snpqc an R pipeline for quality control of Illumina SNP data
|
|
|
- Rhoda Parrish
- 5 years ago
- Views:
Transcription
1 snpqc an R pipeline for quality control of Illumina SNP data 1. In a nutshell snpqc is a series of R scripts to perform quality control analysis on Illumina SNP data. The objective of the program is to run a fully automated QC pipeline from raw data through to filtered data ready for downstream analysis. Briefly, starting with flat files exported from Genome Studio the program will: 1 build an SQLite database to store genotypes, sample information and details on the SNPs. 2 run various QC metrics on the SNPs, samples and mapping information 3 filter SNPs and samples based on user defined QC criteria 4 output filtered data for downstream analysis and various summary files 5 build a genomic relationship matrix (GRM) for genomic selection/prediction 5 generate a fully automated report of the QC results in PDF 6 store QC results metrics in the database and as flat files snpqc is freely available for download from: If you have any questions, problems, bugs, etc, please contact Cedric Gondro at cgondro2@une.edu.au; I ll try to help out as much as I can. If you have any particular needs or would like some extension feel free to get in touch time allowing, I ll try to accommodate. I have not built an R library package for snpqc (but it is quite doable, again if there s real interest I ll package it up). Personally I prefer to keep the plain scripts in a working directory. The idea behind snpqc is more along the lines of an automated workflow and not a collection of functions to be called to perform tasks. The scripts are easy to modify for custom routines and, particularly for the reporting, it can be convenient to be able to edit the files for individual custom jobs or use different database engines. For those of you who have no time to spare, there s also a compiled C# version that bypasses the database creation it s much faster but depending on the size of the dataset it can be quite demanding on memory and will eat up whatever cores are available until the job finishes (the pure R version is slow but the load can be balanced so it should run on just about any machine).
2 2. Files for snpqc To run snpqc you ll need 3 data files that can be exported directly from GenomeStudio (you can use Analysis/Reports/Report Wizard for this). If the genotyping is done externally you service provider can generate the files. The files needed are: A sample file with at least one column with unique identifiers for each sample (can also have phenotypes, covariates, groupings, etc.). The ID column in this file should match the ID column in the genotypes file (essentially just a list of IDs is enough). For example: Sample ID id1_1 id2_1 id3_1 id4_1 id5_1 id6_1 id7_1 id8_1 group A snp map file with SNP names (must match those in the genotypes file), chromosome names and physical location in bp. These files can be downloaded from Illumina s website and can also be exported from GenomeStudio. For example: SNP Name Chr Position BFGL-NGS X BFGL-NGS BFGL-NGS BFGL-NGS And a genotypes file, one SNP per sample per line with SNP name, sample ID, allele 1, allele 2, GC score and X, Y coordinates; e.g.: SNP Name Sample ID Allele1 - AB Allele2 - AB X Y GC Score BFGL-NGS id1_1 A B BFGL-NGS id1_1 A A BFGL-NGS id1_1 A B BFGL-NGS id1_1 A A Files can have any number of header lines (Illumina s output currently has 9 lines plus the column headers) and any number of additional columns in any of the files. The normal separator is TAB, but the user can define it in the Parameters file (details below). In the genotypes file the X and Y are not mandatory fields currently just used to make a figure in the report and not used for QC filtering. The GC score column can also be absent but that s not recommended most of the QC is based on GC score filtering. If GC scores are not available filtering will be based on population parameters only.
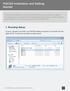
3 3. Parameters file This is the heart of the program and also where you ll need to pay some attention. Here all parameters for the run are defined: input files, output files, filtering criteria and what is what in the files. The main parameters are discussed below. It is important to note that this file is rigid in structure everything needs to be in the exact line, everything after the # symbol on a line gets ignored and if there is more than one criterion in the line it has to be tab delimited. The suggestion is to use the example parameter file as a template and modify for your projects. Figure 1 screenshot of Example parameters file for snpqc. The parameters file has 7 main sections, which are (default values are on the left hand side of the # to illustrate): ############ data ###################### The path and names of the snp map, sample and genotypes files. This can be a fully qualified path or just the names of the files (then the script will look inside R s current working directory). A note on this: paths with spaces cause problems with the C# version better to avoid spaces in the path. Make sure to use the correct separator between folders for Windows (\) and MacOs, Linux (/).
4 ############ data file parameters ######### Basically just to tell the program what to look for. A bit convoluted with many parameters but makes it quite flexible to work with various data sources and formats. 1 # index (column number) of SNP in genotype file - must match names in SNP map file or database column (the latter if the DB already exists and you don t want to recreate it). 2 # index (column number) of sample identifiers in genotype file - must match names in sample map file or database column. 3 # index (column number) of SNP allele1 in genotype file - currently only uses A/B allele calls or database column (make sure to include Allele1 AB and Allele2 AB in the Genome Studio export). Other columns are optional (e.g. forward, top ). 4 # index (column number) of SNP allele2 in genotype file - currently only uses A/B allele calls or database column. 7 # index (column number) of gcscore in genotype file or database column. You can set this to zero if GC scores not available - but not an ideal scenario since most QC metrics are based on this. 1 # index (column number) of sample identifiers in sample map file or database column. 1 # index (column number) of SNP names in SNP map file or database column. 2 # index (column number) of chromosome in SNP map file or database column. 3 # index (column number) of base pair position in SNP map file or database column. 1 # number of lines to skip in genotypes file (headers, etc). 1 # number of lines to skip in sample map file (headers, etc). 1 # number of lines to skip in SNP map file (headers, etc). - # symbols for missing alleles (missing data in the genotypes file). Note that the index must match file or database column means that if you are starting from scratch you tell the program where to find the data in the flat files. If you ve already built the database and want to rerun e.g. the QC the program will use these indices from the database; i.e. the columns to search for data in the database. Generally you don t have to change the mapping will be the same between the files and the database, but the possibility to build or use an already available DB is there. All other data columns found in the files will be added to the database that is created. Initially it will have 3 tables: snpmap with all columns found in the map file, samples with all columns from the samples it s worthwhile to include phenotypes, covariates, etc in this file so that the database is comprehensive (but minimally just needs a column with sample IDs); snps with the genotype data. The database will index SNP and samples in the snps table for rapid data retrieval (also chromosome information in the snpmap table). Indexing can be extremely slow with large datasets.
5 ########### filtering parameters ########### These are the actual parameters used for filtering the data. Filtering is performed across SNP and across samples. SNP filters: 5 # over X percent genotyping fail. If more than X% fail, SNP is rejected. Removes SNP that consistently fail across samples # median call rates (GC scores) smaller than X. If median call rate below this value, the SNP is rejected. 0 # all GC scores zero or NA (do not change). 0.5 # GC < X in Y (next parameter, below) percent of samples. If a Y% of the samples have GC scores below the threshold the SNP is rejected. Note that this value is used to flag individual bad SNP any individual genotypes in a sample below this value is set to missing. 90 # GC < X (above) in Y percent of samples. The percent for Y (for above parameter). 0.0 # MAF zero - snps fixed across all samples, use negative value not to exclude SNP that are not segregating # minor allele frequency MAF < X. SNP with MAF under the cutoff are excluded (set to zero or negative not to exclude on MAF). 3 # heterozygosity deviation in number of standard deviations X. Excludes SNP that show much lower/higher levels of hererozygosity. 10e-16 # Hardy-Weinberg threshold cut off value. Except for GC scores (and not even these entirely), the parameters are somewhat subjective since they are based on population metrics there s some confounding between technical essay problems (which is what we want to identify here) and real population structure in the data. There is a danger of throwing out the baby with the bath but can also end up adding noise to the data which will have a larger or smaller effect depending on the downstream intentions. This is probably the highlight of snpqc because it generates a comprehensive report that is very useful to understand the data structure and it makes it easy to change parameters, rerun and compare results. Sample filters: 0.90 # call rates lower than X. If less than X% genotypes called the whole sample is excluded the DNA might have been of low quality. 3 # heterozygosity deviation in number of standard deviations X # correlation between samples - not used for exclusion, just for listing similar/different samples. Note that the correlation does not exclude missing values the point is to strongly tease apart differences. Do not use this for relationships, use the GRM instead. This just helps identify duplicates and compare correlation between duplicates.
6 ########## map info to remove SNP ##### 0 X # unmapped, X, Y, mitochondrial, etc - specific for the map file. Tab separate whichever SNP should be excluded based on chromosome information (not limited to X, Y, MT can exclude autosomes as well e.g. 1 or 2). Leave blank not to exclude based on mapping information ######### output parameters ############# false # exclude all rejected SNP and samples from reshaped.txt (output files detailed below) file. If false all SNP and samples are maintained in file (sample and snp files will also show only subset). I ve generally found it more practical to keep the full data and then subset based on the list of exclusions. Note that all SNP on a sample below the GC score cutoff will still be set to the missing value. false # set all rejected samples and SNP to missing value (only relevant if above parameter set to 'false'). This keeps all data but sets everything rejected to the missing value. It s also usually preferable to keep the original genotypes and then subset the data. These two parameters work best to accommodate routine workflows were adequate QC parameters are well known and defined already. 9 # symbol for missing/rejected values sets rejected genotypes to this value (could also be NA, -, ) based on GC score cut offs and the parameter above # values for genotypes in output file (does not need to be numeric, could be e.g. AA, AB, BB). Can also separate into allele format by leaving a space between alleles, e.g. 0/0 0/1 1/1. Tab separated. true # build genomic relationship matrix (true/false). Uses van Raden s (2008) approach; SNP that have missing genotypes above the average are excluded. The GRM is built on the subset. The GRM can be used downstream for genomic prediction, principal component analysis or to evaluate relationships in the data. ######## database info ################## ExampleDB # name of database (Note: if already exists genotypes/samples will be appended and map file replaced) by default will be saved in directory databases in the current working directory; output/report results will be stored in new directories 'dbname_output' and 'dbname_report' next to DB. false # use default DB location, under databases in working directory (use false for custom location). Other output files will also be this directory. /Users/joe/myGWASprojects/results # only used if above set to false. Path to where to write the DB and output folders can use a network path to save on a remote machine. \t # file separator for the output files 5 # Column number for X values in genotype file (use zero for missing or to bypass). Simply used to create a picture of good/bad clusters for the report. 6 # Column number for Y values in genotype file (use zero for missing or to bypass).
7 ######## info for report ################# Only used for the report title. Example 50K simulated John Doe and Jane Doe # title of project # authors 4. How to run and R packages needed Before running the program, you ll need to install the R packages: lattice, gplots, gtools, RSQLite, snowfall, gdata, catools, bitops, DBI, snow and compiler. Will also need LaTex installed and available in the path; there are no additional packages needed for LaTex a standard install will suffice (MikTex is a good option). If there is no LaTex, the tex file will still be generated but will have to be converted to pdf manually (MikTex portable works well if you don t want/have LaTex installed). Running the program is very straightforward: 1 setup the parameters file for the run. 2 start R and set the working directory to the folders with the snpqc scripts 3 load the scripts using source. The entry point is the snpqc.r file; in R run: source( snpqc.r ) 4 select the parameters file, e.g. myparam <- C:\mySNPdata\MyParamFile.txt 5 run the full QC with: runqc(myparam) The program has a single function for direct user access (runqc). The parameters are: runqc(runparam,numhits=50000,dbin=true,qc=true,filter=true,report=true,dbout=true) runparam is the path/name of the parameters file. numhits is the number of SNP that are read from the DB in each iteration. The bigger the number, the faster the QC; but more demanding on memory. All other parameters are simply Booleans on whether or not a QC step should be performed. DBin build database of genotypes (TRUE/FALSE) qc run QC metrics (TRUE/FALSE) filter summarize results and build plots for report (TRUE/FALSE) report make PDF report (TRUE/FALSE) DBout write QC results to database (TRUE/FALSE) Note: none of DBin, qc, report or DBout can be run if the previous steps have not been run before (as in never before). You'll need the database to run the QC; you'll need QC results to build the summaries/plots and you'll need summaries/plots to write the report. You can actually skip filter and report steps and go straight from qc to writing (DBout) the results to the database.
8 The database is mandatory for the R version (the C# one can bypass the DB but will be computationally demanding will crash if not enough memory, but it s very fast). If you don t want the report there s no need to run filter/report but this is quite fast, not much time savings to be made on this front (and beats a bit the purpose of this workflow). DBout will store and index all QC metrics in the database, very useful for sub setting on different parameters later on (can query/extract directly with SQL). Population parameters are also included (allelic and genotypic frequencies, HW values ). The database is SQLite which is practical and easily portable but not much in terms of security and may not fit in with other parts of the lab. It s easy to change the scripts to use other engines and integrate with the current system. snpqc is not an R package primarily to simplify this level of customisation. You can e.g. add extra QC checks (family relationships, PCA of sample structures ), personalize the QC report (e.g. add a logo or change the wording), change the parallelization. There are some hidden parameters than can be set once the scripts are loaded:.maxcores=32 # increase max number of cores.cpu=8 # setup number of cores in cluster, can use more or less - if more than 32 need to increase.maxcores.machinename="localhost" # change to run on a remote machine, will probably need to sort out ssh tunneling to get it to work useyates=true # True or false for using Yates correction on the chi-square values 5. Outputs snpqc will generate the DB and two directories: DBname_output and DBname_report. The report folder will contain various images and the full PDF report itself (snpqcreport.pdf). The text file to recreate the pdf is snpqcreport.tex. Just a note on the tex file, you ll need the Sweave.sty style visible to LaTex (it s included with the scripts) for the build to work (will work fine if both files are in the same directory). The DBname_output folder contains all the QC results in flat files. The names should be selfexplanatory but the ones of interest are usually: reshaped.txt this is the main output file with genotypes. There are no column/row names to simplify importing into other programs. The default is to use 0, 1, 2 and 9 for missing, with data separated by space (but can change in parameters file, see above). Number of SNP and samples will depend on parameters selected (i.e. exclude or not bad samples/snp). The format is SNP x sample. animalid.txt ordered list of sample identifiers (matches the reshaped.txt file columns). If the option to exclude bad samples was used, only samples that passed QC are included. snpid.txt is the same as above but just with a single column of ordered SNP that matches columns in reshaped.txt. Numbers will also depend on parameters selected (e.g. exclude bad, excluded some chromosomes). The file orderedmap.txt is the map file ordered by chromosome name and base pair position within chromosomes. SNP in reshaped.txt are ordered according to this map (minus anything excluded). Simply subset orderedmap.txt with snpid.txt and reshaped.txt can be used directly in e.g. GWAS + Manhattan plot or LD analysis.
9 Lists of SNP and samples that did not pass QC are found in the files rejectsamplenames.txt and rejectsnpnames.txt (can be used to exclude from reshaped file after QC). The file rejectmapnames.txt has the SNP names that were removed based on mapping criteria (essentially anything the user decides to exclude). Note: these are always excluded from reshaped.txt. The file rejectmapandsnps.txt combines all SNP that were flagged for exclusion. GRM.txt is the genomic relationship matrix ordered as per reshaped.txt and animalid.txt. Other files contain summaries for the report and the tables for the database (but can also be used externally for additional/new filtering). 6. Where things can go sour Just a few problems that different users have come across: The three main problems you might encounter are incorrect settings in the parameter file; problems in initializing the cluster and out of memory issues (the latter is addressed in the next section). As a reference, if an error occurs right away it s probably a parameter file error, if you get an error after building the database it s probably a cluster problem. The cluster has initiated successfully after you see a message similar to snowfall initialized (using snow ): parallel execution on 8 CPUs. Anything after this is probably a memory issue. The parameters file. The most common source of grief! Make sure that the paths to the 3 data files are correct and with the correct file names. And don t forget to also set the path for the output files correctly. And remember to use / for path delimiters in Linux or Mac and \ in Windows. Also make sure to carefully check the indexes of the data in the data files (e.g. in which column are the SNP names in the genotypes file); it s a bit cumbersome with all those indexes but allows for a lot of flexibility in data formats. Install packages in Linux. Sometimes users do not have permission (actually almost always, unless it s your own machine) to install packages. Create a personal library and point R to it. snow /dosnow for the parallel steps can occasionally be tricky across machines on a network (might need ssh tunnelling to work). On a single machine usually no problems but in Linux you might again not have the necessary permissions to instantiate the cluster; if you get an error here, install the snow/dosnow parallel R packages in a personal library. The program uses simple socket connections which work well in terms of generality but are also slower it s something that can be tinkered with to improve performance (but you need to know what you are doing). You ll need LaTex installed to generate the PDF and it has to be visible in the path. If you don t have LaTex, the PDF can be generated later on using the snpqcreport.tex file (in the report directory). But note that you ll need the Sweave.sty style visible to LaTex (it s included with the scripts) for the build to work (will work fine if both files are in the same directory). You ll also need the images for the PDF (the PDFs in the report folder). In some server settings users do not have permission to build the PDF file programmatically (more of a Windows server issue) even when LaTex is installed (you ll need to do it manually or ask the sys admin to modify permissions). In Windows, MikTex is a good option (or MikTex portable if you don t want to install it).
10 The C# version cannot have space in the path; it s fast but will crash if there s not enough memory (and it s only for the 64 bit Windows OS). If enough demand I ll port to C++ for multi-platform support. 7. Memory management and data sizes snpqc does a reasonable job of handling large datasets with reasonably limited computational resources. But this does come at the cost of speed very large datasets say, ~5,000 samples on 700k arrays will take a full day to index the database (this is the largest bottleneck) but it will give you a useful database to ensure data integrity in the future. If you do not want the database, the compiled version is much faster (the above job runs in less than 30 minutes) but there is no memory management your machine can either run the job or not (and also currently only in Windows). The R version is much more flexible but you cannot bypass the database building step. However you might also run into problems depending on the settings used. To speed up the database, it is built and indexed directly in memory. You should not have any problems with a machine that has 12 or more Gb (it s set to max out memory usage at ~12Gb). But if an error occurs at this point you ll need to edit the file BuilDB.r manually. Try modifying the parameters (you can also tinker with this to improve speed): dbgetquery(dbcon, "pragma cache_size = ") dbgetquery(dbcon, "pragma synchronous = 0") dbgetquery(dbcon, "pragma temp_store = 2") dbgetquery(dbcon, "pragma journal_mode = off") dbgetquery(dbcon, "pragma page_size=65536") dbgetquery(dbcon, "pragma locking_mode=exclusive") Note: you can also achieve very significant speed improvements (almost 2X) by reading in the data from one disk and building the database on another; preferably use fast or ssd disks. If an out of memory error occurs while running the QC steps (after the indexing is complete and the cluster was successfully instantiated, you ll be seeing on the screen something along the lines of evaluating SNPS: ); then try to reduce the number of nodes used in the cluster (R is not very memory efficient in parallelized jobs change the.cpu parameter to e.g..cpu=4, default is 8) or reduce the number of SNP being tested each time (default is numhits=10,000). As a ballpark figure expect each core to use around 80% of the amount of memory used when running as a single threaded job (i.e. 10 cores will use ~8 times more memory). Note: the number of cores does not necessarily mean a faster run with the R version. It will not scale linearly and R can take a lot of time to copy data onto the threads which may cause an inverse effect : more cores makes the run slower. The last memory problem you may encounter is to generate the correlation matrix or the GRM. Here the entire dataset is read into memory (in numeric format, reasonably efficient). For this problem there is no easy fix. If the correlation or GRM cannot be built, the program will bypass the function; this is not a problem for the correlation since it is just a visual aid for diagnostic purposes but you
11 will not be able to build the GRM. At this point I have opted not to read in chunks of data to build the matrices since it would be painfully slow and it has not been an issue up to now (but let me know, datasets are growing rapidly). For reference purposes you d need around 24Gb to handle 20,000 samples on 50k arrays and around 48Gb for 5,000 samples on 700,000k. Figure 2 shows relative memory usage and speed with different numbers of samples (50k genotypes). R4 and R8 refer to the R version with 4 and 8 cores. Note that the C# version scales more linearly than the R version. Values are for reference purposes only and will change completely on different architectures (but the relationships between program versions and cores should be similar). Finally, if none of this works, the last resort is to partition the data into subsets to run the QC (any machine with 8GB will have no problems running 1000 samples on 50k at a time). Just note that you will not able to build a GRM with all samples. 8. More help and citing snpqc If you have any questions or need some help, flick an to Cedric Gondro: cgondro2@une.edu.au (I ll try to help out) also let me know if there are any bugs in the program. If it works for you and you want to cite (hopefully, if it gets accepted): Gondro, C., L.R. Porto-Neto and S.H. Lee (2014). snpqc - an R pipeline for quality control of Illumina SNP data. under review.
Step-by-Step Guide to Relatedness and Association Mapping Contents
 Step-by-Step Guide to Relatedness and Association Mapping Contents OBJECTIVES... 2 INTRODUCTION... 2 RELATEDNESS MEASURES... 2 POPULATION STRUCTURE... 6 Q-K ASSOCIATION ANALYSIS... 10 K MATRIX COMPRESSION...
Step-by-Step Guide to Relatedness and Association Mapping Contents OBJECTIVES... 2 INTRODUCTION... 2 RELATEDNESS MEASURES... 2 POPULATION STRUCTURE... 6 Q-K ASSOCIATION ANALYSIS... 10 K MATRIX COMPRESSION...
Release Note. Agilent Genomic Workbench 6.5 Lite
 Release Note Agilent Genomic Workbench 6.5 Lite Associated Products and Part Number # G3794AA G3799AA - DNA Analytics Software Modules New for the Agilent Genomic Workbench SNP genotype and Copy Number
Release Note Agilent Genomic Workbench 6.5 Lite Associated Products and Part Number # G3794AA G3799AA - DNA Analytics Software Modules New for the Agilent Genomic Workbench SNP genotype and Copy Number
Step-by-Step Guide to Advanced Genetic Analysis
 Step-by-Step Guide to Advanced Genetic Analysis Page 1 Introduction In the previous document, 1 we covered the standard genetic analyses available in JMP Genomics. Here, we cover the more advanced options
Step-by-Step Guide to Advanced Genetic Analysis Page 1 Introduction In the previous document, 1 we covered the standard genetic analyses available in JMP Genomics. Here, we cover the more advanced options
Genetic Analysis. Page 1
 Genetic Analysis Page 1 Genetic Analysis Objectives: 1) Set up Case-Control Association analysis and the Basic Genetics Workflow 2) Use JMP tools to interact with and explore results 3) Learn advanced
Genetic Analysis Page 1 Genetic Analysis Objectives: 1) Set up Case-Control Association analysis and the Basic Genetics Workflow 2) Use JMP tools to interact with and explore results 3) Learn advanced
500K Data Analysis Workflow using BRLMM
 500K Data Analysis Workflow using BRLMM I. INTRODUCTION TO BRLMM ANALYSIS TOOL... 2 II. INSTALLATION AND SET-UP... 2 III. HARDWARE REQUIREMENTS... 3 IV. BRLMM ANALYSIS TOOL WORKFLOW... 3 V. RESULTS/OUTPUT
500K Data Analysis Workflow using BRLMM I. INTRODUCTION TO BRLMM ANALYSIS TOOL... 2 II. INSTALLATION AND SET-UP... 2 III. HARDWARE REQUIREMENTS... 3 IV. BRLMM ANALYSIS TOOL WORKFLOW... 3 V. RESULTS/OUTPUT
Step-by-Step Guide to Basic Genetic Analysis
 Step-by-Step Guide to Basic Genetic Analysis Page 1 Introduction This document shows you how to clean up your genetic data, assess its statistical properties and perform simple analyses such as case-control
Step-by-Step Guide to Basic Genetic Analysis Page 1 Introduction This document shows you how to clean up your genetic data, assess its statistical properties and perform simple analyses such as case-control
Axiom Analysis Suite Release Notes (For research use only. Not for use in diagnostic procedures.)
 Axiom Analysis Suite 4.0.1 Release Notes (For research use only. Not for use in diagnostic procedures.) Axiom Analysis Suite 4.0.1 includes the following changes/updates: 1. For library packages that support
Axiom Analysis Suite 4.0.1 Release Notes (For research use only. Not for use in diagnostic procedures.) Axiom Analysis Suite 4.0.1 includes the following changes/updates: 1. For library packages that support
BICF Nano Course: GWAS GWAS Workflow Development using PLINK. Julia Kozlitina April 28, 2017
 BICF Nano Course: GWAS GWAS Workflow Development using PLINK Julia Kozlitina Julia.Kozlitina@UTSouthwestern.edu April 28, 2017 Getting started Open the Terminal (Search -> Applications -> Terminal), and
BICF Nano Course: GWAS GWAS Workflow Development using PLINK Julia Kozlitina Julia.Kozlitina@UTSouthwestern.edu April 28, 2017 Getting started Open the Terminal (Search -> Applications -> Terminal), and
FVGWAS- 3.0 Manual. 1. Schematic overview of FVGWAS
 FVGWAS- 3.0 Manual Hongtu Zhu @ UNC BIAS Chao Huang @ UNC BIAS Nov 8, 2015 More and more large- scale imaging genetic studies are being widely conducted to collect a rich set of imaging, genetic, and clinical
FVGWAS- 3.0 Manual Hongtu Zhu @ UNC BIAS Chao Huang @ UNC BIAS Nov 8, 2015 More and more large- scale imaging genetic studies are being widely conducted to collect a rich set of imaging, genetic, and clinical
The H3ABioNet GWAS Pipeline
 School of Electrical and UNIVERSITY OF THE WITWATERSRAND, JOHANNESBURG Information Engineering The H3ABioNet GWAS Pipeline https://github.com/h3abionet/h3agwas Scott Hazelhurst 1 Introduction Need to build
School of Electrical and UNIVERSITY OF THE WITWATERSRAND, JOHANNESBURG Information Engineering The H3ABioNet GWAS Pipeline https://github.com/h3abionet/h3agwas Scott Hazelhurst 1 Introduction Need to build
Helpful Galaxy screencasts are available at:
 This user guide serves as a simplified, graphic version of the CloudMap paper for applicationoriented end-users. For more details, please see the CloudMap paper. Video versions of these user guides and
This user guide serves as a simplified, graphic version of the CloudMap paper for applicationoriented end-users. For more details, please see the CloudMap paper. Video versions of these user guides and
D-Optimal Designs. Chapter 888. Introduction. D-Optimal Design Overview
 Chapter 888 Introduction This procedure generates D-optimal designs for multi-factor experiments with both quantitative and qualitative factors. The factors can have a mixed number of levels. For example,
Chapter 888 Introduction This procedure generates D-optimal designs for multi-factor experiments with both quantitative and qualitative factors. The factors can have a mixed number of levels. For example,
SOLOMON: Parentage Analysis 1. Corresponding author: Mark Christie
 SOLOMON: Parentage Analysis 1 Corresponding author: Mark Christie christim@science.oregonstate.edu SOLOMON: Parentage Analysis 2 Table of Contents: Installing SOLOMON on Windows/Linux Pg. 3 Installing
SOLOMON: Parentage Analysis 1 Corresponding author: Mark Christie christim@science.oregonstate.edu SOLOMON: Parentage Analysis 2 Table of Contents: Installing SOLOMON on Windows/Linux Pg. 3 Installing
Recalling Genotypes with BEAGLECALL Tutorial
 Recalling Genotypes with BEAGLECALL Tutorial Release 8.1.4 Golden Helix, Inc. June 24, 2014 Contents 1. Format and Confirm Data Quality 2 A. Exclude Non-Autosomal Markers......................................
Recalling Genotypes with BEAGLECALL Tutorial Release 8.1.4 Golden Helix, Inc. June 24, 2014 Contents 1. Format and Confirm Data Quality 2 A. Exclude Non-Autosomal Markers......................................
Word: Print Address Labels Using Mail Merge
 Word: Print Address Labels Using Mail Merge No Typing! The Quick and Easy Way to Print Sheets of Address Labels Here at PC Knowledge for Seniors we re often asked how to print sticky address labels in
Word: Print Address Labels Using Mail Merge No Typing! The Quick and Easy Way to Print Sheets of Address Labels Here at PC Knowledge for Seniors we re often asked how to print sticky address labels in
Deep Learning for Visual Computing Prof. Debdoot Sheet Department of Electrical Engineering Indian Institute of Technology, Kharagpur
 Deep Learning for Visual Computing Prof. Debdoot Sheet Department of Electrical Engineering Indian Institute of Technology, Kharagpur Lecture - 05 Classification with Perceptron Model So, welcome to today
Deep Learning for Visual Computing Prof. Debdoot Sheet Department of Electrical Engineering Indian Institute of Technology, Kharagpur Lecture - 05 Classification with Perceptron Model So, welcome to today
Exsys RuleBook Selector Tutorial. Copyright 2004 EXSYS Inc. All right reserved. Printed in the United States of America.
 Exsys RuleBook Selector Tutorial Copyright 2004 EXSYS Inc. All right reserved. Printed in the United States of America. This documentation, as well as the software described in it, is furnished under license
Exsys RuleBook Selector Tutorial Copyright 2004 EXSYS Inc. All right reserved. Printed in the United States of America. This documentation, as well as the software described in it, is furnished under license
Spectroscopic Analysis: Peak Detector
 Electronics and Instrumentation Laboratory Sacramento State Physics Department Spectroscopic Analysis: Peak Detector Purpose: The purpose of this experiment is a common sort of experiment in spectroscopy.
Electronics and Instrumentation Laboratory Sacramento State Physics Department Spectroscopic Analysis: Peak Detector Purpose: The purpose of this experiment is a common sort of experiment in spectroscopy.
BEAGLECALL 1.0. Brian L. Browning Department of Medicine Division of Medical Genetics University of Washington. 15 November 2010
 BEAGLECALL 1.0 Brian L. Browning Department of Medicine Division of Medical Genetics University of Washington 15 November 2010 BEAGLECALL 1.0 P a g e i Contents 1 Introduction... 1 1.1 Citing BEAGLECALL...
BEAGLECALL 1.0 Brian L. Browning Department of Medicine Division of Medical Genetics University of Washington 15 November 2010 BEAGLECALL 1.0 P a g e i Contents 1 Introduction... 1 1.1 Citing BEAGLECALL...
Polymorphism and Variant Analysis Lab
 Polymorphism and Variant Analysis Lab Arian Avalos PowerPoint by Casey Hanson Polymorphism and Variant Analysis Matt Hudson 2018 1 Exercise In this exercise, we will do the following:. 1. Gain familiarity
Polymorphism and Variant Analysis Lab Arian Avalos PowerPoint by Casey Hanson Polymorphism and Variant Analysis Matt Hudson 2018 1 Exercise In this exercise, we will do the following:. 1. Gain familiarity
The software comes with 2 installers: (1) SureCall installer (2) GenAligners (contains BWA, BWA- MEM).
 Release Notes Agilent SureCall 4.0 Product Number G4980AA SureCall Client 6-month named license supports installation of one client and server (to host the SureCall database) on one machine. For additional
Release Notes Agilent SureCall 4.0 Product Number G4980AA SureCall Client 6-month named license supports installation of one client and server (to host the SureCall database) on one machine. For additional
Ricopili: Introdution. WCPG Education Day Stephan Ripke / Raymond Walters Toronto, October 2015
 Ricopili: Introdution WCPG Education Day Stephan Ripke / Raymond Walters Toronto, October 2015 What will we offer? Practical: Sorry, no practical sessions today, please refer to the summer school, organized
Ricopili: Introdution WCPG Education Day Stephan Ripke / Raymond Walters Toronto, October 2015 What will we offer? Practical: Sorry, no practical sessions today, please refer to the summer school, organized
Tutorial: De Novo Assembly of Paired Data
 : De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
MacVector for Mac OS X. The online updater for this release is MB in size
 MacVector 17.0.3 for Mac OS X The online updater for this release is 143.5 MB in size You must be running MacVector 15.5.4 or later for this updater to work! System Requirements MacVector 17.0 is supported
MacVector 17.0.3 for Mac OS X The online updater for this release is 143.5 MB in size You must be running MacVector 15.5.4 or later for this updater to work! System Requirements MacVector 17.0 is supported
Gegenees genome format...7. Gegenees comparisons...8 Creating a fragmented all-all comparison...9 The alignment The analysis...
 User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
Release Notes. JMP Genomics. Version 4.0
 JMP Genomics Version 4.0 Release Notes Creativity involves breaking out of established patterns in order to look at things in a different way. Edward de Bono JMP. A Business Unit of SAS SAS Campus Drive
JMP Genomics Version 4.0 Release Notes Creativity involves breaking out of established patterns in order to look at things in a different way. Edward de Bono JMP. A Business Unit of SAS SAS Campus Drive
SSIM Collection & Archiving Infrastructure Scaling & Performance Tuning Guide
 SSIM Collection & Archiving Infrastructure Scaling & Performance Tuning Guide April 2013 SSIM Engineering Team Version 3.0 1 Document revision history Date Revision Description of Change Originator 03/20/2013
SSIM Collection & Archiving Infrastructure Scaling & Performance Tuning Guide April 2013 SSIM Engineering Team Version 3.0 1 Document revision history Date Revision Description of Change Originator 03/20/2013
MAGA: Meta-Analysis of Gene-level Associations
 MAGA: Meta-Analysis of Gene-level Associations SYNOPSIS MAGA [--sfile] [--chr] OPTIONS Option Default Description --sfile specification.txt Select a specification file --chr Select a chromosome DESCRIPTION
MAGA: Meta-Analysis of Gene-level Associations SYNOPSIS MAGA [--sfile] [--chr] OPTIONS Option Default Description --sfile specification.txt Select a specification file --chr Select a chromosome DESCRIPTION
ACTIVANT. Prophet 21 ACTIVANT PROPHET 21. New Features Guide Version 11.0 ADMINISTRATION NEW FEATURES GUIDE (SS, SA, PS) Pre-Release Documentation
 I ACTIVANT ACTIVANT PROPHET 21 Prophet 21 ADMINISTRATION NEW FEATURES GUIDE (SS, SA, PS) New Features Guide Version 11.0 Version 11.5 Pre-Release Documentation This manual contains reference information
I ACTIVANT ACTIVANT PROPHET 21 Prophet 21 ADMINISTRATION NEW FEATURES GUIDE (SS, SA, PS) New Features Guide Version 11.0 Version 11.5 Pre-Release Documentation This manual contains reference information
The fgwas software. Version 1.0. Pennsylvannia State University
 The fgwas software Version 1.0 Zhong Wang 1 and Jiahan Li 2 1 Department of Public Health Science, 2 Department of Statistics, Pennsylvannia State University 1. Introduction Genome-wide association studies
The fgwas software Version 1.0 Zhong Wang 1 and Jiahan Li 2 1 Department of Public Health Science, 2 Department of Statistics, Pennsylvannia State University 1. Introduction Genome-wide association studies
Agilent Genomic Workbench 7.0
 Agilent Genomic Workbench 7.0 Workflow User Guide For Research Use Only. Not for use in diagnostic procedures. Agilent Technologies Notices Agilent Technologies, Inc. 2012, 2015 No part of this manual
Agilent Genomic Workbench 7.0 Workflow User Guide For Research Use Only. Not for use in diagnostic procedures. Agilent Technologies Notices Agilent Technologies, Inc. 2012, 2015 No part of this manual
PRSice: Polygenic Risk Score software - Vignette
 PRSice: Polygenic Risk Score software - Vignette Jack Euesden, Paul O Reilly March 22, 2016 1 The Polygenic Risk Score process PRSice ( precise ) implements a pipeline that has become standard in Polygenic
PRSice: Polygenic Risk Score software - Vignette Jack Euesden, Paul O Reilly March 22, 2016 1 The Polygenic Risk Score process PRSice ( precise ) implements a pipeline that has become standard in Polygenic
ToCatchAThief c ryan campbell & jenn coughlan 7/23/2018
 ToCatchAThief c ryan campbell & jenn coughlan 7/23/2018 Welcome to the To Catch a Thief: With Data! walkthrough! https://bioconductor.org/packages/devel/ bioc/vignettes/snprelate/inst/doc/snprelatetutorial.html
ToCatchAThief c ryan campbell & jenn coughlan 7/23/2018 Welcome to the To Catch a Thief: With Data! walkthrough! https://bioconductor.org/packages/devel/ bioc/vignettes/snprelate/inst/doc/snprelatetutorial.html
XP: Backup Your Important Files for Safety
 XP: Backup Your Important Files for Safety X 380 / 1 Protect Your Personal Files Against Accidental Loss with XP s Backup Wizard Your computer contains a great many important files, but when it comes to
XP: Backup Your Important Files for Safety X 380 / 1 Protect Your Personal Files Against Accidental Loss with XP s Backup Wizard Your computer contains a great many important files, but when it comes to
Running SNAP. The SNAP Team October 2012
 Running SNAP The SNAP Team October 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
Running SNAP The SNAP Team October 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
HOW TO EXPORT BUYER NAMES & ADDRESSES FROM PAYPAL TO A CSV FILE
 HOW TO EXPORT BUYER NAMES & ADDRESSES FROM PAYPAL TO A CSV FILE If your buyers use PayPal to pay for their purchases, you can quickly export all names and addresses to a type of spreadsheet known as a
HOW TO EXPORT BUYER NAMES & ADDRESSES FROM PAYPAL TO A CSV FILE If your buyers use PayPal to pay for their purchases, you can quickly export all names and addresses to a type of spreadsheet known as a
BOSTON USER GROUP. Workflows for Threading. kcura LLC. All rights reserved.
 BOSTON USER GROUP Workflows for Email Threading Jacob Cross Overview The objective of today s session is to give you some ideas that will hopefully increase your knowledge of how email threading works
BOSTON USER GROUP Workflows for Email Threading Jacob Cross Overview The objective of today s session is to give you some ideas that will hopefully increase your knowledge of how email threading works
Running SNAP. The SNAP Team February 2012
 Running SNAP The SNAP Team February 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
Running SNAP The SNAP Team February 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
Importing and Merging Data Tutorial
 Importing and Merging Data Tutorial Release 1.0 Golden Helix, Inc. February 17, 2012 Contents 1. Overview 2 2. Import Pedigree Data 4 3. Import Phenotypic Data 6 4. Import Genetic Data 8 5. Import and
Importing and Merging Data Tutorial Release 1.0 Golden Helix, Inc. February 17, 2012 Contents 1. Overview 2 2. Import Pedigree Data 4 3. Import Phenotypic Data 6 4. Import Genetic Data 8 5. Import and
Maximizing Public Data Sources for Sequencing and GWAS
 Maximizing Public Data Sources for Sequencing and GWAS February 4, 2014 G Bryce Christensen Director of Services Questions during the presentation Use the Questions pane in your GoToWebinar window Agenda
Maximizing Public Data Sources for Sequencing and GWAS February 4, 2014 G Bryce Christensen Director of Services Questions during the presentation Use the Questions pane in your GoToWebinar window Agenda
CMSC 201 Fall 2018 Lab 04 While Loops
 CMSC 201 Fall 2018 Lab 04 While Loops Assignment: Lab 04 While Loops Due Date: During discussion, September 24 th through September 27 th Value: 10 points (8 points during lab, 2 points for Pre Lab quiz)
CMSC 201 Fall 2018 Lab 04 While Loops Assignment: Lab 04 While Loops Due Date: During discussion, September 24 th through September 27 th Value: 10 points (8 points during lab, 2 points for Pre Lab quiz)
Performance issues in Cerm What to check first?
 Performance issues in Cerm What to check first? The Cerm software is built as a client server model. This means the client and the server need the correct specifications, but also the network in between
Performance issues in Cerm What to check first? The Cerm software is built as a client server model. This means the client and the server need the correct specifications, but also the network in between
Before beginning a session, be sure these packages are loaded using the library() or require() commands.
 Instructions for running state-space movement models in WinBUGS via R. This readme file is included with a package of R and WinBUGS scripts included as an online supplement (Ecological Archives EXXX-XXX-03)
Instructions for running state-space movement models in WinBUGS via R. This readme file is included with a package of R and WinBUGS scripts included as an online supplement (Ecological Archives EXXX-XXX-03)
TPC Desktop Series. Least Squares Learning Guide 1/18
 TPC Desktop Series Least Squares Learning Guide 1/18 NOTICE The information in this document is subject to change without notice. TRAVERSE PC. Inc. assumes no responsibility for any errors that may appear
TPC Desktop Series Least Squares Learning Guide 1/18 NOTICE The information in this document is subject to change without notice. TRAVERSE PC. Inc. assumes no responsibility for any errors that may appear
CTL mapping in R. Danny Arends, Pjotr Prins, and Ritsert C. Jansen. University of Groningen Groningen Bioinformatics Centre & GCC Revision # 1
 CTL mapping in R Danny Arends, Pjotr Prins, and Ritsert C. Jansen University of Groningen Groningen Bioinformatics Centre & GCC Revision # 1 First written: Oct 2011 Last modified: Jan 2018 Abstract: Tutorial
CTL mapping in R Danny Arends, Pjotr Prins, and Ritsert C. Jansen University of Groningen Groningen Bioinformatics Centre & GCC Revision # 1 First written: Oct 2011 Last modified: Jan 2018 Abstract: Tutorial
Preparation of alignments for variant calling with GATK: exercise instructions for BioHPC Lab computers
 Preparation of alignments for variant calling with GATK: exercise instructions for BioHPC Lab computers Data used in the exercise We will use D. melanogaster WGS paired-end Illumina data with NCBI accessions
Preparation of alignments for variant calling with GATK: exercise instructions for BioHPC Lab computers Data used in the exercise We will use D. melanogaster WGS paired-end Illumina data with NCBI accessions
Getting Started Guide
 Getting Started Guide for education accounts Setup Manual Edition 7 Last updated: September 15th, 2016 Note: Click on File and select Make a copy to save this to your Google Drive, or select Print, to
Getting Started Guide for education accounts Setup Manual Edition 7 Last updated: September 15th, 2016 Note: Click on File and select Make a copy to save this to your Google Drive, or select Print, to
Affymetrix Genotyping Console 4.2 Release Notes (For research use only. Not for use in diagnostic procedures.)
 Affymetrix Genotyping Console 4.2 Release Notes (For research use only. Not for use in diagnostic procedures.) Genotyping Console 4.2 includes the following changes and enhancements: 1. Edit Calls within
Affymetrix Genotyping Console 4.2 Release Notes (For research use only. Not for use in diagnostic procedures.) Genotyping Console 4.2 includes the following changes and enhancements: 1. Edit Calls within
Automating Digital Downloads
 Automating Digital Downloads (Copyright 2018 Reed Hoffmann, not to be shared without permission) One of the best things you can do to simplify your imaging workflow is to automate the download process.
Automating Digital Downloads (Copyright 2018 Reed Hoffmann, not to be shared without permission) One of the best things you can do to simplify your imaging workflow is to automate the download process.
CPSC 340: Machine Learning and Data Mining. Feature Selection Fall 2016
 CPSC 34: Machine Learning and Data Mining Feature Selection Fall 26 Assignment 3: Admin Solutions will be posted after class Wednesday. Extra office hours Thursday: :3-2 and 4:3-6 in X836. Midterm Friday:
CPSC 34: Machine Learning and Data Mining Feature Selection Fall 26 Assignment 3: Admin Solutions will be posted after class Wednesday. Extra office hours Thursday: :3-2 and 4:3-6 in X836. Midterm Friday:
Visual Streamline FAQ
 Program Overview: Visual Streamline FAQ How does the program Map Import, located in: Inventory > Global Changes, work? This program enables users the flexibility to use their own excel spreadsheet, and
Program Overview: Visual Streamline FAQ How does the program Map Import, located in: Inventory > Global Changes, work? This program enables users the flexibility to use their own excel spreadsheet, and
QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL
 QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL User manual for QIAseq Targeted RNAscan Panel Analysis 0.5.2 beta 1 Windows, Mac OS X and Linux February 5, 2018 This software is for research
QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL User manual for QIAseq Targeted RNAscan Panel Analysis 0.5.2 beta 1 Windows, Mac OS X and Linux February 5, 2018 This software is for research
CSCI 1100L: Topics in Computing Lab Lab 11: Programming with Scratch
 CSCI 1100L: Topics in Computing Lab Lab 11: Programming with Scratch Purpose: We will take a look at programming this week using a language called Scratch. Scratch is a programming language that was developed
CSCI 1100L: Topics in Computing Lab Lab 11: Programming with Scratch Purpose: We will take a look at programming this week using a language called Scratch. Scratch is a programming language that was developed
Emile R. Chimusa Division of Human Genetics Department of Pathology University of Cape Town
 Advanced Genomic data manipulation and Quality Control with plink Emile R. Chimusa (emile.chimusa@uct.ac.za) Division of Human Genetics Department of Pathology University of Cape Town Outlines: 1.Introduction
Advanced Genomic data manipulation and Quality Control with plink Emile R. Chimusa (emile.chimusa@uct.ac.za) Division of Human Genetics Department of Pathology University of Cape Town Outlines: 1.Introduction
1 Installation (briefly)
 Jumpstart Linux Bo Waggoner Updated: 2014-09-15 Abstract A basic, rapid tutorial on Linux and its command line for the absolute beginner. Prerequisites: a computer on which to install, a DVD and/or USB
Jumpstart Linux Bo Waggoner Updated: 2014-09-15 Abstract A basic, rapid tutorial on Linux and its command line for the absolute beginner. Prerequisites: a computer on which to install, a DVD and/or USB
Welcome Back! Without further delay, let s get started! First Things First. If you haven t done it already, download Turbo Lister from ebay.
 Welcome Back! Now that we ve covered the basics on how to use templates and how to customise them, it s time to learn some more advanced techniques that will help you create outstanding ebay listings!
Welcome Back! Now that we ve covered the basics on how to use templates and how to customise them, it s time to learn some more advanced techniques that will help you create outstanding ebay listings!
Optimizing Testing Performance With Data Validation Option
 Optimizing Testing Performance With Data Validation Option 1993-2016 Informatica LLC. No part of this document may be reproduced or transmitted in any form, by any means (electronic, photocopying, recording
Optimizing Testing Performance With Data Validation Option 1993-2016 Informatica LLC. No part of this document may be reproduced or transmitted in any form, by any means (electronic, photocopying, recording
Creating a new form with check boxes, drop-down list boxes, and text box fill-ins. Customizing each of the three form fields.
 In This Chapter Creating a new form with check boxes, drop-down list boxes, and text box fill-ins. Customizing each of the three form fields. Adding help text to any field to assist users as they fill
In This Chapter Creating a new form with check boxes, drop-down list boxes, and text box fill-ins. Customizing each of the three form fields. Adding help text to any field to assist users as they fill
Release Notes. Agilent CytoGenomics 2.7. Product Number. Key new features. Overview
 Release Notes Agilent CytoGenomics 2.7 Product Number G1662AA CytoGenomics Client 1 year named license (including Feature Extraction). This license supports installation of one client and server (to host
Release Notes Agilent CytoGenomics 2.7 Product Number G1662AA CytoGenomics Client 1 year named license (including Feature Extraction). This license supports installation of one client and server (to host
Foxtrot Certified Expert Study Guide
 Foxtrot Certified Expert Study Guide Click for the Practice Exam Useful Terms: Client Machine Typically referred to as a user s machine that points to a License Path. Data Execution Prevention (DEP) A
Foxtrot Certified Expert Study Guide Click for the Practice Exam Useful Terms: Client Machine Typically referred to as a user s machine that points to a License Path. Data Execution Prevention (DEP) A
Intermediate/Advanced Python. Michael Weinstein (Day 1)
 Intermediate/Advanced Python Michael Weinstein (Day 1) Who am I? Most of my experience is on the molecular and animal modeling side I also design computer programs for analyzing biological data, particularly
Intermediate/Advanced Python Michael Weinstein (Day 1) Who am I? Most of my experience is on the molecular and animal modeling side I also design computer programs for analyzing biological data, particularly
ELAI user manual. Yongtao Guan Baylor College of Medicine. Version June Copyright 2. 3 A simple example 2
 ELAI user manual Yongtao Guan Baylor College of Medicine Version 1.0 25 June 2015 Contents 1 Copyright 2 2 What ELAI Can Do 2 3 A simple example 2 4 Input file formats 3 4.1 Genotype file format....................................
ELAI user manual Yongtao Guan Baylor College of Medicine Version 1.0 25 June 2015 Contents 1 Copyright 2 2 What ELAI Can Do 2 3 A simple example 2 4 Input file formats 3 4.1 Genotype file format....................................
RLMM - Robust Linear Model with Mahalanobis Distance Classifier
 RLMM - Robust Linear Model with Mahalanobis Distance Classifier Nusrat Rabbee and Gary Wong June 13, 2018 Contents 1 Introduction 1 2 Instructions for Genotyping Affymetrix Mapping 100K array - Xba set
RLMM - Robust Linear Model with Mahalanobis Distance Classifier Nusrat Rabbee and Gary Wong June 13, 2018 Contents 1 Introduction 1 2 Instructions for Genotyping Affymetrix Mapping 100K array - Xba set
OnCommand Insight 6.3 Data Warehouse Custom Report Hands-on Lab Guide
 OnCommand Insight 6.3 Data Warehouse Custom Report Hands-on Lab Guide How to easily create adhoc and multi-tenancy reports in OnCommand Insight DWH using Business Insight Advanced Dave Collins, Technical
OnCommand Insight 6.3 Data Warehouse Custom Report Hands-on Lab Guide How to easily create adhoc and multi-tenancy reports in OnCommand Insight DWH using Business Insight Advanced Dave Collins, Technical
Retrospect 8 for Windows Reviewer s Guide
 Retrospect 8 for Windows Reviewer s Guide 2012 Retrospect, Inc. About this Reviewer s Guide This document provides a concise guide to understanding Retrospect 8 for Windows. While it is not designed to
Retrospect 8 for Windows Reviewer s Guide 2012 Retrospect, Inc. About this Reviewer s Guide This document provides a concise guide to understanding Retrospect 8 for Windows. While it is not designed to
Package lodgwas. R topics documented: November 30, Type Package
 Type Package Package lodgwas November 30, 2015 Title Genome-Wide Association Analysis of a Biomarker Accounting for Limit of Detection Version 1.0-7 Date 2015-11-10 Author Ahmad Vaez, Ilja M. Nolte, Peter
Type Package Package lodgwas November 30, 2015 Title Genome-Wide Association Analysis of a Biomarker Accounting for Limit of Detection Version 1.0-7 Date 2015-11-10 Author Ahmad Vaez, Ilja M. Nolte, Peter
Matlab for FMRI Module 1: the basics Instructor: Luis Hernandez-Garcia
 Matlab for FMRI Module 1: the basics Instructor: Luis Hernandez-Garcia The goal for this tutorial is to make sure that you understand a few key concepts related to programming, and that you know the basics
Matlab for FMRI Module 1: the basics Instructor: Luis Hernandez-Garcia The goal for this tutorial is to make sure that you understand a few key concepts related to programming, and that you know the basics
Three OPTIMIZING. Your System for Photoshop. Tuning for Performance
 Three OPTIMIZING Your System for Photoshop Tuning for Performance 72 Power, Speed & Automation with Adobe Photoshop This chapter goes beyond speeding up how you can work faster in Photoshop to how to make
Three OPTIMIZING Your System for Photoshop Tuning for Performance 72 Power, Speed & Automation with Adobe Photoshop This chapter goes beyond speeding up how you can work faster in Photoshop to how to make
Chapter One: Getting Started With IBM SPSS for Windows
 Chapter One: Getting Started With IBM SPSS for Windows Using Windows The Windows start-up screen should look something like Figure 1-1. Several standard desktop icons will always appear on start up. Note
Chapter One: Getting Started With IBM SPSS for Windows Using Windows The Windows start-up screen should look something like Figure 1-1. Several standard desktop icons will always appear on start up. Note
V6 Programming Fundamentals: Part 1 Stored Procedures and Beyond David Adams & Dan Beckett. All rights reserved.
 Summit 97 V6 Programming Fundamentals: Part 1 Stored Procedures and Beyond by David Adams & Dan Beckett 1997 David Adams & Dan Beckett. All rights reserved. Content adapted from Programming 4th Dimension:
Summit 97 V6 Programming Fundamentals: Part 1 Stored Procedures and Beyond by David Adams & Dan Beckett 1997 David Adams & Dan Beckett. All rights reserved. Content adapted from Programming 4th Dimension:
Intro to NGS Tutorial
 Intro to NGS Tutorial Release 8.6.0 Golden Helix, Inc. October 31, 2016 Contents 1. Overview 2 2. Import Variants and Quality Fields 3 3. Quality Filters 10 Generate Alternate Read Ratio.........................................
Intro to NGS Tutorial Release 8.6.0 Golden Helix, Inc. October 31, 2016 Contents 1. Overview 2 2. Import Variants and Quality Fields 3 3. Quality Filters 10 Generate Alternate Read Ratio.........................................
Technical White Paper
 Technical White Paper Via Excel (VXL) Item Templates This technical white paper is designed for Spitfire Project Management System users. In this paper, you will learn how to create Via Excel Item Templates
Technical White Paper Via Excel (VXL) Item Templates This technical white paper is designed for Spitfire Project Management System users. In this paper, you will learn how to create Via Excel Item Templates
Module 1: Introduction RStudio
 Module 1: Introduction RStudio Contents Page(s) Installing R and RStudio Software for Social Network Analysis 1-2 Introduction to R Language/ Syntax 3 Welcome to RStudio 4-14 A. The 4 Panes 5 B. Calculator
Module 1: Introduction RStudio Contents Page(s) Installing R and RStudio Software for Social Network Analysis 1-2 Introduction to R Language/ Syntax 3 Welcome to RStudio 4-14 A. The 4 Panes 5 B. Calculator
Bioinformatics - Homework 1 Q&A style
 Bioinformatics - Homework 1 Q&A style Instructions: in this assignment you will test your understanding of basic GWAS concepts and GenABEL functions. The materials needed for the homework (two datasets
Bioinformatics - Homework 1 Q&A style Instructions: in this assignment you will test your understanding of basic GWAS concepts and GenABEL functions. The materials needed for the homework (two datasets
Managing custom montage files Quick montages How custom montage files are applied Markers Adding markers...
 AnyWave Contents What is AnyWave?... 3 AnyWave home directories... 3 Opening a file in AnyWave... 4 Quick re-open a recent file... 4 Viewing the content of a file... 5 Choose what you want to view and
AnyWave Contents What is AnyWave?... 3 AnyWave home directories... 3 Opening a file in AnyWave... 4 Quick re-open a recent file... 4 Viewing the content of a file... 5 Choose what you want to view and
1. Summary statistics test_gwas. This file contains a set of 50K random SNPs of the Subjective Well-being GWAS of the Netherlands Twin Register
 Quality Control for Genome-Wide Association Studies Bart Baselmans & Meike Bartels Boulder 2017 Setting up files and directories To perform a quality control protocol in a Genome-Wide Association Meta
Quality Control for Genome-Wide Association Studies Bart Baselmans & Meike Bartels Boulder 2017 Setting up files and directories To perform a quality control protocol in a Genome-Wide Association Meta
Techniques for Optimizing Reusable Content in LibGuides
 University of Louisville From the SelectedWorks of Terri Holtze April 21, 2017 Techniques for Optimizing Reusable Content in LibGuides Terri Holtze, University of Louisville Available at: https://works.bepress.com/terri-holtze/4/
University of Louisville From the SelectedWorks of Terri Holtze April 21, 2017 Techniques for Optimizing Reusable Content in LibGuides Terri Holtze, University of Louisville Available at: https://works.bepress.com/terri-holtze/4/
USING DRUPAL. Hampshire College Website Editors Guide https://drupal.hampshire.edu
 USING DRUPAL Hampshire College Website Editors Guide 2014 https://drupal.hampshire.edu Asha Kinney Hampshire College Information Technology - 2014 HOW TO GET HELP Your best bet is ALWAYS going to be to
USING DRUPAL Hampshire College Website Editors Guide 2014 https://drupal.hampshire.edu Asha Kinney Hampshire College Information Technology - 2014 HOW TO GET HELP Your best bet is ALWAYS going to be to
Microsoft Excel Level 2
 Microsoft Excel Level 2 Table of Contents Chapter 1 Working with Excel Templates... 5 What is a Template?... 5 I. Opening a Template... 5 II. Using a Template... 5 III. Creating a Template... 6 Chapter
Microsoft Excel Level 2 Table of Contents Chapter 1 Working with Excel Templates... 5 What is a Template?... 5 I. Opening a Template... 5 II. Using a Template... 5 III. Creating a Template... 6 Chapter
Learn how to login to Sitefinity and what possible errors you can get if you do not have proper permissions.
 USER GUIDE This guide is intended for users of all levels of expertise. The guide describes in detail Sitefinity user interface - from logging to completing a project. Use it to learn how to create pages
USER GUIDE This guide is intended for users of all levels of expertise. The guide describes in detail Sitefinity user interface - from logging to completing a project. Use it to learn how to create pages
Welcome to the world of .
 Welcome to the world of e-mail. E-mail, short for electronic mail, allows computer users to easily send messages back and forth between acquaintances around the world. There are a variety of ways to do
Welcome to the world of e-mail. E-mail, short for electronic mail, allows computer users to easily send messages back and forth between acquaintances around the world. There are a variety of ways to do
PISCES Installation and Getting Started 1
 This document will walk you through the PISCES setup process and get you started accessing the suite of available tools. It will begin with what options to choose during the actual installation and the
This document will walk you through the PISCES setup process and get you started accessing the suite of available tools. It will begin with what options to choose during the actual installation and the
Here we will look at some methods for checking data simply using JOSM. Some of the questions we are asking about our data are:
 Validating for Missing Maps Using JOSM This document covers processes for checking data quality in OpenStreetMap, particularly in the context of Humanitarian OpenStreetMap Team and Red Cross Missing Maps
Validating for Missing Maps Using JOSM This document covers processes for checking data quality in OpenStreetMap, particularly in the context of Humanitarian OpenStreetMap Team and Red Cross Missing Maps
Agilent Genomic Workbench 6.5
 Agilent Genomic Workbench 6.5 Product Overview Guide For Research Use Only. Not for use in diagnostic procedures. Agilent Technologies Notices Agilent Technologies, Inc. 2010, 2015 No part of this manual
Agilent Genomic Workbench 6.5 Product Overview Guide For Research Use Only. Not for use in diagnostic procedures. Agilent Technologies Notices Agilent Technologies, Inc. 2010, 2015 No part of this manual
SiteProxy adds security, reduces network traffic on the camera, and improves performance.
 SiteProxy SiteProxy is a cross-platform HTTP server for managing one or more Internet Camera Servers (Canon VB101, VB150, and VB-C10/R, and others) streaming video servers. Location of the SiteProxy Software
SiteProxy SiteProxy is a cross-platform HTTP server for managing one or more Internet Camera Servers (Canon VB101, VB150, and VB-C10/R, and others) streaming video servers. Location of the SiteProxy Software
Very large searches present a number of challenges. These are the topics we will cover during this presentation.
 1 Very large searches present a number of challenges. These are the topics we will cover during this presentation. 2 The smartest way to merge files, like fractions from a MudPIT run, is using Mascot Daemon.
1 Very large searches present a number of challenges. These are the topics we will cover during this presentation. 2 The smartest way to merge files, like fractions from a MudPIT run, is using Mascot Daemon.
NEW FEATURES. Manage. Analyze. Discover.
 Manage. Analyze. Discover. NEW FEATURES Applied Maths proudly presents BioNumerics version 7.5, honoring a good old Applied Maths tradition of supplying its minor upgrade with a new features list worth
Manage. Analyze. Discover. NEW FEATURES Applied Maths proudly presents BioNumerics version 7.5, honoring a good old Applied Maths tradition of supplying its minor upgrade with a new features list worth
SEQGWAS: Integrative Analysis of SEQuencing and GWAS Data
 SEQGWAS: Integrative Analysis of SEQuencing and GWAS Data SYNOPSIS SEQGWAS [--sfile] [--chr] OPTIONS Option Default Description --sfile specification.txt Select a specification file --chr Select a chromosome
SEQGWAS: Integrative Analysis of SEQuencing and GWAS Data SYNOPSIS SEQGWAS [--sfile] [--chr] OPTIONS Option Default Description --sfile specification.txt Select a specification file --chr Select a chromosome
EssEntial indesign skills Brought to you By InDesign
 10 EssEntial indesign skills Brought to you By InDesignSkills 10 Essential InDesign Skills by InDesignSkills www.indesignskills.com Copyright 2014 Illustration: Henry Rivers Contents In this ebook you
10 EssEntial indesign skills Brought to you By InDesignSkills 10 Essential InDesign Skills by InDesignSkills www.indesignskills.com Copyright 2014 Illustration: Henry Rivers Contents In this ebook you
When you first launch CrushFTP you may be notified that port 21 is locked. You will be prompted to fix this.
 This is a quick start guide. Its intent is to help you get up and running with as little configuration as possible. This walk through should take less than 10 minutes until you are able to login with your
This is a quick start guide. Its intent is to help you get up and running with as little configuration as possible. This walk through should take less than 10 minutes until you are able to login with your
Company System Administrator (CSA) User Guide
 BMO HARRIS ONLINE BANKING SM FOR SMALL BUSINESS Company System Administrator (CSA) User Guide Copyright 2011 BMO Harris Bank N.A. TABLE OF CONTENTS WELCOME... 1 Who should use this guide... 1 What it covers...
BMO HARRIS ONLINE BANKING SM FOR SMALL BUSINESS Company System Administrator (CSA) User Guide Copyright 2011 BMO Harris Bank N.A. TABLE OF CONTENTS WELCOME... 1 Who should use this guide... 1 What it covers...
POC Evaluation Guide May 09, 2017
 POC Evaluation Guide May 09, 2017 This page intentionally left blank P r o p r i e t a r y a n d C o n f i d e n t i a l. 2 0 1 7 R F P M o n k e y. c o m L L C Page 2 CONTENTS Read Me First... 4 About
POC Evaluation Guide May 09, 2017 This page intentionally left blank P r o p r i e t a r y a n d C o n f i d e n t i a l. 2 0 1 7 R F P M o n k e y. c o m L L C Page 2 CONTENTS Read Me First... 4 About
GenomeStudio Software Release Notes
 GenomeStudio Software 2009.2 Release Notes 1. GenomeStudio Software 2009.2 Framework... 1 2. Illumina Genome Viewer v1.5...2 3. Genotyping Module v1.5... 4 4. Gene Expression Module v1.5... 6 5. Methylation
GenomeStudio Software 2009.2 Release Notes 1. GenomeStudio Software 2009.2 Framework... 1 2. Illumina Genome Viewer v1.5...2 3. Genotyping Module v1.5... 4 4. Gene Expression Module v1.5... 6 5. Methylation
TUTORIAL FOR IMPORTING OTTAWA FIRE HYDRANT PARKING VIOLATION DATA INTO MYSQL
 TUTORIAL FOR IMPORTING OTTAWA FIRE HYDRANT PARKING VIOLATION DATA INTO MYSQL We have spent the first part of the course learning Excel: importing files, cleaning, sorting, filtering, pivot tables and exporting
TUTORIAL FOR IMPORTING OTTAWA FIRE HYDRANT PARKING VIOLATION DATA INTO MYSQL We have spent the first part of the course learning Excel: importing files, cleaning, sorting, filtering, pivot tables and exporting
Lecture 5. Essential skills for bioinformatics: Unix/Linux
 Lecture 5 Essential skills for bioinformatics: Unix/Linux UNIX DATA TOOLS Text processing with awk We have illustrated two ways awk can come in handy: Filtering data using rules that can combine regular
Lecture 5 Essential skills for bioinformatics: Unix/Linux UNIX DATA TOOLS Text processing with awk We have illustrated two ways awk can come in handy: Filtering data using rules that can combine regular
An introduction to plotting data
 An introduction to plotting data Eric D. Black California Institute of Technology February 25, 2014 1 Introduction Plotting data is one of the essential skills every scientist must have. We use it on a
An introduction to plotting data Eric D. Black California Institute of Technology February 25, 2014 1 Introduction Plotting data is one of the essential skills every scientist must have. We use it on a
MAGMA manual (version 1.06)
 MAGMA manual (version 1.06) TABLE OF CONTENTS OVERVIEW 3 QUICKSTART 4 ANNOTATION 6 OVERVIEW 6 RUNNING THE ANNOTATION 6 ADDING AN ANNOTATION WINDOW AROUND GENES 7 RESTRICTING THE ANNOTATION TO A SUBSET
MAGMA manual (version 1.06) TABLE OF CONTENTS OVERVIEW 3 QUICKSTART 4 ANNOTATION 6 OVERVIEW 6 RUNNING THE ANNOTATION 6 ADDING AN ANNOTATION WINDOW AROUND GENES 7 RESTRICTING THE ANNOTATION TO A SUBSET
FLIR Tools+ and Report Studio
 Creating and Processing Word Templates http://www.infraredtraining.com 09-20-2017 2017, Infrared Training Center. 1 FLIR Report Studio Overview Report Studio is a Microsoft Word Reporting module that is
Creating and Processing Word Templates http://www.infraredtraining.com 09-20-2017 2017, Infrared Training Center. 1 FLIR Report Studio Overview Report Studio is a Microsoft Word Reporting module that is
Exome sequencing. Jong Kyoung Kim
 Exome sequencing Jong Kyoung Kim Genome Analysis Toolkit The GATK is the industry standard for identifying SNPs and indels in germline DNA and RNAseq data. Its scope is now expanding to include somatic
Exome sequencing Jong Kyoung Kim Genome Analysis Toolkit The GATK is the industry standard for identifying SNPs and indels in germline DNA and RNAseq data. Its scope is now expanding to include somatic
User Guide. v Released June Advaita Corporation 2016
 User Guide v. 0.9 Released June 2016 Copyright Advaita Corporation 2016 Page 2 Table of Contents Table of Contents... 2 Background and Introduction... 4 Variant Calling Pipeline... 4 Annotation Information
User Guide v. 0.9 Released June 2016 Copyright Advaita Corporation 2016 Page 2 Table of Contents Table of Contents... 2 Background and Introduction... 4 Variant Calling Pipeline... 4 Annotation Information
