IntegronFinder Documentation
|
|
|
- Chastity Fleming
- 5 years ago
- Views:
Transcription
1 IntegronFinder Documentation Release Jean Cury, Bertrand Néron, Eduardo PC Rocha Feb 16, 2018
2 Contents 1 Introduction 2 2 Installation IntegronFinder dependencies Installation procedure Uninstallation procedure How to install Python Tutorial Basic use Advanced options Mobyle How to use it Results Galaxy How to use it Results Developer guide Getting a local working copy of the repository Send changes to upstream repository Developer installation Tests Documentation References 20 i
3 IntegronFinder is a program that detects integrons in DNA sequences. The program is available on a webserver Mobyle (closing end of 2016), on the Galaxy Pasteur, or by command line (Integron- Finder on github). You already read the paper and want to install it? Click here You did not read the paper (yet) but you would like to have rapid introduction to integrons and the program? click here Contents 1
4 { CHAPTER 1 Introduction Integrons are major genetic element, notorious for their major implication in the spread of antibiotic resistance genes. More generally, integrons are gene-capturing platform, whose broader evolutionary role remains poorly understood. IntegronFinder is able to detect with high accuracy integron in DNA sequences. It is accurate because it combines the use of HMM profiles for the detection of the essential protein, the site-specific integron integrase, and the use of Covariance Models for the detection of the recombination site, the attc site. A 2 inti P inti Pc atti attc { 1 cassette B C D Complete In0 CALIN How does it work? First, IntegronFinder annotates the DNA sequence s CDS with Prodigal. Second, IntegronFinder detects independently integron integrase and attc recombination sites. The Integron integrase is detected by using the intersection of two HMM profiles: 2
5 one specific of tyrosine-recombinase (PF00589) one specific of the integron integrase, near the patch III domain of tyrosine recombinases. The attc recombination site is detected with a covariance model (CM), which models the secondary structure in addition to the few conserved sequence positions. Third, the results are integrated, and IntegronFinder distinguishes 3 types of elements: complete integron (panel B above) Integron with integron integrase nearby attc site(s) In0 element (panel C above) Integron integrase only, without any attc site nearby CALIN element (panel D above) Cluster of attc sites Lacking INtegrase nearby. A rule of thumb to avoid false positive is to filter out singleton of attc site. IntegronFinder can also annotate gene cassettes (CDS nearby attc sites) using Resfams, a database of HMM profiles aiming at annotating antibiotic resistance genes. This database is provided but the user can add any other HMM profiles database of its own interest. When available, IntegronFinder annotates the promoters and atti sites by pattern matching. Input: DNA sequence Fasta CDS annotation Prodigal --func_annot attc detection INFERNAL IntI detection HMMER --local_max Aggregation+ Classification Functional annotation HMMER Promoters and atti annotation Output: Integron Annotation Genbank Mandatory path Optional path 3
6 Does it work? Yes! The estimated sensitivity is 61% on average with the default option and goes up to 88% with the --local_max option. The missing attc sites are usually at the end of the array. The False positive rate with the --local_max option is estimated between 0.03 False Positive per Megabases (FP/Mb) to 0.72 FP/Mb. This leads to a probability of finding 2 consecutive false attc sites within 4kb between 4.10^-6 and 7.10^-9. Overall, the probability of finding an integron in a chromosome (including finding a part of it) is more than 95%. Finally, these parameters do not depend on the G+C percent of the given replicon. See the paper for more information (freely accessible). Default local_max Sensitivity 61.20% 88.03% FP rate 0.02 FP/Mb 0.03 FP/Mb Mean time 2.59 s s FP / Mb TP Sensitivity (%) TP +FN G+C content The time in the table correspond to the average time per run with a pseudogenome having attc sites on a Mac Pro, 2 x 2.4 GHz 6-Core Intel Xeon, 16 Gb RAM, with options cpu 20 and no-proteins. Note: The time does not vary depending of the mode (default or local_max), and is about a couple of second, if the replicon does not contain any attc site. 4
7 CHAPTER 2 Installation 2.1 IntegronFinder dependencies IntegronFinder is built with Python 2.7, and a few libraries are needed: Python 2.7 Pandas (>=0.18.0) Numpy (>=1.9.1) Biopython (>=1.65) Matplotlib (>=1.4.3) From version 1.5.1, integron_finder will check and install theses libraries for you. In addition, IntegronFinder has external dependencies, which have to be installed prior the use of the program (click to access the corresponding website). HMMER 3.1b1 INFERNAL 1.1 Prodigal V2.6.2 After installation of these programs, they should be in your $PATH (i.e. you can type in a terminal hmmsearch, cmsearch, or prodigal and a command not found shall not be displayed). If you have them installed somewhere else, please refer to integron_finder s parameters to give complete path to IntegronFinder. 5
8 2.2 Installation procedure Warning: When installing a new version (up to included) of IntegronFinder, do not forget to uninstall the previous version installed! From Version and after 1. Open a terminal and hit: (sudo) pip install integron_finder 2. To get an updated version (no need to uninstall): (sudo) pip install -U integron_finder For Version 1.5 and before 1. Download the latest release that can be installed like this (v1.5) 2. Uncompress it 3. In a shell (e.g. a terminal), go to the directory and run: (sudo) python setup.py install Note: Super-user privileges (i.e., sudo) are necesserary if you want to install the program in the general file architecture. Note: If you do not have the privileges, or if you do not want to install IntegronFinder in the Python libraries of your system, you can install IntegronFinder in a virtual environment. See virtualenv or if you re using Canopy, see Canopy CLI Warning: The installer does not work with pure setuptools procedure, it does not work in egg. Unless you disable egg by using the --root option. python setup.py install --root /prefix/where/to/install/integron_finder 2.2. Installation procedure 6
9 2.3 Uninstallation procedure From Version and after To uninstall IntegronFinder, run in the following command: (sudo) pip uninstall integron_finder It will uninstall integron_finder executable From Version 1.0 to Version 1.5 Go to the directory from where you installed IntegronFinder (e.g. Integron_Finder-1.5), and run: (sudo) python setup.py uninstall 2.4 How to install Python The purpose of this section is to provide some help about installing python dependencies for IntegronFinder if you never installed any python package. As IntegronFinder has not been test on Windows, we assume Unix-based operating system. For Windows users, the best would be to install a unix virtual machine on your computer. Usually a python distribution is already installed on your machine. However, if you don t know how to install libraries, we recommend to re-install it from a distribution which contains precompiled libraries. There are two main distributions (click to access website): Enthought Canopy Anaconda Download version 2.7 which correspond to your machine, then make sure that python from these distributions is the default one (you can possibly choose that in the preference and/or during installation). They both come with all the needed packages but Biopython. If you have a student adress from a university-delivering degree, you can request an academic licence to Enthough Canopy (see Canopy for Academics) which will allow you to download additional packages including Biopython. Otherwise, you will have to install Biopython manually. pip is recommended as a python packages installer. It works as follow: (sudo) pip install Biopython== Uninstallation procedure 7
10 To install version 1.65 of Biopython (recommended for IntegronFinder). Note: If you don t manage to install all the packages, try googling the error, or don t hesisate to ask a question on stackoverflow How to install Python 8
11 CHAPTER 3 Tutorial We assume here that the program is installed. 3.1 Basic use Note: The different options will be shown separately, but they can be used alltogether unless otherwise stated. You can see all available options with: integron_finder -h You can go to directory containing your sequence, or specify the path to that sequence and call: integron_finder mysequence.fst or: integron_finder path/to/mysequence.fst It will perform a search, and outputs the results in a directory called Results_Integron_Finder_mysequence. Within this directory, you can find: mysequence.integrons A tabular file with the annotations of the different elements 9
12 mysequence.gbk A GenBank file with the sequence annotated with the same annotations from the previous file. mysequence_x.pdf For each complete integron, a simple graphic of the region is depicted other A folder containing outputs of the different step in the program. It includes notably the protein file in fasta (mysequence.prt) Thorough local detection This option allows a more sensitive search. It will be slower if integrons are found, but will be as fast if nothing is detected: integron_finder mysequence.fst --local_max Functional annotation This option allows to annotate cassettes given HMM profiles. As Resfams database is distributed, to annotate antibiotic resistance genes, just use: integron_finder mysequence.fst --func_annot IntegronFinder will look in the directory Integron_Finder-x.x/data/ Functional_annotation and use all.hmm files available to annotate. By default, there is only Resfams.hmm, but one can add any other HMM file here. Alternatively, if one wants to use a database which is present elsewhere on the user s computer without copying it into that directory, one can specify the following option: integron_finder mysequence.fst --path_func_annot bank_hmm where bank_hmm is a file containing one absolute path to a hmm file per line, and you can comment out a line: ~/Downloads/Integron_Finder-x.x/data/Functional_annotation/Resfams.hmm ~/Documents/Data/Pfam-A.hmm # ~/Documents/Data/Pfam-B.hmm Here, annotation will be made using Pfam-A et Resfams, but not Pfam-B. If a protein is hit by 2 different profiles, the one with the best e-value will be kept Parallelization The time limiting part are HMMER and INFERNAL. So IntegronFinder does not have parallel implementation (yet?), but the user can set the number of CPU used by HMMER and INFERNAL: 3.1. Basic use 10
13 integron_finder mysequence.fst --cpu 4 Default is Circularity By default, IntegronFinder assumes your replicon to be circular. However, if they aren t, or if it s PCR fragments or contigs, you can specify that it s a linear fragment: integron_finder mylinearsequence.fst --linear However, if --linear is not used and the replicon is smaller than 4 x dt (where dt is the distance threshold, so 4kb by default), the replicon is considered linear to avoid clustering problem 3.2 Advanced options Clustering of elements attc sites are clustered together if they are on the same strand and if they are less than 4 kb apart. To cluster an array of attc sites and an integron integrase, they also must be less than 4 kb apart. This value has been empirically estimated and is consistent with previous observations showing that biggest gene cassettes are about 2 kb long. This value of 4 kb can be modify though: integron_finder mysequence.fst --distance_thresh or, equivalently: integron_finder mysequence.fst -dt This sets the threshold for clustering to 10 kb. Note: The option --outdir allows you to chose the location of the Results folder (Results_Integron_Finder_mysequence). If this folder already exists, IntegronFinder will not re-run analyses already done, except functional annotation. It allows you to re-run rapidly IntegronFinder with a different --distance_threshold value. Functional annotation needs to re-run each time because depending on the aggregation parameters, the proteins associated with an integron might change Advanced options 11
14 3.2.2 attc evalue The default evalue is 1. Sometimes, degenerated attc sites can have a evalue above 1 and one may want to increase this value to have a better sensitivity, to the cost of a much higher false positive rate. integron_finder mysequence.fst --evalue_attc Palindromes attc sites are more or less palindromic sequences, and sometimes, a single attc site can be detected on the 2 strands. By default, the one with the highest evalue is discarded, but you can choose to keep them with the following option: integron_finder mysequence.fst --keep_palindromes 3.2. Advanced options 12
15 CHAPTER 4 Mobyle You can access IntegronFinder online, on the Mobyle server of the Pasteur institute 4.1 How to use it 1. Copy your sequence or upload it in the appropriate field. 2. Select the options you want 3. Click on Run If you want more options: 3. Click on advanced options (instead of Run) 4. Select the options you want 5. Click on Run You can see the role of the different functions in the tutorial page, or by clicking on the corresponding field. After submitting your job, you may need to enter your . in the 4.2 Results Once the job is finished, you have a result page, which contains: 13
16 integron_finder.out: Log of the run. It tells you how many integrons have been found for each types along with the number of attc sites per type. Schema of complete integron(s) [replicon_x.pdf] Simple representation of one or more complete integrons found. The representation is very basic and a better representation can be obtained from the GenBank file and a software (eg Geneious) to represent it. annotated sequence [replicon.gbk] The GenBank file of the input sequence with the annotation corresponding to the elements found (integrase, attc, promoter, atti, etc... ). putative integrons [replicon.integrons] A tabular file listing all the elements and their caracteristics. Finally, you have your initial sequence of the replicon and the command line used. For each of the aforementionned files, you can save them by clicking on the save button Results 14
17 CHAPTER 5 Galaxy You can access IntegronFinder online, on the Galaxy server of the Pasteur institute 5.1 How to use it Registration on the Galaxy server of the Pasteur institute is not required to use the tool. Yet, if you wish to keep your history, we recommend you to register. 1. Upload your sequence with Get Data - Upload File in the menu on the left 2. Select your file in the Replicon file list of Integron Finder 3. Select the options you want 4. Click on Execute If you want more options: 3. Select Show on advanced parameters 4. Select the options you want 5. Click on Execute You can see the role of the different functions in the tutorial page. 15
18 5.2 Results Once the job is finished, you get your results on right panel. All files contain the log of the run which tells you how many integrons have been found for each types along with the number of attc sites per type. There are 4 different outputs created: Raw results archive: An archive containing all raw results. Integrons annotations: A tabular file listing all the elements and their caracteristics. GenBank: The GenBank file of the input sequence with the annotation corresponding to the elements found (integrase, attc, promoter, atti, etc... ). Graphics: Simple representation of one or more complete integrons found. The representation is very basic and a better representation can be obtained from the GenBank file and a software (eg Geneious) to represent it. For each of the aforementionned files, you can save them by clicking on the download button Results 16
19 CHAPTER 6 Developer guide This part is for developers, who want to work on IntegronFinder scripts. 6.1 Getting a local working copy of the repository If you are not part of the project, start by forking IntegronFinder repository. For that, sign in to your account on github, and go to Then, click on Fork (under your account icone). This will create a copy of the repository, but with your username instead of gem-pasteur. Then, clone it to your computer to have a local working copy, by opening a terminal window, and typing: git clone or, if you linked your ssh key: git clone git@github.com:<your_login>/integron_finder.git Go to the Integron_Finder repository created: cd Integron_Finder (referred here after as the root directory of IntegronFinder ). Then, it is better if you work on your own branch. For that, type: git checkout -b <your branch name> with <your branch name> a descriptive name (e.g. adding-xx-feature, fixing-typos, etc.), so that others understand what your are working on. 17
20 You can then install IntegronFinder as explained above, and do your modifications, with their associated tests. Don t forget to commit your changes at each step. 6.2 Send changes to upstream repository Once you are done, you can push your changes to your forked repository: git push --set-upstream origin <your branch name> Then, create a pull request, to add your changes to the official repository. For that: go to your forked repository on github Click on New pull request Choose your repository and the branch on which you did your changes in head fork (righthand side), and choose gem-pasteur/integron_finder with the branch on which you want to merge (probably master) in base fork (left-hand side). A green Able to merge text should appear if git is able to automatically merge the 2 branches. In that case, click on Create pull request, write your comments on the changes you made, why etc., and save. We will receive the pull request. Warning: Before submitting your pull request, please be sure that you provide the unit tests corresponding to the new features you added. 6.3 Developer installation To be able to work on the scripts and test your changes, you can either work with the scripts without installing the package, either install the package in developer mode. If you do not install the package, you always need to be in the root directory of IntegronFinder to launch it using:./integron_finder If you want to install it in developer mode, run, from the root of IntegronFinder: pip install -e. Then, you will be able to run IntegronFinder using integron_finder from anywhere. In any case, you also need to define the INTEGRON_HOME environment variable, with the path to the root of IntegronFinder directory: 6.2. Send changes to upstream repository 18
21 export INTEGRON_HOME=<path_to_integronfinder_directory> 6.4 Tests IntegronFinder is provided with unit (resp. functional) tests. You can find them in tests/unit (resp. tests/functional) repository. You can use them to check that your changes did not break the previous features, and you can update them, and add your own tests for the new features. Tests are done using unittest Running tests To run the tests (use option corresponding to unit or functional test according to what you want): python tests/run_tests.py [--unit] [--functional] Or, if you also want to get code coverage: coverage run tests/run_tests.py [--unit] [--functional] Add -vvv to get more details on each test passed/failed. The html coverage report will be generated in coverage_html/index.html Adding tests If you want to create a new test file, it must be saved in tests/unit or tests/functional according to the type of test, and the test file must start with test_. Then, write your tests using unittest framework (examples in existing files), and run them. 6.5 Documentation Documentation is done using sphinx. Source files are located in doc/sources. You can complete them. To generate html documentation, go to doc directory, and run: make html 6.4. Tests 19
22 CHAPTER 7 References If you use this software, please cite: Identification and analysis of integrons and cassette arrays in bacterial genomes Jean Cury; Thomas Jove; Marie Touchon; Bertrand Neron; Eduardo PC Rocha. Nucleic Acids Research, 2016; doi: /nar/gkw319 Please cite also the following articles: Nawrocki, E.P. and Eddy, S.R. (2013) Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics, 29, Eddy, S.R. (2011) Accelerated Profile HMM Searches. PLoS Comput Biol, 7, e Hyatt, D., Chen, G.L., Locascio, P.F., Land, M.L., Larimer, F.W. and Hauser, L.J. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11, 119. and if you use ResFams, cite the corresponding articles: Gibson, M.K., Forsberg, K.J. and Dantas, G. (2015) Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J, 9,
IntegronFinder Documentation
 IntegronFinder Documentation Release 2-2018-11-08 Jean Cury, Bertrand Néron, Eduardo PC Rocha Nov 08, 2018 Contents 1 User Guide 2 1.1 User Guide...................................... 2 2 Developer Guide
IntegronFinder Documentation Release 2-2018-11-08 Jean Cury, Bertrand Néron, Eduardo PC Rocha Nov 08, 2018 Contents 1 User Guide 2 1.1 User Guide...................................... 2 2 Developer Guide
COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP. Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas
 COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas First of all connect once again to the CBS system: Open ssh shell client. Press Quick
COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas First of all connect once again to the CBS system: Open ssh shell client. Press Quick
Django MFA Documentation
 Django MFA Documentation Release 1.0 Micro Pyramid Sep 20, 2018 Contents 1 Getting started 3 1.1 Requirements............................................... 3 1.2 Installation................................................
Django MFA Documentation Release 1.0 Micro Pyramid Sep 20, 2018 Contents 1 Getting started 3 1.1 Requirements............................................... 3 1.2 Installation................................................
The Old World. Have you ever had to collaborate on a project by
 What the Git? The Old World Have you ever had to collaborate on a project by Shuttling a USB drive back and forth Using Dropbox E-mailing your document around Have you ever accidentally deleted someone
What the Git? The Old World Have you ever had to collaborate on a project by Shuttling a USB drive back and forth Using Dropbox E-mailing your document around Have you ever accidentally deleted someone
CLC Server. End User USER MANUAL
 CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
TPS Documentation. Release Thomas Roten
 TPS Documentation Release 0.1.0 Thomas Roten Sep 27, 2017 Contents 1 TPS: TargetProcess in Python! 3 2 Installation 5 3 Contributing 7 3.1 Types of Contributions..........................................
TPS Documentation Release 0.1.0 Thomas Roten Sep 27, 2017 Contents 1 TPS: TargetProcess in Python! 3 2 Installation 5 3 Contributing 7 3.1 Types of Contributions..........................................
Python Project Example Documentation
 Python Project Example Documentation Release 0.1.0 Neil Stoddard Mar 22, 2017 Contents 1 Neilvana Example 3 1.1 Features.................................................. 3 1.2 Credits..................................................
Python Project Example Documentation Release 0.1.0 Neil Stoddard Mar 22, 2017 Contents 1 Neilvana Example 3 1.1 Features.................................................. 3 1.2 Credits..................................................
Lecture 3: Processing Language Data, Git/GitHub. LING 1340/2340: Data Science for Linguists Na-Rae Han
 Lecture 3: Processing Language Data, Git/GitHub LING 1340/2340: Data Science for Linguists Na-Rae Han Objectives What do linguistic data look like? Homework 1: What did you process? How does collaborating
Lecture 3: Processing Language Data, Git/GitHub LING 1340/2340: Data Science for Linguists Na-Rae Han Objectives What do linguistic data look like? Homework 1: What did you process? How does collaborating
Job Submitter Documentation
 Job Submitter Documentation Release 0+untagged.133.g5a1e521.dirty Juan Eiros February 27, 2017 Contents 1 Job Submitter 3 1.1 Before you start............................................. 3 1.2 Features..................................................
Job Submitter Documentation Release 0+untagged.133.g5a1e521.dirty Juan Eiros February 27, 2017 Contents 1 Job Submitter 3 1.1 Before you start............................................. 3 1.2 Features..................................................
Git. Charles J. Geyer School of Statistics University of Minnesota. Stat 8054 Lecture Notes
 Git Charles J. Geyer School of Statistics University of Minnesota Stat 8054 Lecture Notes 1 Before Anything Else Tell git who you are. git config --global user.name "Charles J. Geyer" git config --global
Git Charles J. Geyer School of Statistics University of Minnesota Stat 8054 Lecture Notes 1 Before Anything Else Tell git who you are. git config --global user.name "Charles J. Geyer" git config --global
HIPPIE User Manual. (v0.0.2-beta, 2015/4/26, Yih-Chii Hwang, yihhwang [at] mail.med.upenn.edu)
![HIPPIE User Manual. (v0.0.2-beta, 2015/4/26, Yih-Chii Hwang, yihhwang [at] mail.med.upenn.edu) HIPPIE User Manual. (v0.0.2-beta, 2015/4/26, Yih-Chii Hwang, yihhwang [at] mail.med.upenn.edu)](/thumbs/71/65752105.jpg) HIPPIE User Manual (v0.0.2-beta, 2015/4/26, Yih-Chii Hwang, yihhwang [at] mail.med.upenn.edu) OVERVIEW OF HIPPIE o Flowchart of HIPPIE o Requirements PREPARE DIRECTORY STRUCTURE FOR HIPPIE EXECUTION o
HIPPIE User Manual (v0.0.2-beta, 2015/4/26, Yih-Chii Hwang, yihhwang [at] mail.med.upenn.edu) OVERVIEW OF HIPPIE o Flowchart of HIPPIE o Requirements PREPARE DIRECTORY STRUCTURE FOR HIPPIE EXECUTION o
chatterbot-weather Documentation
 chatterbot-weather Documentation Release 0.1.1 Gunther Cox Nov 23, 2018 Contents 1 chatterbot-weather 3 1.1 Installation................................................ 3 1.2 Example.................................................
chatterbot-weather Documentation Release 0.1.1 Gunther Cox Nov 23, 2018 Contents 1 chatterbot-weather 3 1.1 Installation................................................ 3 1.2 Example.................................................
CNRS ANF PYTHON Packaging & Life Cycle
 CNRS ANF PYTHON Packaging & Life Cycle Marc Poinot Numerical Simulation Dept. Outline Package management with Python Concepts Software life cycle Package services Pragmatic approach Practical works Source
CNRS ANF PYTHON Packaging & Life Cycle Marc Poinot Numerical Simulation Dept. Outline Package management with Python Concepts Software life cycle Package services Pragmatic approach Practical works Source
BanzaiDB Documentation
 BanzaiDB Documentation Release 0.3.0 Mitchell Stanton-Cook Jul 19, 2017 Contents 1 BanzaiDB documentation contents 3 2 Indices and tables 11 i ii BanzaiDB is a tool for pairing Microbial Genomics Next
BanzaiDB Documentation Release 0.3.0 Mitchell Stanton-Cook Jul 19, 2017 Contents 1 BanzaiDB documentation contents 3 2 Indices and tables 11 i ii BanzaiDB is a tool for pairing Microbial Genomics Next
Roman Numeral Converter Documentation
 Roman Numeral Converter Documentation Release 0.1.0 Adrian Cruz October 07, 2014 Contents 1 Roman Numeral Converter 3 1.1 Features.................................................. 3 2 Installation 5
Roman Numeral Converter Documentation Release 0.1.0 Adrian Cruz October 07, 2014 Contents 1 Roman Numeral Converter 3 1.1 Features.................................................. 3 2 Installation 5
google-search Documentation
 google-search Documentation Release 1.0.0 Anthony Hseb May 08, 2017 Contents 1 google-search 3 1.1 Features.................................................. 3 1.2 Credits..................................................
google-search Documentation Release 1.0.0 Anthony Hseb May 08, 2017 Contents 1 google-search 3 1.1 Features.................................................. 3 1.2 Credits..................................................
sainsmart Documentation
 sainsmart Documentation Release 0.3.1 Victor Yap Jun 21, 2017 Contents 1 sainsmart 3 1.1 Install................................................... 3 1.2 Usage...................................................
sainsmart Documentation Release 0.3.1 Victor Yap Jun 21, 2017 Contents 1 sainsmart 3 1.1 Install................................................... 3 1.2 Usage...................................................
TangeloHub Documentation
 TangeloHub Documentation Release None Kitware, Inc. September 21, 2015 Contents 1 User s Guide 3 1.1 Managing Data.............................................. 3 1.2 Running an Analysis...........................................
TangeloHub Documentation Release None Kitware, Inc. September 21, 2015 Contents 1 User s Guide 3 1.1 Managing Data.............................................. 3 1.2 Running an Analysis...........................................
django-idioticon Documentation
 django-idioticon Documentation Release 0.0.1 openpolis June 10, 2014 Contents 1 django-idioticon 3 1.1 Documentation.............................................. 3 1.2 Quickstart................................................
django-idioticon Documentation Release 0.0.1 openpolis June 10, 2014 Contents 1 django-idioticon 3 1.1 Documentation.............................................. 3 1.2 Quickstart................................................
RAMMCAP The Rapid Analysis of Multiple Metagenomes with a Clustering and Annotation Pipeline
 RAMMCAP The Rapid Analysis of Multiple Metagenomes with a Clustering and Annotation Pipeline Weizhong Li, liwz@sdsc.edu CAMERA project (http://camera.calit2.net) Contents: 1. Introduction 2. Implementation
RAMMCAP The Rapid Analysis of Multiple Metagenomes with a Clustering and Annotation Pipeline Weizhong Li, liwz@sdsc.edu CAMERA project (http://camera.calit2.net) Contents: 1. Introduction 2. Implementation
Github/Git Primer. Tyler Hague
 Github/Git Primer Tyler Hague Why Use Github? Github keeps all of our code up to date in one place Github tracks changes so we can see what is being worked on Github has issue tracking for keeping up with
Github/Git Primer Tyler Hague Why Use Github? Github keeps all of our code up to date in one place Github tracks changes so we can see what is being worked on Github has issue tracking for keeping up with
Using GitHub to Share with SparkFun a
 Using GitHub to Share with SparkFun a learn.sparkfun.com tutorial Available online at: http://sfe.io/t52 Contents Introduction Gitting Started Forking a Repository Committing, Pushing and Pulling Syncing
Using GitHub to Share with SparkFun a learn.sparkfun.com tutorial Available online at: http://sfe.io/t52 Contents Introduction Gitting Started Forking a Repository Committing, Pushing and Pulling Syncing
Browser Exercises - I. Alignments and Comparative genomics
 Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Python wrapper for Viscosity.app Documentation
 Python wrapper for Viscosity.app Documentation Release Paul Kremer March 08, 2014 Contents 1 Python wrapper for Viscosity.app 3 1.1 Features.................................................. 3 2 Installation
Python wrapper for Viscosity.app Documentation Release Paul Kremer March 08, 2014 Contents 1 Python wrapper for Viscosity.app 3 1.1 Features.................................................. 3 2 Installation
Git Tutorial. André Sailer. ILD Technical Meeting April 24, 2017 CERN-EP-LCD. ILD Technical Meeting, Apr 24, 2017 A. Sailer: Git Tutorial 1/36
 ILD Technical Meeting, Apr 24, 2017 A. Sailer: Git Tutorial 1/36 Git Tutorial André Sailer CERN-EP-LCD ILD Technical Meeting April 24, 2017 LD Technical Meeting, Apr 24, 2017 A. Sailer: Git Tutorial 2/36
ILD Technical Meeting, Apr 24, 2017 A. Sailer: Git Tutorial 1/36 Git Tutorial André Sailer CERN-EP-LCD ILD Technical Meeting April 24, 2017 LD Technical Meeting, Apr 24, 2017 A. Sailer: Git Tutorial 2/36
Python Schema Generator Documentation
 Python Schema Generator Documentation Release 1.0.0 Peter Demin June 26, 2016 Contents 1 Mutant - Python code generator 3 1.1 Project Status............................................... 3 1.2 Design..................................................
Python Schema Generator Documentation Release 1.0.0 Peter Demin June 26, 2016 Contents 1 Mutant - Python code generator 3 1.1 Project Status............................................... 3 1.2 Design..................................................
Python simple arp table reader Documentation
 Python simple arp table reader Documentation Release 0.0.1 David Francos Nov 17, 2017 Contents 1 Python simple arp table reader 3 1.1 Features.................................................. 3 1.2 Usage...................................................
Python simple arp table reader Documentation Release 0.0.1 David Francos Nov 17, 2017 Contents 1 Python simple arp table reader 3 1.1 Features.................................................. 3 1.2 Usage...................................................
Tips on how to set up a GitHub account:
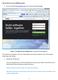 Tips on how to set up a GitHub account: 1. Go to the website https://github.com/, you will see the following page: Figure 1: The GitHub main webpage (before you create an account and sign in) Then choose
Tips on how to set up a GitHub account: 1. Go to the website https://github.com/, you will see the following page: Figure 1: The GitHub main webpage (before you create an account and sign in) Then choose
I2C LCD Documentation
 I2C LCD Documentation Release 0.1.0 Peter Landoll Sep 04, 2017 Contents 1 I2C LCD 3 1.1 Features.................................................. 3 1.2 Credits..................................................
I2C LCD Documentation Release 0.1.0 Peter Landoll Sep 04, 2017 Contents 1 I2C LCD 3 1.1 Features.................................................. 3 1.2 Credits..................................................
PyCRC Documentation. Release 1.0
 PyCRC Documentation Release 1.0 Cristian Năvălici May 12, 2018 Contents 1 PyCRC 3 1.1 Features.................................................. 3 2 Installation 5 3 Usage 7 4 Contributing 9 4.1 Types
PyCRC Documentation Release 1.0 Cristian Năvălici May 12, 2018 Contents 1 PyCRC 3 1.1 Features.................................................. 3 2 Installation 5 3 Usage 7 4 Contributing 9 4.1 Types
Huber & Bulyk, BMC Bioinformatics MS ID , Additional Methods. Installation and Usage of MultiFinder, SequenceExtractor and BlockFilter
 Installation and Usage of MultiFinder, SequenceExtractor and BlockFilter I. Introduction: MultiFinder is a tool designed to combine the results of multiple motif finders and analyze the resulting motifs
Installation and Usage of MultiFinder, SequenceExtractor and BlockFilter I. Introduction: MultiFinder is a tool designed to combine the results of multiple motif finders and analyze the resulting motifs
Proteome Comparison: A fine-grained tool for comparative genomics
 Proteome Comparison: A fine-grained tool for comparative genomics In addition to the Protein Family Sorter that allows researchers to examine up to the protein families from up to 500 genomes at a time,
Proteome Comparison: A fine-grained tool for comparative genomics In addition to the Protein Family Sorter that allows researchers to examine up to the protein families from up to 500 genomes at a time,
About the Tutorial. Audience. Prerequisites. Copyright & Disclaimer. Gerrit
 Gerrit About the Tutorial Gerrit is a web-based code review tool, which is integrated with Git and built on top of Git version control system (helps developers to work together and maintain the history
Gerrit About the Tutorial Gerrit is a web-based code review tool, which is integrated with Git and built on top of Git version control system (helps developers to work together and maintain the history
django simple pagination Documentation
 django simple pagination Documentation Release 1.1.5 Micro Pyramid Nov 08, 2017 Contents 1 Getting started 3 1.1 Requirements............................................... 3 1.2 Installation................................................
django simple pagination Documentation Release 1.1.5 Micro Pyramid Nov 08, 2017 Contents 1 Getting started 3 1.1 Requirements............................................... 3 1.2 Installation................................................
Aircrack-ng python bindings Documentation
 Aircrack-ng python bindings Documentation Release 0.1.1 David Francos Cuartero January 20, 2016 Contents 1 Aircrack-ng python bindings 3 1.1 Features..................................................
Aircrack-ng python bindings Documentation Release 0.1.1 David Francos Cuartero January 20, 2016 Contents 1 Aircrack-ng python bindings 3 1.1 Features..................................................
CONCOCT Documentation. Release 1.0.0
 CONCOCT Documentation Release 1.0.0 Johannes Alneberg, Brynjar Smari Bjarnason, Ino de Bruijn, Melan December 12, 2018 Contents 1 Features 3 2 Installation 5 3 Contribute 7 4 Support 9 5 Licence 11 6
CONCOCT Documentation Release 1.0.0 Johannes Alneberg, Brynjar Smari Bjarnason, Ino de Bruijn, Melan December 12, 2018 Contents 1 Features 3 2 Installation 5 3 Contribute 7 4 Support 9 5 Licence 11 6
1. Which of these Git client commands creates a copy of the repository and a working directory in the client s workspace. (Choose one.
 Multiple-Choice Questions: 1. Which of these Git client commands creates a copy of the repository and a working directory in the client s workspace. (Choose one.) a. update b. checkout c. clone d. import
Multiple-Choice Questions: 1. Which of these Git client commands creates a copy of the repository and a working directory in the client s workspace. (Choose one.) a. update b. checkout c. clone d. import
Introduction to Git and GitHub for Writers Workbook February 23, 2019 Peter Gruenbaum
 Introduction to Git and GitHub for Writers Workbook February 23, 2019 Peter Gruenbaum Table of Contents Preparation... 3 Exercise 1: Create a repository. Use the command line.... 4 Create a repository...
Introduction to Git and GitHub for Writers Workbook February 23, 2019 Peter Gruenbaum Table of Contents Preparation... 3 Exercise 1: Create a repository. Use the command line.... 4 Create a repository...
Python Project Documentation
 Python Project Documentation Release 1.0 Tim Diels Jan 10, 2018 Contents 1 Simple project structure 3 1.1 Code repository usage.......................................... 3 1.2 Versioning................................................
Python Project Documentation Release 1.0 Tim Diels Jan 10, 2018 Contents 1 Simple project structure 3 1.1 Code repository usage.......................................... 3 1.2 Versioning................................................
Poulpe Documentation. Release Edouard Klein
 Poulpe Documentation Release 0.0.5 Edouard Klein Jul 18, 2017 Contents 1 Poulpe 1 1.1 Features.................................................. 1 2 Usage 3 3 Installation 5 4 Contributing 7 4.1 Types
Poulpe Documentation Release 0.0.5 Edouard Klein Jul 18, 2017 Contents 1 Poulpe 1 1.1 Features.................................................. 1 2 Usage 3 3 Installation 5 4 Contributing 7 4.1 Types
CLC Sequence Viewer 6.5 Windows, Mac OS X and Linux
 CLC Sequence Viewer Manual for CLC Sequence Viewer 6.5 Windows, Mac OS X and Linux January 26, 2011 This software is for research purposes only. CLC bio Finlandsgade 10-12 DK-8200 Aarhus N Denmark Contents
CLC Sequence Viewer Manual for CLC Sequence Viewer 6.5 Windows, Mac OS X and Linux January 26, 2011 This software is for research purposes only. CLC bio Finlandsgade 10-12 DK-8200 Aarhus N Denmark Contents
Getting the files for the first time...2. Making Changes, Commiting them and Pull Requests:...5. Update your repository from the upstream master...
 Table of Contents Getting the files for the first time...2 Making Changes, Commiting them and Pull Requests:...5 Update your repository from the upstream master...8 Making a new branch (for leads, do this
Table of Contents Getting the files for the first time...2 Making Changes, Commiting them and Pull Requests:...5 Update your repository from the upstream master...8 Making a new branch (for leads, do this
gunny Documentation Release David Blewett
 gunny Documentation Release 0.1.0 David Blewett December 29, 2013 Contents 1 gunny 3 1.1 Features.................................................. 3 2 Installation 5 2.1 Dependencies...............................................
gunny Documentation Release 0.1.0 David Blewett December 29, 2013 Contents 1 gunny 3 1.1 Features.................................................. 3 2 Installation 5 2.1 Dependencies...............................................
FPLLL. Contributing. Martin R. Albrecht 2017/07/06
 FPLLL Contributing Martin R. Albrecht 2017/07/06 Outline Communication Setup Reporting Bugs Topic Branches and Pull Requests How to Get your Pull Request Accepted Documentation Overview All contributions
FPLLL Contributing Martin R. Albrecht 2017/07/06 Outline Communication Setup Reporting Bugs Topic Branches and Pull Requests How to Get your Pull Request Accepted Documentation Overview All contributions
VIP Documentation. Release Carlos Alberto Gomez Gonzalez, Olivier Wertz & VORTEX team
 VIP Documentation Release 0.8.9 Carlos Alberto Gomez Gonzalez, Olivier Wertz & VORTEX team Feb 17, 2018 Contents 1 Introduction 3 2 Documentation 5 3 Jupyter notebook tutorial 7 4 TL;DR setup guide 9
VIP Documentation Release 0.8.9 Carlos Alberto Gomez Gonzalez, Olivier Wertz & VORTEX team Feb 17, 2018 Contents 1 Introduction 3 2 Documentation 5 3 Jupyter notebook tutorial 7 4 TL;DR setup guide 9
Software Development I
 6.148 Software Development I Two things How to write code for web apps. How to collaborate and keep track of your work. A text editor A text editor A text editor Anything that you re used to using Even
6.148 Software Development I Two things How to write code for web apps. How to collaborate and keep track of your work. A text editor A text editor A text editor Anything that you re used to using Even
dj-libcloud Documentation
 dj-libcloud Documentation Release 0.2.0 Daniel Greenfeld December 19, 2016 Contents 1 dj-libcloud 3 1.1 Documentation.............................................. 3 1.2 Quickstart................................................
dj-libcloud Documentation Release 0.2.0 Daniel Greenfeld December 19, 2016 Contents 1 dj-libcloud 3 1.1 Documentation.............................................. 3 1.2 Quickstart................................................
Google Domain Shared Contacts Client Documentation
 Google Domain Shared Contacts Client Documentation Release 0.1.0 Robert Joyal Mar 31, 2018 Contents 1 Google Domain Shared Contacts Client 3 1.1 Features..................................................
Google Domain Shared Contacts Client Documentation Release 0.1.0 Robert Joyal Mar 31, 2018 Contents 1 Google Domain Shared Contacts Client 3 1.1 Features..................................................
Lab 01 How to Survive & Introduction to Git. Web Programming DataLab, CS, NTHU
 Lab 01 How to Survive & Introduction to Git Web Programming DataLab, CS, NTHU Notice These slides will focus on how to submit you code by using Git command line You can also use other Git GUI tool or built-in
Lab 01 How to Survive & Introduction to Git Web Programming DataLab, CS, NTHU Notice These slides will focus on how to submit you code by using Git command line You can also use other Git GUI tool or built-in
Introduction to Git and Github Repositories
 Introduction to Git and Github Repositories Benjamin Audren École Polytechnique Fédérale de Lausanne 29/10/2014 Benjamin Audren (EPFL) CLASS/MP MP runs 29/10/2014 1 / 16 Version Control survey Survey Who
Introduction to Git and Github Repositories Benjamin Audren École Polytechnique Fédérale de Lausanne 29/10/2014 Benjamin Audren (EPFL) CLASS/MP MP runs 29/10/2014 1 / 16 Version Control survey Survey Who
open-helpdesk Documentation
 open-helpdesk Documentation Release 0.9.9 Simone Dalla Nov 16, 2017 Contents 1 Overview 3 1.1 Dependencies............................................... 3 1.2 Documentation..............................................
open-helpdesk Documentation Release 0.9.9 Simone Dalla Nov 16, 2017 Contents 1 Overview 3 1.1 Dependencies............................................... 3 1.2 Documentation..............................................
Simple libtorrent streaming module Documentation
 Simple libtorrent streaming module Documentation Release 0.1.0 David Francos August 31, 2015 Contents 1 Simple libtorrent streaming module 3 1.1 Dependences...............................................
Simple libtorrent streaming module Documentation Release 0.1.0 David Francos August 31, 2015 Contents 1 Simple libtorrent streaming module 3 1.1 Dependences...............................................
ChIP-Seq Tutorial on Galaxy
 1 Introduction ChIP-Seq Tutorial on Galaxy 2 December 2010 (modified April 6, 2017) Rory Stark The aim of this practical is to give you some experience handling ChIP-Seq data. We will be working with data
1 Introduction ChIP-Seq Tutorial on Galaxy 2 December 2010 (modified April 6, 2017) Rory Stark The aim of this practical is to give you some experience handling ChIP-Seq data. We will be working with data
Archan. Release 2.0.1
 Archan Release 2.0.1 Jul 30, 2018 Contents 1 Archan 1 1.1 Features.................................................. 1 1.2 Installation................................................ 1 1.3 Documentation..............................................
Archan Release 2.0.1 Jul 30, 2018 Contents 1 Archan 1 1.1 Features.................................................. 1 1.2 Installation................................................ 1 1.3 Documentation..............................................
pygenbank Documentation
 pygenbank Documentation Release 0.0.1 Matthieu Bruneaux February 06, 2017 Contents 1 Description 1 2 Contents 3 2.1 Installation................................................ 3 2.2 genbank module.............................................
pygenbank Documentation Release 0.0.1 Matthieu Bruneaux February 06, 2017 Contents 1 Description 1 2 Contents 3 2.1 Installation................................................ 3 2.2 genbank module.............................................
699DR git/github Tutorial
 699DR git/github Tutorial Sep 20 2017 This tutorial gives a high-level introduction into basic usage of the version control software git in combination with the online platform Github. The git commands
699DR git/github Tutorial Sep 20 2017 This tutorial gives a high-level introduction into basic usage of the version control software git in combination with the online platform Github. The git commands
Release Nicholas A. Del Grosso
 wavefront r eaderdocumentation Release 0.1.0 Nicholas A. Del Grosso Apr 12, 2017 Contents 1 wavefront_reader 3 1.1 Features.................................................. 3 1.2 Credits..................................................
wavefront r eaderdocumentation Release 0.1.0 Nicholas A. Del Grosso Apr 12, 2017 Contents 1 wavefront_reader 3 1.1 Features.................................................. 3 1.2 Credits..................................................
SAP GUI 7.30 for Windows Computer
 SAP GUI 7.30 for Windows Computer Student and Faculty Installation Instructions Table of Contents Caution:... 2 System Requirements:... 2 System Memory (RAM) requirements:... 2 Disk Space requirements:...
SAP GUI 7.30 for Windows Computer Student and Faculty Installation Instructions Table of Contents Caution:... 2 System Requirements:... 2 System Memory (RAM) requirements:... 2 Disk Space requirements:...
Lecture 2: Data in Linguistics, Git/GitHub, Jupyter Notebook. LING 1340/2340: Data Science for Linguists Na-Rae Han
 Lecture 2: Data in Linguistics, Git/GitHub, Jupyter Notebook LING 1340/2340: Data Science for Linguists Na-Rae Han Objectives What do linguistic data look like? Tools: You should be taking NOTES! Git and
Lecture 2: Data in Linguistics, Git/GitHub, Jupyter Notebook LING 1340/2340: Data Science for Linguists Na-Rae Han Objectives What do linguistic data look like? Tools: You should be taking NOTES! Git and
projecto Documentation
 projecto Documentation Release 0.0.1 Projecto Team September 08, 2014 Contents 1 Part I: Projecto Overview 3 1.1 Features.................................................. 3 1.2 Project Layout..............................................
projecto Documentation Release 0.0.1 Projecto Team September 08, 2014 Contents 1 Part I: Projecto Overview 3 1.1 Features.................................................. 3 1.2 Project Layout..............................................
DNS Zone Test Documentation
 DNS Zone Test Documentation Release 1.1.3 Maarten Diemel Dec 02, 2017 Contents 1 DNS Zone Test 3 1.1 Features.................................................. 3 1.2 Credits..................................................
DNS Zone Test Documentation Release 1.1.3 Maarten Diemel Dec 02, 2017 Contents 1 DNS Zone Test 3 1.1 Features.................................................. 3 1.2 Credits..................................................
xmljson Documentation
 xmljson Documentation Release 0.1.9 S Anand Aug 01, 2017 Contents 1 About 3 2 Convert data to XML 5 3 Convert XML to data 7 4 Conventions 9 5 Options 11 6 Installation 13 7 Roadmap 15 8 More information
xmljson Documentation Release 0.1.9 S Anand Aug 01, 2017 Contents 1 About 3 2 Convert data to XML 5 3 Convert XML to data 7 4 Conventions 9 5 Options 11 6 Installation 13 7 Roadmap 15 8 More information
Git better. Collaborative project management using Git and GitHub. Matteo Sostero March 13, Sant Anna School of Advanced Studies
 Git better Collaborative project management using Git and GitHub Matteo Sostero March 13, 2018 Sant Anna School of Advanced Studies Let s Git it done! These slides are a brief primer to Git, and how it
Git better Collaborative project management using Git and GitHub Matteo Sostero March 13, 2018 Sant Anna School of Advanced Studies Let s Git it done! These slides are a brief primer to Git, and how it
[v1.2] SIMBA docs LGCM Federal University of Minas Gerais 2015
![[v1.2] SIMBA docs LGCM Federal University of Minas Gerais 2015 [v1.2] SIMBA docs LGCM Federal University of Minas Gerais 2015](/thumbs/81/84357512.jpg) [v1.2] SIMBA docs LGCM Federal University of Minas Gerais 2015 2 Summary 1. Introduction... 3 1.1 Why use SIMBA?... 3 1.2 How does SIMBA work?... 4 1.3 How to download SIMBA?... 4 1.4 SIMBA VM... 4 2.
[v1.2] SIMBA docs LGCM Federal University of Minas Gerais 2015 2 Summary 1. Introduction... 3 1.1 Why use SIMBA?... 3 1.2 How does SIMBA work?... 4 1.3 How to download SIMBA?... 4 1.4 SIMBA VM... 4 2.
Poetaster. Release 0.1.1
 Poetaster Release 0.1.1 September 21, 2016 Contents 1 Overview 1 1.1 Installation................................................ 1 1.2 Documentation.............................................. 1 1.3
Poetaster Release 0.1.1 September 21, 2016 Contents 1 Overview 1 1.1 Installation................................................ 1 1.2 Documentation.............................................. 1 1.3
8:15 Introduction/Overview Michelle Giglio. 8:45 CloVR background W. Florian Fricke. 9:15 Hands-on: Start CloVR W. Florian Fricke
 Hands-On Exercises 2016 1 Agenda 8:15 Introduction/Overview Michelle Giglio 8:45 CloVR background W. Florian Fricke 9:15 Hands-on: Start CloVR W. Florian Fricke 9:45 Break 9:55 Hands-on: Start CloVR-Microbe
Hands-On Exercises 2016 1 Agenda 8:15 Introduction/Overview Michelle Giglio 8:45 CloVR background W. Florian Fricke 9:15 Hands-on: Start CloVR W. Florian Fricke 9:45 Break 9:55 Hands-on: Start CloVR-Microbe
Python AutoTask Web Services Documentation
 Python AutoTask Web Services Documentation Release 0.5.1 Matt Parr May 15, 2018 Contents 1 Python AutoTask Web Services 3 1.1 Features.................................................. 3 1.2 Credits..................................................
Python AutoTask Web Services Documentation Release 0.5.1 Matt Parr May 15, 2018 Contents 1 Python AutoTask Web Services 3 1.1 Features.................................................. 3 1.2 Credits..................................................
CROWDCOIN MASTERNODE SETUP COLD WALLET ON WINDOWS WITH LINUX VPS
 CROWDCOIN MASTERNODE SETUP COLD WALLET ON WINDOWS WITH LINUX VPS This tutorial shows the steps required to setup your Crowdcoin Masternode on a Linux server and run your wallet on a Windows operating system
CROWDCOIN MASTERNODE SETUP COLD WALLET ON WINDOWS WITH LINUX VPS This tutorial shows the steps required to setup your Crowdcoin Masternode on a Linux server and run your wallet on a Windows operating system
Getting the Source Code
 Getting the Source Code The CORD source code is available from our Gerrit system at gerrit.opencord.org. Setting up a Gerrit account and ssh access will also enable you to submit your own changes to CORD
Getting the Source Code The CORD source code is available from our Gerrit system at gerrit.opencord.org. Setting up a Gerrit account and ssh access will also enable you to submit your own changes to CORD
eventbrite-sdk-python Documentation
 eventbrite-sdk-python Documentation Release 3.3.4 Eventbrite December 18, 2016 Contents 1 eventbrite-sdk-python 3 1.1 Installation from PyPI.......................................... 3 1.2 Usage...................................................
eventbrite-sdk-python Documentation Release 3.3.4 Eventbrite December 18, 2016 Contents 1 eventbrite-sdk-python 3 1.1 Installation from PyPI.......................................... 3 1.2 Usage...................................................
Git, the magical version control
 Git, the magical version control Git is an open-source version control system (meaning, it s free!) that allows developers to track changes made on their code files throughout the lifetime of a project.
Git, the magical version control Git is an open-source version control system (meaning, it s free!) that allows developers to track changes made on their code files throughout the lifetime of a project.
Frontier Documentation
 Frontier Documentation Release 0.1.3-dev Sam Nicholls August 14, 2014 Contents 1 Frontier 3 1.1 Requirements............................................... 3 1.2 Installation................................................
Frontier Documentation Release 0.1.3-dev Sam Nicholls August 14, 2014 Contents 1 Frontier 3 1.1 Requirements............................................... 3 1.2 Installation................................................
Getting Started with Python
 Getting Started with Python A beginner course to Python Ryan Leung Updated: 2018/01/30 yanyan.ryan.leung@gmail.com Links Tutorial Material on GitHub: http://goo.gl/grrxqj 1 Learning Outcomes Python as
Getting Started with Python A beginner course to Python Ryan Leung Updated: 2018/01/30 yanyan.ryan.leung@gmail.com Links Tutorial Material on GitHub: http://goo.gl/grrxqj 1 Learning Outcomes Python as
flask-dynamo Documentation
 flask-dynamo Documentation Release 0.1.2 Randall Degges January 22, 2018 Contents 1 User s Guide 3 1.1 Quickstart................................................ 3 1.2 Getting Help...............................................
flask-dynamo Documentation Release 0.1.2 Randall Degges January 22, 2018 Contents 1 User s Guide 3 1.1 Quickstart................................................ 3 1.2 Getting Help...............................................
FEEG Applied Programming 3 - Version Control and Git II
 FEEG6002 - Applied Programming 3 - Version Control and Git II Richard Boardman, Sam Sinayoko 2016-10-19 Outline Learning outcomes Working with a single repository (review) Working with multiple versions
FEEG6002 - Applied Programming 3 - Version Control and Git II Richard Boardman, Sam Sinayoko 2016-10-19 Outline Learning outcomes Working with a single repository (review) Working with multiple versions
Conda Documentation. Release latest
 Conda Documentation Release latest August 09, 2015 Contents 1 Installation 3 2 Getting Started 5 3 Building Your Own Packages 7 4 Getting Help 9 5 Contributing 11 i ii Conda Documentation, Release latest
Conda Documentation Release latest August 09, 2015 Contents 1 Installation 3 2 Getting Started 5 3 Building Your Own Packages 7 4 Getting Help 9 5 Contributing 11 i ii Conda Documentation, Release latest
doconv Documentation Release Jacob Mourelos
 doconv Documentation Release 0.1.6 Jacob Mourelos October 17, 2016 Contents 1 Introduction 3 2 Features 5 2.1 Available Format Conversions...................................... 5 3 Installation 7 3.1
doconv Documentation Release 0.1.6 Jacob Mourelos October 17, 2016 Contents 1 Introduction 3 2 Features 5 2.1 Available Format Conversions...................................... 5 3 Installation 7 3.1
pyldavis Documentation
 pyldavis Documentation Release 2.1.2 Ben Mabey Feb 06, 2018 Contents 1 pyldavis 3 1.1 Installation................................................ 3 1.2 Usage...................................................
pyldavis Documentation Release 2.1.2 Ben Mabey Feb 06, 2018 Contents 1 pyldavis 3 1.1 Installation................................................ 3 1.2 Usage...................................................
Merge Conflicts p. 92 More GitHub Workflows: Forking and Pull Requests p. 97 Using Git to Make Life Easier: Working with Past Commits p.
 Preface p. xiii Ideology: Data Skills for Robust and Reproducible Bioinformatics How to Learn Bioinformatics p. 1 Why Bioinformatics? Biology's Growing Data p. 1 Learning Data Skills to Learn Bioinformatics
Preface p. xiii Ideology: Data Skills for Robust and Reproducible Bioinformatics How to Learn Bioinformatics p. 1 Why Bioinformatics? Biology's Growing Data p. 1 Learning Data Skills to Learn Bioinformatics
When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame
 1 When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame translation on the fly. That is, 3 reading frames from
1 When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame translation on the fly. That is, 3 reading frames from
git-flow Documentation
 git-flow Documentation Release 1.0 Johan Cwiklinski Jul 14, 2017 Contents 1 Presentation 3 1.1 Conventions............................................... 4 1.2 Pre-requisites...............................................
git-flow Documentation Release 1.0 Johan Cwiklinski Jul 14, 2017 Contents 1 Presentation 3 1.1 Conventions............................................... 4 1.2 Pre-requisites...............................................
TFM-Explorer user manual
 TFM-Explorer user manual Laurie Tonon January 27, 2010 1 Contents 1 Introduction 3 1.1 What is TFM-Explorer?....................... 3 1.2 Versions................................ 3 1.3 Licence................................
TFM-Explorer user manual Laurie Tonon January 27, 2010 1 Contents 1 Introduction 3 1.1 What is TFM-Explorer?....................... 3 1.2 Versions................................ 3 1.3 Licence................................
Annotating a single sequence
 BioNumerics Tutorial: Annotating a single sequence 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn how
BioNumerics Tutorial: Annotating a single sequence 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn how
About SJTUG. SJTU *nix User Group SJTU Joyful Techie User Group
 About SJTUG SJTU *nix User Group SJTU Joyful Techie User Group Homepage - https://sjtug.org/ SJTUG Mirrors - https://mirrors.sjtug.sjtu.edu.cn/ GitHub - https://github.com/sjtug Git Basic Tutorial Zhou
About SJTUG SJTU *nix User Group SJTU Joyful Techie User Group Homepage - https://sjtug.org/ SJTUG Mirrors - https://mirrors.sjtug.sjtu.edu.cn/ GitHub - https://github.com/sjtug Git Basic Tutorial Zhou
CSC 2700: Scientific Computing
 CSC 2700: Scientific Computing Record and share your work: revision control systems Dr Frank Löffler Center for Computation and Technology Louisiana State University, Baton Rouge, LA Feb 13 2014 Overview
CSC 2700: Scientific Computing Record and share your work: revision control systems Dr Frank Löffler Center for Computation and Technology Louisiana State University, Baton Rouge, LA Feb 13 2014 Overview
umicount Documentation
 umicount Documentation Release 1.0 Mickael June 30, 2015 Contents 1 Introduction 3 2 Recommendations 5 3 Install 7 4 How to use umicount 9 4.1 Working with a single bed file......................................
umicount Documentation Release 1.0 Mickael June 30, 2015 Contents 1 Introduction 3 2 Recommendations 5 3 Install 7 4 How to use umicount 9 4.1 Working with a single bed file......................................
smartfilesorter Documentation
 smartfilesorter Documentation Release 0.2.0 Jason Short September 14, 2014 Contents 1 Smart File Sorter 3 1.1 Features.................................................. 3 2 Installation 5 3 Usage Example
smartfilesorter Documentation Release 0.2.0 Jason Short September 14, 2014 Contents 1 Smart File Sorter 3 1.1 Features.................................................. 3 2 Installation 5 3 Usage Example
AMath 483/583 Lecture 2
 AMath 483/583 Lecture 2 Outline: Binary storage, floating point numbers Version control main ideas Client-server version control, e.g., CVS, Subversion Distributed version control, e.g., git, Mercurial
AMath 483/583 Lecture 2 Outline: Binary storage, floating point numbers Version control main ideas Client-server version control, e.g., CVS, Subversion Distributed version control, e.g., git, Mercurial
Windows. Everywhere else
 Git version control Enable native scrolling Git is a tool to manage sourcecode Never lose your coding progress again An empty folder 1/30 Windows Go to your programs overview and start Git Bash Everywhere
Git version control Enable native scrolling Git is a tool to manage sourcecode Never lose your coding progress again An empty folder 1/30 Windows Go to your programs overview and start Git Bash Everywhere
django-konfera Documentation
 django-konfera Documentation Release 0.1 SPy o.z. Mar 21, 2017 Contents 1 Installation 3 1.1 Using Pip................................................. 3 1.2 Using the Source.............................................
django-konfera Documentation Release 0.1 SPy o.z. Mar 21, 2017 Contents 1 Installation 3 1.1 Using Pip................................................. 3 1.2 Using the Source.............................................
AMath 483/583 Lecture 2. Notes: Notes: Homework #1. Class Virtual Machine. Notes: Outline:
 AMath 483/583 Lecture 2 Outline: Binary storage, floating point numbers Version control main ideas Client-server version control, e.g., CVS, Subversion Distributed version control, e.g., git, Mercurial
AMath 483/583 Lecture 2 Outline: Binary storage, floating point numbers Version control main ideas Client-server version control, e.g., CVS, Subversion Distributed version control, e.g., git, Mercurial
Release Ralph Offinger
 nagios c heck p aloaltodocumentation Release 0.3.2 Ralph Offinger May 30, 2017 Contents 1 nagios_check_paloalto: a Nagios/Icinga Plugin 3 1.1 Documentation..............................................
nagios c heck p aloaltodocumentation Release 0.3.2 Ralph Offinger May 30, 2017 Contents 1 nagios_check_paloalto: a Nagios/Icinga Plugin 3 1.1 Documentation..............................................
MacVector for Mac OS X. The online updater for this release is MB in size
 MacVector 17.0.3 for Mac OS X The online updater for this release is 143.5 MB in size You must be running MacVector 15.5.4 or later for this updater to work! System Requirements MacVector 17.0 is supported
MacVector 17.0.3 for Mac OS X The online updater for this release is 143.5 MB in size You must be running MacVector 15.5.4 or later for this updater to work! System Requirements MacVector 17.0 is supported
Tizen/Artik IoT Practice Part 4 Open Source Development
 1 Tizen/Artik IoT Practice Part 4 Open Source Development Sungkyunkwan University Contents 2 SCM Tool: Git Version Management Local & Remote Repository Branch Management Github Contribution Process Issue
1 Tizen/Artik IoT Practice Part 4 Open Source Development Sungkyunkwan University Contents 2 SCM Tool: Git Version Management Local & Remote Repository Branch Management Github Contribution Process Issue
DNASIS MAX V2.0. Tutorial Booklet
 Sequence Analysis Software DNASIS MAX V2.0 Tutorial Booklet CONTENTS Introduction...2 1. DNASIS MAX...5 1-1: Protein Translation & Function...5 1-2: Nucleic Acid Alignments(BLAST Search)...10 1-3: Vector
Sequence Analysis Software DNASIS MAX V2.0 Tutorial Booklet CONTENTS Introduction...2 1. DNASIS MAX...5 1-1: Protein Translation & Function...5 1-2: Nucleic Acid Alignments(BLAST Search)...10 1-3: Vector
Rubix Documentation. Release Qubole
 Rubix Documentation Release 0.2.12 Qubole Jul 02, 2018 Contents: 1 RubiX 3 1.1 Usecase.................................................. 3 1.2 Supported Engines and Cloud Stores..................................
Rubix Documentation Release 0.2.12 Qubole Jul 02, 2018 Contents: 1 RubiX 3 1.1 Usecase.................................................. 3 1.2 Supported Engines and Cloud Stores..................................
Release Fulfil.IO Inc.
 api a idocumentation Release 0.1.0 Fulfil.IO Inc. July 29, 2016 Contents 1 api_ai 3 1.1 Features.................................................. 3 1.2 Installation................................................
api a idocumentation Release 0.1.0 Fulfil.IO Inc. July 29, 2016 Contents 1 api_ai 3 1.1 Features.................................................. 3 1.2 Installation................................................
django-reinhardt Documentation
 django-reinhardt Documentation Release 0.1.0 Hyuntak Joo December 02, 2016 Contents 1 django-reinhardt 3 1.1 Installation................................................ 3 1.2 Usage...................................................
django-reinhardt Documentation Release 0.1.0 Hyuntak Joo December 02, 2016 Contents 1 django-reinhardt 3 1.1 Installation................................................ 3 1.2 Usage...................................................
Introduction to Genome Browsers
 Introduction to Genome Browsers Rolando Garcia-Milian, MLS, AHIP (Rolando.milian@ufl.edu) Department of Biomedical and Health Information Services Health Sciences Center Libraries, University of Florida
Introduction to Genome Browsers Rolando Garcia-Milian, MLS, AHIP (Rolando.milian@ufl.edu) Department of Biomedical and Health Information Services Health Sciences Center Libraries, University of Florida
Revision Control. An Introduction Using Git 1/15
 Revision Control An Introduction Using Git 1/15 Overview 1. What is revision control? 2. 30,000 foot view 3. Software - git and gitk 4. Setting up your own repository on onyx 2/15 What is version control?
Revision Control An Introduction Using Git 1/15 Overview 1. What is revision control? 2. 30,000 foot view 3. Software - git and gitk 4. Setting up your own repository on onyx 2/15 What is version control?
