Boosting k-nearest Neighbor Queries Estimating Suitable Query Radii
|
|
|
- Harvey Oswin Leonard
- 5 years ago
- Views:
Transcription
1 Boosting k-nearest Neighbor Queries Estimating Suitable Query Radii Marcos R. Vieira, Caetano Traina Jr., Agma J.M. Traina, Adriano Arantes, Christos Faloutsos Department of Computer Science, George Mason University, Fairfax, VA - USA Computer Science Department, University of Sao Paulo at Sao Carlos, SP - Brazil Department of Computer Science, Carnegie Mellon University, Pittsburgh, PA - USA mrvieira@gmu.edu, {caetano agma arantes}@icmc.usp.br, christos@cs.cmu.edu Abstract This paper proposes novel and effective techniques to estimate a radius to answer k-nearest neighbor queries. The first technique targets datasets where it is possible to learn the distribution about the pairwise distances between the elements, generating a global estimation that applies to the whole dataset. The second technique targets datasets where the first technique cannot be employed, generating estimations that depend on where the query center is located. The proposed k-nnf () algorithm combines both techniques, achieving remarkable speedups. Experiments performed on both real and synthetic datasets have shown that the proposed algorithm can accelerate k-nn queries more than 26 times compared with the incremental algorithm and spends half of the total time compared with the traditional k-n N() algorithms. 1 Introduction Complex data, such as genomic sequences and spatial and multimedia data, are searched mainly by similarity criteria, which motivated the development of algorithms to retrieve data based on similarity. Comparing two elements based on their similarity relies on a measurement, often calculated by a distance function δ : S S R +, which compares a pair of elements of the data domain S and returns a numerical value that is smaller as the elements become more similar. A distance function is called a metric when for any elements s 1, s 2, s 3 S it satisfies the following properties: δ(s 1, s 1 ) = 0 and δ(s 1, s 2 ) > 0, s 1 s 2 (non-negativeness); δ(s 1, s 2 ) = δ(s 2, s 1 ) (symmetry); δ(s 1, s 3 ) δ(s 1, s 2 ) + δ(s 2, s 3 ) (triangular inequality). Metrics are fundamental to create access methods to index a dataset S S, the so-called Metric Access Methods (MAM), which is true when S is a complex data domain. A MAM, such as the M-tree [2] and the Slimtree [15], can accelerate similarity queries of complex data by orders of magnitude. There are basically two types of similarity queries, both of which search a dataset S S for elements s i S regarding their similarity to a query center s q S: the Similarity Range Query (RQ) and the k-nearest Neighbor Query (knnq). The first retrieves every element in the dataset nearer than a given radius r q to the query center. A range query example on a dataset S of genomic sequences is: Q1: Choose the polypeptide chains that are different from the chain p by at most 5 codons. The second retrieves the k elements in the dataset nearest to the query center. An example of a k-nn query on S is: Q2: Choose the 10 polypeptide chains closer to polypeptide chains p. Due to the high computational cost to calculate distances between pairs of elements in complex domains, similarity queries often take advantage of index structures. Trees are the most common index structures for metric domains, where each node typically stores a data element called the representative, a subset of elements, and a covering radius, so that no element stored in the node or in any of its subtrees is farther from the representative than the covering radius. In non-leaf nodes, each element stored is the representative of a node of the next tree level. Index structures employ the triangular inequality property of metric domains to prune subtrees, using a limiting radius: a node whose representative is farther than the limiting radius added to its covering radius never contributes to the answer. An algorithm Range(s q, r q ) executes range queries using r q as the limiting radius, thus the pruning ability of the Range algorithms using index structures is typically high. However, there is no a priori limiting radius to perform a k-nn query. A k-nn(s q, k) algorithm starts collecting a list L of k elements, sorted by the distance δ(s q, s i ) of each element s i S to the query center s q. The limiting radius dynamically keeps track of the largest distance from the query center to the elements in L. Whenever a nearer element is found, it is included in the list, removing the farthest element and reducing the limiting radius accordingly. The limiting radius must start with a value larger than the maximum distance between any pair of elements in the dataset (or simply with infinity). Until at least k elements have been found, no pruning can be executed. Moreover, while r L is larger than the final distance r k of the true k-th element nearest to s q, the false answers 1
2 included in L cannot be discarded. If a proper limiting radius can be set from the beginning, a significant reduction of distance calculations can be achieved. Therefore, the problem posed is : How to estimate a suitable radius r k to execute a Range Query - RQ that returns the same elements of a k-nnq? This paper proposes two novel techniques to precisely estimate the final limiting radius r f of a k-nn query, taking advantage of the intrinsic dimensionality of the dataset. The first technique targets datasets where the global data distribution can be obtained by estimating their intrinsic dimensionality, which is the large class of self-similar datasets. The second technique targets datasets where it is not possible to assume a global data distribution, therefore local estimations are generated for each query, depending on where the query center is located. Based on those estimates, we developed a new algorithm, called k-nnf (s q, k), which uses those estimations to accelerate k-nn queries. In the experiments performed, our technique decreased the number of distance calculations up to 37%, the number of disk accesses up to 18%, bringing an overwhelming gain in time, as it reduced the total time demanded by the query processing up to 26 times. The remainder of this paper is structured as follows: Section 2 discusses the related works; Section 3 provides a short description of the Correlation Fractal Dimension concept, which is employed to estimate the intrinsic dimension of a dataset; Section 4 introduces the proposed algorithms and the mathematical support used to define the estimated radius r k and the k-nnf () algorithm; Section 5 presents the experiments results; and Section 6 gives the conclusions of this paper. 2 Related Work The development of algorithms to answer similarity queries has motivated much research based on index structures. A common approach is the branch-andbound where an indexing tree is traversed from the root, and at each step a heuristic is used to determine which branches can be pruned from the search and which one must be traversed next. One of the most influential algorithms in this category was proposed by Roussopoulos et al. [10] to find the k-nearest neighbors in multidimensional datasets stored in R-trees [4]. This algorithm has also been used in MAM to perform similarity searching following the branch-and-bound approach, which we call here the df (depth-first) k-n N() algorithm. The heuristic of this algorithm involves choosing in a node the next subtree to be explored, which is the subtree closest to the query object. Thus, one priority queue is used for each node visited. In [2], another algorithm is proposed that creates a unique global list (i.e. it is employed only one priority queue for the whole query) of nodes not yet traversed. When a new subtree must be traversed, it proceeds from the node closest to the s q, regardless of its level in the tree. Using the globally closest node generates an algorithm considered better than the df k-n N() algorithm, which is called the bf (best-first) k-nn() algorithm. Another approach uses incremental techniques to answer similarity queries, such as the work of Hjaltason and Samet [5], which retrieves the (k +1) nearest neighbor after the first k have been found. This algorithm employs the triangular inequality property to delay the real distance calculations as much as possible, using a priority queue to keep track of which objects or subtrees must be analyzed next. The triangular inequality limits the minimum and the maximum distance allowable for each element s i to s q, using the distance from s q to a node representative s rep. A dynamic limiting radius can be derived by composing the minima and maxima distances of several nodes, even before performing the first real distance calculation between the s q and a stored element. Metric trees, such as the Slim-tree, store the distance from s rep to each subtree (or to the stored elements in leaf nodes). This algorithm delays pruning subtrees and objects until no improvement can be made to the minima and maxima limits, making its priority queue management very costly. The performance of the priority queue is highly affected by the dataset intrinsic dimensionality, as we show in the experiments in Section 5. We call this the inc k-nn() algorithm, and together with the df and bf, this is one of the k-nn() algorithms that we compare to our proposed algorithm. Previous works trying to estimate a limiting radius for k-nn queries were presented in [12, 6, 9]. Their approaches are based on a probability density function to estimate a limiting radius, using a histogram of distances based on chosen pivots. The drawbacks are that it requires a costly construction of a histogram of the distribution of distances between the dataset objects and a set of pivots and it does not capture distinct distributions over different parts of the data space. Moreover, the histogram must be updated if the dataset is modified, as well as the number of pivots. Also, the pivots to be used in this approach need to be carefully chosen. Our approach differs from the previous works because it does not need sampling as the others do; it only uses the intrinsic dimensionality of the dataset. Moreover, by using the intrinsic dimensionality of the dataset instead of the number of attributes (even in spatial datasets), our approach avoids the problem of object dispersion in high-dimensional spaces [7], allowing it to deal with the varying density of elements in each query region. 2
3 3 Background Uniformity and independence assumptions have long been discredited to model data distributions [1]. Real data overwhelmingly disobey these assumptions because they typically are skewed and have subtle dependencies between attributes, leading the majority of estimates and cost models to deliver inaccurate, pessimistic figures [8]. On the other side, some experimental evidence has shown that the distribution of distances between pairs of elements, in the majority of real datasets, presents a fractal behavior or self-similarity; the properties of parts of the dataset in a usable range of scales being similar to the properties of the whole dataset. In self-similar datasets the distribution of distances between elements follows power laws [11, 3] as follows: Given a set of N objects in a dataset with a distance function δ(), the average number k of neighbors within a given distance r is proportional to r raised to D, where D is the correlation fractal dimension of the dataset [3]. Thus, the pair-count P C(r) of pairs of elements within distance r follows the power law: P C(r) = K p r D, (1) where K p is a proportionality constant. Whenever a metric between pairs of elements is defined for a dataset, a graph depicting Equation 1 can be drawn, even when the dataset is not in a dimensional domain. For the majority of real datasets, this graph, called the Distance Plot, is plotted in log-log scales results in an almost straight line for a significant range of distances. The slope of the line in the Distance Plot is the exponent D in Equation 1, so it is called the Distance Exponent [14]. The results are independent of the base of the logarithm used (in this paper we use the natural logarithm log e (x)). It is interesting to note that D closely approximates the correlation fractal dimension of a dataset, and therefore its intrinsic dimensionality [11]. Hence, D can be seen as a measurement of the distribution of distances between elements, even for non-spatial datasets. Figure 1 shows the distance plot of a dataset whose elements are the geographical coordinates 1 of streets and roads in Montgomery County, MD, USA (MGCounty). As can be seen, the plots are linear for the most requested range sizes (typically we are not interested in radius much smaller or larger than the typical distances involved in the dataset). The Distance Exponent D of any dataset can be calculated as the slope of the line that best fits the resulting curve in the Distance Plot, as in Figure 1. Therefore, Equation 1 can be expressed as log(p C(r)) = D log(r) + K d, K d = log(k p ). (2) D has many interesting properties derived from the Correlation Fractal Dimension. The main property is 1 This dataset is in the US Census Bureau Tiger format Fractal Dimension of MGCounty D= Log(r) Figure 1. Distance Plot of the MGCounty. that D is invariant to the size of the dataset, provided that a reasonable number of elements exists [3]. Therefore, the slopes of the lines corresponding to distinct datasets S from the same data domain S are always the same, regardless of the S cardinality. This property enables us to know D of a dataset even if the dataset had been updated. 4 How to estimate a suitable initial radius for k-nn queries In this section we present the main contributions of this paper. Our goal is to take advantage of the intrinsic dimension of a dataset S to speed up k-nn(s q, k) queries estimating a final radius r f, therefore the majority of the elements unlikely to pertain to the answer set are pruned right away, even before k elements have been inserted in L. The improved algorithm, which we call k-nnf (s q, k), performs three steps: (1) Estimates the final radius r f, using a global estimate for r k, (2) Performs a radius-limited k-nn() algorithm using r f, (3) Refines the search procedure, using a local estimate for r f, if the required number of objects k was not retrieved. The main ideas here are: (a) use the fractal dimension to estimate the ideal radius r f ; (b) over-estimating r f in a systematic way; and (c) react when the estimate r q is lower than the real final radius. The first and third steps require calculating respectively a global and a local estimate for the final radius r f. In the next subsections we present techniques to calculate them. The second step can be performed by a new algorithm kandrange(s q, k, r), defined as follows: Definition 1 k-nearest and Range Query: Given an element s q S, a quantity k and a distance r R +, a k-nearest and Range algorithm kandrange(s q, k, r) retrieves at most k elements s i S δ(s q, s i ) r, such that for every non-retrieved element s j, δ(s q, s j ) δ(s q, s i ) for any retrieved element s i. The algorithm kandrange(s q, k, r) can be implemented by modifying an existing k-nn(s q, k) algorithm changing the dynamic query radius initialization from infinity to r f (obtained in the first step). The resulting algorithm answers a composite query, equivalent to the intersection of a Range(s q, r) and a k-nn(s q, k) using the same s q. That is, considering the dataset S, the following queries are better expressed in relational algebra, 3
4 as: Fractal Dimension of MGCounty = = Log(r) Figure 2. How to use the Distance Plot to obtain the global estimate r f = GEst(k). σ ( Range(s q,r q) k NN(s q,k)) S σ Range(sq,r q)s σ k NN(sq,k)S σ kandrange(sq,k,r q)s. For example, this algorithm alone could answer queries such as: Q3: Select the 10 nearest restaurants not farther than a mile from here kandrange(here, 10, 1mile). Notice that the algorithm will recover less than 10 restaurants if the range given is not sufficiently large. 4.1 Calculating a global estimate for the final query radius The first step of the proposed k-nnf (s q, k) algorithm requires a method to estimate the final radius r f of the k- NN query, so it can be used to call kandrange(s q, k, r f ). It supposes that the elements in the dataset follow a mapped distribution regarding the distances between the elements (see Equation 1). Therefore, the resulting estimate is global for the whole dataset, independent of where the query is centered. The global estimate is defined as follows: Definition 2 Global Estimate GEst(k): Given the number of neighbors k to be retrieved, GEst(k) returns the estimated distance r f of the equivalent range query that return k elements, based on the assumption that the dataset follows a self-similar distribution. Figure 2 illustrates the main idea to obtain GEst(k) using the Distance Plot of the dataset. To perform the estimate, an adequate line with slope D, depicted as Line 1, must be chosen to convert the number of elements k into the corresponding estimated radius r f. The GEst(k) method assumes that, as long as the general characteristics of the dataset regarding its distance distribution are preserved when the dataset is updated (i.e. the updates are performed randomly), the distance exponent remains the same. This means that the line best fitting the slope of the Distance Plot can go up or down as elements are inserted or deleted, but its slope is preserved. However, besides knowing the slope, one must define a particular line for the current dataset, using a known point in the graph. We propose using the total number of objects N currently in the dataset and the diameter R of the dataset. This pair of values defines a point in the graph, which we call Point 0. The line of slope D including Point 0 estimates how to convert from number of elements to radius. The number N is easily obtained (it is the number of elements in the dataset), and the diameter R can be estimated from the indexing structure, as the diameter of the root node of the indexing tree. Point 0 can be calculated through Equation 2. However, the equation uses the number of pairs P C(r) within a distance r, and we are interested in the number of elements k involved. Therefore we must convert numbers of elements into numbers of pairs within a distance. The number of pairs in a subset of k elements (counting each pair only once) is P airs(k) = k(k 1)/2. Thus, given a dataset with cardinality N, the number of pairs separated by distances less than the diameter of the dataset is P C(R) = P airs(n) = N(N 1)/2, and a line specific to the dataset when a query is issued can be found considering Point 0 to be log(r), log(p airs(n)) on the Distance Plot of the dataset. Using this line, the number of elements k that form pairs at a distance less or equal r can be estimated using P C(r) = P airs(k). Line 0 in Figure 2 is the one originally employed to calculate the intrinsic dimension D of the dataset (the line that best fits the Distance Plot). It approximates the average number of points (in log scale) over the full range of distances that occur in the dataset. Notice that real datasets usually have fewer distances with values near the diameter of the dataset, therefore the number of the largest distances increases at a slower pace than those of medium or small distances. This tendency explains why there is a typical flattening of pair-counting plots at large radius of real datasets, as shown in Figure 1. Now let us consider Line 1 passing at Point 0 = log(r), log(p airs(n)) (see Figure 2). As it represents the relationship between the radii and the number of element pairs within each radius in the whole dataset, it is adequate to estimate the final radius r f of a k-nn query. The constant K d from Equation 2 applied over Line 1 can be calculated as: K d = log(p airs(n)) D log(r) = log (N(N 1)/2) D log(r). (3) Notice that Equation 2 calculates the number of pairs in a set of k elements within a given distance. If we want to know the number of distances between a subset of k elements contained in S and the set of all N elements stored in the dataset S, then Equation 2 turns into: P airs(k) = N(k 1)/2. (4) 4
5 Combining Equation 3 with 4 and 2 we obtain: log(r f ) = [log(p C(r)) K d ]/D = [log (N(k 1)/2) log (N(N 1)/2)+D log(r)]/d r f = R exp ([log (k 1) log (N 1)]/D) (5) Fractal Dimension of MGCounty Equation 5 shows how to calculate the global estimate for the final radius r f = GEst(k) of a k-nearest neighbor query. The final radius r f estimated by GEst(k) is an approximation, as the exact radius of a query also depends on the local density of elements around the s q. Therefore, if the query is centered where the density of elements is similar to or higher than the average density of the whole dataset, then calling kandrange(s q, k, r f ) in Step 2 of algorithm k-nnf (s q, k) using r f estimated by GEst(k) answers the k-nn query with a much better performance than the traditional k-nn(s q, k) algorithms. However, if r f is under-estimated, then the kandrange(s q, k, r f ) algorithm will return fewer elements than required (that is, a quantity k < k of elements), and another call to the kandrange() algorithm with a larger radius is required. As calling this algorithm a second time reduces the overall performance gain, it would help to slightly augment the estimate r f to reduce the odds of having to call the kandrange() algorithm more than once. In fact, an augment is already embodied in Equation 5, when we estimate the size of the dataset by looking at the indexing structure: as the covering radius of the nodes of metric trees needs to guarantee that every element stored at each subtree is covered by that radius, they are always equal to or larger than the minimum required radius. 4.2 Calculating a local estimate for the final query radius The third step of the proposed k-nnf (s q, k) algorithm is executed when r f reveals to be under-estimated, despite the inflated diameter obtained from the root node of the metric tree. In this case, another search is required with a larger value r f > r f. Calculating r f must take into account the local density of elements around the s q, hence a local estimate LEst(), defined as follows, must be performed. Definition 3 Local Estimate LEst(): Knowing that the local data distribution around the s q retrieves k elements with a radius r f, then LEst(k, k, r f ) returns the local estimate r f required to retrieve the required number of neighbors k. To develop the LEst() algorithm, we take advantage of the Distance Plot again. We know that the first call to algorithm kandrange() using the estimated radius r f = GEst(k) returned an insufficient quantity k of elements. Therefore, that first call was in fact a probe of the 5 = = = Log(r) Figure 3. How to use the Distance Plot to obtain the local estimate r f = LEst(k, k, r f ). space around the s q, and the value k can be used to estimate the local density of the dataset around the s q. Let us reproduce Figure 2 in Figure 3, marking the point given by log(r f ), log(p airs(k )) as Point 1. This point corresponds to the actual amount of elements k within distance r f around the s q. Now, let us draw another line with the same slope D containing this point, and call it Line 2. Line 2 reflects the local density of the region where the query is centered, consequently it is useful to calculate the local estimate. The radius r f can be estimated by using equation 5, now using Line 2 and Point 1. This leads to: r f = r f exp ([log (k 1) log (k 1)]/D) (6) Equation 6 shows how to calculate the local estimate for the final radius r f = LEst(k, k, r f ) of a k-nn query considering the local density of elements around the s q. Once the local estimate r f has been calculated, another incursion in the dataset must be performed to retrieve the additional k k elements. This operation can be performed by another algorithm, defined as follows: Definition 4 kringrange: Given an element s q S, the inner radius r f, the outer radius r f and the maximum number of elements k k still to be retrieved, the kringrange(s q, k k, r f, r f ) algorithm searches the dataset to find at most k k elements s i in the ring centered at s q, so that r f < δ(s q, s i ) r f. The kringrange() algorithm is basically a kandrange() algorithm modified to prune not only elements and nodes that are outside the ball limited by the outer radius r f, but also the nodes and elements that are inside the ball defined by the inner radius r f. The radius r f can be reduced whenever the required number of objects are found. The first call to LEst() and to the kringrange() algorithm retrieves k elements, where k k k. It is not expected to have k < k, but it can. In this case, the point log(r f ), log(p airs(k )) can be used to estimate another radius r f. This new estimate must then be used 5
6 in another call to the kringrange() algorithm, repeating this last step until the desired number of elements k are retrieved. The k-nnf () algorithm, shown as Algorithm 1, uses kandrange(), kringrange(), GEst() and LEst() to perform the same duty of a usual k-n N() algorithm, receiving the same parameters and producing the same results, but with a better performance, as we show in the next section. Algorithm 1 k-nnf (s q, k) Output: The list L of k pairs OId, δ(s ki, s q ). 1: Obtain N as the number of elements in the dataset 2: Obtain R as the diameter of the dataset indexed 3: Clear list L 4: Set r f = GEst(k) 5: Execute KAndRange(s q, k, r f ), store the answer in L, and set k as the number of retrieved elements 6: while k k do 7: Set r f = LEst(k, k, r f ) 8: Execute kringrange(s q, r f, r f, k k ), store the answer in L, and set k to the number of elements in L 9: set r f =r f 10: Return L 5 Experimental Results To evaluate the effectiveness of the presented approach, we worked on a variety of synthetic and real datasets, experimenting with a variety of metrics, including vectorial and non-vectorial ones. As it is not possible to discuss all of them here, we selected three real and one synthetic datasets that represent and summarize the behavior of the proposed technique. The experimental results show that the estimates given by GEst() and LEst() are accurate, leading to the building of an efficient k-nnf () algorithm. The datasets are described as follows. The Corel- Histo is a dataset of color histograms extracted from 68,040 images from the Corel Image Collection ( This is a real world dataset, where each element is a 32-dimensional vector (we consider it as a high-dimensional dataset). The MetricHisto is a real world dataset consisting of 40,000 metric histograms of medical gray-level images. It does not have a predefined embedded dimension, since the number of buckets (instead of bins in traditional histograms) varies from one image to another. Its elements are compared using the M HD() metric function [13]. The MG- County consists of a 2-dimensional coordinates dataset with 27,282 road intersections in Montgomery County, MD, Figure 4(a). The Sierpinsky is a synthetic dataset with 531,441 points of a Sierpinsky triangle, Figure 4(b). These are points from a perfect fractal, whose behavior is predictable, so it is useful to corroborate the correctness of the theoretical assumptions made in this work. Figure 4. The two-dimensional vector datasets used in the experiments: (a) MGCounty and (b) Sierpinsky triangle. Table 1. Summary of the datasets. Dataset D #Attrib. # Objs Metric CorelHisto ,040 Manhattan MetricHisto ,000 MHD() MGCounty ,282 Euclidean Sierpinsky ,441 Euclidean SieDVar 1.56 (up to 256) 531,441 Euclidean We also used several variations of the Sierpinsky dataset, adding up to 254 new attributes (SieDVar), calculated by polynomial expressions of the first two, to evaluate the behavior of the algorithms for datasets with varying embedded dimensionality and the same intrinsic dimensionality. Only 10% of each dataset were used to calculate its D, as it is close to the one obtained using the whole dataset. Table 1 summarizes the main features of the datasets, including their intrinsic dimension D, number of attributes (for the spatial datasets), number of elements, and the metric employed. All algorithms were implemented in C++ using the GNU gcc compiler. The kringrange() and kandrange() algorithms were implemented following the approach given by the bf k-nn() algorithm. We implemented the inc k-n N() algorithm following the description and the best parameters given in [5]. The experiments were performed on an Intel Pentium IV 1.6GHz machine with 128 MB of RAM memory, using the Linux operating system. Each measured point in a plot corresponds to 500 k- NN queries asking for the same number of neighbors at different query centers. The 500 query centers were sampled from the respective datasets, where half of them were removed from the dataset. Therefore 250 queries ask for neighbors from centers stored in the dataset, and 250 queries ask for neighbors from centers that are not in the dataset, but that correspond to centers likely to be used in real queries. The size of each disk page of the Slim-tree was 16KB for the CorelHisto, 4KB for the MG- County and the MetricHisto, and 1KB for the Sierpinsky datasets. We used the default configuration parameters to build each Slim-tree. Tests performed with different page sizes obtained similar results. 5.1 Performance improvement This section discusses the results of experiments comparing the proposed k-nnf () algorithm with the most 6
7 representatives of the three existing classes (as explained in Section 2): the df k-nn(), the bf k-nn() and the inc k-nn() algorithms. Moreover, if the distance r k from the query center to the farthest element in the final list of k nearest neighbors could be known in advance, then a Range() limited by r k could be used in place of the k-n N() algorithm because the processing of Range() is more efficient. Therefore, we also compared the k-nn() algorithms to the standard Range() algorithm, using the limiting radius r k previously discovered by one of the k-n N() algorithms. This later test provides a theoretical lower bound of the best possible algorithm to find the nearest neighbors. All five algorithms were implemented using the Slim-tree as the indexing method. In this experiment, the number of neighbors in the k-nn queries varies from 5 to 100 in steps of 5 elements. For each dataset, we measured the average number of distance calculations, the average number of disk accesses and the average total time (in seconds) required by each algorithm to perform each set of 500 k-nn queries. The number of distance calculations is important because comparing complex elements can be very costly. The reported number of disk accesses were obtained counting the number of requests generated by the algorithm, therefore issues regarding uses of cache memory were not taken into account. The objective of these measurements is to provide clues about the amount of data required by the algorithm, as the same index structure is used for every algorithm. However, the most important measurement is the total time, as it reports the global complexity of the algorithm. Thus, reducing the processing time is the ultimate objective of improving the performance of any algorithm. Figure 5 shows the results of experiments on the four datasets. The first column shows the results from the CorelHisto dataset, the second from the MetricHisto, the third from the MGCounty, and the fourth column from the Sierpinsky dataset. The first row corresponds to the average number of disk accesses required to answer each query for the number of neighbors corresponding to the abscissas, the second row gives the average number of distance calculations, and the third row presents the average total time for 500 queries at each point in the plots. Figure 5 shows that, as expected, no k-n N() algorithm can perform better than the Range() algorithm, as it is the theoretical lower bound. It also shows that the average number of disk accesses and distance calculations required by the inc k-nn() algorithm follows very closely those required by the theoretical best algorithm (the Range() one), as both plots are practically coincidental for every dataset. However, the inc k-nn() algorithm achieves these results at the expenses of a heavy internal processing required to maintain the minima and maxima distances for the elements retrieved from the index structure. This processing turns the inc k-nn() al- Table 2. Total time (s) for k=10 and k=100. Dataset df k-nn() bf k-nn() k-nnf () Range() inc k-nn() k=10 k=10 k=10 k=10 k=10 CorelHisto MetricHisto MGCounty k=100 k=100 k=100 k=100 k=100 CorelHisto MetricHisto MGCounty Table 3. AvgOper for all queues (k=20 and k=40). Dataset df k-nn() bf k-nn() k-nnf () inc k-nn() k=20 k=40 k=20 k=40 k=20 k=40 k=20 k=40 CorelHisto ,601 2,856 MetricHisto ,997 3,354 MGCounty ,210 Sierpinsky ,304 1,482 gorithm the slowest for every dataset, as shown in the third row of Figure 5. To better compare the other algorithms regarding time, Table 2 reproduces the values for k=10 and k=100 of the third row of Figure 5 for the first three datasets. The measurements show that the inc k-nn() algorithm is up to 26 times slower than the others for the real world datasets (i.e., the CorelHisto, MetricHisto and MG- County datasets). Therefore, the inc k-n N() algorithm is valuable regarding the theoretical aspects of reducing the number of disk accesses and distance calculations, but it is impractical, as what really matters is the total time. Figure 5 shows that, whereas the average number of disk accesses and distance calculations required by every algorithm present a sub-linear behavior for increasing number of neighbors, the average total time presents a slightly super-linear behavior. This is due to the increasing complexity to maintain the internal structures of the algorithms. In fact, the k-nn() algorithms use a priority queue whose management has super-linear complexity regarding the number of elements managed. On the other side, the complexity of the proposed k-nnf () algorithm is much smaller, reducing the processing time accordingly. Table 3 shows the total number of queue operations (add/remove elements to/from it) (AvgOper) averaged for 500 queries, for the four k-nn() algorithms, using k = 20 and k = 40. As we see, the queue cost for the k-nnf () algorithm is smaller than any other, contributing to making it the fast algorithm among the k-nn() algorithms. Three aspects from this table enforce our finds: (1) the larger the embedded dimension of the dataset, the worse the performance of the inc k-nn() algorithm; (2) the cost of the k-nnf () queue is much smaller than the cost of the inc k-nn() queue (up to 50 times smaller for the AvgOper); and (3) the k-nnf () algorithm has smaller queue cost regarding any other algorithm. Comparing the df and the bf k-nn() algorithms in Table 2 and in the first and second rows of Figure 5, we can see that the bf k-nn() is slightly better than the df k-n N(). However, comparing the df and the bf k-nn() with the proposed k-nnf (), it can be seen that the k-n N F () algorithm significantly reduces the three 7
8 Figure 5. Comparing the proposed k-nnf () with the other algorithms to answer 500 queries. parameters measured (disk, distance, and time), for every dataset. The k-nnf () algorithm decreases up to 37% (MGCounty and Sierpinsky - df k-nn) the need for distance calculations, decreases up to 18% the number of disk accesses (CorelHisto and Sierpinsky - df k-nn) and it spends half of total time than the traditional k-nn() algorithms (Sierpinsky). Comparing the k-n N F () algorithm with the inc k-nn() algorithm (the best theoretical algorithm), it is up to 26 times faster than the later algorithm (MGCounty). The k-n N F () algorithm achieves its best performance for the metric dataset MetricHisto, mainly considering that the Range() algorithm is a lower bound for improvements. As can be seen in Table 2, it can answer k-nn queries, spending at most 5% more time than the Range() algorithm, whereas its closest competitor requires at least 17% more time. The Range() algorithm is the fastest because its internal structure does not require a priority queue to order by distance all subtrees to be visited, nor does it require the adjustment of the dynamic radius. As the k-nnf () algorithm is based on the kandrange() algorithm, it does need a priority queue, but it remains smaller than the others as a limiting query radius is available from the beginning. Excluding the Range() algorithm which requires a previous knowledge of the maximum radius, the proposed k-nnf () algorithm is always the fastest one for every dataset evaluated. 5.2 Internal behavior of the k-n N F () algorithm In this section we present measurements about the internal behavior of the k-n N F () algorithm, showing statistics about the estimates GEst() and LEst(). Figure 6 shows the Distance Plot of the MGCounty dataset superimposed with the final radius r f from the query center to its k-th nearest neighbor for the various values of k employed to generate the third column of plots from Figure 5 (with k varying from 5 to 100 in steps of 5 elements). Each point shown as a represents the point ln(r f ), ln(p airs(k)) in the log-log scale of the graph, which is used as parameters of the kandrange() algorithms. It is clearly seen that the set of points are over a line passing close to ln(r), ln(p airs(n)) ( Point 0 in Figure 3), the rightmost point of the Distance Plot used to obtain the fractal dimension of the dataset. Therefore, these points represent the global estimate GEst(k) for each number of neighbors k asked in the queries. Figure 6 also shows the average distance of r f and the minimum/maximum radii for each value of k, measured from the set of 500 queries. As can be seen, the averages are roughly over the Distance Plot curve, confirming that it gives a suitable estimation of the final distance r k. Moreover, it also shows that a sample of the dataset, such as the centers of the 500 queries, can be used to estimate 8
9 Figure 7. Scalability tests for k-nnf (). Figure 6. Global estimations for the distances from the query centers to their k-th NN and the average and minimum/maximum of the real distances measured, for the MGCounty dataset. the intrinsic dimension D of the dataset, as shown in the Figure 6. It also shows that the increase in the estimation of r f (provided by the flattening of the curve and the super-estimation of the dataset radius) is enough to lead the number of required callings to kringrange() to a very small value, as the estimated values of r f are always close to the maximum value r k. Therefore, Figure 6 confirms that the global estimations GEst() is accurate. An thought-provoking point to check is determining when the kringrange() algorithm needs to be called, that is, when a local estimation LEst() is required. To verify this point, we measured the number of times that kringrange() is called at each set of 500 queries using the same number k of required elements (step 8 of Algorithm 1). It turned out that the kringrange() algorithm was never called a second time. In fact, the algorithm is seldom called. Only for k = 100 the kringrange() was called 5 out of 500 queries for the CorelHisto dataset, but was never called for the other datasets, showing that the GEst() estimation is accurate. The fact that the kringrange() algorithm was never called twice confirms that the local estimation LEst() is also very accurate. 5.3 Scalability and Dimensionality In this section we present results of experiments performed to evaluate the behavior of the proposed k-nnf () algorithm when varying the dataset size. To perform these experiments, we shuffled each dataset randomly and divided it in 10 parts of equal size. We created the index structure by inserting the first part and measured the first point of the plots. Thereafter the next part was inserted and the second point was taken, and so on, until having the measurements for the complete dataset. As before, each point in the plots corresponds to the average total time of 500 queries with different query centers. We performed experiments asking for 10, 20, 30, and 40 nearest neighbors (500 queries for each value of k). Figure 7 presents the plots for the average total time to answer 500 queries for the MetricHisto (a) and Sierpinsky (b) dataset. As can be seen, the results reveal an essentially linear behavior for every measured parameter. Figure 7(b) presents the plots for the Sierpinsky dataset. As can be seen, the results reveal sub-linear behavior. The plots for average number of disk accesses and distance calculations follow a similar pattern (not shown here). The experiments with other datasets revealed that the general behavior is at least linear for high dimensional and non-dimensional (pure metric) datasets and is sub-linear for the low dimensional datasets. We also performed experiments to evaluate the behavior of the proposed k-nnf () algorithm when varying the embedded dimension. To evaluate these experiments, we generated several versions of the SieDVar dataset, increasing the embedded dimension and keeping its intrinsic dimension constant. The extra dimensions were generated as linear combinations of the previous dimensions to increase the embedded dimension while keeping the fractal dimension unchanged. We created one index structure for each version, having the same height, node occupation and other parameters, keeping the same properties for every structure of each dataset, aiming at performing a fair comparison. As before, each point in the plots correspond to the average total time of 500 queries with different query centers, asking for 10 nearest neighbors. Figure 8 presents the plots for the various versions of the SieDVar dataset, varying the embedded dimension from 2 (original dataset) to 256. As can be seen, the results reveal that our proposed technique works very well for datasets with high embedded dimensionality. On the other hand, the inc k-n N() algorithm degenerates very fast when increasing the embedded dimensionality. From this experiment, we can conclude that as the dimensionality grows, the performance of the inc k-nn() algorithm deteriorates. On the other hand, the k-nnf () algorithm maintains its good performance when the dimensionality of the dataset increases, maintaining its position as the best of all algorithms. 6 Conclusions This paper aims to develop techniques to explore the intrinsic dimensionality of a dataset, measured by its correlation fractal dimension, to estimate the final radius of 9
10 real improvement in answering k-nn queries, the proposed algorithm is simple to implement, consumes very little additional memory and requires less computational power than the competing algorithms in the literature. As additional research problems, we are interested in using the global and local estimates to improve clustering and classification algorithms, and also interested in employing the results of traditional k-nn and other similarity queries to calculate the intrinsic dimensionality D of a dataset. Figure 8. The embedded dimensionality test. a k-nearest neighbor query to accelerate the query processing. Its main contributions are the following: (1) Two methods to estimate the final radius were proposed and implemented. The first one assumes the elements in the dataset following a known distribution (given by D obtained from the Distance Plot) regarding the distances between the elements and generates a global estimate GEst(). The second method takes into account the local density of elements around the query center, and generates a local estimate LEst(); (2) Using the two estimates provided by GEst() and LEst(), the paper presents the new k-n N F () algorithm, which improves processing k-nn queries over both spatial and metric datasets; (3) The two methods are independent of the underlying MAM employed; (4) The experiments compared the proposed k-nnf () algorithm with the three most representative existing ones from their corresponding classes: the df k-nn(), the bf k-nn() and the inc k-nn() algorithms. We also compared k-nnf () to the standard range query, using the previously-known limiting radius r f. This later test provides a theoretical lower bound of the best possible algorithm to find the nearest neighbors. The experiments were performed on both real and synthetic datasets. Although our approach requires more distance calculations and disk accesses than the best incremental algorithm, it requires very few calculations to estimate the final radius and few extra operations besides the element comparisons to perform the search procedure, as it requires smaller queues than the others to manage its subtrees. Therefore, the proposed k-n N F () algorithm was the fastest for every dataset evaluated and required almost no extra memory to achieve such performance. The experiments conducted to evaluate the algorithms showed that the k-nnf () algorithm can reduce at least one order of magnitude the time demanded to answer a query and is up to 26 times faster than the inc k-nn() algorithm. Considering the non-incremental algorithms, our propose algorithm requires up to 37% less distance calculations, up to 18% less disk accesses, and spends half of the total time of its closest competitor. Besides presenting a Acknowledgement This work has been supported by CNPq (Brazilian National Council for Research Supporting) and FAPESP (São Paulo State Research Foundation) grants, and a CAPES (Brazilian Federal Agency for Post-Graduate Education)/Fulbright Ph.D. fellowship for the first author. References [1] S. Christodoulakis. Implications of certain assumptions in database performance evaluation. TODS, [2] P. Ciaccia, M. Patella, and P. Zezula. M-tree: An efficient access method for similarity search in metric spaces. In VLDB, [3] C. Faloutsos, B. Seeger, A. J. M. Traina, and C. Traina Jr. Spatial join selectivity using power laws. In SIGMOD, [4] A. Guttman. R-tree: A dynamic index structure for spatial searching. In SIGMOD, [5] G. R. Hjaltason and H. Samet. Index-driven similarity search in metric spaces. TODS, [6] L. Jin, N. Koudas, and C. Li. Nnh: Improving performance of nearest-neighbor searches using histograms. In EDBT, [7] N. Katayama and S. Satoh. Distinctiveness-sensitive nearest-neighbor search for efficient similarity retrieval of multimedia information. In ICDE, [8] F. Korn, B. Pagel, and C. Faloutsos. On the dimensionality curse and the self-similarity blessing. TKDE, [9] C. A. Lang and A. K. Singh. Accelerating highdimensional nearest neighbor queries. In SSDBM, [10] N. Roussopoulos, S. Kelley, and F. Vincent. Nearest neighbor queries. In SIGMOD, [11] M. Schroeder. Fractals, Chaos, Power Laws [12] M. Tasan and Z. M. Özsoyoglu. Improvements in distance-based indexing. In SSDBM, [13] A. J. M. Traina, C. Traina Jr., J. Bueno, F. Chino, and P. M. Marques. Efficient content-based image retrieval through metric histograms. WWWJ, [14] C. Traina Jr., A. J. M. Traina, and C. Faloutsos. Distance exponent: a new concept for selectivity estimation in metric trees. In ICDE, [15] C. Traina Jr., A. J. M. Traina, C. Faloutsos, and B. Seeger. Fast indexing and visualization of metric datasets using slim-trees. TKDE,
Bulk-loading Dynamic Metric Access Methods
 Bulk-loading Dynamic Metric Access Methods Thiago Galbiatti Vespa 1, Caetano Traina Jr 1, Agma Juci Machado Traina 1 1 ICMC - Institute of Mathematics and Computer Sciences USP - University of São Paulo
Bulk-loading Dynamic Metric Access Methods Thiago Galbiatti Vespa 1, Caetano Traina Jr 1, Agma Juci Machado Traina 1 1 ICMC - Institute of Mathematics and Computer Sciences USP - University of São Paulo
DBM-Tree: A Dynamic Metric Access Method Sensitive to Local Density Data
 DB: A Dynamic Metric Access Method Sensitive to Local Density Data Marcos R. Vieira, Caetano Traina Jr., Fabio J. T. Chino, Agma J. M. Traina Department of Computer Science, University of São Paulo at
DB: A Dynamic Metric Access Method Sensitive to Local Density Data Marcos R. Vieira, Caetano Traina Jr., Fabio J. T. Chino, Agma J. M. Traina Department of Computer Science, University of São Paulo at
How to Use the Fractal Dimension to Find Correlations between Attributes
 How to Use the Fractal Dimension to Find Correlations between Attributes Elaine Parros Machado de Sousa, Caetano Traina Jr., Agma J. M. Traina University of Sao Paulo at Sao Carlos - Department of Computer
How to Use the Fractal Dimension to Find Correlations between Attributes Elaine Parros Machado de Sousa, Caetano Traina Jr., Agma J. M. Traina University of Sao Paulo at Sao Carlos - Department of Computer
Branch and Bound. Algorithms for Nearest Neighbor Search: Lecture 1. Yury Lifshits
 Branch and Bound Algorithms for Nearest Neighbor Search: Lecture 1 Yury Lifshits http://yury.name Steklov Institute of Mathematics at St.Petersburg California Institute of Technology 1 / 36 Outline 1 Welcome
Branch and Bound Algorithms for Nearest Neighbor Search: Lecture 1 Yury Lifshits http://yury.name Steklov Institute of Mathematics at St.Petersburg California Institute of Technology 1 / 36 Outline 1 Welcome
Nearest Neighbor Search by Branch and Bound
 Nearest Neighbor Search by Branch and Bound Algorithmic Problems Around the Web #2 Yury Lifshits http://yury.name CalTech, Fall 07, CS101.2, http://yury.name/algoweb.html 1 / 30 Outline 1 Short Intro to
Nearest Neighbor Search by Branch and Bound Algorithmic Problems Around the Web #2 Yury Lifshits http://yury.name CalTech, Fall 07, CS101.2, http://yury.name/algoweb.html 1 / 30 Outline 1 Short Intro to
Similarity Search without Tears: the OMNI-Family of All-Purpose Access Methods
 Similarity Search without Tears: the OMNI-Family of All-Purpose Access Methods Roberto Figueira Santos Filho 1 Agma Traina 1 Caetano Traina Jr. 1 Christos Faloutsos 2 1 Department of Computer Science and
Similarity Search without Tears: the OMNI-Family of All-Purpose Access Methods Roberto Figueira Santos Filho 1 Agma Traina 1 Caetano Traina Jr. 1 Christos Faloutsos 2 1 Department of Computer Science and
DBM-Tree: A Dynamic Metric Access Method Sensitive to Local Density Data
 DB: A Dynamic Metric Access Method Sensitive to Local Density Data Marcos R. Vieira, Caetano Traina Jr., Fabio J. T. Chino, Agma J. M. Traina ICMC Institute of Mathematics and Computer Sciences USP University
DB: A Dynamic Metric Access Method Sensitive to Local Density Data Marcos R. Vieira, Caetano Traina Jr., Fabio J. T. Chino, Agma J. M. Traina ICMC Institute of Mathematics and Computer Sciences USP University
Experimental Evaluation of Spatial Indices with FESTIval
 Experimental Evaluation of Spatial Indices with FESTIval Anderson Chaves Carniel 1, Ricardo Rodrigues Ciferri 2, Cristina Dutra de Aguiar Ciferri 1 1 Department of Computer Science University of São Paulo
Experimental Evaluation of Spatial Indices with FESTIval Anderson Chaves Carniel 1, Ricardo Rodrigues Ciferri 2, Cristina Dutra de Aguiar Ciferri 1 1 Department of Computer Science University of São Paulo
Fast Similarity Search for High-Dimensional Dataset
 Fast Similarity Search for High-Dimensional Dataset Quan Wang and Suya You Computer Science Department University of Southern California {quanwang,suyay}@graphics.usc.edu Abstract This paper addresses
Fast Similarity Search for High-Dimensional Dataset Quan Wang and Suya You Computer Science Department University of Southern California {quanwang,suyay}@graphics.usc.edu Abstract This paper addresses
doc. RNDr. Tomáš Skopal, Ph.D. Department of Software Engineering, Faculty of Information Technology, Czech Technical University in Prague
 Praha & EU: Investujeme do vaší budoucnosti Evropský sociální fond course: Searching the Web and Multimedia Databases (BI-VWM) Tomáš Skopal, 2011 SS2010/11 doc. RNDr. Tomáš Skopal, Ph.D. Department of
Praha & EU: Investujeme do vaší budoucnosti Evropský sociální fond course: Searching the Web and Multimedia Databases (BI-VWM) Tomáš Skopal, 2011 SS2010/11 doc. RNDr. Tomáš Skopal, Ph.D. Department of
Pivoting M-tree: A Metric Access Method for Efficient Similarity Search
 Pivoting M-tree: A Metric Access Method for Efficient Similarity Search Tomáš Skopal Department of Computer Science, VŠB Technical University of Ostrava, tř. 17. listopadu 15, Ostrava, Czech Republic tomas.skopal@vsb.cz
Pivoting M-tree: A Metric Access Method for Efficient Similarity Search Tomáš Skopal Department of Computer Science, VŠB Technical University of Ostrava, tř. 17. listopadu 15, Ostrava, Czech Republic tomas.skopal@vsb.cz
NNH: Improving Performance of Nearest-Neighbor Searches Using Histograms (Full Version. UCI Technical Report, Dec. 2003)
 NNH: Improving Performance of Nearest-Neighbor Searches Using Histograms (Full Version. UCI Technical Report, Dec. 2003) Liang Jin 1, Nick Koudas 2,andChenLi 1 1 School of Information and Computer Science,
NNH: Improving Performance of Nearest-Neighbor Searches Using Histograms (Full Version. UCI Technical Report, Dec. 2003) Liang Jin 1, Nick Koudas 2,andChenLi 1 1 School of Information and Computer Science,
Search Space Reductions for Nearest-Neighbor Queries
 Search Space Reductions for Nearest-Neighbor Queries Micah Adler 1 and Brent Heeringa 2 1 Department of Computer Science, University of Massachusetts, Amherst 140 Governors Drive Amherst, MA 01003 2 Department
Search Space Reductions for Nearest-Neighbor Queries Micah Adler 1 and Brent Heeringa 2 1 Department of Computer Science, University of Massachusetts, Amherst 140 Governors Drive Amherst, MA 01003 2 Department
Reducing the Complexity of k-nearest Diverse Neighbor Queries in Medical Image Datasets through Fractal Analysis
 Reducing the Complexity of k-nearest Diverse Neighbor Queries in Medical Image Datasets through Fractal Analysis Rafael L. Dias, Renato Bueno, Marcela X. Ribeiro Department of Computing Federal University
Reducing the Complexity of k-nearest Diverse Neighbor Queries in Medical Image Datasets through Fractal Analysis Rafael L. Dias, Renato Bueno, Marcela X. Ribeiro Department of Computing Federal University
Slim-trees: High Performance Metric Trees Minimizing Overlap Between Nodes
 Slim-trees: High Performance Metric Trees Minimizing Overlap Between Nodes Caetano Traina Jr. 1, Agma Traina 2, Bernhard Seeger 3, Christos Faloutsos 4 October, 1999 CMU-CS-99-170 School of Computer Science
Slim-trees: High Performance Metric Trees Minimizing Overlap Between Nodes Caetano Traina Jr. 1, Agma Traina 2, Bernhard Seeger 3, Christos Faloutsos 4 October, 1999 CMU-CS-99-170 School of Computer Science
Speeding up Queries in a Leaf Image Database
 1 Speeding up Queries in a Leaf Image Database Daozheng Chen May 10, 2007 Abstract We have an Electronic Field Guide which contains an image database with thousands of leaf images. We have a system which
1 Speeding up Queries in a Leaf Image Database Daozheng Chen May 10, 2007 Abstract We have an Electronic Field Guide which contains an image database with thousands of leaf images. We have a system which
Nearest neighbors. Focus on tree-based methods. Clément Jamin, GUDHI project, Inria March 2017
 Nearest neighbors Focus on tree-based methods Clément Jamin, GUDHI project, Inria March 2017 Introduction Exact and approximate nearest neighbor search Essential tool for many applications Huge bibliography
Nearest neighbors Focus on tree-based methods Clément Jamin, GUDHI project, Inria March 2017 Introduction Exact and approximate nearest neighbor search Essential tool for many applications Huge bibliography
Nearest Neighbor Search on Vertically Partitioned High-Dimensional Data
 Nearest Neighbor Search on Vertically Partitioned High-Dimensional Data Evangelos Dellis, Bernhard Seeger, and Akrivi Vlachou Department of Mathematics and Computer Science, University of Marburg, Hans-Meerwein-Straße,
Nearest Neighbor Search on Vertically Partitioned High-Dimensional Data Evangelos Dellis, Bernhard Seeger, and Akrivi Vlachou Department of Mathematics and Computer Science, University of Marburg, Hans-Meerwein-Straße,
Geometric data structures:
 Geometric data structures: Machine Learning for Big Data CSE547/STAT548, University of Washington Sham Kakade Sham Kakade 2017 1 Announcements: HW3 posted Today: Review: LSH for Euclidean distance Other
Geometric data structures: Machine Learning for Big Data CSE547/STAT548, University of Washington Sham Kakade Sham Kakade 2017 1 Announcements: HW3 posted Today: Review: LSH for Euclidean distance Other
Clustering Billions of Images with Large Scale Nearest Neighbor Search
 Clustering Billions of Images with Large Scale Nearest Neighbor Search Ting Liu, Charles Rosenberg, Henry A. Rowley IEEE Workshop on Applications of Computer Vision February 2007 Presented by Dafna Bitton
Clustering Billions of Images with Large Scale Nearest Neighbor Search Ting Liu, Charles Rosenberg, Henry A. Rowley IEEE Workshop on Applications of Computer Vision February 2007 Presented by Dafna Bitton
High Dimensional Indexing by Clustering
 Yufei Tao ITEE University of Queensland Recall that, our discussion so far has assumed that the dimensionality d is moderately high, such that it can be regarded as a constant. This means that d should
Yufei Tao ITEE University of Queensland Recall that, our discussion so far has assumed that the dimensionality d is moderately high, such that it can be regarded as a constant. This means that d should
Benchmarking Access Structures for High-Dimensional Multimedia Data
 Benchmarking Access Structures for High-Dimensional Multimedia Data by Nathan G. Colossi and Mario A. Nascimento Technical Report TR 99-05 December 1999 DEPARTMENT OF COMPUTING SCIENCE University of Alberta
Benchmarking Access Structures for High-Dimensional Multimedia Data by Nathan G. Colossi and Mario A. Nascimento Technical Report TR 99-05 December 1999 DEPARTMENT OF COMPUTING SCIENCE University of Alberta
Voronoi-Based K Nearest Neighbor Search for Spatial Network Databases
 Voronoi-Based K Nearest Neighbor Search for Spatial Network Databases Mohammad Kolahdouzan and Cyrus Shahabi Department of Computer Science University of Southern California Los Angeles, CA, 90089, USA
Voronoi-Based K Nearest Neighbor Search for Spatial Network Databases Mohammad Kolahdouzan and Cyrus Shahabi Department of Computer Science University of Southern California Los Angeles, CA, 90089, USA
The Metric Histogram: A New and Efficient Approach for Content-based Image Retrieval
 The Metric Histogram: A New and Efficient Approach for Content-based Image Retrieval Agma J. M. Traina, Caetano Traina Jr., Josiane M. Bueno Computer Science Department University of Sao Paulo at Sao Carlos
The Metric Histogram: A New and Efficient Approach for Content-based Image Retrieval Agma J. M. Traina, Caetano Traina Jr., Josiane M. Bueno Computer Science Department University of Sao Paulo at Sao Carlos
9/23/2009 CONFERENCES CONTINUOUS NEAREST NEIGHBOR SEARCH INTRODUCTION OVERVIEW PRELIMINARY -- POINT NN QUERIES
 CONFERENCES Short Name SIGMOD Full Name Special Interest Group on Management Of Data CONTINUOUS NEAREST NEIGHBOR SEARCH Yufei Tao, Dimitris Papadias, Qiongmao Shen Hong Kong University of Science and Technology
CONFERENCES Short Name SIGMOD Full Name Special Interest Group on Management Of Data CONTINUOUS NEAREST NEIGHBOR SEARCH Yufei Tao, Dimitris Papadias, Qiongmao Shen Hong Kong University of Science and Technology
Class-Constraint Similarity Queries
 Class-Constraint Similarity Queries Jessica A. de Souza Institute of Mathematics and Computer Sciences University of Sao Paulo Sao Carlos, SP, Brazil jessicasouza@usp.br Agma J. M. Traina Institute of
Class-Constraint Similarity Queries Jessica A. de Souza Institute of Mathematics and Computer Sciences University of Sao Paulo Sao Carlos, SP, Brazil jessicasouza@usp.br Agma J. M. Traina Institute of
Benchmarking the UB-tree
 Benchmarking the UB-tree Michal Krátký, Tomáš Skopal Department of Computer Science, VŠB Technical University of Ostrava, tř. 17. listopadu 15, Ostrava, Czech Republic michal.kratky@vsb.cz, tomas.skopal@vsb.cz
Benchmarking the UB-tree Michal Krátký, Tomáš Skopal Department of Computer Science, VŠB Technical University of Ostrava, tř. 17. listopadu 15, Ostrava, Czech Republic michal.kratky@vsb.cz, tomas.skopal@vsb.cz
Structure-Based Similarity Search with Graph Histograms
 Structure-Based Similarity Search with Graph Histograms Apostolos N. Papadopoulos and Yannis Manolopoulos Data Engineering Lab. Department of Informatics, Aristotle University Thessaloniki 5006, Greece
Structure-Based Similarity Search with Graph Histograms Apostolos N. Papadopoulos and Yannis Manolopoulos Data Engineering Lab. Department of Informatics, Aristotle University Thessaloniki 5006, Greece
Chapter 4: Non-Parametric Techniques
 Chapter 4: Non-Parametric Techniques Introduction Density Estimation Parzen Windows Kn-Nearest Neighbor Density Estimation K-Nearest Neighbor (KNN) Decision Rule Supervised Learning How to fit a density
Chapter 4: Non-Parametric Techniques Introduction Density Estimation Parzen Windows Kn-Nearest Neighbor Density Estimation K-Nearest Neighbor (KNN) Decision Rule Supervised Learning How to fit a density
DB-GNG: A constructive Self-Organizing Map based on density
 DB-GNG: A constructive Self-Organizing Map based on density Alexander Ocsa, Carlos Bedregal, Ernesto Cuadros-Vargas Abstract Nowadays applications require efficient and fast techniques due to the growing
DB-GNG: A constructive Self-Organizing Map based on density Alexander Ocsa, Carlos Bedregal, Ernesto Cuadros-Vargas Abstract Nowadays applications require efficient and fast techniques due to the growing
Nearest Neighbor Queries
 Nearest Neighbor Queries Nick Roussopoulos Stephen Kelley Frederic Vincent University of Maryland May 1995 Problem / Motivation Given a point in space, find the k NN classic NN queries (find the nearest
Nearest Neighbor Queries Nick Roussopoulos Stephen Kelley Frederic Vincent University of Maryland May 1995 Problem / Motivation Given a point in space, find the k NN classic NN queries (find the nearest
Chapter 2 Basic Structure of High-Dimensional Spaces
 Chapter 2 Basic Structure of High-Dimensional Spaces Data is naturally represented geometrically by associating each record with a point in the space spanned by the attributes. This idea, although simple,
Chapter 2 Basic Structure of High-Dimensional Spaces Data is naturally represented geometrically by associating each record with a point in the space spanned by the attributes. This idea, although simple,
Integers & Absolute Value Properties of Addition Add Integers Subtract Integers. Add & Subtract Like Fractions Add & Subtract Unlike Fractions
 Unit 1: Rational Numbers & Exponents M07.A-N & M08.A-N, M08.B-E Essential Questions Standards Content Skills Vocabulary What happens when you add, subtract, multiply and divide integers? What happens when
Unit 1: Rational Numbers & Exponents M07.A-N & M08.A-N, M08.B-E Essential Questions Standards Content Skills Vocabulary What happens when you add, subtract, multiply and divide integers? What happens when
Spatial Queries. Nearest Neighbor Queries
 Spatial Queries Nearest Neighbor Queries Spatial Queries Given a collection of geometric objects (points, lines, polygons,...) organize them on disk, to answer efficiently point queries range queries k-nn
Spatial Queries Nearest Neighbor Queries Spatial Queries Given a collection of geometric objects (points, lines, polygons,...) organize them on disk, to answer efficiently point queries range queries k-nn
Using Novel Method ProMiSH Search Nearest keyword Set In Multidimensional Dataset
 Using Novel Method ProMiSH Search Nearest keyword Set In Multidimensional Dataset Miss. Shilpa Bhaskar Thakare 1, Prof. Jayshree.V.Shinde 2 1 Department of Computer Engineering, Late G.N.Sapkal C.O.E,
Using Novel Method ProMiSH Search Nearest keyword Set In Multidimensional Dataset Miss. Shilpa Bhaskar Thakare 1, Prof. Jayshree.V.Shinde 2 1 Department of Computer Engineering, Late G.N.Sapkal C.O.E,
Dimension Induced Clustering
 Dimension Induced Clustering Aris Gionis Alexander Hinneburg Spiros Papadimitriou Panayiotis Tsaparas HIIT, University of Helsinki Martin Luther University, Halle Carnegie Melon University HIIT, University
Dimension Induced Clustering Aris Gionis Alexander Hinneburg Spiros Papadimitriou Panayiotis Tsaparas HIIT, University of Helsinki Martin Luther University, Halle Carnegie Melon University HIIT, University
Improving Range Query Performance on Historic Web Page Data
 Improving Range Query Performance on Historic Web Page Data Geng LI Lab of Computer Networks and Distributed Systems, Peking University Beijing, China ligeng@net.pku.edu.cn Bo Peng Lab of Computer Networks
Improving Range Query Performance on Historic Web Page Data Geng LI Lab of Computer Networks and Distributed Systems, Peking University Beijing, China ligeng@net.pku.edu.cn Bo Peng Lab of Computer Networks
Introduction to Indexing R-trees. Hong Kong University of Science and Technology
 Introduction to Indexing R-trees Dimitris Papadias Hong Kong University of Science and Technology 1 Introduction to Indexing 1. Assume that you work in a government office, and you maintain the records
Introduction to Indexing R-trees Dimitris Papadias Hong Kong University of Science and Technology 1 Introduction to Indexing 1. Assume that you work in a government office, and you maintain the records
Striped Grid Files: An Alternative for Highdimensional
 Striped Grid Files: An Alternative for Highdimensional Indexing Thanet Praneenararat 1, Vorapong Suppakitpaisarn 2, Sunchai Pitakchonlasap 1, and Jaruloj Chongstitvatana 1 Department of Mathematics 1,
Striped Grid Files: An Alternative for Highdimensional Indexing Thanet Praneenararat 1, Vorapong Suppakitpaisarn 2, Sunchai Pitakchonlasap 1, and Jaruloj Chongstitvatana 1 Department of Mathematics 1,
Chapter 5 Hashing. Introduction. Hashing. Hashing Functions. hashing performs basic operations, such as insertion,
 Introduction Chapter 5 Hashing hashing performs basic operations, such as insertion, deletion, and finds in average time 2 Hashing a hash table is merely an of some fixed size hashing converts into locations
Introduction Chapter 5 Hashing hashing performs basic operations, such as insertion, deletion, and finds in average time 2 Hashing a hash table is merely an of some fixed size hashing converts into locations
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION
 CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
Trajectory Similarity Search in Spatial Networks
 Trajectory Similarity Search in Spatial Networks Eleftherios Tiakas Apostolos N. Papadopoulos Alexandros Nanopoulos Yannis Manolopoulos Department of Informatics, Aristotle University 54124 Thessaloniki,
Trajectory Similarity Search in Spatial Networks Eleftherios Tiakas Apostolos N. Papadopoulos Alexandros Nanopoulos Yannis Manolopoulos Department of Informatics, Aristotle University 54124 Thessaloniki,
Supplementary Material for The Generalized PatchMatch Correspondence Algorithm
 Supplementary Material for The Generalized PatchMatch Correspondence Algorithm Connelly Barnes 1, Eli Shechtman 2, Dan B Goldman 2, Adam Finkelstein 1 1 Princeton University, 2 Adobe Systems 1 Overview
Supplementary Material for The Generalized PatchMatch Correspondence Algorithm Connelly Barnes 1, Eli Shechtman 2, Dan B Goldman 2, Adam Finkelstein 1 1 Princeton University, 2 Adobe Systems 1 Overview
Seminar on. A Coarse-Grain Parallel Formulation of Multilevel k-way Graph Partitioning Algorithm
 Seminar on A Coarse-Grain Parallel Formulation of Multilevel k-way Graph Partitioning Algorithm Mohammad Iftakher Uddin & Mohammad Mahfuzur Rahman Matrikel Nr: 9003357 Matrikel Nr : 9003358 Masters of
Seminar on A Coarse-Grain Parallel Formulation of Multilevel k-way Graph Partitioning Algorithm Mohammad Iftakher Uddin & Mohammad Mahfuzur Rahman Matrikel Nr: 9003357 Matrikel Nr : 9003358 Masters of
THE EFFECT OF JOIN SELECTIVITIES ON OPTIMAL NESTING ORDER
 THE EFFECT OF JOIN SELECTIVITIES ON OPTIMAL NESTING ORDER Akhil Kumar and Michael Stonebraker EECS Department University of California Berkeley, Ca., 94720 Abstract A heuristic query optimizer must choose
THE EFFECT OF JOIN SELECTIVITIES ON OPTIMAL NESTING ORDER Akhil Kumar and Michael Stonebraker EECS Department University of California Berkeley, Ca., 94720 Abstract A heuristic query optimizer must choose
Local Dimensionality Reduction: A New Approach to Indexing High Dimensional Spaces
 Local Dimensionality Reduction: A New Approach to Indexing High Dimensional Spaces Kaushik Chakrabarti Department of Computer Science University of Illinois Urbana, IL 61801 kaushikc@cs.uiuc.edu Sharad
Local Dimensionality Reduction: A New Approach to Indexing High Dimensional Spaces Kaushik Chakrabarti Department of Computer Science University of Illinois Urbana, IL 61801 kaushikc@cs.uiuc.edu Sharad
Parallel Performance Studies for a Clustering Algorithm
 Parallel Performance Studies for a Clustering Algorithm Robin V. Blasberg and Matthias K. Gobbert Naval Research Laboratory, Washington, D.C. Department of Mathematics and Statistics, University of Maryland,
Parallel Performance Studies for a Clustering Algorithm Robin V. Blasberg and Matthias K. Gobbert Naval Research Laboratory, Washington, D.C. Department of Mathematics and Statistics, University of Maryland,
External Memory Algorithms and Data Structures Fall Project 3 A GIS system
 External Memory Algorithms and Data Structures Fall 2003 1 Project 3 A GIS system GSB/RF November 17, 2003 1 Introduction The goal of this project is to implement a rudimentary Geographical Information
External Memory Algorithms and Data Structures Fall 2003 1 Project 3 A GIS system GSB/RF November 17, 2003 1 Introduction The goal of this project is to implement a rudimentary Geographical Information
Exploratory Data Analysis
 Chapter 10 Exploratory Data Analysis Definition of Exploratory Data Analysis (page 410) Definition 12.1. Exploratory data analysis (EDA) is a subfield of applied statistics that is concerned with the investigation
Chapter 10 Exploratory Data Analysis Definition of Exploratory Data Analysis (page 410) Definition 12.1. Exploratory data analysis (EDA) is a subfield of applied statistics that is concerned with the investigation
Analytical Modeling of Parallel Systems. To accompany the text ``Introduction to Parallel Computing'', Addison Wesley, 2003.
 Analytical Modeling of Parallel Systems To accompany the text ``Introduction to Parallel Computing'', Addison Wesley, 2003. Topic Overview Sources of Overhead in Parallel Programs Performance Metrics for
Analytical Modeling of Parallel Systems To accompany the text ``Introduction to Parallel Computing'', Addison Wesley, 2003. Topic Overview Sources of Overhead in Parallel Programs Performance Metrics for
Chapter 11: Indexing and Hashing
 Chapter 11: Indexing and Hashing Basic Concepts Ordered Indices B + -Tree Index Files B-Tree Index Files Static Hashing Dynamic Hashing Comparison of Ordered Indexing and Hashing Index Definition in SQL
Chapter 11: Indexing and Hashing Basic Concepts Ordered Indices B + -Tree Index Files B-Tree Index Files Static Hashing Dynamic Hashing Comparison of Ordered Indexing and Hashing Index Definition in SQL
Search Algorithms for Discrete Optimization Problems
 Search Algorithms for Discrete Optimization Problems Ananth Grama, Anshul Gupta, George Karypis, and Vipin Kumar To accompany the text ``Introduction to Parallel Computing'', Addison Wesley, 2003. Topic
Search Algorithms for Discrete Optimization Problems Ananth Grama, Anshul Gupta, George Karypis, and Vipin Kumar To accompany the text ``Introduction to Parallel Computing'', Addison Wesley, 2003. Topic
An Experiment in Visual Clustering Using Star Glyph Displays
 An Experiment in Visual Clustering Using Star Glyph Displays by Hanna Kazhamiaka A Research Paper presented to the University of Waterloo in partial fulfillment of the requirements for the degree of Master
An Experiment in Visual Clustering Using Star Glyph Displays by Hanna Kazhamiaka A Research Paper presented to the University of Waterloo in partial fulfillment of the requirements for the degree of Master
Top-K Ranking Spatial Queries over Filtering Data
 Top-K Ranking Spatial Queries over Filtering Data 1 Lakkapragada Prasanth, 2 Krishna Chaitanya, 1 Student, 2 Assistant Professor, NRL Instiute of Technology,Vijayawada Abstract: A spatial preference query
Top-K Ranking Spatial Queries over Filtering Data 1 Lakkapragada Prasanth, 2 Krishna Chaitanya, 1 Student, 2 Assistant Professor, NRL Instiute of Technology,Vijayawada Abstract: A spatial preference query
Optimal k-anonymity with Flexible Generalization Schemes through Bottom-up Searching
 Optimal k-anonymity with Flexible Generalization Schemes through Bottom-up Searching Tiancheng Li Ninghui Li CERIAS and Department of Computer Science, Purdue University 250 N. University Street, West
Optimal k-anonymity with Flexible Generalization Schemes through Bottom-up Searching Tiancheng Li Ninghui Li CERIAS and Department of Computer Science, Purdue University 250 N. University Street, West
Search K Nearest Neighbors on Air
 Search K Nearest Neighbors on Air Baihua Zheng 1, Wang-Chien Lee 2, and Dik Lun Lee 1 1 Hong Kong University of Science and Technology Clear Water Bay, Hong Kong {baihua,dlee}@cs.ust.hk 2 The Penn State
Search K Nearest Neighbors on Air Baihua Zheng 1, Wang-Chien Lee 2, and Dik Lun Lee 1 1 Hong Kong University of Science and Technology Clear Water Bay, Hong Kong {baihua,dlee}@cs.ust.hk 2 The Penn State
Outline. Other Use of Triangle Inequality Algorithms for Nearest Neighbor Search: Lecture 2. Orchard s Algorithm. Chapter VI
 Other Use of Triangle Ineuality Algorithms for Nearest Neighbor Search: Lecture 2 Yury Lifshits http://yury.name Steklov Institute of Mathematics at St.Petersburg California Institute of Technology Outline
Other Use of Triangle Ineuality Algorithms for Nearest Neighbor Search: Lecture 2 Yury Lifshits http://yury.name Steklov Institute of Mathematics at St.Petersburg California Institute of Technology Outline
Clustering For Similarity Search And Privacyguaranteed Publishing Of Hi-Dimensional Data Ashwini.R #1, K.Praveen *2, R.V.
 Clustering For Similarity Search And Privacyguaranteed Publishing Of Hi-Dimensional Data Ashwini.R #1, K.Praveen *2, R.V.Krishnaiah *3 #1 M.Tech, Computer Science Engineering, DRKIST, Hyderabad, Andhra
Clustering For Similarity Search And Privacyguaranteed Publishing Of Hi-Dimensional Data Ashwini.R #1, K.Praveen *2, R.V.Krishnaiah *3 #1 M.Tech, Computer Science Engineering, DRKIST, Hyderabad, Andhra
Themes in the Texas CCRS - Mathematics
 1. Compare real numbers. a. Classify numbers as natural, whole, integers, rational, irrational, real, imaginary, &/or complex. b. Use and apply the relative magnitude of real numbers by using inequality
1. Compare real numbers. a. Classify numbers as natural, whole, integers, rational, irrational, real, imaginary, &/or complex. b. Use and apply the relative magnitude of real numbers by using inequality
HOT asax: A Novel Adaptive Symbolic Representation for Time Series Discords Discovery
 HOT asax: A Novel Adaptive Symbolic Representation for Time Series Discords Discovery Ninh D. Pham, Quang Loc Le, Tran Khanh Dang Faculty of Computer Science and Engineering, HCM University of Technology,
HOT asax: A Novel Adaptive Symbolic Representation for Time Series Discords Discovery Ninh D. Pham, Quang Loc Le, Tran Khanh Dang Faculty of Computer Science and Engineering, HCM University of Technology,
Introduction. hashing performs basic operations, such as insertion, better than other ADTs we ve seen so far
 Chapter 5 Hashing 2 Introduction hashing performs basic operations, such as insertion, deletion, and finds in average time better than other ADTs we ve seen so far 3 Hashing a hash table is merely an hashing
Chapter 5 Hashing 2 Introduction hashing performs basic operations, such as insertion, deletion, and finds in average time better than other ADTs we ve seen so far 3 Hashing a hash table is merely an hashing
A Pivot-based Index Structure for Combination of Feature Vectors
 A Pivot-based Index Structure for Combination of Feature Vectors Benjamin Bustos Daniel Keim Tobias Schreck Department of Computer and Information Science, University of Konstanz Universitätstr. 10 Box
A Pivot-based Index Structure for Combination of Feature Vectors Benjamin Bustos Daniel Keim Tobias Schreck Department of Computer and Information Science, University of Konstanz Universitätstr. 10 Box
Chapter 12: Indexing and Hashing. Basic Concepts
 Chapter 12: Indexing and Hashing! Basic Concepts! Ordered Indices! B+-Tree Index Files! B-Tree Index Files! Static Hashing! Dynamic Hashing! Comparison of Ordered Indexing and Hashing! Index Definition
Chapter 12: Indexing and Hashing! Basic Concepts! Ordered Indices! B+-Tree Index Files! B-Tree Index Files! Static Hashing! Dynamic Hashing! Comparison of Ordered Indexing and Hashing! Index Definition
Z-KNN Join for the Swiss Feed Database: a feasibility study
 Z-KNN Join for the Swiss Feed Database: a feasibility study Francesco Luminati University Of Zurich, Switzerland francesco.luminati@uzh.ch 1 Introduction K-nearest neighbor query (knn) and k-nearest neighbor
Z-KNN Join for the Swiss Feed Database: a feasibility study Francesco Luminati University Of Zurich, Switzerland francesco.luminati@uzh.ch 1 Introduction K-nearest neighbor query (knn) and k-nearest neighbor
9/24/ Hash functions
 11.3 Hash functions A good hash function satis es (approximately) the assumption of SUH: each key is equally likely to hash to any of the slots, independently of the other keys We typically have no way
11.3 Hash functions A good hash function satis es (approximately) the assumption of SUH: each key is equally likely to hash to any of the slots, independently of the other keys We typically have no way
Exploring intersection trees for indexing metric spaces
 Exploring intersection trees for indexing metric spaces Zineddine KOUAHLA Laboratoire d informatique de Nantes Atlantique (LINA, UMR CNRS 6241) Polytechnic School of the University of Nantes, Nantes, France
Exploring intersection trees for indexing metric spaces Zineddine KOUAHLA Laboratoire d informatique de Nantes Atlantique (LINA, UMR CNRS 6241) Polytechnic School of the University of Nantes, Nantes, France
A Scalable Index Mechanism for High-Dimensional Data in Cluster File Systems
 A Scalable Index Mechanism for High-Dimensional Data in Cluster File Systems Kyu-Woong Lee Hun-Soon Lee, Mi-Young Lee, Myung-Joon Kim Abstract We address the problem of designing index structures that
A Scalable Index Mechanism for High-Dimensional Data in Cluster File Systems Kyu-Woong Lee Hun-Soon Lee, Mi-Young Lee, Myung-Joon Kim Abstract We address the problem of designing index structures that
Chapter 12: Indexing and Hashing
 Chapter 12: Indexing and Hashing Basic Concepts Ordered Indices B+-Tree Index Files B-Tree Index Files Static Hashing Dynamic Hashing Comparison of Ordered Indexing and Hashing Index Definition in SQL
Chapter 12: Indexing and Hashing Basic Concepts Ordered Indices B+-Tree Index Files B-Tree Index Files Static Hashing Dynamic Hashing Comparison of Ordered Indexing and Hashing Index Definition in SQL
Processing Rank-Aware Queries in P2P Systems
 Processing Rank-Aware Queries in P2P Systems Katja Hose, Marcel Karnstedt, Anke Koch, Kai-Uwe Sattler, and Daniel Zinn Department of Computer Science and Automation, TU Ilmenau P.O. Box 100565, D-98684
Processing Rank-Aware Queries in P2P Systems Katja Hose, Marcel Karnstedt, Anke Koch, Kai-Uwe Sattler, and Daniel Zinn Department of Computer Science and Automation, TU Ilmenau P.O. Box 100565, D-98684
3 Nonlinear Regression
 3 Linear models are often insufficient to capture the real-world phenomena. That is, the relation between the inputs and the outputs we want to be able to predict are not linear. As a consequence, nonlinear
3 Linear models are often insufficient to capture the real-world phenomena. That is, the relation between the inputs and the outputs we want to be able to predict are not linear. As a consequence, nonlinear
PATMaths Fourth Edition alignment to the NSW mathematics syllabus
 Fourth Edition alignment to the NSW mathematics syllabus http://acer.ac/patmnsw Australian Council for Educational Research Fourth Edition alignment to the NSW mathematics syllabus Test 1 item descriptors
Fourth Edition alignment to the NSW mathematics syllabus http://acer.ac/patmnsw Australian Council for Educational Research Fourth Edition alignment to the NSW mathematics syllabus Test 1 item descriptors
Theorem 2.9: nearest addition algorithm
 There are severe limits on our ability to compute near-optimal tours It is NP-complete to decide whether a given undirected =(,)has a Hamiltonian cycle An approximation algorithm for the TSP can be used
There are severe limits on our ability to compute near-optimal tours It is NP-complete to decide whether a given undirected =(,)has a Hamiltonian cycle An approximation algorithm for the TSP can be used
An index structure for efficient reverse nearest neighbor queries
 An index structure for efficient reverse nearest neighbor queries Congjun Yang Division of Computer Science, Department of Mathematical Sciences The University of Memphis, Memphis, TN 38152, USA yangc@msci.memphis.edu
An index structure for efficient reverse nearest neighbor queries Congjun Yang Division of Computer Science, Department of Mathematical Sciences The University of Memphis, Memphis, TN 38152, USA yangc@msci.memphis.edu
Comparing Implementations of Optimal Binary Search Trees
 Introduction Comparing Implementations of Optimal Binary Search Trees Corianna Jacoby and Alex King Tufts University May 2017 In this paper we sought to put together a practical comparison of the optimality
Introduction Comparing Implementations of Optimal Binary Search Trees Corianna Jacoby and Alex King Tufts University May 2017 In this paper we sought to put together a practical comparison of the optimality
Bumptrees for Efficient Function, Constraint, and Classification Learning
 umptrees for Efficient Function, Constraint, and Classification Learning Stephen M. Omohundro International Computer Science Institute 1947 Center Street, Suite 600 erkeley, California 94704 Abstract A
umptrees for Efficient Function, Constraint, and Classification Learning Stephen M. Omohundro International Computer Science Institute 1947 Center Street, Suite 600 erkeley, California 94704 Abstract A
Small-World Overlay P2P Networks: Construction and Handling Dynamic Flash Crowd
 Small-World Overlay P2P Networks: Construction and Handling Dynamic Flash Crowd Ken Y.K. Hui John C. S. Lui David K.Y. Yau Dept. of Computer Science & Engineering Computer Science Department The Chinese
Small-World Overlay P2P Networks: Construction and Handling Dynamic Flash Crowd Ken Y.K. Hui John C. S. Lui David K.Y. Yau Dept. of Computer Science & Engineering Computer Science Department The Chinese
Evaluating Classifiers
 Evaluating Classifiers Charles Elkan elkan@cs.ucsd.edu January 18, 2011 In a real-world application of supervised learning, we have a training set of examples with labels, and a test set of examples with
Evaluating Classifiers Charles Elkan elkan@cs.ucsd.edu January 18, 2011 In a real-world application of supervised learning, we have a training set of examples with labels, and a test set of examples with
Search. The Nearest Neighbor Problem
 3 Nearest Neighbor Search Lab Objective: The nearest neighbor problem is an optimization problem that arises in applications such as computer vision, pattern recognition, internet marketing, and data compression.
3 Nearest Neighbor Search Lab Objective: The nearest neighbor problem is an optimization problem that arises in applications such as computer vision, pattern recognition, internet marketing, and data compression.
Detecting Clusters and Outliers for Multidimensional
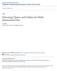 Kennesaw State University DigitalCommons@Kennesaw State University Faculty Publications 2008 Detecting Clusters and Outliers for Multidimensional Data Yong Shi Kennesaw State University, yshi5@kennesaw.edu
Kennesaw State University DigitalCommons@Kennesaw State University Faculty Publications 2008 Detecting Clusters and Outliers for Multidimensional Data Yong Shi Kennesaw State University, yshi5@kennesaw.edu
Week 7 Picturing Network. Vahe and Bethany
 Week 7 Picturing Network Vahe and Bethany Freeman (2005) - Graphic Techniques for Exploring Social Network Data The two main goals of analyzing social network data are identification of cohesive groups
Week 7 Picturing Network Vahe and Bethany Freeman (2005) - Graphic Techniques for Exploring Social Network Data The two main goals of analyzing social network data are identification of cohesive groups
Spatial Index Keyword Search in Multi- Dimensional Database
 Spatial Index Keyword Search in Multi- Dimensional Database Sushma Ahirrao M. E Student, Department of Computer Engineering, GHRIEM, Jalgaon, India ABSTRACT: Nearest neighbor search in multimedia databases
Spatial Index Keyword Search in Multi- Dimensional Database Sushma Ahirrao M. E Student, Department of Computer Engineering, GHRIEM, Jalgaon, India ABSTRACT: Nearest neighbor search in multimedia databases
An Efficient Single Chord-based Accumulation Technique (SCA) to Detect More Reliable Corners
 An Efficient Single Chord-based Accumulation Technique (SCA) to Detect More Reliable Corners Mohammad Asiful Hossain, Abdul Kawsar Tushar, and Shofiullah Babor Computer Science and Engineering Department,
An Efficient Single Chord-based Accumulation Technique (SCA) to Detect More Reliable Corners Mohammad Asiful Hossain, Abdul Kawsar Tushar, and Shofiullah Babor Computer Science and Engineering Department,
Leveraging Set Relations in Exact Set Similarity Join
 Leveraging Set Relations in Exact Set Similarity Join Xubo Wang, Lu Qin, Xuemin Lin, Ying Zhang, and Lijun Chang University of New South Wales, Australia University of Technology Sydney, Australia {xwang,lxue,ljchang}@cse.unsw.edu.au,
Leveraging Set Relations in Exact Set Similarity Join Xubo Wang, Lu Qin, Xuemin Lin, Ying Zhang, and Lijun Chang University of New South Wales, Australia University of Technology Sydney, Australia {xwang,lxue,ljchang}@cse.unsw.edu.au,
A Study on Reverse Top-K Queries Using Monochromatic and Bichromatic Methods
 A Study on Reverse Top-K Queries Using Monochromatic and Bichromatic Methods S.Anusuya 1, M.Balaganesh 2 P.G. Student, Department of Computer Science and Engineering, Sembodai Rukmani Varatharajan Engineering
A Study on Reverse Top-K Queries Using Monochromatic and Bichromatic Methods S.Anusuya 1, M.Balaganesh 2 P.G. Student, Department of Computer Science and Engineering, Sembodai Rukmani Varatharajan Engineering
Locality- Sensitive Hashing Random Projections for NN Search
 Case Study 2: Document Retrieval Locality- Sensitive Hashing Random Projections for NN Search Machine Learning for Big Data CSE547/STAT548, University of Washington Sham Kakade April 18, 2017 Sham Kakade
Case Study 2: Document Retrieval Locality- Sensitive Hashing Random Projections for NN Search Machine Learning for Big Data CSE547/STAT548, University of Washington Sham Kakade April 18, 2017 Sham Kakade
Efficient Construction of Safe Regions for Moving knn Queries Over Dynamic Datasets
 Efficient Construction of Safe Regions for Moving knn Queries Over Dynamic Datasets Mahady Hasan, Muhammad Aamir Cheema, Xuemin Lin, Ying Zhang The University of New South Wales, Australia {mahadyh,macheema,lxue,yingz}@cse.unsw.edu.au
Efficient Construction of Safe Regions for Moving knn Queries Over Dynamic Datasets Mahady Hasan, Muhammad Aamir Cheema, Xuemin Lin, Ying Zhang The University of New South Wales, Australia {mahadyh,macheema,lxue,yingz}@cse.unsw.edu.au
Inital Starting Point Analysis for K-Means Clustering: A Case Study
 lemson University TigerPrints Publications School of omputing 3-26 Inital Starting Point Analysis for K-Means lustering: A ase Study Amy Apon lemson University, aapon@clemson.edu Frank Robinson Vanderbilt
lemson University TigerPrints Publications School of omputing 3-26 Inital Starting Point Analysis for K-Means lustering: A ase Study Amy Apon lemson University, aapon@clemson.edu Frank Robinson Vanderbilt
Incorporating Metric Access Methods for Similarity Searching on Oracle Database
 Incorporating Metric Access Methods for Similarity Searching on Oracle Database Daniel S. Kaster 1,2,, Pedro H. Bugatti 2, Agma J. M. Traina 2, Caetano Traina Jr. 2 1 Department of Computer Science, University
Incorporating Metric Access Methods for Similarity Searching on Oracle Database Daniel S. Kaster 1,2,, Pedro H. Bugatti 2, Agma J. M. Traina 2, Caetano Traina Jr. 2 1 Department of Computer Science, University
On the Relationships between Zero Forcing Numbers and Certain Graph Coverings
 On the Relationships between Zero Forcing Numbers and Certain Graph Coverings Fatemeh Alinaghipour Taklimi, Shaun Fallat 1,, Karen Meagher 2 Department of Mathematics and Statistics, University of Regina,
On the Relationships between Zero Forcing Numbers and Certain Graph Coverings Fatemeh Alinaghipour Taklimi, Shaun Fallat 1,, Karen Meagher 2 Department of Mathematics and Statistics, University of Regina,
Cache-Oblivious Traversals of an Array s Pairs
 Cache-Oblivious Traversals of an Array s Pairs Tobias Johnson May 7, 2007 Abstract Cache-obliviousness is a concept first introduced by Frigo et al. in [1]. We follow their model and develop a cache-oblivious
Cache-Oblivious Traversals of an Array s Pairs Tobias Johnson May 7, 2007 Abstract Cache-obliviousness is a concept first introduced by Frigo et al. in [1]. We follow their model and develop a cache-oblivious
Hierarchical Intelligent Cuttings: A Dynamic Multi-dimensional Packet Classification Algorithm
 161 CHAPTER 5 Hierarchical Intelligent Cuttings: A Dynamic Multi-dimensional Packet Classification Algorithm 1 Introduction We saw in the previous chapter that real-life classifiers exhibit structure and
161 CHAPTER 5 Hierarchical Intelligent Cuttings: A Dynamic Multi-dimensional Packet Classification Algorithm 1 Introduction We saw in the previous chapter that real-life classifiers exhibit structure and
DeLiClu: Boosting Robustness, Completeness, Usability, and Efficiency of Hierarchical Clustering by a Closest Pair Ranking
 In Proc. 10th Pacific-Asian Conf. on Advances in Knowledge Discovery and Data Mining (PAKDD'06), Singapore, 2006 DeLiClu: Boosting Robustness, Completeness, Usability, and Efficiency of Hierarchical Clustering
In Proc. 10th Pacific-Asian Conf. on Advances in Knowledge Discovery and Data Mining (PAKDD'06), Singapore, 2006 DeLiClu: Boosting Robustness, Completeness, Usability, and Efficiency of Hierarchical Clustering
A note on an L-approach for solving the manufacturer s pallet loading problem
 A note on an L-approach for solving the manufacturer s pallet loading problem Ernesto G. Birgin Reinaldo Morabito Fabio H. Nishihara November 19 th, 2004. Abstract An L-approach for packing (l, w)-rectangles
A note on an L-approach for solving the manufacturer s pallet loading problem Ernesto G. Birgin Reinaldo Morabito Fabio H. Nishihara November 19 th, 2004. Abstract An L-approach for packing (l, w)-rectangles
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS
 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
( ) = Y ˆ. Calibration Definition A model is calibrated if its predictions are right on average: ave(response Predicted value) = Predicted value.
 Calibration OVERVIEW... 2 INTRODUCTION... 2 CALIBRATION... 3 ANOTHER REASON FOR CALIBRATION... 4 CHECKING THE CALIBRATION OF A REGRESSION... 5 CALIBRATION IN SIMPLE REGRESSION (DISPLAY.JMP)... 5 TESTING
Calibration OVERVIEW... 2 INTRODUCTION... 2 CALIBRATION... 3 ANOTHER REASON FOR CALIBRATION... 4 CHECKING THE CALIBRATION OF A REGRESSION... 5 CALIBRATION IN SIMPLE REGRESSION (DISPLAY.JMP)... 5 TESTING
MVAPICH2 vs. OpenMPI for a Clustering Algorithm
 MVAPICH2 vs. OpenMPI for a Clustering Algorithm Robin V. Blasberg and Matthias K. Gobbert Naval Research Laboratory, Washington, D.C. Department of Mathematics and Statistics, University of Maryland, Baltimore
MVAPICH2 vs. OpenMPI for a Clustering Algorithm Robin V. Blasberg and Matthias K. Gobbert Naval Research Laboratory, Washington, D.C. Department of Mathematics and Statistics, University of Maryland, Baltimore
Clustered Pivot Tables for I/O-optimized Similarity Search
 Clustered Pivot Tables for I/O-optimized Similarity Search Juraj Moško Charles University in Prague, Faculty of Mathematics and Physics, SIRET research group mosko.juro@centrum.sk Jakub Lokoč Charles University
Clustered Pivot Tables for I/O-optimized Similarity Search Juraj Moško Charles University in Prague, Faculty of Mathematics and Physics, SIRET research group mosko.juro@centrum.sk Jakub Lokoč Charles University
Chapter 6 Memory 11/3/2015. Chapter 6 Objectives. 6.2 Types of Memory. 6.1 Introduction
 Chapter 6 Objectives Chapter 6 Memory Master the concepts of hierarchical memory organization. Understand how each level of memory contributes to system performance, and how the performance is measured.
Chapter 6 Objectives Chapter 6 Memory Master the concepts of hierarchical memory organization. Understand how each level of memory contributes to system performance, and how the performance is measured.
Unsupervised Learning
 Outline Unsupervised Learning Basic concepts K-means algorithm Representation of clusters Hierarchical clustering Distance functions Which clustering algorithm to use? NN Supervised learning vs. unsupervised
Outline Unsupervised Learning Basic concepts K-means algorithm Representation of clusters Hierarchical clustering Distance functions Which clustering algorithm to use? NN Supervised learning vs. unsupervised
A visual framework to understand similarity queries and explore data in Metric Access Methods
 370 Int. J. Business Intelligence and Data Mining, Vol. 5, No. 4, 2010 A visual framework to understand similarity queries and explore data in Metric Access Methods Marcos R. Vieira*, Fabio J.T. Chino,
370 Int. J. Business Intelligence and Data Mining, Vol. 5, No. 4, 2010 A visual framework to understand similarity queries and explore data in Metric Access Methods Marcos R. Vieira*, Fabio J.T. Chino,
