UNIVERSITY OF PUERTO RICO RIO PIEDRAS CAMPUS COLLEGE OF NATURAL SCIENCE DEPARTMENT OF MATHEMATICS
|
|
|
- Elizabeth Clarke
- 5 years ago
- Views:
Transcription
1 UNIVERSITY OF PUERTO RICO RIO PIEDRAS CAMPUS COLLEGE OF NATURAL SCIENCE DEPARTMENT OF MATHEMATICS Spectral Graph Algorithms: Applications in Neural Images and Social Networks By: Richard García-Lebrón A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE MAY, 2014
2 APPROVED BY THE MASTER THESIS COMMITTEE IN PARTIAL FULLFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE IN MATHEMATICS AT THE UNIVERSITY OF PUERTO RICO RIO PIEDRAS CAMPUS ADVISOR: Ioannis Koutis, Ph.D. Assistant Professor of Computer Science University of Puerto Rico, Rio Piedras READERS: Patricia Ordoñez, Ph.D. Assistant Professor of Computer Science University of Puerto Rico, Rio Piedras Edusmildo Orozco, Ph.D. Associate Professor of Computer Science University of Puerto Rico, Rio Piedras Eduardo Rosa-Molinar, Ph.D. Associate Professor of Biology University of Puerto Rico, Rio Piedras
3 You can t connect the dots looking forward; you can only connect them looking backwards. So you have to trust that the dots will somehow connect in your future. You have to trust in something your gut, destiny, life, karma, whatever. This approach has never let me down, and it has made all the difference in my life. Stay hungry. Stay foolish. Steve Jobs
4 Acknowledgments To my grandparents: Eugenio Lebrón Negrón ( ) & Carmen L. Milian De León Thanks to my wife Wilnelia Antuna Camacho, my mother Angie I. Lebrón Milian and my family for all the love and support. Moreover I would like to thank my advisor Ioannis Koutis, my mentor Ivelisse Rubio, the commitee, the Biological Imaging Group UPR and Puerto Rico Louis Stokes Alliance for Minority Participation thanks for the experiences, knowledge and support. I am because you are. iv
5 Spectral Graph Algorithms: Applications in Neural Images and Social Networks Richard García Lebrón Department of Mathematics University of Puerto Rico Rio Piedras, San Juan 2014 ABSTRACT Algorithms based on spectral graph theory have brought up powerful advances in the analysis of complex networks. We present spectral algorithms for data mining and image segmentation. The core of the algorithms is a recently discovered very fast linear system solver for the important class of symmetric diagonally dominant matrices. Our first contribution is the Fast Effective Resistance Library FastER, for computing the effective resistances of graph, viewed as an electrical network. We also present the Graph Clustering Library GraphCL which applies FastER to the community detection problem. A further application of FastER is in the analysis of edge importance. Electrical and combinatorial edge importance measures were compared using an information propagation model. The effective resistance measure performed better in identifying more influential edges in the graph. Our second contribution is the irandom-walker algorithm, a modified version of the Grady s Random-Walker algorithm for image segmentation. We use the irandom- Walker to build i3d-segmentation, a framework for semi-automated segmentation of neurons in their three-dimensional space, implemented as an Imaris MATLAB extension.
6 Contents 1 Introduction Problems and Solutions Fast computation of effective resistances in electrical networks: a MATLAB implementation Introduction Background Graphs and Laplacians Graphs as electrical resistive circuits Implementation Experimental Accuracy Analysis Experimental Computational Time Analysis Edge-Importance Introduction Background SpanningTree centrality The CurrentFlow centrality Edge Importance Tool Computing SpanningTree Tools Speedups Experiments Experiments for SpanningTree Edge-importance measures and information propagation Results vi
7 4 Spectral and Electrical Clustering Introduction Background Algorithms Graph Embeddings Implementation Experiments and Results Random Walks: For Image Segmentation Introduction Background Random Walker Iterative Random Walk Solving Three-dimensional Segmentation of Neurons Using Spectral Graph Algorithms Introduction Frame Work Results Bibliography Index Glossary
8 List of Figures 2.1 A graph: dots are vertices and connections are edges An example graph G = (V, E, w) Circuit representation for the graph in Figure Average and standard deviation of the computed effective resistances for ɛ = 0.1 and tolerance Average and standard deviation of the computed effective resistances for ɛ = 0.01 and tolerance Standard Deviation summary Average and standard deviation of the relative error for ɛ = 0.1 and tolerance Average and standard deviation of the relative error for ɛ = 0.01 and tolerance Relative error, standard deviation summary The C graph Computational time for query and static versions using C n n n with n from 9 to A network, viewed as an electrical resistive circuit. The thickness of an edge represents the amount of current it carries, if a battery is attached to nodes s and t Accuracy-efficiency tradeoff; y-axis (logarithmic scale): running time of the Fast-TreeC algorithm; x-axis: error parameter ɛ Limiting the computation on the 2-core shows a measurable improvement in the running time of Fast-TreeC The values k as a function of k for the different importance measures The number of components as a function of x The number of infected nodes as a function of x The number of components as a function of x viii
9 3.8 The number of infected nodes as a function of x Graph with communities. Each community is circled by a line A social network graph where dots nodes are user and users that are friends are connected by a line edges An example graph G = (V, E, w) Summary of the ratio R on the communities obtained by Algorithm 6 and Analysis of 80% best clusters Visualization for k = 6: Each node represents a community, the ratio of the node is proportional to the number of individuals in the community and the edge weight is the number of connections between the communities Visualization for k = 12: Each node represents a community, the ratio of the node is proportional to the number of individuals in the community and the edge weight is the number of connections between the communities Visualization of twelve communities using the Force Atlas 2 layout from Gephi Visualization of eighteen communities using the Force Atlas 2 layout from Gephi The pixels at column one and row three are labeled as out pixel and the pixel at column two and row two is labeled as in. The other pixels remain unlabeled Solution Left: the pre-labeled pixels at the second iteration. Right: the solution after two iterations of the irandom-walker Left: The pre-labeled pixels (input for irandom-walker). Right: the solution after one iteration of the irandom-walker A) Registered serial three-dimensional (3D) image stack of high contrast images of nervous system; B) Selectively labeled neuron with Alexa FluorR 594 biocytin; C) Example of expert input A) Registered serial three-dimensional (3D) image stack of high contrast images of nervous system; B) Selectively labeled neuron with Alexa FluorR 594 biocytin; C) Example of expert input
10 6.3 A) Contour on image with folding artifact; B) Contour on image with a chatter artifacts Images correspond to a sequence of frames in order: A) Image with out artifact. B) Example of image with artifact in and its contour. C) Image with out artifact Surface comparison: A) Manual segmentation. B) Semi-automated segmentation (smoother surface)
11 List of Tables 2.1 Experimental relative error analysis of the solver precision Experimental Relative Error Analysis Social Networks time results Statistics of the collection of datasets used in our experiments xi
12 Chapter 1 Introduction The last decade has seen an explosion in digital data and information. Socials Networks such as Facebook, Twitter and Google+, are central platforms for communication, content sharing, news, picture, videos and marketing advertising. Indeed this is the motivation of many research problems in computer science and applied mathematics. Consider for example the following problem: How should Facebook choose to recommend new friends for its users? A natural way to address this problem is by creating a graph where users are represented by vertices, and edges represent friendship relationships. Given this framework one solution would be to find communities in the graph and suggest as new friends people that appear to be in the same community. Questions of the same type naturally suggest the use of graphs to encode the information. Graph encodings can be used in many other significant but less obvious ways. A great example is computer vision where pixels can be represented as vertices and neighboring pixels are connected with weighted edges. In this case, community detection can be used as a metaphor for object and image segmentation. Graph-based problems can thus vary from the analysis of social networks to the reconstruction of the connectome (network of all neurons in the central nervous system) from 3D images. In general, graphs encode individuals as vertices and the relation between individuals as edges. Graph themselves can be encoded in many other ways, including as algebraic objects known as matrices, such as the Laplacian or the adjacency matrix. The Lapla- 1
13 cian matrix captures many combinatorial properties of the graph in algebraic terms. For example there are known connections with sparse graph cuts, with commute times in random walks as well as with the computation of the effective resistances. The powerful properties of Laplacians were well understood for decades, but they have received special attention after the work of Spielman and Teng [ST04a]. In 2004 Spielman and Teng described the first nearly-linear time solver for matrix of the class Symmetric Diagonal Dominant (SDD). Because Laplacians are in the class of SDD matrices many existing algorithm based on Laplacians instantly became feasible for huge data sets. Further work on SDD solvers [KMP10, KMP11a] has brought the running time down to near-o(m log m) where m is the number of non-zeros entries in the system. Recent works present a simple algorithm but with a slightly higher running time: O(m log 2 n log log n) [KOSZ13]. Based on recent research on SDD solvers is also the Combinatorial Multigrid solver (CMGSolve) which empirically exhibits a linear running time [KMT11]. The emerging of this technology makes possible the analysis of graph with millions of connection in time almost proportional to the number of connection in the graph. 1.1 Problems and Solutions In this work a graph G = (V, E, w) is composed by the set of vertices V, the set of edges E and the list of edge weights w. The set of vertices V in general represent individuals from a network. Depending on the network type, an individual can be a user (in social networks), a pixel (in image segmentation problems), an intersection of streets (in maps) etc. The edges represent the relationship of the vertices. For example, for the case of image related problems, edges connect spatially adjacent pixels. The edge weight quantifies how similar are the two pixels. 2
14 In the context of this very general framework we study the following problems. Edge-Importance: Measures that quantify the importance of edges are valuable in the analysis of various types of network data, including social networks, biological networks, computer networks and many more. With rare exceptions, the applicability of such measures to large data sets is hindered by the lack of fast algorithms for their computation. In this thesis, we present tools that enable the computation of significant edge importance measures on very large networks. These measures fall in the broad class of electrical measures. At a high level, we quantify the importance of an edge as a function of the electrical flow that passes through it when electric sources are applied to different nodes on the network. For this task we propose the use of effective resistances as a metric for edge importance also known as spanning-tree centrality. Clustering: Clustering refers to the detection of disjoint or overlapping sets of vertices in a graph. In this work we focus on disjoint clusters. The task is to find clusters that are of good quality, according to some predetermined measure. We focus in minimizing the edge-expansion value which refers to the proportion of edges leaving the cluster compared with the number of edges inside the cluster. Another measure of interest is to maximize the conductance of the sub-graph induced by the cluster. We also propose using a combination of the two measures as a metric for quantifying the quality of the cluster. We present implementations and comparisons between three of the most recent clustering tools [KC12a, NJW01, LRTV12] based on electrical and spectral measures. Three-dimensional Segmentation of Neurons: The semi-automatic segmentation of individual neurons in electron microscopic (EM) images is crucial in the acquisition and analysis of connectomes. EM images are three-dimensional. The nature of the problem suggests the use of all images as one object. However it has been commonly thought that approaches which use contextual information from distant parts 3
15 of the image to make local decisions, should be computationally infeasible. Combined with the topological complexity of three-dimensional (3D) space, this belief has been deterring the development of algorithms that work genuinely in 3D. However, recent breakthrough results in spectral graph theory show that this intuition is wrong. It is in fact possible to solve linear systems of matrices associated with the affinity graphs derived from the images in time that essentially scales with their size. This renders feasible a multitude of previously proposed algorithms for image segmentation, and in particular algorithms based on the computation of fundamental spectral properties of the graphs, which encode information valuable for segmentation. In this problem we adapt Grady s Random Walker method [Gra06] to expand a rough shape of the neuron into a significantly more precise segmentation. Supplemented with the recently discovered linear system solvers our algorithms make efficient use of 3D contextual information to generate noise-insensitive neuron segmentation that delivers the surface of the neuron as whole, rather than as a stack of 2D boundaries. 4
16 Chapter 2 Fast computation of effective resistances in electrical networks: a MATLAB implementation 2.1 Introduction Graphs are frequently used to model data. The most well understood example is that of a social network graph: persons are represented by vertices and their mutual relationships are encoded in edges. Of course, there are many other examples from a variety of disciplines including ecology networks, protein networks, electrical networks, street networks etc. The common theme in these representations is that entities are represented as nodes, and relationships between individual entities are represented as edges; edges can have weights indicating the degree of relationship. For example in an ecology network, species in the ecosystem are represented as the nodes in the graph and the weight of an edge between two species can be a function of the number of common prey between the species. Another perhaps less obvious example is digital images where pixels are represented as nodes and the weight of edges encodes the similarity in brightness or color of the corresponding pixels. Besides a systematic framework for approaching data analysis, graph representations offer the opportunity to transfer notions, algorithms and ideas between the different contexts and disciplines. These connections can be quite surprising. For example, 5
17 we can view graphs as electrical resistive networks where edges become resistors of capacitance equal to their weight. The effective resistance between two nodes i and j is the inverse of the current that will flow between i and j if we apply the two poles of a 1Volt battery to the two nodes. On the other hand, we can define the commute time between i and j, as the expected time between two visits to the vertex i under the constraint that vertex j has been visited in the mean time. These two seemingly different notions are in fact equivalent; the effective resistance is proportional to the commute time [CRR + 89]. There has recently been significant interest in using commute time/effective resistances for data mining, as it has been observed that they behave well both locally and globally as a similarity measure especially in the presence of noise in the data. In [KC10] the commute time was employed in the detection of outliers in a set of points embedded in the Euclidean space. In particular, after constructing a graph from the points, it was shown that applying k-means clustering with respect to commute distance over the graph instead of the Euclidean distance, improves the detection of local and global outliers. In [FPS05] an effective resistance-based approach was used for the problem of recommending movies to the users of a user-movies database. Commute times have also been in used in combination with k-means and fuzzy k-means algorithms, for graph clustering applications [YFD + 07]. As we will see, computing the effective resistance for even one pair of vertices requires by definition the solution of a symmetric diagonally dominant linear (SDD) system of equations. The running time required to solve one such linear system was prohibitive to using commute distance-based methods on large data, at least until the work of Spielman and Teng [ST04b] who gave a near-linear time algorithm for the problem. Further work on SDD solvers [KMP10, KMP11a] has brought the running time down to near-o(m log m) where m is the number of non-zero entries in the 6
18 system. Based on recent research on SDD solvers is also the Combinatorial Multigrid solver (CMGSolve) which empirically exhibits a linear running time [KMT11]. Still, the availability of fast solvers doesn t answer the problem of computing many, potentially O(m), effective resistances in a given graph. A very fast approach to compute good approximations of many effective resistances was given by Spielman and Srivastava in [SS08]; they essentially show that after a preprocessing phase which consists of solving O(log n) linear systems where n is the number node in the graph, one can approximately compute any effective resistance in O(log n) time. In this section we present and discuss a MATLAB implementation of the Spielman- Srivastava algorithm. The algorithm is based on CMGSolve. We actually present two variants of the algorithm. The query version outputs a function R(i, j) which returns an approximation to the effective resistance between the vertices i and j. The static version of the algorithm gives an alternative, more space-efficient and faster implementation for the case when the list of needed effective resistances is known before the preprocessing phase; the code outputs directly approximate values for the requested list. We complement these two variants with code that computes the exact effective resistances for smaller networks. Our goal is to observe in practice the accuracy of the method for various types of networks and recommend safe choices for the parameters of the algorithm. We are aware of only one prior implementation [WC12], which was used to accelerate the method in [KC10]; this code is not publicly available. 7
19 2.2 Background Graphs and Laplacians A graph is formally defined as a triple G = (V, E, w) where: (i) V is the set of vertices (also called nodes), (ii) E is a set of edges, with each edge being an unordered pair of vertices, (iii) w is a weight function that maps each edge in E to a real value. Graphs can be visualized as in Figure 2.1. Figure 2.1: A graph: dots are vertices and connections are edges. To handle graphs computationally we need to define some type of encoding for them. We will find it useful to assign to each vertex a unique integer in [1, n], where n is the number of vertices in the graph. We will do the same for edges, and denote by m the number of edges in the graph, and A common graph encoding is the adjacency matrix. Definition 1. The adjacency matrix A of a graph G = (V, E, w) is defined as: w i,j if (i, j) E A i,j = 0 otherwise 8
20 We will make crucial use of a related matrix the graph Laplacian. Definition 2. The Laplacian matrix L of a graph G = (V, E, w) is defined as: L i,j = w i,j if (i, j) E i j w i,j if i = j 0 otherwise An example of a graph and its Laplacian is given in Figure 2.2. Definition 3. For an arbitrary orientation of the graph G the signed edge-vertex incidence matrix B with size m n is defined as: Let e an edge, 1 k m and 1 h n then: B k,h = 1 if e k = (i, j) and h = i for 1 k m 1 if e k = (i, j) and h = j for 1 k m 0 otherwise Consider the graph on Figure 2.1 its signed edge-vertex incident matrix is as follows: B =
21 L G = C A Figure 2.2: An example graph G = (V, E, w) Let W be the m m diagonal matrix containing the weights of the edges. The Laplacian and edge-incident matrices are connected through the following identity L = B T W B (2.1) Laplacians matrices are symmetric and diagonally dominant - SDD - linear systems of the form Ax = b, where A is SDD can be solved in near-linear time O(m log c n), where c is constant [ST04a]. There are practical implementations that are been use in [KMT11] Graphs as electrical resistive circuits Every graph can be viewed as a resistive electrical network, where each edge (i, j) is a resistor with resistance R i,j = 1/w i,j. An illustration is given in Figure Figure 2.3: Circuit representation for the graph in Figure 2.2. Laplacians arise naturally in the context of resistive electrical networks. For example, 10
22 consider the quadratic form of the Laplacian: x T Lx = (x i x j ) 2 w i,j (2.2) (i,j) E To see the natural meaning of the above equality, imagine that the voltages at the nodes of the circuit/graph are given in the corresponding entry of a vector x. The energy dissipated by this setting on edge (i, j) is equal to w i,j (x i x j ) 2. Then it can be seen that the quadratic form x T Lx is equal to the total dissipation in the circuit. For a second use of the Laplacian, consider the case when two nodes s and t are hooked to the poles of a battery and a unit of current (1A) flow from s to t. In order to compute the voltages needed in the resistive electrical network for this to happen, we solve the linear system Lv = b, where b is a zero vector with the exception of b s = 1 and b t = 1. Solving the linear system Lv = b comes directly from the Kirchoff s and Ohm s law. From Ohm s law the physical meaning of R is equal to the voltage difference that has to be applied to its two endpoints in order to drive one unit of current flow across the wire V = RI. Moreover it is known from Kirchoff s law that the net current is equal to zeros - I 1 + I 2 + = 0-. Example 1. The effective resistance of the first and last node in a serial circuit with two wires when driving 1A through the circuit is V 3 V 1 by Kirchoff s law. The voltages needed to drive a one unit of current over the circuit when a battery is hooked to the first and last node are computed by solving the following system equations: V 1 I 1 = 1 R 1 V 2 I 2 = 0 R 2 V 3 I 3 = +1 11
23 V 1 V 2 R 1 = 1 V 2 V 1 R 1 + V 2 V 3 R 2 = 0 V 3 V 2 R 2 = +1 1 R 1 1 R R 1 1 R R 2 1 R R 2 1 R 2 V 1 V 2 V 3 = By Kirchoff s law the current in the second node must be zero. Notice the matrix of the system is the Laplacian matrix of the graph equivalent to the two wire circuit. Therefore the effective resistance is R 1 +R 2, this is the well known formula to compute the effective resistance in a parallel circuit. Viewing the computation of the effective resistances as the solution of Lv = b motivates the following algorithm to compute all the effective resistances. Algorithm 1 Exact computation of the effective resistances, this algorithm is implemented as ExactER or exact version see 2.3 Input: Laplacian matrix L. Output: R(e) for e = (s, t) 1: let b k be a column vector with all zeros except the k row. 2: b s,t = b s b t 3: solve Lv = b s,t 4: report R((s, t)) = b T s,t v A fast approximation for effective resistances 1 Computing even one effective resistance using Algorithm 1 requires the solution of a linear system with a Laplacian matrix, so computing the effective resistancesfor all edges of a network seems to require at least quadratic time. Can something better be done if we settle for approximations of the effective resistances? Before answering the question, we observe that the effective resistances can be expressed as a distance between vectors: 12
24 Definition 4. Effective resistances as distance between vectors, let L by the pseudo inverse of the Laplacian matrix then: R(s, t) = (b s,t ) T L (b s,t ) = (b s b t ) T L (b s b t ) = (b s b t ) T L LL (b s b t ) = (b s b t ) T L B T W BL (b s b t ) = (W 1 2 BL (b s b t )) T (W 1 2 BL (b s b t )) = (W 1 2 BL (b s b t )) 2 2. (2.3) This expression of the effective resistances as a L 2 2 distance make possible the use of the well known Johnson-Lindenstrauss Lemma, which roughly states that randomly projecting n vectors to a space of dimension O(log n) preserves their distances [JL84]. More precisely, it is known that in order to achieve accuracy 1 ± ɛ we need at most C log n/ɛ 2 dimensions, where C is some constant independent of the parameters. Because the distances are taken from the columns of W 1 2 BL we can use the Johnson Lindenstrauss projection in every column of W 1 2 BL to reduce the number of linear system needed to be solved. This idea was first proposed by Spielman and Srivastava [SS08]. Let us be more concrete. Theorem 1. (Johnson-Lindenstrauss) Let ɛ > 0, n be an integer and k be a positive integer such that k k 0 = O( log n ). For every set P of n points in R d there exists ɛ f : R d R k such that for all u, v P. (1 ɛ) u v 2 f(u) f(v) 2 (1 + ɛ) u v 2 13
25 In practice a random matrix Q with dimension O(log n) n is used to project n vectors (the points) into a space to a space of dimension O(log n). The particular construction below is due to Arriaga et al. [AV06] the performance analysis of this construction was introduced in [Ach03]. Lemma 1. Given fixed vectors v 1... v n R d and ɛ > 0, let Q k d be a random ± 1 k matrix (i.e. independent Bernoulli entries) with k > 24 log n/ɛ 2. Then with probability at least 1 1, we have: n (1 ɛ) v i v j 2 2 Qv i Qv j 2 2 (1 + ɛ) v i v j 2 2 for all pairs i, j n. Algorithm 2 Computing approximations for effective resistances Input: G = (V, E). Output: R(e) for every e i E. 1: Construct the projection matrix Q shown in Lemma 1. 2: Compute Y = Q W 1 2 B 3: Construct Z as follows: for each i-th row y i of Y solve Lz i = y i. 4: Report R((i, j)) = Z(:, i) Z(:, j) 2 2, the distance between the i-th column and the j-th column of Z. The above algorithm assumes that the linear systems are solved exactly. However, the fast linear solvers we use are iterative, i.e. they only compute an approximate solution, up to a tolerance specified by the user. Spielman and Srivastava [SS08] show that in order to guarantee an 1 ± ɛ precision in the effective resistance we can set the tolerance to ɛ/n 2, impacting the running time of the solver by an O(log(n/ɛ)) factor. Koutis et al. [KLP12] show that it actually suffices to set the tolerance to ɛ, saving an O(log n) factor from the running time for most values of ɛ. We now comment on the running time of the algorithm, omitting the O(log(1/ɛ)) factor we discussed above. It is easy to see that Step 1 takes time O(m log n). More 14
26 interesting are Steps 2 and 3 where we project the columns of the matrix W 1/2 BL into the columns of the matrix Z. Because B is sparse and has 2m non zero entries, Step 2 takes time O(m log n). As we state before the Laplacian matrix is SDD and systems of the form Lz i = y i can be solved fast using the Koutis et al. [KMT11] solver which runs in O(m log n) time, therefore Step 3 takes O(m log 2 n). Finally Step 4 can be done in time O(log n) per edge. This shows that for computing approximation of the effective resistances we only need time O(m log 2 n). In the current implementation we use CMGSolve by Koutis et al. [KMT11], which in practice has a linear running time for sparse graphs and saves another O(log n) factor for most practical problems. 2.3 Implementation Different kinds of problems involve the effective resistances computation as shown in 2.1. There are (i) problems where the user knows exactly the edges for which the effective resistances need to be computed (ii) problems where the user doesn t know ahead of time which effective resistances are going to be needed. For these two kinds of problems we came up with two implementations called static for the fist type of problems and query for the second type of problems. These implementations have been developed on MATLAB and the code can be accessed from EVKKLF. In the query version the user is able to query for the effective resistance between any pair of nodes (i, j). This version is an exact implementation of Algorithm 2. The Z is constructed and stored in the background allowing the user to reuse the matrix for further queries. The query version is suggested for users that are not interested in a specific small set of edges or pairs of vertices, but they want to have access to any pair of vertices, as need arises. 15
27 In comparison with the query version, the static version returns the effective resistances of a list of edges given by the user as an input. This version doesn t save the matrix Z for future queries. Indeed the static version is faster and suitable for huge systems. The question that arises naturally is why is the static version faster if both version are based on the same algorithm? The key in Algorithm 2 is how the random vectors and the projection matrix are created. In the static version we combine the four steps to avoid the use of Q, this approach reduces the space needed in memory. As a result reducing the memory speedups the implementation by reducing the overhead time it takes for the operating system to provide access to the memory and by reducing the use of virtual memory Experimental Accuracy Analysis In this section we experimentally compare the accuracy of the two different implementations, the query and the static version, against the exact effective resistances computations. Experiment 1: For the lattices graphs L and the L we run the exact version to get the exact effective resistances then we compare the relative error using the approximation of query version with a fixed ɛ and varying the solver tolerance. The result of the experiments are summarized in Table 2.1. In agreement with the result in [KLP12], the tolerance in the accuracy of the linear system solver does not affect the accuracy in the approximation of effective resistances, if it is set to be sufficiently small. So in the following experiments we will be setting uniformly the tolerance to a small value, for each type of experiment. Experiment 2: We compute the effective resistances for the path graph P 1000 with one thousand vertex and the lattice graph L with eight hundred vertex and 16
28 Graph solver tolerance Average relative error MSE L L L L L L Table 2.1: Experimental relative error analysis of the solver precision Error / Version query static Maximum Median Mean Quantile 25% Quantile 75% Quantile 95% MSE (a) Analysis for P 1000 Error / Version query static Maximum Median Mean Quantile 25% Quantile 75% Quantile 95% MSE (b) Analysis for L Table 2.2: Experimental Relative Error Analysis compute the relative error. The tolerance for the solver was set to and the algorithm precision to ɛ = 0.1. In Table 2.2a we present common statistical measures of the relative error to compare the behavior between the implementations. Experiment 3: In this experiment we use the collaboration network for General Relativity and Quantum Cosmology (ca-grqc). The effective resistances were computed with both versions, for accuracy ɛ = 0.1 and ɛ = 0.01, and solver tolerance set to The computation is repeated five times, from which we compute the average µ and the standard deviation σ of the computed effective resistances. In Figures 2.4, 2.5 and 2.6 we plot µ ± σ and the summary of the standard deviation respectively. Experiment 4: For this experiment we compute the relative error of both implementations using the ca-grqc data set. The average and standard deviation of five computations are plotted in Figures 2.7 and 2.8. The summary of the standard deviation is in Figure
29 ca GrQc (epsilon=0.1) 0.6 ER (mean) 0.4 type Exact Query (eps = 0.1) Static (eps = 0.1) Sorted edge index Figure 2.4: Average and standard deviation of the computed effective resistances for ɛ = 0.1 and tolerance Version Time (s) query static (a) LiveJournal social network Version Time (s) query 12.1 static 9.78 (b) Facebook social network Table 2.3: Social Networks time results. Since the static version is nearly identical to the query version, as expected similar results were observed in Experiments 2, 3 and 4. The small differences occurs because the random generator changes between experiments Experimental Computational Time Analysis The goal of this section is to compare the speed of the implementations for the query and static versions. The hardware used in the experiment was a Server with Two Xeon 2.3 Ghz and 96Gb of RAM. 18
30 Sorted edge index ER (mean) type Exact Query (eps = 0.01) Static (eps = 0.01) ca GrQc (epsilon=0.01) Figure 2.5: Average and standard deviation of the computed effective resistances for ɛ = 0.01 and tolerance Query (eps = 0.01) Query (eps = 0.1) Static (eps = 0.01) Static (eps = 0.1) Algorithm Version Standart Deviation type Query (eps = 0.01) Query (eps = 0.1) Static (eps = 0.01) Static (eps = 0.1) ca GrQc Figure 2.6: Standard Deviation summary 19
31 0.4 ca GrQc (epsilon=0.1) 0.3 Relative error (mean) type Query (eps = 0.1) Static (eps = 0.1) Edge index Figure 2.7: Average and standard deviation of the relative error for ɛ = 0.1 and tolerance 10 8 ca GrQc (epsilon=0.01) Relative error (mean) type Query (eps = 0.01) Static (eps = 0.01) Edge index Figure 2.8: Average and standard deviation of the relative error for ɛ = 0.01 and tolerance
32 Query (eps = 0.01) Query (eps = 0.1) Static (eps = 0.01) Static (eps = 0.1) Algorithm Version Relative error Standart Deviation type Query (eps = 0.01) Query (eps = 0.1) Static (eps = 0.01) Static (eps = 0.1) ca GrQc Figure 2.9: Relative error, standard deviation summary Experiment 1: The graph used in this experiment is the three-dimensional grid graph C n n n with 3n 2 (n 1) edges, illustrated in Figure We use n from 9 to 300. In Figure 2.11 we show the time in seconds for both implementations. We see that the static version is slightly faster than the query version. We observe that using the query version with n > 300 we have space (memory on hardware 96GB) limitations in comparison to the static version we can go up to C which has more edges than C Experiment 2: The graphs in this experiment are the LiveJournal social network described in [BHKL06] and [LLDM08] and the Facebook social network described in [ML12]. Both networks where converted to undirected weighted graphs using the number of common friends in the social network as the weights. The LiveJournal graph is disconnected to convert the graph to a connected graph we choose five nodes at random for each connected component and add edges between them. The results are summarized in Tables 2.3a and 2.3b. 21
33 The C 7x7x7 graph Figure 2.10: The C graph query time V.S. static time Time (s) static query Number of edges in the graph x 10 7 Figure 2.11: Computational time for query and static versions using C n n n with n from 9 to
34 Chapter 3 Edge-Importance 3.1 Introduction Which streets should one monitor, in order to accurately track the traffic in a city? Or, if one wants to block the spread of information in a network by severing a small percentage of its links, which ones should we choose in order to maximize the impact? Questions like these have motivated the definition of importance measures, for both the nodes and the edges of a network. They have been studied for various types of data, ranging from social and media networks, the Internet and the Web, to transportation networks, biological networks and other domains. In principle, all reasonable importance measures can reveal useful information about a network. But computation is a serious issue. As the size of the network increases to contain billions of nodes and edges, the usability of such importance scores depends on the time and memory resources required for their computation. Measures whose computation can be carried out even approximately for networks with millions of nodes are the rare exception [DMFFR12, BV]. In this work we present a method for computing an edge importance measure for very large networks, in time nearly proportional to their size. More specifically, we focus on one electrical measure, namely the SpanningTree centrality, that arise when viewing the network as an electrical resistive network, where edges correspond to resistors. The effectiveness of this measure has been demonstrated in the context 23
35 s t Figure 3.1: A network, viewed as an electrical resistive circuit. The thickness of an edge represents the amount of current it carries, if a battery is attached to nodes s and t. of applications in phylogenetic trees [TMC + 13], social networks [BF05] and proteinprotein interaction networks [MLZ + 09]. Electrical measures have the potential for providing significant network information in many other contexts, making them a powerful tool for network analysis. We provide evidence in support of this claim by experimentally studying electrical measures with respect to an information propagation process over the network. Electrical measures: To get a better understanding of electrical measures, consider Figure 3.1. Suppose that we hook the poles of a battery to nodes s and t and apply a voltage difference sufficient to drive one unit of current (1A) from s to t. Doing that, each node in the network will get a voltage value and electrical current will flow essentially everywhere. At a high level, the electrical measures quantify the importance of an edge by aggregating the value of the flows that pass through it over different applications of voltage differences across pairs of nodes s and t. In fact, different aggregation schemes and acceptable battery placements lead to different definitions of edge-importance measures. 24
36 Contributions: We provide fast implementations for the computation of one concrete measure of edge importance: SpanningTree centrality. Our computations are randomized approximate, and in the case of SpanningTree, they also come with strict theoretical guarantees, which are completely under our control. For example, for a network consisting of 1.5 million nodes, we are able to compute the SpanningTree centrality values that are within 5% of the original ones in 30 minutes. Note that 5% variations of the values can be produced even with very small local perturbations (additions and removal of local links) in the given graphs. The core component of our implementation is a fast linear system solver for Laplacian matrices [KMT11]. The computation of SpanningTree centrality also uses the remarkable work of Spielman and Srivastava [SS11]. While we leverage all these algorithmic tools, our ability to experiment with large-scale networks are possible in time proportional to the number of node in the network. Finally, in a thorough experimental evaluation we demonstrate the ability of our algorithms to compute the proposed centrality measures for very large graphs. In addition to demonstrating the computational efficiency of our algorithms, we also investigate the meaning of importance as defined by the different measures. We quantify this abstract notion by measuring the effect of the addition/deletion of important edges as selected by the different measures on the result of an information propagation process over the underlying network. As we have already discussed, there exists a plethora of measures for quantifying the importance of network entities (i.e., nodes and edges) [Ant71, Bav48, Bor05, Bra01, BF05, DMFFR12, Fre77, FBW91, IETB12, KPST11, New05, Shi53]. Here, we limit our review to edge-importance measures. A widely used measure of edge importance is betweenness centrality. Originally de- 25
37 fined for nodes [Fre77], betweenness centrality has a natural extension to edges. Specifically, the betweenness centrality of an edge e is defined as the fraction of shortest paths between all pairs of nodes that pass through e. From the structural perspective, the main problem with betweenness centrality is that the addition of even one shortcut link joining two nodes that were previously at distance two (a very common occurrence in social networks), can change dramatically the betweenness scores and the ranking of links according to it [New05]. On the contrary, electrical measures alleviate this lack of robustness and appear to be more appropriate for several applications including the analysis of protein-protein interaction networks [MLZ + 09]. From the computational point of view, betweenness centrality can be computed in time O(mn) or O(nm + n 2 log n), where n (resp. m) is the number of nodes (resp. edges) of the network [Bra01]. The main bottleneck of that computation lies in finding the all-pairs shortest paths. Existing algorithms for speeding up this computation rely on reducing the number of such shortest-path computations. For example, Brandes and Pich [BP07] propose sampling pairs of source-destination pairs as a means for alleviating this computational problem. Then, they experimentally evaluate the accuracy of different source-destination sampling schemes, including random sampling. Geisberger et al. [GSS08] also propose sampling of source destination pairs. The only difference is that, in their case, the contribution of every sampled pair to the centrality of a node depends on the distance of the node to the nodes of the selected pair. Instead of sampling random source-destination pairs, Bader et al. [BKMM07] sample only sources from which they form a DFS traversal of the input graph. Therefore the shortest-paths from the selected source to all other nodes are retrieved. The key of their method is that the sampling of such sources is adaptive based on the exploration (through DFS trees) that has already been made. Although all the above 26
38 provide significant speedups, they do not provide any approximation guarantees and the resulting algorithms remain quadratic. The algorithms we present here for our measures are almost linear and thus they can be adopted for the analysis of very large datasets. Electrical measures of edge importance, like the one we study here, are also not new. For example, the SpanningTree centrality have been previously proposed to capture central edges in applications like phylogenetic trees [TMC + 13]. Although we use the same definitions in our work, we also provide a near linear-time algorithms to compute it. 3.2 Background In this section, we provide the definitions of several electrical measures of edge centrality and their combinatorial interpretation in terms of the input graph as well as their electrical interpretation in terms of the resistive network defined by the input graph SpanningTree centrality The SpanningTree centrality assigns edge importance based on the number of spanning trees an edge participates in. That is, important edges participate in many spanning trees. Formally, the measure has been defined recently by Teixeira et al. [TMC + 13] as follows: Definition 5 (SpanningTree). Given a connected, unweighted and undirected graph G = (V, E) the SpanningTree centrality of an edge e, denoted by StC(e, G) is defined as the fraction of spanning trees of G that contain edge e. 27
39 By definition, StC(e) (0, 1] and the larger the StC score of edge e, the more spanning trees e participates in. In cases where we want to specify the graph G used for the computation of the StC of an edge e, we extend the set of arguments of StC with an extra argument: StC(e, G). Intuition: In order to develop some intuition, it is interesting to discuss which edges are assigned high StC scores: the only edges that achieve the highest possible StC score of 1 are bridges, i.e., edges that once removed make G disconnected. This is means that they participate in all possible spanning trees. The extreme case of bridges helps demonstrate the notion of importance captured by the StC scores for the rest of the edges: assuming that spanning trees encode candidate pathways through which information propagates in the network, then edges with high StC are those that, once removed, would incur a significant burden on the rest of the edges. SpanningTree centrality as an electrical measure: Our algorithms for computing the SpanningTree centrality efficiently rely on the connection between the StC scores and the effective resistances of edges. The notion of effective resistance of edges comes from viewing the input graph as an electrical circuit [DS84] in which each edge is a resistor with unit resistance. The effective resistance R(u, v) between two nodes u, v of the graph (that may or may not be connected) is equal to the potential difference between nodes u and v when a unit of current is injected in one vertex (e.g., u) and extracted at the other (e.g., v). In fact, it can be shown [Bol98, DS84] that for any graph G = (V, E) and edge e E, the effective resistance of e, denoted by R(e), is equal to the probability that edge e appears in a random spanning tree of G = (V, E). That is, StC(e) = R(e). This fact makes the theory of resistive electrical networks [DS84] available to us. The details of these computations are given in Section
40 3.2.2 The CurrentFlow centrality Instead of using the spanning trees that edge e participates in as an indicator of e s importance, one can alternatively harness the paths that include e to quantify its importance. This is the intuition behind the CurrentFlow centrality, which was first proposed by Brandes and Fleisher[BF05]. Edge e is considered important if it participates in many paths that connect any two random nodes in the network. This intuition is best captured via a definition based on electrical flows in the resistive network defined by the graph G. More specifically, consider two fixed nodes s and t on which we apply a voltage difference sufficient to drive one unit of current (1A) from s to t. When we apply this, every node in the network has some voltage values and electric current flows through every edge. The (s, t)-flow of edge e = {u, v}, denoted by f st (u, v), is the flow that passes through edge e in this configuration. From this, we define the CurrentFlow centrality of an edge as follows: Definition 6 (CurrentFlow centrality). Given a connected and undirected network G = (V, E) the CurrentFlow centrality of edge e E (denoted by CfC(e)) is the average (s, t)-flow of edge e, when considering all distinct pairs of nodes (s, t). That is, CfC(e = {u, v}) ( 1 n ) f st (u, v). (3.1) 2 (s,t) CurrentFlow vs. SpanningTree centrality: From the combinatorial perspective, the CurrentFlow considers an edge as important if it participates in many paths, while SpanningTree deems an edge important if it participates in many spanning trees. From electrical network point of view, the SpanningTree centrality 29
41 of edge e, can be defined as follows ( notation as in Definition 6) StC(e = {u, v}) = f uv (u, v). (3.2) Thus, while CurrentFlow takes into consideration multiple placements of batteries on all pairs of nodes (u, v), SpanningTree only considers a single battery placement. CurrentFlow vs. betweenness centrality: One should think of the CfC(e) of edge e as a measure of participation of the edge in paths that go from s to t for all (s, t) pairs. The idea of counting participation of edges is also central in the definition of betweenness centrality [Bra01]. However, the betweenness centrality takes into consideration only the shortest paths between all source-destination pairs. Therefore, an edge may have low betweenness score if it does not participate in many short paths, even if it participates in a lot of short paths. More importantly, the betweenness score of edges may change by the addition of a small number of edges (e.g., edges that create triangles) [New05]. Clearly, the CurrentFlow centrality does not suffer from such unstable behavior since it takes into account the importance of the edge in all paths that connect all source-destination pairs. 3.3 Edge Importance Tool 3.4 Computing SpanningTree In this section, we present our algorithm for evaluating the SpanningTree centrality of all the edges of the input graph. For that, we first discuss existing tools and how they are currently used. Then, we show how the SDD solvers by Koutis et al. [KMP12, KMP11b] can speed up existing algorithms. Finally, we present a set of speedups that 30
42 we can apply to these tools towards an efficient and practical implementation Tools Matrix-tree theorem for SpanningTree centralities: Existing algorithms for evaluating the SpanningTree scores of edges are based on the celebrated Kirchoff s matrix-tree theorem [HHM08, Tut01]. The best known algorithm based on the matrixtree theorem has running time O(mn 3/2 ) [TMC + 13], which makes it impossible to use even for networks that consist of a few thousands of nodes and edges. Random projections for SpanningTree centralities: The equivalence between the effective resistance of edge e, denoted by R(e), and the StC(e) has the following consequence: the effective resistances of all edges {u, v} are the diagonal elements of the m m matrix R computed as [DS84]: R = BL B T, (3.3) where B is the incidence matrix and L is the pseudoinverse of the Laplacian matrix L of G. Unfortunately, this computations requires O(n 3 ) time. Equation (3.3) provides us with a useful intuition: the effective resistance of an edge e = {u, v} can be re-written as the distance between two vectors that only depend on nodes u and v. To see this consider the following notation: for node v V assume an n 1 unit vector e v such that e v has value one in its v-th position and value zero in all other positions (i.e., e v (v) = 1 and e v (v ) = 0 for v v ). Using Equation (3.3) as our guide, we can write the effective resistance R (e) between nodes u, v V as 31
43 follows: R (e) = (e u e v ) T L (e u e v ) = BL (e u e v ) 2 2. Thus, the effective resistances of edges e = {u, v} can be interpreted as the pairwise distance between vectors {BL e v } v V. This viewpoint of effective resistances as the L 2 2 distances of this vectors, allows us to use the Johnson-Lindenstraus Lemma [JL82]. Thus, these distances are preserved if we project the vectors into a lower-dimensional space spanned by O(log n) random vectors. This observation has led to Algorithm 3, which was first proposed by Spielman and Srivastava [SS11]. We refer to this algorithm with the name TreeC. Algorithm 3 The TreeC algorithm. Input: G = (V, E). Output: R(e) for every e = {u, v} E 1: Z = [ ], L = Laplacian of G 2: Construct random projection matrix Q of size k m 3: Compute Y = QB 4: for i = 1... k do 5: Approximate z i by solving: Lz i = Y (:, i) 6: Z = [Z; z T i ] 7: return R (e) = Z(:, u) Z(:, v) 2 2 In Line 2, a random ±1/ k matrix Q of size k m is created. This is the projection matrix, for which, using the Johnson-Lindenstrauss Lemma, k = O(log n). After forming the projection matrix Q, we could simply project matrix BL on the k random vectors defined by the rows of Q, i.e., we could compute QBL. However, this would not help in terms of running time since such a computation would require computing L which requires time O(n 3 ). The steps shown in Lines 3 and 5 try exactly to approximate QBL, without computing the pseudoinverse of the Laplacian. This is 32
44 achieved as follows: first, Y = QB is computed in time O(2m log n) this is because B is sparse and has only 2m non-zero entries. Then, the execution of Line 5 results in the approximation of the rows z i of matrix QBL by (approximately) solving the system Lz i = y i, where y i is the i-th row of Y. Therefore, the result of the TreeC algorithm is the set of rows of matrix Z = [z1 T,..., zk T ], which is an approximation of QBL. By the Johnson-Lindenstrauss lemma we know that, if k = O(log n), the TreeC algorithm will guarantee that the estimates R(e) of R(e) satisfy (1 ɛ)r(e) R(e) (1 + ɛ)r(e), with probability at least 1 1/n. We call ɛ, the error parameter of the algorithm. Now, if the running time required to solve the linear system in Line 5 is I(n, m), then the total running time of the TreeC algorithm is O(I(n, m) log n). Incorporating SDD solvers: Now, if we settle for approximate solutions to the linear systems solved by TreeC and we deploy the SDD solver proposed by Koutis et al. [KMP12, KMP11b], then we have that I(n, m) = Õ(m log n) and, therefore, the version of ß that deploys such solvers runs in time Õ ( m log 2 n log ( 1 ɛ )) and with probability (1 1/n) the estimates R(e) of R(e) satisfy (1 ɛ) 2 R(e) R(e) (1 + ɛ) 2 R(e). (3.4) We refer to the version of the TreeC algorithm that uses such solvers as the Fast- TreeC algorithm. The running time of Fast-TreeC increases linearly with the number of edges and logarithmically with the number of nodes. This dependency manifests itself clearly in our experiments in Section
45 3.4.2 Speedups We now describe three observations that lead to significant space and speed savings over the version of Fast-TreeC shown in Algorithm 3. Space-efficient implementation: First we observe that intermediate variables Y and Z of Algorithm 3 need not be stored in k n matrices but that vectors y and z of size 1 n are sufficient. The pseudocode that implements this observation is shown in Algorithm 4 In this case, the algorithm proceeds in k = O(log n) iterations. In each iteration a single random vector q (i.e., a row of the matrix Q used in Algorithm 3) is created and the projection of all nodes on this vector are computed. The effective resistance of edge e = {u, v} is computed additively in each iteration the portion of the effective resistance score that is due to the particular dimension is added to the total effective resistance score (Line 6 of Algorithm 4). Algorithm 4 The space-efficient version of the Fast-TreeC algorithm. Input: G = (V, E). Output: R(e) for every e = {u, v} E 1: L = Laplacian of G 2: for i = 1... k do 3: Construct a vector q of size 1 m 4: Compute y = qb 5: Approximate z by solving: Lz = y 6: return R (e) = R (e) + z(u) z(v) 2 2 Parallel implementation: Algorithm 4 reveals also that Fast-TreeC is amenable to parallelization. That is, the execution of every iteration of the for loop (Line 2 of Algorithm 4) can be done independently and in parallel in different cores and the results can be combined. This observation leads to a further running-improvement: in a parallel system with O(log n) cores, the running time of a parallel version of 34
46 the Fast-TreeC algorithm is Õ ( m log n log ( 1 ɛ )). In all our experiments we use this parallelization. Reducing the size of the input to the 2-core: As it has been observed in Section 3.2.1, the bridges of a graph participate in all the spanning trees of the graph and thus have StC score equal to 1. Although we know how to extract bridges efficiently [Tar74], assigning to those edges StC score of 1 and applying the Fast- TreeC algorithm on each disconnected component would not give us the correct result since it is not clear how to combine the StC scores computed in different connected components. However, we observe that this can be done for a subset of the bridges. Let us first provide some intuition before making a more general statement. Consider an input graph G = (V, E) and an edge e = {u, v} connecting node v of degree one to the rest of the network via node u. Clearly e participates in all spanning trees of G and therefore StC(e, G) = 1 1. Now assume that edge e and node v are removed from G, resulting into graph G = (V \ {v}, E \ {e}). Since e was connecting a node of degree 1 to the rest of the network, then the number of spanning trees in G is equal to the number of spanning trees in G. Thus, for every edge e E \ {e} it will hold that StC(e, G ) = StC(e, G). Now the key observation is that the above argument can be applied recursively. Formally, consider the input graph G = (V, E) and let C 2 (G) = (V, E ) be the 2-core of G, i.e., the subgraph of G that has the following property: the degree of every node v V in C 2 (G) is at least 2. Then, we have the following observation: Lemma 2. If G = (V, E) is a connected graph and C 2 (G) = (V, E ) is its 2-core, 1 For this discussion we use two arguments for StC in order to specify the graph on which the StC score of an edge is computed. 35
47 then for every edge e E StC(e, C 2 (G)) = StC(e, G). The above observation suggests the following speedup for Fast-TreeC: given graph G = (V, E), first extract the 2-core C 2 (G) = (V, E ). Then for every edge e E compute StC(e) using the Fast-TreeC algorithm with input C 2 (G). For every e E \ E, set StC(e) = 1. The computational savings of such scheme depend on the time required to extract C 2 (G) from G. At a high level this can be done by recursively removing from G nodes with degree 1 and their incident edges. This algorithm, which we call Extract2Core, runs in time O(m) [BZ03]. Our experiments (Section 3.5) indicate that extracting C 2 (G) and applying Fast-TreeC on this subgraph is more efficient than applying Fast-TreeC on the original graph, i.e., the time required for running Extract2Core is much less than the speedup achieved by reducing the input size. By default, we use this speedup in all our experiments. 3.5 Experiments In this section, we experimentally evaluate our method for computing the SpanningTree centrality and we study their effect on information propagation, the size and the number of clusters produced when removing or adding edges ranked by the electrical measure. For the evaluation, we use a large collection of datasets of different sizes, which come from a diverse set of domains. Experimental setup: We implemented Fast-TreeC using a combination of Matlab and C code. The CMG solver[kmt11] is written mostly in C and can be invoked 36
48 Table 3.1: Statistics of the collection of datasets used in our experiments. Dataset #Nodes in #Edges in #Nodes in #Edges in name LCC LCC the 2-Core the 2-Core GrQc p2p-gnutella Oregon HepTh wiki-vote p2p-gnutella Epinions Slashdot Amazon DBLP roadnet-tx Youtube as-skitter Patents from Matlab. We ran all our experiments on a machine with 4 Intel Xeon 2.9GHz, with 512GB of memory. We need to note here that none of our algorithms pushed the memory of the machine near its limit. Finally, we used 12 hardware threads to take advantage of the parallelizability of both Fast-TreeC and Fast-FlowC. Datasets: In order to demonstrate the applicability of our algorithm on different types of data, we used a large collection of real-world datasets of varying sizes, coming from different application domains. Table 3.1 provides a comprehensive description of these datasets shown in increasing size (in terms of the number of edges). The smallest dataset consists of approximately nodes, while the largest one has almost nodes. For each dataset, the first two columns of Table 3.1 report the number of nodes and the number of edges in the Largest Connected Component (LCC) of the graph that correspond to this dataset. The third and fourth columns report the number of nodes and edges in the 2-core of each dataset. The 2-core of a graph is extracted using the 37
49 algorithm of Batagelj et al. [BZ03]. The statistics of these last two columns will be revisited when we explore the significance of applying Extract2Core in the running time of Fast-TreeC. In addition to their varying sizes, the datasets also come from a wide set of application domains, including collaboration networks (HepTh, GrQc, DBLP and Patents), social networks (wiki-vote, Slashdot, Epinions, Orkut and Youtube), communication networks, (p2p-gnutella08, p2p-gnutella31, Oregon-1 and as-skitter) and road networks (roadnet-tx). All the above datasets are publicly available through the Stanford Large Network Dataset Collection (SNAP). 2 For consistency, we maintain the names of the datasets from the original SNAP website. Since our methods only apply to undirected graphs, if the original graphs were directed or had self-loops, we ignored the directions of the edges as well as the self loops Experiments for SpanningTree Accuracy-efficiency tradeoff: Our first experiment aims to convey the practical semantics of the accuracy-efficiency tradeoff offered by the Fast-TreeC algorithm. For this, we recorded the running time of the Fast-TreeC algorithm for different values of the error parameter ɛ (see Equation (3.4)) and for different datasets. Note that the running times reported for this experiment are obtained after applying all the three speedups that we discuss in Section The results are shown in Figure 3.2; for better readability the figure shows the results we obtained only for a subset of our datasets (from different applications and with different sizes); the trends in other datasets are very similar. As expected, the running time of Fast-TreeC decreases as the value of ɛ, the error parameter, increases. Given
50 Runtime 8 hours 4 hours 2 hours 1 hour 30 mins 15 mins 5 mins p2p-gnutella31 Epinions Slashdot DBLP Youtube roadnet-tx 1 min as-skitter Error parameter ǫ Figure 3.2: Accuracy-efficiency tradeoff; y-axis (logarithmic scale): running time of the Fast-TreeC algorithm; x-axis: error parameter ɛ. 39
51 that the y-axis in Figure 3.2 is logarithmic, this decrease in the running time is as expected exponential. Even for our largest datasets (e.g., as-skitter and roadnet- TX), the running time of Fast-TreeC even for very small values of ɛ (e.g., ɛ = 0.05) was never more than 8 hours. Also, for ɛ = 0.15, which is a very reasonable accuracy requirement, Fast-TreeC calculates the StC scores of all the edges in the graphs in less than 1 hour. Also, despite the fact that the roadnet-tx and as-skitter datasets have almost the same number of nodes, as-skitter runs significantly faster than roadnet-tx for the same value of ɛ. This is due to the fact that as-skitter has approximately 5 times more edges than the corresponding graph of roadnet-tx and that the running time of Fast-TreeC is linear to the number of edges yet logarithmic to the number of nodes of the input graph. Effect of the 2-core speedup: Here, we explore the impact of reducing the size of the input to the 2-core of the original graph on the running time of Fast-TreeC. For this, we fix the value of the error parameter ɛ = 0.1, and run the Fast-TreeC algorithm twice; once using as input the original graph G and then using as input the 2-core of the same graph, denoted by C 2 (G). Then, we report the running times of both these executions. We separately also compute the time required to extract C 2 (G) from G using the Extract2Core routine described in Section Figure 3.3 shows the times required for all the above operations. In the figure, we use Fast-TreeC(G) (resp. Fast-TreeC(C 2 (G))) to denote the running time of Fast- TreeC on input G (resp. C 2 (G)). We also use Extract2Core to denote the running time of Extract2Core for the corresponding input. For each of these datasets, we computed the StC scores of the edges, before(left) and after(right), and we report the running time in the y-axis using logarithmic scale. 40
52 Runtime 16 hours 8 hours 4 hours 2 hours 1 hour 30 mins 15 mins 5 mins 1 min Fast-TreeC(G) Extract2Core Fast-TreeC(C 2 (G)) GrQc HepTh p2p-gnutella08 Epinions p2p-gnutella31 wiki-vote Youtube roadnet-tx Slashdot Patents as-skitter Figure 3.3: Limiting the computation on the 2-core shows a measurable improvement in the running time of Fast-TreeC. Note that on top of the box that represents the runtime of Fast-TreeC(C 2 (G)), we have also stacked a box with size relative to the time it took us to find that 2-core subgraph. It is hard to discern this box, because the time spent for Extract2Core is minimal compared to the time it took to compute the StC scores. Only in the case of smaller graphs is this box visible, but again, in these cases, the total runtime does not exceed a minute. For instance, for Patents (our largest graph) spending less than 5 minutes to find the 2-core of the graph lowered the runtime of Fast-TreeC down to less than 8 hours, which is less than half of the original. Moreover, the difference between the height of the left and the right bar for the different datasets behaves similarly. Hence, as the size of the dataset and the runtime of Fast-TreeC grow exponentially, so does the speedup. 41
53 k 100 k k k (a) SpanningTree k k (b) CurrentFlow k (c) betweenness k (d) random Figure 3.4: The values k as a function of k for the different importance measures Edge-importance measures and information propagation A natural question to consider is the following: what do all the different edgeimportance measures capture and how do they relate to each other? Here, we describe an experiment that allows us to quantitatively answer this question. On a high level, on Experiments 1, 2 and 3 we do so by investigating how edges ranked as important (or less important) affect the result of an information-propagation process in the network. Moreover, in the experiments we verify if the edges ranked as important work as bridges between clusters. To evaluate this we compute the number and the size of the clusters produced by removing those edges. Methodology: Experiment 1. Given a network G = (V, E), we compute the spread of an information propagation process, by picking 5% of the nodes of the graph, running the popular independent cascade model [KKT03] using these nodes as seeds and comput- 42
54 ing the expected number of infected nodes in the end of this process. By repeating the experiment 20 times and taking the average we compute the Spread(G). For any edge-importance measure, we compute the scores of all edges according to this measure, then edges are ranked in decreasing order using the given score and finally a set of l edges is chosen, where l = 0.02 E, such that they are at positions from ((k 1)l + 1) to kl, for k = 1,..., E /l. We refer to the set of edges picked for any k as E k. For every k, we then remove the edges in E k, forming graph G k, and then compute Spread(G k ). In order to quantify the influence that the set E k has on the information-propagation process we compute: k = Spread(G) Spread(G k ). Clearly, k 0 and the larger the values of k the larger the effect of the removed edges E k on the propagation process. We experiment with the following four measures of importance: SpanningTree, CurrentFlow, betweenness and random. Recall that the betweenness score of an edge is the fraction of all pairs shortest paths that go through this edge [Fre77]. random simply assigns a random order on the edges in E. Experiment 2. (Deterministic) Part A: Given a network G = (V, E), we compute the spread of an information propagation process, by removing the top x% of the edges in the graph G to produce the graph Ĝ, running the popular independent cascade model [KKT03] on Ĝ to compute the number of infected nodes. By repeating the experiment 20 times we compute the expected number of infected nodes. Part B: Starting from an empty graph we compute the number of clusters generated when adding the top x% ranked edges. 43
55 ca GrQc (deterministic) 3000 Number of WCC (mean) type cb CFC STB % of edges in the graph (a) GrQc ca HepTh (deterministic) 6000 Number of WCC (mean) type cb CFC STB % of edges in the graph (b) HepTh p2p Gnutella08 (deterministic) 4000 Number of WCC (mean) type cb CFC STB % of edges in the graph (c) p2p-gnutella08 Figure 3.5: The number of components as a function of x. 44
56 ca GrQc (deterministic) Number of Infected Nodes (mean) type cb CFC STB % of edges removed from the graph (a) GrQc ca HepTh (deterministic) 7000 Number of Infected Nodes (mean) type cb CFC STB % of edges removed from the graph (b) HepTh p2p_gnutella08 (deterministic) Number of Infected Nodes (mean) type cb CFC STB % of edges removed from the graph (c) p2p-gnutella08 Figure 3.6: The number of infected nodes as a function of x. 45
57 Steps: For every edge-importance measure, we compute the scores of all edges according to this measure, we rank the edges in decreasing order of this score and then remove a x% of the edges choosing from the top edges. We increase x by 2% and 0% x 70%. We experiment with the following four measures of importance: SpanningTree, CurrentFlow and betweenness. We evaluate the rate of information propagation lost. Experiment 3. (Probabilistic) Part A: Let M(0, 1) be a probability distribution proportional to an edge-important measure. Then given a network G = (V, E), we compute the spread of an information propagation process, by removing x% M(0, 1) of the nodes in the graph G to produce Ĝ, running 20 trials of the popular independent cascade model [KKT03] on the graph Ĝ we compute the expected number of infected nodes. Part B: Starting from an empty graph we compute the number of clusters generated when adding the top x% ranked edges. Steps: For any edge-importance measures, we compute the scores of all edges according to this measure, rank the edges in decreasing order of this score and then pick a set of with x% M(0, 1). We increase x by 2% and 0% x 70%. We experiment with the following four measures of importance: SpanningTree, CurrentFlow and betweenness. We evaluate the rate of information propagation lost. 46
58 ca GrQc (probabilistic) 3000 Number of WCC (mean) type cb CFC STB % of edges in the graph (a) GrQc ca HepTh (probabilistic) 6000 Number of WCC (mean) type cb CFC STB % of edges in the graph (b) HepTh p2p Gnutella08 (probabilistic) 4000 Number of WCC (mean) type cb CFC STB % of edges in the graph (c) p2p-gnutella08 Figure 3.7: The number of components as a function of x. 47
59 3500 ca GrQc (probabilistic) Number of Infected Nodes (mean) type cb CFC STB % of edges removed from the graph (a) GrQc ca HepTh (probabilistic) 7000 Number of Infected Nodes (mean) type cb CFC STB % of edges removed from the graph (b) HepTh p2p Gnutella08 (probabilistic) 4500 Number of Infected Nodes (mean) type cb CFC STB % of edges removed from the graph (c) p2p-gnutella08 Figure 3.8: The number of infected nodes as a function of x. 48
60 3.6 Results Results: Figure 3.4 shows the values of k in the case of the HepTh network, for k = 1,..., 49 when sets E k are determined by the different importance measures. Overall we observe that the trend of k varies across measurements. More specifically, in the case of SpanningTree centrality (Figure 3.4a), k takes larger values for small k and appears to drop consistently until k = 30. This behavior can be explained by the definition of the SpanningTree centrality an edge is important if it is part of many spanning trees in the network and the fact that the propagation of information in a graph can be represented as a spanning tree. CurrentFlow and betweenness (Figures 3.4b and 3.4c) behave differently. They appear to give medium importance scores to edges that have high impact on the spread. For CfC, these are the edges in E k for k [38, 43] and, for betweenness, the edges for k [13, 18]. These edges correspond to the peaks we see in Figures 3.4b and 3.4c. Observe that, for betweenness, this peak appears for smaller values of k, indicating that in this particular graph, there are edges that participate in relatively many shortest paths and, once removed, they disconnect the network, hindering the propagation process. Finally, the results for random show no specific pattern, indicating that what we observed in Figures 3.4a 3.4c is statistically significant. On Experiment 2 the loss of information propagation corresponding to the edges ranked using the SpanningTree centrality occurs at a higher rate compared with the other measures see Figure 3.6, a similar behavior occurs on Experiment 3 see Figure 3.8 except for the data set GrQc on Figure 3.8a where after 42% the other measures show higher rate. However this shows how the SpanningTree centrality capture more influent edges. Moreover when computing the number of connected components the SpanningTree centrality connects the graph faster than the other measurements. This confirm again the performance of the SpanningTree to chose important edges on the network see 49
61 Figure 3.5 and
62 Chapter 4 Spectral and Electrical Clustering 4.1 Introduction Studies of social networks have been an important topic in sociology at least since the 1930 s [Sco91]. More recently, Internet s growth induced the creation of virtual groups, that live on the WEB as on-line social communities, further increasing efforts on social network studies. We are all empirically aware of the fact that social networks are organized in communities, e.g. groups of friends, professional communities, interest groups, or groups of people that are related in space or time. Furthermore, communities often exhibit a hierarchical organization: a larger community includes several smaller ones. Detecting communities in a large network is a problem with significant applications. For example, detecting a group of patients with similar biological features may enable targeted drug discovery or delivery. Or, companies on purchase relationship networks can find communities of customers with similar product interests and demographical features. By finding this communities companies can effectively recommend products to its customers [RKSR02]. The computational detection of communities requires two general steps. The first step is a concrete definition or quantification of the community notion. For example, a community can be defined as a cluster of nodes with a high density of edges inside the cluster, and a relatively lower number of edges leaving the cluster. This is 51
63 illustrated in Figure 4.1. This type of clustering will be our main focus in the rest of this Section. The second step is the actual computation of clusters that are good in terms of the underlying definition, a task that can be very demanding given the sheer size of modern networks. Figure 4.1: Graph with communities. Each community is circled by a line.. There is a vast literature on both general steps outlined in the previous paragraph, and surveying them is out the scope of this work. The purpose of this Section is to contribute and experiment with computational methods for clustering that can benefit from the availability of very fast linear system solvers [KMP11a], and in particular our work on computing effective resistances. Clustering Challenges. Computing clusters is an NP-hard problem with respect to most natural clustering optimization criteria. So, for even small networks the use of exact clustering algorithms is pointless. In practice the huge size of networks renders impractical the use of any algorithm that is not designed to run in nearly-linear time. The common approach is to use algorithms that are faster, in exchange for some error in the solution, which is not approximate. In general approximation algorithms are non-deterministic, their output can slightly vary between trials with the same input parameters, and they may have a small probability of failure, in which case they can be 52
64 repeated until they succeed. Heuristic algorithms are also approximation algorithms, but they don t strictly bound the probability of failure and so they are not guaranteed to eventually succeed. In what follows we will include and study both approximation algorithms and heuristics. A constraint of many algorithms is that they require the reciprocal interaction of the node in the graph, in which case the graph is said to be undirected. In this work we concentrate on undirected graphs. However the relationship between the nodes in the network can be non reciprocal. As an example consider the social network Twitter where the user u 1 can follow user u 2, but this doesn t implies that user u 2 follows user u 1, as in the case for the social network Facebook. Graphs with non reciprocal relationships are said to be directed. Another classic directed graph is the WWW where hyper-links represent the relation between the websites, from which only 10% are reciprocal [AJB99]. The analysis of asymmetric system tends to be more complex and finding communities on directed graph is a difficult task. A common solution is to relax the graph and assume a reciprocal relationship. So, even though some researchers have shown that this is not always a good practice [RB08], studying the undirected case bears applications to the directed case as well. Yet another difficulty is when nodes can be part of more that one community. Nodes on overlapping communities can be part of multiple clusters complicating the process of clustering. For this work we restrict to networks with reciprocal interactions and non overlapping communities. 4.2 Background The goal of this section is to provide some basic definitions about graphs and theirs encoding data structures. In addition we review two metrics used to evaluate the 53
65 quality of communities. A graph is formally defined as a triple G = (V, E, w) where: (i) V is the set of vertices (also called nodes), (ii) E is a set of edges, with each edge being an unordered pair of nodes, (iii) w is a weight function that maps each edge in E to a real value. Consider the Facebook friendship relationship system where nodes are the users, the friendship relationship between two users is represented as an edge between the two users as shown in Figure 4.2. Figure 4.2: A social network graph where dots nodes are user and users that are friends are connected by a line edges. We will assign to each node a unique integer in [1, n], where n is the number of nodes in the graph. We will do the same for edges, and denote by m the number of edges in the graph. Graph Encoding: A useful graph encoding is the well know adjacency matrix where adjacent nodes have value equal one and zero otherwise. Definition 7. The adjacency matrix A of a graph G = (V, E, w) is defined as: w i,j if (i, j) E A i,j = 0 otherwise 54
66 L G = C A Figure 4.3: An example graph G = (V, E, w) The core of our work is based on the graph Laplacian matrix. Definition 8. The Laplacian matrix L of a graph G = (V, E, w) is defined as: L i,j = w i,j if (i, j) E i j w i,j if i = j 0 otherwise An example of a graph and its Laplacian is given in Figure 4.3. Definition 9. For an arbitrary orientation of the graph G the signed edge-vertex incidence matrix B with size m n is defined as: Let e be an edge, 1 k m and 1 h n then B k,h = 1 if e k = (i, j) and h = i for 1 k m 1 if e k = (i, j) and h = j for 1 k m 0 otherwise Let W be the m m diagonal matrix containing the weights of the edges. The Laplacian and edge-incident matrices are connected through the following identity: L = B T W B (4.1) 55
67 Furthermore the Laplacian matrix is related to the adjacency matrix through the following identity L = D A, where D is a matrix with diagonal equals to the nodes degree for weighted graph use the nodes volume instead. Definition 10. Let d i = j V \i w i,j be the degree of the node i and let D 1 2 be the diagonal matrix with entries D 1 2 (i, i) = 1 di, then the normalized Laplacian matrix ˆL of a graph G = (V, E, w) is defined as: ˆL = D 1 2 LD 1 2 (4.2) Graph Clustering: We will find it useful to define graph clustering as the procedure of finding sets V 1, V 2,... V k such that V = k i=1v i where the pairwise intersection between any two clusters is empty, V i V j =. Ideally we want to find communities with good quality. To measure the quality of a cluster many measures have been introduced [Ste04, TSK05, Jac12, Mog10]. In this work, we focus on the edgeexpansion and cluster-conductance. Before we define them, we introduce some intermediate quantities. Graph-cut is defined as: cut(v a, V b ) = i V a,j V b w i,j and the cluster-association: assoc(v a, V ) = i V w a,j V i,j. Definition 11. Let V a V then: edge expansion(v a ) = cut(v a, V \ V a ) assoc(v a, V a ). (4.3) Definition 12. The conductance of a graph G = (V, E, w) is defined as follows. φ(g) = min V a V φ(v a) 56
68 φ(v a ) = cut(v a, V a \ V ) min(assoc(v a, V \ V a ), assoc(v \ V a, V a )) Definition 13. Let V a V and G a be the subgraph of G induced by the nodes in V a. The cluster-conductance of the cluster V a is defined as φ(g a ). Our implementation is a combination of heuristic implementation of the k-means algorithm also know as Hartigan and Wong algorithm [HW79] and spectral or electrical embeddings [NJW01, KC12b]. After computing the clusters we will measure the quality of the clusters using the ratio: R a = edge expansion(v a) φ(g a ) V a (4.4) Intuitively, this measure favors clusters that are well connected in their interior, and therefore have a high cluster-conductance, and have a small connection to the exterior. So, the quality of the cluster V a is inversely proportional to R a. Furthermore cluster conductance(v a ) can be approximate through the second smallest eigenvalue of the normalize Laplacian for the graph induced by V a. 4.3 Algorithms Graph Embeddings Certain graph clustering methods in general follows two steps: (i) form an embedding of the graph into a geometric space, i.e. a one-to-one map between vertices and points in the space and (ii) apply the k-means algorithm. Here we are going to focus on the second step and how two different embeddings affect the quality of the clusters. Spectral Embedding: Let ν i be the eigenvector that corresponds to the eigenvalue 57
69 λ i of the normalized Laplacian matrix. Node i is embedded to the i-th row of: Π = ν 2... ν k+1 (4.5) where k is the number of cluster wanted. This embedding was first introduced by Ng et al. [NJW01]. The recent implementation of fast SDD linear solver [KMT11], can be combined to find such embedding in time nearly proportional to the number of edges in the graph O(m log m). Effective Resistance Embedding: In Chapter 2, we saw that we can efficiently compute all effective resistances of a graph. The algorithm has two steps: (i) the vertices of the graph are embedded to points (vectors) in the Euclidean space of dimension O(log n). (ii) the effective resistance of a an edge (i, j) is approximated by the squared distance between the corresponding points for i and j. In this chapter we will be using the space embedding proposed by Spielman and Srivastava [SS08]. Naturally, we can associate a n O(log n) matrix Z with this embedding. The use of this embedding for clustering has recently been explored by Khoa and Chawla [KC12b] Implementation In this section we expose two clustering algorithms: Algorithm 5: k-means spectral clustering (Sc) introduced by Ng et al. [NJW01]. In the first step, the graph is encoded using the normalized Laplacian matrix, the spectral embedding is constructed in steps two and three. In step four the k-means algorithm is used with the spectral embedding to find the communities. The second algorithm: Algorithm 6 k-means effective resistance clustering (ErC) 58
70 Algorithm 5 k-means Spectral Clustering. Input: G = (V, E). Output: l i for every v i V 1: Encode the graph in the normalized Laplacian L 2: Find the k eigenvectors ν 2,..., ν k+1 of L 3: Construct Π 4: Compute l = kmeans(π) 5: return l introduced by Khoa and Chawla [KC12b]. Algorithm 6 k-means Effective Resistance Clustering. Input: G = (V, E). Output: l i for every v i V 1: Construct the embedding matrix Z 2: Compute l = kmeans(z) 3: return l The graphs is encoded using the Laplacian matrix in step one, the electrical embedding is constructed in step two, and in step four the k-means algorithm is used with the electrical embedding to find the communities. 4.4 Experiments and Results We have implemented Algorithm 5 and Algorithm 6 using the CMGSolve [KMT11] to approximate the eigenvectors and the effective resistances embedding. The code for both implementations can be found at: To test our implementations and compare their performance we use data that comes from the Stanford Network Analysis Project (SNAP). We run Algorithms 5 and 6 for k equal to 2, 3, 6, 12, 18, 24. For smaller communities i.e. k = {2, 3, 6} the spectral embedding perform better than the electrical embedding see Figure 4.5. However the electrical embedding perform better using more communities i.e. k = {12, 18, 24}. Moreover on Figure 4.5a the 0.8-quantile of the list R = {R 1,... R k } on the different commu- 59
71 R R E12 E18 E2 E3 E6 embedding (a) Community analysis with Algorithm 6, and k values 2, 3, 6, 12, S12 S18 S2 S3 S6 embedding (b) Community analysis with Algorithm 5, and k values 2, 3, 6, 12, 18. Figure 4.4: Summary of the ratio R on the communities obtained by Algorithm 6 and 5. nities are similar between the spectral and electrical embeddings, except for k = 3. Compared with Figure 4.5b where the performance difference between embeddings is higher. To visualize the communities we plot the graph using the Force Atlas 2 layout from Gephi and overlap labels produced by the Algorithms 5 and 6 respectively on Figure 4.6b and Figure 4.6a. We also analyze the community structure and interaction on Figure 4.7 where the communities produced using the electrical embedding to communicate with more communities around the network see Figure 4.7a when compared with the spectral embedding see Figure 4.7b. Even thought the number of edges between the communities is smaller using the electrical embedding, the volume of edges between communities is less compare to the spectral embedding. 60
72 quantile of R E2 E3 E6 S2 S3 S6 R E12 E18 E24 S12 S18 S E2 E3 E6 S2 S3 S6 (a) The 0.8-quantile of R in the communities obtained with k values equal to: 2, 3, 6. E12 E18 E24 S12 S18 S24 (b) The 0.8-quantile of R in the communities obtained with k values equal to: 12, 18, 24. Figure 4.5: Analysis of 80% best clusters (a) Community vizulization: Algorithm 6, k = 6. (b) Community vizulization: Algorithm 5, k = 6. Figure 4.6: Visualization for k = 6: Each node represents a community, the ratio of the node is proportional to the number of individuals in the community and the edge weight is the number of connections between the communities. 61
73 (a) Community visualization: Algorithm 6, k = 12. (b) Community visualization: Algorithm 5, k = 12. Figure 4.7: Visualization for k = 12: Each node represents a community, the ratio of the node is proportional to the number of individuals in the community and the edge weight is the number of connections between the communities. (a) Community visualization: Algorithm 6, k = 12. (b) Community visualization: Algorithm 5, k = 12. Figure 4.8: Visualization of twelve communities using the Force Atlas 2 layout from Gephi. 62
74 (a) Community visualization: Algorithm 6, k = 18 (b) Community visualization: Algorithm 5, k = 18 Figure 4.9: Visualization of eighteen communities using the Force Atlas 2 layout from Gephi 63
75 Chapter 5 Random Walks: For Image Segmentation 5.1 Introduction Solutions for automatic image segmentation problems have been a popular application of spectral graph theory, following the work by Shi and Malik [SM00], where the use of the normalized cut measure was proposed to segment images, instead of the well known minimum cut on graphs. Mathematically the algorithm considers a relaxed version of the binary graph cut problem, using the solution of the generalized eigenvalue problem. The goal of finding the normalized cut, is to avoid the segmentation of irrelevant small partitions that may be obtained when computing the minimum cut. Two solutions for k-way partitioning where proposed by Shi and Malik [SM00]. The first and more intuitive is the recursive use of the two-way partitioning method. The second is the use of multiple (k) eigenvectors. The k-way partitioning was further explored by Ng et al. [NJW01, CH53] and Yu and Shi [YS03]. The last work motivated the contribution proposed by Tolliver and Miller [TM06] where a spectral method was iteratively used to adjust the edges of the graph until the graph gets disconnected. It is not surprising that even the best automatic image segmentation algorithms are far from perfect. An approach that can help obtain improved results is semi-automated or guided image segmentation. An image segmentation algorithm is consider guided or semi-automated when information provided by the user or previous segmentation 64
76 process (i.e. an automatic image segmentation algorithm) is incorporated in the algorithm. A case of guided image segmentation algorithms is the family of active contours and levels sets [MS96, KWT88], where the user inputs a contour within the desired boundary, and an optimization algorithm runs locally on the user input to get the boundary. Another member of the guided image segmentation algorithms is the intelligent scissor algorithm [MB98]. The core of the algorithm is Dijkstra s shortest path algorithm. The user inputs points along the boundary and the algorithm finds the shortest path from point to point until the boundary is traced. The quality of the segmentation is proportional to the number of points the user assign along the boundary. Similar to automated image segmentation approaches, guided algorithms have been formulated as graph cuts problems [BJ01]. The intuition in this algorithm is to find the min-cut/maxflow between the sink and source vertex given by the user. Indeed this algorithm is easy to implement and generalizes in three-dimensions. A novel guided algorithm is the random walker proposed by Grady [Gra06]. The random walker algorithm takes its name by its implicity use of random walks in deciding the cluster or segment of each pixel. More concretely, the input of the user is a labeling of some pixels, where each such pixel receives one of k possible levels, indicating which segment they are part of. For each unlabeled pixel, one can consider a random walk starting at that pixel and propagating in the graph until it first reaches a labeled pixel. The algorithm computes the probability that the random walk will end to each given type of label, and it assigns to the pixel the label which has the highest such probability. Prior to Grady the first applications of random walks in vision was the work by Wechsler and Kidode [WK79], based on texture discrimination. Another use of random walks is in the isoperimetrical graph partitioning algorithm, that can be view as a computation of hitting times of random walks [GS06]. Also the 65
77 use hitting time of random walks was used for object characterization algorithms by Golerick et al. [GGS + 04]. Other related applications for clustering were proposed by Yen et al. [YVW + 05] where the use of commute time was used as a distance in the k-mean algorithm. Here we propose a variant of the random walker where pixels are labeled iteratively, as some unlabeled pixels remain unlabeled during the iterations. This framework is combined with the recent nearly-linear time solvers of Koutis et al. [KMP10, KMT11], and a nearly-linear time implementation is presented here. 5.2 Background In general a random walk consists of a list obtained by traversing the graph, randomly. Assuming that at step t of the walk is at vertex u, the next vertex (at step t + 1) is selected by randomly picking an edge adjacent to u and following it. The probability that an edge is traversed is proportional to its weight. Definition 14. Random Walk: Let P be a transition matrix with entries P i,j f being the probabilities of going from vertex i to vertex j. Also let s 0 be a zero vector except a one at the starting vertex. The probability that a random walker is at vertex j after t steps, is given by the j entry in the vector s t where: s t = s t 1 P (5.1) The image segmentation problem we consider here can be defined as assigning the same label to the pixels that are within the same region in a given image. After encoding the image in an affinity graph we can formulate the problem as a graph partitioning problem. To form the affinity graph, pixels are taken to be vertices and 66
78 each edge represents the similarity between the pixel and its adjacent pixels 1. Affinity graphs are formally defined as a triple G = (V, E, w) in where V and E are the usual vertex and edges sets and w : E R is an affinity function that maps edges to real numbers. There exist many affinity functions. Here we are going to focus in two type of affinity functions called the exponential difference and the exponential minimum see Definitions 15 and 16. Our affinity function definitions are expressed for gray scale images. In the work by Shi and Malik [SM00] the exponential difference affinity function is presented for images in general including color and textured images. Definition 15. Let i(v) be the color intensity of the vertex v. The exponential difference affinity function w dif is defined as: w dif : E R ( ) 1 w dif (s, t) = exp [i(s) i(t)] 2 Definition 16. Let i(v) be the color intensity of the vertex v. The exponential minimum affinity function w min is defined as: w min : E R ( ) 1 w min (s, t) = exp min(i(s), i(t)) We will find it useful assign to each vertex a unique integer in [1, n], where n is the number of vertices in the graph. We will do the same for edges, and denote by m the number of edges in the graph, and a common graph encoding is the Laplacian matrix. 1 For this work pixels are consider adjacent if the Mahnattan distance between the two pixels is one. Other studies suggest as adjacency a vertex with distance at most two but those implementations are left as future work 67
79 Definition 17. The Laplacian matrix L of a graph G = (V, E, w) is defined as: L i,j = w i,j if (i, j) E i j w i,j if i = j 0 otherwise Definition 18. For an arbitrary orientation of the graph G the signed edge-vertex incidence matrix B with size m n is defined as: Let e be an edge 1 k m and 1 h n then: B k,h = 1 if e k = (i, j) and h = i for 1 k m 1 if e k = (i, j) and h = j for 1 k m 0 otherwise Let W be the m m diagonal matrix containing the weights of the edges. The Laplacian and edge-incident matrices are connected through the following identity: L = B T W B (5.2) Random Walker In the class on guided image segmentation algorithms is the Random Walker by Grady [Gra06] that classify the pixels to regions in the image. The user input some prelabeled pixel, that are classified with two or more labels. Therefore the Random Walker algorithm classify all the unlabeled pixel u on the graph by starting a random walk at u and finding the probability of passing through a pre-label pixel l before 68
80 passing through u. The combinatorial formulation is as follows: D[x] = 1 2 (Bx)T W (Bx) = 1 2 xt Lx = 1 w i,j (x i x j ) 2 (5.3) 2 e i,j E Laplacian matrices are semi-definite, as a result the critical points in D[x] are minimal. Let V = V l V u for V l V u = where V l contains the vertex with labels assign by the user and V u contains the unlabeled vertex. Assuming an order with labeled vertex first we can decompose Equation 5.3 as follows: D[x u ] = 1 2 [xt l x T u ] L l U T U L u x l x u = 1 2 (xt l L l x l + 2x T u U T x l + x t ul u x u ) (5.4) where x l correspond to the potentials (probabilities) of the labeled vertex and x u the potential of the unlabeled vertex. The system in Equation 5.5 with V u unknowns is obtained after differentiating D[x u ] respect x u and finding its minimal points. L u x u = U T x l (5.5) Considering that our graph is connected, the Equation 5.5 will be nonsingular [Big93]. Let x l i be the potentials assumed at vertex v i for each label l. Then define the vector m l : V l 1 with entries equal to 1 if the label of v i is equal to l m l i = 0 otherwise (5.6) 69
81 for every vertex v i in V l. Consequently the solution of the combinatorial Dirichlet problem for the label l is given by solving the system: L u x l = U T m l (5.7) For more than one label let M, be the matrix with columns equal to m l for each label and X with columns taken to be each x l, then solve the following system: L u X = U T M (5.8) Solving for x in either case, find the potentials (probability) for all the unlabeled vertices. In In In Out In Out Out In Out Out In Figure 5.1: The pixels at column one and row three are labeled as out pixel and the pixel at column two and row two is labeled as in. The other pixels remain unlabeled. Example 2. Lets consider the Laplacian matrix L I for the image in Figure 5.1 with pixels values zero and one, represented as black and white respectively. 70
82 2 L I = Assuming a column mayor 2 indexing, in where the pixels in 3the first column correspond to the indexes: 1, 2, 13, second 0 3 column 0 1to: 4, 0 5, 60and the third column: L u = , 8, 9. Solving the Equation we0can find 1 0the1.37 probabilities of 7 the unlabeled pixels V u = {1, 2, 4, 6, 7, 8, 9} been label as out or been label as in. Moreover L u, U, m in and m out have the following values: U = L I = m in = 0 m out = L u = Therefore by solving for x in the following systems 1 L u x out = U T m out and L u x in = U T m in the results are: x in = [ ] T 71
83 x out = [ ] T Final labeling are visualized in Figure 5.2. In In In Out In Out Out In Out Figure 5.2: Solution The steps used in the Example 2 follows from the Random Walker Algorithm 7. The core of the algorithm is the solution of Equation 5.7, notice that this linear system Out In is a SDD system therefore the recent nearly-linear time algorithm [KMP10] to solve this system can be used. Algorithm 7 Random Walker RW Input: G = (V l V u, E, w) and it Laplacian matrix L Output: the label of every pixel. 1: Compute L u and U. 2: For every label l compute m l. 3: For every label l solve L u x l = U T m l. 4: Report for each vertex (row) the maximum entry on X = (x 1 x 2... x K ), where K is the number of labels. In Step 1 computing the sub matrix L u comes from taking the columns and rows corresponding to the vertex in V u and computing the sub matrix U comes from taking the row and columns corresponding to the vertex on V l and V u respectively. Step 2 comes from Equation 5.6. In Steps 3 the CMGSolve [KMP11a] can be use to solve 72
84 the linear system and find the probabilities in almost linear time. Assigning the label with the highest probability to unlabeled pixel occurs on Step Iterative Random Walk Some guided algorithm for image segmentation can be constrained by the quality of the user input. There results are usually reflected in the quantity and quality of the user input. This was the case for the intelligent scissor algorithm, where the quality of the segmentation was proportional with the number of points inputs across the boundary. Similarly happens to the RW, where the result improves proportional to the number of pixels that are labeled by the user, for convenience also known as pre-labeled pixels or seeds. Understanding this limitation motivates the following algorithm that automatically increase the number of pre-labeled pixels and iteratively repeat the RW algorithm. Algorithm 8 Iterative Random Walker irandom-walker Input: G = (V l V u, E, w), its Laplacian matrix L and t Output: the label of every pixel. 1: Repeat t times; 2: Compute L u and U; 3: For every label l compute m l ; 4: For every label l solve L u x l = U T m l ; 5: Update V u and V l ; 6: Report for each vertex (row) the maximum entry on X = (x 1 x 2... x K ), where K is the number of labels; The steps from Step 2 to Step 4 come directly from the RW algorithm. The key step is Step 5 where sets V u and V l get updated, where the following criterion is used. Update Criterion A: To increase the number of pre-labeled pixels we will assign high probability labels to the vertex at each iteration. In details for each label k 73
85 repeat: pick q such (1 1 ) < q < 1, therefore a unlabel vertex v K i ˆV q k and has label k if x k (i) > q. Then update V l = V l ˆV k q. Corollary 1. Choosing q such (1 1 ) < q < 1 where K 2 is the number of unique K labels, guaranty a unique label to each pre-label vertex v i. Proof: Let K 2 and l be the set of unique labels. For an arbitrary vertex v i the h l xh (i) = 1 by probability. Then if the vertex v i has label k is because x k (i) q, thus the h l\k xh (i) (1 q) by probability, as consequent the vertex v i get assigned only the label. Update Criterion B: To increase the number of pre-labeled pixels we will assign high probability labels to the vertex at each iteration. In details for each label k repeat: pick q such (1 1 K ) < q < 1, therefore a unlabel vertex v i ˆV k q and has label k if x k (i) > q quantile of x k. Then update V l = V l ˆV k q. Corollary 2. Choosing q-quantile such that (1 1 ) < q < 1 where K 2 is the K number of unique labels, guaranty a unique label to each pre-label vertex v i. Proof: Let K 2 and l be the set of unique labels. For an arbitrary vertex v i the h l xh (i) = 1 by probability. Then if the vertex v i has label k is because x k (i) q- quantile, thus the h l\k xh (i) (1 q)-quantile by probability, as consequent the vertex v i get assigned only the label. Example 3. Continuing Example 2, where the results miss-label one of the pixels, the resultant probabilities on Example 2 fails one pixel. This labeling error maybe could be resolved using more pixels labels as out or by choosing different pixels. By using the Algorithm 8 in this example with q = 0.51 > 1 2 and the inputs from Example 2. The results are improved where the 8 pixel gets the correct label, see Figure 5.2. The values for L u, V l, V u, U, m out and m in on the first iteration are described in Example 74
86 2 and the values on the second iterations are given as follows: ˆV in 0.51 = {1, 4, 7} ˆV out 0.51 = {2, 6, 9} the sets ˆV k 0.6 are been computed using the 0.6-quantile equal to 0.70 and 0.42 for x in and x out respectively. V u = {8} V l = {1, 2, 3, 4, 5, 6, 7, 9} U = [ ] L u = 1.74 m in = [ ] T m out = [ ] T In In In Out In Out Out Out In In In In In In Out In Out In Out Out Out Out Out Out Out Figure 5.3: Left: the pre-labeled pixels at the second iteration. Right: the solution after two iterations of the irandom-walker. In In In The Matlab implementation for the irandom-walker algorithm can be found in link. Out In Out Out Out Example 4. Let run the irandom-walker on Figure 5.4 where pixels 3, 6, 11 are label as L 1, L 2, L 3 respectively and q = 0.67 > 1. After one iteration the irandom- 3 Out Walker get correct labels on the image see Figure 5.4. In this example both the Random-Walker and the irandom-walker will produce the same results. 75
87 Figure 5.4: Left: The pre-labeled pixels (input for irandom-walker). solution after one iteration of the irandom-walker. Right: the 76
88 Chapter 6 Solving Three-dimensional Segmentation of Neurons Using Spectral Graph Algorithms 6.1 Introduction Segmenting individual neurons is a crucial step for the construction of connectomes and synaptomes, comprehensive maps of neural connections. Manual segmentation is an extremely laborious task requiring a great deal of expertise and experience. Automating segmentation is desirable but it poses a crucial and difficult problem. The large amount of contextual information within the data high contrast images of CNS tissues, places significant constraints on algorithms and heuristics used for automating segmentation. As a result, algorithmic approaches that use contextual information from distant parts of the image to make local decisions are often perceived as being infeasible. However, very fast linear system solvers that recently became available, make it technically possible to solve optimization problems that incorporate information from distant parts of the image. Here we discuss the engineering and implementation of the i3d-segmentation [GLKTV + 14], a segmentation framework based on random walk in graphs. In a preprocessing phase that relies on some human assistance, the system focuses on a shallow initial part of the neuron and classifies it with certainty a number of pixels being in or out the neuron. The system then uses random walk to classify the uncertain pixels and trace the neurons in their 77
89 three-dimensional space. Significance: Connectomics and synaptomics are new research efforts in neuroscience, dedicated to mapping the global dendritic and axonal branching topology and fine details of dendritic spine geometry, all of which are crucial determinants of neuronal connectivity, function, plasticity, and pathology. Connectomics and synaptomics potential in advancing understanding of the neuronal connectivity, has led to the development and refinement of tools and methods for generating usable multiscale and multi-modal large-scale data sets. Still, the sheer size of serial neural imaging data imposes a challenging constraint on developing and implementing algorithms for high- throughput, highly-accurate, automated image segmentation. The described work to develop algorithms based on recent breakthroughs in spectral graph theory will enable large-scale high- throughput generations of accurate and reliable 3D models of neuronal morphologies. Such models are crucial to neuroscientists ability to accurately and reliably reconstruct, quantify, and model dendritic processes at highresolution, an essential prerequisite to understanding the structural determinants of neuronal function. The work described is in keeping with the Brain Research Advancing Innovative Neurotechnologies (BRAIN) initiative that seeks to accelerate the development and applications of innovative new tools and technologies needed to construct a dynamic picture of the brain structure and function. Related Work: Image segmentation is a well-studied problem and hundreds, if not thousands, of publications have been devoted to it. Nevertheless, the sheer size of serial neural imaging data imposes a challenging constraint that is not present in prior image segmentation studies. Any algorithm should perform a small number of operations per pixel to be considered reasonable or feasible, meaning the almost overwhelming size of neural data poses what seems to be an inevitable design constraint: the detection of pixels on the boundary should mostly be based on local procedures 78
90 that consider the pixel brightness in a small neighborhood around each candidate boundary pixel [LDT + 11]. Most researchers consider the inclusion of distant contextual information impossible and in practice avoid such inclusion [JST10]. As a result, several studies have focused on approaches based on machine learning and, more specifically, convolutional networks (e.g. [JST10, TMJ + 10]). Such approaches train an algorithm to learn the appropriate function and thus have the potential of including distant contextual information. Such training is completely opaque to human understanding. In addition, in order to learn the algorithm must be able to use manually segmented images. Consequently, these approaches require significant human labor. Moreover, the training/learning phase of the algorithm can take weeks even on large workstations. Another significant problem with current methods is that in most cases they work by segmenting each two-dimensional (2D) scan separately and then following the boundaries through the three-dimensional (3D) stack to construct a surface [KSH13]. There are two underlying problems with the methods: (i) running time of the methods used does not scale with the size of the data, and thus breaking the data into smaller chunks improves their applicability; (ii) the topological properties of 3D space are much more complex than those of 2D space and several of the approaches do not generalize easily, including certain machine learning-based ones [JST10]; Thus, the 3D implementations are perpetually left as future work. As a result, only a small number of methods work genuinely in 3D, and often they are concerned with constructing only a 1D skeleton of the neuron [ZCB + 10, REHW09, PLM11], or with segmenting only the neuron axons [SZM + 07, SLZ + 10]. Attempts to segment the surface of a neuron require strong properties that are simply not present in the data [REHW06]. Engineering methods that work genuinely in 3D and use contextual information from distant parts of the volume would overcome a significant obstacle to constructing connectomes. 79
91 In principle, we view machine learning-based approaches as complementary to explicit 3D algorithm approaches because 3D pre-processing has the potential to reduce learning time and perhaps assist in the design of unsupervised learning algorithms. 6.2 Frame Work Images Acquisition: Here we provide some advantages of using explicit 3D algorithms to segment and reconstruct spinal motor neurons in 3D. Spinal motor neurons were labeled by combining Alexa FluorR 594 biocytin (Invitrogen Corporation, Carlsbad, CA) retrograde neuronal labeling with whole-mount immunohistochemistry using the avidin-biotin-peroxidase complex (ABP complex method (Vectastain R Elite Kit, Vector Laboratories, Burlingame, CA) and by further enhancing the ABP complex with EnzMet (Nanoprobes Inc. Yaphank, NY); This was followed by in bloc staining using heavy metal ions; spinal motor neurons were imaged using whole-mount reflection laser scanning confocal microscopy (LSCM). In the protocol for whole-mount reflection LSCM, Alexa Fluor R 594 fluorescence can be combined with biocytin immunoreactive staining, thus allowing double-labeling of neurons and their processes. Once the samples were imaged using whole-mount reflection LSCM, the samples were flat embedded in epoxy plastic resin (EMbed 812; Electron Microscopy Sciences, Hatfield, PA). After polymerization, the blocks containing the embedded tissue were glued onto a sample stub using conductive silver paint nm ultrathin-sections were serially cut using a modified diamond knife in a transverse plane. After the removal of each ultrathin-section, the sectioning process was paused and the freshly cut blockface surface was imaged with a low energy acceleration potential ( 2keV) using the in-column through The-Lens (TLD) detector. All ultrathin-sections were cut using the 3ViewTM (Gatan Inc., Pleasanton, CA) ultramicrotome within a FEI QuantaTM 80
92 200 vpfesem (FEI, Eindhoven, The Netherlands). For detecting the backscattered electrons, the TLD was set to repel all low energy electrons by setting the retardation voltage higher than 50eV. Serially imaging the block-face surface rather than individual sections and the stability of 3View s stage assures excellent registration between all images within an image stack (see Figure 6.1A) Figure 6.1: A) Registered serial three-dimensional (3D) image stack of high contrast images of nervous system; B) Selectively labeled neuron with Alexa FluorR 594 biocytin; C) Example of expert input. Engineering our system: We initially applied some computationally inexpensive filters on the electron microscopic image, primarily to increase even more the contrast of the ABP complex. We have found that doing so, increases the accuracy of subse- 81
93 quent steps. The following step of the process required expert intervention only for a very small portion of the image (10 out of 200 frames in the 3D stack) segmentation and reconstruction. The expert identified and outlined the outermost boundary of a retrogradely labeled neuron (see blue line in Figure 6.1C). The pixel intensity within the boundary is higher than that of the surrounding area because of the ABP complex. The algorithm learns to distinguish between the ABP complex pixel intensity and the pixel intensity of the surrounding area (see asterisk in Figure 6.1C). The experts compilation of all the segmented images within the image data stack results in a fully segmented and 3D rendered neuron, as illustrated in Figure 6.2A. Crucial to the algorithm is the representation of image as an affinity graph. Each pixel corresponds to a node and neighboring pixels are joined by weighted edges; The weight encodes the similarity between two pixels. Based on the expert input and simple thresholding, the nodes in the random-walk-on graph are classified as inside, outside and uncertain. Given the relatively small volume of one neuron relative to the whole image, most pixels near the area of expert input are classified as inside or outside. Thus, for pixels that were classified as uncertain, we ask the following question once we start a Random-Walker on the graph: what is the probability that we will first reach an inside pixel rather than an outside pixel? This question was first proposed and studied by Grady [Gra06]. We actually applied the irandom-walker a variant of the Gradys method [Gra06], in which we iteratively expand the area inside the 3D volume. 6.3 Results The irandom-walker can be used on a computer of reasonable cost and is very fast. The method allows the segmentation of one neuron in about 20 minutes (whole time) 82
94 as compared to 60 minutes per neuron with manual segmentation. We believe that this time can be reduced further by optimizing the code. Moreover the algorithm have demonstrated been invulnerable to image artifacts. When using a 3-dimensional graph from a stack of images the irandom-walker algorithm gets global information content of images with artifacts and images without artifacts within the same stack, this global information allows the algorithm identify the correct labels on images with artifacts, since the information of the images without artifacts help to reduce the effects of the artifacts on the segmentation as shown [KGLFZ + 11] an example of this is in Figure 6.3 and Figure 6.5. In Figure 6.2 we compare the output of segmentations obtained manually (see Figure 6.2A, 6.2D) to our semi-automated 3D algorithm (see Figure 6.2B, 6.2E). The comparison between the manual and semiautomated segmentation are almost identical. Moreover smoothness of the surface obtained is better compared with the manual segmentation as shown on Figure??. Thus, algorithms based on recent breakthroughs in spectral graph theory combined with improved staining and multi-scale and multi-modal imaging (i.e. reflection laser scanning confocal microscopy combined with serial block-face imaging technologies) will enable large- scale high-throughput generation of accurate and reliable 3D models of neuronal morphologies. The work described here is in keeping with the Brain Research Advancing Innovative Neurotechnologies (BRAIN) initiative that specifically seeks to accelerate the development and application of innovative new tools and technologies needed to construct a dynamic picture of brain structure and function. Examples of the i3d-segmentation tool is available at 83
95 Figure 6.2: A) Registered serial three-dimensional (3D) image stack of high contrast images of nervous system; B) Selectively labeled neuron with Alexa FluorR 594 biocytin; C) Example of expert input. Figure 6.3: A) Contour on image with folding artifact; B) Contour on image with a chatter artifacts. 84
96 Figure 6.4: Images correspond to a sequence of frames in order: A) Image with out artifact. B) Example of image with artifact in and its contour. C) Image with out artifact. Figure 6.5: Surface comparison: A) Manual segmentation. B) Semi-automated segmentation (smoother surface). 85
Spectral Graph Sparsification: overview of theory and practical methods. Yiannis Koutis. University of Puerto Rico - Rio Piedras
 Spectral Graph Sparsification: overview of theory and practical methods Yiannis Koutis University of Puerto Rico - Rio Piedras Graph Sparsification or Sketching Compute a smaller graph that preserves some
Spectral Graph Sparsification: overview of theory and practical methods Yiannis Koutis University of Puerto Rico - Rio Piedras Graph Sparsification or Sketching Compute a smaller graph that preserves some
SDD Solvers: Bridging theory and practice
 Yiannis Koutis University of Puerto Rico, Rio Piedras joint with Gary Miller, Richard Peng Carnegie Mellon University SDD Solvers: Bridging theory and practice The problem: Solving very large Laplacian
Yiannis Koutis University of Puerto Rico, Rio Piedras joint with Gary Miller, Richard Peng Carnegie Mellon University SDD Solvers: Bridging theory and practice The problem: Solving very large Laplacian
Mining Social Network Graphs
 Mining Social Network Graphs Analysis of Large Graphs: Community Detection Rafael Ferreira da Silva rafsilva@isi.edu http://rafaelsilva.com Note to other teachers and users of these slides: We would be
Mining Social Network Graphs Analysis of Large Graphs: Community Detection Rafael Ferreira da Silva rafsilva@isi.edu http://rafaelsilva.com Note to other teachers and users of these slides: We would be
Combinatorial meets Convex Leveraging combinatorial structure to obtain faster algorithms, provably
 Combinatorial meets Convex Leveraging combinatorial structure to obtain faster algorithms, provably Lorenzo Orecchia Department of Computer Science 6/30/15 Challenges in Optimization for Data Science 1
Combinatorial meets Convex Leveraging combinatorial structure to obtain faster algorithms, provably Lorenzo Orecchia Department of Computer Science 6/30/15 Challenges in Optimization for Data Science 1
Social-Network Graphs
 Social-Network Graphs Mining Social Networks Facebook, Google+, Twitter Email Networks, Collaboration Networks Identify communities Similar to clustering Communities usually overlap Identify similarities
Social-Network Graphs Mining Social Networks Facebook, Google+, Twitter Email Networks, Collaboration Networks Identify communities Similar to clustering Communities usually overlap Identify similarities
Spanning Edge Centrality: Large-scale Computation and Applications
 Spanning Edge Centrality: Large-scale Computation and Applications ABSTRACT Charalampos Mavroforakis Boston University cmav@cs.bu.edu Ioannis Koutis University of Puerto Rico Rio Piedras ioannis.koutis@upr.edu
Spanning Edge Centrality: Large-scale Computation and Applications ABSTRACT Charalampos Mavroforakis Boston University cmav@cs.bu.edu Ioannis Koutis University of Puerto Rico Rio Piedras ioannis.koutis@upr.edu
Algorithms for Grid Graphs in the MapReduce Model
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Computer Science and Engineering: Theses, Dissertations, and Student Research Computer Science and Engineering, Department
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Computer Science and Engineering: Theses, Dissertations, and Student Research Computer Science and Engineering, Department
Lecture 20 : Trees DRAFT
 CS/Math 240: Introduction to Discrete Mathematics 4/12/2011 Lecture 20 : Trees Instructor: Dieter van Melkebeek Scribe: Dalibor Zelený DRAFT Last time we discussed graphs. Today we continue this discussion,
CS/Math 240: Introduction to Discrete Mathematics 4/12/2011 Lecture 20 : Trees Instructor: Dieter van Melkebeek Scribe: Dalibor Zelený DRAFT Last time we discussed graphs. Today we continue this discussion,
Lecture 11: Clustering and the Spectral Partitioning Algorithm A note on randomized algorithm, Unbiased estimates
 CSE 51: Design and Analysis of Algorithms I Spring 016 Lecture 11: Clustering and the Spectral Partitioning Algorithm Lecturer: Shayan Oveis Gharan May nd Scribe: Yueqi Sheng Disclaimer: These notes have
CSE 51: Design and Analysis of Algorithms I Spring 016 Lecture 11: Clustering and the Spectral Partitioning Algorithm Lecturer: Shayan Oveis Gharan May nd Scribe: Yueqi Sheng Disclaimer: These notes have
An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization
 An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization Pedro Ribeiro (DCC/FCUP & CRACS/INESC-TEC) Part 1 Motivation and emergence of Network Science
An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization Pedro Ribeiro (DCC/FCUP & CRACS/INESC-TEC) Part 1 Motivation and emergence of Network Science
Graph Theory Review. January 30, Network Science Analytics Graph Theory Review 1
 Graph Theory Review Gonzalo Mateos Dept. of ECE and Goergen Institute for Data Science University of Rochester gmateosb@ece.rochester.edu http://www.ece.rochester.edu/~gmateosb/ January 30, 2018 Network
Graph Theory Review Gonzalo Mateos Dept. of ECE and Goergen Institute for Data Science University of Rochester gmateosb@ece.rochester.edu http://www.ece.rochester.edu/~gmateosb/ January 30, 2018 Network
Big Data Management and NoSQL Databases
 NDBI040 Big Data Management and NoSQL Databases Lecture 10. Graph databases Doc. RNDr. Irena Holubova, Ph.D. holubova@ksi.mff.cuni.cz http://www.ksi.mff.cuni.cz/~holubova/ndbi040/ Graph Databases Basic
NDBI040 Big Data Management and NoSQL Databases Lecture 10. Graph databases Doc. RNDr. Irena Holubova, Ph.D. holubova@ksi.mff.cuni.cz http://www.ksi.mff.cuni.cz/~holubova/ndbi040/ Graph Databases Basic
Graph drawing in spectral layout
 Graph drawing in spectral layout Maureen Gallagher Colleen Tygh John Urschel Ludmil Zikatanov Beginning: July 8, 203; Today is: October 2, 203 Introduction Our research focuses on the use of spectral graph
Graph drawing in spectral layout Maureen Gallagher Colleen Tygh John Urschel Ludmil Zikatanov Beginning: July 8, 203; Today is: October 2, 203 Introduction Our research focuses on the use of spectral graph
Visual Representations for Machine Learning
 Visual Representations for Machine Learning Spectral Clustering and Channel Representations Lecture 1 Spectral Clustering: introduction and confusion Michael Felsberg Klas Nordberg The Spectral Clustering
Visual Representations for Machine Learning Spectral Clustering and Channel Representations Lecture 1 Spectral Clustering: introduction and confusion Michael Felsberg Klas Nordberg The Spectral Clustering
1 Homophily and assortative mixing
 1 Homophily and assortative mixing Networks, and particularly social networks, often exhibit a property called homophily or assortative mixing, which simply means that the attributes of vertices correlate
1 Homophily and assortative mixing Networks, and particularly social networks, often exhibit a property called homophily or assortative mixing, which simply means that the attributes of vertices correlate
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION
 CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
Symmetric Product Graphs
 Rochester Institute of Technology RIT Scholar Works Theses Thesis/Dissertation Collections 5-20-2015 Symmetric Product Graphs Evan Witz Follow this and additional works at: http://scholarworks.rit.edu/theses
Rochester Institute of Technology RIT Scholar Works Theses Thesis/Dissertation Collections 5-20-2015 Symmetric Product Graphs Evan Witz Follow this and additional works at: http://scholarworks.rit.edu/theses
CS224W: Analysis of Networks Jure Leskovec, Stanford University
 CS224W: Analysis of Networks Jure Leskovec, Stanford University http://cs224w.stanford.edu 11/13/17 Jure Leskovec, Stanford CS224W: Analysis of Networks, http://cs224w.stanford.edu 2 Observations Models
CS224W: Analysis of Networks Jure Leskovec, Stanford University http://cs224w.stanford.edu 11/13/17 Jure Leskovec, Stanford CS224W: Analysis of Networks, http://cs224w.stanford.edu 2 Observations Models
Copyright 2000, Kevin Wayne 1
 Chapter 3 - Graphs Undirected Graphs Undirected graph. G = (V, E) V = nodes. E = edges between pairs of nodes. Captures pairwise relationship between objects. Graph size parameters: n = V, m = E. Directed
Chapter 3 - Graphs Undirected Graphs Undirected graph. G = (V, E) V = nodes. E = edges between pairs of nodes. Captures pairwise relationship between objects. Graph size parameters: n = V, m = E. Directed
Introduction III. Graphs. Motivations I. Introduction IV
 Introduction I Graphs Computer Science & Engineering 235: Discrete Mathematics Christopher M. Bourke cbourke@cse.unl.edu Graph theory was introduced in the 18th century by Leonhard Euler via the Königsberg
Introduction I Graphs Computer Science & Engineering 235: Discrete Mathematics Christopher M. Bourke cbourke@cse.unl.edu Graph theory was introduced in the 18th century by Leonhard Euler via the Königsberg
Lecture #3: PageRank Algorithm The Mathematics of Google Search
 Lecture #3: PageRank Algorithm The Mathematics of Google Search We live in a computer era. Internet is part of our everyday lives and information is only a click away. Just open your favorite search engine,
Lecture #3: PageRank Algorithm The Mathematics of Google Search We live in a computer era. Internet is part of our everyday lives and information is only a click away. Just open your favorite search engine,
Spectral Clustering and Community Detection in Labeled Graphs
 Spectral Clustering and Community Detection in Labeled Graphs Brandon Fain, Stavros Sintos, Nisarg Raval Machine Learning (CompSci 571D / STA 561D) December 7, 2015 {btfain, nisarg, ssintos} at cs.duke.edu
Spectral Clustering and Community Detection in Labeled Graphs Brandon Fain, Stavros Sintos, Nisarg Raval Machine Learning (CompSci 571D / STA 561D) December 7, 2015 {btfain, nisarg, ssintos} at cs.duke.edu
Foundations of Computing
 Foundations of Computing Darmstadt University of Technology Dept. Computer Science Winter Term 2005 / 2006 Copyright c 2004 by Matthias Müller-Hannemann and Karsten Weihe All rights reserved http://www.algo.informatik.tu-darmstadt.de/
Foundations of Computing Darmstadt University of Technology Dept. Computer Science Winter Term 2005 / 2006 Copyright c 2004 by Matthias Müller-Hannemann and Karsten Weihe All rights reserved http://www.algo.informatik.tu-darmstadt.de/
Centrality Book. cohesion.
 Cohesion The graph-theoretic terms discussed in the previous chapter have very specific and concrete meanings which are highly shared across the field of graph theory and other fields like social network
Cohesion The graph-theoretic terms discussed in the previous chapter have very specific and concrete meanings which are highly shared across the field of graph theory and other fields like social network
Advanced Algorithms Class Notes for Monday, October 23, 2012 Min Ye, Mingfu Shao, and Bernard Moret
 Advanced Algorithms Class Notes for Monday, October 23, 2012 Min Ye, Mingfu Shao, and Bernard Moret Greedy Algorithms (continued) The best known application where the greedy algorithm is optimal is surely
Advanced Algorithms Class Notes for Monday, October 23, 2012 Min Ye, Mingfu Shao, and Bernard Moret Greedy Algorithms (continued) The best known application where the greedy algorithm is optimal is surely
REDUCING GRAPH COLORING TO CLIQUE SEARCH
 Asia Pacific Journal of Mathematics, Vol. 3, No. 1 (2016), 64-85 ISSN 2357-2205 REDUCING GRAPH COLORING TO CLIQUE SEARCH SÁNDOR SZABÓ AND BOGDÁN ZAVÁLNIJ Institute of Mathematics and Informatics, University
Asia Pacific Journal of Mathematics, Vol. 3, No. 1 (2016), 64-85 ISSN 2357-2205 REDUCING GRAPH COLORING TO CLIQUE SEARCH SÁNDOR SZABÓ AND BOGDÁN ZAVÁLNIJ Institute of Mathematics and Informatics, University
Lecture 27: Fast Laplacian Solvers
 Lecture 27: Fast Laplacian Solvers Scribed by Eric Lee, Eston Schweickart, Chengrun Yang November 21, 2017 1 How Fast Laplacian Solvers Work We want to solve Lx = b with L being a Laplacian matrix. Recall
Lecture 27: Fast Laplacian Solvers Scribed by Eric Lee, Eston Schweickart, Chengrun Yang November 21, 2017 1 How Fast Laplacian Solvers Work We want to solve Lx = b with L being a Laplacian matrix. Recall
Theorem 2.9: nearest addition algorithm
 There are severe limits on our ability to compute near-optimal tours It is NP-complete to decide whether a given undirected =(,)has a Hamiltonian cycle An approximation algorithm for the TSP can be used
There are severe limits on our ability to compute near-optimal tours It is NP-complete to decide whether a given undirected =(,)has a Hamiltonian cycle An approximation algorithm for the TSP can be used
The Simplex Algorithm
 The Simplex Algorithm Uri Feige November 2011 1 The simplex algorithm The simplex algorithm was designed by Danzig in 1947. This write-up presents the main ideas involved. It is a slight update (mostly
The Simplex Algorithm Uri Feige November 2011 1 The simplex algorithm The simplex algorithm was designed by Danzig in 1947. This write-up presents the main ideas involved. It is a slight update (mostly
Random projection for non-gaussian mixture models
 Random projection for non-gaussian mixture models Győző Gidófalvi Department of Computer Science and Engineering University of California, San Diego La Jolla, CA 92037 gyozo@cs.ucsd.edu Abstract Recently,
Random projection for non-gaussian mixture models Győző Gidófalvi Department of Computer Science and Engineering University of California, San Diego La Jolla, CA 92037 gyozo@cs.ucsd.edu Abstract Recently,
On the Relationships between Zero Forcing Numbers and Certain Graph Coverings
 On the Relationships between Zero Forcing Numbers and Certain Graph Coverings Fatemeh Alinaghipour Taklimi, Shaun Fallat 1,, Karen Meagher 2 Department of Mathematics and Statistics, University of Regina,
On the Relationships between Zero Forcing Numbers and Certain Graph Coverings Fatemeh Alinaghipour Taklimi, Shaun Fallat 1,, Karen Meagher 2 Department of Mathematics and Statistics, University of Regina,
Detecting and Analyzing Communities in Social Network Graphs for Targeted Marketing
 Detecting and Analyzing Communities in Social Network Graphs for Targeted Marketing Gautam Bhat, Rajeev Kumar Singh Department of Computer Science and Engineering Shiv Nadar University Gautam Buddh Nagar,
Detecting and Analyzing Communities in Social Network Graphs for Targeted Marketing Gautam Bhat, Rajeev Kumar Singh Department of Computer Science and Engineering Shiv Nadar University Gautam Buddh Nagar,
Stanford University CS359G: Graph Partitioning and Expanders Handout 1 Luca Trevisan January 4, 2011
 Stanford University CS359G: Graph Partitioning and Expanders Handout 1 Luca Trevisan January 4, 2011 Lecture 1 In which we describe what this course is about. 1 Overview This class is about the following
Stanford University CS359G: Graph Partitioning and Expanders Handout 1 Luca Trevisan January 4, 2011 Lecture 1 In which we describe what this course is about. 1 Overview This class is about the following
Fast Fuzzy Clustering of Infrared Images. 2. brfcm
 Fast Fuzzy Clustering of Infrared Images Steven Eschrich, Jingwei Ke, Lawrence O. Hall and Dmitry B. Goldgof Department of Computer Science and Engineering, ENB 118 University of South Florida 4202 E.
Fast Fuzzy Clustering of Infrared Images Steven Eschrich, Jingwei Ke, Lawrence O. Hall and Dmitry B. Goldgof Department of Computer Science and Engineering, ENB 118 University of South Florida 4202 E.
An Experiment in Visual Clustering Using Star Glyph Displays
 An Experiment in Visual Clustering Using Star Glyph Displays by Hanna Kazhamiaka A Research Paper presented to the University of Waterloo in partial fulfillment of the requirements for the degree of Master
An Experiment in Visual Clustering Using Star Glyph Displays by Hanna Kazhamiaka A Research Paper presented to the University of Waterloo in partial fulfillment of the requirements for the degree of Master
Lecture 26: Graphs: Traversal (Part 1)
 CS8 Integrated Introduction to Computer Science Fisler, Nelson Lecture 6: Graphs: Traversal (Part ) 0:00 AM, Apr, 08 Contents Introduction. Definitions........................................... Representations.......................................
CS8 Integrated Introduction to Computer Science Fisler, Nelson Lecture 6: Graphs: Traversal (Part ) 0:00 AM, Apr, 08 Contents Introduction. Definitions........................................... Representations.......................................
Economics of Information Networks
 Economics of Information Networks Stephen Turnbull Division of Policy and Planning Sciences Lecture 4: December 7, 2017 Abstract We continue discussion of the modern economics of networks, which considers
Economics of Information Networks Stephen Turnbull Division of Policy and Planning Sciences Lecture 4: December 7, 2017 Abstract We continue discussion of the modern economics of networks, which considers
TELCOM2125: Network Science and Analysis
 School of Information Sciences University of Pittsburgh TELCOM2125: Network Science and Analysis Konstantinos Pelechrinis Spring 2015 2 Part 4: Dividing Networks into Clusters The problem l Graph partitioning
School of Information Sciences University of Pittsburgh TELCOM2125: Network Science and Analysis Konstantinos Pelechrinis Spring 2015 2 Part 4: Dividing Networks into Clusters The problem l Graph partitioning
Spectral Clustering X I AO ZE N G + E L HA M TA BA S SI CS E CL A S S P R ESENTATION MA RCH 1 6,
 Spectral Clustering XIAO ZENG + ELHAM TABASSI CSE 902 CLASS PRESENTATION MARCH 16, 2017 1 Presentation based on 1. Von Luxburg, Ulrike. "A tutorial on spectral clustering." Statistics and computing 17.4
Spectral Clustering XIAO ZENG + ELHAM TABASSI CSE 902 CLASS PRESENTATION MARCH 16, 2017 1 Presentation based on 1. Von Luxburg, Ulrike. "A tutorial on spectral clustering." Statistics and computing 17.4
The Encoding Complexity of Network Coding
 The Encoding Complexity of Network Coding Michael Langberg Alexander Sprintson Jehoshua Bruck California Institute of Technology Email: mikel,spalex,bruck @caltech.edu Abstract In the multicast network
The Encoding Complexity of Network Coding Michael Langberg Alexander Sprintson Jehoshua Bruck California Institute of Technology Email: mikel,spalex,bruck @caltech.edu Abstract In the multicast network
Algorithms. Graphs. Algorithms
 Algorithms Graphs Algorithms Graphs Definition: A graph is a collection of edges and vertices. Each edge connects two vertices. Algorithms 1 Graphs Vertices: Nodes, points, computers, users, items,...
Algorithms Graphs Algorithms Graphs Definition: A graph is a collection of edges and vertices. Each edge connects two vertices. Algorithms 1 Graphs Vertices: Nodes, points, computers, users, items,...
Challenges in large-scale graph processing on HPC platforms and the Graph500 benchmark. by Nkemdirim Dockery
 Challenges in large-scale graph processing on HPC platforms and the Graph500 benchmark by Nkemdirim Dockery High Performance Computing Workloads Core-memory sized Floating point intensive Well-structured
Challenges in large-scale graph processing on HPC platforms and the Graph500 benchmark by Nkemdirim Dockery High Performance Computing Workloads Core-memory sized Floating point intensive Well-structured
Clustering. SC4/SM4 Data Mining and Machine Learning, Hilary Term 2017 Dino Sejdinovic
 Clustering SC4/SM4 Data Mining and Machine Learning, Hilary Term 2017 Dino Sejdinovic Clustering is one of the fundamental and ubiquitous tasks in exploratory data analysis a first intuition about the
Clustering SC4/SM4 Data Mining and Machine Learning, Hilary Term 2017 Dino Sejdinovic Clustering is one of the fundamental and ubiquitous tasks in exploratory data analysis a first intuition about the
Stanford University CS359G: Graph Partitioning and Expanders Handout 18 Luca Trevisan March 3, 2011
 Stanford University CS359G: Graph Partitioning and Expanders Handout 8 Luca Trevisan March 3, 20 Lecture 8 In which we prove properties of expander graphs. Quasirandomness of Expander Graphs Recall that
Stanford University CS359G: Graph Partitioning and Expanders Handout 8 Luca Trevisan March 3, 20 Lecture 8 In which we prove properties of expander graphs. Quasirandomness of Expander Graphs Recall that
Lecture 25: Element-wise Sampling of Graphs and Linear Equation Solving, Cont. 25 Element-wise Sampling of Graphs and Linear Equation Solving,
 Stat260/CS294: Randomized Algorithms for Matrices and Data Lecture 25-12/04/2013 Lecture 25: Element-wise Sampling of Graphs and Linear Equation Solving, Cont. Lecturer: Michael Mahoney Scribe: Michael
Stat260/CS294: Randomized Algorithms for Matrices and Data Lecture 25-12/04/2013 Lecture 25: Element-wise Sampling of Graphs and Linear Equation Solving, Cont. Lecturer: Michael Mahoney Scribe: Michael
Topic: Local Search: Max-Cut, Facility Location Date: 2/13/2007
 CS880: Approximations Algorithms Scribe: Chi Man Liu Lecturer: Shuchi Chawla Topic: Local Search: Max-Cut, Facility Location Date: 2/3/2007 In previous lectures we saw how dynamic programming could be
CS880: Approximations Algorithms Scribe: Chi Man Liu Lecturer: Shuchi Chawla Topic: Local Search: Max-Cut, Facility Location Date: 2/3/2007 In previous lectures we saw how dynamic programming could be
Algorithms: Graphs. Amotz Bar-Noy. Spring 2012 CUNY. Amotz Bar-Noy (CUNY) Graphs Spring / 95
 Algorithms: Graphs Amotz Bar-Noy CUNY Spring 2012 Amotz Bar-Noy (CUNY) Graphs Spring 2012 1 / 95 Graphs Definition: A graph is a collection of edges and vertices. Each edge connects two vertices. Amotz
Algorithms: Graphs Amotz Bar-Noy CUNY Spring 2012 Amotz Bar-Noy (CUNY) Graphs Spring 2012 1 / 95 Graphs Definition: A graph is a collection of edges and vertices. Each edge connects two vertices. Amotz
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS
 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
Prepare a stem-and-leaf graph for the following data. In your final display, you should arrange the leaves for each stem in increasing order.
 Chapter 2 2.1 Descriptive Statistics A stem-and-leaf graph, also called a stemplot, allows for a nice overview of quantitative data without losing information on individual observations. It can be a good
Chapter 2 2.1 Descriptive Statistics A stem-and-leaf graph, also called a stemplot, allows for a nice overview of quantitative data without losing information on individual observations. It can be a good
2.3 Algorithms Using Map-Reduce
 28 CHAPTER 2. MAP-REDUCE AND THE NEW SOFTWARE STACK one becomes available. The Master must also inform each Reduce task that the location of its input from that Map task has changed. Dealing with a failure
28 CHAPTER 2. MAP-REDUCE AND THE NEW SOFTWARE STACK one becomes available. The Master must also inform each Reduce task that the location of its input from that Map task has changed. Dealing with a failure
Topological Classification of Data Sets without an Explicit Metric
 Topological Classification of Data Sets without an Explicit Metric Tim Harrington, Andrew Tausz and Guillaume Troianowski December 10, 2008 A contemporary problem in data analysis is understanding the
Topological Classification of Data Sets without an Explicit Metric Tim Harrington, Andrew Tausz and Guillaume Troianowski December 10, 2008 A contemporary problem in data analysis is understanding the
Collaborative filtering based on a random walk model on a graph
 Collaborative filtering based on a random walk model on a graph Marco Saerens, Francois Fouss, Alain Pirotte, Luh Yen, Pierre Dupont (UCL) Jean-Michel Renders (Xerox Research Europe) Some recent methods:
Collaborative filtering based on a random walk model on a graph Marco Saerens, Francois Fouss, Alain Pirotte, Luh Yen, Pierre Dupont (UCL) Jean-Michel Renders (Xerox Research Europe) Some recent methods:
Non Overlapping Communities
 Non Overlapping Communities Davide Mottin, Konstantina Lazaridou HassoPlattner Institute Graph Mining course Winter Semester 2016 Acknowledgements Most of this lecture is taken from: http://web.stanford.edu/class/cs224w/slides
Non Overlapping Communities Davide Mottin, Konstantina Lazaridou HassoPlattner Institute Graph Mining course Winter Semester 2016 Acknowledgements Most of this lecture is taken from: http://web.stanford.edu/class/cs224w/slides
CSE 417: Algorithms and Computational Complexity. 3.1 Basic Definitions and Applications. Goals. Chapter 3. Winter 2012 Graphs and Graph Algorithms
 Chapter 3 CSE 417: Algorithms and Computational Complexity Graphs Reading: 3.1-3.6 Winter 2012 Graphs and Graph Algorithms Slides by Kevin Wayne. Copyright 2005 Pearson-Addison Wesley. All rights reserved.
Chapter 3 CSE 417: Algorithms and Computational Complexity Graphs Reading: 3.1-3.6 Winter 2012 Graphs and Graph Algorithms Slides by Kevin Wayne. Copyright 2005 Pearson-Addison Wesley. All rights reserved.
Algorithms: Lecture 7. Chalmers University of Technology
 Algorithms: Lecture 7 Chalmers University of Technology Today s Lecture Divide & Conquer Counting Inversions Closest Pair of Points Multiplication of large integers Intro to the forthcoming problems Graphs:
Algorithms: Lecture 7 Chalmers University of Technology Today s Lecture Divide & Conquer Counting Inversions Closest Pair of Points Multiplication of large integers Intro to the forthcoming problems Graphs:
Electrical Circuits and Random Walks: Challenge Problems
 Electrical Circuits and Random Walks: Challenge Problems LA Math Circle, High School II December 9, 2018 1 Effective Resistance The most efficient way of calculating electrical properties of a circuit
Electrical Circuits and Random Walks: Challenge Problems LA Math Circle, High School II December 9, 2018 1 Effective Resistance The most efficient way of calculating electrical properties of a circuit
Lecture 9 - Matrix Multiplication Equivalences and Spectral Graph Theory 1
 CME 305: Discrete Mathematics and Algorithms Instructor: Professor Aaron Sidford (sidford@stanfordedu) February 6, 2018 Lecture 9 - Matrix Multiplication Equivalences and Spectral Graph Theory 1 In the
CME 305: Discrete Mathematics and Algorithms Instructor: Professor Aaron Sidford (sidford@stanfordedu) February 6, 2018 Lecture 9 - Matrix Multiplication Equivalences and Spectral Graph Theory 1 In the
Algorithms: Lecture 10. Chalmers University of Technology
 Algorithms: Lecture 10 Chalmers University of Technology Today s Topics Basic Definitions Path, Cycle, Tree, Connectivity, etc. Graph Traversal Depth First Search Breadth First Search Testing Bipartatiness
Algorithms: Lecture 10 Chalmers University of Technology Today s Topics Basic Definitions Path, Cycle, Tree, Connectivity, etc. Graph Traversal Depth First Search Breadth First Search Testing Bipartatiness
Math 7 Glossary Terms
 Math 7 Glossary Terms Absolute Value Absolute value is the distance, or number of units, a number is from zero. Distance is always a positive value; therefore, absolute value is always a positive value.
Math 7 Glossary Terms Absolute Value Absolute value is the distance, or number of units, a number is from zero. Distance is always a positive value; therefore, absolute value is always a positive value.
st-orientations September 29, 2005
 st-orientations September 29, 2005 Introduction Let G = (V, E) be an undirected biconnected graph of n nodes and m edges. The main problem this chapter deals with is different algorithms for orienting
st-orientations September 29, 2005 Introduction Let G = (V, E) be an undirected biconnected graph of n nodes and m edges. The main problem this chapter deals with is different algorithms for orienting
Lecture 2 September 3
 EE 381V: Large Scale Optimization Fall 2012 Lecture 2 September 3 Lecturer: Caramanis & Sanghavi Scribe: Hongbo Si, Qiaoyang Ye 2.1 Overview of the last Lecture The focus of the last lecture was to give
EE 381V: Large Scale Optimization Fall 2012 Lecture 2 September 3 Lecturer: Caramanis & Sanghavi Scribe: Hongbo Si, Qiaoyang Ye 2.1 Overview of the last Lecture The focus of the last lecture was to give
Homework 4: Clustering, Recommenders, Dim. Reduction, ML and Graph Mining (due November 19 th, 2014, 2:30pm, in class hard-copy please)
 Virginia Tech. Computer Science CS 5614 (Big) Data Management Systems Fall 2014, Prakash Homework 4: Clustering, Recommenders, Dim. Reduction, ML and Graph Mining (due November 19 th, 2014, 2:30pm, in
Virginia Tech. Computer Science CS 5614 (Big) Data Management Systems Fall 2014, Prakash Homework 4: Clustering, Recommenders, Dim. Reduction, ML and Graph Mining (due November 19 th, 2014, 2:30pm, in
Chapter 3. Graphs. Slides by Kevin Wayne. Copyright 2005 Pearson-Addison Wesley. All rights reserved.
 Chapter 3 Graphs Slides by Kevin Wayne. Copyright 2005 Pearson-Addison Wesley. All rights reserved. 1 3.1 Basic Definitions and Applications Undirected Graphs Undirected graph. G = (V, E) V = nodes. E
Chapter 3 Graphs Slides by Kevin Wayne. Copyright 2005 Pearson-Addison Wesley. All rights reserved. 1 3.1 Basic Definitions and Applications Undirected Graphs Undirected graph. G = (V, E) V = nodes. E
Knowledge Discovery and Data Mining 1 (VO) ( )
 Knowledge Discovery and Data Mining 1 (VO) (707.003) Data Matrices and Vector Space Model Denis Helic KTI, TU Graz Nov 6, 2014 Denis Helic (KTI, TU Graz) KDDM1 Nov 6, 2014 1 / 55 Big picture: KDDM Probability
Knowledge Discovery and Data Mining 1 (VO) (707.003) Data Matrices and Vector Space Model Denis Helic KTI, TU Graz Nov 6, 2014 Denis Helic (KTI, TU Graz) KDDM1 Nov 6, 2014 1 / 55 Big picture: KDDM Probability
Clustering: Classic Methods and Modern Views
 Clustering: Classic Methods and Modern Views Marina Meilă University of Washington mmp@stat.washington.edu June 22, 2015 Lorentz Center Workshop on Clusters, Games and Axioms Outline Paradigms for clustering
Clustering: Classic Methods and Modern Views Marina Meilă University of Washington mmp@stat.washington.edu June 22, 2015 Lorentz Center Workshop on Clusters, Games and Axioms Outline Paradigms for clustering
CSE 255 Lecture 6. Data Mining and Predictive Analytics. Community Detection
 CSE 255 Lecture 6 Data Mining and Predictive Analytics Community Detection Dimensionality reduction Goal: take high-dimensional data, and describe it compactly using a small number of dimensions Assumption:
CSE 255 Lecture 6 Data Mining and Predictive Analytics Community Detection Dimensionality reduction Goal: take high-dimensional data, and describe it compactly using a small number of dimensions Assumption:
Leveraging Set Relations in Exact Set Similarity Join
 Leveraging Set Relations in Exact Set Similarity Join Xubo Wang, Lu Qin, Xuemin Lin, Ying Zhang, and Lijun Chang University of New South Wales, Australia University of Technology Sydney, Australia {xwang,lxue,ljchang}@cse.unsw.edu.au,
Leveraging Set Relations in Exact Set Similarity Join Xubo Wang, Lu Qin, Xuemin Lin, Ying Zhang, and Lijun Chang University of New South Wales, Australia University of Technology Sydney, Australia {xwang,lxue,ljchang}@cse.unsw.edu.au,
Course Introduction / Review of Fundamentals of Graph Theory
 Course Introduction / Review of Fundamentals of Graph Theory Hiroki Sayama sayama@binghamton.edu Rise of Network Science (From Barabasi 2010) 2 Network models Many discrete parts involved Classic mean-field
Course Introduction / Review of Fundamentals of Graph Theory Hiroki Sayama sayama@binghamton.edu Rise of Network Science (From Barabasi 2010) 2 Network models Many discrete parts involved Classic mean-field
CS246: Mining Massive Datasets Jure Leskovec, Stanford University
 CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu SPAM FARMING 2/11/2013 Jure Leskovec, Stanford C246: Mining Massive Datasets 2 2/11/2013 Jure Leskovec, Stanford
CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu SPAM FARMING 2/11/2013 Jure Leskovec, Stanford C246: Mining Massive Datasets 2 2/11/2013 Jure Leskovec, Stanford
Chapter 2 Basic Structure of High-Dimensional Spaces
 Chapter 2 Basic Structure of High-Dimensional Spaces Data is naturally represented geometrically by associating each record with a point in the space spanned by the attributes. This idea, although simple,
Chapter 2 Basic Structure of High-Dimensional Spaces Data is naturally represented geometrically by associating each record with a point in the space spanned by the attributes. This idea, although simple,
Figure 1: A directed graph.
 1 Graphs A graph is a data structure that expresses relationships between objects. The objects are called nodes and the relationships are called edges. For example, social networks can be represented as
1 Graphs A graph is a data structure that expresses relationships between objects. The objects are called nodes and the relationships are called edges. For example, social networks can be represented as
Lecture 5: Graphs. Rajat Mittal. IIT Kanpur
 Lecture : Graphs Rajat Mittal IIT Kanpur Combinatorial graphs provide a natural way to model connections between different objects. They are very useful in depicting communication networks, social networks
Lecture : Graphs Rajat Mittal IIT Kanpur Combinatorial graphs provide a natural way to model connections between different objects. They are very useful in depicting communication networks, social networks
Massachusetts Institute of Technology Department of Electrical Engineering and Computer Science Algorithms For Inference Fall 2014
 Suggested Reading: Massachusetts Institute of Technology Department of Electrical Engineering and Computer Science 6.438 Algorithms For Inference Fall 2014 Probabilistic Modelling and Reasoning: The Junction
Suggested Reading: Massachusetts Institute of Technology Department of Electrical Engineering and Computer Science 6.438 Algorithms For Inference Fall 2014 Probabilistic Modelling and Reasoning: The Junction
Efficient Second-Order Iterative Methods for IR Drop Analysis in Power Grid
 Efficient Second-Order Iterative Methods for IR Drop Analysis in Power Grid Yu Zhong Martin D. F. Wong Dept. of Electrical and Computer Engineering Dept. of Electrical and Computer Engineering Univ. of
Efficient Second-Order Iterative Methods for IR Drop Analysis in Power Grid Yu Zhong Martin D. F. Wong Dept. of Electrical and Computer Engineering Dept. of Electrical and Computer Engineering Univ. of
An Empirical Analysis of Communities in Real-World Networks
 An Empirical Analysis of Communities in Real-World Networks Chuan Sheng Foo Computer Science Department Stanford University csfoo@cs.stanford.edu ABSTRACT Little work has been done on the characterization
An Empirical Analysis of Communities in Real-World Networks Chuan Sheng Foo Computer Science Department Stanford University csfoo@cs.stanford.edu ABSTRACT Little work has been done on the characterization
GRAPHS (Undirected) Graph: Set of objects with pairwise connections. Why study graph algorithms?
 GRAPHS (Undirected) Graph: Set of objects with pairwise connections. Why study graph algorithms? Interesting and broadly useful abstraction. Challenging branch of computer science and discrete math. Hundreds
GRAPHS (Undirected) Graph: Set of objects with pairwise connections. Why study graph algorithms? Interesting and broadly useful abstraction. Challenging branch of computer science and discrete math. Hundreds
MVAPICH2 vs. OpenMPI for a Clustering Algorithm
 MVAPICH2 vs. OpenMPI for a Clustering Algorithm Robin V. Blasberg and Matthias K. Gobbert Naval Research Laboratory, Washington, D.C. Department of Mathematics and Statistics, University of Maryland, Baltimore
MVAPICH2 vs. OpenMPI for a Clustering Algorithm Robin V. Blasberg and Matthias K. Gobbert Naval Research Laboratory, Washington, D.C. Department of Mathematics and Statistics, University of Maryland, Baltimore
E-Companion: On Styles in Product Design: An Analysis of US. Design Patents
 E-Companion: On Styles in Product Design: An Analysis of US Design Patents 1 PART A: FORMALIZING THE DEFINITION OF STYLES A.1 Styles as categories of designs of similar form Our task involves categorizing
E-Companion: On Styles in Product Design: An Analysis of US Design Patents 1 PART A: FORMALIZING THE DEFINITION OF STYLES A.1 Styles as categories of designs of similar form Our task involves categorizing
The Encoding Complexity of Network Coding
 The Encoding Complexity of Network Coding Michael Langberg Alexander Sprintson Jehoshua Bruck California Institute of Technology Email mikel,spalex,bruck @caltech.edu Abstract In the multicast network
The Encoding Complexity of Network Coding Michael Langberg Alexander Sprintson Jehoshua Bruck California Institute of Technology Email mikel,spalex,bruck @caltech.edu Abstract In the multicast network
PERFECT MATCHING THE CENTRALIZED DEPLOYMENT MOBILE SENSORS THE PROBLEM SECOND PART: WIRELESS NETWORKS 2.B. SENSOR NETWORKS OF MOBILE SENSORS
 SECOND PART: WIRELESS NETWORKS 2.B. SENSOR NETWORKS THE CENTRALIZED DEPLOYMENT OF MOBILE SENSORS I.E. THE MINIMUM WEIGHT PERFECT MATCHING 1 2 ON BIPARTITE GRAPHS Prof. Tiziana Calamoneri Network Algorithms
SECOND PART: WIRELESS NETWORKS 2.B. SENSOR NETWORKS THE CENTRALIZED DEPLOYMENT OF MOBILE SENSORS I.E. THE MINIMUM WEIGHT PERFECT MATCHING 1 2 ON BIPARTITE GRAPHS Prof. Tiziana Calamoneri Network Algorithms
3.1 Basic Definitions and Applications. Chapter 3. Graphs. Undirected Graphs. Some Graph Applications
 Chapter 3 31 Basic Definitions and Applications Graphs Slides by Kevin Wayne Copyright 2005 Pearson-Addison Wesley All rights reserved 1 Undirected Graphs Some Graph Applications Undirected graph G = (V,
Chapter 3 31 Basic Definitions and Applications Graphs Slides by Kevin Wayne Copyright 2005 Pearson-Addison Wesley All rights reserved 1 Undirected Graphs Some Graph Applications Undirected graph G = (V,
From Smith-Waterman to BLAST
 From Smith-Waterman to BLAST Jeremy Buhler July 23, 2015 Smith-Waterman is the fundamental tool that we use to decide how similar two sequences are. Isn t that all that BLAST does? In principle, it is
From Smith-Waterman to BLAST Jeremy Buhler July 23, 2015 Smith-Waterman is the fundamental tool that we use to decide how similar two sequences are. Isn t that all that BLAST does? In principle, it is
Lecture and notes by: Nate Chenette, Brent Myers, Hari Prasad November 8, Property Testing
 Property Testing 1 Introduction Broadly, property testing is the study of the following class of problems: Given the ability to perform (local) queries concerning a particular object (e.g., a function,
Property Testing 1 Introduction Broadly, property testing is the study of the following class of problems: Given the ability to perform (local) queries concerning a particular object (e.g., a function,
6. Lecture notes on matroid intersection
 Massachusetts Institute of Technology 18.453: Combinatorial Optimization Michel X. Goemans May 2, 2017 6. Lecture notes on matroid intersection One nice feature about matroids is that a simple greedy algorithm
Massachusetts Institute of Technology 18.453: Combinatorial Optimization Michel X. Goemans May 2, 2017 6. Lecture notes on matroid intersection One nice feature about matroids is that a simple greedy algorithm
Parallel Architecture & Programing Models for Face Recognition
 Parallel Architecture & Programing Models for Face Recognition Submitted by Sagar Kukreja Computer Engineering Department Rochester Institute of Technology Agenda Introduction to face recognition Feature
Parallel Architecture & Programing Models for Face Recognition Submitted by Sagar Kukreja Computer Engineering Department Rochester Institute of Technology Agenda Introduction to face recognition Feature
Copyright 2007 Pearson Addison-Wesley. All rights reserved. A. Levitin Introduction to the Design & Analysis of Algorithms, 2 nd ed., Ch.
 Iterative Improvement Algorithm design technique for solving optimization problems Start with a feasible solution Repeat the following step until no improvement can be found: change the current feasible
Iterative Improvement Algorithm design technique for solving optimization problems Start with a feasible solution Repeat the following step until no improvement can be found: change the current feasible
Clustering. CE-717: Machine Learning Sharif University of Technology Spring Soleymani
 Clustering CE-717: Machine Learning Sharif University of Technology Spring 2016 Soleymani Outline Clustering Definition Clustering main approaches Partitional (flat) Hierarchical Clustering validation
Clustering CE-717: Machine Learning Sharif University of Technology Spring 2016 Soleymani Outline Clustering Definition Clustering main approaches Partitional (flat) Hierarchical Clustering validation
Basic Idea. The routing problem is typically solved using a twostep
 Global Routing Basic Idea The routing problem is typically solved using a twostep approach: Global Routing Define the routing regions. Generate a tentative route for each net. Each net is assigned to a
Global Routing Basic Idea The routing problem is typically solved using a twostep approach: Global Routing Define the routing regions. Generate a tentative route for each net. Each net is assigned to a
Cache-Oblivious Traversals of an Array s Pairs
 Cache-Oblivious Traversals of an Array s Pairs Tobias Johnson May 7, 2007 Abstract Cache-obliviousness is a concept first introduced by Frigo et al. in [1]. We follow their model and develop a cache-oblivious
Cache-Oblivious Traversals of an Array s Pairs Tobias Johnson May 7, 2007 Abstract Cache-obliviousness is a concept first introduced by Frigo et al. in [1]. We follow their model and develop a cache-oblivious
CSE 258 Lecture 6. Web Mining and Recommender Systems. Community Detection
 CSE 258 Lecture 6 Web Mining and Recommender Systems Community Detection Dimensionality reduction Goal: take high-dimensional data, and describe it compactly using a small number of dimensions Assumption:
CSE 258 Lecture 6 Web Mining and Recommender Systems Community Detection Dimensionality reduction Goal: take high-dimensional data, and describe it compactly using a small number of dimensions Assumption:
CS246: Mining Massive Datasets Jure Leskovec, Stanford University
 CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu HITS (Hypertext Induced Topic Selection) Is a measure of importance of pages or documents, similar to PageRank
CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu HITS (Hypertext Induced Topic Selection) Is a measure of importance of pages or documents, similar to PageRank
Network Traffic Measurements and Analysis
 DEIB - Politecnico di Milano Fall, 2017 Introduction Often, we have only a set of features x = x 1, x 2,, x n, but no associated response y. Therefore we are not interested in prediction nor classification,
DEIB - Politecnico di Milano Fall, 2017 Introduction Often, we have only a set of features x = x 1, x 2,, x n, but no associated response y. Therefore we are not interested in prediction nor classification,
MCL. (and other clustering algorithms) 858L
 MCL (and other clustering algorithms) 858L Comparing Clustering Algorithms Brohee and van Helden (2006) compared 4 graph clustering algorithms for the task of finding protein complexes: MCODE RNSC Restricted
MCL (and other clustering algorithms) 858L Comparing Clustering Algorithms Brohee and van Helden (2006) compared 4 graph clustering algorithms for the task of finding protein complexes: MCODE RNSC Restricted
Graph Theory S 1 I 2 I 1 S 2 I 1 I 2
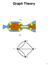 Graph Theory S I I S S I I S Graphs Definition A graph G is a pair consisting of a vertex set V (G), and an edge set E(G) ( ) V (G). x and y are the endpoints of edge e = {x, y}. They are called adjacent
Graph Theory S I I S S I I S Graphs Definition A graph G is a pair consisting of a vertex set V (G), and an edge set E(G) ( ) V (G). x and y are the endpoints of edge e = {x, y}. They are called adjacent
Extracting Information from Complex Networks
 Extracting Information from Complex Networks 1 Complex Networks Networks that arise from modeling complex systems: relationships Social networks Biological networks Distinguish from random networks uniform
Extracting Information from Complex Networks 1 Complex Networks Networks that arise from modeling complex systems: relationships Social networks Biological networks Distinguish from random networks uniform
Math 443/543 Graph Theory Notes
 Math 443/543 Graph Theory Notes David Glickenstein September 8, 2014 1 Introduction We will begin by considering several problems which may be solved using graphs, directed graphs (digraphs), and networks.
Math 443/543 Graph Theory Notes David Glickenstein September 8, 2014 1 Introduction We will begin by considering several problems which may be solved using graphs, directed graphs (digraphs), and networks.
Lecture 2 - Graph Theory Fundamentals - Reachability and Exploration 1
 CME 305: Discrete Mathematics and Algorithms Instructor: Professor Aaron Sidford (sidford@stanford.edu) January 11, 2018 Lecture 2 - Graph Theory Fundamentals - Reachability and Exploration 1 In this lecture
CME 305: Discrete Mathematics and Algorithms Instructor: Professor Aaron Sidford (sidford@stanford.edu) January 11, 2018 Lecture 2 - Graph Theory Fundamentals - Reachability and Exploration 1 In this lecture
Chapter 10. Fundamental Network Algorithms. M. E. J. Newman. May 6, M. E. J. Newman Chapter 10 May 6, / 33
 Chapter 10 Fundamental Network Algorithms M. E. J. Newman May 6, 2015 M. E. J. Newman Chapter 10 May 6, 2015 1 / 33 Table of Contents 1 Algorithms for Degrees and Degree Distributions Degree-Degree Correlation
Chapter 10 Fundamental Network Algorithms M. E. J. Newman May 6, 2015 M. E. J. Newman Chapter 10 May 6, 2015 1 / 33 Table of Contents 1 Algorithms for Degrees and Degree Distributions Degree-Degree Correlation
Singular Value Decomposition, and Application to Recommender Systems
 Singular Value Decomposition, and Application to Recommender Systems CSE 6363 Machine Learning Vassilis Athitsos Computer Science and Engineering Department University of Texas at Arlington 1 Recommendation
Singular Value Decomposition, and Application to Recommender Systems CSE 6363 Machine Learning Vassilis Athitsos Computer Science and Engineering Department University of Texas at Arlington 1 Recommendation
MITOCW ocw f99-lec12_300k
 MITOCW ocw-18.06-f99-lec12_300k This is lecture twelve. OK. We've reached twelve lectures. And this one is more than the others about applications of linear algebra. And I'll confess. When I'm giving you
MITOCW ocw-18.06-f99-lec12_300k This is lecture twelve. OK. We've reached twelve lectures. And this one is more than the others about applications of linear algebra. And I'll confess. When I'm giving you
