A SAS based solution for define.xml
|
|
|
- Preston Watkins
- 6 years ago
- Views:
Transcription
1 Paper RA02 A SAS based solution for define.xml Monika Kawohl, Accovion GmbH, Marburg, Germany ABSTRACT When submitting data to the FDA, a data definition file, describing the structure and contents of the data, is a mandatory deliverable. For data submitted in the CDISC SDTM format the define.xml as published by the CDISC define.xml team is the preferred type of a data definition file. Compared to define.pdf, the previous data definition file format, define.xml is more suitable for providing the different types of metadata required to adequately describe data in the SDTM format. An additional benefit of define.xml is its machine-readability. After Accovion successfully implemented an automated process for define.pdf in SAS, building a SAS based automation process for define.xml was the logical consequence. This paper illustrates the process chosen by Accovion to compile all the necessary metadata and build a define.xml as described in the CDISC Case Report Tabulation Data Definition Specification (define.xml) V1.0 Standard [1]. And as this standard will evolve over the next couple of years, it will briefly touch on the expected enhancements for define.xml. INTRODUCTION A data definition file, formally called Case Report Tabulation Data Definitions (CRT DD), is necessary to facilitate the review of the study data submitted to a regulatory authority. Well-defined, standardized metadata minimizes the time needed to familiarize with the data, which can speed up the overall review process. The Case Report Tabulation Data Definition Specification (define.xml) V1.0 Standard as prepared by the CDISC define.xml team in 2005 has been referenced in the FDA s ectd Study Data Specifications [6] as the preferred data definition file when data are submitted in the SDTM format. The most important benefit of define.xml compared to define.pdf, the previous data definition file format, is that an XML file is both human- and machine-readable. Whereas the human-readability helps the reviewer to understand and work with the data, machine-readable metadata can be exploited when transferring data between different systems. They also provide the basis for automation of transposing the data submitted into different structures. Even the generation of define.xml can be automated, taken into account that a lot of the metadata required for define.xml are already inherent in the datasets themselves. The SAS based automation solution for define.xml presented in this paper refers to the Case Report Tabulation Data Definition Specification (define.xml) V1.0 Standard and the sample define.xml as prepared by the CDISC define.xml team in End of July 2007, the CDISC Submission Data Standards (SDS) Metadata team has released a draft version of the Metadata Submission Guidelines [2], Appendix to the Study Data Tabulation Model Implementation Guide [3], for review. A sample electronic submission with a new sample define.xml is included with this Appendix. These new documents provide some further guidance on how to describe the metadata of the data submitted. The navigation through define.xml has improved via additional hyperlinks and bookmarks. Therefore, relevant differences between the new (2007) and the old (2005) define.xml sample will be highlighted throughout this paper, where necessary. XML BASICS In order to build define.xml with SAS, a basic understanding of the XML technology is required. In essence there are three different file types that work together to achieve the human and machine readable characteristic of the XML technology: 1
2 Schema (extension:.xsd) Definition and declaration of elements and their attributes (prerequisite for machine-readability) XML file (extension:.xml) Description of data and metadata in machine-readable format using the elements defined in the schema Style sheet (extension:.xsl) Definition of the layout in a browser tool for the human readable representation of the XML file Example: When opening a define.xml file with a text editor, it looks like this: <?xml version="1.0" encoding="iso "?> <?xml-stylesheet type="text/xsl" href="define1-0-0.xsl"?> <ODM <ItemGroupDef OID="DM" Name="DM" Repeating="No" IsReferenceData="No" Purpose="Tabulation" def:label="demographics" def:structure="one record per subject" def:domainkeys=" STUDYID USUBJID" def:class="special Purpose" def:archivelocationid="location.dm"> The style sheet reference define1-0-0.xsl in the second line (style sheet used in the 2005 define.xml sample) leads to the following representation of define.xml in the Internet Explorer: Datasets for Study <XY> Dataset Description Structure Purpose Keys Location DM Demographics Special Purpose - One record per subject Tabulation STUDYID, USUBJID crt/datasets/<xy>/dm.xpt (Underlined texts indicate hyperlinks) The style sheet defines how the define.xml elements (e.g., ItemGroupDef) and their attributes (e.g., Name=) are displayed in a browser tool. The elements and their attributes themselves are declared in the schema file(s) and referenced both in the XML file and the XSL file. Some attributes are relevant for the machine-readability mainly (e.g., Repeating=). If these attributes are not described in the style sheet, they are omitted from the human readable representation of define.xml. When a layout change is desired, only the style sheet and not the XML file itself has to be modified. Applying the style sheet included in the 2007 define.xml sample to this specific piece of information, the layout looks slightly different, and a bookmark panel (not displayed below) is also included in the new browser representation: Datasets for Study <XY> Dataset Description Class Structure Purpose Keys Location DM Demographics SPECIAL PURPOSE One record per subject Tabulation STUDYID, USUBJID Dm.xpt 2
3 The define.xml schema, an extension to the CDISC ODM (Operational Data Model) [4], and the style sheet define1-0-0.xsl (either the 2005 or the 2007 version) can be used for the define.xml generation as prepared by the CDISC teams. Only the contents of the define.xml file has to be adapted to the respective data. DEFINE.XML SECTIONS (ELEMENTS) The define.xml as displayed via the style sheet define1-0-0.xsl provided by the CDISC define.xml team in 2005 has the following sections: Data Metadata (TOC) Variable Metadata Variable Value Level Metadata Computational Algorithms Controlled Terminology/Code Lists Additionally the following external and internal hyperlinks are included: External links to the first page of the annotated CRF External links to a supplemental data definitions document External links to the respective SAS Version 5 transport files Internal links to the top of the define.xml document (=TOC) Several internal links between different define.xml sections Their purpose and their contents are described in the following subsections. Comparing this layout with the layout obtained via the style sheet used in the 2007 define.xml sample, especially the hyperlinking capability has been greatly extended to better suit a reviewer s needs. Comments on notable improvements when using the new style sheet will be included in the following subsections under Notable Changes. DATA METADATA (TOC) The Data Metadata Section serves as a table of contents (TOC). It describes each dataset in a standardized fashion. Excerpt of selected datasets: Datasets for Study <XY> Dataset Description Structure Purpose Keys Location DM Demographics Special Purpose - One record per subject Tabulation STUDYID, USUBJID crt/datasets/<xy>/dm.xpt LB Laboratory Tests Findings - One record per lab test per time point per visit per subject Tabulation STUDYID, USUBJID, LBTESTCD, VISITNUM, LBTPTNUM crt/datasets/<xy>/lb.xpt SUPPDM Supplemental Qualifiers for DM Related - One record per variable value per subject Tabulation STUDYID, RDOMAIN, USUBJID, QNAM crt/datasets/<xy>/suppdm.xpt When clicking on the location link the SAS V5 transport file of the respective dataset opens up in the associated program, e.g., SAS System Viewer. The description link leads to the Variable Metadata of the selected dataset. Notable changes Except for splitting the contents displayed in the column Structure into the two columns Class and Structure, no further changes have been performed on the TOC section. 3
4 However, the Draft Metadata Submission Guidelines recommend a specific order for the display of the datasets. The datasets should be listed alphabetically within their domain class. Sponsor-defined domains should appear after all of the CDISC domains in the respective class. The domain class order is: Trial Design, Special Purpose, Interventions, Events, Findings, Relationships (including Supplemental Qualifiers). VARIABLE METADATA All variables included in a certain dataset are described per dataset in the Variable Metadata Section. Excerpt of selected variables: Laboratory Tests Dataset (LB) Variable Label Type Controlled Terms or Format Origin Role Comment USUBJID Unique Subject Identifier text Sponsor Defined Identifier Concatenation of Study, Site and Subject Identifiers separated by '-' LBTESTCD LAB Test or Examination Short Name text CRF Topic CRF pages 5, 10, 15, 20 LBTEST LAB Test or Examination Name text CRF Synonym Qualifier CRF pages 5, 10, 15, 20 LBORRES Result or Finding in Original Units text CRF Result Qualifier CRF pages 5, 10, 15, 20 LBORRESU Original Units text LBORU CRF Variable Qualifier CRF pages 5, 10, 15, 20 LBSTRESN Numeric Result/Finding in Standard Units float Derived Result Qualifier LBSTRESN = numeric representation of LBORRES converted to standard unit LBSTRESU Standard Units text LBSTU Sponsor Defined Variable Qualifier Sponsor defined standard unit for analysis LBBLFL Baseline Flag text YF Derived Record Qualifier See Computational Method: COMPMETHOD.LBBLFL LBDTC Date/Time of Specimen Collection datetime CRF Timing CRF pages 5, 10, 15, 20 The columns Variable and Label are self-explaining. The Type conforms to the data type definitions of the ODM schema. Valid values are: text, integer, float, datetime, date and time. For variables with a discrete list of values attached (e.g., No/Yes questions) a format name is provided in the Controlled Terms or Format column that links to the Controlled Terms/Code Lists Section. In the target section the possible values and their decodes are listed. The Origin of a variable may be Sponsor Defined, CRF or Derived. The Role values correspond to the metadata definition of the SDTM. The Comment column contains CRF page references for variable originated from the CRF, a short explanation of Sponsor Defined variables or, for derived variables, the derivation rule or a computational method reference. This reference links to the computational algorithms section for complex derivations or derivations used more than once. In normalized datasets of vertical format, e.g., findings datasets built with test short name, test name and test result variables, the variable Test Short Name links to the Variable Value Level Metadata Section. Notable changes In the 2007 define.xml sample the same information is displayed like this: 4
5 Excerpt of selected variables: Laboratory Tests Dataset (LB) lb.xpt (1.) Variable Label Type Controlled Terms or Format Origin Role Comment USUBJID Unique Subject Identifier text Sponsor Defined IDENTIFIER Concatenation of Study, Site and Subject Identifiers separated by '-' LBTESTCD LAB Test or Examination Short Name text CRF Pages 5, 10, 15, 20 TOPIC (2.) LBORRES Result or Finding in Original Units text CRF Pages 5, 10, 15, 20 (3.) RESULT QUALIFIER LBSTRESN Numeric Result/Finding in Standard Units float Derived RESULT QUALIFIER LBSTRESN = numeric representation of LBORRES converted to standard unit LBBLFL Baseline Flag text YF Derived RECORD QUALIFIER See Computational Method: COMPMETHOD.LBBLFL LBDTC Date/Time of Specimen Collection text CRF Pages 5, 10, 15, 20 TIMING Supplemental Qualifier Dataset (ValueList.SUPPLB.QNAM) (4.) The style sheet improvements include: 1. Link to respective SAS transport file 2. Link to the Role Codelist details in the Controlled Terminology / Codelists section 3. Links to specific Annotated CRF pages 4. Link to the Variable Value Level Metadata of QNAM if a supplemental qualifier dataset exist for the respective domain. In order to make use of the specific CRF page link functionality, the list of respective CRF pages have to appear in the Origin column following the keywords CRF Page or CRF Pages, respectively. The page numbers listed have to be identical with the physical page numbers in the Annotated CRF PDF document. It is also noteworthy that the Draft Metadata Submission Guidelines clearly state, that the type description of date or datetime variables should be text as long as data are submitted in the SAS V5 transport format. Additionally, the new keyword EDT (Electronic Data Transfer) is introduced as code of origin, if data have not been collected on the CRF but have been received electronically. VARIABLE VALUE LEVEL METADATA The Variable Value Level Metadata describes each unique value of a certain test short name similarly to the variables described in the Variable Metadata. For review and analysis a distinction between each of the single tests is required. The list of short names allows for a quick selection. Attributes like origin, data type and format may differ between the single tests available in a dataset, and the data type and format information of the respective test result is especially useful for transposing the vertical datasets to horizontal datasets, i.e. variables per test, if required. 5
6 Excerpt of selected variable values: Source Variable Value Label Type Controlled Terms or Format Origin Role Comment LBTESTCD ALB Albumin float CRF CRF pages 5, 15 LBTESTCD OPIATES Opiates text POSNEGF CRF CRF pages 10, 20 QNAM ITT Intent to Treat Population Flag text YNF Derived See Computational Method: COMPMETHOD.ITT QNAM PPROT Per Protocol Set Flag text YNF Derived See Computational Method: COMPMETHOD.PPROT Notable changes In the 2007 define.xml sample, there are separate bookmarked sections per variable explained in the Variable Value Level Metadata. This is particularly helpful to link the information for supplemental qualifier datasets back to their parent dataset (e.g., section title: Value Level Metadata (ValueList.SUPPDM.QNAM)). For the ease of navigation, a link to the Variable Metadata of the parent dataset is included in these cases (e.g., link via table footnote Dataset (DM) ). As mentioned above, the CRF page references are best to be placed into the Origin column in order to use the CRF page link functionality. For Variable Value Level Metadata there is another option to express the Origin of a variable value, i.e., specifying the name of the result variable in the parent dataset. The variable displayed is the variable which holds the value that would become the new variable value when transposing the vertical SDTM dataset to a horizontal structure. Applying this option to the excerpt above, the Origin of the variable value ALB becomes LBSTRESN whereas LBSTRESC is the Origin for OPIATES. This option focuses on the machine-readability. However, for the review of the metadata at the regulatory authority the CRF page references or the derivation descriptions seem preferable. COMPUTATIONAL ALGORITHMS All computational methods referred to by either the variable metadata or the variable value level metadata are shown in the Computational Algorithms Section. It includes the computational method name and the derivation rule described in a kind of pseudo code. Excerpt of selected computational algorithms: Computational Algorithms Section Reference Name Computation Method COMPMETHOD.LBBLFL Derive mean of pre-treatment measurements. Create new record with result and flag LBBLFL= Y. Notable changes None CONTROLLED TERMINOLOGY / CODE LISTS SECTION All format names referred to by either the variable metadata or the variable value level metadata, their values and their decodes are displayed in the Controlled Terminology/Code Lists Section. As the code values correspond to the values stored in the datasets, they may be equal to the decode values, e.g., result codes of NEGATIVE or POSITIVE. 6
7 Excerpt of selected code lists: Controlled Terminology (Code Lists) Section Code Value Code Text POSNEGF, Reference Name (POSNEGF) NEGATIVE POSITIVE NEGATIVE POSITIVE YF, Reference Name (YF) Y YES YNF, Reference Name (YNF) N Y NO YES Notable changes Each referenced code list is bookmarked separately. The 2007 define.xml sample additionally includes a subsection for external code list references like medical dictionaries. Example: Controlled Terminology (External Dictionaries) ADVERSE EVENT DICTIONARY, Reference Name (AEDICT) External Dictionary Dictionary Version MEDDRA 8.0 DRUG DICTIONARY, Reference Name (CMDICT) External Dictionary Dictionary Version WHODRUG HYPERLINKS A standard footer is included at the end of each of the single metadata tables/sections of the 2005 define.xml sample, that links to the annotated CRF, the supplemental data definitions document and the top of define.xml (TOC). Footer example: Annotated Case Report Form (blankcrf.pdf) Supplemental Data Definitions Document (supplementaldatadefinitions.pdf) Go to the top of the define.xml Date of document generation ( T16:59:49) 7
8 Notable changes Since the stylesheet used in the 2007 define.xml sample includes bookmarks for the annotated CRF and the supplemental data definitions document, if available, the footer in this version is reduced to the link to the Top of define.xml and to the Date of document generation. ANNOTATED CRF According to the FDA s ectd Study Data Specifications the annotated CRF has to be named blankcrf and has to be provided in PDF format. All variables captured on the CRF (i.e. with Origin CRF) should appear in the annotated CRF. Annotations for the test short names as described in the variable value level metadata are also helpful. Information on controlled terminology, if attached to a variable, can also be included. Excerpt of an annotated CRF: Notable changes The Draft Metadata Submission Guidelines include detailed recommendations on Annotating CRFs. A blankcrf.pdf file is part in the sample electronic submission, too. Two especially noteworthy recommendations are to Annotate repeat CRF pages with references to the first appearance of the page only, e.g., see Page 5 Annotate data fields not included in the case report tabulation datasets, e.g., monitoring questions like Check if no AE occurred with [NOT SUBMITTED]. SUPPLEMENTAL DATA DEFINITIONS DOCUMENT The supplemental data definitions document is another PDF document. It includes any additional information, that is deemed necessary to facilitate the review of the submitted data, and that do not fit into the metadata structures described above, e.g., general assumptions or flow charts to illustrate derivation dependencies. Notable changes A supplemental data definition document is not included in the latest submission sample provided by CDISC, but the functionality to include such a document is still available. 8
9 DEFINE.XML GENERATION PROCESS After a look at what is required for define.xml, it becomes apparent that a lot of the metadata to be described is already included in the datasets. Thus, rather than creating duplicate information manually, which might be necessary when using other tools, an automated solution for the generation of define.xml based on SAS seems preferable, especially when taking into account the good experiences with the SAS based define.pdf generation. EXPERIENCES WITH DEFINE.PDF AUTOMATION Data definition files for the documentation of submitted datasets have already been requested by the FDA before the CDISC initiative. In the 1999 FDA s guidance document, Providing Regulatory Submissions in Electronic Format [7], define.pdf was described. Since a lot of information required for define.pdf is inherent in the SAS data themselves, an automated process for the generation of define.pdf using SAS was developed at Accovion as presented at PHARMASUG in 2003 [8]. The main ideas of this process are: Use all the metadata already available in SAS Provide the additional information required (basically the contents of the comment column describing each variable) in an Excel spreadsheet Combine metadata and additional information in SAS Create RTF code for the define document in SAS Convert define.rtf into define.pdf with Adobe Acrobat Distiller Comparing the contents of define.xml with the contents of define.pdf as described by the FDA in 1999, more additional information, not already included in the SAS dictionary tables, is required for define.xml. Whereas for define.pdf, only the contents of the comments column has to be provided from an external source, more than one look-up file for additional information is required for define.xml. Nevertheless, a similar approach for creating define.xml is feasible. This time XML code instead of RTF has to be written in SAS. The details on the required XML code can be picked up from the CDISC Case Report Tabulation Data Specification (define.xml), V1.0. The sample define.xml prepared by the CDISC define.xml team also proved to be extremely helpful when familiarizing with the task of define.xml code generation. METADATA ALREADY AVAILABLE IN SAS As mentioned before, a lot of information required to describe the data submitted is inherent in the data themselves. The following information, required for the different define.xml sections/elements, can either be extracted from the SAS dictionary tables, the contents of certain variables of a dataset, or a format catalog. DEFINE.XML SECTION TARGET COLUMN/ELEMENT ATTRIBUTE IN DEFINE.XML Data Metadata Dataset Description Location (= standard path + dataset name) Variable Metadata Variable Label Type (derivation required) Controlled Terms or Format SOURCE DICTIONARY.COLUMNS/ SASHELP.VCOLUMN DICTIONARY.COLUMNS/ SASHELP.VCOLUMN Element attributes not displayed but required for machine-readability: Length Significant Digits Display Format 9
10 DEFINE.XML SECTION TARGET COLUMN/ELEMENT ATTRIBUTE IN DEFINE.XML Variable Value Level Metadata Source Variable Value Label Controlled Terminology/Code Lists Reference Name Code Value Code Text SOURCE Value = Value of source variable in source dataset (e.g., --TESTCD) Label = Value of corresponding test name variable in source dataset (e.g., --TEST) SAS format library Element attributes not displayed but required for machine-readability: Data Type EXCEL FILES FOR SUPPLEMENTARY INFORMATION In order to build the full define.xml, external look-up tables for the information that is not inherent in the SAS data are required. Excel is one possibility to capture the supplementary information because the contents of those files can be easily transferred to SAS. Several Excel files or worksheets are necessary to provide the extra information required, basically at least one file per define.xml section, i.e., Data Metadata (List of Datasets), Variable Metadata, Variable Value Level Metadata and Computational Algorithms. Only the Controlled Terminology/Code Lists Section can be created without additional look-up files. Data Metadata (List of Datasets) The Excel file structure chosen to capture the information for the Data Metadata looks very much like the Data Metadata Section of define.xml in a browser tool. It has the following columns: Dataset (dataset name) Description (dataset label) CDISC Domain Class (e.g., Findings, Interventions, Events, Special Purpose) Structure (e.g., One record per lab test per time point per visit per subject) Purpose ( Tabulation for all SDTM datasets) Keys (e.g., STUDYID, USUBJID, LBTESTCD, VISITNUM, LBTPTNUM for dataset LB) A list of datasets is often generated at the design and specification phase of a programming project. If this list is already prepared in the format mentioned above, redundancies and inconsistencies can be avoided. Variable Metadata When looking for an Excel file structure for the Variable Metadata, the SDTM Domain Models as set up in the CDISC SDTM Implementation Guide, Version 3.1.1, are the ideal starting point. This structure is also worth considering when creating specifications for SDTM datasets. To link the SDTM Domain Model to a specific study, just one additional column is required that contains the contents of comment column for define.xml. 10
11 Excerpt of selected variables from Excel file for Variable Metadata: Except for the addition of the study specific comment column, the following edits might be necessary: Controlled Terms and Formats: The * indicates that there is a controlled terminology for a variable, which will be explained via format codes and decodes in the Controlled Terminology/Code Lists Section. For test short name and test name variables that are further explained by the variable value level metadata this column should be blank. Origin: The contents of this column in the CDISC SDTM Domain Model might include choices, e.g., CRF or Derived. The unique study specific origin is required for define.xml. Study specific comment = Not needed : Not all variables included in the SDTM Domain Models might be required for a study. Permissible variables can be omitted. Rather than deleting these rows from the dataset specification files, one might agree on a certain keyword, e.g., Not needed, in the new study specific comment column. If the structure and contents of the SDTM Domain Models are retained as much as possible, the data that are redundant to the metadata taken from the datasets themselves, e.g., Label, Type and whether a variable is formatted can be used for SDTM conformance checks of the data to be described by define.xml. The extra information for the variable metadata can be organized in either one big file or separate files per domain/dataset. Separate files are preferable when the look-up file preparation is divided between several programmers. Variable Value Level Metadata The Excel look-up file for the variable value level metadata can either be created manually or in SAS using the SDTM datasets contents and the data of the study specific comment column of the variable metadata/dataset specification files. We decided on the automated generation of a draft in SAS as this ensures consistency with the data submitted without any extra checks required. Nevertheless, this Excel file requires manual adaptation, as type, length, significant digits, display format, origin, role and comment might differ between the unique values of a source variable, e.g., numeric lab test results vs. character lab test results. 11
12 Variable value level metadata is required for the trial summary dataset, each findings and each supplemental qualifiers dataset. The columns of the variable value level metadata Excel file, their source and contents, and whether editing might be required after the initial draft has been created, can be depicted from the following table. Column Source/Contents Editing required? Source Dataset Name of source dataset (applicable for findings and No supplemental qualifier datasets) Source Variable Variable --TESTCD for findings datasets, No variable QNAM for supplemental qualifiers datasets Value Each unique value of TSPARMCD, --TESTCD or QNAM, No respectively Label Corresponding value of TSPARM, --TEST or QLABEL No Type Defaulted to text No Length*) Defaulted to: 200 Yes Significant Digits*) Defaulted to: blank Yes Display Format*) Defaulted to: blank Yes Controlled Terms or Format Defaulted to: blank Yes Origin Origin of --ORRES or QVAL according to variable Yes metadata/dataset specification files Comment Comment for --ORRES or QVAL according to variable metadata/dataset specification files Yes *) Not displayed via style sheet define1-0-0.xsl but required according to schema to ensure machine-readability. Computational Algorithms The Excel file structure chosen to capture the information for the Computational Algorithms conforms to the browser representation of the Computational Algorithms Section in define.xml. It has to following columns: Reference Name: Name specified in the comment column of the variable metadata or the variable value level metadata Computation Method: Complex derivation rule expressed in pseudo code A computation method reference rather than the derivation description itself is put into the comment columns of either the variable metadata or the variable value level metadata when the size of the derivation description compromises the tabular structure. PROCESS FLOW The define.xml implementation process chosen at Accovion, using SAS internal metadata and metadata compiled externally, can be summarized as follows. 12
13 1. Create Excel file Dataset Metadata (List of Datasets) 2. Create Excel files Variable Metadata/Dataset Specifications per dataset 3. Create Excel file Computational Algorithms 4. Create permanent formats required for the variables/variable values with a discrete list of values attached (e.g., proc format library=library; value $SEXF M = Male F = Female ;) 5. Create datasets according to the specifications - Assign defined formats to variables with a discrete list of values - Assign numeric formats to variables of type float, e.g., format LBSTRESN 9.3; 6. Create Preliminary Excel file Variable Value Level Metadata in SAS 7. Edit Excel file Variable Value Level Metadata as necessary 8. Create define.xml in SAS - Retrieve metadata available in SAS - Import Excel files with additional information - Combine and structure as required for the define.xml generation - Check metadata inherent in SAS for SDTM conformity - Create required XML code 9. Provide annotated CRF (blankcrf.pdf) for hyperlinking 10. Provide Supplemental Data Definitions Document in PDF format for hyperlinking 11. Provide datasets in SAS V5 transport format for hyperlinking 12. Place define.xml, referenced style sheet, blankcrf.pdf, supplementaldatadefinitions.pdf and the XPT files of the datasets in the same target directory XML CODE GENERATION IN SAS The XML code for define.xml is generated as described in the CDISC define.xml V1.0 standard and shown in the sample define.xml. The XML elements are written to define.xml in the following order. XML header including style sheet reference ODM open tag + schema references + ODM attributes incl. FileOID= <STUDY ID> Study open tag with OID= <STUDY ID> GlobalVariables defining study attributes MetaDataVersion open tag def:annotatedcrf for link to Annotated CRF def:supplementaldoc for link to supplemental data definition file 13
14 def:computationmethod per computational algorithm available (Attribute OID is set to reference name, e.g., COMPMETHOD.LBBLFL ) def:valuelistdef per variable to be explained by variable value level metadata (Attribute OID is set to ValueList.<dataset name>.<variable name>, e.g., ValueList.LB.LBTESTCD ) o ItemRef per variable value of variable listed in def:valuelistdef (Attribute ItemOID is set to <dataset name>.<variable name>.<variable value>, e.g., LB.LBTESTCD.ALB ) ItemGroupDef per dataset for data metadata (Attribute OID is set to dataset name, e.g., LB ) o ItemRef per variable included in the dataset (Attribute ItemOID is set to <dataset name>.<variable name>, e.g., LB.LBTESTCD ) ItemDef per variable referenced in ItemGroupDef (Attribute OID is set to <dataset name>.<variable name>, e.g., LB.LBTESTCD, to link with ItemRef attribute ItemOID for dataset variables; attribute def:computationmethodoid is set to reference name of computational algorithm, if applicable, to link with def:computationmethod attribute OID) o def:valuelistref for link to variable value level metadata, if applicable (Attribute ValueListOID is set to ValueList.<dataset name>.<variable name>, e.g., ValueList.LB.LBTESTCD ) o CodeListRef for link to controlled teminology/code lists, if applicable (Attribute CodeListOID is set to format name, e.g., YF ) ItemDef per variable value referenced in def:valuelistdef (Attribute OID is set to <dataset name>.<variable name>.<variable value>, e.g., LB.LBTESTCD.ALB, to link with ItemRef attribute ItemOID for variable values; attribute def:computationmethodoid and child element CodeListRef as for dataset variables ItemDefs, if applicable) CodeList per controlled terminology/code list name (Attribute OID is set to format name, e.g., YF, to link with the CodeListOID referenced in ItemDef) o CodeListItem per coded value of parent code list MetaDataVersion close tag Study close tag ODM close tag Following the order of elements described above, define.xml can easily be created by several data steps, putting the contents of the combined define.xml metadata datasets into XML code. The dependencies/interactions between the single define.xml elements are illustrated in the following excerpt of define.xml as displayed in a text editor. These dependencies are facilitated in the define1-0-0.xsl style sheet to create the hyperlinks in define.xml when displayed in a browser tool. <def:computationmethod OID="COMPMETHOD.LBBLFL"> Target of internal link 1 Derive mean of pre-treatment measurements. Create new record with result and flag LBBLFL='Y' </def:computationmethod> <def:valuelistdef OID="ValueList.LB.LBTESTCD"> Target of internal link 2 <ItemRef ItemOID="LB.LBTESTCD.ALB" Internal link 3 OrderNumber="1" Mandatory="No"/> </def:valuelistdef> <ItemGroupDef OID="LB" Name="LB" Repeating="Yes" IsReferenceData="No" Purpose="Tabulation" def:label="laboratory Tests" def:structure="one record per lab test per time point per visit per subject" def:domainkeys="studyid USUBJID LBTESTCD VISITNUM LBTPTNUM" def:class="findings" def:archivelocationid="location.lb"> Internal link 4 <ItemRef ItemOID="LB.LBTESTCD" Internal link 5 OrderNumber="5" Mandatory="Yes" Role="Topic"/> 14
15 <ItemRef ItemOID="LB.LBBLFL" Internal link 6 OrderNumber="22" Mandatory="No" Role="Record Qualifier"/> <def:leaf ID="Location.LB" xlink:href="lb.xpt"> Target of internal link 4 + External link <def:title>crt/datasets/<study>/lb.xpt </def:title> </def:leaf> </ItemGroupDef> <ItemDef OID="LB.LBTESTCD" Target of internal link 5 Name="LBTESTCD" DataType="text" Length="15" Origin="CRF" Comment="CRF page 98" def:label="lab Test or Examination Short Name"> <def:valuelistref ValueListOID="ValueList.LB.LBTESTCD"/> Internal link 2 </ItemDef> <ItemDef OID="LB.LBBLFL" Target of internal link 6 Name="LBBLFL" DataType="text" Length="1" Origin="Derived" def:label="baseline Flag" def:computationmethodoid="compmethod.lbblfl"> Internal link 1 <CodeListRef CodeListOID="YF"/> Internal link 7 </ItemDef> <ItemDef OID="LB.LBTESTCD.ALB" Target of internal link 3 Name="ALB" DataType="float" Length="8" SignificantDigits="1" Origin="CRF" Comment="CRF pages 5, 15" def:label="albumin" def:displayformat="5.1"/> <CodeList OID="YF" Target of internal link 7 Name="YF" DataType="text"> <CodeListItem CodedValue="Y"> <Decode> <TranslatedText xml:lang="en">yes</translatedtext> </Decode> </CodeListItem> </CodeList> Special characters like ampersand, apostrophe, quote, less than, greater than must be treated specially. They are replaced with &, ', ", <, >, respectively, when creating define.xml in SAS to ensure XML conformity. OIDs must be unique CONSISTENCY CHECKS When using the CDISC SDTM Domain Models as look-up files for the additional information required for define.xml, their contents can also be used for a SDTM adherence check of the datasets. The following SDTM adherence checks are possible: Availability of datasets and variables Order of variables in a dataset Dataset label and variable labels Variable data type Variables with controlled terminology (a SAS format should be attached) Since the information required for define.xml comes from different sources, it is also important to include define.xml specific consistency checks to ensure conformance to the CRT DD Specification and provision of a well-formed XML file. Those checks include: COMPMETHOD reference but no details provided and vice versa Format reference but format not available in format catalog and vice versa Variable value level metadata link but no variable value level metadata provided and vice versa 15
16 Consistency of data metadata with variable metadata is available through the SAS metadata themselves. ADAPTATION TO THE NEW CDISC DEFINE.XML SAMPLE The define.xml automation process described above was built according to the sample define.xml provided by the CDISC team in For new submissions, the adaptation to the new sample define.xml as included in the Draft Metadata Submission Guidelines Review Package should be considered. When applying the formatting of the new style sheet to a define.xml file constructed according to the 2005 define.xml sample, the majority of the increased functionality (e.g., bookmarks, hyperlinks to SAS transport files from variable metadata, hyperlinks to supplemental qualifiers, if applicable) is already available, because the style sheet enhancements are only built on the information already inherent in the XML file and the SDTM naming conventions (e.g., supplemental qualifier dataset for DM is called SUPPDM). In order to exploit the full functionality of the new style sheet, there are just some minor straightforward changes required to the SAS macro generating the define.xml code. Links to the referenced annotated CRF pages only work when the list of CRF page references is assigned to the Origin attribute of the ItemDef element. Thus, whenever the contents of the comment column from the variable metadata look-up file starts with CRF Page the information has to be assigned to the Origin attribute rather than the Comment attribute. However, this should only be done, if the page numbers printed on the CRF equal the physical page numbers of the blankcrf.pdf. For the links from the Role column values of the variable metadata to the Role Code List the attribute RoleCodeListOID has to be added to each ItemRef, which includes the attribute Role. A new permanent format has to be created for each possible Role value, which will be described in the Controlled Terminology/Code Lists section. The format name becomes the value referenced by RoleCodeListOID. In the CDISC define.xml sample of 2005, -DTC variables were shown as date or datetime variables. According to the Draft Metadata Submission Guidelines, the data type of these variables should be text. Thus, the SAS algorithms to derive the corresponding ODM data type of each variable for the Type attribute of the element ItemDef can even be simplified. The pilot define.xml implementation at Accovion did not include external dictionary references via the Controlled Terms or Format column. The respective coding dictionary and its version were only described in the Comment column of the dictionary coded variables, e.g., AEDECOD. To comply with the latest CDISC define.xml sample, the define.xml code generation could be extended to handle predefined external dictionary reference names, e.g., MEDDRA and WHODD as required. The Draft Metadata Submission Guidelines introduce a specific way to describe the metadata of multiple questionnaire CRFs included in one QS dataset. In order to describe the variable values per questionnaire CRF, a first level of variable value level metadata is created for the QSCAT variable, which contains the different questionnaire names. A second level of variable value level metadata is provided for the QSTESTCD variable per questionnaire, i.e., per unique value of QSCAT. The SAS algorithm has to be extended to describe these data appropriately in the XML code. Finally, when using the style sheet define1-0-0.xsl of the electronic submission sample from 2007, the files define.css, icon1.gif, icon2.gif, and icon3.gif, referenced by this style sheet have to be placed into the submission data folder. EXPECTED DEFINE.XML ENHANCEMENTS The define.xml according to the CDISC Case Report Tabulation Data Definition Specification (define.xml) V1.0 Standard was constructed to meet or exceed the minimum FDA requirements for a data definition file. End of June 2007 the Draft Metadata Submission Guidelines were released for review. This document provides detailed recommendations on how to describe the metadata of SDTM datasets efficiently for regulatory review. It clarifies a lot of the areas not yet unambiguously defined in the CDISC define.xml V1.0 standard. The new define.xml sample referenced in the Draft Metadata Submission Guidelines, includes most of the functionality missed in the first implementation sample of define.xml, e.g., links to single CRF pages. Nevertheless, some further enhancements might come up. CORRECTION OF SOFTWARE ISSUES WITH LATEST DEFINE.XML SAMPLE The internal and external hyperlinks implemented in the style sheet of the 2007 define.xml sample are quite extensive. However, depending on the Software versions used, not all links might work as expected. For correct linking to specific CRF pages Adobe Versions other than Adobe 7 should be used. With Adobe 7 the links will always lead to the first page of the blankcrf.pdf. In some cases the back arrows do not work when navigating within define.xml. This issue might be attributable to the Internet Explorer Version used. 16
17 ADAPTATION TO LATEST CDISC ODM STANDARD The current CDISC define.xml standard is based on the CDISC ODM Version 1.2. The current CDISC ODM Version is Version 1.3. New versions of the define.xml standard will probably be based on the latest available CDISC ODM standard to make use of the new functionality available. IMPROVED PRINTABILITY The human readable browser representation of define.xml works well when reviewing the contents on the screen, but in order to print it properly, a workaround has to be used. The sample electronic submission referenced in the Draft Metadata Submission Guidelines includes a file define.xml_printable.pdf. The final version of the guidelines document might include further information on how this file was created and whether it should generally be included in an electronic submission. EXTENSION FOR CDISC ADAM SPECIFIC METADATA The CDISC define.xml V1.0 standard focuses on the documentation of Case Report Tabulation Datasets in SDTM format. For the statistical review of the study results, more analysis ready datasets constructed according to the Analysis Data Model (ADaM) [5] are also required. These datasets and at least the most important analysis results themselves also have to be documented via standard metadata concepts. The Metadata tables described in the CDISC Analysis Data Model, Version 2.0, are similar but not equal to the metadata sections of the current define.xml. The Analysis Metadata, for example, which describe the most important analysis results and link them to the respective datasets, can only be included in define.xml via a sponsor specific schema extension. However, a standard format is desirable. The CDISC define.xml team is currently working on an integration of the ADaM specific metadata as one of the results from the FDA-CDISC Pilot, a mock submission to the FDA to test the interoperability of the CDISC standards and to demonstrate that these standards meet the needs of the FDA reviewers. EXECUTABLE COMPUTATIONAL ALGORITHMS Currently, the Computational Algorithms Section of define.xml is just a human readable description of the computations or derivations performed. One of the CDISC visions is to also embed some executable code to allow for reproduction of the results submitted. However, this is probably the least likely standard enhancement to be seen in the near future. CONCLUSIONS The SAS based solution for the automated generation of define.xml presented in this paper is just one possibility to implement define.xml. People with more XML experience might chose XML tools rather than SAS to perform this task. For people with a SAS programming background, on the other hand, XML code can easily be created in SAS following the well-documented CDISC define.xml standard and the recommendations provided in the Draft Metadata Submission Guidelines. The more challenging part of the define.xml implementation process is to compile the necessary additional, non-sas inherent, information and to organize them in a format ready for input into SAS. When deciding for a SAS based solution, there are several options to integrate the define.xml generation into the submission datasets preparation process. Structuring the design and specification documents as needed for the define.xml generation increases consistency and avoids redundancy. Facilitating the CDISC SDTM domain model tables as input files, also allows for a SDTM adherence check of the data submitted. When using a SAS based implementation process, no cross checks between the metadata described in define.xml and metadata inherent in the datasets submitted are necessary as there is built-in consistency with the data. Finally, SAS PROC CDISC will probably offer an alternative solution for the define.xml generation in the future. So before starting a define.xml implementation, it might be worth checking on the PROC CDISC status. However, according to the latest development notes on PROC CDISC, SAS has deferred the work on define.xml until the CDISC define team progresses further in their discussion of a new version that supports data submission via XML. REFERENCES 1. CDISC Case Report Tabulation Data Definition Specification (define.xml), Version 1.0, February 9, 2005 ( 2. CDISC Metadata Submission Guidelines, Appendix to the SDTM IG V3.1.1, Draft Version 0.9, July 25, 2007 ( 3. CDISC Study Data Tabulation Model (SDTM) Implementation Guide, Final Version 3.1.1, September 8, 2005 ( 4. CDISC Operational Data Model (ODM), Version 1.3 ( 17
18 5. CDISC Analysis Data Model, Final Version 2.0, November 8, 2006 ( 6. FDA Guidance: electronic Common Technical Document (ectd) ( Study Data Specifications, Version 1.4 ( 7. Guidance for Industry: Providing Regulatory Submissions to the Center for Biologics Evaluation and Research (CBER) in Electronic Format Biologics Marketing Applications by U.S. Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research (CBER), November 1999 ( 8. Using RTF, EPS, PDFMARK to Automate the Creation of Your DEFINE.PDF Document for Electronic Submissions, Dirk Spruck, PharmaSUG 2003 ( ACKNOWLEDGMENTS The author would like to thank Beate Hientzsch, Beate Jakobi-Plöhn, Michael Ludwig, Cordula Massion and Dirk Spruck from Accovion for their valuable help and input. CONTACT INFORMATION Your comments and questions are valued and encouraged. Contact the author at: Monika Kawohl Accovion GmbH Softwarecenter Marburg, Germany Work Phone: Fax: Monika.Kawohl@accovion.com Web: Brand and product names are trademarks of their respective companies. 18
Introduction to define.xml
 Introduction to define.xml Dave Iberson-Hurst 27 th May 2010 ESUG Webinar 1 Outline 2 Outline Introduction Purpose of define.xml XML How define works FAQ Q&A 3 Introduction 4 Introduction 5 Purpose 6 Purpose
Introduction to define.xml Dave Iberson-Hurst 27 th May 2010 ESUG Webinar 1 Outline 2 Outline Introduction Purpose of define.xml XML How define works FAQ Q&A 3 Introduction 4 Introduction 5 Purpose 6 Purpose
Introduction to define.xml
 Introduction to define.xml Dave Iberson-Hurst 22 nd March 2012 (based on a presentation given on 27 th May 2010) ESUG Webinar 1 Outline Introduction Purpose of define.xml XML How define works FAQ Q&A 2
Introduction to define.xml Dave Iberson-Hurst 22 nd March 2012 (based on a presentation given on 27 th May 2010) ESUG Webinar 1 Outline Introduction Purpose of define.xml XML How define works FAQ Q&A 2
Aquila's Lunch And Learn CDISC The FDA Data Standard. Disclosure Note 1/17/2014. Host: Josh Boutwell, MBA, RAC CEO Aquila Solutions, LLC
 Aquila's Lunch And Learn CDISC The FDA Data Standard Host: Josh Boutwell, MBA, RAC CEO Aquila Solutions, LLC Disclosure Note This free training session will be placed on Aquila s website after the session
Aquila's Lunch And Learn CDISC The FDA Data Standard Host: Josh Boutwell, MBA, RAC CEO Aquila Solutions, LLC Disclosure Note This free training session will be placed on Aquila s website after the session
Introduction to Define.xml
 Introduction to Define.xml Bay Area CDISC Implementation Network 4 April 2008 John Brega, PharmaStat LLC Presentation Objectives 1. Introduce the concept and purpose of define.xml 2. Introduce the published
Introduction to Define.xml Bay Area CDISC Implementation Network 4 April 2008 John Brega, PharmaStat LLC Presentation Objectives 1. Introduce the concept and purpose of define.xml 2. Introduce the published
Define.xml - Tips and Techniques for Creating CRT - DDS
 Paper CD09 Define.xml - Tips and Techniques for Creating CRT - DDS Julie Maddox, SAS Institute Inc., Cary, NC, USA Mark Lambrecht, SAS Institute Inc., Tervuren, Belgium ABSTRACT The Case Report Tabulation
Paper CD09 Define.xml - Tips and Techniques for Creating CRT - DDS Julie Maddox, SAS Institute Inc., Cary, NC, USA Mark Lambrecht, SAS Institute Inc., Tervuren, Belgium ABSTRACT The Case Report Tabulation
Study Data Reviewer s Guide Completion Guideline
 Study Data Reviewer s Guide Completion Guideline 22-Feb-2013 Revision History Date Version Summary 02-Nov-2012 0.1 Draft 20-Nov-2012 0.2 Added Finalization Instructions 10-Jan-2013 0.3 Updated based on
Study Data Reviewer s Guide Completion Guideline 22-Feb-2013 Revision History Date Version Summary 02-Nov-2012 0.1 Draft 20-Nov-2012 0.2 Added Finalization Instructions 10-Jan-2013 0.3 Updated based on
Lex Jansen Octagon Research Solutions, Inc.
 Converting the define.xml to a Relational Database to enable Printing and Validation Lex Jansen Octagon Research Solutions, Inc. Leading the Electronic Transformation of Clinical R&D PhUSE 2009, Basel,
Converting the define.xml to a Relational Database to enable Printing and Validation Lex Jansen Octagon Research Solutions, Inc. Leading the Electronic Transformation of Clinical R&D PhUSE 2009, Basel,
Submission-Ready Define.xml Files Using SAS Clinical Data Integration Melissa R. Martinez, SAS Institute, Cary, NC USA
 PharmaSUG 2016 - Paper SS12 Submission-Ready Define.xml Files Using SAS Clinical Data Integration Melissa R. Martinez, SAS Institute, Cary, NC USA ABSTRACT SAS Clinical Data Integration simplifies the
PharmaSUG 2016 - Paper SS12 Submission-Ready Define.xml Files Using SAS Clinical Data Integration Melissa R. Martinez, SAS Institute, Cary, NC USA ABSTRACT SAS Clinical Data Integration simplifies the
CS05 Creating define.xml from a SAS program
 CS05 Creating define.xml from a SAS program Jørgen Mangor Iversen LEO Pharma A/S 21 November 2013 LEO Pharma About LEO Pharma A/S Independent, research-based pharmaceutical company Founded in 1908 Employs
CS05 Creating define.xml from a SAS program Jørgen Mangor Iversen LEO Pharma A/S 21 November 2013 LEO Pharma About LEO Pharma A/S Independent, research-based pharmaceutical company Founded in 1908 Employs
How to handle different versions of SDTM & DEFINE generation in a Single Study?
 Paper CD15 How to handle different versions of SDTM & DEFINE generation in a Single Study? Edwin Ponraj Thangarajan, PRA Health Sciences, Chennai, India Giri Balasubramanian, PRA Health Sciences, Chennai,
Paper CD15 How to handle different versions of SDTM & DEFINE generation in a Single Study? Edwin Ponraj Thangarajan, PRA Health Sciences, Chennai, India Giri Balasubramanian, PRA Health Sciences, Chennai,
Creating Define-XML v2 with the SAS Clinical Standards Toolkit 1.6 Lex Jansen, SAS
 Creating Define-XML v2 with the SAS Clinical Standards Toolkit 1.6 Lex Jansen, SAS Agenda Introduction to the SAS Clinical Standards Toolkit (CST) Define-XML History and Background What is Define-XML?
Creating Define-XML v2 with the SAS Clinical Standards Toolkit 1.6 Lex Jansen, SAS Agenda Introduction to the SAS Clinical Standards Toolkit (CST) Define-XML History and Background What is Define-XML?
Define 2.0: What is new? How to switch from 1.0 to 2.0? Presented by FH-Prof.Dr. Jozef Aerts University of Applied Sciences FH Joanneum Graz, Austria
 1 Define 2.0: What is new? How to switch from 1.0 to 2.0? Presented by FH-Prof.Dr. Jozef Aerts University of Applied Sciences FH Joanneum Graz, Austria 2 What's new in define.xml 2.0? Based on ODM 1.3(.2)
1 Define 2.0: What is new? How to switch from 1.0 to 2.0? Presented by FH-Prof.Dr. Jozef Aerts University of Applied Sciences FH Joanneum Graz, Austria 2 What's new in define.xml 2.0? Based on ODM 1.3(.2)
Beyond OpenCDISC: Using Define.xml Metadata to Ensure End-to-End Submission Integrity. John Brega Linda Collins PharmaStat LLC
 Beyond OpenCDISC: Using Define.xml Metadata to Ensure End-to-End Submission Integrity John Brega Linda Collins PharmaStat LLC Topics Part 1: A Standard with Many Uses Status of the Define.xml Standard
Beyond OpenCDISC: Using Define.xml Metadata to Ensure End-to-End Submission Integrity John Brega Linda Collins PharmaStat LLC Topics Part 1: A Standard with Many Uses Status of the Define.xml Standard
Understanding the define.xml and converting it to a relational database. Lex Jansen, Octagon Research Solutions, Wayne, PA
 Paper HW02 Understanding the define.xml and converting it to a relational database Lex Jansen, Octagon Research Solutions, Wayne, PA ABSTRACT When submitting clinical study data in electronic format to
Paper HW02 Understanding the define.xml and converting it to a relational database Lex Jansen, Octagon Research Solutions, Wayne, PA ABSTRACT When submitting clinical study data in electronic format to
MethodDefs, ValueLists and WhereClauses:
 MethodDefs, ValueLists and WhereClauses: How to Define Data Traceability in an MDR DH03 Judith Goud, Nurocor Agenda Data Traceability Standards Traceability in Clinical Development Systems Conclusion Data
MethodDefs, ValueLists and WhereClauses: How to Define Data Traceability in an MDR DH03 Judith Goud, Nurocor Agenda Data Traceability Standards Traceability in Clinical Development Systems Conclusion Data
Lex Jansen Octagon Research Solutions, Inc.
 Converting the define.xml to a Relational Database to Enable Printing and Validation Lex Jansen Octagon Research Solutions, Inc. Leading the Electronic Transformation of Clinical R&D * PharmaSUG 2009,
Converting the define.xml to a Relational Database to Enable Printing and Validation Lex Jansen Octagon Research Solutions, Inc. Leading the Electronic Transformation of Clinical R&D * PharmaSUG 2009,
It s All About Getting the Source and Codelist Implementation Right for ADaM Define.xml v2.0
 PharmaSUG 2018 - Paper SS-15 It s All About Getting the Source and Codelist Implementation Right for ADaM Define.xml v2.0 ABSTRACT Supriya Davuluri, PPD, LLC, Morrisville, NC There are some obvious challenges
PharmaSUG 2018 - Paper SS-15 It s All About Getting the Source and Codelist Implementation Right for ADaM Define.xml v2.0 ABSTRACT Supriya Davuluri, PPD, LLC, Morrisville, NC There are some obvious challenges
Generating Define.xml from Pinnacle 21 Community
 PharmaSUG 2018 - Paper AD-29 ABSTRACT Generating Define.xml from Pinnacle 21 Community Pinky Anandani Dutta, Inclin, Inc Define.xml is an XML document that describes the structure and contents (metadata
PharmaSUG 2018 - Paper AD-29 ABSTRACT Generating Define.xml from Pinnacle 21 Community Pinky Anandani Dutta, Inclin, Inc Define.xml is an XML document that describes the structure and contents (metadata
Mapping Corporate Data Standards to the CDISC Model. David Parker, AstraZeneca UK Ltd, Manchester, UK
 Mapping Corporate Data Standards to the CDISC Model David Parker, AstraZeneca UK Ltd, Manchester, UK Introduction Discuss CDISC Case Report Tabulations Data Definition, Study Data Tabulation Model, Operational
Mapping Corporate Data Standards to the CDISC Model David Parker, AstraZeneca UK Ltd, Manchester, UK Introduction Discuss CDISC Case Report Tabulations Data Definition, Study Data Tabulation Model, Operational
Planning to Pool SDTM by Creating and Maintaining a Sponsor-Specific Controlled Terminology Database
 PharmaSUG 2017 - Paper DS13 Planning to Pool SDTM by Creating and Maintaining a Sponsor-Specific Controlled Terminology Database ABSTRACT Cori Kramer, Ragini Hari, Keith Shusterman, Chiltern When SDTM
PharmaSUG 2017 - Paper DS13 Planning to Pool SDTM by Creating and Maintaining a Sponsor-Specific Controlled Terminology Database ABSTRACT Cori Kramer, Ragini Hari, Keith Shusterman, Chiltern When SDTM
Edwin Ponraj Thangarajan, PRA Health Sciences, Chennai, India Giri Balasubramanian, PRA Health Sciences, Chennai, India
 Paper CD15 PhUSE 2016 How to handle different versions of SDTM & DEFINE generation in a Single Study? Edwin Ponraj Thangarajan, PRA Health Sciences, Chennai, India Giri Balasubramanian, PRA Health Sciences,
Paper CD15 PhUSE 2016 How to handle different versions of SDTM & DEFINE generation in a Single Study? Edwin Ponraj Thangarajan, PRA Health Sciences, Chennai, India Giri Balasubramanian, PRA Health Sciences,
Customer oriented CDISC implementation
 Paper CD10 Customer oriented CDISC implementation Edelbert Arnold, Accovion GmbH, Eschborn, Germany Ulrike Plank, Accovion GmbH, Eschborn, Germany ABSTRACT The Clinical Data Interchange Standards Consortium
Paper CD10 Customer oriented CDISC implementation Edelbert Arnold, Accovion GmbH, Eschborn, Germany Ulrike Plank, Accovion GmbH, Eschborn, Germany ABSTRACT The Clinical Data Interchange Standards Consortium
Study Data Reviewer s Guide
 Revision History Date Study Data Reviewer s Guide Completion Guideline: Nonclinical (nnsdrg) Version Summary V1.1 03 March 2016 1.0 First Public Version: posted for Public Comment 1.1 Update from Public
Revision History Date Study Data Reviewer s Guide Completion Guideline: Nonclinical (nnsdrg) Version Summary V1.1 03 March 2016 1.0 First Public Version: posted for Public Comment 1.1 Update from Public
Implementing CDISC Using SAS. Full book available for purchase here.
 Implementing CDISC Using SAS. Full book available for purchase here. Contents About the Book... ix About the Authors... xv Chapter 1: Implementation Strategies... 1 The Case for Standards... 1 Which Models
Implementing CDISC Using SAS. Full book available for purchase here. Contents About the Book... ix About the Authors... xv Chapter 1: Implementation Strategies... 1 The Case for Standards... 1 Which Models
The Wonderful World of Define.xml.. Practical Uses Today. Mark Wheeldon, CEO, Formedix DC User Group, Washington, 9 th December 2008
 The Wonderful World of Define.xml.. Practical Uses Today Mark Wheeldon, CEO, Formedix DC User Group, Washington, 9 th December 2008 Agenda Introduction to Formedix What is Define.xml? Features and Benefits
The Wonderful World of Define.xml.. Practical Uses Today Mark Wheeldon, CEO, Formedix DC User Group, Washington, 9 th December 2008 Agenda Introduction to Formedix What is Define.xml? Features and Benefits
Doctor's Prescription to Re-engineer Process of Pinnacle 21 Community Version Friendly ADaM Development
 PharmaSUG 2018 - Paper DS-15 Doctor's Prescription to Re-engineer Process of Pinnacle 21 Community Version Friendly ADaM Development Aakar Shah, Pfizer Inc; Tracy Sherman, Ephicacy Consulting Group, Inc.
PharmaSUG 2018 - Paper DS-15 Doctor's Prescription to Re-engineer Process of Pinnacle 21 Community Version Friendly ADaM Development Aakar Shah, Pfizer Inc; Tracy Sherman, Ephicacy Consulting Group, Inc.
Creating Define-XML version 2 including Analysis Results Metadata with the SAS Clinical Standards Toolkit
 Creating Define-XML version 2 including Analysis Results Metadata with the SAS Clinical Standards Toolkit Lex Jansen Principal Software Developer @ SAS PharmaSUG 2016 Agenda Why Analysis Results Metadata?
Creating Define-XML version 2 including Analysis Results Metadata with the SAS Clinical Standards Toolkit Lex Jansen Principal Software Developer @ SAS PharmaSUG 2016 Agenda Why Analysis Results Metadata?
Data Integrity through DEFINE.PDF and DEFINE.XML
 Data Integrity through DEFINE.PDF and DEFINE.XML Sy Truong, Meta Xceed, Inc, Fremont, CA ABSTRACT One of the key questions asked in determining if an analysis dataset is valid is simply, what did you do
Data Integrity through DEFINE.PDF and DEFINE.XML Sy Truong, Meta Xceed, Inc, Fremont, CA ABSTRACT One of the key questions asked in determining if an analysis dataset is valid is simply, what did you do
SDTM Implementation Guide Clear as Mud: Strategies for Developing Consistent Company Standards
 Paper CD02 SDTM Implementation Guide Clear as Mud: Strategies for Developing Consistent Company Standards Brian Mabe, UCB Biosciences, Raleigh, USA ABSTRACT Many pharmaceutical companies are now entrenched
Paper CD02 SDTM Implementation Guide Clear as Mud: Strategies for Developing Consistent Company Standards Brian Mabe, UCB Biosciences, Raleigh, USA ABSTRACT Many pharmaceutical companies are now entrenched
CDASH Standards and EDC CRF Library. Guang-liang Wang September 18, Q3 DCDISC Meeting
 CDASH Standards and EDC CRF Library Guang-liang Wang September 18, 2014 2014 Q3 DCDISC Meeting 1 Disclaimer The content of this presentation does not represent the views of my employer or any of its affiliates.
CDASH Standards and EDC CRF Library Guang-liang Wang September 18, 2014 2014 Q3 DCDISC Meeting 1 Disclaimer The content of this presentation does not represent the views of my employer or any of its affiliates.
DIA 11234: CDER Data Standards Common Issues Document webinar questions
 Q: What is the preferred data definition format for ADaM analysis data, define.xml or define.pdf? 1 ADaM Define File Q: The CRTDDS does not describe how to submit a define.xml for ADaM. Does CDER expect
Q: What is the preferred data definition format for ADaM analysis data, define.xml or define.pdf? 1 ADaM Define File Q: The CRTDDS does not describe how to submit a define.xml for ADaM. Does CDER expect
SDTM-ETL 3.2 User Manual and Tutorial
 SDTM-ETL 3.2 User Manual and Tutorial Author: Jozef Aerts, XML4Pharma Last update: 2017-07-29 Adding mappings for the Vital Signs domain After loading and validating the ODM file with metadata, your load
SDTM-ETL 3.2 User Manual and Tutorial Author: Jozef Aerts, XML4Pharma Last update: 2017-07-29 Adding mappings for the Vital Signs domain After loading and validating the ODM file with metadata, your load
Linking Metadata from CDASH to ADaM Author: João Gonçalves Business & Decision Life Sciences, Brussels, Belgium
 PhUSE 2016 Paper CD10 Linking Metadata from CDASH to ADaM Author: João Gonçalves Business & Decision Life Sciences, Brussels, Belgium ABSTRACT CDISC standards include instructions describing how variables
PhUSE 2016 Paper CD10 Linking Metadata from CDASH to ADaM Author: João Gonçalves Business & Decision Life Sciences, Brussels, Belgium ABSTRACT CDISC standards include instructions describing how variables
Best Practice for Explaining Validation Results in the Study Data Reviewer s Guide
 Paper DS06 Best Practice for Explaining Validation Results in the Study Data Reviewer s Guide Kristin Kelly, Pinnacle 21 LLC, Plymouth Meeting, PA, USA Michael Beers, Pinnacle 21 LLC, Plymouth Meeting,
Paper DS06 Best Practice for Explaining Validation Results in the Study Data Reviewer s Guide Kristin Kelly, Pinnacle 21 LLC, Plymouth Meeting, PA, USA Michael Beers, Pinnacle 21 LLC, Plymouth Meeting,
esubmission - Are you really Compliant?
 ABSTRACT PharmaSUG 2018 - Paper SS21 esubmission - Are you really Compliant? Majdoub Haloui, Merck & Co., Inc., Upper Gwynedd, PA, USA Suhas R. Sanjee, Merck & Co., Inc., Upper Gwynedd, PA, USA Pinnacle
ABSTRACT PharmaSUG 2018 - Paper SS21 esubmission - Are you really Compliant? Majdoub Haloui, Merck & Co., Inc., Upper Gwynedd, PA, USA Suhas R. Sanjee, Merck & Co., Inc., Upper Gwynedd, PA, USA Pinnacle
Material covered in the Dec 2014 FDA Binding Guidances
 Accenture Accelerated R&D Services Rethink Reshape Restructure for better patient outcomes Sandra Minjoe Senior ADaM Consultant Preparing ADaM and Related Files for Submission Presentation Focus Material
Accenture Accelerated R&D Services Rethink Reshape Restructure for better patient outcomes Sandra Minjoe Senior ADaM Consultant Preparing ADaM and Related Files for Submission Presentation Focus Material
Define.xml 2.0: More Functional, More Challenging
 Define.xml 2.0: More Functional, More Challenging Bay Area CDISC Implementation Network 24 July 2013 John Brega, PharmaStat LLC Presentation Objectives 1. Introduce the new features in Define.xml 2.0 with
Define.xml 2.0: More Functional, More Challenging Bay Area CDISC Implementation Network 24 July 2013 John Brega, PharmaStat LLC Presentation Objectives 1. Introduce the new features in Define.xml 2.0 with
Accessing and using the metadata from the define.xml. Lex Jansen, Octagon Research Solutions, Wayne, PA
 PharmaSUG2010 - Paper HW05 Accessing and using the metadata from the define.xml Lex Jansen, Octagon Research Solutions, Wayne, PA ABSTRACT When submitting clinical study data in electronic format to the
PharmaSUG2010 - Paper HW05 Accessing and using the metadata from the define.xml Lex Jansen, Octagon Research Solutions, Wayne, PA ABSTRACT When submitting clinical study data in electronic format to the
Harmonizing CDISC Data Standards across Companies: A Practical Overview with Examples
 PharmaSUG 2017 - Paper DS06 Harmonizing CDISC Data Standards across Companies: A Practical Overview with Examples Keith Shusterman, Chiltern; Prathima Surabhi, AstraZeneca; Binoy Varghese, Medimmune ABSTRACT
PharmaSUG 2017 - Paper DS06 Harmonizing CDISC Data Standards across Companies: A Practical Overview with Examples Keith Shusterman, Chiltern; Prathima Surabhi, AstraZeneca; Binoy Varghese, Medimmune ABSTRACT
From Implementing CDISC Using SAS. Full book available for purchase here. About This Book... xi About The Authors... xvii Acknowledgments...
 From Implementing CDISC Using SAS. Full book available for purchase here. Contents About This Book... xi About The Authors... xvii Acknowledgments... xix Chapter 1: Implementation Strategies... 1 Why CDISC
From Implementing CDISC Using SAS. Full book available for purchase here. Contents About This Book... xi About The Authors... xvii Acknowledgments... xix Chapter 1: Implementation Strategies... 1 Why CDISC
SDTM-ETL 4.0 Preview of New Features
 SDTM-ETL 4.0 Preview of New Features Author: Jozef Aerts, XML4Pharma Last update: 2018-12-01 Automated installation of new or additional SDTM/SEND templates When new versions of the SDTM-IG or SEND-IG
SDTM-ETL 4.0 Preview of New Features Author: Jozef Aerts, XML4Pharma Last update: 2018-12-01 Automated installation of new or additional SDTM/SEND templates When new versions of the SDTM-IG or SEND-IG
Generating Define.xml Using SAS By Element-by-Element And Domain-by-Domian Mechanism Lina Qin, Beijing, China
 PharmaSUG China 2015-30 Generating Define.xml Using SAS By Element-by-Element And Domain-by-Domian Mechanism Lina Qin, Beijing, China ABSTRACT An element-by-element and domain-by-domain mechanism is introduced
PharmaSUG China 2015-30 Generating Define.xml Using SAS By Element-by-Element And Domain-by-Domian Mechanism Lina Qin, Beijing, China ABSTRACT An element-by-element and domain-by-domain mechanism is introduced
Updates on CDISC Standards Validation
 Updates on CDISC Standards Validation NJ CDISC User Group September 19, 2013 Topics CDISC standards validation initiative FDA update on SEND checks OpenCDISC v1.4.1 release OpenCDISC plans 2 CDISC validation
Updates on CDISC Standards Validation NJ CDISC User Group September 19, 2013 Topics CDISC standards validation initiative FDA update on SEND checks OpenCDISC v1.4.1 release OpenCDISC plans 2 CDISC validation
CDASH MODEL 1.0 AND CDASHIG 2.0. Kathleen Mellars Special Thanks to the CDASH Model and CDASHIG Teams
 CDASH MODEL 1.0 AND CDASHIG 2.0 Kathleen Mellars Special Thanks to the CDASH Model and CDASHIG Teams 1 What is CDASH? Clinical Data Acquisition Standards Harmonization (CDASH) Standards for the collection
CDASH MODEL 1.0 AND CDASHIG 2.0 Kathleen Mellars Special Thanks to the CDASH Model and CDASHIG Teams 1 What is CDASH? Clinical Data Acquisition Standards Harmonization (CDASH) Standards for the collection
Helping The Define.xml User
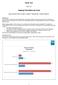 Paper TT01 Helping The Define.xml User Dave Iberson-Hurst, Assero Limited, Teignmouth, United Kingdom ABSTRACT The FDA often comment at industry gatherings on the quality of define.xml files received as
Paper TT01 Helping The Define.xml User Dave Iberson-Hurst, Assero Limited, Teignmouth, United Kingdom ABSTRACT The FDA often comment at industry gatherings on the quality of define.xml files received as
CDISC SDTM and ADaM Real World Issues
 CDISC SDTM and ADaM Real World Issues Washington DC CDISC Data Standards User Group Meeting Sy Truong President MXI, Meta-Xceed, Inc. http://www.meta-x.com Agenda CDISC SDTM and ADaM Fundamentals CDISC
CDISC SDTM and ADaM Real World Issues Washington DC CDISC Data Standards User Group Meeting Sy Truong President MXI, Meta-Xceed, Inc. http://www.meta-x.com Agenda CDISC SDTM and ADaM Fundamentals CDISC
Adding, editing and managing links to external documents in define.xml
 SDTM-ETL 3.1 User Manual and Tutorial Author: Jozef Aerts, XML4Pharma Last update: 2015-05-25 Adding, editing and managing links to external documents in define.xml Define.xml contains the metadata of
SDTM-ETL 3.1 User Manual and Tutorial Author: Jozef Aerts, XML4Pharma Last update: 2015-05-25 Adding, editing and managing links to external documents in define.xml Define.xml contains the metadata of
Preparing the Office of Scientific Investigations (OSI) Requests for Submissions to FDA
 PharmaSUG 2018 - Paper EP15 Preparing the Office of Scientific Investigations (OSI) Requests for Submissions to FDA Ellen Lin, Wei Cui, Ran Li, and Yaling Teng Amgen Inc, Thousand Oaks, CA ABSTRACT The
PharmaSUG 2018 - Paper EP15 Preparing the Office of Scientific Investigations (OSI) Requests for Submissions to FDA Ellen Lin, Wei Cui, Ran Li, and Yaling Teng Amgen Inc, Thousand Oaks, CA ABSTRACT The
Global Checklist to QC SDTM Lab Data Murali Marneni, PPD, LLC, Morrisville, NC Sekhar Badam, PPD, LLC, Morrisville, NC
 PharmaSUG 2018 Paper DS-13 Global Checklist to QC SDTM Lab Data Murali Marneni, PPD, LLC, Morrisville, NC Sekhar Badam, PPD, LLC, Morrisville, NC ABSTRACT Laboratory data is one of the primary datasets
PharmaSUG 2018 Paper DS-13 Global Checklist to QC SDTM Lab Data Murali Marneni, PPD, LLC, Morrisville, NC Sekhar Badam, PPD, LLC, Morrisville, NC ABSTRACT Laboratory data is one of the primary datasets
SAS Application to Automate a Comprehensive Review of DEFINE and All of its Components
 PharmaSUG 2017 - Paper AD19 SAS Application to Automate a Comprehensive Review of DEFINE and All of its Components Walter Hufford, Vincent Guo, and Mijun Hu, Novartis Pharmaceuticals Corporation ABSTRACT
PharmaSUG 2017 - Paper AD19 SAS Application to Automate a Comprehensive Review of DEFINE and All of its Components Walter Hufford, Vincent Guo, and Mijun Hu, Novartis Pharmaceuticals Corporation ABSTRACT
Experience of electronic data submission via Gateway to PMDA
 PharmaSUG 2018 - Paper EP-21 ABSTRACT Experience of electronic data submission via Gateway to PMDA Iori Sakakibara, Kumiko Kimura, Amgen Astellas BioPharma K.K. and Laurence Carpenter, Amgen Ltd PMDA started
PharmaSUG 2018 - Paper EP-21 ABSTRACT Experience of electronic data submission via Gateway to PMDA Iori Sakakibara, Kumiko Kimura, Amgen Astellas BioPharma K.K. and Laurence Carpenter, Amgen Ltd PMDA started
Dealing with changing versions of SDTM and Controlled Terminology (CT)
 CDISC UK Network Breakout session Notes 07/06/16 Afternoon Session 1: Dealing with changing versions of SDTM and Controlled Terminology (CT) How do people manage this? Is this managed via a sponsor Standards
CDISC UK Network Breakout session Notes 07/06/16 Afternoon Session 1: Dealing with changing versions of SDTM and Controlled Terminology (CT) How do people manage this? Is this managed via a sponsor Standards
Dataset-XML - A New CDISC Standard
 Dataset-XML - A New CDISC Standard Lex Jansen Principal Software Developer @ SAS CDISC XML Technologies Team Single Day Event CDISC Tools and Optimization September 29, 2014, Cary, NC Agenda Dataset-XML
Dataset-XML - A New CDISC Standard Lex Jansen Principal Software Developer @ SAS CDISC XML Technologies Team Single Day Event CDISC Tools and Optimization September 29, 2014, Cary, NC Agenda Dataset-XML
Robust approach to create Define.xml v2.0. Vineet Jain
 Robust approach to create Define.xml v2.0 Vineet Jain DEFINE SYSTEM GOALS Generic Powerful Reliable Integrable Efficient Works across SDTM, ADaM & SEND Create define.xml, annotated CRF & define.pdf Create
Robust approach to create Define.xml v2.0 Vineet Jain DEFINE SYSTEM GOALS Generic Powerful Reliable Integrable Efficient Works across SDTM, ADaM & SEND Create define.xml, annotated CRF & define.pdf Create
How to write ADaM specifications like a ninja.
 Poster PP06 How to write ADaM specifications like a ninja. Caroline Francis, Independent SAS & Standards Consultant, Torrevieja, Spain ABSTRACT To produce analysis datasets from CDISC Study Data Tabulation
Poster PP06 How to write ADaM specifications like a ninja. Caroline Francis, Independent SAS & Standards Consultant, Torrevieja, Spain ABSTRACT To produce analysis datasets from CDISC Study Data Tabulation
Paper FC02. SDTM, Plus or Minus. Barry R. Cohen, Octagon Research Solutions, Wayne, PA
 Paper FC02 SDTM, Plus or Minus Barry R. Cohen, Octagon Research Solutions, Wayne, PA ABSTRACT The CDISC Study Data Tabulation Model (SDTM) has become the industry standard for the regulatory submission
Paper FC02 SDTM, Plus or Minus Barry R. Cohen, Octagon Research Solutions, Wayne, PA ABSTRACT The CDISC Study Data Tabulation Model (SDTM) has become the industry standard for the regulatory submission
Codelists Here, Versions There, Controlled Terminology Everywhere Shelley Dunn, Regulus Therapeutics, San Diego, California
 ABSTRACT PharmaSUG 2016 - Paper DS16 lists Here, Versions There, Controlled Terminology Everywhere Shelley Dunn, Regulus Therapeutics, San Diego, California Programming SDTM and ADaM data sets for a single
ABSTRACT PharmaSUG 2016 - Paper DS16 lists Here, Versions There, Controlled Terminology Everywhere Shelley Dunn, Regulus Therapeutics, San Diego, California Programming SDTM and ADaM data sets for a single
XML4Pharma's ODM Study Designer New features of version 2010-R1 and 2010-R2
 XML4Pharma's ODM Study Designer New features of version 2010-R1 and 2010-R2 Author: Jozef Aerts, XML4Pharma Last Version: September 7, 2010 New features of version 2010-R2 Search panel further improved
XML4Pharma's ODM Study Designer New features of version 2010-R1 and 2010-R2 Author: Jozef Aerts, XML4Pharma Last Version: September 7, 2010 New features of version 2010-R2 Search panel further improved
Introduction to ADaM and What s new in ADaM
 Introduction to ADaM and What s new in ADaM Italian CDISC UN Day - Milan 27 th October 2017 Silvia Faini Principal Statistical Programmer CROS NT - Verona ADaM Purpose Why are standards needed in analysis
Introduction to ADaM and What s new in ADaM Italian CDISC UN Day - Milan 27 th October 2017 Silvia Faini Principal Statistical Programmer CROS NT - Verona ADaM Purpose Why are standards needed in analysis
Mapping and Terminology. English Speaking CDISC User Group Meeting on 13-Mar-08
 Mapping and Terminology English Speaking CDISC User Group Meeting on 13-Mar-08 Statement of the Problem GSK has a large drug portfolio, therefore there are many drug project teams GSK has standards 8,200
Mapping and Terminology English Speaking CDISC User Group Meeting on 13-Mar-08 Statement of the Problem GSK has a large drug portfolio, therefore there are many drug project teams GSK has standards 8,200
PharmaSUG Paper PO21
 PharmaSUG 2015 - Paper PO21 Evaluating SDTM SUPP Domain For AdaM - Trash Can Or Buried Treasure Xiaopeng Li, Celerion, Lincoln, NE Yi Liu, Celerion, Lincoln, NE Chun Feng, Celerion, Lincoln, NE ABSTRACT
PharmaSUG 2015 - Paper PO21 Evaluating SDTM SUPP Domain For AdaM - Trash Can Or Buried Treasure Xiaopeng Li, Celerion, Lincoln, NE Yi Liu, Celerion, Lincoln, NE Chun Feng, Celerion, Lincoln, NE ABSTRACT
Study Composer: a CRF design tool enabling the re-use of CDISC define.xml metadata
 Paper SD02 Study Composer: a CRF design tool enabling the re-use of CDISC define.xml metadata Dr. Philippe Verplancke, XClinical GmbH, Munich, Germany ABSTRACT define.xml is often created at the end of
Paper SD02 Study Composer: a CRF design tool enabling the re-use of CDISC define.xml metadata Dr. Philippe Verplancke, XClinical GmbH, Munich, Germany ABSTRACT define.xml is often created at the end of
Sandra Minjoe, Accenture Life Sciences John Brega, PharmaStat. PharmaSUG Single Day Event San Francisco Bay Area
 Sandra Minjoe, Accenture Life Sciences John Brega, PharmaStat PharmaSUG Single Day Event San Francisco Bay Area 2015-02-10 What is the Computational Sciences Symposium? CSS originally formed to help FDA
Sandra Minjoe, Accenture Life Sciences John Brega, PharmaStat PharmaSUG Single Day Event San Francisco Bay Area 2015-02-10 What is the Computational Sciences Symposium? CSS originally formed to help FDA
Improving Metadata Compliance and Assessing Quality Metrics with a Standards Library
 PharmaSUG 2018 - Paper SS-12 Improving Metadata Compliance and Assessing Quality Metrics with a Standards Library Veena Nataraj, Erica Davis, Shire ABSTRACT Establishing internal Data Standards helps companies
PharmaSUG 2018 - Paper SS-12 Improving Metadata Compliance and Assessing Quality Metrics with a Standards Library Veena Nataraj, Erica Davis, Shire ABSTRACT Establishing internal Data Standards helps companies
Now let s take a look
 1 2 3 4 Manage assets across the end to end life cycle of your studies This includes forms, datasets, terminologies, files, links and more, for example: - Studies may contain the protocol, a set of Forms,
1 2 3 4 Manage assets across the end to end life cycle of your studies This includes forms, datasets, terminologies, files, links and more, for example: - Studies may contain the protocol, a set of Forms,
OpenCDISC Validator 1.4 What s New?
 OpenCDISC Validator 1.4 What s New? Bay Area CDISC Implementation Network 23 May 2013 David Borbas Sr Director, Data Management Jazz Pharmaceuticals, Inc. Disclaimers The opinions expressed in this presentation
OpenCDISC Validator 1.4 What s New? Bay Area CDISC Implementation Network 23 May 2013 David Borbas Sr Director, Data Management Jazz Pharmaceuticals, Inc. Disclaimers The opinions expressed in this presentation
PharmaSUG Paper DS24
 PharmaSUG 2017 - Paper DS24 ADQRS: Basic Principles for Building Questionnaire, Rating and Scale Datasets Nancy Brucken, inventiv Health, Ann Arbor, MI Karin LaPann, Shire, Lexington, MA ABSTRACT Questionnaires,
PharmaSUG 2017 - Paper DS24 ADQRS: Basic Principles for Building Questionnaire, Rating and Scale Datasets Nancy Brucken, inventiv Health, Ann Arbor, MI Karin LaPann, Shire, Lexington, MA ABSTRACT Questionnaires,
Paper DS07. Generating Define.xml and Analysis Result Metadata using Specifications, Datasets and TFL Annotation
 Paper DS07 Generating Define.xml and Analysis Result Metadata using Specifications, s and TFL Annotation Mark Crangle, ICON Clinical Research, Marlow, United Kingdom ABSTRACT The most recent addition to
Paper DS07 Generating Define.xml and Analysis Result Metadata using Specifications, s and TFL Annotation Mark Crangle, ICON Clinical Research, Marlow, United Kingdom ABSTRACT The most recent addition to
SDTM Attribute Checking Tool Ellen Xiao, Merck & Co., Inc., Rahway, NJ
 PharmaSUG2010 - Paper CC20 SDTM Attribute Checking Tool Ellen Xiao, Merck & Co., Inc., Rahway, NJ ABSTRACT Converting clinical data into CDISC SDTM format is a high priority of many pharmaceutical/biotech
PharmaSUG2010 - Paper CC20 SDTM Attribute Checking Tool Ellen Xiao, Merck & Co., Inc., Rahway, NJ ABSTRACT Converting clinical data into CDISC SDTM format is a high priority of many pharmaceutical/biotech
PhUSE EU Connect Paper PP15. Stop Copying CDISC Standards. Craig Parry, SyneQuaNon, Diss, England
 Paper PP15 Abstract Stop Copying CDISC Standards Craig Parry, SyneQuaNon, Diss, England We repeatedly see repositories which require a large amount of front loading, a lot of duplicating of the Clinical
Paper PP15 Abstract Stop Copying CDISC Standards Craig Parry, SyneQuaNon, Diss, England We repeatedly see repositories which require a large amount of front loading, a lot of duplicating of the Clinical
SAS, XML, and CDISC. Anthony T Friebel XML Development Manager, SAS XML Libname Engine Architect SAS Institute Inc.
 SAS, XML, and CDISC Anthony T Friebel XML Development Manager, SAS XML Libname Engine Architect SAS Institute Inc. SAS is a registered trademark or trademark of SAS Institute Inc. in the USA and other
SAS, XML, and CDISC Anthony T Friebel XML Development Manager, SAS XML Libname Engine Architect SAS Institute Inc. SAS is a registered trademark or trademark of SAS Institute Inc. in the USA and other
ADaM Compliance Starts with ADaM Specifications
 PharmaSUG 2017 - Paper DS16 ADaM Compliance Starts with ADaM Specifications Trevor Mankus, Kent Letourneau, PRA Health Sciences ABSTRACT As of December 17th, 2016, the FDA and PMDA require that all new
PharmaSUG 2017 - Paper DS16 ADaM Compliance Starts with ADaM Specifications Trevor Mankus, Kent Letourneau, PRA Health Sciences ABSTRACT As of December 17th, 2016, the FDA and PMDA require that all new
What is high quality study metadata?
 What is high quality study metadata? Sergiy Sirichenko PhUSE Annual Conference Barcelona, 2016 What is study metadata? Trial Design domains Reviewer s Guides acrf Define.xml Conclusion Topics What is study
What is high quality study metadata? Sergiy Sirichenko PhUSE Annual Conference Barcelona, 2016 What is study metadata? Trial Design domains Reviewer s Guides acrf Define.xml Conclusion Topics What is study
ABSTRACT INTRODUCTION WHERE TO START? 1. DATA CHECK FOR CONSISTENCIES
 Developing Integrated Summary of Safety Database using CDISC Standards Rajkumar Sharma, Genentech Inc., A member of the Roche Group, South San Francisco, CA ABSTRACT Most individual trials are not powered
Developing Integrated Summary of Safety Database using CDISC Standards Rajkumar Sharma, Genentech Inc., A member of the Roche Group, South San Francisco, CA ABSTRACT Most individual trials are not powered
Automate Clinical Trial Data Issue Checking and Tracking
 PharmaSUG 2018 - Paper AD-31 ABSTRACT Automate Clinical Trial Data Issue Checking and Tracking Dale LeSueur and Krishna Avula, Regeneron Pharmaceuticals Inc. Well organized and properly cleaned data are
PharmaSUG 2018 - Paper AD-31 ABSTRACT Automate Clinical Trial Data Issue Checking and Tracking Dale LeSueur and Krishna Avula, Regeneron Pharmaceuticals Inc. Well organized and properly cleaned data are
Legacy to SDTM Conversion Workshop: Tools and Techniques
 Legacy to SDTM Conversion Workshop: Tools and Techniques Mike Todd President Nth Analytics Legacy Data Old studies never die Legacy studies are often required for submissions or pharmacovigilence. Often
Legacy to SDTM Conversion Workshop: Tools and Techniques Mike Todd President Nth Analytics Legacy Data Old studies never die Legacy studies are often required for submissions or pharmacovigilence. Often
Streamline SDTM Development and QC
 Paper SI09 Streamline SDTM Development and QC Stephen Gormley, Amgen, United Kingdom ABSTRACT Amgen s Global Statistical Programming ( GSP ) function have one centralised team (The CDISC Consultancy and
Paper SI09 Streamline SDTM Development and QC Stephen Gormley, Amgen, United Kingdom ABSTRACT Amgen s Global Statistical Programming ( GSP ) function have one centralised team (The CDISC Consultancy and
An Efficient Solution to Efficacy ADaM Design and Implementation
 PharmaSUG 2017 - Paper AD05 An Efficient Solution to Efficacy ADaM Design and Implementation Chengxin Li, Pfizer Consumer Healthcare, Madison, NJ, USA Zhongwei Zhou, Pfizer Consumer Healthcare, Madison,
PharmaSUG 2017 - Paper AD05 An Efficient Solution to Efficacy ADaM Design and Implementation Chengxin Li, Pfizer Consumer Healthcare, Madison, NJ, USA Zhongwei Zhou, Pfizer Consumer Healthcare, Madison,
Automating the Creation of Data Definition Tables (Define.pdf) Using SAS Version 8.2
 Automating the Creation of Data Definition Tables (Define.pdf) Using SAS Version 8.2 Eugene Yeh, PharmaNet, Inc., Cary, NC Syamala Kasichainula, PharmaNet, Inc., Cary, NC Katie Lanier, PharmaNet, Inc.,
Automating the Creation of Data Definition Tables (Define.pdf) Using SAS Version 8.2 Eugene Yeh, PharmaNet, Inc., Cary, NC Syamala Kasichainula, PharmaNet, Inc., Cary, NC Katie Lanier, PharmaNet, Inc.,
SDTM-ETL 3.1 User Manual and Tutorial
 SDTM-ETL 3.1 User Manual and Tutorial Author: Jozef Aerts, XML4Pharma Last update: 2014-07-19 Creating mappings for the AE domain Now that we have created (and executed) mappings for several domains, let
SDTM-ETL 3.1 User Manual and Tutorial Author: Jozef Aerts, XML4Pharma Last update: 2014-07-19 Creating mappings for the AE domain Now that we have created (and executed) mappings for several domains, let
ADaM Reviewer s Guide Interpretation and Implementation
 Paper CD13 ADaM Reviewer s Guide Interpretation and Implementation Steve Griffiths, GlaxoSmithKline, Stockley Park, UK ABSTRACT Throughout the course of a study, teams will make a lot of decisions about
Paper CD13 ADaM Reviewer s Guide Interpretation and Implementation Steve Griffiths, GlaxoSmithKline, Stockley Park, UK ABSTRACT Throughout the course of a study, teams will make a lot of decisions about
Define.xml tools supporting SEND/SDTM data process
 Define.xml tools supporting SEND/SDTM data process - Using SAS Clinical Standards Toolkit PRESENTER / Kirsten Walther Langendorf DATE / 11. Oct 2017 OUTLINE Business Process High-level SEND process Creating
Define.xml tools supporting SEND/SDTM data process - Using SAS Clinical Standards Toolkit PRESENTER / Kirsten Walther Langendorf DATE / 11. Oct 2017 OUTLINE Business Process High-level SEND process Creating
Business & Decision Life Sciences
 Business & Decision Life Sciences Moving to Define.xml v2.0.0 for CDISC FSUG Anne-Sophie Bekx / 22 May 2014 Introduction Differences Impact Introduction History of Define.xml February 2005: CRT-DDS Final
Business & Decision Life Sciences Moving to Define.xml v2.0.0 for CDISC FSUG Anne-Sophie Bekx / 22 May 2014 Introduction Differences Impact Introduction History of Define.xml February 2005: CRT-DDS Final
PharmaSUG Paper AD03
 PharmaSUG 2017 - Paper AD03 Three Issues and Corresponding Work-Around Solution for Generating Define.xml 2.0 Using Pinnacle 21 Enterprise Jeff Xia, Merck & Co., Inc., Rahway, NJ, USA Lugang (Larry) Xie,
PharmaSUG 2017 - Paper AD03 Three Issues and Corresponding Work-Around Solution for Generating Define.xml 2.0 Using Pinnacle 21 Enterprise Jeff Xia, Merck & Co., Inc., Rahway, NJ, USA Lugang (Larry) Xie,
The Benefits of Traceability Beyond Just From SDTM to ADaM in CDISC Standards Maggie Ci Jiang, Teva Pharmaceuticals, Great Valley, PA
 PharmaSUG 2017 - Paper DS23 The Benefits of Traceability Beyond Just From SDTM to ADaM in CDISC Standards Maggie Ci Jiang, Teva Pharmaceuticals, Great Valley, PA ABSTRACT Since FDA released the Analysis
PharmaSUG 2017 - Paper DS23 The Benefits of Traceability Beyond Just From SDTM to ADaM in CDISC Standards Maggie Ci Jiang, Teva Pharmaceuticals, Great Valley, PA ABSTRACT Since FDA released the Analysis
Comparison of FDA and PMDA Requirements for Electronic Submission of Study Data
 Comparison of FDA and PMDA Requirements for Electronic Submission of Study Data Monika Kawohl Statistical Programming Accovion CDISC GSUG Meeting 15-Sep-2015 1 References FDA Website: Study Data Standards
Comparison of FDA and PMDA Requirements for Electronic Submission of Study Data Monika Kawohl Statistical Programming Accovion CDISC GSUG Meeting 15-Sep-2015 1 References FDA Website: Study Data Standards
NCI/CDISC or User Specified CT
 NCI/CDISC or User Specified CT Q: When to specify CT? CT should be provided for every variable with a finite set of valid values (e.g., the variable AESEV in ADAE can have the values MILD, MODERATE or
NCI/CDISC or User Specified CT Q: When to specify CT? CT should be provided for every variable with a finite set of valid values (e.g., the variable AESEV in ADAE can have the values MILD, MODERATE or
Automated Creation of Submission-Ready Artifacts Silas McKee, Accenture, Pennsylvania, USA Lourdes Devenney, Accenture, Pennsylvania, USA
 Paper DH06 Automated Creation of Submission-Ready Artifacts Silas McKee, Accenture, Pennsylvania, USA Lourdes Devenney, Accenture, Pennsylvania, USA ABSTRACT Despite significant progress towards the standardization
Paper DH06 Automated Creation of Submission-Ready Artifacts Silas McKee, Accenture, Pennsylvania, USA Lourdes Devenney, Accenture, Pennsylvania, USA ABSTRACT Despite significant progress towards the standardization
Considerations on creation of SDTM datasets for extended studies
 13/May/2016 As one of the activities of CDISC Japan User Group (CJUG), a small group, "Extension study team" was organized in 2015. The team discussed what kind of approach works better for SDTM creation
13/May/2016 As one of the activities of CDISC Japan User Group (CJUG), a small group, "Extension study team" was organized in 2015. The team discussed what kind of approach works better for SDTM creation
SAS 9.3 CDISC Procedure
 SAS 9.3 CDISC Procedure User s Guide SAS Documentation The correct bibliographic citation for this manual is as follows: SAS Institute Inc. 2011. SAS 9.3 CDISC Procedure: User s Guide. Cary, NC: SAS Institute
SAS 9.3 CDISC Procedure User s Guide SAS Documentation The correct bibliographic citation for this manual is as follows: SAS Institute Inc. 2011. SAS 9.3 CDISC Procedure: User s Guide. Cary, NC: SAS Institute
define.xml: A Crash Course Frank DiIorio
 sponsor requests ODM extensions XSL-FO schema/xsd define.xml define.xml: A Crash Course metadata tables XML4Pharma metadata interface (the other) define.pdf ODM itext Frank DiIorio CodeCrafters, Inc. Philadelphia
sponsor requests ODM extensions XSL-FO schema/xsd define.xml define.xml: A Crash Course metadata tables XML4Pharma metadata interface (the other) define.pdf ODM itext Frank DiIorio CodeCrafters, Inc. Philadelphia
%check_codelist: A SAS macro to check SDTM domains against controlled terminology
 Paper CS02 %check_codelist: A SAS macro to check SDTM domains against controlled terminology Guido Wendland, UCB Biosciences GmbH, Monheim, Germany ABSTRACT The SAS macro %check_codelist allows programmers
Paper CS02 %check_codelist: A SAS macro to check SDTM domains against controlled terminology Guido Wendland, UCB Biosciences GmbH, Monheim, Germany ABSTRACT The SAS macro %check_codelist allows programmers
Power Data Explorer (PDE) - Data Exploration in an All-In-One Dynamic Report Using SAS & EXCEL
 Power Data Explorer (PDE) - Data Exploration in an All-In-One Dynamic Report Using SAS & EXCEL ABSTRACT Harry Chen, Qian Zhao, Janssen R&D China Lisa Lyons, Janssen R&D US Getting to know your data is
Power Data Explorer (PDE) - Data Exploration in an All-In-One Dynamic Report Using SAS & EXCEL ABSTRACT Harry Chen, Qian Zhao, Janssen R&D China Lisa Lyons, Janssen R&D US Getting to know your data is
Revision of Technical Conformance Guide on Electronic Study Data Submissions
 Notification No. 0824001 August 24, 2016 To: Prefectural Health Department (Bureau) Director of the Advanced Review with Electronic Data Promotion Group, Pharmaceuticals and Medical Devices Agency Revision
Notification No. 0824001 August 24, 2016 To: Prefectural Health Department (Bureau) Director of the Advanced Review with Electronic Data Promotion Group, Pharmaceuticals and Medical Devices Agency Revision
Advantages of a real end-to-end approach with CDISC standards
 Advantages of a real end-to-end approach with CDISC standards Dr. Philippe Verplancke CEO XClinical GmbH 26th Annual EuroMeeting 25-27 March 2014 ACV, Vienna Austria Disclaimer The views and opinions expressed
Advantages of a real end-to-end approach with CDISC standards Dr. Philippe Verplancke CEO XClinical GmbH 26th Annual EuroMeeting 25-27 March 2014 ACV, Vienna Austria Disclaimer The views and opinions expressed
IS03: An Introduction to SDTM Part II. Jennie Mc Guirk
 IS03: An Introduction to SDTM Part II Jennie Mc Guirk SDTM Framework 1. Where should the data go? 3. What is the minimum information needed? 2. What type of information should it contain? SDTM Framework:
IS03: An Introduction to SDTM Part II Jennie Mc Guirk SDTM Framework 1. Where should the data go? 3. What is the minimum information needed? 2. What type of information should it contain? SDTM Framework:
Optimization of the traceability when applying an ADaM Parallel Conversion Method
 Paper SI04 Optimization of the traceability when applying an ADaM Parallel Conversion Method DEBRUS Roxane, Business & Decision Life Sciences, Brussels, Belgium ABSTRACT One of the methods to create CDISC
Paper SI04 Optimization of the traceability when applying an ADaM Parallel Conversion Method DEBRUS Roxane, Business & Decision Life Sciences, Brussels, Belgium ABSTRACT One of the methods to create CDISC
Making a List, Checking it Twice (Part 1): Techniques for Specifying and Validating Analysis Datasets
 PharmaSUG2011 Paper CD17 Making a List, Checking it Twice (Part 1): Techniques for Specifying and Validating Analysis Datasets Elizabeth Li, PharmaStat LLC, Newark, California Linda Collins, PharmaStat
PharmaSUG2011 Paper CD17 Making a List, Checking it Twice (Part 1): Techniques for Specifying and Validating Analysis Datasets Elizabeth Li, PharmaStat LLC, Newark, California Linda Collins, PharmaStat
SAS offers technology to facilitate working with CDISC standards : the metadata perspective.
 SAS offers technology to facilitate working with CDISC standards : the metadata perspective. Mark Lambrecht, PhD Principal Consultant, Life Sciences SAS Agenda SAS actively supports CDISC standards Tools
SAS offers technology to facilitate working with CDISC standards : the metadata perspective. Mark Lambrecht, PhD Principal Consultant, Life Sciences SAS Agenda SAS actively supports CDISC standards Tools
A Taste of SDTM in Real Time
 A Taste of SDTM in Real Time Changhong Shi, Merck & Co., Inc., Rahway, NJ Beilei Xu, Merck & Co., Inc., Rahway, NJ ABSTRACT The Study Data Tabulation Model (SDTM) is a Clinical Data Interchange Standards
A Taste of SDTM in Real Time Changhong Shi, Merck & Co., Inc., Rahway, NJ Beilei Xu, Merck & Co., Inc., Rahway, NJ ABSTRACT The Study Data Tabulation Model (SDTM) is a Clinical Data Interchange Standards
