A methodology for biomedical ontology reuse
|
|
|
- Poppy Lang
- 6 years ago
- Views:
Transcription
1 A methodology for biomedical ontology reuse Zulkarnain, NZ and Meziane, F _1 Title Authors Type URL A methodology for biomedical ontology reuse Zulkarnain, NZ and Meziane, F Article Published Date 2016 This version is available at: USIR is a digital collection of the research output of the University of Salford. Where copyright permits, full text material held in the repository is made freely available online and can be read, downloaded and copied for non commercial private study or research purposes. Please check the manuscript for any further copyright restrictions. For more information, including our policy and submission procedure, please contact the Repository Team at: usir@salford.ac.uk.
2 A Methodology for Biomedical Ontology Reuse Nur Zareen Zulkarnain 1, Farid Meziane 1 and Gillian Crofts 2 1 Informatics Research Centre, School of Computing, Science and Engineering, University of Salford, M5 4WT, UK N.Z.Zulkarnain@edu.salford.ac.uk,F.Meziane@salford.ac.uk, 2 School of Health Science, University of Salford, M6 6PU, UK G.Crofts@salford.ac.uk Abstract The abundance of biomedical ontologies is beneficial to the development of biomedical related systems. However, existing biomedical ontologies such as the National Cancer Institute Thesaurus (NCIT), Foundational Model of Anatomy (FMA) and Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT) are often too large to be implemented in a particular system and cause unnecessary high usage of memory and slow down the system s processing time. Developing a new ontology from scratch just for the use of a particular system is deemed as inefficient since it requires additional time and causes redundancy. Thus, a potentially better method is by reusing existing ontologies. However, currently there are no specific methods or tools for reusing ontologies. This paper aims to provide readers with a step by step method in reusing ontologies together with the tools that can be used to ease the process. Keywords: Ontology, Ontology Reuse, Biomedical Ontology, BioPortal 1 Introduction Biomedical systems are an integral part of today s medical world. Systems such as electronic patient records and clinical decision support systems (CDSS) have played an important role in assisting the works of medical personnel. One area that could benefit from the development of biomedical systems is ultrasound reporting. In ultrasound, reports generated have more value compared to the image captured during the examination [2]. Variations in ultrasound reporting impacts the way a report is interpreted as well as in decision making. Thus, the standardization of these reports is important. In order to achieve this goal, ontologies are used to understand the reports and structure them according to a certain format [16] as well as recognizing the relationships between the parts of the text composing the report. General and established domains such as medicine have existing ontologies that cover the general concepts in the domain. Examples of these ontologies include the National Cancer Institute Thesaurus (NCIT), Foundational Model of Anatomy (FMA) and Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT). These ontologies however are often too large to be manipulated
3 2 Nur Zareen Zulkarnain, Farid Meziane and Gillian Crofts or processed in a specific application. Thus, a domain specific ontology is needed to solve this problem. Building a new domain specific ontology from scratch would not be efficient since this will cause redundancy and takes a lot of time. Thus, ontology reuse has been potentially seen as a better alternative. This paper discusses how ontology reuse has been done before and proposes a methodology to reuse ontologies together with the existing tools that can be used to ease the reuse process. The development of the Abdominal Ultrasound Ontology (AUO) as the knowledge base for an ultrasound reporting system developed by Zulkarnain et al. [16] is used in this paper to explain the proposed ontology reuse methodology. 2 Related Work Ontology reuse can be defined as a process where a small portion of existing ontologies is taken as an input to build a new one [3]. The process of reusing large existing ontologies allows their use without slowing down the process of an application. Ontology reuse also increases interoperability [14]. Indeed, when an ontology is reused by several other new ontologies, interoperability between these ontologies can be achieved much easier since they share several features such as classes naming method and concept modelling. Even though ontology reuse brings a lot of benefits, there are currently no tools that provide adequate support for the ontology reuse process [8,14] which hinders the effort of ontology reuse. There is also no one specific method agreed in reusing ontologies. Even so, most ontology reuse methodologies that have been used in previous works [1,4,5,11,12,14,15] falls along the line of these four steps: (i) Ontology selection for reuse, (ii) Concept selection, (iii) Concept customization and (iv) Ontology integration. The first step for ontology reuse is to select the ontology to be reused. Ontology selection is done according to several criteria according to the needs of the new ontology, for example the language of the ontology, its comprehensiveness and its reasoning capabilities. Once the ontology for reuse is chosen, the next step would be to select the concepts that would be reused. One or several ontologies can be selected for reuse depending on the needs of the new ontology. Russ et al. [11] in their work merged two aircraft ontologies where most of its concepts were selected to develop a broader aircraft ontology. Shah et al. [12] on the other hand reused just one ontology; SNOMED CT where he selected the concepts needed then adds other relevant concepts not included in SNOMED CT. Concepts selected are then translated into the same semantic language and then merged. In Caldarola et al. s work [4] this includes manually translating metadata to better understand concepts. Alani [1] in developing his ontology has merged several ontologies that contain different properties for one same concept which resulted in additional knowledge representation. Several different concepts have also been selected from different ontologies which are then compared and merged. Finally, the ontology will be integrated into the system or application. In this research, these four steps serve as a guideline in developing an ontology reuse methodology for the biomedical domain. The methodology proposed in this
4 A Methodology for Biomedical Ontology Reuse 3 research will allow for the ontology to be reused from multiple existing ontologies and suggest tools that would help in each step of the methodology. The ontology developed, Abdominal Ultrasound Ontology (AUO), will serve two purposes in this research: (i) it will be used to standardize the development of ultrasound reports and enforce the use of standard terminology and (ii) to analyse the reports written in Natural Language (English free-text) with the aim of automatically transforming them into a structured format. 3 The Proposed Methodology In developing a new ontology by reusing existing biomedical ones, proper planning and execution are important in order to ensure the modularity of the concepts reused. Thus, the ontology reuse methodology developed in this paper, adopted the general four steps mentioned in section 2 and summarised in Figure 1. Figure 1. Ontology Reuse Methodology 3.1 Term Extraction The first step in ontology reuse or even in developing one from scratch is to decide on its scope and domain. In this case of developing the Abdominal Ultrasound Ontology (AUO), the scope and domain of the ontology is abdominal ultrasound. 49 sample ultrasound reports have been collected and used as the basis of our ontology corpus. These sample reports were obtained from the Radiology Departments in a large NHS Trust incorporating 4 regionally based hospitals in Manchester and Salford. Once we have our corpus, the next step is to extract relevant terms from the corpus to generate a list of terms for reuse. Two biomedical term extraction applications; (i) TerMine 1 and (ii) BioTex 2 have been used for
5 4 Nur Zareen Zulkarnain, Farid Meziane and Gillian Crofts the extraction. All 49 sample reports were submitted to both applications and the results are shown in Table 1. Table 1. Comparison of biomedical term extraction using TerMine and BioTex TerMine BioTex Language English English License Open Open POS Tagger GENIA Tagger / Tree Tagger Tree Tagger Terms Found 241 (GENIA Tagger) (Tree Tagger) Extraction Type Multi-word extraction Multi-word and Single-word extraction From this comparison, BioTex was chosen as the better biomedical term extractor in this research because of its ability to extract more terms compared to TerMine. BioTex is an automatic term recognition and extraction application that allows for both multi-word and single-word extraction [7]. It is important that the term extractor is able to extract not only multi-word but also single-word terms. For example, if the sentence Unremarkable appearances of the liver with no intrahepatic lesions was submitted to both applications, TerMine will only extract two multi-word terms Unremarkable appearance and intrahepatic lesion while BioTex will extract not only the two multi-word terms but also liver which is a single word term. If single-word terms such as liver, kidney and spleen were not extracted, the ontology developed would be incomplete. Terms which are extracted from BioTex were also validated using the Unified Medical Language System (UMLS) [7] which is a set of documents containing health and biomedical vocabularies and standards. 3.2 Ontology Recommendation The next step after obtaining a list of terms for ontology reuse would be to select the suitable ontology to be reused. Three important criteria were used for selecting the ontology in this research: (i) Ontology coverage - To which extend does the ontology covers the terms extracted from the corpus? (ii) Ontology acceptance - Is the ontology being accepted in the medical field and how often is it used? and (iii) Ontology language - Is the ontology written in OWL, OBO or other semantic languages? Initial review resulted in choosing FMA, SNOMED-CT and RadLex as suitable candidates because of their domain coverage, acceptance in the biomedical community and language which is OWL. In order to verify this, an ontology recommender was developed using BioPortal s ontology recommender
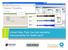
6 A Methodology for Biomedical Ontology Reuse 5 API 3 which is an open ontology library that contains ontologies with domains that range from anatomy, phenotype and chemistry to experimental conditions [10]. Figure 2. BioPortal s Ontology Recommender BioPortal has an ontology recommender available on its portal that can be used to obtain suggestions on suitable ontology to be reused for certain corpus. The ontology recommender makes a decision according to the following three criteria: (i) Coverage - Which ontology provides most coverage to the input text?, (ii) Connectivity - How often the ontology is mapped by other ontologies? and (iii) Size - Number of concepts in the ontology [6]. When a list of terms is submitted to the recommender, it will give a recommendation of 25 ontologies which are ranked from the highest to lowest scores (see Figure 2). The final score is calculated based on the following formula: F inalscore = (CoverageScore 0.55) + (AcceptanceScore 0.15) + (KnowledgeDetailScore 0.15) + (SpecializationScore 0.15) (1) The coverage score is given based on the number of terms in the input that are covered by the ontology. The acceptance score indicates how well-known and trusted the ontology is in the biomedical field. Knowledge detail score on the other hand indicates the level of details in the ontology; i.e. does the ontology have definitions, synonym or other details. Specialization score is given based on how well the ontology covers the domain of the input. An example is given in Fig. 2 where 21 terms where submitted. There is however a limitation in using it on the portal whereby it only allows for 500 words to be submitted. This limitation 3
7 6 Nur Zareen Zulkarnain, Farid Meziane and Gillian Crofts has prompted us to develop our own recommender by manipulating the data from BioPortal s ontology recommender API. We first develop the recommender that would give 25 ontology recommendations just like how it would be in the BioPortal s recommender. However, it seems that 761 terms were too big for the recommender s server to handle. Because of this, a recommender that would suggest ontology for each term was developed. A list of terms, in this case the 761 terms that have been extracted, are submitted to the algorithm that would submit each term to BioPortal s recommender and get ontology recommendations for each term. Then, the frequency of each ontology recommended will be counted and sorted from highest to lowest. The recommender has ranked NCI Thesaurus as the ontology with the highest frequency (341) followed by SNOMED CT (140) and RadLex (37). Figure 3 shows an excerpt of the result from processing 761 terms using the recommender we have developed. Figure 3. (a) Ontology recommendation for each term (b) Ranking of ontology recommended 3.3 Term to Concept Mapping Once the ontology for reuse has been selected, the next step in building the abdominal ultrasound ontology is to map the terms extracted to concepts in the ontology which is done by referring to the result from BioPortal s Search API. The API allows us to insert several parameters to perform concept search which in this case, the parameters used are q to specify the term that we would like to search for, and ontologies which specifies the ontology where we would like to look for the term. Once these parameters have been submitted, the API will return a concept if there is a match with the term submitted. The concept will be returned with several other properties such as the preferred label, definition, synonym, match type and the terms relationship with its children, descendant, parents and ancestors.
8 A Methodology for Biomedical Ontology Reuse 7 In previous works by Mejino et al. [9] and Shah et al. [13], term to concept mapping was done by referring to the existing ontology and mapping it into the new one by deleting and adding concepts in the ontology to make it complete. Using BioPortal s API consumes less time and work as the terms are queried according to the provided parameters. This will also ensure the accuracy of the relationship between concepts and its children, descendant, parents and ancestors since there are links that can be clearly seen in the API result. There was an intention to auto populate these data into Protègè (the OWL editor that was used in this research) by taking advantage of the option of saving the results in XML compared to JSON. However, there are two reasons why this is not possible at the moment. The first reason was that data from the API does not give the complete properties of a concept. For example, parents and ancestors were provided as links which makes it hard for the data to be manipulated since the properties of the parents and ancestors can only be obtained after the link is visited. The second reason is there are terms which matched several concepts in the ontology. For example, the term calculus could mean branch of mathematics concerned with calculation or an abnormal concretion occurring mostly in the urinary and biliary tracts, usually composed of mineral salts. Thus, it is important to know in which context it is being used in order to adopt the correct meaning. Figure 4. Term to concept mapping guide In deciding whether a term should be reused or not, the term to concept mapping guide (see Fig. 4) was used. Firstly, a term from the term list will be queried using the Search API in the ontology with the highest frequency which in this case is NCIT. If there is a match, we will see whether the match is a preferred label, synonym or partial match. A preferred label (PrefLabel) match means that the API found a concept that has an exact match to the term while
9 8 Nur Zareen Zulkarnain, Farid Meziane and Gillian Crofts synonym match means that the term is found as a synonym to the concept. Partial match on the other hand means that there is no exact match for the term but there are at least two concepts that match the term. For example, for the term intrahepatic biliary, there are no concepts that match the term exactly. However, there is the concept intrahepatic which is an anatomy qualifier in NCIT and the concept duct in NCIT which is an organ that matches. If the match is a PrefLabel or synonym match, the concept will be reused. If the match is partial, the concepts that make up the term will also be reused. However, the term would still remain in the term list so that it could be compared to concepts in other ontologies. After the concept has been reused, we will find out if the term has a parent or ancestors. If there is, the parent or ancestors will also be reused. Once all terms have been searched, this process will then be repeated for the remaining recommended ontologies. In this research, all terms are first searched in NCIT followed by SNOMED CT and RadLex. The Abdominal Ultrasound Ontology modelling follows the modelling of NCIT since it is the main ontology being reused. When merging ontologies from SNOMED CT and RadLex into the ontologies reused from NCIT, we would first find a parent that would be suitable for the concept. If no such parent exists, the parent and ancestors of the concept will then be reused. This is done to ensure the modularity of the ontology developed. If no match is found in any of these ontologies, a new concept will then be created with the help of domain experts. The main objective of using this ontology reuse methodology is to achieve as much coverage as possible and reduce the need for domain experts in developing the ontology. 3.4 Ontology Evaluation by Domain Expert Figure 5. Snapshot of the Abdominal Ultrasound Ontology
10 A Methodology for Biomedical Ontology Reuse 9 Once a complete Abdominal Ultrasound Ontology has been developed using the ontology reuse methodology, it is important that the ontology be evaluated by a domain expert in order to verify that the relationship between the terms as well as their definitions are correct. In evaluating this ontology, we have sat down together with a domain expert and went through the ontology. There are some corrections that need to be done but overall, the domain expert believes that the 92.6% ontology coverage is enough to cover all the important concepts that an abdominal ultrasound report would need. For the other 7.4% terms that have no match in the ontology, some of it were caused by human error whereby spelling mistakes were made by the reporter. As for the rest of it, the domain expert will help in giving definitions and suggestions on where it would fit in the ontology. Out of the 7.4% terms that have no match in the ontology, there are also several terms that the domain expert believes we can omit since these words should not be in an ultrasound report for good practice. Examples of such words are comet tail, NAD, and hepato petal. Figure 5 shows a snapshot of the complete Abdominal Ultrasound Ontology. 4 Result and Discussion The ontology reuse methodology used to develop the Abdominal Ultrasound Ontology (AUO) has given the highest number of concept match compared to using only one ontology. This can be proved by performing a term to concept matching using the 761 terms extracted from the sample ultrasound report corpus. Figure 6 shows the comparison of total matches according to type (PrefLabel match, synonym match, partial match and no match) between NCIT, SNOMED CT and AUO. Between NCIT and SNOMED CT, NCIT has the higher concept match total with 151 PrefLabel matches, 79 synonyms matches and 438 partial matches. SNOMED CT on the other hand has only 98 PrefLabel matches, 104 synonyms matches and 431 partial matches. The reason SNOMED CT has lower PrefLabel matches compared to synonyms is because of its naming convention. For example, the preferred label for kidney is kidney structure and entire gallbladder for gallbladder. When writing report, radiologist often used simpler words like kidney and gallbladder instead of kidney structure and entire gallbladder thus, when term to concept matching was performed, SNOMED CT returned more synonym matches compared to PrefLabel. Compared to NCIT and SNOMED CT, AUO returns the highest total match where it has 176 PrefLabel matches, 111 synonym matches and 418 partial matches. The reason AUO returns the most number of matches is because the ontology reuse methodology selects the best match from different ontologies and merge it into the AUO. Its exhaustive mapping in several ontologies based on the ontology rank has ensured that almost all terms in the corpus are covered by AUO. Whenever possible, a PrefLabel match will be inserted in the ontology. If not, a synonym match will be added then only partial matches are included to ensure the ontology has a wide coverage of the corpus.
11 10 Nur Zareen Zulkarnain, Farid Meziane and Gillian Crofts Figure 6. Breakdown of total match according to type against NCIT, SNOMED CT and Abdominal Ultrasound Ontology (AUO) From the analysis, it can be concluded that it is better to reuse from several ontologies compared to just one. This is because reusing several ontologies offers better term coverage compared to reusing just one. Fig. 7 shows the percentage of total match and no match in all three ontologies. If ontology reuse was done by mapping the 761 terms against NCIT, there will only be an 87.8% of coverage. If the mapping were done against SNOMED CT, the percentage of coverage would be only 83.2% which is lower than NCIT. However, the percentage of coverage increases to 92.6% when several ontologies were reused; which in this case are NCIT, SNOMED CT, and RadLex. Figure 7. Percentage of total match and no match in NCIT, SNOMED CT and AUO
12 A Methodology for Biomedical Ontology Reuse 11 The percentage of no match is also very small (7.4%) which means that the AUO covers almost all the terms in the corpus. After ontology evaluation with domain expert, the percentage of no match has been reduced to only 5% after the domain expert included new concepts which before this have no match in any of the other ontologies being reused. The reason there is still 5% of no match is because there are several term in the corpus that the domain experts believe are poor usage of terms to describe findings in an ultrasound report. The domain expert believes that this is bad practice and the medical ultrasound experts are now slowly cutting down the usage of such words thus making it irrelevant to be in the AUO. Another reason for the 5% of no match is spelling errors made by ultrasound reporters. This is not a concern for now but for future work, we could consider using the ontology to also correct and understand these errors. NCIT has a total of 113,794 classes while SNOMED CT has 316,031 classes. However, there are only 668 and 633 matches respectively for each NCIT and SNOMED CT regarding abdominal ultrasound terminology. On the other hand, AUO has only 509 classes which is less than 0.5% of either NCIT or SNOMED CT but still managed to have 705 matches which is more than the matches NCIT and SNOMED CT each gets. This is because of the specialization of the ontology. Since the ontology has an intended purpose in an application, it is much better and more efficient to build a domain specific ontology through reuse. It definitely would not be efficient to store a large ontology such as NCIT and SNOMED CT and use only less than 0.3% of it. This is because it would take a lot of storage space and it will also slow down the application since the application will need to go through the whole ontology to find a match. Thus the better way to develop an ontology based application is to build a new domain specific ontology through ontology reuse methodology. 5 Conclusion Ontology reuse can be beneficial in developing domain specific ontologies for application system whereby it reduces development time and redundancy. The lack of proper methodology and tools in reusing ontology has hindered this effort. Thus, this paper proposed a methodology to reuse ontology together with supporting tools that would make the ontology reuse process much easier. The development of AUO using this methodology has proven that ontology reuse is beneficial in developing a small domain specific ontology which has wide coverage of the terminology used in the application system compared to using a large general domain ontology. It is hoped that the proposed ontology reuse methodology would encourage more usage of ontology in medical system without the development of similar domain ontologies that would cause redundancy. References 1. Harith Alani. Position paper: Ontology construction from online ontologies. In Proceedings of the 15th International Conference on World Wide Web, WWW 06, pages , New York, NY, USA, ACM.
13 12 Nur Zareen Zulkarnain, Farid Meziane and Gillian Crofts 2. G Boland. Enhancing the radiology product: the value of voice-recognition technology. Clinical radiology, 62(11):1127, Elena Paslaru Bontas, Malgorzata Mochol, and Robert Tolksdorf. Case studies on ontology reuse. In Proceedings of the 5th International Conference on Knowledge Management IKNOW05, Enrico G Caldarola, Antonio Picariello, and Antonio M Rinaldi. An approach to ontology integration for ontology reuse in knowledge based digital ecosystems. In Proceedings of the 7th International Conference on Management of computational and collective intelligence in Digital EcoSystems, pages 1 8. ACM, Maria Angeles Capellades. Assessment of reusability of ontologies: a practical example. In Proceedings of AAAI1999 Workshop on Ontology Management, AAAI Press, pages 74 79, Clement Jonquet, Mark A Musen, and Nigam H Shah. Building a biomedical ontology recommender web service. Journal of Biomedical Semantics, 1(S1), Juan Antonio Lossio-Ventura, Clement Jonquet, Mathieu Roche, and Maguelonne Teisseire. Biotex: A system for biomedical terminology extraction, ranking, and validation. In 13th International Semantic Web Conference, A. Maedche, B. Motik, L. Stojanovic, R. Studer, and R. Volz. An infrastructure for searching, reusing and evolving distributed ontologies. In Proceedings of the 12th International Conference on World Wide Web, WWW 03, pages , New York, NY, USA, ACM. 9. Jose LV Mejino Jr, Daniel L Rubin, and James F Brinkley. Fma-radlex: An application ontology of radiological anatomy derived from the foundational model of anatomy reference ontology. In AMIA Annual Symposium Proceedings, volume 2008, page 465. American Medical Informatics Association, Natalya F Noy, Nigam H Shah, Patricia L Whetzel, Benjamin Dai, Michael Dorf, Nicholas Griffith, Clement Jonquet, Daniel L Rubin, Margaret-Anne Storey, Christopher G Chute, et al. Bioportal: ontologies and integrated data resources at the click of a mouse. Nucleic acids research, pages W170 W173, Thomas Russ, Andre Valente, Robert MacGregor, and William Swartout. Practical experiences in trading off ontology usability and reusability. In Proceedings of the 12th Workshop on Knowledge Acquisition, Modeling and Management (KAW 99, pages 16 21, Tejal Shah, Fethi Rabhi, and Priyadip Ray. Oshco: A cross-domain ontology for semantic interoperability across medical and oral health domains. In e-health Networking, Applications Services (Healthcom), 2013 IEEE 15th International Conference on, pages , Oct Tejal Shah, Fethi Rabhi, Priyadip Ray, and Kerry Taylor. A guiding framework for ontology reuse in the biomedical domain. In System Sciences (HICSS), th Hawaii International Conference on, pages IEEE, Elena Simperl. Reusing ontologies on the semantic web: A feasibility study. Data Knowl. Eng., 68(10): , October Mike Uschold, Peter Clark, Mike Healy, Keith Williamson, and Steven Woods. An experiment in ontology reuse. In Proc. of the 11th Knowledge Acquisition Workshop, Nur Zareen Zulkarnain, Gillian Crofts, and Farid Meziane. An architecture to support ultrasound report generation and standardisation. In Proceedings of the International Conference on Health Informatics, pages , All links were last followed on January 25, 2016.
Prototyping a Biomedical Ontology Recommender Service
 Prototyping a Biomedical Ontology Recommender Service Clement Jonquet Nigam H. Shah Mark A. Musen jonquet@stanford.edu 1 Ontologies & data & annota@ons (1/2) Hard for biomedical researchers to find the
Prototyping a Biomedical Ontology Recommender Service Clement Jonquet Nigam H. Shah Mark A. Musen jonquet@stanford.edu 1 Ontologies & data & annota@ons (1/2) Hard for biomedical researchers to find the
Acquiring Experience with Ontology and Vocabularies
 Acquiring Experience with Ontology and Vocabularies Walt Melo Risa Mayan Jean Stanford The author's affiliation with The MITRE Corporation is provided for identification purposes only, and is not intended
Acquiring Experience with Ontology and Vocabularies Walt Melo Risa Mayan Jean Stanford The author's affiliation with The MITRE Corporation is provided for identification purposes only, and is not intended
Community-based ontology development, alignment, and evaluation. Natasha Noy Stanford Center for Biomedical Informatics Research Stanford University
 Community-based ontology development, alignment, and evaluation Natasha Noy Stanford Center for Biomedical Informatics Research Stanford University Community-based Ontology... Everything Development and
Community-based ontology development, alignment, and evaluation Natasha Noy Stanford Center for Biomedical Informatics Research Stanford University Community-based Ontology... Everything Development and
warwick.ac.uk/lib-publications
 Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
A Semantic Web-Based Approach for Harvesting Multilingual Textual. definitions from Wikipedia to support ICD-11 revision
 A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
ORES-2010 Ontology Repositories and Editors for the Semantic Web
 Vol-596 urn:nbn:de:0074-596-3 Copyright 2010 for the individual papers by the papers' authors. Copying permitted only for private and academic purposes. This volume is published and copyrighted by its
Vol-596 urn:nbn:de:0074-596-3 Copyright 2010 for the individual papers by the papers' authors. Copying permitted only for private and academic purposes. This volume is published and copyrighted by its
A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES
 A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES 1 Yuxin Mao 1 School of Computer and Information Engineering, Zhejiang Gongshang University, Hangzhou 310018, P.R. China E-mail: 1 maoyuxin@zjgsu.edu.cn ABSTRACT
A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES 1 Yuxin Mao 1 School of Computer and Information Engineering, Zhejiang Gongshang University, Hangzhou 310018, P.R. China E-mail: 1 maoyuxin@zjgsu.edu.cn ABSTRACT
NCI Thesaurus, managing towards an ontology
 NCI Thesaurus, managing towards an ontology CENDI/NKOS Workshop October 22, 2009 Gilberto Fragoso Outline Background on EVS The NCI Thesaurus BiomedGT Editing Plug-in for Protege Semantic Media Wiki supports
NCI Thesaurus, managing towards an ontology CENDI/NKOS Workshop October 22, 2009 Gilberto Fragoso Outline Background on EVS The NCI Thesaurus BiomedGT Editing Plug-in for Protege Semantic Media Wiki supports
Case Studies on Ontology Reuse
 Case Studies on Ontology Reuse Elena Paslaru Bontas, Malgorzata Mochol, Robert Tolksdorf (Freie Universität Berlin, Germany paslaru, mochol, tolk@inf.fu-berlin.de) Abstract: The development of new ontologies
Case Studies on Ontology Reuse Elena Paslaru Bontas, Malgorzata Mochol, Robert Tolksdorf (Freie Universität Berlin, Germany paslaru, mochol, tolk@inf.fu-berlin.de) Abstract: The development of new ontologies
NCBO Technology: Powering semantically aware applications
 JOURNAL OF BIOMEDICAL SEMANTICS PROCEEDINGS Open Access NCBO Technology: Powering semantically aware applications Patricia L Whetzel 1*, NCBO Team 1,2,3,4 From Bio-Ontologies 2012 Long Beach, CA, USA.
JOURNAL OF BIOMEDICAL SEMANTICS PROCEEDINGS Open Access NCBO Technology: Powering semantically aware applications Patricia L Whetzel 1*, NCBO Team 1,2,3,4 From Bio-Ontologies 2012 Long Beach, CA, USA.
Knowledge Representations. How else can we represent knowledge in addition to formal logic?
 Knowledge Representations How else can we represent knowledge in addition to formal logic? 1 Common Knowledge Representations Formal Logic Production Rules Semantic Nets Schemata and Frames 2 Production
Knowledge Representations How else can we represent knowledge in addition to formal logic? 1 Common Knowledge Representations Formal Logic Production Rules Semantic Nets Schemata and Frames 2 Production
Practical Experiences in Building Ontology-based Retrieval Systems
 Practical Experiences in Building Ontology-based Retrieval Systems Elena Paslaru Bontas Freie Universität Berlin Institut für Informatik Takustr. 9, D-14195 Berlin, Germany paslaru@inf.fu-berlin.de Abstract.
Practical Experiences in Building Ontology-based Retrieval Systems Elena Paslaru Bontas Freie Universität Berlin Institut für Informatik Takustr. 9, D-14195 Berlin, Germany paslaru@inf.fu-berlin.de Abstract.
NEO: Systematic Non-Lattice Embedding of Ontologies for Comparing the Subsumption Relationship in SNOMED CT and in FMA Using MapReduce
 NEO: Systematic Non-Lattice Embedding of Ontologies for Comparing the Subsumption Relationship in SNOMED CT and in FMA Using MapReduce Wei Zhu, Guo-Qiang Zhang, PhD, Shiqiang Tao, MS, Mengmeng Sun, Licong
NEO: Systematic Non-Lattice Embedding of Ontologies for Comparing the Subsumption Relationship in SNOMED CT and in FMA Using MapReduce Wei Zhu, Guo-Qiang Zhang, PhD, Shiqiang Tao, MS, Mengmeng Sun, Licong
A comparison of computer science and software engineering programmes in English universities
 A comparison of computer science and software engineering programmes in English universities Meziane, F and Vadera, S Title Authors Type URL Published Date 2004 A comparison of computer science and software
A comparison of computer science and software engineering programmes in English universities Meziane, F and Vadera, S Title Authors Type URL Published Date 2004 A comparison of computer science and software
From Lexicon To Mammographic Ontology: Experiences and Lessons
 From Lexicon To Mammographic : Experiences and Lessons Bo Hu, Srinandan Dasmahapatra and Nigel Shadbolt Department of Electronics and Computer Science University of Southampton United Kingdom Email: {bh,
From Lexicon To Mammographic : Experiences and Lessons Bo Hu, Srinandan Dasmahapatra and Nigel Shadbolt Department of Electronics and Computer Science University of Southampton United Kingdom Email: {bh,
A few contributions of the SIFR project
 A few contributions of the SIFR project Semantic Indexing of French biomedical Resources Data seminar- December 10th 2015 LIRMM University of Montpellier Clement Jonquet, Mathieu Roche, Sandra Bringay
A few contributions of the SIFR project Semantic Indexing of French biomedical Resources Data seminar- December 10th 2015 LIRMM University of Montpellier Clement Jonquet, Mathieu Roche, Sandra Bringay
Utilizing NCBO Tools to Develop & Use an ECG Ontology
 Utilizing NCBO Tools to Develop & Use an ECG Ontology Stephen J. Granite, MS, MBA The Johns Hopkins University Institute for Computational Medicine (sgranite at jhu dot edu) The CardioVascular Research
Utilizing NCBO Tools to Develop & Use an ECG Ontology Stephen J. Granite, MS, MBA The Johns Hopkins University Institute for Computational Medicine (sgranite at jhu dot edu) The CardioVascular Research
WebProtégé. Protégé going Web. Tania Tudorache, Jennifer Vendetti, Natasha Noy. Stanford Center for Biomedical Informatics
 WebProtégé Protégé going Web Tania Tudorache, Jennifer Vendetti, Natasha Noy Stanford Center for Biomedical Informatics Protégé conference 2009 Amsterdam, June 24, 2009 WebProtégé quick overview WebProtégé
WebProtégé Protégé going Web Tania Tudorache, Jennifer Vendetti, Natasha Noy Stanford Center for Biomedical Informatics Protégé conference 2009 Amsterdam, June 24, 2009 WebProtégé quick overview WebProtégé
Languages and tools for building and using ontologies. Simon Jupp, James Malone
 An overview of ontology technology Languages and tools for building and using ontologies Simon Jupp, James Malone jupp@ebi.ac.uk, malone@ebi.ac.uk Outline Languages OWL and OBO classes, individuals, relations,
An overview of ontology technology Languages and tools for building and using ontologies Simon Jupp, James Malone jupp@ebi.ac.uk, malone@ebi.ac.uk Outline Languages OWL and OBO classes, individuals, relations,
SEMANTIC SUPPORT FOR MEDICAL IMAGE SEARCH AND RETRIEVAL
 SEMANTIC SUPPORT FOR MEDICAL IMAGE SEARCH AND RETRIEVAL Wang Wei, Payam M. Barnaghi School of Computer Science and Information Technology The University of Nottingham Malaysia Campus {Kcy3ww, payam.barnaghi}@nottingham.edu.my
SEMANTIC SUPPORT FOR MEDICAL IMAGE SEARCH AND RETRIEVAL Wang Wei, Payam M. Barnaghi School of Computer Science and Information Technology The University of Nottingham Malaysia Campus {Kcy3ww, payam.barnaghi}@nottingham.edu.my
Humboldt-University of Berlin
 Humboldt-University of Berlin Exploiting Link Structure to Discover Meaningful Associations between Controlled Vocabulary Terms exposé of diploma thesis of Andrej Masula 13th October 2008 supervisor: Louiqa
Humboldt-University of Berlin Exploiting Link Structure to Discover Meaningful Associations between Controlled Vocabulary Terms exposé of diploma thesis of Andrej Masula 13th October 2008 supervisor: Louiqa
How orthogonal are the OBO Foundry ontologies?
 JOURNAL OF BIOMEDICAL SEMANTICS PROCEEDINGS Open Access How orthogonal are the OBO Foundry ontologies? Amir Ghazvinian *, Natalya F Noy *, Mark A Musen From Bio-Ontologies 2010: Semantic Applications in
JOURNAL OF BIOMEDICAL SEMANTICS PROCEEDINGS Open Access How orthogonal are the OBO Foundry ontologies? Amir Ghazvinian *, Natalya F Noy *, Mark A Musen From Bio-Ontologies 2010: Semantic Applications in
Development of an Environment for Data Annotation in Bioengineering
 Development of an Environment for Data Annotation in Bioengineering Renato Galina Barbosa Azambuja Correia Instituto Superior Técnico, Universidade Técnica de Lisboa, Portugal renato_correia1@hotmail.com
Development of an Environment for Data Annotation in Bioengineering Renato Galina Barbosa Azambuja Correia Instituto Superior Técnico, Universidade Técnica de Lisboa, Portugal renato_correia1@hotmail.com
This is the author s version of a work that was submitted/accepted for publication in the following source:
 This is the author s version of a work that was submitted/accepted for publication in the following source: Koopman, Bevan, Bruza, Peter, Sitbon, Laurianne, & Lawley, Michael (2011) AEHRC & QUT at TREC
This is the author s version of a work that was submitted/accepted for publication in the following source: Koopman, Bevan, Bruza, Peter, Sitbon, Laurianne, & Lawley, Michael (2011) AEHRC & QUT at TREC
ResPubliQA 2010
 SZTAKI @ ResPubliQA 2010 David Mark Nemeskey Computer and Automation Research Institute, Hungarian Academy of Sciences, Budapest, Hungary (SZTAKI) Abstract. This paper summarizes the results of our first
SZTAKI @ ResPubliQA 2010 David Mark Nemeskey Computer and Automation Research Institute, Hungarian Academy of Sciences, Budapest, Hungary (SZTAKI) Abstract. This paper summarizes the results of our first
Maintaining a Catalog of Manually-Indexed, Clinically-Oriented World Wide Web Content
 Maintaining a Catalog of Manually-Indexed, Clinically-Oriented World Wide Web Content William Hersh, M.D., Andrea Ball, M.L.S., Bikram Day, M.S., Mary Masterson, M.P.H., Li Zhang, M.S., Lynetta Sacherek,
Maintaining a Catalog of Manually-Indexed, Clinically-Oriented World Wide Web Content William Hersh, M.D., Andrea Ball, M.L.S., Bikram Day, M.S., Mary Masterson, M.P.H., Li Zhang, M.S., Lynetta Sacherek,
Ontology-Assisted Analysis of Web Queries to Determine the Knowledge Radiologists Seek
 Ontology-Assisted Analysis of Web Queries to Determine the Knowledge Radiologists Seek Daniel L. Rubin, 1 Adam Flanders, 2 Woojin Kim, 3 Khan M. Siddiqui, 4,5 and Charles E. Kahn Jr 6 Radiologists frequently
Ontology-Assisted Analysis of Web Queries to Determine the Knowledge Radiologists Seek Daniel L. Rubin, 1 Adam Flanders, 2 Woojin Kim, 3 Khan M. Siddiqui, 4,5 and Charles E. Kahn Jr 6 Radiologists frequently
What Four Million Mappings Can Tell You about Two Hundred Ontologies
 What Four Million Mappings Can Tell You about Two Hundred Ontologies Amir Ghazvinian, Natasha Noy, Clement Jonquet, Nigam Shah, Mark Musen To cite this version: Amir Ghazvinian, Natasha Noy, Clement Jonquet,
What Four Million Mappings Can Tell You about Two Hundred Ontologies Amir Ghazvinian, Natasha Noy, Clement Jonquet, Nigam Shah, Mark Musen To cite this version: Amir Ghazvinian, Natasha Noy, Clement Jonquet,
Towards Ease of Building Legos in Assessing ehealth Language Technologies
 Towards Ease of Building Legos in Assessing ehealth Language Technologies A RESTful Laboratory for Data and Software Hanna Suominen 1, 2, Karl Kreiner 3, Mike Wu 1, Leif Hanlen 1, 2 1 NICTA, National ICT
Towards Ease of Building Legos in Assessing ehealth Language Technologies A RESTful Laboratory for Data and Software Hanna Suominen 1, 2, Karl Kreiner 3, Mike Wu 1, Leif Hanlen 1, 2 1 NICTA, National ICT
Ontology Refinement and Evaluation based on is-a Hierarchy Similarity
 Ontology Refinement and Evaluation based on is-a Hierarchy Similarity Takeshi Masuda The Institute of Scientific and Industrial Research, Osaka University Abstract. Ontologies are constructed in fields
Ontology Refinement and Evaluation based on is-a Hierarchy Similarity Takeshi Masuda The Institute of Scientific and Industrial Research, Osaka University Abstract. Ontologies are constructed in fields
Collaborative Ontology Construction using Template-based Wiki for Semantic Web Applications
 2009 International Conference on Computer Engineering and Technology Collaborative Ontology Construction using Template-based Wiki for Semantic Web Applications Sung-Kooc Lim Information and Communications
2009 International Conference on Computer Engineering and Technology Collaborative Ontology Construction using Template-based Wiki for Semantic Web Applications Sung-Kooc Lim Information and Communications
SOFTWARE REQUIREMENT REUSE MODEL BASED ON LEVENSHTEIN DISTANCES
 SOFTWARE REQUIREMENT REUSE MODEL BASED ON LEVENSHTEIN DISTANCES 1 WONG PO HUI, 2,* WAN MOHD NAZMEE WAN ZAINON School of Computer Sciences, Universiti Sains Malaysia, 118 Penang, Malaysia 1 wphui.ucom12@student.usm.my,
SOFTWARE REQUIREMENT REUSE MODEL BASED ON LEVENSHTEIN DISTANCES 1 WONG PO HUI, 2,* WAN MOHD NAZMEE WAN ZAINON School of Computer Sciences, Universiti Sains Malaysia, 118 Penang, Malaysia 1 wphui.ucom12@student.usm.my,
A System for Ontology-Based Annotation of Biomedical Data
 A System for Ontology-Based Annotation of Biomedical Data Clement Jonquet, Mark A. Musen, and Nigam Shah Stanford Center for Biomedical Informatics Research Stanford University School of Medicine Medical
A System for Ontology-Based Annotation of Biomedical Data Clement Jonquet, Mark A. Musen, and Nigam Shah Stanford Center for Biomedical Informatics Research Stanford University School of Medicine Medical
Ontology-Driven Software: What We Learned From Using Ontologies As Infrastructure For Software Or How Does It Taste to Eat Our Own Dogfood
 Ontology-Driven Software: What We Learned From Using Ontologies As Infrastructure For Software Or How Does It Taste to Eat Our Own Dogfood Csongor Nyulas, Natalya F. Noy, Michael Dorf, Nicholas Griffith,
Ontology-Driven Software: What We Learned From Using Ontologies As Infrastructure For Software Or How Does It Taste to Eat Our Own Dogfood Csongor Nyulas, Natalya F. Noy, Michael Dorf, Nicholas Griffith,
Context-enhanced Ontology Reuse
 Context-enhanced Ontology Reuse Elena Paslaru Bontas Free University of Berlin Institute of Computer Science Takustr. 9 14195 Berlin, Germany paslaru@inf.fu-berlin.de Abstract. Though the importance of
Context-enhanced Ontology Reuse Elena Paslaru Bontas Free University of Berlin Institute of Computer Science Takustr. 9 14195 Berlin, Germany paslaru@inf.fu-berlin.de Abstract. Though the importance of
Collaborative Ontology Development on the (Semantic) Web
 Collaborative Ontology Development on the (Semantic) Web Natalya F. Noy and Tania Tudorache Stanford Center for Biomedical Informatics Research Stanford University Stanford, CA 94305 {noy,tudorache}@stanford.edu
Collaborative Ontology Development on the (Semantic) Web Natalya F. Noy and Tania Tudorache Stanford Center for Biomedical Informatics Research Stanford University Stanford, CA 94305 {noy,tudorache}@stanford.edu
Collaborative & WebProtégé
 Collaborative & WebProtégé Tania Tudorache Stanford Center for Biomedical Informatics Research Joint Ontolog-OOR Panel Session July 16, 2009 1 Collaborative Ontology Development Collaboration: several
Collaborative & WebProtégé Tania Tudorache Stanford Center for Biomedical Informatics Research Joint Ontolog-OOR Panel Session July 16, 2009 1 Collaborative Ontology Development Collaboration: several
POMap results for OAEI 2017
 POMap results for OAEI 2017 Amir Laadhar 1, Faiza Ghozzi 2, Imen Megdiche 1, Franck Ravat 1, Olivier Teste 1, and Faiez Gargouri 2 1 Paul Sabatier University, IRIT (CNRS/UMR 5505) 118 Route de Narbonne
POMap results for OAEI 2017 Amir Laadhar 1, Faiza Ghozzi 2, Imen Megdiche 1, Franck Ravat 1, Olivier Teste 1, and Faiez Gargouri 2 1 Paul Sabatier University, IRIT (CNRS/UMR 5505) 118 Route de Narbonne
Interoperability of Protégé 2.0 beta and OilEd 3.5 in the Domain Knowledge of Osteoporosis
 EXPERIMENT: Interoperability of Protégé 2.0 beta and OilEd 3.5 in the Domain Knowledge of Osteoporosis Franz Calvo, MD fcalvo@u.washington.edu and John H. Gennari, PhD gennari@u.washington.edu Department
EXPERIMENT: Interoperability of Protégé 2.0 beta and OilEd 3.5 in the Domain Knowledge of Osteoporosis Franz Calvo, MD fcalvo@u.washington.edu and John H. Gennari, PhD gennari@u.washington.edu Department
Definition of the evaluation protocol and goals for competition 2
 www.visceral.eu Definition of the evaluation protocol and goals for competition 2 Deliverable number D4.2 Dissemination level Public Delivery date 7 February 2014 Status Author(s) Final Georg Langs, Bjoern
www.visceral.eu Definition of the evaluation protocol and goals for competition 2 Deliverable number D4.2 Dissemination level Public Delivery date 7 February 2014 Status Author(s) Final Georg Langs, Bjoern
Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR
 Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR Guoqian Jiang 1, Eric Prud Hommeaux 2, Guohui Xiao 3, and Harold R. Solbrig 1 1 Mayo Clinic, Rochester,
Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR Guoqian Jiang 1, Eric Prud Hommeaux 2, Guohui Xiao 3, and Harold R. Solbrig 1 1 Mayo Clinic, Rochester,
Available online at ScienceDirect. Procedia Computer Science 52 (2015 )
 Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 52 (2015 ) 1071 1076 The 5 th International Symposium on Frontiers in Ambient and Mobile Systems (FAMS-2015) Health, Food
Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 52 (2015 ) 1071 1076 The 5 th International Symposium on Frontiers in Ambient and Mobile Systems (FAMS-2015) Health, Food
PRIOR System: Results for OAEI 2006
 PRIOR System: Results for OAEI 2006 Ming Mao, Yefei Peng University of Pittsburgh, Pittsburgh, PA, USA {mingmao,ypeng}@mail.sis.pitt.edu Abstract. This paper summarizes the results of PRIOR system, which
PRIOR System: Results for OAEI 2006 Ming Mao, Yefei Peng University of Pittsburgh, Pittsburgh, PA, USA {mingmao,ypeng}@mail.sis.pitt.edu Abstract. This paper summarizes the results of PRIOR system, which
Health Information Exchange Content Model Architecture Building Block HISO
 Health Information Exchange Content Model Architecture Building Block HISO 10040.2 To be used in conjunction with HISO 10040.0 Health Information Exchange Overview and Glossary HISO 10040.1 Health Information
Health Information Exchange Content Model Architecture Building Block HISO 10040.2 To be used in conjunction with HISO 10040.0 Health Information Exchange Overview and Glossary HISO 10040.1 Health Information
Context Ontology Construction For Cricket Video
 Context Ontology Construction For Cricket Video Dr. Sunitha Abburu Professor& Director, Department of Computer Applications Adhiyamaan College of Engineering, Hosur, pin-635109, Tamilnadu, India Abstract
Context Ontology Construction For Cricket Video Dr. Sunitha Abburu Professor& Director, Department of Computer Applications Adhiyamaan College of Engineering, Hosur, pin-635109, Tamilnadu, India Abstract
Daniel L. Rubin and Kaustubh Supekar, Stanford University
 S e m a n t i c S c i e n t i f i c K n o w l e d g e I n t e g r a t i o n Annotation and Image Markup: Accessing and Interoperating with the Semantic Content in Medical Imaging The Annotation Daniel
S e m a n t i c S c i e n t i f i c K n o w l e d g e I n t e g r a t i o n Annotation and Image Markup: Accessing and Interoperating with the Semantic Content in Medical Imaging The Annotation Daniel
An Annotation Tool for Semantic Documents
 An Annotation Tool for Semantic Documents (System Description) Henrik Eriksson Dept. of Computer and Information Science Linköping University SE-581 83 Linköping, Sweden her@ida.liu.se Abstract. Document
An Annotation Tool for Semantic Documents (System Description) Henrik Eriksson Dept. of Computer and Information Science Linköping University SE-581 83 Linköping, Sweden her@ida.liu.se Abstract. Document
Software re-use assessment for quality M. Ramachandran School of Computing and Mathematical Sciences, Jo/m Moores C/mrerszZ?/,
 Software re-use assessment for quality M. Ramachandran School of Computing and Mathematical Sciences, Jo/m Moores C/mrerszZ?/, ABSTRACT Reuse of software components can improve software quality and productivity
Software re-use assessment for quality M. Ramachandran School of Computing and Mathematical Sciences, Jo/m Moores C/mrerszZ?/, ABSTRACT Reuse of software components can improve software quality and productivity
A Study on Website Quality Models
 International Journal of Scientific and Research Publications, Volume 4, Issue 12, December 2014 1 A Study on Website Quality Models R.Anusha Department of Information Systems Management, M.O.P Vaishnav
International Journal of Scientific and Research Publications, Volume 4, Issue 12, December 2014 1 A Study on Website Quality Models R.Anusha Department of Information Systems Management, M.O.P Vaishnav
Enhancing Wrapper Usability through Ontology Sharing and Large Scale Cooperation
 Enhancing Wrapper Usability through Ontology Enhancing Sharing Wrapper and Large Usability Scale Cooperation through Ontology Sharing and Large Scale Cooperation Christian Schindler, Pranjal Arya, Andreas
Enhancing Wrapper Usability through Ontology Enhancing Sharing Wrapper and Large Usability Scale Cooperation through Ontology Sharing and Large Scale Cooperation Christian Schindler, Pranjal Arya, Andreas
Medical Imaging on the Semantic Web: Annotation and Image Markup
 Medical Imaging on the Semantic Web: Annotation and Image Markup Daniel L. Rubin, 1 Pattanasak Mongkolwat, 2 Vladimir Kleper, 2 Kaustubh Supekar, 1 and David S. Channin 2 1 Department of Radiology and
Medical Imaging on the Semantic Web: Annotation and Image Markup Daniel L. Rubin, 1 Pattanasak Mongkolwat, 2 Vladimir Kleper, 2 Kaustubh Supekar, 1 and David S. Channin 2 1 Department of Radiology and
Ontology Creation and Development Model
 Ontology Creation and Development Model Pallavi Grover, Sonal Chawla Research Scholar, Department of Computer Science & Applications, Panjab University, Chandigarh, India Associate. Professor, Department
Ontology Creation and Development Model Pallavi Grover, Sonal Chawla Research Scholar, Department of Computer Science & Applications, Panjab University, Chandigarh, India Associate. Professor, Department
Utilizing Semantic Web Technologies in Healthcare
 Utilizing Semantic Web Technologies in Healthcare Vassileios D. Kolias, John Stoitsis, Spyretta Golemati and Konstantina S. Nikita Abstract The technological breakthrough in biomedical engineering and
Utilizing Semantic Web Technologies in Healthcare Vassileios D. Kolias, John Stoitsis, Spyretta Golemati and Konstantina S. Nikita Abstract The technological breakthrough in biomedical engineering and
Interoperability and Semantics in Use- Application of UML, XMI and MDA to Precision Medicine and Cancer Research
 Interoperability and Semantics in Use- Application of UML, XMI and MDA to Precision Medicine and Cancer Research Ian Fore, D.Phil. Associate Director, Biorepository and Pathology Informatics Senior Program
Interoperability and Semantics in Use- Application of UML, XMI and MDA to Precision Medicine and Cancer Research Ian Fore, D.Phil. Associate Director, Biorepository and Pathology Informatics Senior Program
Controlled Medical Vocabulary in the CPR Generations
 Tutorials, B. Hieb, M.D. Research Note 5 November 2003 Controlled Medical Vocabulary in the CPR Generations A CMV capability becomes progressively more important as computer-based patient record systems
Tutorials, B. Hieb, M.D. Research Note 5 November 2003 Controlled Medical Vocabulary in the CPR Generations A CMV capability becomes progressively more important as computer-based patient record systems
Semantic Annotation for Semantic Social Networks. Using Community Resources
 Semantic Annotation for Semantic Social Networks Using Community Resources Lawrence Reeve and Hyoil Han College of Information Science and Technology Drexel University, Philadelphia, PA 19108 lhr24@drexel.edu
Semantic Annotation for Semantic Social Networks Using Community Resources Lawrence Reeve and Hyoil Han College of Information Science and Technology Drexel University, Philadelphia, PA 19108 lhr24@drexel.edu
APPLYING HUMAN FACTORS ENGINEERING TO IMPROVE USABILITY AND WORKFLOW IN PATHOLOGY INFORMATICS
 Proceedings of the 2017 International Symposium on Human Factors and Ergonomics in Health Care 23 APPLYING HUMAN FACTORS ENGINEERING TO IMPROVE USABILITY AND WORKFLOW IN PATHOLOGY INFORMATICS Austin F.
Proceedings of the 2017 International Symposium on Human Factors and Ergonomics in Health Care 23 APPLYING HUMAN FACTORS ENGINEERING TO IMPROVE USABILITY AND WORKFLOW IN PATHOLOGY INFORMATICS Austin F.
Specification of a Cardiovascular Metadata Model: A Consensus Standard
 Specification of a Cardiovascular Metadata Model: A Consensus Standard Rebecca Wilgus, RN MSN 1 ; Dana Pinchotti 2 ; Salvatore Mungal 3 ; David F Kong, MD AM 1 ; James E Tcheng, MD 1 ; Brian McCourt 1
Specification of a Cardiovascular Metadata Model: A Consensus Standard Rebecca Wilgus, RN MSN 1 ; Dana Pinchotti 2 ; Salvatore Mungal 3 ; David F Kong, MD AM 1 ; James E Tcheng, MD 1 ; Brian McCourt 1
A COMPARATIVE STUDY OF BYG SEARCH ENGINES
 American Journal of Engineering Research (AJER) e-issn: 2320-0847 p-issn : 2320-0936 Volume-2, Issue-4, pp-39-43 www.ajer.us Research Paper Open Access A COMPARATIVE STUDY OF BYG SEARCH ENGINES Kailash
American Journal of Engineering Research (AJER) e-issn: 2320-0847 p-issn : 2320-0936 Volume-2, Issue-4, pp-39-43 www.ajer.us Research Paper Open Access A COMPARATIVE STUDY OF BYG SEARCH ENGINES Kailash
Effective Mapping Composition for Biomedical Ontologies
 Effective Mapping Composition for Biomedical Ontologies Michael Hartung 1,2, Anika Gross 1,2, Toralf Kirsten 2, and Erhard Rahm 1,2 1 Department of Computer Science, University of Leipzig 2 Interdisciplinary
Effective Mapping Composition for Biomedical Ontologies Michael Hartung 1,2, Anika Gross 1,2, Toralf Kirsten 2, and Erhard Rahm 1,2 1 Department of Computer Science, University of Leipzig 2 Interdisciplinary
Sheffield University and the TREC 2004 Genomics Track: Query Expansion Using Synonymous Terms
 Sheffield University and the TREC 2004 Genomics Track: Query Expansion Using Synonymous Terms Yikun Guo, Henk Harkema, Rob Gaizauskas University of Sheffield, UK {guo, harkema, gaizauskas}@dcs.shef.ac.uk
Sheffield University and the TREC 2004 Genomics Track: Query Expansion Using Synonymous Terms Yikun Guo, Henk Harkema, Rob Gaizauskas University of Sheffield, UK {guo, harkema, gaizauskas}@dcs.shef.ac.uk
Transforming Enterprise Ontologies into SBVR formalizations
 Transforming Enterprise Ontologies into SBVR formalizations Frederik Gailly Faculty of Economics and Business Administration Ghent University Frederik.Gailly@ugent.be Abstract In 2007 the Object Management
Transforming Enterprise Ontologies into SBVR formalizations Frederik Gailly Faculty of Economics and Business Administration Ghent University Frederik.Gailly@ugent.be Abstract In 2007 the Object Management
ServOMap and ServOMap-lt Results for OAEI 2012
 ServOMap and ServOMap-lt Results for OAEI 2012 Mouhamadou Ba 1, Gayo Diallo 1 1 LESIM/ISPED, Univ. Bordeaux Segalen, F-33000, France first.last@isped.u-bordeaux2.fr Abstract. We present the results obtained
ServOMap and ServOMap-lt Results for OAEI 2012 Mouhamadou Ba 1, Gayo Diallo 1 1 LESIM/ISPED, Univ. Bordeaux Segalen, F-33000, France first.last@isped.u-bordeaux2.fr Abstract. We present the results obtained
Comment-based Keyword Programming
 Comment-based Keyword Programming Yusuke Sakamoto, Haruhiko Sato, Satoshi Oyama, Masahito Kurihara Abstract Keyword programming is a technique to generate code fragments automatically from the several
Comment-based Keyword Programming Yusuke Sakamoto, Haruhiko Sato, Satoshi Oyama, Masahito Kurihara Abstract Keyword programming is a technique to generate code fragments automatically from the several
UMLS-Query: A Perl Module for Querying the UMLS
 UMLS-Query: A Perl Module for Querying the UMLS Nigam H. Shah, MBBS, PhD, Mark A. Musen, MD, PhD Center for Biomedical Informatics Research, Stanford University, Stanford, CA Abstract The Metathesaurus
UMLS-Query: A Perl Module for Querying the UMLS Nigam H. Shah, MBBS, PhD, Mark A. Musen, MD, PhD Center for Biomedical Informatics Research, Stanford University, Stanford, CA Abstract The Metathesaurus
Modeling Systems Using Design Patterns
 Modeling Systems Using Design Patterns Jaroslav JAKUBÍK Slovak University of Technology Faculty of Informatics and Information Technologies Ilkovičova 3, 842 16 Bratislava, Slovakia jakubik@fiit.stuba.sk
Modeling Systems Using Design Patterns Jaroslav JAKUBÍK Slovak University of Technology Faculty of Informatics and Information Technologies Ilkovičova 3, 842 16 Bratislava, Slovakia jakubik@fiit.stuba.sk
Collaborative Editing and Linking of Astronomy Vocabularies Using Semantic Mediawiki
 Collaborative Editing and Linking of Astronomy Vocabularies Using Semantic Mediawiki Stuart Chalmers 1, Norman Gray 2, Iadh Ounis 1, and Alasdair Gray 3 1 Computing Science, University of Glasgow, Glasgow,
Collaborative Editing and Linking of Astronomy Vocabularies Using Semantic Mediawiki Stuart Chalmers 1, Norman Gray 2, Iadh Ounis 1, and Alasdair Gray 3 1 Computing Science, University of Glasgow, Glasgow,
WHO ICD11 Wiki LexWiki, Semantic MediaWiki and the International Classification of Diseases
 WHO ICD11 Wiki LexWiki, Semantic MediaWiki and the International Classification of Diseases Guoqian Jiang, PhD Harold Solbrig Division of Biomedical Statistics and Informatics Mayo Clinic College of Medicine
WHO ICD11 Wiki LexWiki, Semantic MediaWiki and the International Classification of Diseases Guoqian Jiang, PhD Harold Solbrig Division of Biomedical Statistics and Informatics Mayo Clinic College of Medicine
Component-Based Software Engineering TIP
 Component-Based Software Engineering TIP X LIU, School of Computing, Napier University This chapter will present a complete picture of how to develop software systems with components and system integration.
Component-Based Software Engineering TIP X LIU, School of Computing, Napier University This chapter will present a complete picture of how to develop software systems with components and system integration.
BORN Ontario s Data Quality Framework
 BORN Ontario s Data Quality Framework At BORN Ontario we make Data Privacy and Data Quality our highest priority. We recognize that the quality of the data directly impacts use of the data. With addition
BORN Ontario s Data Quality Framework At BORN Ontario we make Data Privacy and Data Quality our highest priority. We recognize that the quality of the data directly impacts use of the data. With addition
Domain-specific Concept-based Information Retrieval System
 Domain-specific Concept-based Information Retrieval System L. Shen 1, Y. K. Lim 1, H. T. Loh 2 1 Design Technology Institute Ltd, National University of Singapore, Singapore 2 Department of Mechanical
Domain-specific Concept-based Information Retrieval System L. Shen 1, Y. K. Lim 1, H. T. Loh 2 1 Design Technology Institute Ltd, National University of Singapore, Singapore 2 Department of Mechanical
OWL/ZIP: Distributing Large and Modular Ontologies
 OWL/ZIP: Distributing Large and Modular Ontologies Nicolas Matentzoglu and Bijan Parsia The University of Manchester Oxford Road, Manchester, M13 9PL, UK {bparsia,matentzn}@cs.manchester.ac.uk Abstract.
OWL/ZIP: Distributing Large and Modular Ontologies Nicolas Matentzoglu and Bijan Parsia The University of Manchester Oxford Road, Manchester, M13 9PL, UK {bparsia,matentzn}@cs.manchester.ac.uk Abstract.
UNIFYING SEMANTIC ANNOTATION AND QUERYING IN BIOMEDICAL IMAGE REPOSITORIES One Solution for Two Problems of Medical Knowledge Engineering
 UNIFYING SEMANTIC ANNOTATION AND QUERYING IN BIOMEDICAL IMAGE REPOSITORIES One Solution for Two Problems of Medical Knowledge Engineering Daniel Sonntag German Research Center for Artificial Intelligence,
UNIFYING SEMANTIC ANNOTATION AND QUERYING IN BIOMEDICAL IMAGE REPOSITORIES One Solution for Two Problems of Medical Knowledge Engineering Daniel Sonntag German Research Center for Artificial Intelligence,
FCA-Map Results for OAEI 2016
 FCA-Map Results for OAEI 2016 Mengyi Zhao 1 and Songmao Zhang 2 1,2 Institute of Mathematics, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, P. R. China 1 myzhao@amss.ac.cn,
FCA-Map Results for OAEI 2016 Mengyi Zhao 1 and Songmao Zhang 2 1,2 Institute of Mathematics, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, P. R. China 1 myzhao@amss.ac.cn,
Classification and retrieval of biomedical literatures: SNUMedinfo at CLEF QA track BioASQ 2014
 Classification and retrieval of biomedical literatures: SNUMedinfo at CLEF QA track BioASQ 2014 Sungbin Choi, Jinwook Choi Medical Informatics Laboratory, Seoul National University, Seoul, Republic of
Classification and retrieval of biomedical literatures: SNUMedinfo at CLEF QA track BioASQ 2014 Sungbin Choi, Jinwook Choi Medical Informatics Laboratory, Seoul National University, Seoul, Republic of
Ontology Mapping based on Similarity Measure and Fuzzy Logic
 Ontology Mapping based on Similarity Measure and Fuzzy Logic Author Niwattanakul, S., Martin, Philippe, Eboueya, M., Khaimook, K. Published 2007 Conference Title Proceedings of E-Learn 2007 Copyright Statement
Ontology Mapping based on Similarity Measure and Fuzzy Logic Author Niwattanakul, S., Martin, Philippe, Eboueya, M., Khaimook, K. Published 2007 Conference Title Proceedings of E-Learn 2007 Copyright Statement
Precise Medication Extraction using Agile Text Mining
 Precise Medication Extraction using Agile Text Mining Chaitanya Shivade *, James Cormack, David Milward * The Ohio State University, Columbus, Ohio, USA Linguamatics Ltd, Cambridge, UK shivade@cse.ohio-state.edu,
Precise Medication Extraction using Agile Text Mining Chaitanya Shivade *, James Cormack, David Milward * The Ohio State University, Columbus, Ohio, USA Linguamatics Ltd, Cambridge, UK shivade@cse.ohio-state.edu,
Identification and Classification of A/E/C Web Sites and Pages
 Construction Informatics Digital Library http://itc.scix.net/ paper w78-2002-34.content Theme: Title: Author(s): Institution(s): E-mail(s): Abstract: Keywords: Identification and Classification of A/E/C
Construction Informatics Digital Library http://itc.scix.net/ paper w78-2002-34.content Theme: Title: Author(s): Institution(s): E-mail(s): Abstract: Keywords: Identification and Classification of A/E/C
Introduction to The Cochrane Library
 Introduction to The Cochrane Library What is The Cochrane Library? A collection of databases that include high quality, independent, reliable evidence from Cochrane and other systematic reviews, clinical
Introduction to The Cochrane Library What is The Cochrane Library? A collection of databases that include high quality, independent, reliable evidence from Cochrane and other systematic reviews, clinical
From Passages into Elements in XML Retrieval
 From Passages into Elements in XML Retrieval Kelly Y. Itakura David R. Cheriton School of Computer Science, University of Waterloo 200 Univ. Ave. W. Waterloo, ON, Canada yitakura@cs.uwaterloo.ca Charles
From Passages into Elements in XML Retrieval Kelly Y. Itakura David R. Cheriton School of Computer Science, University of Waterloo 200 Univ. Ave. W. Waterloo, ON, Canada yitakura@cs.uwaterloo.ca Charles
XETA: extensible metadata System
 XETA: extensible metadata System Abstract: This paper presents an extensible metadata system (XETA System) which makes it possible for the user to organize and extend the structure of metadata. We discuss
XETA: extensible metadata System Abstract: This paper presents an extensible metadata system (XETA System) which makes it possible for the user to organize and extend the structure of metadata. We discuss
WEB SEARCH, FILTERING, AND TEXT MINING: TECHNOLOGY FOR A NEW ERA OF INFORMATION ACCESS
 1 WEB SEARCH, FILTERING, AND TEXT MINING: TECHNOLOGY FOR A NEW ERA OF INFORMATION ACCESS BRUCE CROFT NSF Center for Intelligent Information Retrieval, Computer Science Department, University of Massachusetts,
1 WEB SEARCH, FILTERING, AND TEXT MINING: TECHNOLOGY FOR A NEW ERA OF INFORMATION ACCESS BRUCE CROFT NSF Center for Intelligent Information Retrieval, Computer Science Department, University of Massachusetts,
A framework for multi-level semantic trace abstraction
 A framework for multi-level semantic trace abstraction Manuel Striani 1 1 PhD Candidate Department of Computer Science, University of Torino Corso Svizzera 185, 10149 Torino, Italy striani@di.unito.it
A framework for multi-level semantic trace abstraction Manuel Striani 1 1 PhD Candidate Department of Computer Science, University of Torino Corso Svizzera 185, 10149 Torino, Italy striani@di.unito.it
A Model for Interactive Web Information Retrieval
 A Model for Interactive Web Information Retrieval Orland Hoeber and Xue Dong Yang University of Regina, Regina, SK S4S 0A2, Canada {hoeber, yang}@uregina.ca Abstract. The interaction model supported by
A Model for Interactive Web Information Retrieval Orland Hoeber and Xue Dong Yang University of Regina, Regina, SK S4S 0A2, Canada {hoeber, yang}@uregina.ca Abstract. The interaction model supported by
Linked Data: Fast, low cost semantic interoperability for health care?
 Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
Keywords Repository, Retrieval, Component, Reusability, Query.
 Volume 4, Issue 3, March 2014 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com Multiple Search
Volume 4, Issue 3, March 2014 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com Multiple Search
MeSH: A Thesaurus for PubMed
 Resources and tools for bibliographic research MeSH: A Thesaurus for PubMed October 24, 2012 What is MeSH? Who uses MeSH? Why use MeSH? Searching by using the MeSH Database What is MeSH? Acronym for Medical
Resources and tools for bibliographic research MeSH: A Thesaurus for PubMed October 24, 2012 What is MeSH? Who uses MeSH? Why use MeSH? Searching by using the MeSH Database What is MeSH? Acronym for Medical
Mapping Composition for Matching Large Life Science Ontologies
 Mapping Composition for Matching Large Life Science Ontologies Anika Groß 1,2, Michael Hartung 1,2, Toralf Kirsten 2,3, Erhard Rahm 1,2 1 Department of Computer Science, University of Leipzig 2 Interdisciplinary
Mapping Composition for Matching Large Life Science Ontologies Anika Groß 1,2, Michael Hartung 1,2, Toralf Kirsten 2,3, Erhard Rahm 1,2 1 Department of Computer Science, University of Leipzig 2 Interdisciplinary
Tania Tudorache Stanford University. - Ontolog forum invited talk04. October 2007
 Collaborative Ontology Development in Protégé Tania Tudorache Stanford University - Ontolog forum invited talk04. October 2007 Outline Introduction and Background Tools for collaborative knowledge development
Collaborative Ontology Development in Protégé Tania Tudorache Stanford University - Ontolog forum invited talk04. October 2007 Outline Introduction and Background Tools for collaborative knowledge development
Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology (DDI)
 ICBO 2013 Workshop on Vaccine and Drug Ontology Studies Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology (DDI) Zhe He 1 Christopher Ochs 1 Larisa
ICBO 2013 Workshop on Vaccine and Drug Ontology Studies Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology (DDI) Zhe He 1 Christopher Ochs 1 Larisa
Citation for published version (APA): Jorritsma, W. (2016). Human-computer interaction in radiology [Groningen]: Rijksuniversiteit Groningen
![Citation for published version (APA): Jorritsma, W. (2016). Human-computer interaction in radiology [Groningen]: Rijksuniversiteit Groningen Citation for published version (APA): Jorritsma, W. (2016). Human-computer interaction in radiology [Groningen]: Rijksuniversiteit Groningen](/thumbs/83/88406495.jpg) University of Groningen Human-computer interaction in radiology Jorritsma, Wiard IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
University of Groningen Human-computer interaction in radiology Jorritsma, Wiard IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
Customisable Curation Workflows in Argo
 Customisable Curation Workflows in Argo Rafal Rak*, Riza Batista-Navarro, Andrew Rowley, Jacob Carter and Sophia Ananiadou National Centre for Text Mining, University of Manchester, UK *Corresponding author:
Customisable Curation Workflows in Argo Rafal Rak*, Riza Batista-Navarro, Andrew Rowley, Jacob Carter and Sophia Ananiadou National Centre for Text Mining, University of Manchester, UK *Corresponding author:
Introduction to Ovid. As a Clinical Librarían tool! Masoud Mohammadi Golestan University of Medical Sciences
 Introduction to Ovid As a Clinical Librarían tool! Masoud Mohammadi Golestan University of Medical Sciences Overview Ovid helps researchers, librarians, clinicians, and other healthcare professionals find
Introduction to Ovid As a Clinical Librarían tool! Masoud Mohammadi Golestan University of Medical Sciences Overview Ovid helps researchers, librarians, clinicians, and other healthcare professionals find
Designing a System Engineering Environment in a structured way
 Designing a System Engineering Environment in a structured way Anna Todino Ivo Viglietti Bruno Tranchero Leonardo-Finmeccanica Aircraft Division Torino, Italy Copyright held by the authors. Rubén de Juan
Designing a System Engineering Environment in a structured way Anna Todino Ivo Viglietti Bruno Tranchero Leonardo-Finmeccanica Aircraft Division Torino, Italy Copyright held by the authors. Rubén de Juan
Workshop 2. > Interoperability <
 Workshop 2 21 / 08 / 2011 > Interoperability < Heiko Zimmermann R&D Engineer, AHI CR Santec Heiko.Zimmermann@tudor.lu Interoperability definition Picture from NCI-Wiki (https://wiki.nci.nih.gov) 2 Interoperability
Workshop 2 21 / 08 / 2011 > Interoperability < Heiko Zimmermann R&D Engineer, AHI CR Santec Heiko.Zimmermann@tudor.lu Interoperability definition Picture from NCI-Wiki (https://wiki.nci.nih.gov) 2 Interoperability
REDCap under FHIR Enhancing electronic data capture with FHIR capability
 REDCap under FHIR Enhancing electronic data capture with FHIR capability Hugo Leroux Research Scientist 09 August 2017 THE AUSTRALIAN E-HEALTH RESEARCH CENTRE, HEALTH AND BIOSECURITY Rationale Clinical
REDCap under FHIR Enhancing electronic data capture with FHIR capability Hugo Leroux Research Scientist 09 August 2017 THE AUSTRALIAN E-HEALTH RESEARCH CENTRE, HEALTH AND BIOSECURITY Rationale Clinical
Open Research Online The Open University s repository of research publications and other research outputs
 Open Research Online The Open University s repository of research publications and other research outputs The Smart Book Recommender: An Ontology-Driven Application for Recommending Editorial Products
Open Research Online The Open University s repository of research publications and other research outputs The Smart Book Recommender: An Ontology-Driven Application for Recommending Editorial Products
3.4 Data-Centric workflow
 3.4 Data-Centric workflow One of the most important activities in a S-DWH environment is represented by data integration of different and heterogeneous sources. The process of extract, transform, and load
3.4 Data-Centric workflow One of the most important activities in a S-DWH environment is represented by data integration of different and heterogeneous sources. The process of extract, transform, and load
Protégé-2000: A Flexible and Extensible Ontology-Editing Environment
 Protégé-2000: A Flexible and Extensible Ontology-Editing Environment Natalya F. Noy, Monica Crubézy, Ray W. Fergerson, Samson Tu, Mark A. Musen Stanford Medical Informatics Stanford University Stanford,
Protégé-2000: A Flexible and Extensible Ontology-Editing Environment Natalya F. Noy, Monica Crubézy, Ray W. Fergerson, Samson Tu, Mark A. Musen Stanford Medical Informatics Stanford University Stanford,
Ontrez Project Report National Center for Biomedical Ontology November, 2007
 Ontrez Project Report National Center for Biomedical Ontology November, 2007 Executive summary Currently, genomics data and data repositories in the public domain are expanding at an explosive pace. 1
Ontrez Project Report National Center for Biomedical Ontology November, 2007 Executive summary Currently, genomics data and data repositories in the public domain are expanding at an explosive pace. 1
