Package batman. Installation and Testing
|
|
|
- Janice O’Brien’
- 5 years ago
- Views:
Transcription
1 Package batman Installation and Testing Table of Contents 1. INSTALLATION INSTRUCTIONS TESTING... 3 Test 1: Single spectrum from designed mixture data... 3 Test 2: Multiple spectra from designed mixture data... 4 Test 3: Multiple spectra from designed mixture data (adjusted relative intensities)... 7 Test 4: Multiple spectra from bacterial supernatants data CHEMICAL SHIFT SORTING Installation instructions For all platforms: Install R , or higher (note restriction for Mac binary below). Install R packages dosnow and plotrix using the following command in R: install.packages("dosnow") install.packages("plotrix") User can specify where to install the package in argument lib. For detailed help of install.packages(), type in R:?install.packages Windows & Linux: Install using online repository: R install command: install.packages("batman", repos=" Mac: Download and install from local copy: 1
2 There are two options: 1. Download & install from Mac binary Note that, Mac OS X 10.6 (Snow Leopard) and higher users need to download the binary file and follow the instructions below. It does not support Mac OS X 10.9 (Mavericks) and higher at the moment. Download the Mac binary file batman_ tgz (to user desktop in this example) using svn command in terminal (check the downloaded file size): svn export svn://scm.r-forge.r-project.org/svnroot/batman/pkg/batman_ tgz /home/user/desktop/batman_ tgz And in R type the following command: install.packages( /path/of/batman_ tgz, repos = NULL) The fortan compiler ( may be required in order to run BATMAN in some case. 2. Download & install from source Download the source code file batman_ tar.gz using the following svn command in terminal (check the downloaded file size): svn export svn://scm.r-forge.r-project.org/svnroot/batman/pkg/batman_ tar.gz /home/user/desktop/batman_ tar.gz And in R type the following command (make sure you have a C++ compiler is installed before installing from source code and possibly fortan compiler as well which can be downloaded from install.packages( /path/of/batman_ tar.gz, repos = NULL, type="source") (Note: in this guide, users should replace the italic characters in directory name with the directory in their cases.) If you receive error messages on installation, check if the required packages dosnow and plotrix are installed. Also check the size of downloaded files. If no error messages are produced, proceed to testing below. 2
3 2. Testing Load batman package in R: library(batman) There are 4 Tests included in this guide using, a) designed mixture data [1] and b) bacterial supernatant data [2]: Test 1 is a quick run of batman to check if it is installed properly. Test 2 shows how to run batman on the six designed mixture spectra to estimate levels of 6 metabolites. Test 3 uses the designed mixture data to explain how to modify the properties of the spectral templates, specifically the relative intensities of each multiplet, thus improving the fit. Test 4 is a real biological sample, a bacterial supernatant, with larger number of spectra and possibly more metabolites to fit, which may take longer time to run. The run times stated in this document were obtained on a Mac Pro with a dual 3.0 GHz quad core processors, 13 GB RAM. Test 1: Single spectrum from designed mixture data This is a simple test checking if the package has been installed correctly and can run. The number of iterations is deliberately set very low and one should not expect a particularly good fit. It should take about half minute to run. Run batman with included test dataset (designed mixture data) in user specified directory: bm<-batman(runbatmandir = /user/specified/directory ) Or just use the current directory: bm<-batman() Output from R: Running batman... Number of burn-in iterations: 200 Number of post-burn-in iterations: 100 The template file used is 2: The user input template of multiplets in multi_data_user.csv file. Loading multi_data_user.csv... 0% Size of each spectrum is 682. Size of metabolite list is 6. of which 6 have resonances in/near the specified region and will be fit. Constructing chain data structure... time used is 1 seconds. Running MCMC... =========================================================== 67% time used for burnin is 16 seconds. ========================================================================================= 100% time used is 23 seconds. saving posteriors... 3
4 Standardized Intensity time elapsed second. Reading in saved data in folder /user/specified/directory/runbatman/batmanoutput/31_oct_16_03_38 Completed. The green lines shown above may only show when using R from terminal (in Mac and Linux) or CMD (in Windows). Time elapsed will vary depending on the machine power. The mean posterior fit result similar to the one below will be shown in a figure window. A PDF file of this plot with the name specfit_2_.pdf can be found in folder /user/specified/directory/runbatman/batmanoutput/date_time. N M R S pectrum 2: X 2 Original Spectrum Metabolites Fit Wavelet Fit Fit Sum ppm Test 2: Multiple spectra from designed mixture data This test fits the six designed mixture spectra and uses the parallel processes function to process a separate spectrum on each core. The final example output has been processed with a large number of iterations, which takes about 11 minutes to run (using six cores, thus about the same time needed to run one spectrum), but the user can try first with less iterations. Below is a screen shot of the multiplet template file /user/specified/directory/runbatman/batmaninput/multi_data_user.csv used for this test. The user does not need to change anything in this file at the moment. In the next test, details will be given on how to change the relative intensities of multiplets to improve the fit. 4
5 Refer to batman documentation for more detailed information on the parameters in this file. Chemical shift for each multiplet can be changed here to a more accurate position specific to the dataset analysed. Set to 0 to exclude the corresponding multiplet from the fit. Further details are given in the batman documentation which can be found at Open file batmanoptions.txt in folder /user/specified/directory/runbatman/batmaninput, find the following parameter lines: specno - Ranges of spectra number to be included (e.g. 1,3-4 etc.): 2 paraproc - No of parallel processes (multicores) (only 1 core will be used for single spectrum): 1 nitburnin - Number of burn-in iterations: 200 nitpostburnin - Number of post-burn-in iterations: 100 And change their parameters to: specno - Ranges of spectra number to be included (e.g. 1,3-4 etc.): 1-6 paraproc - No of parallel processes (multicores) (only 1 core will be used for single spectrum): 6 nitburnin - Number of burn-in iterations: 7000 nitpostburnin - Number of post-burn-in iterations: 1000 Save the options file. This will change the input from spectrum number 2 to all the 6 (1 to 6) spectra in test dataset. Note: paraproc parameter: This depends on the number of cores on the user s machine. nitburnin & nitpostburnin parameters: user can test with less iterations which will need less time to run. Now run the same command used in Test 1: Run batman with included test dataset (designed mixture data) in user specified directory: bm<-batman(runbatmandir = /user/specified/directory ) Or just use the current directory: 5
6 Standardized Intensity bm<-batman() Output from R: Running batman... Number of burn-in iterations: 7000 Number of post-burn-in iterations: 1000 The template file used is 2: The user input template of multiplets in multi_data_user.csv file. Loading multi_data_user.csv... Number of parallel processes (multicores) used to run the multi-spectra analysis: 6 Parallel processing of multi spectra currently cannot display the progress bar (or any words), please be patient for the results :) time elapsed second. Reading in saved data in folder /user/specified/directory/runbatman/batmanoutput/date_time Completed. The 3 figures with the results for all 6 spectra (not shown here) will be generated. Alternatively to inspect the results, the user can plot all 6 spectra in a single stack plot as follows: plotbatmanfitstack(bm,offset = 0.8,placeLegend = 'topleft',yto = 5) A stack plot of the fit result is shown below: (pdf file with the name specfit_stack_.pdf can be found in folder /user/specified/directory/runbatman/batmanoutput/date_time ) N M R S pectra Original Spectrum Metabolites Fit Mirrored Wavelet Fit Fit Sum ppm 6
7 Test 3: Multiple spectra from designed mixture data (adjusted relative intensities) Often, the relative intensities of resonances do not reflect the theoretical ratios expected from the molecular structure. This could be due to, for example, incomplete relaxation of different spin systems, and varies according to the pulse sequence and sample type. The effect may lead to poor fitting. This test uses the same spectra as in Test 2 but with some adjustment of relative intensities of the multiplet template used to attempt to improve the fit. It should take about the same time as of Test 2. Below is a screen shot of multiplet template file /user/specified/directory/runbatman/batmaninput/multi_data_user.csv used for this test. The user can either change the indicated column to the same value as shown, or rename the file multi_data_user.csv used in Test 2 to a different one, and then change the file name from /user/specified/directory/runbatman/batmaninput/multi_data_user_adj.csv to /user/specified/directory/runbatman/batmaninput/multi_data_user.csv, so that batman will use the adjusted template parameters. Adjust the relative_intensity column to improve multiplet fitting. Refer to documentation for more detail. Plot all 6 spectra in a single stack plot use the same command as in Test 2: plotbatmanfitstack(bm,offset = 0.8,placeLegend = 'topleft',yto = 5) A stack plot of the fit result is shown below: (pdf file with the name specfit_stack_.pdf can be found in folder /user/specified/directory/runbatman/batmanoutput/date_time ) 7
8 Standardized Intensity N M R S pectra Original Spectrum Metabolites Fit Mirrored Wavelet Fit Fit Sum ppm It can be observed that the fit for some metabolites is improved (particularly nicotinic acid resonances around 8ppm). If you are able to generate the above results, you have correctly and successfully installed BATMAN! Test 4: Multiple spectra from bacterial supernatants data This test uses 31 spectra from bacterial supernatants. To reduce run time we fit a small ppm range corresponding to resonances from the metabolites glycine, valine, threonine and glucose. It will take about 1 minute per spectrum. The total run time depends on the number of cores (parallel processes) batman can use. The chemical shift sorting information is also provided with this test, details of how to get this information are explained in section 3: Chemical Shift Sorting. The test dataset and input files for batman are zipped in folder Test4.zip, which can be downloaded from Unzip this file, then: 1. Backup the 4 files: batmanoptions.txt, chemshiftperspec.csv, metaboliteslist.csv, and multi_data_user.csv in folder /user/specified/directory/runbatman/batmaninput, for example by giving them another name. (chemshiftperspec.csv may not exist if you haven t run this option before). 2. Move all the 5 files from the unzipped folder Test4 (include: batmanoptions.txt, chemshiftperspec.csv, metaboliteslist.csv, multi_data_user.csv, NMRdata_Test4.txt) into the input folder /user/specified/directory/runbatman/batmaninput. The user should check if the input for paraproc parameter in batmanoptions.txt is appropriate for your machine, which is set to 7 in the default file. In this case the 31 spectra take around 4 minutes to process. 8
9 Standardized Intensity paraproc - No of parallel processes (multicores) (only 1 core will be used for single spectrum): 7 Now run the same command used in previous tests with two additional input arguments: bm<-batman(runbatmandir = /user/specified/directory, txtfile = /user/specified/directory/runbatman/batmaninput/nmrdata_test4.txt', figbatmanfit = FALSE) Or bm<-batman(txtfile = './runbatman/batmaninput/nmrdata_test4.txt',figbatmanfit = FALSE) The user can leave the setting of figbatmanfit to its default (TRUE) by removing that argument from input, in which case 16 figures will be produced when batman fitting finishes. Plot all 31 spectra in a single stack plot as follows: plotbatmanfitstack(bm, offset = 0.4, yto = 15, overwrite = T, placelegend = 'topleft', orientation = "P") A stack plot of the fit result is shown below: (pdf file with the name specfit_stack_.pdf can be found in folder /user/specified/directory/runbatman/batmanoutput/date_time ) N M R S pectra Original Spectrum Metabolites Fit Mirrored Wavelet Fit Fit Sum ppm 3. Chemical Shift Sorting The SplineFitBATMAN is a MATLAB tool developed to work with BATMAN. The idea is to plot a stack of spectra, where the order is controlled by the chemical shift of a shifting peak. This often 9
10 shows a coherent shift across many peaks in the spectra allowing their positions to be estimated more clearly. Then a spline fitting tool allows the user to quickly interactively specify the position of each multiplet across a large number of spectra. This gives BATMAN accurate prior knowledge on the position of each resonance in each spectrum and can be extremely beneficial with peaks that shift a lot. Note that, when BATMAN is used with this information, peaks are still allowed to shift, according to the parameter rdelta (which can be controlled for individual multiplets in mulit_data.csv). The tool gets input from chemshiftperspec.csv and, NMRdata.txt (generated by BATMAN). The codes can be found in in SplineFitBATMAN_MATLABcode folder from: The csflag in batman options file should be set to 1 in order to use this tool. csflag - Specify chemical shift for each multiplet in each spectrum? (chemshiftperspectra.csv file) (Yes - 1 / No - 0): 1 At this stage, you should have finished Test 4, which uses the provided file chemshiftperspec.csv. Rename that file to a different name, and run batman again. The following error message (in red) will show. > bm<-batman(txtfile = './runbatman/batmaninput/nmrdata_test4.txt',figbatmanfit = FALSE) Running batman... Number of burn-in iterations: 6000 Number of post-burn-in iterations: 1000 The template file used is 2: The user input template of multiplets in multi_data_user.csv file. Loading multi_data_user.csv... Copying multiplet list from multi_data_user.csv to chemshiftperspec.csv... Error in batman(txtfile = "./runbatman/batmaninput/nmrdata_test4.txt", : No chemshiftperspec.csv file found in BatmanInput folder. Creating one now, please modify the values. A new chemshiftperspec.csv file should be created in batman input folder. Keep this file closed when using SplineFitBATMAN to write in it. Run the SplineFitBATMAN tool in Matlab, the following GUI should appear. 10
11 11
12 The figures below show how the tool can be used to fit a spline curve through the sorted stack plot to estimate the positions of shifting multiplets. The cross is where the cursor is. The red pentagrams are where a cursor clicked, the blue squares are the chemical shifts which will be output for use in BATMAN. Step 1: 1.1 The input for NMR spectra file should be.txt file generated by BATMAN. Calling "batman" once in R should generate NMRdata.txt in../runbatman/batmaninput folder which can be used here. 1.2 The input for chemical shift per spectra file, chemshiftperspec.csv can also be found in folder../runbatman/batmaninput. Keep this file closed when writing to file. 1.3 "Sort spectra using peak in ppm range": choose a peak to sort on, suggest an isolated but shifting peak. In this example (test 4 data) we suggest sorting on the histidine peak in the region ppm. 1.4 "Stack plot offset": sets the offset for stack plotting NMR spectra. Adjust the offset so that the multiplets of interest do not intersect with each other between spectra. This parameter can be modified for different multiplets. 1.5 Click Sort and Plot to plot the stack of spectra sorted according to chemical shift of the selected peak. Repeat steps until satisfied with the stack plot. 12
13 Step 2: 2.1 Choose a multiplet from the drop down menu, and then zoom in to the range containing that multiplet on the figure. This does not have to be the multiplet used in step 1.3 above. 2.2 The algorithm can output ppm positions found by a) simply fitting a spline between points clicked on the figure, b) finding the maximum intensity in the vicinity of the spline position (e.g. to find position of a singlet or triplet) or c) finding the minimum intensity in the vicinity of the spline position (e.g. to find position of a doublet). The parameter "ppm range for spline intersection" sets the search range on the left and right (+/-) of the intersection between spline and each spectrum. 2.3 Click "Select and Run". 2.4 Click on the points at which you wish to place the spline. You do not need to click on every spectrum in the stack. For example, a peak, which shifts smoothly across the stack, may only require 2 or 3 clicks to define the spline. Make sure to click on at least the FIRST and LAST spectra in stack plot each time. At the end, press return on the keyboard and the tool will draw the spline and the chemical shift positions to be output for use in BATMAN (blue squares). 2.5 If you are not satisfied with results, try changing the parameters including ppm range for spline intersection and method for locating ppm, and then click "Relocate chemical shift per spectrum" (no need to re-select points), or "Select and Run" to re-select points. 2.6 If satisfied with estimated positions, click "Save and Next", to save and go to next multiplet. 2.7 When finished, click "Save and Write to File" to save results to file chemshiftperspec.csv. Note that you do not need to do this for all multiplets or all metabolites. If no spectrum-specific chemical shifts are available for a given multiplet, BATMAN will use the default parameters specified in multi_data.csv (or multi_data_user.csv, as selected by the user). Troubleshooting: If not satisfied with results try: checking if the FIRST and LAST spectra were clicked, or choosing different method for locating ppm, or clicking on figure at more accurate points, or clicking more spectra on figure. Sometimes MATLAB does not register all clicks on the plot. Try leaving more time between clicks. If satisfied with fitting results, return to Test 4 to use the newly estimated chemical shifts in batman. References: 1. Viant, M.R., et al., International NMR-based environmental metabolomics intercomparison exercise. Environ Sci Technol, (1): p Behrends, V., et al., Metabolite profiling to characterize disease-related bacteria: gluconate excretion by Pseudomonas aeruginosa mutants and clinical isolates from cystic fibrosis patients. J Biol Chem, (21): p
Package batman. June 19, 2016
 Version 1.2.1.08 Author Jie Hao, William Astle, Maria De Iorio, Timothy Ebbels Maintainer Jie Hao Package batman June 19, 2016 Title Bayesian AuTomated Metabolite Analyser for NMR spectra
Version 1.2.1.08 Author Jie Hao, William Astle, Maria De Iorio, Timothy Ebbels Maintainer Jie Hao Package batman June 19, 2016 Title Bayesian AuTomated Metabolite Analyser for NMR spectra
Processing data with Bruker TopSpin
 Processing data with Bruker TopSpin This exercise has three parts: a 1D 1 H spectrum to baseline correct, integrate, peak-pick, and plot; a 2D spectrum to plot with a 1 H spectrum as a projection; and
Processing data with Bruker TopSpin This exercise has three parts: a 1D 1 H spectrum to baseline correct, integrate, peak-pick, and plot; a 2D spectrum to plot with a 1 H spectrum as a projection; and
How to analyze complex NMR signals through spin simulation/iteration
 How to analyze complex NMR signals through spin simulation/iteration 1. Introduction When a NMR signal s multiplicity exceeds the common doublet, triplet, quartet or doublet of doublet structure, analysis
How to analyze complex NMR signals through spin simulation/iteration 1. Introduction When a NMR signal s multiplicity exceeds the common doublet, triplet, quartet or doublet of doublet structure, analysis
NMR Users Guide Organic Chemistry Laboratory
 NMR Users Guide Organic Chemistry Laboratory Introduction The chemistry department is fortunate to have a high field (400 MHz) Nuclear Magnetic Resonance (NMR) spectrometer. You will be using this instrument
NMR Users Guide Organic Chemistry Laboratory Introduction The chemistry department is fortunate to have a high field (400 MHz) Nuclear Magnetic Resonance (NMR) spectrometer. You will be using this instrument
Chemical Shift Perturbation Tutorial
 CcpNmr Analysis Version 3 Chemical Shift Perturbation Tutorial Written by: Luca Mureddu CCPN application developer, PhD Student MCB, University of Leicester, Lancaster Road, Leicester, LE1 9HN, UK 1 t:
CcpNmr Analysis Version 3 Chemical Shift Perturbation Tutorial Written by: Luca Mureddu CCPN application developer, PhD Student MCB, University of Leicester, Lancaster Road, Leicester, LE1 9HN, UK 1 t:
NMR Assignments using NMRView I: Introduction to NMRView
 NMR Assignments using NMRView I: Introduction to NMRView For Mac OS X, you should have a subdirectory nmrview. At UGA this is /Users/bcmb8190/nmrview. The contents should look like this: -in the file hsqc.tcl
NMR Assignments using NMRView I: Introduction to NMRView For Mac OS X, you should have a subdirectory nmrview. At UGA this is /Users/bcmb8190/nmrview. The contents should look like this: -in the file hsqc.tcl
PROCESSING 2D SPECTRA USING VNMRJ JB Stothers NMR Facility Materials Science Addition 0216 Department of Chemistry Western University
 PROCESSING 2D SPECTRA USING VNMRJ JB Stothers NMR Facility Materials Science Addition 0216 Department of Chemistry Western University 1. INTRODUCTION...1 1.1. About this Worksheet... 1 1.2. A Very Brief
PROCESSING 2D SPECTRA USING VNMRJ JB Stothers NMR Facility Materials Science Addition 0216 Department of Chemistry Western University 1. INTRODUCTION...1 1.1. About this Worksheet... 1 1.2. A Very Brief
NMR Assignments using NMRView I: Introduction to NMRView
 NMR Assignments using NMRView I: Introduction to NMRView In your home directory, you should have a subdirectory called nmrview. The contents of that directory should be as shown below (subdirectories are
NMR Assignments using NMRView I: Introduction to NMRView In your home directory, you should have a subdirectory called nmrview. The contents of that directory should be as shown below (subdirectories are
An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data
 Supplementary material: User manual An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data Juan Prada, Manju
Supplementary material: User manual An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data Juan Prada, Manju
VNMRJ 4.2 INSTRUCTIONS: QANUC 500 FOR CHEMISTS
 VNMRJ 4.2 INSTRUCTIONS: QANUC 500 FOR CHEMISTS April 16, 2018 1. Sample preparation a. Tubes of any length can be used b. Make your samples 4 cm deep. They will not shim as well if they are shorter; you
VNMRJ 4.2 INSTRUCTIONS: QANUC 500 FOR CHEMISTS April 16, 2018 1. Sample preparation a. Tubes of any length can be used b. Make your samples 4 cm deep. They will not shim as well if they are shorter; you
Using OPUS to Process Evolved Gas Data (8/12/15 edits highlighted)
 Using OPUS to Process Evolved Gas Data (8/12/15 edits highlighted) Once FTIR data has been acquired for the gases evolved during your DSC/TGA run, you will process using the OPUS software package. Select
Using OPUS to Process Evolved Gas Data (8/12/15 edits highlighted) Once FTIR data has been acquired for the gases evolved during your DSC/TGA run, you will process using the OPUS software package. Select
Import and preprocessing of raw spectrum data
 BioNumerics Tutorial: Import and preprocessing of raw spectrum data 1 Aim Comprehensive tools for the import of spectrum data, both raw spectrum data as processed spectrum data are incorporated into BioNumerics.
BioNumerics Tutorial: Import and preprocessing of raw spectrum data 1 Aim Comprehensive tools for the import of spectrum data, both raw spectrum data as processed spectrum data are incorporated into BioNumerics.
TopSpin. Multiplet Analysis Tutorial
 TopSpin Multiplet Analysis Tutorial Contents Automatic Multiplet Definition Manual Single-Level Multiplet Definition Manual Multi-Level Multiplet Definition Working with Multiplets Report dialog and export
TopSpin Multiplet Analysis Tutorial Contents Automatic Multiplet Definition Manual Single-Level Multiplet Definition Manual Multi-Level Multiplet Definition Working with Multiplets Report dialog and export
North Carolina State University Department of Chemistry Varian NMR Training Manual
 North Carolina State University Department of Chemistry Varian NMR Training Manual by J.B. Clark IV & Dr. S. Sankar 1 st Edition 05/15/2009 Section 1: Essential Operations for Basic 1D Spectra Preparing
North Carolina State University Department of Chemistry Varian NMR Training Manual by J.B. Clark IV & Dr. S. Sankar 1 st Edition 05/15/2009 Section 1: Essential Operations for Basic 1D Spectra Preparing
GETTING STARTED. Welcome To Trilian!
 GETTING STARTED Welcome To Trilian! GETTING STARTED Quickstart This Quick Start provides an overview of the essential things you will need to know in order to get started using Trilian. Trilian is a software
GETTING STARTED Welcome To Trilian! GETTING STARTED Quickstart This Quick Start provides an overview of the essential things you will need to know in order to get started using Trilian. Trilian is a software
MRMPROBS tutorial. Hiroshi Tsugawa RIKEN Center for Sustainable Resource Science
 MRMPROBS tutorial Edited in 2014/10/6 Introduction MRMPROBS is a tool for the analysis of data from multiple reaction monitoring (MRM)- or selected reaction monitoring (SRM)-based metabolomics studies.
MRMPROBS tutorial Edited in 2014/10/6 Introduction MRMPROBS is a tool for the analysis of data from multiple reaction monitoring (MRM)- or selected reaction monitoring (SRM)-based metabolomics studies.
Instruction for Mass Spectra Viewer
 Instruction for Mass Spectra Viewer 1. A general description Basic functions Setup and Pre-process View, browser and undo Grouping, highlighting and labeling 2. Setup and Pre-process These procedures must
Instruction for Mass Spectra Viewer 1. A general description Basic functions Setup and Pre-process View, browser and undo Grouping, highlighting and labeling 2. Setup and Pre-process These procedures must
MestReC Cheat Sheet. by Monika Ivancic, July 1 st 2005
 MestReC Cheat Sheet by Monika Ivancic, July 1 st 2005 This Cheat Sheet is to be used at UW-Madison as a quick guide to processing using the MestReC NMR software. You may find more help at the MestReC homepage
MestReC Cheat Sheet by Monika Ivancic, July 1 st 2005 This Cheat Sheet is to be used at UW-Madison as a quick guide to processing using the MestReC NMR software. You may find more help at the MestReC homepage
Recording Your Audio and Creating Your MP3 File using Audacity
 http://www.larkin.net.au/ Page 1 Recording Your Audio and Creating Your MP3 File using Audacity Many people who are working with digital audio are choosing a program called Audacity for many reasons: 1.
http://www.larkin.net.au/ Page 1 Recording Your Audio and Creating Your MP3 File using Audacity Many people who are working with digital audio are choosing a program called Audacity for many reasons: 1.
IMSERC NMR MANUAL 02: Basic Processing of Varian 1D NMR Data
 IMSERC NMR MANUAL 02: Basic Processing of Varian 1D NMR Data Last updated: July 15, 2011 by Josh Kurutz This manual describes how to process NMR data on the offline processing computer in the IMSERC lab
IMSERC NMR MANUAL 02: Basic Processing of Varian 1D NMR Data Last updated: July 15, 2011 by Josh Kurutz This manual describes how to process NMR data on the offline processing computer in the IMSERC lab
CSImage Tutorial v August 2000
 CSImage Tutorial v 1.0 8 August 2000 To use CSImage you need to have Java2 installed. Development was done using JDK 1.2.2 from Sun Microsystems. The program has been run on Digital Unix (Compaq Tru64
CSImage Tutorial v 1.0 8 August 2000 To use CSImage you need to have Java2 installed. Development was done using JDK 1.2.2 from Sun Microsystems. The program has been run on Digital Unix (Compaq Tru64
UNIFIT - Spectrum Processing, Peak Fitting, Analysis and Presentation Software for XPS, AES, XAS and RAMAN Spectroscopy Based on WINDOWS
 UNIFIT - Spectrum Processing, Peak Fitting, Analysis and Presentation Software for XPS, AES, XAS and RAMAN Spectroscopy Based on WINDOWS UNIFIT FOR WINDOWS is an universal processing, analysis and presentation
UNIFIT - Spectrum Processing, Peak Fitting, Analysis and Presentation Software for XPS, AES, XAS and RAMAN Spectroscopy Based on WINDOWS UNIFIT FOR WINDOWS is an universal processing, analysis and presentation
Enhanced material contrast by dual-energy microct imaging
 Enhanced material contrast by dual-energy microct imaging Method note Page 1 of 12 2 Method note: Dual-energy microct analysis 1. Introduction 1.1. The basis for dual energy imaging Micro-computed tomography
Enhanced material contrast by dual-energy microct imaging Method note Page 1 of 12 2 Method note: Dual-energy microct analysis 1. Introduction 1.1. The basis for dual energy imaging Micro-computed tomography
User Guide for ModuLand Cytoscape plug-in
 User Guide for ModuLand Cytoscape plug-in Created for the ModuLand plug-in version 1.3 (April 2012) This user guide is based on the following publications, where the ModuLand method and its versions have
User Guide for ModuLand Cytoscape plug-in Created for the ModuLand plug-in version 1.3 (April 2012) This user guide is based on the following publications, where the ModuLand method and its versions have
Introduction to R. base -> R win32.exe (this will change depending on the latest version)
 Dr Raffaella Calabrese, Essex Business School 1. GETTING STARTED Introduction to R R is a powerful environment for statistical computing which runs on several platforms. R is available free of charge.
Dr Raffaella Calabrese, Essex Business School 1. GETTING STARTED Introduction to R R is a powerful environment for statistical computing which runs on several platforms. R is available free of charge.
Diffusion Ordered Spectroscopy (DOSY) Overview BRUKER last edit 5/14/12
 Diffusion Ordered Spectroscopy (DOSY) Overview BRUKER last edit 5/14/12 DOSY Diffusion Ordered SpectroscopY. NMR diffusion experiments, like DOSY, are used to determine the diffusion coefficients of solute
Diffusion Ordered Spectroscopy (DOSY) Overview BRUKER last edit 5/14/12 DOSY Diffusion Ordered SpectroscopY. NMR diffusion experiments, like DOSY, are used to determine the diffusion coefficients of solute
Package ASICS. January 23, 2018
 Type Package Package ASICS January 23, 2018 Title Automatic Statistical Identification in Complex Spectra Version 1.0.1 With a set of pure metabolite spectra, ASICS quantifies metabolites concentration
Type Package Package ASICS January 23, 2018 Title Automatic Statistical Identification in Complex Spectra Version 1.0.1 With a set of pure metabolite spectra, ASICS quantifies metabolites concentration
ClaNC: The Manual (v1.1)
 ClaNC: The Manual (v1.1) Alan R. Dabney June 23, 2008 Contents 1 Installation 3 1.1 The R programming language............................... 3 1.2 X11 with Mac OS X....................................
ClaNC: The Manual (v1.1) Alan R. Dabney June 23, 2008 Contents 1 Installation 3 1.1 The R programming language............................... 3 1.2 X11 with Mac OS X....................................
SMART NOTEBOOK: A semi-automated approach for the rapid assignment of triple resonance NMR spectra. Valerie Booth Brian Sykes
 SMART NOTEBOOK: A semi-automated approach for the rapid assignment of triple resonance NMR spectra. Programmer: Consultants: Robert Boyko Carolyn Slupsky Valerie Booth Brian Sykes Protein Engineering Network
SMART NOTEBOOK: A semi-automated approach for the rapid assignment of triple resonance NMR spectra. Programmer: Consultants: Robert Boyko Carolyn Slupsky Valerie Booth Brian Sykes Protein Engineering Network
Installing the Software
 Logic User s Guide Note: this is a new version to support the 1.1.0+ software. Please let us know if you can t find what you re looking for http://www.saleae.com/contact Contents Installing the Software...
Logic User s Guide Note: this is a new version to support the 1.1.0+ software. Please let us know if you can t find what you re looking for http://www.saleae.com/contact Contents Installing the Software...
NMRProcFlow Macro-command Reference Guide
 NMRProcFlow Macro-command Reference Guide This document is the reference guide of the macro-commands Daniel Jacob UMR 1332 BFP, Metabolomics Facility CGFB Bordeaux, MetaboHUB - 2018 1 NMRProcFlow - Macro-command
NMRProcFlow Macro-command Reference Guide This document is the reference guide of the macro-commands Daniel Jacob UMR 1332 BFP, Metabolomics Facility CGFB Bordeaux, MetaboHUB - 2018 1 NMRProcFlow - Macro-command
ProShake 2.0. The first step in this tutorial exercise is to start the ProShake 2.0 program. Click on the ProShake 2.0 icon to start the program.
 PROSHAKE 2.0 TUTORIAL The easiest way to learn the basics of ProShake s organization and operation is to complete the tutorial exercise detailed in this section. This tutorial will take you through nearly
PROSHAKE 2.0 TUTORIAL The easiest way to learn the basics of ProShake s organization and operation is to complete the tutorial exercise detailed in this section. This tutorial will take you through nearly
T 1 Relaxation Measurement: The Inversion-Recovery Experiment (Using IconNMR) Revised
 T 1 Relaxation Measurement: The Inversion-Recovery Experiment (Using IconNMR) Revised 4-21-2015 Relaxation times are divided into two types: longitudinal, which concerns change in magnetization along the
T 1 Relaxation Measurement: The Inversion-Recovery Experiment (Using IconNMR) Revised 4-21-2015 Relaxation times are divided into two types: longitudinal, which concerns change in magnetization along the
Geneious Microsatellite Plugin. Biomatters Ltd
 Geneious Microsatellite Plugin Biomatters Ltd November 24, 2018 2 Introduction This plugin imports ABI fragment analysis files and allows you to visualize traces, fit ladders, call peaks, predict bins,
Geneious Microsatellite Plugin Biomatters Ltd November 24, 2018 2 Introduction This plugin imports ABI fragment analysis files and allows you to visualize traces, fit ladders, call peaks, predict bins,
Tutorial for the CCPN Metabolomics project
 Tutorial for the CCPN Metabolomics project Francesca Chignola 1, Silvia Mari 1, Tim J. Stevens 2, Rasmus H. Fogh 2, Valeria Mannella 1, Wayne Boucher 2, Giovanna Musco 1 1. Biomolecular NMR Laboratory,
Tutorial for the CCPN Metabolomics project Francesca Chignola 1, Silvia Mari 1, Tim J. Stevens 2, Rasmus H. Fogh 2, Valeria Mannella 1, Wayne Boucher 2, Giovanna Musco 1 1. Biomolecular NMR Laboratory,
FIFI-LS: Basic Cube Analysis using SOSPEX
 FIFI-LS: Basic Cube Analysis using SOSPEX Date: 1 Oct 2018 Revision: - CONTENTS 1 INTRODUCTION... 1 2 INGREDIENTS... 1 3 INSPECTING THE CUBE... 3 4 COMPARING TO A REFERENCE IMAGE... 5 5 REFERENCE VELOCITY
FIFI-LS: Basic Cube Analysis using SOSPEX Date: 1 Oct 2018 Revision: - CONTENTS 1 INTRODUCTION... 1 2 INGREDIENTS... 1 3 INSPECTING THE CUBE... 3 4 COMPARING TO A REFERENCE IMAGE... 5 5 REFERENCE VELOCITY
USB 2.0 DISPLAY ADAPTER INSTALLATION GUIDE ON MAC
 USB 2.0 DISPLAY ADAPTER INSTALLATION GUIDE ON MAC 1 SYSTEM REQUIREMENTS Available USB 2.0 port Minimum requirement to use one USB 2.0 DISPLAY ADAPTER: CPU: Intel Core 2 Duo 2.4GHz RAM: 2GB memory 2~3 Displays
USB 2.0 DISPLAY ADAPTER INSTALLATION GUIDE ON MAC 1 SYSTEM REQUIREMENTS Available USB 2.0 port Minimum requirement to use one USB 2.0 DISPLAY ADAPTER: CPU: Intel Core 2 Duo 2.4GHz RAM: 2GB memory 2~3 Displays
Tutorial: De Novo Assembly of Paired Data
 : De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
Rat 2D EPSI Dual Band Variable Flip Angle 13 C Dynamic Spectroscopy
 Rat 2D EPSI Dual Band Variable Flip Angle 13 C Dynamic Spectroscopy In this example you will load a dynamic MRS animal data set acquired on a GE 3T scanner. This data was acquired with an EPSI sequence
Rat 2D EPSI Dual Band Variable Flip Angle 13 C Dynamic Spectroscopy In this example you will load a dynamic MRS animal data set acquired on a GE 3T scanner. This data was acquired with an EPSI sequence
R-software multims-toolbox. (User Guide)
 R-software multims-toolbox (User Guide) Pavel Cejnar 1, Štěpánka Kučková 2 1 Department of Computing and Control Engineering, Institute of Chemical Technology, Technická 3, 166 28 Prague 6, Czech Republic,
R-software multims-toolbox (User Guide) Pavel Cejnar 1, Štěpánka Kučková 2 1 Department of Computing and Control Engineering, Institute of Chemical Technology, Technická 3, 166 28 Prague 6, Czech Republic,
Tutorial 2: Analysis of DIA/SWATH data in Skyline
 Tutorial 2: Analysis of DIA/SWATH data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantification.
Tutorial 2: Analysis of DIA/SWATH data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantification.
ALGORITHM USER GUIDE FOR RVD
 ALGORITHM USER GUIDE FOR RVD The RVD program takes BAM files of deep sequencing reads in as input. Using a Beta-Binomial model, the algorithm estimates the error rate at each base position in the reference
ALGORITHM USER GUIDE FOR RVD The RVD program takes BAM files of deep sequencing reads in as input. Using a Beta-Binomial model, the algorithm estimates the error rate at each base position in the reference
Using Eclipse for Java. Using Eclipse for Java 1 / 1
 Using Eclipse for Java Using Eclipse for Java 1 / 1 Using Eclipse IDE for Java Development Download the latest version of Eclipse (Eclipse for Java Developers or the Standard version) from the website:
Using Eclipse for Java Using Eclipse for Java 1 / 1 Using Eclipse IDE for Java Development Download the latest version of Eclipse (Eclipse for Java Developers or the Standard version) from the website:
VNMRJ 4.2 INSTRUCTIONS: VARIAN MERCURY 400 AND VARIAN VNMRS 500
 VNMRJ 4.2 INSTRUCTIONS: VARIAN MERCURY 400 AND VARIAN VNMRS 500 August 9, 2016 1. Sample preparation a. Tubes of any length can be used b. Make your samples 4 cm deep. They will not shim as well if they
VNMRJ 4.2 INSTRUCTIONS: VARIAN MERCURY 400 AND VARIAN VNMRS 500 August 9, 2016 1. Sample preparation a. Tubes of any length can be used b. Make your samples 4 cm deep. They will not shim as well if they
Desktop & Laptop Edition
 Desktop & Laptop Edition USER MANUAL For Mac OS X Copyright Notice & Proprietary Information Redstor Limited, 2016. All rights reserved. Trademarks - Mac, Leopard, Snow Leopard, Lion and Mountain Lion
Desktop & Laptop Edition USER MANUAL For Mac OS X Copyright Notice & Proprietary Information Redstor Limited, 2016. All rights reserved. Trademarks - Mac, Leopard, Snow Leopard, Lion and Mountain Lion
Part 2 Downloading and installing templates and dummy patient records into Medical Director
 Part 2 Downloading and installing templates and dummy patient records into Medical Director Preparing a folder for the patient files 1. Close any programs and folders so that you have a clear view of your
Part 2 Downloading and installing templates and dummy patient records into Medical Director Preparing a folder for the patient files 1. Close any programs and folders so that you have a clear view of your
SIVIC GUI Overview. SIVIC GUI Layout Overview
 SIVIC GUI Overview SIVIC GUI Layout Overview At the top of the SIVIC GUI is a row of buttons called the Toolbar. It is a quick interface for loading datasets, controlling how the mouse manipulates the
SIVIC GUI Overview SIVIC GUI Layout Overview At the top of the SIVIC GUI is a row of buttons called the Toolbar. It is a quick interface for loading datasets, controlling how the mouse manipulates the
Setting up Eclipse (Windows), or GCC (Mac) Setting up Atmel Studio (Windows & Mac)
 ECE3411 Fall 2015 Lecture # 0 Setting up Eclipse (Windows), or GCC (Mac) Setting up Atmel Studio (Windows & Mac) Marten van Dijk, Syed Kamran Haider Department of Electrical & Computer Engineering University
ECE3411 Fall 2015 Lecture # 0 Setting up Eclipse (Windows), or GCC (Mac) Setting up Atmel Studio (Windows & Mac) Marten van Dijk, Syed Kamran Haider Department of Electrical & Computer Engineering University
14 - Multiple Files and Folders Dragging and dropping File name collisions revisited
 14 - Multiple Files and Folders In the last lesson, we saw how to use the context menu or the ribbon to copy and move files on our hard drive. In this lesson, we will review and build on those skills as
14 - Multiple Files and Folders In the last lesson, we saw how to use the context menu or the ribbon to copy and move files on our hard drive. In this lesson, we will review and build on those skills as
Tutorial to QuotationFinder_0.6
 Tutorial to QuotationFinder_0.6 What is QuotationFinder, and for which purposes can it be used? QuotationFinder is a tool for the automatic comparison of fully digitized texts. It can detect quotations,
Tutorial to QuotationFinder_0.6 What is QuotationFinder, and for which purposes can it be used? QuotationFinder is a tool for the automatic comparison of fully digitized texts. It can detect quotations,
Characterizing dissolved organic matter fluorescence with parallel factor analysis: a tutorial
 Limnol. Oceanogr.: Methods 6, 2008 2008, by the American Society of Limnology and Oceanography, Inc. Characterizing dissolved organic matter fluorescence with parallel factor analysis: a tutorial Colin
Limnol. Oceanogr.: Methods 6, 2008 2008, by the American Society of Limnology and Oceanography, Inc. Characterizing dissolved organic matter fluorescence with parallel factor analysis: a tutorial Colin
USB 3.0 DISPLAY ADAPTER INSTALLATION GUIDE ON MAC
 USB 3.0 DISPLAY ADAPTER INSTALLATION GUIDE ON MAC 1 SYSTEM REQUIREMENTS Available USB 2.0/ USB 3.0 port (USB 3.0 is recommended) Minimum requirement to use one USB Display Adapter: CPU: Intel Core 2 Duo
USB 3.0 DISPLAY ADAPTER INSTALLATION GUIDE ON MAC 1 SYSTEM REQUIREMENTS Available USB 2.0/ USB 3.0 port (USB 3.0 is recommended) Minimum requirement to use one USB Display Adapter: CPU: Intel Core 2 Duo
Appendix I. TACTICS Toolbox v3.x. Interactive MATLAB Platform For Bioimaging informatics. User Guide TRACKING MODULE
 TACTICS Toolbox v3.x Interactive MATLAB Platform For Bioimaging informatics User Guide TRACKING MODULE -17- Cell Tracking Module 1 (user interface) Once the images were successfully segmented, the next
TACTICS Toolbox v3.x Interactive MATLAB Platform For Bioimaging informatics User Guide TRACKING MODULE -17- Cell Tracking Module 1 (user interface) Once the images were successfully segmented, the next
Spectrum Assignment using Ansig Ansig Setup
 Spectrum Assignment using Ansig Ansig Setup First you will go through the backbone assignment procedure. Go into the ansig directory. This contains several files: 1. The embo_back.ctr defines the al procedure
Spectrum Assignment using Ansig Ansig Setup First you will go through the backbone assignment procedure. Go into the ansig directory. This contains several files: 1. The embo_back.ctr defines the al procedure
Tutorial. De Novo Assembly of Paired Data. Sample to Insight. November 21, 2017
 De Novo Assembly of Paired Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
De Novo Assembly of Paired Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Agilent MicroLab Quant Calibration Software: Measure Oil in Water using Method IP 426
 Agilent MicroLab Quant Calibration Software: Measure Oil in Water using Method IP 426 Application Note Environmental Authors John Seelenbinder and Dipak Mainali Agilent Technologies, Inc. Introduction
Agilent MicroLab Quant Calibration Software: Measure Oil in Water using Method IP 426 Application Note Environmental Authors John Seelenbinder and Dipak Mainali Agilent Technologies, Inc. Introduction
Creating a DVD with menu from an existing DVD using DVDLab Pro
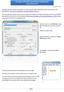 Creating a DVD with menu from an existing DVD using DVDLab Pro September 24, 2010 This guide has been tested with DVDLab Pro 2.51 (compiler 4.215). Evaluation version can be found in the following link:
Creating a DVD with menu from an existing DVD using DVDLab Pro September 24, 2010 This guide has been tested with DVDLab Pro 2.51 (compiler 4.215). Evaluation version can be found in the following link:
1.0 The System Architecture and Design Features
 1.0 The System Architecture and Design Features Figure 1. System Architecture The overall guiding design philosophy behind the Data Capture and Logging System Architecture is to have a clean design that
1.0 The System Architecture and Design Features Figure 1. System Architecture The overall guiding design philosophy behind the Data Capture and Logging System Architecture is to have a clean design that
MNova Version Installation Instructions.
 MNova Version 12.0.0 Installation Instructions. Note: This Document contains information for a new NMR user. Carefully read and accomplish each section before moving to the next. If you can t finish a
MNova Version 12.0.0 Installation Instructions. Note: This Document contains information for a new NMR user. Carefully read and accomplish each section before moving to the next. If you can t finish a
OCT Segmentation Manual Jens Kowal, Pascal Dufour This manual gives a quick introduction into the OCT Segmentation application in particular it
 OCT Segmentation Manual Jens Kowal, Pascal Dufour This manual gives a quick introduction into the OCT Segmentation application in particular it explains how to prepare the OCT data in order to be loaded
OCT Segmentation Manual Jens Kowal, Pascal Dufour This manual gives a quick introduction into the OCT Segmentation application in particular it explains how to prepare the OCT data in order to be loaded
Adobe illustrator Introduction
 Adobe illustrator Introduction This document was prepared by Luke Easterbrook 2013 1 Summary This document is an introduction to using adobe illustrator for scientific illustration. The document is a filleable
Adobe illustrator Introduction This document was prepared by Luke Easterbrook 2013 1 Summary This document is an introduction to using adobe illustrator for scientific illustration. The document is a filleable
GPR Analyzer version 1.23 User s Manual
 GPR Analyzer version 1.23 User s Manual GPR Analyzer is a tool to quickly analyze multi- species microarrays, especially designed for use with the MIDTAL (Microarray Detection of Toxic ALgae) chip. It
GPR Analyzer version 1.23 User s Manual GPR Analyzer is a tool to quickly analyze multi- species microarrays, especially designed for use with the MIDTAL (Microarray Detection of Toxic ALgae) chip. It
OCAP: An R package for analysing itraq data.
 OCAP: An R package for analysing itraq data. Penghao Wang, Pengyi Yang, Yee Hwa Yang 10, January 2012 Contents 1. Introduction... 2 2. Getting started... 2 2.1 Download... 2 2.2 Install to R... 2 2.3 Load
OCAP: An R package for analysing itraq data. Penghao Wang, Pengyi Yang, Yee Hwa Yang 10, January 2012 Contents 1. Introduction... 2 2. Getting started... 2 2.1 Download... 2 2.2 Install to R... 2 2.3 Load
English. Image management software for the EyeSpecial C-II
 English Image management software for the EyeSpecial C-II SureFile Overview... 1 How to activate SureFile... 3 Instructions for use... 4 Reference... 11 Specifications... 14 Ver.1.3 SureFile Overview SureFile
English Image management software for the EyeSpecial C-II SureFile Overview... 1 How to activate SureFile... 3 Instructions for use... 4 Reference... 11 Specifications... 14 Ver.1.3 SureFile Overview SureFile
To get started download the dataset, unzip files, start Maven, and follow steps below.
 Getting Started. This document provides basic overview of Maven functionality. For the purpose of demonstration we will use an example CMV Viral Infection time course dataset. (Dataset Download). There
Getting Started. This document provides basic overview of Maven functionality. For the purpose of demonstration we will use an example CMV Viral Infection time course dataset. (Dataset Download). There
Spectroscopic Analysis: Peak Detector
 Electronics and Instrumentation Laboratory Sacramento State Physics Department Spectroscopic Analysis: Peak Detector Purpose: The purpose of this experiment is a common sort of experiment in spectroscopy.
Electronics and Instrumentation Laboratory Sacramento State Physics Department Spectroscopic Analysis: Peak Detector Purpose: The purpose of this experiment is a common sort of experiment in spectroscopy.
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
Performer to DP2 Hot Folder Reference Manual Rev There is only one file involved with installing the Performer to DP2 Hot Folder.
 Performer to DP2 Hot Folder Reference Manual Rev. 07.11.05 Install Files: There is only one file involved with installing the Performer to DP2 Hot Folder. The installer file is named PP2DP2_1.x.x.EXE.
Performer to DP2 Hot Folder Reference Manual Rev. 07.11.05 Install Files: There is only one file involved with installing the Performer to DP2 Hot Folder. The installer file is named PP2DP2_1.x.x.EXE.
WaveAR User s Manual. Blake J. Landry and Matthew J. Hancock. WaveAR User s Manual page 1 of 19
 WaveAR User s Manual by Blake J. Landry and Matthew J. Hancock WaveAR User s Manual page 1 of 19 WaveAR User s Manual Contents 1. Introduction... 3 2. Installation... 3 2.1 Standalone executable... 3 2.2
WaveAR User s Manual by Blake J. Landry and Matthew J. Hancock WaveAR User s Manual page 1 of 19 WaveAR User s Manual Contents 1. Introduction... 3 2. Installation... 3 2.1 Standalone executable... 3 2.2
In this exercise, you will convert labels into geodatabase annotation so you can edit the text features.
 Instructions: Use the provided data stored in a USB. For the report: 1. Start a new word document. 2. Follow an exercise step as given below. 3. Describe what you did in that step in the word document
Instructions: Use the provided data stored in a USB. For the report: 1. Start a new word document. 2. Follow an exercise step as given below. 3. Describe what you did in that step in the word document
Protein Deconvolution Quick Start Guide
 Protein Deconvolution Quick Start Guide The electrospray ionization (ESI) of intact peptides and proteins produces mass spectra containing a series of multiply charged ions with associated mass-to-charge
Protein Deconvolution Quick Start Guide The electrospray ionization (ESI) of intact peptides and proteins produces mass spectra containing a series of multiply charged ions with associated mass-to-charge
SELECTION OF A MULTIVARIATE CALIBRATION METHOD
 SELECTION OF A MULTIVARIATE CALIBRATION METHOD 0. Aim of this document Different types of multivariate calibration methods are available. The aim of this document is to help the user select the proper
SELECTION OF A MULTIVARIATE CALIBRATION METHOD 0. Aim of this document Different types of multivariate calibration methods are available. The aim of this document is to help the user select the proper
Fitting NMR peaks for N,N DMA
 Fitting NMR peaks for N,N DMA Importing the FID file to your local system Any ftp program may be used to transfer the FID file from the NMR computer. The description below will take you through the process
Fitting NMR peaks for N,N DMA Importing the FID file to your local system Any ftp program may be used to transfer the FID file from the NMR computer. The description below will take you through the process
Icare CLINIC Icare EXPORT Icare PATIENT app INSTRUCTION MANUAL FOR HEALTHCARE PROFESSIONALS ENGLISH
 Icare CLINIC Icare EXPORT Icare PATIENT app INSTRUCTION MANUAL FOR HEALTHCARE PROFESSIONALS ENGLISH Icare CLINIC (Type: TS02), Icare EXPORT (Type: TS03) and Icare PATIENT (Type: TS04) app - Instruction
Icare CLINIC Icare EXPORT Icare PATIENT app INSTRUCTION MANUAL FOR HEALTHCARE PROFESSIONALS ENGLISH Icare CLINIC (Type: TS02), Icare EXPORT (Type: TS03) and Icare PATIENT (Type: TS04) app - Instruction
REF2020 open access policy
 REF2020 open access policy File formats required for the evidence of the acceptance date and how to produce them REF2020 open access policy requires authors to upload their final peer reviewed and accepted
REF2020 open access policy File formats required for the evidence of the acceptance date and how to produce them REF2020 open access policy requires authors to upload their final peer reviewed and accepted
TraceFinder Analysis Quick Reference Guide
 TraceFinder Analysis Quick Reference Guide This quick reference guide describes the Analysis mode tasks assigned to the Technician role in the Thermo TraceFinder 3.0 analytical software. For detailed descriptions
TraceFinder Analysis Quick Reference Guide This quick reference guide describes the Analysis mode tasks assigned to the Technician role in the Thermo TraceFinder 3.0 analytical software. For detailed descriptions
Multivariate Calibration Quick Guide
 Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Tutorial 7: Automated Peak Picking in Skyline
 Tutorial 7: Automated Peak Picking in Skyline Skyline now supports the ability to create custom advanced peak picking and scoring models for both selected reaction monitoring (SRM) and data-independent
Tutorial 7: Automated Peak Picking in Skyline Skyline now supports the ability to create custom advanced peak picking and scoring models for both selected reaction monitoring (SRM) and data-independent
25 October :28. User Manual
 25 October 2018 11:28 User Manual Requirements Requirements Software and hardware requirements Windows PC OS version Window 7, Windows 8, Windows 10 CPU 2GhHz with SSE (2.5GHz multicore system recommended)
25 October 2018 11:28 User Manual Requirements Requirements Software and hardware requirements Windows PC OS version Window 7, Windows 8, Windows 10 CPU 2GhHz with SSE (2.5GHz multicore system recommended)
Gegenees genome format...7. Gegenees comparisons...8 Creating a fragmented all-all comparison...9 The alignment The analysis...
 User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
How To Changing Album Name On Macbook Pro Home Folder
 How To Changing Album Name On Macbook Pro Home Folder One or more iphoto album names are not listed in the Photos tab in itunes. Check to see if you've accidentally moved your iphoto Library file to another
How To Changing Album Name On Macbook Pro Home Folder One or more iphoto album names are not listed in the Photos tab in itunes. Check to see if you've accidentally moved your iphoto Library file to another
1. Open up PRO-DESKTOP from your programmes menu. Then click on the file menu > new> design.
 Radio Tutorial Draw your spatula shape by:- 1. Open up PRO-DESKTOP from your programmes menu. Then click on the file menu > new> design. 2. The new design window will now open. Double click on design 1
Radio Tutorial Draw your spatula shape by:- 1. Open up PRO-DESKTOP from your programmes menu. Then click on the file menu > new> design. 2. The new design window will now open. Double click on design 1
USB DISPLAY ADAPTER QUICK INSTALLATION GUIDE JUA210 / JUA230 / JUA250
 USB DISPLAY ADAPTER QUICK INSTALLATION GUIDE JUA210 / JUA230 / JUA250 GETTING STARTED Installation on Windows 1. Insert the Driver CD into the drive; the setup program should launch automatically. Make
USB DISPLAY ADAPTER QUICK INSTALLATION GUIDE JUA210 / JUA230 / JUA250 GETTING STARTED Installation on Windows 1. Insert the Driver CD into the drive; the setup program should launch automatically. Make
The power of centralized computing at your fingertips
 Pinnacle 3 Professional The power of centralized computing at your fingertips Philips Pinnacle 3 Professional specifications The power of centralized computing in a scalable offering for mid-size clinics
Pinnacle 3 Professional The power of centralized computing at your fingertips Philips Pinnacle 3 Professional specifications The power of centralized computing in a scalable offering for mid-size clinics
Basic 1D Processing. Opening Saved Data. Start the VnmrJ software. Click File > Open.
 Basic 1D Processing Opening Saved Data Start the VnmrJ software. Click File > Open. This will open a pop-up window. Clicking Home will take you to the data directory within your account. You can create
Basic 1D Processing Opening Saved Data Start the VnmrJ software. Click File > Open. This will open a pop-up window. Clicking Home will take you to the data directory within your account. You can create
MAC ACS CLIENT SOFTWARE USER MANUAL
 MAC ACS CLIENT SOFTWARE USER MANUAL 1 MAC ACS USER GUIDE 1.1 System Requirement Recommended System Requirement OS CPU VGA RAM HDD OS CPU VGA RAM HDD Mac OS X 10.6(snow leopard) Built-in Intel CPU MAC Recommended
MAC ACS CLIENT SOFTWARE USER MANUAL 1 MAC ACS USER GUIDE 1.1 System Requirement Recommended System Requirement OS CPU VGA RAM HDD OS CPU VGA RAM HDD Mac OS X 10.6(snow leopard) Built-in Intel CPU MAC Recommended
PLEASE NOTE THAT THERE ARE QUESTIONS IN SOME OF THESE SECTIONS THAT ARE TO BE TURNED IN AS HOMEWORK.
 Seismic Reflection Lab for Geology 594 BEFORE YOU DO ANYTHING INSERT THE DONGLE ON THE LEFT SIDE OF THE KEYBOARD (LIGHT SIDE UP). The little green light will come on showing the dongle is activated. You
Seismic Reflection Lab for Geology 594 BEFORE YOU DO ANYTHING INSERT THE DONGLE ON THE LEFT SIDE OF THE KEYBOARD (LIGHT SIDE UP). The little green light will come on showing the dongle is activated. You
TOPSPINPLOT(XWINPLOT)
 Mississippi NMR Workshop TOPSPINPLOT(XWINPLOT) Easy and Simple Plotting Written by: Mike Brown Version 01092006.meb Bruker South Training Center Topics Covered Definitions Starting XWINPLOT Layouts Plot
Mississippi NMR Workshop TOPSPINPLOT(XWINPLOT) Easy and Simple Plotting Written by: Mike Brown Version 01092006.meb Bruker South Training Center Topics Covered Definitions Starting XWINPLOT Layouts Plot
EPBscore user guide. Requirements:
 EPBscore user guide Requirements: Up to Matlab 2014a. Windows-only versions (both 32- and 64-bit) were tested. Toolboxes: Control, Images, Local, Matlab, Optim, Shared, Signal, Simulink, Stats, Wavelet
EPBscore user guide Requirements: Up to Matlab 2014a. Windows-only versions (both 32- and 64-bit) were tested. Toolboxes: Control, Images, Local, Matlab, Optim, Shared, Signal, Simulink, Stats, Wavelet
ADVANCED DATA PROCESSING
 ADVANCED DATA PROCESSING Making Backups The NMR lab periodically backs up data on CD-ROMs for your convenience ([Files] [Oldfids]). If you would like your own personal backups, you can do this via the
ADVANCED DATA PROCESSING Making Backups The NMR lab periodically backs up data on CD-ROMs for your convenience ([Files] [Oldfids]). If you would like your own personal backups, you can do this via the
Issues in MCMC use for Bayesian model fitting. Practical Considerations for WinBUGS Users
 Practical Considerations for WinBUGS Users Kate Cowles, Ph.D. Department of Statistics and Actuarial Science University of Iowa 22S:138 Lecture 12 Oct. 3, 2003 Issues in MCMC use for Bayesian model fitting
Practical Considerations for WinBUGS Users Kate Cowles, Ph.D. Department of Statistics and Actuarial Science University of Iowa 22S:138 Lecture 12 Oct. 3, 2003 Issues in MCMC use for Bayesian model fitting
Code::Blocks Student Manual
 Code::Blocks Student Manual Lawrence Goetz, Network Administrator Yedidyah Langsam, Professor and Theodore Raphan, Distinguished Professor Dept. of Computer and Information Science Brooklyn College of
Code::Blocks Student Manual Lawrence Goetz, Network Administrator Yedidyah Langsam, Professor and Theodore Raphan, Distinguished Professor Dept. of Computer and Information Science Brooklyn College of
ECO375 Tutorial 1 Introduction to Stata
 ECO375 Tutorial 1 Introduction to Stata Matt Tudball University of Toronto Mississauga September 14, 2017 Matt Tudball (University of Toronto) ECO375H5 September 14, 2017 1 / 25 What Is Stata? Stata is
ECO375 Tutorial 1 Introduction to Stata Matt Tudball University of Toronto Mississauga September 14, 2017 Matt Tudball (University of Toronto) ECO375H5 September 14, 2017 1 / 25 What Is Stata? Stata is
10kTrees - Exercise #2. Viewing Trees Downloaded from 10kTrees: FigTree, R, and Mesquite
 10kTrees - Exercise #2 Viewing Trees Downloaded from 10kTrees: FigTree, R, and Mesquite The goal of this worked exercise is to view trees downloaded from 10kTrees, including tree blocks. You may wish to
10kTrees - Exercise #2 Viewing Trees Downloaded from 10kTrees: FigTree, R, and Mesquite The goal of this worked exercise is to view trees downloaded from 10kTrees, including tree blocks. You may wish to
ANALYSIS OF PULMONARY FIBROSIS IN MRI, USING AN ELASTIC REGISTRATION TECHNIQUE IN A MODEL OF FIBROSIS: Scleroderma
 ANALYSIS OF PULMONARY FIBROSIS IN MRI, USING AN ELASTIC REGISTRATION TECHNIQUE IN A MODEL OF FIBROSIS: Scleroderma ORAL DEFENSE 8 th of September 2017 Charlotte MARTIN Supervisor: Pr. MP REVEL M2 Bio Medical
ANALYSIS OF PULMONARY FIBROSIS IN MRI, USING AN ELASTIC REGISTRATION TECHNIQUE IN A MODEL OF FIBROSIS: Scleroderma ORAL DEFENSE 8 th of September 2017 Charlotte MARTIN Supervisor: Pr. MP REVEL M2 Bio Medical
Year 8. Revision Exercise April Computer Science CARDINAL NEWMAN CATHOLIC SCHOOL. Student Name : Subject Teacher : Tutor Group:
 CARDINAL NEWMAN CATHOLIC SCHOOL Computer Science Year 8 Revision Exercise April 2017 Student Name : Subject Teacher : Tutor Group: Page 1 of 17 Section A: File Management 1. Develop some materials to advise
CARDINAL NEWMAN CATHOLIC SCHOOL Computer Science Year 8 Revision Exercise April 2017 Student Name : Subject Teacher : Tutor Group: Page 1 of 17 Section A: File Management 1. Develop some materials to advise
Living Image. Release Notes Version Purpose. 2. New Features. FLIT Algorithm Improvements. Single-Click FLIT and DLIT Reconstructions
 Living Image Release Notes Version 4.5 1. Purpose This document provides a brief overview of new features and improvements in Living Image 4.5. This release serves as an update for Living Image 4.4 on
Living Image Release Notes Version 4.5 1. Purpose This document provides a brief overview of new features and improvements in Living Image 4.5. This release serves as an update for Living Image 4.4 on
Direct Infusion Mass Spectrometry Processing (DIMaSP) Instructions for use
 Direct Infusion Mass Spectrometry Processing (DIMaSP) Instructions for use The following instructions assume the user has a batch of paired sample and blank spectra that have been exported to a comma-separated
Direct Infusion Mass Spectrometry Processing (DIMaSP) Instructions for use The following instructions assume the user has a batch of paired sample and blank spectra that have been exported to a comma-separated
Rubik s Cube in SketchUp
 This project shows how to start with one cube, and use it to build a Rubik s cube, which you can spin and try to solve. For this project, it helps to have some basic knowledge of SketchUp (though detailed
This project shows how to start with one cube, and use it to build a Rubik s cube, which you can spin and try to solve. For this project, it helps to have some basic knowledge of SketchUp (though detailed
GIS LAB 1. Basic GIS Operations with ArcGIS. Calculating Stream Lengths and Watershed Areas.
 GIS LAB 1 Basic GIS Operations with ArcGIS. Calculating Stream Lengths and Watershed Areas. ArcGIS offers some advantages for novice users. The graphical user interface is similar to many Windows packages
GIS LAB 1 Basic GIS Operations with ArcGIS. Calculating Stream Lengths and Watershed Areas. ArcGIS offers some advantages for novice users. The graphical user interface is similar to many Windows packages
