From Smith-Waterman to BLAST
|
|
|
- Garry Houston
- 6 years ago
- Views:
Transcription
1 From Smith-Waterman to BLAST Jeremy Buhler July 23, 2015 Smith-Waterman is the fundamental tool that we use to decide how similar two sequences are. Isn t that all that BLAST does? In principle, it is possible to build a biosequence database search tool that uses only Smith-Waterman (and associated Karlin-Altschul significance testing) to do its work. If you want to try such a program, Bill Pearson s ssearch is a good example. If you really, really want to use pure Smith-Waterman on a large database, a company named TimeLogic will sell you an application called DeCypherSW, which uses specialized hardware to make the search go faster. It costs only tens of thousands of dollars. However, most people don t want to buy specialized hardware and find that pure Smith-Waterman in software is much too slow for their search needs, and so they use BLAST or a related tool (FASTA, BLAT, etc.). There is one other important limitation of Smith-Waterman: given two sequences, it returns only one alignment between them. For genomic applications, one would of like to find all sufficiently good matches between the query and (say) a chromosome. For example, we d like to see both of the alignments in the following comparison: Query sequence Query Paralogous features in genome Genome 1
2 We may want to see all good feature matches, even if they don t all have the optimal score. Today, we ll talk about how BLAST (and friends) address the two key limitations of Smith-Waterman: speed and returning multiple hits from one database sequence. 1 Beyond Dynamic Programming? The key idea of BLAST is to do some fast preprocessing up front to strictly limit the fraction of the database that is subjected to more sensitive (but expensive) Smith-Waterman alignment. This strategy is called generate and filter. 1. Generate a list of candidate patterns that may indicate the presence of a high-scoring alignment nearby. 2. Filter the candidate list by searching for evidence of high-scoring alignments near each one. Discard candidates that do not yield such evidence. 3. Repeat step 2 as needed on the remaining candidates, using progressively more sensitive filters, until the candidate set is small enough that we can just run Smith-Waterman on each candidate. It may help to visualize this strategy as running inside Smith-Waterman s dynamic programming matrix: Initial candidates Refined candidates Final Alignments QUERY DATABASE Filtering Smith-Waterman To realize this strategy, we need to answer two questions: 1. What kind of candidate patterns should we look for? 2. How do we find the best alignment near a candidate? 2
3 2 Candidate Generation We ll describe the approach that NCBI BLASTN takes to candidate generation. What might be evidence that two sequences are locally similar? Think about DNA in particular... Observe that a high-scoring alignment between DNA sequences contains many matching bases, so it is likely to exhibit several matches in a row. Defn: a k-mer match ( word match ) between two sequences is a string of k contiguous residues that occurs somewhere in both sequences. (k = 4) atacatcactacgatcc-a agacatg--tgcaatccca We propose to treat every word match of sufficient length ( some fixed k) as a candidate. The first question to ask is: can we locate k-mer matches between two sequences more efficiently than finding high-scoring alignments? Here s a quick-and-dirty strategy that works well. First, make a list of all the k-mers in the query sequence. Then, create a table T with 4 k entries, one for each possible k-mer. If a k-mer w is present in the query, then entry T [w] lists its position(s) in the query. When searching a database D, for each position d into the database, check whether the k-mer w d = D[d..d + k 1] appears in T. If so, each query position in T [w d ] begins a k-mer match between query and the database at position d. 3
4 BLASTN works this way, except that it uses a more space-efficient data structure called a hash table to store T in space proportional to the query size, which may be much less than 4 k. In general, the build a table abstraction lets us spend time proportional to one sequence s length to construct the table, and then spend time proportional to the other sequence s length to search it. This yields an algorithm whose cost is proportional to the sum, not the product, of lengths. If we want to compare a bunch of queries to the same database, it might be faster to build a table from the k-mers in the database once and scan each query against this table, rather than scanning the whole database for each query. This is exactly what BLAT does to support large numbers of queries against the UCSC Genome Database. The table is stored in memory or on other fast storage such as an SSD. There are also much fancier, more space-efficient index structures than a simple table, such as the virtual suffix tree used by short-read alignment tools like Bowtie, but the principle is the same. Generating k-mer word matches can be done very fast! But is it any good? We have one parameter to play with the word length k. By changing this parameter, we can trade off between sensitivity (ability to find high-scoring alignments) and specificity (ability to discard sequences without such alignments). First, let s quantify specificity. Given two unrelated DNA sequences S, T, how long a match is likely to occur between them by chance alone? To make this question concrete, we assume that S and T are i.i.d. random with equal base frequencies. Pr(S[i] = T [i]) = 1 4 ( ) 1 k Pr(S[i... i + k 1] = T [j... j + k 1]) =. 4 4
5 Let M = S T, and let k = log 4 M. Then E[# matches of length = k] = i,j Pr(S[i... i + k 1] = T [j... j + k 1]) ( 1 k M 4) = M 4 log 4 M = M M = 1. For example, if we search a 1000-base pattern against a 1 gigabase database, we shouldn t be surprised to see a word match of length log bases. The number of expected matches grows exponentially as k decreases. Roughly S T /4 k k-mer matches are expected by chance alone. For the above sequence sizes, that s about 1, mer matches, 60, mer matches, or 1 million 10-mer matches. In practice, nonuniform and non-i.i.d. sequences exhibit a higher rate of chance matches. All these matches are junk they do not arise from an interesting alignment. Now, let s think about how to quantify the sensitivity of word matching. This is harder to analyze what is the frequency of k-mer matches in pairs of conserved biological sequence features? To get a rough idea, we can study the set of all alignments of some length L that contain exactly f L matching residue pairs. Such alignments have 100f percent identity. Assume that the non-matching residue pairs are distributed uniformly at random throughout the alignment. (This is actually a more challenging model than many real biological sequence alignments.) Our strategy can detect these alignments only if they contain at least one k-mer match. 5
6 How large a value of k is likely to detect most alignments? We can address this question through fancy math or via simulation. The following plot, produced by simulation, shows the probability that a random alignment of length L = 100 from the above model contains a word match, as a function of the word length. The solid line is for alignments with f = 0.8; the dashed line is for f = % identity 67% identity 0.8 probability of occurrence (L = 100) word length In practice, a match length of around k = 12 bases catches most alignments with 75-80% identity. However, to discover most matches with 67% identity, a better length would be 8-9 bases. NCBI BLASTN uses a match length of 11 bases. Hence, we expect it to find most medium- to high-similarity alignments in DNA, but almost all candidates it generates will be false positives. RepeatMasker, which tries hard to find very diverged repeat alignments, uses match lengths as small as 8 bases. BLAT, which runs super-fast but does not try very hard to find distant homologies, uses matches of 15 bases or more. Lest you think that we ve said all there is to say about word-matching algorithms, the strategy used for protein is slightly different because we can t rely on seeing many exact residue matches. Also, there are tricks we 6
7 can (must?) do to avoid generating many candidates that will lead to the same alignment. But for now, let s move on. 3 Using the Smith-Waterman Algorithm as a Filter Each candidate match indicates that we should search the dynamic programming matrix nearby for high-scoring local alignments. This idea raises some technical issues. First, what shape of region do we use when searching for alignments around a candidate? Second, how large should this region be? The third issue is a bit more subtle. Recall that Smith-Waterman only returns a single alignment (in this case, per candidate, since we run it once for each candidate). Suppose some alignment A generates a candidate, but the region we search contains both A and some other alignment A with a higher score than A. If we return only a single alignment per region, we will see A but not A. This is called the shadowing problem A is said to shadow A. Lower-scoring A is shadowed by higher-scoring A A Query A A Query A Genome Genome 7
8 Examples of shadowed features might include part of a tandem duplication, as at left, or all but one exon of a multi-exon gene, as at right. There are many ways to address these issues. I ll sketch one consistent approach, which isn t quite what NCBI BLAST uses but can be found in other BLAST-like tools. Suppose our candidate match is centered on bases S[i], T [j] in sequences S and T, respectively. We will compute an optimal local alignment between S and T that passes through entry (i, j) of the DP matrix. This is not hard to do just compute two half-anchored alignments. The first alignment A f is a best alignment starting from (i, j) and ending anywhere below and to the right of it. The second alignment A b is a best alignment ending at (i, j) and starting anywhere above and to the left of it. The desired alignment A is just the concatenation of A f with A b. A b Sequence 1 A f A Sequence 2 (Note: to compute A b, we can run the dynamic programming algorithm backwards, starting from the lower-right corner of the matrix.) The above strategy is said to pin the alignment to position (i, j). Why might it be a good idea? Pinning anchors the alignment window near the candidate. 8
9 More precisely, it nails down at least one endpoint of each of two dynamic programming problems for that candidate. Pinning addresses shadowing: nearby alignments that do not pass through the candidate will not be found. The one thing pinning doesn t tell us is: how large a region must be searched by each of the two dynamic programming problems? In theory, we could simply bound the two search regions by the ends of the two sequences. But as the following figure shows, this would cost about as much as not filtering at all! DP fill region half of matrix Moreover, if we do not limit the search region, we can have the opposite problem to shadowing. Consider aligning two sequences that contain two nearby matching features (for example, genomic fragments with two adjacent homologous exons). Seq 2 Seq 1 Junk! Seq 2 Feature A Feature B Seq
10 Suppose we generate a candidate match in the first feature and then search for high-scoring alignments passing through this candidate. An optimal alignment might subsume both features, even if it includes a completely spurious and biologically meaningless alignment of the sequence between them! In the example above, the penalty ( 39) for the junk alignment between the two features is less than the bonus (+40) for the second feature match, so the second feature s alignment is linked to that of the first feature. This is called the chaining problem. To avoid chaining and limit the size of the region to be explored by dynamic programming, we need a little more secret sauce. 4 Banded Alignment with X-Drop The key idea for bounding the search region will be to ignore alignments that are not promising. That is, don t pursue alignments that are unlikely to be the best one passing through the candidate. One heuristic that is intuitively plausible is don t consider alignments that contain very large gaps. Firstly, gaps are expensive under most common scoring systems, so an alignment with a large gap will likely have a poor score. Secondly, if we have to create a large gap to form an alignment, we should be suspicious that there is chaining going on. It would be better to stop before the gap and discover each of the two parts separately. How can we limit the size of gaps in an alignment? For any entry (i, j) in the dynamic programming matrix, call the difference j i its diagonal. Diagonals correspond (surprise!) matrix. to diagonal lines of entries in the 10
11 Our heuristic is: if an alignment is pinned to entry (i, j), do not let it pass through any entry (i, j ) for which (j i ) (j i) > b. The region of all DP entries that satisfy this constraint for a given (i, j) form a diagonal band in the matrix: 2b+1 The parameter b is called the bandwidth of the search, and it is typically at most a few tens of bases. The band itself is actually 2b + 1 entries wide. If we restrict alignments to lie completely inside the band, it is impossible to have a contiguous gap, or a series of adjacent gaps, of length more than 2b residues in one sequence. The following figure shows a few such impossible alignments for bandwidth b: > b (iꞌ,jꞌ) > b (iꞌ,jꞌ) bb 1 + bb 2 > bb b 1 b b 2 (iꞌ,jꞌ) Some alignments with jj ii jj ii > bb > 2b (iꞌ,jꞌ) 11
12 The variant of the Smith-Waterman algorithm that restricts alignments to a diagonal band is known as banded alignment. Banded alignment guarantees that the number of DP entries computed is at most proportional to the length of the shorter sequence (times b), rather than the product of the sequence lengths. Banded alignment already provides a huge speedup over the naive approach, but we can do even better. Suppose that (say) the DP search for the forward alignment A f starting from entry (i, j) has at some point discovered an entry (i, j ) with score σ. Now suppose that we compute another entry (u, v) and find that its score is < σ x. If an optimal alignment starting at (i, j) passes through (u, v), then the portion induced by the path starting at (u, v) must have score greater than x otherwise, it couldn t be optimal! A f (u,v) (i*,j*) MM ii jj = σσ MM uuuu < σσ xx If total score of A f is σ, score of this piece must be > x If x is a big number (typically a few tens for DNA alignment), we think it unlikely that an alignment scoring less than σ x will improve enough to become optimal after further extension. Moreover, if it were to improve, then we would worry about chaining the part after (u, v) might be a distinct feature. Hence, we would be justified in suppressing any alignments starting from (u, v). This observation leads to a heuristic called the X-drop. 12
13 Track the best score σ for any entry seen so far in the current dynamic programming search. (Initially, σ = 0.) If an entry (u, v) receives a score < σ x, for some value x chosen a priori, then replace this score with. Every alignment path passing through this entry will now receive score, so Smith-Waterman will never return such a path as optimal (since we can always return an empty alignment with score 0 instead!). If, when filling in the matrix, we find that every entry on a given row of the band is, then all future rows will also be, and we we can stop the alignment algorithm altogether. The combination of banded alignment and X-drop typically limits the search area for filtering to just a small region around each candidate. BLAST uses a slightly different X-drop strategy that does not rely on banding, but the basic idea is the same. 13
BIOL591: Introduction to Bioinformatics Alignment of pairs of sequences
 BIOL591: Introduction to Bioinformatics Alignment of pairs of sequences Reading in text (Mount Bioinformatics): I must confess that the treatment in Mount of sequence alignment does not seem to me a model
BIOL591: Introduction to Bioinformatics Alignment of pairs of sequences Reading in text (Mount Bioinformatics): I must confess that the treatment in Mount of sequence alignment does not seem to me a model
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1. Database Searching
 COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
BLAST. Basic Local Alignment Search Tool. Used to quickly compare a protein or DNA sequence to a database.
 BLAST Basic Local Alignment Search Tool Used to quickly compare a protein or DNA sequence to a database. There is no such thing as a free lunch BLAST is fast and highly sensitive compared to competitors.
BLAST Basic Local Alignment Search Tool Used to quickly compare a protein or DNA sequence to a database. There is no such thing as a free lunch BLAST is fast and highly sensitive compared to competitors.
Exercise 2: Browser-Based Annotation and RNA-Seq Data
 Exercise 2: Browser-Based Annotation and RNA-Seq Data Jeremy Buhler July 24, 2018 This exercise continues your introduction to practical issues in comparative annotation. You ll be annotating genomic sequence
Exercise 2: Browser-Based Annotation and RNA-Seq Data Jeremy Buhler July 24, 2018 This exercise continues your introduction to practical issues in comparative annotation. You ll be annotating genomic sequence
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014
 Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST
 An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
Data Structures and Algorithms Dr. Naveen Garg Department of Computer Science and Engineering Indian Institute of Technology, Delhi.
 Data Structures and Algorithms Dr. Naveen Garg Department of Computer Science and Engineering Indian Institute of Technology, Delhi Lecture 18 Tries Today we are going to be talking about another data
Data Structures and Algorithms Dr. Naveen Garg Department of Computer Science and Engineering Indian Institute of Technology, Delhi Lecture 18 Tries Today we are going to be talking about another data
As of August 15, 2008, GenBank contained bases from reported sequences. The search procedure should be
 48 Bioinformatics I, WS 09-10, S. Henz (script by D. Huson) November 26, 2009 4 BLAST and BLAT Outline of the chapter: 1. Heuristics for the pairwise local alignment of two sequences 2. BLAST: search and
48 Bioinformatics I, WS 09-10, S. Henz (script by D. Huson) November 26, 2009 4 BLAST and BLAT Outline of the chapter: 1. Heuristics for the pairwise local alignment of two sequences 2. BLAST: search and
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, This lecture is based on the following papers, which are all recommended reading:
 24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
BLAST, Profile, and PSI-BLAST
 BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
Running SNAP. The SNAP Team October 2012
 Running SNAP The SNAP Team October 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
Running SNAP The SNAP Team October 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio CS 466 Saurabh Sinha
 BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio. 1990. CS 466 Saurabh Sinha Motivation Sequence homology to a known protein suggest function of newly sequenced protein Bioinformatics
BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio. 1990. CS 466 Saurabh Sinha Motivation Sequence homology to a known protein suggest function of newly sequenced protein Bioinformatics
Running SNAP. The SNAP Team February 2012
 Running SNAP The SNAP Team February 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
Running SNAP The SNAP Team February 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:
 FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:56 4001 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem
FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:56 4001 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem
Brief review from last class
 Sequence Alignment Brief review from last class DNA is has direction, we will use only one (5 -> 3 ) and generate the opposite strand as needed. DNA is a 3D object (see lecture 1) but we will model it
Sequence Alignment Brief review from last class DNA is has direction, we will use only one (5 -> 3 ) and generate the opposite strand as needed. DNA is a 3D object (see lecture 1) but we will model it
Computational Molecular Biology
 Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
CS313 Exercise 4 Cover Page Fall 2017
 CS313 Exercise 4 Cover Page Fall 2017 Due by the start of class on Thursday, October 12, 2017. Name(s): In the TIME column, please estimate the time you spent on the parts of this exercise. Please try
CS313 Exercise 4 Cover Page Fall 2017 Due by the start of class on Thursday, October 12, 2017. Name(s): In the TIME column, please estimate the time you spent on the parts of this exercise. Please try
Biology 644: Bioinformatics
 Find the best alignment between 2 sequences with lengths n and m, respectively Best alignment is very dependent upon the substitution matrix and gap penalties The Global Alignment Problem tries to find
Find the best alignment between 2 sequences with lengths n and m, respectively Best alignment is very dependent upon the substitution matrix and gap penalties The Global Alignment Problem tries to find
FastA & the chaining problem
 FastA & the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 1 Sources for this lecture: Lectures by Volker Heun, Daniel Huson and Knut Reinert,
FastA & the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 1 Sources for this lecture: Lectures by Volker Heun, Daniel Huson and Knut Reinert,
Lecture Overview. Sequence search & alignment. Searching sequence databases. Sequence Alignment & Search. Goals: Motivations:
 Lecture Overview Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating
Lecture Overview Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating
Sequence Alignment & Search
 Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating the first version
Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating the first version
Algorithms in Bioinformatics: A Practical Introduction. Database Search
 Algorithms in Bioinformatics: A Practical Introduction Database Search Biological databases Biological data is double in size every 15 or 16 months Increasing in number of queries: 40,000 queries per day
Algorithms in Bioinformatics: A Practical Introduction Database Search Biological databases Biological data is double in size every 15 or 16 months Increasing in number of queries: 40,000 queries per day
Chapter 2 Basic Structure of High-Dimensional Spaces
 Chapter 2 Basic Structure of High-Dimensional Spaces Data is naturally represented geometrically by associating each record with a point in the space spanned by the attributes. This idea, although simple,
Chapter 2 Basic Structure of High-Dimensional Spaces Data is naturally represented geometrically by associating each record with a point in the space spanned by the attributes. This idea, although simple,
Bioinformatics for Biologists
 Bioinformatics for Biologists Sequence Analysis: Part I. Pairwise alignment and database searching Fran Lewitter, Ph.D. Director Bioinformatics & Research Computing Whitehead Institute Topics to Cover
Bioinformatics for Biologists Sequence Analysis: Part I. Pairwise alignment and database searching Fran Lewitter, Ph.D. Director Bioinformatics & Research Computing Whitehead Institute Topics to Cover
L4: Blast: Alignment Scores etc.
 L4: Blast: Alignment Scores etc. Why is Blast Fast? Silly Question Prove or Disprove: There are two people in New York City with exactly the same number of hairs. Large database search Database (n) Query
L4: Blast: Alignment Scores etc. Why is Blast Fast? Silly Question Prove or Disprove: There are two people in New York City with exactly the same number of hairs. Large database search Database (n) Query
Lectures by Volker Heun, Daniel Huson and Knut Reinert, in particular last years lectures
 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 4.1 Sources for this lecture Lectures by Volker Heun, Daniel Huson and Knut
4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 4.1 Sources for this lecture Lectures by Volker Heun, Daniel Huson and Knut
TCCAGGTG-GAT TGCAAGTGCG-T. Local Sequence Alignment & Heuristic Local Aligners. Review: Probabilistic Interpretation. Chance or true homology?
 Local Sequence Alignment & Heuristic Local Aligners Lectures 18 Nov 28, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall
Local Sequence Alignment & Heuristic Local Aligners Lectures 18 Nov 28, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall
Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA.
 Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA. Fasta is used to compare a protein or DNA sequence to all of the
Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA. Fasta is used to compare a protein or DNA sequence to all of the
Sequence Alignment. part 2
 Sequence Alignment part 2 Dynamic programming with more realistic scoring scheme Using the same initial sequences, we ll look at a dynamic programming example with a scoring scheme that selects for matches
Sequence Alignment part 2 Dynamic programming with more realistic scoring scheme Using the same initial sequences, we ll look at a dynamic programming example with a scoring scheme that selects for matches
Basic Local Alignment Search Tool (BLAST)
 BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
BLAST & Genome assembly
 BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies May 15, 2014 1 BLAST What is BLAST? The algorithm 2 Genome assembly De novo assembly Mapping assembly 3
BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies May 15, 2014 1 BLAST What is BLAST? The algorithm 2 Genome assembly De novo assembly Mapping assembly 3
Lecture 3: February Local Alignment: The Smith-Waterman Algorithm
 CSCI1820: Sequence Alignment Spring 2017 Lecture 3: February 7 Lecturer: Sorin Istrail Scribe: Pranavan Chanthrakumar Note: LaTeX template courtesy of UC Berkeley EECS dept. Notes are also adapted from
CSCI1820: Sequence Alignment Spring 2017 Lecture 3: February 7 Lecturer: Sorin Istrail Scribe: Pranavan Chanthrakumar Note: LaTeX template courtesy of UC Berkeley EECS dept. Notes are also adapted from
Sequence Alignment. GBIO0002 Archana Bhardwaj University of Liege
 Sequence Alignment GBIO0002 Archana Bhardwaj University of Liege 1 What is Sequence Alignment? A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity.
Sequence Alignment GBIO0002 Archana Bhardwaj University of Liege 1 What is Sequence Alignment? A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity.
Reconstructing long sequences from overlapping sequence fragment. Searching databases for related sequences and subsequences
 SEQUENCE ALIGNMENT ALGORITHMS 1 Why compare sequences? Reconstructing long sequences from overlapping sequence fragment Searching databases for related sequences and subsequences Storing, retrieving and
SEQUENCE ALIGNMENT ALGORITHMS 1 Why compare sequences? Reconstructing long sequences from overlapping sequence fragment Searching databases for related sequences and subsequences Storing, retrieving and
Bioinformatics explained: Smith-Waterman
 Bioinformatics Explained Bioinformatics explained: Smith-Waterman May 1, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com
Bioinformatics Explained Bioinformatics explained: Smith-Waterman May 1, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com
Sequence alignment theory and applications Session 3: BLAST algorithm
 Sequence alignment theory and applications Session 3: BLAST algorithm Introduction to Bioinformatics online course : IBT Sonal Henson Learning Objectives Understand the principles of the BLAST algorithm
Sequence alignment theory and applications Session 3: BLAST algorithm Introduction to Bioinformatics online course : IBT Sonal Henson Learning Objectives Understand the principles of the BLAST algorithm
FASTA. Besides that, FASTA package provides SSEARCH, an implementation of the optimal Smith- Waterman algorithm.
 FASTA INTRODUCTION Definition (by David J. Lipman and William R. Pearson in 1985) - Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence
FASTA INTRODUCTION Definition (by David J. Lipman and William R. Pearson in 1985) - Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence
Bioinformatics explained: BLAST. March 8, 2007
 Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Notes on Dynamic-Programming Sequence Alignment
 Notes on Dynamic-Programming Sequence Alignment Introduction. Following its introduction by Needleman and Wunsch (1970), dynamic programming has become the method of choice for rigorous alignment of DNA
Notes on Dynamic-Programming Sequence Alignment Introduction. Following its introduction by Needleman and Wunsch (1970), dynamic programming has become the method of choice for rigorous alignment of DNA
Alignment of Long Sequences
 Alignment of Long Sequences BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2009 Mark Craven craven@biostat.wisc.edu Pairwise Whole Genome Alignment: Task Definition Given a pair of genomes (or other large-scale
Alignment of Long Sequences BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2009 Mark Craven craven@biostat.wisc.edu Pairwise Whole Genome Alignment: Task Definition Given a pair of genomes (or other large-scale
PROTEIN MULTIPLE ALIGNMENT MOTIVATION: BACKGROUND: Marina Sirota
 Marina Sirota MOTIVATION: PROTEIN MULTIPLE ALIGNMENT To study evolution on the genetic level across a wide range of organisms, biologists need accurate tools for multiple sequence alignment of protein
Marina Sirota MOTIVATION: PROTEIN MULTIPLE ALIGNMENT To study evolution on the genetic level across a wide range of organisms, biologists need accurate tools for multiple sequence alignment of protein
CISC 636 Computational Biology & Bioinformatics (Fall 2016)
 CISC 636 Computational Biology & Bioinformatics (Fall 2016) Sequence pairwise alignment Score statistics: E-value and p-value Heuristic algorithms: BLAST and FASTA Database search: gene finding and annotations
CISC 636 Computational Biology & Bioinformatics (Fall 2016) Sequence pairwise alignment Score statistics: E-value and p-value Heuristic algorithms: BLAST and FASTA Database search: gene finding and annotations
Pairwise Sequence Alignment. Zhongming Zhao, PhD
 Pairwise Sequence Alignment Zhongming Zhao, PhD Email: zhongming.zhao@vanderbilt.edu http://bioinfo.mc.vanderbilt.edu/ Sequence Similarity match mismatch A T T A C G C G T A C C A T A T T A T G C G A T
Pairwise Sequence Alignment Zhongming Zhao, PhD Email: zhongming.zhao@vanderbilt.edu http://bioinfo.mc.vanderbilt.edu/ Sequence Similarity match mismatch A T T A C G C G T A C C A T A T T A T G C G A T
Alignments BLAST, BLAT
 Alignments BLAST, BLAT Genome Genome Gene vs Built of DNA DNA Describes Organism Protein gene Stored as Circular/ linear Single molecule, or a few of them Both (depending on the species) Part of genome
Alignments BLAST, BLAT Genome Genome Gene vs Built of DNA DNA Describes Organism Protein gene Stored as Circular/ linear Single molecule, or a few of them Both (depending on the species) Part of genome
B L A S T! BLAST: Basic local alignment search tool. Copyright notice. February 6, Pairwise alignment: key points. Outline of tonight s lecture
 February 6, 2008 BLAST: Basic local alignment search tool B L A S T! Jonathan Pevsner, Ph.D. Introduction to Bioinformatics pevsner@jhmi.edu 4.633.0 Copyright notice Many of the images in this powerpoint
February 6, 2008 BLAST: Basic local alignment search tool B L A S T! Jonathan Pevsner, Ph.D. Introduction to Bioinformatics pevsner@jhmi.edu 4.633.0 Copyright notice Many of the images in this powerpoint
Dynamic Programming. Lecture Overview Introduction
 Lecture 12 Dynamic Programming 12.1 Overview Dynamic Programming is a powerful technique that allows one to solve many different types of problems in time O(n 2 ) or O(n 3 ) for which a naive approach
Lecture 12 Dynamic Programming 12.1 Overview Dynamic Programming is a powerful technique that allows one to solve many different types of problems in time O(n 2 ) or O(n 3 ) for which a naive approach
Tutorial 1: Exploring the UCSC Genome Browser
 Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 04: Variations of sequence alignments http://www.pitt.edu/~mcs2/teaching/biocomp/tutorials/global.html Slides adapted from Dr. Shaojie Zhang (University
EECS730: Introduction to Bioinformatics Lecture 04: Variations of sequence alignments http://www.pitt.edu/~mcs2/teaching/biocomp/tutorials/global.html Slides adapted from Dr. Shaojie Zhang (University
Mouse, Human, Chimpanzee
 More Alignments 1 Mouse, Human, Chimpanzee Mouse to Human Chimpanzee to Human 2 Mouse v.s. Human Chromosome X of Mouse to Human 3 Local Alignment Given: two sequences S and T Find: substrings of S and
More Alignments 1 Mouse, Human, Chimpanzee Mouse to Human Chimpanzee to Human 2 Mouse v.s. Human Chromosome X of Mouse to Human 3 Local Alignment Given: two sequences S and T Find: substrings of S and
BLAST & Genome assembly
 BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies November 17, 2012 1 Introduction Introduction 2 BLAST What is BLAST? The algorithm 3 Genome assembly De
BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies November 17, 2012 1 Introduction Introduction 2 BLAST What is BLAST? The algorithm 3 Genome assembly De
Principles of Bioinformatics. BIO540/STA569/CSI660 Fall 2010
 Principles of Bioinformatics BIO540/STA569/CSI660 Fall 2010 Lecture 11 Multiple Sequence Alignment I Administrivia Administrivia The midterm examination will be Monday, October 18 th, in class. Closed
Principles of Bioinformatics BIO540/STA569/CSI660 Fall 2010 Lecture 11 Multiple Sequence Alignment I Administrivia Administrivia The midterm examination will be Monday, October 18 th, in class. Closed
USING AN EXTENDED SUFFIX TREE TO SPEED-UP SEQUENCE ALIGNMENT
 IADIS International Conference Applied Computing 2006 USING AN EXTENDED SUFFIX TREE TO SPEED-UP SEQUENCE ALIGNMENT Divya R. Singh Software Engineer Microsoft Corporation, Redmond, WA 98052, USA Abdullah
IADIS International Conference Applied Computing 2006 USING AN EXTENDED SUFFIX TREE TO SPEED-UP SEQUENCE ALIGNMENT Divya R. Singh Software Engineer Microsoft Corporation, Redmond, WA 98052, USA Abdullah
In this section we describe how to extend the match refinement to the multiple case and then use T-Coffee to heuristically compute a multiple trace.
 5 Multiple Match Refinement and T-Coffee In this section we describe how to extend the match refinement to the multiple case and then use T-Coffee to heuristically compute a multiple trace. This exposition
5 Multiple Match Refinement and T-Coffee In this section we describe how to extend the match refinement to the multiple case and then use T-Coffee to heuristically compute a multiple trace. This exposition
NGS Data and Sequence Alignment
 Applications and Servers SERVER/REMOTE Compute DB WEB Data files NGS Data and Sequence Alignment SSH WEB SCP Manpreet S. Katari App Aug 11, 2016 Service Terminal IGV Data files Window Personal Computer/Local
Applications and Servers SERVER/REMOTE Compute DB WEB Data files NGS Data and Sequence Alignment SSH WEB SCP Manpreet S. Katari App Aug 11, 2016 Service Terminal IGV Data files Window Personal Computer/Local
A BANDED SMITH-WATERMAN FPGA ACCELERATOR FOR MERCURY BLASTP
 A BANDED SITH-WATERAN FPGA ACCELERATOR FOR ERCURY BLASTP Brandon Harris*, Arpith C. Jacob*, Joseph. Lancaster*, Jeremy Buhler*, Roger D. Chamberlain* *Dept. of Computer Science and Engineering, Washington
A BANDED SITH-WATERAN FPGA ACCELERATOR FOR ERCURY BLASTP Brandon Harris*, Arpith C. Jacob*, Joseph. Lancaster*, Jeremy Buhler*, Roger D. Chamberlain* *Dept. of Computer Science and Engineering, Washington
The levels of a memory hierarchy. Main. Memory. 500 By 1MB 4GB 500GB 0.25 ns 1ns 20ns 5ms
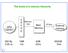 The levels of a memory hierarchy CPU registers C A C H E Memory bus Main Memory I/O bus External memory 500 By 1MB 4GB 500GB 0.25 ns 1ns 20ns 5ms 1 1 Some useful definitions When the CPU finds a requested
The levels of a memory hierarchy CPU registers C A C H E Memory bus Main Memory I/O bus External memory 500 By 1MB 4GB 500GB 0.25 ns 1ns 20ns 5ms 1 1 Some useful definitions When the CPU finds a requested
Lecture 1: Overview
 15-150 Lecture 1: Overview Lecture by Stefan Muller May 21, 2018 Welcome to 15-150! Today s lecture was an overview that showed the highlights of everything you re learning this semester, which also meant
15-150 Lecture 1: Overview Lecture by Stefan Muller May 21, 2018 Welcome to 15-150! Today s lecture was an overview that showed the highlights of everything you re learning this semester, which also meant
Acceleration of Ungapped Extension in Mercury BLAST. Joseph Lancaster Jeremy Buhler Roger Chamberlain
 Acceleration of Ungapped Extension in Mercury BLAST Joseph Lancaster Jeremy Buhler Roger Chamberlain Joseph Lancaster, Jeremy Buhler, and Roger Chamberlain, Acceleration of Ungapped Extension in Mercury
Acceleration of Ungapped Extension in Mercury BLAST Joseph Lancaster Jeremy Buhler Roger Chamberlain Joseph Lancaster, Jeremy Buhler, and Roger Chamberlain, Acceleration of Ungapped Extension in Mercury
15-451/651: Design & Analysis of Algorithms January 26, 2015 Dynamic Programming I last changed: January 28, 2015
 15-451/651: Design & Analysis of Algorithms January 26, 2015 Dynamic Programming I last changed: January 28, 2015 Dynamic Programming is a powerful technique that allows one to solve many different types
15-451/651: Design & Analysis of Algorithms January 26, 2015 Dynamic Programming I last changed: January 28, 2015 Dynamic Programming is a powerful technique that allows one to solve many different types
Database Searching Using BLAST
 Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Genome 373: Mapping Short Sequence Reads I. Doug Fowler
 Genome 373: Mapping Short Sequence Reads I Doug Fowler Two different strategies for parallel amplification BRIDGE PCR EMULSION PCR Two different strategies for parallel amplification BRIDGE PCR EMULSION
Genome 373: Mapping Short Sequence Reads I Doug Fowler Two different strategies for parallel amplification BRIDGE PCR EMULSION PCR Two different strategies for parallel amplification BRIDGE PCR EMULSION
Preliminary Syllabus. Genomics. Introduction & Genome Assembly Sequence Comparison Gene Modeling Gene Function Identification
 Preliminary Syllabus Sep 30 Oct 2 Oct 7 Oct 9 Oct 14 Oct 16 Oct 21 Oct 25 Oct 28 Nov 4 Nov 8 Introduction & Genome Assembly Sequence Comparison Gene Modeling Gene Function Identification OCTOBER BREAK
Preliminary Syllabus Sep 30 Oct 2 Oct 7 Oct 9 Oct 14 Oct 16 Oct 21 Oct 25 Oct 28 Nov 4 Nov 8 Introduction & Genome Assembly Sequence Comparison Gene Modeling Gene Function Identification OCTOBER BREAK
A Study On Pair-Wise Local Alignment Of Protein Sequence For Identifying The Structural Similarity
 A Study On Pair-Wise Local Alignment Of Protein Sequence For Identifying The Structural Similarity G. Pratyusha, Department of Computer Science & Engineering, V.R.Siddhartha Engineering College(Autonomous)
A Study On Pair-Wise Local Alignment Of Protein Sequence For Identifying The Structural Similarity G. Pratyusha, Department of Computer Science & Engineering, V.R.Siddhartha Engineering College(Autonomous)
CAP BLAST. BIOINFORMATICS Su-Shing Chen CISE. 8/20/2005 Su-Shing Chen, CISE 1
 CAP 5510-6 BLAST BIOINFORMATICS Su-Shing Chen CISE 8/20/2005 Su-Shing Chen, CISE 1 BLAST Basic Local Alignment Prof Search Su-Shing Chen Tool A Fast Pair-wise Alignment and Database Searching Tool 8/20/2005
CAP 5510-6 BLAST BIOINFORMATICS Su-Shing Chen CISE 8/20/2005 Su-Shing Chen, CISE 1 BLAST Basic Local Alignment Prof Search Su-Shing Chen Tool A Fast Pair-wise Alignment and Database Searching Tool 8/20/2005
Worst-case running time for RANDOMIZED-SELECT
 Worst-case running time for RANDOMIZED-SELECT is ), even to nd the minimum The algorithm has a linear expected running time, though, and because it is randomized, no particular input elicits the worst-case
Worst-case running time for RANDOMIZED-SELECT is ), even to nd the minimum The algorithm has a linear expected running time, though, and because it is randomized, no particular input elicits the worst-case
Darwin-WGA. A Co-processor Provides Increased Sensitivity in Whole Genome Alignments with High Speedup
 Darwin-WGA A Co-processor Provides Increased Sensitivity in Whole Genome Alignments with High Speedup Yatish Turakhia*, Sneha D. Goenka*, Prof. Gill Bejerano, Prof. William J. Dally * Equal contribution
Darwin-WGA A Co-processor Provides Increased Sensitivity in Whole Genome Alignments with High Speedup Yatish Turakhia*, Sneha D. Goenka*, Prof. Gill Bejerano, Prof. William J. Dally * Equal contribution
Hashing and sketching
 Hashing and sketching 1 The age of big data An age of big data is upon us, brought on by a combination of: Pervasive sensing: so much of what goes on in our lives and in the world at large is now digitally
Hashing and sketching 1 The age of big data An age of big data is upon us, brought on by a combination of: Pervasive sensing: so much of what goes on in our lives and in the world at large is now digitally
Long Read RNA-seq Mapper
 UNIVERSITY OF ZAGREB FACULTY OF ELECTRICAL ENGENEERING AND COMPUTING MASTER THESIS no. 1005 Long Read RNA-seq Mapper Josip Marić Zagreb, February 2015. Table of Contents 1. Introduction... 1 2. RNA Sequencing...
UNIVERSITY OF ZAGREB FACULTY OF ELECTRICAL ENGENEERING AND COMPUTING MASTER THESIS no. 1005 Long Read RNA-seq Mapper Josip Marić Zagreb, February 2015. Table of Contents 1. Introduction... 1 2. RNA Sequencing...
Project and Production Management Prof. Arun Kanda Department of Mechanical Engineering Indian Institute of Technology, Delhi
 Project and Production Management Prof. Arun Kanda Department of Mechanical Engineering Indian Institute of Technology, Delhi Lecture - 8 Consistency and Redundancy in Project networks In today s lecture
Project and Production Management Prof. Arun Kanda Department of Mechanical Engineering Indian Institute of Technology, Delhi Lecture - 8 Consistency and Redundancy in Project networks In today s lecture
Dynamic Programming Part I: Examples. Bioinfo I (Institut Pasteur de Montevideo) Dynamic Programming -class4- July 25th, / 77
 Dynamic Programming Part I: Examples Bioinfo I (Institut Pasteur de Montevideo) Dynamic Programming -class4- July 25th, 2011 1 / 77 Dynamic Programming Recall: the Change Problem Other problems: Manhattan
Dynamic Programming Part I: Examples Bioinfo I (Institut Pasteur de Montevideo) Dynamic Programming -class4- July 25th, 2011 1 / 77 Dynamic Programming Recall: the Change Problem Other problems: Manhattan
Multiple Sequence Alignment: Multidimensional. Biological Motivation
 Multiple Sequence Alignment: Multidimensional Dynamic Programming Boston University Biological Motivation Compare a new sequence with the sequences in a protein family. Proteins can be categorized into
Multiple Sequence Alignment: Multidimensional Dynamic Programming Boston University Biological Motivation Compare a new sequence with the sequences in a protein family. Proteins can be categorized into
Data Mining Technologies for Bioinformatics Sequences
 Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it.
 Sequence Alignments Overview Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it. Sequence alignment means arranging
Sequence Alignments Overview Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it. Sequence alignment means arranging
Introduction to BLAST with Protein Sequences. Utah State University Spring 2014 STAT 5570: Statistical Bioinformatics Notes 6.2
 Introduction to BLAST with Protein Sequences Utah State University Spring 2014 STAT 5570: Statistical Bioinformatics Notes 6.2 1 References Chapter 2 of Biological Sequence Analysis (Durbin et al., 2001)
Introduction to BLAST with Protein Sequences Utah State University Spring 2014 STAT 5570: Statistical Bioinformatics Notes 6.2 1 References Chapter 2 of Biological Sequence Analysis (Durbin et al., 2001)
Highly Scalable and Accurate Seeds for Subsequence Alignment
 Highly Scalable and Accurate Seeds for Subsequence Alignment Abhijit Pol Tamer Kahveci Department of Computer and Information Science and Engineering, University of Florida, Gainesville, FL, USA, 32611
Highly Scalable and Accurate Seeds for Subsequence Alignment Abhijit Pol Tamer Kahveci Department of Computer and Information Science and Engineering, University of Florida, Gainesville, FL, USA, 32611
CS246: Mining Massive Datasets Jure Leskovec, Stanford University
 CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu 2/24/2014 Jure Leskovec, Stanford CS246: Mining Massive Datasets, http://cs246.stanford.edu 2 High dim. data
CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu 2/24/2014 Jure Leskovec, Stanford CS246: Mining Massive Datasets, http://cs246.stanford.edu 2 High dim. data
Research Article International Journals of Advanced Research in Computer Science and Software Engineering ISSN: X (Volume-7, Issue-6)
 International Journals of Advanced Research in Computer Science and Software Engineering ISSN: 77-18X (Volume-7, Issue-6) Research Article June 017 DDGARM: Dotlet Driven Global Alignment with Reduced Matrix
International Journals of Advanced Research in Computer Science and Software Engineering ISSN: 77-18X (Volume-7, Issue-6) Research Article June 017 DDGARM: Dotlet Driven Global Alignment with Reduced Matrix
BLAST MCDB 187. Friday, February 8, 13
 BLAST MCDB 187 BLAST Basic Local Alignment Sequence Tool Uses shortcut to compute alignments of a sequence against a database very quickly Typically takes about a minute to align a sequence against a database
BLAST MCDB 187 BLAST Basic Local Alignment Sequence Tool Uses shortcut to compute alignments of a sequence against a database very quickly Typically takes about a minute to align a sequence against a database
CS5112: Algorithms and Data Structures for Applications
 CS5112: Algorithms and Data Structures for Applications Lecture 3: Hashing Ramin Zabih Some figures from Wikipedia/Google image search Administrivia Web site is: https://github.com/cornelltech/cs5112-f18
CS5112: Algorithms and Data Structures for Applications Lecture 3: Hashing Ramin Zabih Some figures from Wikipedia/Google image search Administrivia Web site is: https://github.com/cornelltech/cs5112-f18
CS 284A: Algorithms for Computational Biology Notes on Lecture: BLAST. The statistics of alignment scores.
 CS 284A: Algorithms for Computational Biology Notes on Lecture: BLAST. The statistics of alignment scores. prepared by Oleksii Kuchaiev, based on presentation by Xiaohui Xie on February 20th. 1 Introduction
CS 284A: Algorithms for Computational Biology Notes on Lecture: BLAST. The statistics of alignment scores. prepared by Oleksii Kuchaiev, based on presentation by Xiaohui Xie on February 20th. 1 Introduction
BLAST. NCBI BLAST Basic Local Alignment Search Tool
 BLAST NCBI BLAST Basic Local Alignment Search Tool http://www.ncbi.nlm.nih.gov/blast/ Global versus local alignments Global alignments: Attempt to align every residue in every sequence, Most useful when
BLAST NCBI BLAST Basic Local Alignment Search Tool http://www.ncbi.nlm.nih.gov/blast/ Global versus local alignments Global alignments: Attempt to align every residue in every sequence, Most useful when
Combinatorial Pattern Matching. CS 466 Saurabh Sinha
 Combinatorial Pattern Matching CS 466 Saurabh Sinha Genomic Repeats Example of repeats: ATGGTCTAGGTCCTAGTGGTC Motivation to find them: Genomic rearrangements are often associated with repeats Trace evolutionary
Combinatorial Pattern Matching CS 466 Saurabh Sinha Genomic Repeats Example of repeats: ATGGTCTAGGTCCTAGTGGTC Motivation to find them: Genomic rearrangements are often associated with repeats Trace evolutionary
6. Relational Algebra (Part II)
 6. Relational Algebra (Part II) 6.1. Introduction In the previous chapter, we introduced relational algebra as a fundamental model of relational database manipulation. In particular, we defined and discussed
6. Relational Algebra (Part II) 6.1. Introduction In the previous chapter, we introduced relational algebra as a fundamental model of relational database manipulation. In particular, we defined and discussed
Introduction to Computational Molecular Biology
 18.417 Introduction to Computational Molecular Biology Lecture 13: October 21, 2004 Scribe: Eitan Reich Lecturer: Ross Lippert Editor: Peter Lee 13.1 Introduction We have been looking at algorithms to
18.417 Introduction to Computational Molecular Biology Lecture 13: October 21, 2004 Scribe: Eitan Reich Lecturer: Ross Lippert Editor: Peter Lee 13.1 Introduction We have been looking at algorithms to
Similarity Searches on Sequence Databases
 Similarity Searches on Sequence Databases Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Zürich, October 2004 Swiss Institute of Bioinformatics Swiss EMBnet node Outline Importance of
Similarity Searches on Sequence Databases Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Zürich, October 2004 Swiss Institute of Bioinformatics Swiss EMBnet node Outline Importance of
Algorithmic Approaches for Biological Data, Lecture #20
 Algorithmic Approaches for Biological Data, Lecture #20 Katherine St. John City University of New York American Museum of Natural History 20 April 2016 Outline Aligning with Gaps and Substitution Matrices
Algorithmic Approaches for Biological Data, Lecture #20 Katherine St. John City University of New York American Museum of Natural History 20 April 2016 Outline Aligning with Gaps and Substitution Matrices
C E N T R. Introduction to bioinformatics 2007 E B I O I N F O R M A T I C S V U F O R I N T. Lecture 13 G R A T I V. Iterative homology searching,
 C E N T R E F O R I N T E G R A T I V E B I O I N F O R M A T I C S V U Introduction to bioinformatics 2007 Lecture 13 Iterative homology searching, PSI (Position Specific Iterated) BLAST basic idea use
C E N T R E F O R I N T E G R A T I V E B I O I N F O R M A T I C S V U Introduction to bioinformatics 2007 Lecture 13 Iterative homology searching, PSI (Position Specific Iterated) BLAST basic idea use
Genome 559: Introduction to Statistical and Computational Genomics. Lecture15a Multiple Sequence Alignment Larry Ruzzo
 Genome 559: Introduction to Statistical and Computational Genomics Lecture15a Multiple Sequence Alignment Larry Ruzzo 1 Multiple Alignment: Motivations Common structure, function, or origin may be only
Genome 559: Introduction to Statistical and Computational Genomics Lecture15a Multiple Sequence Alignment Larry Ruzzo 1 Multiple Alignment: Motivations Common structure, function, or origin may be only
Lecture 5: Multiple sequence alignment
 Lecture 5: Multiple sequence alignment Introduction to Computational Biology Teresa Przytycka, PhD (with some additions by Martin Vingron) Why do we need multiple sequence alignment Pairwise sequence alignment
Lecture 5: Multiple sequence alignment Introduction to Computational Biology Teresa Przytycka, PhD (with some additions by Martin Vingron) Why do we need multiple sequence alignment Pairwise sequence alignment
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Rochester Institute of Technology. Making personalized education scalable using Sequence Alignment Algorithm
 Rochester Institute of Technology Making personalized education scalable using Sequence Alignment Algorithm Submitted by: Lakhan Bhojwani Advisor: Dr. Carlos Rivero 1 1. Abstract There are many ways proposed
Rochester Institute of Technology Making personalized education scalable using Sequence Alignment Algorithm Submitted by: Lakhan Bhojwani Advisor: Dr. Carlos Rivero 1 1. Abstract There are many ways proposed
Test Requirement Catalog. Generic Clues, Developer Version
 Version 4..0 PART 1: DATA TYPES Test Requirement Catalog SIMPLE DATA TYPES... 4 BOOLEAN... 4 CASES... 4 COUNTS... 5 INTERVALS... 5 [LOW, HIGH] an interval that contains both LOW and HIGH... 6 [LOW, HIGH)
Version 4..0 PART 1: DATA TYPES Test Requirement Catalog SIMPLE DATA TYPES... 4 BOOLEAN... 4 CASES... 4 COUNTS... 5 INTERVALS... 5 [LOW, HIGH] an interval that contains both LOW and HIGH... 6 [LOW, HIGH)
CS246: Mining Massive Datasets Jure Leskovec, Stanford University
 CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu 3/6/2012 Jure Leskovec, Stanford CS246: Mining Massive Datasets, http://cs246.stanford.edu 2 In many data mining
CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu 3/6/2012 Jure Leskovec, Stanford CS246: Mining Massive Datasets, http://cs246.stanford.edu 2 In many data mining
CS246: Mining Massive Datasets Jure Leskovec, Stanford University
 CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu 2/25/2013 Jure Leskovec, Stanford CS246: Mining Massive Datasets, http://cs246.stanford.edu 3 In many data mining
CS246: Mining Massive Datasets Jure Leskovec, Stanford University http://cs246.stanford.edu 2/25/2013 Jure Leskovec, Stanford CS246: Mining Massive Datasets, http://cs246.stanford.edu 3 In many data mining
Today s Lecture. Edit graph & alignment algorithms. Local vs global Computational complexity of pairwise alignment Multiple sequence alignment
 Today s Lecture Edit graph & alignment algorithms Smith-Waterman algorithm Needleman-Wunsch algorithm Local vs global Computational complexity of pairwise alignment Multiple sequence alignment 1 Sequence
Today s Lecture Edit graph & alignment algorithms Smith-Waterman algorithm Needleman-Wunsch algorithm Local vs global Computational complexity of pairwise alignment Multiple sequence alignment 1 Sequence
GSNAP: Fast and SNP-tolerant detection of complex variants and splicing in short reads by Thomas D. Wu and Serban Nacu
 GSNAP: Fast and SNP-tolerant detection of complex variants and splicing in short reads by Thomas D. Wu and Serban Nacu Matt Huska Freie Universität Berlin Computational Methods for High-Throughput Omics
GSNAP: Fast and SNP-tolerant detection of complex variants and splicing in short reads by Thomas D. Wu and Serban Nacu Matt Huska Freie Universität Berlin Computational Methods for High-Throughput Omics
BLAST Exercise 2: Using mrna and EST Evidence in Annotation Adapted by W. Leung and SCR Elgin from Annotation Using mrna and ESTs by Dr. J.
 BLAST Exercise 2: Using mrna and EST Evidence in Annotation Adapted by W. Leung and SCR Elgin from Annotation Using mrna and ESTs by Dr. J. Buhler Prerequisites: BLAST Exercise: Detecting and Interpreting
BLAST Exercise 2: Using mrna and EST Evidence in Annotation Adapted by W. Leung and SCR Elgin from Annotation Using mrna and ESTs by Dr. J. Buhler Prerequisites: BLAST Exercise: Detecting and Interpreting
Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD
 Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD Assumptions: Biological sequences evolved by evolution. Micro scale changes: For short sequences (e.g. one
Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD Assumptions: Biological sequences evolved by evolution. Micro scale changes: For short sequences (e.g. one
6.338 Final Paper: Parallel Huffman Encoding and Move to Front Encoding in Julia
 6.338 Final Paper: Parallel Huffman Encoding and Move to Front Encoding in Julia Gil Goldshlager December 2015 1 Introduction 1.1 Background The Burrows-Wheeler transform (BWT) is a string transform used
6.338 Final Paper: Parallel Huffman Encoding and Move to Front Encoding in Julia Gil Goldshlager December 2015 1 Introduction 1.1 Background The Burrows-Wheeler transform (BWT) is a string transform used
A Scalable Coprocessor for Bioinformatic Sequence Alignments
 A Scalable Coprocessor for Bioinformatic Sequence Alignments Scott F. Smith Department of Electrical and Computer Engineering Boise State University Boise, ID, U.S.A. Abstract A hardware coprocessor for
A Scalable Coprocessor for Bioinformatic Sequence Alignments Scott F. Smith Department of Electrical and Computer Engineering Boise State University Boise, ID, U.S.A. Abstract A hardware coprocessor for
