The lmekin function. Terry Therneau Mayo Clinic. May 11, 2018
|
|
|
- Kelly Jacobs
- 5 years ago
- Views:
Transcription
1 The lmekin function Terry Therneau Mayo Clinic May 11, Background The original kinship library had an implementation of linear mixed effects models using the matrix code found in coxme. Since the primary motivation for the functions in that library was to fit models with random family effects, i.e., using a kinship matrix for the correlation, the name lmekin was chosen. The reason for the program was entirely to check our arithmetic: the result of the matrix manipulations contained in it should give exactly the same answer as lme, and since the underlying routines were shared with coxme that gave a validity check for parts of coxme. With more time and a larger test suite the routine is no longer necessary for this purpose, however, it became popular with users (they often do unanticipated things) since it can fit a few models that lme cannot. Let me emphasis this: most models that can be fit with the lmekin function can also be fit with lme and/or lmer. For any such model the lme/lmer functions will be faster and have superior support routines (residuals, printing, plotting, etc.) The solution code for lmer is likely also more reliable since it has been exercised on a much wider variety of data sets. However, there are models that lmekin will fit which lme will not. The most obvious of these are models with a random genetic effect, e.g. a kinship matrix. The second class will be models for which the user has written their own variance extension, as described in the variance vignette. The follow-up methods for lmekin are limited, which reflects the fact that linear mixed effects models are not a primary focus for me, the author of the coxme package. A primary reason to update lmekin at all is a desire to depreciate the original kinship package; this routine was the last bit of functionality that is not otherwise available. The set of models fit by lmekin was also extended to include all of the random effects structures supported by coxme, which should make the routine more valuable. Contributions by others with deeper interest will be warmly received. Nevertheless, the core code is solid and reliable to the best of my ability and will be actively maintained. 2 Simple Models The control code for lmekin is identical to coxme with respect to specifying the random effects, and both are modeled on the methods used in lmer. Here is a simple example using one of the data sets from Pinheiro and Bates. 1
2 > library(coxme) > require(nlme) > fit1 <- lme(effort~type, random= ~ 1 Subject,data=ergoStool, method="ml") > fit2 <- lmekin(effort ~ Type + (1 Subject), data=ergostool, method="ml") > print(fit1) Linear mixed-effects model fit by maximum likelihood Data: ergostool Log-likelihood: Fixed: effort ~ Type (Intercept) TypeT2 TypeT3 TypeT : Formula: ~1 Subject (Intercept) Residual StdDev: Number of Observations: 36 Number of Groups: 9 > print(fit2) Data: ergostool Log-likelihood = n= 36 Model: effort ~ Type + (1 Subject) (Intercept) e+00 TypeT e-15 TypeT e-06 TypeT e-01 Group Variable Std Dev Variance Subject Intercept Residual error= And here is a slightly more complex one based on data from J. Cortinas [2]. There are 37 centers of varying size, and the simulated data set has both random intercepts and treatment effects per center. 2
3 > tdata <-eortc > tdata$center2 <- factor(tdata$center) > fit3 <- lme(y ~ trt, random= ~ trt center2, data=tdata, method="ml") > fit3 Linear mixed-effects model fit by maximum likelihood Data: tdata Log-likelihood: Fixed: y ~ trt (Intercept) trt : Formula: ~trt center2 Structure: General positive-definite, Log-Cholesky parametrization StdDev Corr (Intercept) (Intr) trt Residual Number of Observations: 2323 Number of Groups: 37 > fit4 <- lmekin(y ~ trt + (1+ trt center), tdata) > fit4 Data: tdata Log-likelihood = n= 2323 Model: y ~ trt + (1 + trt center) (Intercept) trt Group Variable Std Dev Variance Corr center Intercept e e e+08 trt e e+04 Residual error= > all.equal(fit3$loglik, fit4$loglik) [1] TRUE 3
4 First note that the two fits give identical log-likelihoods, even though the coefficients differ. The log-likelihood function is somewhat flat on top, and because of different default starting estimates the two programs do not end up at exactly the same place. One small difference above is that lmekin is a little more forgiving with respect to groups. The center variable in the eortc data set is numeric, when it appears on the right hand side of the vertical bar (1 + trt center) the program assumes it is a grouping effect. The lme routine insists that the grouping variable be a factor. (In defense of lme, if one were to accidentally put a continuous variable on the right such as age, which has no business being there, the error message is welcome.) A more important difference from lme (and lmer) is the inclusion of random intercepts. In lmer a random term like (age group will actually fit the model (1+age group), i.e., an intercept term is assumed unless it is specifically removed by adding -1 to the model. In lmekin an intercept is not assumed, the random effect you type is the one that you get. The primary reason for this is that lmer mimics lm, which also adds an intercept unless it is explicitly suppressed. The coxme function mimics coxph, which does not add an intercept. Since lmekin is built on the same routines as coxme it also follows that convention. (In Cox models there is not an intercept term for the fixed effects since this is absorbed into the baseline hazard). 3 GAW example The following examples use data from one of the Genetic Analaysis Workshops (I don t remeber which year). First read in the saved data, create the pedigrees, and create the kinship matrix. Figure 1 shows a plot of the smallest of the 23 families in the file. > require(kinship2) > load("gaw.rda") > gped <- with(gdata, pedigree(id, father, mother, sex=sex, famid=famid)) > kmat <- kinship(gped) > plot(gped[9]) > gfit0 <- lm(age ~ q1, gdata) > summary(gfit0) Call: lm(formula = age ~ q1, data = gdata) Residuals: Min 1Q Median 3Q Max Coefficients: Estimate Std. Error t value Pr(> t ) (Intercept) q <2e-16 Residual standard error: on 998 degrees of freedom (497 observations deleted due to missingness) 4
5 Figure 1: Pedigree 9 from the GAW data. 5
6 Multiple R-squared: , Adjusted R-squared: F-statistic: on 1 and 998 DF, p-value: < 2.2e-16 > gfit1 <- lmekin(age ~ q1 + (1 id), data=gdata, varlist=kmat*2) > gfit1 Data: gdata Log-likelihood = n=1000 (497 observations deleted due to missingness) Model: age ~ q1 + (1 id) (Intercept) q Group Variable Std Dev Variance id Vmat Residual error= The fit predicts age at onset using one quantitative trait along with a familial affect. The residual error is decreased when we include a familial effect, and the familial effects is substantial. The kinship matrix has diagonal elements of.5 (if there is no inbreeding); it is traditional to use a scaled version with elements of 1 in genetics models. A next step is to look at the effect of a particular locus. The saved rda file also contains the results of a single SOLAR run at locus 6.90 along with the pedindex file created by SOLAR. We need to convert these into sparse matrix form, and add appropriate labels. (When there are kinship or ibd matrices, the coxme routine uses the matrix labels to match the proper row/col to the proper subject). The SOLAR package may reorder subjects in the data set; the pedindex matrix contains the new subject and family numbers in colums 1 and 6, and the original family and subject values in the last two columns. In this data set each subject has a unique identifier, so we do not need to include the family id in the matrix dimnames to obtain correct matches. > sid <- pedindex[,9] > ibd6.90 <- with(solar6.90, sparsematrix(id2, id1, x=x, symmetric=true, dimnames=list(sid, sid))) > gfit2 <- lmekin(age ~ (1 id), data=gdata, varlist= list(kmat, ibd6.90)) > print(gfit2) Data: gdata Log-likelihood = n=1000 (497 observations deleted due to missingness) 6
7 Model: age ~ (1 id) (Intercept) Group Variable Std Dev Variance id Vmat Vmat Residual error= The specific effect is modest for this locus: it partitions the familial effect found above into about 1/3 locus specific and 2/3 multifactorial. Another possible fit is to assume a common environmental effect for each family. (For pedigrees this large I have serious doubts about the relevance of the model below, but it serves as an illustration). When there are multiple random terms the varlist argument is matched up to them one by one, with the default choice used for any remaining, so in the model below the first will be a kinship effect and the second an uncorrelated random intercept per family. > gfit3 <- lmekin(age ~ q1 + (1 id) + (1 famid), data=gdata, varlist=kmat) > gfit3 Data: gdata Log-likelihood = n=1000 (497 observations deleted due to missingness) Model: age ~ q1 + (1 id) + (1 famid) (Intercept) q Group Variable Std Dev Variance id Vmat famid Intercept Residual error= If one wanted to be specific the above model could be written as below, to identify the actual variance functions used for each. > lmekin(age ~ q1 + (1 id) + (1 famid), data=gdata, varlist=list(coxmemlist(kmat), coxmefull)) 7
8 4 Computation The random effects linear model is y = Xβ + Zb + ɛ (1) b N(0, σ 2 A(θ) (2) ɛ = N(0, σ 2 ) (3) Here β are the fixed and b the random coefficients, and the variance matrix A of the random effects depends on some arbitrary vector of parameters θ. For any fixed value of θ the solution for the remaining parameters is based on a QR decomposition, exactly as is laid out in section 2.2 of Pinheiro and Bates ([1]), leading also a profile likelihood value L(θ). For known A, this is solved as an augmented least squares problem with ( ) ( ) ( ) y y = X X = Z Z = 0 0 where = A 1. The dummy rows of data have y = 0, X = 0 and as the predictor variables. With known, this gives the solution to all the other parameters as an ordinary least squares problem, which is solved using a QR decompostion. The Z matrix is often sparse, so the QR computations are done using the Matrix library to take advantage of this. Maximization of L(θ) with respect to θ is accomplished with the optim() function. Thus, during the solution process A will contain relative variances for components of b, something that Pinheiro and Bates refer to as the precision matrix. When the results of a fit are printed out A is multiplied by σ 2 to give the variance of b directly. This decomposition will be invisible to most users, unless they either set initial values or retrieve variances directly from the coxme object. Initial values are for the parameters θ of A, and the results of the VarCorr function will also be terms of θ, not multiplied by the residual variance. This causes a complication if a user wanted to fix the the overall variance of the random effect at some constant; no solution to this is yet in place. For comparison see section of Pinheiro and Bates. They also use the values of the Cholesky decomposition directly as the unknowns for the optim function. This has the advantage of further numerical precision, avoids computing the Cholesky decompostion anew at each iteration, and guarrantees that the variance matrix is positive definite. However, though it works well for an unstructured variance, the lme default, the common genetic models do not have a simple representation in the Cholesky space and so we work directly with A. References [1] José C. Pinheiro and Douglas M. Bates, Mixed-Effects Models in S and S-PLUS, Springer, [2] Cortinas Abrahantes, Jose; Burzykowski, Tomasz, A version of the EM algorithm for proportional hazards models with random effects, Lecture Notes of the ICB Seminars, p ,
Package coxme. R topics documented: May 13, Title Mixed Effects Cox Models Priority optional Version
 Title Mixed Effects Cox Models Priority optional Version 2.2-10 Package coxme May 13, 2018 Depends survival (>= 2.36.14), methods, bdsmatrix(>= 1.3) Imports nlme, Matrix (>= 1.0) Suggests mvtnorm, kinship2
Title Mixed Effects Cox Models Priority optional Version 2.2-10 Package coxme May 13, 2018 Depends survival (>= 2.36.14), methods, bdsmatrix(>= 1.3) Imports nlme, Matrix (>= 1.0) Suggests mvtnorm, kinship2
9.1 Random coefficients models Constructed data Consumer preference mapping of carrots... 10
 St@tmaster 02429/MIXED LINEAR MODELS PREPARED BY THE STATISTICS GROUPS AT IMM, DTU AND KU-LIFE Module 9: R 9.1 Random coefficients models...................... 1 9.1.1 Constructed data........................
St@tmaster 02429/MIXED LINEAR MODELS PREPARED BY THE STATISTICS GROUPS AT IMM, DTU AND KU-LIFE Module 9: R 9.1 Random coefficients models...................... 1 9.1.1 Constructed data........................
lme for SAS PROC MIXED Users
 lme for SAS PROC MIXED Users Douglas M. Bates Department of Statistics University of Wisconsin Madison José C. Pinheiro Bell Laboratories Lucent Technologies 1 Introduction The lme function from the nlme
lme for SAS PROC MIXED Users Douglas M. Bates Department of Statistics University of Wisconsin Madison José C. Pinheiro Bell Laboratories Lucent Technologies 1 Introduction The lme function from the nlme
Section 2.2: Covariance, Correlation, and Least Squares
 Section 2.2: Covariance, Correlation, and Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 A Deeper
Section 2.2: Covariance, Correlation, and Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 A Deeper
Practical 4: Mixed effect models
 Practical 4: Mixed effect models This practical is about how to fit (generalised) linear mixed effects models using the lme4 package. You may need to install it first (using either the install.packages
Practical 4: Mixed effect models This practical is about how to fit (generalised) linear mixed effects models using the lme4 package. You may need to install it first (using either the install.packages
lme4: Mixed-effects modeling with R
 Douglas M. Bates lme4: Mixed-effects modeling with R February 17, 2010 Springer Page: 1 job: lmmwr macro: svmono.cls date/time: 17-Feb-2010/14:23 Page: 2 job: lmmwr macro: svmono.cls date/time: 17-Feb-2010/14:23
Douglas M. Bates lme4: Mixed-effects modeling with R February 17, 2010 Springer Page: 1 job: lmmwr macro: svmono.cls date/time: 17-Feb-2010/14:23 Page: 2 job: lmmwr macro: svmono.cls date/time: 17-Feb-2010/14:23
Linear Methods for Regression and Shrinkage Methods
 Linear Methods for Regression and Shrinkage Methods Reference: The Elements of Statistical Learning, by T. Hastie, R. Tibshirani, J. Friedman, Springer 1 Linear Regression Models Least Squares Input vectors
Linear Methods for Regression and Shrinkage Methods Reference: The Elements of Statistical Learning, by T. Hastie, R. Tibshirani, J. Friedman, Springer 1 Linear Regression Models Least Squares Input vectors
The linear mixed model: modeling hierarchical and longitudinal data
 The linear mixed model: modeling hierarchical and longitudinal data Analysis of Experimental Data AED The linear mixed model: modeling hierarchical and longitudinal data 1 of 44 Contents 1 Modeling Hierarchical
The linear mixed model: modeling hierarchical and longitudinal data Analysis of Experimental Data AED The linear mixed model: modeling hierarchical and longitudinal data 1 of 44 Contents 1 Modeling Hierarchical
Multiple Linear Regression
 Multiple Linear Regression Rebecca C. Steorts, Duke University STA 325, Chapter 3 ISL 1 / 49 Agenda How to extend beyond a SLR Multiple Linear Regression (MLR) Relationship Between the Response and Predictors
Multiple Linear Regression Rebecca C. Steorts, Duke University STA 325, Chapter 3 ISL 1 / 49 Agenda How to extend beyond a SLR Multiple Linear Regression (MLR) Relationship Between the Response and Predictors
Organizing data in R. Fitting Mixed-Effects Models Using the lme4 Package in R. R packages. Accessing documentation. The Dyestuff data set
 Fitting Mixed-Effects Models Using the lme4 Package in R Deepayan Sarkar Fred Hutchinson Cancer Research Center 18 September 2008 Organizing data in R Standard rectangular data sets (columns are variables,
Fitting Mixed-Effects Models Using the lme4 Package in R Deepayan Sarkar Fred Hutchinson Cancer Research Center 18 September 2008 Organizing data in R Standard rectangular data sets (columns are variables,
Simulating power in practice
 Simulating power in practice Author: Nicholas G Reich This material is part of the statsteachr project Made available under the Creative Commons Attribution-ShareAlike 3.0 Unported License: http://creativecommons.org/licenses/by-sa/3.0/deed.en
Simulating power in practice Author: Nicholas G Reich This material is part of the statsteachr project Made available under the Creative Commons Attribution-ShareAlike 3.0 Unported License: http://creativecommons.org/licenses/by-sa/3.0/deed.en
This is called a linear basis expansion, and h m is the mth basis function For example if X is one-dimensional: f (X) = β 0 + β 1 X + β 2 X 2, or
 STA 450/4000 S: February 2 2005 Flexible modelling using basis expansions (Chapter 5) Linear regression: y = Xβ + ɛ, ɛ (0, σ 2 ) Smooth regression: y = f (X) + ɛ: f (X) = E(Y X) to be specified Flexible
STA 450/4000 S: February 2 2005 Flexible modelling using basis expansions (Chapter 5) Linear regression: y = Xβ + ɛ, ɛ (0, σ 2 ) Smooth regression: y = f (X) + ɛ: f (X) = E(Y X) to be specified Flexible
The Lander-Green Algorithm in Practice. Biostatistics 666
 The Lander-Green Algorithm in Practice Biostatistics 666 Last Lecture: Lander-Green Algorithm More general definition for I, the "IBD vector" Probability of genotypes given IBD vector Transition probabilities
The Lander-Green Algorithm in Practice Biostatistics 666 Last Lecture: Lander-Green Algorithm More general definition for I, the "IBD vector" Probability of genotypes given IBD vector Transition probabilities
Gelman-Hill Chapter 3
 Gelman-Hill Chapter 3 Linear Regression Basics In linear regression with a single independent variable, as we have seen, the fundamental equation is where ŷ bx 1 b0 b b b y 1 yx, 0 y 1 x x Bivariate Normal
Gelman-Hill Chapter 3 Linear Regression Basics In linear regression with a single independent variable, as we have seen, the fundamental equation is where ŷ bx 1 b0 b b b y 1 yx, 0 y 1 x x Bivariate Normal
GAMs semi-parametric GLMs. Simon Wood Mathematical Sciences, University of Bath, U.K.
 GAMs semi-parametric GLMs Simon Wood Mathematical Sciences, University of Bath, U.K. Generalized linear models, GLM 1. A GLM models a univariate response, y i as g{e(y i )} = X i β where y i Exponential
GAMs semi-parametric GLMs Simon Wood Mathematical Sciences, University of Bath, U.K. Generalized linear models, GLM 1. A GLM models a univariate response, y i as g{e(y i )} = X i β where y i Exponential
D-Optimal Designs. Chapter 888. Introduction. D-Optimal Design Overview
 Chapter 888 Introduction This procedure generates D-optimal designs for multi-factor experiments with both quantitative and qualitative factors. The factors can have a mixed number of levels. For example,
Chapter 888 Introduction This procedure generates D-optimal designs for multi-factor experiments with both quantitative and qualitative factors. The factors can have a mixed number of levels. For example,
Package GWAF. March 12, 2015
 Type Package Package GWAF March 12, 2015 Title Genome-Wide Association/Interaction Analysis and Rare Variant Analysis with Family Data Version 2.2 Date 2015-03-12 Author Ming-Huei Chen
Type Package Package GWAF March 12, 2015 Title Genome-Wide Association/Interaction Analysis and Rare Variant Analysis with Family Data Version 2.2 Date 2015-03-12 Author Ming-Huei Chen
- 1 - Fig. A5.1 Missing value analysis dialog box
 WEB APPENDIX Sarstedt, M. & Mooi, E. (2019). A concise guide to market research. The process, data, and methods using SPSS (3 rd ed.). Heidelberg: Springer. Missing Value Analysis and Multiple Imputation
WEB APPENDIX Sarstedt, M. & Mooi, E. (2019). A concise guide to market research. The process, data, and methods using SPSS (3 rd ed.). Heidelberg: Springer. Missing Value Analysis and Multiple Imputation
Regression Analysis and Linear Regression Models
 Regression Analysis and Linear Regression Models University of Trento - FBK 2 March, 2015 (UNITN-FBK) Regression Analysis and Linear Regression Models 2 March, 2015 1 / 33 Relationship between numerical
Regression Analysis and Linear Regression Models University of Trento - FBK 2 March, 2015 (UNITN-FBK) Regression Analysis and Linear Regression Models 2 March, 2015 1 / 33 Relationship between numerical
Package lmesplines. R topics documented: February 20, Version
 Version 1.1-10 Package lmesplines February 20, 2015 Title Add smoothing spline modelling capability to nlme. Author Rod Ball Maintainer Andrzej Galecki
Version 1.1-10 Package lmesplines February 20, 2015 Title Add smoothing spline modelling capability to nlme. Author Rod Ball Maintainer Andrzej Galecki
Week 4: Simple Linear Regression II
 Week 4: Simple Linear Regression II Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Algebraic properties
Week 4: Simple Linear Regression II Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Algebraic properties
Section 2.1: Intro to Simple Linear Regression & Least Squares
 Section 2.1: Intro to Simple Linear Regression & Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 Regression:
Section 2.1: Intro to Simple Linear Regression & Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 Regression:
Lecture 26: Missing data
 Lecture 26: Missing data Reading: ESL 9.6 STATS 202: Data mining and analysis December 1, 2017 1 / 10 Missing data is everywhere Survey data: nonresponse. 2 / 10 Missing data is everywhere Survey data:
Lecture 26: Missing data Reading: ESL 9.6 STATS 202: Data mining and analysis December 1, 2017 1 / 10 Missing data is everywhere Survey data: nonresponse. 2 / 10 Missing data is everywhere Survey data:
A short explanation of Linear Mixed Models (LMM)
 A short explanation of Linear Mixed Models (LMM) DO NOT TRUST M ENGLISH! This PDF is downloadable at "My learning page" of http://www.lowtem.hokudai.ac.jp/plantecol/akihiro/sumida-index.html ver 20121121e
A short explanation of Linear Mixed Models (LMM) DO NOT TRUST M ENGLISH! This PDF is downloadable at "My learning page" of http://www.lowtem.hokudai.ac.jp/plantecol/akihiro/sumida-index.html ver 20121121e
Generalized Additive Models
 :p Texts in Statistical Science Generalized Additive Models An Introduction with R Simon N. Wood Contents Preface XV 1 Linear Models 1 1.1 A simple linear model 2 Simple least squares estimation 3 1.1.1
:p Texts in Statistical Science Generalized Additive Models An Introduction with R Simon N. Wood Contents Preface XV 1 Linear Models 1 1.1 A simple linear model 2 Simple least squares estimation 3 1.1.1
Salary 9 mo : 9 month salary for faculty member for 2004
 22s:52 Applied Linear Regression DeCook Fall 2008 Lab 3 Friday October 3. The data Set In 2004, a study was done to examine if gender, after controlling for other variables, was a significant predictor
22s:52 Applied Linear Regression DeCook Fall 2008 Lab 3 Friday October 3. The data Set In 2004, a study was done to examine if gender, after controlling for other variables, was a significant predictor
Lecture 7: Linear Regression (continued)
 Lecture 7: Linear Regression (continued) Reading: Chapter 3 STATS 2: Data mining and analysis Jonathan Taylor, 10/8 Slide credits: Sergio Bacallado 1 / 14 Potential issues in linear regression 1. Interactions
Lecture 7: Linear Regression (continued) Reading: Chapter 3 STATS 2: Data mining and analysis Jonathan Taylor, 10/8 Slide credits: Sergio Bacallado 1 / 14 Potential issues in linear regression 1. Interactions
Online Supplementary Appendix for. Dziak, Nahum-Shani and Collins (2012), Multilevel Factorial Experiments for Developing Behavioral Interventions:
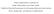 Online Supplementary Appendix for Dziak, Nahum-Shani and Collins (2012), Multilevel Factorial Experiments for Developing Behavioral Interventions: Power, Sample Size, and Resource Considerations 1 Appendix
Online Supplementary Appendix for Dziak, Nahum-Shani and Collins (2012), Multilevel Factorial Experiments for Developing Behavioral Interventions: Power, Sample Size, and Resource Considerations 1 Appendix
Model selection. Peter Hoff. 560 Hierarchical modeling. Statistics, University of Washington 1/41
 1/41 Model selection 560 Hierarchical modeling Peter Hoff Statistics, University of Washington /41 Modeling choices Model: A statistical model is a set of probability distributions for your data. In HLM,
1/41 Model selection 560 Hierarchical modeling Peter Hoff Statistics, University of Washington /41 Modeling choices Model: A statistical model is a set of probability distributions for your data. In HLM,
Contrasts. 1 An experiment with two factors. Chapter 5
 Chapter 5 Contrasts 5 Contrasts 1 1 An experiment with two factors................. 1 2 The data set........................... 2 3 Interpretation of coefficients for one factor........... 5 3.1 Without
Chapter 5 Contrasts 5 Contrasts 1 1 An experiment with two factors................. 1 2 The data set........................... 2 3 Interpretation of coefficients for one factor........... 5 3.1 Without
Estimating Variance Components in MMAP
 Last update: 6/1/2014 Estimating Variance Components in MMAP MMAP implements routines to estimate variance components within the mixed model. These estimates can be used for likelihood ratio tests to compare
Last update: 6/1/2014 Estimating Variance Components in MMAP MMAP implements routines to estimate variance components within the mixed model. These estimates can be used for likelihood ratio tests to compare
DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R
 DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R Lee, Rönnegård & Noh LRN@du.se Lee, Rönnegård & Noh HGLM book 1 / 24 Overview 1 Background to the book 2 Crack growth example 3 Contents
DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R Lee, Rönnegård & Noh LRN@du.se Lee, Rönnegård & Noh HGLM book 1 / 24 Overview 1 Background to the book 2 Crack growth example 3 Contents
Chapters 5-6: Statistical Inference Methods
 Chapters 5-6: Statistical Inference Methods Chapter 5: Estimation (of population parameters) Ex. Based on GSS data, we re 95% confident that the population mean of the variable LONELY (no. of days in past
Chapters 5-6: Statistical Inference Methods Chapter 5: Estimation (of population parameters) Ex. Based on GSS data, we re 95% confident that the population mean of the variable LONELY (no. of days in past
STEPHEN WOLFRAM MATHEMATICADO. Fourth Edition WOLFRAM MEDIA CAMBRIDGE UNIVERSITY PRESS
 STEPHEN WOLFRAM MATHEMATICADO OO Fourth Edition WOLFRAM MEDIA CAMBRIDGE UNIVERSITY PRESS Table of Contents XXI a section new for Version 3 a section new for Version 4 a section substantially modified for
STEPHEN WOLFRAM MATHEMATICADO OO Fourth Edition WOLFRAM MEDIA CAMBRIDGE UNIVERSITY PRESS Table of Contents XXI a section new for Version 3 a section new for Version 4 a section substantially modified for
610 R12 Prof Colleen F. Moore Analysis of variance for Unbalanced Between Groups designs in R For Psychology 610 University of Wisconsin--Madison
 610 R12 Prof Colleen F. Moore Analysis of variance for Unbalanced Between Groups designs in R For Psychology 610 University of Wisconsin--Madison R is very touchy about unbalanced designs, partly because
610 R12 Prof Colleen F. Moore Analysis of variance for Unbalanced Between Groups designs in R For Psychology 610 University of Wisconsin--Madison R is very touchy about unbalanced designs, partly because
Mixed Models with R: Non-linear Models Asymptotic Functions of Time
 SCS 2017: Longitudinal and Nested Data Mixed Models with R: Non-linear Models Asymptotic Functions of Time Georges Monette, March 2017 2 2 3 3 4 4 5 5 6 6 7 7 Modeling individual trajectories A good strategy
SCS 2017: Longitudinal and Nested Data Mixed Models with R: Non-linear Models Asymptotic Functions of Time Georges Monette, March 2017 2 2 3 3 4 4 5 5 6 6 7 7 Modeling individual trajectories A good strategy
Pine Trees and Comas: Asymptotic Functions of Time Part II
 SPIDA 2009 Mixed Models with R Pine Trees and Comas: Asymptotic Functions of Time Part II Georges Monette 1 June 2009 e-mail: georges@yorku.ca web page: http://wiki.math.yorku.ca/spida_2009 1 with thanks
SPIDA 2009 Mixed Models with R Pine Trees and Comas: Asymptotic Functions of Time Part II Georges Monette 1 June 2009 e-mail: georges@yorku.ca web page: http://wiki.math.yorku.ca/spida_2009 1 with thanks
Introduction to Mixed Models: Multivariate Regression
 Introduction to Mixed Models: Multivariate Regression EPSY 905: Multivariate Analysis Spring 2016 Lecture #9 March 30, 2016 EPSY 905: Multivariate Regression via Path Analysis Today s Lecture Multivariate
Introduction to Mixed Models: Multivariate Regression EPSY 905: Multivariate Analysis Spring 2016 Lecture #9 March 30, 2016 EPSY 905: Multivariate Regression via Path Analysis Today s Lecture Multivariate
Using BLUPF90 UGA
 Using BLUPF90 UGA 05-2018 BLUPF90 family programs All programs are controled by the SAME paramenter file. Extra options could be used to set non-default behaviour of each program Understanding parameter
Using BLUPF90 UGA 05-2018 BLUPF90 family programs All programs are controled by the SAME paramenter file. Extra options could be used to set non-default behaviour of each program Understanding parameter
Exercise 2.23 Villanova MAT 8406 September 7, 2015
 Exercise 2.23 Villanova MAT 8406 September 7, 2015 Step 1: Understand the Question Consider the simple linear regression model y = 50 + 10x + ε where ε is NID(0, 16). Suppose that n = 20 pairs of observations
Exercise 2.23 Villanova MAT 8406 September 7, 2015 Step 1: Understand the Question Consider the simple linear regression model y = 50 + 10x + ε where ε is NID(0, 16). Suppose that n = 20 pairs of observations
Workshop 5: Linear mixed-effects models
 Workshop 5: Linear mixed-effects models In this workshop we will fit linear mixed-effects models to data in R. Linear mixed-effects models are used when you have random effects, which occurs when multiple
Workshop 5: Linear mixed-effects models In this workshop we will fit linear mixed-effects models to data in R. Linear mixed-effects models are used when you have random effects, which occurs when multiple
DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R
 DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R Lee, Rönnegård & Noh LRN@du.se Lee, Rönnegård & Noh HGLM book 1 / 25 Overview 1 Background to the book 2 A motivating example from my own
DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R Lee, Rönnegård & Noh LRN@du.se Lee, Rönnegård & Noh HGLM book 1 / 25 Overview 1 Background to the book 2 A motivating example from my own
CSSS 510: Lab 2. Introduction to Maximum Likelihood Estimation
 CSSS 510: Lab 2 Introduction to Maximum Likelihood Estimation 2018-10-12 0. Agenda 1. Housekeeping: simcf, tile 2. Questions about Homework 1 or lecture 3. Simulating heteroskedastic normal data 4. Fitting
CSSS 510: Lab 2 Introduction to Maximum Likelihood Estimation 2018-10-12 0. Agenda 1. Housekeeping: simcf, tile 2. Questions about Homework 1 or lecture 3. Simulating heteroskedastic normal data 4. Fitting
BIOL 458 BIOMETRY Lab 10 - Multiple Regression
 BIOL 458 BIOMETRY Lab 0 - Multiple Regression Many problems in biology science involve the analysis of multivariate data sets. For data sets in which there is a single continuous dependent variable, but
BIOL 458 BIOMETRY Lab 0 - Multiple Regression Many problems in biology science involve the analysis of multivariate data sets. For data sets in which there is a single continuous dependent variable, but
Chapter 6: Linear Model Selection and Regularization
 Chapter 6: Linear Model Selection and Regularization As p (the number of predictors) comes close to or exceeds n (the sample size) standard linear regression is faced with problems. The variance of the
Chapter 6: Linear Model Selection and Regularization As p (the number of predictors) comes close to or exceeds n (the sample size) standard linear regression is faced with problems. The variance of the
Regression. Dr. G. Bharadwaja Kumar VIT Chennai
 Regression Dr. G. Bharadwaja Kumar VIT Chennai Introduction Statistical models normally specify how one set of variables, called dependent variables, functionally depend on another set of variables, called
Regression Dr. G. Bharadwaja Kumar VIT Chennai Introduction Statistical models normally specify how one set of variables, called dependent variables, functionally depend on another set of variables, called
Goals of the Lecture. SOC6078 Advanced Statistics: 9. Generalized Additive Models. Limitations of the Multiple Nonparametric Models (2)
 SOC6078 Advanced Statistics: 9. Generalized Additive Models Robert Andersen Department of Sociology University of Toronto Goals of the Lecture Introduce Additive Models Explain how they extend from simple
SOC6078 Advanced Statistics: 9. Generalized Additive Models Robert Andersen Department of Sociology University of Toronto Goals of the Lecture Introduce Additive Models Explain how they extend from simple
Generalized Additive Models
 Generalized Additive Models Statistics 135 Autumn 2005 Copyright c 2005 by Mark E. Irwin Generalized Additive Models GAMs are one approach to non-parametric regression in the multiple predictor setting.
Generalized Additive Models Statistics 135 Autumn 2005 Copyright c 2005 by Mark E. Irwin Generalized Additive Models GAMs are one approach to non-parametric regression in the multiple predictor setting.
Package EBglmnet. January 30, 2016
 Type Package Package EBglmnet January 30, 2016 Title Empirical Bayesian Lasso and Elastic Net Methods for Generalized Linear Models Version 4.1 Date 2016-01-15 Author Anhui Huang, Dianting Liu Maintainer
Type Package Package EBglmnet January 30, 2016 Title Empirical Bayesian Lasso and Elastic Net Methods for Generalized Linear Models Version 4.1 Date 2016-01-15 Author Anhui Huang, Dianting Liu Maintainer
Section 3.4: Diagnostics and Transformations. Jared S. Murray The University of Texas at Austin McCombs School of Business
 Section 3.4: Diagnostics and Transformations Jared S. Murray The University of Texas at Austin McCombs School of Business 1 Regression Model Assumptions Y i = β 0 + β 1 X i + ɛ Recall the key assumptions
Section 3.4: Diagnostics and Transformations Jared S. Murray The University of Texas at Austin McCombs School of Business 1 Regression Model Assumptions Y i = β 0 + β 1 X i + ɛ Recall the key assumptions
Stat 8053, Fall 2013: Additive Models
 Stat 853, Fall 213: Additive Models We will only use the package mgcv for fitting additive and later generalized additive models. The best reference is S. N. Wood (26), Generalized Additive Models, An
Stat 853, Fall 213: Additive Models We will only use the package mgcv for fitting additive and later generalized additive models. The best reference is S. N. Wood (26), Generalized Additive Models, An
Statistics Lab #7 ANOVA Part 2 & ANCOVA
 Statistics Lab #7 ANOVA Part 2 & ANCOVA PSYCH 710 7 Initialize R Initialize R by entering the following commands at the prompt. You must type the commands exactly as shown. options(contrasts=c("contr.sum","contr.poly")
Statistics Lab #7 ANOVA Part 2 & ANCOVA PSYCH 710 7 Initialize R Initialize R by entering the following commands at the prompt. You must type the commands exactly as shown. options(contrasts=c("contr.sum","contr.poly")
Introduction to Programming in C Department of Computer Science and Engineering. Lecture No. #44. Multidimensional Array and pointers
 Introduction to Programming in C Department of Computer Science and Engineering Lecture No. #44 Multidimensional Array and pointers In this video, we will look at the relation between Multi-dimensional
Introduction to Programming in C Department of Computer Science and Engineering Lecture No. #44 Multidimensional Array and pointers In this video, we will look at the relation between Multi-dimensional
Predictive Checking. Readings GH Chapter 6-8. February 8, 2017
 Predictive Checking Readings GH Chapter 6-8 February 8, 2017 Model Choice and Model Checking 2 Questions: 1. Is my Model good enough? (no alternative models in mind) 2. Which Model is best? (comparison
Predictive Checking Readings GH Chapter 6-8 February 8, 2017 Model Choice and Model Checking 2 Questions: 1. Is my Model good enough? (no alternative models in mind) 2. Which Model is best? (comparison
Comparing Fitted Models with the fit.models Package
 Comparing Fitted Models with the fit.models Package Kjell Konis Acting Assistant Professor Computational Finance and Risk Management Dept. Applied Mathematics, University of Washington History of fit.models
Comparing Fitted Models with the fit.models Package Kjell Konis Acting Assistant Professor Computational Finance and Risk Management Dept. Applied Mathematics, University of Washington History of fit.models
Expectation Maximization (EM) and Gaussian Mixture Models
 Expectation Maximization (EM) and Gaussian Mixture Models Reference: The Elements of Statistical Learning, by T. Hastie, R. Tibshirani, J. Friedman, Springer 1 2 3 4 5 6 7 8 Unsupervised Learning Motivation
Expectation Maximization (EM) and Gaussian Mixture Models Reference: The Elements of Statistical Learning, by T. Hastie, R. Tibshirani, J. Friedman, Springer 1 2 3 4 5 6 7 8 Unsupervised Learning Motivation
Introduction to the R Statistical Computing Environment R Programming: Exercises
 Introduction to the R Statistical Computing Environment R Programming: Exercises John Fox (McMaster University) ICPSR 2014 1. A straightforward problem: Write an R function for linear least-squares regression.
Introduction to the R Statistical Computing Environment R Programming: Exercises John Fox (McMaster University) ICPSR 2014 1. A straightforward problem: Write an R function for linear least-squares regression.
Lasso. November 14, 2017
 Lasso November 14, 2017 Contents 1 Case Study: Least Absolute Shrinkage and Selection Operator (LASSO) 1 1.1 The Lasso Estimator.................................... 1 1.2 Computation of the Lasso Solution............................
Lasso November 14, 2017 Contents 1 Case Study: Least Absolute Shrinkage and Selection Operator (LASSO) 1 1.1 The Lasso Estimator.................................... 1 1.2 Computation of the Lasso Solution............................
Package simr. April 30, 2018
 Type Package Package simr April 30, 2018 Title Power Analysis for Generalised Linear Mixed Models by Simulation Calculate power for generalised linear mixed models, using simulation. Designed to work with
Type Package Package simr April 30, 2018 Title Power Analysis for Generalised Linear Mixed Models by Simulation Calculate power for generalised linear mixed models, using simulation. Designed to work with
NEURAL NETWORKS. Cement. Blast Furnace Slag. Fly Ash. Water. Superplasticizer. Coarse Aggregate. Fine Aggregate. Age
 NEURAL NETWORKS As an introduction, we ll tackle a prediction task with a continuous variable. We ll reproduce research from the field of cement and concrete manufacturing that seeks to model the compressive
NEURAL NETWORKS As an introduction, we ll tackle a prediction task with a continuous variable. We ll reproduce research from the field of cement and concrete manufacturing that seeks to model the compressive
Package citools. October 20, 2018
 Type Package Package citools October 20, 2018 Title Confidence or Prediction Intervals, Quantiles, and Probabilities for Statistical Models Version 0.5.0 Maintainer John Haman Functions
Type Package Package citools October 20, 2018 Title Confidence or Prediction Intervals, Quantiles, and Probabilities for Statistical Models Version 0.5.0 Maintainer John Haman Functions
Bayes Estimators & Ridge Regression
 Bayes Estimators & Ridge Regression Readings ISLR 6 STA 521 Duke University Merlise Clyde October 27, 2017 Model Assume that we have centered (as before) and rescaled X o (original X) so that X j = X o
Bayes Estimators & Ridge Regression Readings ISLR 6 STA 521 Duke University Merlise Clyde October 27, 2017 Model Assume that we have centered (as before) and rescaled X o (original X) so that X j = X o
STENO Introductory R-Workshop: Loading a Data Set Tommi Suvitaival, Steno Diabetes Center June 11, 2015
 STENO Introductory R-Workshop: Loading a Data Set Tommi Suvitaival, tsvv@steno.dk, Steno Diabetes Center June 11, 2015 Contents 1 Introduction 1 2 Recap: Variables 2 3 Data Containers 2 3.1 Vectors................................................
STENO Introductory R-Workshop: Loading a Data Set Tommi Suvitaival, tsvv@steno.dk, Steno Diabetes Center June 11, 2015 Contents 1 Introduction 1 2 Recap: Variables 2 3 Data Containers 2 3.1 Vectors................................................
DESIGN OF EXPERIMENTS and ROBUST DESIGN
 DESIGN OF EXPERIMENTS and ROBUST DESIGN Problems in design and production environments often require experiments to find a solution. Design of experiments are a collection of statistical methods that,
DESIGN OF EXPERIMENTS and ROBUST DESIGN Problems in design and production environments often require experiments to find a solution. Design of experiments are a collection of statistical methods that,
Applied Statistics and Econometrics Lecture 6
 Applied Statistics and Econometrics Lecture 6 Giuseppe Ragusa Luiss University gragusa@luiss.it http://gragusa.org/ March 6, 2017 Luiss University Empirical application. Data Italian Labour Force Survey,
Applied Statistics and Econometrics Lecture 6 Giuseppe Ragusa Luiss University gragusa@luiss.it http://gragusa.org/ March 6, 2017 Luiss University Empirical application. Data Italian Labour Force Survey,
Poisson Regression and Model Checking
 Poisson Regression and Model Checking Readings GH Chapter 6-8 September 27, 2017 HIV & Risk Behaviour Study The variables couples and women_alone code the intervention: control - no counselling (both 0)
Poisson Regression and Model Checking Readings GH Chapter 6-8 September 27, 2017 HIV & Risk Behaviour Study The variables couples and women_alone code the intervention: control - no counselling (both 0)
Repeated Measures Part 4: Blood Flow data
 Repeated Measures Part 4: Blood Flow data /* bloodflow.sas */ options linesize=79 pagesize=100 noovp formdlim='_'; title 'Two within-subjecs factors: Blood flow data (NWK p. 1181)'; proc format; value
Repeated Measures Part 4: Blood Flow data /* bloodflow.sas */ options linesize=79 pagesize=100 noovp formdlim='_'; title 'Two within-subjecs factors: Blood flow data (NWK p. 1181)'; proc format; value
Random coefficients models
 enote 9 1 enote 9 Random coefficients models enote 9 INDHOLD 2 Indhold 9 Random coefficients models 1 9.1 Introduction.................................... 2 9.2 Example: Constructed data...........................
enote 9 1 enote 9 Random coefficients models enote 9 INDHOLD 2 Indhold 9 Random coefficients models 1 9.1 Introduction.................................... 2 9.2 Example: Constructed data...........................
MACAU User Manual. Xiang Zhou. March 15, 2017
 MACAU User Manual Xiang Zhou March 15, 2017 Contents 1 Introduction 2 1.1 What is MACAU...................................... 2 1.2 How to Cite MACAU................................... 2 1.3 The Model.........................................
MACAU User Manual Xiang Zhou March 15, 2017 Contents 1 Introduction 2 1.1 What is MACAU...................................... 2 1.2 How to Cite MACAU................................... 2 1.3 The Model.........................................
Lecture 20: Outliers and Influential Points
 Lecture 20: Outliers and Influential Points An outlier is a point with a large residual. An influential point is a point that has a large impact on the regression. Surprisingly, these are not the same
Lecture 20: Outliers and Influential Points An outlier is a point with a large residual. An influential point is a point that has a large impact on the regression. Surprisingly, these are not the same
Lab #13 - Resampling Methods Econ 224 October 23rd, 2018
 Lab #13 - Resampling Methods Econ 224 October 23rd, 2018 Introduction In this lab you will work through Section 5.3 of ISL and record your code and results in an RMarkdown document. I have added section
Lab #13 - Resampling Methods Econ 224 October 23rd, 2018 Introduction In this lab you will work through Section 5.3 of ISL and record your code and results in an RMarkdown document. I have added section
Least Squares and SLAM Pose-SLAM
 Least Squares and SLAM Pose-SLAM Giorgio Grisetti Part of the material of this course is taken from the Robotics 2 lectures given by G.Grisetti, W.Burgard, C.Stachniss, K.Arras, D. Tipaldi and M.Bennewitz
Least Squares and SLAM Pose-SLAM Giorgio Grisetti Part of the material of this course is taken from the Robotics 2 lectures given by G.Grisetti, W.Burgard, C.Stachniss, K.Arras, D. Tipaldi and M.Bennewitz
Chapter 6: DESCRIPTIVE STATISTICS
 Chapter 6: DESCRIPTIVE STATISTICS Random Sampling Numerical Summaries Stem-n-Leaf plots Histograms, and Box plots Time Sequence Plots Normal Probability Plots Sections 6-1 to 6-5, and 6-7 Random Sampling
Chapter 6: DESCRIPTIVE STATISTICS Random Sampling Numerical Summaries Stem-n-Leaf plots Histograms, and Box plots Time Sequence Plots Normal Probability Plots Sections 6-1 to 6-5, and 6-7 Random Sampling
Linear Equation Systems Iterative Methods
 Linear Equation Systems Iterative Methods Content Iterative Methods Jacobi Iterative Method Gauss Seidel Iterative Method Iterative Methods Iterative methods are those that produce a sequence of successive
Linear Equation Systems Iterative Methods Content Iterative Methods Jacobi Iterative Method Gauss Seidel Iterative Method Iterative Methods Iterative methods are those that produce a sequence of successive
COMP 558 lecture 19 Nov. 17, 2010
 COMP 558 lecture 9 Nov. 7, 2 Camera calibration To estimate the geometry of 3D scenes, it helps to know the camera parameters, both external and internal. The problem of finding all these parameters is
COMP 558 lecture 9 Nov. 7, 2 Camera calibration To estimate the geometry of 3D scenes, it helps to know the camera parameters, both external and internal. The problem of finding all these parameters is
Section 4.1: Time Series I. Jared S. Murray The University of Texas at Austin McCombs School of Business
 Section 4.1: Time Series I Jared S. Murray The University of Texas at Austin McCombs School of Business 1 Time Series Data and Dependence Time-series data are simply a collection of observations gathered
Section 4.1: Time Series I Jared S. Murray The University of Texas at Austin McCombs School of Business 1 Time Series Data and Dependence Time-series data are simply a collection of observations gathered
Today. Lecture 4: Last time. The EM algorithm. We examine clustering in a little more detail; we went over it a somewhat quickly last time
 Today Lecture 4: We examine clustering in a little more detail; we went over it a somewhat quickly last time The CAD data will return and give us an opportunity to work with curves (!) We then examine
Today Lecture 4: We examine clustering in a little more detail; we went over it a somewhat quickly last time The CAD data will return and give us an opportunity to work with curves (!) We then examine
Regression III: Lab 4
 Regression III: Lab 4 This lab will work through some model/variable selection problems, finite mixture models and missing data issues. You shouldn t feel obligated to work through this linearly, I would
Regression III: Lab 4 This lab will work through some model/variable selection problems, finite mixture models and missing data issues. You shouldn t feel obligated to work through this linearly, I would
Standard Errors in OLS Luke Sonnet
 Standard Errors in OLS Luke Sonnet Contents Variance-Covariance of ˆβ 1 Standard Estimation (Spherical Errors) 2 Robust Estimation (Heteroskedasticity Constistent Errors) 4 Cluster Robust Estimation 7
Standard Errors in OLS Luke Sonnet Contents Variance-Covariance of ˆβ 1 Standard Estimation (Spherical Errors) 2 Robust Estimation (Heteroskedasticity Constistent Errors) 4 Cluster Robust Estimation 7
The kinship Package. February 13, 2006
 The kinship Package February 13, 2006 Version 1.1.0-11 Date 2006-2-13 Title mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees Author Beth Atkinson (atkinson@mayo.edu) for
The kinship Package February 13, 2006 Version 1.1.0-11 Date 2006-2-13 Title mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees Author Beth Atkinson (atkinson@mayo.edu) for
Lecture 13: Model selection and regularization
 Lecture 13: Model selection and regularization Reading: Sections 6.1-6.2.1 STATS 202: Data mining and analysis October 23, 2017 1 / 17 What do we know so far In linear regression, adding predictors always
Lecture 13: Model selection and regularization Reading: Sections 6.1-6.2.1 STATS 202: Data mining and analysis October 23, 2017 1 / 17 What do we know so far In linear regression, adding predictors always
Lecture 24: Generalized Additive Models Stat 704: Data Analysis I, Fall 2010
 Lecture 24: Generalized Additive Models Stat 704: Data Analysis I, Fall 2010 Tim Hanson, Ph.D. University of South Carolina T. Hanson (USC) Stat 704: Data Analysis I, Fall 2010 1 / 26 Additive predictors
Lecture 24: Generalized Additive Models Stat 704: Data Analysis I, Fall 2010 Tim Hanson, Ph.D. University of South Carolina T. Hanson (USC) Stat 704: Data Analysis I, Fall 2010 1 / 26 Additive predictors
Resources for statistical assistance. Quantitative covariates and regression analysis. Methods for predicting continuous outcomes.
 Resources for statistical assistance Quantitative covariates and regression analysis Carolyn Taylor Applied Statistics and Data Science Group (ASDa) Department of Statistics, UBC January 24, 2017 Department
Resources for statistical assistance Quantitative covariates and regression analysis Carolyn Taylor Applied Statistics and Data Science Group (ASDa) Department of Statistics, UBC January 24, 2017 Department
The kinship Package. October 2, 2007
 The kinship Package October 2, 2007 Version 1.1.0-18 Date 2007-10-1 Title mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees Author Beth Atkinson (atkinson@mayo.edu) for
The kinship Package October 2, 2007 Version 1.1.0-18 Date 2007-10-1 Title mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees Author Beth Atkinson (atkinson@mayo.edu) for
Package PedCNV. February 19, 2015
 Type Package Package PedCNV February 19, 2015 Title An implementation for association analysis with CNV data. Version 0.1 Date 2013-08-03 Author, Sungho Won and Weicheng Zhu Maintainer
Type Package Package PedCNV February 19, 2015 Title An implementation for association analysis with CNV data. Version 0.1 Date 2013-08-03 Author, Sungho Won and Weicheng Zhu Maintainer
Advanced Techniques for Mobile Robotics Graph-based SLAM using Least Squares. Wolfram Burgard, Cyrill Stachniss, Kai Arras, Maren Bennewitz
 Advanced Techniques for Mobile Robotics Graph-based SLAM using Least Squares Wolfram Burgard, Cyrill Stachniss, Kai Arras, Maren Bennewitz SLAM Constraints connect the poses of the robot while it is moving
Advanced Techniques for Mobile Robotics Graph-based SLAM using Least Squares Wolfram Burgard, Cyrill Stachniss, Kai Arras, Maren Bennewitz SLAM Constraints connect the poses of the robot while it is moving
An introduction to SPSS
 An introduction to SPSS To open the SPSS software using U of Iowa Virtual Desktop... Go to https://virtualdesktop.uiowa.edu and choose SPSS 24. Contents NOTE: Save data files in a drive that is accessible
An introduction to SPSS To open the SPSS software using U of Iowa Virtual Desktop... Go to https://virtualdesktop.uiowa.edu and choose SPSS 24. Contents NOTE: Save data files in a drive that is accessible
Multiple Regression White paper
 +44 (0) 333 666 7366 Multiple Regression White paper A tool to determine the impact in analysing the effectiveness of advertising spend. Multiple Regression In order to establish if the advertising mechanisms
+44 (0) 333 666 7366 Multiple Regression White paper A tool to determine the impact in analysing the effectiveness of advertising spend. Multiple Regression In order to establish if the advertising mechanisms
Generalized Least Squares (GLS) and Estimated Generalized Least Squares (EGLS)
 Generalized Least Squares (GLS) and Estimated Generalized Least Squares (EGLS) Linear Model in matrix notation for the population Y = Xβ + Var ( ) = In GLS, the error covariance matrix is known In EGLS
Generalized Least Squares (GLS) and Estimated Generalized Least Squares (EGLS) Linear Model in matrix notation for the population Y = Xβ + Var ( ) = In GLS, the error covariance matrix is known In EGLS
Non-Linear Regression. Business Analytics Practice Winter Term 2015/16 Stefan Feuerriegel
 Non-Linear Regression Business Analytics Practice Winter Term 2015/16 Stefan Feuerriegel Today s Lecture Objectives 1 Understanding the need for non-parametric regressions 2 Familiarizing with two common
Non-Linear Regression Business Analytics Practice Winter Term 2015/16 Stefan Feuerriegel Today s Lecture Objectives 1 Understanding the need for non-parametric regressions 2 Familiarizing with two common
Estimation of Item Response Models
 Estimation of Item Response Models Lecture #5 ICPSR Item Response Theory Workshop Lecture #5: 1of 39 The Big Picture of Estimation ESTIMATOR = Maximum Likelihood; Mplus Any questions? answers Lecture #5:
Estimation of Item Response Models Lecture #5 ICPSR Item Response Theory Workshop Lecture #5: 1of 39 The Big Picture of Estimation ESTIMATOR = Maximum Likelihood; Mplus Any questions? answers Lecture #5:
CDAA No. 4 - Part Two - Multiple Regression - Initial Data Screening
 CDAA No. 4 - Part Two - Multiple Regression - Initial Data Screening Variables Entered/Removed b Variables Entered GPA in other high school, test, Math test, GPA, High school math GPA a Variables Removed
CDAA No. 4 - Part Two - Multiple Regression - Initial Data Screening Variables Entered/Removed b Variables Entered GPA in other high school, test, Math test, GPA, High school math GPA a Variables Removed
Lecture 22 The Generalized Lasso
 Lecture 22 The Generalized Lasso 07 December 2015 Taylor B. Arnold Yale Statistics STAT 312/612 Class Notes Midterm II - Due today Problem Set 7 - Available now, please hand in by the 16th Motivation Today
Lecture 22 The Generalized Lasso 07 December 2015 Taylor B. Arnold Yale Statistics STAT 312/612 Class Notes Midterm II - Due today Problem Set 7 - Available now, please hand in by the 16th Motivation Today
Package snm. July 20, 2018
 Type Package Title Supervised Normalization of Microarrays Version 1.28.0 Package snm July 20, 2018 Author Brig Mecham and John D. Storey SNM is a modeling strategy especially designed
Type Package Title Supervised Normalization of Microarrays Version 1.28.0 Package snm July 20, 2018 Author Brig Mecham and John D. Storey SNM is a modeling strategy especially designed
Statistical Analysis of Series of N-of-1 Trials Using R. Artur Araujo
 Statistical Analysis of Series of N-of-1 Trials Using R Artur Araujo March 2016 Acknowledgements I would like to thank Boehringer Ingelheim GmbH for having paid my tuition fees at the University of Sheffield
Statistical Analysis of Series of N-of-1 Trials Using R Artur Araujo March 2016 Acknowledgements I would like to thank Boehringer Ingelheim GmbH for having paid my tuition fees at the University of Sheffield
Splines and penalized regression
 Splines and penalized regression November 23 Introduction We are discussing ways to estimate the regression function f, where E(y x) = f(x) One approach is of course to assume that f has a certain shape,
Splines and penalized regression November 23 Introduction We are discussing ways to estimate the regression function f, where E(y x) = f(x) One approach is of course to assume that f has a certain shape,
Section 2.3: Simple Linear Regression: Predictions and Inference
 Section 2.3: Simple Linear Regression: Predictions and Inference Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.4 1 Simple
Section 2.3: Simple Linear Regression: Predictions and Inference Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.4 1 Simple
Regression on the trees data with R
 > trees Girth Height Volume 1 8.3 70 10.3 2 8.6 65 10.3 3 8.8 63 10.2 4 10.5 72 16.4 5 10.7 81 18.8 6 10.8 83 19.7 7 11.0 66 15.6 8 11.0 75 18.2 9 11.1 80 22.6 10 11.2 75 19.9 11 11.3 79 24.2 12 11.4 76
> trees Girth Height Volume 1 8.3 70 10.3 2 8.6 65 10.3 3 8.8 63 10.2 4 10.5 72 16.4 5 10.7 81 18.8 6 10.8 83 19.7 7 11.0 66 15.6 8 11.0 75 18.2 9 11.1 80 22.6 10 11.2 75 19.9 11 11.3 79 24.2 12 11.4 76
S CHAPTER return.data S CHAPTER.Data S CHAPTER
 1 S CHAPTER return.data S CHAPTER.Data MySwork S CHAPTER.Data 2 S e > return ; return + # 3 setenv S_CLEDITOR emacs 4 > 4 + 5 / 3 ## addition & divison [1] 5.666667 > (4 + 5) / 3 ## using parentheses [1]
1 S CHAPTER return.data S CHAPTER.Data MySwork S CHAPTER.Data 2 S e > return ; return + # 3 setenv S_CLEDITOR emacs 4 > 4 + 5 / 3 ## addition & divison [1] 5.666667 > (4 + 5) / 3 ## using parentheses [1]
Predict Outcomes and Reveal Relationships in Categorical Data
 PASW Categories 18 Specifications Predict Outcomes and Reveal Relationships in Categorical Data Unleash the full potential of your data through predictive analysis, statistical learning, perceptual mapping,
PASW Categories 18 Specifications Predict Outcomes and Reveal Relationships in Categorical Data Unleash the full potential of your data through predictive analysis, statistical learning, perceptual mapping,
