Genomics - Problem Set 2 Part 1 due Friday, 1/25/2019 by 9:00am Part 2 due Friday, 2/1/2019 by 9:00am
|
|
|
- Oswald Miller
- 5 years ago
- Views:
Transcription
1 Genomics - Part 1 due Friday, 1/25/2019 by 9:00am Part 2 due Friday, 2/1/2019 by 9:00am One major aspect of functional genomics is measuring the transcript abundance of all genes simultaneously. This was initially done using DNA microarrays, although RNA sequencing (RNAseq) and its variants are now widely used due to their greater reproducibility and utility in exploring other biological questions. For this problem set, you will use existing tools as well as your own custom scripts to extract biological information from a raw mrna expression dataset. Problem: You are exploring a budding yeast gene, YPL267W. You decide to explore published datasets to see how the relative abundance of RNA from YPL267W changes, and to see what other genes show similar patterns of expression, hoping to gain insight into the role of YPL267W. A standard method of defining groups of co-expressed genes in a gene expression dataset is hierarchical clustering. We have not covered clustering in class yet, but in general, clustering constructs a tree in which genes with similar expression patterns are correlated (or clustered together ) using either a neighbor joining or UPGMA-type algorithm applied to a distance matrix. If you would like more information, a good starting point is a paper by Eisen, et al. There are several software products that perform clustering; we will use a program written by Gavin Sherlock, called XCluster, which does centroid-based clustering (to be discussed in lecture). We will work with a data file that is a subset of the data described in: Spellman P.T., Sherlock, G., et al. (1998). Comprehensive identification of cell cycleregulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9(12): To do this problem set, you will use FarmShare servers (e.g. rice.stanford.edu). Use this tab-delimited input data file for your analysis: /afs/ir.stanford.edu/class/gene211/misc/cellcycle.pcl Start by making a copy of the.pcl to your own directory and examine it using any of the Unix utilities we have worked with (e.g. more or less ). The first row of this file (the header) contains the experiment names (here representing a single experimental replicate). The experiments described in this file compared yeast cells synchronized in different phases of the cell cycle with asynchronous yeast cells. The first two columns of the file contain the gene unique identifier (ORF systematic name) and the gene name (somethings called gene symbol), and the third column contains a gene weight, you will not need this for your analysis. The remaining columns contain the log2-transformed expression ratios (that is, 2-fold differences in RNA expression based on the amount of RNA observed) for that gene in the corresponding experiment.
2 Part 1 (due January 25): Question 1: Why is it convenient to represent the change in expression for each gene in each experiment as a log2 ratio as opposed to the raw expression measurements? Describe any mathematical and biological reasons why this might be advantageous. Cluster the cell cycle dataset by executing the XCluster program as follows (you must be in the directory containing your local copy of the cellcycle.pcl file, and you must be logged onto one of the Farmshare2 servers): /afs/ir.stanford.edu/class/gene211/bin/cluster -f cellcycle.pcl If you want more information about the clustering software or the file formats it uses see: Question 2: What metrics does XCluster allow you to use to determine the distances between genes? Which is used by default? Does it matter which one you use? If yes, explain and provide a concrete example of a case where there is a difference between the two methods. If no, explain why the choice of the distance metric does not matter. Executing the clustering command above produces two output files: cellcycle.cdt and cellcycle.gtr (both explained below). To view the data, download cellcycle.cdt and cellcycle.gtr to your laptop and open the cellcycle.cdt file with MapleTree. MapleTree is a tool available from For MacOS X, after you download, click on the MapleTree.jar file, probably in Downloads/MapleTree, while holding down the 'control' key. Select Open, and then in the resulting dialog box click Open. To load the.cdt file in MapleTree select Load -> Hierarchical Clustering Data -> Browse... -> Open -> Load. If you haven t previously used Java on your computer you may need to install a Java Development Kit (JDK). Using MapleTree to explore the data, you should see that plenty of genes show similar expression to your gene of interest, YPL267W. You can Find the gene, and then use the arrow keys to expand the set of genes in the detailed view. Similar to the analysis we performed in PS1, download the SGD features file to your laptop. /afs/ir.stanford.edu/class/gene211/misc/sgd_features.tab Now use Python to find information on genes with similar expression to YPL267W. Programming Exercise: Write a Python script, extract_info_better.py, that accepts as input any number of yeast gene names, feature names, gene aliases, or primary/secondary SGDIDs
3 (or a mixture thereof) on the command line and will extract information about those genes from the SGD_features.tab. The script s output should include the following information for each gene, one gene per line, in a tab-delimited format: 1 Primary SGDID (SGD s database identifier) 2 Feature Type 3 Feature Qualifier 4 Feature Name 5 Standard Gene Name 6 Aliases (when more than one, delimited by a character) 7 Parent feature name 8 Secondary SGDID (when more than one, delimited by the character) 9 Chromosome 10 Start coordinate 11 Stop coordinate 12 Strand (W or C) 13 Description As a reminder the SGD_features.tab file contains the following information: 1 Primary SGDID 2 Feature Type 3 Feature Qualifier* 4 Feature Name* 5 Standard Gene Name* 6 Alias* (when more than one, delimited by the character) 7 Parent Feature Name* 8 Secondary SGDID* (when more than one, delimited by the character) 9 Chromosome* 10 Start coordinate* 11 Stop coordinate* 12 Strand* 13 Genetic position* 14 Coordinate version* 15 Sequence version* 16 Description* * Within the file these fields are optional Algorithms and Methods: Your script should be executable on the command line as follows: python3 extract_info_better.py gene1 gene2 genen < SGD_features.tab
4 The < directs the contents of SGD_features.tab to your Python script via standard input (sys.stdin). In your script, access each line of SGD_features.tab within a for loop as follows: import sys for line in sys.stdin: # process str variable line Within this for loop, your program will read through SGD_features.tab one line at a time, and the loop will exit when there are no more lines to be read, i.e. the end of the file. We covered how to split a str variable based on the tab character in Problem Set 1, which you can re-use. You will also need to further split the alias and secondary SGDID fields based on the vertical bar character ( ), use the follows. # convert str aliases into list aliases_list aliases_list = aliases.split(" ") You can now check each line of the SGD_features.tab file for each of the gene IDs in the input list from the command line. The command line arguments are found in the list slice sys.argv[1:] gene_names = sys.argv[1:] Remember that many genes have similar names and some gene names may be subsets of other gene names as is found sometimes with alias names, please design your code accordingly. Be sure to make your gene matching case-insensitive. Note that in SGD_features.tab for each protein coding genes, open-reading frame (ORF), there are a minimum of two lines, one for the ORF (analogous to a gene in the yeast world), and one for the coding sequence (CDS, a.k.a. the exonic portion). In the case of spliced genes there will be more than one CDS, as well as one or more introns. It is most useful to print out only the top-level part of the feature these have a parent feature name that beings with chromosome such as chromosome 1. To test whether a string begins with chromosome, you can use the startswith() method of the string object in this fashion, e.g. for the str variable named parent in PS1: if parent.startswith("chromosome"): # something useful Finally, save the output of your program to a file. Use command line redirection as covered in PS0 and PS1. Additionally, your script should be case-insensitive this means that the terms cdc6 and CDC6 should be treated identically so that the user does not have to worry about the correct casing of any gene identifier. Finally, your code should be well commented.
5 Submission of Part 1 (due January 25): Write out answers to Questions 1 and 2 above in a text file called README and submit this with your script, extract_info_better.py. You should also submit a file, output.txt, generated by your script with the following input identifiers (all on one command line): S L YAL030W TRN1 ssu21 ZRG11 CBP80 SUT1 PAT1 YPK1 YIH1 CDL13 ARS309 YNL067W-B R0010W LSR1 Put all three files in a temporary directory on FarmShare and run the following script. /afs/ir.stanford.edu/class/gene211/bin/submit.pl Part 2 (due February 1): The program you wrote in Part 1 proves useful, but it needs to be augmented. As your analysis continues you find yourself looking at lots of datasets, and then cluster after cluster in a viewer, typing many gene names on the command line, and iterating for your search of all genes in the experiments of interest. Apart from being error prone this process becomes quite tedious. It is important to be consistent from one dataset to the next, so that your definition of similar expression doesn t change over time. You realize you can take advantage of the structure of the cluster itself you re really interested in extracting sub-clusters that include your gene of interest. There is a better way than looking at each cluster by hand. Examine the output files that were created earlier, the cluster data file cellcycle.cdt and the gene tree file cellcycle.gtr. The.cdt file is in the same format as the.pcl file except: (1) In the.cdt file, the genes (rows) are ordered such that genes that cluster together are adjacent (however note that at the boundary of two clusters, genes on adjacent rows are not necessarily clustered together) (2) There is an additional column at the start of each row with a new (unique) gene identifier. The.gtr file consists of a number of rows, each of which describes a node. This is an example tree and its corresponding.gtr file information.
6 The first column is the name of a node. The next two columns are the children (branches) of that node. In this case, NODE2X has two branches, one leading to the gene (leaf) GENE1X and the other leading to another node, NODE1X. The last column is the level of correlation between the data of GENE1X and the (average) data of NODE1X, 0.6. The columns are tab-delimited.
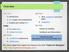
7 Goal of Part 2: You resolve to write a script named parse_cluster.py that takes the output from XCluster, for the gene (YPL267W) and a correlation cutoff (0.86) as input and prints out all genes (i.e., their ORF name) that cluster with the input gene up to the input correlation cutoff. Algorithms and Methods: Your script should be designed to execute like this from the command line on FarmShare: python3 parse_cluster.py cellcycle.cdt cellcycle.gtr gene_name cutoff_value In this program, you will have two files open for reading. To do this, you cannot read both files using standard input like was done in Part 1. In order to have two files open, you use file I/O. An example of this is shown below for the cdt file: with open("cellcycle.cdt", "r") as cdt_file: for line in cdt_file: # process line NOTE: The problem outlined in this exercise can be solved using a variety of algorithms and implementations of those algorithms feel free to explore any possibilities that you like, with the only requirements being: Your script must use sys.argv to read all command-line parameters Your script must include at least one function. The function must be passed at least one argument, return at least one value, and must be used directly by your script to identify the gene(s) that cluster with the gene of interest Your program should write the output using the print() function or similar (ensure the output goes to stdout) for easy command line redirection. Within the output there is only one gene name per line. Also generate an output file called coexpressed_genes.txt that contains just the ORF name for each of the genes, one name per line. We supply a description of one possible algorithm at the end of this document, which uses two functions. You may ignore it and design your own algorithm. Finally, in addition to answering the questions below, use your script to find all genes in the cluster that are co-expressed with ORF YPL267W with a correlation greater than Note: You are to consider the highest order branches that have correlations greater than 0.86; i.e. if a higher order node has a correlation >.86 and a lower order node has a correlation =<.86, still consider the higher order node and all of its sub nodes to be clustered together as in case C below. There should be about 40 genes listed in an output file called coexpressed_genes.txt. Consider these example images:
8 In the cases above, we want to return all of the genes highlighted in red if the input is Gene 1 and the cutoff is In case A, return all three genes connected to nodes with correlations >0.86. In case B, no node above Gene 1 has a correlation higher than the cutoff, so we should only return Gene 1. In case C, the highest order node with correlation greater than 0.86 contains genes 1 through 4. We should return Genes 1 through 4, even though there is a lower order node with a correlation below the cutoff. Question 3: Use the GO::TermFinder tool provided by SGD to determine information about the localization of the genes co-expressed with ORF YPL267W. Based on the Gene Ontology annotations, which Cellular Component is enriched for these genes? What do the differentcolored boxes displayed by GO::TermFinder represent? The URL for this tool is: For more information on GO::TermFinder check out: Note: It may take the GO::TermFinder a minute or two to return its results depending on the size of the gene list and the number of annotations associated with those genes.
9 Question 4: Now that you know where the gene products tend to be located in the cell, repeat the search using the Biological Process aspect of GO. Why are the p-values so different between the two ontologies? How are the p-values calculated? What is the difference between the p-value and the False Discovery Rate (FDR)? Now with the list of co-expressed genes and with some clues as to the biological processes in which these genes participate, you would like to learn more about the regulation of these genes. Question 5: Consider the experiments that were used to generate the data for the list of genes along with your clustering results. What type of gene regulation would explain the co-expression of those genes? For what biological reasons might these genes be clustered together? Submission of Part 2 (due February 1): Submit your answers to the questions in a file named README. Also submit parse_cluster.py and coexpressed_genes.txt. These files can be submitted by putting them all in a temporary directory and running the following script, then following the instructions: /afs/ir.stanford.edu/class/gene211/bin/submit.pl Algorithm Hints, Traversing the Gene Cluster Tree 1. Read in the cdt file. Create a dictionary indexing the UID identifier (e.g. YDL037C) with the XCluster generated GID (e.g. GENE93X). 2. Read in the gtr file. For each node and gene in the file make an entry into a parent dictionary with the node ID or gene ID as the key and the node ID of its parent node as the value. Create two children dictionaries, where each node ID is entered as a key in both children dictionaries and indexed with the other IDs (every node should have exactly two children, which are either another node or a gene). You will also want to store the correlations within another dictionary. 3. To find the co-expressed genes, you should now be able to write two simple functions. The first will find the node ID for the highest order node on the tree above a gene where the correlation is above an arbitrary cutoff. The second will find all gene IDs for the constituents of a given node. You must then find a way to combine the use of these functions to solve the problem. Note: This is a naturally recursive problem, but you may also be able to solve it without recursion if that's more intuitive to you. Wikipedia has some good information about recursion and tree traversal:
Genomics - Problem Set 2 Part 1 due Friday, 1/26/2018 by 9:00am Part 2 due Friday, 2/2/2018 by 9:00am
 Genomics - Part 1 due Friday, 1/26/2018 by 9:00am Part 2 due Friday, 2/2/2018 by 9:00am One major aspect of functional genomics is measuring the transcript abundance of all genes simultaneously. This was
Genomics - Part 1 due Friday, 1/26/2018 by 9:00am Part 2 due Friday, 2/2/2018 by 9:00am One major aspect of functional genomics is measuring the transcript abundance of all genes simultaneously. This was
Contents. ! Data sets. ! Distance and similarity metrics. ! K-means clustering. ! Hierarchical clustering. ! Evaluation of clustering results
 Statistical Analysis of Microarray Data Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation of clustering results Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be
Statistical Analysis of Microarray Data Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation of clustering results Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be
Clustering Jacques van Helden
 Statistical Analysis of Microarray Data Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation
Statistical Analysis of Microarray Data Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation
Supervised Clustering of Yeast Gene Expression Data
 Supervised Clustering of Yeast Gene Expression Data In the DeRisi paper five expression profile clusters were cited, each containing a small number (7-8) of genes. In the following examples we apply supervised
Supervised Clustering of Yeast Gene Expression Data In the DeRisi paper five expression profile clusters were cited, each containing a small number (7-8) of genes. In the following examples we apply supervised
CompClustTk Manual & Tutorial
 CompClustTk Manual & Tutorial Brandon King Copyright c California Institute of Technology Version 0.1.10 May 13, 2004 Contents 1 Introduction 1 1.1 Purpose.............................................
CompClustTk Manual & Tutorial Brandon King Copyright c California Institute of Technology Version 0.1.10 May 13, 2004 Contents 1 Introduction 1 1.1 Purpose.............................................
Tutorial:OverRepresentation - OpenTutorials
 Tutorial:OverRepresentation From OpenTutorials Slideshow OverRepresentation (about 12 minutes) (http://opentutorials.rbvi.ucsf.edu/index.php?title=tutorial:overrepresentation& ce_slide=true&ce_style=cytoscape)
Tutorial:OverRepresentation From OpenTutorials Slideshow OverRepresentation (about 12 minutes) (http://opentutorials.rbvi.ucsf.edu/index.php?title=tutorial:overrepresentation& ce_slide=true&ce_style=cytoscape)
Tutorial: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and Expression measures
 : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
An Efficient Optimal Leaf Ordering for Hierarchical Clustering in Microarray Gene Expression Data Analysis
 An Efficient Optimal Leaf Ordering for Hierarchical Clustering in Microarray Gene Expression Data Analysis Jianting Zhang Le Gruenwald School of Computer Science The University of Oklahoma Norman, Oklahoma,
An Efficient Optimal Leaf Ordering for Hierarchical Clustering in Microarray Gene Expression Data Analysis Jianting Zhang Le Gruenwald School of Computer Science The University of Oklahoma Norman, Oklahoma,
/ Computational Genomics. Normalization
 10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
Radmacher, M, McShante, L, Simon, R (2002) A paradigm for Class Prediction Using Expression Profiles, J Computational Biol 9:
 Microarray Statistics Module 3: Clustering, comparison, prediction, and Go term analysis Johanna Hardin and Laura Hoopes Worksheet to be handed in the week after discussion Name Clustering algorithms:
Microarray Statistics Module 3: Clustering, comparison, prediction, and Go term analysis Johanna Hardin and Laura Hoopes Worksheet to be handed in the week after discussion Name Clustering algorithms:
Tutorial for the Exon Ontology website
 Tutorial for the Exon Ontology website Table of content Outline Step-by-step Guide 1. Preparation of the test-list 2. First analysis step (without statistical analysis) 2.1. The output page is composed
Tutorial for the Exon Ontology website Table of content Outline Step-by-step Guide 1. Preparation of the test-list 2. First analysis step (without statistical analysis) 2.1. The output page is composed
Bioinformatics? Reads, assembly, annotation, comparative genomics and a bit of phylogeny.
 Bioinformatics? Reads, assembly, annotation, comparative genomics and a bit of phylogeny stefano.gaiarsa@unimi.it Linux and the command line PART 1 Survival kit for the bash environment Purpose of the
Bioinformatics? Reads, assembly, annotation, comparative genomics and a bit of phylogeny stefano.gaiarsa@unimi.it Linux and the command line PART 1 Survival kit for the bash environment Purpose of the
Differential Expression Analysis at PATRIC
 Differential Expression Analysis at PATRIC The following step- by- step workflow is intended to help users learn how to upload their differential gene expression data to their private workspace using Expression
Differential Expression Analysis at PATRIC The following step- by- step workflow is intended to help users learn how to upload their differential gene expression data to their private workspace using Expression
MSCBIO 2070/02-710: Computational Genomics, Spring A4: spline, HMM, clustering, time-series data analysis, RNA-folding
 MSCBIO 2070/02-710:, Spring 2015 A4: spline, HMM, clustering, time-series data analysis, RNA-folding Due: April 13, 2015 by email to Silvia Liu (silvia.shuchang.liu@gmail.com) TA in charge: Silvia Liu
MSCBIO 2070/02-710:, Spring 2015 A4: spline, HMM, clustering, time-series data analysis, RNA-folding Due: April 13, 2015 by email to Silvia Liu (silvia.shuchang.liu@gmail.com) TA in charge: Silvia Liu
How do microarrays work
 Lecture 3 (continued) Alvis Brazma European Bioinformatics Institute How do microarrays work condition mrna cdna hybridise to microarray condition Sample RNA extract labelled acid acid acid nucleic acid
Lecture 3 (continued) Alvis Brazma European Bioinformatics Institute How do microarrays work condition mrna cdna hybridise to microarray condition Sample RNA extract labelled acid acid acid nucleic acid
Tutorial. RNA-Seq Analysis of Breast Cancer Data. Sample to Insight. November 21, 2017
 RNA-Seq Analysis of Breast Cancer Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
RNA-Seq Analysis of Breast Cancer Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Introduction to Bioinformatics AS Laboratory Assignment 2
 Introduction to Bioinformatics AS 250.265 Laboratory Assignment 2 Last week, we discussed several high-throughput methods for the analysis of gene expression in cells. Of those methods, microarray technologies
Introduction to Bioinformatics AS 250.265 Laboratory Assignment 2 Last week, we discussed several high-throughput methods for the analysis of gene expression in cells. Of those methods, microarray technologies
Tutorial 1: Exploring the UCSC Genome Browser
 Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Genome Browsers - The UCSC Genome Browser
 Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Automated Bioinformatics Analysis System on Chip ABASOC. version 1.1
 Automated Bioinformatics Analysis System on Chip ABASOC version 1.1 Phillip Winston Miller, Priyam Patel, Daniel L. Johnson, PhD. University of Tennessee Health Science Center Office of Research Molecular
Automated Bioinformatics Analysis System on Chip ABASOC version 1.1 Phillip Winston Miller, Priyam Patel, Daniel L. Johnson, PhD. University of Tennessee Health Science Center Office of Research Molecular
m6aviewer Version Documentation
 m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
Gene Survey: FAQ. Gene Survey: FAQ Tod Casasent DRAFT
 Gene Survey: FAQ Tod Casasent 2016-02-22-1245 DRAFT 1 What is this document? This document is intended for use by internal and external users of the Gene Survey package, results, and output. This document
Gene Survey: FAQ Tod Casasent 2016-02-22-1245 DRAFT 1 What is this document? This document is intended for use by internal and external users of the Gene Survey package, results, and output. This document
Clustering Techniques
 Clustering Techniques Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 16 Lopresti Fall 2007 Lecture 16-1 - Administrative notes Your final project / paper proposal is due on Friday,
Clustering Techniques Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 16 Lopresti Fall 2007 Lecture 16-1 - Administrative notes Your final project / paper proposal is due on Friday,
Click Trust to launch TableView.
 Visualizing Expression data using the Co-expression Tool Web service and TableView Introduction. TableView was written by James (Jim) E. Johnson and colleagues at the University of Minnesota Center for
Visualizing Expression data using the Co-expression Tool Web service and TableView Introduction. TableView was written by James (Jim) E. Johnson and colleagues at the University of Minnesota Center for
Exercise 2: Browser-Based Annotation and RNA-Seq Data
 Exercise 2: Browser-Based Annotation and RNA-Seq Data Jeremy Buhler July 24, 2018 This exercise continues your introduction to practical issues in comparative annotation. You ll be annotating genomic sequence
Exercise 2: Browser-Based Annotation and RNA-Seq Data Jeremy Buhler July 24, 2018 This exercise continues your introduction to practical issues in comparative annotation. You ll be annotating genomic sequence
Getting Started. April Strand Life Sciences, Inc All rights reserved.
 Getting Started April 2015 Strand Life Sciences, Inc. 2015. All rights reserved. Contents Aim... 3 Demo Project and User Interface... 3 Downloading Annotations... 4 Project and Experiment Creation... 6
Getting Started April 2015 Strand Life Sciences, Inc. 2015. All rights reserved. Contents Aim... 3 Demo Project and User Interface... 3 Downloading Annotations... 4 Project and Experiment Creation... 6
User s Guide. Using the R-Peridot Graphical User Interface (GUI) on Windows and GNU/Linux Systems
 User s Guide Using the R-Peridot Graphical User Interface (GUI) on Windows and GNU/Linux Systems Pitágoras Alves 01/06/2018 Natal-RN, Brazil Index 1. The R Environment Manager...
User s Guide Using the R-Peridot Graphical User Interface (GUI) on Windows and GNU/Linux Systems Pitágoras Alves 01/06/2018 Natal-RN, Brazil Index 1. The R Environment Manager...
HymenopteraMine Documentation
 HymenopteraMine Documentation Release 1.0 Aditi Tayal, Deepak Unni, Colin Diesh, Chris Elsik, Darren Hagen Apr 06, 2017 Contents 1 Welcome to HymenopteraMine 3 1.1 Overview of HymenopteraMine.....................................
HymenopteraMine Documentation Release 1.0 Aditi Tayal, Deepak Unni, Colin Diesh, Chris Elsik, Darren Hagen Apr 06, 2017 Contents 1 Welcome to HymenopteraMine 3 1.1 Overview of HymenopteraMine.....................................
Comparisons and validation of statistical clustering techniques for microarray gene expression data. Outline. Microarrays.
 Comparisons and validation of statistical clustering techniques for microarray gene expression data Susmita Datta and Somnath Datta Presented by: Jenni Dietrich Assisted by: Jeffrey Kidd and Kristin Wheeler
Comparisons and validation of statistical clustering techniques for microarray gene expression data Susmita Datta and Somnath Datta Presented by: Jenni Dietrich Assisted by: Jeffrey Kidd and Kristin Wheeler
Introduction to GE Microarray data analysis Practical Course MolBio 2012
 Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
GenViewer Tutorial / Manual
 GenViewer Tutorial / Manual Table of Contents Importing Data Files... 2 Configuration File... 2 Primary Data... 4 Primary Data Format:... 4 Connectivity Data... 5 Module Declaration File Format... 5 Module
GenViewer Tutorial / Manual Table of Contents Importing Data Files... 2 Configuration File... 2 Primary Data... 4 Primary Data Format:... 4 Connectivity Data... 5 Module Declaration File Format... 5 Module
Long Read RNA-seq Mapper
 UNIVERSITY OF ZAGREB FACULTY OF ELECTRICAL ENGENEERING AND COMPUTING MASTER THESIS no. 1005 Long Read RNA-seq Mapper Josip Marić Zagreb, February 2015. Table of Contents 1. Introduction... 1 2. RNA Sequencing...
UNIVERSITY OF ZAGREB FACULTY OF ELECTRICAL ENGENEERING AND COMPUTING MASTER THESIS no. 1005 Long Read RNA-seq Mapper Josip Marić Zagreb, February 2015. Table of Contents 1. Introduction... 1 2. RNA Sequencing...
CompClustTk Manual & Tutorial
 CompClustTk Manual & Tutorial Brandon King Diane Trout Copyright c California Institute of Technology Version 0.2.0 May 16, 2005 Contents 1 Introduction 1 1.1 Purpose.............................................
CompClustTk Manual & Tutorial Brandon King Diane Trout Copyright c California Institute of Technology Version 0.2.0 May 16, 2005 Contents 1 Introduction 1 1.1 Purpose.............................................
Practical Bioinformatics
 5/12/2015 Gotchas Strings are quoted, names of things are not. mystring = mystring Gotchas Strings are quoted, names of things are not. mystring = mystring Case matters for variable names: mystring MyString
5/12/2015 Gotchas Strings are quoted, names of things are not. mystring = mystring Gotchas Strings are quoted, names of things are not. mystring = mystring Case matters for variable names: mystring MyString
day one day four today day five day three Python for Biologists
 Overview day one today 0. introduction 1. text output and manipulation 2. reading and writing files 3. lists and loops 4. writing functions day three 5. conditional statements 6. dictionaries day four
Overview day one today 0. introduction 1. text output and manipulation 2. reading and writing files 3. lists and loops 4. writing functions day three 5. conditional statements 6. dictionaries day four
Week - 01 Lecture - 04 Downloading and installing Python
 Programming, Data Structures and Algorithms in Python Prof. Madhavan Mukund Department of Computer Science and Engineering Indian Institute of Technology, Madras Week - 01 Lecture - 04 Downloading and
Programming, Data Structures and Algorithms in Python Prof. Madhavan Mukund Department of Computer Science and Engineering Indian Institute of Technology, Madras Week - 01 Lecture - 04 Downloading and
Tutorial: chloroplast genomes
 Tutorial: chloroplast genomes Stacia Wyman Department of Computer Sciences Williams College Williamstown, MA 01267 March 10, 2005 ASSUMPTIONS: You are using Internet Explorer under OS X on the Mac. You
Tutorial: chloroplast genomes Stacia Wyman Department of Computer Sciences Williams College Williamstown, MA 01267 March 10, 2005 ASSUMPTIONS: You are using Internet Explorer under OS X on the Mac. You
java cp PATH_to_minorthird.jar edu.cmu.minorthird.text.gui.textbaseeditor DATA_DIRECTORY LabelsFlName urlmap.txt
 The labeling tool can help us label protein and cell names, and also specify some properties of the labels(i also call them annotations). For example, your confidence for labeling this as a protein name
The labeling tool can help us label protein and cell names, and also specify some properties of the labels(i also call them annotations). For example, your confidence for labeling this as a protein name
implementing the breadth-first search algorithm implementing the depth-first search algorithm
 Graph Traversals 1 Graph Traversals representing graphs adjacency matrices and adjacency lists 2 Implementing the Breadth-First and Depth-First Search Algorithms implementing the breadth-first search algorithm
Graph Traversals 1 Graph Traversals representing graphs adjacency matrices and adjacency lists 2 Implementing the Breadth-First and Depth-First Search Algorithms implementing the breadth-first search algorithm
SHARPR (Systematic High-resolution Activation and Repression Profiling with Reporter-tiling) User Manual (v1.0.2)
 SHARPR (Systematic High-resolution Activation and Repression Profiling with Reporter-tiling) User Manual (v1.0.2) Overview Email any questions to Jason Ernst (jason.ernst@ucla.edu) SHARPR is software for
SHARPR (Systematic High-resolution Activation and Repression Profiling with Reporter-tiling) User Manual (v1.0.2) Overview Email any questions to Jason Ernst (jason.ernst@ucla.edu) SHARPR is software for
Tutorial: RNA-Seq analysis part I: Getting started
 : RNA-Seq analysis part I: Getting started August 9, 2012 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com : RNA-Seq analysis
: RNA-Seq analysis part I: Getting started August 9, 2012 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com : RNA-Seq analysis
QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL
 QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL User manual for QIAseq Targeted RNAscan Panel Analysis 0.5.2 beta 1 Windows, Mac OS X and Linux February 5, 2018 This software is for research
QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL User manual for QIAseq Targeted RNAscan Panel Analysis 0.5.2 beta 1 Windows, Mac OS X and Linux February 5, 2018 This software is for research
When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame
 1 When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame translation on the fly. That is, 3 reading frames from
1 When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame translation on the fly. That is, 3 reading frames from
Lecture 5. Functional Analysis with Blast2GO Enriched functions. Kegg Pathway Analysis Functional Similarities B2G-Far. FatiGO Babelomics.
 Lecture 5 Functional Analysis with Blast2GO Enriched functions FatiGO Babelomics FatiScan Kegg Pathway Analysis Functional Similarities B2G-Far 1 Fisher's Exact Test One Gene List (A) The other list (B)
Lecture 5 Functional Analysis with Blast2GO Enriched functions FatiGO Babelomics FatiScan Kegg Pathway Analysis Functional Similarities B2G-Far 1 Fisher's Exact Test One Gene List (A) The other list (B)
7.36/7.91/20.390/20.490/6.802/6.874 PROBLEM SET 3. Gibbs Sampler, RNA secondary structure, Protein Structure with PyRosetta, Connections (25 Points)
 7.36/7.91/20.390/20.490/6.802/6.874 PROBLEM SET 3. Gibbs Sampler, RNA secondary structure, Protein Structure with PyRosetta, Connections (25 Points) Due: Thursday, April 3 th at noon. Python Scripts All
7.36/7.91/20.390/20.490/6.802/6.874 PROBLEM SET 3. Gibbs Sampler, RNA secondary structure, Protein Structure with PyRosetta, Connections (25 Points) Due: Thursday, April 3 th at noon. Python Scripts All
Genetics 211 Genomics Winter 2014 Problem Set 4
 Genomics - Part 1 due Friday, 2/21/2014 by 9:00am Part 2 due Friday, 3/7/2014 by 9:00am For this problem set, we re going to use real data from a high-throughput sequencing project to look for differential
Genomics - Part 1 due Friday, 2/21/2014 by 9:00am Part 2 due Friday, 3/7/2014 by 9:00am For this problem set, we re going to use real data from a high-throughput sequencing project to look for differential
CLC Server. End User USER MANUAL
 CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
Browser Exercises - I. Alignments and Comparative genomics
 Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Database Repository and Tools
 Database Repository and Tools John Matese May 9, 2008 What is the Repository? Save and exchange retrieved and analyzed datafiles Perform datafile manipulations (averaging and annotations) Run specialized
Database Repository and Tools John Matese May 9, 2008 What is the Repository? Save and exchange retrieved and analyzed datafiles Perform datafile manipulations (averaging and annotations) Run specialized
Week - 04 Lecture - 01 Merge Sort. (Refer Slide Time: 00:02)
 Programming, Data Structures and Algorithms in Python Prof. Madhavan Mukund Department of Computer Science and Engineering Indian Institute of Technology, Madras Week - 04 Lecture - 01 Merge Sort (Refer
Programming, Data Structures and Algorithms in Python Prof. Madhavan Mukund Department of Computer Science and Engineering Indian Institute of Technology, Madras Week - 04 Lecture - 01 Merge Sort (Refer
Homework Assignment #3
 CS 540-2: Introduction to Artificial Intelligence Homework Assignment #3 Assigned: Monday, February 20 Due: Saturday, March 4 Hand-In Instructions This assignment includes written problems and programming
CS 540-2: Introduction to Artificial Intelligence Homework Assignment #3 Assigned: Monday, February 20 Due: Saturday, March 4 Hand-In Instructions This assignment includes written problems and programming
ChIP-seq Analysis Practical
 ChIP-seq Analysis Practical Vladimir Teif (vteif@essex.ac.uk) An updated version of this document will be available at http://generegulation.info/index.php/teaching In this practical we will learn how
ChIP-seq Analysis Practical Vladimir Teif (vteif@essex.ac.uk) An updated version of this document will be available at http://generegulation.info/index.php/teaching In this practical we will learn how
Eval: A Gene Set Comparison System
 Masters Project Report Eval: A Gene Set Comparison System Evan Keibler evan@cse.wustl.edu Table of Contents Table of Contents... - 2 - Chapter 1: Introduction... - 5-1.1 Gene Structure... - 5-1.2 Gene
Masters Project Report Eval: A Gene Set Comparison System Evan Keibler evan@cse.wustl.edu Table of Contents Table of Contents... - 2 - Chapter 1: Introduction... - 5-1.1 Gene Structure... - 5-1.2 Gene
Lecture 3. Essential skills for bioinformatics: Unix/Linux
 Lecture 3 Essential skills for bioinformatics: Unix/Linux RETRIEVING DATA Overview Whether downloading large sequencing datasets or accessing a web application hundreds of times to download specific files,
Lecture 3 Essential skills for bioinformatics: Unix/Linux RETRIEVING DATA Overview Whether downloading large sequencing datasets or accessing a web application hundreds of times to download specific files,
BovineMine Documentation
 BovineMine Documentation Release 1.0 Deepak Unni, Aditi Tayal, Colin Diesh, Christine Elsik, Darren Hag Oct 06, 2017 Contents 1 Tutorial 3 1.1 Overview.................................................
BovineMine Documentation Release 1.0 Deepak Unni, Aditi Tayal, Colin Diesh, Christine Elsik, Darren Hag Oct 06, 2017 Contents 1 Tutorial 3 1.1 Overview.................................................
Tutorial. Introduction. Starting GenePattern. Prerequisites. Scientific Scenario
 Tutorial The GenePattern Tutorial introduces you to GenePattern by providing step-by-step instructions for analyzing gene expression. It takes approximately 40 minutes to complete. All of the information
Tutorial The GenePattern Tutorial introduces you to GenePattern by providing step-by-step instructions for analyzing gene expression. It takes approximately 40 minutes to complete. All of the information
Practical Bioinformatics
 4/25/2017 Mean def mean ( x ) : s = 0. 0 f o r i i n x : s += i return s / len ( x ) def mean ( x ) : return sum( x )/ f l o a t ( len ( x ) ) Standard Deviation σ x = N i (x i x) 2 N 1 Standard Deviation
4/25/2017 Mean def mean ( x ) : s = 0. 0 f o r i i n x : s += i return s / len ( x ) def mean ( x ) : return sum( x )/ f l o a t ( len ( x ) ) Standard Deviation σ x = N i (x i x) 2 N 1 Standard Deviation
Analyzing ChIP- Seq Data in Galaxy
 Analyzing ChIP- Seq Data in Galaxy Lauren Mills RISS ABSTRACT Step- by- step guide to basic ChIP- Seq analysis using the Galaxy platform. Table of Contents Introduction... 3 Links to helpful information...
Analyzing ChIP- Seq Data in Galaxy Lauren Mills RISS ABSTRACT Step- by- step guide to basic ChIP- Seq analysis using the Galaxy platform. Table of Contents Introduction... 3 Links to helpful information...
Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides
 1 Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides ways to flexibly merge your Mascot search and quantitation
1 Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides ways to flexibly merge your Mascot search and quantitation
Package ctc. R topics documented: August 2, Version Date Depends amap. Title Cluster and Tree Conversion.
 Package ctc August 2, 2013 Version 1.35.0 Date 2005-11-16 Depends amap Title Cluster and Tree Conversion. Author Antoine Lucas , Laurent Gautier biocviews Microarray,
Package ctc August 2, 2013 Version 1.35.0 Date 2005-11-16 Depends amap Title Cluster and Tree Conversion. Author Antoine Lucas , Laurent Gautier biocviews Microarray,
Application of Hierarchical Clustering to Find Expression Modules in Cancer
 Application of Hierarchical Clustering to Find Expression Modules in Cancer T. M. Murali August 18, 2008 Innovative Application of Hierarchical Clustering A module map showing conditional activity of expression
Application of Hierarchical Clustering to Find Expression Modules in Cancer T. M. Murali August 18, 2008 Innovative Application of Hierarchical Clustering A module map showing conditional activity of expression
Creating and Using Genome Assemblies Tutorial
 Creating and Using Genome Assemblies Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Create a Genome Assembly for Danio rerio 2 2. Building Annotation Sources 5 A. Creating a Reference
Creating and Using Genome Assemblies Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Create a Genome Assembly for Danio rerio 2 2. Building Annotation Sources 5 A. Creating a Reference
MetScape User Manual
 MetScape 2.3.2 User Manual A Plugin for Cytoscape National Center for Integrative Biomedical Informatics July 2012 2011 University of Michigan This work is supported by the National Center for Integrative
MetScape 2.3.2 User Manual A Plugin for Cytoscape National Center for Integrative Biomedical Informatics July 2012 2011 University of Michigan This work is supported by the National Center for Integrative
Exercises. Biological Data Analysis Using InterMine workshop exercises with answers
 Exercises Biological Data Analysis Using InterMine workshop exercises with answers Exercise1: Faceted Search Use HumanMine for this exercise 1. Search for one or more of the following using the keyword
Exercises Biological Data Analysis Using InterMine workshop exercises with answers Exercise1: Faceted Search Use HumanMine for this exercise 1. Search for one or more of the following using the keyword
CLUSTERING IN BIOINFORMATICS
 CLUSTERING IN BIOINFORMATICS CSE/BIMM/BENG 8 MAY 4, 0 OVERVIEW Define the clustering problem Motivation: gene expression and microarrays Types of clustering Clustering algorithms Other applications of
CLUSTERING IN BIOINFORMATICS CSE/BIMM/BENG 8 MAY 4, 0 OVERVIEW Define the clustering problem Motivation: gene expression and microarrays Types of clustering Clustering algorithms Other applications of
Identiyfing splice junctions from RNA-Seq data
 Identiyfing splice junctions from RNA-Seq data Joseph K. Pickrell pickrell@uchicago.edu October 4, 2010 Contents 1 Motivation 2 2 Identification of potential junction-spanning reads 2 3 Calling splice
Identiyfing splice junctions from RNA-Seq data Joseph K. Pickrell pickrell@uchicago.edu October 4, 2010 Contents 1 Motivation 2 2 Identification of potential junction-spanning reads 2 3 Calling splice
The Allen Human Brain Atlas offers three types of searches to allow a user to: (1) obtain gene expression data for specific genes (or probes) of
 Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Missing Data Estimation in Microarrays Using Multi-Organism Approach
 Missing Data Estimation in Microarrays Using Multi-Organism Approach Marcel Nassar and Hady Zeineddine Progress Report: Data Mining Course Project, Spring 2008 Prof. Inderjit S. Dhillon April 02, 2008
Missing Data Estimation in Microarrays Using Multi-Organism Approach Marcel Nassar and Hady Zeineddine Progress Report: Data Mining Course Project, Spring 2008 Prof. Inderjit S. Dhillon April 02, 2008
Step-by-Step Guide to Relatedness and Association Mapping Contents
 Step-by-Step Guide to Relatedness and Association Mapping Contents OBJECTIVES... 2 INTRODUCTION... 2 RELATEDNESS MEASURES... 2 POPULATION STRUCTURE... 6 Q-K ASSOCIATION ANALYSIS... 10 K MATRIX COMPRESSION...
Step-by-Step Guide to Relatedness and Association Mapping Contents OBJECTIVES... 2 INTRODUCTION... 2 RELATEDNESS MEASURES... 2 POPULATION STRUCTURE... 6 Q-K ASSOCIATION ANALYSIS... 10 K MATRIX COMPRESSION...
Gene expression & Clustering (Chapter 10)
 Gene expression & Clustering (Chapter 10) Determining gene function Sequence comparison tells us if a gene is similar to another gene, e.g., in a new species Dynamic programming Approximate pattern matching
Gene expression & Clustering (Chapter 10) Determining gene function Sequence comparison tells us if a gene is similar to another gene, e.g., in a new species Dynamic programming Approximate pattern matching
Public Repositories Tutorial: Bulk Downloads
 Public Repositories Tutorial: Bulk Downloads Almost all of the public databases, genome browsers, and other tools you have explored so far offer some form of access to rapidly download all or large chunks
Public Repositories Tutorial: Bulk Downloads Almost all of the public databases, genome browsers, and other tools you have explored so far offer some form of access to rapidly download all or large chunks
Lecture 5. Essential skills for bioinformatics: Unix/Linux
 Lecture 5 Essential skills for bioinformatics: Unix/Linux UNIX DATA TOOLS Text processing with awk We have illustrated two ways awk can come in handy: Filtering data using rules that can combine regular
Lecture 5 Essential skills for bioinformatics: Unix/Linux UNIX DATA TOOLS Text processing with awk We have illustrated two ways awk can come in handy: Filtering data using rules that can combine regular
Clustering. Lecture 6, 1/24/03 ECS289A
 Clustering Lecture 6, 1/24/03 What is Clustering? Given n objects, assign them to groups (clusters) based on their similarity Unsupervised Machine Learning Class Discovery Difficult, and maybe ill-posed
Clustering Lecture 6, 1/24/03 What is Clustering? Given n objects, assign them to groups (clusters) based on their similarity Unsupervised Machine Learning Class Discovery Difficult, and maybe ill-posed
Import GEO Experiment into Partek Genomics Suite
 Import GEO Experiment into Partek Genomics Suite This tutorial will illustrate how to: Import a gene expression experiment from GEO SOFT files Specify annotations Import RAW data from GEO for gene expression
Import GEO Experiment into Partek Genomics Suite This tutorial will illustrate how to: Import a gene expression experiment from GEO SOFT files Specify annotations Import RAW data from GEO for gene expression
Fusion Detection Using QIAseq RNAscan Panels
 Fusion Detection Using QIAseq RNAscan Panels June 11, 2018 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com ts-bioinformatics@qiagen.com
Fusion Detection Using QIAseq RNAscan Panels June 11, 2018 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com ts-bioinformatics@qiagen.com
6.00 Introduction to Computer Science and Programming Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu 6.00 Introduction to Computer Science and Programming Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu 6.00 Introduction to Computer Science and Programming Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
(Refer Slide Time 6:48)
 Digital Circuits and Systems Prof. S. Srinivasan Department of Electrical Engineering Indian Institute of Technology Madras Lecture - 8 Karnaugh Map Minimization using Maxterms We have been taking about
Digital Circuits and Systems Prof. S. Srinivasan Department of Electrical Engineering Indian Institute of Technology Madras Lecture - 8 Karnaugh Map Minimization using Maxterms We have been taking about
11/8/2017 Trinity De novo Transcriptome Assembly Workshop trinityrnaseq/rnaseq_trinity_tuxedo_workshop Wiki GitHub
 trinityrnaseq / RNASeq_Trinity_Tuxedo_Workshop Trinity De novo Transcriptome Assembly Workshop Brian Haas edited this page on Oct 17, 2015 14 revisions De novo RNA-Seq Assembly and Analysis Using Trinity
trinityrnaseq / RNASeq_Trinity_Tuxedo_Workshop Trinity De novo Transcriptome Assembly Workshop Brian Haas edited this page on Oct 17, 2015 14 revisions De novo RNA-Seq Assembly and Analysis Using Trinity
Analysis of ChIP-seq data
 Before we start: 1. Log into tak (step 0 on the exercises) 2. Go to your lab space and create a folder for the class (see separate hand out) 3. Connect to your lab space through the wihtdata network and
Before we start: 1. Log into tak (step 0 on the exercises) 2. Go to your lab space and create a folder for the class (see separate hand out) 3. Connect to your lab space through the wihtdata network and
Locality-sensitive hashing and biological network alignment
 Locality-sensitive hashing and biological network alignment Laura LeGault - University of Wisconsin, Madison 12 May 2008 Abstract Large biological networks contain much information about the functionality
Locality-sensitive hashing and biological network alignment Laura LeGault - University of Wisconsin, Madison 12 May 2008 Abstract Large biological networks contain much information about the functionality
COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP. Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas
 COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas First of all connect once again to the CBS system: Open ssh shell client. Press Quick
COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas First of all connect once again to the CBS system: Open ssh shell client. Press Quick
6.001 Notes: Section 15.1
 6.001 Notes: Section 15.1 Slide 15.1.1 Our goal over the next few lectures is to build an interpreter, which in a very basic sense is the ultimate in programming, since doing so will allow us to define
6.001 Notes: Section 15.1 Slide 15.1.1 Our goal over the next few lectures is to build an interpreter, which in a very basic sense is the ultimate in programming, since doing so will allow us to define
Lab 7c: Rainfall patterns and drainage density
 Lab 7c: Rainfall patterns and drainage density This is the third of a four-part handout for class the last two weeks before spring break. Due: Be done with this by class on 11/3. Task: Extract your watersheds
Lab 7c: Rainfall patterns and drainage density This is the third of a four-part handout for class the last two weeks before spring break. Due: Be done with this by class on 11/3. Task: Extract your watersheds
Data Structures and Algorithms Dr. Naveen Garg Department of Computer Science and Engineering Indian Institute of Technology, Delhi.
 Data Structures and Algorithms Dr. Naveen Garg Department of Computer Science and Engineering Indian Institute of Technology, Delhi Lecture 18 Tries Today we are going to be talking about another data
Data Structures and Algorithms Dr. Naveen Garg Department of Computer Science and Engineering Indian Institute of Technology, Delhi Lecture 18 Tries Today we are going to be talking about another data
Design and Annotation Files
 Design and Annotation Files Release Notes SeqCap EZ Exome Target Enrichment System The design and annotation files provide information about genomic regions covered by the capture probes and the genes
Design and Annotation Files Release Notes SeqCap EZ Exome Target Enrichment System The design and annotation files provide information about genomic regions covered by the capture probes and the genes
Intermediate/Advanced Python. Michael Weinstein (Day 1)
 Intermediate/Advanced Python Michael Weinstein (Day 1) Who am I? Most of my experience is on the molecular and animal modeling side I also design computer programs for analyzing biological data, particularly
Intermediate/Advanced Python Michael Weinstein (Day 1) Who am I? Most of my experience is on the molecular and animal modeling side I also design computer programs for analyzing biological data, particularly
Computational Theory MAT542 (Computational Methods in Genomics) - Part 2 & 3 -
 Computational Theory MAT542 (Computational Methods in Genomics) - Part 2 & 3 - Benjamin King Mount Desert Island Biological Laboratory bking@mdibl.org Overview of 4 Lectures Introduction to Computation
Computational Theory MAT542 (Computational Methods in Genomics) - Part 2 & 3 - Benjamin King Mount Desert Island Biological Laboratory bking@mdibl.org Overview of 4 Lectures Introduction to Computation
University of Wisconsin-Madison Spring 2018 BMI/CS 776: Advanced Bioinformatics Homework #2
 Assignment goals Use mutual information to reconstruct gene expression networks Evaluate classifier predictions Examine Gibbs sampling for a Markov random field Control for multiple hypothesis testing
Assignment goals Use mutual information to reconstruct gene expression networks Evaluate classifier predictions Examine Gibbs sampling for a Markov random field Control for multiple hypothesis testing
GPU Accelerated PK-means Algorithm for Gene Clustering
 GPU Accelerated PK-means Algorithm for Gene Clustering Wuchao Situ, Yau-King Lam, Yi Xiao, P.W.M. Tsang, and Chi-Sing Leung Department of Electronic Engineering, City University of Hong Kong, Hong Kong,
GPU Accelerated PK-means Algorithm for Gene Clustering Wuchao Situ, Yau-King Lam, Yi Xiao, P.W.M. Tsang, and Chi-Sing Leung Department of Electronic Engineering, City University of Hong Kong, Hong Kong,
Unix tutorial, tome 5: deep-sequencing data analysis
 Unix tutorial, tome 5: deep-sequencing data analysis by Hervé December 8, 2008 Contents 1 Input files 2 2 Data extraction 3 2.1 Overview, implicit assumptions.............................. 3 2.2 Usage............................................
Unix tutorial, tome 5: deep-sequencing data analysis by Hervé December 8, 2008 Contents 1 Input files 2 2 Data extraction 3 2.1 Overview, implicit assumptions.............................. 3 2.2 Usage............................................
Expander Online Documentation
 Expander Online Documentation Table of Contents Introduction...1 Starting EXPANDER...2 Input Data...4 Preprocessing GE Data...8 Viewing Data Plots...12 Clustering GE Data...14 Biclustering GE Data...17
Expander Online Documentation Table of Contents Introduction...1 Starting EXPANDER...2 Input Data...4 Preprocessing GE Data...8 Viewing Data Plots...12 Clustering GE Data...14 Biclustering GE Data...17
Overview. Dataset: testpos DNA: CCCATGGTCGGGGGGGGGGAGTCCATAACCC Num exons: 2 strand: + RNA (from file): AUGGUCAGUCCAUAA peptide (from file): MVSP*
 Overview In this homework, we will write a program that will print the peptide (a string of amino acids) from four pieces of information: A DNA sequence (a string). The strand the gene appears on (a string).
Overview In this homework, we will write a program that will print the peptide (a string of amino acids) from four pieces of information: A DNA sequence (a string). The strand the gene appears on (a string).
CSCI 204 Introduction to Computer Science II
 CSCI 204 Project 2 Maze Assigned: Wednesday 09/27/2017 First Phase (Recursion) Due Friday, 10/06/2017 Second Phase (Stack) Due Monday, 10/16/2017 1 Objective The purpose of this assignment is to give you
CSCI 204 Project 2 Maze Assigned: Wednesday 09/27/2017 First Phase (Recursion) Due Friday, 10/06/2017 Second Phase (Stack) Due Monday, 10/16/2017 1 Objective The purpose of this assignment is to give you
Week - 01 Lecture - 03 Euclid's Algorithm for gcd. Let us continue with our running example of gcd to explore more issues involved with program.
 Programming, Data Structures and Algorithms in Python Prof. Madhavan Mukund Department of Computer Science and Engineering Indian Institute of Technology, Madras Week - 01 Lecture - 03 Euclid's Algorithm
Programming, Data Structures and Algorithms in Python Prof. Madhavan Mukund Department of Computer Science and Engineering Indian Institute of Technology, Madras Week - 01 Lecture - 03 Euclid's Algorithm
Supplementary information: Detection of differentially expressed segments in tiling array data
 Supplementary information: Detection of differentially expressed segments in tiling array data Christian Otto 1,2, Kristin Reiche 3,1,4, Jörg Hackermüller 3,1,4 July 1, 212 1 Bioinformatics Group, Department
Supplementary information: Detection of differentially expressed segments in tiling array data Christian Otto 1,2, Kristin Reiche 3,1,4, Jörg Hackermüller 3,1,4 July 1, 212 1 Bioinformatics Group, Department
CMSC 201 Spring 2017 Lab 05 Lists
 CMSC 201 Spring 2017 Lab 05 Lists Assignment: Lab 05 Lists Due Date: During discussion, February 27th through March 2nd Value: 10 points (8 points during lab, 2 points for Pre Lab quiz) This week s lab
CMSC 201 Spring 2017 Lab 05 Lists Assignment: Lab 05 Lists Due Date: During discussion, February 27th through March 2nd Value: 10 points (8 points during lab, 2 points for Pre Lab quiz) This week s lab
Algorithms for Bounded-Error Correlation of High Dimensional Data in Microarray Experiments
 Algorithms for Bounded-Error Correlation of High Dimensional Data in Microarray Experiments Mehmet Koyutürk, Ananth Grama, and Wojciech Szpankowski Department of Computer Sciences, Purdue University West
Algorithms for Bounded-Error Correlation of High Dimensional Data in Microarray Experiments Mehmet Koyutürk, Ananth Grama, and Wojciech Szpankowski Department of Computer Sciences, Purdue University West
Supplementary Materials for. A gene ontology inferred from molecular networks
 Supplementary Materials for A gene ontology inferred from molecular networks Janusz Dutkowski, Michael Kramer, Michal A Surma, Rama Balakrishnan, J Michael Cherry, Nevan J Krogan & Trey Ideker 1. Supplementary
Supplementary Materials for A gene ontology inferred from molecular networks Janusz Dutkowski, Michael Kramer, Michal A Surma, Rama Balakrishnan, J Michael Cherry, Nevan J Krogan & Trey Ideker 1. Supplementary
CTL mapping in R. Danny Arends, Pjotr Prins, and Ritsert C. Jansen. University of Groningen Groningen Bioinformatics Centre & GCC Revision # 1
 CTL mapping in R Danny Arends, Pjotr Prins, and Ritsert C. Jansen University of Groningen Groningen Bioinformatics Centre & GCC Revision # 1 First written: Oct 2011 Last modified: Jan 2018 Abstract: Tutorial
CTL mapping in R Danny Arends, Pjotr Prins, and Ritsert C. Jansen University of Groningen Groningen Bioinformatics Centre & GCC Revision # 1 First written: Oct 2011 Last modified: Jan 2018 Abstract: Tutorial
Tutorial. Identification of Variants in a Tumor Sample. Sample to Insight. November 21, 2017
 Identification of Variants in a Tumor Sample November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Identification of Variants in a Tumor Sample November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
