Principles of Bioinformatics. BIO540/STA569/CSI660 Fall 2010
|
|
|
- Jodie Moore
- 5 years ago
- Views:
Transcription
1 Principles of Bioinformatics BIO540/STA569/CSI660 Fall 2010
2 Lecture 11 Multiple Sequence Alignment I
3 Administrivia
4 Administrivia The midterm examination will be Monday, October 18 th, in class. Closed book and notes. More details soon. Fall 2010 BIO540/STA569/CSI660 4
5 Administrivia I ve put up a set of exercises on pairwise sequence alignment on the class web page. They are not a formal homework, but rather a resource to help you study for the midterm exam. Fall 2010 BIO540/STA569/CSI660 5
6 Today s Content
7 Readings MSA Basics 4.5, 6.4 Blocks, Motifs and Patterns 4.8, 4.9, 6.1, 6.3 HMMs 6.2 Alternatives 4.10, 6.5, 6.6 Fall 2009 BIO540/CSI660 7
8 Multiple Sequence Alignment (MSA) Just as two sequences can be aligned to maximize the common elements of the pair, three or more sequences can be aligned in the same manner. This is multiple sequence alignment (MSA). Fall 2009 BIO540/CSI660 8
9 Multiple Sequence Alignment MSA has many uses: Detect the overall similarity of a set of sequences. Find similar regions in sequences. As the starting point of a phylogenetic analysis to determine evolutionary relatedness. Find overlapping DNA fragments as part of genome sequencing efforts. Fall 2009 BIO540/CSI660 9
10 From Pairwise to Multiple Sequence Alignment In principle, MSA can be done as an extension of the dynamic programming used for pairwise sequence alignment. However, each sequence adds a new dimension to the alignment matrix. This causes the matrix to grow too quickly to either store it or compute using it. Fall 2009 BIO540/CSI660 10
11 From Pairwise to Multiple Sequence Alignment In a pairwise alignment, the size of the matrix is approximately l 2 where l is the length of the longer of the two sequences. So, for two sequences of length 300, the matrix has 90,000 elements. Fall 2009 BIO540/CSI660 11
12 From Pairwise to Multiple Sequence Alignment For a MSA, the size of the matrix will be approximately l n where l is the length of the longest of the sequences to be aligned, and n is the number of sequences. Fall 2009 BIO540/CSI660 12
13 From Pairwise to Multiple Sequence Alignment Using the l n formula, and assuming we have length 300 sequences, we get the following matrix sizes: 3 sequences: = 27,000, sequences: = 2,430,000,000, sequences: = x = 5,904,900,000,000,000,000,000, sequences: = x sequences: = x Fall 2009 BIO540/CSI660 13
14 From Pairwise to Multiple Sequence Alignment Clearly, a straight-forward generalization of the dynamic programming algorithm is impractical for MSA. Fall 2009 BIO540/CSI660 14
15 Defining Multiple Sequence Alignment Before we look at how MSA is done in practice, we need to define just what it is we want to do. Fall 2009 BIO540/CSI660 15
16 Scoring Multiple Sequence Alignments A tractable way of doing MSA starts with the sum of pairs (SP) scoring function. Instead of, for example, an unconstrained combination of all the symbols, just look at the scores of the pairwise combinations. Fall 2009 BIO540/CSI660 16
17 Scoring Multiple Sequence Alignments The SP score is the sum of all pairwise scores of symbols in the alignment column. For example: S S - C - Fall 2009 BIO540/CSI660 17
18 Scoring Multiple Sequence Alignments The SP score is the sum of all pairwise scores of symbols in the alignment column. For example: S S - C - Here the SP-score = p(s,s) +p(s,-) + p(s,c) + p(s,-) + p(s,-) + p(s,c) + p(s,-) + p(-,c) + p(-,-) + p(c,-) Fall 2009 BIO540/CSI660 18
19 Scoring Multiple Sequence Alignments The SP score is the sum of all pairwise scores of symbols in the alignment column. For example: S S - C - Here the SP-score = p(s,s) +p(s,-) + p(s,c) + p(s,-) + p(s,-) + p(s,c) + p(s,-) + p(-,c) + p(-,-) + p(c,-) Fall 2009 BIO540/CSI660 19
20 Scoring Multiple Sequence Alignments The SP score is the sum of all pairwise scores of symbols in the alignment column. For example: S S - C - Here the SP-score = p(s,s) +p(s,-) + p(s,c) + p(s,-) + p(s,-) + p(s,c) + p(s,-) + p(-,c) + p(-,-) + p(c,-) Fall 2009 BIO540/CSI660 20
21 Scoring Multiple Sequence Alignments The SP score is the sum of all pairwise scores of symbols in the alignment column. For example: S S - C - Here the SP-score = p(s,s) +p(s,-) + p(s,c) + p(s,-) + p(s,-) + p(s,c) + p(s,-) + p(-,c) + p(-,-) + p(c,-) Fall 2009 BIO540/CSI660 21
22 Scoring Multiple Sequence Alignments The SP score is the sum of all pairwise scores of symbols in the alignment column. For example: S S - C - Here the SP-score = p(s,s) +p(s,-) + p(s,c) + p(s,-) + p(s,-) + p(s,c) + p(s,-) + p(-,c) + p(-,-) + p(c,-) Fall 2009 BIO540/CSI660 22
23 Scoring Multiple Sequence Alignments How do we score p(-,-)? Generally, p(-, -) = 0. Fall 2009 BIO540/CSI660 23
24 Scoring Multiple Sequence Alignments The rationale for this is that, if we set p(-, -) = 0, then if we pick out a pair of sequences where we have spaces for both in the column, then the pairwise score of them is the same as the SP-score for the two. Fall 2009 BIO540/CSI660 24
25 Scoring Multiple Sequence Alignments The SP score is the sum of all pairwise scores of symbols in the alignment column. For example: S S - C - So, the example s SP-score = = -13 Fall 2009 BIO540/CSI660 25
26 Scoring Multiple Sequence Alignments Picking a pairwise alignment out of a multiple alignment is known as an induced pairwise alignment. In an induced pairwise alignment, a column that has a gap in both sequences ( -,- ) is removed. Fall 2009 BIO540/CSI660 26
27 Scoring Multiple Sequence Alignments In general, the SP-score for the entire multiple sequence list, α ( alpha ), is the same as the sum of the pairwise alignment scores. SP-score(α) = Σ i < j score(α ij ) For this to be true, p(-, -) must equal zero. Fall 2009 BIO540/CSI660 27
28 Scoring Multiple Sequence Alignments Now that we have a way to evaluate multiple alignments, we can now try and determine the alignment of a set of sequences that gives us a maximum alignment score. These alignments of maximum score are known as optimal alignments. Fall 2009 BIO540/CSI660 28
29 From Pairwise to Multiple Sequence Alignment In practice, a dynamic programming MSA algorithm will only construct and examine a very small portion of the multidimensional alignment matrix. Glossing over some details, the algorithm sets a threshold, and it will not build alignments beyond elements that score worse than the threshold. Fall 2009 BIO540/CSI660 29
30 From Pairwise to Multiple Sequence Alignment By analogy to mining, this algorithm only tunnels into portions of the matrix that have rich veins of mineral - in this case high partial alignment values. Fall 2009 BIO540/CSI660 30
31 From Pairwise to Multiple Sequence Alignment Fall 2009 BIO540/CSI660 31
32 Progressive Methods of MSA Even so, these dynamic programming MSA methods have some serious problems. They are still limited to alignments of about a half-dozen sequences. They do not guarantee an optimal solution. Although it is very likely that they will find an alignment that is at least very close to optimal. Fall 2009 BIO540/CSI660 32
33 Progressive Methods of MSA However, a different use of dynamic programming can be used for practical MSA. This technique, progressive alignment, starts by aligning the most similar sequences, and then progressively adds the other, less similar, sequences to the overall alignment. Fall 2009 BIO540/CSI660 33
34 Progressive Methods of MSA Progressive alignment is usually done as follows: Make an assessment of the phylogenetic relatedness of the sequences and their branching. Progressively align more and more sequences, starting with the closest ones. At each point, either align a sequence to a subalignment, or two sub-alignments, depending on what's closest. Fall 2009 BIO540/CSI660 34
35 Progressive Methods of MSA The method is heuristic, that is, it is not guaranteed to produce optimal alignments. Fall 2009 BIO540/CSI660 35
36 Progressive Methods of MSA The relatedness of the sequences is done by creating a tree. The tree shows how closely related the sequences to be aligned are. The tree can be created by doing pairwise comparisons of all the sequences. Fall 2009 BIO540/CSI660 36
37 Progressive Methods of MSA Trees can be created either using a full Needleman-Wunsch dynamic programming global alignment of each pair of sequences, or a simplified, faster comparison that only looks at identical amino acids in k-tuples in the sequences. k-tuples are patterns of characters of length k occurring in the two sequences. Fall 2009 BIO540/CSI660 37
38 Progressive Methods of MSA Using the tree as a guide, the closest two sequences are aligned. This creates a sub-alignment. Fall 2009 BIO540/CSI660 38
39 Progressive Methods of MSA Then, the next two closest sequences/subalignments are aligned. Fall 2009 BIO540/CSI660 39
40 Progressive Methods of MSA This continues until all the sequences have been aligned. Fall 2009 BIO540/CSI660 40
41 Progressive Methods of MSA Seq1 Seq2 Seq3 Seq4 Seq5 Seq6 Fall 2009 BIO540/CSI660 41
42 Progressive Methods of MSA Seq2 Seq3 Seq4 Seq5 Seq6 Seq1 Fall 2009 BIO540/CSI660 42
43 Progressive Methods of MSA Seq2 Seq3 Seq4 Seq5 Seq1 Seq6 Fall 2009 BIO540/CSI660 43
44 Progressive Methods of MSA Seq3 Seq4 Seq5 Seq1 Seq6 Seq2 Fall 2009 BIO540/CSI660 44
45 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 45
46 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 46
47 Progressive Methods of MSA Seq3 Seq4-43 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 47
48 Progressive Methods of MSA Seq3 45 Seq4 Seq Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 48
49 Progressive Methods of MSA Seq3 45 Seq4 25 Seq Seq Seq2-25 Seq5 Fall 2009 BIO540/CSI660 49
50 Progressive Methods of MSA Seq3 45 Seq4 25 Seq Seq Seq2-25 Seq5 Fall 2009 BIO540/CSI660 50
51 Progressive Methods of MSA Seq3 Seq4 Seq Seq Seq2-25 Seq5 Fall 2009 BIO540/CSI660 51
52 Progressive Methods of MSA Seq3 Seq4 Seq Seq Seq2-25 Seq5 Fall 2009 BIO540/CSI660 52
53 Progressive Methods of MSA Seq3 Seq4 Seq Seq Seq2-25 Seq5 Fall 2009 BIO540/CSI660 53
54 Progressive Methods of MSA Seq3 Seq Seq6 Seq1-37 Seq2 Seq5 Fall 2009 BIO540/CSI660 54
55 Progressive Methods of MSA Seq3 Seq4 17 Seq Seq5 Seq6 Seq2 Fall 2009 BIO540/CSI660 55
56 Progressive Methods of MSA Seq3 Seq4 17 Seq Seq5 Seq6 Seq2 Fall 2009 BIO540/CSI660 56
57 Progressive Methods of MSA Seq3 Seq4 17 Seq6 Seq1 Seq2 Seq5 Fall 2009 BIO540/CSI660 57
58 Progressive Methods of MSA Seq3 Seq4 Seq Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 58
59 Progressive Methods of MSA Seq3 Seq4 Seq Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 59
60 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 60
61 Progressive Methods of MSA Seq3 Seq4 7 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 61
62 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 62
63 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 63
64 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 64
65 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 65
66 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq6 Seq2 Seq5 Fall 2009 BIO540/CSI660 66
67 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq2 Seq6 Seq5 Fall 2009 BIO540/CSI660 67
68 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq2 Seq6 Seq5 Fall 2009 BIO540/CSI660 68
69 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq2 Seq5 Seq6 Fall 2009 BIO540/CSI660 69
70 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq2 Seq5 Seq6 Fall 2009 BIO540/CSI660 70
71 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq2 Seq5 Seq6 Fall 2009 BIO540/CSI660 71
72 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq2 Seq5 Seq6 Fall 2009 BIO540/CSI660 72
73 Progressive Methods of MSA Seq3 Seq4 Seq1 Seq2 Seq5 Seq6 Fall 2009 BIO540/CSI660 73
74 Clustal The most popular program to do progressive multiple sequence alignment is Clustal. The current version is ClustalW. If you re using a UNIX computer with X- Windows, there is a ClustalX version. Fall 2009 BIO540/CSI660 74
75 Clustal The relatedness of any two sequences is determined by a genetic distance. This distance is the number of mismatched positions divided by the number of matched positions. Gaps are not counted in this value. Fall 2009 BIO540/CSI660 75
76 Clustal Genetic Distance Example: LQLLKDE--QWIDVPPMRHSIVVNLGDQ LQATRDGGRTWITVQPVEGAFVVNLGDH Number of plusses: 12 Number of minuses: 13: Genetic Distance = 13/12 = Fall 2009 BIO540/CSI660 76
77 Clustal The W in ClustalW stands for weighting. Different sequences are given different weights, or significance values in the alignment. Fall 2009 BIO540/CSI660 77
78 Clustal In the weighting scheme for ClustalW, distant sequences are more heavily weighted in the alignment than more closely related ones. Fall 2009 BIO540/CSI660 78
79 Clustal The rationale for this is that closely related sequences provide little extra information for alignment purposes. This is similar in spirit to PSI-BLAST s removing almost identical sequences. Fall 2009 BIO540/CSI660 79
80 Clustal How gaps are placed is very important in determining the overall quality of the final multiple sequence alignment. Researchers have found patterns in gaps in related proteins - gaps avoid secondary structure elements, and conserved regions in the aligned sequences. ClustalW tries to model these patterns. Fall 2009 BIO540/CSI660 80
81 Clustal ClustalW uses an affine gap model, with a gap opening penalty and a further penalty for each position in the gap. The cost of the gap opening penalty changes in different circumstances. Fall 2009 BIO540/CSI660 81
82 Clustal First, the basic Gap Opening Penalty (GOP) and a Gap Extension Penalty (GEP) are chosen. Then, they are modified as follows for each pair of sequences/ sub-alignments to be aligned: GOP A B (GOP initial + log (min(m, N))) GEP GEP initial (1.0 + log (min(m, N))) where A is the average value for a mismatch in the amino acid weight matrix, B is the percent identity of the two sequences, and M and N are the lengths of the sequences to be aligned, Fall 2009 BIO540/CSI660 82
83 Clustal Then for each position in the alignment, they are further modified as follows: Use lower gap penalties at positions where gaps already occur, Increase gap penalties adjacent to positions where gaps already occur, Reduce gap penalties where stretches of hydrophilic residues occur, and Increase or decrease gap penalties using tables of the observed frequencies of gaps adjacent to each of the 20 amino acids. Fall 2009 BIO540/CSI660 83
84 MSA with Clustal
85 MSA with Clustal There are several things that should be taken into account when doing MSA with any program. Here are some things to be aware of when doing MSA with ClustalW. Fall 2009 BIO540/CSI660 85
86 Clustal & Substitution Matrices
87 MSA with Clustal The first decision is what substitution matrix to use. For example, the ClustalW server at EBI ( has PAM BLOSUM Gonnet, and DNA identity matrices. Fall 2009 BIO540/CSI660 87
88 MSA with Clustal We are familiar with the amino acid substitution matrices. The EBI web site has the following comments: The Blosum matrices are best for detecting local alignments. The Blosum62 matrix is the best for detecting the majority of weak protein similarities. The Blosum45 matrix is the best for detecting long and weak alignments. Fall 2009 BIO540/CSI660 88
89 PAM and BLOSUM According to the EBI web site help, the following matrices are roughly equivalent: PAM100 and Blosum90 PAM120 and Blosum80 PAM160 and Blosum60 PAM200 and Blosum52 PAM250 and Blosum45 Fall 2009 BIO540/CSI660 89
90 Gonnet Matrices Gonnet, Cohen and Benner (1992) used exhaustive pairwise alignments of the protein databases then available to create a distance matrix. This matrix was used iteratively to refine the alignment, and then refine the matrix. They propose that their matrix should be used for an initial alignment, and that a PAM matrix suitable for the task s distance then be used for subsequent ones. Fall 2009 BIO540/CSI660 90
91 DNA Identity Matrices The last class of matrices for use with Clustal at EBI is the DNA identity matrix. Compared to the others, it is a simple matrix. Each DNA base match is worth +1, and Each mismatch is worth -10,000. This virtually ensures aligning no mismatched positions, creating gaps in preference to mismatches. Depending on the gap penalties, of course. Fall 2009 BIO540/CSI660 91
92 Other Factors
93 MSA with ClustalW Some things to keep in mind when aligning with ClustalW. If the input to Clustalw is already aligned, any subsequent alignment will not remove existing gaps. The order of the sequences in the input is significant. Different orders of the same sequences can give different results. Fall 2009 BIO540/CSI660 93
94 MSA with ClustalW Some things to keep in mind when aligning with ClustalW. ClustalW has many parameters (cf. the server at EBI). Some of them are often not worthwhile to adjust (e.g. the gap penalties). If you are using ClustalW extensively, you should read the papers on it by the authors. Fall 2009 BIO540/CSI660 94
95 MSA with ClustalW If you are using ClustalW extensively, you should read the papers on it by the authors. Higgins, D.G., Thompson, J.D., and Gibson, T.J Using CLUSTAL for Multiple Sequence Alignments. Methods Enzymol. 266: Several older references. Fall 2009 BIO540/CSI660 95
96 MSA with ClustalW Additional things to keep in mind when aligning with ClustalW. If you know the secondary structures of one of the sequences in the alignment, that information can be used to bias the alignment. Gaps will be penalized more highly in secondary structure regions. Fall 2009 BIO540/CSI660 96
PROTEIN MULTIPLE ALIGNMENT MOTIVATION: BACKGROUND: Marina Sirota
 Marina Sirota MOTIVATION: PROTEIN MULTIPLE ALIGNMENT To study evolution on the genetic level across a wide range of organisms, biologists need accurate tools for multiple sequence alignment of protein
Marina Sirota MOTIVATION: PROTEIN MULTIPLE ALIGNMENT To study evolution on the genetic level across a wide range of organisms, biologists need accurate tools for multiple sequence alignment of protein
CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment
 CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment Courtesy of jalview 1 Motivations Collective statistic Protein families Identification and representation of conserved sequence features
CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment Courtesy of jalview 1 Motivations Collective statistic Protein families Identification and representation of conserved sequence features
Multiple Sequence Alignment. Mark Whitsitt - NCSA
 Multiple Sequence Alignment Mark Whitsitt - NCSA What is a Multiple Sequence Alignment (MA)? GMHGTVYANYAVDSSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHV GMHGTVYANYAVEHSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKTPHV
Multiple Sequence Alignment Mark Whitsitt - NCSA What is a Multiple Sequence Alignment (MA)? GMHGTVYANYAVDSSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHV GMHGTVYANYAVEHSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKTPHV
GLOBEX Bioinformatics (Summer 2015) Multiple Sequence Alignment
 GLOBEX Bioinformatics (Summer 2015) Multiple Sequence Alignment Scoring Dynamic Programming algorithms Heuristic algorithms CLUSTAL W Courtesy of jalview Motivations Collective (or aggregate) statistic
GLOBEX Bioinformatics (Summer 2015) Multiple Sequence Alignment Scoring Dynamic Programming algorithms Heuristic algorithms CLUSTAL W Courtesy of jalview Motivations Collective (or aggregate) statistic
3.4 Multiple sequence alignment
 3.4 Multiple sequence alignment Why produce a multiple sequence alignment? Using more than two sequences results in a more convincing alignment by revealing conserved regions in ALL of the sequences Aligned
3.4 Multiple sequence alignment Why produce a multiple sequence alignment? Using more than two sequences results in a more convincing alignment by revealing conserved regions in ALL of the sequences Aligned
Multiple sequence alignment. November 20, 2018
 Multiple sequence alignment November 20, 2018 Why do multiple alignment? Gain insight into evolutionary history Can assess time of divergence by looking at the number of mutations needed to change one
Multiple sequence alignment November 20, 2018 Why do multiple alignment? Gain insight into evolutionary history Can assess time of divergence by looking at the number of mutations needed to change one
Lecture 5: Multiple sequence alignment
 Lecture 5: Multiple sequence alignment Introduction to Computational Biology Teresa Przytycka, PhD (with some additions by Martin Vingron) Why do we need multiple sequence alignment Pairwise sequence alignment
Lecture 5: Multiple sequence alignment Introduction to Computational Biology Teresa Przytycka, PhD (with some additions by Martin Vingron) Why do we need multiple sequence alignment Pairwise sequence alignment
Multiple Sequence Alignment (MSA)
 I519 Introduction to Bioinformatics, Fall 2013 Multiple Sequence Alignment (MSA) Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Outline Multiple sequence alignment (MSA) Generalize
I519 Introduction to Bioinformatics, Fall 2013 Multiple Sequence Alignment (MSA) Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Outline Multiple sequence alignment (MSA) Generalize
Bioinformatics for Biologists
 Bioinformatics for Biologists Sequence Analysis: Part I. Pairwise alignment and database searching Fran Lewitter, Ph.D. Director Bioinformatics & Research Computing Whitehead Institute Topics to Cover
Bioinformatics for Biologists Sequence Analysis: Part I. Pairwise alignment and database searching Fran Lewitter, Ph.D. Director Bioinformatics & Research Computing Whitehead Institute Topics to Cover
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Basic Local Alignment Search Tool (BLAST)
 BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
Algorithmic Approaches for Biological Data, Lecture #20
 Algorithmic Approaches for Biological Data, Lecture #20 Katherine St. John City University of New York American Museum of Natural History 20 April 2016 Outline Aligning with Gaps and Substitution Matrices
Algorithmic Approaches for Biological Data, Lecture #20 Katherine St. John City University of New York American Museum of Natural History 20 April 2016 Outline Aligning with Gaps and Substitution Matrices
BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio CS 466 Saurabh Sinha
 BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio. 1990. CS 466 Saurabh Sinha Motivation Sequence homology to a known protein suggest function of newly sequenced protein Bioinformatics
BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio. 1990. CS 466 Saurabh Sinha Motivation Sequence homology to a known protein suggest function of newly sequenced protein Bioinformatics
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 06: Multiple Sequence Alignment https://upload.wikimedia.org/wikipedia/commons/thumb/7/79/rplp0_90_clustalw_aln.gif/575px-rplp0_90_clustalw_aln.gif Slides
EECS730: Introduction to Bioinformatics Lecture 06: Multiple Sequence Alignment https://upload.wikimedia.org/wikipedia/commons/thumb/7/79/rplp0_90_clustalw_aln.gif/575px-rplp0_90_clustalw_aln.gif Slides
Multiple Sequence Alignment Sum-of-Pairs and ClustalW. Ulf Leser
 Multiple Sequence Alignment Sum-of-Pairs and ClustalW Ulf Leser This Lecture Multiple Sequence Alignment The problem Theoretical approach: Sum-of-Pairs scores Practical approach: ClustalW Ulf Leser: Bioinformatics,
Multiple Sequence Alignment Sum-of-Pairs and ClustalW Ulf Leser This Lecture Multiple Sequence Alignment The problem Theoretical approach: Sum-of-Pairs scores Practical approach: ClustalW Ulf Leser: Bioinformatics,
Lecture Overview. Sequence search & alignment. Searching sequence databases. Sequence Alignment & Search. Goals: Motivations:
 Lecture Overview Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating
Lecture Overview Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST
 An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
Reconstructing long sequences from overlapping sequence fragment. Searching databases for related sequences and subsequences
 SEQUENCE ALIGNMENT ALGORITHMS 1 Why compare sequences? Reconstructing long sequences from overlapping sequence fragment Searching databases for related sequences and subsequences Storing, retrieving and
SEQUENCE ALIGNMENT ALGORITHMS 1 Why compare sequences? Reconstructing long sequences from overlapping sequence fragment Searching databases for related sequences and subsequences Storing, retrieving and
Sequence Alignment & Search
 Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating the first version
Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating the first version
Today s Lecture. Multiple sequence alignment. Improved scoring of pairwise alignments. Affine gap penalties Profiles
 Today s Lecture Multiple sequence alignment Improved scoring of pairwise alignments Affine gap penalties Profiles 1 The Edit Graph for a Pair of Sequences G A C G T T G A A T G A C C C A C A T G A C G
Today s Lecture Multiple sequence alignment Improved scoring of pairwise alignments Affine gap penalties Profiles 1 The Edit Graph for a Pair of Sequences G A C G T T G A A T G A C C C A C A T G A C G
Stephen Scott.
 1 / 33 sscott@cse.unl.edu 2 / 33 Start with a set of sequences In each column, residues are homolgous Residues occupy similar positions in 3D structure Residues diverge from a common ancestral residue
1 / 33 sscott@cse.unl.edu 2 / 33 Start with a set of sequences In each column, residues are homolgous Residues occupy similar positions in 3D structure Residues diverge from a common ancestral residue
Profiles and Multiple Alignments. COMP 571 Luay Nakhleh, Rice University
 Profiles and Multiple Alignments COMP 571 Luay Nakhleh, Rice University Outline Profiles and sequence logos Profile hidden Markov models Aligning profiles Multiple sequence alignment by gradual sequence
Profiles and Multiple Alignments COMP 571 Luay Nakhleh, Rice University Outline Profiles and sequence logos Profile hidden Markov models Aligning profiles Multiple sequence alignment by gradual sequence
Biology 644: Bioinformatics
 Find the best alignment between 2 sequences with lengths n and m, respectively Best alignment is very dependent upon the substitution matrix and gap penalties The Global Alignment Problem tries to find
Find the best alignment between 2 sequences with lengths n and m, respectively Best alignment is very dependent upon the substitution matrix and gap penalties The Global Alignment Problem tries to find
AlignMe Manual. Version 1.1. Rene Staritzbichler, Marcus Stamm, Kamil Khafizov and Lucy R. Forrest
 AlignMe Manual Version 1.1 Rene Staritzbichler, Marcus Stamm, Kamil Khafizov and Lucy R. Forrest Max Planck Institute of Biophysics Frankfurt am Main 60438 Germany 1) Introduction...3 2) Using AlignMe
AlignMe Manual Version 1.1 Rene Staritzbichler, Marcus Stamm, Kamil Khafizov and Lucy R. Forrest Max Planck Institute of Biophysics Frankfurt am Main 60438 Germany 1) Introduction...3 2) Using AlignMe
Computational Molecular Biology
 Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
BLAST MCDB 187. Friday, February 8, 13
 BLAST MCDB 187 BLAST Basic Local Alignment Sequence Tool Uses shortcut to compute alignments of a sequence against a database very quickly Typically takes about a minute to align a sequence against a database
BLAST MCDB 187 BLAST Basic Local Alignment Sequence Tool Uses shortcut to compute alignments of a sequence against a database very quickly Typically takes about a minute to align a sequence against a database
Comparison and Evaluation of Multiple Sequence Alignment Tools In Bininformatics
 IJCSNS International Journal of Computer Science and Network Security, VOL.9 No.7, July 2009 51 Comparison and Evaluation of Multiple Sequence Alignment Tools In Bininformatics Asieh Sedaghatinia, Dr Rodziah
IJCSNS International Journal of Computer Science and Network Security, VOL.9 No.7, July 2009 51 Comparison and Evaluation of Multiple Sequence Alignment Tools In Bininformatics Asieh Sedaghatinia, Dr Rodziah
Lecture 5 Advanced BLAST
 Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1. Database Searching
 COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
Sequence alignment theory and applications Session 3: BLAST algorithm
 Sequence alignment theory and applications Session 3: BLAST algorithm Introduction to Bioinformatics online course : IBT Sonal Henson Learning Objectives Understand the principles of the BLAST algorithm
Sequence alignment theory and applications Session 3: BLAST algorithm Introduction to Bioinformatics online course : IBT Sonal Henson Learning Objectives Understand the principles of the BLAST algorithm
Biochemistry 324 Bioinformatics. Multiple Sequence Alignment (MSA)
 Biochemistry 324 Bioinformatics Multiple Sequence Alignment (MSA) Big- Οh notation Greek omicron symbol Ο The Big-Oh notation indicates the complexity of an algorithm in terms of execution speed and storage
Biochemistry 324 Bioinformatics Multiple Sequence Alignment (MSA) Big- Οh notation Greek omicron symbol Ο The Big-Oh notation indicates the complexity of an algorithm in terms of execution speed and storage
Salvador Capella-Gutiérrez, Jose M. Silla-Martínez and Toni Gabaldón
 trimal: a tool for automated alignment trimming in large-scale phylogenetics analyses Salvador Capella-Gutiérrez, Jose M. Silla-Martínez and Toni Gabaldón Version 1.2b Index of contents 1. General features
trimal: a tool for automated alignment trimming in large-scale phylogenetics analyses Salvador Capella-Gutiérrez, Jose M. Silla-Martínez and Toni Gabaldón Version 1.2b Index of contents 1. General features
Multiple Sequence Alignment. With thanks to Eric Stone and Steffen Heber, North Carolina State University
 Multiple Sequence Alignment With thanks to Eric Stone and Steffen Heber, North Carolina State University Definition: Multiple sequence alignment Given a set of sequences, a multiple sequence alignment
Multiple Sequence Alignment With thanks to Eric Stone and Steffen Heber, North Carolina State University Definition: Multiple sequence alignment Given a set of sequences, a multiple sequence alignment
Multiple Sequence Alignment II
 Multiple Sequence Alignment II Lectures 20 Dec 5, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall (JHN) 022 1 Outline
Multiple Sequence Alignment II Lectures 20 Dec 5, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall (JHN) 022 1 Outline
Multiple sequence alignment. November 2, 2017
 Multiple sequence alignment November 2, 2017 Why do multiple alignment? Gain insight into evolutionary history Can assess time of divergence by looking at the number of mutations needed to change one sequence
Multiple sequence alignment November 2, 2017 Why do multiple alignment? Gain insight into evolutionary history Can assess time of divergence by looking at the number of mutations needed to change one sequence
CS313 Exercise 4 Cover Page Fall 2017
 CS313 Exercise 4 Cover Page Fall 2017 Due by the start of class on Thursday, October 12, 2017. Name(s): In the TIME column, please estimate the time you spent on the parts of this exercise. Please try
CS313 Exercise 4 Cover Page Fall 2017 Due by the start of class on Thursday, October 12, 2017. Name(s): In the TIME column, please estimate the time you spent on the parts of this exercise. Please try
.. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar..
 .. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar.. PAM and BLOSUM Matrices Prepared by: Jason Banich and Chris Hoover Background As DNA sequences change and evolve, certain amino acids are more
.. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar.. PAM and BLOSUM Matrices Prepared by: Jason Banich and Chris Hoover Background As DNA sequences change and evolve, certain amino acids are more
Genome 559: Introduction to Statistical and Computational Genomics. Lecture15a Multiple Sequence Alignment Larry Ruzzo
 Genome 559: Introduction to Statistical and Computational Genomics Lecture15a Multiple Sequence Alignment Larry Ruzzo 1 Multiple Alignment: Motivations Common structure, function, or origin may be only
Genome 559: Introduction to Statistical and Computational Genomics Lecture15a Multiple Sequence Alignment Larry Ruzzo 1 Multiple Alignment: Motivations Common structure, function, or origin may be only
Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA.
 Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA. Fasta is used to compare a protein or DNA sequence to all of the
Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA. Fasta is used to compare a protein or DNA sequence to all of the
Bioinformatics explained: Smith-Waterman
 Bioinformatics Explained Bioinformatics explained: Smith-Waterman May 1, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com
Bioinformatics Explained Bioinformatics explained: Smith-Waterman May 1, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com
Alignment ABC. Most slides are modified from Serafim s lectures
 Alignment ABC Most slides are modified from Serafim s lectures Complete genomes Evolution Evolution at the DNA level C ACGGTGCAGTCACCA ACGTTGCAGTCCACCA SEQUENCE EDITS REARRANGEMENTS Sequence conservation
Alignment ABC Most slides are modified from Serafim s lectures Complete genomes Evolution Evolution at the DNA level C ACGGTGCAGTCACCA ACGTTGCAGTCCACCA SEQUENCE EDITS REARRANGEMENTS Sequence conservation
Sequence analysis Pairwise sequence alignment
 UMF11 Introduction to bioinformatics, 25 Sequence analysis Pairwise sequence alignment 1. Sequence alignment Lecturer: Marina lexandersson 12 September, 25 here are two types of sequence alignments, global
UMF11 Introduction to bioinformatics, 25 Sequence analysis Pairwise sequence alignment 1. Sequence alignment Lecturer: Marina lexandersson 12 September, 25 here are two types of sequence alignments, global
Heuristic methods for pairwise alignment:
 Bi03c_1 Unit 03c: Heuristic methods for pairwise alignment: k-tuple-methods k-tuple-methods for alignment of pairs of sequences Bi03c_2 dynamic programming is too slow for large databases Use heuristic
Bi03c_1 Unit 03c: Heuristic methods for pairwise alignment: k-tuple-methods k-tuple-methods for alignment of pairs of sequences Bi03c_2 dynamic programming is too slow for large databases Use heuristic
Lecture 4: January 1, Biological Databases and Retrieval Systems
 Algorithms for Molecular Biology Fall Semester, 1998 Lecture 4: January 1, 1999 Lecturer: Irit Orr Scribe: Irit Gat and Tal Kohen 4.1 Biological Databases and Retrieval Systems In recent years, biological
Algorithms for Molecular Biology Fall Semester, 1998 Lecture 4: January 1, 1999 Lecturer: Irit Orr Scribe: Irit Gat and Tal Kohen 4.1 Biological Databases and Retrieval Systems In recent years, biological
In this section we describe how to extend the match refinement to the multiple case and then use T-Coffee to heuristically compute a multiple trace.
 5 Multiple Match Refinement and T-Coffee In this section we describe how to extend the match refinement to the multiple case and then use T-Coffee to heuristically compute a multiple trace. This exposition
5 Multiple Match Refinement and T-Coffee In this section we describe how to extend the match refinement to the multiple case and then use T-Coffee to heuristically compute a multiple trace. This exposition
Pairwise Sequence Alignment: Dynamic Programming Algorithms. COMP Spring 2015 Luay Nakhleh, Rice University
 Pairwise Sequence Alignment: Dynamic Programming Algorithms COMP 571 - Spring 2015 Luay Nakhleh, Rice University DP Algorithms for Pairwise Alignment The number of all possible pairwise alignments (if
Pairwise Sequence Alignment: Dynamic Programming Algorithms COMP 571 - Spring 2015 Luay Nakhleh, Rice University DP Algorithms for Pairwise Alignment The number of all possible pairwise alignments (if
A New Approach For Tree Alignment Based on Local Re-Optimization
 A New Approach For Tree Alignment Based on Local Re-Optimization Feng Yue and Jijun Tang Department of Computer Science and Engineering University of South Carolina Columbia, SC 29063, USA yuef, jtang
A New Approach For Tree Alignment Based on Local Re-Optimization Feng Yue and Jijun Tang Department of Computer Science and Engineering University of South Carolina Columbia, SC 29063, USA yuef, jtang
BLAST, Profile, and PSI-BLAST
 BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
FASTA. Besides that, FASTA package provides SSEARCH, an implementation of the optimal Smith- Waterman algorithm.
 FASTA INTRODUCTION Definition (by David J. Lipman and William R. Pearson in 1985) - Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence
FASTA INTRODUCTION Definition (by David J. Lipman and William R. Pearson in 1985) - Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence
MULTIPLE SEQUENCE ALIGNMENT SOLUTIONS AND APPLICATIONS
 MULTIPLE SEQUENCE ALIGNMENT SOLUTIONS AND APPLICATIONS By XU ZHANG A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE
MULTIPLE SEQUENCE ALIGNMENT SOLUTIONS AND APPLICATIONS By XU ZHANG A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE
Programming assignment for the course Sequence Analysis (2006)
 Programming assignment for the course Sequence Analysis (2006) Original text by John W. Romein, adapted by Bart van Houte (bart@cs.vu.nl) Introduction Please note: This assignment is only obligatory for
Programming assignment for the course Sequence Analysis (2006) Original text by John W. Romein, adapted by Bart van Houte (bart@cs.vu.nl) Introduction Please note: This assignment is only obligatory for
As of August 15, 2008, GenBank contained bases from reported sequences. The search procedure should be
 48 Bioinformatics I, WS 09-10, S. Henz (script by D. Huson) November 26, 2009 4 BLAST and BLAT Outline of the chapter: 1. Heuristics for the pairwise local alignment of two sequences 2. BLAST: search and
48 Bioinformatics I, WS 09-10, S. Henz (script by D. Huson) November 26, 2009 4 BLAST and BLAT Outline of the chapter: 1. Heuristics for the pairwise local alignment of two sequences 2. BLAST: search and
Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods
 Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods Khaddouja Boujenfa, Nadia Essoussi, and Mohamed Limam International Science Index, Computer and Information Engineering waset.org/publication/482
Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods Khaddouja Boujenfa, Nadia Essoussi, and Mohamed Limam International Science Index, Computer and Information Engineering waset.org/publication/482
Global Alignment Scoring Matrices Local Alignment Alignment with Affine Gap Penalties
 Global Alignment Scoring Matrices Local Alignment Alignment with Affine Gap Penalties From LCS to Alignment: Change the Scoring The Longest Common Subsequence (LCS) problem the simplest form of sequence
Global Alignment Scoring Matrices Local Alignment Alignment with Affine Gap Penalties From LCS to Alignment: Change the Scoring The Longest Common Subsequence (LCS) problem the simplest form of sequence
1. R. Durbin, S. Eddy, A. Krogh und G. Mitchison: Biological sequence analysis, Cambridge, 1998
 7 Multiple Sequence Alignment The exposition was prepared by Clemens Gröpl, based on earlier versions by Daniel Huson, Knut Reinert, and Gunnar Klau. It is based on the following sources, which are all
7 Multiple Sequence Alignment The exposition was prepared by Clemens Gröpl, based on earlier versions by Daniel Huson, Knut Reinert, and Gunnar Klau. It is based on the following sources, which are all
C E N T R. Introduction to bioinformatics 2007 E B I O I N F O R M A T I C S V U F O R I N T. Lecture 13 G R A T I V. Iterative homology searching,
 C E N T R E F O R I N T E G R A T I V E B I O I N F O R M A T I C S V U Introduction to bioinformatics 2007 Lecture 13 Iterative homology searching, PSI (Position Specific Iterated) BLAST basic idea use
C E N T R E F O R I N T E G R A T I V E B I O I N F O R M A T I C S V U Introduction to bioinformatics 2007 Lecture 13 Iterative homology searching, PSI (Position Specific Iterated) BLAST basic idea use
1. R. Durbin, S. Eddy, A. Krogh und G. Mitchison: Biological sequence analysis, Cambridge, 1998
 7 Multiple Sequence Alignment The exposition was prepared by Clemens GrÃP pl, based on earlier versions by Daniel Huson, Knut Reinert, and Gunnar Klau. It is based on the following sources, which are all
7 Multiple Sequence Alignment The exposition was prepared by Clemens GrÃP pl, based on earlier versions by Daniel Huson, Knut Reinert, and Gunnar Klau. It is based on the following sources, which are all
Parsimony-Based Approaches to Inferring Phylogenetic Trees
 Parsimony-Based Approaches to Inferring Phylogenetic Trees BMI/CS 576 www.biostat.wisc.edu/bmi576.html Mark Craven craven@biostat.wisc.edu Fall 0 Phylogenetic tree approaches! three general types! distance:
Parsimony-Based Approaches to Inferring Phylogenetic Trees BMI/CS 576 www.biostat.wisc.edu/bmi576.html Mark Craven craven@biostat.wisc.edu Fall 0 Phylogenetic tree approaches! three general types! distance:
Lecture 10: Local Alignments
 Lecture 10: Local Alignments Study Chapter 6.8-6.10 1 Outline Edit Distances Longest Common Subsequence Global Sequence Alignment Scoring Matrices Local Sequence Alignment Alignment with Affine Gap Penalties
Lecture 10: Local Alignments Study Chapter 6.8-6.10 1 Outline Edit Distances Longest Common Subsequence Global Sequence Alignment Scoring Matrices Local Sequence Alignment Alignment with Affine Gap Penalties
JET 2 User Manual 1 INSTALLATION 2 EXECUTION AND FUNCTIONALITIES. 1.1 Download. 1.2 System requirements. 1.3 How to install JET 2
 JET 2 User Manual 1 INSTALLATION 1.1 Download The JET 2 package is available at www.lcqb.upmc.fr/jet2. 1.2 System requirements JET 2 runs on Linux or Mac OS X. The program requires some external tools
JET 2 User Manual 1 INSTALLATION 1.1 Download The JET 2 package is available at www.lcqb.upmc.fr/jet2. 1.2 System requirements JET 2 runs on Linux or Mac OS X. The program requires some external tools
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014
 Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
BLAST - Basic Local Alignment Search Tool
 Lecture for ic Bioinformatics (DD2450) April 11, 2013 Searching 1. Input: Query Sequence 2. Database of sequences 3. Subject Sequence(s) 4. Output: High Segment Pairs (HSPs) Sequence Similarity Measures:
Lecture for ic Bioinformatics (DD2450) April 11, 2013 Searching 1. Input: Query Sequence 2. Database of sequences 3. Subject Sequence(s) 4. Output: High Segment Pairs (HSPs) Sequence Similarity Measures:
Data Mining Technologies for Bioinformatics Sequences
 Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Similarity Searches on Sequence Databases
 Similarity Searches on Sequence Databases Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Zürich, October 2004 Swiss Institute of Bioinformatics Swiss EMBnet node Outline Importance of
Similarity Searches on Sequence Databases Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Zürich, October 2004 Swiss Institute of Bioinformatics Swiss EMBnet node Outline Importance of
Pairwise Sequence Alignment: Dynamic Programming Algorithms COMP 571 Luay Nakhleh, Rice University
 1 Pairwise Sequence Alignment: Dynamic Programming Algorithms COMP 571 Luay Nakhleh, Rice University DP Algorithms for Pairwise Alignment 2 The number of all possible pairwise alignments (if gaps are allowed)
1 Pairwise Sequence Alignment: Dynamic Programming Algorithms COMP 571 Luay Nakhleh, Rice University DP Algorithms for Pairwise Alignment 2 The number of all possible pairwise alignments (if gaps are allowed)
B L A S T! BLAST: Basic local alignment search tool. Copyright notice. February 6, Pairwise alignment: key points. Outline of tonight s lecture
 February 6, 2008 BLAST: Basic local alignment search tool B L A S T! Jonathan Pevsner, Ph.D. Introduction to Bioinformatics pevsner@jhmi.edu 4.633.0 Copyright notice Many of the images in this powerpoint
February 6, 2008 BLAST: Basic local alignment search tool B L A S T! Jonathan Pevsner, Ph.D. Introduction to Bioinformatics pevsner@jhmi.edu 4.633.0 Copyright notice Many of the images in this powerpoint
A multiple alignment tool in 3D
 Outline Department of Computer Science, Bioinformatics Group University of Leipzig TBI Winterseminar Bled, Slovenia February 2005 Outline Outline 1 Multiple Alignments Problems Goal Outline Outline 1 Multiple
Outline Department of Computer Science, Bioinformatics Group University of Leipzig TBI Winterseminar Bled, Slovenia February 2005 Outline Outline 1 Multiple Alignments Problems Goal Outline Outline 1 Multiple
Computational Molecular Biology
 Computational Molecular Biology Erwin M. Bakker Lecture 2 Materials used from R. Shamir [2] and H.J. Hoogeboom [4]. 1 Molecular Biology Sequences DNA A, T, C, G RNA A, U, C, G Protein A, R, D, N, C E,
Computational Molecular Biology Erwin M. Bakker Lecture 2 Materials used from R. Shamir [2] and H.J. Hoogeboom [4]. 1 Molecular Biology Sequences DNA A, T, C, G RNA A, U, C, G Protein A, R, D, N, C E,
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 04: Variations of sequence alignments http://www.pitt.edu/~mcs2/teaching/biocomp/tutorials/global.html Slides adapted from Dr. Shaojie Zhang (University
EECS730: Introduction to Bioinformatics Lecture 04: Variations of sequence alignments http://www.pitt.edu/~mcs2/teaching/biocomp/tutorials/global.html Slides adapted from Dr. Shaojie Zhang (University
Chapter 6. Multiple sequence alignment (week 10)
 Course organization Introduction ( Week 1,2) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 3)» Algorithm complexity analysis
Course organization Introduction ( Week 1,2) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 3)» Algorithm complexity analysis
Cost Partitioning Techniques for Multiple Sequence Alignment. Mirko Riesterer,
 Cost Partitioning Techniques for Multiple Sequence Alignment Mirko Riesterer, 10.09.18 Agenda. 1 Introduction 2 Formal Definition 3 Solving MSA 4 Combining Multiple Pattern Databases 5 Cost Partitioning
Cost Partitioning Techniques for Multiple Sequence Alignment Mirko Riesterer, 10.09.18 Agenda. 1 Introduction 2 Formal Definition 3 Solving MSA 4 Combining Multiple Pattern Databases 5 Cost Partitioning
MULTIPLE SEQUENCE ALIGNMENT
 MULTIPLE SEQUENCE ALIGNMENT Multiple Alignment versus Pairwise Alignment Up until now we have only tried to align two sequences. What about more than two? A faint similarity between two sequences becomes
MULTIPLE SEQUENCE ALIGNMENT Multiple Alignment versus Pairwise Alignment Up until now we have only tried to align two sequences. What about more than two? A faint similarity between two sequences becomes
Sequence alignment algorithms
 Sequence alignment algorithms Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, February 23 rd 27 After this lecture, you can decide when to use local and global sequence alignments
Sequence alignment algorithms Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, February 23 rd 27 After this lecture, you can decide when to use local and global sequence alignments
- G T G T A C A C
 Name Student ID.. Sequence alignment 1. Globally align sequence V (GTGTACAC) and sequence W (GTACC) by hand using dynamic programming algorithm. The alignment will be performed based on match premium of
Name Student ID.. Sequence alignment 1. Globally align sequence V (GTGTACAC) and sequence W (GTACC) by hand using dynamic programming algorithm. The alignment will be performed based on match premium of
Distributed Protein Sequence Alignment
 Distributed Protein Sequence Alignment ABSTRACT J. Michael Meehan meehan@wwu.edu James Hearne hearne@wwu.edu Given the explosive growth of biological sequence databases and the computational complexity
Distributed Protein Sequence Alignment ABSTRACT J. Michael Meehan meehan@wwu.edu James Hearne hearne@wwu.edu Given the explosive growth of biological sequence databases and the computational complexity
Biologically significant sequence alignments using Boltzmann probabilities
 Biologically significant sequence alignments using Boltzmann probabilities P Clote Department of Biology, Boston College Gasson Hall 16, Chestnut Hill MA 0267 clote@bcedu Abstract In this paper, we give
Biologically significant sequence alignments using Boltzmann probabilities P Clote Department of Biology, Boston College Gasson Hall 16, Chestnut Hill MA 0267 clote@bcedu Abstract In this paper, we give
Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD
 Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD Assumptions: Biological sequences evolved by evolution. Micro scale changes: For short sequences (e.g. one
Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD Assumptions: Biological sequences evolved by evolution. Micro scale changes: For short sequences (e.g. one
Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it.
 Sequence Alignments Overview Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it. Sequence alignment means arranging
Sequence Alignments Overview Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it. Sequence alignment means arranging
Multiple Sequence Alignment Gene Finding, Conserved Elements
 Multiple Sequence Alignment Gene Finding, Conserved Elements Definition Given N sequences x 1, x 2,, x N : Insert gaps (-) in each sequence x i, such that All sequences have the same length L Score of
Multiple Sequence Alignment Gene Finding, Conserved Elements Definition Given N sequences x 1, x 2,, x N : Insert gaps (-) in each sequence x i, such that All sequences have the same length L Score of
Brief review from last class
 Sequence Alignment Brief review from last class DNA is has direction, we will use only one (5 -> 3 ) and generate the opposite strand as needed. DNA is a 3D object (see lecture 1) but we will model it
Sequence Alignment Brief review from last class DNA is has direction, we will use only one (5 -> 3 ) and generate the opposite strand as needed. DNA is a 3D object (see lecture 1) but we will model it
Bioinformatics explained: BLAST. March 8, 2007
 Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Research Article Aligning Sequences by Minimum Description Length
 Hindawi Publishing Corporation EURASIP Journal on Bioinformatics and Systems Biology Volume 2007, Article ID 72936, 14 pages doi:10.1155/2007/72936 Research Article Aligning Sequences by Minimum Description
Hindawi Publishing Corporation EURASIP Journal on Bioinformatics and Systems Biology Volume 2007, Article ID 72936, 14 pages doi:10.1155/2007/72936 Research Article Aligning Sequences by Minimum Description
Sequence comparison: Local alignment
 Sequence comparison: Local alignment Genome 559: Introuction to Statistical an Computational Genomics Prof. James H. Thomas http://faculty.washington.eu/jht/gs559_217/ Review global alignment en traceback
Sequence comparison: Local alignment Genome 559: Introuction to Statistical an Computational Genomics Prof. James H. Thomas http://faculty.washington.eu/jht/gs559_217/ Review global alignment en traceback
Alignment of Pairs of Sequences
 Bi03a_1 Unit 03a: Alignment of Pairs of Sequences Partners for alignment Bi03a_2 Protein 1 Protein 2 =amino-acid sequences (20 letter alphabeth + gap) LGPSSKQTGKGS-SRIWDN LN-ITKSAGKGAIMRLGDA -------TGKG--------
Bi03a_1 Unit 03a: Alignment of Pairs of Sequences Partners for alignment Bi03a_2 Protein 1 Protein 2 =amino-acid sequences (20 letter alphabeth + gap) LGPSSKQTGKGS-SRIWDN LN-ITKSAGKGAIMRLGDA -------TGKG--------
Chapter 8 Multiple sequence alignment. Chaochun Wei Spring 2018
 1896 1920 1987 2006 Chapter 8 Multiple sequence alignment Chaochun Wei Spring 2018 Contents 1. Reading materials 2. Multiple sequence alignment basic algorithms and tools how to improve multiple alignment
1896 1920 1987 2006 Chapter 8 Multiple sequence alignment Chaochun Wei Spring 2018 Contents 1. Reading materials 2. Multiple sequence alignment basic algorithms and tools how to improve multiple alignment
Approaches to Efficient Multiple Sequence Alignment and Protein Search
 Approaches to Efficient Multiple Sequence Alignment and Protein Search Thesis statements of the PhD dissertation Adrienn Szabó Supervisor: István Miklós Eötvös Loránd University Faculty of Informatics
Approaches to Efficient Multiple Sequence Alignment and Protein Search Thesis statements of the PhD dissertation Adrienn Szabó Supervisor: István Miklós Eötvös Loránd University Faculty of Informatics
CS273: Algorithms for Structure Handout # 4 and Motion in Biology Stanford University Thursday, 8 April 2004
 CS273: Algorithms for Structure Handout # 4 and Motion in Biology Stanford University Thursday, 8 April 2004 Lecture #4: 8 April 2004 Topics: Sequence Similarity Scribe: Sonil Mukherjee 1 Introduction
CS273: Algorithms for Structure Handout # 4 and Motion in Biology Stanford University Thursday, 8 April 2004 Lecture #4: 8 April 2004 Topics: Sequence Similarity Scribe: Sonil Mukherjee 1 Introduction
Lab 4: Multiple Sequence Alignment (MSA)
 Lab 4: Multiple Sequence Alignment (MSA) The objective of this lab is to become familiar with the features of several multiple alignment and visualization tools, including the data input and output, basic
Lab 4: Multiple Sequence Alignment (MSA) The objective of this lab is to become familiar with the features of several multiple alignment and visualization tools, including the data input and output, basic
PyMod Documentation (Version 2.1, September 2011)
 PyMod User s Guide PyMod Documentation (Version 2.1, September 2011) http://schubert.bio.uniroma1.it/pymod/ Emanuele Bramucci & Alessandro Paiardini, Francesco Bossa, Stefano Pascarella, Department of
PyMod User s Guide PyMod Documentation (Version 2.1, September 2011) http://schubert.bio.uniroma1.it/pymod/ Emanuele Bramucci & Alessandro Paiardini, Francesco Bossa, Stefano Pascarella, Department of
Lecture 10. Sequence alignments
 Lecture 10 Sequence alignments Alignment algorithms: Overview Given a scoring system, we need to have an algorithm for finding an optimal alignment for a pair of sequences. We want to maximize the score
Lecture 10 Sequence alignments Alignment algorithms: Overview Given a scoring system, we need to have an algorithm for finding an optimal alignment for a pair of sequences. We want to maximize the score
Sequence clustering. Introduction. Clustering basics. Hierarchical clustering
 Sequence clustering Introduction Data clustering is one of the key tools used in various incarnations of data-mining - trying to make sense of large datasets. It is, thus, natural to ask whether clustering
Sequence clustering Introduction Data clustering is one of the key tools used in various incarnations of data-mining - trying to make sense of large datasets. It is, thus, natural to ask whether clustering
Database Searching Using BLAST
 Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
LAGAN and Multi-LAGAN: Efficient Tools for Large-Scale Multiple Alignment of Genomic DNA
 LAGAN and Multi-LAGAN: Efficient Tools for Large-Scale Multiple Alignment of Genomic DNA Michael Brudno, Chuong B. Do, Gregory M. Cooper, et al. Presented by Xuebei Yang About Alignments Pairwise Alignments
LAGAN and Multi-LAGAN: Efficient Tools for Large-Scale Multiple Alignment of Genomic DNA Michael Brudno, Chuong B. Do, Gregory M. Cooper, et al. Presented by Xuebei Yang About Alignments Pairwise Alignments
Dynamic Programming in 3-D Progressive Alignment Profile Progressive Alignment (ClustalW) Scoring Multiple Alignments Entropy Sum of Pairs Alignment
 Dynamic Programming in 3-D Progressive Alignment Profile Progressive Alignment (ClustalW) Scoring Multiple Alignments Entropy Sum of Pairs Alignment Partial Order Alignment (POA) A-Bruijin (ABA) Approach
Dynamic Programming in 3-D Progressive Alignment Profile Progressive Alignment (ClustalW) Scoring Multiple Alignments Entropy Sum of Pairs Alignment Partial Order Alignment (POA) A-Bruijin (ABA) Approach
Sequence Comparison: Dynamic Programming. Genome 373 Genomic Informatics Elhanan Borenstein
 Sequence omparison: Dynamic Programming Genome 373 Genomic Informatics Elhanan Borenstein quick review: hallenges Find the best global alignment of two sequences Find the best global alignment of multiple
Sequence omparison: Dynamic Programming Genome 373 Genomic Informatics Elhanan Borenstein quick review: hallenges Find the best global alignment of two sequences Find the best global alignment of multiple
Notes on Dynamic-Programming Sequence Alignment
 Notes on Dynamic-Programming Sequence Alignment Introduction. Following its introduction by Needleman and Wunsch (1970), dynamic programming has become the method of choice for rigorous alignment of DNA
Notes on Dynamic-Programming Sequence Alignment Introduction. Following its introduction by Needleman and Wunsch (1970), dynamic programming has become the method of choice for rigorous alignment of DNA
Scoring and heuristic methods for sequence alignment CG 17
 Scoring and heuristic methods for sequence alignment CG 17 Amino Acid Substitution Matrices Used to score alignments. Reflect evolution of sequences. Unitary Matrix: M ij = 1 i=j { 0 o/w Genetic Code Matrix:
Scoring and heuristic methods for sequence alignment CG 17 Amino Acid Substitution Matrices Used to score alignments. Reflect evolution of sequences. Unitary Matrix: M ij = 1 i=j { 0 o/w Genetic Code Matrix:
DNA Alignment With Affine Gap Penalties
 DNA Alignment With Affine Gap Penalties Laurel Schuster Why Use Affine Gap Penalties? When aligning two DNA sequences, one goal may be to infer the mutations that made them different. Though it s impossible
DNA Alignment With Affine Gap Penalties Laurel Schuster Why Use Affine Gap Penalties? When aligning two DNA sequences, one goal may be to infer the mutations that made them different. Though it s impossible
Sequence Alignment AGGCTATCACCTGACCTCCAGGCCGATGCCC TAGCTATCACGACCGCGGTCGATTTGCCCGAC -AGGCTATCACCTGACCTCCAGGCCGA--TGCCC--
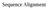 Sequence Alignment Sequence Alignment AGGCTATCACCTGACCTCCAGGCCGATGCCC TAGCTATCACGACCGCGGTCGATTTGCCCGAC -AGGCTATCACCTGACCTCCAGGCCGA--TGCCC-- TAG-CTATCAC--GACCGC--GGTCGATTTGCCCGAC Distance from sequences
Sequence Alignment Sequence Alignment AGGCTATCACCTGACCTCCAGGCCGATGCCC TAGCTATCACGACCGCGGTCGATTTGCCCGAC -AGGCTATCACCTGACCTCCAGGCCGA--TGCCC-- TAG-CTATCAC--GACCGC--GGTCGATTTGCCCGAC Distance from sequences
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, This lecture is based on the following papers, which are all recommended reading:
 24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
