Supplementary Figure 1
|
|
|
- Charles McBride
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 Experimental unmodified 2D and astigmatic 3D PSFs. (a) The averaged experimental unmodified 2D PSF used in this study. PSFs at axial positions from -800 nm to 800 nm are shown. The x-z reconstruction represents a vertical cross-section along the line depicted in the image corresponding to the axial position 0 nm. (b) Same as (a), but for the averaged experimental astigmatic 3D PSF. Scale bars 1 µm.
2 Supplementary Figure 2 The localization precision of the fitter achieves minimum uncertainty with the cspline interpolated experimental astigmatic PSF. Single molecule images were simulated using an experimental astigmatic PSF model with 5000 photons/localization (panel a) or 500 photons/localization (panel b) and 10 background photons/pixel and fitted with the interpolated cubic spline model. We then evaluated the x, y and z localization precisions at different axial positions as the standard deviation of the error between fitted 3D positions and ground truth of the simulated molecules at each axial position (symbols). Additionally, we calculated the root-mean-square error between fitted positions and ground truth (dots). The localization precision as well as the localization accuracy of the fitted positions achieves the estimated Cramér Rao lower bound (CRLB) (denoted by lines) in all 3 dimensions. Only for dimmer fluorophores and large defocus, a small fraction (<5%) of fluorophores that converged to a z position close to the starting z position (focus), result in a deteriorated average localization accuracy. The CRLB is evaluated as the diagonal element of the inverse of the Fisher information matrix with the fitted parameters and cspline interpolated experimental PSF model.
3 Supplementary Figure 3 Experimental localization precision compared with the Cramér Rao lower bound (CRLB), measured by repeated imaging of a single bead. For each z-position, spaced 50 nm apart, 50 images were acquired. The upper panel shows the fitted z-position vs. the objective z position and the lower panel compares the experimental localization precision (standard deviation of the fitted z position) with the CRLB estimated by the software. (a): The EM gain was switched off, median photons per localization were (b): The EM gain was switched off, median photons per localization were For fitting of a, b, the CCD readout noise was taken into account by fitting the data with the scmos noise model, using a pixel-independent noise calibrated from a stack of dark images (21.7 photoelectrons). (c): An EM gain of 200 was used, median photons per localization were The beads were fitted with a noise model that approximates the EM excess noise by Poisson noise 1. To this end, we divided the photon numbers prior to fitting by the noise excess factor, which approaches 2 for large EM gain. This approximation seems to overestimate the noise; thus, the experimental uncertainty is slightly below the CRLB. 1. Fullerton, S., Bennett, K., Toda, E. & Takahashi, T. Optimization of precision localization microscopy using CMOS camera technology. Proc of SPIE 8228: 82280T 1 (2012).
4 Supplementary Figure 4 Experimental profiles and localization precisions for the data used in this paper. The insets denote the regions where the profiles were taken. Dotted lines: Fit with two Gaussians with a distance d and a standard deviation s in nm. Scale bars 100 nm.
5 Supplementary Figure 5 Comparison of our fitter to previous work, using the example of immunolabeled microtubules. (a) Front view reconstructions of immunolabeled microtubules taken from the literature 2 4. The hollow structure of the microtubules can only be resolved in the interferometric 4-Pi methods. (b) Front view reconstructions of microtubules presented in this work. We are able to resolve hollow, cylinder-like structure of microtubules in both DNA-PAINT and dstorm imaging. (c) Sketch of a microtubule labeled with antibodies to illustrate its dimensions. Scale bars 100 nm. (a, top) Adapted with permission from Ref 2, Springer Customer Service Centre GmbH: Springer Nature, Nature Photonics, copyright (a, middle and bottom) Adapted with permission from Refs 3 and 4, respectively, under creative commons license CC BY 4.0 ( cropped from original and added scale bar. 2. Verdeny-Vilanova, I. et al. 3D motion of vesicles along microtubules helps them to circumvent obstacles in cells. J. Cell Sci. 130, (2017). 3. Bourg, N. et al. Direct optical nanoscopy with axially localized detection. Nat Phot. 9, (2015). 4. Huang, F. et al. Ultra-High Resolution 3D Imaging of Whole Cells. Cell 166, (2016).
6 Supplementary Figure 6 3D astigmatic dstorm image of microtubules. Microtubules in U-2 OS cells, labeled with anti-alpha tubulin primary and Alexa Fluor 647-coupled secondary antibodies, were imaged using dstorm with a cylindrical lens. Upper panel: x-y top view. Lines denote regions for which 250 nm x-z (side view) cross-sections are shown in the lower panels. Corresponding localization precisions and profiles can be found in Supplementary Figure 4. Scale bars: 1 µm (top view), 100 nm (side views).
7 Supplementary Figure 7 Convergence and robustness of Levenberg Marquardt versus the Newton iterative method. (a), (b): Levenberg-Marquardt converges faster than Newton. We compared for the L-M and Newton method the change of the MLE cost function at each iteration. is defined as in ref. 5. The iteration stops when the relative change of decreases by less than The number of iterations is 35±4 for the Newton method and 12±1 for the L-M method, which indicates the L-M method is more efficient than the custom Newton solver. (a) Update of during each iteration. (b) Relative change of during each iteration. Newton iterative scheme as in Smith et al 6. (c): The Levenberg-Marquardt algorithm is more robust with respect to starting parameters than the Newton method. The localization precision is plotted in dependence on the photons/localization. Both algorithms can achieve the CRLB for molecules with more than 100 photons when the centroid of the fitting window is used as a start parameter. We then purposely offset the starting parameter for x, y by 2 pixels. Whereas the final localization precision for the L-M algorithm is robust with respect to the start parameters, the Newton method often fails to converge for a wrong starting parameter estimate. Simulation parameters: Elliptical Gaussian PSF model with a sigma 1.5 and 2.5 pixels in x and y, respectively. 5 background photons per pixel. 500 photons per localization was used for (a) and (b). Fitting parameters: ROI size pixels, 1000 molecules per data point, maximum number of iterations was Laurence, T. A. & Chromy, B. A. Efficient maximum likelihood estimator fitting of histograms. Nat. Methods 7, (2010). 6. Smith, C. S., Joseph, N., Rieger, B. & Lidke, K. a. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods 7, (2010).
8 Supplementary Figure 8 Localization precision and accuracy for a Gaussian versus an experimental PSF model. Single molecule images were simulated using an experimental PSF model with 5000 photons/localization and 10 background photons/pixel. For the cspline 3D and elliptical Gaussian fit, an experimental astigmatic PSF was used for the simulation. For the cspline 2D fit, an unmodified experimental PSF was used. We then evaluated both localization precision and accuracy in 3D at different axial positions from 1000 simulated molecules at each axial position. The localization precision is calculated as the standard deviation of the difference between the fitted positions and ground truth positions, and the localization accuracy is calculated as the root mean square error between the fitted positions and ground truth positions. (a) Localization accuracy and (b) localization precision in z for different models at different axial positions. For the elliptical Gaussian PSF model, two algorithms were used for z localization as described in Online Methods. (c, d) are the same as (a, b) but in lateral direction.
9 Supplementary Figure 9 Comparison of the experimental PSF fit with a Gaussian fit for astigmatic data on the nuclear complex. (a) Nup107-SNAP-BG-AF647 in U-2 OS cells, imaged using dstorm with a cylindrical lens (top view). (b) 500 nm side view cross-section of the region denoted in (a) using the experimental PSF fit. (c) Side view reconstruction of the region denoted in (a) using the Gaussian z fit 6. Scale bars 1 µm. 6. Smith, C. S., Joseph, N., Rieger, B. & Lidke, K. a. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods 7, (2010).
10 Supplementary Figure 10 Photon count underestimation by Gaussian fit of experimental PSFs. Single molecule images were simulated using an experimental astigmatic PSF model with 5000 photons/localization and 10 background photons/pixel. The simulated single molecule images were fitted with a cspline interpolated PSF model (a) and an elliptical Gaussian model (b). Accuracy (root mean square error) and precision (standard deviation) of fitted photons/localization at different axial positions for (a) cspline fit and (b) elliptical Gaussian fit. Though the returned photon precision of the Gaussian fit is close to the CRLB, the returned accuracy is much larger than the precision, which indicates a strong systematic error in estimating the photons using the Gaussian PSF model. In contrast, for the cspline interpolated PSF model the accuracy, precision and CRLB of the photons/localization agree well with each other.
11 Supplementary Figure 11 A noisy PSF model leads to stripe artifacts. (a) Overview image of microtubules in a U-2 OS cell. The axial positions were color coded according to the color scale bar. (b) y-z plot of the boxed region in (a) when the PSF was modeled using only a single bead without smoothing. (c) y-z plot of the boxed region in (a) when PSF was robustly modeled with many beads across different fields of view and with regularization, as described in Online Methods. (d) z-profiles of (b) and (c). The stripe artifact is avoided using the proper averaging and regularization. Scale bars: (a) 1 µm, (b), (c) 100 nm.
12 Supplementary Figure 12 Computational speed of different fitting routines. (a) Comparison of the speed of the fitter presented in this work and previously published fitters. A pixel window size was used for benchmarking. The fitting speeds of Babcock s code and Kirshner s code were measured using the example data provided in the software package by the authors. The fitting speed of ThunderSTORM was estimated using 1,000 frames of dstorm data from Supplementary Figure 6. We measured the overall time difference between an analysis with a stringent detection cutoff and one with a lower threshold. We then divided the difference in number of fitted localization by the time difference to estimate the number of fits per second. (b) Comparison of the speed of the Newton method and the L-M algorithm using an elliptical Gaussian PSF model. The effect of the readout noise variance for scmos camera data was also evaluated. Even though the L- M algorithm has higher complexity as it evaluates every iteration to determine the damping factor, it is still slightly faster than Newton method due to the fact that less iterations are needed. (c) Comparison of the speed of astigmatic Gauss z fit and cspline z fit. The GPU code is overall more than 100 times faster than the CPU code running on a single thread. It is interesting to notice that the spline fit is even faster than the Gauss z fit in the CPU code while it is slower than the Gauss z fit in GPU code. This is probably because the spline coefficients are accessed much more frequently in the GPU than the CPU, which slows down the performance.
13 Supplementary Figure 13 The scmos noise model avoids readout-noise-induced bias. Single-molecule images were simulated considering pixel-dependent readout noise, as is present in scmos cameras. The readout noise of the overall pixels is set as a Gaussian distribution with a mean of 1.4 e - and a sigma with 0.2 e -. (a) The map of the readout noise. The readout noise of the pixel located 2 pixels to the up of the center is set as 30 e -. (b) Simulated single molecule with the readout noise as shown in (a). Each molecule was simulated as an experimental PSF, 200 photons/localization and 2 background photons/pixel. The positions of the molecules were all placed in the center (6, 6) of the window. (c) Scatter plot of the 1000 fitted positions of the molecules as shown in (b) fitted with the EMCCD noise model. (d) Scatter plot of the 1000 fitted positions of the molecules as shown in (b) fitted with the scmos noise model.
14 Supplementary Figure 14 The cspline fitter works on reference data, as well as with commercial microscopes, advanced PSF engineering approaches, and highly aberrated PSFs. (a), (b) Fit of the 2016 SMLM Challenge ( training data set with our fitter for experimental PSFs. a) Bi-directional fitting of the 2D training dataset (MT0.N1.LD-2D). The panels show topview and side-view reconstructions and slices through the PSF used for fitting. (b) Fitting of the Double-Helix PSF training dataset (MT0.N1.LD-DH). Panels as for (a). Scale bars: 1 µm. (c)-(h) Spline interpolated PSF models were derived from bead stacks and used to fit 6 individual beads from a different stack. (c) Zeiss Elyra, using a phase-ramp PSF 7. (d) Leica SR GSD 3D with astigmatic PSF. (e) Double-Helix PSF from the SMLM challenge (MT0.N1.LD-DH). (f)-(h) Experimental aberrated PSFs
15 generated with a deformable mirror on a custom microscope. 7. Baddeley, D., Cannell, M. B. & Soeller, C. Three-dimensional sub-100 nm super-resolution imaging of biological samples using a phase ramp in the objective pupil. Nano Res. 4, (2011).
16 Supplementary Figure 15 The cspline fitter can improve the resolution on commercial microscopes. Nup107-SNAP-BG-AlexaFluor647 in U-2 OS cells, imaged using dstorm on a commercial Leica SR GSD 3D microscope. (a) overview, (b) side-view reconstruction using the cspline fitter with an experimentally derived PSF model, (c) as (b), but using the coordinates fitted with the Leica LAS X software.
Faster STORM using compressed sensing. Supplementary Software. Nature Methods. Nature Methods: doi: /nmeth.1978
 Nature Methods Faster STORM using compressed sensing Lei Zhu, Wei Zhang, Daniel Elnatan & Bo Huang Supplementary File Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3 Supplementary
Nature Methods Faster STORM using compressed sensing Lei Zhu, Wei Zhang, Daniel Elnatan & Bo Huang Supplementary File Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3 Supplementary
Live-cell 3D super-resolution imaging in thick biological samples
 Nature Methods Live-cell 3D super-resolution imaging in thick biological samples Francesca Cella Zanacchi, Zeno Lavagnino, Michela Perrone Donnorso, Alessio Del Bue, Lauria Furia, Mario Faretta & Alberto
Nature Methods Live-cell 3D super-resolution imaging in thick biological samples Francesca Cella Zanacchi, Zeno Lavagnino, Michela Perrone Donnorso, Alessio Del Bue, Lauria Furia, Mario Faretta & Alberto
Vutara 350. Innovation with Integrity. The Fastest, Super-Resolution Microscope Deep 3D Imaging on Live Cells, Quickly and Easily
 Vutara 350 The Fastest, Super-Resolution Microscope Deep 3D Imaging on Live Cells, Quickly and Easily Innovation with Integrity Fluorescence Microscopy Vutara 350 Don t Get Left Behind Bruker s Vutara
Vutara 350 The Fastest, Super-Resolution Microscope Deep 3D Imaging on Live Cells, Quickly and Easily Innovation with Integrity Fluorescence Microscopy Vutara 350 Don t Get Left Behind Bruker s Vutara
Supporting Information for Azimuthal Polarization Filtering for Accurate, Precise, and Robust Single-Molecule Localization Microscopy
 Nano Letters Supporting Information for Azimuthal Polarization Filtering for Accurate, Precise, and Robust Single-Molecule Localization Microscopy Matthew D. Lew, and W. E. Moerner *, Departments of Chemistry
Nano Letters Supporting Information for Azimuthal Polarization Filtering for Accurate, Precise, and Robust Single-Molecule Localization Microscopy Matthew D. Lew, and W. E. Moerner *, Departments of Chemistry
1. ABOUT INSTALLATION COMPATIBILITY SURESIM WORKFLOWS a. Workflow b. Workflow SURESIM TUTORIAL...
 SuReSim manual 1. ABOUT... 2 2. INSTALLATION... 2 3. COMPATIBILITY... 2 4. SURESIM WORKFLOWS... 2 a. Workflow 1... 3 b. Workflow 2... 4 5. SURESIM TUTORIAL... 5 a. Import Data... 5 b. Parameter Selection...
SuReSim manual 1. ABOUT... 2 2. INSTALLATION... 2 3. COMPATIBILITY... 2 4. SURESIM WORKFLOWS... 2 a. Workflow 1... 3 b. Workflow 2... 4 5. SURESIM TUTORIAL... 5 a. Import Data... 5 b. Parameter Selection...
Gpufit: An open-source toolkit for GPU-accelerated curve fitting
 Gpufit: An open-source toolkit for GPU-accelerated curve fitting Adrian Przybylski, Björn Thiel, Jan Keller-Findeisen, Bernd Stock, and Mark Bates Supplementary Information Table of Contents Calculating
Gpufit: An open-source toolkit for GPU-accelerated curve fitting Adrian Przybylski, Björn Thiel, Jan Keller-Findeisen, Bernd Stock, and Mark Bates Supplementary Information Table of Contents Calculating
Effect of OTF attenuation on single-slice SR-SIM reconstruction
 Effect of OTF attenuation on single-slice SR-SIM reconstruction Supplementary Figure 1: Actin filaments in U2OS cells, labelled with Phalloidin-Atto488, measured on a DeltaVision OMX, excited at 488 nm
Effect of OTF attenuation on single-slice SR-SIM reconstruction Supplementary Figure 1: Actin filaments in U2OS cells, labelled with Phalloidin-Atto488, measured on a DeltaVision OMX, excited at 488 nm
BSI scmos. High Quantum Efficiency. Cooled Scientific CMOS Camera
 95 BSI scmos High Quantum Efficiency Cooled Scientific CMOS Camera Backside-illuminated scmos technology Opening a new era of high sensitivity imaging applications! The Dhyana 95 is a highly sensitive
95 BSI scmos High Quantum Efficiency Cooled Scientific CMOS Camera Backside-illuminated scmos technology Opening a new era of high sensitivity imaging applications! The Dhyana 95 is a highly sensitive
Single Molecule Light Microscopy ImageJ Plugins
 Single Molecule Light Microscopy ImageJ Plugins Alex Herbert MRC Genome Damage and Stability Centre School of Life Sciences University of Sussex Science Road Falmer BN1 9RQ a.herbert@sussex.ac.uk Page
Single Molecule Light Microscopy ImageJ Plugins Alex Herbert MRC Genome Damage and Stability Centre School of Life Sciences University of Sussex Science Road Falmer BN1 9RQ a.herbert@sussex.ac.uk Page
COLOCALISATION. Alexia Ferrand. Imaging Core Facility Biozentrum Basel
 COLOCALISATION Alexia Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to analyse
COLOCALISATION Alexia Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to analyse
MetaMorph Super-Resolution Software. Localization-based super-resolution microscopy
 MetaMorph Super-Resolution Software Localization-based super-resolution microscopy KEY BENEFITS Lateral Resolution up to 20 nm & Axial Resolution up to 40nm Real-time and offline superresolution image
MetaMorph Super-Resolution Software Localization-based super-resolution microscopy KEY BENEFITS Lateral Resolution up to 20 nm & Axial Resolution up to 40nm Real-time and offline superresolution image
SUPPLEMENTARY INFORMATION
 Multicolour localization microscopy by point-spread-function engineering Yoav Shechtman 1, Lucien E. Weiss 1, Adam S. Backer 1,2, Maurice Y. Lee 1,3, and W.E. Moerner 1 1 Department of Chemistry, Stanford
Multicolour localization microscopy by point-spread-function engineering Yoav Shechtman 1, Lucien E. Weiss 1, Adam S. Backer 1,2, Maurice Y. Lee 1,3, and W.E. Moerner 1 1 Department of Chemistry, Stanford
Transmitted light Illuminator VIS-LED, color temperature. Epifluorescence lamp module X-Cite 120PC Q (Excelitas
 ELYRA PS.1 Multi-functional fluorescence inverted widefield microscope enabling live-cell imaging, TIRF or HILO illuminion and two super-resolution techniques: structured illuminion microscopy (SIM) and
ELYRA PS.1 Multi-functional fluorescence inverted widefield microscope enabling live-cell imaging, TIRF or HILO illuminion and two super-resolution techniques: structured illuminion microscopy (SIM) and
Automatic Partiicle Tracking Software USE ER MANUAL Update: May 2015
 Automatic Particle Tracking Software USER MANUAL Update: May 2015 File Menu The micrograph below shows the panel displayed when a movie is opened, including a playback menu where most of the parameters
Automatic Particle Tracking Software USER MANUAL Update: May 2015 File Menu The micrograph below shows the panel displayed when a movie is opened, including a playback menu where most of the parameters
Chao, J., Ram, S., Ward, E. S., and Ober, R. J. Investigating the usage of point spread functions in point source and microsphere localization.
 Chao, J., Ram, S., Ward, E. S., and Ober, R. J. Investigating the usage of point spread functions in point source and microsphere localization. Proceedings of the SPIE, Three-Dimensional and Multidimensional
Chao, J., Ram, S., Ward, E. S., and Ober, R. J. Investigating the usage of point spread functions in point source and microsphere localization. Proceedings of the SPIE, Three-Dimensional and Multidimensional
Swammerdam Institute for Life Sciences (Universiteit van Amsterdam), 1098 XH Amsterdam, The Netherland
 Sparse deconvolution of high-density super-resolution images (SPIDER) Siewert Hugelier 1, Johan J. de Rooi 2,4, Romain Bernex 1, Sam Duwé 3, Olivier Devos 1, Michel Sliwa 1, Peter Dedecker 3, Paul H. C.
Sparse deconvolution of high-density super-resolution images (SPIDER) Siewert Hugelier 1, Johan J. de Rooi 2,4, Romain Bernex 1, Sam Duwé 3, Olivier Devos 1, Michel Sliwa 1, Peter Dedecker 3, Paul H. C.
CS 395T Lecture 12: Feature Matching and Bundle Adjustment. Qixing Huang October 10 st 2018
 CS 395T Lecture 12: Feature Matching and Bundle Adjustment Qixing Huang October 10 st 2018 Lecture Overview Dense Feature Correspondences Bundle Adjustment in Structure-from-Motion Image Matching Algorithm
CS 395T Lecture 12: Feature Matching and Bundle Adjustment Qixing Huang October 10 st 2018 Lecture Overview Dense Feature Correspondences Bundle Adjustment in Structure-from-Motion Image Matching Algorithm
COLOCALISATION. Alexia Loynton-Ferrand. Imaging Core Facility Biozentrum Basel
 COLOCALISATION Alexia Loynton-Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to
COLOCALISATION Alexia Loynton-Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to
Virtual Frap User Guide
 Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Supporting Information. Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking
 Supporting Information Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking Daan van der Zwaag 1,2, Nane Vanparijs 3, Sjors Wijnands 1,4, Riet De Rycke 5, Bruno G. De Geest 2* and
Supporting Information Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking Daan van der Zwaag 1,2, Nane Vanparijs 3, Sjors Wijnands 1,4, Riet De Rycke 5, Bruno G. De Geest 2* and
Super-Resolution on Moving Objects and Background
 Super-Resolution on Moving Objects and Background A. van Eekeren K. Schutte J. Dijk D.J.J. de Lange L.J. van Vliet TNO Defence, Security and Safety, P.O. Box 96864, 2509 JG, The Hague, The Netherlands
Super-Resolution on Moving Objects and Background A. van Eekeren K. Schutte J. Dijk D.J.J. de Lange L.J. van Vliet TNO Defence, Security and Safety, P.O. Box 96864, 2509 JG, The Hague, The Netherlands
Back Illuminated Scientific CMOS
 Prime 95B Scientific CMOS Camera Datasheet CMOS, EMCCD AND CCD CAMERAS FOR LIFE SCIENCES Back Illuminated Scientific CMOS Discovery depends on every photon Primary applications: Super-Resolution Microscopy
Prime 95B Scientific CMOS Camera Datasheet CMOS, EMCCD AND CCD CAMERAS FOR LIFE SCIENCES Back Illuminated Scientific CMOS Discovery depends on every photon Primary applications: Super-Resolution Microscopy
Single Particle Tracking Analyses of Hyperspectral Fluorescent Images. FACSS October 20 th, 2010 Pat Cutler, University of New Mexico
 Single Particle Tracking Analyses of Hyperspectral Fluorescent Images FACSS October 20 th, 2010 Pat Cutler, University of New Mexico Labeling proteins Biomolecule (SA) Cell with QDs QD-IgE Polymer Coating
Single Particle Tracking Analyses of Hyperspectral Fluorescent Images FACSS October 20 th, 2010 Pat Cutler, University of New Mexico Labeling proteins Biomolecule (SA) Cell with QDs QD-IgE Polymer Coating
Localization Accuracy in Single-Molecule Microscopy
 Biophysical Journal Volume 86 February 004 85 00 85 Localization Accuracy in Single-Molecule Microscopy Raimund J. Ober,* y Sripad Ram, z and E. Sally Ward yz *Department of Electrical Engineering, University
Biophysical Journal Volume 86 February 004 85 00 85 Localization Accuracy in Single-Molecule Microscopy Raimund J. Ober,* y Sripad Ram, z and E. Sally Ward yz *Department of Electrical Engineering, University
4.2 Megapixel Scientific CMOS Camera
 CMOS, EMCCD AND CCD CAMERAS FOR LIFE SCIENCES 4.2 Megapixel Scientific CMOS Camera Photometrics Prime is the first intelligent scientific CMOS (scmos) camera to incorporate a powerful FPGA-based Embedded
CMOS, EMCCD AND CCD CAMERAS FOR LIFE SCIENCES 4.2 Megapixel Scientific CMOS Camera Photometrics Prime is the first intelligent scientific CMOS (scmos) camera to incorporate a powerful FPGA-based Embedded
Digital Volume Correlation for Materials Characterization
 19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
Searching for Handedness in Large Data
 Searching for Handedness in Large Data Summer project Mark Jenei, UCL [4] Supervisors: Prof. Ian Robinson, Chritopher Lynch Summer of 2015 1 Abstract Many biological systems have a well-defined chirality
Searching for Handedness in Large Data Summer project Mark Jenei, UCL [4] Supervisors: Prof. Ian Robinson, Chritopher Lynch Summer of 2015 1 Abstract Many biological systems have a well-defined chirality
doi: /
 Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Detector systems for light microscopy
 Detector systems for light microscopy The human eye the perfect detector? Resolution: 0.1-0.3mm @25cm object distance Spectral sensitivity ~400-700nm Has a dynamic range of 10 decades Two detectors: rods
Detector systems for light microscopy The human eye the perfect detector? Resolution: 0.1-0.3mm @25cm object distance Spectral sensitivity ~400-700nm Has a dynamic range of 10 decades Two detectors: rods
Time-resolved wavelet- based acquisitions using a single pixel camera
 Florian Rousset 1,2, Nicolas Ducros 1, Andrea Farina 2, Gianluca Valentini 2, Cosimo D Andrea 2, Françoise Peyrin 1 1 Univ Lyon, INSA Lyon, CNRS 5220, INSERM U1206, CREATIS Lyon, France 2 Politecnico di
Florian Rousset 1,2, Nicolas Ducros 1, Andrea Farina 2, Gianluca Valentini 2, Cosimo D Andrea 2, Françoise Peyrin 1 1 Univ Lyon, INSA Lyon, CNRS 5220, INSERM U1206, CREATIS Lyon, France 2 Politecnico di
Accurate extraction of reciprocal space information from transmission electron microscopy images. Edward Rosten and Susan Cox LA-UR
 Accurate extraction of reciprocal space information from transmission electron microscopy images Edward Rosten and Susan Cox LA-UR-06-8287 Diffraction patterns Diffraction pattern is the Power Spectral
Accurate extraction of reciprocal space information from transmission electron microscopy images Edward Rosten and Susan Cox LA-UR-06-8287 Diffraction patterns Diffraction pattern is the Power Spectral
Detection of Sub-resolution Dots in Microscopy Images
 Detection of Sub-resolution Dots in Microscopy Images Karel Štěpka, 2012 Centre for Biomedical Image Analysis, FI MU supervisor: prof. RNDr. Michal Kozubek, Ph.D. Outline Introduction Existing approaches
Detection of Sub-resolution Dots in Microscopy Images Karel Štěpka, 2012 Centre for Biomedical Image Analysis, FI MU supervisor: prof. RNDr. Michal Kozubek, Ph.D. Outline Introduction Existing approaches
Image Coding with Active Appearance Models
 Image Coding with Active Appearance Models Simon Baker, Iain Matthews, and Jeff Schneider CMU-RI-TR-03-13 The Robotics Institute Carnegie Mellon University Abstract Image coding is the task of representing
Image Coding with Active Appearance Models Simon Baker, Iain Matthews, and Jeff Schneider CMU-RI-TR-03-13 The Robotics Institute Carnegie Mellon University Abstract Image coding is the task of representing
Thomas Cajgfinger, Eric Chabanat, Agnes Dominjon, Quang T. Doan, Cyrille Guerin, Julien Houles, Remi Barbier.
 Thomas Cajgfinger, Eric Chabanat, Agnes Dominjon, Quang T. Doan, Cyrille Guerin, Julien Houles, Remi Barbier. IPNL, Université de Lyon, Université Lyon 1, CNRS/IN2P3, 4 rue E. Fermi 69622 Villeurbanne
Thomas Cajgfinger, Eric Chabanat, Agnes Dominjon, Quang T. Doan, Cyrille Guerin, Julien Houles, Remi Barbier. IPNL, Université de Lyon, Université Lyon 1, CNRS/IN2P3, 4 rue E. Fermi 69622 Villeurbanne
Edges, interpolation, templates. Nuno Vasconcelos ECE Department, UCSD (with thanks to David Forsyth)
 Edges, interpolation, templates Nuno Vasconcelos ECE Department, UCSD (with thanks to David Forsyth) Gradients and edges edges are points of large gradient magnitude edge detection strategy 1. determine
Edges, interpolation, templates Nuno Vasconcelos ECE Department, UCSD (with thanks to David Forsyth) Gradients and edges edges are points of large gradient magnitude edge detection strategy 1. determine
Locally Weighted Least Squares Regression for Image Denoising, Reconstruction and Up-sampling
 Locally Weighted Least Squares Regression for Image Denoising, Reconstruction and Up-sampling Moritz Baecher May 15, 29 1 Introduction Edge-preserving smoothing and super-resolution are classic and important
Locally Weighted Least Squares Regression for Image Denoising, Reconstruction and Up-sampling Moritz Baecher May 15, 29 1 Introduction Edge-preserving smoothing and super-resolution are classic and important
High Resolution BSI Scientific CMOS
 CMOS, EMCCD AND CCD CAMERAS FOR LIFE SCIENCES High Resolution BSI Scientific CMOS Prime BSI delivers the perfect balance between high resolution imaging and sensitivity with an optimized pixel design and
CMOS, EMCCD AND CCD CAMERAS FOR LIFE SCIENCES High Resolution BSI Scientific CMOS Prime BSI delivers the perfect balance between high resolution imaging and sensitivity with an optimized pixel design and
45 µm polystyrene bead embedded in scattering tissue phantom. (a,b) raw images under oblique
 Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
Application of Proximal Algorithms to Three Dimensional Deconvolution Microscopy
 Application of Proximal Algorithms to Three Dimensional Deconvolution Microscopy Paroma Varma Stanford University paroma@stanford.edu Abstract In microscopy, shot noise dominates image formation, which
Application of Proximal Algorithms to Three Dimensional Deconvolution Microscopy Paroma Varma Stanford University paroma@stanford.edu Abstract In microscopy, shot noise dominates image formation, which
Intensity-based axial localization approaches for multifocal plane microscopy
 Vol. 25, No. 4 Feb 17 OPTICS EXPRESS 3394 Intensity-based axial localization approaches for multifocal plane microscopy RAMRAJ VELMURUGAN, 1,2,3 JERRY CHAO, 1,4 SRIPAD RAM, 5 E. SALLY WARD, 1,2 AND RAIMUND
Vol. 25, No. 4 Feb 17 OPTICS EXPRESS 3394 Intensity-based axial localization approaches for multifocal plane microscopy RAMRAJ VELMURUGAN, 1,2,3 JERRY CHAO, 1,4 SRIPAD RAM, 5 E. SALLY WARD, 1,2 AND RAIMUND
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10934 Supplementary Methods Mathematical implementation of the EST method. The EST method begins with padding each projection with zeros (that is, embedding
SUPPLEMENTARY INFORMATION doi:10.1038/nature10934 Supplementary Methods Mathematical implementation of the EST method. The EST method begins with padding each projection with zeros (that is, embedding
Raster Image Correlation Spectroscopy and Number and Brightness Analysis
 Raster Image Correlation Spectroscopy and Number and Brightness Analysis 15 Principles of Fluorescence Course Paolo Annibale, PhD On behalf of Dr. Enrico Gratton lfd Raster Image Correlation Spectroscopy
Raster Image Correlation Spectroscopy and Number and Brightness Analysis 15 Principles of Fluorescence Course Paolo Annibale, PhD On behalf of Dr. Enrico Gratton lfd Raster Image Correlation Spectroscopy
Broad field that includes low-level operations as well as complex high-level algorithms
 Image processing About Broad field that includes low-level operations as well as complex high-level algorithms Low-level image processing Computer vision Computational photography Several procedures and
Image processing About Broad field that includes low-level operations as well as complex high-level algorithms Low-level image processing Computer vision Computational photography Several procedures and
Supplementary Figure 1. Decoding results broken down for different ROIs
 Supplementary Figure 1 Decoding results broken down for different ROIs Decoding results for areas V1, V2, V3, and V1 V3 combined. (a) Decoded and presented orientations are strongly correlated in areas
Supplementary Figure 1 Decoding results broken down for different ROIs Decoding results for areas V1, V2, V3, and V1 V3 combined. (a) Decoded and presented orientations are strongly correlated in areas
Guided Image Super-Resolution: A New Technique for Photogeometric Super-Resolution in Hybrid 3-D Range Imaging
 Guided Image Super-Resolution: A New Technique for Photogeometric Super-Resolution in Hybrid 3-D Range Imaging Florin C. Ghesu 1, Thomas Köhler 1,2, Sven Haase 1, Joachim Hornegger 1,2 04.09.2014 1 Pattern
Guided Image Super-Resolution: A New Technique for Photogeometric Super-Resolution in Hybrid 3-D Range Imaging Florin C. Ghesu 1, Thomas Köhler 1,2, Sven Haase 1, Joachim Hornegger 1,2 04.09.2014 1 Pattern
Colocalisation/Correlation
 Colocalisation/Correlation The past: I see yellow - therefore there is colocalisation but published images look over exposed. No colocalisation definition + No stats = No Science. From Now On: 3D. Quantification.
Colocalisation/Correlation The past: I see yellow - therefore there is colocalisation but published images look over exposed. No colocalisation definition + No stats = No Science. From Now On: 3D. Quantification.
Tomographic Algorithm for Industrial Plasmas
 Tomographic Algorithm for Industrial Plasmas More info about this article: http://www.ndt.net/?id=22342 1 Sudhir K. Chaudhary, 1 Kavita Rathore, 2 Sudeep Bhattacharjee, 1 Prabhat Munshi 1 Nuclear Engineering
Tomographic Algorithm for Industrial Plasmas More info about this article: http://www.ndt.net/?id=22342 1 Sudhir K. Chaudhary, 1 Kavita Rathore, 2 Sudeep Bhattacharjee, 1 Prabhat Munshi 1 Nuclear Engineering
EPFL SV PTBIOP BIOP COURSE 2015 OPTICAL SLICING METHODS
 BIOP COURSE 2015 OPTICAL SLICING METHODS OPTICAL SLICING METHODS Scanning Methods Wide field Methods Point Scanning Deconvolution Line Scanning Multiple Beam Scanning Single Photon Multiple Photon Total
BIOP COURSE 2015 OPTICAL SLICING METHODS OPTICAL SLICING METHODS Scanning Methods Wide field Methods Point Scanning Deconvolution Line Scanning Multiple Beam Scanning Single Photon Multiple Photon Total
Experimental Data and Training
 Modeling and Control of Dynamic Systems Experimental Data and Training Mihkel Pajusalu Alo Peets Tartu, 2008 1 Overview Experimental data Designing input signal Preparing data for modeling Training Criterion
Modeling and Control of Dynamic Systems Experimental Data and Training Mihkel Pajusalu Alo Peets Tartu, 2008 1 Overview Experimental data Designing input signal Preparing data for modeling Training Criterion
Supplementary Information
 Supplementary Information Interferometric scattering microscopy with polarization-selective dual detection scheme: Capturing the orientational information of anisotropic nanometric objects Il-Buem Lee
Supplementary Information Interferometric scattering microscopy with polarization-selective dual detection scheme: Capturing the orientational information of anisotropic nanometric objects Il-Buem Lee
Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences
 Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences W. J. Godinez 1, M. Lampe 2, S. Wörz 1, B. Müller 2, R. Eils 1 and K. Rohr 1 1 University of Heidelberg, IPMB, and DKFZ
Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences W. J. Godinez 1, M. Lampe 2, S. Wörz 1, B. Müller 2, R. Eils 1 and K. Rohr 1 1 University of Heidelberg, IPMB, and DKFZ
Renishaw invia Raman Microscope (April 2006)
 Renishaw invia Raman Microscope (April 2006) I. Starting the System 1. The main system unit is ON all the time. 2. Switch on the Leica microscope and light source for reflective bright field (BF) imaging.
Renishaw invia Raman Microscope (April 2006) I. Starting the System 1. The main system unit is ON all the time. 2. Switch on the Leica microscope and light source for reflective bright field (BF) imaging.
Comparison of fiber orientation analysis methods in Avizo
 Comparison of fiber orientation analysis methods in Avizo More info about this article: http://www.ndt.net/?id=20865 Abstract Rémi Blanc 1, Peter Westenberger 2 1 FEI, 3 Impasse Rudolf Diesel, Bât A, 33708
Comparison of fiber orientation analysis methods in Avizo More info about this article: http://www.ndt.net/?id=20865 Abstract Rémi Blanc 1, Peter Westenberger 2 1 FEI, 3 Impasse Rudolf Diesel, Bât A, 33708
On the quality of measured optical aberration coefficients using phase wheel monitor
 On the quality of measured optical aberration coefficients using phase wheel monitor Lena V. Zavyalova *, Aaron R. Robinson, Anatoly Bourov, Neal V. Lafferty, and Bruce W. Smith Center for Nanolithography
On the quality of measured optical aberration coefficients using phase wheel monitor Lena V. Zavyalova *, Aaron R. Robinson, Anatoly Bourov, Neal V. Lafferty, and Bruce W. Smith Center for Nanolithography
x' = c 1 x + c 2 y + c 3 xy + c 4 y' = c 5 x + c 6 y + c 7 xy + c 8
 1. Explain about gray level interpolation. The distortion correction equations yield non integer values for x' and y'. Because the distorted image g is digital, its pixel values are defined only at integer
1. Explain about gray level interpolation. The distortion correction equations yield non integer values for x' and y'. Because the distorted image g is digital, its pixel values are defined only at integer
2, the coefficient of variation R 2, and properties of the photon counts traces
 Supplementary Figure 1 Quality control of FCS traces. (a) Typical trace that passes the quality control (QC) according to the parameters shown in f. The QC is based on thresholds applied to fitting parameters
Supplementary Figure 1 Quality control of FCS traces. (a) Typical trace that passes the quality control (QC) according to the parameters shown in f. The QC is based on thresholds applied to fitting parameters
Contents. 1 Introduction Background Organization Features... 7
 Contents 1 Introduction... 1 1.1 Background.... 1 1.2 Organization... 2 1.3 Features... 7 Part I Fundamental Algorithms for Computer Vision 2 Ellipse Fitting... 11 2.1 Representation of Ellipses.... 11
Contents 1 Introduction... 1 1.1 Background.... 1 1.2 Organization... 2 1.3 Features... 7 Part I Fundamental Algorithms for Computer Vision 2 Ellipse Fitting... 11 2.1 Representation of Ellipses.... 11
Structured Light II. Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov
 Structured Light II Johannes Köhler Johannes.koehler@dfki.de Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov Introduction Previous lecture: Structured Light I Active Scanning Camera/emitter
Structured Light II Johannes Köhler Johannes.koehler@dfki.de Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov Introduction Previous lecture: Structured Light I Active Scanning Camera/emitter
Software Reference Manual June, 2015 revision 3.1
 Software Reference Manual June, 2015 revision 3.1 Innovations Foresight 2015 Powered by Alcor System 1 For any improvement and suggestions, please contact customerservice@innovationsforesight.com Some
Software Reference Manual June, 2015 revision 3.1 Innovations Foresight 2015 Powered by Alcor System 1 For any improvement and suggestions, please contact customerservice@innovationsforesight.com Some
Assembly dynamics of microtubules at molecular resolution
 Supplementary Information with: Assembly dynamics of microtubules at molecular resolution Jacob W.J. Kerssemakers 1,2, E. Laura Munteanu 1, Liedewij Laan 1, Tim L. Noetzel 2, Marcel E. Janson 1,3, and
Supplementary Information with: Assembly dynamics of microtubules at molecular resolution Jacob W.J. Kerssemakers 1,2, E. Laura Munteanu 1, Liedewij Laan 1, Tim L. Noetzel 2, Marcel E. Janson 1,3, and
Hartley - Zisserman reading club. Part I: Hartley and Zisserman Appendix 6: Part II: Zhengyou Zhang: Presented by Daniel Fontijne
 Hartley - Zisserman reading club Part I: Hartley and Zisserman Appendix 6: Iterative estimation methods Part II: Zhengyou Zhang: A Flexible New Technique for Camera Calibration Presented by Daniel Fontijne
Hartley - Zisserman reading club Part I: Hartley and Zisserman Appendix 6: Iterative estimation methods Part II: Zhengyou Zhang: A Flexible New Technique for Camera Calibration Presented by Daniel Fontijne
DeltaVision OMX SR super-resolution microscope. Super-resolution doesn t need to be complicated
 DeltaVision OMX SR super-resolution microscope Super-resolution doesn t need to be complicated Live-cell super-resolution microscopy DeltaVision OMX SR is a compact super-resolution microscope system optimized
DeltaVision OMX SR super-resolution microscope Super-resolution doesn t need to be complicated Live-cell super-resolution microscopy DeltaVision OMX SR is a compact super-resolution microscope system optimized
Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration
 Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration Thomas Kilgus, Thiago R. dos Santos, Alexander Seitel, Kwong Yung, Alfred M. Franz, Anja Groch, Ivo Wolf, Hans-Peter Meinzer,
Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration Thomas Kilgus, Thiago R. dos Santos, Alexander Seitel, Kwong Yung, Alfred M. Franz, Anja Groch, Ivo Wolf, Hans-Peter Meinzer,
Phasor based single-molecule localization microscopy in 3D. (psmlm-3d): an algorithm for MHz localization rates using
 Phasor based single-molecule localization microscopy in 3D (psmlm-3d): an algorithm for MHz localization rates using standard CPUs Koen J.A. Martens 1,2, Arjen N. Bader 1,3, Sander Baas 1, Bernd Rieger
Phasor based single-molecule localization microscopy in 3D (psmlm-3d): an algorithm for MHz localization rates using standard CPUs Koen J.A. Martens 1,2, Arjen N. Bader 1,3, Sander Baas 1, Bernd Rieger
OPTICAL COHERENCE TOMOGRAPHY:SIGNAL PROCESSING AND ALGORITHM
 OPTICAL COHERENCE TOMOGRAPHY:SIGNAL PROCESSING AND ALGORITHM OCT Medical imaging modality with 1-10 µ m resolutions and 1-2 mm penetration depths High-resolution, sub-surface non-invasive or minimally
OPTICAL COHERENCE TOMOGRAPHY:SIGNAL PROCESSING AND ALGORITHM OCT Medical imaging modality with 1-10 µ m resolutions and 1-2 mm penetration depths High-resolution, sub-surface non-invasive or minimally
Cluster Analysis of Super Resolution Fluorescence Images Ki Woong Sung
 NSERC USRA Report Summer 2015 Cluster Analysis of Super Resolution Fluorescence Images Ki Woong Sung 1 Biological Backgound Direct Stochastic Optical Reconstruction Microscopy (dstorm) is a novel, high
NSERC USRA Report Summer 2015 Cluster Analysis of Super Resolution Fluorescence Images Ki Woong Sung 1 Biological Backgound Direct Stochastic Optical Reconstruction Microscopy (dstorm) is a novel, high
Leica Microsystems Intelligent Structured Illumination Microscopy
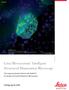 Widefield Mouse kidney section. Maximum projection of a stack containing 65 planes. Green: glomeruli and convoluted tubules (wheat germ agglutinin Alexa Fluor 488) Blue: Nuclei (DAPI). Structured Illumination
Widefield Mouse kidney section. Maximum projection of a stack containing 65 planes. Green: glomeruli and convoluted tubules (wheat germ agglutinin Alexa Fluor 488) Blue: Nuclei (DAPI). Structured Illumination
On the uncertainty of Astigmatic Particle Tracking Velocimetry in the depth direction
 On the uncertainty of Astigmatic Particle Tracking Velocimetry in the depth direction Massimiliano Rossi 1,*, Christian J. Kähler 1 1: Institute for Fluid Mechanics and Aerodynamics, Bundeswehr University
On the uncertainty of Astigmatic Particle Tracking Velocimetry in the depth direction Massimiliano Rossi 1,*, Christian J. Kähler 1 1: Institute for Fluid Mechanics and Aerodynamics, Bundeswehr University
Super-Resolution from Image Sequences A Review
 Super-Resolution from Image Sequences A Review Sean Borman, Robert L. Stevenson Department of Electrical Engineering University of Notre Dame 1 Introduction Seminal work by Tsai and Huang 1984 More information
Super-Resolution from Image Sequences A Review Sean Borman, Robert L. Stevenson Department of Electrical Engineering University of Notre Dame 1 Introduction Seminal work by Tsai and Huang 1984 More information
Optical Sectioning. Bo Huang. Pharmaceutical Chemistry
 Optical Sectioning Bo Huang Pharmaceutical Chemistry Approaches to 3D imaging Physical cutting Technical difficulty Highest resolution Highest sensitivity Optical sectioning Simple sample prep. No physical
Optical Sectioning Bo Huang Pharmaceutical Chemistry Approaches to 3D imaging Physical cutting Technical difficulty Highest resolution Highest sensitivity Optical sectioning Simple sample prep. No physical
Independent Resolution Test of
 Independent Resolution Test of as conducted and published by Dr. Adam Puche, PhD University of Maryland June 2005 as presented by (formerly Thales Optem Inc.) www.qioptiqimaging.com Independent Resolution
Independent Resolution Test of as conducted and published by Dr. Adam Puche, PhD University of Maryland June 2005 as presented by (formerly Thales Optem Inc.) www.qioptiqimaging.com Independent Resolution
Dense Tracking and Mapping for Autonomous Quadrocopters. Jürgen Sturm
 Computer Vision Group Prof. Daniel Cremers Dense Tracking and Mapping for Autonomous Quadrocopters Jürgen Sturm Joint work with Frank Steinbrücker, Jakob Engel, Christian Kerl, Erik Bylow, and Daniel Cremers
Computer Vision Group Prof. Daniel Cremers Dense Tracking and Mapping for Autonomous Quadrocopters Jürgen Sturm Joint work with Frank Steinbrücker, Jakob Engel, Christian Kerl, Erik Bylow, and Daniel Cremers
Digital Image Processing. Prof. P. K. Biswas. Department of Electronic & Electrical Communication Engineering
 Digital Image Processing Prof. P. K. Biswas Department of Electronic & Electrical Communication Engineering Indian Institute of Technology, Kharagpur Lecture - 21 Image Enhancement Frequency Domain Processing
Digital Image Processing Prof. P. K. Biswas Department of Electronic & Electrical Communication Engineering Indian Institute of Technology, Kharagpur Lecture - 21 Image Enhancement Frequency Domain Processing
Quantifying Caveolin-1 Molecular Domains via Machine Learning and Computational Network Analysis of Super-resolution Microscopy Data
 Quantifying Caveolin-1 Molecular Domains via Machine Learning and Computational Network Analysis of Super-resolution Microscopy Data Ismail M. Khater1, Fanrui Meng2, Ivan Robert Nabi2, Ghassan Hamarneh1
Quantifying Caveolin-1 Molecular Domains via Machine Learning and Computational Network Analysis of Super-resolution Microscopy Data Ismail M. Khater1, Fanrui Meng2, Ivan Robert Nabi2, Ghassan Hamarneh1
ANALYSIS OF GEOPHYSICAL POTENTIAL FIELDS A Digital Signal Processing Approach
 ADVANCES IN EXPLORATION GEOPHYSICS 5 ANALYSIS OF GEOPHYSICAL POTENTIAL FIELDS A Digital Signal Processing Approach PRABHAKAR S. NAIDU Indian Institute of Science, Bangalore 560012, India AND M.P. MATHEW
ADVANCES IN EXPLORATION GEOPHYSICS 5 ANALYSIS OF GEOPHYSICAL POTENTIAL FIELDS A Digital Signal Processing Approach PRABHAKAR S. NAIDU Indian Institute of Science, Bangalore 560012, India AND M.P. MATHEW
arxiv: v1 [cs.cv] 28 Sep 2018
![arxiv: v1 [cs.cv] 28 Sep 2018 arxiv: v1 [cs.cv] 28 Sep 2018](/thumbs/93/113542646.jpg) Camera Pose Estimation from Sequence of Calibrated Images arxiv:1809.11066v1 [cs.cv] 28 Sep 2018 Jacek Komorowski 1 and Przemyslaw Rokita 2 1 Maria Curie-Sklodowska University, Institute of Computer Science,
Camera Pose Estimation from Sequence of Calibrated Images arxiv:1809.11066v1 [cs.cv] 28 Sep 2018 Jacek Komorowski 1 and Przemyslaw Rokita 2 1 Maria Curie-Sklodowska University, Institute of Computer Science,
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror
 Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
3-D. Here red spheres show the location of gold nanoparticles inside/around a cell nucleus.
 3-D The CytoViva 3-D System allows the user can locate objects of interest in a 3-D space. It does this by acquiring multiple Z planes and performing our custom software routines to locate and observe
3-D The CytoViva 3-D System allows the user can locate objects of interest in a 3-D space. It does this by acquiring multiple Z planes and performing our custom software routines to locate and observe
Product Information Version 1.1. ZEISS Axiocam 503 mono Your Flexible and Sensitive 3 Megapixel Microscope Camera for Live Cell Imaging
 Product Information Version 1.1 ZEISS Axiocam 503 mono Your Flexible and Sensitive 3 Megapixel Microscope Camera for Live Cell Imaging ZEISS Axiocam 503 mono Sensor Model Sensor Pixel Count Pixel Size
Product Information Version 1.1 ZEISS Axiocam 503 mono Your Flexible and Sensitive 3 Megapixel Microscope Camera for Live Cell Imaging ZEISS Axiocam 503 mono Sensor Model Sensor Pixel Count Pixel Size
3D Computer Vision. Structured Light II. Prof. Didier Stricker. Kaiserlautern University.
 3D Computer Vision Structured Light II Prof. Didier Stricker Kaiserlautern University http://ags.cs.uni-kl.de/ DFKI Deutsches Forschungszentrum für Künstliche Intelligenz http://av.dfki.de 1 Introduction
3D Computer Vision Structured Light II Prof. Didier Stricker Kaiserlautern University http://ags.cs.uni-kl.de/ DFKI Deutsches Forschungszentrum für Künstliche Intelligenz http://av.dfki.de 1 Introduction
Adaptive Multiple-Frame Image Super- Resolution Based on U-Curve
 Adaptive Multiple-Frame Image Super- Resolution Based on U-Curve IEEE Transaction on Image Processing, Vol. 19, No. 12, 2010 Qiangqiang Yuan, Liangpei Zhang, Huanfeng Shen, and Pingxiang Li Presented by
Adaptive Multiple-Frame Image Super- Resolution Based on U-Curve IEEE Transaction on Image Processing, Vol. 19, No. 12, 2010 Qiangqiang Yuan, Liangpei Zhang, Huanfeng Shen, and Pingxiang Li Presented by
Super-Resolution. Many slides from Miki Elad Technion Yosi Rubner RTC and more
 Super-Resolution Many slides from Mii Elad Technion Yosi Rubner RTC and more 1 Example - Video 53 images, ratio 1:4 2 Example Surveillance 40 images ratio 1:4 3 Example Enhance Mosaics 4 5 Super-Resolution
Super-Resolution Many slides from Mii Elad Technion Yosi Rubner RTC and more 1 Example - Video 53 images, ratio 1:4 2 Example Surveillance 40 images ratio 1:4 3 Example Enhance Mosaics 4 5 Super-Resolution
Advanced Image Combine Techniques
 Advanced Image Combine Techniques Ray Gralak November 16 2008 AIC 2008 Important Equation 1 of 22 (Joke! ) Outline Some History The Imager s Enemies artifacts! The Basic Raw Image Types Image Calibration
Advanced Image Combine Techniques Ray Gralak November 16 2008 AIC 2008 Important Equation 1 of 22 (Joke! ) Outline Some History The Imager s Enemies artifacts! The Basic Raw Image Types Image Calibration
Correcting Radial Distortion of Cameras With Wide Angle Lens Using Point Correspondences
 Correcting Radial istortion of Cameras With Wide Angle Lens Using Point Correspondences Leonardo Romero and Cuauhtemoc Gomez Universidad Michoacana de San Nicolas de Hidalgo Morelia, Mich., 58000, Mexico
Correcting Radial istortion of Cameras With Wide Angle Lens Using Point Correspondences Leonardo Romero and Cuauhtemoc Gomez Universidad Michoacana de San Nicolas de Hidalgo Morelia, Mich., 58000, Mexico
pco.edge electrons 2048 x 1536 pixel 50 fps :1 > 60 % pco. low noise high resolution high speed high dynamic range
 edge 3.1 scientific CMOS camera high resolution 2048 x 1536 pixel low noise 1.1 electrons global shutter USB 3.0 small form factor high dynamic range 27 000:1 high speed 50 fps high quantum efficiency
edge 3.1 scientific CMOS camera high resolution 2048 x 1536 pixel low noise 1.1 electrons global shutter USB 3.0 small form factor high dynamic range 27 000:1 high speed 50 fps high quantum efficiency
SUPPLEMENTARY INFORMATION
 Removing orientation-induced localization biases in single-molecule microscopy using a broadband metasurface mask Mikael P. Backlund, Amir Arbabi, Petar N. Petrov, Ehsan Arbabi, Saumya Saurabh, Andrei
Removing orientation-induced localization biases in single-molecule microscopy using a broadband metasurface mask Mikael P. Backlund, Amir Arbabi, Petar N. Petrov, Ehsan Arbabi, Saumya Saurabh, Andrei
TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy
 TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy Introduction The OptiGrid converts the illumination system of a conventional wide
TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy Introduction The OptiGrid converts the illumination system of a conventional wide
Neo scmos. Specifications Summary. Pixel size (W x H) 6.5 µm. Pixel well depth (typical) 30,000 e - Pixel readout rate (MHz) 560, 200
 Interline CCD 5.5 e - read noise Low Light Imaging Ultra Sensitive Imaging Andor scmos 1 e - read noise Features and Benefits TE cooling to -40 C Minimization of dark current and pixel blemish 1 e - read
Interline CCD 5.5 e - read noise Low Light Imaging Ultra Sensitive Imaging Andor scmos 1 e - read noise Features and Benefits TE cooling to -40 C Minimization of dark current and pixel blemish 1 e - read
arxiv: v1 [physics.optics] 13 Oct 2017
![arxiv: v1 [physics.optics] 13 Oct 2017 arxiv: v1 [physics.optics] 13 Oct 2017](/thumbs/90/101585266.jpg) Aberration-free calibration for 3D single molecule localization microscopy Clément Cabriel 1,*, Nicolas Bourg 1, Guillaume Dupuis 2, and Sandrine Lévêque-Fort 1,** arxiv:1710.04957v1 [physics.optics] 13
Aberration-free calibration for 3D single molecule localization microscopy Clément Cabriel 1,*, Nicolas Bourg 1, Guillaume Dupuis 2, and Sandrine Lévêque-Fort 1,** arxiv:1710.04957v1 [physics.optics] 13
Iterative Closest Point Algorithm in the Presence of Anisotropic Noise
 Iterative Closest Point Algorithm in the Presence of Anisotropic Noise L. Maier-Hein, T. R. dos Santos, A. M. Franz, H.-P. Meinzer German Cancer Research Center, Div. of Medical and Biological Informatics
Iterative Closest Point Algorithm in the Presence of Anisotropic Noise L. Maier-Hein, T. R. dos Santos, A. M. Franz, H.-P. Meinzer German Cancer Research Center, Div. of Medical and Biological Informatics
ProEM -HS:1KBX3-10µm
 The ProEM-HS:1KBX3-1µm is the fastest EMCCD camera on the market utilizing the latest low-noise readout electronics and a 124 x 124 EMCCD. This camera delivers single photon sensitivity and the best fringe
The ProEM-HS:1KBX3-1µm is the fastest EMCCD camera on the market utilizing the latest low-noise readout electronics and a 124 x 124 EMCCD. This camera delivers single photon sensitivity and the best fringe
ProEM -HS:1024BX3 FEATURES BENEFITS
 ProEM -HS:124BX3 The ProEM-HS: 124BX3 is the most advanced EMCCD camera on the market utilizing the latest low-noise readout electronics and a 124 x 124 EMCCD. This camera delivers single photon sensitivity
ProEM -HS:124BX3 The ProEM-HS: 124BX3 is the most advanced EMCCD camera on the market utilizing the latest low-noise readout electronics and a 124 x 124 EMCCD. This camera delivers single photon sensitivity
Local Readjustment for High-Resolution 3D Reconstruction: Supplementary Material
 Local Readjustment for High-Resolution 3D Reconstruction: Supplementary Material Siyu Zhu 1, Tian Fang 2, Jianxiong Xiao 3, and Long Quan 4 1,2,4 The Hong Kong University of Science and Technology 3 Princeton
Local Readjustment for High-Resolution 3D Reconstruction: Supplementary Material Siyu Zhu 1, Tian Fang 2, Jianxiong Xiao 3, and Long Quan 4 1,2,4 The Hong Kong University of Science and Technology 3 Princeton
Bioimage Informatics
 Bioimage Informatics Lecture 10, Spring 2012 Bioimage Data Analysis (II): Applications of Point Feature Detection Techniques: Super Resolution Fluorescence Microscopy Bioimage Data Analysis (III): Edge
Bioimage Informatics Lecture 10, Spring 2012 Bioimage Data Analysis (II): Applications of Point Feature Detection Techniques: Super Resolution Fluorescence Microscopy Bioimage Data Analysis (III): Edge
Status of PSF Reconstruction at Lick
 Status of PSF Reconstruction at Lick Mike Fitzgerald Workshop on AO PSF Reconstruction May 10-12, 2004 Quick Outline Recap Lick AO system's features Reconstruction approach Implementation issues Calibration
Status of PSF Reconstruction at Lick Mike Fitzgerald Workshop on AO PSF Reconstruction May 10-12, 2004 Quick Outline Recap Lick AO system's features Reconstruction approach Implementation issues Calibration
TAU-POGRAPHY-FLIM Software
 TAU-POGRAPHY-FLIM Software P FLIM VERSION 3.1 Authors : Christian Brière, Fabien Esteveny et Alain Jauneau Contacts : briere@lrsv.ups-tlse.fr ; jauneau@lrsv.ups-tlse.fr imageriefraib@lrsv.ups-tlse.fr 2017/03/01
TAU-POGRAPHY-FLIM Software P FLIM VERSION 3.1 Authors : Christian Brière, Fabien Esteveny et Alain Jauneau Contacts : briere@lrsv.ups-tlse.fr ; jauneau@lrsv.ups-tlse.fr imageriefraib@lrsv.ups-tlse.fr 2017/03/01
Multi-Input Cardiac Image Super-Resolution using Convolutional Neural Networks
 Multi-Input Cardiac Image Super-Resolution using Convolutional Neural Networks Ozan Oktay, Wenjia Bai, Matthew Lee, Ricardo Guerrero, Konstantinos Kamnitsas, Jose Caballero, Antonio de Marvao, Stuart Cook,
Multi-Input Cardiac Image Super-Resolution using Convolutional Neural Networks Ozan Oktay, Wenjia Bai, Matthew Lee, Ricardo Guerrero, Konstantinos Kamnitsas, Jose Caballero, Antonio de Marvao, Stuart Cook,
Level-set MCMC Curve Sampling and Geometric Conditional Simulation
 Level-set MCMC Curve Sampling and Geometric Conditional Simulation Ayres Fan John W. Fisher III Alan S. Willsky February 16, 2007 Outline 1. Overview 2. Curve evolution 3. Markov chain Monte Carlo 4. Curve
Level-set MCMC Curve Sampling and Geometric Conditional Simulation Ayres Fan John W. Fisher III Alan S. Willsky February 16, 2007 Outline 1. Overview 2. Curve evolution 3. Markov chain Monte Carlo 4. Curve
Chapter 3 Image Registration. Chapter 3 Image Registration
 Chapter 3 Image Registration Distributed Algorithms for Introduction (1) Definition: Image Registration Input: 2 images of the same scene but taken from different perspectives Goal: Identify transformation
Chapter 3 Image Registration Distributed Algorithms for Introduction (1) Definition: Image Registration Input: 2 images of the same scene but taken from different perspectives Goal: Identify transformation
SPATIAL DENSITY ESTIMATION BASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES
 SPATIAL DENSITY ESTIMATION ASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES Kuan-Chieh Jackie Chen 1,2,3, Ge Yang 1,2, and Jelena Kovačević 3,1,2 1 Dept. of iomedical Eng., 2 Center
SPATIAL DENSITY ESTIMATION ASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES Kuan-Chieh Jackie Chen 1,2,3, Ge Yang 1,2, and Jelena Kovačević 3,1,2 1 Dept. of iomedical Eng., 2 Center
