Tutorial: Jump Start on the Human Epigenome Browser at Washington University
|
|
|
- Tamsin Mills
- 5 years ago
- Views:
Transcription
1 Tutorial: Jump Start on the Human Epigenome Browser at Washington University This brief tutorial aims to introduce some of the basic features of the Human Epigenome Browser, allowing users to navigate the browser and interact with data tracks in a meaningful way. In order to access the public site, please visit In this tutorial, heatmap will be used to refer to genome heatmap (panel A in Supplementary Figure 2), and control panel will be used to refer to panel G in Supplementary Figure 2. Help desk Help desk for the Epigenome browser is at Please check the site for most updated tutorial and video demo. Fine-tuning genome heatmap display The color of individual heatmap cells is determined by the numerical data associated with a cell and a threshold value for that data track. To understand how the threshold value is computed, go to the tool box which floats on the web page (it can be moved by dragging the banner on the top): The drop-down menu for the track threshold gives options for computing the track threshold score. If max is selected, the maximum value for each track at the currently displayed region will be used as the threshold value. Otherwise, the user can select a percentile value to be the threshold. Once a method is selected to compute track thresholds, the heatmap cell color will be decided accordingly. This can be visualized using the context menu accessed by right-clicking on a single data track in the heatmap: 1
2 Each row of colored bars represents a different color scheme that can be assigned to a data track by the user. If the cell value is below the threshold score, the color of the cell will be determined according to the color gradient leading up to the threshold. As a value approaches the threshold, the color assigned to the cell will be progressively darker. If a value is above the threshold by any amount, the cell will be assigned a distinct color. The height of each row in the heatmap can be changed using the track height option in the toolbox. If track height is greater than 20 pixels, the heatmap display will be replaced by a wiggle plot, where vertical lines indicate the relative heatmap cell value: Finally, tracks in the heatmap can be rearranged in various ways to make visualization more intuitive for the individual user. Click and drag the striped bar corresponding to a data track, found to the right of metadata heatmap, in order to move a single track. Group of tracks can be moved together by dragging the colored blocks in metadata heatmap. Tracks can be sorted according to metadata terms by clicking on the column labels in the metadata heatmap. When the correlation function is turned on, tracks can be sorted according to correlation coeffients by clicking on the coefficients image to the left of the heatmap. Navigation To view a wider genomic region in the heatmap, click the zoom out button in the toolbox; to zoom in, click on the chromosome ideogram below the heatmap and drag the cursor to the desired genomic coordinate. The heatmap will zoom to display the selected region down to single nucleotide views. To move left of right within the current zoom level, click, hold and drag the heatmap in either direction. Please wait for the loading image to disappear before dragging the heatmap further. Depending on the speed of internet connection and the number of tracks displayed, this waiting time can be somewhere between several milliseconds to two seconds. Customizing metadata color-coding area Columns in the metadata heatmap correspond to the metadata terms. In order to add/remove terms or browse the collection of terms, go to the Metadata tab in the control panel. Terms 2
3 with / icon can be expanded or collapsed by clicking to show or hide children terms. Use the check boxes to add or remove columns from the metadata heatmap: As an example, the metadata terms chosen above will produce the following metadata heatmap: Clicking on term names above the heatmap will sort tracks according to the annotation of that attribute. Drag the term name to re-arrange metadata columns. Track selection Open the Track management tab in the control panel to access the track selection grid (Supplementary Figure 2). By default, rows of the grid correspond to sequencing experiment sample names, and columns correspond to the type of assay (epigenetic marks). Name with / icon means the marks can be clicked on to show/hide detailed terms. Numbers in the grid occur in pairs, indicating the number of tracks annotated by the combination of row/column terms, and number of tracks in this group currently on display. Click a pair of numbers, and a panel will appear in toolbox allowing the user to add/remove tracks with check boxes: 3
4 In this Choose track panel, tracks currently on display will be labeled with a red check. Click on it and the track will be removed from display. Click on Remove all to remove all tracks. Tracks that are available for but not on display will have a check box on the left of the track name. Check the box will result in the following panel: In this Track to be added panel, click on Add these tracks button to add the tracks for browsing. The user can make selections on multiple number pairs and add tracks all together. To view detailed information for each track, click on the blue letter i following the track name and an additional panel with detailed track information will appear in toolbox. By clicking the GEO icon, the user will be taken to the record to this track in the GEO database: 4
5 Short cuts are available to quickly select/remove all tracks annotated by one term. Scroll mouse over a term in the track selection box and a small menu will appear with options Select all and Un-select all. Click on either option to take effect: The track display can be cleared in a single click, using buttons at the bottom of the track selection grid. Using the Advanced options tab on top of the track selection grid, the grid layout can be customized. The track selection panel can be reorganized using a single criteria. In this case, an indented text tree will be generated, and the user can click on the pair of numbers to select all tracks annotated by that term: 5
6 Genomic feature tracks Many types of genomic feature annotation are available under the Genomic feature tab in the control panel. Three plotting modes can be selected from (thin, full, density), and the gene structure will be plotted if the gene body track is turned to full mode. Once a genomic feature track is displayed, the user can right click on the track image to display a context menu for additional visualization options. An example using genomic feature track display is shown below. Gene body is in full mode, and various repetitive elements are in thin mode. The region surrounding the NSUN6 gene is shown. While SINE, LTR, LINE, DNA transposons are relatively uniformly distributed in this region, satellite DNA only exists inside the 8 th intron of NSUN6, and that region has strong methylation signal (MeDIP experiment tracks). 6
7 Correlation analysis Open the Correlation tab in the control panel for correlation analysis options. Track scores can be correlated with genomic feature density data at the currently displayed region and an inter-track correlation can also be computed. Similar operations can also be applied to custom bigbed tracks. To activate these options, check the corresponding button in the control panel. Click the Turn off correlation function button to cancel correlation analysis. Hypothesis test Open the Hypothesis test tab in the control panel for options on this function. At least two groups of tracks must be defined to run a hypothesis test. In the panel shown below, GROUP 1 and GROUP 2 are two group holders provided by default, which can be modified or removed by the user. To assign a new track to a group, the user can use the drop-down menus in the box on the right side of screen shot. Entries denoting tracks will appear in group boxes. To remove a track from selection, click the red cross image in the group box. A track can be assigned to a group using the context menu by right-clicking on the heatmap. Likewise, a group of tracks can be assigned to a group by right-clicking on the metadata heatmap. Click the clear button to remove all track assignments: 7
8 Click the Run hypothesis test button and the server will run the test. Once done, vertical bars corresponding to P-values will be plotted in a track below the chromosome ideogram. P- values less than or equal to the significance cutoff are plotted in red: Multiple test correction can be applied to correct P-values. Select a correction option using the drop-down menu and click the Run hypothesis test button to run test again to generate corrected P-values. Custom bigwig track Custom bigwig tracks can be used to display numerical data across the genome. The bigwig format file can be generated using the wigtobigwig and bedgraphtobigwig utility programs available in the Kent Source Tree. The user needs to set up a HTTP web server and place this file on the web server directory. To submit the custom bigwig track, open the Custom track tab in the control panel, click on Submit bigwig button, enter the bigwig file URL and a track name, and press Submit button to submit without specifying metadata for this track: 8
9 The track will be displayed in the heatmap in the default color, blue. To change the color of the track, right click on the track to display the context menu and select a color scheme by clicking a row of colored bars. The track will also be assigned a row in the metadata heatmap. If no metadata for the track is provided, the cell will be colored gray. To annotate custom bigwig tracks with metadata, the user can use the metadata vocabulary provided by the browser. Open this Metadata box to add annotation terms to the track using the drop-down menu: If suitable terms are not available in the existing vocabulary, the user can create his/her own metadata term with a number of attributes, and annotate custom bigwig track using it. Open the corresponding box, click Add new term and a new panel will be displayed: 9
10 Enter the term name and attributes for this term to create a custom metadata term. Once added, the term will show up in a table ready to be used: Following submission, the custom metadata term as well as the custom bigwig track will be displayed like the following: Custom bigbed track Users can display their collection of genomic features using a custom bigbed track. Genomic locations must be specified for the features, and the name/strand can be optionally specified. A special requirement is id on the 5 th field of the bed file, in order for it to be compatible the drag-move feature of the Human Epigenome Browser. Open the bigbed custom track submission panel and enter the file URL and track name to submit. 10
11 Above is an example showing the appearance of a custom bigbed track with name featureless genebody. Submitted custom tracks will occupy entries in the table seen at the bottom of screen shot, and clicking the red cross image will remove that track. Data collation Data collation is a unique feature of the Human Epigenome Browser, allowing the user to reorganize the heatmap view to concentrate on specific genomic features. To collate track data on a specific genomic feature, open the Data collation tab in the control panel. Under the functional genomic features tab, click the button next to the genomic feature to be used for collation. If this genomic feature is already on display, check the Collate option in the context menu by right-clicking on the track: Following collation, the browser view will be similar to the following: 11
12 In the above example, data collation is performed on genes, and all the intergenic regions are removed as seen in the gene body track. A gray line is drawn between adjacent and nonoverlapping genes, and will not be drawn between overlapping genes. All navigation operations still apply to this view (move/zoom). Click the cancel collation button to return to the genome view. Gene set view Data collation on genomic features will use features from part of the genome to organize the view. The features' original orientation and relative positions are all preserved. Conversely, using the gene set view, a set of genes can be used to organize the view irrespective of their original positions in genome. Open the Gene set view tab, click the Custom gene set button, and the following form will appear: A list of gene names or genomic coordinates can be used as input. By default, a short list of cytochrome P450 genes is provided in no particular order. Following submission, the heatmap view will look like the following: In this view, the chromosome ideogram below the heatmap is replaced by boxes. Each box represents one item (gene or genomic coordinate) submitted in the gene set, with the name inside of or adjacent to the box. The width of each box represents the relative length of the item. The Zoom in operation can be performed on this image by clicking on the image and dragging the cursor over the region of interest. When submitting gene sets, the user can define which gene features parts should be displayed using the drop-down menu. Ready-made gene features can be selected for viewing, including 3 kb promoter (with or without gene body) and 5' or 3' UTRs, of the user can define a custom region. This allows the user to select a generic region around relevant gene features (i.e. transcription start/stop sites, or gene body) by defining the upstream and downstream sequence lengths. Upon choosing this option, a small control panel will appear. Drag the colored gliders to define the length of upstream/downstream regions to display: 12
13 Following gene set submission, the control panel will contain the list of submitted genes and a brief detail for review. Also shown is a gene set editing panel. The user can change which gene features to display of sort the gene set. Clicking the remove all genes button will clear this gene list and return to the genome view: The gene set can be sorted according to the mean track signal. By selecting the by average track score option in the sort gene set drop down menu, an additional drop-down menu of track names will appear. The user can then select a track and click the Update button to sort by the average score of the selected track: More conveniently, the user can right click on the heatmap to select a track for sorting (after selection, the user must click the Update button to sort): 13
14 The following is the view after sorting the gene set by the average track score of the last track (in green): KEGG pathway view The Kyoto Encylopedia of Genes and Genomes (KEGG, database contains carefully curated information on metabolic pathways, enzyme-coding genes, and metabolites in all genome-sequenced species. With the Human Epigenome Browser users can view track data on genes in KEGG pathways with a few simple steps. Go to the Data collation panel, and click KEGG pathway button. The following panel will appear: 14
15 The user can directly enter KEGG pathway identifiers (e.g. path:hsa00010) and view data; however, the user will usually need to do keyword search for pathways of interest. By searching for the keyword glyco, the following pathways will be found: Click the pathway icon to view pathway details in the KEGG database. Click on the pathway link to perform data collation. The following is the view produced by data collation on the 65 genes in the glycolysis pathway (half of entire gene set is shown): Session Users can save their entire browsing status, including their custom tracks and metadata entries using the session function. Go to the Session tab, click the Save status button and the current browsing status will be saved under current session. Multiple statuses can be saved under one session, and an optional short text is available to describe each status: 15
16 Save the session ID and use it to retrieve saved statuses later or for sharing with colleagues. Enter the session ID into the second text field in the panel and click the Retrieve status button. All saved statuses will be displayed in the table. Previous browsing statuses can be restored by clicking on the status link. 16
W ASHU E PI G ENOME B ROWSER
 W ASHU E PI G ENOME B ROWSER Keystone Symposium on DNA and RNA Methylation January 23 rd, 2018 Fairmont Hotel Vancouver, Vancouver, British Columbia, Canada Presenter: Renee Sears and Josh Jang Tutorial
W ASHU E PI G ENOME B ROWSER Keystone Symposium on DNA and RNA Methylation January 23 rd, 2018 Fairmont Hotel Vancouver, Vancouver, British Columbia, Canada Presenter: Renee Sears and Josh Jang Tutorial
epigenomegateway.wustl.edu
 Everything can be found at epigenomegateway.wustl.edu REFERENCES 1. Zhou X, et al., Nature Methods 8, 989-990 (2011) 2. Zhou X & Wang T, Current Protocols in Bioinformatics Unit 10.10 (2012) 3. Zhou X,
Everything can be found at epigenomegateway.wustl.edu REFERENCES 1. Zhou X, et al., Nature Methods 8, 989-990 (2011) 2. Zhou X & Wang T, Current Protocols in Bioinformatics Unit 10.10 (2012) 3. Zhou X,
W ASHU E PI G ENOME B ROWSER
 Roadmap Epigenomics Workshop W ASHU E PI G ENOME B ROWSER SOT 2016 Satellite Meeting March 17 th, 2016 Ernest N. Morial Convention Center, New Orleans, LA Presenter: Ting Wang Tutorial Overview: WashU
Roadmap Epigenomics Workshop W ASHU E PI G ENOME B ROWSER SOT 2016 Satellite Meeting March 17 th, 2016 Ernest N. Morial Convention Center, New Orleans, LA Presenter: Ting Wang Tutorial Overview: WashU
WASHU EPIGENOME BROWSER 2018 epigenomegateway.wustl.edu
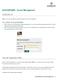
 WASHU EPIGENOME BROWSER 2018 epigenomegateway.wustl.edu 3 BROWSER MAP 14 15 16 17 18 19 20 21 22 23 24 1 2 5 4 8 9 10 12 6 11 7 13 Key 1 = Go to this page number to learn about the browser feature TABLE
WASHU EPIGENOME BROWSER 2018 epigenomegateway.wustl.edu 3 BROWSER MAP 14 15 16 17 18 19 20 21 22 23 24 1 2 5 4 8 9 10 12 6 11 7 13 Key 1 = Go to this page number to learn about the browser feature TABLE
A short Introduction to UCSC Genome Browser
 A short Introduction to UCSC Genome Browser Elodie Girard, Nicolas Servant Institut Curie/INSERM U900 Bioinformatics, Biostatistics, Epidemiology and computational Systems Biology of Cancer 1 Why using
A short Introduction to UCSC Genome Browser Elodie Girard, Nicolas Servant Institut Curie/INSERM U900 Bioinformatics, Biostatistics, Epidemiology and computational Systems Biology of Cancer 1 Why using
Integrative Genomics Viewer. Prat Thiru
 Integrative Genomics Viewer Prat Thiru 1 Overview User Interface Basics Browsing the Data Data Formats IGV Tools Demo Outline Based on ISMB 2010 Tutorial by Robinson and Thorvaldsdottir 2 Why IGV? IGV
Integrative Genomics Viewer Prat Thiru 1 Overview User Interface Basics Browsing the Data Data Formats IGV Tools Demo Outline Based on ISMB 2010 Tutorial by Robinson and Thorvaldsdottir 2 Why IGV? IGV
The Fleet page provides you with the tools needed to display, find, and manage your equipment. The page views and elements include:
 Using the Fleet Page The Fleet page provides you with the tools needed to display, find, and manage your equipment. The page views and elements include: Assets tab Components tab Asset Details view Add/Modify
Using the Fleet Page The Fleet page provides you with the tools needed to display, find, and manage your equipment. The page views and elements include: Assets tab Components tab Asset Details view Add/Modify
m6aviewer Version Documentation
 m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
You will be re-directed to the following result page.
 ENCODE Element Browser Goal: to navigate the candidate DNA elements predicted by the ENCODE consortium, including gene expression, DNase I hypersensitive sites, TF binding sites, and candidate enhancers/promoters.
ENCODE Element Browser Goal: to navigate the candidate DNA elements predicted by the ENCODE consortium, including gene expression, DNase I hypersensitive sites, TF binding sites, and candidate enhancers/promoters.
Genomic Analysis with Genome Browsers.
 Genomic Analysis with Genome Browsers http://barc.wi.mit.edu/hot_topics/ 1 Outline Genome browsers overview UCSC Genome Browser Navigating: View your list of regions in the browser Available tracks (eg.
Genomic Analysis with Genome Browsers http://barc.wi.mit.edu/hot_topics/ 1 Outline Genome browsers overview UCSC Genome Browser Navigating: View your list of regions in the browser Available tracks (eg.
MetScape User Manual
 MetScape 2.3.2 User Manual A Plugin for Cytoscape National Center for Integrative Biomedical Informatics July 2012 2011 University of Michigan This work is supported by the National Center for Integrative
MetScape 2.3.2 User Manual A Plugin for Cytoscape National Center for Integrative Biomedical Informatics July 2012 2011 University of Michigan This work is supported by the National Center for Integrative
EGAN Tutorial: A Basic Use-case
 EGAN Tutorial: A Basic Use-case July 2010 Jesse Paquette Biostatistics and Computational Biology Core Helen Diller Family Comprehensive Cancer Center University of California, San Francisco (AKA BCBC HDFCCC
EGAN Tutorial: A Basic Use-case July 2010 Jesse Paquette Biostatistics and Computational Biology Core Helen Diller Family Comprehensive Cancer Center University of California, San Francisco (AKA BCBC HDFCCC
Tutorial. RNA-Seq Analysis of Breast Cancer Data. Sample to Insight. November 21, 2017
 RNA-Seq Analysis of Breast Cancer Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
RNA-Seq Analysis of Breast Cancer Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
SEEK User Manual. Introduction
 SEEK User Manual Introduction SEEK is a computational gene co-expression search engine. It utilizes a vast human gene expression compendium to deliver fast, integrative, cross-platform co-expression analyses.
SEEK User Manual Introduction SEEK is a computational gene co-expression search engine. It utilizes a vast human gene expression compendium to deliver fast, integrative, cross-platform co-expression analyses.
Genome Browsers - The UCSC Genome Browser
 Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Roxen Content Provider
 Roxen Content Provider Generation 3 Templates Purpose This workbook is designed to provide a training and reference tool for placing University of Alaska information on the World Wide Web (WWW) using the
Roxen Content Provider Generation 3 Templates Purpose This workbook is designed to provide a training and reference tool for placing University of Alaska information on the World Wide Web (WWW) using the
Table of contents Genomatix AG 1
 Table of contents! Introduction! 3 Getting started! 5 The Genome Browser window! 9 The toolbar! 9 The general annotation tracks! 12 Annotation tracks! 13 The 'Sequence' track! 14 The 'Position' track!
Table of contents! Introduction! 3 Getting started! 5 The Genome Browser window! 9 The toolbar! 9 The general annotation tracks! 12 Annotation tracks! 13 The 'Sequence' track! 14 The 'Position' track!
SPAR outputs and report page
 SPAR outputs and report page Landing results page (full view) Landing results / outputs page (top) Input files are listed Job id is shown Download all tables, figures, tracks as zip Percentage of reads
SPAR outputs and report page Landing results page (full view) Landing results / outputs page (top) Input files are listed Job id is shown Download all tables, figures, tracks as zip Percentage of reads
BANNER 9 QUICK NAVIGATION GUIDE
 Application Navigator Application Navigator provides a single interface to navigate the Banner 9 JAVA pages. It is a tool that allows you to go back and forth between the current Banner forms and the new
Application Navigator Application Navigator provides a single interface to navigate the Banner 9 JAVA pages. It is a tool that allows you to go back and forth between the current Banner forms and the new
Wilson Leung 01/03/2018 An Introduction to NCBI BLAST. Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment
 An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
Tutorial. Variant Detection. Sample to Insight. November 21, 2017
 Resequencing: Variant Detection November 21, 2017 Map Reads to Reference and Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Resequencing: Variant Detection November 21, 2017 Map Reads to Reference and Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
BLACKBOARD: Course Management
 BLACKBOARD: Course Management LOGGING IN Note: you will use your Babson username and password to access Blackboard. TO LOGIN TO BLACKBOARD: 1. Open a web browser and type http://blackboard.babson.edu.
BLACKBOARD: Course Management LOGGING IN Note: you will use your Babson username and password to access Blackboard. TO LOGIN TO BLACKBOARD: 1. Open a web browser and type http://blackboard.babson.edu.
Differential Expression Analysis at PATRIC
 Differential Expression Analysis at PATRIC The following step- by- step workflow is intended to help users learn how to upload their differential gene expression data to their private workspace using Expression
Differential Expression Analysis at PATRIC The following step- by- step workflow is intended to help users learn how to upload their differential gene expression data to their private workspace using Expression
mirnet Tutorial Starting with expression data
 mirnet Tutorial Starting with expression data Computer and Browser Requirements A modern web browser with Java Script enabled Chrome, Safari, Firefox, and Internet Explorer 9+ For best performance and
mirnet Tutorial Starting with expression data Computer and Browser Requirements A modern web browser with Java Script enabled Chrome, Safari, Firefox, and Internet Explorer 9+ For best performance and
How to use earray to create custom content for the SureSelect Target Enrichment platform. Page 1
 How to use earray to create custom content for the SureSelect Target Enrichment platform Page 1 Getting Started Access earray Access earray at: https://earray.chem.agilent.com/earray/ Log in to earray,
How to use earray to create custom content for the SureSelect Target Enrichment platform Page 1 Getting Started Access earray Access earray at: https://earray.chem.agilent.com/earray/ Log in to earray,
Oracle General Navigation Overview
 Oracle 11.5.9 General Navigation Overview 1 Logging On to Oracle Applications You may access Oracle, by logging onto the ATC Applications Login System Status page located at www.atc.caltech.edu/support/index.php
Oracle 11.5.9 General Navigation Overview 1 Logging On to Oracle Applications You may access Oracle, by logging onto the ATC Applications Login System Status page located at www.atc.caltech.edu/support/index.php
Faculty Guide to Grade Center in Blackboard
 Faculty Guide to Grade Center in Blackboard Grade Center, formally known as Gradebook, is a central repository for assessment data, student information, and instructor notes. Although it includes items
Faculty Guide to Grade Center in Blackboard Grade Center, formally known as Gradebook, is a central repository for assessment data, student information, and instructor notes. Although it includes items
GraphWorX64 Productivity Tips
 Description: Overview of the most important productivity tools in GraphWorX64 General Requirement: Basic knowledge of GraphWorX64. Introduction GraphWorX64 has a very powerful development environment in
Description: Overview of the most important productivity tools in GraphWorX64 General Requirement: Basic knowledge of GraphWorX64. Introduction GraphWorX64 has a very powerful development environment in
Coach s Office Playbook Tutorial Playbook i
 Playbook i The Playbook... 1 Overview... 1 Open the Playbook... 1 The Playbook Window... 2 Name the Chapter... 2 Insert the First Page... 3 Page Templates... 3 Define the Template Boxes... 4 Text on the
Playbook i The Playbook... 1 Overview... 1 Open the Playbook... 1 The Playbook Window... 2 Name the Chapter... 2 Insert the First Page... 3 Page Templates... 3 Define the Template Boxes... 4 Text on the
Appleworks 6.0 Word Processing
 Appleworks 6.0 Word Processing AppleWorks 6.0 Starting Points What s New in AppleWorks 6.0 AppleWorks 6.0 is a versatile and powerful program that integrates the best of everything you need - word processing,
Appleworks 6.0 Word Processing AppleWorks 6.0 Starting Points What s New in AppleWorks 6.0 AppleWorks 6.0 is a versatile and powerful program that integrates the best of everything you need - word processing,
Word Select New in the left pane. 3. Select Blank document in the Available Templates pane. 4. Click the Create button.
 Microsoft QUICK Word 2010 Source Getting Started The Word Window u v w x z Opening a Document 2. Select Open in the left pane. 3. In the Open dialog box, locate and select the file you want to open. 4.
Microsoft QUICK Word 2010 Source Getting Started The Word Window u v w x z Opening a Document 2. Select Open in the left pane. 3. In the Open dialog box, locate and select the file you want to open. 4.
Supplementary Figure 1. Fast read-mapping algorithm of BrowserGenome.
 Supplementary Figure 1 Fast read-mapping algorithm of BrowserGenome. (a) Indexing strategy: The genome sequence of interest is divided into non-overlapping 12-mers. A Hook table is generated that contains
Supplementary Figure 1 Fast read-mapping algorithm of BrowserGenome. (a) Indexing strategy: The genome sequence of interest is divided into non-overlapping 12-mers. A Hook table is generated that contains
Adding Text and Images. IMCOM Enterprise Web CMS Tutorial 1 Version 2
 Adding Text and Images IMCOM Enterprise Web CMS Tutorial 1 Version 2 Contents and general instructions PAGE: 3. First steps: Open a page and a block to edit 4. Edit text / The menu bar 5. Link to sites,
Adding Text and Images IMCOM Enterprise Web CMS Tutorial 1 Version 2 Contents and general instructions PAGE: 3. First steps: Open a page and a block to edit 4. Edit text / The menu bar 5. Link to sites,
Tutorial: De Novo Assembly of Paired Data
 : De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
Genome Browsers Guide
 Genome Browsers Guide Take a Class This guide supports the Galter Library class called Genome Browsers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
Genome Browsers Guide Take a Class This guide supports the Galter Library class called Genome Browsers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
Latvijas Banka Statistical Database
 Latvijas Banka Statistical Database User Manual Version 2.03 Riga, July 2015 Latvijas Banka Statistical Database. User Manual 2 Table of contents 1. GENERAL PRINCIPLES... 3 1.1. LAYOUT OF THE USER INTERFACE...
Latvijas Banka Statistical Database User Manual Version 2.03 Riga, July 2015 Latvijas Banka Statistical Database. User Manual 2 Table of contents 1. GENERAL PRINCIPLES... 3 1.1. LAYOUT OF THE USER INTERFACE...
BANNER 9 QUICK NAVIGATION GUIDE
 MARCH 2017 Application Navigator Application Navigator provides a single interface to seamlessly navigate between Banner 9 JAVA pages and Banner 8 Oracle forms. It is a tool that allows you to go back
MARCH 2017 Application Navigator Application Navigator provides a single interface to seamlessly navigate between Banner 9 JAVA pages and Banner 8 Oracle forms. It is a tool that allows you to go back
ChIP-seq practical: peak detection and peak annotation. Mali Salmon-Divon Remco Loos Myrto Kostadima
 ChIP-seq practical: peak detection and peak annotation Mali Salmon-Divon Remco Loos Myrto Kostadima March 2012 Introduction The goal of this hands-on session is to perform some basic tasks in the analysis
ChIP-seq practical: peak detection and peak annotation Mali Salmon-Divon Remco Loos Myrto Kostadima March 2012 Introduction The goal of this hands-on session is to perform some basic tasks in the analysis
Faculty Guide to Grade Center in Blackboard 9.1
 Faculty Guide to Grade Center in Blackboard 9.1 Grade Center, formally known as Gradebook, is a central repository for assessment data, student information, and instructor notes. Although it includes items
Faculty Guide to Grade Center in Blackboard 9.1 Grade Center, formally known as Gradebook, is a central repository for assessment data, student information, and instructor notes. Although it includes items
SPREADSHEET (Excel 2007)
 SPREADSHEET (Excel 2007) 1 U N I T 0 4 BY I F T I K H A R H U S S A I N B A B U R Spreadsheet Microsoft Office Excel 2007 (or Excel) is a computer program used to enter, analyze, and present quantitative
SPREADSHEET (Excel 2007) 1 U N I T 0 4 BY I F T I K H A R H U S S A I N B A B U R Spreadsheet Microsoft Office Excel 2007 (or Excel) is a computer program used to enter, analyze, and present quantitative
Genome Environment Browser (GEB) user guide
 Genome Environment Browser (GEB) user guide GEB is a Java application developed to provide a dynamic graphical interface to visualise the distribution of genome features and chromosome-wide experimental
Genome Environment Browser (GEB) user guide GEB is a Java application developed to provide a dynamic graphical interface to visualise the distribution of genome features and chromosome-wide experimental
Tutorial: chloroplast genomes
 Tutorial: chloroplast genomes Stacia Wyman Department of Computer Sciences Williams College Williamstown, MA 01267 March 10, 2005 ASSUMPTIONS: You are using Internet Explorer under OS X on the Mac. You
Tutorial: chloroplast genomes Stacia Wyman Department of Computer Sciences Williams College Williamstown, MA 01267 March 10, 2005 ASSUMPTIONS: You are using Internet Explorer under OS X on the Mac. You
VectorBase Web Apollo April Web Apollo 1
 Web Apollo 1 Contents 1. Access points: Web Apollo, Genome Browser and BLAST 2. How to identify genes that need to be annotated? 3. Gene manual annotations 4. Metadata 1. Access points Web Apollo tool
Web Apollo 1 Contents 1. Access points: Web Apollo, Genome Browser and BLAST 2. How to identify genes that need to be annotated? 3. Gene manual annotations 4. Metadata 1. Access points Web Apollo tool
Tutorial 1: Getting Started with Excel
 Tutorial 1: Getting Started with Excel Microsoft Excel 2010 Objectives Understand the use of spreadsheets and Excel Learn the parts of the Excel window Scroll through a worksheet and navigate between worksheets
Tutorial 1: Getting Started with Excel Microsoft Excel 2010 Objectives Understand the use of spreadsheets and Excel Learn the parts of the Excel window Scroll through a worksheet and navigate between worksheets
Unified Classroom: Class Pages
 Unified Classroom: Class Pages Unified Classroom April 2018 Release Unified Classroom: Class Pages Basics Contents Unified Classroom: Class Pages Basics... 3 Building Class Pages... 3 Access Class Pages...
Unified Classroom: Class Pages Unified Classroom April 2018 Release Unified Classroom: Class Pages Basics Contents Unified Classroom: Class Pages Basics... 3 Building Class Pages... 3 Access Class Pages...
Tutorial: RNA-Seq analysis part I: Getting started
 : RNA-Seq analysis part I: Getting started August 9, 2012 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com : RNA-Seq analysis
: RNA-Seq analysis part I: Getting started August 9, 2012 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com : RNA-Seq analysis
Integrating Sintelix and ANB. Learn how to access and explore Sintelix networks in IBM i2 Analyst s Notebook
 Integrating Sintelix and ANB Learn how to access and explore Sintelix networks in IBM i2 Analyst s Notebook 2 Integrating Sintelix and ANB By the end of this tutorial you will know how to: 1 2 3 Install
Integrating Sintelix and ANB Learn how to access and explore Sintelix networks in IBM i2 Analyst s Notebook 2 Integrating Sintelix and ANB By the end of this tutorial you will know how to: 1 2 3 Install
The Allen Human Brain Atlas offers three types of searches to allow a user to: (1) obtain gene expression data for specific genes (or probes) of
 Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
BLAST Exercise 2: Using mrna and EST Evidence in Annotation Adapted by W. Leung and SCR Elgin from Annotation Using mrna and ESTs by Dr. J.
 BLAST Exercise 2: Using mrna and EST Evidence in Annotation Adapted by W. Leung and SCR Elgin from Annotation Using mrna and ESTs by Dr. J. Buhler Prerequisites: BLAST Exercise: Detecting and Interpreting
BLAST Exercise 2: Using mrna and EST Evidence in Annotation Adapted by W. Leung and SCR Elgin from Annotation Using mrna and ESTs by Dr. J. Buhler Prerequisites: BLAST Exercise: Detecting and Interpreting
Microsoft Excel 2010 Part 2: Intermediate Excel
 CALIFORNIA STATE UNIVERSITY, LOS ANGELES INFORMATION TECHNOLOGY SERVICES Microsoft Excel 2010 Part 2: Intermediate Excel Spring 2014, Version 1.0 Table of Contents Introduction...3 Working with Rows and
CALIFORNIA STATE UNIVERSITY, LOS ANGELES INFORMATION TECHNOLOGY SERVICES Microsoft Excel 2010 Part 2: Intermediate Excel Spring 2014, Version 1.0 Table of Contents Introduction...3 Working with Rows and
Océ Engineering Exec. Electronic Job Ticket
 Océ Engineering Exec Electronic Job Ticket Océ-Technologies B.V. Copyright 2004, Océ-Technologies B.V. Venlo, The Netherlands All rights reserved. No part of this work may be reproduced, copied, adapted,
Océ Engineering Exec Electronic Job Ticket Océ-Technologies B.V. Copyright 2004, Océ-Technologies B.V. Venlo, The Netherlands All rights reserved. No part of this work may be reproduced, copied, adapted,
User Guide Product Design Version 1.7
 User Guide Product Design Version 1.7 1 INTRODUCTION 3 Guide 3 USING THE SYSTEM 4 Accessing the System 5 Logging In Using an Access Email 5 Normal Login 6 Resetting a Password 6 Logging Off 6 Home Page
User Guide Product Design Version 1.7 1 INTRODUCTION 3 Guide 3 USING THE SYSTEM 4 Accessing the System 5 Logging In Using an Access Email 5 Normal Login 6 Resetting a Password 6 Logging Off 6 Home Page
This Tutorial is for Word 2007 but 2003 instructions are included in [brackets] after of each step.
![This Tutorial is for Word 2007 but 2003 instructions are included in [brackets] after of each step. This Tutorial is for Word 2007 but 2003 instructions are included in [brackets] after of each step.](/thumbs/89/98497436.jpg) This Tutorial is for Word 2007 but 2003 instructions are included in [brackets] after of each step. Table of Contents Get Organized... 1 Create the Home Page... 1 Save the Home Page as a Word Document...
This Tutorial is for Word 2007 but 2003 instructions are included in [brackets] after of each step. Table of Contents Get Organized... 1 Create the Home Page... 1 Save the Home Page as a Word Document...
Click Trust to launch TableView.
 Visualizing Expression data using the Co-expression Tool Web service and TableView Introduction. TableView was written by James (Jim) E. Johnson and colleagues at the University of Minnesota Center for
Visualizing Expression data using the Co-expression Tool Web service and TableView Introduction. TableView was written by James (Jim) E. Johnson and colleagues at the University of Minnesota Center for
Tutorial. De Novo Assembly of Paired Data. Sample to Insight. November 21, 2017
 De Novo Assembly of Paired Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
De Novo Assembly of Paired Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
TEACHER: CREATE PRACTICE QUIZ
 TEACHER: CREATE PRACTICE QUIZ Table of Contents Select Questions... 3 Create Practice Test: 1. Select questions... 4 Create Practice Test: 2. Add to an Existing Test... 6 Create versions & Editing Tabs...
TEACHER: CREATE PRACTICE QUIZ Table of Contents Select Questions... 3 Create Practice Test: 1. Select questions... 4 Create Practice Test: 2. Add to an Existing Test... 6 Create versions & Editing Tabs...
Mockup Step-by-Step Guide
 Guide CONTENTS Contents... 1 Overview... 2 Key Takeaways... 2 Mockup User Interface... 3 Mockup Toolbar... 3 Options... 3 General Options... 4 Float Properties Popup... 4 Creating a Mockup... 6 Opening
Guide CONTENTS Contents... 1 Overview... 2 Key Takeaways... 2 Mockup User Interface... 3 Mockup Toolbar... 3 Options... 3 General Options... 4 Float Properties Popup... 4 Creating a Mockup... 6 Opening
LONG TERM CARE HOSPITAL ASSESSMENT SUBMISSION ENTRY & REPORTING TOOL (LASER) USER GUIDE IN SUPPORT OF VERSION (APRIL 2017)
 LONG TERM CARE HOSPITAL ASSESSMENT SUBMISSION ENTRY & REPORTING TOOL (LASER) USER GUIDE IN SUPPORT OF VERSION 1.3.0 (APRIL 2017) Updated 3/2017 TABLE OF CONTENT INTRODUCTION... 4 INSTALLATION... 4 Installers
LONG TERM CARE HOSPITAL ASSESSMENT SUBMISSION ENTRY & REPORTING TOOL (LASER) USER GUIDE IN SUPPORT OF VERSION 1.3.0 (APRIL 2017) Updated 3/2017 TABLE OF CONTENT INTRODUCTION... 4 INSTALLATION... 4 Installers
Tutorial: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and Expression measures
 : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
Annotating a single sequence
 BioNumerics Tutorial: Annotating a single sequence 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn how
BioNumerics Tutorial: Annotating a single sequence 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn how
GeneSpring Tutorial version 3.5
 version 3.5 Release date, 28 December 2000 Copyright 1998-2000 Silicon Genetics. All rights reserved. All rights reserved. GeneSpring, GeneSpider, GenEx, GeNet, and MicroSift are trademarks of Silicon
version 3.5 Release date, 28 December 2000 Copyright 1998-2000 Silicon Genetics. All rights reserved. All rights reserved. GeneSpring, GeneSpider, GenEx, GeNet, and MicroSift are trademarks of Silicon
Agilent Genomic Workbench 7.0
 Agilent Genomic Workbench 7.0 Data Viewing User Guide Agilent Technologies Notices Agilent Technologies, Inc. 2012, 2015 No part of this manual may be reproduced in any form or by any means (including
Agilent Genomic Workbench 7.0 Data Viewing User Guide Agilent Technologies Notices Agilent Technologies, Inc. 2012, 2015 No part of this manual may be reproduced in any form or by any means (including
GenomeStudio Software Release Notes
 GenomeStudio Software 2009.2 Release Notes 1. GenomeStudio Software 2009.2 Framework... 1 2. Illumina Genome Viewer v1.5...2 3. Genotyping Module v1.5... 4 4. Gene Expression Module v1.5... 6 5. Methylation
GenomeStudio Software 2009.2 Release Notes 1. GenomeStudio Software 2009.2 Framework... 1 2. Illumina Genome Viewer v1.5...2 3. Genotyping Module v1.5... 4 4. Gene Expression Module v1.5... 6 5. Methylation
Browser Exercises - I. Alignments and Comparative genomics
 Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Useful software utilities for computational genomics. Shamith Samarajiwa CRUK Autumn School in Bioinformatics September 2017
 Useful software utilities for computational genomics Shamith Samarajiwa CRUK Autumn School in Bioinformatics September 2017 Overview Search and download genomic datasets: GEOquery, GEOsearch and GEOmetadb,
Useful software utilities for computational genomics Shamith Samarajiwa CRUK Autumn School in Bioinformatics September 2017 Overview Search and download genomic datasets: GEOquery, GEOsearch and GEOmetadb,
CLC Sequence Viewer 6.5 Windows, Mac OS X and Linux
 CLC Sequence Viewer Manual for CLC Sequence Viewer 6.5 Windows, Mac OS X and Linux January 26, 2011 This software is for research purposes only. CLC bio Finlandsgade 10-12 DK-8200 Aarhus N Denmark Contents
CLC Sequence Viewer Manual for CLC Sequence Viewer 6.5 Windows, Mac OS X and Linux January 26, 2011 This software is for research purposes only. CLC bio Finlandsgade 10-12 DK-8200 Aarhus N Denmark Contents
AutoCAD 2009 User InterfaceChapter1:
 AutoCAD 2009 User InterfaceChapter1: Chapter 1 The AutoCAD 2009 interface has been enhanced to make AutoCAD even easier to use, while making as much screen space available as possible. In this chapter,
AutoCAD 2009 User InterfaceChapter1: Chapter 1 The AutoCAD 2009 interface has been enhanced to make AutoCAD even easier to use, while making as much screen space available as possible. In this chapter,
Fusion Detection Using QIAseq RNAscan Panels
 Fusion Detection Using QIAseq RNAscan Panels June 11, 2018 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com ts-bioinformatics@qiagen.com
Fusion Detection Using QIAseq RNAscan Panels June 11, 2018 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com ts-bioinformatics@qiagen.com
CyKEGGParser User Manual
 CyKEGGParser User Manual Table of Contents Introduction... 3 Development... 3 Citation... 3 License... 3 Getting started... 4 Pathway loading... 4 Laoding KEGG pathways from local KGML files... 4 Importing
CyKEGGParser User Manual Table of Contents Introduction... 3 Development... 3 Citation... 3 License... 3 Getting started... 4 Pathway loading... 4 Laoding KEGG pathways from local KGML files... 4 Importing
Wilson Leung 05/27/2008 A Simple Introduction to NCBI BLAST
 A Simple Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at http://www.ncbi.nih.gov/blast/
A Simple Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at http://www.ncbi.nih.gov/blast/
HOSPICE ABSTRACTION REPORTING TOOL (HART) USER GUIDE
 HOSPICE ABSTRACTION REPORTING TOOL (HART) USER GUIDE IN SUPPORT OF VERSION 1.5.0 (APRIL 2019) Page 1 of 60 TABLE OF CONTENT TABLE OF CONTENT... 2 INTRODUCTION... 4 INSTALLATION... 4 Installers Access Rights...
HOSPICE ABSTRACTION REPORTING TOOL (HART) USER GUIDE IN SUPPORT OF VERSION 1.5.0 (APRIL 2019) Page 1 of 60 TABLE OF CONTENT TABLE OF CONTENT... 2 INTRODUCTION... 4 INSTALLATION... 4 Installers Access Rights...
Click on "+" button Select your VCF data files (see #Input Formats->1 above) Remove file from files list:
 CircosVCF: CircosVCF is a web based visualization tool of genome-wide variant data described in VCF files using circos plots. The provided visualization capabilities, gives a broad overview of the genomic
CircosVCF: CircosVCF is a web based visualization tool of genome-wide variant data described in VCF files using circos plots. The provided visualization capabilities, gives a broad overview of the genomic
Guide to KI-ELN, downloaded/remote desktop client
 Guide to KI-ELN, downloaded/remote desktop client Conventions used in this tutorial Bold a button Underline selection from a menu Italics Right mouse-click menu item You can access the system in three
Guide to KI-ELN, downloaded/remote desktop client Conventions used in this tutorial Bold a button Underline selection from a menu Italics Right mouse-click menu item You can access the system in three
FrontPage 2000 Tutorial -- Advanced
 FrontPage 2000 Tutorial -- Advanced Shared Borders Shared Borders are parts of the web page that share content with the other pages in the web. They are located at the top, bottom, left side, or right
FrontPage 2000 Tutorial -- Advanced Shared Borders Shared Borders are parts of the web page that share content with the other pages in the web. They are located at the top, bottom, left side, or right
Blackboard 1: Building a Course Site
 Blackboard 1: Building a Course Site This QuickStart outlines the material covered in the first of four workshops on teaching with Blackboard. It will help you to begin creating your Blackboard course
Blackboard 1: Building a Course Site This QuickStart outlines the material covered in the first of four workshops on teaching with Blackboard. It will help you to begin creating your Blackboard course
Report Designer Report Types Table Report Multi-Column Report Label Report Parameterized Report Cross-Tab Report Drill-Down Report Chart with Static
 Table of Contents Report Designer Report Types Table Report Multi-Column Report Label Report Parameterized Report Cross-Tab Report Drill-Down Report Chart with Static Series Chart with Dynamic Series Master-Detail
Table of Contents Report Designer Report Types Table Report Multi-Column Report Label Report Parameterized Report Cross-Tab Report Drill-Down Report Chart with Static Series Chart with Dynamic Series Master-Detail
Data Grid Utilities - Searching Data Grids
 Data Grid Utilities - Searching Data Grids Step 1 : To search any column on selected data grids begin by right mouse-clicking the column header for the column you want to search. This feature works on
Data Grid Utilities - Searching Data Grids Step 1 : To search any column on selected data grids begin by right mouse-clicking the column header for the column you want to search. This feature works on
Integrated Genome browser (IGB) installation
 Integrated Genome browser (IGB) installation Navigate to the IGB download page http://bioviz.org/igb/download.html You will see three icons for download: The three icons correspond to different memory
Integrated Genome browser (IGB) installation Navigate to the IGB download page http://bioviz.org/igb/download.html You will see three icons for download: The three icons correspond to different memory
Beyond 20/20. Browser - English. Version 7.0, SP3
 Beyond 20/20 Browser - English Version 7.0, SP3 Notice of Copyright Beyond 20/20 Desktop Browser Version 7.0, SP3 Copyright 1992-2006 Beyond 20/20 Inc. All rights reserved. This document forms part of
Beyond 20/20 Browser - English Version 7.0, SP3 Notice of Copyright Beyond 20/20 Desktop Browser Version 7.0, SP3 Copyright 1992-2006 Beyond 20/20 Inc. All rights reserved. This document forms part of
Website Management with the CMS
 Website Management with the CMS In Class Step-by-Step Guidebook Updated 12/22/2010 Quick Reference Links CMS Login http://staging.montgomerycollege.edu/cmslogin.aspx Sample Department Site URLs (staging
Website Management with the CMS In Class Step-by-Step Guidebook Updated 12/22/2010 Quick Reference Links CMS Login http://staging.montgomerycollege.edu/cmslogin.aspx Sample Department Site URLs (staging
Anima-LP. Version 2.1alpha. User's Manual. August 10, 1992
 Anima-LP Version 2.1alpha User's Manual August 10, 1992 Christopher V. Jones Faculty of Business Administration Simon Fraser University Burnaby, BC V5A 1S6 CANADA chris_jones@sfu.ca 1992 Christopher V.
Anima-LP Version 2.1alpha User's Manual August 10, 1992 Christopher V. Jones Faculty of Business Administration Simon Fraser University Burnaby, BC V5A 1S6 CANADA chris_jones@sfu.ca 1992 Christopher V.
Overview of Adobe Fireworks
 Adobe Fireworks Overview of Adobe Fireworks In this guide, you ll learn how to do the following: Work with the Adobe Fireworks workspace: tools, Document windows, menus, and panels. Customize the workspace.
Adobe Fireworks Overview of Adobe Fireworks In this guide, you ll learn how to do the following: Work with the Adobe Fireworks workspace: tools, Document windows, menus, and panels. Customize the workspace.
Electrical System Functional Definition
 Electrical System Functional Definition Preface What's New? Getting Started Basic Tasks Advanced Tasks Workbench Description Customizing Glossary Index Dassault Systèmes 1994-2000. All rights reserved.
Electrical System Functional Definition Preface What's New? Getting Started Basic Tasks Advanced Tasks Workbench Description Customizing Glossary Index Dassault Systèmes 1994-2000. All rights reserved.
The Text scrap sheet is also needed for copying image and webpage URLs for easy access. Additionally you will use it to look at HTML when needed.
 epro 2 Hints Always have open Word and Text scrap sheets so you can write your text, spell check etc. in Word. You MUST copy and paste your writings onto Text to remove formatting. The Text scrap sheet
epro 2 Hints Always have open Word and Text scrap sheets so you can write your text, spell check etc. in Word. You MUST copy and paste your writings onto Text to remove formatting. The Text scrap sheet
panda Documentation Release 1.0 Daniel Vera
 panda Documentation Release 1.0 Daniel Vera February 12, 2014 Contents 1 mat.make 3 1.1 Usage and option summary....................................... 3 1.2 Arguments................................................
panda Documentation Release 1.0 Daniel Vera February 12, 2014 Contents 1 mat.make 3 1.1 Usage and option summary....................................... 3 1.2 Arguments................................................
AgWare ClickFORMS 7.0 Introductory Tutorial
 AgWare ClickFORMS 7.0 Introductory Tutorial MAIN WINDOW If you have used Office 2007, our new layout will look familiar. Here is a screenshot of the new and improved AgWare ClickFORMS window: The Forms
AgWare ClickFORMS 7.0 Introductory Tutorial MAIN WINDOW If you have used Office 2007, our new layout will look familiar. Here is a screenshot of the new and improved AgWare ClickFORMS window: The Forms
Working with Mailbox Manager
 Working with Mailbox Manager A user guide for Mailbox Manager supporting the Message Storage Server component of the Avaya S3400 Message Server Mailbox Manager Version 5.0 February 2003 Copyright 2003
Working with Mailbox Manager A user guide for Mailbox Manager supporting the Message Storage Server component of the Avaya S3400 Message Server Mailbox Manager Version 5.0 February 2003 Copyright 2003
Tutorial: Resequencing Analysis using Tracks
 : Resequencing Analysis using Tracks September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : Resequencing
: Resequencing Analysis using Tracks September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : Resequencing
EXERCISE: GETTING STARTED WITH SAV
 Sequencing Analysis Viewer (SAV) Overview 1 EXERCISE: GETTING STARTED WITH SAV Purpose This exercise explores the following topics: How to load run data into SAV How to explore run metrics with SAV Getting
Sequencing Analysis Viewer (SAV) Overview 1 EXERCISE: GETTING STARTED WITH SAV Purpose This exercise explores the following topics: How to load run data into SAV How to explore run metrics with SAV Getting
SERVICE MANU SER VICE MANU
 SERVICE MANUAL H ow t o U s e Table of Contents 1. 2. 3. 4. 5. 6. Launching the Service Manual Setting up the Service Manual icon Shutting down the Service Manual Basic controls Basic controls of Acrobat
SERVICE MANUAL H ow t o U s e Table of Contents 1. 2. 3. 4. 5. 6. Launching the Service Manual Setting up the Service Manual icon Shutting down the Service Manual Basic controls Basic controls of Acrobat
General Help & Instructions to use with Examples
 General Help & Instructions to use with Examples Contents Types of Searches and their Purposes... 2 Basic Search:... 2 Advance search option... 6 List Search:... 7 Details Page... 8 Results Grid functionalities:...
General Help & Instructions to use with Examples Contents Types of Searches and their Purposes... 2 Basic Search:... 2 Advance search option... 6 List Search:... 7 Details Page... 8 Results Grid functionalities:...
Table Basics. The structure of an table
 TABLE -FRAMESET Table Basics A table is a grid of rows and columns that intersect to form cells. Two different types of cells exist: Table cell that contains data, is created with the A cell that
TABLE -FRAMESET Table Basics A table is a grid of rows and columns that intersect to form cells. Two different types of cells exist: Table cell that contains data, is created with the A cell that
Tutorial 1: Exploring the UCSC Genome Browser
 Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Introduction Accessing MICS Compiler Learning MICS Compiler CHAPTER 1: Searching for Data Surveys Indicators...
 Acknowledgement MICS Compiler is a web application that has been developed by UNICEF to provide access to Multiple Indicator Cluster Survey data. The system is built on DevInfo technology. 3 Contents Introduction...
Acknowledgement MICS Compiler is a web application that has been developed by UNICEF to provide access to Multiple Indicator Cluster Survey data. The system is built on DevInfo technology. 3 Contents Introduction...
Easy Edit Editing the Public Website
 Easy Edit Editing the Public Website Recommended browser for using the easy edit suite: Firefox Editing a Live Page 2. Click on the edit button 3. Place the page in to safe edit mode Either by clicking
Easy Edit Editing the Public Website Recommended browser for using the easy edit suite: Firefox Editing a Live Page 2. Click on the edit button 3. Place the page in to safe edit mode Either by clicking
The ATLAS.ti Interface
 28 STARTING ATLAS.TI Importing Copy Bundle Files (Versions 5, 6 Or 7) To import a copy bundle file, open ATLAS.ti 8 and select the option IMPORT LEGACY PROJECT. If ATLAS.ti is already open, select File
28 STARTING ATLAS.TI Importing Copy Bundle Files (Versions 5, 6 Or 7) To import a copy bundle file, open ATLAS.ti 8 and select the option IMPORT LEGACY PROJECT. If ATLAS.ti is already open, select File
User Guide for TASKE Contact Web Interface
 User Guide for TASKE Contact Web Interface For Avaya Communication Manager with Application Enablement Services Version: 8.9 Date: 2011-06 This document is provided to you for informational purposes only.
User Guide for TASKE Contact Web Interface For Avaya Communication Manager with Application Enablement Services Version: 8.9 Date: 2011-06 This document is provided to you for informational purposes only.
Warping & Blending AP
 Warping & Blending AP Operation about AP This AP provides three major functions including Warp, Edge Blending and Black Level. If the AP is already installed, please remove previous version before installing
Warping & Blending AP Operation about AP This AP provides three major functions including Warp, Edge Blending and Black Level. If the AP is already installed, please remove previous version before installing
User Guide. v7.5. September 4, For the most recent version of this document, visit kcura's Documentation Site.
 User Guide v7.5 September 4, 2013 For the most recent version of this document, visit kcura's Documentation Site. Table of Contents 1 User guide overview 4 2 Relativity objects 4 3 Workspace 6 3.1 Workspaces
User Guide v7.5 September 4, 2013 For the most recent version of this document, visit kcura's Documentation Site. Table of Contents 1 User guide overview 4 2 Relativity objects 4 3 Workspace 6 3.1 Workspaces
SmartView. User Guide - Analysis. Version 2.0
 SmartView User Guide - Analysis Version 2.0 Table of Contents Page i Table of Contents Table Of Contents I Introduction 1 Dashboard Layouts 2 Dashboard Mode 2 Story Mode 3 Dashboard Controls 4 Dashboards
SmartView User Guide - Analysis Version 2.0 Table of Contents Page i Table of Contents Table Of Contents I Introduction 1 Dashboard Layouts 2 Dashboard Mode 2 Story Mode 3 Dashboard Controls 4 Dashboards
