BIOM Documentation. Release dev. The BIOM Project
|
|
|
- Austin Norton
- 6 years ago
- Views:
Transcription
1 BIOM Documentation Release dev The BIOM Project Dec 13, 2017
2
3 Contents 1 Projects using the BIOM format 3 2 Contents BIOM Documentation The biom file format Tips and FAQs regarding the BIOM file format Converting between file formats Adding sample and observation metadata to biom files Summarizing BIOM tables The BIOM Format License BIOM version 35 4 Installing the biom-format Python package 37 5 Installing the biom R package 39 6 Citing the BIOM project 41 7 Development team 43 i
4 ii
5 The BIOM file format (canonically pronounced biome) is designed to be a general-use format for representing biological sample by observation contingency tables. BIOM is a recognized standard for the Earth Microbiome Project and is a Genomics Standards Consortium supported project. The BIOM format is designed for general use in broad areas of comparative -omics. For example, in marker-gene surveys, the primary use of this format is to represent OTU tables: the observations in this case are OTUs and the matrix contains counts corresponding to the number of times each OTU is observed in each sample. With respect to metagenome data, this format would be used to represent metagenome tables: the observations in this case might correspond to SEED subsystems, and the matrix would contain counts corresponding to the number of times each subsystem is observed in each metagenome. Similarly, with respect to genome data, this format may be used to represent a set of genomes: the observations in this case again might correspond to SEED subsystems, and the counts would correspond to the number of times each subsystem is observed in each genome. The BIOM project consists of the following components: definition of the BIOM file format; command line interface (CLI) for working with BIOM files, including converting between file formats, adding metadata to BIOM files, and summarizing BIOM files (run biom to see the full list of commands); application programming interface (API) for working with BIOM files in multiple programming languages (including Python and R). The biom-format package provides a command line interface and Python API for working with BIOM files. The rest of this site contains details about the BIOM file format (which is independent of the API) and the Python biom-format package. For more details about the R API, please see the CRAN biom package. Contents 1
6 2 Contents
7 CHAPTER 1 Projects using the BIOM format QIIME MG-RAST PICRUSt Mothur phyloseq MEGAN VAMPS metagenomeseq Phinch RDP Classifier USEARCH PhyloToAST EBI Metagenomics GCModeller MetaPhlAn 2 If you are using BIOM in your project, and would like your project to be listed, please submit a pull request to the BIOM project. More information on submitting pull requests can be found here. 3
8 4 Chapter 1. Projects using the BIOM format
9 CHAPTER 2 Contents 2.1 BIOM Documentation These pages provide format specifications and API information for the BIOM table objects The biom file format The BIOM project consists of two independent tools: the biom-format software package, which contains software tools for working with BIOM-formatted files and the tables they represent; and the BIOM file format. As of the software version and the 1.0 file format version, the version of the software and the file format are independent of one another. Version specific documentation of the file formats can be found on the following pages. The biom file format: Version 1.0 The biom format is based on JSON to provide the overall structure for the format. JSON is a widely supported format with native parsers available within many programming languages. Required top-level fields: id null) format format_url type : <string or null> a field that can be used to id a table (or : <string> The name and version of the current biom format : <url> A string with a static URL providing format details : <string> Table type (a controlled vocabulary) Acceptable values: "OTU table" "Pathway table" "Function table" "Ortholog table" "Gene table" "Metabolite table" "Taxon table" 5
10 generated_by : <string> Package and revision that built the table date : <datetime> Date the table was built (ISO 8601 format) rows : <list of objects> An ORDERED list of obj describing the rows (explained in detail below) columns : <list of objects> An ORDERED list of obj describing the columns (explained in detail below) matrix_type : <string> Type of matrix data representation (a controlled vocabulary) Acceptable values: "sparse" : only non-zero values are specified "dense" : every element must be specified matrix_element_type : Value type in matrix (a controlled vocabulary) Acceptable values: "int" : integer "float" : floating point "unicode" : unicode string shape : <list of ints>, the number of rows and number of columns in data data : <list of lists>, counts of observations by sample if matrix_type is "sparse", [[row, column, value], [row, column, value],...] if matrix_type is "dense", [[value, value, value,...], [value, value, value,...],...] Optional top-level fields: comment : <string> A free text field containing any information that you feel is relevant (or just feel like sharing) The rows value is an ORDERED list of objects where each object corresponds to a single row in the matrix. Each object can currently store arbitrary keys, although this might become restricted based on table type. Each object must provide, at the minimum: id metadata pairs : <string> an arbitrary UNIQUE identifier : <an object or null> A object containing key, value metadata The columns value is an ORDERED list of objects where each object corresponds to a single column in the matrix. Each object can currently store arbitrary keys, although this might become restricted based on table type. Each object must provide, at the minimum: id metadata pairs : <string> an arbitrary UNIQUE identifier : <an object or null> A object containing key, value metadata Example biom files Below are examples of minimal and rich biom files in both sparse and dense formats. To decide which of these you should generate for new data types, see the section on Tips and FAQs regarding the BIOM file format. 6 Chapter 2. Contents
11 Minimal sparse OTU table "id":null, "format": "Biological Observation Matrix dev", "format_url": " ", "type": "OTU table", "generated_by": "QIIME revision dev", "date": " T19:00:00", "rows":[ "id":"gg_otu_1", "metadata":null, "id":"gg_otu_2", "metadata":null, "id":"gg_otu_3", "metadata":null, "id":"gg_otu_4", "metadata":null, "id":"gg_otu_5", "metadata":null ], "columns": [ "id":"sample1", "metadata":null, "id":"sample2", "metadata":null, "id":"sample3", "metadata":null, "id":"sample4", "metadata":null, "id":"sample5", "metadata":null, "id":"sample6", "metadata":null ], "matrix_type": "sparse", "matrix_element_type": "int", "shape": [5, 6], "data":[[0,2,1], [1,0,5], [1,1,1], [1,3,2], [1,4,3], [1,5,1], [2,2,1], [2,3,4], [2,4,2], [3,0,2], [3,1,1], [3,2,1], [3,5,1], [4,1,1], [4,2,1] ] Minimal dense OTU table "id":null, "format": "Biological Observation Matrix dev", "format_url": " ", "type": "OTU table", "generated_by": "QIIME revision dev", "date": " T19:00:00", 2.1. BIOM Documentation 7
12 "rows":[ "id":"gg_otu_1", "metadata":null, "id":"gg_otu_2", "metadata":null, "id":"gg_otu_3", "metadata":null, "id":"gg_otu_4", "metadata":null, "id":"gg_otu_5", "metadata":null ], "columns": [ "id":"sample1", "metadata":null, "id":"sample2", "metadata":null, "id":"sample3", "metadata":null, "id":"sample4", "metadata":null, "id":"sample5", "metadata":null, "id":"sample6", "metadata":null ], "matrix_type": "dense", "matrix_element_type": "int", "shape": [5,6], "data": [[0,0,1,0,0,0], [5,1,0,2,3,1], [0,0,1,4,2,0], [2,1,1,0,0,1], [0,1,1,0,0,0]] Rich sparse OTU table "id":null, "format": "Biological Observation Matrix dev", "format_url": " "type": "OTU table", "generated_by": "QIIME revision dev", "date": " T19:00:00", "rows":[ "id":"gg_otu_1", "metadata":"taxonomy":["k Bacteria", "p Proteobacteria", "c Gammaproteobacteria", "o Enterobacteriales", "f Enterobacteriaceae", "g Escherichia", "s "], "id":"gg_otu_2", "metadata":"taxonomy":["k Bacteria", "p Cyanobacteria", "c Nostocophycideae", "o Nostocales", "f Nostocaceae", "g Dolichospermum", "s "], "id":"gg_otu_3", "metadata":"taxonomy":["k Archaea", "p Euryarchaeota", "c Methanomicrobia", "o Methanosarcinales", "f Methanosarcinaceae", "g Methanosarcina", "s "], "id":"gg_otu_4", "metadata":"taxonomy":["k Bacteria", "p Firmicutes", "c Clostridia", "o Halanaerobiales", "f Halanaerobiaceae", "g Halanaerobium", "s Halanaerobiumsaccharolyticum"], "id":"gg_otu_5", "metadata":"taxonomy":["k Bacteria", "p Proteobacteria", "c Gammaproteobacteria", "o Enterobacteriales", "f Enterobacteriaceae", "g Escherichia", "s "] ], "columns":[ "id":"sample1", "metadata": "BarcodeSequence":"CGCTTATCGAGA", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"gut", "Description":"human gut", 8 Chapter 2. Contents
13 "id":"sample2", "metadata": "BarcodeSequence":"CATACCAGTAGC", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"gut", "Description":"human gut", "id":"sample3", "metadata": "BarcodeSequence":"CTCTCTACCTGT", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"gut", "Description":"human gut", "id":"sample4", "metadata": "BarcodeSequence":"CTCTCGGCCTGT", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"skin", "Description":"human skin", "id":"sample5", "metadata": "BarcodeSequence":"CTCTCTACCAAT", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"skin", "Description":"human skin", "id":"sample6", "metadata": "BarcodeSequence":"CTAACTACCAAT", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"skin", "Description":"human skin" ], "matrix_type": "sparse", "matrix_element_type": "int", "shape": [5, 6], "data":[[0,2,1], [1,0,5], [1,1,1], [1,3,2], [1,4,3], [1,5,1], [2,2,1], [2,3,4], [2,5,2], [3,0,2], [3,1,1], [3,2,1], [3,5,1], [4,1,1], [4,2,1] ] Rich dense OTU table "id":null, "format": "Biological Observation Matrix dev", "format_url": " "type": "OTU table", "generated_by": "QIIME revision dev", "date": " T19:00:00", 2.1. BIOM Documentation 9
14 "rows":[ "id":"gg_otu_1", "metadata":"taxonomy":["k Bacteria", "p Proteobacteria", "c Gammaproteobacteria", "o Enterobacteriales", "f Enterobacteriaceae", "g Escherichia", "s "], "id":"gg_otu_2", "metadata":"taxonomy":["k Bacteria", "p Cyanobacteria", "c Nostocophycideae", "o Nostocales", "f Nostocaceae", "g Dolichospermum", "s "], "id":"gg_otu_3", "metadata":"taxonomy":["k Archaea", "p Euryarchaeota", "c Methanomicrobia", "o Methanosarcinales", "f Methanosarcinaceae", "g Methanosarcina", "s "], "id":"gg_otu_4", "metadata":"taxonomy":["k Bacteria", "p Firmicutes", "c Clostridia", "o Halanaerobiales", "f Halanaerobiaceae", "g Halanaerobium", "s Halanaerobiumsaccharolyticum"], "id":"gg_otu_5", "metadata":"taxonomy":["k Bacteria", "p Proteobacteria", "c Gammaproteobacteria", "o Enterobacteriales", "f Enterobacteriaceae", "g Escherichia", "s "] ], "columns":[ "id":"sample1", "metadata": "BarcodeSequence":"CGCTTATCGAGA", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"gut", "Description":"human gut", "id":"sample2", "metadata": "BarcodeSequence":"CATACCAGTAGC", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"gut", "Description":"human gut", "id":"sample3", "metadata": "BarcodeSequence":"CTCTCTACCTGT", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"gut", "Description":"human gut", "id":"sample4", "metadata": "BarcodeSequence":"CTCTCGGCCTGT", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"skin", "Description":"human skin", "id":"sample5", "metadata": "BarcodeSequence":"CTCTCTACCAAT", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"skin", "Description":"human skin", "id":"sample6", "metadata": "BarcodeSequence":"CTAACTACCAAT", "LinkerPrimerSequence":"CATGCTGCCTCCCGTAGGAGT", "BODY_SITE":"skin", "Description":"human skin" ], "matrix_type": "dense", "matrix_element_type": "int", "shape": [5,6], "data": [[0,0,1,0,0,0], [5,1,0,2,3,1], [0,0,1,4,2,0], [2,1,1,0,0,1], [0,1,1,0,0,0]] 10 Chapter 2. Contents
15 The biom file format: Version 2.0 The biom format is based on HDF5 to provide the overall structure for the format. HDF5 is a widely supported format with native parsers available within many programming languages. Required top-level attributes: id null) format format-url type generated-by creation-date nnz shape : <string or null> a field that can be used to id a table (or : <string> The name and version of the current biom format : <url> A string with a static URL providing format details : <string> Table type (a controlled vocabulary) Acceptable values: "OTU table" "Pathway table" "Function table" "Ortholog table" "Gene table" "Metabolite table" "Taxon table" : <string> Package and revision that built the table : <datetime> Date the table was built (ISO 8601 format) : <int> The number of non-zero elements in the table : <list of ints>, the number of rows and number of columns in data Required groups: observation/ : The HDF5 group that contains observation specific information and an observation oriented view of the data observation/matrix : The HDF5 group that contains matrix data oriented for observation-wise operations (e.g., in compressed sparse row format) sample/ : The HDF5 group that contains sample specific information and a sample oriented data oriented view of the data sample/matrix : The HDF5 group that contains matrix data oriented for sample- wise operations (e.g., in compressed sparse column format) Required datasets: observation/ids : <string> or <variable length string> A (N,) dataset of the observation IDs, where N is the total number of IDs observation/matrix/data : <float64> A (nnz,) dataset containing the actual matrix data observation/matrix/indices : <int32> A (nnz,) dataset containing the column indices (e.g., maps into samples/ids) observation/matrix/indptr : <int32> A (M+1,) dataset containing the compressed row offsets sample/ids : <string> or <variable length string> A (M,) dataset of the sample IDs, where M is the total number of IDs sample/matrix/data : <float64> A (nnz,) dataset containing the actual matrix data sample/matrix/indices : <int32> A (nnz,) dataset containing the row indices (e.g., maps into observation/ids) sample/matrix/indptr : <int32> A (N+1,) dataset containing the compressed column offsets Optional datasets: 2.1. BIOM Documentation 11
16 observation/metadata : <variable length string or null> If specified, a (1,) dataset containing a JSON-string representation of the metadata sample/metadata : <variable length string or null> If specified, a (1,) dataset containing a JSON-string representation of the metadata The metadata for each axis (observation and sample) are described with JSON. The required structure, if the metadata are specified, is a list of objects, where the list is in index order with respect to the axis (e.g, the object at element 0 corresponds to ID 0 for the given axis). Any metadata that corresponds to the ID, such as taxonomy, can be represented in the object. For instance, the following JSON string describes taxonomy for three IDs: Metadata description: [ "taxonomy": ["k Bacteria", "p Proteobacteria", "c Gammaproteobacteria", "o Enterobacteriales", "f Enterobacteriaceae", "g Escherichia", "s "], "taxonomy": ["k Bacteria", "p Cyanobacteria", "c Nostocophycideae", "o Nostocales", "f Nostocaceae", "g Dolichospermum", "s "], "taxonomy": ["k Archaea", "p Euryarchaeota", "c Methanomicrobia", "o Methanosarcinales", "f Methanosarcinaceae", "g Methanosarcina", "s "] ] Example biom files Below are examples of minimal and rich biom files. BIOM 2.0 OTU table in the HDF5 data description langauge (DDL) HDF5 "rich_sparse_otu_table_hdf5.biom" GROUP "/" ATTRIBUTE "creation-date" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SCALAR (0): " T14:50: " ATTRIBUTE "format-url" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SCALAR (0): " ATTRIBUTE "format-version" 12 Chapter 2. Contents
17 DATATYPE H5T_STD_I64LE DATASPACE SIMPLE ( 2 ) / ( 2 ) (0): 2, 0 ATTRIBUTE "generated-by" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE (0): "example" SCALAR ATTRIBUTE "id" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SCALAR (0): "No Table ID" ATTRIBUTE "nnz" DATATYPE H5T_STD_I64LE DATASPACE SCALAR (0): 15 ATTRIBUTE "shape" DATATYPE H5T_STD_I64LE DATASPACE SIMPLE ( 2 ) / ( 2 ) (0): 5, 6 ATTRIBUTE "type" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SCALAR (0): "otu table" GROUP "observation" DATASET "ids" 2.1. BIOM Documentation 13
18 DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 5 ) / ( 5 ) (0): "GG_OTU_1", "GG_OTU_2", "GG_OTU_3", "GG_OTU_4", "GG_OTU_5" GROUP "matrix" DATASET "data" DATATYPE H5T_IEEE_F64LE DATASPACE SIMPLE ( 15 ) / ( 15 ) (0): 1, 5, 1, 2, 3, 1, 1, 4, 2, 2, 1, 1, 1, 1, 1 DATASET "indices" DATATYPE H5T_STD_I32LE DATASPACE SIMPLE ( 15 ) / ( 15 ) (0): 2, 0, 1, 3, 4, 5, 2, 3, 5, 0, 1, 2, 5, 1, 2 DATASET "indptr" DATATYPE H5T_STD_I32LE DATASPACE SIMPLE ( 6 ) / ( 6 ) (0): 0, 1, 6, 9, 13, 15 DATASET "metadata" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 1 ) / ( 1 ) (0): "["taxonomy": ["k Bacteria", "p Proteobacteria", "c Gammaproteobacteria", "o Enterobacteriales", "f Enterobacteriaceae", "g Escherichia", "s "], "taxonomy": ["k Bacteria", "p Cyanobacteria", "c Nostocophycideae", "o Nostocales", "f Nostocaceae", "g Dolichospermum", "s "], "taxonomy": ["k Archaea", "p Euryarchaeota", "c Methanomicrobia", "o Methanosarcinales", "f Methanosarcinaceae", "g Methanosarcina", "s "], "taxonomy": ["k Bacteria", "p Firmicutes", "c Clostridia", "o Halanaerobiales", "f Halanaerobiaceae", "g Halanaerobium", "s Halanaerobiumsaccharolyticum"], "taxonomy": ["k Bacteria", "p Proteobacteria", "c Gammaproteobacteria", "o Enterobacteriales", "f Enterobacteriaceae", "g Escherichia", "s "]]" GROUP "sample" DATASET "ids" 14 Chapter 2. Contents
19 DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 6 ) / ( 6 ) (0): "Sample1", "Sample2", "Sample3", "Sample4", "Sample5", (5): "Sample6" GROUP "matrix" DATASET "data" DATATYPE H5T_IEEE_F64LE DATASPACE SIMPLE ( 15 ) / ( 15 ) (0): 5, 2, 1, 1, 1, 1, 1, 1, 1, 2, 4, 3, 1, 2, 1 DATASET "indices" DATATYPE H5T_STD_I32LE DATASPACE SIMPLE ( 15 ) / ( 15 ) (0): 1, 3, 1, 3, 4, 0, 2, 3, 4, 1, 2, 1, 1, 2, 3 DATASET "indptr" DATATYPE H5T_STD_I32LE DATASPACE SIMPLE ( 7 ) / ( 7 ) (0): 0, 2, 5, 9, 11, 12, 15 DATASET "metadata" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 1 ) / ( 1 ) (0): "["LinkerPrimerSequence": "CATGCTGCCTCCCGTAGGAGT", "BarcodeSequence": "CGCTTATCGAGA", "Description": "human gut", "BODY_SITE": "gut", "LinkerPrimerSequence": "CATGCTGCCTCCCGTAGGAGT", "BarcodeSequence": "CATACCAGTAGC", "Description": "human gut", "BODY_SITE": "gut", "LinkerPrimerSequence": "CATGCTGCCTCCCGTAGGAGT", "BarcodeSequence": "CTCTCTACCTGT", "Description": "human gut", "BODY_SITE": "gut", "LinkerPrimerSequence": "CATGCTGCCTCCCGTAGGAGT", "BarcodeSequence": "CTCTCGGCCTGT", "Description": "human skin", "BODY_SITE": "skin", "LinkerPrimerSequence": "CATGCTGCCTCCCGTAGGAGT", "BarcodeSequence": "CTCTCTACCAAT ", "Description": "human skin", "BODY_SITE": "skin", "LinkerPrimerSequence": "CATGCTGCCTCCCGTAGGAGT", "BarcodeSequence": "CTAACTACCAAT", "Description": "human skin", "BODY_SITE": "skin"]" 2.1. BIOM Documentation 15
20 The biom file format: Version 2.1 The biom format is based on HDF5 to provide the overall structure for the format. HDF5 is a widely supported format with native parsers available within many programming languages. Required top-level attributes: id null) type format-url format-version generated-by creation-date shape data nnz : <string or null> a field that can be used to id a table (or : <string> Table type (a controlled vocabulary) Acceptable values: "OTU table" "Pathway table" "Function table" "Ortholog table" "Gene table" "Metabolite table" "Taxon table" : <url> A string with a static URL providing format details : <tuple> The version of the current biom format, major and minor : <string> Package and revision that built the table : <datetime> Date the table was built (ISO 8601 format) : <list of ints>, the number of rows and number of columns in : <int> The number of non-zero elements in the table Required groups: observation/ : The HDF5 group that contains observation specific information and an observation oriented view of the data observation/matrix : The HDF5 group that contains matrix data oriented for observation-wise operations (e.g., in compressed sparse row format) observation/metadata : The HDF5 group that contains observation specific metadata information observation/group-metadata : The HDF5 group that contains observation specific group metadata information (e.g., phylogenetic tree) sample/ : The HDF5 group that contains sample specific information and a sample oriented data oriented view of the data sample/matrix : The HDF5 group that contains matrix data oriented for sample-wise operations (e.g., in compressed sparse column format) sample/metadata : The HDF5 group that contains sample specific metadata information sample/group-metadata : The HDF5 group that contains sample specific group metadata information (e.g., relationships between samples) Required datasets: observation/ids : <string> or <variable length string> A (N,) dataset of the observation IDs, where N is the total number of IDs observation/matrix/data : <float64> A (nnz,) dataset containing the actual matrix data observation/matrix/indices : <int32> A (nnz,) dataset containing the column indices (e.g., maps into samples/ids) observation/matrix/indptr : <int32> A (M+1,) dataset containing the compressed row offsets 16 Chapter 2. Contents
21 sample/ids : <string> or <variable length string> A (M,) dataset of the sample IDs, where M is the total number of IDs sample/matrix/data : <float64> A (nnz,) dataset containing the actual matrix data sample/matrix/indices : <int32> A (nnz,) dataset containing the row indices (e.g., maps into observation/ids) sample/matrix/indptr : <int32> A (N+1,) dataset containing the compressed column offsets Under the observation/metadata and sample/metadata groups, the user can specify an arbitrary number of datasets that represents a metadata category for that axis. The expected structure for each of these metadata datasets is a list of atomic type objects (int, float, str,... ) where the index order of the list corresponds to the index order of the relevant axis IDs. Special complex metadata fields have been defined, and they are stored in a specific way. Currently, the available special metadata fields are: observation/metadata/taxonomy : <string> or <variable length string> A (N,?) dataset containing the taxonomy names assigned to the observation observation/metadata/kegg_pathways : <string> or <variable length string> A (N,?) dataset containing the KEGG Pathways assigned to the observation observation/metadata/collapsed_ids : <string> or <variable length string> A (N,?) dataset containing the observation ids of the original table that have been collapsed in the given observation sample/metadata/collapsed_ids : <string> or <variable length string> A (M,?) dataset containing the sample ids of the original table that have been collapsed in the given sample Under the observation/group-metadata and sample/group-metadata groups, the user can specify an arbitrary number of datasets that represents a relationship between the ids for that axis. The expected structure for each of these group metadata datasets is a single string or variable length string. Each of these datasets should have defined an attribute called data_type, which specifies how the string should be interpreted. One example of such group metadata dataset is observation/group-metadata/phylogeny, with the attribute observation/ group-metadata/phylogeny.attrs['data_type'] = "newick", which stores a single string with the newick format of the phylogenetic tree for the observations. Example biom files Below is an example of a rich biom file. BIOM 2.1 OTU table in the HDF5 data description language (DDL) HDF5 "examples/rich_sparse_otu_table_hdf5.biom" GROUP "/" ATTRIBUTE "creation-date" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SCALAR (0): " T16:16: " 2.1. BIOM Documentation 17
22 ATTRIBUTE "format-url" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SCALAR (0): " ATTRIBUTE "format-version" DATATYPE H5T_STD_I64LE DATASPACE SIMPLE ( 2 ) / ( 2 ) (0): 2, 1 ATTRIBUTE "generated-by" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE (0): "example" SCALAR ATTRIBUTE "id" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SCALAR (0): "No Table ID" ATTRIBUTE "nnz" DATATYPE H5T_STD_I64LE DATASPACE SCALAR (0): 15 ATTRIBUTE "shape" DATATYPE H5T_STD_I64LE DATASPACE SIMPLE ( 2 ) / ( 2 ) (0): 5, 6 ATTRIBUTE "type" 18 Chapter 2. Contents
23 DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SCALAR (0): "otu table" GROUP "observation" GROUP "group-metadata" DATASET "ids" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 5 ) / ( 5 ) (0): "GG_OTU_1", "GG_OTU_2", "GG_OTU_3", "GG_OTU_4", "GG_OTU_5" GROUP "matrix" DATASET "data" DATATYPE H5T_IEEE_F64LE DATASPACE SIMPLE ( 15 ) / ( 15 ) (0): 1, 5, 1, 2, 3, 1, 1, 4, 2, 2, 1, 1, 1, 1, 1 DATASET "indices" DATATYPE H5T_STD_I32LE DATASPACE SIMPLE ( 15 ) / ( 15 ) (0): 2, 0, 1, 3, 4, 5, 2, 3, 5, 0, 1, 2, 5, 1, 2 DATASET "indptr" DATATYPE H5T_STD_I32LE DATASPACE SIMPLE ( 6 ) / ( 6 ) (0): 0, 1, 6, 9, 13, 15 GROUP "metadata" DATASET "taxonomy" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 5, 7 ) / ( 5, 7 ) 2.1. BIOM Documentation 19
24 (0,0): "k Bacteria", "p Proteobacteria", (0,2): "c Gammaproteobacteria", "o Enterobacteriales", (0,4): "f Enterobacteriaceae", "g Escherichia", "s ", (1,0): "k Bacteria", "p Cyanobacteria", "c Nostocophycideae", (1,3): "o Nostocales", "f Nostocaceae", "g Dolichospermum", (1,6): "s ", (2,0): "k Archaea", "p Euryarchaeota", "c Methanomicrobia", (2,3): "o Methanosarcinales", "f Methanosarcinaceae", (2,5): "g Methanosarcina", "s ", (3,0): "k Bacteria", "p Firmicutes", "c Clostridia", (3,3): "o Halanaerobiales", "f Halanaerobiaceae", (3,5): "g Halanaerobium", "s Halanaerobiumsaccharolyticum", (4,0): "k Bacteria", "p Proteobacteria", (4,2): "c Gammaproteobacteria", "o Enterobacteriales", (4,4): "f Enterobacteriaceae", "g Escherichia", "s " GROUP "sample" GROUP "group-metadata" DATASET "ids" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 6 ) / ( 6 ) (0): "Sample1", "Sample2", "Sample3", "Sample4", "Sample5", (5): "Sample6" GROUP "matrix" DATASET "data" DATATYPE H5T_IEEE_F64LE DATASPACE SIMPLE ( 15 ) / ( 15 ) (0): 5, 2, 1, 1, 1, 1, 1, 1, 1, 2, 4, 3, 1, 2, 1 DATASET "indices" DATATYPE H5T_STD_I32LE DATASPACE SIMPLE ( 15 ) / ( 15 ) (0): 1, 3, 1, 3, 4, 0, 2, 3, 4, 1, 2, 1, 1, 2, 3 DATASET "indptr" DATATYPE H5T_STD_I32LE DATASPACE SIMPLE ( 7 ) / ( 7 ) (0): 0, 2, 5, 9, 11, 12, Chapter 2. Contents
25 GROUP "metadata" DATASET "BODY_SITE" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_UTF8; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 6 ) / ( 6 ) (0): "gut", "gut", "gut", "skin", "skin", "skin" DATASET "BarcodeSequence" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_UTF8; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 6 ) / ( 6 ) (0): "CGCTTATCGAGA", "CATACCAGTAGC", "CTCTCTACCTGT", (3): "CTCTCGGCCTGT", "CTCTCTACCAAT", "CTAACTACCAAT" DATASET "Description" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_UTF8; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 6 ) / ( 6 ) (0): "human gut", "human gut", "human gut", "human skin", (4): "human skin", "human skin" DATASET "LinkerPrimerSequence" DATATYPE H5T_STRING STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_UTF8; CTYPE H5T_C_S1; DATASPACE SIMPLE ( 6 ) / ( 6 ) (0): "CATGCTGCCTCCCGTAGGAGT", "CATGCTGCCTCCCGTAGGAGT", (2): "CATGCTGCCTCCCGTAGGAGT", "CATGCTGCCTCCCGTAGGAGT", (4): "CATGCTGCCTCCCGTAGGAGT", "CATGCTGCCTCCCGTAGGAGT" 2.1. BIOM Documentation 21
26 Release versions contain three integers in the following format: major-version.minor-version. micro-version. When -dev is appended to the end of a version string that indicates a development (or betweenrelease version). For example, dev would refer to the development version following the release Tips and FAQs regarding the BIOM file format Motivation for the BIOM format The BIOM format was motivated by several goals. First, to facilitate efficient handling and storage of large, sparse biological contingency tables; second, to support encapsulation of core study data (contingency table data and sample/observation metadata) in a single file; and third, to facilitate the use of these tables between tools that support this format (e.g., passing of data between QIIME, MG-RAST, and VAMPS.). Efficient handling and storage of very large tables In QIIME, we began hitting limitations with OTU table objects when working with thousands of samples and hundreds of thousands of OTUs. In the near future we expect that we ll be dealing with hundreds of thousands of samples in single analyses. The OTU table format up to QIIME involved a dense matrix: if an OTU was not observed in a given sample, that would be indicated with a zero. We now primarily represent OTU tables in a sparse format: if an OTU is not observed in a sample, there is no count for that OTU. The two ways of representing this data are exemplified here. A dense representation of an OTU table: OTU ID PC.354 PC.355 PC.356 OTU OTU OTU OTU A sparse representation of an OTU table: PC.354 OTU1 6 PC.354 OTU2 1 PC.356 OTU0 4 PC.356 OTU2 7 PC.356 OTU3 3 OTU table data tends to be sparse (e.g., greater than 90% of counts are zero, and frequently as many as 99% of counts are zero) in which case the latter format is more convenient to work with as it has a smaller memory footprint. In biom-format 1.0.0, both of these representations are supported in the biom-format project via dense and sparse Table types. In biom-format 2.x, only sparse is supported as in practice, dense was not useful especially with improved study designs that utilize increasing numbers of samples. Encapsulation of core study data (OTU table data and sample/otu metadata) in a single file Formats, such as JSON and HDF5, made more efficient storage of highly sparse data and allowed for storage of arbitrary amounts of sample and OTU metadata in a single file. Sample metadata corresponds to what is generally found in QIIME mapping files. At this stage inclusion of this information in the OTU table file is optional, but it may be useful for sharing these files with other QIIME users and for publishing or archiving results of analyses. OTU metadata (generally a taxonomic assignment for an OTU) is also optional. In contrast to the previous OTU table 22 Chapter 2. Contents
27 format, you can now store more than one OTU metadata value in this field, so for example you can score taxonomic assignments based on two different taxonomic assignment approaches. Facilitating the use of tables between tools that support this format Different tools, such as QIIME, MG-RAST, and VAMPS work with similar data structures that represent different types of data. An example of this is a metagenome table that could be generated by MG-RAST (where for example, columns are metagenomes and rows are functional categories). Exporting this data from MG-RAST in a suitable format will allow for the application of many of the QIIME tools to this data (such as generation of alpha rarefaction plots or beta diversity ordination plots). This new format is far more general than previous formats, so will support adoption by groups working with different data types and is already being integrated to support transfer of data between QIIME, MG-RAST, and VAMPS. File extension We recommend that BIOM files use the.biom extension Converting between file formats The convert command in the biom-format project can be used to convert between biom and tab-delimited table formats. This converting biom format tables to tab-delimited tables for easy viewing in programs such as Excel converting between sparse and dense biom formats (note: dense is only supported in biom-format 1.0.0) Note: The tab-delimited tables are commonly referred to as the classic format tables, while BIOM formatted tables are referred to as biom tables. General usage examples Convert a tab-delimited table to a HDF5 or JSON biom format. Note that you must specify the type of table here: biom convert -i table.txt -o table.from_txt_json.biom --table-type="otu table" --to- json biom convert -i table.txt -o table.from_txt_hdf5.biom --table-type="otu table" --to- hdf5 Convert biom format to tab-delimited table format: biom convert -i table.biom -o table.from_biom.txt --to-tsv Convert biom format to classic format, including the taxonomy observation metadata as the last column of the classic format table. Because the BIOM format can support an arbitrary number of observation (or sample) metadata entries, and the classic format can support only a single observation metadata entry, you must specify which of the observation metadata entries you want to include in the output table: biom convert -i table.biom -o table.from_biom_w_taxonomy.txt --to-tsv --header-key taxonomy 2.1. BIOM Documentation 23
28 Convert biom format to classic format, including the taxonomy observation metadata as the last column of the classic format table, but renaming that column as ConsensusLineage. This is useful when using legacy tools that require a specific name for the observation metadata column.: biom convert -i table.biom -o table.from_biom_w_consensuslineage.txt --to-tsv -- header-key taxonomy --output-metadata-id "ConsensusLineage" Special case usage examples Round-tripping between biom and tsv In specific cases, see this comment, it is still useful to convert our biom table to tsv so we can open in Excel, make some changes to the file and then convert back to biom. For this cases you should follow this steps: Convert from biom to txt: biom convert -i otu_table.biom -o otu_table.txt --to-tsv --header-key taxonomy Make your changes in Excel. Convert back to biom: biom convert -i otu_table.txt -o new_otu_table.biom --to-hdf5 --table-type="otu table" --process-obs-metadata taxonomy Converting QIIME and earlier OTU tables to BIOM format If you are converting a QIIME or earlier OTU table to BIOM format, there are a few steps to go through. First, for convenience, you might want to rename the ConsensusLineage column taxonomy. You can do this with the following command: sed 's/consensus Lineage/ConsensusLineage/' < otu_table.txt sed 's/consensuslineage/ taxonomy/' > otu_table.taxonomy.txt Then, you ll want to perform the conversion including a step to convert the taxonomy string from the classic OTU table to a taxonomy list, as it s represented in QIIME dev and later: biom convert -i otu_table.taxonomy.txt -o otu_table.from_txt.biom --table-type="otu table" --process-obs-metadata taxonomy --to-hdf Adding sample and observation metadata to biom files Frequently you ll have an existing BIOM file and want to add sample and/or observation metadata to it. For samples, metadata is frequently environmental or technical details about your samples: the subject that a sample was collected from, the ph of the sample, the PCR primers used to amplify DNA from the samples, etc. For observations, metadata is frequently a categorization of the observation: the taxonomy of an OTU, or the EC hierarchy of a gene. You can use the biom add-metadata command to add this information to an existing BIOM file. To get help with add-metadata you can call: biom add-metadata -h 24 Chapter 2. Contents
29 This command takes a BIOM file, and corresponding sample and/or observation mapping files. The following examples are used in the commands below. You can find these files in the biom-format/examples directory. Your BIOM file might look like the following: "id":null, "format": "1.0.0", "format_url": " "type": "OTU table", "generated_by": "some software package", "date": " T19:00:00", "rows":[ "id":"gg_otu_1", "metadata":null, "id":"gg_otu_2", "metadata":null, "id":"gg_otu_3", "metadata":null, "id":"gg_otu_4", "metadata":null, "id":"gg_otu_5", "metadata":null ], "columns": [ "id":"sample1", "metadata":null, "id":"sample2", "metadata":null, "id":"sample3", "metadata":null, "id":"sample4", "metadata":null, "id":"sample5", "metadata":null, "id":"sample6", "metadata":null ], "matrix_type": "sparse", "matrix_element_type": "int", "shape": [5, 6], "data":[[0,2,1], [1,0,5], [1,1,1], [1,3,2], [1,4,3], [1,5,1], [2,2,1], [2,3,4], [2,5,2], [3,0,2], [3,1,1], [3,2,1], [3,5,1], [4,1,1], [4,2,1] ] A sample metadata mapping file could then look like the following. Notice that there is an extra sample in here with respect to the above BIOM table. Any samples in the mapping file that are not in the BIOM file are ignored. #SampleID BarcodeSequence DOB # Some optional # comment lines... Sample1 AGCACGAGCCTA Sample2 AACTCGTCGATG Sample3 ACAGACCACTCA Sample4 ACCAGCGACTAG Sample5 AGCAGCACTTGT BIOM Documentation 25
30 Sample6 AGCAGCACAACT An observation metadata mapping file might look like the following. Notice that there is an extra observation in here with respect to the above BIOM table. Any observations in the mapping file that are not in the BIOM file are ignored. #OTUID taxonomy confidence # Some optional # comment lines GG_OTU_0 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f GG_OTU_1 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae GG_OTU_2 Root;k Bacteria GG_OTU_3 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae GG_OTU_4 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae GG_OTU_5 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae Adding metadata To add sample metadata to a BIOM file, you can run the following: biom add-metadata -i min_sparse_otu_table.biom -o table.w_smd.biom --sample-metadata- fp sam_md.txt To add observation metadata to a BIOM file, you can run the following: biom add-metadata -i min_sparse_otu_table.biom -o table.w_omd.biom --observation- metadata-fp obs_md.txt You can also combine these in a single command to add both observation and sample metadata: biom add-metadata -i min_sparse_otu_table.biom -o table.w_md.biom --observation- metadata-fp obs_md.txt --sample-metadata-fp sam_md.txt In the last case, the resulting BIOM file will look like the following: "columns": [ "id": "Sample1", "metadata": "BarcodeSequence": "AGCACGAGCCTA", "DOB": " ", "id": "Sample2", "metadata": "BarcodeSequence": "AACTCGTCGATG", "DOB": " ", 26 Chapter 2. Contents
31 ,,, "id": "Sample3", "metadata": "BarcodeSequence": "ACAGACCACTCA", "DOB": " " "id": "Sample4", "metadata": "BarcodeSequence": "ACCAGCGACTAG", "DOB": " " "id": "Sample5", "metadata": "BarcodeSequence": "AGCAGCACTTGT", "DOB": " " "id": "Sample6", "metadata": "BarcodeSequence": "AGCAGCACAACT", "DOB": " " ], "data": [ [0, 2, 1.0], [1, 0, 5.0], [1, 1, 1.0], [1, 3, 2.0], [1, 4, 3.0], [1, 5, 1.0], [2, 2, 1.0], [2, 3, 4.0], [2, 5, 2.0], [3, 0, 2.0], [3, 1, 1.0], [3, 2, 1.0], [3, 5, 1.0], [4, 1, 1.0], [4, 2, 1.0] ], "date": " T07:36: ", "format": "Biological Observation Matrix 1.0.0", "format_url": " "generated_by": "some software package", "id": null, "matrix_element_type": "float", "matrix_type": "sparse", "rows": [ "id": "GG_OTU_1", "metadata": "confidence": "0.665", "taxonomy": "Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae" 2.1. BIOM Documentation 27
32 ,, "id": "GG_OTU_2", "metadata": "confidence": "0.980", "taxonomy": "Root;k Bacteria" "id": "GG_OTU_3", "metadata": "confidence": "1.000", "taxonomy": "Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae", "id": "GG_OTU_4", "metadata": "confidence": "0.842", "taxonomy": "Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae", "id": "GG_OTU_5", "metadata": "confidence": "1.000", "taxonomy": "Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae" ], "shape": [5, 6], "type": "OTU table" Processing metadata while adding There are some additional parameters you can pass to this command for more complex processing. You can tell the command to process certain metadata column values as integers (--int-fields), floating point (i.e., decimal or real) numbers (--float-fields), or as hierarchical semicolon-delimited data (--sc-separated). biom add-metadata -i min_sparse_otu_table.biom -o table.w_md.biom --observation- metadata-fp obs_md.txt --sample-metadata-fp sam_md.txt --int-fields DOB --sc- separated taxonomy --float-fields confidence Here your resulting BIOM file will look like the following, where DOB values are now integers (compare to the above: they re not quoted now), confidence values are now floating point numbers (again, not quoted now), and taxonomy values are now lists where each entry is a taxonomy level, opposed to above where they appear as a single semi-colon-separated string. 28 Chapter 2. Contents
33 "columns": [ "id": "Sample1", "metadata": "BarcodeSequence": "AGCACGAGCCTA", "DOB": , "id": "Sample2", "metadata": "BarcodeSequence": "AACTCGTCGATG", "DOB": , "id": "Sample3", "metadata": "BarcodeSequence": "ACAGACCACTCA", "DOB": , "id": "Sample4", "metadata": "BarcodeSequence": "ACCAGCGACTAG", "DOB": , "id": "Sample5", "metadata": "BarcodeSequence": "AGCAGCACTTGT", "DOB": , "id": "Sample6", "metadata": "BarcodeSequence": "AGCAGCACAACT", "DOB": ], "data": [ [0, 2, 1.0], [1, 0, 5.0], [1, 1, 1.0], [1, 3, 2.0], [1, 4, 3.0], [1, 5, 1.0], [2, 2, 1.0], [2, 3, 4.0], [2, 5, 2.0], [3, 0, 2.0], [3, 1, 1.0], [3, 2, 1.0], 2.1. BIOM Documentation 29
34 [3, 5, 1.0], [4, 1, 1.0], [4, 2, 1.0] ], "date": " T07:30: ", "format": "Biological Observation Matrix 1.0.0", "format_url": " "generated_by": "some software package", "id": null, "matrix_element_type": "float", "matrix_type": "sparse", "rows": [ "id": "GG_OTU_1", "metadata": "confidence": 0.665, "taxonomy": ["Root", "k Bacteria", "p Firmicutes", "c Clostridia", "o Clostridiales", "f Lachnospiraceae"], "id": "GG_OTU_2", "metadata": "confidence": 0.98, "taxonomy": ["Root", "k Bacteria"], "id": "GG_OTU_3", "metadata": "confidence": 1.0, "taxonomy": ["Root", "k Bacteria", "p Firmicutes", "c Clostridia", "o Clostridiales", "f Lachnospiraceae"], "id": "GG_OTU_4", "metadata": "confidence": 0.842, "taxonomy": ["Root", "k Bacteria", "p Firmicutes", "c Clostridia", "o Clostridiales", "f Lachnospiraceae"], "id": "GG_OTU_5", "metadata": "confidence": 1.0, "taxonomy": ["Root", "k Bacteria", "p Firmicutes", "c Clostridia", "o Clostridiales", "f Lachnospiraceae"] ], "shape": [5, 6], "type": "OTU table" If you have multiple fields that you d like processed in one of these ways, you can pass a comma-separated list of field names (e.g., --float-fields confidence,ph). 30 Chapter 2. Contents
35 Renaming (or naming) metadata columns while adding You can also override the names of the metadata fields provided in the mapping files with the --observation-header and --sample-header parameters. This is useful if you want to rename metadata columns, or if metadata column headers aren t present in your metadata mapping file. If you pass either of these parameters, you must name all columns in order. If there are more columns in the metadata mapping file then there are headers, extra columns will be ignored (so this is also a useful way to select only the first n columns from your mapping file). For example, if you want to rename the DOB column in the sample metadata mapping you could do the following: biom add-metadata -i min_sparse_otu_table.biom -o table.w_smd.biom --sample-metadata- fp sam_md.txt --sample-header SampleID,BarcodeSequence,DateOfBirth If you have a mapping file without headers such as the following: GG_OTU_0 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f GG_OTU_1 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae GG_OTU_2 Root;k Bacteria GG_OTU_3 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae GG_OTU_4 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae GG_OTU_5 Root;k Bacteria;p Firmicutes;c Clostridia;o Clostridiales;f Lachnospiraceae you could name these while adding them as follows: biom add-metadata -i min_sparse_otu_table.biom -o table.w_omd.biom --observation- metadata-fp obs_md.txt --observation-header OTUID,taxonomy,confidence As a variation on the last command, if you only want to include the taxonomy column and exclude the confidence column, you could run: biom add-metadata -i min_sparse_otu_table.biom -o table.w_omd.biom --observation- metadata-fp obs_md.txt --observation-header OTUID,taxonomy --sc-separated taxonomy Summarizing BIOM tables If you have an existing BIOM file and want to compile a summary of the information in that table, you can use the biom summarize-table command. To get help with biom summarize-table you can call: biom summarize-table -h This command takes a BIOM file or gzipped BIOM file as input, and will print a summary of the count information on a per-sample basis to the new file specified by the -o parameter. The example file used in the commands below can be found in the biom-format/examples directory. Summarizing sample data To summarize the per-sample data in a BIOM file, you can run: 2.1. BIOM Documentation 31
36 biom summarize-table -i rich_sparse_otu_table.biom -o rich_sparse_otu_table_summary. txt The following information will be written to rich_sparse_otu_table_summary.txt: Num samples: 6 Num observations: 5 Total count: 27 Table density (fraction of non-zero values): Counts/sample summary: Min: 3.0 Max: 7.0 Median: Mean: Std. dev.: Sample Metadata Categories: LinkerPrimerSequence; BarcodeSequence; Description; BODY_ SITE Observation Metadata Categories: taxonomy Counts/sample detail: Sample5: 3.0 Sample2: 3.0 Sample6: 4.0 Sample3: 4.0 Sample4: 6.0 Sample1: 7.0 As you can see, general summary information about the table is provided, including the number of samples, the number of observations, the total count (i.e., the sum of all values in the table), and so on, followed by the per-sample counts. Summarizing sample data qualitatively To summarize the per-sample data in a BIOM file qualitatively, where the number of unique observations per sample (rather than the total count of observations per sample) are provided, you can run: biom summarize-table -i rich_sparse_otu_table.biom --qualitative -o rich_sparse_otu_ table_qual_summary.txt The following information will be written to rich_sparse_otu_table_qual_summary.txt: Num samples: 6 Num observations: 5 Observations/sample summary: Min: 1 Max: 4 Median: Mean: Std. dev.: Sample Metadata Categories: LinkerPrimerSequence; BarcodeSequence; Description; BODY_ SITE Observation Metadata Categories: taxonomy Observations/sample detail: Sample5: 1 Sample4: 2 32 Chapter 2. Contents
37 Sample1: 2 Sample6: 3 Sample2: The BIOM Format License The BIOM Format project is licensed under the terms of the Modified BSD License (also known as New or Revised BSD), as follows: Copyright (c) , The BIOM Format Development Team <gregcaporaso@gmail.com> All rights reserved. Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met: * Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer. * Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. * Neither the name of the BIOM Format Development Team nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission. THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE BIOM FORMAT DEVELOPMENT TEAM BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. The following banner should be used in any source code file to indicate the copyright and license terms: # # Copyright (c) , The BIOM Format Development Team. # # Distributed under the terms of the Modified BSD License. # # The full license is in the file COPYING.txt, distributed with this software. # The BIOM Format License 33
ASDF Definition. Release Lion Krischer, James Smith, Jeroen Tromp
 ASDF Definition Release 1.0.0 Lion Krischer, James Smith, Jeroen Tromp March 22, 2016 Contents 1 Introduction 2 1.1 Why introduce a new seismic data format?............................. 2 2 Big Picture
ASDF Definition Release 1.0.0 Lion Krischer, James Smith, Jeroen Tromp March 22, 2016 Contents 1 Introduction 2 1.1 Why introduce a new seismic data format?............................. 2 2 Big Picture
Package fst. December 18, 2017
 Type Package Package fst December 18, 2017 Title Lightning Fast Serialization of Data Frames for R Multithreaded serialization of compressed data frames using the 'fst' format. The 'fst' format allows
Type Package Package fst December 18, 2017 Title Lightning Fast Serialization of Data Frames for R Multithreaded serialization of compressed data frames using the 'fst' format. The 'fst' format allows
Introduction to HDF5
 Introduction to HDF5 Dr. Shelley L. Knuth Research Computing, CU-Boulder December 11, 2014 h/p://researchcompu7ng.github.io/meetup_fall_2014/ Download data used today from: h/p://neondataskills.org/hdf5/exploring-
Introduction to HDF5 Dr. Shelley L. Knuth Research Computing, CU-Boulder December 11, 2014 h/p://researchcompu7ng.github.io/meetup_fall_2014/ Download data used today from: h/p://neondataskills.org/hdf5/exploring-
Table of Contents Overview...2 Selecting Post-Processing: ColorMap...3 Overview of Options Copyright, license, warranty/disclaimer...
 1 P a g e ColorMap Post-Processing Plugin for OpenPolScope software ColorMap processing with Pol-Acquisition and Pol-Analyzer plugin v. 2.0, Last Modified: April 16, 2013; Revision 1.00 Copyright, license,
1 P a g e ColorMap Post-Processing Plugin for OpenPolScope software ColorMap processing with Pol-Acquisition and Pol-Analyzer plugin v. 2.0, Last Modified: April 16, 2013; Revision 1.00 Copyright, license,
ProgressBar Abstract
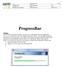 Doc type here 1(21) ProgressBar Abstract The WireFlow progressbar module is an easy way to add progress bars to an application. It is easy to customize the look of the displayed progress window, since
Doc type here 1(21) ProgressBar Abstract The WireFlow progressbar module is an easy way to add progress bars to an application. It is easy to customize the look of the displayed progress window, since
HYDRODESKTOP VERSION 1.1 BETA QUICK START GUIDE
 HYDRODESKTOP VERSION 1.1 BETA QUICK START GUIDE A guide to help you get started using this free and open source desktop application for discovering, accessing, and using hydrologic data. September 15,
HYDRODESKTOP VERSION 1.1 BETA QUICK START GUIDE A guide to help you get started using this free and open source desktop application for discovering, accessing, and using hydrologic data. September 15,
Intel Stress Bitstreams and Encoder (Intel SBE) 2017 AVS2 Release Notes (Version 2.3)
 Intel Stress Bitstreams and Encoder (Intel SBE) 2017 AVS2 Release Notes (Version 2.3) Overview Changes History Installation Package Contents Known Limitations Attributions Legal Information Overview The
Intel Stress Bitstreams and Encoder (Intel SBE) 2017 AVS2 Release Notes (Version 2.3) Overview Changes History Installation Package Contents Known Limitations Attributions Legal Information Overview The
HALCoGen TMS570LS31x Help: example_sci_uart_9600.c
 Page 1 of 6 example_sci_uart_9600.c This example code configures SCI and transmits a set of characters. An UART receiver can be used to receive this data. The scilin driver files should be generated with
Page 1 of 6 example_sci_uart_9600.c This example code configures SCI and transmits a set of characters. An UART receiver can be used to receive this data. The scilin driver files should be generated with
Open Source Used In TSP
 Open Source Used In TSP 3.5.11 Cisco Systems, Inc. www.cisco.com Cisco has more than 200 offices worldwide. Addresses, phone numbers, and fax numbers are listed on the Cisco website at www.cisco.com/go/offices.
Open Source Used In TSP 3.5.11 Cisco Systems, Inc. www.cisco.com Cisco has more than 200 offices worldwide. Addresses, phone numbers, and fax numbers are listed on the Cisco website at www.cisco.com/go/offices.
Package biomformat. April 11, 2018
 Version 1.7.0 Date 2016-04-16 Package biomformat April 11, 2018 Maintainer Paul J. McMurdie License GPL-2 Title An interface package for the BIOM file format Type Package Author
Version 1.7.0 Date 2016-04-16 Package biomformat April 11, 2018 Maintainer Paul J. McMurdie License GPL-2 Title An interface package for the BIOM file format Type Package Author
Package fst. June 7, 2018
 Type Package Package fst June 7, 2018 Title Lightning Fast Serialization of Data Frames for R Multithreaded serialization of compressed data frames using the 'fst' format. The 'fst' format allows for random
Type Package Package fst June 7, 2018 Title Lightning Fast Serialization of Data Frames for R Multithreaded serialization of compressed data frames using the 'fst' format. The 'fst' format allows for random
MUMPS IO Documentation
 MUMPS IO Documentation Copyright (c) 1999, 2000, 2001, 2002, 2003 Raymond Douglas Newman. All rights reserved. Redistribution and use in source and binary forms, with or without modification, are permitted
MUMPS IO Documentation Copyright (c) 1999, 2000, 2001, 2002, 2003 Raymond Douglas Newman. All rights reserved. Redistribution and use in source and binary forms, with or without modification, are permitted
IETF TRUST. Legal Provisions Relating to IETF Documents. Approved November 6, Effective Date: November 10, 2008
 IETF TRUST Legal Provisions Relating to IETF Documents Approved November 6, 2008 Effective Date: November 10, 2008 1. Background The IETF Trust was formed on December 15, 2005, for, among other things,
IETF TRUST Legal Provisions Relating to IETF Documents Approved November 6, 2008 Effective Date: November 10, 2008 1. Background The IETF Trust was formed on December 15, 2005, for, among other things,
Data Deduplication Metadata Extension
 Data Deduplication Metadata Extension Version 1.1c ABSTRACT: This document describes a proposed extension to the SNIA Cloud Data Management Interface (CDMI) International Standard. Publication of this
Data Deduplication Metadata Extension Version 1.1c ABSTRACT: This document describes a proposed extension to the SNIA Cloud Data Management Interface (CDMI) International Standard. Publication of this
ANZ TRANSACTIVE MOBILE for ipad
 ANZ TRANSACTIVE MOBILE for ipad CORPORATE CASH AND TRADE MANAGEMENT ON THE GO QUICK REFERENCE GUIDE April 2016 HOME SCREEN The home screen provides immediate visibility of your favourite accounts and transactions
ANZ TRANSACTIVE MOBILE for ipad CORPORATE CASH AND TRADE MANAGEMENT ON THE GO QUICK REFERENCE GUIDE April 2016 HOME SCREEN The home screen provides immediate visibility of your favourite accounts and transactions
IETF TRUST. Legal Provisions Relating to IETF Documents. February 12, Effective Date: February 15, 2009
 IETF TRUST Legal Provisions Relating to IETF Documents February 12, 2009 Effective Date: February 15, 2009 1. Background The IETF Trust was formed on December 15, 2005, for, among other things, the purpose
IETF TRUST Legal Provisions Relating to IETF Documents February 12, 2009 Effective Date: February 15, 2009 1. Background The IETF Trust was formed on December 15, 2005, for, among other things, the purpose
iwrite technical manual iwrite authors and contributors Revision: 0.00 (Draft/WIP)
 iwrite technical manual iwrite authors and contributors Revision: 0.00 (Draft/WIP) June 11, 2015 Chapter 1 Files This section describes the files iwrite utilizes. 1.1 report files An iwrite report consists
iwrite technical manual iwrite authors and contributors Revision: 0.00 (Draft/WIP) June 11, 2015 Chapter 1 Files This section describes the files iwrite utilizes. 1.1 report files An iwrite report consists
Explaining & Accessing the SPDX License List
 Explaining & Accessing the SPDX License List SOFTWARE PACKAGE DATA EXCHANGE Gary O Neall Source Auditor Inc. Jilayne Lovejoy ARM August, 2014 Copyright Linux Foundation 2014 1 The SPDX License List 2 The
Explaining & Accessing the SPDX License List SOFTWARE PACKAGE DATA EXCHANGE Gary O Neall Source Auditor Inc. Jilayne Lovejoy ARM August, 2014 Copyright Linux Foundation 2014 1 The SPDX License List 2 The
HYDRODESKTOP VERSION 1.4 QUICK START GUIDE
 HYDRODESKTOP VERSION 1.4 QUICK START GUIDE A guide to using this free and open source application for discovering, accessing, and using hydrologic data February 8, 2012 by: Tim Whiteaker Center for Research
HYDRODESKTOP VERSION 1.4 QUICK START GUIDE A guide to using this free and open source application for discovering, accessing, and using hydrologic data February 8, 2012 by: Tim Whiteaker Center for Research
System Log NextAge Consulting Pete Halsted
 System Log NextAge Consulting Pete Halsted 110 East Center St. #1035 Madison, SD 57042 pete@thenextage.com www.thenextage.com www.thenextage.com/wordpress Table of Contents Table of Contents BSD 3 License
System Log NextAge Consulting Pete Halsted 110 East Center St. #1035 Madison, SD 57042 pete@thenextage.com www.thenextage.com www.thenextage.com/wordpress Table of Contents Table of Contents BSD 3 License
Ecma International Policy on Submission, Inclusion and Licensing of Software
 Ecma International Policy on Submission, Inclusion and Licensing of Software Experimental TC39 Policy This Ecma International Policy on Submission, Inclusion and Licensing of Software ( Policy ) is being
Ecma International Policy on Submission, Inclusion and Licensing of Software Experimental TC39 Policy This Ecma International Policy on Submission, Inclusion and Licensing of Software ( Policy ) is being
PyWin32ctypes Documentation
 PyWin32ctypes Documentation Release 0.1.3.dev1 David Cournapeau, Ioannis Tziakos Sep 01, 2017 Contents 1 Usage 3 2 Development setup 5 3 Reference 7 3.1 PyWin32 Compatibility Layer......................................
PyWin32ctypes Documentation Release 0.1.3.dev1 David Cournapeau, Ioannis Tziakos Sep 01, 2017 Contents 1 Usage 3 2 Development setup 5 3 Reference 7 3.1 PyWin32 Compatibility Layer......................................
HIS document 2 Loading Observations Data with the ODDataLoader (version 1.0)
 HIS document 2 Loading Observations Data with the ODDataLoader (version 1.0) A guide to using CUAHSI s ODDataLoader tool for loading observations data into an Observations Data Model compliant database
HIS document 2 Loading Observations Data with the ODDataLoader (version 1.0) A guide to using CUAHSI s ODDataLoader tool for loading observations data into an Observations Data Model compliant database
The Cron service allows you to register STAF commands that will be executed at a specified time interval(s).
 Cron Service User's Guide Version 1.2.6 Last updated: March 29, 2006 Overview The Cron service allows you to register STAF commands that will be executed at a specified time interval(s). Note that Cron
Cron Service User's Guide Version 1.2.6 Last updated: March 29, 2006 Overview The Cron service allows you to register STAF commands that will be executed at a specified time interval(s). Note that Cron
Preface. Audience. Cisco IOS Software Documentation. Organization
 This preface describes the audience, organization, and conventions of this publication, and provides information on how to obtain related documentation. Cisco documentation and additional literature are
This preface describes the audience, organization, and conventions of this publication, and provides information on how to obtain related documentation. Cisco documentation and additional literature are
Ecma International Policy on Submission, Inclusion and Licensing of Software
 Ecma International Policy on Submission, Inclusion and Licensing of Software Experimental TC39 Policy This Ecma International Policy on Submission, Inclusion and Licensing of Software ( Policy ) is being
Ecma International Policy on Submission, Inclusion and Licensing of Software Experimental TC39 Policy This Ecma International Policy on Submission, Inclusion and Licensing of Software ( Policy ) is being
Open Source and Standards: A Proposal for Collaboration
 ETSI Workshop on Open Source and ization: Legal Interactions September 16, 2016 Sophia Antipolis Open Source and s: A Proposal for Collaboration David Marr VP & Legal Counsel Open Source Group Qualcomm
ETSI Workshop on Open Source and ization: Legal Interactions September 16, 2016 Sophia Antipolis Open Source and s: A Proposal for Collaboration David Marr VP & Legal Counsel Open Source Group Qualcomm
Definiens. Image Miner bit and 64-bit Edition. Release Notes
 Definiens Image Miner 2.1.1 32-bit and 64-bit Edition Release Notes Definiens Documentation: Image Miner 2.1.1 Release Notes Imprint 2013 Definiens AG. All rights reserved. This document may be copied
Definiens Image Miner 2.1.1 32-bit and 64-bit Edition Release Notes Definiens Documentation: Image Miner 2.1.1 Release Notes Imprint 2013 Definiens AG. All rights reserved. This document may be copied
User Manual. Date Aug 30, Enertrax DAS Download Client
 EnertraxDL - DAS Download Client User Manual Date Aug 30, 2004 Page 1 Copyright Information Copyright 2004, Obvius Holdings, LLC. All rights reserved. Redistribution and use in source and binary forms,
EnertraxDL - DAS Download Client User Manual Date Aug 30, 2004 Page 1 Copyright Information Copyright 2004, Obvius Holdings, LLC. All rights reserved. Redistribution and use in source and binary forms,
Static analysis for quality mobile applications
 Static analysis for quality mobile applications Julia Perdigueiro MOTODEV Studio for Android Project Manager Instituto de Pesquisas Eldorado Eric Cloninger Product Line Manager Motorola Mobility Life.
Static analysis for quality mobile applications Julia Perdigueiro MOTODEV Studio for Android Project Manager Instituto de Pesquisas Eldorado Eric Cloninger Product Line Manager Motorola Mobility Life.
An Easy Way to Split a SAS Data Set into Unique and Non-Unique Row Subsets Thomas E. Billings, MUFG Union Bank, N.A., San Francisco, California
 An Easy Way to Split a SAS Data Set into Unique and Non-Unique Row Subsets Thomas E. Billings, MUFG Union Bank, N.A., San Francisco, California This work by Thomas E. Billings is licensed (2017) under
An Easy Way to Split a SAS Data Set into Unique and Non-Unique Row Subsets Thomas E. Billings, MUFG Union Bank, N.A., San Francisco, California This work by Thomas E. Billings is licensed (2017) under
pyserial-asyncio Documentation
 pyserial-asyncio Documentation Release 0.4 pyserial-team Feb 12, 2018 Contents 1 Short introduction 3 2 pyserial-asyncio API 5 2.1 asyncio.................................................. 5 3 Appendix
pyserial-asyncio Documentation Release 0.4 pyserial-team Feb 12, 2018 Contents 1 Short introduction 3 2 pyserial-asyncio API 5 2.1 asyncio.................................................. 5 3 Appendix
Open Source Used In Cisco Configuration Professional for Catalyst 1.0
 Open Source Used In Cisco Configuration Professional for Catalyst 1.0 Cisco Systems, Inc. www.cisco.com Cisco has more than 200 offices worldwide. Addresses, phone numbers, and fax numbers are listed on
Open Source Used In Cisco Configuration Professional for Catalyst 1.0 Cisco Systems, Inc. www.cisco.com Cisco has more than 200 offices worldwide. Addresses, phone numbers, and fax numbers are listed on
ColdFusion Builder 3.2 Third Party Software Notices and/or Additional Terms and Conditions
 ColdFusion Builder 3.2 Third Party Software Notices and/or Additional Terms and Conditions Date Generated: 2018/09/10 Apache Tomcat ID: 306 Apache Foundation and Contributors This product includes software
ColdFusion Builder 3.2 Third Party Software Notices and/or Additional Terms and Conditions Date Generated: 2018/09/10 Apache Tomcat ID: 306 Apache Foundation and Contributors This product includes software
Flask-Sitemap Documentation
 Flask-Sitemap Documentation Release 0.3.0 CERN May 06, 2018 Contents 1 Contents 3 2 Installation 5 2.1 Requirements............................................... 5 3 Usage 7 3.1 Simple Example.............................................
Flask-Sitemap Documentation Release 0.3.0 CERN May 06, 2018 Contents 1 Contents 3 2 Installation 5 2.1 Requirements............................................... 5 3 Usage 7 3.1 Simple Example.............................................
Graphic Inspector 2 User Guide
 www.zevrix.com support@zevrix.com Graphic Inspector 2 User Guide Installation & System Requirements 2 Scanning Files and Folders 2 Checkup Presets 3 File Table and Info Panel 4 Export Data 5 Support 6
www.zevrix.com support@zevrix.com Graphic Inspector 2 User Guide Installation & System Requirements 2 Scanning Files and Folders 2 Checkup Presets 3 File Table and Info Panel 4 Export Data 5 Support 6
Open Source Used In c1101 and c1109 Cisco IOS XE Fuji
 Open Source Used In c1101 and c1109 Cisco IOS XE Fuji 16.8.1 Cisco Systems, Inc. www.cisco.com Cisco has more than 200 offices worldwide. Addresses, phone numbers, and fax numbers are listed on the Cisco
Open Source Used In c1101 and c1109 Cisco IOS XE Fuji 16.8.1 Cisco Systems, Inc. www.cisco.com Cisco has more than 200 offices worldwide. Addresses, phone numbers, and fax numbers are listed on the Cisco
Copyright PFU LIMITED 2016
 -------------------------------------------------------- PaperStream Capture Lite 1.0.1 README File -------------------------------------------------------- Copyright PFU LIMITED 2016 This file contains
-------------------------------------------------------- PaperStream Capture Lite 1.0.1 README File -------------------------------------------------------- Copyright PFU LIMITED 2016 This file contains
Documentation Roadmap for Cisco Prime LAN Management Solution 4.2
 Documentation Roadmap for Cisco Prime LAN Thank you for purchasing Cisco Prime LAN Management Solution (LMS) 4.2. This document provides an introduction to the Cisco Prime LMS and lists the contents of
Documentation Roadmap for Cisco Prime LAN Thank you for purchasing Cisco Prime LAN Management Solution (LMS) 4.2. This document provides an introduction to the Cisco Prime LMS and lists the contents of
SORRENTO MANUAL. by Aziz Gulbeden
 SORRENTO MANUAL by Aziz Gulbeden September 13, 2004 Table of Contents SORRENTO SELF-ORGANIZING CLUSTER FOR PARALLEL DATA- INTENSIVE APPLICATIONS... 1 OVERVIEW... 1 COPYRIGHT... 1 SORRENTO WALKTHROUGH -
SORRENTO MANUAL by Aziz Gulbeden September 13, 2004 Table of Contents SORRENTO SELF-ORGANIZING CLUSTER FOR PARALLEL DATA- INTENSIVE APPLICATIONS... 1 OVERVIEW... 1 COPYRIGHT... 1 SORRENTO WALKTHROUGH -
Flask Gravatar. Release 0.5.0
 Flask Gravatar Release 0.5.0 Jan 05, 2018 Contents 1 Contents 3 1.1 Installation................................................ 3 1.2 Usage................................................... 3 2 Parameters
Flask Gravatar Release 0.5.0 Jan 05, 2018 Contents 1 Contents 3 1.1 Installation................................................ 3 1.2 Usage................................................... 3 2 Parameters
Ryft REST API - Swagger.io
 Ryft REST API - Swagger.io User Guide Ryft Document Number: 1192 Document Version: 1.1.0 Revision Date: June 2017 2017 Ryft Systems, Inc. All Rights in this documentation are reserved. RYFT SYSTEMS, INC.
Ryft REST API - Swagger.io User Guide Ryft Document Number: 1192 Document Version: 1.1.0 Revision Date: June 2017 2017 Ryft Systems, Inc. All Rights in this documentation are reserved. RYFT SYSTEMS, INC.
HYDROOBJECTS VERSION 1.1
 o HYDROOBJECTS VERSION 1.1 July, 2008 by: Tim Whiteaker Center for Research in Water Resources The University of Texas at Austin Distribution The HydroObjects software, source code, and documentation are
o HYDROOBJECTS VERSION 1.1 July, 2008 by: Tim Whiteaker Center for Research in Water Resources The University of Texas at Austin Distribution The HydroObjects software, source code, and documentation are
Denkh XML Reporter. Web Based Report Generation Software. Written By Scott Auge Amduus Information Works, Inc.
 Denkh XML Reporter Web Based Report Generation Software Written By Scott Auge sauge@amduus.com Page 1 of 13 Table of Contents License 3 What is it? 4 Basic Software Requirements 5 Basic Report Designer
Denkh XML Reporter Web Based Report Generation Software Written By Scott Auge sauge@amduus.com Page 1 of 13 Table of Contents License 3 What is it? 4 Basic Software Requirements 5 Basic Report Designer
NemHandel Referenceklient 2.3.1
 OIO Service Oriented Infrastructure NemHandel Referenceklient 2.3.1 Release Notes Contents 1 Introduction... 3 2 Release Content... 3 3 What is changed?... 4 3.1 NemHandel Referenceklient version 2.3.1...
OIO Service Oriented Infrastructure NemHandel Referenceklient 2.3.1 Release Notes Contents 1 Introduction... 3 2 Release Content... 3 3 What is changed?... 4 3.1 NemHandel Referenceklient version 2.3.1...
ASDF Definition. Release Lion Krischer, James Smith, Jeroen Tromp
 ASDF Definition Release 1.0.2 Lion Krischer, James Smith, Jeroen Tromp Mar 26, 2018 Contents 1 ASDF Format Changelog 3 1.1 Introduction............................................. 3 1.2 Big Picture..............................................
ASDF Definition Release 1.0.2 Lion Krischer, James Smith, Jeroen Tromp Mar 26, 2018 Contents 1 ASDF Format Changelog 3 1.1 Introduction............................................. 3 1.2 Big Picture..............................................
Lax-Hopf-based LWR solver. Matlab implementation: Manual
 Lax-Hopf-based LWR solver Matlab implementation: Manual Pierre-Emmanuel Mazaré, Christian Claudel, Alexandre Bayen June 15, 2010 This document describes the sample implementation of an exact, grid-free
Lax-Hopf-based LWR solver Matlab implementation: Manual Pierre-Emmanuel Mazaré, Christian Claudel, Alexandre Bayen June 15, 2010 This document describes the sample implementation of an exact, grid-free
Bar Code Discovery. Administrator's Guide
 Bar Code Discovery Administrator's Guide November 2012 www.lexmark.com Contents 2 Contents Overview...3 Configuring the application...4 Configuring the application...4 Configuring Bar Code Discovery...4
Bar Code Discovery Administrator's Guide November 2012 www.lexmark.com Contents 2 Contents Overview...3 Configuring the application...4 Configuring the application...4 Configuring Bar Code Discovery...4
AccuTerm 7 Internet Edition Connection Designer Help. Copyright Schellenbach & Assoc., Inc.
 AccuTerm 7 Internet Edition Connection Designer Help Contents 3 Table of Contents Foreword 0 Part I AccuTerm 7 Internet Edition 6 1 Description... 6 2 Connection... Designer 6 3 Internet... Client 6 4
AccuTerm 7 Internet Edition Connection Designer Help Contents 3 Table of Contents Foreword 0 Part I AccuTerm 7 Internet Edition 6 1 Description... 6 2 Connection... Designer 6 3 Internet... Client 6 4
Definiens. Image Miner bit and 64-bit Editions. Release Notes
 Definiens Image Miner 2.0.2 32-bit and 64-bit Editions Release Notes Definiens Documentation: Image Miner 2.0.2 Release Notes Imprint 2012 Definiens AG. All rights reserved. This document may be copied
Definiens Image Miner 2.0.2 32-bit and 64-bit Editions Release Notes Definiens Documentation: Image Miner 2.0.2 Release Notes Imprint 2012 Definiens AG. All rights reserved. This document may be copied
Business Rules NextAge Consulting Pete Halsted
 Business Rules NextAge Consulting Pete Halsted 110 East Center St. #1035 Madison, SD 57042 pete@thenextage.com www.thenextage.com www.thenextage.com/wordpress Table of Contents Table of Contents BSD 3
Business Rules NextAge Consulting Pete Halsted 110 East Center St. #1035 Madison, SD 57042 pete@thenextage.com www.thenextage.com www.thenextage.com/wordpress Table of Contents Table of Contents BSD 3
DAP Controller FCO
 Release Note DAP Controller 6.40.0412 FCO 2016.046 System : Business Mobility IP DECT Date : 30 June 2016 Category : Maintenance Product Identity : DAP Controller 6.40.0412 Queries concerning this document
Release Note DAP Controller 6.40.0412 FCO 2016.046 System : Business Mobility IP DECT Date : 30 June 2016 Category : Maintenance Product Identity : DAP Controller 6.40.0412 Queries concerning this document
LabVIEW Driver. User guide Version
 LabVIEW Driver User guide Version 1.0.0 2016 Table of Contents Version History...3 Copyright...4 Software License...5 Operational Safety...6 Warranty and Support...7 Introduction...8 Requirements...9 How
LabVIEW Driver User guide Version 1.0.0 2016 Table of Contents Version History...3 Copyright...4 Software License...5 Operational Safety...6 Warranty and Support...7 Introduction...8 Requirements...9 How
Grouper UI csrf xsrf prevention
 Grouper UI csrf xsrf prevention Wiki Home Download Grouper Grouper Guides Community Contributions Developer Resources Grouper Website This is in Grouper 2.2 UI. btw, Ive heard this does not work with IE8.
Grouper UI csrf xsrf prevention Wiki Home Download Grouper Grouper Guides Community Contributions Developer Resources Grouper Website This is in Grouper 2.2 UI. btw, Ive heard this does not work with IE8.
Copyright PFU LIMITED
 -------------------------------------------------------- PaperStream Capture 1.0.12 README File -------------------------------------------------------- Copyright PFU LIMITED 2013-2015 This file contains
-------------------------------------------------------- PaperStream Capture 1.0.12 README File -------------------------------------------------------- Copyright PFU LIMITED 2013-2015 This file contains
MagicInfo Express Content Creator
 MagicInfo Express Content Creator MagicInfo Express Content Creator User Guide MagicInfo Express Content Creator is a program that allows you to conveniently create LFD content using a variety of templates.
MagicInfo Express Content Creator MagicInfo Express Content Creator User Guide MagicInfo Express Content Creator is a program that allows you to conveniently create LFD content using a variety of templates.
NEC Display Solutions MultiProfiler for Linux x64 Versions Installation Guide
 NEC Display Solutions MultiProfiler for Linux x64 Versions 1.3.60 Installation Guide 1 ABOUT MULTIPROFILER... 4 2 SYSTEM REQUIREMENTS... 5 2.1 SUPPORTED NEC DISPLAY MODELS AND DESCRIPTION... 5 2.1.1 Minimum
NEC Display Solutions MultiProfiler for Linux x64 Versions 1.3.60 Installation Guide 1 ABOUT MULTIPROFILER... 4 2 SYSTEM REQUIREMENTS... 5 2.1 SUPPORTED NEC DISPLAY MODELS AND DESCRIPTION... 5 2.1.1 Minimum
Small Logger File System
 Small Logger File System (http://www.tnkernel.com/) Copyright 2011 Yuri Tiomkin Document Disclaimer The information in this document is subject to change without notice. While the information herein is
Small Logger File System (http://www.tnkernel.com/) Copyright 2011 Yuri Tiomkin Document Disclaimer The information in this document is subject to change without notice. While the information herein is
CSCE Inspection Activity Name(s):
 CSCE 747 - Inspection Activity Name(s): You are inspecting the source code for the Graduate Record and Data System (GRADS) - a system that graduate students can log into and use to view their transcript
CSCE 747 - Inspection Activity Name(s): You are inspecting the source code for the Graduate Record and Data System (GRADS) - a system that graduate students can log into and use to view their transcript
About This Guide. and with the Cisco Nexus 1010 Virtual Services Appliance: N1K-C1010
 This guide describes how to use Cisco Network Analysis Module Traffic Analyzer 4.2 (NAM 4.2) software. This preface has the following sections: Chapter Overview, page xvi Audience, page xvii Conventions,
This guide describes how to use Cisco Network Analysis Module Traffic Analyzer 4.2 (NAM 4.2) software. This preface has the following sections: Chapter Overview, page xvi Audience, page xvii Conventions,
@list = bsd_glob('*.[ch]'); $homedir = bsd_glob('~gnat', GLOB_TILDE GLOB_ERR);
![@list = bsd_glob('*.[ch]'); $homedir = bsd_glob('~gnat', GLOB_TILDE GLOB_ERR); @list = bsd_glob('*.[ch]'); $homedir = bsd_glob('~gnat', GLOB_TILDE GLOB_ERR);](/thumbs/87/97210082.jpg) NAME File::Glob - Perl extension for BSD glob routine SYNOPSIS use File::Glob ':bsd_glob'; @list = bsd_glob('*.[ch]'); $homedir = bsd_glob('~gnat', GLOB_TILDE GLOB_ERR); if (GLOB_ERROR) { # an error occurred
NAME File::Glob - Perl extension for BSD glob routine SYNOPSIS use File::Glob ':bsd_glob'; @list = bsd_glob('*.[ch]'); $homedir = bsd_glob('~gnat', GLOB_TILDE GLOB_ERR); if (GLOB_ERROR) { # an error occurred
Intercloud Federation using via Semantic Resource Federation API and Dynamic SDN Provisioning
 Intercloud Federation using via Semantic Resource Federation API and Dynamic SDN Provisioning David Bernstein Deepak Vij Copyright 2013, 2014 IEEE. All rights reserved. Redistribution and use in source
Intercloud Federation using via Semantic Resource Federation API and Dynamic SDN Provisioning David Bernstein Deepak Vij Copyright 2013, 2014 IEEE. All rights reserved. Redistribution and use in source
Trimble. ecognition. Release Notes
 Trimble ecognition Release Notes Trimble Documentation: ecognition 8.9 Release Notes Imprint and Version Document Version 8.9 Copyright 2013 Trimble Germany GmbH. All rights reserved. This document may
Trimble ecognition Release Notes Trimble Documentation: ecognition 8.9 Release Notes Imprint and Version Document Version 8.9 Copyright 2013 Trimble Germany GmbH. All rights reserved. This document may
NemHandel Referenceklient 2.3.0
 OIO Service Oriented Infrastructure OIO Service Oriented Infrastructure NemHandel Referenceklient 2.3.0 Release Notes Contents 1 Introduction... 3 2 Release Content... 3 3 What is changed?... 4 3.1 NemHandel
OIO Service Oriented Infrastructure OIO Service Oriented Infrastructure NemHandel Referenceklient 2.3.0 Release Notes Contents 1 Introduction... 3 2 Release Content... 3 3 What is changed?... 4 3.1 NemHandel
LGR Toolset (beta) User Guide. IDN Program 24 October 2017
 LGR Toolset (beta) User Guide IDN Program 24 October 2017 1 Introduction to LGR Toolset (beta) Label Generation Rulesets (LGRs) specify metadata, code point repertoire, variant rules and Whole Label Evaluation
LGR Toolset (beta) User Guide IDN Program 24 October 2017 1 Introduction to LGR Toolset (beta) Label Generation Rulesets (LGRs) specify metadata, code point repertoire, variant rules and Whole Label Evaluation
TWAIN driver User s Guide
 4037-9571-05 TWAIN driver User s Guide Contents 1 Introduction 1.1 System requirements...1-1 2 Installing the TWAIN Driver 2.1 Installation procedure...2-1 To install the software...2-1 2.2 Uninstalling...2-1
4037-9571-05 TWAIN driver User s Guide Contents 1 Introduction 1.1 System requirements...1-1 2 Installing the TWAIN Driver 2.1 Installation procedure...2-1 To install the software...2-1 2.2 Uninstalling...2-1
US Government Users Restricted Rights - Use, duplication or disclosure restricted by GSA ADP Schedule Contract with IBM Corp.
 Service Data Objects (SDO) DFED Sample Application README Copyright IBM Corporation, 2012, 2013 US Government Users Restricted Rights - Use, duplication or disclosure restricted by GSA ADP Schedule Contract
Service Data Objects (SDO) DFED Sample Application README Copyright IBM Corporation, 2012, 2013 US Government Users Restricted Rights - Use, duplication or disclosure restricted by GSA ADP Schedule Contract
ODM 1.1. An application Hydrologic. June Prepared by: Jeffery S. Horsburgh Justin Berger Utah Water
 ODM M STREAMING VERSION DATAA LOADER 1.1 An application for loading streaming sensor data into the CUAHSI Hydrologic Information System Observations Data Model June 2008 Prepared by: Jeffery S. Horsburgh
ODM M STREAMING VERSION DATAA LOADER 1.1 An application for loading streaming sensor data into the CUAHSI Hydrologic Information System Observations Data Model June 2008 Prepared by: Jeffery S. Horsburgh
SMS2CMDB Project Summary v1.6
 SMS2CMDB Project Summary v1.6 Project Abstract SMS2CMDB provides the capability to integrate Microsoft Systems Management Server (MS- SMS) data with BMC Atrium CMDB (Atrium CMDB) and the BMC Remedy Asset
SMS2CMDB Project Summary v1.6 Project Abstract SMS2CMDB provides the capability to integrate Microsoft Systems Management Server (MS- SMS) data with BMC Atrium CMDB (Atrium CMDB) and the BMC Remedy Asset
NTLM NTLM. Feature Description
 Feature Description VERSION: 6.0 UPDATED: JULY 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP Technologies logo are registered
Feature Description VERSION: 6.0 UPDATED: JULY 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP Technologies logo are registered
cs50.c /**************************************************************************** * CS50 Library 6 *
 cs50.c 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. 22. 23. 24. 25. 26. 27. 28. 29. 30. 31. 32. 33. 34. 35. 36. 37. 38. 39. 40. 41. 42. 43. 44. 45. 46. 47. 48. / CS50 Library
cs50.c 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. 22. 23. 24. 25. 26. 27. 28. 29. 30. 31. 32. 33. 34. 35. 36. 37. 38. 39. 40. 41. 42. 43. 44. 45. 46. 47. 48. / CS50 Library
Internet Connection Guide
 Internet Connection Guide v1.10 CVP-509/505/503/501 PSR-S910/S710 Enjoy your instrument with Internet Direct Connection This instrument can be directly connected to the Internet, conveniently letting you
Internet Connection Guide v1.10 CVP-509/505/503/501 PSR-S910/S710 Enjoy your instrument with Internet Direct Connection This instrument can be directly connected to the Internet, conveniently letting you
ODM STREAMING DATA LOADER
 ODM STREAMING DATA LOADER An application for loading streaming sensor data into the CUAHSI Hydrologic Information System Observations Data Model March 2012 Prepared by: Jeffery S. Horsburgh Utah Water
ODM STREAMING DATA LOADER An application for loading streaming sensor data into the CUAHSI Hydrologic Information System Observations Data Model March 2012 Prepared by: Jeffery S. Horsburgh Utah Water
Cassette Documentation
 Cassette Documentation Release 0.3.8 Charles-Axel Dein October 01, 2015 Contents 1 User s Guide 3 1.1 Foreword................................................. 3 1.2 Quickstart................................................
Cassette Documentation Release 0.3.8 Charles-Axel Dein October 01, 2015 Contents 1 User s Guide 3 1.1 Foreword................................................. 3 1.2 Quickstart................................................
Distinction Import Module User Guide. DISTINCTION.CO.UK
 Distinction Import Module User Guide. Distinction Import Module. Licence: Copyright (c) 2018, Distinction Limited. All rights reserved. Redistribution and use in source and binary forms, with or without
Distinction Import Module User Guide. Distinction Import Module. Licence: Copyright (c) 2018, Distinction Limited. All rights reserved. Redistribution and use in source and binary forms, with or without
License, Rules, and Application Form
 Generic Interface for Cameras License, Rules, and Application Form GenICam_License.doc Page 1 of 11 Table of Contents 1 OVERVIEW... 4 2 SUBJECT OF THE GENICAM LICENSE... 4 3 RULES FOR STANDARD COMPLIANCY...
Generic Interface for Cameras License, Rules, and Application Form GenICam_License.doc Page 1 of 11 Table of Contents 1 OVERVIEW... 4 2 SUBJECT OF THE GENICAM LICENSE... 4 3 RULES FOR STANDARD COMPLIANCY...
DoJSON Documentation. Release Invenio collaboration
 DoJSON Documentation Release 1.2.0 Invenio collaboration March 21, 2016 Contents 1 About 1 2 Installation 3 3 Documentation 5 4 Testing 7 5 Example 9 5.1 User s Guide...............................................
DoJSON Documentation Release 1.2.0 Invenio collaboration March 21, 2016 Contents 1 About 1 2 Installation 3 3 Documentation 5 4 Testing 7 5 Example 9 5.1 User s Guide...............................................
Definiens. Tissue Studio Release Notes
 Definiens Tissue Studio 3.0.1 Release Notes Definiens Documentation: Definiens Tissue Studio 3.0.1 Release Notes Imprint 2012 Definiens AG. All rights reserved. This document may be copied and printed
Definiens Tissue Studio 3.0.1 Release Notes Definiens Documentation: Definiens Tissue Studio 3.0.1 Release Notes Imprint 2012 Definiens AG. All rights reserved. This document may be copied and printed
Navigator Documentation
 Navigator Documentation Release 1.0.0 Simon Holywell February 18, 2016 Contents 1 Installation 3 1.1 Packagist with Composer........................................ 3 1.2 git Clone or Zip Package.........................................
Navigator Documentation Release 1.0.0 Simon Holywell February 18, 2016 Contents 1 Installation 3 1.1 Packagist with Composer........................................ 3 1.2 git Clone or Zip Package.........................................
Migration Tool. Migration Tool (Beta) Technical Note
 Migration Tool (Beta) Technical Note VERSION: 6.0 UPDATED: MARCH 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP Technologies logo
Migration Tool (Beta) Technical Note VERSION: 6.0 UPDATED: MARCH 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP Technologies logo
Scott Auge
 Scott Auge sauge@amduus.com Amduus Information Works, Inc. http://www.amduus.com Page 1 of 14 LICENSE This is your typical BSD license. Basically it says do what you want with it - just don't sue me. Written
Scott Auge sauge@amduus.com Amduus Information Works, Inc. http://www.amduus.com Page 1 of 14 LICENSE This is your typical BSD license. Basically it says do what you want with it - just don't sue me. Written
SW MAPS TEMPLATE BUILDER. User s Manual
 SW MAPS TEMPLATE BUILDER User s Manual Copyright (c) 2017 SOFTWEL (P) Ltd All rights reserved. Redistribution and use in binary forms, without modification, are permitted provided that the following conditions
SW MAPS TEMPLATE BUILDER User s Manual Copyright (c) 2017 SOFTWEL (P) Ltd All rights reserved. Redistribution and use in binary forms, without modification, are permitted provided that the following conditions
openresty / array-var-nginx-module
 1 of 6 2/17/2015 11:20 AM Explore Gist Blog Help itpp16 + openresty / array-var-nginx-module 4 22 4 Add support for array variables to nginx config files 47 commits 1 branch 4 releases 2 contributors array-var-nginx-module
1 of 6 2/17/2015 11:20 AM Explore Gist Blog Help itpp16 + openresty / array-var-nginx-module 4 22 4 Add support for array variables to nginx config files 47 commits 1 branch 4 releases 2 contributors array-var-nginx-module
Simba Cassandra ODBC Driver with SQL Connector
 Simba Cassandra ODBC Driver with SQL Connector Last Revised: March 26, 2013 Simba Technologies Inc. Copyright 2012-2013 Simba Technologies Inc. All Rights Reserved. Information in this document is subject
Simba Cassandra ODBC Driver with SQL Connector Last Revised: March 26, 2013 Simba Technologies Inc. Copyright 2012-2013 Simba Technologies Inc. All Rights Reserved. Information in this document is subject
Informatica Cloud Spring Microsoft Azure Blob Storage V2 Connector Guide
 Informatica Cloud Spring 2017 Microsoft Azure Blob Storage V2 Connector Guide Informatica Cloud Microsoft Azure Blob Storage V2 Connector Guide Spring 2017 October 2017 Copyright Informatica LLC 2017 This
Informatica Cloud Spring 2017 Microsoft Azure Blob Storage V2 Connector Guide Informatica Cloud Microsoft Azure Blob Storage V2 Connector Guide Spring 2017 October 2017 Copyright Informatica LLC 2017 This
Fujitsu ScandAll PRO V2.1.5 README
 -------------------------------------------------------- Fujitsu ScandAll PRO V2.1.5 README -------------------------------------------------------- Copyright PFU Limited 2007-2017 This file contains information
-------------------------------------------------------- Fujitsu ScandAll PRO V2.1.5 README -------------------------------------------------------- Copyright PFU Limited 2007-2017 This file contains information
 xxxx xxxx xxxx xxxx xxxx xxxx xxxx xxxx Android Calendar Syn (http://github.com/mukesh4u/android-calendar-sync/), Apache License 2.0 Android NDK (https://developer.android.com/tools/sdk/ndk/index.html),
xxxx xxxx xxxx xxxx xxxx xxxx xxxx xxxx Android Calendar Syn (http://github.com/mukesh4u/android-calendar-sync/), Apache License 2.0 Android NDK (https://developer.android.com/tools/sdk/ndk/index.html),
KEMP Driver for Red Hat OpenStack. KEMP LBaaS Red Hat OpenStack Driver. Installation Guide
 KEMP LBaaS Red Hat OpenStack Driver Installation Guide VERSION: 2.0 UPDATED: AUGUST 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP
KEMP LBaaS Red Hat OpenStack Driver Installation Guide VERSION: 2.0 UPDATED: AUGUST 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP
Encrypted Object Extension
 Encrypted Object Extension ABSTRACT: "Publication of this Working Draft for review and comment has been approved by the Cloud Storage Technical Working Group. This draft represents a "best effort" attempt
Encrypted Object Extension ABSTRACT: "Publication of this Working Draft for review and comment has been approved by the Cloud Storage Technical Working Group. This draft represents a "best effort" attempt
calio / form-input-nginx-module
 https://github.com/ 1 of 5 2/17/2015 11:27 AM Explore Gist Blog Help itpp16 + calio / form-input-nginx-module 5 46 9 This is a nginx module that reads HTTP POST and PUT request body encoded in "application/x-www-formurlencoded",
https://github.com/ 1 of 5 2/17/2015 11:27 AM Explore Gist Blog Help itpp16 + calio / form-input-nginx-module 5 46 9 This is a nginx module that reads HTTP POST and PUT request body encoded in "application/x-www-formurlencoded",
Splunk. Splunk. Deployment Guide
 Deployment Guide VERSION: 1.0 UPDATED: JULY 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP Technologies logo are registered trademarks
Deployment Guide VERSION: 1.0 UPDATED: JULY 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP Technologies logo are registered trademarks
AT03262: SAM D/R/L/C System Pin Multiplexer (SYSTEM PINMUX) Driver. Introduction. SMART ARM-based Microcontrollers APPLICATION NOTE
 SMART ARM-based Microcontrollers AT03262: SAM D/R/L/C System Pin Multiplexer (SYSTEM PINMUX) Driver APPLICATION NOTE Introduction This driver for Atmel SMART ARM -based microcontrollers provides an interface
SMART ARM-based Microcontrollers AT03262: SAM D/R/L/C System Pin Multiplexer (SYSTEM PINMUX) Driver APPLICATION NOTE Introduction This driver for Atmel SMART ARM -based microcontrollers provides an interface
StorageGRID Webscale NAS Bridge Management API Guide
 StorageGRID Webscale NAS Bridge 2.0.3 Management API Guide January 2018 215-12414_B0 doccomments@netapp.com Table of Contents 3 Contents Understanding the NAS Bridge management API... 4 RESTful web services
StorageGRID Webscale NAS Bridge 2.0.3 Management API Guide January 2018 215-12414_B0 doccomments@netapp.com Table of Contents 3 Contents Understanding the NAS Bridge management API... 4 RESTful web services
Grouping Nodes. X3D Graphics for Web Authors. Chapter 3
 X3D Graphics for Web Authors Chapter 3 Grouping Nodes A Working Group is a technical committee that researches and proposes solutions to specific technical problems relating to X3D. Web3D Consortium Contents
X3D Graphics for Web Authors Chapter 3 Grouping Nodes A Working Group is a technical committee that researches and proposes solutions to specific technical problems relating to X3D. Web3D Consortium Contents
CMS Content Management System
 CMS Content Management System Written by Scott Auge sauge@amduus.com scott_auge@yahoo.com Copyright 2002 Amduus Information Works, Inc. http://www.amduus.com Page 1 of 31 Contents License... 5 Purpose...
CMS Content Management System Written by Scott Auge sauge@amduus.com scott_auge@yahoo.com Copyright 2002 Amduus Information Works, Inc. http://www.amduus.com Page 1 of 31 Contents License... 5 Purpose...
notify2 Documentation
 notify2 Documentation Release 0.3 Thomas Kluyver Nov 13, 2017 Contents 1 License and Contributors 3 2 Creating and showing notifications 5 3 Extra parameters 7 4 Callbacks 9 5 Constants 11 6 Indices and
notify2 Documentation Release 0.3 Thomas Kluyver Nov 13, 2017 Contents 1 License and Contributors 3 2 Creating and showing notifications 5 3 Extra parameters 7 4 Callbacks 9 5 Constants 11 6 Indices and
iphone/ipad Connection Manual
 For Electone users / Connection Manual By connecting your, or ipod touch to a compatible Electone and using the various dedicated applications, you can expand the potential of the Electone and make it
For Electone users / Connection Manual By connecting your, or ipod touch to a compatible Electone and using the various dedicated applications, you can expand the potential of the Electone and make it
TheGreenBow VPN Client ios User Guide
 www.thegreenbow.com TheGreenBow VPN Client ios User Guide Property of TheGreenBow 2018 Table of Contents 1 Presentation... 3 1.1 TheGreenBow VPN Client... 3 1.2 TheGreenBow VPN Client main features...
www.thegreenbow.com TheGreenBow VPN Client ios User Guide Property of TheGreenBow 2018 Table of Contents 1 Presentation... 3 1.1 TheGreenBow VPN Client... 3 1.2 TheGreenBow VPN Client main features...
AMPHORA2 User Manual. An Automated Phylogenomic Inference Pipeline for Bacterial and Archaeal Sequences. COPYRIGHT 2011 by Martin Wu
 AMPHORA2 User Manual An Automated Phylogenomic Inference Pipeline for Bacterial and Archaeal Sequences. COPYRIGHT 2011 by Martin Wu AMPHORA2 is free software: you may redistribute it and/or modify its
AMPHORA2 User Manual An Automated Phylogenomic Inference Pipeline for Bacterial and Archaeal Sequences. COPYRIGHT 2011 by Martin Wu AMPHORA2 is free software: you may redistribute it and/or modify its
VMware vcenter Log Insight Manager. Deployment Guide
 VMware vcenter Log Insight Manager Deployment Guide VERSION: 6.0 UPDATED: JULY 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP Technologies
VMware vcenter Log Insight Manager Deployment Guide VERSION: 6.0 UPDATED: JULY 2016 Copyright Notices Copyright 2002-2016 KEMP Technologies, Inc.. All rights reserved.. KEMP Technologies and the KEMP Technologies
