Comparison of Centralities for Biological Networks
|
|
|
- Piers Sanders
- 5 years ago
- Views:
Transcription
1 Comparison of Centralities for Biological Networks Dirk Koschützki and Falk Schreiber Bioinformatics Center Gatersleben-Halle Institute of Plant Genetics and Crop Plant Research Corrensstraße Gatersleben, Germany. koschuet,schreibe@ipk-gatersleben.de Abstract: The analysis of biological networks involves the evaluation of the vertices within the connection structure of the network. To support this analysis we discuss five centrality measures and demonstrate their applicability on two example networks, a protein-protein-interaction network and a transcriptional regulation network. We show that all five centrality measures result in different valuations of the vertices and that for the analysis of biological networks all five measures are of interest. 1 Introduction Centrality analysis is particularly useful in analyzing biological networks and hence in helping to understand the underlying biological processes. It has been shown that central vertices in protein-protein interaction networks are often functionally important and the deletion of such vertices is related to lethality [JMBO01]. In [WS03] three different types of centralities are defined and applied to metabolic, protein-protein interaction and domain sequence networks. Fell and Wagner discuss the possibility that metabolites with highest degree may belong to the oldest part of the metabolism [FW00]. However, it has also been shown that the degree of a vertex alone, as a specific centrality measure, is not sufficient to distinguish lethal proteins clearly from viable ones [Wu02], that in protein networks there is no relation between network connectivity and robustness against amino-acid substitutions [HCW02], and that for biological network analysis several centrality measures have to be considered [WS03]. To assist scientists in the exploration of biological networks, we discuss and compare five different centrality measures. Some of these are already known in biological sciences, others are transferred from different fields of sciences such as social network analysis. The application of these measures shows that some correlate strongly in one network and weakly in another. As a result, we conclude that for the analysis of biological networks several measures should be considered. This work was supported by the German Ministry of Education and Research (BMBF) under grant A. 199
2 This paper is organized as follows: in Sect. 2 we define the graph model on which we operate. Section 3 introduces the five centralities in networks, all are explained using one example graph. These measures are applied to typical biological networks in Sect Definitions A undirected graph G = (V, E) consists of a finite set V of vertices (n = V ) and a finite set E V V of edges (m = E ). An edge e = (u, v) E connects two vertices u and v. The vertices u and v are said to be incident with the edge e and adjacent to each other. The set of all vertices which are adjacent to u is called the neighborhood N(u) of u (N(u) = {v : (u, v) E}). A graph is called loop-free if no edge connects a vertex to itself. An adjacency matrix A of a graph G = (V, E) is a (n n) matrix, where a ij = 1 if and only if (i, j) E and a ij = 0 otherwise. The adjacency matrix of any undirected graph is symmetric. The degree d(v) of a vertex v is the number of its incident edges. Let (e 1,..., e k ) be a sequence of edges in a graph G = (V, E). This sequence is called a walk if there are vertices v 0,..., v k such that e i = (v i 1, v i ) for i = 1,..., k. If the edges e i are pairwise distinct and the vertices v i are pairwise distinct the walk is called a path. The length of a walk or path is given by its number of edges, k = (e 1,..., e k ). A shortest path between two vertices u, v is a path with minimal length, all shortest paths between u, v are called geodesics. The distance (dist(u, v)) between two vertices u, v is the length of a shortest path between them. Two vertices u, v of a graph G = (V, E) are called connected if there exists a walk from vertex u to vertex v. If any pair of different vertices of the graph is connected, the graph G = (V, E) is called connected. If a walk starts at vertex u, chooses uniformly at random one of the incident edges of the current vertex until it finally reaches the target v then we call this walk a random walk between u and v. In the remainder of this paper we consider only non-trivial 1 undirected loop-free connected graphs. This restriction is required for a common definition of all centrality measures covered in this paper. Some of the centralities can easily be expanded to cover directed or unconnected graphs, even an extension towards weighted edges is possible. 3 Centralities in Networks Formally a centrality is a function C which assigns every vertex v V of a given graph G a value C(v) R. As we are interested in the ranking of the vertices of the given graph G we choose the convention that a vertex u is more important than another vertex v iff C(u) > C(v). In the following sections we explain five different centrality measures and show an example graph and the corresponding centrality values. 1 Graphs of at least two vertices and one edge. 200
3 3.1 Degree An obvious order of the vertices of a graph can be established by sorting them according to their degree. The corresponding centrality measure degree-centrality (C d ) is defined as C d (v) := d(v). See the work of Freeman [Fr79] for a long list of references to the usage of degree-centrality in social network analysis. For biological network analysis degreecentrality for example is used in [JMBO01] to correlate the degree of a protein in the network with the lethality of its removal. See Fig. 1 for an example graph and Table 1 for the corresponding centrality values. 3.2 Eccentricity The next three definitions of centralities all operate on the concepts of paths within the given graph. The simplest definition uses the distance between vertices. The eccentricity ecc of a vertex u is defined as ecc(u) := max v V dist(u, v) and the eccentricity-centrality. The reciprocal of ecc(u) is used to assure that more central vertices have a higher value of C e, because such central vertices are the ones with the smallest eccentricity value. An application of eccentricity within the biological context is shown in [WS03]. Again, as for degree-centrality, Fig. 1 and Table 1 show an example. (C e ) as C e (u) := 1 ecc(u) 3.3 Closeness In contrast to eccentricity, closeness-centrality uses not only the maximum distance between the vertex of interest and all other vertices but uses the sum of the distances of this 1 vertex and all other vertices. The closeness-centrality is defined as C c (u) := sumdist(u) with sumdist(u) = v V dist(u, v). Closeness-based centrality measures are cited extensively in the work of Freeman [Fr79]. Wuchty et al. [WS03] apply this centrality to different biological networks and show the correspondence with the service facility location problem. 3.4 Random Walk Betweenness Within networks a communication between two vertices u, v may be visible to a third vertex w if this vertex lies in the path of the communication between these vertices. To measure the centrality of a vertex the ability to observe communication is a feasible approach. Different methods to model communication are conceivable. There are for example communications over shortest paths, paths with maximum flow and random walks. All of these are potential models for betweenness and are covered in the literature [Fr77, FBW91, Ne03]. 201
4 Figure 1: A graph to show the five different centrality measures Newman s random walk approach models information transmission and therefore matches problems often modelled in biological networks. For the random-walk betweenness centrality (C r ) the centrality of a vertex w is equal to the number of times that a random walk from u to v goes through w, averaged over all u and v [Ne03]. A detailed coverage of the required calculations for the random-walk betweenness is beyond the scope of this paper and the reader is directed toward [Ne03]. 3.5 Bonacich s Eigenvector Centrality A different approach to order the vertices of a graph was suggested by Bonacich [Bo72]. His idea is based on the assumption that the value of a single vertex is determined by the values of the neighboring vertices. In contrast to the previous measures not only the position of a vertex within the graph is considered but also the centrality values of its neighbors. Bonacich suggested the following definition: C λ (u) := v N(u) C λ(v). If we consider the adjacency matrix representation of the graph, this is equivalent to C λ (v i ) := n j=1 a ijc λ (v j ). This leads directly to the well known problem of eigenvector computation λs = AS and the eigenvector of the largest eigenvalue is the eigenvector-centrality (C λ := S) [Bo72]. 3.6 Centralities shown on an example graph To demonstrate that all of the explained centrality measures usually give different results we use the example graph shown in Fig. 1. For this graph Table 1 shows all centrality values for the five centrality measures. 202
5 Vertex C d Vertex C e Vertex C c Vertex C r Vertex C λ Table 1: The centrality values for the example graph. The vertices are ordered by descending centrality value 4 Application and Discussion We applied the presented centrality measures to two biological networks, a protein-proteininteraction (PPI) network and a transcriptional regulation (TR) network. The PPI network is based on the April 2004 release of the Homo sapiens network from the DIP- Database [SMS + 04]. It models proteins as vertices and interactions as edges. The TR network of Escherichia coli models operons as vertices and regulation between transcription factors and operons as edges, see [SOMMA02] for details and the link to the data. For the PPI-network we removed 51 self interactions as the graph has to be loop-free for the calculation of the eigenvector-centrality C λ. Furthermore, we removed vertices and edges not connected to the giant component, and the resulting graph consisted of 563 vertices and 870 edges. For the TR network we used the data from [SOMMA02] and applied the same strategy: we removed self regulation and used the giant component for the analysis. As the regulation data is directed we used the underlying undirected graph. The activation and repression information at the edges was not used as our framework is based on unweighted graphs. Finally, the graph for the TR network consisted of 325 vertices and 453 edges. The calculation of the five centrality measures for both graphs was done on a modern desktop PC (3 GHz, 2 GB Ram, Linux 2.6.x, Java 1.4.2) and took between less than a second (C d ) and several minutes (C r ). This was expected as the complexity of the different algorithms ranges from O(n) to O((n + m)n 2 ). To analyse a network we calculated the presented centralities for all vertices. Then for each centrality all vertices were ordered by descending centrality value and, for vertices with the same centrality value, by ascending vertex-label. The vertices were enumerated 203
6 from 1 to n, this number is the position of the vertex according to the centrality. Finally, we build a new table which for every vertex and every centrality contains the position of the vertex according to the centrality. With the calculated positions we made several analyses of the correlation. It shows that some of the measures, e.g. eccentricity C e and eigenvector C λ, are highly correlated in the PPI-network (see Fig. 2) while others are only weakly correlated. Within the TR network a strong correlation between eigenvector C λ and closeness C c was observed (see Fig. 3), but in contrast to the PPI network there is only a weak correlation between C e and C λ. Tables 2 and 3 show all correlation coefficients based on Pearson s method. Note that the strong correlation of degree C d and random-walk betweenness C r is discussed in [Ne03]. In conclusion, the analysis of biological networks clearly benefits from the application of several centrality measures. Our next step lies in the comparison of different centralities measures with biological information Degree Eccentricity Closeness RWB Eigenvector Figure 2: Scatter plot matrix of the centrality positions for the PPI network C d C e C c C r C λ Degree Eccentricity Closeness RWB Eigenvector C d C e C c C r C λ Table 2: Correlation coefficients for the centrality positions for the PPI-network 204
7 Degree Eccentricity Closeness RWB Eigenvector Figure 3: Scatter plot matrix of the centrality positions for the TR network C d C e C c C r C λ Degree Eccentricity Closeness RWB Eigenvector C d C e C c C r C λ Table 3: Correlation coefficients for the centrality positions for the TR network References [Bo72] Bonacich, P.: Factoring and weighting approaches to status scores and clique identification. Journal of Mathematical Sociology. 2: [FBW91] Freeman, L. S., Borgatti, S. P., and White, D. R.: Centrality in valued graphs: a measure of betweenness based on network flow. Social Networks. 13: [Fr77] [Fr79] [FW00] Freeman, L. C.: A Set of Measures of Centrality Based on Betweenness. Sociometry. 40(6): Freeman, L. S.: Centrality in social networks: Conceptual clarification. Social Networks. 1: Fell, D. A. and Wagner, A.: The small world of metabolism. Nature Biotechnology. 18:
8 [HCW02] [JMBO01] Hahn, M. W., Conant, G., and Wagner, A.: Molecular evolution in large genetic networks: connectivity does not equal importance. Technical report. Santa Fe Institute Jeong, H., Mason, S. P., Barabási, A. L., and Oltvai, Z. N.: Lethality and centrality in protein networks. Nature. 411: [Ne03] Newman, M. E. J.: A measure of betweenness centrality based on random walks. arxiv cond-mat/ [SMS + 04] Salwinski, L., Miller, C. S., Smith, A. J., Pettit, F. K., Bowie, J. U., and Eisenberg, D.: The database of interacting proteins: 2004 update. Nucleic Acids Res. 32(1): [SOMMA02] Shen-Orr, S. S., Milo, R., Mangan, S., and Alon, U.: Network motifs in the transcriptional regulation network of escherichia coli. Nature Genetics. 31(1): [WS03] Wuchty, S. and Stadler, P. F.: Centers of complex networks. J Theor Biol. 223(1): [Wu02] Wuchty, S.: Interaction and domain networks of yeast. Proteomics. 2:
Special-topic lecture bioinformatics: Mathematics of Biological Networks
 Special-topic lecture bioinformatics: Leistungspunkte/Credit points: 5 (V2/Ü1) This course is taught in English language. The material (from books and original literature) are provided online at the course
Special-topic lecture bioinformatics: Leistungspunkte/Credit points: 5 (V2/Ü1) This course is taught in English language. The material (from books and original literature) are provided online at the course
Advanced Algorithms and Models for Computational Biology -- a machine learning approach
 Advanced Algorithms and Models for Computational Biology -- a machine learning approach Biological Networks & Network Evolution Eric Xing Lecture 22, April 10, 2006 Reading: Molecular Networks Interaction
Advanced Algorithms and Models for Computational Biology -- a machine learning approach Biological Networks & Network Evolution Eric Xing Lecture 22, April 10, 2006 Reading: Molecular Networks Interaction
Response Network Emerging from Simple Perturbation
 Journal of the Korean Physical Society, Vol 44, No 3, March 2004, pp 628 632 Response Network Emerging from Simple Perturbation S-W Son, D-H Kim, Y-Y Ahn and H Jeong Department of Physics, Korea Advanced
Journal of the Korean Physical Society, Vol 44, No 3, March 2004, pp 628 632 Response Network Emerging from Simple Perturbation S-W Son, D-H Kim, Y-Y Ahn and H Jeong Department of Physics, Korea Advanced
Coordinated Perspectives and Enhanced Force-Directed Layout for the Analysis of Network Motifs
 Coordinated Perspectives and Enhanced Force-Directed Layout for the Analysis of Network Motifs Christian Klukas Falk Schreiber Henning Schwöbbermeyer Leibniz Institute of Plant Genetics and Crop Plant
Coordinated Perspectives and Enhanced Force-Directed Layout for the Analysis of Network Motifs Christian Klukas Falk Schreiber Henning Schwöbbermeyer Leibniz Institute of Plant Genetics and Crop Plant
An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization
 An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization Pedro Ribeiro (DCC/FCUP & CRACS/INESC-TEC) Part 1 Motivation and emergence of Network Science
An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization Pedro Ribeiro (DCC/FCUP & CRACS/INESC-TEC) Part 1 Motivation and emergence of Network Science
Exploration of biological network centralities with CentiBiN
 Exploration of biological network centralities with CentiBiN Björn H. Junker 1, Dirk Koschützki 1 and Falk Schreiber 1 1 Department of Molecular Genetics, Leibniz Institute of Plant Genetics and Crop Plant
Exploration of biological network centralities with CentiBiN Björn H. Junker 1, Dirk Koschützki 1 and Falk Schreiber 1 1 Department of Molecular Genetics, Leibniz Institute of Plant Genetics and Crop Plant
Incoming, Outgoing Degree and Importance Analysis of Network Motifs
 Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology IJCSMC, Vol. 4, Issue. 6, June 2015, pg.758
Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology IJCSMC, Vol. 4, Issue. 6, June 2015, pg.758
Centrality Measures to Identify Traffic Congestion on Road Networks: A Case Study of Sri Lanka
 IOSR Journal of Mathematics (IOSR-JM) e-issn: 2278-5728, p-issn: 2319-765X. Volume 13, Issue 2 Ver. I (Mar. - Apr. 2017), PP 13-19 www.iosrjournals.org Centrality Measures to Identify Traffic Congestion
IOSR Journal of Mathematics (IOSR-JM) e-issn: 2278-5728, p-issn: 2319-765X. Volume 13, Issue 2 Ver. I (Mar. - Apr. 2017), PP 13-19 www.iosrjournals.org Centrality Measures to Identify Traffic Congestion
Graph Theory for Network Science
 Graph Theory for Network Science Dr. Natarajan Meghanathan Professor Department of Computer Science Jackson State University, Jackson, MS E-mail: natarajan.meghanathan@jsums.edu Networks or Graphs We typically
Graph Theory for Network Science Dr. Natarajan Meghanathan Professor Department of Computer Science Jackson State University, Jackson, MS E-mail: natarajan.meghanathan@jsums.edu Networks or Graphs We typically
Graph Theory Review. January 30, Network Science Analytics Graph Theory Review 1
 Graph Theory Review Gonzalo Mateos Dept. of ECE and Goergen Institute for Data Science University of Rochester gmateosb@ece.rochester.edu http://www.ece.rochester.edu/~gmateosb/ January 30, 2018 Network
Graph Theory Review Gonzalo Mateos Dept. of ECE and Goergen Institute for Data Science University of Rochester gmateosb@ece.rochester.edu http://www.ece.rochester.edu/~gmateosb/ January 30, 2018 Network
A Generating Function Approach to Analyze Random Graphs
 A Generating Function Approach to Analyze Random Graphs Presented by - Vilas Veeraraghavan Advisor - Dr. Steven Weber Department of Electrical and Computer Engineering Drexel University April 8, 2005 Presentation
A Generating Function Approach to Analyze Random Graphs Presented by - Vilas Veeraraghavan Advisor - Dr. Steven Weber Department of Electrical and Computer Engineering Drexel University April 8, 2005 Presentation
V2: Measures and Metrics (II)
 - Betweenness Centrality V2: Measures and Metrics (II) - Groups of Vertices - Transitivity - Reciprocity - Signed Edges and Structural Balance - Similarity - Homophily and Assortative Mixing 1 Betweenness
- Betweenness Centrality V2: Measures and Metrics (II) - Groups of Vertices - Transitivity - Reciprocity - Signed Edges and Structural Balance - Similarity - Homophily and Assortative Mixing 1 Betweenness
Graph Theory for Network Science
 Graph Theory for Network Science Dr. Natarajan Meghanathan Professor Department of Computer Science Jackson State University, Jackson, MS E-mail: natarajan.meghanathan@jsums.edu Networks or Graphs We typically
Graph Theory for Network Science Dr. Natarajan Meghanathan Professor Department of Computer Science Jackson State University, Jackson, MS E-mail: natarajan.meghanathan@jsums.edu Networks or Graphs We typically
CentiBiN Centralities in Biological Networks
 CentiBiN Centralities in Biological Networks Dirk Koschützki Research Group Network Analysis Leibniz Institute of Plant Genetics and Crop Plant Research, 06466 Gatersleben, Germany 23rd March 2006 1 Idea
CentiBiN Centralities in Biological Networks Dirk Koschützki Research Group Network Analysis Leibniz Institute of Plant Genetics and Crop Plant Research, 06466 Gatersleben, Germany 23rd March 2006 1 Idea
ALTERNATIVES TO BETWEENNESS CENTRALITY: A MEASURE OF CORRELATION COEFFICIENT
 ALTERNATIVES TO BETWEENNESS CENTRALITY: A MEASURE OF CORRELATION COEFFICIENT Xiaojia He 1 and Natarajan Meghanathan 2 1 University of Georgia, GA, USA, 2 Jackson State University, MS, USA 2 natarajan.meghanathan@jsums.edu
ALTERNATIVES TO BETWEENNESS CENTRALITY: A MEASURE OF CORRELATION COEFFICIENT Xiaojia He 1 and Natarajan Meghanathan 2 1 University of Georgia, GA, USA, 2 Jackson State University, MS, USA 2 natarajan.meghanathan@jsums.edu
1 Homophily and assortative mixing
 1 Homophily and assortative mixing Networks, and particularly social networks, often exhibit a property called homophily or assortative mixing, which simply means that the attributes of vertices correlate
1 Homophily and assortative mixing Networks, and particularly social networks, often exhibit a property called homophily or assortative mixing, which simply means that the attributes of vertices correlate
Localized Bridging Centrality for Distributed Network Analysis
 Localized Bridging Centrality for Distributed Network Analysis Soumendra Nanda Department of Computer Science Dartmouth College David Kotz Institute for Security Technology Studies Dartmouth College Abstract
Localized Bridging Centrality for Distributed Network Analysis Soumendra Nanda Department of Computer Science Dartmouth College David Kotz Institute for Security Technology Studies Dartmouth College Abstract
Graphs. Pseudograph: multiple edges and loops allowed
 Graphs G = (V, E) V - set of vertices, E - set of edges Undirected graphs Simple graph: V - nonempty set of vertices, E - set of unordered pairs of distinct vertices (no multiple edges or loops) Multigraph:
Graphs G = (V, E) V - set of vertices, E - set of edges Undirected graphs Simple graph: V - nonempty set of vertices, E - set of unordered pairs of distinct vertices (no multiple edges or loops) Multigraph:
Course Introduction / Review of Fundamentals of Graph Theory
 Course Introduction / Review of Fundamentals of Graph Theory Hiroki Sayama sayama@binghamton.edu Rise of Network Science (From Barabasi 2010) 2 Network models Many discrete parts involved Classic mean-field
Course Introduction / Review of Fundamentals of Graph Theory Hiroki Sayama sayama@binghamton.edu Rise of Network Science (From Barabasi 2010) 2 Network models Many discrete parts involved Classic mean-field
Properties of Biological Networks
 Properties of Biological Networks presented by: Ola Hamud June 12, 2013 Supervisor: Prof. Ron Pinter Based on: NETWORK BIOLOGY: UNDERSTANDING THE CELL S FUNCTIONAL ORGANIZATION By Albert-László Barabási
Properties of Biological Networks presented by: Ola Hamud June 12, 2013 Supervisor: Prof. Ron Pinter Based on: NETWORK BIOLOGY: UNDERSTANDING THE CELL S FUNCTIONAL ORGANIZATION By Albert-László Barabási
2. CONNECTIVITY Connectivity
 2. CONNECTIVITY 70 2. Connectivity 2.1. Connectivity. Definition 2.1.1. (1) A path in a graph G = (V, E) is a sequence of vertices v 0, v 1, v 2,..., v n such that {v i 1, v i } is an edge of G for i =
2. CONNECTIVITY 70 2. Connectivity 2.1. Connectivity. Definition 2.1.1. (1) A path in a graph G = (V, E) is a sequence of vertices v 0, v 1, v 2,..., v n such that {v i 1, v i } is an edge of G for i =
FastA & the chaining problem
 FastA & the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 1 Sources for this lecture: Lectures by Volker Heun, Daniel Huson and Knut Reinert,
FastA & the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 1 Sources for this lecture: Lectures by Volker Heun, Daniel Huson and Knut Reinert,
FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:
 FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:56 4001 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem
FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:56 4001 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem
Centralities (4) By: Ralucca Gera, NPS. Excellence Through Knowledge
 Centralities (4) By: Ralucca Gera, NPS Excellence Through Knowledge Some slide from last week that we didn t talk about in class: 2 PageRank algorithm Eigenvector centrality: i s Rank score is the sum
Centralities (4) By: Ralucca Gera, NPS Excellence Through Knowledge Some slide from last week that we didn t talk about in class: 2 PageRank algorithm Eigenvector centrality: i s Rank score is the sum
Preliminaries: networks and graphs
 978--52-8795-7 - Dynamical Processes on Complex Networks Preliminaries: networks and graphs In this chapter we introduce the reader to the basic definitions of network and graph theory. We define metrics
978--52-8795-7 - Dynamical Processes on Complex Networks Preliminaries: networks and graphs In this chapter we introduce the reader to the basic definitions of network and graph theory. We define metrics
Summary: What We Have Learned So Far
 Summary: What We Have Learned So Far small-world phenomenon Real-world networks: { Short path lengths High clustering Broad degree distributions, often power laws P (k) k γ Erdös-Renyi model: Short path
Summary: What We Have Learned So Far small-world phenomenon Real-world networks: { Short path lengths High clustering Broad degree distributions, often power laws P (k) k γ Erdös-Renyi model: Short path
arxiv:cond-mat/ v1 [cond-mat.other] 2 Feb 2004
![arxiv:cond-mat/ v1 [cond-mat.other] 2 Feb 2004 arxiv:cond-mat/ v1 [cond-mat.other] 2 Feb 2004](/thumbs/93/112340418.jpg) A measure of centrality based on the network efficiency arxiv:cond-mat/0402050v1 [cond-mat.other] 2 Feb 2004 Vito Latora a and Massimo Marchiori b,c a Dipartimento di Fisica e Astronomia, Università di
A measure of centrality based on the network efficiency arxiv:cond-mat/0402050v1 [cond-mat.other] 2 Feb 2004 Vito Latora a and Massimo Marchiori b,c a Dipartimento di Fisica e Astronomia, Università di
Graph Theory and Network Measurment
 Graph Theory and Network Measurment Social and Economic Networks MohammadAmin Fazli Social and Economic Networks 1 ToC Network Representation Basic Graph Theory Definitions (SE) Network Statistics and
Graph Theory and Network Measurment Social and Economic Networks MohammadAmin Fazli Social and Economic Networks 1 ToC Network Representation Basic Graph Theory Definitions (SE) Network Statistics and
Structure of biological networks. Presentation by Atanas Kamburov
 Structure of biological networks Presentation by Atanas Kamburov Seminar Gute Ideen in der theoretischen Biologie / Systembiologie 08.05.2007 Overview Motivation Definitions Large-scale properties of cellular
Structure of biological networks Presentation by Atanas Kamburov Seminar Gute Ideen in der theoretischen Biologie / Systembiologie 08.05.2007 Overview Motivation Definitions Large-scale properties of cellular
Lecture 5: Graphs. Rajat Mittal. IIT Kanpur
 Lecture : Graphs Rajat Mittal IIT Kanpur Combinatorial graphs provide a natural way to model connections between different objects. They are very useful in depicting communication networks, social networks
Lecture : Graphs Rajat Mittal IIT Kanpur Combinatorial graphs provide a natural way to model connections between different objects. They are very useful in depicting communication networks, social networks
Basics of Network Analysis
 Basics of Network Analysis Hiroki Sayama sayama@binghamton.edu Graph = Network G(V, E): graph (network) V: vertices (nodes), E: edges (links) 1 Nodes = 1, 2, 3, 4, 5 2 3 Links = 12, 13, 15, 23,
Basics of Network Analysis Hiroki Sayama sayama@binghamton.edu Graph = Network G(V, E): graph (network) V: vertices (nodes), E: edges (links) 1 Nodes = 1, 2, 3, 4, 5 2 3 Links = 12, 13, 15, 23,
Centrality. Peter Hoff. 567 Statistical analysis of social networks. Statistics, University of Washington 1/36
 1/36 Centrality 567 Statistical analysis of social networks Peter Hoff Statistics, University of Washington 2/36 Centrality A common goal in SNA is to identify the central nodes of a network. What does
1/36 Centrality 567 Statistical analysis of social networks Peter Hoff Statistics, University of Washington 2/36 Centrality A common goal in SNA is to identify the central nodes of a network. What does
Centralities for undirected graphs
 Chapter 4 Centralities for undirected graphs The next step in network analysis is to define, using mathematical formalisms, the different features we want to compute. So we present indexes, called centralities,
Chapter 4 Centralities for undirected graphs The next step in network analysis is to define, using mathematical formalisms, the different features we want to compute. So we present indexes, called centralities,
Lectures by Volker Heun, Daniel Huson and Knut Reinert, in particular last years lectures
 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 4.1 Sources for this lecture Lectures by Volker Heun, Daniel Huson and Knut
4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 4.1 Sources for this lecture Lectures by Volker Heun, Daniel Huson and Knut
BACKGROUND: A BRIEF INTRODUCTION TO GRAPH THEORY
 BACKGROUND: A BRIEF INTRODUCTION TO GRAPH THEORY General definitions; Representations; Graph Traversals; Topological sort; Graphs definitions & representations Graph theory is a fundamental tool in sparse
BACKGROUND: A BRIEF INTRODUCTION TO GRAPH THEORY General definitions; Representations; Graph Traversals; Topological sort; Graphs definitions & representations Graph theory is a fundamental tool in sparse
Introduction aux Systèmes Collaboratifs Multi-Agents
 M1 EEAII - Découverte de la Recherche (ViRob) Introduction aux Systèmes Collaboratifs Multi-Agents UPJV, Département EEA Fabio MORBIDI Laboratoire MIS Équipe Perception et Robotique E-mail: fabio.morbidi@u-picardie.fr
M1 EEAII - Découverte de la Recherche (ViRob) Introduction aux Systèmes Collaboratifs Multi-Agents UPJV, Département EEA Fabio MORBIDI Laboratoire MIS Équipe Perception et Robotique E-mail: fabio.morbidi@u-picardie.fr
Analysis of Biological Networks. 1. Clustering 2. Random Walks 3. Finding paths
 Analysis of Biological Networks 1. Clustering 2. Random Walks 3. Finding paths Problem 1: Graph Clustering Finding dense subgraphs Applications Identification of novel pathways, complexes, other modules?
Analysis of Biological Networks 1. Clustering 2. Random Walks 3. Finding paths Problem 1: Graph Clustering Finding dense subgraphs Applications Identification of novel pathways, complexes, other modules?
Why is a power law interesting? 2. it begs a question about mechanism: How do networks come to have power-law degree distributions in the first place?
 you ve got the power Why is a power law interesting? 1. it is scale-free 2. it begs a question about mechanism: How do networks come to have power-law degree distributions in the first place? powerful
you ve got the power Why is a power law interesting? 1. it is scale-free 2. it begs a question about mechanism: How do networks come to have power-law degree distributions in the first place? powerful
Structured prediction using the network perceptron
 Structured prediction using the network perceptron Ta-tsen Soong Joint work with Stuart Andrews and Prof. Tony Jebara Motivation A lot of network-structured data Social networks Citation networks Biological
Structured prediction using the network perceptron Ta-tsen Soong Joint work with Stuart Andrews and Prof. Tony Jebara Motivation A lot of network-structured data Social networks Citation networks Biological
Introduction to network metrics
 Universitat Politècnica de Catalunya Version 0.5 Complex and Social Networks (2018-2019) Master in Innovation and Research in Informatics (MIRI) Instructors Argimiro Arratia, argimiro@cs.upc.edu, http://www.cs.upc.edu/~argimiro/
Universitat Politècnica de Catalunya Version 0.5 Complex and Social Networks (2018-2019) Master in Innovation and Research in Informatics (MIRI) Instructors Argimiro Arratia, argimiro@cs.upc.edu, http://www.cs.upc.edu/~argimiro/
THE KNOWLEDGE MANAGEMENT STRATEGY IN ORGANIZATIONS. Summer semester, 2016/2017
 THE KNOWLEDGE MANAGEMENT STRATEGY IN ORGANIZATIONS Summer semester, 2016/2017 SOCIAL NETWORK ANALYSIS: THEORY AND APPLICATIONS 1. A FEW THINGS ABOUT NETWORKS NETWORKS IN THE REAL WORLD There are four categories
THE KNOWLEDGE MANAGEMENT STRATEGY IN ORGANIZATIONS Summer semester, 2016/2017 SOCIAL NETWORK ANALYSIS: THEORY AND APPLICATIONS 1. A FEW THINGS ABOUT NETWORKS NETWORKS IN THE REAL WORLD There are four categories
The Generalized Topological Overlap Matrix in Biological Network Analysis
 The Generalized Topological Overlap Matrix in Biological Network Analysis Andy Yip, Steve Horvath Email: shorvath@mednet.ucla.edu Depts Human Genetics and Biostatistics, University of California, Los Angeles
The Generalized Topological Overlap Matrix in Biological Network Analysis Andy Yip, Steve Horvath Email: shorvath@mednet.ucla.edu Depts Human Genetics and Biostatistics, University of California, Los Angeles
Random Graph Model; parameterization 2
 Agenda Random Graphs Recap giant component and small world statistics problems: degree distribution and triangles Recall that a graph G = (V, E) consists of a set of vertices V and a set of edges E V V.
Agenda Random Graphs Recap giant component and small world statistics problems: degree distribution and triangles Recall that a graph G = (V, E) consists of a set of vertices V and a set of edges E V V.
Lesson 4. Random graphs. Sergio Barbarossa. UPC - Barcelona - July 2008
 Lesson 4 Random graphs Sergio Barbarossa Graph models 1. Uncorrelated random graph (Erdős, Rényi) N nodes are connected through n edges which are chosen randomly from the possible configurations 2. Binomial
Lesson 4 Random graphs Sergio Barbarossa Graph models 1. Uncorrelated random graph (Erdős, Rényi) N nodes are connected through n edges which are chosen randomly from the possible configurations 2. Binomial
A New Measure of Linkage Between Two Sub-networks 1
 CONNECTIONS 26(1): 62-70 2004 INSNA http://www.insna.org/connections-web/volume26-1/6.flom.pdf A New Measure of Linkage Between Two Sub-networks 1 2 Peter L. Flom, Samuel R. Friedman, Shiela Strauss, Alan
CONNECTIONS 26(1): 62-70 2004 INSNA http://www.insna.org/connections-web/volume26-1/6.flom.pdf A New Measure of Linkage Between Two Sub-networks 1 2 Peter L. Flom, Samuel R. Friedman, Shiela Strauss, Alan
A SOCIAL NETWORK ANALYSIS APPROACH TO ANALYZE ROAD NETWORKS INTRODUCTION
 A SOCIAL NETWORK ANALYSIS APPROACH TO ANALYZE ROAD NETWORKS Kyoungjin Park Alper Yilmaz Photogrammetric and Computer Vision Lab Ohio State University park.764@osu.edu yilmaz.15@osu.edu ABSTRACT Depending
A SOCIAL NETWORK ANALYSIS APPROACH TO ANALYZE ROAD NETWORKS Kyoungjin Park Alper Yilmaz Photogrammetric and Computer Vision Lab Ohio State University park.764@osu.edu yilmaz.15@osu.edu ABSTRACT Depending
Locality-sensitive hashing and biological network alignment
 Locality-sensitive hashing and biological network alignment Laura LeGault - University of Wisconsin, Madison 12 May 2008 Abstract Large biological networks contain much information about the functionality
Locality-sensitive hashing and biological network alignment Laura LeGault - University of Wisconsin, Madison 12 May 2008 Abstract Large biological networks contain much information about the functionality
Characterizing Graphs (3) Characterizing Graphs (1) Characterizing Graphs (2) Characterizing Graphs (4)
 S-72.2420/T-79.5203 Basic Concepts 1 S-72.2420/T-79.5203 Basic Concepts 3 Characterizing Graphs (1) Characterizing Graphs (3) Characterizing a class G by a condition P means proving the equivalence G G
S-72.2420/T-79.5203 Basic Concepts 1 S-72.2420/T-79.5203 Basic Concepts 3 Characterizing Graphs (1) Characterizing Graphs (3) Characterizing a class G by a condition P means proving the equivalence G G
Networks in economics and finance. Lecture 1 - Measuring networks
 Networks in economics and finance Lecture 1 - Measuring networks What are networks and why study them? A network is a set of items (nodes) connected by edges or links. Units (nodes) Individuals Firms Banks
Networks in economics and finance Lecture 1 - Measuring networks What are networks and why study them? A network is a set of items (nodes) connected by edges or links. Units (nodes) Individuals Firms Banks
Some Graph Theory for Network Analysis. CS 249B: Science of Networks Week 01: Thursday, 01/31/08 Daniel Bilar Wellesley College Spring 2008
 Some Graph Theory for Network Analysis CS 9B: Science of Networks Week 0: Thursday, 0//08 Daniel Bilar Wellesley College Spring 008 Goals this lecture Introduce you to some jargon what we call things in
Some Graph Theory for Network Analysis CS 9B: Science of Networks Week 0: Thursday, 0//08 Daniel Bilar Wellesley College Spring 008 Goals this lecture Introduce you to some jargon what we call things in
Graph. Vertex. edge. Directed Graph. Undirected Graph
 Module : Graphs Dr. Natarajan Meghanathan Professor of Computer Science Jackson State University Jackson, MS E-mail: natarajan.meghanathan@jsums.edu Graph Graph is a data structure that is a collection
Module : Graphs Dr. Natarajan Meghanathan Professor of Computer Science Jackson State University Jackson, MS E-mail: natarajan.meghanathan@jsums.edu Graph Graph is a data structure that is a collection
Module 11. Directed Graphs. Contents
 Module 11 Directed Graphs Contents 11.1 Basic concepts......................... 256 Underlying graph of a digraph................ 257 Out-degrees and in-degrees.................. 258 Isomorphism..........................
Module 11 Directed Graphs Contents 11.1 Basic concepts......................... 256 Underlying graph of a digraph................ 257 Out-degrees and in-degrees.................. 258 Isomorphism..........................
1 More configuration model
 1 More configuration model In the last lecture, we explored the definition of the configuration model, a simple method for drawing networks from the ensemble, and derived some of its mathematical properties.
1 More configuration model In the last lecture, we explored the definition of the configuration model, a simple method for drawing networks from the ensemble, and derived some of its mathematical properties.
Varying Applications (examples)
 Graph Theory Varying Applications (examples) Computer networks Distinguish between two chemical compounds with the same molecular formula but different structures Solve shortest path problems between cities
Graph Theory Varying Applications (examples) Computer networks Distinguish between two chemical compounds with the same molecular formula but different structures Solve shortest path problems between cities
Introduction to Mathematical Programming IE406. Lecture 16. Dr. Ted Ralphs
 Introduction to Mathematical Programming IE406 Lecture 16 Dr. Ted Ralphs IE406 Lecture 16 1 Reading for This Lecture Bertsimas 7.1-7.3 IE406 Lecture 16 2 Network Flow Problems Networks are used to model
Introduction to Mathematical Programming IE406 Lecture 16 Dr. Ted Ralphs IE406 Lecture 16 1 Reading for This Lecture Bertsimas 7.1-7.3 IE406 Lecture 16 2 Network Flow Problems Networks are used to model
Centrality Estimation in Large Networks
 Centrality Estimation in Large Networks Ulrik Brandes Christian Pich Department of Computer and Information Science University of Konstanz August 18, 26 Abstract Centrality indices are an essential concept
Centrality Estimation in Large Networks Ulrik Brandes Christian Pich Department of Computer and Information Science University of Konstanz August 18, 26 Abstract Centrality indices are an essential concept
CSE101: Design and Analysis of Algorithms. Ragesh Jaiswal, CSE, UCSD
 Recap. Growth rates: Arrange the following functions in ascending order of growth rate: n 2 log n n log n 2 log n n/ log n n n Introduction Algorithm: A step-by-step way of solving a problem. Design of
Recap. Growth rates: Arrange the following functions in ascending order of growth rate: n 2 log n n log n 2 log n n/ log n n n Introduction Algorithm: A step-by-step way of solving a problem. Design of
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, This lecture is based on the following papers, which are all recommended reading:
 24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
Critical Phenomena in Complex Networks
 Critical Phenomena in Complex Networks Term essay for Physics 563: Phase Transitions and the Renormalization Group University of Illinois at Urbana-Champaign Vikyath Deviprasad Rao 11 May 2012 Abstract
Critical Phenomena in Complex Networks Term essay for Physics 563: Phase Transitions and the Renormalization Group University of Illinois at Urbana-Champaign Vikyath Deviprasad Rao 11 May 2012 Abstract
V 1 Introduction! Mon, Oct 15, 2012! Bioinformatics 3 Volkhard Helms!
 V 1 Introduction! Mon, Oct 15, 2012! Bioinformatics 3 Volkhard Helms! How Does a Cell Work?! A cell is a crowded environment! => many different proteins,! metabolites, compartments,! On a microscopic level!
V 1 Introduction! Mon, Oct 15, 2012! Bioinformatics 3 Volkhard Helms! How Does a Cell Work?! A cell is a crowded environment! => many different proteins,! metabolites, compartments,! On a microscopic level!
V4 Matrix algorithms and graph partitioning
 V4 Matrix algorithms and graph partitioning - Community detection - Simple modularity maximization - Spectral modularity maximization - Division into more than two groups - Other algorithms for community
V4 Matrix algorithms and graph partitioning - Community detection - Simple modularity maximization - Spectral modularity maximization - Division into more than two groups - Other algorithms for community
Chapter 10. Fundamental Network Algorithms. M. E. J. Newman. May 6, M. E. J. Newman Chapter 10 May 6, / 33
 Chapter 10 Fundamental Network Algorithms M. E. J. Newman May 6, 2015 M. E. J. Newman Chapter 10 May 6, 2015 1 / 33 Table of Contents 1 Algorithms for Degrees and Degree Distributions Degree-Degree Correlation
Chapter 10 Fundamental Network Algorithms M. E. J. Newman May 6, 2015 M. E. J. Newman Chapter 10 May 6, 2015 1 / 33 Table of Contents 1 Algorithms for Degrees and Degree Distributions Degree-Degree Correlation
Centrality Book. cohesion.
 Cohesion The graph-theoretic terms discussed in the previous chapter have very specific and concrete meanings which are highly shared across the field of graph theory and other fields like social network
Cohesion The graph-theoretic terms discussed in the previous chapter have very specific and concrete meanings which are highly shared across the field of graph theory and other fields like social network
Paths, Circuits, and Connected Graphs
 Paths, Circuits, and Connected Graphs Paths and Circuits Definition: Let G = (V, E) be an undirected graph, vertices u, v V A path of length n from u to v is a sequence of edges e i = {u i 1, u i} E for
Paths, Circuits, and Connected Graphs Paths and Circuits Definition: Let G = (V, E) be an undirected graph, vertices u, v V A path of length n from u to v is a sequence of edges e i = {u i 1, u i} E for
Graph Theory S 1 I 2 I 1 S 2 I 1 I 2
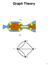 Graph Theory S I I S S I I S Graphs Definition A graph G is a pair consisting of a vertex set V (G), and an edge set E(G) ( ) V (G). x and y are the endpoints of edge e = {x, y}. They are called adjacent
Graph Theory S I I S S I I S Graphs Definition A graph G is a pair consisting of a vertex set V (G), and an edge set E(G) ( ) V (G). x and y are the endpoints of edge e = {x, y}. They are called adjacent
Signal Processing for Big Data
 Signal Processing for Big Data Sergio Barbarossa 1 Summary 1. Networks 2.Algebraic graph theory 3. Random graph models 4. OperaGons on graphs 2 Networks The simplest way to represent the interaction between
Signal Processing for Big Data Sergio Barbarossa 1 Summary 1. Networks 2.Algebraic graph theory 3. Random graph models 4. OperaGons on graphs 2 Networks The simplest way to represent the interaction between
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1. Database Searching
 COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Graph Theory. Graph Theory. COURSE: Introduction to Biological Networks. Euler s Solution LECTURE 1: INTRODUCTION TO NETWORKS.
 Graph Theory COURSE: Introduction to Biological Networks LECTURE 1: INTRODUCTION TO NETWORKS Arun Krishnan Koenigsberg, Russia Is it possible to walk with a route that crosses each bridge exactly once,
Graph Theory COURSE: Introduction to Biological Networks LECTURE 1: INTRODUCTION TO NETWORKS Arun Krishnan Koenigsberg, Russia Is it possible to walk with a route that crosses each bridge exactly once,
Introduction to Graph Theory
 Introduction to Graph Theory Tandy Warnow January 20, 2017 Graphs Tandy Warnow Graphs A graph G = (V, E) is an object that contains a vertex set V and an edge set E. We also write V (G) to denote the vertex
Introduction to Graph Theory Tandy Warnow January 20, 2017 Graphs Tandy Warnow Graphs A graph G = (V, E) is an object that contains a vertex set V and an edge set E. We also write V (G) to denote the vertex
Local higher-order graph clustering
 Local higher-order graph clustering Hao Yin Stanford University yinh@stanford.edu Austin R. Benson Cornell University arb@cornell.edu Jure Leskovec Stanford University jure@cs.stanford.edu David F. Gleich
Local higher-order graph clustering Hao Yin Stanford University yinh@stanford.edu Austin R. Benson Cornell University arb@cornell.edu Jure Leskovec Stanford University jure@cs.stanford.edu David F. Gleich
Centralities (4) Ralucca Gera, Applied Mathematics Dept. Naval Postgraduate School Monterey, California Excellence Through Knowledge
 Centralities (4) Ralucca Gera, Applied Mathematics Dept. Naval Postgraduate School Monterey, California rgera@nps.edu Excellence Through Knowledge Different types of centralities: Betweenness Centrality
Centralities (4) Ralucca Gera, Applied Mathematics Dept. Naval Postgraduate School Monterey, California rgera@nps.edu Excellence Through Knowledge Different types of centralities: Betweenness Centrality
CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment
 CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment Courtesy of jalview 1 Motivations Collective statistic Protein families Identification and representation of conserved sequence features
CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment Courtesy of jalview 1 Motivations Collective statistic Protein families Identification and representation of conserved sequence features
Social Network Analysis
 Social Network Analysis Mathematics of Networks Manar Mohaisen Department of EEC Engineering Adjacency matrix Network types Edge list Adjacency list Graph representation 2 Adjacency matrix Adjacency matrix
Social Network Analysis Mathematics of Networks Manar Mohaisen Department of EEC Engineering Adjacency matrix Network types Edge list Adjacency list Graph representation 2 Adjacency matrix Adjacency matrix
Unit 2: Graphs and Matrices. ICPSR University of Michigan, Ann Arbor Summer 2015 Instructor: Ann McCranie
 Unit 2: Graphs and Matrices ICPSR University of Michigan, Ann Arbor Summer 2015 Instructor: Ann McCranie Four main ways to notate a social network There are a variety of ways to mathematize a social network,
Unit 2: Graphs and Matrices ICPSR University of Michigan, Ann Arbor Summer 2015 Instructor: Ann McCranie Four main ways to notate a social network There are a variety of ways to mathematize a social network,
Social Networks. Slides by : I. Koutsopoulos (AUEB), Source:L. Adamic, SN Analysis, Coursera course
 Social Networks Slides by : I. Koutsopoulos (AUEB), Source:L. Adamic, SN Analysis, Coursera course Introduction Political blogs Organizations Facebook networks Ingredient networks SN representation Networks
Social Networks Slides by : I. Koutsopoulos (AUEB), Source:L. Adamic, SN Analysis, Coursera course Introduction Political blogs Organizations Facebook networks Ingredient networks SN representation Networks
Weighted networks. 9.1 The strength of weak ties
 Weighted networks 9 In this lecture we consider networks in which links differ from each other. The links are made individual by ascribing a positive number a weight to each of them. These weighted networks
Weighted networks 9 In this lecture we consider networks in which links differ from each other. The links are made individual by ascribing a positive number a weight to each of them. These weighted networks
W4231: Analysis of Algorithms
 W4231: Analysis of Algorithms 10/21/1999 Definitions for graphs Breadth First Search and Depth First Search Topological Sort. Graphs AgraphG is given by a set of vertices V and a set of edges E. Normally
W4231: Analysis of Algorithms 10/21/1999 Definitions for graphs Breadth First Search and Depth First Search Topological Sort. Graphs AgraphG is given by a set of vertices V and a set of edges E. Normally
A PRIME FACTOR THEOREM FOR A GENERALIZED DIRECT PRODUCT
 Discussiones Mathematicae Graph Theory 26 (2006 ) 135 140 A PRIME FACTOR THEOREM FOR A GENERALIZED DIRECT PRODUCT Wilfried Imrich Department of Mathematics and Information Technology Montanuniversität
Discussiones Mathematicae Graph Theory 26 (2006 ) 135 140 A PRIME FACTOR THEOREM FOR A GENERALIZED DIRECT PRODUCT Wilfried Imrich Department of Mathematics and Information Technology Montanuniversität
Extracting Information from Complex Networks
 Extracting Information from Complex Networks 1 Complex Networks Networks that arise from modeling complex systems: relationships Social networks Biological networks Distinguish from random networks uniform
Extracting Information from Complex Networks 1 Complex Networks Networks that arise from modeling complex systems: relationships Social networks Biological networks Distinguish from random networks uniform
Supplementary material to Epidemic spreading on complex networks with community structures
 Supplementary material to Epidemic spreading on complex networks with community structures Clara Stegehuis, Remco van der Hofstad, Johan S. H. van Leeuwaarden Supplementary otes Supplementary ote etwork
Supplementary material to Epidemic spreading on complex networks with community structures Clara Stegehuis, Remco van der Hofstad, Johan S. H. van Leeuwaarden Supplementary otes Supplementary ote etwork
TELCOM2125: Network Science and Analysis
 School of Information Sciences University of Pittsburgh TELCOM2125: Network Science and Analysis Konstantinos Pelechrinis Spring 2015 2 Part 4: Dividing Networks into Clusters The problem l Graph partitioning
School of Information Sciences University of Pittsburgh TELCOM2125: Network Science and Analysis Konstantinos Pelechrinis Spring 2015 2 Part 4: Dividing Networks into Clusters The problem l Graph partitioning
Data mining --- mining graphs
 Data mining --- mining graphs University of South Florida Xiaoning Qian Today s Lecture 1. Complex networks 2. Graph representation for networks 3. Markov chain 4. Viral propagation 5. Google s PageRank
Data mining --- mining graphs University of South Florida Xiaoning Qian Today s Lecture 1. Complex networks 2. Graph representation for networks 3. Markov chain 4. Viral propagation 5. Google s PageRank
On subgraphs of Cartesian product graphs
 On subgraphs of Cartesian product graphs Sandi Klavžar 1 Department of Mathematics, PEF, University of Maribor, Koroška cesta 160, 2000 Maribor, Slovenia sandi.klavzar@uni-lj.si Alenka Lipovec Department
On subgraphs of Cartesian product graphs Sandi Klavžar 1 Department of Mathematics, PEF, University of Maribor, Koroška cesta 160, 2000 Maribor, Slovenia sandi.klavzar@uni-lj.si Alenka Lipovec Department
On Page Rank. 1 Introduction
 On Page Rank C. Hoede Faculty of Electrical Engineering, Mathematics and Computer Science University of Twente P.O.Box 217 7500 AE Enschede, The Netherlands Abstract In this paper the concept of page rank
On Page Rank C. Hoede Faculty of Electrical Engineering, Mathematics and Computer Science University of Twente P.O.Box 217 7500 AE Enschede, The Netherlands Abstract In this paper the concept of page rank
MCL. (and other clustering algorithms) 858L
 MCL (and other clustering algorithms) 858L Comparing Clustering Algorithms Brohee and van Helden (2006) compared 4 graph clustering algorithms for the task of finding protein complexes: MCODE RNSC Restricted
MCL (and other clustering algorithms) 858L Comparing Clustering Algorithms Brohee and van Helden (2006) compared 4 graph clustering algorithms for the task of finding protein complexes: MCODE RNSC Restricted
Online Social Networks and Media
 Online Social Networks and Media Absorbing Random Walks Link Prediction Why does the Power Method work? If a matrix R is real and symmetric, it has real eigenvalues and eigenvectors: λ, w, λ 2, w 2,, (λ
Online Social Networks and Media Absorbing Random Walks Link Prediction Why does the Power Method work? If a matrix R is real and symmetric, it has real eigenvalues and eigenvectors: λ, w, λ 2, w 2,, (λ
A graph is finite if its vertex set and edge set are finite. We call a graph with just one vertex trivial and all other graphs nontrivial.
 2301-670 Graph theory 1.1 What is a graph? 1 st semester 2550 1 1.1. What is a graph? 1.1.2. Definition. A graph G is a triple (V(G), E(G), ψ G ) consisting of V(G) of vertices, a set E(G), disjoint from
2301-670 Graph theory 1.1 What is a graph? 1 st semester 2550 1 1.1. What is a graph? 1.1.2. Definition. A graph G is a triple (V(G), E(G), ψ G ) consisting of V(G) of vertices, a set E(G), disjoint from
A new general family of deterministic hierarchical networks
 A new general family of deterministic hierarchical networks arxiv:1608.02197v1 [math.co] 7 Aug 2016 C. Dalfó a, M.A. Fiol a,b a Departament de Matemàtica Aplicada IV Universitat Politècnica de Catalunya
A new general family of deterministic hierarchical networks arxiv:1608.02197v1 [math.co] 7 Aug 2016 C. Dalfó a, M.A. Fiol a,b a Departament de Matemàtica Aplicada IV Universitat Politècnica de Catalunya
Bioinformatics explained: BLAST. March 8, 2007
 Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Mathematics of Networks II
 Mathematics of Networks II 26.10.2016 1 / 30 Definition of a network Our definition (Newman): A network (graph) is a collection of vertices (nodes) joined by edges (links). More precise definition (Bollobàs):
Mathematics of Networks II 26.10.2016 1 / 30 Definition of a network Our definition (Newman): A network (graph) is a collection of vertices (nodes) joined by edges (links). More precise definition (Bollobàs):
CS224W: Analysis of Networks Jure Leskovec, Stanford University
 CS224W: Analysis of Networks Jure Leskovec, Stanford University http://cs224w.stanford.edu 11/13/17 Jure Leskovec, Stanford CS224W: Analysis of Networks, http://cs224w.stanford.edu 2 Observations Models
CS224W: Analysis of Networks Jure Leskovec, Stanford University http://cs224w.stanford.edu 11/13/17 Jure Leskovec, Stanford CS224W: Analysis of Networks, http://cs224w.stanford.edu 2 Observations Models
CS473-Algorithms I. Lecture 13-A. Graphs. Cevdet Aykanat - Bilkent University Computer Engineering Department
 CS473-Algorithms I Lecture 3-A Graphs Graphs A directed graph (or digraph) G is a pair (V, E), where V is a finite set, and E is a binary relation on V The set V: Vertex set of G The set E: Edge set of
CS473-Algorithms I Lecture 3-A Graphs Graphs A directed graph (or digraph) G is a pair (V, E), where V is a finite set, and E is a binary relation on V The set V: Vertex set of G The set E: Edge set of
3.1 Basic Definitions and Applications. Chapter 3. Graphs. Undirected Graphs. Some Graph Applications
 Chapter 3 31 Basic Definitions and Applications Graphs Slides by Kevin Wayne Copyright 2005 Pearson-Addison Wesley All rights reserved 1 Undirected Graphs Some Graph Applications Undirected graph G = (V,
Chapter 3 31 Basic Definitions and Applications Graphs Slides by Kevin Wayne Copyright 2005 Pearson-Addison Wesley All rights reserved 1 Undirected Graphs Some Graph Applications Undirected graph G = (V,
Robustness of Centrality Measures for Small-World Networks Containing Systematic Error
 Robustness of Centrality Measures for Small-World Networks Containing Systematic Error Amanda Lannie Analytical Systems Branch, Air Force Research Laboratory, NY, USA Abstract Social network analysis is
Robustness of Centrality Measures for Small-World Networks Containing Systematic Error Amanda Lannie Analytical Systems Branch, Air Force Research Laboratory, NY, USA Abstract Social network analysis is
Chordal deletion is fixed-parameter tractable
 Chordal deletion is fixed-parameter tractable Dániel Marx Institut für Informatik, Humboldt-Universität zu Berlin, Unter den Linden 6, 10099 Berlin, Germany. dmarx@informatik.hu-berlin.de Abstract. It
Chordal deletion is fixed-parameter tractable Dániel Marx Institut für Informatik, Humboldt-Universität zu Berlin, Unter den Linden 6, 10099 Berlin, Germany. dmarx@informatik.hu-berlin.de Abstract. It
Spectral Clustering X I AO ZE N G + E L HA M TA BA S SI CS E CL A S S P R ESENTATION MA RCH 1 6,
 Spectral Clustering XIAO ZENG + ELHAM TABASSI CSE 902 CLASS PRESENTATION MARCH 16, 2017 1 Presentation based on 1. Von Luxburg, Ulrike. "A tutorial on spectral clustering." Statistics and computing 17.4
Spectral Clustering XIAO ZENG + ELHAM TABASSI CSE 902 CLASS PRESENTATION MARCH 16, 2017 1 Presentation based on 1. Von Luxburg, Ulrike. "A tutorial on spectral clustering." Statistics and computing 17.4
Simplicial Complexes of Networks and Their Statistical Properties
 Simplicial Complexes of Networks and Their Statistical Properties Slobodan Maletić, Milan Rajković*, and Danijela Vasiljević Institute of Nuclear Sciences Vinča, elgrade, Serbia *milanr@vin.bg.ac.yu bstract.
Simplicial Complexes of Networks and Their Statistical Properties Slobodan Maletić, Milan Rajković*, and Danijela Vasiljević Institute of Nuclear Sciences Vinča, elgrade, Serbia *milanr@vin.bg.ac.yu bstract.
Graph and Digraph Glossary
 1 of 15 31.1.2004 14:45 Graph and Digraph Glossary A B C D E F G H I-J K L M N O P-Q R S T U V W-Z Acyclic Graph A graph is acyclic if it contains no cycles. Adjacency Matrix A 0-1 square matrix whose
1 of 15 31.1.2004 14:45 Graph and Digraph Glossary A B C D E F G H I-J K L M N O P-Q R S T U V W-Z Acyclic Graph A graph is acyclic if it contains no cycles. Adjacency Matrix A 0-1 square matrix whose
Section 7.12: Similarity. By: Ralucca Gera, NPS
 Section 7.12: Similarity By: Ralucca Gera, NPS Motivation We talked about global properties Average degree, average clustering, ave path length We talked about local properties: Some node centralities
Section 7.12: Similarity By: Ralucca Gera, NPS Motivation We talked about global properties Average degree, average clustering, ave path length We talked about local properties: Some node centralities
