Package REMP. January 16, 2019
|
|
|
- Blaze Williamson
- 5 years ago
- Views:
Transcription
1 Type Package Title Repetitive Element Methylation Prediction Version Package REMP January 16, 2019 Machine learning-based tools to predict DNA methylation of locus-specific repetitive elements (RE) by learning surrounding genetic and epigenetic information. These tools provide genomewide and single-base resolution of DNA methylation prediction on RE that are difficult to measure using array-based or sequencing-based platforms, which enables epigenomewide association study (EWAS) and differentially methylated region (DMR) analysis on RE. License GPL-3 Depends R (>= 3.4), SummarizedExperiment(>= 1.1.6), minfi (>= ), IlluminaHumanMethylation450kanno.ilmn12.hg19, IlluminaHumanMethylationEPICanno.ilm10b2.hg19 Imports graphics, stats, utils, methods, settings, BiocGenerics, S4Vectors, Biostrings, GenomicRanges, IRanges, BiocParallel, doparallel, parallel, foreach, caret, kernlab, ranger, BSgenome, AnnotationHub, BSgenome.Hsapiens.UCSC.hg19, org.hs.eg.db, impute, iterators Suggests knitr, rmarkdown, minfidataepic Collate REMParcel-class.R REMProduct-class.R generics.r REMParcel-methods.R REMProduct-methods.R options.r utils.r REMPtools.R getgm12878.r groomethy.r initremp.r remp.r DemoData.R REMP-package.R URL BugReports Encoding UTF-8 LazyData true biocviews ImmunoOncology, DNAMethylation, Microarray, MethylationArray, GenePrediction, Sequencing, Alignment, Epigenetics, Normalization, Preprocessing, MultiChannel, TwoChannel, DifferentialMethylation, QualityControl, DataImport RoxygenNote git_url git_branch master git_last_commit a23f642 1
2 2 REMP-package git_last_commit_date Date/Publication Author Yinan Zheng [aut, cre], Lei Liu [aut], Wei Zhang [aut], Warren Kibbe [aut], Lifang Hou [aut, cph] Maintainer Yinan Zheng R topics documented: REMP-package Alu.demo fetchrefseqgene fetchrmsk findrecpg getbackend getgm GRannot groomethy initremp remp REMParcel-class REMProduct-class remprofile remp_options Index 21 REMP-package Repetitive Element Methylation Prediction Machine learning-based tools to predict DNA methylation of locus-specific repetitive elements (RE) by learning surrounding genetic and epigenetic information. These tools provide genomewide and single-base resolution of DNA methylation prediction on RE that are difficult to measure using array-based or sequencing-based platforms, which enables epigenome-wide association study (EWAS) and differentially methylated region (DMR) analysis on RE. Overview - standard procedure Step 1 Start out generating required dataset for prediction using initremp. The datasets include RE information, RE-CpG (i.e. CpGs located in RE region) information, and gene annotation, which are maintained in a REMParcel object. It is recommended to save these generated data to the working directory so they can be used in the future. Step 2 Clean Illumina methylation dataset using groomethy. This function can help identify and fix abnormal values and automatically impute missing values, which are essential for downstream prediction. Step 3 Run remp to predict genome-wide locus specific RE methylation.
3 Alu.demo 3 Step 4 Use the built-in accessors and utilities in REMProduct object to get or refine the prediction results. Author(s) Yinan Zheng <y-zheng@northwestern.edu>, Lei Liu <lei.liu@northwestern.edu>, Wei Zhang <wei.zhang1@northwestern.edu>, Warren Kibbe <warren.kibbe@nih.gov>, Lifang Hou <l-hou@northwestern.ed Maintainer: Yinan Zheng <y-zheng@northwestern.edu> References Zheng Y, Joyce BT, Liu L, Zhang Z, Kibbe WA, Zhang W, Hou L. Prediction of genome-wide DNA methylation in repetitive elements. Nucleic Acids Res. 2017;45(15): PubMed PMID: ; PMCID: PMC Alu.demo Subset of Alu sequence dataset Format Details Source A GRanges dataset containing 500 Alu sequences that have CpGs profiled by both Illumina 450k and EPIC array. The variables are as follows: Alu.demo A GRanges object. seqnames chromosome number ranges hg19 genomic position strand DNA strand name Alu subfamily score Smith Waterman (SW) alignment score Index internal index Scripts for generating this object are contained in the scripts directory. A GRanges object with 500 ranges and 3 metadata columns. RepeatMasker database provided by package AnnotationHub.
4 4 fetchrmsk fetchrefseqgene Get RefSeq gene database fetchrefseqgene is used to obtain refseq gene database provided by AnnotationHub. fetchrefseqgene(ah, mainonly = FALSE, verbose = FALSE) ah mainonly verbose An AnnotationHub object. Use AnnotationHub() to retrive information about all records in the hub. Logical parameter. See details. Logical parameter. Should the function be verbose? Details When mainonly = FALSE, only the transcript location information will be returned, Otherwise, a GRangesList object containing gene regions information will be added. Gene regions include: 2000 base pair upstream of the transcript start site ($tss)), 5 UTR ($fiveutr)), coding sequence ($cds)), exon ($exon)), and 3 UTR ($threeutr)). The index column is an internal index that is used to facilitate data referral, which is meaningless for external use. A single GRanges (for main refgene data) object or a list incorporating both GRanges object (for main refgene data) and GRangesList object (for gene regions data). ah <- AnnotationHub::AnnotationHub() refgene <- fetchrefseqgene(ah, mainonly = TRUE, verbose = TRUE) refgene fetchrmsk Get RE database from RepeatMasker fetchrmsk is used to obtain specified RE database from RepeatMasker Database provided by AnnotationHub. fetchrmsk(ah, REtype, verbose = FALSE)
5 findrecpg 5 ah REtype verbose An AnnotationHub object. Use AnnotationHub() to retrive information about all records in the hub. Type of RE. Currently "Alu" and "L1" are supported. Logical parameter. Should the function be verbose? A GRanges object containing RE database. name column indicates the RE subfamily; score column indicates the SW score; Index is an internal index for RE to facilitate data referral, which is meaningless for external use. ah <- AnnotationHub::AnnotationHub() L1 <- fetchrmsk(ah, 'L1', verbose = TRUE) L1 findrecpg Find RE-CpG genomic location given RE ranges information findrecpg is used to obtain RE-CpG genomic location data. findrecpg(re.hg19, REtype = c("alu", "L1"), be = NULL, verbose = FALSE) RE.hg19 REtype be verbose an GRanges object of RE genomic location database. This can be obtained by fetchrmsk. Type of RE. Currently "Alu" and "L1" are supported. A BiocParallel object containing back-end information that is ready for paralle computing. This can be obtained by getbackend. logical parameter. Should the function be verbose? Details CpG site is defined as 5 -C-p-G-3. It is reasonable to assume that the methylation status across all CpG/CpG dyads are concordant. Maintenance methyltransferase exhibits a preference for hemimethylated CpG/CpG dyads (methylated on one strand only). As a result, methyaltion status of CpG sites in both forward and reverse strands are usually consistent. Therefore, to accommodate the cytosine loci in both strands, the returned genomic ranges cover the CG sequence with width of 2. The stand information indicates the strand of the RE.
6 6 getbackend A GRanges object containing identified RE-CpG genomic location data. data(alu.demo) be <- getbackend(1, verbose = TRUE) RE.CpG <- findrecpg(alu.demo, 'Alu', be, verbose = TRUE) RE.CpG getbackend Get BiocParallel back-end getbackend is used to obtain BiocParallel Back-end to allow parallel computing. getbackend(ncore, BPPARAM = NULL, verbose = FALSE) ncore BPPARAM verbose Number of cores to run parallel computation. By default max number of cores available in the machine will be utilized. If ncore = 1, no parallel computation is allowed. An optional BiocParallelParam instance determining the parallel back-end to be used during evaluation. If not specified, default back-end in the machine will be used. Logical parameter. Should the function be verbose? A BiocParallel object that can be used for parallel computing. # Non-parallel mode be <- getbackend(1, verbose = TRUE) be # parallel mode (2 workers) be <- getbackend(2, verbose = TRUE) be
7 getgm getgm12878 Get methylation data of HapMap LCL sample GM12878 profiled by Illumina 450k array or EPIC array getgm12878 is used to obtain public available methylation profiling data of HapMap LCL sample GM getgm12878(arraytype = c("450k", "EPIC"), mapgenome = FALSE) arraytype mapgenome Illumina methylation array type. Currently "450k" and "EPIC" are supported. Default = "450k". Logical parameter. If TRUE, function will return a GenomicRatioSet object instead of a link{ratioset} object. Details Illumina 450k data were sourced and curated from ENCODE edu/goldenpath/hg19/encodedcc/wgencodehaibmethyl450/wgencodehaibmethyl450gm12878sitesrep1. bed.gz. Illumina EPIC data were obtained from data package minfidataepic. A RatioSet or GenomicRatioSet containing beta value and M value of the methylation data. # Get GM12878 methylation data (EPIC array) GM12878_EPIC <- getgm12878('epic') GM12878_EPIC GRannot Annotate genomic ranges data with gene region information. GRannot is used to annotate a GRanges dataset with gene region information using refseq gene database GRannot(object.GR, refgene, symbol = FALSE, verbose = FALSE)
8 8 groomethy object.gr refgene symbol verbose An GRanges object of a genomic location database. A complete refgene annotation database returned by fetchrefseqgene (with parameter mainonly = FALSE). Logical parameter. Should the annotation return gene symbol? Logical parameter. Should the function be verbose? Details The annotated gene region information includes: protein coding gene (InNM), noncoding RNA gene (InNR), 2000 base pair upstream of the transcript start site (InTSS), 5 UTR (In5UTR), coding sequence (InCDS), exon (InExon), and 3 UTR (In3UTR). The intergenic and intron regions can then be represented by the combination of these region data. The number shown in these columns represent the row number or index column in the main refgene database obtained by fetchrefseqgene. A GRanges or a GRangesList object containing refseq Gene database. data(alu.demo) ah <- AnnotationHub::AnnotationHub() refgene.hg19 <- fetchrefseqgene(ah, verbose = TRUE) Alu.demo.refGene <- GRannot(Alu.demo, refgene.hg19, verbose = TRUE) Alu.demo.refGene groomethy Groom methylation data to fix potential data issues groomethy is used to automatically detect and fix data issues including zero beta value, missing value, and infinite value. groomethy(methydat, impute = TRUE, imputebyrow = TRUE, mapgenome = FALSE, verbose = FALSE) methydat impute imputebyrow A RatioSet, GenomicRatioSet, DataFrame, data.table, data.frame, or matrix of Illumina BeadChip methylation data (450k or EPIC array). If TRUE, K-Nearest Neighbouring imputation will be applied to fill the missing values. Default = TRUE. See Details. If TRUE, missing values will be imputed using similar values in row (i.e., across samples); if FALSE, missing values will be imputed using similar values in column (i.e., across CpGs). Default is TRUE.
9 initremp 9 mapgenome verbose Logical parameter. If TRUE, function will return a GenomicRatioSet object instead of a RatioSet. Logical parameter. Should the function be verbose? Details For methylation data in beta value, if zero value exists, the logit transformation from beta to M value will produce negative infinite value. Therefore, zero beta value will be replaced with the smallest non-zero beta value found in the dataset. groomethy can also handle missing value (i.e. NA or NaN) using KNN imputation (see impute.knn). The infinite value will be also treated as missing value for imputation. If the original dataset is in beta value, groomethy will first transform it to M value before imputation is carried out. If the imputed value is out of the original range (which is possible when imputebyrow = FALSE), mean value will be used instead. Warning: imputed values for multimodal distributed CpGs (across samples) may not be correct. Please check package ENmix to identify the CpGs with multimodal distribution. Please note that groomethy is also embedded in remp so the user can run remp directly without explicitly running groomethy. A RatioSet or GenomicRatioSet containing beta value and M value of the methylation data. GM12878_450k <- getgm12878('450k') # Get GM12878 methylation data (450k array) groomethy(gm12878_450k, verbose = TRUE) groomethy(minfi::getbeta(gm12878_450k), verbose = TRUE) initremp RE Annotation Database Initialization initremp is used to initialize annotation database for RE methylation prediction. Two major RE types in human, Alu element (Alu) and LINE-1 (L1) are available. initremp(arraytype = c("450k", "EPIC"), REtype = c("alu", "L1"), RE = NULL, ncore = NULL, BPPARAM = NULL, export = FALSE, work.dir = tempdir(), verbose = FALSE) arraytype REtype RE Illumina methylation array type. Currently "450k" and "EPIC" are supported. Default = "450k". Type of RE. Currently "Alu" and "L1" are supported. A GRanges object containing user-specified RE genomic location information. If NULL, the function will retrive RepeatMasker RE database from AnnotationHub (build hg19).
10 10 remp ncore BPPARAM export work.dir verbose Number of cores to run parallel computation. By default max number of cores available in the machine will be utilized. If ncore = 1, no parallel computation is allowed. An optional BiocParallelParam instance determining the parallel back-end to be used during evaluation. If not specified, default back-end in the machine will be used. Logical. Should the returned REMParcel object be saved to local machine? See Details. Path to the directory where the generated data will be saved. Valid when export = TRUE. If not specified and export = TRUE, temporary directory tempdir() will be used. Logical parameter. Should the function be verbose? Details Currently, we support two major types of RE in human, Alu and L1. The main purpose of initremp is to generate and annotate CpG/RE data using the refseq Gene annotation database (provided by AnnotationHub). These annotation data are crucial to RE methylation prediction in remp. Once generated, the data can be reused in the future (data can be very large). Therefore, we recommend user to save the output from initremp to the local machine, so that user only need to run this function once as long as there is no change to the RE database. To minimize the size of resulting data file, the generated annotation data are only for REs that contain RE-CpGs with neighboring profiled CpGs. By default, the neighboring CpGs are confined within 1200 bp flanking window. This window size can be modified using remp_options. An REMParcel object containing data needed for RE methylation prediction. See Also See remp for RE methylation prediction. data(alu.demo) remparcel <- initremp(arraytype = '450k', REtype = 'Alu', RE = Alu.demo, ncore = 1) remparcel # Save the data for later use saveparcel(remparcel) remp Repetitive element methylation prediction remp is used to predict genomewide methylation levels of locus-specific repetitive elements (RE). Two major RE types in human, Alu element (Alu) and LINE-1 (L1) are available.
11 remp 11 remp(methydat, REtype = c("alu", "L1"), parcel = NULL, work.dir = tempdir(), win = 1000, method = c("rf", "svmlinear", "svmradial", "naive"), autotune = TRUE, param = NULL, seed = NULL, ncore = NULL, BPPARAM = NULL, verbose = FALSE) methydat REtype parcel work.dir win method autotune param seed ncore BPPARAM verbose A RatioSet, GenomicRatioSet, DataFrame, data.table, data.frame, or matrix of Illumina BeadChip methylation data (450k or EPIC array). See Details. Type of RE. Currently "Alu" and "L1" are supported. An REMParcel object containing necessary data to carry out the prediction. If NULL, the function will search the.rds data file in work.dir exported by initremp (with export = TRUE) or saveparcel. Path to the directory where the annotation data generated by initremp are saved. Valid when the argument parcel is missing. If not specified, temporary directory tempdir() will be used. If specified, the directory path has to be the same as the one specified in initremp or in saveparcel. An integer specifying window size to confine the upstream and downstream flanking region centered on the predicted CpG in RE for prediction. Default = See Details. Name of model/approach for prediction. Currently "rf" (Random Forest), "svmlinear" (SVM with linear kernel), "svmradial" (SVM with linear kernel), and "naive" (carrying over methylation values of the closest CpG site) are available. Default = "rf" (Random Forest). See Details. Logical parameter. If TRUE, a 3-time repeated 5-fold cross validation will be performed to determine the best model parameter. If FALSE, the param option (see below) must be specified. Default = TRUE. Auto-tune will be disabled using Random Forest. See Details. A number or a vector specifying the model tuning parameter(s) (not applicable for Random Forest). For SVM, param represents Cost (for linear kernel) or Sigma and Cost (for radial basis function kernel). This parameter is valid only when autotune = FALSE. Random seed for Random Forest model for reproducible prediction results. Default is NULL, which generates a seed. Number of cores to run parallel computation. By default, max number of cores available in the machine will be utilized. If ncore = 1, no parallel computation is allowed. An optional BiocParallelParam instance determining the parallel back-end to be used during evaluation. If not specified, default back-end in the machine will be used. Logical parameter. Should the function be verbose? Details Before running remp, user should make sure the methylation data have gone through proper quality control, background correction, and normalization procedures. Both beta value and M value are allowed. Rows represents probes and columns represents samples. Please make sure to have row
12 12 remp names that specify the Illumina probe ID (i.e. cg ). Parameter win = 1000 is based on previous findings showing that neighboring CpGs are more likely to be co-modified within 1000 bp. User can specify narrower window size for slight improvement of prediction accuracy at the cost of less predicted RE. Window size greater than 1000 is not recommended as the machine learning models would not be able to learn much userful information for prediction but introduce noise. Random Forest model (method = "rf") is recommented as it offers more accurate prediction and it also enables prediction reliability functionality. Prediction reliability is estimated by conditional standard deviation using Quantile Regression Forest. Please note that if parallel computing is allowed, parallel Random Forest (powered by package ranger) will be used automatically. The performance of Random Forest model is often relatively insensitive to the choice of mtry. Therefore, auto-tune will be turned off using Random Forest and mtry will be set to one third of the total number of predictors. For SVM, if autotune = TRUE, preset tuning parameter search grid can be access and modified using remp_options. See Also A REMProduct object containing predicted RE methylation results. See initremp to prepare necessary annotation database before running remp. # Obtain example Illumina example data (450k) GM12878_450k <- getgm12878('450k') # Make sure you have run 'initremp'. See?initREMP. # Run prediction remp.res <- remp(gm12878_450k, REtype = 'Alu', ncore = 1) remp.res details(remp.res) rempb(remp.res) # Methylation data (beta value) # Extract CpG location information (inherit from class 'RangedSummarizedExperiment') rowranges(remp.res) # RE annotation information rempannot(remp.res) # Add gene annotation remp.res <- decodeannot(remp.res, type = "symbol") rempannot(remp.res) # (Recommended) Trim off less reliable prediction remp.res <- remptrim(remp.res) # (Recommended) Obtain RE-level methylation (aggregate by mean) remp.res <- rempaggregate(remp.res) rempb(remp.res) # Methylation data (beta value) # Extract RE location information rowranges(remp.res) # Density plot across predicted RE
13 REMParcel-class 13 plot(remp.res) REMParcel-class REMParcel instances REMParcel is a container class to organize required datasets for RE methylation prediction generated from initremp and used in remp. REMParcel(REtype = "Unknown", platform = "Unknown", RefGene = GRanges(), RE = GRanges(), RECpG = GRanges(), ILMN = GRanges()) getrefgene(object) getre(object) getrecpg(object) getilmn(object,...) saveparcel(object,...) ## S4 method for signature 'REMParcel' saveparcel(object, work.dir = tempdir(), verbose = FALSE,...) ## S4 method for signature 'REMParcel' getrefgene(object) ## S4 method for signature 'REMParcel' getre(object) ## S4 method for signature 'REMParcel' getrecpg(object) ## S4 method for signature 'REMParcel' getilmn(object, REonly = FALSE) REtype platform RefGene RE Type of RE ("Alu" or "L1"). Illumina methylation profiling platform ("450k" or "EPIC"). refseq gene annotation data, which can be obtained by fetchrefseqgene. Annotated RE genomic range data, which can be obtained by fetchrmsk and annotated by GRannot.
14 14 REMProduct-class RECpG ILMN object Genomic range data of annotated CpG site identified in RE DNA sequence, which can be obtained by findrecpg and annotated by GRannot. Illumina CpG probe genomic range data. A REMParcel object.... For saveparcel: other parameters to be passed to the saverds method. See saverds. work.dir verbose REonly For saveparcel: path to the directory where the generated data will be saved. If not specified, temporary directory tempdir() will be used. For saveparcel: logical parameter. Should the function be verbose? For getilmn: see Accessors. An object of class REMParcel for the constructor. Accessors getrefgene(object) Return RefSeq gene annotation data. getre(object) Return RE genomic location data for prediction (annotated by refseq gene database). getrecpg(object) Return RE-CpG genomic location data for prediction. getilmn(object, REonly = FALSE) Return Illumina CpG probe genomic location data for prediction (annotated by refseq gene database). If REonly = TRUE, only probes within RE region are returned. Utilities saveparcel(object, work.dir = tempdir(), verbose = FALSE,...) Save the object to local machine. showclass("remparcel") REMProduct-class REMProduct instances Class REMProduct is to maintain RE methylation prediction results. REMProduct inherits Bioconductor s RangedSummarizedExperiment class.
15 REMProduct-class 15 REMProduct(REtype = "Unknown", platform = "Unknown", win = "Unknown", predictmodel = "Unknown", QCModel = "Unknown", rempm = NULL, rempb = NULL, rempqc = NULL, cpgranges = GRanges(), sampleinfo = DataFrame(), REannotation = GRanges(), RECpG = GRanges(), regioncode = DataFrame(), refgene = GRanges(), varimp = DataFrame(), REStats = DataFrame(), GeneStats = DataFrame(), Seed = NULL) rempm(object) rempb(object) rempqc(object) rempannot(object) rempimp(object) rempstats(object) details(object) decodeannot(object,...) remptrim(object,...) rempaggregate(object,...) rempcombine(object1, object2) ## S4 method for signature 'REMProduct' rempm(object) ## S4 method for signature 'REMProduct' rempb(object) ## S4 method for signature 'REMProduct' rempqc(object) ## S4 method for signature 'REMProduct' rempimp(object) ## S4 method for signature 'REMProduct' rempannot(object) ## S4 method for signature 'REMProduct' rempstats(object) ## S4 method for signature 'REMProduct,missing' plot(x, type = c("individual", "overall"),...)
16 16 REMProduct-class ## S4 method for signature 'REMProduct' details(object) ## S4 method for signature 'REMProduct' decodeannot(object, type = c("symbol", "entrez"), ncore = 1, BPPARAM = NULL) ## S4 method for signature 'REMProduct' remptrim(object, threshold = 1.7, missingrate = 0.2) ## S4 method for signature 'REMProduct' rempaggregate(object, NCpG = 2, ncore = 1, BPPARAM = NULL) ## S4 method for signature 'REMProduct,REMProduct' rempcombine(object1, object2) REtype platform win predictmodel QCModel rempm rempb rempqc cpgranges sampleinfo REannotation RECpG regioncode refgene varimp REStats GeneStats Seed object Type of RE ("Alu" or "L1"). Illumina methylation profiling platform ("450k" or "EPIC"). Flanking window size of the predicting RE-CpG. Name of the model used for prediction. Name of the model used for prediction quality evaluation. Predicted methylation level in M value. Predicted methylation level in beta value (optional). Prediction quality scores, which is available only when Random Forest model is used in remp. Genomic ranges of the predicting RE-CpG. Sample information. Annotation data for the predicting RE. Annotation data for the RE-CpG profiled by Illumina platform. Internal index code defined in refgene for gene region indicators. refseq gene annotation data, which can be obtained by fetchrefseqgene. Importance of the predictors. RE coverage statistics, which is internally generated in remp. Gene coverage statistics, which is internally generated in remp. Random seed for Random Forest model for reproducible prediction results. A REMProduct object.... For plot: graphical parameters to be passed to the plot method. object1 object2 x type A REMProduct object. A REMProduct object. For plot: an REMProduct object. For plot and decodeannot: see Utilities.
17 REMProduct-class 17 ncore BPPARAM threshold missingrate NCpG For decodeannot and rempaggregate: number of cores to run parallel computation. By default no parallel computation is allowed (ncore = 1). For decodeannot and rempaggregate: an optional BiocParallelParam instance determining the parallel back-end to be used during evaluation. If not specified, default back-end in the machine will be used. For remptrim: see Utilities. For remptrim: see Utilities. For rempaggregate: see Utilities. An object of class REMProduct for the constructor. Accessors rempm(object) Return M value of the prediction. rempb(object) Return beta value of the prediction. rempqc(object) Return prediction quality scores. rempimp(object) Return relative importance of predictors. rempstats(object) Return RE and gene coverage statistics. rempannot(object) Return annotation data for the predicted RE. Utilities plot(x, type = c("individual", "overall"),...) Make a density plot of predicted methylation (beta values) in the REMProduct object x. If type = "individual", density curves will be plotted for each of the samples; If type = "overall", one density curve of the mean methylation level across the samples will be plotted. Default type = "individual". details(object) Display detailed descriptive statistics of the predicion results. decodeannot(object, type = c("symbol", "entrez")), ncore = NULL, BPPARAM = NULL Decode the RE annotation data by Gene Symbol (when type = "Symbol") or Entrez Gene (when type = "Entrez").Default type = "Symbol". Annotation data are provided by org.hs.eg.db. remptrim(object, threshold = 1.7, missingrate = 0.2) Any predicted CpG values with quality score < threshold (default = 1.7, specified by threshold = 1.7) will be replaced with NA. CpGs contain more than missingrate * 100 rate across samples will be discarded. Relavant statistics will be re-evaluated. rempaggregate(object, NCpG = 2, ncore = NULL, BPPARAM = NULL) Aggregate the predicted RE- CpG methylation by RE using mean. To ensure the reliability of the aggregation, by default only RE with at least 2 predicted CpG sites (specified by NCpG = 2) will be aggregated. rempcombine(object1, object2) Combine two REMProduct objects by column. showclass("remproduct")
18 18 remprofile remprofile Extract DNA methylation data profiled in RE remprofile is used to extract profiled methylation of CpG sites in RE. remprofile(methydat, REtype = c("alu", "L1"), RE = NULL, impute = TRUE, imputebyrow = TRUE, verbose = FALSE) methydat REtype RE impute imputebyrow verbose A RatioSet, GenomicRatioSet, DataFrame, data.table, data.frame, or matrix of Illumina BeadChip methylation data (450k or EPIC array). Type of RE. Currently "Alu" and "L1" are supported. A GRanges object containing user-specified RE genomic location information. If NULL, the function will retrive RepeatMasker RE database from AnnotationHub (build hg19). Parameter used by groomethy. If TRUE, K-Nearest Neighbouring imputation will be applied to fill the missing values. Default = TRUE. Parameter used by groomethy. If TRUE, missing values will be imputed using similar values in row (i.e., across samples); if FALSE, missing values will be imputed using similar values in column (i.e., across CpGs). Default is TRUE. Logical parameter. Should the function be verbose? A REMProduct object containing profiled RE methylation results. data(alu.demo) GM12878_450k <- getgm12878('450k') remp.res <- remprofile(gm12878_450k, REtype = "Alu", RE = Alu.demo) details(remp.res) rempb(remp.res) # Methylation data (beta value) remp.res <- rempaggregate(remp.res) details(remp.res) rempb(remp.res) # Methylation data (beta value)
19 remp_options 19 remp_options Set or get options for REMP package Tools to manage global setting options for REMP package. remp_options(...) remp_reset()... Option names to retrieve option values or [key]=[value] pairs to set options. NULL Supported options The following options are supported.default.alufamily A list of Alu subfamily to be included in the prediction..default.l1family A list of L1 subfamily to be included in the prediction..default.gm k.url URL to download GM k methylation profiling data..default.ah.repeatmasker.hg19 AnnotationHub data ID linked to RepeatMasker database (build hg19).default.ah.refgene.hg19 AnnotationHub data ID linked to refseq gene database (build hg19).default.tss.upstream Define the upstream range of transcription start site region..default.tss.downstream Define the downstream range of transcription start site region..default.max.flankwindow Define the max size of the flanking window surrounding the predicted RE-CpG..default.450k.total.probes Total number of probes designed in Illumina 450k array..default.epic.total.probes Total number of probes designed in Illumina EPIC array..default.450k.annotation A character string associated with the Illumina 450k array annotation dataset..default.epic.annotation A character string associated with the Illumina EPIC array annotation dataset..default.genomicregioncolnames Define the names of the genomic regions for prediction..default.predictors Define the names of predictors for RE methylation prediction..default.c.svmlinear.tune Define the default C (Cost) parameter for Support Vector Machine (SVM) using linear kernel..default.sigma.svmradial.tune Define the default sigma parameter for SVM using Radial basis function kernel..default.c.svmradial.tune Define the default C (Cost) parameter for SVM using Radial basis function kernel.
20 20 remp_options # Display all default settings remp_options() # Display a specified setting remp_options(".default.max.flankwindow") # Change default maximum flanking window size to 2000 remp_options(.default.max.flankwindow = 2000) # Reset all options remp_reset()
21 Index Topic datasets Alu.demo, 3 Topic package REMP-package, 2 Alu.demo, 3 AnnotationHub, 5, 9, 10, 18 BiocParallel, 5, 6 BiocParallelParam, 10, 11, 17 DataFrame, 8, 11, 18 decodeannot decodeannot,remproduct-method details details,remproduct-method fetchrefseqgene, 4, 8, 13, 16 fetchrmsk, 4, 5, 13 findrecpg, 5, 14 GenomicRatioSet, 7 9, 11, 18 getbackend, 5, 6 getgm12878, 7 getilmn (REMParcel-class), 13 getilmn,remparcel-method (REMParcel-class), 13 getre (REMParcel-class), 13 getre,remparcel-method (REMParcel-class), 13 getrecpg (REMParcel-class), 13 getrecpg,remparcel-method (REMParcel-class), 13 getrefgene (REMParcel-class), 13 getrefgene,remparcel-method (REMParcel-class), 13 GRanges, 4 6, 8, 9, 18 GRangesList, 4, 8 GRannot, 7, 13, 14 graphical parameters, 16 groomethy, 2, 8, 18 impute.knn, 9 initremp, 2, 9, org.hs.eg.db, 17 plot,remproduct,missing-method ranger, 12 RatioSet, 7 9, 11, 18 REMP (REMP-package), 2 remp, 2, 9, 10, 10, 13, 16 REMP-package, 2 remp_options, 10, 12, 19 remp_reset (remp_options), 19 rempaggregate rempaggregate,remproduct-method rempannot rempannot,remproduct-method REMParcel, 2, 10, 11 REMParcel (REMParcel-class), 13 REMParcel-class, 13 rempb rempb,remproduct-method rempcombine rempcombine,remproduct,remproduct-method rempimp rempimp,remproduct-method rempm rempm,remproduct-method rempqc rempqc,remproduct-method REMProduct, 3, 12, 18 REMProduct REMProduct-class, 14 remprofile, 18 rempstats 21
22 22 INDEX rempstats,remproduct-method remptrim remptrim,remproduct-method saveparcel, 11 saveparcel (REMParcel-class), 13 saveparcel,remparcel-method (REMParcel-class), 13 saverds, 14
Package dmrseq. September 14, 2018
 Type Package Package dmrseq September 14, 2018 Title Detection and inference of differentially methylated regions from Whole Genome Bisulfite Sequencing Version 1.1.15 Author Keegan Korthauer ,
Type Package Package dmrseq September 14, 2018 Title Detection and inference of differentially methylated regions from Whole Genome Bisulfite Sequencing Version 1.1.15 Author Keegan Korthauer ,
Package DMCHMM. January 19, 2019
 Package DMCHMM January 19, 2019 Type Package Title Differentially Methylated CpG using Hidden Markov Model Version 1.4.0 Author Farhad Shokoohi Maintainer A pipeline for identifying differentially methylated
Package DMCHMM January 19, 2019 Type Package Title Differentially Methylated CpG using Hidden Markov Model Version 1.4.0 Author Farhad Shokoohi Maintainer A pipeline for identifying differentially methylated
Package MEAL. August 16, 2018
 Title Perform methylation analysis Version 1.11.6 Package MEAL August 16, 2018 Package to integrate methylation and expression data. It can also perform methylation or expression analysis alone. Several
Title Perform methylation analysis Version 1.11.6 Package MEAL August 16, 2018 Package to integrate methylation and expression data. It can also perform methylation or expression analysis alone. Several
Package Organism.dplyr
 Package Organism.dplyr January 9, 2019 Title dplyr-based Access to Bioconductor Annotation Resources Version 1.11.0 Description This package provides an alternative interface to Bioconductor 'annotation'
Package Organism.dplyr January 9, 2019 Title dplyr-based Access to Bioconductor Annotation Resources Version 1.11.0 Description This package provides an alternative interface to Bioconductor 'annotation'
Package MIRA. October 16, 2018
 Version 1.2.0 Date 2018-04-17 Package MIRA October 16, 2018 Title Methylation-Based Inference of Regulatory Activity MIRA measures the degree of ``dip'' in methylation level surrounding a regulatory site
Version 1.2.0 Date 2018-04-17 Package MIRA October 16, 2018 Title Methylation-Based Inference of Regulatory Activity MIRA measures the degree of ``dip'' in methylation level surrounding a regulatory site
Package metagene. November 1, 2018
 Version 2.15.0 Date 2017-10-13 Title A package to produce metagene plots Author Charles Joly Beauparlant Package metagene November 1, 2018 , Fabien Claude Lamaze
Version 2.15.0 Date 2017-10-13 Title A package to produce metagene plots Author Charles Joly Beauparlant Package metagene November 1, 2018 , Fabien Claude Lamaze
Package conumee. February 17, 2018
 Package conumee February 17, 2018 Title Enhanced copy-number variation analysis using Illumina DNA methylation arrays Version 1.13.0 Author Volker Hovestadt, Marc Zapatka Maintainer Address Division of
Package conumee February 17, 2018 Title Enhanced copy-number variation analysis using Illumina DNA methylation arrays Version 1.13.0 Author Volker Hovestadt, Marc Zapatka Maintainer Address Division of
Package methyvim. April 5, 2019
 Package methyvim April 5, 2019 Title Targeted, Robust, and Model-free Differential Methylation Analysis Version 1.4.0 Maintainer Nima Hejazi Author Nima Hejazi [aut, cre, cph], Mark
Package methyvim April 5, 2019 Title Targeted, Robust, and Model-free Differential Methylation Analysis Version 1.4.0 Maintainer Nima Hejazi Author Nima Hejazi [aut, cre, cph], Mark
Package BiocNeighbors
 Version 0.99.22 Date 2018-10-15 Package BiocNeighbors October 16, 2018 Title Nearest Neighbor Detection for Bioconductor Packages Depends R (>= 3.5), BiocParallel Imports Rcpp, S4Vectors, stats, methods
Version 0.99.22 Date 2018-10-15 Package BiocNeighbors October 16, 2018 Title Nearest Neighbor Detection for Bioconductor Packages Depends R (>= 3.5), BiocParallel Imports Rcpp, S4Vectors, stats, methods
Package DMRcate. May 1, 2018
 Package DMRcate May 1, 2018 Title Methylation array and sequencing spatial analysis methods Version 1.16.0 Date 2017-07-02 Author Tim Peters Maintainer Tim Peters De novo identification
Package DMRcate May 1, 2018 Title Methylation array and sequencing spatial analysis methods Version 1.16.0 Date 2017-07-02 Author Tim Peters Maintainer Tim Peters De novo identification
Package ENmix. September 17, 2015
 Version 1.0.0 Date 2015-04-15 Package ENmix September 17, 2015 Title Data preprocessing and quality control for Illumina HumanMethylation450 BeadChip Type Package Illumina HumanMethylation450 BeadChip
Version 1.0.0 Date 2015-04-15 Package ENmix September 17, 2015 Title Data preprocessing and quality control for Illumina HumanMethylation450 BeadChip Type Package Illumina HumanMethylation450 BeadChip
Package seqcat. March 25, 2019
 Package seqcat March 25, 2019 Title High Throughput Sequencing Cell Authentication Toolkit Version 1.4.1 The seqcat package uses variant calling data (in the form of VCF files) from high throughput sequencing
Package seqcat March 25, 2019 Title High Throughput Sequencing Cell Authentication Toolkit Version 1.4.1 The seqcat package uses variant calling data (in the form of VCF files) from high throughput sequencing
Package compartmap. November 10, Type Package
 Type Package Package compartmap November 10, 2018 Title A/B compartment inference from ATAC-seq and methylation array data Compartmap performs shrunken A/B compartment inference from ATACseq and methylation
Type Package Package compartmap November 10, 2018 Title A/B compartment inference from ATAC-seq and methylation array data Compartmap performs shrunken A/B compartment inference from ATACseq and methylation
Package ChIPseeker. July 12, 2018
 Type Package Package ChIPseeker July 12, 2018 Title ChIPseeker for ChIP peak Annotation, Comparison, and Visualization Version 1.16.0 Encoding UTF-8 Maintainer Guangchuang Yu
Type Package Package ChIPseeker July 12, 2018 Title ChIPseeker for ChIP peak Annotation, Comparison, and Visualization Version 1.16.0 Encoding UTF-8 Maintainer Guangchuang Yu
Package scmap. March 29, 2019
 Type Package Package scmap March 29, 2019 Title A tool for unsupervised projection of single cell RNA-seq data Version 1.4.1 Author Vladimir Kiselev Maintainer Vladimir Kiselev
Type Package Package scmap March 29, 2019 Title A tool for unsupervised projection of single cell RNA-seq data Version 1.4.1 Author Vladimir Kiselev Maintainer Vladimir Kiselev
Package RcisTarget. July 17, 2018
 Type Package Package RcisTarget July 17, 2018 Title RcisTarget: Identify transcription factor binding motifs enriched on a gene list Version 1.1.2 Date 2018-05-26 Author Sara Aibar, Gert Hulselmans, Stein
Type Package Package RcisTarget July 17, 2018 Title RcisTarget: Identify transcription factor binding motifs enriched on a gene list Version 1.1.2 Date 2018-05-26 Author Sara Aibar, Gert Hulselmans, Stein
Package RTCGAToolbox
 Type Package Package RTCGAToolbox November 27, 2017 Title A new tool for exporting TCGA Firehose data Version 2.9.2 Author Mehmet Kemal Samur Maintainer Marcel Ramos Managing
Type Package Package RTCGAToolbox November 27, 2017 Title A new tool for exporting TCGA Firehose data Version 2.9.2 Author Mehmet Kemal Samur Maintainer Marcel Ramos Managing
Package bnbc. R topics documented: April 1, Version 1.4.0
 Version 1.4.0 Package bnbc April 1, 2019 Title Bandwise normalization and batch correction of Hi-C data Tools to normalize (several) Hi-C data from replicates. Depends R (>= 3.4.0), methods, BiocGenerics,
Version 1.4.0 Package bnbc April 1, 2019 Title Bandwise normalization and batch correction of Hi-C data Tools to normalize (several) Hi-C data from replicates. Depends R (>= 3.4.0), methods, BiocGenerics,
Package TFutils. October 11, 2018
 Title TFutils Package to work with TF data. Version 1.0.0 Depends R (>= 3.5.0) Package TFutils October 11, 2018 Imports methods, GenomicRanges, IRanges, S4Vectors, GSEABase, shiny, miniui, data.table,
Title TFutils Package to work with TF data. Version 1.0.0 Depends R (>= 3.5.0) Package TFutils October 11, 2018 Imports methods, GenomicRanges, IRanges, S4Vectors, GSEABase, shiny, miniui, data.table,
Package multihiccompare
 Package multihiccompare September 23, 2018 Title Normalize and detect differences between Hi-C datasets when replicates of each experimental condition are available Version 0.99.44 multihiccompare provides
Package multihiccompare September 23, 2018 Title Normalize and detect differences between Hi-C datasets when replicates of each experimental condition are available Version 0.99.44 multihiccompare provides
Package decomptumor2sig
 Type Package Package February 8, 2019 Title Decomposition of individual tumors into mutational signatures by signature refitting Depends R(>= 3.5), ggplot2 Imports methods, Matrix, quadprog(>= 1.5-5),
Type Package Package February 8, 2019 Title Decomposition of individual tumors into mutational signatures by signature refitting Depends R(>= 3.5), ggplot2 Imports methods, Matrix, quadprog(>= 1.5-5),
Package M3D. March 21, 2019
 Type Package Package M3D March 21, 2019 Title Identifies differentially methylated regions across testing groups Version 1.16.0 Date 2016-06-22 Author Tom Mayo Maintainer This package identifies statistically
Type Package Package M3D March 21, 2019 Title Identifies differentially methylated regions across testing groups Version 1.16.0 Date 2016-06-22 Author Tom Mayo Maintainer This package identifies statistically
Handling genomic data using Bioconductor II: GenomicRanges and GenomicFeatures
 Handling genomic data using Bioconductor II: GenomicRanges and GenomicFeatures Motivating examples Genomic Features (e.g., genes, exons, CpG islands) on the genome are often represented as intervals, e.g.,
Handling genomic data using Bioconductor II: GenomicRanges and GenomicFeatures Motivating examples Genomic Features (e.g., genes, exons, CpG islands) on the genome are often represented as intervals, e.g.,
Package methylgsa. March 7, 2019
 Package methylgsa March 7, 2019 Type Package Title Gene Set Analysis Using the Outcome of Differential Methylation Version 1.1.3 The main functions for methylgsa are methylglm and methylrra. methylgsa
Package methylgsa March 7, 2019 Type Package Title Gene Set Analysis Using the Outcome of Differential Methylation Version 1.1.3 The main functions for methylgsa are methylglm and methylrra. methylgsa
Package roar. August 31, 2018
 Type Package Package roar August 31, 2018 Title Identify differential APA usage from RNA-seq alignments Version 1.16.0 Date 2016-03-21 Author Elena Grassi Maintainer Elena Grassi Identify
Type Package Package roar August 31, 2018 Title Identify differential APA usage from RNA-seq alignments Version 1.16.0 Date 2016-03-21 Author Elena Grassi Maintainer Elena Grassi Identify
IRanges and GenomicRanges An introduction
 IRanges and GenomicRanges An introduction Kasper Daniel Hansen CSAMA, Brixen 2011 1 / 28 Why you should care IRanges and GRanges are data structures I use often to solve a variety of
IRanges and GenomicRanges An introduction Kasper Daniel Hansen CSAMA, Brixen 2011 1 / 28 Why you should care IRanges and GRanges are data structures I use often to solve a variety of
Package EventPointer
 Type Package Package EventPointer September 5, 2018 Title An effective identification of alternative splicing events using junction arrays and RNA-Seq data Version 1.4.0 Author Juan Pablo Romero, Ander
Type Package Package EventPointer September 5, 2018 Title An effective identification of alternative splicing events using junction arrays and RNA-Seq data Version 1.4.0 Author Juan Pablo Romero, Ander
Package LOLA. December 24, 2018
 Package LOLA December 24, 2018 Version 1.13.0 Date 2015-06-26 Title Locus overlap analysis for enrichment of genomic ranges Provides functions for testing overlap of sets of genomic regions with public
Package LOLA December 24, 2018 Version 1.13.0 Date 2015-06-26 Title Locus overlap analysis for enrichment of genomic ranges Provides functions for testing overlap of sets of genomic regions with public
Package preprocomb. June 26, 2016
 Type Package Title Tools for Preprocessing Combinations Version 0.3.0 Date 2016-6-26 Author Markus Vattulainen Package preprocomb June 26, 2016 Maintainer Markus Vattulainen
Type Package Title Tools for Preprocessing Combinations Version 0.3.0 Date 2016-6-26 Author Markus Vattulainen Package preprocomb June 26, 2016 Maintainer Markus Vattulainen
Package rgreat. June 19, 2018
 Type Package Title Client for GREAT Analysis Version 1.12.0 Date 2018-2-20 Author Zuguang Gu Maintainer Zuguang Gu Package rgreat June 19, 2018 Depends R (>= 3.1.2), GenomicRanges, IRanges,
Type Package Title Client for GREAT Analysis Version 1.12.0 Date 2018-2-20 Author Zuguang Gu Maintainer Zuguang Gu Package rgreat June 19, 2018 Depends R (>= 3.1.2), GenomicRanges, IRanges,
Package DRIMSeq. December 19, 2018
 Type Package Package DRIMSeq December 19, 2018 Title Differential transcript usage and tuqtl analyses with Dirichlet-multinomial model in RNA-seq Version 1.10.0 Date 2017-05-24 Description The package
Type Package Package DRIMSeq December 19, 2018 Title Differential transcript usage and tuqtl analyses with Dirichlet-multinomial model in RNA-seq Version 1.10.0 Date 2017-05-24 Description The package
Package CAGEr. September 4, 2018
 Package CAGEr September 4, 2018 Title Analysis of CAGE (Cap Analysis of Gene Expression) sequencing data for precise mapping of transcription start sites and promoterome mining Version 1.23.4 Date 2018-08-29
Package CAGEr September 4, 2018 Title Analysis of CAGE (Cap Analysis of Gene Expression) sequencing data for precise mapping of transcription start sites and promoterome mining Version 1.23.4 Date 2018-08-29
segmentseq: methods for detecting methylation loci and differential methylation
 segmentseq: methods for detecting methylation loci and differential methylation Thomas J. Hardcastle October 30, 2018 1 Introduction This vignette introduces analysis methods for data from high-throughput
segmentseq: methods for detecting methylation loci and differential methylation Thomas J. Hardcastle October 30, 2018 1 Introduction This vignette introduces analysis methods for data from high-throughput
Package igc. February 10, 2018
 Type Package Package igc February 10, 2018 Title An integrated analysis package of Gene expression and Copy number alteration Version 1.8.0 This package is intended to identify differentially expressed
Type Package Package igc February 10, 2018 Title An integrated analysis package of Gene expression and Copy number alteration Version 1.8.0 This package is intended to identify differentially expressed
Package gmapr. January 22, 2019
 Package gmapr January 22, 2019 Maintainer Michael Lawrence License Artistic-2.0 Title An R interface to the GMAP/GSNAP/GSTRUCT suite Type Package Author Cory Barr, Thomas Wu,
Package gmapr January 22, 2019 Maintainer Michael Lawrence License Artistic-2.0 Title An R interface to the GMAP/GSNAP/GSTRUCT suite Type Package Author Cory Barr, Thomas Wu,
segmentseq: methods for detecting methylation loci and differential methylation
 segmentseq: methods for detecting methylation loci and differential methylation Thomas J. Hardcastle October 13, 2015 1 Introduction This vignette introduces analysis methods for data from high-throughput
segmentseq: methods for detecting methylation loci and differential methylation Thomas J. Hardcastle October 13, 2015 1 Introduction This vignette introduces analysis methods for data from high-throughput
Package MethylSeekR. January 26, 2019
 Type Package Title Segmentation of Bis-seq data Version 1.22.0 Date 2014-7-1 Package MethylSeekR January 26, 2019 Author Lukas Burger, Dimos Gaidatzis, Dirk Schubeler and Michael Stadler Maintainer Lukas
Type Package Title Segmentation of Bis-seq data Version 1.22.0 Date 2014-7-1 Package MethylSeekR January 26, 2019 Author Lukas Burger, Dimos Gaidatzis, Dirk Schubeler and Michael Stadler Maintainer Lukas
Package cattonum. R topics documented: May 2, Type Package Version Title Encode Categorical Features
 Type Package Version 0.0.2 Title Encode Categorical Features Package cattonum May 2, 2018 Functions for dummy encoding, frequency encoding, label encoding, leave-one-out encoding, mean encoding, median
Type Package Version 0.0.2 Title Encode Categorical Features Package cattonum May 2, 2018 Functions for dummy encoding, frequency encoding, label encoding, leave-one-out encoding, mean encoding, median
Package GEM. R topics documented: January 31, Type Package
 Type Package Package GEM January 31, 2018 Title GEM: fast association study for the interplay of Gene, Environment and Methylation Version 1.5.0 Date 2015-12-05 Author Hong Pan, Joanna D Holbrook, Neerja
Type Package Package GEM January 31, 2018 Title GEM: fast association study for the interplay of Gene, Environment and Methylation Version 1.5.0 Date 2015-12-05 Author Hong Pan, Joanna D Holbrook, Neerja
Package RNASeqR. January 8, 2019
 Type Package Package RNASeqR January 8, 2019 Title RNASeqR: RNA-Seq workflow for case-control study Version 1.1.3 Date 2018-8-7 Author Maintainer biocviews Genetics, Infrastructure,
Type Package Package RNASeqR January 8, 2019 Title RNASeqR: RNA-Seq workflow for case-control study Version 1.1.3 Date 2018-8-7 Author Maintainer biocviews Genetics, Infrastructure,
Package gpart. November 19, 2018
 Package gpart November 19, 2018 Title Human genome partitioning of dense sequencing data by identifying haplotype blocks Version 1.0.0 Depends R (>= 3.5.0), grid, Homo.sapiens, TxDb.Hsapiens.UCSC.hg38.knownGene,
Package gpart November 19, 2018 Title Human genome partitioning of dense sequencing data by identifying haplotype blocks Version 1.0.0 Depends R (>= 3.5.0), grid, Homo.sapiens, TxDb.Hsapiens.UCSC.hg38.knownGene,
Package genomeintervals
 Version 1.38.0 Date 2017-04-05 Type Package Title Operations on genomic intervals Package genomeintervals April 16, 2019 Author Julien Gagneur , Joern Toedling, Richard Bourgon,
Version 1.38.0 Date 2017-04-05 Type Package Title Operations on genomic intervals Package genomeintervals April 16, 2019 Author Julien Gagneur , Joern Toedling, Richard Bourgon,
Package SC3. September 29, 2018
 Type Package Title Single-Cell Consensus Clustering Version 1.8.0 Author Vladimir Kiselev Package SC3 September 29, 2018 Maintainer Vladimir Kiselev A tool for unsupervised
Type Package Title Single-Cell Consensus Clustering Version 1.8.0 Author Vladimir Kiselev Package SC3 September 29, 2018 Maintainer Vladimir Kiselev A tool for unsupervised
epigenomegateway.wustl.edu
 Everything can be found at epigenomegateway.wustl.edu REFERENCES 1. Zhou X, et al., Nature Methods 8, 989-990 (2011) 2. Zhou X & Wang T, Current Protocols in Bioinformatics Unit 10.10 (2012) 3. Zhou X,
Everything can be found at epigenomegateway.wustl.edu REFERENCES 1. Zhou X, et al., Nature Methods 8, 989-990 (2011) 2. Zhou X & Wang T, Current Protocols in Bioinformatics Unit 10.10 (2012) 3. Zhou X,
Goldmine: Integrating information to place sets of genomic ranges into biological contexts Jeff Bhasin
 Goldmine: Integrating information to place sets of genomic ranges into biological contexts Jeff Bhasin 2016-04-22 Contents Purpose 1 Prerequisites 1 Installation 2 Example 1: Annotation of Any Set of Genomic
Goldmine: Integrating information to place sets of genomic ranges into biological contexts Jeff Bhasin 2016-04-22 Contents Purpose 1 Prerequisites 1 Installation 2 Example 1: Annotation of Any Set of Genomic
Package HTSeqGenie. April 16, 2019
 Package HTSeqGenie April 16, 2019 Imports BiocGenerics (>= 0.2.0), S4Vectors (>= 0.9.25), IRanges (>= 1.21.39), GenomicRanges (>= 1.23.21), Rsamtools (>= 1.8.5), Biostrings (>= 2.24.1), chipseq (>= 1.6.1),
Package HTSeqGenie April 16, 2019 Imports BiocGenerics (>= 0.2.0), S4Vectors (>= 0.9.25), IRanges (>= 1.21.39), GenomicRanges (>= 1.23.21), Rsamtools (>= 1.8.5), Biostrings (>= 2.24.1), chipseq (>= 1.6.1),
Package AnnotationHub
 Type Package Package AnnotationHub Title Client to access AnnotationHub resources Version 2.11.2 November 27, 2017 biocviews Infrastructure, DataImport, GUI, ThirdPartyClient Maintainer Bioconductor Package
Type Package Package AnnotationHub Title Client to access AnnotationHub resources Version 2.11.2 November 27, 2017 biocviews Infrastructure, DataImport, GUI, ThirdPartyClient Maintainer Bioconductor Package
Package customprodb. September 9, 2018
 Type Package Package customprodb September 9, 2018 Title Generate customized protein database from NGS data, with a focus on RNA-Seq data, for proteomics search Version 1.20.2 Date 2018-08-08 Author Maintainer
Type Package Package customprodb September 9, 2018 Title Generate customized protein database from NGS data, with a focus on RNA-Seq data, for proteomics search Version 1.20.2 Date 2018-08-08 Author Maintainer
Package mimager. March 7, 2019
 Package mimager March 7, 2019 Version 1.6.0 Type Package Title mimager: The Microarray Imager Easily visualize and inspect microarrays for spatial artifacts. License MIT + file LICENSE LazyData TRUE Depends
Package mimager March 7, 2019 Version 1.6.0 Type Package Title mimager: The Microarray Imager Easily visualize and inspect microarrays for spatial artifacts. License MIT + file LICENSE LazyData TRUE Depends
Package STROMA4. October 22, 2018
 Version 1.5.2 Date 2017-03-23 Title Assign Properties to TNBC Patients Package STROMA4 October 22, 2018 This package estimates four stromal properties identified in TNBC patients in each patient of a gene
Version 1.5.2 Date 2017-03-23 Title Assign Properties to TNBC Patients Package STROMA4 October 22, 2018 This package estimates four stromal properties identified in TNBC patients in each patient of a gene
Package JunctionSeq. November 1, 2018
 Package JunctionSeq November 1, 2018 Version 1.12.0 Title JunctionSeq: A Utility for Detection of Differential Exon and Splice-Junction Usage in RNA-Seq data Depends R (>= 3.2.2), methods, SummarizedExperiment
Package JunctionSeq November 1, 2018 Version 1.12.0 Title JunctionSeq: A Utility for Detection of Differential Exon and Splice-Junction Usage in RNA-Seq data Depends R (>= 3.2.2), methods, SummarizedExperiment
Package bisect. April 16, 2018
 Package bisect April 16, 2018 Title Estimating Cell Type Composition from Methylation Sequencing Data Version 0.9.0 Maintainer Eyal Fisher Author Eyal Fisher [aut, cre] An implementation
Package bisect April 16, 2018 Title Estimating Cell Type Composition from Methylation Sequencing Data Version 0.9.0 Maintainer Eyal Fisher Author Eyal Fisher [aut, cre] An implementation
Package ChIPanalyser
 Type Package Package ChIPanalyser December 7, 2018 Title ChIPanalyser: Predicting Transcription Factor Binding Sites Version 1.4.0 Date 2017-09-01 Author Patrick C.N.Martin & Nicolae Radu Zabet Maintainer
Type Package Package ChIPanalyser December 7, 2018 Title ChIPanalyser: Predicting Transcription Factor Binding Sites Version 1.4.0 Date 2017-09-01 Author Patrick C.N.Martin & Nicolae Radu Zabet Maintainer
Package methyanalysis
 Type Package Package methyanalysis June 19, 2018 Title DNA methylation data analysis and visualization Version 1.22.0 Date 2017-01-09 Depends R (>= 2.10), grid, BiocGenerics, IRanges, GenomeInfoDb, GenomicRanges,
Type Package Package methyanalysis June 19, 2018 Title DNA methylation data analysis and visualization Version 1.22.0 Date 2017-01-09 Depends R (>= 2.10), grid, BiocGenerics, IRanges, GenomeInfoDb, GenomicRanges,
BioConductor Overviewr
 BioConductor Overviewr 2016-09-28 Contents Installing Bioconductor 1 Bioconductor basics 1 ExressionSet 2 assaydata (gene expression)........................................ 2 phenodata (sample annotations).....................................
BioConductor Overviewr 2016-09-28 Contents Installing Bioconductor 1 Bioconductor basics 1 ExressionSet 2 assaydata (gene expression)........................................ 2 phenodata (sample annotations).....................................
Package bumphunter. September 22, 2018
 Package bumphunter September 22, 2018 Version 1.22.0 Title Bump Hunter Tools for finding bumps in genomic data Depends R (>= 3.4), S4Vectors (>= 0.9.25), IRanges (>= 2.3.23), GenomeInfoDb, GenomicRanges,
Package bumphunter September 22, 2018 Version 1.22.0 Title Bump Hunter Tools for finding bumps in genomic data Depends R (>= 3.4), S4Vectors (>= 0.9.25), IRanges (>= 2.3.23), GenomeInfoDb, GenomicRanges,
Package ClusterSignificance
 Package ClusterSignificance May 11, 2018 Title The ClusterSignificance package provides tools to assess if class clusters in dimensionality reduced data representations have a separation different from
Package ClusterSignificance May 11, 2018 Title The ClusterSignificance package provides tools to assess if class clusters in dimensionality reduced data representations have a separation different from
Package dupradar. R topics documented: July 12, Type Package
 Type Package Package dupradar July 12, 2018 Title Assessment of duplication rates in RNA-Seq datasets Version 1.10.0 Date 2015-09-26 Author Sergi Sayols , Holger Klein
Type Package Package dupradar July 12, 2018 Title Assessment of duplication rates in RNA-Seq datasets Version 1.10.0 Date 2015-09-26 Author Sergi Sayols , Holger Klein
Package CoGAPS. January 24, 2019
 Version 3.2.2 Date 2018-04-24 Title Coordinated Gene Activity in Pattern Sets Package CoGAPS January 24, 2019 Author Thomas Sherman, Wai-shing Lee, Conor Kelton, Ondrej Maxian, Jacob Carey, Genevieve Stein-O'Brien,
Version 3.2.2 Date 2018-04-24 Title Coordinated Gene Activity in Pattern Sets Package CoGAPS January 24, 2019 Author Thomas Sherman, Wai-shing Lee, Conor Kelton, Ondrej Maxian, Jacob Carey, Genevieve Stein-O'Brien,
Package sigqc. June 13, 2018
 Title Quality Control Metrics for Gene Signatures Version 0.1.20 Package sigqc June 13, 2018 Description Provides gene signature quality control metrics in publication ready plots. Namely, enables the
Title Quality Control Metrics for Gene Signatures Version 0.1.20 Package sigqc June 13, 2018 Description Provides gene signature quality control metrics in publication ready plots. Namely, enables the
Package TnT. November 22, 2017
 Package TnT November 22, 2017 Title Interactive Visualization for Genomic Features Version 1.0.1 A R interface to the TnT javascript library (https://github.com/ tntvis) to provide interactive and flexible
Package TnT November 22, 2017 Title Interactive Visualization for Genomic Features Version 1.0.1 A R interface to the TnT javascript library (https://github.com/ tntvis) to provide interactive and flexible
Package ENmix. December 27, 2018
 Version 1.18.0 Date 2017-09-05 Package ENmix December 27, 2018 Title Data preprocessing and quality control for Illumina HumanMethylation450 and MethylationEPIC BeadChip Type Package The ENmix package
Version 1.18.0 Date 2017-09-05 Package ENmix December 27, 2018 Title Data preprocessing and quality control for Illumina HumanMethylation450 and MethylationEPIC BeadChip Type Package The ENmix package
Package icnv. R topics documented: March 8, Title Integrated Copy Number Variation detection Version Author Zilu Zhou, Nancy Zhang
 Title Integrated Copy Number Variation detection Version 1.2.1 Author Zilu Zhou, Nancy Zhang Package icnv March 8, 2019 Maintainer Zilu Zhou Integrative copy number variation
Title Integrated Copy Number Variation detection Version 1.2.1 Author Zilu Zhou, Nancy Zhang Package icnv March 8, 2019 Maintainer Zilu Zhou Integrative copy number variation
Package GOTHiC. November 22, 2017
 Title Binomial test for Hi-C data analysis Package GOTHiC November 22, 2017 This is a Hi-C analysis package using a cumulative binomial test to detect interactions between distal genomic loci that have
Title Binomial test for Hi-C data analysis Package GOTHiC November 22, 2017 This is a Hi-C analysis package using a cumulative binomial test to detect interactions between distal genomic loci that have
Package ChIC. R topics documented: May 4, Title Quality Control Pipeline for ChIP-Seq Data Version Author Carmen Maria Livi
 Title Quality Control Pipeline for ChIP-Seq Data Version 1.0.0 Author Carmen Maria Livi Package ChIC May 4, 2018 Maintainer Carmen Maria Livi Quality control pipeline for ChIP-seq
Title Quality Control Pipeline for ChIP-Seq Data Version 1.0.0 Author Carmen Maria Livi Package ChIC May 4, 2018 Maintainer Carmen Maria Livi Quality control pipeline for ChIP-seq
Package preprosim. July 26, 2016
 Package preprosim July 26, 2016 Type Package Title Lightweight Data Quality Simulation for Classification Version 0.2.0 Date 2016-07-26 Data quality simulation can be used to check the robustness of data
Package preprosim July 26, 2016 Type Package Title Lightweight Data Quality Simulation for Classification Version 0.2.0 Date 2016-07-26 Data quality simulation can be used to check the robustness of data
Package DNAshapeR. February 9, 2018
 Type Package Package DNAshapeR February 9, 2018 Title High-throughput prediction of DNA shape features Version 1.7.0 Date 2017-10-24 Author Tsu-Pei Chiu and Federico Comoglio Maintainer Tsu-Pei Chiu
Type Package Package DNAshapeR February 9, 2018 Title High-throughput prediction of DNA shape features Version 1.7.0 Date 2017-10-24 Author Tsu-Pei Chiu and Federico Comoglio Maintainer Tsu-Pei Chiu
Package SGSeq. December 31, 2018
 Type Package Package SGSeq December 31, 2018 Title Splice event prediction and quantification from RNA-seq data Version 1.16.2 Author Maintainer SGSeq is a software package for analyzing
Type Package Package SGSeq December 31, 2018 Title Splice event prediction and quantification from RNA-seq data Version 1.16.2 Author Maintainer SGSeq is a software package for analyzing
ChromHMM: automating chromatin-state discovery and characterization
 Nature Methods ChromHMM: automating chromatin-state discovery and characterization Jason Ernst & Manolis Kellis Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3 Supplementary Figure
Nature Methods ChromHMM: automating chromatin-state discovery and characterization Jason Ernst & Manolis Kellis Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3 Supplementary Figure
Package FunciSNP. November 16, 2018
 Type Package Package FunciSNP November 16, 2018 Title Integrating Functional Non-coding Datasets with Genetic Association Studies to Identify Candidate Regulatory SNPs Version 1.26.0 Date 2013-01-19 Author
Type Package Package FunciSNP November 16, 2018 Title Integrating Functional Non-coding Datasets with Genetic Association Studies to Identify Candidate Regulatory SNPs Version 1.26.0 Date 2013-01-19 Author
Package wavcluster. January 23, 2019
 Type Package Package wavcluster January 23, 2019 Title Sensitive and highly resolved identification of RNA-protein interaction sites in PAR-CLIP data Version 2.16.1 Date 2015-05-07 Depends R (>= 3.2),
Type Package Package wavcluster January 23, 2019 Title Sensitive and highly resolved identification of RNA-protein interaction sites in PAR-CLIP data Version 2.16.1 Date 2015-05-07 Depends R (>= 3.2),
Package mzid. April 23, 2019
 Package mzid April 23, 2019 Type Package Title An mzidentml parser for R Version 1.21.1 Date 2016-04-21 Author Thomas Lin Pedersen, Vladislav A Petyuk with contributions from Laurent Gatto and Sebastian
Package mzid April 23, 2019 Type Package Title An mzidentml parser for R Version 1.21.1 Date 2016-04-21 Author Thomas Lin Pedersen, Vladislav A Petyuk with contributions from Laurent Gatto and Sebastian
Package BDMMAcorrect
 Type Package Package BDMMAcorrect March 6, 2019 Title Meta-analysis for the metagenomic read counts data from different cohorts Version 1.0.1 Author ZHENWEI DAI Maintainer ZHENWEI
Type Package Package BDMMAcorrect March 6, 2019 Title Meta-analysis for the metagenomic read counts data from different cohorts Version 1.0.1 Author ZHENWEI DAI Maintainer ZHENWEI
Package SCAN.UPC. October 9, Type Package. Title Single-channel array normalization (SCAN) and University Probability of expression Codes (UPC)
 Package SCAN.UPC October 9, 2013 Type Package Title Single-channel array normalization (SCAN) and University Probability of expression Codes (UPC) Version 2.0.2 Author Stephen R. Piccolo and W. Evan Johnson
Package SCAN.UPC October 9, 2013 Type Package Title Single-channel array normalization (SCAN) and University Probability of expression Codes (UPC) Version 2.0.2 Author Stephen R. Piccolo and W. Evan Johnson
Package TxRegInfra. March 17, 2019
 Package TxRegInfra March 17, 2019 Title Metadata management for multiomic specification of transcriptional regulatory networks This package provides interfaces to genomic metadata employed in regulatory
Package TxRegInfra March 17, 2019 Title Metadata management for multiomic specification of transcriptional regulatory networks This package provides interfaces to genomic metadata employed in regulatory
Package GreyListChIP
 Type Package Version 1.14.0 Package GreyListChIP November 2, 2018 Title Grey Lists -- Mask Artefact Regions Based on ChIP Inputs Date 2018-01-21 Author Gord Brown Maintainer Gordon
Type Package Version 1.14.0 Package GreyListChIP November 2, 2018 Title Grey Lists -- Mask Artefact Regions Based on ChIP Inputs Date 2018-01-21 Author Gord Brown Maintainer Gordon
Useful software utilities for computational genomics. Shamith Samarajiwa CRUK Autumn School in Bioinformatics September 2017
 Useful software utilities for computational genomics Shamith Samarajiwa CRUK Autumn School in Bioinformatics September 2017 Overview Search and download genomic datasets: GEOquery, GEOsearch and GEOmetadb,
Useful software utilities for computational genomics Shamith Samarajiwa CRUK Autumn School in Bioinformatics September 2017 Overview Search and download genomic datasets: GEOquery, GEOsearch and GEOmetadb,
Package EMDomics. November 11, 2018
 Package EMDomics November 11, 2018 Title Earth Mover's Distance for Differential Analysis of Genomics Data The EMDomics algorithm is used to perform a supervised multi-class analysis to measure the magnitude
Package EMDomics November 11, 2018 Title Earth Mover's Distance for Differential Analysis of Genomics Data The EMDomics algorithm is used to perform a supervised multi-class analysis to measure the magnitude
Package fastdummies. January 8, 2018
 Type Package Package fastdummies January 8, 2018 Title Fast Creation of Dummy (Binary) Columns and Rows from Categorical Variables Version 1.0.0 Description Creates dummy columns from columns that have
Type Package Package fastdummies January 8, 2018 Title Fast Creation of Dummy (Binary) Columns and Rows from Categorical Variables Version 1.0.0 Description Creates dummy columns from columns that have
Package scruff. November 6, 2018
 Package scruff November 6, 2018 Title Single Cell RNA-Seq UMI Filtering Facilitator (scruff) Version 1.0.0 Date 2018-08-29 A pipeline which processes single cell RNA-seq (scrna-seq) reads from CEL-seq
Package scruff November 6, 2018 Title Single Cell RNA-Seq UMI Filtering Facilitator (scruff) Version 1.0.0 Date 2018-08-29 A pipeline which processes single cell RNA-seq (scrna-seq) reads from CEL-seq
R topics documented: 2 R topics documented:
 Package cn.mops March 20, 2019 Maintainer Guenter Klambauer Author Guenter Klambauer License LGPL (>= 2.0) Type Package Title cn.mops - Mixture of Poissons for CNV detection in
Package cn.mops March 20, 2019 Maintainer Guenter Klambauer Author Guenter Klambauer License LGPL (>= 2.0) Type Package Title cn.mops - Mixture of Poissons for CNV detection in
Package gggenes. R topics documented: November 7, Title Draw Gene Arrow Maps in 'ggplot2' Version 0.3.2
 Title Draw Gene Arrow Maps in 'ggplot2' Version 0.3.2 Package gggenes November 7, 2018 Provides a 'ggplot2' geom and helper functions for drawing gene arrow maps. Depends R (>= 3.3.0) Imports grid (>=
Title Draw Gene Arrow Maps in 'ggplot2' Version 0.3.2 Package gggenes November 7, 2018 Provides a 'ggplot2' geom and helper functions for drawing gene arrow maps. Depends R (>= 3.3.0) Imports grid (>=
Package MultiDataSet
 Type Package Package MultiDataSet August 16, 2018 Title Implementation of MultiDataSet and ResultSet Version 1.8.0 Date 2018-02-23 Implementation of the BRGE's (Bioinformatic Research Group in Epidemiology
Type Package Package MultiDataSet August 16, 2018 Title Implementation of MultiDataSet and ResultSet Version 1.8.0 Date 2018-02-23 Implementation of the BRGE's (Bioinformatic Research Group in Epidemiology
Package RTNduals. R topics documented: March 7, Type Package
 Type Package Package RTNduals March 7, 2019 Title Analysis of co-regulation and inference of 'dual regulons' Version 1.7.0 Author Vinicius S. Chagas, Clarice S. Groeneveld, Gordon Robertson, Kerstin B.
Type Package Package RTNduals March 7, 2019 Title Analysis of co-regulation and inference of 'dual regulons' Version 1.7.0 Author Vinicius S. Chagas, Clarice S. Groeneveld, Gordon Robertson, Kerstin B.
Package SingleCellExperiment
 Version 1.5.0 Date 2018-10-26 Package SingleCellExperiment Title S4 Classes for Single Cell Data Depends R (>= 3.5), SummarizedExperiment December 13, 2018 Imports S4Vectors, methods, BiocGenerics, utils,
Version 1.5.0 Date 2018-10-26 Package SingleCellExperiment Title S4 Classes for Single Cell Data Depends R (>= 3.5), SummarizedExperiment December 13, 2018 Imports S4Vectors, methods, BiocGenerics, utils,
Package BASiNET. October 2, 2018
 Package BASiNET October 2, 2018 Title Classification of RNA Sequences using Complex Network Theory Version 0.0.4 Author Maintainer It makes the creation of networks from sequences
Package BASiNET October 2, 2018 Title Classification of RNA Sequences using Complex Network Theory Version 0.0.4 Author Maintainer It makes the creation of networks from sequences
Package IsoCorrectoR. R topics documented:
 Package IsoCorrectoR March 19, 2019 Title Correction for natural isotope abundance and tracer purity in MS and MS/MS data from stable isotope labeling experiments Version 1.0.5 Imports dplyr, magrittr,
Package IsoCorrectoR March 19, 2019 Title Correction for natural isotope abundance and tracer purity in MS and MS/MS data from stable isotope labeling experiments Version 1.0.5 Imports dplyr, magrittr,
A quick introduction to GRanges and GRangesList objects
 A quick introduction to GRanges and GRangesList objects Hervé Pagès hpages@fredhutch.org Michael Lawrence lawrence.michael@gene.com July 2015 GRanges objects The GRanges() constructor GRanges accessors
A quick introduction to GRanges and GRangesList objects Hervé Pagès hpages@fredhutch.org Michael Lawrence lawrence.michael@gene.com July 2015 GRanges objects The GRanges() constructor GRanges accessors
Package FastqCleaner
 Type Package Package FastqCleaner April 8, 2019 Title A Shiny Application for Quality Control, Filtering and Trimming of FASTQ Files Version 1.1.0 Date 2018-05-19 Maintainer An interactive web application
Type Package Package FastqCleaner April 8, 2019 Title A Shiny Application for Quality Control, Filtering and Trimming of FASTQ Files Version 1.1.0 Date 2018-05-19 Maintainer An interactive web application
Package SCAN.UPC. July 17, 2018
 Type Package Package SCAN.UPC July 17, 2018 Title Single-channel array normalization (SCAN) and Universal expression Codes (UPC) Version 2.22.0 Author and Andrea H. Bild and W. Evan Johnson Maintainer
Type Package Package SCAN.UPC July 17, 2018 Title Single-channel array normalization (SCAN) and Universal expression Codes (UPC) Version 2.22.0 Author and Andrea H. Bild and W. Evan Johnson Maintainer
Package OmaDB. R topics documented: December 19, Title R wrapper for the OMA REST API Version Author Klara Kaleb
 Title R wrapper for the OMA REST API Version 1.2.0 Author Klara Kaleb Package OmaDB December 19, 2018 Maintainer Klara Kaleb , Adrian Altenhoff A
Title R wrapper for the OMA REST API Version 1.2.0 Author Klara Kaleb Package OmaDB December 19, 2018 Maintainer Klara Kaleb , Adrian Altenhoff A
Package biomformat. April 11, 2018
 Version 1.7.0 Date 2016-04-16 Package biomformat April 11, 2018 Maintainer Paul J. McMurdie License GPL-2 Title An interface package for the BIOM file format Type Package Author
Version 1.7.0 Date 2016-04-16 Package biomformat April 11, 2018 Maintainer Paul J. McMurdie License GPL-2 Title An interface package for the BIOM file format Type Package Author
Tutorial: Jump Start on the Human Epigenome Browser at Washington University
 Tutorial: Jump Start on the Human Epigenome Browser at Washington University This brief tutorial aims to introduce some of the basic features of the Human Epigenome Browser, allowing users to navigate
Tutorial: Jump Start on the Human Epigenome Browser at Washington University This brief tutorial aims to introduce some of the basic features of the Human Epigenome Browser, allowing users to navigate
WASHU EPIGENOME BROWSER 2018 epigenomegateway.wustl.edu
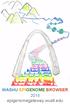 WASHU EPIGENOME BROWSER 2018 epigenomegateway.wustl.edu 3 BROWSER MAP 14 15 16 17 18 19 20 21 22 23 24 1 2 5 4 8 9 10 12 6 11 7 13 Key 1 = Go to this page number to learn about the browser feature TABLE
WASHU EPIGENOME BROWSER 2018 epigenomegateway.wustl.edu 3 BROWSER MAP 14 15 16 17 18 19 20 21 22 23 24 1 2 5 4 8 9 10 12 6 11 7 13 Key 1 = Go to this page number to learn about the browser feature TABLE
Package ccmap. June 17, 2018
 Type Package Title Combination Connectivity Mapping Version 1.6.0 Author Alex Pickering Package ccmap June 17, 2018 Maintainer Alex Pickering Finds drugs and drug combinations
Type Package Title Combination Connectivity Mapping Version 1.6.0 Author Alex Pickering Package ccmap June 17, 2018 Maintainer Alex Pickering Finds drugs and drug combinations
featuredb storing and querying genomic annotation
 featuredb storing and querying genomic annotation Work in progress Arne Müller Preclinical Safety Informatics, Novartis, December 14th 2012 Acknowledgements: Florian Hahne + Bioconductor Community featuredb
featuredb storing and querying genomic annotation Work in progress Arne Müller Preclinical Safety Informatics, Novartis, December 14th 2012 Acknowledgements: Florian Hahne + Bioconductor Community featuredb
m6aviewer Version Documentation
 m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
Package FitHiC. October 16, 2018
 Type Package Package FitHiC October 16, 2018 Title Confidence estimation for intra-chromosomal contact maps Version 1.6.0 Date 2017-04-12 Description Fit-Hi-C is a tool for assigning statistical confidence
Type Package Package FitHiC October 16, 2018 Title Confidence estimation for intra-chromosomal contact maps Version 1.6.0 Date 2017-04-12 Description Fit-Hi-C is a tool for assigning statistical confidence
Package RNAseqNet. May 16, 2017
 Type Package Package RNAseqNet May 16, 2017 Title Log-Linear Poisson Graphical Model with Hot-Deck Multiple Imputation Version 0.1.1 Date 2017-05-16 Maintainer Nathalie Villa-Vialaneix
Type Package Package RNAseqNet May 16, 2017 Title Log-Linear Poisson Graphical Model with Hot-Deck Multiple Imputation Version 0.1.1 Date 2017-05-16 Maintainer Nathalie Villa-Vialaneix
Rsubread package: high-performance read alignment, quantification and mutation discovery
 Rsubread package: high-performance read alignment, quantification and mutation discovery Wei Shi 14 September 2015 1 Introduction This vignette provides a brief description to the Rsubread package. For
Rsubread package: high-performance read alignment, quantification and mutation discovery Wei Shi 14 September 2015 1 Introduction This vignette provides a brief description to the Rsubread package. For
