Parallel Network Motif Finding Michael Schatz, Elliott Cooper-Balis, Adam Bazinet CMSC 714 High Performance Computing
|
|
|
- Paula Lindsey
- 6 years ago
- Views:
Transcription
1 Parallel Network Motif Finding Michael Schatz, Elliott Cooper-Balis, Adam Bazinet CMSC 714 High Performance Computing Abstract Network motifs are over-represented patterns within a network, and signify the fundamental building blocks of that network. The process of finding network motifs is closely related to the traditional subgraph isomorphism problem in computer science, which finds instances of a particular subgraph in a graph. This problem has been proven NP-complete, and thus even for relatively small subgraphs and graphs, today s most efficient approaches require a large amount of computation time. Here we present parallel algorithms for network motif finding, including both query parallelization, where different subgraphs are searched for in parallel, and network parallelization, where the original network is partitioned into overlapping regions, and a single subgraph is searched for in parallel among the different regions. The network partitioning algorithm is a novel application of hierarchical clustering and divides the network into strongly connected components, while minimizing the overlap between them. Our results show query parallel network motif finding speeds up the computation time linearly with the number of workers, and allows us to systematically search for larger motifs than ever before. In addition, our novel network partitioning algorithms are effective at partitioning the network, which enables network parallelization to effectively reduce the time required to count instances of a particular subgraph. 1. Introduction In 2002, Milo et al. spurred interest in the field of network motif analysis [1] by discovering that the same motifs are present in networks that originate from highly diverse sources. A network motif is a pattern (subgraph) of a larger network that occurs at a much higher frequency than is expected by chance. These patterns often have specific functional roles, and form basic building blocks of all complex networks. Milo et al. searched for overrepresented motifs in a variety of biological and technological networks, including gene regulation networks in yeast, neural connections within the nematode worm, food webs in different environments, electronic circuit designs, inter-page hyperlinks on the World Wide Web, and connections between routers on the Internet. The specific network motifs discovered in the different networks varied, and the authors hypothesize the particular networks motifs discovered in each network reveal the fundamental properties of that complex network. For example, the feed-forward loop between 3 nodes: A->B, B->C, A->C (configuration 5 in Figure 1), was commonly found in electronic circuit design, as it requires the signal at A to be persistent to activate C via B, and thus dampens noise at A. Surprisingly, though, it was also overrepresented in gene regulation networks, suggesting gene regulation is tolerant to noise using the same mechanism as in an electronic circuit. Recent work has searched for motifs in networks called protein-protein interaction (PPI) networks, in which nodes represent proteins in an organism, and (undirected) edges connect proteins that interact within an organism. These networks typically have several thousand nodes and tens of thousands of edges, but the exact numbers vary according to the complexity of the organism. PPI networks are publicly available for several model organisms and were built using high throughput laboratory methods that test for molecular interactions between all pairs of proteins. This work has discovered several statistically and biologically important
2 motifs [2]. For example, signaling pathways within a cell will be present as long chains of connections, while protein complexes will be represented as a clique. The method Milo et al. used for discovering overrepresented network motifs was to count the number of instances of all possible 3 and 4 node subgraphs in the target network, and then do the same in randomized versions of the target network. Afterwards, they report the motifs that were statistically overrepresented in the target network when compared to the random networks. For example, there are 13 possible 3-node subgraphs to search for in a directed graph (Figure 1). Milo s algorithm for counting possible motifs was a brute force search of all 3 and 4 node subgraphs in the target networks. This algorithm is so computationally expensive that only very small motifs (n<=4) could be analyzed in a reasonable amount of time, but large motifs are also of great interest. Figure 1. All possible 3 node directed graphs In a follow-up paper published in 2007, Grochow and Kellis [3] present a dramatically faster algorithm for network motif discovery. This is the fastest known serial algorithm for network motif discovery. Unlike the Milo algorithm, which counts instances of all possible 3 or 4 node graphs simultaneously, the Grochow and Kellis algorithm takes as input a single query graph and counts the number of instances of it in the network with a very efficient method. To reproduce Milo s analysis, one would simply run the Grochow and Kellis algorithm on all possible query motifs up to a given size in the target network and in several random networks. Their algorithm directly addresses the classic subgraph isomorphism problem, which is known to be NP-complete. Even so, their algorithm achieves an exponential speedup over the brute force approach using algorithmic techniques they call subgraph enumeration and symmetry breaking. These optimizations help to avoid rediscovering automorphisms of a previously discovered subgraph, such as the rotation of labels in a clique. Despite the exponential speedup over the brute force approach, the running time for the algorithm is still exponential in the number of nodes in the query graph, and searching all possible query graphs up to 7 nodes requires hours of computation per network. Consequently, there had been no attempt to exhaustively study larger motifs. However, since their algorithm counts instances of a single query motif in the larger network, the authors state their algorithm is amenable to parallelization by having different processing units search for instances of different motifs. We explored their remark and implemented both query parallelization, where a single processing unit processes a single query subgraph in the full target network as was proposed by the authors, and network parallelization, where the target network is partitioned into overlapping regions which are searched in parallel for a single query subgraph. Query parallelization is embarrassingly parallel, since the different queries are completely independent and require no inter-processor communication. In contrast, network parallelization requires coordination to partition and distribute the network intelligently, and to merge the results without over-counting instances of a subgraph in the overlapping regions. This is especially challenging for the biological networks, which are non-planar and have low effective diameter. 2. Methods Grochow and Kellis made their source code available under the condition that we did not distribute their code without their permission. The subgraph isomorphism code is approximately 10,000 lines of Java, not including the graph and ancillary libraries. Therefore, we decided to implement our solution in Java given the time available for the project. We selected Parallel Java [4] for inter-processor communication for our project, since it appears to be the most actively developed message passing library for Java. It supports both OpenMP-style fork-join parallelism for SMP machines, and MPIstyle SPMD parallelism for clusters of machines.
3 Parallel Java is packaged as a single JAR file to be included in the CLASSPATH of the parallel application, and uses ssh to launch processes from a frontend processor. Inter-node communication is handled through a Comm class that provides MPIstyle primitives such as send/receive, and size/ rank. Primitive data types are transferred natively, and Java objects can be transferred using serialization techniques. 2.1 Query Parallelization Query parallelization is useful for performing a high throughput search for network motifs by counting instances of every possible subgraph up to a given size. Every processing unit gets a full copy of the target network, and searches for 1 or more queries. The queries are independent, but the search time per query varies considerably depending on the number of nodes and edges in the query subgraph. Therefore, the total runtime depends on how the queries are scheduled. To address this, we implemented 2 scheduling policies: naïve and first-fit. Under the naïve policy, the full list of S possible subgraphs is precomputed and each of the P processing units is assigned a block of size S/P subgraphs to evaluate. This scheduler has very low overhead, and the master node simply sends a range of queries to each processing unit. However, this scheduler could have large load imbalance if a given worker gets a block of unusually complicated subgraphs. As an alternative, we also implemented a first-fit scheduler, where the master distributes queries one at a time to workers. This ensures each work is always busy, (except possibly for the last P-1 queries), but has higher overhead since workers must request work after every subgraph. 2.2 Network Parallelization In contrast to query parallelization, network parallelization is primarily useful for speeding up the search of a single subgraph. It may even be necessary to use network parallelization to make the problem feasible, such as when the target network is too large to fit into main memory on a single processing unit. Even when the entire graph would fit into memory, network parallelization may have significant cache performance benefits from increased locality in the search. Network parallelization, however, also has additional overhead. It has to partition the network into different regions, distribute the partitions and merge the results from each worker. The partitioning can potentially be reused for several query subgraphs, so the time to partition can be amortized over the full execution time. However, the regions of the network are not independent, and a particular instance of a subgraph may be present in the overlap between multiple regions. Therefore each instance of the subgraph must be examined to avoid over-counting the total number of instances Network Partitioning The success of network parallelization depends in large part on how well the search network is divided. There are several algorithms for partitioning a graph into multiple regions called a multi-cut of the graph [5,6,7]. These algorithms typically attempt to balance the number of nodes in each partition, or minimize the number of edges that are cut. However, none of the known algorithms are suitable for subgraph isomorphism, since we also require partitions to overlap by the radius of the query to guarantee that every instance of the query will be found. Otherwise the subgraph isomorphism algorithm would have to be modified extensively to consider and merge partial matches, which is well beyond the scope of our project. We implemented a variation of hierarchical clustering [8] for partitioning the target network for a query with radius r. The radius of a graph is the minimum distance to reach every node in the graph from the center node of the graph. For example, for a linear chain of 7 nodes, the maximal distance between any two nodes, the diameter of the graph, is 6, from the leftmost to the rightmost, but the radius is only 3, from the center to either the leftmost or rightmost. For a circular ring of 7 nodes, both the diameter and radius are 3. For a clique of any size, both the diameter and radius are 1. The partitioning algorithm works as follows. First, the connected components of the target network are extracted and processed separately, since a query subgraph cannot span multiple disconnected components. Then, for each node n of a connected component of size N > MIN_COMPONENT_SIZE, the algorithm first creates a partition centered at n that includes all nodes that are within r hops from n. This creates N overlapping subgraphs that are
4 highly redundant, but are guaranteed to fully contain every query of radius r (Figure 2). This is easily demonstrated by considering a connected component W, and a subgraph Q with radius r, i.e. from some node n c in Q, every other node in Q is reachable within r hops. Since the partitioning algorithm creates a partition centered at every node containing every node within r hops, we are guaranteed to consider every center node n c of Q with all nodes within r hops and thus count every instance of Q. After the first phase, if there are more partitions than desired, the partitions are merged together. The number of partitions can be controlled directly, or based on a characteristic of the graph, such as the desired size per partition. The algorithm finds merges the pair of partitions with the highest overlap, defined by the fraction of nodes in common compared to the sum of their sizes (Figure 4). Merging two partitions may make other partitions redundant, so the remaining partitions are checked and redundant partitions are discarded. This merging process is repeated until just the target number of partitions remains. This process of merging partitions with the most overlap is analogous to a traditional hierarchical clustering algorithm, where the most similar data points are merged together into the same cluster. This procedure is thus effective at keeping highly connected regions of the network in the same partition and separating regions that are relatively disjoint, i.e., those which have a low degree of overlap. Figure 2. A partition of radius=1 is formed around each node in the original graph, shown in the same color as the center node. The original partitions may be redundant, so the algorithm next removes all partitions that are completely contained in another partitions (Figure 3). The result of this first phase is a set of nonredundant, but overlapping partitions. Figure 3. Redundant partitions are discarded, leaving just the set of non-redundant, but overlapping partitions. Figure 4. The pair of partitions with the highest overlap is merged into a single partition. The original network partitioning algorithm forms partitions from the most highly connected regions of the network and attempts to minimize the amount of overlap between the partitions, but it makes no attempt to balance the size of the partitions. Minimizing the amount of overlap is important for avoiding duplicate work in the search phase, but may lead to widely varying partition sizes and thus high load imbalance. Therefore, we implemented a second partitioning algorithm that uses a different rule for merging partitions. Instead of merging the pair with greatest overlap, the algorithm merges the smallest partition with the partition it that overlaps the most. This ensures the small partitions will be
5 merged into larger partitions, but at the cost of having potentially more overlap in the aggregate Query Distribution At initialization, all processing units master and worker read both the target graph and query graph files using calls from the supplied graphtools library. With both the master and workers containing the graphs in memory, the issues of sending large pieces of data via message passing are circumvented. Since the partitioning is based on the radius of the query graph, the master node precomputes all possible partitions based on the various radii of all queries to be performed. The correct partition is fetched and used when searching for a particular query. In this way we avoid recomputing the partitioning for each query graph. The input to the program is a file containing all query graphs, and each query in the file is sequentially searched for in the target graph. Basic communication between master and workers consists of the master sending compact graph representations to the workers, and the workers sending back the total instances found in their respective chunk of the target graph. For generality, handshaking logic is done using messages containing these same data types: at the beginning and end of a query, the master sends a null graph object to alert the workers, and similarly the workers report their status by sending various negative values -1 for ready for query and -2 for completed final query. The target graph is made up of two types of components: those that are large enough to be partitioned into smaller sub-components and those that are not. When searching for a particular query, the master creates a compact graph object of each component and sends it to a worker that is awaiting data. Regardless of whether or not this component has been partitioned into smaller sub-components, the entire component is sent. If the component has been partitioned, a particular sub-component is flagged so that the worker knows that is the one to search. When a worker has completed searching a given component, it reports back an integer value of the number of instances found, using the ownership rule explained below to avoid over-counting. Upon receiving the count, the master updates its running tally for the query subgraph and sends a new component to the now idle worker Counting Queries Correctly Since the partitions of a component overlap, it may be possible for the same instance of a query to be found multiple times in the overlapping region, and thus the number of instances would be overcounted. One solution is to store each instance in a hash table and count only the unique instances. This method has high communication and memory overhead since a query may be present hundreds of thousands of times in a network. Instead we use an ownership rule to determine the partition that should count an instance of a query. As explained above, each worker is sent the query subgraph, and an array containing all of the partitions with one partition flagged to search. For each instance of the query found in its partition, the worker checks the array to see if another partition with lower index could also contain the query. If there is another partition, the instance is not counted; otherwise, that partition owns the instance, and adds it to the count. This distributes the problem of over-counting results to the individual workers and has minimal overhead. 3. Results Our main focus is to search for motifs in biological networks, specifically the yeast protein-protein interaction network examined in the Grochow and Kellis paper. This network records 2493 interactions between 1379 proteins. The network has 162 connected components, although it is dominated by a few large components, the largest having 778 nodes. The diameter of large connected component is 25, but the effective diameter (the average distance between any 2 nodes) is only 9.4, indicating the network is relatively dense and thus hard to partition. Prior work on this network had systematically explored motifs with 7 or fewer nodes, but we used the nauty [9,10] graph tools to enumerate all possible graphs up to 8 nodes (Figure 5), each with radius between 1 and 4. Computation was performed using the 64 node CBCB wren cluster. Each node has a dual core 3.2 GHz Xeon processor with 4 GB of RAM, several TB of disk space shared over NFS, and allows direct ssh access. Thus, it was a suitable resource for Parallel Java and for our programs, although we
6 did have to contend with minimal interference from other people s Condor jobs. Figure 5. The number of possible graphs of a given size. Figure 6. The running time of both naïve block scheduling and the better first-fit scheduling decreases as the number of processors increases. 3.1 Query Parallelization Generally speaking, we expected first-fit Query Parallelization to do better than naïve Query Parallelization on any significantly large query set. Indeed, this is what Figure 6 shows: first-fit outperforms naïve QP in the sweet spot, that is, where message passing overhead is offset by continuous computation. This particular run was for a set of all 6-node queries, and did not run for a particularly long time. No doubt the sweet spot would be larger and more pronounced for even larger query sets, which leads us to believe that first-fit QP is appropriate in the majority of cases. Next, we conducted exhaustive timings of first-fit QP on all N-node query sets where N went from 2 to 8. For each query set, we varied the number of processors from 1 to 64, as appropriate. The results are presented in Figure 7. Speedup was as expected, generally linear with a small amount of message passing overhead. Using our parallelization technique, we were able to compute counts for all 8- node queries in a reasonable amount of time, thus making exhaustive analysis of this next order of magnitude possible. Figure 7. Query parallelization is effective over a wide variety of query sizes and worker configurations. 3.2 Network Partitioning The network partitioning algorithms are designed to partition the network at regions with relatively few edges, its bottlenecks, and leave regions with small overlap in separate partitions. The success of the
7 algorithms depends on the number and placement of bottlenecks in the network. Very regular networks, such as a regular grid of 20x20 nodes, with each node connected to its 8 neighbors, may have no natural bottlenecks. Nevertheless, both the hierarchical partitioning algorithm (HIERARCHICAL), and the variant that merges the partition with minimum size (MIN_SIZE), can partition the grid into 2 roughly equal sized partitions in less than 2 seconds (Figure 8). partitions of the 778 node large connected component. As expected, the hierarchical partitioning had very few duplicated instances, since it attempts to minimize the overlap between partitions. In contrast, the minimum size variant often had a large amount of overlap, and as much as 225% of the instances were redundant. For cliques with between 3 and 8 nodes, each with radius 1, there were very few duplicated instances for the different algorithms, except for MIN_SIZE_50, which had between 15% and 54% duplicates (results not shown). Figure 8. Network partitioning of a 20x20 grid for a query of radius 1. The black nodes in the middle are shared by both the top and bottom partitions. In contrast, biological networks have an irregular scale-free structure, such that the degree distribution follows a power law, with most nodes having low degree, but a few nodes having very high degree. In these networks, it may be possible to find true bottlenecks that can be used to partition the network and with small overlap. However, if the bottlenecks are not evenly spaced, the partition sizes may be unbalanced. We explored the overlap between partitions by computing the percent of total number of instances of a query found in the large component of the yeast PPI network that were duplicates for the different partitioning algorithms. Figure 9 shows the results for linear chains of nodes between 3 and 8 nodes long, each having radius=len/2. For the 2 partitioning algorithms, we set the partitioning algorithms to create partitions of approximately 50 or 100 nodes each, corresponding to 15 or 7 Figure 9. The fraction of duplicated instances for query chains of different lengths. Figure 10 shows the standard deviation in partition size for queries with radius between 1 and 4. As expected, the minimum size partitioning creates partitions that are relatively uniform in size, and have consistently low standard deviation in size. In contrast, hierarchical partitioning creates partitions with a very large standard deviation in size. This is because it has a tendency to create partitions of the relatively small but densely connected regions on the periphery of the yeast PPI network and leave the bulk of the network in a few larger partitions. 3.3 Network Parallelization Network parallelization is primarily useful for speeding up the search for a particular query subgraph. We explored the performance of network parallelization by choosing a random sample of eight 7-node queries and recording the computation time while varying the number of workers and partitioning algorithms. The partitioning algorithm
8 was set to create on average 25, 50 or 100 node partitions, corresponding to 30, 15, or 7 partitions for the large connected component, respectively. If the connected component had fewer nodes than the requested size, it was not partitioned but sent to a worker as a whole. between the partitions, so it takes a high number of workers before the overall runtime is faster than the serial search on the monolithic graph. Figure 11. Average runtime for 7 node queries using network parallelization. Figure 10. Standard deviation in partition sizes. Hierarchical partitions vary widely in size, but minimum size variation creates relatively uniform partitions. Figure 11 shows the results for a serial monolithic search on a 1 processor, and a parallel execution with 4, 8, 16 and 32 workers. The hierarchical partitioning algorithm had relatively uniform runtime regardless of the number of worker threads because the partitioning was only marginally effective in this network (HIERARCHICAL_25 and HIERARCHCAL_50 are not shown, but have similar results). As explained in the previous section the majority of the nodes were placed in a single partition, and this partition dominated the overall runtime. The overall runtime was between 10% to 15% slower than the serial search, suggesting a very high message passing overhead with Parallel Java, since the hierarchical partitioning algorithm has minimal overlap between partitions. The minimum size partitioning algorithm showed good speedup between 4, 8, 16, and 32 workers. The most effective partitioning was MIN_SIZE_25, which creates approximately the same number of partitions as workers from the large component, and thus had the best load balance. It had nearly linear speedup between 4 and 32 workers. This partitioning also has the highest level of overlap 4. Conclusions Network motifs reveal the building blocks of complex networks and can show surprising similarity between networks from diverse sources. Searching for network motifs requires a large amount of independent computation and is thus a good candidate for parallel computing. Our results show that we can dramatically reduce the runtime needed for finding motifs, by searching for occurrences of different query subgraphs in parallel using a technique called query parallelization. This technique has almost perfect linear speedup in the overall computation. Furthermore, we have shown that we can speed up the computation for a single query using a technique called network parallelization. At the core of this technique is a novel algorithm for decomposing an irregular network into overlapping regions with minimal overlap, inspired by hierarchical clustering. We also presented a second algorithm for partitioning a network that creates partitions of uniform size by merging the smallest partition with the partition it overlaps the most. We demonstrated that network parallelization can have near linear speed up for searching for individual queries when used in conjunction with the minimize size partitioning algorithm.
9 In the future, we plan to explore larger networks to give the partitioning algorithms more opportunity to partition the network, and to have more work for the individual workers and thus offset the message passing overhead. The yeast PPI graph only has interactions for 1379 proteins, but some organisms have many more proteins. Unfortunately, only a small number of organisms have PPI networks available at this time, and the networks are of generally lower quality. The low effective diameter and relatively few bottlenecks of the yeast PPI network makes it intrinsically difficult to partition, but other networks may have more structure to exploit. We also plan to explore variants of the partitioning algorithms that can both minimize overlap and create uniform sized partitions. (CSUR), Volume 31, Issue 3, ACM Press, New York [9] Nauty Graph Tools [10] B. D. McKay, Practical graph isomorphism, 10th. Manitoba Conference on Numerical Mathematics and Computing (Winnipeg, 1980); Congressus Numerantium, 30 (1981) References [1] Milo et al. Network motifs: simple building blocks of complex networks. Science 298: , [2] Zhang et al. Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J Biology 4:6, [3] Grochow and Kellis. Network motif discovery using subgraph enumeration and symmetrybreaking. RECOMB [4] Parallel Java Library. [5] Golovin, D., Nagarajan, V., and Singh, M Approximating the k-multicut problem. In Proceedings of the Seventeenth Annual ACM- SIAM Symposium on Discrete Algorithm (Miami, Florida, January 22-26, 2006). SODA '06. ACM, New York, NY, [6] E. Dahlhaus, D. S. Johnson, C. H. Papadimitriou, P. D. Seymour, and M. Yannakakis. Complexity of Multiterminal Cuts. SIAM Journal On Computing 23: , 1994 [7] N. Garg, V.V Vazirani, M. Yannakais.Multiway Cuts in Directed Graphs and Node Weighted Graphs.Proceedings of ICALP, 1994, PP [8] A. K. Jain, M. N. Murty, P. J. Flynn Data Clustering: A Review. ACM Computing Surveys
Biology, Physics, Mathematics, Sociology, Engineering, Computer Science, Etc
 Motivation Motifs Algorithms G-Tries Parallelism Complex Networks Networks are ubiquitous! Biology, Physics, Mathematics, Sociology, Engineering, Computer Science, Etc Images: UK Highways Agency, Uriel
Motivation Motifs Algorithms G-Tries Parallelism Complex Networks Networks are ubiquitous! Biology, Physics, Mathematics, Sociology, Engineering, Computer Science, Etc Images: UK Highways Agency, Uriel
Parameterized graph separation problems
 Parameterized graph separation problems Dániel Marx Department of Computer Science and Information Theory, Budapest University of Technology and Economics Budapest, H-1521, Hungary, dmarx@cs.bme.hu Abstract.
Parameterized graph separation problems Dániel Marx Department of Computer Science and Information Theory, Budapest University of Technology and Economics Budapest, H-1521, Hungary, dmarx@cs.bme.hu Abstract.
Arabesque. A system for distributed graph mining. Mohammed Zaki, RPI
 rabesque system for distributed graph mining Mohammed Zaki, RPI Carlos Teixeira, lexandre Fonseca, Marco Serafini, Georgos Siganos, shraf boulnaga, Qatar Computing Research Institute (QCRI) 1 Big Data
rabesque system for distributed graph mining Mohammed Zaki, RPI Carlos Teixeira, lexandre Fonseca, Marco Serafini, Georgos Siganos, shraf boulnaga, Qatar Computing Research Institute (QCRI) 1 Big Data
Layer-Based Scheduling Algorithms for Multiprocessor-Tasks with Precedence Constraints
 Layer-Based Scheduling Algorithms for Multiprocessor-Tasks with Precedence Constraints Jörg Dümmler, Raphael Kunis, and Gudula Rünger Chemnitz University of Technology, Department of Computer Science,
Layer-Based Scheduling Algorithms for Multiprocessor-Tasks with Precedence Constraints Jörg Dümmler, Raphael Kunis, and Gudula Rünger Chemnitz University of Technology, Department of Computer Science,
Complexity Results on Graphs with Few Cliques
 Discrete Mathematics and Theoretical Computer Science DMTCS vol. 9, 2007, 127 136 Complexity Results on Graphs with Few Cliques Bill Rosgen 1 and Lorna Stewart 2 1 Institute for Quantum Computing and School
Discrete Mathematics and Theoretical Computer Science DMTCS vol. 9, 2007, 127 136 Complexity Results on Graphs with Few Cliques Bill Rosgen 1 and Lorna Stewart 2 1 Institute for Quantum Computing and School
A Hybrid Recursive Multi-Way Number Partitioning Algorithm
 Proceedings of the Twenty-Second International Joint Conference on Artificial Intelligence A Hybrid Recursive Multi-Way Number Partitioning Algorithm Richard E. Korf Computer Science Department University
Proceedings of the Twenty-Second International Joint Conference on Artificial Intelligence A Hybrid Recursive Multi-Way Number Partitioning Algorithm Richard E. Korf Computer Science Department University
Network Design Considerations for Grid Computing
 Network Design Considerations for Grid Computing Engineering Systems How Bandwidth, Latency, and Packet Size Impact Grid Job Performance by Erik Burrows, Engineering Systems Analyst, Principal, Broadcom
Network Design Considerations for Grid Computing Engineering Systems How Bandwidth, Latency, and Packet Size Impact Grid Job Performance by Erik Burrows, Engineering Systems Analyst, Principal, Broadcom
1 Homophily and assortative mixing
 1 Homophily and assortative mixing Networks, and particularly social networks, often exhibit a property called homophily or assortative mixing, which simply means that the attributes of vertices correlate
1 Homophily and assortative mixing Networks, and particularly social networks, often exhibit a property called homophily or assortative mixing, which simply means that the attributes of vertices correlate
Automatic Compiler-Based Optimization of Graph Analytics for the GPU. Sreepathi Pai The University of Texas at Austin. May 8, 2017 NVIDIA GTC
 Automatic Compiler-Based Optimization of Graph Analytics for the GPU Sreepathi Pai The University of Texas at Austin May 8, 2017 NVIDIA GTC Parallel Graph Processing is not easy 299ms HD-BFS 84ms USA Road
Automatic Compiler-Based Optimization of Graph Analytics for the GPU Sreepathi Pai The University of Texas at Austin May 8, 2017 NVIDIA GTC Parallel Graph Processing is not easy 299ms HD-BFS 84ms USA Road
Introduction to Parallel Computing
 Portland State University ECE 588/688 Introduction to Parallel Computing Reference: Lawrence Livermore National Lab Tutorial https://computing.llnl.gov/tutorials/parallel_comp/ Copyright by Alaa Alameldeen
Portland State University ECE 588/688 Introduction to Parallel Computing Reference: Lawrence Livermore National Lab Tutorial https://computing.llnl.gov/tutorials/parallel_comp/ Copyright by Alaa Alameldeen
Mitigating Data Skew Using Map Reduce Application
 Ms. Archana P.M Mitigating Data Skew Using Map Reduce Application Mr. Malathesh S.H 4 th sem, M.Tech (C.S.E) Associate Professor C.S.E Dept. M.S.E.C, V.T.U Bangalore, India archanaanil062@gmail.com M.S.E.C,
Ms. Archana P.M Mitigating Data Skew Using Map Reduce Application Mr. Malathesh S.H 4 th sem, M.Tech (C.S.E) Associate Professor C.S.E Dept. M.S.E.C, V.T.U Bangalore, India archanaanil062@gmail.com M.S.E.C,
Parallel Algorithm Design. Parallel Algorithm Design p. 1
 Parallel Algorithm Design Parallel Algorithm Design p. 1 Overview Chapter 3 from Michael J. Quinn, Parallel Programming in C with MPI and OpenMP Another resource: http://www.mcs.anl.gov/ itf/dbpp/text/node14.html
Parallel Algorithm Design Parallel Algorithm Design p. 1 Overview Chapter 3 from Michael J. Quinn, Parallel Programming in C with MPI and OpenMP Another resource: http://www.mcs.anl.gov/ itf/dbpp/text/node14.html
Analyzing Dshield Logs Using Fully Automatic Cross-Associations
 Analyzing Dshield Logs Using Fully Automatic Cross-Associations Anh Le 1 1 Donald Bren School of Information and Computer Sciences University of California, Irvine Irvine, CA, 92697, USA anh.le@uci.edu
Analyzing Dshield Logs Using Fully Automatic Cross-Associations Anh Le 1 1 Donald Bren School of Information and Computer Sciences University of California, Irvine Irvine, CA, 92697, USA anh.le@uci.edu
Principles of Parallel Algorithm Design: Concurrency and Mapping
 Principles of Parallel Algorithm Design: Concurrency and Mapping John Mellor-Crummey Department of Computer Science Rice University johnmc@rice.edu COMP 422/534 Lecture 3 17 January 2017 Last Thursday
Principles of Parallel Algorithm Design: Concurrency and Mapping John Mellor-Crummey Department of Computer Science Rice University johnmc@rice.edu COMP 422/534 Lecture 3 17 January 2017 Last Thursday
Locality-sensitive hashing and biological network alignment
 Locality-sensitive hashing and biological network alignment Laura LeGault - University of Wisconsin, Madison 12 May 2008 Abstract Large biological networks contain much information about the functionality
Locality-sensitive hashing and biological network alignment Laura LeGault - University of Wisconsin, Madison 12 May 2008 Abstract Large biological networks contain much information about the functionality
GPU ACCELERATED SELF-JOIN FOR THE DISTANCE SIMILARITY METRIC
 GPU ACCELERATED SELF-JOIN FOR THE DISTANCE SIMILARITY METRIC MIKE GOWANLOCK NORTHERN ARIZONA UNIVERSITY SCHOOL OF INFORMATICS, COMPUTING & CYBER SYSTEMS BEN KARSIN UNIVERSITY OF HAWAII AT MANOA DEPARTMENT
GPU ACCELERATED SELF-JOIN FOR THE DISTANCE SIMILARITY METRIC MIKE GOWANLOCK NORTHERN ARIZONA UNIVERSITY SCHOOL OF INFORMATICS, COMPUTING & CYBER SYSTEMS BEN KARSIN UNIVERSITY OF HAWAII AT MANOA DEPARTMENT
Serial. Parallel. CIT 668: System Architecture 2/14/2011. Topics. Serial and Parallel Computation. Parallel Computing
 CIT 668: System Architecture Parallel Computing Topics 1. What is Parallel Computing? 2. Why use Parallel Computing? 3. Types of Parallelism 4. Amdahl s Law 5. Flynn s Taxonomy of Parallel Computers 6.
CIT 668: System Architecture Parallel Computing Topics 1. What is Parallel Computing? 2. Why use Parallel Computing? 3. Types of Parallelism 4. Amdahl s Law 5. Flynn s Taxonomy of Parallel Computers 6.
A Parallel Implementation of a Higher-order Self Consistent Mean Field. Effectively solving the protein repacking problem is a key step to successful
 Karl Gutwin May 15, 2005 18.336 A Parallel Implementation of a Higher-order Self Consistent Mean Field Effectively solving the protein repacking problem is a key step to successful protein design. Put
Karl Gutwin May 15, 2005 18.336 A Parallel Implementation of a Higher-order Self Consistent Mean Field Effectively solving the protein repacking problem is a key step to successful protein design. Put
More Efficient Classification of Web Content Using Graph Sampling
 More Efficient Classification of Web Content Using Graph Sampling Chris Bennett Department of Computer Science University of Georgia Athens, Georgia, USA 30602 bennett@cs.uga.edu Abstract In mining information
More Efficient Classification of Web Content Using Graph Sampling Chris Bennett Department of Computer Science University of Georgia Athens, Georgia, USA 30602 bennett@cs.uga.edu Abstract In mining information
FASCIA. Fast Approximate Subgraph Counting and Enumeration. 2 Oct Scalable Computing Laboratory The Pennsylvania State University 1 / 28
 FASCIA Fast Approximate Subgraph Counting and Enumeration George M. Slota Kamesh Madduri Scalable Computing Laboratory The Pennsylvania State University 2 Oct. 2013 1 / 28 Overview Background Motivation
FASCIA Fast Approximate Subgraph Counting and Enumeration George M. Slota Kamesh Madduri Scalable Computing Laboratory The Pennsylvania State University 2 Oct. 2013 1 / 28 Overview Background Motivation
High-Performance and Parallel Computing
 9 High-Performance and Parallel Computing 9.1 Code optimization To use resources efficiently, the time saved through optimizing code has to be weighed against the human resources required to implement
9 High-Performance and Parallel Computing 9.1 Code optimization To use resources efficiently, the time saved through optimizing code has to be weighed against the human resources required to implement
Performance of general graph isomorphism algorithms Sara Voss Coe College, Cedar Rapids, IA
 Performance of general graph isomorphism algorithms Sara Voss Coe College, Cedar Rapids, IA I. Problem and Motivation Graphs are commonly used to provide structural and/or relational descriptions. A node
Performance of general graph isomorphism algorithms Sara Voss Coe College, Cedar Rapids, IA I. Problem and Motivation Graphs are commonly used to provide structural and/or relational descriptions. A node
Seminar on. A Coarse-Grain Parallel Formulation of Multilevel k-way Graph Partitioning Algorithm
 Seminar on A Coarse-Grain Parallel Formulation of Multilevel k-way Graph Partitioning Algorithm Mohammad Iftakher Uddin & Mohammad Mahfuzur Rahman Matrikel Nr: 9003357 Matrikel Nr : 9003358 Masters of
Seminar on A Coarse-Grain Parallel Formulation of Multilevel k-way Graph Partitioning Algorithm Mohammad Iftakher Uddin & Mohammad Mahfuzur Rahman Matrikel Nr: 9003357 Matrikel Nr : 9003358 Masters of
2. TOPOLOGICAL PATTERN ANALYSIS
 Methodology for analyzing and quantifying design style changes and complexity using topological patterns Jason P. Cain a, Ya-Chieh Lai b, Frank Gennari b, Jason Sweis b a Advanced Micro Devices, 7171 Southwest
Methodology for analyzing and quantifying design style changes and complexity using topological patterns Jason P. Cain a, Ya-Chieh Lai b, Frank Gennari b, Jason Sweis b a Advanced Micro Devices, 7171 Southwest
An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization
 An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization Pedro Ribeiro (DCC/FCUP & CRACS/INESC-TEC) Part 1 Motivation and emergence of Network Science
An Exploratory Journey Into Network Analysis A Gentle Introduction to Network Science and Graph Visualization Pedro Ribeiro (DCC/FCUP & CRACS/INESC-TEC) Part 1 Motivation and emergence of Network Science
Introduction to Bioinformatics
 Introduction to Bioinformatics Biological Networks Department of Computing Imperial College London Spring 2010 1. Motivation Large Networks model many real-world phenomena technological: www, internet,
Introduction to Bioinformatics Biological Networks Department of Computing Imperial College London Spring 2010 1. Motivation Large Networks model many real-world phenomena technological: www, internet,
On Smart Query Routing: For Distributed Graph Querying with Decoupled Storage
 On Smart Query Routing: For Distributed Graph Querying with Decoupled Storage Arijit Khan Nanyang Technological University (NTU), Singapore Gustavo Segovia ETH Zurich, Switzerland Donald Kossmann Microsoft
On Smart Query Routing: For Distributed Graph Querying with Decoupled Storage Arijit Khan Nanyang Technological University (NTU), Singapore Gustavo Segovia ETH Zurich, Switzerland Donald Kossmann Microsoft
EE/CSCI 451: Parallel and Distributed Computation
 EE/CSCI 451: Parallel and Distributed Computation Lecture #11 2/21/2017 Xuehai Qian Xuehai.qian@usc.edu http://alchem.usc.edu/portal/xuehaiq.html University of Southern California 1 Outline Midterm 1:
EE/CSCI 451: Parallel and Distributed Computation Lecture #11 2/21/2017 Xuehai Qian Xuehai.qian@usc.edu http://alchem.usc.edu/portal/xuehaiq.html University of Southern California 1 Outline Midterm 1:
Definition of RAID Levels
 RAID The basic idea of RAID (Redundant Array of Independent Disks) is to combine multiple inexpensive disk drives into an array of disk drives to obtain performance, capacity and reliability that exceeds
RAID The basic idea of RAID (Redundant Array of Independent Disks) is to combine multiple inexpensive disk drives into an array of disk drives to obtain performance, capacity and reliability that exceeds
SWsoft ADVANCED VIRTUALIZATION AND WORKLOAD MANAGEMENT ON ITANIUM 2-BASED SERVERS
 SWsoft ADVANCED VIRTUALIZATION AND WORKLOAD MANAGEMENT ON ITANIUM 2-BASED SERVERS Abstract Virtualization and workload management are essential technologies for maximizing scalability, availability and
SWsoft ADVANCED VIRTUALIZATION AND WORKLOAD MANAGEMENT ON ITANIUM 2-BASED SERVERS Abstract Virtualization and workload management are essential technologies for maximizing scalability, availability and
Parallel Motif Search Using ParSeq
 Parallel Motif Search Using ParSeq Jun Qin 1, Simon Pinkenburg 2 and Wolfgang Rosenstiel 2 1 Distributed and Parallel Systems Group University of Innsbruck Innsbruck, Austria 2 Department of Computer Engineering
Parallel Motif Search Using ParSeq Jun Qin 1, Simon Pinkenburg 2 and Wolfgang Rosenstiel 2 1 Distributed and Parallel Systems Group University of Innsbruck Innsbruck, Austria 2 Department of Computer Engineering
A Graph Theoretic Approach to Image Database Retrieval
 A Graph Theoretic Approach to Image Database Retrieval Selim Aksoy and Robert M. Haralick Intelligent Systems Laboratory Department of Electrical Engineering University of Washington, Seattle, WA 98195-2500
A Graph Theoretic Approach to Image Database Retrieval Selim Aksoy and Robert M. Haralick Intelligent Systems Laboratory Department of Electrical Engineering University of Washington, Seattle, WA 98195-2500
Optimizing Parallel Access to the BaBar Database System Using CORBA Servers
 SLAC-PUB-9176 September 2001 Optimizing Parallel Access to the BaBar Database System Using CORBA Servers Jacek Becla 1, Igor Gaponenko 2 1 Stanford Linear Accelerator Center Stanford University, Stanford,
SLAC-PUB-9176 September 2001 Optimizing Parallel Access to the BaBar Database System Using CORBA Servers Jacek Becla 1, Igor Gaponenko 2 1 Stanford Linear Accelerator Center Stanford University, Stanford,
Design of Parallel Algorithms. Models of Parallel Computation
 + Design of Parallel Algorithms Models of Parallel Computation + Chapter Overview: Algorithms and Concurrency n Introduction to Parallel Algorithms n Tasks and Decomposition n Processes and Mapping n Processes
+ Design of Parallel Algorithms Models of Parallel Computation + Chapter Overview: Algorithms and Concurrency n Introduction to Parallel Algorithms n Tasks and Decomposition n Processes and Mapping n Processes
Unit 6 Chapter 15 EXAMPLES OF COMPLEXITY CALCULATION
 DESIGN AND ANALYSIS OF ALGORITHMS Unit 6 Chapter 15 EXAMPLES OF COMPLEXITY CALCULATION http://milanvachhani.blogspot.in EXAMPLES FROM THE SORTING WORLD Sorting provides a good set of examples for analyzing
DESIGN AND ANALYSIS OF ALGORITHMS Unit 6 Chapter 15 EXAMPLES OF COMPLEXITY CALCULATION http://milanvachhani.blogspot.in EXAMPLES FROM THE SORTING WORLD Sorting provides a good set of examples for analyzing
Distributed Computations MapReduce. adapted from Jeff Dean s slides
 Distributed Computations MapReduce adapted from Jeff Dean s slides What we ve learnt so far Basic distributed systems concepts Consistency (sequential, eventual) Fault tolerance (recoverability, availability)
Distributed Computations MapReduce adapted from Jeff Dean s slides What we ve learnt so far Basic distributed systems concepts Consistency (sequential, eventual) Fault tolerance (recoverability, availability)
JULIA ENABLED COMPUTATION OF MOLECULAR LIBRARY COMPLEXITY IN DNA SEQUENCING
 JULIA ENABLED COMPUTATION OF MOLECULAR LIBRARY COMPLEXITY IN DNA SEQUENCING Larson Hogstrom, Mukarram Tahir, Andres Hasfura Massachusetts Institute of Technology, Cambridge, Massachusetts, USA 18.337/6.338
JULIA ENABLED COMPUTATION OF MOLECULAR LIBRARY COMPLEXITY IN DNA SEQUENCING Larson Hogstrom, Mukarram Tahir, Andres Hasfura Massachusetts Institute of Technology, Cambridge, Massachusetts, USA 18.337/6.338
Hashing for searching
 Hashing for searching Consider searching a database of records on a given key. There are three standard techniques: Searching sequentially start at the first record and look at each record in turn until
Hashing for searching Consider searching a database of records on a given key. There are three standard techniques: Searching sequentially start at the first record and look at each record in turn until
Running SNAP. The SNAP Team February 2012
 Running SNAP The SNAP Team February 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
Running SNAP The SNAP Team February 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
What is Parallel Computing?
 What is Parallel Computing? Parallel Computing is several processing elements working simultaneously to solve a problem faster. 1/33 What is Parallel Computing? Parallel Computing is several processing
What is Parallel Computing? Parallel Computing is several processing elements working simultaneously to solve a problem faster. 1/33 What is Parallel Computing? Parallel Computing is several processing
MASSACHUSETTS INSTITUTE OF TECHNOLOGY Database Systems: Fall 2008 Quiz II
 Department of Electrical Engineering and Computer Science MASSACHUSETTS INSTITUTE OF TECHNOLOGY 6.830 Database Systems: Fall 2008 Quiz II There are 14 questions and 11 pages in this quiz booklet. To receive
Department of Electrical Engineering and Computer Science MASSACHUSETTS INSTITUTE OF TECHNOLOGY 6.830 Database Systems: Fall 2008 Quiz II There are 14 questions and 11 pages in this quiz booklet. To receive
High Performance Computing. Introduction to Parallel Computing
 High Performance Computing Introduction to Parallel Computing Acknowledgements Content of the following presentation is borrowed from The Lawrence Livermore National Laboratory https://hpc.llnl.gov/training/tutorials
High Performance Computing Introduction to Parallel Computing Acknowledgements Content of the following presentation is borrowed from The Lawrence Livermore National Laboratory https://hpc.llnl.gov/training/tutorials
Lecture 5: Search Algorithms for Discrete Optimization Problems
 Lecture 5: Search Algorithms for Discrete Optimization Problems Definitions Discrete optimization problem (DOP): tuple (S, f), S finite set of feasible solutions, f : S R, cost function. Objective: find
Lecture 5: Search Algorithms for Discrete Optimization Problems Definitions Discrete optimization problem (DOP): tuple (S, f), S finite set of feasible solutions, f : S R, cost function. Objective: find
Lecture Note: Computation problems in social. network analysis
 Lecture Note: Computation problems in social network analysis Bang Ye Wu CSIE, Chung Cheng University, Taiwan September 29, 2008 In this lecture note, several computational problems are listed, including
Lecture Note: Computation problems in social network analysis Bang Ye Wu CSIE, Chung Cheng University, Taiwan September 29, 2008 In this lecture note, several computational problems are listed, including
Lecture 4: Principles of Parallel Algorithm Design (part 4)
 Lecture 4: Principles of Parallel Algorithm Design (part 4) 1 Mapping Technique for Load Balancing Minimize execution time Reduce overheads of execution Sources of overheads: Inter-process interaction
Lecture 4: Principles of Parallel Algorithm Design (part 4) 1 Mapping Technique for Load Balancing Minimize execution time Reduce overheads of execution Sources of overheads: Inter-process interaction
Performance of Multicore LUP Decomposition
 Performance of Multicore LUP Decomposition Nathan Beckmann Silas Boyd-Wickizer May 3, 00 ABSTRACT This paper evaluates the performance of four parallel LUP decomposition implementations. The implementations
Performance of Multicore LUP Decomposition Nathan Beckmann Silas Boyd-Wickizer May 3, 00 ABSTRACT This paper evaluates the performance of four parallel LUP decomposition implementations. The implementations
A DISCUSSION ON SSP STRUCTURE OF PAN, HELM AND CROWN GRAPHS
 VOL. 10, NO. 9, MAY 015 ISSN 1819-6608 A DISCUSSION ON SSP STRUCTURE OF PAN, HELM AND CROWN GRAPHS R. Mary Jeya Jothi Department of Mathematics, Sathyabama University, Chennai, India E-Mail: jeyajothi31@gmail.com
VOL. 10, NO. 9, MAY 015 ISSN 1819-6608 A DISCUSSION ON SSP STRUCTURE OF PAN, HELM AND CROWN GRAPHS R. Mary Jeya Jothi Department of Mathematics, Sathyabama University, Chennai, India E-Mail: jeyajothi31@gmail.com
Complementary Graph Coloring
 International Journal of Computer (IJC) ISSN 2307-4523 (Print & Online) Global Society of Scientific Research and Researchers http://ijcjournal.org/ Complementary Graph Coloring Mohamed Al-Ibrahim a*,
International Journal of Computer (IJC) ISSN 2307-4523 (Print & Online) Global Society of Scientific Research and Researchers http://ijcjournal.org/ Complementary Graph Coloring Mohamed Al-Ibrahim a*,
Parallelization of Depth-First Traversal using Efficient Load Balancing
 Parallelization of Depth-First Traversal using Efficient Load Balancing VSN Harish Rayasam* IIT Madras, India rayasam@cse.iitm.ac.in Rupesh Nasre IIT Madras, India rupesh@cse.iitm.ac.in Abstract Graphs
Parallelization of Depth-First Traversal using Efficient Load Balancing VSN Harish Rayasam* IIT Madras, India rayasam@cse.iitm.ac.in Rupesh Nasre IIT Madras, India rupesh@cse.iitm.ac.in Abstract Graphs
Unsupervised Learning : Clustering
 Unsupervised Learning : Clustering Things to be Addressed Traditional Learning Models. Cluster Analysis K-means Clustering Algorithm Drawbacks of traditional clustering algorithms. Clustering as a complex
Unsupervised Learning : Clustering Things to be Addressed Traditional Learning Models. Cluster Analysis K-means Clustering Algorithm Drawbacks of traditional clustering algorithms. Clustering as a complex
Chapter 12: Indexing and Hashing. Basic Concepts
 Chapter 12: Indexing and Hashing! Basic Concepts! Ordered Indices! B+-Tree Index Files! B-Tree Index Files! Static Hashing! Dynamic Hashing! Comparison of Ordered Indexing and Hashing! Index Definition
Chapter 12: Indexing and Hashing! Basic Concepts! Ordered Indices! B+-Tree Index Files! B-Tree Index Files! Static Hashing! Dynamic Hashing! Comparison of Ordered Indexing and Hashing! Index Definition
Clustering Algorithm (DBSCAN) VISHAL BHARTI Computer Science Dept. GC, CUNY
 Clustering Algorithm (DBSCAN) VISHAL BHARTI Computer Science Dept. GC, CUNY Clustering Algorithm Clustering is an unsupervised machine learning algorithm that divides a data into meaningful sub-groups,
Clustering Algorithm (DBSCAN) VISHAL BHARTI Computer Science Dept. GC, CUNY Clustering Algorithm Clustering is an unsupervised machine learning algorithm that divides a data into meaningful sub-groups,
Optimal Sequential Multi-Way Number Partitioning
 Optimal Sequential Multi-Way Number Partitioning Richard E. Korf, Ethan L. Schreiber, and Michael D. Moffitt Computer Science Department University of California, Los Angeles Los Angeles, CA 90095 IBM
Optimal Sequential Multi-Way Number Partitioning Richard E. Korf, Ethan L. Schreiber, and Michael D. Moffitt Computer Science Department University of California, Los Angeles Los Angeles, CA 90095 IBM
Distributed computing: index building and use
 Distributed computing: index building and use Distributed computing Goals Distributing computation across several machines to Do one computation faster - latency Do more computations in given time - throughput
Distributed computing: index building and use Distributed computing Goals Distributing computation across several machines to Do one computation faster - latency Do more computations in given time - throughput
Chapter 12: Indexing and Hashing
 Chapter 12: Indexing and Hashing Basic Concepts Ordered Indices B+-Tree Index Files B-Tree Index Files Static Hashing Dynamic Hashing Comparison of Ordered Indexing and Hashing Index Definition in SQL
Chapter 12: Indexing and Hashing Basic Concepts Ordered Indices B+-Tree Index Files B-Tree Index Files Static Hashing Dynamic Hashing Comparison of Ordered Indexing and Hashing Index Definition in SQL
Advanced Databases: Parallel Databases A.Poulovassilis
 1 Advanced Databases: Parallel Databases A.Poulovassilis 1 Parallel Database Architectures Parallel database systems use parallel processing techniques to achieve faster DBMS performance and handle larger
1 Advanced Databases: Parallel Databases A.Poulovassilis 1 Parallel Database Architectures Parallel database systems use parallel processing techniques to achieve faster DBMS performance and handle larger
Properties of Biological Networks
 Properties of Biological Networks presented by: Ola Hamud June 12, 2013 Supervisor: Prof. Ron Pinter Based on: NETWORK BIOLOGY: UNDERSTANDING THE CELL S FUNCTIONAL ORGANIZATION By Albert-László Barabási
Properties of Biological Networks presented by: Ola Hamud June 12, 2013 Supervisor: Prof. Ron Pinter Based on: NETWORK BIOLOGY: UNDERSTANDING THE CELL S FUNCTIONAL ORGANIZATION By Albert-László Barabási
6. Results. This section describes the performance that was achieved using the RAMA file system.
 6. Results This section describes the performance that was achieved using the RAMA file system. The resulting numbers represent actual file data bytes transferred to/from server disks per second, excluding
6. Results This section describes the performance that was achieved using the RAMA file system. The resulting numbers represent actual file data bytes transferred to/from server disks per second, excluding
Algorithm Engineering with PRAM Algorithms
 Algorithm Engineering with PRAM Algorithms Bernard M.E. Moret moret@cs.unm.edu Department of Computer Science University of New Mexico Albuquerque, NM 87131 Rome School on Alg. Eng. p.1/29 Measuring and
Algorithm Engineering with PRAM Algorithms Bernard M.E. Moret moret@cs.unm.edu Department of Computer Science University of New Mexico Albuquerque, NM 87131 Rome School on Alg. Eng. p.1/29 Measuring and
Accelerating K-Means Clustering with Parallel Implementations and GPU computing
 Accelerating K-Means Clustering with Parallel Implementations and GPU computing Janki Bhimani Electrical and Computer Engineering Dept. Northeastern University Boston, MA Email: bhimani@ece.neu.edu Miriam
Accelerating K-Means Clustering with Parallel Implementations and GPU computing Janki Bhimani Electrical and Computer Engineering Dept. Northeastern University Boston, MA Email: bhimani@ece.neu.edu Miriam
Milind Kulkarni Research Statement
 Milind Kulkarni Research Statement With the increasing ubiquity of multicore processors, interest in parallel programming is again on the upswing. Over the past three decades, languages and compilers researchers
Milind Kulkarni Research Statement With the increasing ubiquity of multicore processors, interest in parallel programming is again on the upswing. Over the past three decades, languages and compilers researchers
Leveraging Set Relations in Exact Set Similarity Join
 Leveraging Set Relations in Exact Set Similarity Join Xubo Wang, Lu Qin, Xuemin Lin, Ying Zhang, and Lijun Chang University of New South Wales, Australia University of Technology Sydney, Australia {xwang,lxue,ljchang}@cse.unsw.edu.au,
Leveraging Set Relations in Exact Set Similarity Join Xubo Wang, Lu Qin, Xuemin Lin, Ying Zhang, and Lijun Chang University of New South Wales, Australia University of Technology Sydney, Australia {xwang,lxue,ljchang}@cse.unsw.edu.au,
Dynamic Design of Cellular Wireless Networks via Self Organizing Mechanism
 Dynamic Design of Cellular Wireless Networks via Self Organizing Mechanism V.Narasimha Raghavan, M.Venkatesh, Divya Sridharabalan, T.Sabhanayagam, Nithin Bharath Abstract In our paper, we are utilizing
Dynamic Design of Cellular Wireless Networks via Self Organizing Mechanism V.Narasimha Raghavan, M.Venkatesh, Divya Sridharabalan, T.Sabhanayagam, Nithin Bharath Abstract In our paper, we are utilizing
motifs In the context of networks, the term motif may refer to di erent notions. Subgraph motifs Coloured motifs { }
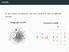 motifs In the context of networks, the term motif may refer to di erent notions. Subgraph motifs Coloured motifs G M { } 2 subgraph motifs 3 motifs Find interesting patterns in a network. 4 motifs Find
motifs In the context of networks, the term motif may refer to di erent notions. Subgraph motifs Coloured motifs G M { } 2 subgraph motifs 3 motifs Find interesting patterns in a network. 4 motifs Find
Introduction to parallel computing
 Introduction to parallel computing 3. Parallel Software Zhiao Shi (modifications by Will French) Advanced Computing Center for Education & Research Vanderbilt University Last time Parallel hardware Multi-core
Introduction to parallel computing 3. Parallel Software Zhiao Shi (modifications by Will French) Advanced Computing Center for Education & Research Vanderbilt University Last time Parallel hardware Multi-core
Parallel Programming Concepts. Parallel Algorithms. Peter Tröger
 Parallel Programming Concepts Parallel Algorithms Peter Tröger Sources: Ian Foster. Designing and Building Parallel Programs. Addison-Wesley. 1995. Mattson, Timothy G.; S, Beverly A.; ers,; Massingill,
Parallel Programming Concepts Parallel Algorithms Peter Tröger Sources: Ian Foster. Designing and Building Parallel Programs. Addison-Wesley. 1995. Mattson, Timothy G.; S, Beverly A.; ers,; Massingill,
Fractal: A Software Toolchain for Mapping Applications to Diverse, Heterogeneous Architecures
 Fractal: A Software Toolchain for Mapping Applications to Diverse, Heterogeneous Architecures University of Virginia Dept. of Computer Science Technical Report #CS-2011-09 Jeremy W. Sheaffer and Kevin
Fractal: A Software Toolchain for Mapping Applications to Diverse, Heterogeneous Architecures University of Virginia Dept. of Computer Science Technical Report #CS-2011-09 Jeremy W. Sheaffer and Kevin
Parallel Exact Inference on the Cell Broadband Engine Processor
 Parallel Exact Inference on the Cell Broadband Engine Processor Yinglong Xia and Viktor K. Prasanna {yinglonx, prasanna}@usc.edu University of Southern California http://ceng.usc.edu/~prasanna/ SC 08 Overview
Parallel Exact Inference on the Cell Broadband Engine Processor Yinglong Xia and Viktor K. Prasanna {yinglonx, prasanna}@usc.edu University of Southern California http://ceng.usc.edu/~prasanna/ SC 08 Overview
ECE 7650 Scalable and Secure Internet Services and Architecture ---- A Systems Perspective
 ECE 7650 Scalable and Secure Internet Services and Architecture ---- A Systems Perspective Part II: Data Center Software Architecture: Topic 3: Programming Models Piccolo: Building Fast, Distributed Programs
ECE 7650 Scalable and Secure Internet Services and Architecture ---- A Systems Perspective Part II: Data Center Software Architecture: Topic 3: Programming Models Piccolo: Building Fast, Distributed Programs
Multi-Way Number Partitioning
 Proceedings of the Twenty-First International Joint Conference on Artificial Intelligence (IJCAI-09) Multi-Way Number Partitioning Richard E. Korf Computer Science Department University of California,
Proceedings of the Twenty-First International Joint Conference on Artificial Intelligence (IJCAI-09) Multi-Way Number Partitioning Richard E. Korf Computer Science Department University of California,
Process size is independent of the main memory present in the system.
 Hardware control structure Two characteristics are key to paging and segmentation: 1. All memory references are logical addresses within a process which are dynamically converted into physical at run time.
Hardware control structure Two characteristics are key to paging and segmentation: 1. All memory references are logical addresses within a process which are dynamically converted into physical at run time.
Clustering CS 550: Machine Learning
 Clustering CS 550: Machine Learning This slide set mainly uses the slides given in the following links: http://www-users.cs.umn.edu/~kumar/dmbook/ch8.pdf http://www-users.cs.umn.edu/~kumar/dmbook/dmslides/chap8_basic_cluster_analysis.pdf
Clustering CS 550: Machine Learning This slide set mainly uses the slides given in the following links: http://www-users.cs.umn.edu/~kumar/dmbook/ch8.pdf http://www-users.cs.umn.edu/~kumar/dmbook/dmslides/chap8_basic_cluster_analysis.pdf
Fast Fuzzy Clustering of Infrared Images. 2. brfcm
 Fast Fuzzy Clustering of Infrared Images Steven Eschrich, Jingwei Ke, Lawrence O. Hall and Dmitry B. Goldgof Department of Computer Science and Engineering, ENB 118 University of South Florida 4202 E.
Fast Fuzzy Clustering of Infrared Images Steven Eschrich, Jingwei Ke, Lawrence O. Hall and Dmitry B. Goldgof Department of Computer Science and Engineering, ENB 118 University of South Florida 4202 E.
A Comparison of Three Document Clustering Algorithms: TreeCluster, Word Intersection GQF, and Word Intersection Hierarchical Agglomerative Clustering
 A Comparison of Three Document Clustering Algorithms:, Word Intersection GQF, and Word Intersection Hierarchical Agglomerative Clustering Abstract Kenrick Mock 9/23/1998 Business Applications Intel Architecture
A Comparison of Three Document Clustering Algorithms:, Word Intersection GQF, and Word Intersection Hierarchical Agglomerative Clustering Abstract Kenrick Mock 9/23/1998 Business Applications Intel Architecture
School of Computer and Information Science
 School of Computer and Information Science CIS Research Placement Report Multiple threads in floating-point sort operations Name: Quang Do Date: 8/6/2012 Supervisor: Grant Wigley Abstract Despite the vast
School of Computer and Information Science CIS Research Placement Report Multiple threads in floating-point sort operations Name: Quang Do Date: 8/6/2012 Supervisor: Grant Wigley Abstract Despite the vast
Treewidth and graph minors
 Treewidth and graph minors Lectures 9 and 10, December 29, 2011, January 5, 2012 We shall touch upon the theory of Graph Minors by Robertson and Seymour. This theory gives a very general condition under
Treewidth and graph minors Lectures 9 and 10, December 29, 2011, January 5, 2012 We shall touch upon the theory of Graph Minors by Robertson and Seymour. This theory gives a very general condition under
Running SNAP. The SNAP Team October 2012
 Running SNAP The SNAP Team October 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
Running SNAP The SNAP Team October 2012 1 Introduction SNAP is a tool that is intended to serve as the read aligner in a gene sequencing pipeline. Its theory of operation is described in Faster and More
Performance Evaluations for Parallel Image Filter on Multi - Core Computer using Java Threads
 Performance Evaluations for Parallel Image Filter on Multi - Core Computer using Java s Devrim Akgün Computer Engineering of Technology Faculty, Duzce University, Duzce,Turkey ABSTRACT Developing multi
Performance Evaluations for Parallel Image Filter on Multi - Core Computer using Java s Devrim Akgün Computer Engineering of Technology Faculty, Duzce University, Duzce,Turkey ABSTRACT Developing multi
Implementation of Parallel Path Finding in a Shared Memory Architecture
 Implementation of Parallel Path Finding in a Shared Memory Architecture David Cohen and Matthew Dallas Department of Computer Science Rensselaer Polytechnic Institute Troy, NY 12180 Email: {cohend4, dallam}
Implementation of Parallel Path Finding in a Shared Memory Architecture David Cohen and Matthew Dallas Department of Computer Science Rensselaer Polytechnic Institute Troy, NY 12180 Email: {cohend4, dallam}
! Parallel machines are becoming quite common and affordable. ! Databases are growing increasingly large
 Chapter 20: Parallel Databases Introduction! Introduction! I/O Parallelism! Interquery Parallelism! Intraquery Parallelism! Intraoperation Parallelism! Interoperation Parallelism! Design of Parallel Systems!
Chapter 20: Parallel Databases Introduction! Introduction! I/O Parallelism! Interquery Parallelism! Intraquery Parallelism! Intraoperation Parallelism! Interoperation Parallelism! Design of Parallel Systems!
Chapter 20: Parallel Databases
 Chapter 20: Parallel Databases! Introduction! I/O Parallelism! Interquery Parallelism! Intraquery Parallelism! Intraoperation Parallelism! Interoperation Parallelism! Design of Parallel Systems 20.1 Introduction!
Chapter 20: Parallel Databases! Introduction! I/O Parallelism! Interquery Parallelism! Intraquery Parallelism! Intraoperation Parallelism! Interoperation Parallelism! Design of Parallel Systems 20.1 Introduction!
Chapter 20: Parallel Databases. Introduction
 Chapter 20: Parallel Databases! Introduction! I/O Parallelism! Interquery Parallelism! Intraquery Parallelism! Intraoperation Parallelism! Interoperation Parallelism! Design of Parallel Systems 20.1 Introduction!
Chapter 20: Parallel Databases! Introduction! I/O Parallelism! Interquery Parallelism! Intraquery Parallelism! Intraoperation Parallelism! Interoperation Parallelism! Design of Parallel Systems 20.1 Introduction!
A Study of High Performance Computing and the Cray SV1 Supercomputer. Michael Sullivan TJHSST Class of 2004
 A Study of High Performance Computing and the Cray SV1 Supercomputer Michael Sullivan TJHSST Class of 2004 June 2004 0.1 Introduction A supercomputer is a device for turning compute-bound problems into
A Study of High Performance Computing and the Cray SV1 Supercomputer Michael Sullivan TJHSST Class of 2004 June 2004 0.1 Introduction A supercomputer is a device for turning compute-bound problems into
Systems Infrastructure for Data Science. Web Science Group Uni Freiburg WS 2014/15
 Systems Infrastructure for Data Science Web Science Group Uni Freiburg WS 2014/15 Lecture X: Parallel Databases Topics Motivation and Goals Architectures Data placement Query processing Load balancing
Systems Infrastructure for Data Science Web Science Group Uni Freiburg WS 2014/15 Lecture X: Parallel Databases Topics Motivation and Goals Architectures Data placement Query processing Load balancing
Designing Parallel Programs. This review was developed from Introduction to Parallel Computing
 Designing Parallel Programs This review was developed from Introduction to Parallel Computing Author: Blaise Barney, Lawrence Livermore National Laboratory references: https://computing.llnl.gov/tutorials/parallel_comp/#whatis
Designing Parallel Programs This review was developed from Introduction to Parallel Computing Author: Blaise Barney, Lawrence Livermore National Laboratory references: https://computing.llnl.gov/tutorials/parallel_comp/#whatis
A Comparative Study on Exact Triangle Counting Algorithms on the GPU
 A Comparative Study on Exact Triangle Counting Algorithms on the GPU Leyuan Wang, Yangzihao Wang, Carl Yang, John D. Owens University of California, Davis, CA, USA 31 st May 2016 L. Wang, Y. Wang, C. Yang,
A Comparative Study on Exact Triangle Counting Algorithms on the GPU Leyuan Wang, Yangzihao Wang, Carl Yang, John D. Owens University of California, Davis, CA, USA 31 st May 2016 L. Wang, Y. Wang, C. Yang,
Maximum Clique Problem. Team Bushido bit.ly/parallel-computing-fall-2014
 Maximum Clique Problem Team Bushido bit.ly/parallel-computing-fall-2014 Agenda Problem summary Research Paper 1 Research Paper 2 Research Paper 3 Software Design Demo of Sequential Program Summary Of the
Maximum Clique Problem Team Bushido bit.ly/parallel-computing-fall-2014 Agenda Problem summary Research Paper 1 Research Paper 2 Research Paper 3 Software Design Demo of Sequential Program Summary Of the
Principles of Parallel Algorithm Design: Concurrency and Mapping
 Principles of Parallel Algorithm Design: Concurrency and Mapping John Mellor-Crummey Department of Computer Science Rice University johnmc@rice.edu COMP 422/534 Lecture 3 28 August 2018 Last Thursday Introduction
Principles of Parallel Algorithm Design: Concurrency and Mapping John Mellor-Crummey Department of Computer Science Rice University johnmc@rice.edu COMP 422/534 Lecture 3 28 August 2018 Last Thursday Introduction
Pregel. Ali Shah
 Pregel Ali Shah s9alshah@stud.uni-saarland.de 2 Outline Introduction Model of Computation Fundamentals of Pregel Program Implementation Applications Experiments Issues with Pregel 3 Outline Costs of Computation
Pregel Ali Shah s9alshah@stud.uni-saarland.de 2 Outline Introduction Model of Computation Fundamentals of Pregel Program Implementation Applications Experiments Issues with Pregel 3 Outline Costs of Computation
MICRO-SPECIALIZATION IN MULTIDIMENSIONAL CONNECTED-COMPONENT LABELING CHRISTOPHER JAMES LAROSE
 MICRO-SPECIALIZATION IN MULTIDIMENSIONAL CONNECTED-COMPONENT LABELING By CHRISTOPHER JAMES LAROSE A Thesis Submitted to The Honors College In Partial Fulfillment of the Bachelors degree With Honors in
MICRO-SPECIALIZATION IN MULTIDIMENSIONAL CONNECTED-COMPONENT LABELING By CHRISTOPHER JAMES LAROSE A Thesis Submitted to The Honors College In Partial Fulfillment of the Bachelors degree With Honors in
Graphs and Network Flows IE411. Lecture 21. Dr. Ted Ralphs
 Graphs and Network Flows IE411 Lecture 21 Dr. Ted Ralphs IE411 Lecture 21 1 Combinatorial Optimization and Network Flows In general, most combinatorial optimization and integer programming problems are
Graphs and Network Flows IE411 Lecture 21 Dr. Ted Ralphs IE411 Lecture 21 1 Combinatorial Optimization and Network Flows In general, most combinatorial optimization and integer programming problems are
Let s say I give you a homework assignment today with 100 problems. Each problem takes 2 hours to solve. The homework is due tomorrow.
 Let s say I give you a homework assignment today with 100 problems. Each problem takes 2 hours to solve. The homework is due tomorrow. Big problems and Very Big problems in Science How do we live Protein
Let s say I give you a homework assignment today with 100 problems. Each problem takes 2 hours to solve. The homework is due tomorrow. Big problems and Very Big problems in Science How do we live Protein
Compressing Pattern Databases
 Compressing Pattern Databases Ariel Felner and Ram Meshulam Computer Science Department Bar-Ilan University Ramat-Gan, Israel 92500 Email: felner,meshulr1 @cs.biu.ac.il Robert C. Holte Computing Science
Compressing Pattern Databases Ariel Felner and Ram Meshulam Computer Science Department Bar-Ilan University Ramat-Gan, Israel 92500 Email: felner,meshulr1 @cs.biu.ac.il Robert C. Holte Computing Science
Analysis of Biological Networks. 1. Clustering 2. Random Walks 3. Finding paths
 Analysis of Biological Networks 1. Clustering 2. Random Walks 3. Finding paths Problem 1: Graph Clustering Finding dense subgraphs Applications Identification of novel pathways, complexes, other modules?
Analysis of Biological Networks 1. Clustering 2. Random Walks 3. Finding paths Problem 1: Graph Clustering Finding dense subgraphs Applications Identification of novel pathways, complexes, other modules?
Introduction to Parallel & Distributed Computing Parallel Graph Algorithms
 Introduction to Parallel & Distributed Computing Parallel Graph Algorithms Lecture 16, Spring 2014 Instructor: 罗国杰 gluo@pku.edu.cn In This Lecture Parallel formulations of some important and fundamental
Introduction to Parallel & Distributed Computing Parallel Graph Algorithms Lecture 16, Spring 2014 Instructor: 罗国杰 gluo@pku.edu.cn In This Lecture Parallel formulations of some important and fundamental
Approximating Fault-Tolerant Steiner Subgraphs in Heterogeneous Wireless Networks
 Approximating Fault-Tolerant Steiner Subgraphs in Heterogeneous Wireless Networks Ambreen Shahnaz and Thomas Erlebach Department of Computer Science University of Leicester University Road, Leicester LE1
Approximating Fault-Tolerant Steiner Subgraphs in Heterogeneous Wireless Networks Ambreen Shahnaz and Thomas Erlebach Department of Computer Science University of Leicester University Road, Leicester LE1
Reducing Disk Latency through Replication
 Gordon B. Bell Morris Marden Abstract Today s disks are inexpensive and have a large amount of capacity. As a result, most disks have a significant amount of excess capacity. At the same time, the performance
Gordon B. Bell Morris Marden Abstract Today s disks are inexpensive and have a large amount of capacity. As a result, most disks have a significant amount of excess capacity. At the same time, the performance
Virtuozzo Containers
 Parallels Virtuozzo Containers White Paper An Introduction to Operating System Virtualization and Parallels Containers www.parallels.com Table of Contents Introduction... 3 Hardware Virtualization... 3
Parallels Virtuozzo Containers White Paper An Introduction to Operating System Virtualization and Parallels Containers www.parallels.com Table of Contents Introduction... 3 Hardware Virtualization... 3
Analysis of parallel suffix tree construction
 168 Analysis of parallel suffix tree construction Malvika Singh 1 1 (Computer Science, Dhirubhai Ambani Institute of Information and Communication Technology, Gandhinagar, Gujarat, India. Email: malvikasingh2k@gmail.com)
168 Analysis of parallel suffix tree construction Malvika Singh 1 1 (Computer Science, Dhirubhai Ambani Institute of Information and Communication Technology, Gandhinagar, Gujarat, India. Email: malvikasingh2k@gmail.com)
Distributed Data Structures and Algorithms for Disjoint Sets in Computing Connected Components of Huge Network
 Distributed Data Structures and Algorithms for Disjoint Sets in Computing Connected Components of Huge Network Wing Ning Li, CSCE Dept. University of Arkansas, Fayetteville, AR 72701 wingning@uark.edu
Distributed Data Structures and Algorithms for Disjoint Sets in Computing Connected Components of Huge Network Wing Ning Li, CSCE Dept. University of Arkansas, Fayetteville, AR 72701 wingning@uark.edu
