Development of an Environment for Data Annotation in Bioengineering
|
|
|
- Logan Jones
- 5 years ago
- Views:
Transcription
1 Development of an Environment for Data Annotation in Bioengineering Renato Galina Barbosa Azambuja Correia Instituto Superior Técnico, Universidade Técnica de Lisboa, Portugal ABSTRACT In the biological sciences, it is increasingly common to have methodologies that produce large amounts of data. In order to make sense of these data, as well as to enable them to be searched, exploited and reused, the context and provenance of the data must be described in the form of metadata. These metadata are usually expressed through annotations, which are a standardized and semantically rich way of expressing them, and always structured as ontologies. The slowness of this process, in the search for the most appropriate ontology for the annotation of a given data and in the identification of the intended concept within this ontology is a pressing problem so it is critical that there is an informatic support to facilitate this process. The objective is the development of an infrastructure to support this type of tasks taking as reference the data repositories BioPortal, AgroPortal and GeoNames and other tools already developed such as OntoMaton, an add-on to google spreadsheets and the Agreement Maker Light (AML), a light ontologies mapper. The solution consists of the design, development and validation of a web environment which supports the data annotation tasks as automated as possible in time and efficiency. Keywords Environment for data annotation, Biology, Bioengineering, BioPortal, AgroPortal, Geonames, OntoMaton, AgreementMakerLight. 1. INTRODUCTION Data annotation in Biology and Bioengineering consists on enriching original data structures, produced in concrete projects, with attributes or complementary data structures generally called metadata, which describe and relate them to more generic contexts. Typically, annotation tasks consist on defining the attributes or data structures to use, and then assign them values, which, ideally, should exist in controlled vocabularies. For that purpose, many annotation structures in Biology exist as ontologies. [1] Annotation of data for ontologies is a timeconsuming and difficult task. The slowness of this process, in the search for the most appropriate ontology for the annotation of a given data and the identification of the desired data / concept within this ontology is a pressing problem. The computer support for this kind of data annotation task is important, especially if it can take advantage of other already existing tools. This paper presents the design, development and validation of a web environment that supports the annotation tasks, taking as reference the data repositories BioPortal[4], AgroPortal 1 and GeoNames 2 and reusing some tools already developed such as OntoMaton ("add -on" to "google spreadsheets")[5] and AgreementMakerLight (a lightweight system for mapping ontologies)[6]. The final solution provides an environment for annotating data that allows annotation in a more automated and accurate way, with possibilities that allow an improvement of the conditions in which these tasks are performed. The remaining parts of this paper describes the challenges of data annotation in plant phenotyping [2], the solution we propose for that, and the results we were able to reach. 2. RELATED WORK The recent methodological evolution in plant phenotyping, as well as the growing importance of its applications in plant science and its reproduction, has produced a significant increase of multidimensional data with great potential for later discoveries and applications. The collection and storage of phenotypic observations is not yet sufficiently patterned. The lack of standardization results mainly from the great variability of the protocols, the multiplicity of measured phenotypic characteristics and the Summary of dissertation for Master degree, Instituto Superior Técnico, Lisboa, October
2 dependence of these characteristics of the environment.[2] A detailed description of the environmental characteristics in which the experiments occur should be expressed through a standardization, to be valid in a community and to ensure interoperability, avoiding misunderstandings among contributors. The interoperability of observations (phenotypic and environmental variables) is achieved by the use of ontologies that are linked and in constant development. In the phenotype domain, it has only recently begun to join to the annotation paradigm with ontologies while in other domains of biology this is already more common. In most cases, the phenotype data and corresponding metadata are managed in spreadsheets (for example CSV or Excel) with non-uniform structure and terminology. The annotation of these data is difficult because there are many different ontologies to describe some aspects, and few ontologies or even none to describe others. The goal is to develop a data annotation environment. In general, this environment receives data structured in CSV or Excel and an annotation scheme or specification, which along with a simple profile of the domain of plant biology will return an annotation to the data. The main benefits are the possible automation of the tool, its speed, efficiency, ease of use and inference ability to return the results. The specification that the data structure in CSV or Excel obeys is the Minimum Information About Plant Phenotyping Experiment (MIAPPE), a decided pattern among plant breeders and modelers of the plant phenotyping community, which defines a list of attributes that may be required to fully describe a set of phenotypic observations, built on the basis of the Minimum Information (MI) documents already created for other branches of research in Biology. These attributes are classified taking into account the aspects common to all major cases of plant phenotyping. An MI document should therefore be considered as a checklist so that researchers who describe and record the data ensure that all the characteristics of important data are included in an experimental process, i.e. what is meaningful for their correct interpretation, evaluation, review and potential replication of experimentation or research.[3] The main technologies analyzed and chosen to build the solution were the OntoMaton and the AgreementMakerLight. OntoMaton is a general purpose tool and one of the most recent components of the ISA software package. It is an open source solution that brings ontology search and markup capabilities in a collaborative cloud-based environment, leveraging Google Spreadsheets and NCBO BioPortal Web Services. OntoMaton can also be used to support the ontology development process. OntoMaton facilitates the search for ontologies and the marking of text-free ontologies from biological experiments directly in Google Sheets. It allows searching for biological terms in NCBO BioPortal Web Services. Lets you search terms in other domains with Linked Open Vocabularies. The provenance of the annotated terms is preserved for downstream processing with URIs and links to definitions. It is implemented in JavaScript using the Google Apps Script API.[5] This tool is, however, limited to the search of NCBO BioPortal Web Services ontologies and only searches for literal terms i.e. if the target data is not written exactly as it is in the ontology, no match is found. AgreementMakerLight is a lightweight ontology mapping system, specialized in the biomedical domain, but applicable to any ontology, which can be used to generate alignments automatically, as a platform for alignment review or as a repair system.[6] This system is relevant for this work since the ontology mapping tasks are very similar to annotation tasks. This implemented environment offers potentialities that contribute more to solving the problems and difficulties encountered by researchers who describe observations of experiments in the field of plant biology. The implementation of this project contributes to the optimization, acceleration, and possible automation of the annotation process of experiments in the phenotype domain of the Elixir's Portuguese Node. 3. PROPOSED TECHNICAL SOLUTION Our solution, named DatAPP (Data Annotation in Plant Phenotyping), was inspired by OntoMaton. DatAPP uses AgreementMakerLight as a base for the creation of a Java back-end that will be run by a Web Service. Also a system of restrictions was implemented, that is, a system that restricts the ontologies where the search will be carried out. DatAPP receives a Google Spreadsheet according to the MIAPPE specification, containing in the first column the data fields, and, in the second column, the restrictions associated with each field. The annotation process starts by reading the Google Spreadsheet line selected by the user with the data to be annotated. Then it is checked whether the field present in this row contains any restrictions saved from the MIAPPE specification. In affirmative case, it will check if the restriction is GeoNames, if it is the search is performed using the GeoNames API. If the restriction is not GeoNames Summary of dissertation for Master degree, Instituto Superior Técnico, Lisboa, October
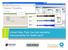
3 the Google Apps Script sends a GET request to the Web Service that will run the Java back-end with the term to be annotated and the restriction/ontology URI as parameters. This Java back-end was created in the Eclipse IDE based on an existing project called AgreementMakerLight (AML) that performs an alignment of ontologies that is similar to ontology matching. This Java back-end works with the ontologies themselves using OWL API for this purpose, constructing the ontology from an URI, storing the classes in a lexicon and the relationships between them in a relationship map. It also contains three Annotator classes, one matching for literal terms, another matching by strings, and another matching by words. Each Annotator returns a set of Annotation Candidates (that are candidates for the annotation, that is, matches found in each Annotator), which are then added to the final set. The Annotation Candidates are only added to the set if there are no equal matches on the set. In the opposite case, they are only added if they contain a larger weight than the one on the set, thus preventing repeated Annotation Candidates. After all the Annotators have ran and the Annotation Candidates added to the final set, this set is organized in descending order of weights and is returned as a JSON file by the Web Service to the Google Apps Script. Then the Google Apps Script will save the first term of the JSON file, that is, the term with the largest weight. And using a function based on OntoMaton, inserts, in the column next to the column containing the terms to be annotated, the URI linked to the term in the ontology specified in the MIAPPE, and in the next column the correspondent term to the URI annotated. The following use case (Figure 1) presents the final requirements of the new tool. Figure 2: Web Service Sequence Diagram Figure 3: Sequence Diagram for GeoNames The architecture of the implemented solution is represented in figure 4 (Figure 4). Figure 4: DatAPP Architecture 4. SOLUTION IMPLEMENTATION Figure 1: Use Case The following figures (Figures 2 and 3) respectively show the Web Service Sequence Diagram and the Sequence Diagram for GeoNames. The annotation tool implemented can be started by turning on the spreadsheets from Google ( Documents; Main Menu; Spreadsheets). A worksheet with data to be annotated is received from the user, in this case a Google Spreadsheet (Figure 5). Summary of dissertation for Master degree, Instituto Superior Técnico, Lisboa, October
4 Now the user only has to select the field and the term intended for the annotation and click on the button Tag terms that is in the User Interface and will be annotated to the data, the URI for the class found by DatAPP to annotate the term, and the name of the class that was found (Figure 8). Figure 5: Google Spreadsheet with data to annotate Then a worksheet is received with a MIAPPE specification, which will serve to restrict the fields of the data to be annotated (Figure 6). Figure 8: Result of annotation/ first line To continue to note the rest of the terms shown in the figure above, the user only has to select the next field and the next term, i.e. the next line of the Google Spreadsheet and click on "Tag terms" as we can see in the following example (Figure 9). Figure 6: Google Spreadsheet with the MIAPPE Specification Providing these two documents, the tool can be started as follows: in the upper bar of Google Spreadsheet select Add-ons; in the options of Addons select DatAPP; in DatAPP select Run Annotator, and on the right side of the Google Spreadsheet will appear the DatAPP user interface (Figure 7). Figure 9: Result of annotation/ second line The process continues in the same way until the last line of the Google Spreadsheet is noted. 5. EVALUATION Figure 7: DatAPP User Interface In order to assess if the main objectives were met, i.e. if the tool performs better with regard to automation, efficiency and accuracy than the methods used to date, the evaluation was done using the same Google Spreadsheet with the annotated data mentioned above in the DatAPP and OntoMaton for the features already mentioned in this document. Summary of dissertation for Master degree, Instituto Superior Técnico, Lisboa, October
5 Thus, two evaluations were carried out, one using OntoMaton and the other using the DatAPP, noting an evaluation dataset consisting of fifty (50) cases to be annotated, that is, fifty (50) lines in a Google Spreadsheet, containing several recommended ontologies and various terms to annotate. A further validation Google Spreadsheet was also used which contained the terms that should be noted for each case. The aspects to be taken into account in the evaluation are directly linked to the main objectives to be achieved. Thus, in each of the evaluations (DatAPP and OntoMaton) the accuracy and precision of the tools response were taken into account. We used a measurement system that allows us to evaluate the quality and accuracy with which a system returns the relevant data requested by the user. These measures are called Recall, Precision and F-Measure and are defined as follows: - Precision = Total number of relevant data returned / Total number of data that is returned. - Recall = Total number of data returned that is relevant / Total number of relevant data. F-Measure (Precision and Recall Harmonic Average) = 2 * Precision * Recall / Precision + Recall. Precision and Recall have an opposing relationship i.e. when we intend to increase the Recall, Precision tends to decrease and vice versa. In this case, we intend to measure Recall, Recall Extended, Precision, F-Measure and F-Measure Extended to evaluate the performance of DatAPP compared to OntoMaton. It is intended that the DatAPP will perform better with regard to its automated ability to return results by increasing the percentage of relevant results without sacrificing precision. Recall considers the percentage of success of the tool when finding and writing down the desired term. For Recall Extended, it is considered the percentage of success of the tool when finding the desired term contained in the terms found during the match process, and for Precision is considered the percentage success of the tool when finding any term in response to the request made by the user. OntoMaton Evaluation For the evaluation of OntoMaton it was necessary to access its definitions and add the restrictions of the ontologies for each field, since in this tool it is necessary to add the restrictions manually. Then, the evaluation dataset was row line by line and the term to be annotated inserted in the search engine of OntoMaton. In the return of the results, the first term returned as the annotated term is considered, since in OntoMaton there is no specific function for ordering the terms while the developed annotation tool adds the term of greater weight of the list of candidates for annotation. The procedures were repeated until the end of the dataset. DatAPP Evaluation For the evaluation of this tool, the line was selected to note as is exemplified in the point above "Solution Implemented" and began the process of request / demand. As already mentioned in this document the tool notes the term with greater weight found in the match process. This was done until the end of the evaluation dataset. In order to analyze the Recall Extended of the developed annotation tool, the URI that Google Apps Service uses to make the GET request to the Web Service directly in an internet browser has been sent, so that all annotation candidates returned by the Web Service to that request are presented, including the one of greater weight that is annotated. In this way we can verify if the desired term is in this list returned by the Web Service so that it can be validated successfully. Precision counts as successful whenever a response is returned. The procedures were repeated until the end of the dataset. 6. RESULTS AND DISCUSSION Before presenting the results of Recall, Recall Extended, Precision and F-Measure we will perform an analysis of the performance of each tool during the evaluation procedures. We can see that regarding automation, DatAPP shows an improvement over OntoMaton. Before starting the annotation process in OntoMaton, it is necessary to access the settings to manually enter the restrictions; DatAPP runs this function automatically. In the case of OntoMaton, the user needs to enter the term to annotate in the search bar and click the search button; OntoMaton returns a list of annotation candidates leaving to the user the task of deciding which one is the most relevant. Regarding DatAPP, you only need to select the line you want to annotate by selecting the first two columns of the Google Spreadsheet (the field and the term), and then clicking the annotate terms button; DatAPP returns an annotation to the data, inferring the relevant term from a list of annotation candidates ordered by weight; As for the results of Recall, Recall Extended and Precision they were accounted for and analyzed and Summary of dissertation for Master degree, Instituto Superior Técnico, Lisboa, October
6 are represented in the following figures (Figure 10 and 11): Resultados MEDIDAS AVALIAÇÃO ONTOMATON RECALL RECALL EXTENDED PRECISION 50% 56% 92% 75% 65% 80% 70% OntoMaton AVALIAÇÃO DATAPP 64% 72% 90% DatAPP Figure 10: Evaluation Results F- Measure F-Measure Extended Figure 12: Evaluation Graphic 50% 64% Resultados OntoMaton 56% 72% DatAPP Figure 11: Evaluation Graphic 92% 90% Recall Recall Extended Precision Given these results we can conclude that DatAPP achieved better results to those of OntoMaton. It maintains precision (with a difference of only 2%) that is OntoMaton in 92% of cases and DatAPP in 90% of cases return a term in response to the request made by the user. It shows an improvement over Recall and Recall Extended (increase of 14% and 16% respectively), which means that it performs better than the previously used tool, increasing the percentage of success in the last two mentioned parameters without, however, a significant decrease precision percentage. Although it does not change the presented results, it is important to note that OntoMaton presents more relevant answers than DatAPP in cases where the text to be annotated corresponds to a synonym and not to the main name of the class of the ontology. The F-Measure and F-Measure Extended were calculated based on the values found for Precision, Recall and Recall Extended. The results are shown in the following figure. (Figure 12). The results of F-Measure show a 10% improvement of DatAPP over OntoMaton and clearly demonstrate the superiority of the developed tool. 7. CONCLUSIONS Data annotation in Biology and Bioengineering consists of enriching original data structures, produced in concrete projects, with attributes or complementary data structures, generally called metadata, which describe and relate them to more generic contexts. Typically, annotation tasks consist of defining the attributes or data structures to use, and then filling them by assigning them values. Attributes, complementary data structures, and values that are assigned to them must belong to controlled vocabularies or domains. Annotation of data for ontologies is a timeconsuming and difficult task. The slowness of this process, in the search for the most appropriate ontology for the annotation of a given data and in the identification of the desired data / concept within this ontology is a pressing problem. In the phenotype domain, it has only recently begun to join to the annotation paradigm with ontologies while in other domains of biology this is already more common. The annotation of phenotyping data is difficult because there are many different ontologies to describe some aspects, and few ontologies or even none to describe others. Also the collection and storage of phenotypic observations is not yet sufficiently patterned. A detailed description of the environmental characteristics in which the experiments occur should be expressed through a standardization, to be valid in a community and to ensure interoperability, avoiding misunderstandings among contributors. Summary of dissertation for Master degree, Instituto Superior Técnico, Lisboa, October
7 The web environment addresses this problem by printing more automation and accuracy than previously used tools. However, there is still a lot of work to do. Improvements can be made to increase accuracy and automation to access heavy ontologies without the need of user interference. This is a challenge yet to come. 8. FUTURE WORK After measuring the results of the evaluations and analyzing the performance of the tool, it was concluded that there are still potentialities in which worth betting and possibilities for intervention in the future. It is worth mentioning in this point two aspects that we would like to see improved in an update of the tool developed in this project: During the evaluation it was noticed that OntoMaton obtained more relevant answers in cases where the text to be annotated corresponds to a synonym and not to the main name of the class of the ontology than DatAPP, which tells us that the tool developed gives greater weight to the similarity between terms than to synonyms. A future work would be to adjust the weight system used in DatAPP so that it presents similar relevance to that of OntoMaton in relation to the synonyms. This adjustment can balance or even improve accuracy values. Consider introducing the option to display a list of all returned annotation candidates, so that the user can compare them with the response returned automatically by the tool, and can choose another term to annotate, risking the loss of some automation in favor of precision and accuracy. It is considered essential that DatAPP be used and evaluated by a large group of users so that suggestions can be made that will lead to future work to be developed. It is also considered a field to explore the adaptation and reuse of the tool for users with different needs but involving matching or query / demand tasks. 9. ACKNOWLEDGEMENTS gratitude to António Higgs for their constant help and advice given. This work was developed in the scope of ELIXIR- PT, supported by the ELIXIR-EXCELERATE project funded by the European Commission (grant agreement ). The researchers from INESC- ID had financial support from the multiannual funding from the PIDDAC programme (UID/CEC/50021/2013). 10. REFERENCES [1] Bose, R., Buneman, P., & Ecklund, D. (2006). Annotating scientific data: why it is important and why it is difficult. In Proceedings of the 2006 UK e-science all hands meeting. (pp ). NeSC. [2] White, J. W. et al. (2012). Field-based phenomics for plant genetics research. Field Crops Research, 133, [3] Ćwiek-Kupczyńska et al. (2016). Measures for interoperability of phenotypic data: minimum information requirements and formatting. Plant Methods. [4] Whetzel, P. L., Noy, N. F., Shah, N. H., Alexander, P. R., Nyulas, C., Tudorache, T., & Musen, M. A. (2011). BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Research, 39 (Web Server issue), W541 W [5] Maguire, E., González-Beltrán, A., Whetzel, P. L., Sansone, S.-A., & Rocca-Serra, P. (2013). OntoMaton: a Bioportal powered ontology widget for Google Spreadsheets. Bioinformatics, 29(4), [6] Faria D., Pesquita C., Santos E., Palmonari M., Cruz I. F. e Couto F. M. (2013). The AgreementMakerLight Ontology Matching System. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). vol LNCS Firstly, I would like to thank Professor José Borbinha for the opportunity to carry out this work, as well as for his guidance during this year and its course. I would like to express a special thanks to Dr. Daniel Faria and João Cardoso for all the help, support and effort. Finally, I would like to show Summary of dissertation for Master degree, Instituto Superior Técnico, Lisboa, October
warwick.ac.uk/lib-publications
 Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
WebProtégé. Protégé going Web. Tania Tudorache, Jennifer Vendetti, Natasha Noy. Stanford Center for Biomedical Informatics
 WebProtégé Protégé going Web Tania Tudorache, Jennifer Vendetti, Natasha Noy Stanford Center for Biomedical Informatics Protégé conference 2009 Amsterdam, June 24, 2009 WebProtégé quick overview WebProtégé
WebProtégé Protégé going Web Tania Tudorache, Jennifer Vendetti, Natasha Noy Stanford Center for Biomedical Informatics Protégé conference 2009 Amsterdam, June 24, 2009 WebProtégé quick overview WebProtégé
Embracing Semantic Technology for Better Metadata Authoring in Biomedicine
 Embracing Semantic Technology for Better Metadata Authoring in Biomedicine Attila L. Egyedi, Martin J. O Connor, Marcos Martínez-Romero, Debra Willrett, Josef Hardi, John Graybeal, and Mark A. Musen Stanford
Embracing Semantic Technology for Better Metadata Authoring in Biomedicine Attila L. Egyedi, Martin J. O Connor, Marcos Martínez-Romero, Debra Willrett, Josef Hardi, John Graybeal, and Mark A. Musen Stanford
AML Results for OAEI 2015
 AML Results for OAEI 2015 Daniel Faria 1, Catarina Martins 2, Amruta Nanavaty 3, Daniela Oliveira 2, Booma S. Balasubramani 3, Aynaz Taheri 3, Catia Pesquita 2, Francisco M. Couto 2, and Isabel F. Cruz
AML Results for OAEI 2015 Daniel Faria 1, Catarina Martins 2, Amruta Nanavaty 3, Daniela Oliveira 2, Booma S. Balasubramani 3, Aynaz Taheri 3, Catia Pesquita 2, Francisco M. Couto 2, and Isabel F. Cruz
I. Background and objectives
 Meeting EMPHASIS ELIXIR 15 May 2018 : Data standards and Information Systems: strategies of the European infrastructures EMPHASIS and ELIXIR Participants : See appendix Main authors: C. Pommier and F.
Meeting EMPHASIS ELIXIR 15 May 2018 : Data standards and Information Systems: strategies of the European infrastructures EMPHASIS and ELIXIR Participants : See appendix Main authors: C. Pommier and F.
A Semantic Web-Based Approach for Harvesting Multilingual Textual. definitions from Wikipedia to support ICD-11 revision
 A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
Semantic Annotation and Linking of Medical Educational Resources
 5 th European IFMBE MBEC, Budapest, September 14-18, 2011 Semantic Annotation and Linking of Medical Educational Resources N. Dovrolis 1, T. Stefanut 2, S. Dietze 3, H.Q. Yu 3, C. Valentine 3 & E. Kaldoudi
5 th European IFMBE MBEC, Budapest, September 14-18, 2011 Semantic Annotation and Linking of Medical Educational Resources N. Dovrolis 1, T. Stefanut 2, S. Dietze 3, H.Q. Yu 3, C. Valentine 3 & E. Kaldoudi
NCBO Technology: Powering semantically aware applications
 JOURNAL OF BIOMEDICAL SEMANTICS PROCEEDINGS Open Access NCBO Technology: Powering semantically aware applications Patricia L Whetzel 1*, NCBO Team 1,2,3,4 From Bio-Ontologies 2012 Long Beach, CA, USA.
JOURNAL OF BIOMEDICAL SEMANTICS PROCEEDINGS Open Access NCBO Technology: Powering semantically aware applications Patricia L Whetzel 1*, NCBO Team 1,2,3,4 From Bio-Ontologies 2012 Long Beach, CA, USA.
Collaborative & WebProtégé
 Collaborative & WebProtégé Tania Tudorache Stanford Center for Biomedical Informatics Research Joint Ontolog-OOR Panel Session July 16, 2009 1 Collaborative Ontology Development Collaboration: several
Collaborative & WebProtégé Tania Tudorache Stanford Center for Biomedical Informatics Research Joint Ontolog-OOR Panel Session July 16, 2009 1 Collaborative Ontology Development Collaboration: several
AML Results for OAEI 2015
 AML Results for OAEI 2015 Daniel Faria 1, Catarina Martins 2, Amruta Nanavaty 4, Daniela Oliveira 2, Booma Sowkarthiga 4, Aynaz Taheri 4, Catia Pesquita 2,3, Francisco M. Couto 2,3, and Isabel F. Cruz
AML Results for OAEI 2015 Daniel Faria 1, Catarina Martins 2, Amruta Nanavaty 4, Daniela Oliveira 2, Booma Sowkarthiga 4, Aynaz Taheri 4, Catia Pesquita 2,3, Francisco M. Couto 2,3, and Isabel F. Cruz
An Annotation Tool for Semantic Documents
 An Annotation Tool for Semantic Documents (System Description) Henrik Eriksson Dept. of Computer and Information Science Linköping University SE-581 83 Linköping, Sweden her@ida.liu.se Abstract. Document
An Annotation Tool for Semantic Documents (System Description) Henrik Eriksson Dept. of Computer and Information Science Linköping University SE-581 83 Linköping, Sweden her@ida.liu.se Abstract. Document
BLOOMS on AgreementMaker: results for OAEI 2010
 BLOOMS on AgreementMaker: results for OAEI 2010 Catia Pesquita 1, Cosmin Stroe 2, Isabel F. Cruz 2, Francisco M. Couto 1 1 Faculdade de Ciencias da Universidade de Lisboa, Portugal cpesquita at xldb.di.fc.ul.pt,
BLOOMS on AgreementMaker: results for OAEI 2010 Catia Pesquita 1, Cosmin Stroe 2, Isabel F. Cruz 2, Francisco M. Couto 1 1 Faculdade de Ciencias da Universidade de Lisboa, Portugal cpesquita at xldb.di.fc.ul.pt,
ORES-2010 Ontology Repositories and Editors for the Semantic Web
 Vol-596 urn:nbn:de:0074-596-3 Copyright 2010 for the individual papers by the papers' authors. Copying permitted only for private and academic purposes. This volume is published and copyrighted by its
Vol-596 urn:nbn:de:0074-596-3 Copyright 2010 for the individual papers by the papers' authors. Copying permitted only for private and academic purposes. This volume is published and copyrighted by its
Using Linked Data and taxonomies to create a quick-start smart thesaurus
 7) MARJORIE HLAVA Using Linked Data and taxonomies to create a quick-start smart thesaurus 1. About the Case Organization The two current applications of this approach are a large scientific publisher
7) MARJORIE HLAVA Using Linked Data and taxonomies to create a quick-start smart thesaurus 1. About the Case Organization The two current applications of this approach are a large scientific publisher
The Final Updates. Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University of Oxford, UK
 The Final Updates Supported by the NIH grant 1U24 AI117966-01 to UCSD PI, Co-Investigators at: Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University
The Final Updates Supported by the NIH grant 1U24 AI117966-01 to UCSD PI, Co-Investigators at: Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University
OAEI 2016 Results of AML
 OAEI 2016 Results of AML Daniel Faria 1, Catia Pesquita 2, Booma S. Balasubramani 3, Catarina Martins 2, João Cardoso 4, Hugo Curado 2, Francisco M. Couto 2, and Isabel F. Cruz 3 1 Instituto Gulbenkian
OAEI 2016 Results of AML Daniel Faria 1, Catia Pesquita 2, Booma S. Balasubramani 3, Catarina Martins 2, João Cardoso 4, Hugo Curado 2, Francisco M. Couto 2, and Isabel F. Cruz 3 1 Instituto Gulbenkian
Linked Data: Fast, low cost semantic interoperability for health care?
 Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
New Approach to Graph Databases
 Paper PP05 New Approach to Graph Databases Anna Berg, Capish, Malmö, Sweden Henrik Drews, Capish, Malmö, Sweden Catharina Dahlbo, Capish, Malmö, Sweden ABSTRACT Graph databases have, during the past few
Paper PP05 New Approach to Graph Databases Anna Berg, Capish, Malmö, Sweden Henrik Drews, Capish, Malmö, Sweden Catharina Dahlbo, Capish, Malmö, Sweden ABSTRACT Graph databases have, during the past few
NOMAD Metadata for all
 EMMC Workshop on Interoperability NOMAD Metadata for all Cambridge, 8 Nov 2017 Fawzi Mohamed FHI Berlin NOMAD Center of excellence goals 200,000 materials known to exist basic properties for very few highly
EMMC Workshop on Interoperability NOMAD Metadata for all Cambridge, 8 Nov 2017 Fawzi Mohamed FHI Berlin NOMAD Center of excellence goals 200,000 materials known to exist basic properties for very few highly
Ontology Servers and Metadata Vocabulary Repositories
 Ontology Servers and Metadata Vocabulary Repositories Dr. Manjula Patel Technical Research and Development m.patel@ukoln.ac.uk http://www.ukoln.ac.uk/ Overview agentcities.net deployment grant Background
Ontology Servers and Metadata Vocabulary Repositories Dr. Manjula Patel Technical Research and Development m.patel@ukoln.ac.uk http://www.ukoln.ac.uk/ Overview agentcities.net deployment grant Background
Long-term preservation for INSPIRE: a metadata framework and geo-portal implementation
 Long-term preservation for INSPIRE: a metadata framework and geo-portal implementation INSPIRE 2010, KRAKOW Dr. Arif Shaon, Dr. Andrew Woolf (e-science, Science and Technology Facilities Council, UK) 3
Long-term preservation for INSPIRE: a metadata framework and geo-portal implementation INSPIRE 2010, KRAKOW Dr. Arif Shaon, Dr. Andrew Woolf (e-science, Science and Technology Facilities Council, UK) 3
Utilizing NCBO Tools to Develop & Use an ECG Ontology
 Utilizing NCBO Tools to Develop & Use an ECG Ontology Stephen J. Granite, MS, MBA The Johns Hopkins University Institute for Computational Medicine (sgranite at jhu dot edu) The CardioVascular Research
Utilizing NCBO Tools to Develop & Use an ECG Ontology Stephen J. Granite, MS, MBA The Johns Hopkins University Institute for Computational Medicine (sgranite at jhu dot edu) The CardioVascular Research
Information Retrieval (IR) through Semantic Web (SW): An Overview
 Information Retrieval (IR) through Semantic Web (SW): An Overview Gagandeep Singh 1, Vishal Jain 2 1 B.Tech (CSE) VI Sem, GuruTegh Bahadur Institute of Technology, GGS Indraprastha University, Delhi 2
Information Retrieval (IR) through Semantic Web (SW): An Overview Gagandeep Singh 1, Vishal Jain 2 1 B.Tech (CSE) VI Sem, GuruTegh Bahadur Institute of Technology, GGS Indraprastha University, Delhi 2
ELIXIR Human Data Use Case
 ELIXIR Human Data Use Case Mikael Borg, ELIXIR Sweden ELIXIR-EXCELERATE is funded by the European Commission within the Research Infrastructures programme of Horizon 2020, grant agreement number 676559.
ELIXIR Human Data Use Case Mikael Borg, ELIXIR Sweden ELIXIR-EXCELERATE is funded by the European Commission within the Research Infrastructures programme of Horizon 2020, grant agreement number 676559.
Community-based ontology development, alignment, and evaluation. Natasha Noy Stanford Center for Biomedical Informatics Research Stanford University
 Community-based ontology development, alignment, and evaluation Natasha Noy Stanford Center for Biomedical Informatics Research Stanford University Community-based Ontology... Everything Development and
Community-based ontology development, alignment, and evaluation Natasha Noy Stanford Center for Biomedical Informatics Research Stanford University Community-based Ontology... Everything Development and
Access Control in Rich Domain Model Web Applications
 Access Control in Rich Domain Model Web Applications Extended Abstract João de Albuquerque Penha Pereira joao.pereira@ist.utl.pt Instituto Superior Técnico November 25, 2010 Abstract Rich Domain Model
Access Control in Rich Domain Model Web Applications Extended Abstract João de Albuquerque Penha Pereira joao.pereira@ist.utl.pt Instituto Superior Técnico November 25, 2010 Abstract Rich Domain Model
PROJECT PERIODIC REPORT
 PROJECT PERIODIC REPORT Grant Agreement number: 257403 Project acronym: CUBIST Project title: Combining and Uniting Business Intelligence and Semantic Technologies Funding Scheme: STREP Date of latest
PROJECT PERIODIC REPORT Grant Agreement number: 257403 Project acronym: CUBIST Project title: Combining and Uniting Business Intelligence and Semantic Technologies Funding Scheme: STREP Date of latest
Languages and tools for building and using ontologies. Simon Jupp, James Malone
 An overview of ontology technology Languages and tools for building and using ontologies Simon Jupp, James Malone jupp@ebi.ac.uk, malone@ebi.ac.uk Outline Languages OWL and OBO classes, individuals, relations,
An overview of ontology technology Languages and tools for building and using ontologies Simon Jupp, James Malone jupp@ebi.ac.uk, malone@ebi.ac.uk Outline Languages OWL and OBO classes, individuals, relations,
Korea Institute of Oriental Medicine, South Korea 2 Biomedical Knowledge Engineering Laboratory,
 A Medical Treatment System based on Traditional Korean Medicine Ontology Sang-Kyun Kim 1, SeJin Nam 2, Dong-Hun Park 1, Yong-Taek Oh 1, Hyunchul Jang 1 1 Literature & Informatics Research Division, Korea
A Medical Treatment System based on Traditional Korean Medicine Ontology Sang-Kyun Kim 1, SeJin Nam 2, Dong-Hun Park 1, Yong-Taek Oh 1, Hyunchul Jang 1 1 Literature & Informatics Research Division, Korea
IMAGENOTION - Collaborative Semantic Annotation of Images and Image Parts and Work Integrated Creation of Ontologies
 IMAGENOTION - Collaborative Semantic Annotation of Images and Image Parts and Work Integrated Creation of Ontologies Andreas Walter, awalter@fzi.de Gabor Nagypal, nagypal@disy.net Abstract: In this paper,
IMAGENOTION - Collaborative Semantic Annotation of Images and Image Parts and Work Integrated Creation of Ontologies Andreas Walter, awalter@fzi.de Gabor Nagypal, nagypal@disy.net Abstract: In this paper,
ONTOLOGY ENGINEERING IN VIRTUAL BREEDING ENVIRONMENTS
 14 ONTOLOGY ENGINEERING IN VIRTUAL BREEDING ENVIRONMENTS Dora Simões 1,3, Hugo Ferreira 1, António Lucas Soares 1,2 INESC Porto 1, Faculty of Engineering Univ. Porto 2, ISCA Univ. Aveiro 3 dora.simoes@isca.ua.pt,
14 ONTOLOGY ENGINEERING IN VIRTUAL BREEDING ENVIRONMENTS Dora Simões 1,3, Hugo Ferreira 1, António Lucas Soares 1,2 INESC Porto 1, Faculty of Engineering Univ. Porto 2, ISCA Univ. Aveiro 3 dora.simoes@isca.ua.pt,
The ELIXIR of Linked Data
 The ELIXIR of Linked Data Professor Carole Goble (UK node) Barend Mons (NL node), Helen Parkinson (EMBL-EBI node) The Interoperability Services Backbone Team European Life Sciences Infrastructure for Biological
The ELIXIR of Linked Data Professor Carole Goble (UK node) Barend Mons (NL node), Helen Parkinson (EMBL-EBI node) The Interoperability Services Backbone Team European Life Sciences Infrastructure for Biological
On the use of Abstract Workflows to Capture Scientific Process Provenance
 On the use of Abstract Workflows to Capture Scientific Process Provenance Paulo Pinheiro da Silva, Leonardo Salayandia, Nicholas Del Rio, Ann Q. Gates The University of Texas at El Paso CENTER OF EXCELLENCE
On the use of Abstract Workflows to Capture Scientific Process Provenance Paulo Pinheiro da Silva, Leonardo Salayandia, Nicholas Del Rio, Ann Q. Gates The University of Texas at El Paso CENTER OF EXCELLENCE
ServOMap and ServOMap-lt Results for OAEI 2012
 ServOMap and ServOMap-lt Results for OAEI 2012 Mouhamadou Ba 1, Gayo Diallo 1 1 LESIM/ISPED, Univ. Bordeaux Segalen, F-33000, France first.last@isped.u-bordeaux2.fr Abstract. We present the results obtained
ServOMap and ServOMap-lt Results for OAEI 2012 Mouhamadou Ba 1, Gayo Diallo 1 1 LESIM/ISPED, Univ. Bordeaux Segalen, F-33000, France first.last@isped.u-bordeaux2.fr Abstract. We present the results obtained
AMNH Gerstner Scholars in Bioinformatics & Computational Biology Application Instructions
 PURPOSE AMNH Gerstner Scholars in Bioinformatics & Computational Biology Application Instructions The seeks highly qualified applicants for its Gerstner postdoctoral fellowship program in Bioinformatics
PURPOSE AMNH Gerstner Scholars in Bioinformatics & Computational Biology Application Instructions The seeks highly qualified applicants for its Gerstner postdoctoral fellowship program in Bioinformatics
biowes Scientific Data Management Solution Dr. Petr Cisar Institute of complex systems, FFPW, USB Czech Republic.
 biowes Scientific Data Management Solution Dr. Petr Cisar Institute of complex systems, FFPW, USB Czech Republic. Reproducibility Reproducibility An independent researcher should be able to replicate the
biowes Scientific Data Management Solution Dr. Petr Cisar Institute of complex systems, FFPW, USB Czech Republic. Reproducibility Reproducibility An independent researcher should be able to replicate the
Data Curation Profile Plant Genetics / Corn Breeding
 Profile Author Author s Institution Contact Researcher(s) Interviewed Researcher s Institution Katherine Chiang Cornell University Library ksc3@cornell.edu Withheld Cornell University Date of Creation
Profile Author Author s Institution Contact Researcher(s) Interviewed Researcher s Institution Katherine Chiang Cornell University Library ksc3@cornell.edu Withheld Cornell University Date of Creation
SWRL RULE EDITOR: A WEB APPLICATION AS RICH AS DESKTOP BUSINESS RULE EDITORS
 SWRL RULE EDITOR: A WEB APPLICATION AS RICH AS DESKTOP BUSINESS RULE EDITORS João Paulo Orlando 1, Adriano Rívolli 1, Saeed Hassanpour 2, Martin J. O'Connor 2, Amar Das 2, and Dilvan A. Moreira 1 1 Dept.
SWRL RULE EDITOR: A WEB APPLICATION AS RICH AS DESKTOP BUSINESS RULE EDITORS João Paulo Orlando 1, Adriano Rívolli 1, Saeed Hassanpour 2, Martin J. O'Connor 2, Amar Das 2, and Dilvan A. Moreira 1 1 Dept.
A Formal Definition of RESTful Semantic Web Services. Antonio Garrote Hernández María N. Moreno García
 A Formal Definition of RESTful Semantic Web Services Antonio Garrote Hernández María N. Moreno García Outline Motivation Resources and Triple Spaces Resources and Processes RESTful Semantic Resources Example
A Formal Definition of RESTful Semantic Web Services Antonio Garrote Hernández María N. Moreno García Outline Motivation Resources and Triple Spaces Resources and Processes RESTful Semantic Resources Example
Linking library data: contributions and role of subject data. Nuno Freire The European Library
 Linking library data: contributions and role of subject data Nuno Freire The European Library Outline Introduction to The European Library Motivation for Linked Library Data The European Library Open Dataset
Linking library data: contributions and role of subject data Nuno Freire The European Library Outline Introduction to The European Library Motivation for Linked Library Data The European Library Open Dataset
Publishing Linked Statistical Data: Aragón, a case study.
 Publishing Linked Statistical Data: Aragón, a case study. Oscar Corcho 1, Idafen Santana-Pérez 1, Hugo Lafuente 2, David Portolés 3, César Cano 4, Alfredo Peris 4, and José María Subero 4 1 Ontology Engineering
Publishing Linked Statistical Data: Aragón, a case study. Oscar Corcho 1, Idafen Santana-Pérez 1, Hugo Lafuente 2, David Portolés 3, César Cano 4, Alfredo Peris 4, and José María Subero 4 1 Ontology Engineering
A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES
 A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES 1 Yuxin Mao 1 School of Computer and Information Engineering, Zhejiang Gongshang University, Hangzhou 310018, P.R. China E-mail: 1 maoyuxin@zjgsu.edu.cn ABSTRACT
A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES 1 Yuxin Mao 1 School of Computer and Information Engineering, Zhejiang Gongshang University, Hangzhou 310018, P.R. China E-mail: 1 maoyuxin@zjgsu.edu.cn ABSTRACT
Linked Data and RDF. COMP60421 Sean Bechhofer
 Linked Data and RDF COMP60421 Sean Bechhofer sean.bechhofer@manchester.ac.uk Building a Semantic Web Annotation Associating metadata with resources Integration Integrating information sources Inference
Linked Data and RDF COMP60421 Sean Bechhofer sean.bechhofer@manchester.ac.uk Building a Semantic Web Annotation Associating metadata with resources Integration Integrating information sources Inference
Data Curation Profile Human Genomics
 Data Curation Profile Human Genomics Profile Author Profile Author Institution Name Contact J. Carlson N. Brown Purdue University J. Carlson, jrcarlso@purdue.edu Date of Creation October 27, 2009 Date
Data Curation Profile Human Genomics Profile Author Profile Author Institution Name Contact J. Carlson N. Brown Purdue University J. Carlson, jrcarlso@purdue.edu Date of Creation October 27, 2009 Date
Acquiring Experience with Ontology and Vocabularies
 Acquiring Experience with Ontology and Vocabularies Walt Melo Risa Mayan Jean Stanford The author's affiliation with The MITRE Corporation is provided for identification purposes only, and is not intended
Acquiring Experience with Ontology and Vocabularies Walt Melo Risa Mayan Jean Stanford The author's affiliation with The MITRE Corporation is provided for identification purposes only, and is not intended
The European Commission s science and knowledge service. Joint Research Centre
 The European Commission s science and knowledge service Joint Research Centre GeoDCAT-AP The story so far Andrea Perego, Antonio Rotundo, Lieven Raes GeoDCAT-AP Webinar 6 June 2018 What is GeoDCAT-AP Geospatial
The European Commission s science and knowledge service Joint Research Centre GeoDCAT-AP The story so far Andrea Perego, Antonio Rotundo, Lieven Raes GeoDCAT-AP Webinar 6 June 2018 What is GeoDCAT-AP Geospatial
Prototyping a Biomedical Ontology Recommender Service
 Prototyping a Biomedical Ontology Recommender Service Clement Jonquet Nigam H. Shah Mark A. Musen jonquet@stanford.edu 1 Ontologies & data & annota@ons (1/2) Hard for biomedical researchers to find the
Prototyping a Biomedical Ontology Recommender Service Clement Jonquet Nigam H. Shah Mark A. Musen jonquet@stanford.edu 1 Ontologies & data & annota@ons (1/2) Hard for biomedical researchers to find the
Financial Dataspaces: Challenges, Approaches and Trends
 Financial Dataspaces: Challenges, Approaches and Trends Finance and Economics on the Semantic Web (FEOSW), ESWC 27 th May, 2012 Seán O Riain ebusiness Copyright 2009. All rights reserved. Motivation Changing
Financial Dataspaces: Challenges, Approaches and Trends Finance and Economics on the Semantic Web (FEOSW), ESWC 27 th May, 2012 Seán O Riain ebusiness Copyright 2009. All rights reserved. Motivation Changing
SKOS. COMP62342 Sean Bechhofer
 SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Ontologies Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Ontologies Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
For each use case, the business need, usage scenario and derived requirements are stated. 1.1 USE CASE 1: EXPLORE AND SEARCH FOR SEMANTIC ASSESTS
 1 1. USE CASES For each use case, the business need, usage scenario and derived requirements are stated. 1.1 USE CASE 1: EXPLORE AND SEARCH FOR SEMANTIC ASSESTS Business need: Users need to be able to
1 1. USE CASES For each use case, the business need, usage scenario and derived requirements are stated. 1.1 USE CASE 1: EXPLORE AND SEARCH FOR SEMANTIC ASSESTS Business need: Users need to be able to
SEXTANT 1. Purpose of the Application
 SEXTANT 1. Purpose of the Application Sextant has been used in the domains of Earth Observation and Environment by presenting its browsing and visualization capabilities using a number of link geospatial
SEXTANT 1. Purpose of the Application Sextant has been used in the domains of Earth Observation and Environment by presenting its browsing and visualization capabilities using a number of link geospatial
Ontology Extraction from Heterogeneous Documents
 Vol.3, Issue.2, March-April. 2013 pp-985-989 ISSN: 2249-6645 Ontology Extraction from Heterogeneous Documents Kirankumar Kataraki, 1 Sumana M 2 1 IV sem M.Tech/ Department of Information Science & Engg
Vol.3, Issue.2, March-April. 2013 pp-985-989 ISSN: 2249-6645 Ontology Extraction from Heterogeneous Documents Kirankumar Kataraki, 1 Sumana M 2 1 IV sem M.Tech/ Department of Information Science & Engg
XML in the bipharmaceutical
 XML in the bipharmaceutical sector XML holds out the opportunity to integrate data across both the enterprise and the network of biopharmaceutical alliances - with little technological dislocation and
XML in the bipharmaceutical sector XML holds out the opportunity to integrate data across both the enterprise and the network of biopharmaceutical alliances - with little technological dislocation and
Ontologies SKOS. COMP62342 Sean Bechhofer
 Ontologies SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
Ontologies SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform
 Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform 1. Exploring the IDR This current IDR web user interface (WUI) is based on the open source
Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform 1. Exploring the IDR This current IDR web user interface (WUI) is based on the open source
Linked Open Europeana: Semantics for the Digital Humanities
 Linked Open Europeana: Semantics for the Digital Humanities Prof. Dr. Stefan Gradmann Humboldt-Universität zu Berlin / School of Library and Information Science stefan.gradmann@ibi.hu-berlin.de 1 Overview
Linked Open Europeana: Semantics for the Digital Humanities Prof. Dr. Stefan Gradmann Humboldt-Universität zu Berlin / School of Library and Information Science stefan.gradmann@ibi.hu-berlin.de 1 Overview
Exploring and Using the Semantic Web
 Exploring and Using the Semantic Web Mathieu d Aquin KMi, The Open University m.daquin@open.ac.uk What?? Exploring the Semantic Web Vocabularies Ontologies Linked Data RDF documents Example: Exploring
Exploring and Using the Semantic Web Mathieu d Aquin KMi, The Open University m.daquin@open.ac.uk What?? Exploring the Semantic Web Vocabularies Ontologies Linked Data RDF documents Example: Exploring
APPLYING KNOWLEDGE BASED AI TO MODERN DATA MANAGEMENT. Mani Keeran, CFA Gi Kim, CFA Preeti Sharma
 APPLYING KNOWLEDGE BASED AI TO MODERN DATA MANAGEMENT Mani Keeran, CFA Gi Kim, CFA Preeti Sharma 2 What we are going to discuss During last two decades, majority of information assets have been digitized
APPLYING KNOWLEDGE BASED AI TO MODERN DATA MANAGEMENT Mani Keeran, CFA Gi Kim, CFA Preeti Sharma 2 What we are going to discuss During last two decades, majority of information assets have been digitized
Global Agricultural Concept Scheme The collaborative integration of three thesauri
 Global Agricultural Concept Scheme The collaborative integration of three thesauri Prof Dr Thomas Baker [1] Dr Osma Suominen [2] Dini Jahrestagung Linked Data Vision und Wirklichkeit Frankfurt, 28. Oktober
Global Agricultural Concept Scheme The collaborative integration of three thesauri Prof Dr Thomas Baker [1] Dr Osma Suominen [2] Dini Jahrestagung Linked Data Vision und Wirklichkeit Frankfurt, 28. Oktober
SEAD Data Services. Jim Best Practices in Data Infrastructure Workshop. Cooperative agreement #OCI
 SEAD Data Services Jim Myers(myersjd@umich.edu), Best Practices in Data Infrastructure Workshop Cooperative agreement #OCI0940824 SEAD: Sustainable Environment - Actionable Data An NSF DataNet project
SEAD Data Services Jim Myers(myersjd@umich.edu), Best Practices in Data Infrastructure Workshop Cooperative agreement #OCI0940824 SEAD: Sustainable Environment - Actionable Data An NSF DataNet project
The Submission Data File System Automating the Creation of CDISC SDTM and ADaM Datasets
 Paper AD-08 The Submission Data File System Automating the Creation of CDISC SDTM and ADaM Datasets Marcus Bloom, Amgen Inc, Thousand Oaks, CA David Edwards, Amgen Inc, Thousand Oaks, CA ABSTRACT From
Paper AD-08 The Submission Data File System Automating the Creation of CDISC SDTM and ADaM Datasets Marcus Bloom, Amgen Inc, Thousand Oaks, CA David Edwards, Amgen Inc, Thousand Oaks, CA ABSTRACT From
Towards a joint service catalogue for e-infrastructure services
 Towards a joint service catalogue for e-infrastructure services Dr British Library 1 DI4R 2016 Workshop Joint service catalogue for research 29 September 2016 15/09/15 Goal A framework for creating a Catalogue
Towards a joint service catalogue for e-infrastructure services Dr British Library 1 DI4R 2016 Workshop Joint service catalogue for research 29 September 2016 15/09/15 Goal A framework for creating a Catalogue
Knowledge-Driven Video Information Retrieval with LOD
 Knowledge-Driven Video Information Retrieval with LOD Leslie F. Sikos, Ph.D., Flinders University ESAIR 15, 23 October 2015 Melbourne, VIC, Australia Knowledge-Driven Video IR Outline Video Retrieval Challenges
Knowledge-Driven Video Information Retrieval with LOD Leslie F. Sikos, Ph.D., Flinders University ESAIR 15, 23 October 2015 Melbourne, VIC, Australia Knowledge-Driven Video IR Outline Video Retrieval Challenges
Conducting a Self-Assessment of a Long-Term Archive for Interdisciplinary Scientific Data as a Trustworthy Digital Repository
 Conducting a Self-Assessment of a Long-Term Archive for Interdisciplinary Scientific Data as a Trustworthy Digital Repository Robert R. Downs and Robert S. Chen Center for International Earth Science Information
Conducting a Self-Assessment of a Long-Term Archive for Interdisciplinary Scientific Data as a Trustworthy Digital Repository Robert R. Downs and Robert S. Chen Center for International Earth Science Information
An Archiving System for Managing Evolution in the Data Web
 An Archiving System for Managing Evolution in the Web Marios Meimaris *, George Papastefanatos and Christos Pateritsas * Institute for the Management of Information Systems, Research Center Athena, Greece
An Archiving System for Managing Evolution in the Web Marios Meimaris *, George Papastefanatos and Christos Pateritsas * Institute for the Management of Information Systems, Research Center Athena, Greece
Design & Manage Persistent URIs
 Training Module 2.3 OPEN DATA SUPPORT Design & Manage Persistent URIs PwC firms help organisations and individuals create the value they re looking for. We re a network of firms in 158 countries with close
Training Module 2.3 OPEN DATA SUPPORT Design & Manage Persistent URIs PwC firms help organisations and individuals create the value they re looking for. We re a network of firms in 158 countries with close
Semantic Web Fundamentals
 Semantic Web Fundamentals Web Technologies (706.704) 3SSt VU WS 2018/19 with acknowledgements to P. Höfler, V. Pammer, W. Kienreich ISDS, TU Graz January 7 th 2019 Overview What is Semantic Web? Technology
Semantic Web Fundamentals Web Technologies (706.704) 3SSt VU WS 2018/19 with acknowledgements to P. Höfler, V. Pammer, W. Kienreich ISDS, TU Graz January 7 th 2019 Overview What is Semantic Web? Technology
Metadata. Week 4 LBSC 671 Creating Information Infrastructures
 Metadata Week 4 LBSC 671 Creating Information Infrastructures Muddiest Points Memory madness Hard drives, DVD s, solid state disks, tape, Digitization Images, audio, video, compression, file names, Where
Metadata Week 4 LBSC 671 Creating Information Infrastructures Muddiest Points Memory madness Hard drives, DVD s, solid state disks, tape, Digitization Images, audio, video, compression, file names, Where
Core Technology Development Team Meeting
 Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
University of Bath. Publication date: Document Version Publisher's PDF, also known as Version of record. Link to publication
 Citation for published version: Patel, M & Duke, M 2004, 'Knowledge Discovery in an Agents Environment' Paper presented at European Semantic Web Symposium 2004, Heraklion, Crete, UK United Kingdom, 9/05/04-11/05/04,.
Citation for published version: Patel, M & Duke, M 2004, 'Knowledge Discovery in an Agents Environment' Paper presented at European Semantic Web Symposium 2004, Heraklion, Crete, UK United Kingdom, 9/05/04-11/05/04,.
Annotation for the Semantic Web During Website Development
 Annotation for the Semantic Web During Website Development Peter Plessers and Olga De Troyer Vrije Universiteit Brussel, Department of Computer Science, WISE, Pleinlaan 2, 1050 Brussel, Belgium {Peter.Plessers,
Annotation for the Semantic Web During Website Development Peter Plessers and Olga De Troyer Vrije Universiteit Brussel, Department of Computer Science, WISE, Pleinlaan 2, 1050 Brussel, Belgium {Peter.Plessers,
Taxonomy Tools: Collaboration, Creation & Integration. Dow Jones & Company
 Taxonomy Tools: Collaboration, Creation & Integration Dave Clarke Global Taxonomy Director dave.clarke@dowjones.com Dow Jones & Company Introduction Software Tools for Taxonomy 1. Collaboration 2. Creation
Taxonomy Tools: Collaboration, Creation & Integration Dave Clarke Global Taxonomy Director dave.clarke@dowjones.com Dow Jones & Company Introduction Software Tools for Taxonomy 1. Collaboration 2. Creation
CHAPTER 6 PROPOSED HYBRID MEDICAL IMAGE RETRIEVAL SYSTEM USING SEMANTIC AND VISUAL FEATURES
 188 CHAPTER 6 PROPOSED HYBRID MEDICAL IMAGE RETRIEVAL SYSTEM USING SEMANTIC AND VISUAL FEATURES 6.1 INTRODUCTION Image representation schemes designed for image retrieval systems are categorized into two
188 CHAPTER 6 PROPOSED HYBRID MEDICAL IMAGE RETRIEVAL SYSTEM USING SEMANTIC AND VISUAL FEATURES 6.1 INTRODUCTION Image representation schemes designed for image retrieval systems are categorized into two
D WSMO Data Grounding Component
 Project Number: 215219 Project Acronym: SOA4All Project Title: Instrument: Thematic Priority: Service Oriented Architectures for All Integrated Project Information and Communication Technologies Activity
Project Number: 215219 Project Acronym: SOA4All Project Title: Instrument: Thematic Priority: Service Oriented Architectures for All Integrated Project Information and Communication Technologies Activity
Ontology Based Prediction of Difficult Keyword Queries
 Ontology Based Prediction of Difficult Keyword Queries Lubna.C*, Kasim K Pursuing M.Tech (CSE)*, Associate Professor (CSE) MEA Engineering College, Perinthalmanna Kerala, India lubna9990@gmail.com, kasim_mlp@gmail.com
Ontology Based Prediction of Difficult Keyword Queries Lubna.C*, Kasim K Pursuing M.Tech (CSE)*, Associate Professor (CSE) MEA Engineering College, Perinthalmanna Kerala, India lubna9990@gmail.com, kasim_mlp@gmail.com
IJCSC Volume 5 Number 1 March-Sep 2014 pp ISSN
 Movie Related Information Retrieval Using Ontology Based Semantic Search Tarjni Vyas, Hetali Tank, Kinjal Shah Nirma University, Ahmedabad tarjni.vyas@nirmauni.ac.in, tank92@gmail.com, shahkinjal92@gmail.com
Movie Related Information Retrieval Using Ontology Based Semantic Search Tarjni Vyas, Hetali Tank, Kinjal Shah Nirma University, Ahmedabad tarjni.vyas@nirmauni.ac.in, tank92@gmail.com, shahkinjal92@gmail.com
Semantic Web Fundamentals
 Semantic Web Fundamentals Web Technologies (706.704) 3SSt VU WS 2017/18 Vedran Sabol with acknowledgements to P. Höfler, V. Pammer, W. Kienreich ISDS, TU Graz December 11 th 2017 Overview What is Semantic
Semantic Web Fundamentals Web Technologies (706.704) 3SSt VU WS 2017/18 Vedran Sabol with acknowledgements to P. Höfler, V. Pammer, W. Kienreich ISDS, TU Graz December 11 th 2017 Overview What is Semantic
Reference Requirements for Records and Documents Management
 Reference Requirements for Records and Documents Management Ricardo Jorge Seno Martins ricardosenomartins@gmail.com Instituto Superior Técnico, Lisboa, Portugal May 2015 Abstract When information systems
Reference Requirements for Records and Documents Management Ricardo Jorge Seno Martins ricardosenomartins@gmail.com Instituto Superior Técnico, Lisboa, Portugal May 2015 Abstract When information systems
Enabling Open Science: Data Discoverability, Access and Use. Jo McEntyre Head of Literature Services
 Enabling Open Science: Data Discoverability, Access and Use Jo McEntyre Head of Literature Services www.ebi.ac.uk About EMBL-EBI Part of the European Molecular Biology Laboratory International, non-profit
Enabling Open Science: Data Discoverability, Access and Use Jo McEntyre Head of Literature Services www.ebi.ac.uk About EMBL-EBI Part of the European Molecular Biology Laboratory International, non-profit
ISA Action 1.17: A Reusable INSPIRE Reference Platform (ARE3NA)
 ISA Action 1.17: A Reusable INSPIRE Reference Platform (ARE3NA) Expert contract supporting the Study on RDF and PIDs for INSPIRE Deliverable D.EC.3.2 RDF in INSPIRE Open issues, tools, and implications
ISA Action 1.17: A Reusable INSPIRE Reference Platform (ARE3NA) Expert contract supporting the Study on RDF and PIDs for INSPIRE Deliverable D.EC.3.2 RDF in INSPIRE Open issues, tools, and implications
Integration of INSPIRE & SDMX data infrastructures for the 2021 population and housing census
 Integration of INSPIRE & SDMX data infrastructures for the 2021 population and housing census Nadezhda VLAHOVA, Fabian BACH, Ekkehard PETRI *, Vlado CETL, Hannes REUTER European Commission (*ekkehard.petri@ec.europa.eu
Integration of INSPIRE & SDMX data infrastructures for the 2021 population and housing census Nadezhda VLAHOVA, Fabian BACH, Ekkehard PETRI *, Vlado CETL, Hannes REUTER European Commission (*ekkehard.petri@ec.europa.eu
A common metadata approach to support egovernment interoperability
 ISWC 2011 10 th International Semantic Web Conference October, 2011 Rue Froissart 36, Brussels - 1040, Belgium A common metadata approach to support egovernment interoperability Makx Dekkers makx@makxdekkers.com
ISWC 2011 10 th International Semantic Web Conference October, 2011 Rue Froissart 36, Brussels - 1040, Belgium A common metadata approach to support egovernment interoperability Makx Dekkers makx@makxdekkers.com
The MEG Metadata Schemas Registry Schemas and Ontologies: building a Semantic Infrastructure for GRIDs and digital libraries Edinburgh, 16 May 2003
 The MEG Metadata Schemas Registry Schemas and Ontologies: building a Semantic Infrastructure for GRIDs and digital libraries Edinburgh, 16 May 2003 Pete Johnston UKOLN, University of Bath Bath, BA2 7AY
The MEG Metadata Schemas Registry Schemas and Ontologies: building a Semantic Infrastructure for GRIDs and digital libraries Edinburgh, 16 May 2003 Pete Johnston UKOLN, University of Bath Bath, BA2 7AY
Exploring the Generation and Integration of Publishable Scientific Facts Using the Concept of Nano-publications
 Exploring the Generation and Integration of Publishable Scientific Facts Using the Concept of Nano-publications Amanda Clare 1,3, Samuel Croset 2,3 (croset@ebi.ac.uk), Christoph Grabmueller 2,3, Senay
Exploring the Generation and Integration of Publishable Scientific Facts Using the Concept of Nano-publications Amanda Clare 1,3, Samuel Croset 2,3 (croset@ebi.ac.uk), Christoph Grabmueller 2,3, Senay
Horizon 2020 and the Open Research Data pilot. Sarah Jones Digital Curation Centre, Glasgow
 Horizon 2020 and the Open Research Data pilot Sarah Jones Digital Curation Centre, Glasgow sarah.jones@glasgow.ac.uk Twitter: @sjdcc Data Management Plan (DMP) workshop, e-infrastructures Austria, Vienna,
Horizon 2020 and the Open Research Data pilot Sarah Jones Digital Curation Centre, Glasgow sarah.jones@glasgow.ac.uk Twitter: @sjdcc Data Management Plan (DMP) workshop, e-infrastructures Austria, Vienna,
Unstructured Text in Big Data The Elephant in the Room
 Unstructured Text in Big Data The Elephant in the Room David Milward ICIC, October 2013 Click Unstructured to to edit edit Master Master Big title Data style title style Big Data Volume, Variety, Velocity
Unstructured Text in Big Data The Elephant in the Room David Milward ICIC, October 2013 Click Unstructured to to edit edit Master Master Big title Data style title style Big Data Volume, Variety, Velocity
Enterprise Data Catalog for Microsoft Azure Tutorial
 Enterprise Data Catalog for Microsoft Azure Tutorial VERSION 10.2 JANUARY 2018 Page 1 of 45 Contents Tutorial Objectives... 4 Enterprise Data Catalog Overview... 5 Overview... 5 Objectives... 5 Enterprise
Enterprise Data Catalog for Microsoft Azure Tutorial VERSION 10.2 JANUARY 2018 Page 1 of 45 Contents Tutorial Objectives... 4 Enterprise Data Catalog Overview... 5 Overview... 5 Objectives... 5 Enterprise
Building the NNEW Weather Ontology
 Building the NNEW Weather Ontology Kelly Moran Kajal Claypool 5 May 2010 1 Outline Introduction Ontology Development Methods & Tools NNEW Weather Ontology Design Application: Semantic Search Summary 2
Building the NNEW Weather Ontology Kelly Moran Kajal Claypool 5 May 2010 1 Outline Introduction Ontology Development Methods & Tools NNEW Weather Ontology Design Application: Semantic Search Summary 2
SELF-SERVICE SEMANTIC DATA FEDERATION
 SELF-SERVICE SEMANTIC DATA FEDERATION WE LL MAKE YOU A DATA SCIENTIST Contact: IPSNP Computing Inc. Chris Baker, CEO Chris.Baker@ipsnp.com (506) 721 8241 BIG VISION: SELF-SERVICE DATA FEDERATION Biomedical
SELF-SERVICE SEMANTIC DATA FEDERATION WE LL MAKE YOU A DATA SCIENTIST Contact: IPSNP Computing Inc. Chris Baker, CEO Chris.Baker@ipsnp.com (506) 721 8241 BIG VISION: SELF-SERVICE DATA FEDERATION Biomedical
Developing a Research Data Policy
 Developing a Research Data Policy Core Elements of the Content of a Research Data Management Policy This document may be useful for defining research data, explaining what RDM is, illustrating workflows,
Developing a Research Data Policy Core Elements of the Content of a Research Data Management Policy This document may be useful for defining research data, explaining what RDM is, illustrating workflows,
FIBO Operational Ontologies Briefing for the Object Management Group
 FIBO Operational Ontologies Briefing for the Object Management Group March 20, 2013, Reston, VA David Newman Strategic Planning Manager, Senior Vice President, Enterprise Architecture Chair, Semantic Technology
FIBO Operational Ontologies Briefing for the Object Management Group March 20, 2013, Reston, VA David Newman Strategic Planning Manager, Senior Vice President, Enterprise Architecture Chair, Semantic Technology
EUDAT B2FIND A Cross-Discipline Metadata Service and Discovery Portal
 EUDAT B2FIND A Cross-Discipline Metadata Service and Discovery Portal Heinrich Widmann, DKRZ DI4R 2016, Krakow, 28 September 2016 www.eudat.eu EUDAT receives funding from the European Union's Horizon 2020
EUDAT B2FIND A Cross-Discipline Metadata Service and Discovery Portal Heinrich Widmann, DKRZ DI4R 2016, Krakow, 28 September 2016 www.eudat.eu EUDAT receives funding from the European Union's Horizon 2020
Just in time and relevant knowledge thanks to recommender systems and Semantic Web.
 Just in time and relevant knowledge thanks to recommender systems and Semantic Web. Plessers, Ben (1); Van Hyfte, Dirk (2); Schreurs, Jeanne (1) Organization(s): 1 Hasselt University, Belgium; 2 i.know,
Just in time and relevant knowledge thanks to recommender systems and Semantic Web. Plessers, Ben (1); Van Hyfte, Dirk (2); Schreurs, Jeanne (1) Organization(s): 1 Hasselt University, Belgium; 2 i.know,
When Communities of Interest Collide: Harmonizing Vocabularies Across Operational Areas C. L. Connors, The MITRE Corporation
 When Communities of Interest Collide: Harmonizing Vocabularies Across Operational Areas C. L. Connors, The MITRE Corporation Three recent trends have had a profound impact on data standardization within
When Communities of Interest Collide: Harmonizing Vocabularies Across Operational Areas C. L. Connors, The MITRE Corporation Three recent trends have had a profound impact on data standardization within
THE ROLE OF STANDARDS IN B2B COMMUNICATION
 THE ROLE OF STANDARDS IN B2B COMMUNICATION Eva Söderström School of Humanities and Informatics, University of Skoevde Box 408, 541 28 Skoevde, Sweden ABSTRACT Recent developments in e.g. technology have
THE ROLE OF STANDARDS IN B2B COMMUNICATION Eva Söderström School of Humanities and Informatics, University of Skoevde Box 408, 541 28 Skoevde, Sweden ABSTRACT Recent developments in e.g. technology have
Mappings from BPEL to PMR for Business Process Registration
 Mappings from BPEL to PMR for Business Process Registration Jingwei Cheng 1, Chong Wang 1 +, Keqing He 1, Jinxu Jia 2, Peng Liang 1 1 State Key Lab. of Software Engineering, Wuhan University, China cinfiniter@gmail.com,
Mappings from BPEL to PMR for Business Process Registration Jingwei Cheng 1, Chong Wang 1 +, Keqing He 1, Jinxu Jia 2, Peng Liang 1 1 State Key Lab. of Software Engineering, Wuhan University, China cinfiniter@gmail.com,
The DIGMAP Virtual Digital Library
 José Borbinha *, Gilberto Pedrosa *, João Luzio *, Hugo Manguinhas *, Bruno Martins * The DIGMAP Virtual Digital Library Keywords: Geographic information; cartographic heritage; information systems architectures;
José Borbinha *, Gilberto Pedrosa *, João Luzio *, Hugo Manguinhas *, Bruno Martins * The DIGMAP Virtual Digital Library Keywords: Geographic information; cartographic heritage; information systems architectures;
Design and Implementation of Cost Effective MIS for Universities
 Fourth LACCEI International Latin American and Caribbean Conference for Engineering and Technology (LACCET 2006) Breaking Frontiers and Barriers in Engineering: Education, Research and Practice 21-23 June
Fourth LACCEI International Latin American and Caribbean Conference for Engineering and Technology (LACCET 2006) Breaking Frontiers and Barriers in Engineering: Education, Research and Practice 21-23 June
Enrichment of Sensor Descriptions and Measurements Using Semantic Technologies. Student: Alexandra Moraru Mentor: Prof. Dr.
 Enrichment of Sensor Descriptions and Measurements Using Semantic Technologies Student: Alexandra Moraru Mentor: Prof. Dr. Dunja Mladenić Environmental Monitoring automation Traffic Monitoring integration
Enrichment of Sensor Descriptions and Measurements Using Semantic Technologies Student: Alexandra Moraru Mentor: Prof. Dr. Dunja Mladenić Environmental Monitoring automation Traffic Monitoring integration
Building an Ontology Catalogue for Smart Cities
 Building an Ontology Catalogue for Smart Cities M. Poveda-Villalón, R. García-Castro and A. Gómez-Pérez Ontology Engineering Group, Escuela Técnica Superior de Ingenieros Informáticos Universidad Politécnica
Building an Ontology Catalogue for Smart Cities M. Poveda-Villalón, R. García-Castro and A. Gómez-Pérez Ontology Engineering Group, Escuela Técnica Superior de Ingenieros Informáticos Universidad Politécnica
