Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues
|
|
|
- Stuart Carr
- 6 years ago
- Views:
Transcription
1 Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena 4, S. J. Price 4,6 Microsoft Research Cambridge, UK Department of Radiology, University of Pennsylvania, Philadelphia, PA, USA 3 Brown University, Providence, RI, USA 4 Cambridge University Hospitals, Cambridge, UK 5 Department of Radiology, Cambridge University, UK 6 Department of Clinical Neurosciences, Cambridge University, UK Abstract. We describe our submission to the Brain Tumor Segmentation Challenge (BraTS) at MICCAI, which is based on our method for tissue-specific segmentation of high-grade brain tumors [3]. The main idea is to cast the segmentation as a classification task, and use the discriminative power of context information. We realize this idea by equipping a classification forest (CF) with spatially non-local features to represent the data, and by providing the CF with initial probability estimates for the single tissue classes as additional input (along-side the MRI channels). The initial probabilities are patient-specific, and computed at test time based on a learned model of intensity. Through the combination of the initial probabilities and the non-local features, our approach is able to capture the context information for each data point. Our method is fully automatic, with segmentation run times in the range of - minutes per patient. We evaluate the submission by crossvalidation on the real and synthetic, high- and low-grade tumor BraTS data sets. Introduction This BraTS submission is based on our work presented in [3]. We approach the segmentation of the tumor tissues as a classification problem, where each point in the brain is assigned a certain tissue class. The basic building block of our approach is a standard classification forest (CF), which is a discriminative multiclass classification method. Classification forests allow us to describe brain points to be classified by very high-dimensional features, which are able to capture information about the spatial context. These features are based on the multichannel intensities and are spatially non-local. Furthermore, we augment the input data to the classification forest with initial tissue probabilities, which are estimated as posterior probabilities resulting from a generative intensity-based model, parametrized by Guassian Mixture models (GMM). Together with the
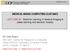
2 D. Zikic et al. Estimation of initial probabilities: posteriors based on tissue-specific intensity-based models (GMM-based) T-gad T T FLAIR (A) input data: MRI Model-B Model-T Model-E p B p T p E (B) initial tissue probabilities Classification Forest spatially non-local, context-sensitive features simultaneous multi-label classification (C) segmentation Fig. : Schematic Method Overview: Based on the input data (A), we first roughly estimate the initial probabilities for the single tissues (B), based on the local intensity information alone. In a second step, we combine the initial probabilities (B) with the input data from (A), resulting in a higher-dimensional multi-channel input for the classification forest. The forest computes the segmentation (C) by a simultaneous multi-label classification, based on non-local and context-sensitive features. context-sensitive features, the initial probabilities as additional input increase the amount of context information and thus improve the classification results. In this paper, we focus on describing our BraTS submission. For more details on motivation for our approach and relation to previous work, please see [3]. Method: Context-sensitive Classification Forests An overview of our approach is given in Figure. We use a standard classification forest [], based on spatially non-local features, and combine it with initial probability estimates for the individual tissue classes. The initial tissue probabilities are based on local intensity information alone. They are estimated with a parametric GMM-based model, as described in Section.. The initial probabilities are then used as additional input channels for the forest, together with the MR image data I. In Section. we give a brief description of classification forests. The types of the context-sensitive features are described in Section.3. We classify three classes C = {B, T, E} for background (B), tumor (T), and edema (E). The MR input data is denoted by I = (I TC, I T, I T, I FLAIR ).. Estimating Initial Tissue Probabilities As the first step of our approach, we estimate the initial class probabilities for a given patient based on the intensity representation in the MRI input data. The initial probabilities are computed as posterior probabilities based on the likelihoods obtained by training a set of GMMs on the training data. For each
3 Context-sensitive Classification Forests for Segmentation of Brain Tumors 3 class c C, we train a single GMM, which captures the likelihood p lik (i c) of the multi-dimensional intensity i R 4 for the class c. With the trained likelihood p lik, for a given test patient data set I, the GMM-based posterior probability p GMM (c p) for the class c is estimated for each point p R 3 by p GMM (c p) = p lik (I(p) c) p(c) c j p lik (I(p) c j ) p(c j ), () with p(c) denoting the prior probability for the class c, computed as a normalized empirical histogram. We can now use the posterior probabilities directly as input for the classification forests, in addition to the multi-channel MR data I. So now, with p GMM c (p):=p GMM (c p), our data for one patient consists of the following channels C = (I T-gad, I T, I T, I FLAIR, p GMM AC, p GMM NC, p GMM E, p GMM B ). () For simplicity, we will denote single channels by C j.. Classification Forests We employ a classification forest (CF) to determine a class c C for a given spatial input point p Ω from a spatial domain Ω of the patient. Our classification forest operates on the representation of a spatial point p by a corresponding feature vector x(p, C), which is based on spatially non-local information from the channels C. CFs are ensembles of (binary) classification trees, indexed and referred to by t [, T ]. As a supervised method, CFs operate in two stages: training and testing. During training, each tree t learns a weak class predictor p t (c x(p, C)). The input training data set is {(x(p, C (k) ), c (k) (p)) : p Ω (k) }, that is, the feature representations of all spatial points p Ω (k), in all training patient data sets k, and the corresponding manual labels c (k) (p). To simplify notation, we will refer to a data point at p by its feature representation x. The set of all data points shall be X. In a classification tree, each node i contains a set of training examples X i, and a class predictor p i t(c x), which is the probability corresponding to the fraction of points with class c in X i (normalized empirical histogram). Starting with the complete training data set X at the root, the training is performed by successively splitting the training examples at every node based on their feature representation, and assigning the partitions X L and X R to the left and right child node. At each node, a number of splits along randomly chosen dimensions of the feature space is considered, and the one maximizing the Information Gain is applied (i.e., an axis-aligned hyperplane is used in the split function). Tree growing is stopped at a certain tree depth D. At testing, a data point x to be classified is pushed through each tree t, by applying the learned split functions. Upon arriving at a leaf node l, the leaf probability is used as the tree probability, i.e. p t (c x) = p l t(c x). The overall probability
4 4 D. Zikic et al. is computed as the average of tree probabilities, i.e. p(c x) = T T t= p t(c x). The actual class estimate ĉ is chosen as the class with the highest probability, i.e. ĉ = arg max c p(c x). For more details on classification forests, see for example []..3 Context-sensitive Feature Types We employ three features types, which are intensity-based and parametrized. Features of these types describe a point to be labeled based on its non-local neighborhood, such that they are context-sensitive. The first two of these feature types are quite generic, while the third one is designed with the intuition of detecting structure changes. We denote the parametrized feature types by x type params. Each combination of type and parameter settings generates one dimension in the feature space, that is x i = x type i params i. Theoretically, the number of possible combinations of type and parameter settings is infinite, and even with exhaustive discrete sampling it remains substantial. In practice, a certain predefined number d of combinations of feature types and parameter settings is randomly drawn for training. In our experiments, we use d =. We use the following notation: Again, p is a spatial point, to be assigned a class, and C j is an input channel. Rj s (p) denotes an p-centered and axis aligned 3D cuboid region in C j with edge lengths l = (l x, l y, l z ), and u R 3 is an offset vector. Feature Type : measures the intensity difference between p in a channel C j and an offset point p + u in a channel C j x t j,j,u(p, C) = C j (p) C j (p + u). (3) Feature Type : measures the difference between intensity means of a cuboid around p in C j, and around an offset point p + u in C j x t j,j,l,l,u(p, C) = µ(r l j (p)) µ(r l j (p + u)). (4) Feature Type 3: captures the intensity range along a 3D line between p and p+u in one channel. This type is designed with the intuition that structure changes can yield a large intensity change, e.g. NC being dark and AC bright in T-gad. x t3 j,u(p, C) = max (C j(p + λu)) min(c j (p + λu)) with λ [, ]. (5) λ λ In the experiments, the types and parameters are drawn uniformly. The offsets u i originate from the range [, ]mm, and the cuboid lengths l i from [, 4]mm.
5 Context-sensitive Classification Forests for Segmentation of Brain Tumors 5 Dice score High-grade (real) Low-grade (real) High-grade (synth) Low-grade (synth) Edema Tumor Edema Tumor Edema Tumor Edema Tumor mean std. dev median Table : Evaluation summary. The Dice scores are computed by the online evaluation tool provided by the organizers of the BraTS challenge. 3 Evaluation We evaluate our approach on the real and synthetic data from the BraTS challenge. Both real and synthetic examples contain separate high-grade (HG) and low-grade (LG) data sets. This results in 4 data sets (Real-HG, Real-LG, Synth- HG, Synth-LG). For each of these data sets, we perform the evaluation independently, i.e., we use only the data from one data set for the training and the testing for this data set. In terms of sizes, Real-HG contains patients, Synth-LG has patients, and the two synthetic data sets contain 5 patients each. For the real data sets, we test our approach on each patient by leave-one-out cross-validation, meaning that for each patient, the training is performed on all other images from the data set, excluding the tested image itself. For the synthetic images, we perform a leave-5-out cross-validation. Pre-processing. We apply bias-field normalization by the ITK N3 implementation from []. Then, we align the mean intensities of the images within each channel by a global multiplicative factor. For speed reasons, we run the evaluation on a down-sampled version of the input images, with isotropic spatial resolution of mm. The computed segmentations are up-sampled back to mm for the evaluation. Settings. In all tests, we employ forests with T = 4 trees of depth D =. Runtime. Our segmentation method is fully automatic, with segmentation run times in the range of - minutes per patient. The training of one tree takes approximately minutes on a single desktop PC. Results. We evaluated our segmentations by the BraTS online evaluation tool, and we summarize the results for the Dice score in Table. Overall, the results indicate a higher segmentation quality for the high-grade tumors than for the low-grade cases, and a better performance on the synthetic data than the real data set.
6 6 D. Zikic et al. Dice Score Dice Score Dice Score Dice Score Specificity Specificity Specificity Specificity Precision Precision Precision Precision Recall (=Sensitivity) Recall (=Sensitivity) Recall (=Sensitivity) Recall (=Sensitivity) SD (mean) [mm] 8 SD (mean) [mm] 8 SD (mean) [mm] 8 SD (mean) [mm] SD (max) [mm] 8 SD (max) [mm] 8 SD (max) [mm] 8 SD (max) [mm] (a) Real-HG (b)real-lg (c) Synth-HG (d) Synth-LG Fig. : Per patient evaluation for the four BraTS data sets (Real-HG, Real- LG, Synth-HG, Synth-LG). We show the results for edema (blue) and tumor tissue (red) per patient, and indicate the respective median results with the horizontal lines. We report the following measures: Dice, Specificity, Precision, Recall(=Sensitivity), Mean Surface Distance (SD), and Maximal SD. Further Evaluation. Furthermore, we reproduce most of the BraTS measures (except Kappa) by our own evaluation in Figure. It can be seen in Figure, that the Specificity is not a very discriminative measure in this application. Therefore, we rather evaluate Precision, which is similar in nature, but does
7 Context-sensitive Classification Forests for Segmentation of Brain Tumors 7 not take the background class into account (TN), and is thus more sensitive to errors. In order to obtain a better understanding of the data and the performance of our method we perform three further measurements.. In Figure 3, we measure the volumes of the brain, and the edema and tumor tissues for the individual patients. This is done in order to be able to evaluate how target volumes influence the segmentation quality.. In Figure 4, we report the results for the basic types of classification outcomes, i.e. true positives (TP), false positives (FP), and false negatives (FN). It is interesting to note the correlation of the TP values with the tissue volumes (cf. Fig. 3). Also, it seems that for edema, the error of our method consists of more FP estimates (wrongly labeled as edema) than FN estimates (wrongly not labeled as edema), i.e. it performs an over-segmentation. 3. In Figure 5, we report additional measures, which might have an applicationspecific relevance. We compute the overall Error, i.e. the volume of all misclassified points FN + FP, and the corresponding relative version, which relates the error to the target volume T, i.e. (FN + FP)/T. Also, we compute the absolute and the relative Volume Error T (TP + FP), and T (TP + FP) /T, which indicate the potential performance for volumetric measurements. The volume error is less sensitive than the error measure, since it does not require an overlap of segmentations but only that the estimated volume is correct (volume error can be expressed as FN FP ). Acknowledgments S. J. Price is funded by a Clinician Scientist Award from the National Institute for Health Research (NIHR). O. M. Thomas is a Clinical Lecturer supported by the NIHR Cambridge Biomedical Research Centre. References. A. Criminisi, J. Shotton, and E. Konukoglu. Decision forests: A unified framework for classification, regression, density estimation, manifold learning and semisupervised learning. Foundations and Trends in Computer Graphics and Vision, 7(-3),.. N. Tustison and J. Gee. N4ITK: Nick s N3 ITK implementation for MRI bias field correction. The Insight Journal,. 3. D. Zikic, B. Glocker, E. Konukoglu, A. Criminisi, C. Demiralp, J. Shotton, O. M. Thomas, T. Das, R. Jena, and Price S. J. Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel mr. In Proc. Medical Image Computing and Computer Assisted Intervention,.
8 8 D. Zikic et al. Brain Volume [x 3 cm 3 ] Brain Volume [x 3 cm 3 ] Brain Volume [x 3 cm 3 ] Brain Volume [x 3 cm 3 ] Tissue Volumes [cm 3 ] 8 Tissue Volumes [cm 3 ] 8 Tissue Volumes [cm 3 ] 8 Tissue Volumes [cm 3 ] (e) Real-HG (f)real-lg (g) Synth-HG (h) Synth-LG Fig. 3: Volume statistics of the BraTS data sets. We compute the brain volumes (top row), and the volumes of the edema (blue) and tumor (red) tissues per patient.. TP / V. TP / V. TP / V. TP / V FP / V FP / V FP / V FP / V FN / V FN / V FN / V FN / V (a) Real-HG (b)real-lg (c) Synth-HG (d) Synth-LG Fig. 4: We report the values of true positives (TP), false positives (FP), and false negatives (FN), for edema (blue), and tumor (red) tissues. To make the values comparable, we report them as percentage of the patient brain volume (V). Again, horizontal lines represent median values. It is interesting to note the correlation of the TP values with the tissue volumes (cf. Fig. 3). Also, it seems that for edema, the error of our method consists of more FP estimates (wrongly labeled as edema) than FN estimates (wrongly not labeled as edema), i.e. it performs an over-segmentation.
9 Context-sensitive Classification Forests for Segmentation of Brain Tumors 9 Error (abs.) [cm 3 ] Error (abs.) [cm 3 ] Error (abs.) [cm 3 ] Error (abs.) [cm 3 ] Error (rel.) Error (rel.) Error (rel.) Error (rel.) Volume Error (abs.) [cm 3 ] Volume Error (abs.) [cm 3 ] Volume Error (abs.) [cm 3 ] Volume Error (abs.) [cm 3 ] Volume Error (rel.) Volume Error (rel.) Volume Error (rel.) Volume Error (rel.) (a) Real-HG (b)real-lg (c) Synth-HG (d) Synth-LG Fig. 5: We further evaluate additional measures which might have applicationspecific relevance. Again, we have blue=edema, red=tumor, and horizontal line=median. In the two top rows, we compute the Error, i.e. the volume of all misclassified points FN + FP, and the relative version, which relates the error to the target volume T, i.e. (FN + FP)/T. In the bottom two rows, we compute the absolute and the relative Volume Error T (TP+FP), and T (TP+FP) /T.
Multimodal Brain Tumor Segmentation
 MICCAI 212 Challenge on Multimodal Brain Tumor Segmentation Proceedings of MICCAI-BRATS 212 October 1 st, Nice, France. I Preface Because of their unpredictable appearance and shape, segmenting brain tumors
MICCAI 212 Challenge on Multimodal Brain Tumor Segmentation Proceedings of MICCAI-BRATS 212 October 1 st, Nice, France. I Preface Because of their unpredictable appearance and shape, segmenting brain tumors
Supervised Learning for Image Segmentation
 Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests)
 SPRING 2016 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
SPRING 2016 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
NIH Public Access Author Manuscript Proc IEEE Int Symp Biomed Imaging. Author manuscript; available in PMC 2014 November 15.
 NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging. 2013 April ; 2013: 748 751. doi:10.1109/isbi.2013.6556583. BRAIN TUMOR SEGMENTATION WITH SYMMETRIC
NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging. 2013 April ; 2013: 748 751. doi:10.1109/isbi.2013.6556583. BRAIN TUMOR SEGMENTATION WITH SYMMETRIC
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation
 Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation
 Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Ezequiel Geremia 2, Nicholas Ayache 2, and Gabor Szekely 1 1 Computer
Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Ezequiel Geremia 2, Nicholas Ayache 2, and Gabor Szekely 1 1 Computer
Ensemble registration: Combining groupwise registration and segmentation
 PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS
 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
Advanced Video Content Analysis and Video Compression (5LSH0), Module 8B
 Advanced Video Content Analysis and Video Compression (5LSH0), Module 8B 1 Supervised learning Catogarized / labeled data Objects in a picture: chair, desk, person, 2 Classification Fons van der Sommen
Advanced Video Content Analysis and Video Compression (5LSH0), Module 8B 1 Supervised learning Catogarized / labeled data Objects in a picture: chair, desk, person, 2 Classification Fons van der Sommen
Mondrian Forests: Efficient Online Random Forests
 Mondrian Forests: Efficient Online Random Forests Balaji Lakshminarayanan (Gatsby Unit, UCL) Daniel M. Roy (Cambridge Toronto) Yee Whye Teh (Oxford) September 4, 2014 1 Outline Background and Motivation
Mondrian Forests: Efficient Online Random Forests Balaji Lakshminarayanan (Gatsby Unit, UCL) Daniel M. Roy (Cambridge Toronto) Yee Whye Teh (Oxford) September 4, 2014 1 Outline Background and Motivation
A Nonparametric Model for Brain Tumor Segmentation and Volumetry in Longitudinal MR Sequences
 A Nonparametric Model for Brain Tumor Segmentation and Volumetry in Longitudinal MR Sequences Esther Alberts 1,2,6, Guillaume Charpiat 3, Yuliya Tarabalka 4, Thomas Huber 1, Marc-André Weber 5, Jan Bauer
A Nonparametric Model for Brain Tumor Segmentation and Volumetry in Longitudinal MR Sequences Esther Alberts 1,2,6, Guillaume Charpiat 3, Yuliya Tarabalka 4, Thomas Huber 1, Marc-André Weber 5, Jan Bauer
A Workflow for Improving Medical Visualization of Semantically Annotated CT-Images
 A Workflow for Improving Medical Visualization of Semantically Annotated CT-Images Alexander Baranya 1,2, Luis Landaeta 1,2, Alexandra La Cruz 1, and Maria-Esther Vidal 2 1 Biophysic and Bioengeneering
A Workflow for Improving Medical Visualization of Semantically Annotated CT-Images Alexander Baranya 1,2, Luis Landaeta 1,2, Alexandra La Cruz 1, and Maria-Esther Vidal 2 1 Biophysic and Bioengeneering
Applying Supervised Learning
 Applying Supervised Learning When to Consider Supervised Learning A supervised learning algorithm takes a known set of input data (the training set) and known responses to the data (output), and trains
Applying Supervised Learning When to Consider Supervised Learning A supervised learning algorithm takes a known set of input data (the training set) and known responses to the data (output), and trains
Best First and Greedy Search Based CFS and Naïve Bayes Algorithms for Hepatitis Diagnosis
 Best First and Greedy Search Based CFS and Naïve Bayes Algorithms for Hepatitis Diagnosis CHAPTER 3 BEST FIRST AND GREEDY SEARCH BASED CFS AND NAÏVE BAYES ALGORITHMS FOR HEPATITIS DIAGNOSIS 3.1 Introduction
Best First and Greedy Search Based CFS and Naïve Bayes Algorithms for Hepatitis Diagnosis CHAPTER 3 BEST FIRST AND GREEDY SEARCH BASED CFS AND NAÏVE BAYES ALGORITHMS FOR HEPATITIS DIAGNOSIS 3.1 Introduction
Using the Kolmogorov-Smirnov Test for Image Segmentation
 Using the Kolmogorov-Smirnov Test for Image Segmentation Yong Jae Lee CS395T Computational Statistics Final Project Report May 6th, 2009 I. INTRODUCTION Image segmentation is a fundamental task in computer
Using the Kolmogorov-Smirnov Test for Image Segmentation Yong Jae Lee CS395T Computational Statistics Final Project Report May 6th, 2009 I. INTRODUCTION Image segmentation is a fundamental task in computer
Ulas Bagci
 CAP5415-Computer Vision Lecture 14-Decision Forests for Computer Vision Ulas Bagci bagci@ucf.edu 1 Readings Slide Credits: Criminisi and Shotton Z. Tu R.Cipolla 2 Common Terminologies Randomized Decision
CAP5415-Computer Vision Lecture 14-Decision Forests for Computer Vision Ulas Bagci bagci@ucf.edu 1 Readings Slide Credits: Criminisi and Shotton Z. Tu R.Cipolla 2 Common Terminologies Randomized Decision
Hybrid Approach for MRI Human Head Scans Classification using HTT based SFTA Texture Feature Extraction Technique
 Volume 118 No. 17 2018, 691-701 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu ijpam.eu Hybrid Approach for MRI Human Head Scans Classification using HTT
Volume 118 No. 17 2018, 691-701 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu ijpam.eu Hybrid Approach for MRI Human Head Scans Classification using HTT
Towards Grading Gleason Score using Generically Trained Deep convolutional Neural Networks
 Towards Grading Gleason Score using Generically Trained Deep convolutional Neural Networks Källén, Hanna; Molin, Jesper; Heyden, Anders; Lundström, Claes; Åström, Karl Published in: 2016 IEEE 13th International
Towards Grading Gleason Score using Generically Trained Deep convolutional Neural Networks Källén, Hanna; Molin, Jesper; Heyden, Anders; Lundström, Claes; Åström, Karl Published in: 2016 IEEE 13th International
CS 229 Midterm Review
 CS 229 Midterm Review Course Staff Fall 2018 11/2/2018 Outline Today: SVMs Kernels Tree Ensembles EM Algorithm / Mixture Models [ Focus on building intuition, less so on solving specific problems. Ask
CS 229 Midterm Review Course Staff Fall 2018 11/2/2018 Outline Today: SVMs Kernels Tree Ensembles EM Algorithm / Mixture Models [ Focus on building intuition, less so on solving specific problems. Ask
Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images
 Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images Sarah Parisot 1,2,3, Hugues Duffau 4, Stéphane Chemouny 3, Nikos Paragios 1,2 1. Center for Visual Computing, Ecole Centrale
Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images Sarah Parisot 1,2,3, Hugues Duffau 4, Stéphane Chemouny 3, Nikos Paragios 1,2 1. Center for Visual Computing, Ecole Centrale
Bioimage Informatics
 Bioimage Informatics Lecture 13, Spring 2012 Bioimage Data Analysis (IV) Image Segmentation (part 2) Lecture 13 February 29, 2012 1 Outline Review: Steger s line/curve detection algorithm Intensity thresholding
Bioimage Informatics Lecture 13, Spring 2012 Bioimage Data Analysis (IV) Image Segmentation (part 2) Lecture 13 February 29, 2012 1 Outline Review: Steger s line/curve detection algorithm Intensity thresholding
Weka ( )
 Weka ( http://www.cs.waikato.ac.nz/ml/weka/ ) The phases in which classifier s design can be divided are reflected in WEKA s Explorer structure: Data pre-processing (filtering) and representation Supervised
Weka ( http://www.cs.waikato.ac.nz/ml/weka/ ) The phases in which classifier s design can be divided are reflected in WEKA s Explorer structure: Data pre-processing (filtering) and representation Supervised
Global Journal of Engineering Science and Research Management
 ADVANCED K-MEANS ALGORITHM FOR BRAIN TUMOR DETECTION USING NAIVE BAYES CLASSIFIER Veena Bai K*, Dr. Niharika Kumar * MTech CSE, Department of Computer Science and Engineering, B.N.M. Institute of Technology,
ADVANCED K-MEANS ALGORITHM FOR BRAIN TUMOR DETECTION USING NAIVE BAYES CLASSIFIER Veena Bai K*, Dr. Niharika Kumar * MTech CSE, Department of Computer Science and Engineering, B.N.M. Institute of Technology,
Supervoxel Classification Forests for Estimating Pairwise Image Correspondences
 Supervoxel Classification Forests for Estimating Pairwise Image Correspondences Fahdi Kanavati 1, Tong Tong 1, Kazunari Misawa 2, Michitaka Fujiwara 3, Kensaku Mori 4, Daniel Rueckert 1, and Ben Glocker
Supervoxel Classification Forests for Estimating Pairwise Image Correspondences Fahdi Kanavati 1, Tong Tong 1, Kazunari Misawa 2, Michitaka Fujiwara 3, Kensaku Mori 4, Daniel Rueckert 1, and Ben Glocker
Norbert Schuff VA Medical Center and UCSF
 Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Classification and Regression
 Classification and Regression Announcements Study guide for exam is on the LMS Sample exam will be posted by Monday Reminder that phase 3 oral presentations are being held next week during workshops Plan
Classification and Regression Announcements Study guide for exam is on the LMS Sample exam will be posted by Monday Reminder that phase 3 oral presentations are being held next week during workshops Plan
Business Club. Decision Trees
 Business Club Decision Trees Business Club Analytics Team December 2017 Index 1. Motivation- A Case Study 2. The Trees a. What is a decision tree b. Representation 3. Regression v/s Classification 4. Building
Business Club Decision Trees Business Club Analytics Team December 2017 Index 1. Motivation- A Case Study 2. The Trees a. What is a decision tree b. Representation 3. Regression v/s Classification 4. Building
Improving Positron Emission Tomography Imaging with Machine Learning David Fan-Chung Hsu CS 229 Fall
 Improving Positron Emission Tomography Imaging with Machine Learning David Fan-Chung Hsu (fcdh@stanford.edu), CS 229 Fall 2014-15 1. Introduction and Motivation High- resolution Positron Emission Tomography
Improving Positron Emission Tomography Imaging with Machine Learning David Fan-Chung Hsu (fcdh@stanford.edu), CS 229 Fall 2014-15 1. Introduction and Motivation High- resolution Positron Emission Tomography
Chapter 3: Supervised Learning
 Chapter 3: Supervised Learning Road Map Basic concepts Evaluation of classifiers Classification using association rules Naïve Bayesian classification Naïve Bayes for text classification Summary 2 An example
Chapter 3: Supervised Learning Road Map Basic concepts Evaluation of classifiers Classification using association rules Naïve Bayesian classification Naïve Bayes for text classification Summary 2 An example
Transfer Forest Based on Covariate Shift
 Transfer Forest Based on Covariate Shift Masamitsu Tsuchiya SECURE, INC. tsuchiya@secureinc.co.jp Yuji Yamauchi, Takayoshi Yamashita, Hironobu Fujiyoshi Chubu University yuu@vision.cs.chubu.ac.jp, {yamashita,
Transfer Forest Based on Covariate Shift Masamitsu Tsuchiya SECURE, INC. tsuchiya@secureinc.co.jp Yuji Yamauchi, Takayoshi Yamashita, Hironobu Fujiyoshi Chubu University yuu@vision.cs.chubu.ac.jp, {yamashita,
Bayesian model ensembling using meta-trained recurrent neural networks
 Bayesian model ensembling using meta-trained recurrent neural networks Luca Ambrogioni l.ambrogioni@donders.ru.nl Umut Güçlü u.guclu@donders.ru.nl Yağmur Güçlütürk y.gucluturk@donders.ru.nl Julia Berezutskaya
Bayesian model ensembling using meta-trained recurrent neural networks Luca Ambrogioni l.ambrogioni@donders.ru.nl Umut Güçlü u.guclu@donders.ru.nl Yağmur Güçlütürk y.gucluturk@donders.ru.nl Julia Berezutskaya
Automatic Vertebrae Localization in Pathological Spine CT using Decision Forests
 Automatic Vertebrae Localization in Pathological Spine CT using Decision Forests Ana Jiménez-Pastor 1, Esther Tomás-González 1, Ángel Alberich-Bayarri 1,2, Fabio García-Castro 1, David García-Juan 1, Luis
Automatic Vertebrae Localization in Pathological Spine CT using Decision Forests Ana Jiménez-Pastor 1, Esther Tomás-González 1, Ángel Alberich-Bayarri 1,2, Fabio García-Castro 1, David García-Juan 1, Luis
Mondrian Forests: Efficient Online Random Forests
 Mondrian Forests: Efficient Online Random Forests Balaji Lakshminarayanan Joint work with Daniel M. Roy and Yee Whye Teh 1 Outline Background and Motivation Mondrian Forests Randomization mechanism Online
Mondrian Forests: Efficient Online Random Forests Balaji Lakshminarayanan Joint work with Daniel M. Roy and Yee Whye Teh 1 Outline Background and Motivation Mondrian Forests Randomization mechanism Online
Evaluation Metrics. (Classifiers) CS229 Section Anand Avati
 Evaluation Metrics (Classifiers) CS Section Anand Avati Topics Why? Binary classifiers Metrics Rank view Thresholding Confusion Matrix Point metrics: Accuracy, Precision, Recall / Sensitivity, Specificity,
Evaluation Metrics (Classifiers) CS Section Anand Avati Topics Why? Binary classifiers Metrics Rank view Thresholding Confusion Matrix Point metrics: Accuracy, Precision, Recall / Sensitivity, Specificity,
ECLT 5810 Evaluation of Classification Quality
 ECLT 5810 Evaluation of Classification Quality Reference: Data Mining Practical Machine Learning Tools and Techniques, by I. Witten, E. Frank, and M. Hall, Morgan Kaufmann Testing and Error Error rate:
ECLT 5810 Evaluation of Classification Quality Reference: Data Mining Practical Machine Learning Tools and Techniques, by I. Witten, E. Frank, and M. Hall, Morgan Kaufmann Testing and Error Error rate:
Contents. Preface to the Second Edition
 Preface to the Second Edition v 1 Introduction 1 1.1 What Is Data Mining?....................... 4 1.2 Motivating Challenges....................... 5 1.3 The Origins of Data Mining....................
Preface to the Second Edition v 1 Introduction 1 1.1 What Is Data Mining?....................... 4 1.2 Motivating Challenges....................... 5 1.3 The Origins of Data Mining....................
HOG-based Pedestriant Detector Training
 HOG-based Pedestriant Detector Training evs embedded Vision Systems Srl c/o Computer Science Park, Strada Le Grazie, 15 Verona- Italy http: // www. embeddedvisionsystems. it Abstract This paper describes
HOG-based Pedestriant Detector Training evs embedded Vision Systems Srl c/o Computer Science Park, Strada Le Grazie, 15 Verona- Italy http: // www. embeddedvisionsystems. it Abstract This paper describes
Evaluating Classifiers
 Evaluating Classifiers Reading for this topic: T. Fawcett, An introduction to ROC analysis, Sections 1-4, 7 (linked from class website) Evaluating Classifiers What we want: Classifier that best predicts
Evaluating Classifiers Reading for this topic: T. Fawcett, An introduction to ROC analysis, Sections 1-4, 7 (linked from class website) Evaluating Classifiers What we want: Classifier that best predicts
Ischemic Stroke Lesion Segmentation Proceedings 5th October 2015 Munich, Germany
 0111010001110001101000100101010111100111011100100011011101110101101012 Ischemic Stroke Lesion Segmentation www.isles-challenge.org Proceedings 5th October 2015 Munich, Germany Preface Stroke is the second
0111010001110001101000100101010111100111011100100011011101110101101012 Ischemic Stroke Lesion Segmentation www.isles-challenge.org Proceedings 5th October 2015 Munich, Germany Preface Stroke is the second
An Implementation and Discussion of Random Forest with Gaussian Process Leaves
 An Implementation and Discussion of Random Forest with Gaussian Process Leaves 1 2 3 4 5 6 Anonymous Author(s) Affiliation Address email 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29
An Implementation and Discussion of Random Forest with Gaussian Process Leaves 1 2 3 4 5 6 Anonymous Author(s) Affiliation Address email 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29
Evaluation Measures. Sebastian Pölsterl. April 28, Computer Aided Medical Procedures Technische Universität München
 Evaluation Measures Sebastian Pölsterl Computer Aided Medical Procedures Technische Universität München April 28, 2015 Outline 1 Classification 1. Confusion Matrix 2. Receiver operating characteristics
Evaluation Measures Sebastian Pölsterl Computer Aided Medical Procedures Technische Universität München April 28, 2015 Outline 1 Classification 1. Confusion Matrix 2. Receiver operating characteristics
Sampling-Based Ensemble Segmentation against Inter-operator Variability
 Sampling-Based Ensemble Segmentation against Inter-operator Variability Jing Huo 1, Kazunori Okada, Whitney Pope 1, Matthew Brown 1 1 Center for Computer vision and Imaging Biomarkers, Department of Radiological
Sampling-Based Ensemble Segmentation against Inter-operator Variability Jing Huo 1, Kazunori Okada, Whitney Pope 1, Matthew Brown 1 1 Center for Computer vision and Imaging Biomarkers, Department of Radiological
Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation
 Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation Ting Song 1, Elsa D. Angelini 2, Brett D. Mensh 3, Andrew Laine 1 1 Heffner Biomedical Imaging Laboratory Department of Biomedical Engineering,
Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation Ting Song 1, Elsa D. Angelini 2, Brett D. Mensh 3, Andrew Laine 1 1 Heffner Biomedical Imaging Laboratory Department of Biomedical Engineering,
Supervised vs unsupervised clustering
 Classification Supervised vs unsupervised clustering Cluster analysis: Classes are not known a- priori. Classification: Classes are defined a-priori Sometimes called supervised clustering Extract useful
Classification Supervised vs unsupervised clustering Cluster analysis: Classes are not known a- priori. Classification: Classes are defined a-priori Sometimes called supervised clustering Extract useful
Data Clustering. Danushka Bollegala
 Data Clustering Danushka Bollegala Outline Why cluster data? Clustering as unsupervised learning Clustering algorithms k-means, k-medoids agglomerative clustering Brown s clustering Spectral clustering
Data Clustering Danushka Bollegala Outline Why cluster data? Clustering as unsupervised learning Clustering algorithms k-means, k-medoids agglomerative clustering Brown s clustering Spectral clustering
Evaluating Classifiers
 Evaluating Classifiers Charles Elkan elkan@cs.ucsd.edu January 18, 2011 In a real-world application of supervised learning, we have a training set of examples with labels, and a test set of examples with
Evaluating Classifiers Charles Elkan elkan@cs.ucsd.edu January 18, 2011 In a real-world application of supervised learning, we have a training set of examples with labels, and a test set of examples with
Lecture 13 Theory of Registration. ch. 10 of Insight into Images edited by Terry Yoo, et al. Spring (CMU RI) : BioE 2630 (Pitt)
 Lecture 13 Theory of Registration ch. 10 of Insight into Images edited by Terry Yoo, et al. Spring 2018 16-725 (CMU RI) : BioE 2630 (Pitt) Dr. John Galeotti The content of these slides by John Galeotti,
Lecture 13 Theory of Registration ch. 10 of Insight into Images edited by Terry Yoo, et al. Spring 2018 16-725 (CMU RI) : BioE 2630 (Pitt) Dr. John Galeotti The content of these slides by John Galeotti,
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION
 ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
Prostate Detection Using Principal Component Analysis
 Prostate Detection Using Principal Component Analysis Aamir Virani (avirani@stanford.edu) CS 229 Machine Learning Stanford University 16 December 2005 Introduction During the past two decades, computed
Prostate Detection Using Principal Component Analysis Aamir Virani (avirani@stanford.edu) CS 229 Machine Learning Stanford University 16 December 2005 Introduction During the past two decades, computed
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 10: Medical Image Segmentation as an Energy Minimization Problem
 SPRING 07 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
SPRING 07 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
Multi-Sectional Views Textural Based SVM for MS Lesion Segmentation in Multi-Channels MRIs
 56 The Open Biomedical Engineering Journal, 2012, 6, 56-72 Open Access Multi-Sectional Views Textural Based SVM for MS Lesion Segmentation in Multi-Channels MRIs Bassem A. Abdullah*, Akmal A. Younis and
56 The Open Biomedical Engineering Journal, 2012, 6, 56-72 Open Access Multi-Sectional Views Textural Based SVM for MS Lesion Segmentation in Multi-Channels MRIs Bassem A. Abdullah*, Akmal A. Younis and
Naïve Bayes Classification. Material borrowed from Jonathan Huang and I. H. Witten s and E. Frank s Data Mining and Jeremy Wyatt and others
 Naïve Bayes Classification Material borrowed from Jonathan Huang and I. H. Witten s and E. Frank s Data Mining and Jeremy Wyatt and others Things We d Like to Do Spam Classification Given an email, predict
Naïve Bayes Classification Material borrowed from Jonathan Huang and I. H. Witten s and E. Frank s Data Mining and Jeremy Wyatt and others Things We d Like to Do Spam Classification Given an email, predict
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information
 Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Vertebrae Segmentation in 3D CT Images based on a Variational Framework
 Vertebrae Segmentation in 3D CT Images based on a Variational Framework Kerstin Hammernik, Thomas Ebner, Darko Stern, Martin Urschler, and Thomas Pock Abstract Automatic segmentation of 3D vertebrae is
Vertebrae Segmentation in 3D CT Images based on a Variational Framework Kerstin Hammernik, Thomas Ebner, Darko Stern, Martin Urschler, and Thomas Pock Abstract Automatic segmentation of 3D vertebrae is
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
CHAPTER 3 TUMOR DETECTION BASED ON NEURO-FUZZY TECHNIQUE
 32 CHAPTER 3 TUMOR DETECTION BASED ON NEURO-FUZZY TECHNIQUE 3.1 INTRODUCTION In this chapter we present the real time implementation of an artificial neural network based on fuzzy segmentation process
32 CHAPTER 3 TUMOR DETECTION BASED ON NEURO-FUZZY TECHNIQUE 3.1 INTRODUCTION In this chapter we present the real time implementation of an artificial neural network based on fuzzy segmentation process
Evaluation of different biological data and computational classification methods for use in protein interaction prediction.
 Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Yanjun Qi, Ziv Bar-Joseph, Judith Klein-Seetharaman Protein 2006 Motivation Correctly
Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Yanjun Qi, Ziv Bar-Joseph, Judith Klein-Seetharaman Protein 2006 Motivation Correctly
MIT Samberg Center Cambridge, MA, USA. May 30 th June 2 nd, by C. Rea, R.S. Granetz MIT Plasma Science and Fusion Center, Cambridge, MA, USA
 Exploratory Machine Learning studies for disruption prediction on DIII-D by C. Rea, R.S. Granetz MIT Plasma Science and Fusion Center, Cambridge, MA, USA Presented at the 2 nd IAEA Technical Meeting on
Exploratory Machine Learning studies for disruption prediction on DIII-D by C. Rea, R.S. Granetz MIT Plasma Science and Fusion Center, Cambridge, MA, USA Presented at the 2 nd IAEA Technical Meeting on
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION
 CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
ECG782: Multidimensional Digital Signal Processing
 ECG782: Multidimensional Digital Signal Processing Object Recognition http://www.ee.unlv.edu/~b1morris/ecg782/ 2 Outline Knowledge Representation Statistical Pattern Recognition Neural Networks Boosting
ECG782: Multidimensional Digital Signal Processing Object Recognition http://www.ee.unlv.edu/~b1morris/ecg782/ 2 Outline Knowledge Representation Statistical Pattern Recognition Neural Networks Boosting
CS145: INTRODUCTION TO DATA MINING
 CS145: INTRODUCTION TO DATA MINING 08: Classification Evaluation and Practical Issues Instructor: Yizhou Sun yzsun@cs.ucla.edu October 24, 2017 Learnt Prediction and Classification Methods Vector Data
CS145: INTRODUCTION TO DATA MINING 08: Classification Evaluation and Practical Issues Instructor: Yizhou Sun yzsun@cs.ucla.edu October 24, 2017 Learnt Prediction and Classification Methods Vector Data
An ITK Filter for Bayesian Segmentation: itkbayesianclassifierimagefilter
 An ITK Filter for Bayesian Segmentation: itkbayesianclassifierimagefilter John Melonakos 1, Karthik Krishnan 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332, USA {jmelonak,
An ITK Filter for Bayesian Segmentation: itkbayesianclassifierimagefilter John Melonakos 1, Karthik Krishnan 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332, USA {jmelonak,
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Network Traffic Measurements and Analysis
 DEIB - Politecnico di Milano Fall, 2017 Sources Hastie, Tibshirani, Friedman: The Elements of Statistical Learning James, Witten, Hastie, Tibshirani: An Introduction to Statistical Learning Andrew Ng:
DEIB - Politecnico di Milano Fall, 2017 Sources Hastie, Tibshirani, Friedman: The Elements of Statistical Learning James, Witten, Hastie, Tibshirani: An Introduction to Statistical Learning Andrew Ng:
The Basics of Decision Trees
 Tree-based Methods Here we describe tree-based methods for regression and classification. These involve stratifying or segmenting the predictor space into a number of simple regions. Since the set of splitting
Tree-based Methods Here we describe tree-based methods for regression and classification. These involve stratifying or segmenting the predictor space into a number of simple regions. Since the set of splitting
Machine Learning Techniques for Data Mining
 Machine Learning Techniques for Data Mining Eibe Frank University of Waikato New Zealand 10/25/2000 1 PART VII Moving on: Engineering the input and output 10/25/2000 2 Applying a learner is not all Already
Machine Learning Techniques for Data Mining Eibe Frank University of Waikato New Zealand 10/25/2000 1 PART VII Moving on: Engineering the input and output 10/25/2000 2 Applying a learner is not all Already
The Comparative Study of Machine Learning Algorithms in Text Data Classification*
 The Comparative Study of Machine Learning Algorithms in Text Data Classification* Wang Xin School of Science, Beijing Information Science and Technology University Beijing, China Abstract Classification
The Comparative Study of Machine Learning Algorithms in Text Data Classification* Wang Xin School of Science, Beijing Information Science and Technology University Beijing, China Abstract Classification
Bus Detection and recognition for visually impaired people
 Bus Detection and recognition for visually impaired people Hangrong Pan, Chucai Yi, and Yingli Tian The City College of New York The Graduate Center The City University of New York MAP4VIP Outline Motivation
Bus Detection and recognition for visually impaired people Hangrong Pan, Chucai Yi, and Yingli Tian The City College of New York The Graduate Center The City University of New York MAP4VIP Outline Motivation
Decision trees. Decision trees are useful to a large degree because of their simplicity and interpretability
 Decision trees A decision tree is a method for classification/regression that aims to ask a few relatively simple questions about an input and then predicts the associated output Decision trees are useful
Decision trees A decision tree is a method for classification/regression that aims to ask a few relatively simple questions about an input and then predicts the associated output Decision trees are useful
Chapter 5: Outlier Detection
 Ludwig-Maximilians-Universität München Institut für Informatik Lehr- und Forschungseinheit für Datenbanksysteme Knowledge Discovery in Databases SS 2016 Chapter 5: Outlier Detection Lecture: Prof. Dr.
Ludwig-Maximilians-Universität München Institut für Informatik Lehr- und Forschungseinheit für Datenbanksysteme Knowledge Discovery in Databases SS 2016 Chapter 5: Outlier Detection Lecture: Prof. Dr.
Fast Sample Generation with Variational Bayesian for Limited Data Hyperspectral Image Classification
 Fast Sample Generation with Variational Bayesian for Limited Data Hyperspectral Image Classification July 26, 2018 AmirAbbas Davari, Hasan Can Özkan, Andreas Maier, Christian Riess Pattern Recognition
Fast Sample Generation with Variational Bayesian for Limited Data Hyperspectral Image Classification July 26, 2018 AmirAbbas Davari, Hasan Can Özkan, Andreas Maier, Christian Riess Pattern Recognition
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 12 Combining
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 12 Combining
Data mining with Support Vector Machine
 Data mining with Support Vector Machine Ms. Arti Patle IES, IPS Academy Indore (M.P.) artipatle@gmail.com Mr. Deepak Singh Chouhan IES, IPS Academy Indore (M.P.) deepak.schouhan@yahoo.com Abstract: Machine
Data mining with Support Vector Machine Ms. Arti Patle IES, IPS Academy Indore (M.P.) artipatle@gmail.com Mr. Deepak Singh Chouhan IES, IPS Academy Indore (M.P.) deepak.schouhan@yahoo.com Abstract: Machine
Two-phase MRI brain tumor segmentation using Random Forests and Level Set Methods
 Two-phase MRI brain tumor segmentation using Random Forests and Level Set Methods László Lefkovits Sapientia University Department of Electrical Engineering Romania, Tg. Mureș lefkolaci@ms.sapientia.ro
Two-phase MRI brain tumor segmentation using Random Forests and Level Set Methods László Lefkovits Sapientia University Department of Electrical Engineering Romania, Tg. Mureș lefkolaci@ms.sapientia.ro
Bagging for One-Class Learning
 Bagging for One-Class Learning David Kamm December 13, 2008 1 Introduction Consider the following outlier detection problem: suppose you are given an unlabeled data set and make the assumptions that one
Bagging for One-Class Learning David Kamm December 13, 2008 1 Introduction Consider the following outlier detection problem: suppose you are given an unlabeled data set and make the assumptions that one
Support Vector Machines
 Support Vector Machines SVM Discussion Overview. Importance of SVMs. Overview of Mathematical Techniques Employed 3. Margin Geometry 4. SVM Training Methodology 5. Overlapping Distributions 6. Dealing
Support Vector Machines SVM Discussion Overview. Importance of SVMs. Overview of Mathematical Techniques Employed 3. Margin Geometry 4. SVM Training Methodology 5. Overlapping Distributions 6. Dealing
Allstate Insurance Claims Severity: A Machine Learning Approach
 Allstate Insurance Claims Severity: A Machine Learning Approach Rajeeva Gaur SUNet ID: rajeevag Jeff Pickelman SUNet ID: pattern Hongyi Wang SUNet ID: hongyiw I. INTRODUCTION The insurance industry has
Allstate Insurance Claims Severity: A Machine Learning Approach Rajeeva Gaur SUNet ID: rajeevag Jeff Pickelman SUNet ID: pattern Hongyi Wang SUNet ID: hongyiw I. INTRODUCTION The insurance industry has
Atlas of Classifiers for Brain MRI Segmentation
 Atlas of Classifiers for Brain MRI Segmentation B. Kodner 1,2, S. H. Gordon 1,2, J. Goldberger 3 and T. Riklin Raviv 1,2 1 Department of Electrical and Computer Engineering, 2 The Zlotowski Center for
Atlas of Classifiers for Brain MRI Segmentation B. Kodner 1,2, S. H. Gordon 1,2, J. Goldberger 3 and T. Riklin Raviv 1,2 1 Department of Electrical and Computer Engineering, 2 The Zlotowski Center for
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations
 Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
MULTI-REGION SEGMENTATION
 MULTI-REGION SEGMENTATION USING GRAPH-CUTS Johannes Ulén Abstract This project deals with multi-region segmenation using graph-cuts and is mainly based on a paper by Delong and Boykov [1]. The difference
MULTI-REGION SEGMENTATION USING GRAPH-CUTS Johannes Ulén Abstract This project deals with multi-region segmenation using graph-cuts and is mainly based on a paper by Delong and Boykov [1]. The difference
CS6375: Machine Learning Gautam Kunapuli. Mid-Term Review
 Gautam Kunapuli Machine Learning Data is identically and independently distributed Goal is to learn a function that maps to Data is generated using an unknown function Learn a hypothesis that minimizes
Gautam Kunapuli Machine Learning Data is identically and independently distributed Goal is to learn a function that maps to Data is generated using an unknown function Learn a hypothesis that minimizes
Joint Classification-Regression Forests for Spatially Structured Multi-Object Segmentation
 Joint Classification-Regression Forests for Spatially Structured Multi-Object Segmentation Ben Glocker 1, Olivier Pauly 2,3, Ender Konukoglu 1, Antonio Criminisi 1 1 Microsoft Research, Cambridge, UK 2
Joint Classification-Regression Forests for Spatially Structured Multi-Object Segmentation Ben Glocker 1, Olivier Pauly 2,3, Ender Konukoglu 1, Antonio Criminisi 1 1 Microsoft Research, Cambridge, UK 2
Boundary-aware Fully Convolutional Network for Brain Tumor Segmentation
 Boundary-aware Fully Convolutional Network for Brain Tumor Segmentation Haocheng Shen, Ruixuan Wang, Jianguo Zhang, and Stephen J. McKenna Computing, School of Science and Engineering, University of Dundee,
Boundary-aware Fully Convolutional Network for Brain Tumor Segmentation Haocheng Shen, Ruixuan Wang, Jianguo Zhang, and Stephen J. McKenna Computing, School of Science and Engineering, University of Dundee,
The exam is closed book, closed notes except your one-page (two-sided) cheat sheet.
 CS 189 Spring 2015 Introduction to Machine Learning Final You have 2 hours 50 minutes for the exam. The exam is closed book, closed notes except your one-page (two-sided) cheat sheet. No calculators or
CS 189 Spring 2015 Introduction to Machine Learning Final You have 2 hours 50 minutes for the exam. The exam is closed book, closed notes except your one-page (two-sided) cheat sheet. No calculators or
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2014 October 07.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
Evaluation of Fourier Transform Coefficients for The Diagnosis of Rheumatoid Arthritis From Diffuse Optical Tomography Images
 Evaluation of Fourier Transform Coefficients for The Diagnosis of Rheumatoid Arthritis From Diffuse Optical Tomography Images Ludguier D. Montejo *a, Jingfei Jia a, Hyun K. Kim b, Andreas H. Hielscher
Evaluation of Fourier Transform Coefficients for The Diagnosis of Rheumatoid Arthritis From Diffuse Optical Tomography Images Ludguier D. Montejo *a, Jingfei Jia a, Hyun K. Kim b, Andreas H. Hielscher
Evaluating Classifiers
 Evaluating Classifiers Reading for this topic: T. Fawcett, An introduction to ROC analysis, Sections 1-4, 7 (linked from class website) Evaluating Classifiers What we want: Classifier that best predicts
Evaluating Classifiers Reading for this topic: T. Fawcett, An introduction to ROC analysis, Sections 1-4, 7 (linked from class website) Evaluating Classifiers What we want: Classifier that best predicts
Learning-based Neuroimage Registration
 Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Neighbourhood Approximation Forests
 Neighbourhood Approximation Forests Ender Konukoglu, Ben Glocker, Darko Zikic and Antonio Criminisi Microsoft Research Cambridge Abstract. Methods that leverage neighbourhood structures in highdimensional
Neighbourhood Approximation Forests Ender Konukoglu, Ben Glocker, Darko Zikic and Antonio Criminisi Microsoft Research Cambridge Abstract. Methods that leverage neighbourhood structures in highdimensional
Adaptive Dictionary Learning For Competitive Classification Of Multiple Sclerosis Lesions
 Adaptive Dictionary Learning For Competitive Classification Of Multiple Sclerosis Lesions Hrishikesh Deshpande, Pierre Maurel, Christian Barillot To cite this version: Hrishikesh Deshpande, Pierre Maurel,
Adaptive Dictionary Learning For Competitive Classification Of Multiple Sclerosis Lesions Hrishikesh Deshpande, Pierre Maurel, Christian Barillot To cite this version: Hrishikesh Deshpande, Pierre Maurel,
CS 584 Data Mining. Classification 3
 CS 584 Data Mining Classification 3 Today Model evaluation & related concepts Additional classifiers Naïve Bayes classifier Support Vector Machine Ensemble methods 2 Model Evaluation Metrics for Performance
CS 584 Data Mining Classification 3 Today Model evaluation & related concepts Additional classifiers Naïve Bayes classifier Support Vector Machine Ensemble methods 2 Model Evaluation Metrics for Performance
8. Tree-based approaches
 Foundations of Machine Learning École Centrale Paris Fall 2015 8. Tree-based approaches Chloé-Agathe Azencott Centre for Computational Biology, Mines ParisTech chloe agathe.azencott@mines paristech.fr
Foundations of Machine Learning École Centrale Paris Fall 2015 8. Tree-based approaches Chloé-Agathe Azencott Centre for Computational Biology, Mines ParisTech chloe agathe.azencott@mines paristech.fr
Regularization and model selection
 CS229 Lecture notes Andrew Ng Part VI Regularization and model selection Suppose we are trying select among several different models for a learning problem. For instance, we might be using a polynomial
CS229 Lecture notes Andrew Ng Part VI Regularization and model selection Suppose we are trying select among several different models for a learning problem. For instance, we might be using a polynomial
Information Management course
 Università degli Studi di Milano Master Degree in Computer Science Information Management course Teacher: Alberto Ceselli Lecture 20: 10/12/2015 Data Mining: Concepts and Techniques (3 rd ed.) Chapter
Università degli Studi di Milano Master Degree in Computer Science Information Management course Teacher: Alberto Ceselli Lecture 20: 10/12/2015 Data Mining: Concepts and Techniques (3 rd ed.) Chapter
A novel supervised learning algorithm and its use for Spam Detection in Social Bookmarking Systems
 A novel supervised learning algorithm and its use for Spam Detection in Social Bookmarking Systems Anestis Gkanogiannis and Theodore Kalamboukis Department of Informatics Athens University of Economics
A novel supervised learning algorithm and its use for Spam Detection in Social Bookmarking Systems Anestis Gkanogiannis and Theodore Kalamboukis Department of Informatics Athens University of Economics
Equation to LaTeX. Abhinav Rastogi, Sevy Harris. I. Introduction. Segmentation.
 Equation to LaTeX Abhinav Rastogi, Sevy Harris {arastogi,sharris5}@stanford.edu I. Introduction Copying equations from a pdf file to a LaTeX document can be time consuming because there is no easy way
Equation to LaTeX Abhinav Rastogi, Sevy Harris {arastogi,sharris5}@stanford.edu I. Introduction Copying equations from a pdf file to a LaTeX document can be time consuming because there is no easy way
Homework. Gaussian, Bishop 2.3 Non-parametric, Bishop 2.5 Linear regression Pod-cast lecture on-line. Next lectures:
 Homework Gaussian, Bishop 2.3 Non-parametric, Bishop 2.5 Linear regression 3.0-3.2 Pod-cast lecture on-line Next lectures: I posted a rough plan. It is flexible though so please come with suggestions Bayes
Homework Gaussian, Bishop 2.3 Non-parametric, Bishop 2.5 Linear regression 3.0-3.2 Pod-cast lecture on-line Next lectures: I posted a rough plan. It is flexible though so please come with suggestions Bayes
INF 4300 Classification III Anne Solberg The agenda today:
 INF 4300 Classification III Anne Solberg 28.10.15 The agenda today: More on estimating classifier accuracy Curse of dimensionality and simple feature selection knn-classification K-means clustering 28.10.15
INF 4300 Classification III Anne Solberg 28.10.15 The agenda today: More on estimating classifier accuracy Curse of dimensionality and simple feature selection knn-classification K-means clustering 28.10.15
Global Probability of Boundary
 Global Probability of Boundary Learning to Detect Natural Image Boundaries Using Local Brightness, Color, and Texture Cues Martin, Fowlkes, Malik Using Contours to Detect and Localize Junctions in Natural
Global Probability of Boundary Learning to Detect Natural Image Boundaries Using Local Brightness, Color, and Texture Cues Martin, Fowlkes, Malik Using Contours to Detect and Localize Junctions in Natural
