Towards Open Publication of Reusable Scientific Workflows: Abstractions, Standards and Linked Data
|
|
|
- Elijah Bryan
- 6 years ago
- Views:
Transcription
1 Towards Open Publication of Reusable Scientific Workflows: Abstractions, Standards and Linked Data Daniel Garijo OEG-DIA Facultad de Informática Universidad Politécnica de Madrid Yolanda Gil Information Sciences Institute and Department of Computer Science University of Southern California Abstract In recent years, a variety of systems have been developed that export the workflows executed to analyze data and make them part of published articles. We argue that the workflows that are published with current approaches are dependent on the specific codes used for execution, the specific workflow system used, and the specific workflow catalogs where they are published. In this paper, we describe a new approach that addresses these shortcomings and makes workflows more reusable through: 1) the publication of abstract workflows that complement executable workflows and that are reusable when the execution environment is different, 2) the publication of both abstract and executable workflows using standards such as the Open Provenance Model that can then be imported by other workflow systems, 3) the publication of workflows as Linked Data that results in open web accessible workflow repositories. We illustrate this approach using a complex workflow that we re-created from an influential publication that describes the generation of drugomes. Keywords: Workflows, provenance, OPM, Wings, reproducibility 1. Introduction Scientific workflows are products of research in their own right and should be treated as first-class citizens in cyberinfrastructure [11]. Workflows represent computations carried out to obtain scientific results, but these computations are typically only described in the narrative of published scientific articles and only at a very high level. Scientific articles describe computational methods informally, often requiring a significant effort from others to reproduce and to reuse. The reproducibility process can be so costly that it has been referred to as forensic research [1]. Studies have shown that reproducibility is not achievable from the article itself, even when datasets are published [3], [19]. Furthermore reviewers often cannot inspect computational methods in enough detail. As a result, retractions of published articles occur more often than would be desirable [32]. A recent editorial proposed tracking the retraction index of scientific journals to indicate the proportion of published articles that are later found problematic [9]. Publishers themselves are asking the community to end black box science that cannot be easily reproduced [29]. The impact of this issue is well beyond scientific research circles and into society at large. Clinical trials based on erroneous results pose significant threats to patients [18]. The validity of 1
2 scientific research methods has been put in question [22]. The public has neutral to low trust on scientists for important topics such as flu pandemics, depression drugs, and autism causes [30]. To facilitate reproducibility, the idea of enhancing scientific publications with explicit workflows has been proposed [5]. Workflows could be incorporated as supplementary material of scientific publications, much like datasets are included today. This would make scientific results more easily reproducible because articles would have not just a textual description of the computational process used but also a workflow that, as a computational artifact, could be inspected and automatically re-executed. Some systems exist that augment publications with scripts or workflows, such as Weaver and GenePattern [8] [23] [24]. Repositories of shared workflows enable scientists to reuse workflows published by others and reproduce their results [33]. Some research addresses the publication of workflows using semantic web technologies [26]. Many workflow researchers have developed and adopted the Open Provenance Model (OPM) as a shared model for workflow publication that is independent of particular workflow systems [28]. These are all important steps towards reusability in workflow publication. However none of the approaches to workflow publication to date supports reproducing workflows across different execution infrastructures. Each lab has an execution infrastructure that includes software libraries and codes for computations that are different from other labs. Therefore, even if a workflow is published and can be run in other workflow systems, it remains tied to the particular software components and execution environment used in the original workflow and therefore its reusability is severely limited. We see a workflow as a digital instrument that enables scientists to analyze data through the lens of the method that the workflow represents. A technological challenge is how to make such instruments reusable across labs and institutions, since each has a diverse software and hardware infrastructure. Publishing and sharing workflows that can only be run using a particular software platform is useful, but their reusability is severely limited to labs that have the same software platform. What is needed is a mechanism to publish workflows that would give scientists access to such digital instruments at very low cost, and that would facilitate the reuse of the method, i.e., the workflow, in the desired execution infrastructure which may be different from the original one used by the workflow publishers. This paper describes a framework to publish computational workflows used in a research article in a manner that is platform independent and easily reusable in different execution environments. Our work has three major contributions: 1. Publishing an abstract representation of the executed workflow. This abstract workflow captures a conceptual and execution-independent view of the data analysis method. It is a more reusable workflow, providing a better understanding of the method and making every workflow step a separate reusable unit. We use the Wings workflow system 1 [10][12][13], which has an expressive language to represent reusable abstract workflow templates using semantic constraints in OWL and RDF. 2. Publishing both the abstract workflow and the executed workflow using the OPM standard. Although other systems publish the executed workflow in OPM, our work is novel in that the abstract workflow is published as well. As a result, the abstract method is no longer dependent on the particular execution environment used to run the original workflow. We extended OPM with a profile called OPMW that includes terms appropriate to describe abstract workflows. 3. Publishing the workflows as accessible web objects using Linked Data principles. Using the Linked Data principles [4][17] enables direct access to workflows, their components, and the datasets used as web objects with a unique URI and represented in RDF. This would enable other scientists to inspect a workflow without having to ask the investigators for details and without having to reproduce it. We offer an RDF repository of published workflows, as accessible from a SPARQL Endpoint. Other applications can import these workflows. An additional advantage is that the workflows could be linked to other entities that are already published as web resources. We applied our framework to reproduce and publish the method of an influential publication that describes how to derive the drug-target network of an organism, called its drugome [20]. Wings is used to create the workflows (both abstract and executable). We extended Wings to publish the workflow in OPM as Linked Data
3 The rest of the paper is organized as follows. The next section briefly reviews the drugome workflow that we reproduced and use in our examples. Section 3 introduces our approach, defining abstract workflows and discussing the modeling decisions made. Section 4 describes the architecture of the conversion and publication process. Section 5 discusses the advantages of publishing workflows as Linked Data along with query examples. Finally, we present conclusions and future work. 2. Initial Focus: The Drugome Workflow Our initial focus is a method to derive the drugtarget network of an organism (i.e., its drugome) described in [20]. The article describes a computational pipeline that accesses data from the Protein Data Bank (PDB) and carries out a systematic analysis of the proteome of Mycobacterium tuberculosis (TB) against all FDA-approved drugs. The process uncovers protein receptors in the organism that could be targeted by drugs currently in use for other purposes. The result is a drug-target network (a drugome ) that includes all known approved drugs. Although the article focuses on a particular organism (TB), the authors state that the methodology may be applied to other pathogens of interest with results improving as more of their structural proteomes are determined through the continued efforts of structural biology/genomics. That is, the expectation is that others should be able to reuse this method to create other drugomes, and to do so periodically as new proteins and drugs are discovered. The original work did not use a workflow system. Instead, the computational steps were run separately and manually. With the help of the authors of the article, we were able to create the executable workflow that reflects the steps described in the original article. We are able to run it with data used in the original experiments. As is usual in computational biology, the paper has a methods section that describes conceptually what computations were carried out, but we needed clarifications from the authors in order to reproduce the computations. Moreover, although the article had just been published we found that some of the software originally used in the experiments was no longer available in the lab, so some of the steps already needed to be done differently Figure 1: An overview of the TB Drugome workflow, where the software component is specified for each computational step. The method described in the original article is decomposed into four conceptual parts. Figure 1 shows the dataflow diagram of the core steps of the drugome workflow represented in Wings. Datasets are represented as ovals, while computations (codes) are shown as rectangles. The main inputs to the workflow are: 1) a list of binding sites of approved drugs that can be associated with protein crystal structures in PDB, 2) a list of proteins of the TB proteome that have solved structures in PDB, and 4 3
4 3) homology models of annotated comparative protein structure models for TB. First, the SMAP 2 tool is used to compare both the binding sites of protein structures and the homology models against the drug binding sites (box 1 in Figure 1). The results are sorted and merged. Next, the FATCAT 3 tool is used to compare the overall similarity of the global protein structures, and only significant pairs are retained (box 2 in Figure 1). A graph of the resulting interaction network is generated, which can be visualized in tools such as Cytoscape 4 (box 3 Figure 1). Finally, the Autodock Vina 5 tool is used to perform molecular docking, to predict the affinity of drug molecules with the proteins (box 4 in Figure 1). However, in order for this drugome workflow to be widely reusable, we would like to be able to publish not just what software was executed, but also the abstract method in a standard language that many groups can reuse. In addition, having the workflow and supporting data openly available through a public access point would allow anyone gain a better understanding of the experiment even without having to reproduce it. 3. Approach Our approach has three key features. First, publishing an abstract workflow provides the means to separate a method from its implementation as an executable workflow. Second, by transforming both the abstract template and the workflow instance results to OPM we separated the workflow from any workflow system representation. Third, publishing workflows as Linked Data provides the added value of allowing sharing and reusing the templates and results from other workflow systems, as well as being able to link resources from datasets already published as Linked Data. 3.1 Abstract workflows A key feature of our approach is the creation of an abstract workflow in addition to the executable workflow. This addresses several limitations of executable workflows regarding reusability. First, the executable workflow runs codes that may not be available to other researchers. In our case, one of the codes was no longer available in the UCSD lab. In the paper, there is a step that obtains docking results from a tool called ehits 6. However, this tool is proprietary. For our workflow we used alternative tool, Autodock Vina, which obtains docking results too, and it is open source. Another tool, SMAP, had been revised and a new version was available that had a few differences with the one originally used. Note that these changes in the execution environment occurred within the same lab that published the original method just in a few months time. The execution environment in other labs that could reproduce the method would be likely to have many more differences. Therefore, publishing the executable workflow has very limited use. This can be addressed if, in addition to publishing the executable workflow that mentions the software that was used, the authors publish an abstract workflow description. In our case, such abstract workflow would include an abstract docking step with the same input and output datasets as ehits but that can be easily mapped to Autodock Vina as an alternative tool for that abstract step. Second, different labs prefer to use different tools for data analysis. In our case, there is a visualization step that can be done using Cytoscape, a known and well-integrated tool, but the lab preferred using yed 7, which is also very popular. Publishing an abstract workflow that has a more general visualization step and does not mention particular tools facilitates the customization to each lab s software environment. Third, an investigator may not be familiar with the particular implementations used by others. Many investigators prefer to use Matlab 8 because it is a commercial product that has been debugged and verified and do not want to use R 9 because it has not been as thoroughly tested. Other investigators strongly favor R because of its wide availability to other researchers. So having abstract descriptions of steps that are independent of the implementation makes the workflow more understandable and therefore more reusable Types of Abstractions in Workflows In our work, we publish abstract workflows in addition to executed workflows. There are many ways to define abstractions, and we turn to AI planning terminology to describe the distinctions of different approaches [21] index.html 9 4
5 One type of abstraction is skeletal planning [13]. In skeletal planning, the same number and type of steps that appear in the abstract plan are the same as those appearing in the specialized plan. Each of the abstract steps in the skeletal plan is specialized to a more specific step in the specialized plan. We have found this to be a very useful type of abstraction when defining workflows and it is the kind that we use in our work. Another type is step abstraction. Here, the steps of a plan may be abstract classes of steps. These classes may be organized in a taxonomy, for example if the steps share similar constraints on the conditions or effects. There are the same steps in the abstract and the specialized plans, and in this regard this type of abstraction is similar to skeletal plans. Another type of abstraction is predicate abstraction. In this type of abstraction, entire predicates are dropped at any given abstraction level. The intuition is that if a predicate requires less steps to be accomplished then it can be placed in a lower abstraction level so the actions to accomplish it will cause minimal disruption to the plan. Ideally, the abstraction layers are designed to have the downward monotonicity property, which implies that any steps and orderings at higher layers of abstraction are unchanged as the plan is elaborated at lower abstraction levels. In workflows, this would be akin to dropping constraints or even inputs of steps at higher levels of abstraction. Another type is macro abstraction, where several computation steps can be compressed together as one step in the abstract plan. The substeps do not disappear from the plan, they are hidden to the user in the abstract plan and can be shown inside the compressed step. It is a common feature in workflow systems. Finally, layered abstraction represents conceptual levels of detail that may have a very loose correspondence with one another. These abstractions can be seen as employing different lenses to view the workflow depending on what perspective one is interested in. This kind of abstraction is very useful to provide an explanation of a workflow for different purposes. For example, a scientist would be interested in seeing the main steps of the flow, while a developer wants to see exactly the preprocessing and post processing done in each step. In our work, we use the skeletal planning approach to workflow abstraction for its simplicity and usability. Macro abstraction is used in some workflow systems. Step abstraction presents technical challenges that have not been addressed in workflow systems research. Layered abstraction has received recent interest and opens a new line of investigation for sharing, comparing and publishing workflows Abstract Workflows in Wings We describe abstract workflows in Wings formally in [13]. Here we describe abstract workflows informally, showing examples from the drugome workflow. Wings models domains with two main ontologies: one describes components (abstract or executable), and one describes the data used and produced by the components. In Wings, workflow components are organized in an ontology. Component classes are abstract components and are not themselves executable. They have specialized components that are actually associated with implemented codes and are therefore executable. Component classes describe the inputs and outputs of each step as well as any constraints they have. In our example workflow, FATCAT is an implemented component that belongs to the abstract component class CompareDissimilarProteinStructures. Workflow templates do not have associated input datasets, they simply specify a reusable workflow structure. An abstract workflow template is composed of abstract components, though it might include some specialized components. These are akin to skeletal plans. An abstract workflow: a) separates a component description from its actual implementation, making it possible to specialize a given abstract component to different specializations, b) makes the workflow more human readable by providing a general view of the steps executed in the workflow in a way that is independent of the particular tool or code that was executed. A specialized workflow template is composed of specialized components. When input datasets are associated with a specialized workflow template, the result is an execution-ready workflow. Figure 2 contrasts abstract and specialized workflows in Wings, using one of the subworkflows from the Drugome workflow. The subworkflow consists on a comparison of dissimilar protein structures, with a formatting step followed by a checking step, and then a filtering step (post processing) after the main comparison. Figure 2(a) shows a specialized workflow template in Wings, where each component is mapped to an executable code. Figure 2(b) shows an abstract workflow template for that same workflow, which has abstract components for every step. 5
6 (a) (b) Figure 2: Specialized workflow template (a) and abstract workflow template (b) for the protein structure comparison portion of the drugome workflow. Wings uses a generation algorithm to create valid execution-ready workflows from an abstract workflow [13]. This algorithm searches through valid specializations for each of the abstract steps, and if many are available, asks the user to choose one or else the system chooses one automatically. The outcome of this algorithm is an execution-ready workflow that specifies all the specialized components to use. An example of such executionready workflow is shown in Figure 3(b). This is the workflow submitted to an execution engine. Wings is typically configured to run Pegasus 10 as the execution engine. Pegasus makes many transformations to the workflow and executes it in the Condor infrastructure 11. That is, the Wings execution-ready workflow is a high-level plan for the execution (which, in turn, Pegasus calls an abstract workflow because it does not contain execution details), and represents a view of what is executed. The detailed execution provenance records for Pegasus and Condor is exported by Pegasus [25] and is not addressed in this paper. Figure 3(a) shows an example of an executed workflow. It has the same structure as the executionready workflow, and all the intermediate results have files associated with them that are created during execution. If the execution was not successful, then some of these files would not exist. An abstract workflow template can be used to represent the method used to analyze data, which is typically described in the Methods section in the scientific literature. Therefore, publishing an abstract workflow would allow us to publish a method in a way that is reusable. They complement the publication of the executed workflow, but should not replace it. The executable workflow provides data products and details of the code invocations that may be useful to other investigators. In this work, we export the executed workflow along with the abstract workflow used to create it. The executed workflow represents the trace of the run, while the abstract workflow represents the method that was used to analyze the data. Both are important to the publication of scientific results
7 SigR FList ChList1288 coutput09 NonSigResults1 SigResults1 (a) (b) Figure 3: Executed workflow (a) and execution ready workflow (b) for a portion of the drugome workflow. 3.2 OPMW: Modeling abstract workflows and executions with OPM To export the abstract workflows and the executable workflows we use OPM, a widely-used domain-independent provenance model result of the Provenance Challenge Series 12 and years of workflow provenance standardization and exchange in the scientific workflow community. We mapped Wings ontologies to the OPMO- OPMV ontology when possible, and extended OPM core concepts and relationships when needed with a new profile called OPMW. There are several reasons to use OPM. First, OPM has been already used successfully in many scientific workflow systems, thus making our published workflows more reusable [27]. Another advantage is that the core definitions in OPM are domain independent and extensible to accommodate other purposes, in our case abstract workflow representations. In addition, OPM can be considered the basis of the emerging W3C Provenance Interchange Language (PROV), which is currently being developed by the W3C Provenance Working Group 13 as a standard for representing and publishing provenance on the Web. OPM offers several core concepts and relationships to represent provenance. OPM models the resources created (in our case the datasets) as artifacts (immutable pieces of state), the steps used as processes (action or series of actions performed on artifacts), and the entities that control those processes as agents. Their relationships are modeled in a provenance graph with five causal edges: used (a process used some artifact), wascontrolledby (an agent controlled some process), wasgeneratedby (a process generated an artifact), wasderivedfrom (an artifact was derived from another artifact) and wastriggeredby (a process was triggered by another process). It also introduces the concept of roles to assign the type of activity that artifacts, processes or agents played when interacting with one another, and the notion of accounts and provenance graphs. An account represents a particular view on the provenance of an artifact based on what was executed. A provenance graph group sets of related OPM assertions
8 San Diego opmo:cause time opmo: Used location opmo:effect opmo:cause time opmo: wasgeneratedy opmo:effect opmv:artifact artif1 opmv:used opmv:process process1 Opmv:Artifact artif2 opmv:wasgeneratedby Figure 4: A portion of the mapped OPMO-OPMV ontology that illustrates their alternative representations of the used and wasgeneratedby relations of OPM. We use two OPM ontologies that have been developed to represent the OPM core specification: the Open Provenance Model Vocabulary (OPMV) 14 and the Open Provenance Model Ontology (OPMO) 15. OPMV is a lightweight vocabulary implementation of the OPM model that has only a subset of the concepts in OPM but facilitates modeling and query formulation. OPMO covers the full OPM model, but it is more complex. A major difference between both ontologies is the modeling of edges of the provenance graph (control, use and generation). OPMV encodes these edges as binary relationships in OWL. They relate Artifacts, Processes and Agents directly, but they cannot be enriched with additional metadata (time, location of the process, etc.), because they are OWL properties. In contrast, OPMO uses the n-ary pattern 16 [31] for adding the edges as classes in the ontology and be able to link them to additional metadata. The OPMO and OPMV ontologies have been mapped to complement each other in the most recent OPMO ontology release 17. By using this combined ontology, we can benefit from the features of each of them. That is, we can represent the structure of the workflow using basic OPMV relationships, and when adding more information is needed then we can use OPMO classes. Figure 4 shows this idea with a small workflow and preserving the namespaces of both ontologies. Here, suppose an opmv:artifact is used in a opmv:process to generate another opmv:artifact. Then, opmv:used is a direct edge between artifact and process. In contrast, opmo:used is a class linked to the time and location of the use. This decision requires adding two additional properties cause and effect to link the used and wasgeneratedby edges to the process and the artifacts. In this work we use the merged OPMO-OPMV ontology so we can use the entire OPM model in OPMO while using the OPMV for a simpler modeling of the edges of the provenance graph. We adopt OPMV s modeling of the edges for several reasons. First, it produces an easy to understand provenance graph (with binary properties rather than n-ary relationships). Second, the provenance queries over the graph are simpler than in OPMO. Third, it is enough to model the provenance produced by WINGS as we do not publish the time or location of the processes. However, if we decided to provide extra information about the edges, we could do it easily with this approach by just adding the classes for the edges as illustrated in Figure
9 Figure 5: An example to illustrate how OPMW represents a process view of workflow provenance, showing a one-step workflow. The abstract workflow is linked to the executed workflow. Terms from OPMV and OPMO are indicated with their prefixes. The terms for OPMW use the opmw prefix. In OPM, each provenance statement belongs to an Account. An Account represents a view of something that has happened (like a workflow execution record), according to some observer. Different Accounts may overlap, and it is possible for alternative accounts to be inconsistent if the observers make different assertions on the same resources. OPMV doesn t specify how to handle Accounts, (although an idea would be to treat them as named graphs), while OPMO introduces Accounts and ProvenanceGraphs as explicit concepts and properties to group statements. ProvenanceGraphs are a general provenance container of Accounts, statements and other annotations. Since named graphs have not been standardized yet, we use the OPMO concepts of Accounts and ProvenanceGraphs already present in the OPMO-OPMV ontology. Executed workflows are mapped to OPM Accounts, reflecting the fact that they capture a Wings view on the execution (recall that Pegasus and Condor each produce their own views on the execution, each at different levels of detail). Abstract workflow templates are not considered accounts since they represent something more abstract than an execution. Therefore, we represent them as a subclass of OPM ProvenanceGraphs. Figure 5 shows a process view high level diagram of the OPM and OPMW representation of an abstract workflow on the left and a workflow execution on the right. The example workflow shown here has one step (executionnode1), which runs the workflow component (speccomp1) that has one input (execinput1) and one output (executionoutput1). For some of the concepts there is a straightforward mapping: datasets are a subtype of Artifacts, while workflow steps (also called nodes) map to OPM Processes. In the figure, the terms taken from OPMO and OPMV are indicated using their namespaces. The new terms that we defined in our extension profile use the OPMW prefix. To make the distinction between datasets used in the abstract workflows and the workflow executions explicit, we have extended OPM Artifacts with opmw:artifacttemplates (the general artifacts used in the abstract workflow) and opmw:artifactinstances (which are bound to their corresponding opmw:artifacttemplate in the abstract workflow). Likewise, we defined two subclasses of OPM Process as opmw:processtemplates (the abstract steps used in the abstract workflow) and opmw:processinstances (the steps in the executed workflow). 9
10 opmv:artifact dataseta opmv:artifact datasetb opmv: used acwings: knowngenes acwings: genesfound opmv: used opmv:process removingduplicates opmv: wasgeneratedby acwings: discoveredgenes acwings :discarded opmv: wasgeneratedby opmv:artifact datasetc opmv:artifact datasetd rdfs:subpropertyof Figure 6: Encoding roles that artifacts play in processes as subproperties of OPM s used and wasgeneratedby. The process template in Figure 5 (templatenode1) uses one input artifact (artifact1), has one abstract component (abscomp1) and generates an output artifact (outputartifact1). All artifact templates and process templates are linked to a opmw:workflowtemplate through opmw:hasartifacttemplate and opmw:hasartifactprocess respectively. On the right side of the figure we can see how the processinstance (executionnode1) was controlled by a user (user1) which is of type Agent, used execinput1 linked to its corresponding ArtifactTemplate artifact1 and generated the ArtifactInstance executionoutput1. All artifacts, processes and agents are linked to the execution account, which is linked to the abstract workflow shown in the left side of the figure by the explicit relationship opmw:hasworkflowtemplate. Notice that each node has a link to the component that is run in that step, for example the workflow in Figure 1 has two nodes that run the same component SMAPV2. There is no OPM term that can be mapped to components, so these are taken from the Wings ontology of workflow components (represented with the acwings prefix). Each component points to its exact code, constraints and execution requirements. We also found the need to define domain-specific extensions to the used and wasgeneratedby properties. For each workflow component, each input and output dataset has a different role. For example, consider a component for removing duplicates that has two datasets of genes as inputs and two datasets of genes as outputs. One of the inputs has the role of known genes, the other has the role genes found. One of the outputs has the role of discovered genes and the other output is discarded. This example illustrates how all four datasets are lists of genes, but each is labeled with a unique role identifier that describes how the data is related to the process carried out. Thus, the known genes role qualifies the relationship in which the input and the process are involved rather than the input or the process in themselves. In OPMO this qualification is captured through the n-ary relationship pattern, linking an instance of a role to the used or wasgeneratedby edges. In order to avoid using OPMO s approach just for introducing roles, we define a subproperty of opmv:used and opmv:wasgeneratedby for each role. The name of the role can be used as the subproperty name (alternatively it could be added as a description of the subproperty). We use a Wings namespace, acwings, for these subproperties. 10
11 Figure 7: An example to illustrate how OPMW represents an attribution view of workflow provenance. We reuse terms from the Dublin Core (DC) Metadata Vocabulary. Terms from DC and OPMO are indicated with their prefixes. The terms for OPMW use the opmw prefix. Figure 6 shows an overview of this approach, illustrating the previous example: two artifacts (datasets A and B) are used with knowngenes and genesfound roles by a removingduplicates process, which generates two other artifacts (datasets C and D in the figure) with discoveredgenes and discarded roles. All these role subproperties are used to link the respective artifacts with the process. Since Wings does not capture the exact time execution of each of the nodes (Pegasus and Condor do) but only captures the starting and ending time of the whole execution, we have linked this information to the execution account along with additional metadata like if it has been a successful execution. Attribution is also key for scientists who publish the workflow, so we have considered crucial to include metadata from the specification as well as from the execution itself. Figure 7 shows an attribution view on the provenance of the same workflow of Figure 5. The abstract workflow contains information of the authors of the original article, the contributors of the workflow, the version, the date of creation and documentation of the abstract workflow (that may include a pointer to the publication). We reuse terms from the Dublin Core (DC) Metadata Vocabulary 18. The executed workflow links to the workflow systems used to create and execute it, the license, date of start and end of the run and the final status (succeeded or failed execution). It is unusual to use a provenance model, aimed at capturing what has happened, to represent an abstract workflow that represents what was planned. From our perspective, the abstract workflow represents an important perspective on the execution. Often, abstractions on what was executed are represented using complex languages that include control statements. In our case, workflow templates can be expressed with a simple direct acyclic graph, just like workflow executions can. So the extensions to OPM to represent abstract workflows in OPMW are straightforward and greatly facilitated by the ProvenanceGraph concept in OPM
12 Table 1: Summary of the terms of the OPMW profile, showing their relationship to OPM terms OPMW term ExecutionAccount ArtifactInstance ProcessInstance WorkflowTemplate ArtifactTemplate ProcessTemplate hasworkflowtemplate hasartifacttemplate hasprocesstemplate hasspecificcomponent hastemplatecomponent createdinworkflowsystem executedinworkflowsystem hasstatus hasstarttime hasendtime lastupdatedtime hasversion hasdocumentation Description Contains the provenance statements of an execution. It is an opmo:account. Resource used or generated in a workflow execution. It is an opmv:artifact. Step executed in a workflow. It is an opmv:process. Contains the abstract workflow. It is an opmo:opmgraph. Abstract type of artifact that is used or generated in a workflow. It is an opmv:artifact. Abstract step of workflow that corresponds to a specific step in the execution of a workflow. It is an opmv:artifact. Relates an ExecutionAccount to its WorkflowTemplate Relates an ArtifactInstance to its ArtifactTemplate Relates a ProcessInstance to its ProcessTemplate Relates a ProcessInstance to the specific component that implements it. Relates a ProcessTemplate to the abstract component that describes it. Relates an ExecutionAccount to a workflow system that created it. Relates an ExecutionAccount to any workflow system that executed it. Each ExecutionAccount has a status that can be either Success or Failed. The start time of the execution of the workflow. The end time of the execution of the workflow. Indicates when a workflow template was last updated. Indicates a version of a workflow template. Documentation for a workflow template. All artifacts, processes, and edge assertions are exported with their OPMV notation in addition to the OPMO and OPMW assertions. This improves interoperability with other systems publishing core concepts in OPM. This means that other systems are not required to do inference to obtain OPMV assertions. For example, although all artifacts are asserted to be OPMW artifacts, we also assert the following for all artifacts: < Instance/7239D> a dcwings:fatcatlist, opmv:artifact, ompw:artifactinstance ; rdfs:label "Instance artifact with id: 7239d" ; 12
13 Figure 8: Architecture overview and conversion process Note the assertion of the type of the artifact, in this case dcwings:fatcatlist. The type uses the namespace of the Wings data catalog, and it is used to describe the type of the datasets and their semantic constraints, which are used for workflow validation. It is key to export the type of the artifacts so it is available to other systems. Figure 9: Linked Data publication architecture Our system does not publish the execution-ready workflow and the specialized workflow template, illustrated in Figure 3(b) and Figure 2(a) respectively. They can both be inferred (derived) from the executed workflow. They could be published in cases where scientists want to share more layers of detail in the creation of the workflow so others would not have to infer them. Table 1 gives an overview of the terms introduced in the OPMW profile. 13
14 3.3 Exporting workflows as Linked Data Publishing the OPM abstract and execution workflows is a very important step for reproducibility and reuse. In order to be able to reference all the resources properly, we have decided to follow the Linked Data principles. According to them, we should use URIs as names for things, (fully compatible with the expression of OPM in RDF), use HTTP URIs so that people can look up those names (making those URIs dereferenceable and available in any browser), provide useful information when someone looks up a URI (by showing the resources that are related to the URI) and include links to other URIs, so they can discover more things. There are several important advantages of publishing workflows as Linked Data: a) link to available web resources, for instance to refer to proteins in the Protein Data Bank by using their published URI; b) get linked from other applications by pointing to the URIs that we publish, which include both the workflows and the data generated by them; and c) produce interoperable results within different systems without having to define particular catalog structures and access interfaces. The Wings workflows published by any user as Linked Data become publicly accessible. In some domains privacy is a concern (e.g., if the workflow processes genomic data), in those cases the publication as Linked Data would not be appropriate. However, there are many areas of science where privacy is not an issue and that would benefit tremendously of a more open architecture for sharing both data and workflows as Linked Data. All the URIs generated by our system to publish any aspect of the workflow are Cool URIs 19, following W3C style. This means that they are produced under a domain under our control, they are unique, and they are not going to change. Each URI identifies a different resource that can be individually accessed. We have separated the ontology URI triples (terminology or T-Box) from the instances of the workflows (assertions or A-Box) with a different URI base. This is to avoid confusion between the A-Box and the T-Box. The main structure for the A-Box is: rce/idofresource An example of an artifact URI from a workflow generated by the system: actinstance/8eb85ddcf2378b b6ad2 F314DF The main structure of the URIs for the T-Box is: me Camel case notation is used for composing the identifiers of classes and instances. The MD5 encoding is used to generate a unique identifier for each resource. The term opmv was included in the ontology namespace to indicate that OPMW is an OPMV profile. 4. Architecture Figure 8 shows an overview of the architecture. Different users produce their own abstract workflows and execution results, either in their own local installation or in central installations of Wings accessible as web portals. These workflows are RDF files, and are converted through a new Wings module to OPM RDF files. The OPM files are then published as Linked Data (center square of Figure 9). Once the files are published on the cloud, they are ready to be queried through external applications or visualized with Linked Data browsers. Users can import workflows to their own installations of Wings, where they can run or change the workflows. Workflows can also be imported to other systems if they are OPM compatible. Figure 9 gives more details about Linked Data publication. The RDF files are loaded into a Triple Store through its interface, and made public through a public endpoint. We have selected Allegro 20 as our triple store and Pubby 21 for browsing and visualizing the RDF. An additional file store is needed to store the files referred to in the links available in the triple store. The file store is in our local servers ( The endpoint can be browsed through generic visualizing tools like Pubby, but it can also be accessed programmatically from other applications. For example, other workflow systems could access the workflows and import them into their framework. The access point for a workflow is simply a URI (of a workflow template or an execution), and all the components and datasets in the workflow can be accessed from it. Additionally, other workflows systems could publish their own workflows on the public endpoint too. For doing so,
15 the only requirement is to support the OPM export in a compatible way and make a secure connection to the triple store. 5. Accessing Workflows as Linked Data The workflow repository is open and accessible over the web 22. The repository will grow as users publish more workflows using our framework. We show the broad accessibility of the published workflows by illustrating the queries that we can issue to the repository. Recall that both abstract workflows and workflow executions coexist in the same repository. Thus, we can query either of them or a mixture of both representations. The latter is very useful, as it enables cross-indexing of methods (the abstract workflows) and runs (executable workflows). We illustrate this with four queries to exemplify how to extract different kinds of information from the repository. To make the text readable, we have included the following prefix exec: < abst: < opmw: opmv: < The first example query is designed to retrieve the executable workflow step that generated a given artifact and the corresponding abstract workflow step. The query starts with the name of an artifact (artifactname) and finds its type (?type), its artifact template (?atempl), the process which generated that artifact (?process) and then its process template (?templp). The example query is: SELECT DISTINCT?process?type?aTempl?templP WHERE { <exec:artifactname> a?type.<exec:artifactname> <opmw:hasartifacttemplate>?atempl. < exec:artifactname> <opmv:wasgeneratedby>?process.?process <opmw:hasprocesstemplate>?templp.} The second example query retrieves all workflows that have used a given dataset. The query also starts with an artifact (artifactname), but we just ask for the accounts that used such artifact, along with their corresponding workflowtemplate (which is the abstract workflow). The query is: SELECT?account? templ WHERE { <exec:artifactname> <opmo:account>?account.?account <opmw:hasworkflowtemplate>?templ} For the third query example we change the perspective of the query to the abstract workflow, and we ask how many executions were run of a given abstract workflow. For each execution we also query the start time (?startt), end time (?endt) and the status (?stat), which specifies whether the execution failed. The query is: SELECT?acc?startT?endT?stat WHERE {?acc <opmw: hasworkflowtemplate> <abst:templatename>.?acc <opmw:hasstarttime>?startt.?acc <opmw:hasendtime>?endt.?acc <opmw:hasstatus>?stat.} In the fourth example query, we mix the use of the ontology and the instances to extract information about the roles played by an Artifact <artifact> when used by one or more processes in the workflow. Since the roles are specified extending the used relationship in the ontology, we just have to ask for the subproperties of used that link both concepts: SELECT?process?role?description WHERE {?process <opmv:used> <artifact>.?process?role <artifact>.?role <rdfs:subpropertyof> <opmv:used>.?role <rdfs:label>?description.} Once we get these roles, we can obtain the descriptions by asking about the label, comments or any additional information given by the authors
16 Types of search Resource URI (Process instance) Autocomplete search bar Resource Properties Specific component used for this process instance Figure 10. GUI snapshot of the Linked Data application. The information shown refers to part of the executed workflow in Figure 3. As we have demonstrated with these queries, the workflows can be accessed with basic knowledge of the OPM ontologies. However, complex queries would require understanding of OPMW. For this reason, and since navigating through the RDF with Linked Data browsers (such as Pubby) might be tedious, we have designed a small Linked Data application 23 for helping users to browse, search and retrieve the data available in the repository. Figure 10 shows an overview of this application, showing a step of the executable workflow of Figure 10. On top of the figure, users can select what kind of search they are aiming for (workflow search, author search or resource search). Users can enter the word terms of the search, which will be auto completed immediately suggesting any available resources. By selecting one of the resources, all its relations will be displayed on the same page. In the case of workflow authors, all their published workflows can be accessed and browsed in detail. 6. Related Work Our system publishes both the executed workflow and the abstract workflow template that led to it. The executed workflow, illustrated in Figure 3(a), represents a trace of what happened as the components of the workflow were executed and created new artifacts. The abstract workflow template, illustrated in Figure 2(b), represents a reusable description of the method, capturing how the different components are related to each other and what are the constraints on the inputs and outputs in order to be valid workflow. Other workflow systems (e.g., [26], [27]), only have the former available for publication, and if they publish a template of the executed workflow it would be the equivalent of the Wings specialized workflow template illustrated in Figure 2(a). Wings is unique in its ability to represent abstract workflow templates, and to specialized them automatically to generate execution-ready workflows
17 Workflow diagrams Component information Inputs of the component Outputs of the component Available code implamentations Template metadata Workflow executions Figure 11: Extended Linked Data application to browse the contents of the workflow templates. Other work [26] has explored exporting workflows as Linked Data. However, the workflows were not published using the OPM standard. One interesting contribution of that work is to show how the workflow inputs and outputs can be linked to Linked Data. In future work, we could take the protein-drug interaction network created by the workflow and expose it as linked data, adding links to the Protein Data Bank (PDB) 24 that is currently exposed in the Linked Data cloud. In other work, alternative ontologies to OPM are used to describe scientific experiments [7] [6]. These ontologies significantly extend OPM to reflect 24 laboratory experiments described in the Method section of scientific publications. OPMW has a narrower scope because it focuses on computational experiments, although OPM terms could be easily used for some core set of concepts describing the experiment process. OPMW is intended for workflows outside of the life sciences, for example we have published workflows for text analytics [16] and ecology [14]. Other approaches have been proposed to publishing scientific workflows such as research objects [2] and nanopublications [15]. Those approaches aim to improve workflow sharing, avoid workflow decay, and increase interoperability. 17
A New Approach for Publishing Workflows: Abstractions, Standards, and Linked Data
 A New Approach for Publishing Workflows: Abstractions, Standards, and Linked Data Daniel Garijo OEG-DIA Facultad de Informática Universidad Politécnica de Madrid dgarijo@delicias.dia.fi.upm.es Yolanda
A New Approach for Publishing Workflows: Abstractions, Standards, and Linked Data Daniel Garijo OEG-DIA Facultad de Informática Universidad Politécnica de Madrid dgarijo@delicias.dia.fi.upm.es Yolanda
Abstract, Link, Publish, Exploit: An End to End Framework for Workflow Sharing
 Abstract, Link, Publish, Exploit: An End to End Framework for Workflow Sharing Abstract Future Generation Computer Systems, Volume 75, October 2017. Daniel Garijo 1, Yolanda Gil 1 and Oscar Corcho 2 1
Abstract, Link, Publish, Exploit: An End to End Framework for Workflow Sharing Abstract Future Generation Computer Systems, Volume 75, October 2017. Daniel Garijo 1, Yolanda Gil 1 and Oscar Corcho 2 1
Orchestrating Music Queries via the Semantic Web
 Orchestrating Music Queries via the Semantic Web Milos Vukicevic, John Galletly American University in Bulgaria Blagoevgrad 2700 Bulgaria +359 73 888 466 milossmi@gmail.com, jgalletly@aubg.bg Abstract
Orchestrating Music Queries via the Semantic Web Milos Vukicevic, John Galletly American University in Bulgaria Blagoevgrad 2700 Bulgaria +359 73 888 466 milossmi@gmail.com, jgalletly@aubg.bg Abstract
Provenance-aware Faceted Search in Drupal
 Provenance-aware Faceted Search in Drupal Zhenning Shangguan, Jinguang Zheng, and Deborah L. McGuinness Tetherless World Constellation, Computer Science Department, Rensselaer Polytechnic Institute, 110
Provenance-aware Faceted Search in Drupal Zhenning Shangguan, Jinguang Zheng, and Deborah L. McGuinness Tetherless World Constellation, Computer Science Department, Rensselaer Polytechnic Institute, 110
Mapping Semantic Workflows to Alternative Workflow Execution Engines
 Proceedings of the Seventh IEEE International Conference on Semantic Computing (ICSC), Irvine, CA, 2013. Mapping Semantic Workflows to Alternative Workflow Execution Engines Yolanda Gil Information Sciences
Proceedings of the Seventh IEEE International Conference on Semantic Computing (ICSC), Irvine, CA, 2013. Mapping Semantic Workflows to Alternative Workflow Execution Engines Yolanda Gil Information Sciences
Ontology Servers and Metadata Vocabulary Repositories
 Ontology Servers and Metadata Vocabulary Repositories Dr. Manjula Patel Technical Research and Development m.patel@ukoln.ac.uk http://www.ukoln.ac.uk/ Overview agentcities.net deployment grant Background
Ontology Servers and Metadata Vocabulary Repositories Dr. Manjula Patel Technical Research and Development m.patel@ukoln.ac.uk http://www.ukoln.ac.uk/ Overview agentcities.net deployment grant Background
5 RDF and Inferencing
 5 RDF and Inferencing In Chapter 1XXX, we introduced the notion of dumb data, and how a more connected web infrastructure can result in behavior that lets smart applications perform to their potential.
5 RDF and Inferencing In Chapter 1XXX, we introduced the notion of dumb data, and how a more connected web infrastructure can result in behavior that lets smart applications perform to their potential.
University of Bath. Publication date: Document Version Publisher's PDF, also known as Version of record. Link to publication
 Citation for published version: Patel, M & Duke, M 2004, 'Knowledge Discovery in an Agents Environment' Paper presented at European Semantic Web Symposium 2004, Heraklion, Crete, UK United Kingdom, 9/05/04-11/05/04,.
Citation for published version: Patel, M & Duke, M 2004, 'Knowledge Discovery in an Agents Environment' Paper presented at European Semantic Web Symposium 2004, Heraklion, Crete, UK United Kingdom, 9/05/04-11/05/04,.
New Approach to Graph Databases
 Paper PP05 New Approach to Graph Databases Anna Berg, Capish, Malmö, Sweden Henrik Drews, Capish, Malmö, Sweden Catharina Dahlbo, Capish, Malmö, Sweden ABSTRACT Graph databases have, during the past few
Paper PP05 New Approach to Graph Databases Anna Berg, Capish, Malmö, Sweden Henrik Drews, Capish, Malmö, Sweden Catharina Dahlbo, Capish, Malmö, Sweden ABSTRACT Graph databases have, during the past few
Using JBI for Service-Oriented Integration (SOI)
 Using JBI for -Oriented Integration (SOI) Ron Ten-Hove, Sun Microsystems January 27, 2006 2006, Sun Microsystems Inc. Introduction How do you use a service-oriented architecture (SOA)? This is an important
Using JBI for -Oriented Integration (SOI) Ron Ten-Hove, Sun Microsystems January 27, 2006 2006, Sun Microsystems Inc. Introduction How do you use a service-oriented architecture (SOA)? This is an important
On the use of Abstract Workflows to Capture Scientific Process Provenance
 On the use of Abstract Workflows to Capture Scientific Process Provenance Paulo Pinheiro da Silva, Leonardo Salayandia, Nicholas Del Rio, Ann Q. Gates The University of Texas at El Paso CENTER OF EXCELLENCE
On the use of Abstract Workflows to Capture Scientific Process Provenance Paulo Pinheiro da Silva, Leonardo Salayandia, Nicholas Del Rio, Ann Q. Gates The University of Texas at El Paso CENTER OF EXCELLENCE
Exploring the Generation and Integration of Publishable Scientific Facts Using the Concept of Nano-publications
 Exploring the Generation and Integration of Publishable Scientific Facts Using the Concept of Nano-publications Amanda Clare 1,3, Samuel Croset 2,3 (croset@ebi.ac.uk), Christoph Grabmueller 2,3, Senay
Exploring the Generation and Integration of Publishable Scientific Facts Using the Concept of Nano-publications Amanda Clare 1,3, Samuel Croset 2,3 (croset@ebi.ac.uk), Christoph Grabmueller 2,3, Senay
Interactive Knowledge Capture
 Interactive Knowledge Capture Director, Knowledge Technologies Associate Division Director for Research Research Professor, Computer Science Intelligent Systems Division Information Sciences Institute
Interactive Knowledge Capture Director, Knowledge Technologies Associate Division Director for Research Research Professor, Computer Science Intelligent Systems Division Information Sciences Institute
Reducing Consumer Uncertainty
 Spatial Analytics Reducing Consumer Uncertainty Towards an Ontology for Geospatial User-centric Metadata Introduction Cooperative Research Centre for Spatial Information (CRCSI) in Australia Communicate
Spatial Analytics Reducing Consumer Uncertainty Towards an Ontology for Geospatial User-centric Metadata Introduction Cooperative Research Centre for Spatial Information (CRCSI) in Australia Communicate
MarcOnt - Integration Ontology for Bibliographic Description Formats
 MarcOnt - Integration Ontology for Bibliographic Description Formats Sebastian Ryszard Kruk DERI Galway Tel: +353 91-495213 Fax: +353 91-495541 sebastian.kruk @deri.org Marcin Synak DERI Galway Tel: +353
MarcOnt - Integration Ontology for Bibliographic Description Formats Sebastian Ryszard Kruk DERI Galway Tel: +353 91-495213 Fax: +353 91-495541 sebastian.kruk @deri.org Marcin Synak DERI Galway Tel: +353
Linked Open Data: a short introduction
 International Workshop Linked Open Data & the Jewish Cultural Heritage Rome, 20 th January 2015 Linked Open Data: a short introduction Oreste Signore (W3C Italy) Slides at: http://www.w3c.it/talks/2015/lodjch/
International Workshop Linked Open Data & the Jewish Cultural Heritage Rome, 20 th January 2015 Linked Open Data: a short introduction Oreste Signore (W3C Italy) Slides at: http://www.w3c.it/talks/2015/lodjch/
Google indexed 3,3 billion of pages. Google s index contains 8,1 billion of websites
 Access IT Training 2003 Google indexed 3,3 billion of pages http://searchenginewatch.com/3071371 2005 Google s index contains 8,1 billion of websites http://blog.searchenginewatch.com/050517-075657 Estimated
Access IT Training 2003 Google indexed 3,3 billion of pages http://searchenginewatch.com/3071371 2005 Google s index contains 8,1 billion of websites http://blog.searchenginewatch.com/050517-075657 Estimated
Automatic Generation of Workflow Provenance
 Automatic Generation of Workflow Provenance Roger S. Barga 1 and Luciano A. Digiampietri 2 1 Microsoft Research, One Microsoft Way Redmond, WA 98052, USA 2 Institute of Computing, University of Campinas,
Automatic Generation of Workflow Provenance Roger S. Barga 1 and Luciano A. Digiampietri 2 1 Microsoft Research, One Microsoft Way Redmond, WA 98052, USA 2 Institute of Computing, University of Campinas,
JENA: A Java API for Ontology Management
 JENA: A Java API for Ontology Management Hari Rajagopal IBM Corporation Page Agenda Background Intro to JENA Case study Tools and methods Questions Page The State of the Web Today The web is more Syntactic
JENA: A Java API for Ontology Management Hari Rajagopal IBM Corporation Page Agenda Background Intro to JENA Case study Tools and methods Questions Page The State of the Web Today The web is more Syntactic
Executing Evaluations over Semantic Technologies using the SEALS Platform
 Executing Evaluations over Semantic Technologies using the SEALS Platform Miguel Esteban-Gutiérrez, Raúl García-Castro, Asunción Gómez-Pérez Ontology Engineering Group, Departamento de Inteligencia Artificial.
Executing Evaluations over Semantic Technologies using the SEALS Platform Miguel Esteban-Gutiérrez, Raúl García-Castro, Asunción Gómez-Pérez Ontology Engineering Group, Departamento de Inteligencia Artificial.
Resilient Linked Data. Dave Reynolds, Epimorphics
 Resilient Linked Data Dave Reynolds, Epimorphics Ltd @der42 Outline What is Linked Data? Dependency problem Approaches: coalesce the graph link sets and partitioning URI architecture governance and registries
Resilient Linked Data Dave Reynolds, Epimorphics Ltd @der42 Outline What is Linked Data? Dependency problem Approaches: coalesce the graph link sets and partitioning URI architecture governance and registries
Efficient, Scalable, and Provenance-Aware Management of Linked Data
 Efficient, Scalable, and Provenance-Aware Management of Linked Data Marcin Wylot 1 Motivation and objectives of the research The proliferation of heterogeneous Linked Data on the Web requires data management
Efficient, Scalable, and Provenance-Aware Management of Linked Data Marcin Wylot 1 Motivation and objectives of the research The proliferation of heterogeneous Linked Data on the Web requires data management
H1 Spring C. A service-oriented architecture is frequently deployed in practice without a service registry
 1. (12 points) Identify all of the following statements that are true about the basics of services. A. Screen scraping may not be effective for large desktops but works perfectly on mobile phones, because
1. (12 points) Identify all of the following statements that are true about the basics of services. A. Screen scraping may not be effective for large desktops but works perfectly on mobile phones, because
Ontological Modeling: Part 2
 Ontological Modeling: Part 2 Terry Halpin LogicBlox This is the second in a series of articles on ontology-based approaches to modeling. The main focus is on popular ontology languages proposed for the
Ontological Modeling: Part 2 Terry Halpin LogicBlox This is the second in a series of articles on ontology-based approaches to modeling. The main focus is on popular ontology languages proposed for the
Supporting Patient Screening to Identify Suitable Clinical Trials
 Supporting Patient Screening to Identify Suitable Clinical Trials Anca BUCUR a,1, Jasper VAN LEEUWEN a, Njin-Zu CHEN a, Brecht CLAERHOUT b Kristof DE SCHEPPER b, David PEREZ-REY c, Raul ALONSO-CALVO c,
Supporting Patient Screening to Identify Suitable Clinical Trials Anca BUCUR a,1, Jasper VAN LEEUWEN a, Njin-Zu CHEN a, Brecht CLAERHOUT b Kristof DE SCHEPPER b, David PEREZ-REY c, Raul ALONSO-CALVO c,
Analysis Traceability and Provenance for HEP
 Analysis Traceability and Provenance for HEP Jetendr Shamdasani, Richard McClatchey, Andrew Branson and Zsolt Kovács University of the West of England, Coldharbour Lane, Bristol, BS16 1QY jet@cern.ch Abstract.
Analysis Traceability and Provenance for HEP Jetendr Shamdasani, Richard McClatchey, Andrew Branson and Zsolt Kovács University of the West of England, Coldharbour Lane, Bristol, BS16 1QY jet@cern.ch Abstract.
Converting Scripts into Reproducible Workflow Research Objects
 Converting Scripts into Reproducible Workflow Research Objects Lucas A. M. C. Carvalho, Khalid Belhajjame, Claudia Bauzer Medeiros lucas.carvalho@ic.unicamp.br Baltimore, Maryland, USA October 23-26, 2016
Converting Scripts into Reproducible Workflow Research Objects Lucas A. M. C. Carvalho, Khalid Belhajjame, Claudia Bauzer Medeiros lucas.carvalho@ic.unicamp.br Baltimore, Maryland, USA October 23-26, 2016
Semantic Web Company. PoolParty - Server. PoolParty - Technical White Paper.
 Semantic Web Company PoolParty - Server PoolParty - Technical White Paper http://www.poolparty.biz Table of Contents Introduction... 3 PoolParty Technical Overview... 3 PoolParty Components Overview...
Semantic Web Company PoolParty - Server PoolParty - Technical White Paper http://www.poolparty.biz Table of Contents Introduction... 3 PoolParty Technical Overview... 3 PoolParty Components Overview...
Developing a Research Data Policy
 Developing a Research Data Policy Core Elements of the Content of a Research Data Management Policy This document may be useful for defining research data, explaining what RDM is, illustrating workflows,
Developing a Research Data Policy Core Elements of the Content of a Research Data Management Policy This document may be useful for defining research data, explaining what RDM is, illustrating workflows,
The Emerging Data Lake IT Strategy
 The Emerging Data Lake IT Strategy An Evolving Approach for Dealing with Big Data & Changing Environments bit.ly/datalake SPEAKERS: Thomas Kelly, Practice Director Cognizant Technology Solutions Sean Martin,
The Emerging Data Lake IT Strategy An Evolving Approach for Dealing with Big Data & Changing Environments bit.ly/datalake SPEAKERS: Thomas Kelly, Practice Director Cognizant Technology Solutions Sean Martin,
DCMI Abstract Model - DRAFT Update
 1 of 7 9/19/2006 7:02 PM Architecture Working Group > AMDraftUpdate User UserPreferences Site Page Actions Search Title: Text: AttachFile DeletePage LikePages LocalSiteMap SpellCheck DCMI Abstract Model
1 of 7 9/19/2006 7:02 PM Architecture Working Group > AMDraftUpdate User UserPreferences Site Page Actions Search Title: Text: AttachFile DeletePage LikePages LocalSiteMap SpellCheck DCMI Abstract Model
The Final Updates. Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University of Oxford, UK
 The Final Updates Supported by the NIH grant 1U24 AI117966-01 to UCSD PI, Co-Investigators at: Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University
The Final Updates Supported by the NIH grant 1U24 AI117966-01 to UCSD PI, Co-Investigators at: Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University
Semantic Web Technologies
 1/33 Semantic Web Technologies Lecture 11: SWT for the Life Sciences 4: BioRDF and Scientifc Workflows Maria Keet email: keet -AT- inf.unibz.it home: http://www.meteck.org blog: http://keet.wordpress.com/category/computer-science/72010-semwebtech/
1/33 Semantic Web Technologies Lecture 11: SWT for the Life Sciences 4: BioRDF and Scientifc Workflows Maria Keet email: keet -AT- inf.unibz.it home: http://www.meteck.org blog: http://keet.wordpress.com/category/computer-science/72010-semwebtech/
The Semantic Planetary Data System
 The Semantic Planetary Data System J. Steven Hughes 1, Daniel J. Crichton 1, Sean Kelly 1, and Chris Mattmann 1 1 Jet Propulsion Laboratory 4800 Oak Grove Drive Pasadena, CA 91109 USA {steve.hughes, dan.crichton,
The Semantic Planetary Data System J. Steven Hughes 1, Daniel J. Crichton 1, Sean Kelly 1, and Chris Mattmann 1 1 Jet Propulsion Laboratory 4800 Oak Grove Drive Pasadena, CA 91109 USA {steve.hughes, dan.crichton,
Minsoo Ryu. College of Information and Communications Hanyang University.
 Software Reuse and Component-Based Software Engineering Minsoo Ryu College of Information and Communications Hanyang University msryu@hanyang.ac.kr Software Reuse Contents Components CBSE (Component-Based
Software Reuse and Component-Based Software Engineering Minsoo Ryu College of Information and Communications Hanyang University msryu@hanyang.ac.kr Software Reuse Contents Components CBSE (Component-Based
Proposal for Implementing Linked Open Data on Libraries Catalogue
 Submitted on: 16.07.2018 Proposal for Implementing Linked Open Data on Libraries Catalogue Esraa Elsayed Abdelaziz Computer Science, Arab Academy for Science and Technology, Alexandria, Egypt. E-mail address:
Submitted on: 16.07.2018 Proposal for Implementing Linked Open Data on Libraries Catalogue Esraa Elsayed Abdelaziz Computer Science, Arab Academy for Science and Technology, Alexandria, Egypt. E-mail address:
Powering Knowledge Discovery. Insights from big data with Linguamatics I2E
 Powering Knowledge Discovery Insights from big data with Linguamatics I2E Gain actionable insights from unstructured data The world now generates an overwhelming amount of data, most of it written in natural
Powering Knowledge Discovery Insights from big data with Linguamatics I2E Gain actionable insights from unstructured data The world now generates an overwhelming amount of data, most of it written in natural
Semantic Web Test
 Semantic Web Test 24.01.2017 Group 1 No. A B C D 1 X X X 2 X X 3 X X 4 X X 5 X X 6 X X X X 7 X X 8 X X 9 X X X 10 X X X 11 X 12 X X X 13 X X 14 X X 15 X X 16 X X 17 X 18 X X 19 X 20 X X 1. Which statements
Semantic Web Test 24.01.2017 Group 1 No. A B C D 1 X X X 2 X X 3 X X 4 X X 5 X X 6 X X X X 7 X X 8 X X 9 X X X 10 X X X 11 X 12 X X X 13 X X 14 X X 15 X X 16 X X 17 X 18 X X 19 X 20 X X 1. Which statements
An Annotation Tool for Semantic Documents
 An Annotation Tool for Semantic Documents (System Description) Henrik Eriksson Dept. of Computer and Information Science Linköping University SE-581 83 Linköping, Sweden her@ida.liu.se Abstract. Document
An Annotation Tool for Semantic Documents (System Description) Henrik Eriksson Dept. of Computer and Information Science Linköping University SE-581 83 Linköping, Sweden her@ida.liu.se Abstract. Document
A tutorial report for SENG Agent Based Software Engineering. Course Instructor: Dr. Behrouz H. Far. XML Tutorial.
 A tutorial report for SENG 609.22 Agent Based Software Engineering Course Instructor: Dr. Behrouz H. Far XML Tutorial Yanan Zhang Department of Electrical and Computer Engineering University of Calgary
A tutorial report for SENG 609.22 Agent Based Software Engineering Course Instructor: Dr. Behrouz H. Far XML Tutorial Yanan Zhang Department of Electrical and Computer Engineering University of Calgary
The role of vocabularies for estimating carbon footprint for food recipies using Linked Open Data
 The role of vocabularies for estimating carbon footprint for food recipies using Linked Open Data Ahsan Morshed Intelligent Sensing and Systems Laboratory, CSIRO, Hobart, Australia {ahsan.morshed, ritaban.dutta}@csiro.au
The role of vocabularies for estimating carbon footprint for food recipies using Linked Open Data Ahsan Morshed Intelligent Sensing and Systems Laboratory, CSIRO, Hobart, Australia {ahsan.morshed, ritaban.dutta}@csiro.au
METADATA INTERCHANGE IN SERVICE BASED ARCHITECTURE
 UDC:681.324 Review paper METADATA INTERCHANGE IN SERVICE BASED ARCHITECTURE Alma Butkovi Tomac Nagravision Kudelski group, Cheseaux / Lausanne alma.butkovictomac@nagra.com Dražen Tomac Cambridge Technology
UDC:681.324 Review paper METADATA INTERCHANGE IN SERVICE BASED ARCHITECTURE Alma Butkovi Tomac Nagravision Kudelski group, Cheseaux / Lausanne alma.butkovictomac@nagra.com Dražen Tomac Cambridge Technology
The National Cancer Institute's Thésaurus and Ontology
 The National Cancer Institute's Thésaurus and Ontology Jennifer Golbeck 1, Gilberto Fragoso 2, Frank Hartel 2, Jim Hendler 1, Jim Oberthaler 2, Bijan Parsia 1 1 University of Maryland, College Park 2 National
The National Cancer Institute's Thésaurus and Ontology Jennifer Golbeck 1, Gilberto Fragoso 2, Frank Hartel 2, Jim Hendler 1, Jim Oberthaler 2, Bijan Parsia 1 1 University of Maryland, College Park 2 National
Multi-agent and Semantic Web Systems: Linked Open Data
 Multi-agent and Semantic Web Systems: Linked Open Data Fiona McNeill School of Informatics 14th February 2013 Fiona McNeill Multi-agent Semantic Web Systems: *lecture* Date 0/27 Jena Vcard 1: Triples Fiona
Multi-agent and Semantic Web Systems: Linked Open Data Fiona McNeill School of Informatics 14th February 2013 Fiona McNeill Multi-agent Semantic Web Systems: *lecture* Date 0/27 Jena Vcard 1: Triples Fiona
Understandability and Semantic Interoperability of Diverse Rules Systems. Adrian Walker, Reengineering [1]
![Understandability and Semantic Interoperability of Diverse Rules Systems. Adrian Walker, Reengineering [1] Understandability and Semantic Interoperability of Diverse Rules Systems. Adrian Walker, Reengineering [1]](/thumbs/84/89149762.jpg) Understandability and Semantic Interoperability of Diverse Rules Systems Adrian Walker, Reengineering [1] Position Paper for the W3C Workshop on Rule Languages for Interoperability 27-28 April 2005, Washington,
Understandability and Semantic Interoperability of Diverse Rules Systems Adrian Walker, Reengineering [1] Position Paper for the W3C Workshop on Rule Languages for Interoperability 27-28 April 2005, Washington,
Reducing Consumer Uncertainty Towards a Vocabulary for User-centric Geospatial Metadata
 Meeting Host Supporting Partner Meeting Sponsors Reducing Consumer Uncertainty Towards a Vocabulary for User-centric Geospatial Metadata 105th OGC Technical Committee Palmerston North, New Zealand Dr.
Meeting Host Supporting Partner Meeting Sponsors Reducing Consumer Uncertainty Towards a Vocabulary for User-centric Geospatial Metadata 105th OGC Technical Committee Palmerston North, New Zealand Dr.
An Approach to Enhancing Workflows Provenance by Leveraging Web 2.0 to Increase Information Sharing, Collaboration and Reuse
 An Approach to Enhancing Workflows Provenance by Leveraging Web 2.0 to Increase Information Sharing, Collaboration and Reuse Aleksander Slominski Department of Computer Science, Indiana University Bloomington,
An Approach to Enhancing Workflows Provenance by Leveraging Web 2.0 to Increase Information Sharing, Collaboration and Reuse Aleksander Slominski Department of Computer Science, Indiana University Bloomington,
Customisable Curation Workflows in Argo
 Customisable Curation Workflows in Argo Rafal Rak*, Riza Batista-Navarro, Andrew Rowley, Jacob Carter and Sophia Ananiadou National Centre for Text Mining, University of Manchester, UK *Corresponding author:
Customisable Curation Workflows in Argo Rafal Rak*, Riza Batista-Navarro, Andrew Rowley, Jacob Carter and Sophia Ananiadou National Centre for Text Mining, University of Manchester, UK *Corresponding author:
Semantic Web. Ontology Pattern. Gerd Gröner, Matthias Thimm. Institute for Web Science and Technologies (WeST) University of Koblenz-Landau
 Semantic Web Ontology Pattern Gerd Gröner, Matthias Thimm {groener,thimm}@uni-koblenz.de Institute for Web Science and Technologies (WeST) University of Koblenz-Landau July 18, 2013 Gerd Gröner, Matthias
Semantic Web Ontology Pattern Gerd Gröner, Matthias Thimm {groener,thimm}@uni-koblenz.de Institute for Web Science and Technologies (WeST) University of Koblenz-Landau July 18, 2013 Gerd Gröner, Matthias
Effective Audit Trail of Data With PROV-O Scott Henninger, Senior Consultant, MarkLogic
 Effective Audit Trail of Data With PROV-O Scott Henninger, Senior Consultant, MarkLogic COPYRIGHT 13 June 2017MARKLOGIC CORPORATION. ALL RIGHTS RESERVED. EFFECTIVE AUDIT TRAIL WITH PROV-O Operationalizing
Effective Audit Trail of Data With PROV-O Scott Henninger, Senior Consultant, MarkLogic COPYRIGHT 13 June 2017MARKLOGIC CORPORATION. ALL RIGHTS RESERVED. EFFECTIVE AUDIT TRAIL WITH PROV-O Operationalizing
Library of Congress BIBFRAME Pilot. NOTSL Fall Meeting October 30, 2015
 Library of Congress BIBFRAME Pilot NOTSL Fall Meeting October 30, 2015 THE BIBFRAME EDITOR AND THE LC PILOT The Semantic Web and Linked Data : a Recap of the Key Concepts Learning Objectives Describe the
Library of Congress BIBFRAME Pilot NOTSL Fall Meeting October 30, 2015 THE BIBFRAME EDITOR AND THE LC PILOT The Semantic Web and Linked Data : a Recap of the Key Concepts Learning Objectives Describe the
SPARQL Protocol And RDF Query Language
 SPARQL Protocol And RDF Query Language WS 2011/12: XML Technologies John Julian Carstens Department of Computer Science Communication Systems Group Christian-Albrechts-Universität zu Kiel March 1, 2012
SPARQL Protocol And RDF Query Language WS 2011/12: XML Technologies John Julian Carstens Department of Computer Science Communication Systems Group Christian-Albrechts-Universität zu Kiel March 1, 2012
A Knowledge-Based System for the Specification of Variables in Clinical Trials
 A Knowledge-Based System for the Specification of Variables in Clinical Trials Matthias Löbe, Barbara Strotmann, Kai-Uwe Hoop, Roland Mücke Institute for Medical Informatics, Statistics and Epidemiology
A Knowledge-Based System for the Specification of Variables in Clinical Trials Matthias Löbe, Barbara Strotmann, Kai-Uwe Hoop, Roland Mücke Institute for Medical Informatics, Statistics and Epidemiology
Contents. G52IWS: The Semantic Web. The Semantic Web. Semantic web elements. Semantic Web technologies. Semantic Web Services
 Contents G52IWS: The Semantic Web Chris Greenhalgh 2007-11-10 Introduction to the Semantic Web Semantic Web technologies Overview RDF OWL Semantic Web Services Concluding comments 1 See Developing Semantic
Contents G52IWS: The Semantic Web Chris Greenhalgh 2007-11-10 Introduction to the Semantic Web Semantic Web technologies Overview RDF OWL Semantic Web Services Concluding comments 1 See Developing Semantic
3.4 Data-Centric workflow
 3.4 Data-Centric workflow One of the most important activities in a S-DWH environment is represented by data integration of different and heterogeneous sources. The process of extract, transform, and load
3.4 Data-Centric workflow One of the most important activities in a S-DWH environment is represented by data integration of different and heterogeneous sources. The process of extract, transform, and load
> Semantic Web Use Cases and Case Studies
 > Semantic Web Use Cases and Case Studies Case Study: Improving Web Search using Metadata Peter Mika, Yahoo! Research, Spain November 2008 Presenting compelling search results depends critically on understanding
> Semantic Web Use Cases and Case Studies Case Study: Improving Web Search using Metadata Peter Mika, Yahoo! Research, Spain November 2008 Presenting compelling search results depends critically on understanding
Comparing Provenance Data Models for Scientific Workflows: an Analysis of PROV-Wf and ProvOne
 Comparing Provenance Data Models for Scientific Workflows: an Analysis of PROV-Wf and ProvOne Wellington Oliveira 1, 2, Paolo Missier 3, Daniel de Oliveira 1, Vanessa Braganholo 1 1 Instituto de Computação,
Comparing Provenance Data Models for Scientific Workflows: an Analysis of PROV-Wf and ProvOne Wellington Oliveira 1, 2, Paolo Missier 3, Daniel de Oliveira 1, Vanessa Braganholo 1 1 Instituto de Computação,
Publishing Linked Statistical Data: Aragón, a case study.
 Publishing Linked Statistical Data: Aragón, a case study. Oscar Corcho 1, Idafen Santana-Pérez 1, Hugo Lafuente 2, David Portolés 3, César Cano 4, Alfredo Peris 4, and José María Subero 4 1 Ontology Engineering
Publishing Linked Statistical Data: Aragón, a case study. Oscar Corcho 1, Idafen Santana-Pérez 1, Hugo Lafuente 2, David Portolés 3, César Cano 4, Alfredo Peris 4, and José María Subero 4 1 Ontology Engineering
Wings Demo Walkthrough
 Wings Demo Walkthrough For a brief overview of Wings, see http://www.isi.edu/~gil/slides/wingstour-8-08.pdf Last Update: August 22, 2008 " 1 Summary of Wings Demonstration Wings can: Express high-level
Wings Demo Walkthrough For a brief overview of Wings, see http://www.isi.edu/~gil/slides/wingstour-8-08.pdf Last Update: August 22, 2008 " 1 Summary of Wings Demonstration Wings can: Express high-level
Controlled vocabularies, taxonomies, and thesauruses (and ontologies)
 Controlled vocabularies, taxonomies, and thesauruses (and ontologies) When people maintain a vocabulary of terms and sometimes, metadata about these terms they often use different words to refer to this
Controlled vocabularies, taxonomies, and thesauruses (and ontologies) When people maintain a vocabulary of terms and sometimes, metadata about these terms they often use different words to refer to this
SEXTANT 1. Purpose of the Application
 SEXTANT 1. Purpose of the Application Sextant has been used in the domains of Earth Observation and Environment by presenting its browsing and visualization capabilities using a number of link geospatial
SEXTANT 1. Purpose of the Application Sextant has been used in the domains of Earth Observation and Environment by presenting its browsing and visualization capabilities using a number of link geospatial
Revisiting Blank Nodes in RDF to Avoid the Semantic Mismatch with SPARQL
 Revisiting Blank Nodes in RDF to Avoid the Semantic Mismatch with SPARQL Marcelo Arenas 1, Mariano Consens 2, and Alejandro Mallea 1,3 1 Pontificia Universidad Católica de Chile 2 University of Toronto
Revisiting Blank Nodes in RDF to Avoid the Semantic Mismatch with SPARQL Marcelo Arenas 1, Mariano Consens 2, and Alejandro Mallea 1,3 1 Pontificia Universidad Católica de Chile 2 University of Toronto
Data Curation Profile Human Genomics
 Data Curation Profile Human Genomics Profile Author Profile Author Institution Name Contact J. Carlson N. Brown Purdue University J. Carlson, jrcarlso@purdue.edu Date of Creation October 27, 2009 Date
Data Curation Profile Human Genomics Profile Author Profile Author Institution Name Contact J. Carlson N. Brown Purdue University J. Carlson, jrcarlso@purdue.edu Date of Creation October 27, 2009 Date
Cybersecurity Through Nimble Task Allocation: Semantic Workflow Reasoning for Mission-Centered Network Models
 Cybersecurity Through Nimble Task Allocation: Semantic Workflow Reasoning for Mission-Centered Network Models Information Sciences Institute University of Southern California gil@isi.edu http://www.isi.edu/~gil
Cybersecurity Through Nimble Task Allocation: Semantic Workflow Reasoning for Mission-Centered Network Models Information Sciences Institute University of Southern California gil@isi.edu http://www.isi.edu/~gil
Bridging the Gap between Semantic Web and Networked Sensors: A Position Paper
 Bridging the Gap between Semantic Web and Networked Sensors: A Position Paper Xiang Su and Jukka Riekki Intelligent Systems Group and Infotech Oulu, FIN-90014, University of Oulu, Finland {Xiang.Su,Jukka.Riekki}@ee.oulu.fi
Bridging the Gap between Semantic Web and Networked Sensors: A Position Paper Xiang Su and Jukka Riekki Intelligent Systems Group and Infotech Oulu, FIN-90014, University of Oulu, Finland {Xiang.Su,Jukka.Riekki}@ee.oulu.fi
Electronic Health Records with Cleveland Clinic and Oracle Semantic Technologies
 Electronic Health Records with Cleveland Clinic and Oracle Semantic Technologies David Booth, Ph.D., Cleveland Clinic (contractor) Oracle OpenWorld 20-Sep-2010 Latest version of these slides: http://dbooth.org/2010/oow/
Electronic Health Records with Cleveland Clinic and Oracle Semantic Technologies David Booth, Ph.D., Cleveland Clinic (contractor) Oracle OpenWorld 20-Sep-2010 Latest version of these slides: http://dbooth.org/2010/oow/
Joining the BRICKS Network - A Piece of Cake
 Joining the BRICKS Network - A Piece of Cake Robert Hecht and Bernhard Haslhofer 1 ARC Seibersdorf research - Research Studios Studio Digital Memory Engineering Thurngasse 8, A-1090 Wien, Austria {robert.hecht
Joining the BRICKS Network - A Piece of Cake Robert Hecht and Bernhard Haslhofer 1 ARC Seibersdorf research - Research Studios Studio Digital Memory Engineering Thurngasse 8, A-1090 Wien, Austria {robert.hecht
SMARTDATA: Leveraging Blockchain to Securely Capture & Verify Scientific Provenance Data
 UT DALLAS Erik Jonsson School of Engineering & Computer Science SMARTDATA: Leveraging Blockchain to Securely Capture & Verify Scientific Provenance Data Dr.Murat Kantarcioglu Joint work with Aravind Ramachandran
UT DALLAS Erik Jonsson School of Engineering & Computer Science SMARTDATA: Leveraging Blockchain to Securely Capture & Verify Scientific Provenance Data Dr.Murat Kantarcioglu Joint work with Aravind Ramachandran
Extracting reproducible simulation studies from model repositories using the CombineArchive Toolkit
 Extracting reproducible simulation studies from model repositories using the CombineArchive Toolkit Martin Scharm, Dagmar Waltemath Department of Systems Biology and Bioinformatics University of Rostock
Extracting reproducible simulation studies from model repositories using the CombineArchive Toolkit Martin Scharm, Dagmar Waltemath Department of Systems Biology and Bioinformatics University of Rostock
Adaptable and Adaptive Web Information Systems. Lecture 1: Introduction
 Adaptable and Adaptive Web Information Systems School of Computer Science and Information Systems Birkbeck College University of London Lecture 1: Introduction George Magoulas gmagoulas@dcs.bbk.ac.uk October
Adaptable and Adaptive Web Information Systems School of Computer Science and Information Systems Birkbeck College University of London Lecture 1: Introduction George Magoulas gmagoulas@dcs.bbk.ac.uk October
Enrichment of Sensor Descriptions and Measurements Using Semantic Technologies. Student: Alexandra Moraru Mentor: Prof. Dr.
 Enrichment of Sensor Descriptions and Measurements Using Semantic Technologies Student: Alexandra Moraru Mentor: Prof. Dr. Dunja Mladenić Environmental Monitoring automation Traffic Monitoring integration
Enrichment of Sensor Descriptions and Measurements Using Semantic Technologies Student: Alexandra Moraru Mentor: Prof. Dr. Dunja Mladenić Environmental Monitoring automation Traffic Monitoring integration
Semantic Web Fundamentals
 Semantic Web Fundamentals Web Technologies (706.704) 3SSt VU WS 2018/19 with acknowledgements to P. Höfler, V. Pammer, W. Kienreich ISDS, TU Graz January 7 th 2019 Overview What is Semantic Web? Technology
Semantic Web Fundamentals Web Technologies (706.704) 3SSt VU WS 2018/19 with acknowledgements to P. Höfler, V. Pammer, W. Kienreich ISDS, TU Graz January 7 th 2019 Overview What is Semantic Web? Technology
Ontology-based Architecture Documentation Approach
 4 Ontology-based Architecture Documentation Approach In this chapter we investigate how an ontology can be used for retrieving AK from SA documentation (RQ2). We first give background information on the
4 Ontology-based Architecture Documentation Approach In this chapter we investigate how an ontology can be used for retrieving AK from SA documentation (RQ2). We first give background information on the
FIBO Metadata in Ontology Mapping
 FIBO Metadata in Ontology Mapping For Open Ontology Repository OOR Metadata Workshop VIII 02 July 2013 Copyright 2010 EDM Council Inc. 1 Overview The Financial Industry Business Ontology Introduction FIBO
FIBO Metadata in Ontology Mapping For Open Ontology Repository OOR Metadata Workshop VIII 02 July 2013 Copyright 2010 EDM Council Inc. 1 Overview The Financial Industry Business Ontology Introduction FIBO
The Data Web and Linked Data.
 Mustafa Jarrar Lecture Notes, Knowledge Engineering (SCOM7348) University of Birzeit 1 st Semester, 2011 Knowledge Engineering (SCOM7348) The Data Web and Linked Data. Dr. Mustafa Jarrar University of
Mustafa Jarrar Lecture Notes, Knowledge Engineering (SCOM7348) University of Birzeit 1 st Semester, 2011 Knowledge Engineering (SCOM7348) The Data Web and Linked Data. Dr. Mustafa Jarrar University of
D WSMO Data Grounding Component
 Project Number: 215219 Project Acronym: SOA4All Project Title: Instrument: Thematic Priority: Service Oriented Architectures for All Integrated Project Information and Communication Technologies Activity
Project Number: 215219 Project Acronym: SOA4All Project Title: Instrument: Thematic Priority: Service Oriented Architectures for All Integrated Project Information and Communication Technologies Activity
Core Technology Development Team Meeting
 Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
SKOS. COMP62342 Sean Bechhofer
 SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Ontologies Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Ontologies Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
The Trustworthiness of Digital Records
 The Trustworthiness of Digital Records International Congress on Digital Records Preservation Beijing, China 16 April 2010 1 The Concept of Record Record: any document made or received by a physical or
The Trustworthiness of Digital Records International Congress on Digital Records Preservation Beijing, China 16 April 2010 1 The Concept of Record Record: any document made or received by a physical or
Linked data and its role in the semantic web. Dave Reynolds, Epimorphics
 Linked data and its role in the semantic web Dave Reynolds, Epimorphics Ltd @der42 Roadmap What is linked data? Modelling Strengths and weaknesses Examples Access other topics image: Leo Oosterloo @ flickr.com
Linked data and its role in the semantic web Dave Reynolds, Epimorphics Ltd @der42 Roadmap What is linked data? Modelling Strengths and weaknesses Examples Access other topics image: Leo Oosterloo @ flickr.com
The RMap Project: Linking the Products of Research and Scholarly Communication Tim DiLauro
 The RMap Project: Linking the Products of Research and Scholarly Communication 2015 04 22 Tim DiLauro Motivation Compound objects fast becoming the norm for outputs of scholarly communication.
The RMap Project: Linking the Products of Research and Scholarly Communication 2015 04 22 Tim DiLauro Motivation Compound objects fast becoming the norm for outputs of scholarly communication.
Semantics. Matthew J. Graham CACR. Methods of Computational Science Caltech, 2011 May 10. matthew graham
 Semantics Matthew J. Graham CACR Methods of Computational Science Caltech, 2011 May 10 semantic web The future of the Internet (Web 3.0) Decentralized platform for distributed knowledge A web of databases
Semantics Matthew J. Graham CACR Methods of Computational Science Caltech, 2011 May 10 semantic web The future of the Internet (Web 3.0) Decentralized platform for distributed knowledge A web of databases
6. The Document Engineering Approach
 6. The Document Engineering Approach DE + IA (INFO 243) - 11 February 2008 Bob Glushko 1 of 40 Plan for Today's Class Modeling Methodologies The Document Engineering Approach 2 of 40 What Modeling Methodologies
6. The Document Engineering Approach DE + IA (INFO 243) - 11 February 2008 Bob Glushko 1 of 40 Plan for Today's Class Modeling Methodologies The Document Engineering Approach 2 of 40 What Modeling Methodologies
Perspectives on Open Data in Science Open Data in Science: Challenges & Opportunities for Europe
 Perspectives on Open Data in Science Open Data in Science: Challenges & Opportunities for Europe Stephane Berghmans, DVM PhD 31 January 2018 9 When talking about data, we talk about All forms of research
Perspectives on Open Data in Science Open Data in Science: Challenges & Opportunities for Europe Stephane Berghmans, DVM PhD 31 January 2018 9 When talking about data, we talk about All forms of research
Pedigree Management and Assessment Framework (PMAF) Demonstration
 Pedigree Management and Assessment Framework (PMAF) Demonstration Kenneth A. McVearry ATC-NY, Cornell Business & Technology Park, 33 Thornwood Drive, Suite 500, Ithaca, NY 14850 kmcvearry@atcorp.com Abstract.
Pedigree Management and Assessment Framework (PMAF) Demonstration Kenneth A. McVearry ATC-NY, Cornell Business & Technology Park, 33 Thornwood Drive, Suite 500, Ithaca, NY 14850 kmcvearry@atcorp.com Abstract.
The Open Group SOA Ontology Technical Standard. Clive Hatton
 The Open Group SOA Ontology Technical Standard Clive Hatton The Open Group Releases SOA Ontology Standard To Increase SOA Adoption and Success Rates Ontology Fosters Common Understanding of SOA Concepts
The Open Group SOA Ontology Technical Standard Clive Hatton The Open Group Releases SOA Ontology Standard To Increase SOA Adoption and Success Rates Ontology Fosters Common Understanding of SOA Concepts
Grid Resources Search Engine based on Ontology
 based on Ontology 12 E-mail: emiao_beyond@163.com Yang Li 3 E-mail: miipl606@163.com Weiguang Xu E-mail: miipl606@163.com Jiabao Wang E-mail: miipl606@163.com Lei Song E-mail: songlei@nudt.edu.cn Jiang
based on Ontology 12 E-mail: emiao_beyond@163.com Yang Li 3 E-mail: miipl606@163.com Weiguang Xu E-mail: miipl606@163.com Jiabao Wang E-mail: miipl606@163.com Lei Song E-mail: songlei@nudt.edu.cn Jiang
Semantic Web Fundamentals
 Semantic Web Fundamentals Web Technologies (706.704) 3SSt VU WS 2017/18 Vedran Sabol with acknowledgements to P. Höfler, V. Pammer, W. Kienreich ISDS, TU Graz December 11 th 2017 Overview What is Semantic
Semantic Web Fundamentals Web Technologies (706.704) 3SSt VU WS 2017/18 Vedran Sabol with acknowledgements to P. Höfler, V. Pammer, W. Kienreich ISDS, TU Graz December 11 th 2017 Overview What is Semantic
Features and Requirements for an XML View Definition Language: Lessons from XML Information Mediation
 Page 1 of 5 Features and Requirements for an XML View Definition Language: Lessons from XML Information Mediation 1. Introduction C. Baru, B. Ludäscher, Y. Papakonstantinou, P. Velikhov, V. Vianu XML indicates
Page 1 of 5 Features and Requirements for an XML View Definition Language: Lessons from XML Information Mediation 1. Introduction C. Baru, B. Ludäscher, Y. Papakonstantinou, P. Velikhov, V. Vianu XML indicates
Ontologies SKOS. COMP62342 Sean Bechhofer
 Ontologies SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
Ontologies SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
Semantic Web: Core Concepts and Mechanisms. MMI ORR Ontology Registry and Repository
 Semantic Web: Core Concepts and Mechanisms MMI ORR Ontology Registry and Repository Carlos A. Rueda Monterey Bay Aquarium Research Institute Moss Landing, CA ESIP 2016 Summer meeting What s all this about?!
Semantic Web: Core Concepts and Mechanisms MMI ORR Ontology Registry and Repository Carlos A. Rueda Monterey Bay Aquarium Research Institute Moss Landing, CA ESIP 2016 Summer meeting What s all this about?!
Component-Based Software Engineering TIP
 Component-Based Software Engineering TIP X LIU, School of Computing, Napier University This chapter will present a complete picture of how to develop software systems with components and system integration.
Component-Based Software Engineering TIP X LIU, School of Computing, Napier University This chapter will present a complete picture of how to develop software systems with components and system integration.
is easing the creation of new ontologies by promoting the reuse of existing ones and automating, as much as possible, the entire ontology
 Preface The idea of improving software quality through reuse is not new. After all, if software works and is needed, just reuse it. What is new and evolving is the idea of relative validation through testing
Preface The idea of improving software quality through reuse is not new. After all, if software works and is needed, just reuse it. What is new and evolving is the idea of relative validation through testing
Application of Named Graphs Towards Custom Provenance Views
 Application of Named Graphs Towards Custom Provenance Views Tara Gibson, Karen Schuchardt, Eric Stephan Pacific Northwest National Laboratory Abstract Provenance capture as applied to execution oriented
Application of Named Graphs Towards Custom Provenance Views Tara Gibson, Karen Schuchardt, Eric Stephan Pacific Northwest National Laboratory Abstract Provenance capture as applied to execution oriented
Linked Data: Fast, low cost semantic interoperability for health care?
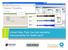 Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
Semantic Web and ehealth
 White Paper Series Semantic Web and ehealth January 2013 SEMANTIC IDENTITY OBSERVE REASON IMAGINE Publication Details Author: Renato Iannella Email: ri@semanticidentity.com Date: January 2013 ISBN: 1 74064
White Paper Series Semantic Web and ehealth January 2013 SEMANTIC IDENTITY OBSERVE REASON IMAGINE Publication Details Author: Renato Iannella Email: ri@semanticidentity.com Date: January 2013 ISBN: 1 74064
VISUALIZATIONS AND INFORMATION WIDGETS
 HORIZON 2020 ICT - INFORMATION AND COMMUNICATION TECHNOLOGIES PRO MOTING FI NANCIAL AWARENESS AND STABILIT Y H2020 687895 VISUALIZATIONS AND INFORMATION WIDGETS WORK PACKAGE NO. WP2 WORK PACKAGE TITLE
HORIZON 2020 ICT - INFORMATION AND COMMUNICATION TECHNOLOGIES PRO MOTING FI NANCIAL AWARENESS AND STABILIT Y H2020 687895 VISUALIZATIONS AND INFORMATION WIDGETS WORK PACKAGE NO. WP2 WORK PACKAGE TITLE
Managing Learning Objects in Large Scale Courseware Authoring Studio 1
 Managing Learning Objects in Large Scale Courseware Authoring Studio 1 Ivo Marinchev, Ivo Hristov Institute of Information Technologies Bulgarian Academy of Sciences, Acad. G. Bonchev Str. Block 29A, Sofia
Managing Learning Objects in Large Scale Courseware Authoring Studio 1 Ivo Marinchev, Ivo Hristov Institute of Information Technologies Bulgarian Academy of Sciences, Acad. G. Bonchev Str. Block 29A, Sofia
Semantic Technologies and CDISC Standards. Frederik Malfait, Information Architect, IMOS Consulting Scott Bahlavooni, Independent
 Semantic Technologies and CDISC Standards Frederik Malfait, Information Architect, IMOS Consulting Scott Bahlavooni, Independent Part I Introduction to Semantic Technology Resource Description Framework
Semantic Technologies and CDISC Standards Frederik Malfait, Information Architect, IMOS Consulting Scott Bahlavooni, Independent Part I Introduction to Semantic Technology Resource Description Framework
ResolutionDefinition - PILIN Team Wiki - Trac. Resolve. Retrieve. Reveal Association. Facets. Indirection. Association data. Retrieval Key.
 Resolve. Retrieve. Reveal Association. Facets. Indirection. Association data. Retrieval Key. ResolutionDefinitionBackground 1. Other definitions XRI definition: Resolution is the function of dereferencing
Resolve. Retrieve. Reveal Association. Facets. Indirection. Association data. Retrieval Key. ResolutionDefinitionBackground 1. Other definitions XRI definition: Resolution is the function of dereferencing
