Automatic Localisation of Vertebrae in DXA Images using Random Forest Regression Voting
|
|
|
- Carmella Daniels
- 6 years ago
- Views:
Transcription
1 Automatic Localisation of Vertebrae in DXA Images using Random Forest Regression Voting Paul A. Bromiley, Judith E. Adams 2, and Timothy F. Cootes Imaging Sciences Research Group, University of Manchester, UK. {paul.bromiley, 2 Radiology & Manchester Academic Health Science Centre, Central Manchester University Hospitals NHS Foundation Trust, UK. Abstract. We describe a method for automatic detection and localisation of vertebrae in clinical images that was designed to avoid making a-priori assumptions of how many vertebrae are visible. Multiple Random Forest regressors were trained to identify vertebral end-plates, providing estimates of both the location and pose of the vertebrae. The highest-weighted responses from each model were combined using a Hough-style voting array. A graphical approach was then used to extract contiguous sets of detections representing neighbouring vertebrae, by finding a path linking modes of high weight, subject to pose constraints. The method was evaluated on 32 lateral dual-energy X-ray absorptiometry spinal images with a high prevalence of osteoporotic vertebral fractures, and detected 92% of the vertebrae between T7 and L4 with a mean localisation error of 2.36mm. When used to initialise a Constrained Local Model segmentation of the vertebrae, the method increased the incidence of fit failures from.5% to 2.% compared to manual initialisation, and produced no difference in fracture classification using a simple classifier. Introduction Osteoporosis is a common skeletal disorder defined by a reduction in bone mineral density (BMD) resulting in a T-score of <2.5 (i.e. more than 2.5 standard deviations below the mean in young adults), measured using dual energy X-ray absorptiometry (DXA) [6]. It significantly increases the risk of fractures, most commonly occurring in the hips, wrists or vertebrae. Approximately 4% of postmenopausal Caucasian women are affected, increasing their lifetime risk of fragility fractures to as much as 4% [6]. Osteoporosis therefore presents a significant public health problem for an ageing population. However, between 3%-6% of vertebral fractures may be asymptomatic and only about one third of those present on images come to clinical attention; they are frequently not noted by radiologists, not entered into medical notes, and do not lead to preventative treatments [6]. Many of these cases involve images acquired for purposes other than assessment for the presence of vertebral fractures, so identification may be opportunistic. However, a recent multi-centre, multinational prospective study [7] found a false negative rate of 34% for reporting vertebral fractures from lateral radiographs of the thoracolumbar spine. The potential utility of computer-aided vertebral fracture identification systems is therefore considerable. Modern clinical imaging is primarily
2 2 P.A. Bromiley et al. digital, with images acquired in Digital Imaging and Communications in Medicine (DI- COM) format and stored on a Picture Archiving and Communication System (PACS). A system that could interface with a PACS, use information from the DICOM header to select images that cover a portion of the spine, automatically segment vertebrae, and detect any abnormal shape would therefore be particularly valuable. Several authors have investigated the use of methods based on statistical shape models to segment vertebrae in both radiographs, e.g. [4], and DXA images e.g. [7]. In recent work [2], the Random Forest Regression Voting Constrained Local Model (RFRV- CLM) [4] was applied to DXA images. This approach uses Random Forest (RF) [] regressors to predict the locations of landmarks that delineate the vertebrae. Each RF predicts one landmark, and fitting is performed subject to a constraint provided by a global shape model. The RFRV-CLM was shown to be more robust, i.e. suffered from a smaller number of fitting failures on more severely fractured vertebrae, than the Active Appearance Model [5], an important consideration in clinical tasks where pathological cases are most significant. High-resolution RFRV-CLMs require initialisation relatively close to the location of the structure being segmented. The use of multi-stage, coarseto-fine models can reduce the required initialisation accuracy. However, fully automatic application of such models still requires an initial estimate of the location of each vertebra. This is problematic due to the repetitive nature of vertebrae and the extensive shape differences between normal and pathological anatomy. A variety of methods that detect potential vertebral candidates have been proposed; see [] for references. However, most assume that the number of vertebrae visible in the image is known a-priori. This is a significant limitation, particularly when using midline sagittal reformatted images from CT scans performed for various clinical indicators and not specifically acquired to view the spine, since the region of the body imaged can vary significantly. DXA images acquired for vertebral fracture assessment (VFA) typically cover the anatomy from T4 to L4. However, confounding bony structures, adipose tissue in the abdomen, or variation in the field of view can result in vertebrae being obscured or omitted, leading to fit failures with models that assume a certain number of vertebrae are present and can be accurately located. Several authors have proposed solutions to the more general problem of vertebra localisation where the number present is not known a-priori. [2] described a framework for fully automatic localisation, identification and segmentation of vertebrae in CT images, producing a triangulated surface mesh and a level label for all visible vertebrae. However, a complex chain of processing steps was required. [] used RF regressors trained on features from arbitrary, unregistered CT images to predict the locations of all visible vertebral centroids simultaneously, followed by a refinement stage using a probabilistic graphical model. Later work [] reported that this approach had problems with images featuring pathology or very narrow fields of view, and instead used RF classifiers to generate a probabilistic label map for vertebral centroids, combined with a shape and appearance model to remove false positives. We describe a method for detecting vertebrae in clinical images, based on RF regressors, that is intended to be robust to obscured or non-present vertebrae. The key idea is to use a set of relatively non-specific regressors trained to predict the location and pose of lower vertebral end-plates. Each regressor detects multiple vertebrae in a query im-
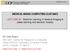
3 Automatic Localisation of Vertebrae using RFRV 3 Fig. : (Left) Example lateral DXA spinal image with manual annotations of 45 points on T7-L4. (Middle) Top detections from each of the lower end-plate detectors, overlaid on the manual annotations. The hue indicates the vertebral level on which the detector was trained, from T7 (green) to L4 (red), and the saturation and line thickness indicate the weighting. (Right) The smoothed Hough voting array produced from the posterior point of the end-plate detections, and the detected modes. age, and so does not require a specific vertebra to be present and visible. The results are combined using a Hough-style voting array, and the modes of the smoothed array represent potential detections. A graphical approach is then used to find the highest-weighted path through the modes subject to pose constraints. An evaluation on 32 DXA VFA images with a high prevalence of osteoporotic fractures is described in Section 3. 2 Method The algorithm described here was based on our own implementation of Hough Forests [8], and used a set of RF regressors, each trained on a different vertebral level, to predict the offset to a distinctive portion of that vertebra given local patches of image features. Figures and 2 show the various stages of the algorithm. Training data consisted of a set of lateral DXA spinal images I with manual annotations x l of N points l =...N on
4 4 P.A. Bromiley et al. Fig. 2: (Left) The highest-weight set of linked modes from the Hough array, overlaid on the original image (see Fig. ). (Middle) Example fitted 99-point RFRV-CLM triplet model initialised with the three lowest automatic detections. (Right) Concatenated points from the central vertebra in all fitted RFRV-CLM triplet models initialised from the automatic vertebra detections. each, outlining the vertebrae (see Fig. ). The two end points of the curve that delineated the lower end plate were extracted. The lower end-plate was used as, of the four sides of the vertebra visible in the lateral view, it tends to exhibit the smallest changes in size and pose when osteoporotic fractures are present. The two reference points were used to calculate the parameters θ of a similarity registration that transformed the image into a standardised reference frame, such that the reference points were transformed to specific coordinates. The image was then resampled into this frame by applying I r (m,n) = I(Tθ (m, n)), where (m, n) specify pixel coordinates. A scaling parameter w f rame set the reference frame width in pixels, allowing variation of the resolution. A set of random displacements d j was then generated by sampling from a uniform distribution in the range[ d max,+d max ] in x and y. Patches of image data of area w 2 patch were extracted from the reference frame images at these displacements, and features f j extracted from them. Haar-like features were used [8], as they have proven effective for a range of applications and can be calculated efficiently from integral images.
5 Automatic Localisation of Vertebrae using RFRV 5 The process was repeated with random perturbations in scale and orientation to make the detector locally pose-invariant. The free parameters were chosen to set w patch to twice the length of the lower end-plate and the reference points to relative coordinates (.75,.25) and(.75,.75) in an undisplaced patch i.e. the patch size was equivalent to an entire vertebra plus a border of (approximately, given that vertebrae are not square) one quarter of the end-plate length around each boundary. The resolution was set to mm per pixel, and d max was set to.4 of the patch size, so that the patches always covered more of the target vertebra than its neighbours. The pose variation was set to. radians, the resolution of the search (see below). A RF was then constructed; each tree was trained on a bootstrap sample of pairs {(f j,d j )} from the training data using a standard, greedy approach. At each node, a random set of n f eat features was chosen by sampling from a uniform distribution of range...n MRS and using the result as skip sizes through the feature list. A feature f i and threshold t that best split the data into two compact groups were chosen by minimising G T (t)=g({d i : f i < t})+g({d i : f i t}) where G({d i })=N s log Σ () G(S) is an entropy measure; N s is the number of samples and Σ is their covariance matrix. Splitting terminated at either a maximum depth, D max, or a minimum number of samples, N min. The process was repeated to generate a forest of size n trees. Free parameters of the RFs (N MRS, D max, N min, n trees ) were set to the values given in [2]. To detect vertebrae in a query image, a grid of points covering the image was defined; the resolution of this grid was set to 3mm. Each of the RF end-plate detectors was applied at each grid location. The required Haar-like features were extracted and passed into the RF, which output a prediction of the displacement to the reference points. The process was repeated with a range of angle and scale variation. Since RF searching is fast, optimisation of free parameters was avoided by using large search ranges: to radians in steps of., and scales from. to 4 in rational/integer steps. For each detector, the predictions were used to vote into a Hough array. Each vote was weighted by the determinant of the covariance matrix of the samples that reached the relevant RF leaf node. The array was then smoothed using a Gaussian kernel of standard deviation (s.d.) mm, to allow mode detection using a 9-way maximum, and the modes detected using exhaustive search. Each mode was a potential end-plate detection. Vertebrae are repeating structures with considerable similarity in shape between neighbours. Therefore, each RF end-plate detector tended to locate multiple vertebrae across a considerable range of the spine (see Fig. ). This provided robustness to variations in exactly which vertebrae were visible in the image. A second stage of Hough voting was performed to combine the results from the multiple models and estimate the location and pose of all detected vertebrae. The highest-weighted N modes modes from each detector were used to cast votes into a single Hough array. Each vote was weighted by the length of the line subtended by the two reference points. This had two effects. First, it acted as a shape prior; vertebrae are the largest structures in spinal images that might be delineated by parallel edges, so the weighting aided in elimination of false detections on ribs, humerus etc. Second, it weighted the vote according to the amount This was performed as a separate step for implementation reasons. We have not yet investigated the possibility of performing the search for all RF regressors using a combined array.
6 6 P.A. Bromiley et al. of information it contained about the vertebral pose. The array was then smoothed with a Gaussian kernel of s.d. σ comb, and modes were detected. Estimating the location and pose simultaneously would require a four-dimensional array, which would be inefficient in terms of memory requirements, and would require a large N modes to avoid problems with sparsity. Therefore, a two stage approach was adopted. The posterior-most point of the detection was used to vote into a 2D array. For each detected mode in this array, all votes within 3σ comb were extracted, and their anterior-most points were used to vote into a second 2D array. This was again smoothed using a Gaussian kernel of s.d. σ comb, and the main mode was detected. σ comb was set to 5mm, and N modes was set to. The result of the combined Hough voting was a single set of N m potential lower end-plate detections (see Fig. ), containing false positives. To initialise a subsequent appearance model, an ordered set of points with no missing vertebrae or false detections was required, and so a graphical method was used to extract the highest-weighted subset of modes subject to pose constraints. This could be performed by optimising over all possible paths through the graph. In practice, it was found to be sufficient to start from the highest-weighted mode in the posterior-point Hough array. The main mode of the corresponding anterior-point Hough array gave an estimate of the vertebral pose. The normals to the vector between these modes specified the local superior and inferior directions of the spine, and a path was extracted by searching in each direction using arg min p i p c s.t. w i > w t, i=...n m,i c rot(a c p c,±π/2).(p i p c ) rot(a c p c,±π/2) (p i p c ) > cosθ t (2) where p i is the location of mode i in the posterior-point Hough array, a i the corresponding anterior point, c specifies the current mode, w i is the weight of mode i, w t and θ t are thresholds, and ± specifies the search direction. θ t was set to twice the angular resolution of the RF search, and w t was set to the 2σ value of a Gaussian distribution with a s.d. of σ comb and a mean scaled to the height of the highest-weighted mode in the posterior-point Hough array. This eliminated any statistically insignificant modes whilst accounting both for the smoothing applied and the typical width of the distribution of votes contributing to the modes. The result of the search was a set of ordered points that specified the posterior-most points on the lower end-plates; an example is given in Fig. 2. The remaining orientation ambiguity was resolved by assuming that the superior-to-inferior axis of the body corresponded more closely to the direction of increasing image y-coordinate. The extracted path might have missing detections, posing a problem for initialisation of subsequent shape and appearance models. Therefore, a second stage of searching through the posterior-point array, equivalent to the use of an adaptive threshold, was included. The lengths of the links in the path were compared to normative vertebral heights learned from the training data, assuming that the lowest detection was on L4, and any heights more than.5 times that expected were referred for the second search. A threshold w t was calculated using the same procedure as w t, but the 3σ point of the distribution. The values in the posterior-point voting array along the link were analysed at a set of rational steps (/2, /3 and 2/3 etc.). The set, if any, that had the highest proportional weight, and individual weights higher than w t, was added to the path. This procedure was sub-optimal as it assumed a vertebral level assignment and, in future work, we intend to replace it with a robust, shape-model based procedure.
7 Automatic Localisation of Vertebrae using RFRV 7 Proportion of points.4.2 Linked modes Linked and filled modes Point-to-point error (mm) Proportion of images.4.2 Linked modes Linked and filled modes Number of points detected Fig. 3: Cumulative distribution function (CDF) of the P-to-P errors (left) and number of points detected (right) for the 32 DXA images, after path extraction and missing point detection. Proportion of Vertebrae T8 T9.4 T T T2.2 L L2 L Mean point-to-curve error (mm) Proportion of Vertebrae T7 T8 T9 T.4 T T2 L.2 L2 L3 L Mean point-to-curve error (mm) Fig. 4: CDFs of the mean P-to-C errors of the 33 points on the central vertebra in each RFRV-CLM triplet model for the 32 DXA images, for each vertebral level, for manual (left) and automatic (right) initialisation. 3 Evaluation The method was evaluated on 32 DXA VFA images scanned on various Hologic (Bedford MA) scanners 2. Manual annotations of 45 landmarks were available for each image, covering the vertebrae from T7 to L4 (see Fig. ). Each of these vertebrae was also classified by an expert radiologist into one of five groups (normal, deformed but not fractured, and grade, 2 and 3 fractures according to the Genant definitions [9]). The automatic initialisation algorithm was applied in a leave-/4-out procedure, using 3/4 of the data to train the detectors and then applying them to the remaining /4. Figure 3 shows the accuracy of the detected lower end-plate points, and the number of such points detected, after the extraction of the linked path and after the additional 2 44 patients from a previous study [5]; 8 female subjects in an epidemiological study of a UK cohort born in 946; 96 females attending a local clinic for DXA BMD measurement, for whom the referring physician had requested VFA (approved by the local ethics committee).
8 8 P.A. Bromiley et al. Proportion of Vertebrae.4 Normal Deformed.2 Grade Grade 2 Grade Mean point-to-curve error (mm) Proportion of Vertebrae.4 Normal Deformed.2 Grade Grade 2 Grade Mean point-to-curve error (mm) Fig. 5: CDFs of the mean P-to-C errors of the 33 points on the central vertebra in each RFRV-CLM triplet model for the 32 DXA images, for each vertebral status, for manual (left) and automatic (right) initialisation. Wegde ratio Biconcavity ratio Sensitivity Manual annotations 5 Manual initialisation Automatic initialisation False Positive Rate Fig. 6: (Left) The whitened biconcavity and wedge ratios used by the simple classifier (black=normal; red=deformed; green,blue,cyan=grade,2,3 fractures, respectively). (Right) ROC curves for classification of the detected vertebrae using the manual annotations, and RFRV-CLM annotations with manual and automatic initialisation. search to detect missing points in the path. A vertebral level was manually assigned to each detection, by finding the closest manual posterior lower end-plate point. Errors are given as the Euclidean distance, in millimetres, between the detected point and the corresponding manual annotation (point-to-point, or P-to-P, error). Additional vertebrae were detected above T7 in many images, and below L4 in some. Since manual annotations were not available for these, detections above the centroid of T7, and below the centroid of L4 plus its height, were removed from the analysis. The proportions of the vertebrae detected at each level are given in Table. There is a reduction in the proportion of detections above T and for L4, where the vertebrae are obscured by other bony structures. However, over 9% of the vertebrae were detected at other levels. The additional search to detect missing points resulted in no significant accuracy loss, but a 4.4% rise in detected points. Nine out of the ten target points were found in 85% of the images, and at least six were found in 99% of the images. RF searching was inefficient
9 Automatic Localisation of Vertebrae using RFRV 9 Table : Statistics of the mean P-to-C errors on each vertebra after RFRV-CLM fitting, using manual and automatic initialisation. Column five gives the %age of vertebrae at each level that had an end-plate detection and, in brackets, a central vertebra from a RFRV-CLM fit assigned to them. Fitting of L4 required L5 to be detected; this is rarely present in DXA VFA images, hence the low percentage of L4 fits. Manual Initialisation Automatic Initialisation Vertebral Level Median (mm) Mean (mm) Errors > 2mm (%) Detected (%) Median (mm) Mean (mm) Errors > 2mm (%) T (46.87) T (7.2) T (88.75) T (95.63) T (98.2) T (99.38) L (99.38) L (98.75) L (79.38).53. L (3.3).53. by design (see Sec. 2) and took 6.7s per regressor per image on a machine with two Xeon X567 processors using a single core; the Hough voting based combination of the results and extraction of the linked modes took.24s per image. The automatically detected lower end-plate points were then used to initialise the fitting of a Random Forest Regression Voting Constrained Local Model (RFRV-CLM) covering a triplet of neighbouring vertebrae, as described in [2], and the results compared to those achieved using manual initialisation of the same points. This model covered 99 points (33 on each of the vertebrae in the triplet). For the automatic initialisation, the RFRV-CLM was applied to all triplets of neighbouring initialisation points. The 33 points on the central vertebra were then extracted; no use was made of the overlap of neighbouring models. A vertebral level was manually assigned by comparing the centroid of these points and of the manual annotations for each vertebra. Additional vertebrae above T7 were again removed from the analysis, and errors were calculated for each point as the minimum Euclidean distance to a piecewise linear curve through the manual annotations (point-to-curve or P-to-C error), to compensate for the aperture problem with dense annotations on extended edges. The mean error over each fitted vertebra was then calculated. The same metric was applied to the manually initialised fits and, for consistency, only those triplets for which all three of the initialisation points were available were included. Therefore, results on T7 and L4 were not available for manual initialisation. Example results from a single triplet model fit, and the concatenated central vertebra points from all triplets, are shown in Fig. 2. Figures 4 and 5 show CDFs of the mean P-to-C error on manually and automatically initialised RFRV-CLM fits for each vertebral level, and for each vertebral status. Tables
10 P.A. Bromiley et al. Table 2: Statistics of the mean P-to-C errors on each vertebra after RFRV-CLM fitting, using manual and automatic initialisation. Manual Initialisation Automatic Initialisation Vertebra %age of Median Mean Errors Detected Median Mean Errors Status Sample (mm) (mm) > 2mm (%) (%) (mm) (mm) > 2mm (%) Normal ± Deformed ± Grade ± Grade ± Grade ± and 2 provide statistics derived from these curves, including the percentage of vertebrae with mean P-to-C errors above 2mm, which gives an indication of the proportion of fit failures. There is little difference between the medians or means for either status or level, indicating that the accuracy of the RFRV-CLM fitting is largely independent of the initialisation as long as it is within the capture range of the model. The percentage of fitting failures on all detected vertebrae between T8 and L3 increased from.5% for manual initialisation to 2.% for automatic, but the increase was larger for fractured vertebrae. However, using the 2mm threshold, at least 86.7% of vertebrae were successfully fitted, regardless of status. Importantly, there was no statistically significant variation in the proportion of vertebrae detected with vertebral status i.e. no evidence of bias against detecting fractured or non-fractured vertebrae. To evaluate the importance of the differences in segmentation accuracy, a simple classifier based on the standard six-point morphometry technique was applied to the results from both manually and automatically initialised RFRV-CLM fits. The anterior H a, middle H m, and posterior H p heights of each vertebra were extracted. A predicted posterior height H p of each vertebra was also calculated by taking the posterior heights of the closest four annotated vertebrae, multiplying by ratios of vertebral heights in normative data obtained from [3], and taking the maximum of the four values (on the basis that fractures reduce vertebral height). This process used the vertebral level assignment derived from the manual annotations for the automatically initialised results. Three ratios were then calculated: the biconcavity ratio H m /H p, the wedge ratio H a /H p, and the crush ratio H p /H p. The data were whitened by subtracting the median and dividing by the square root of the covariance matrix (estimated using the median absolute deviation). The resulting data for the automatically initialised annotations are shown in Fig. 6, showing clear separation between the normal and fractured vertebrae and, in particular, that deformed vertebrae are displaced from the normal class along a different vector through the space than the fractured vertebrae. However, since only a simple classifier was intended, a threshold was applied to the Euclidean distance between the origin and the point defined by the three height ratios, to classify the vertebrae into non-fractured (normal and deformed) and fractured (grade, 2 and 3) classes. Figure 6 shows ROC curves produced by varying the threshold. The simple classifier achieves 9% sensitiv-
11 Automatic Localisation of Vertebrae using RFRV ity at a false positive rate of % when applied to the manual annotations. Using the manually initialised annotations reduced this to 8%, largely due to errors on grade fractures, where shape changes can be quite subtle. However, there was no significant difference in classifier accuracy between manual and automatic initialisation. 4 Conclusion This paper has described a method for automatic detection and localisation of vertebrae in clinical images. The algorithm relies on a set of RF regressors trained to predict the location of vertebral lower end-plates. We have demonstrated that the individual RFs are not specific to the vertebrae they were trained on but instead, due to the similarity of neighbouring vertebrae, respond over a considerable range of the spine. This provides robustness to vertebrae that are obscured or not present. The use of multiple models, and Hough voting to combine their results, provides robustness to fit failures [3]. The failure of any one RF to detect vertebrae will result in responses scattered throughout the voting array. Only locations that result in strong responses from multiple models will result in significant modes. These are detected, and a graphical method applied to find a path through the detections, subject to pose constraints. This can be used to initialise a high-resolution appearance model that provides an accurate segmentation. The method was evaluated on 32 DXA VFA images with a high prevalence of osteoporotic fractures. Other authors who have studied this problem [] [] [2] have used CT images that, since the modality is used for a wider range of clinical purposes, show a much larger variation in the region of the spine being imaged and the number of vertebrae visible. The difference in the dimensionality of the the images prevents any comparison of localisation accuracy. However, we note that the proportions of vertebrae detected in [] and [] are similar to the detection rates presented here. Instead, our evaluation focused on using the automatic annotations, and the equivalent manually annotated points, to initialise a RFRV-CLM. This demonstrated no difference in accuracy when the initialisation was within the capture range of the RFRV-CLM. Automatic initialisation led to more fit failures on grade fractures, where the shape change may be subtle. However, the increase was smaller on grade 2 and 3 fractures. A simple classifier was applied, and showed little difference in performance between RFRV-CLM segmentations using manual and automatic initialisation. One drawback of the algorithm is that it implicitly assumes prior knowledge of an overall image scaling, in the form of the pixel size in mm, although this information is available in the DICOM header for digital clinical images. Another is the relatively simple technique used to extract the path through the modes of the voting array. An appearance model based technique, such as that described in [], might prove more robust. Finally, we have not yet evaluated the technique on images showing wide variation in the number of visible vertebrae. These issues will be addressed in future work. Acknowledgment This publication presents independent research supported by the Health Innovation Challenge Fund (grant no. HICF-R7-44/WT936), a parallel funding partnership between the Depart-
12 2 P.A. Bromiley et al. ment of Health and Wellcome Trust. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health or Wellcome Trust. References. Breiman, L.: Random Forests. Machine Learning 45, 5 32 (2) 2. Bromiley, P.A., Adams, J.E., Cootes, T.F.: Localisation of Vertebrae on DXA Images using Constrained Local Models with Random Forest Regression Voting. In: Recent Advances in Computational Methods and Clinical Applications for Spine Imaging. Lecture Notes in Computational Vision and Biomechanics, vol. 2, pp Springer (25) 3. Bromiley, P.A., Schunke, A.C., Ragheb, H., Thacker, N.A., Tautz, D.: Semi-automatic landmark point annotation for geometric morphometrics. Front Zool (6), 2 (24) 4. de Bruijne, M., Lund, M., Tanko, L., Pettersen, P., Nielsen, M.: Quantitative vertebral morphometry using neighbour-conditional shape models. Med Image Anal, (27) 5. Cootes, T.F., Edwards, G.J., Taylor, C.J.: Active appearance models. IEEE TPAMI 23, (2) 6. Cummings, S.R., Melton, J.: Epidemiology and outcomes of osteoporotic fractures. The Lancet 359(939), (22) 7. Delmas, P.D., van de Langerijt, L., Watts, N.B., Eastell, R., Genant, H.K., Grauer, A., Cahall, D.L.: Underdiagnosis of vertebral fractures is a worldwide problem: the IMPACT study. J Bone Miner Res 2(4), (25) 8. Gall, J., Lempitsky, V.: Class-specific Hough forests for object detection. In: Proc. CVPR. pp IEEE Computer Society (29) 9. Genant, H.K., Wu, C.Y., Kuijk, C.V., Nevitt, M.C.: Vertebral fracture assessment using a semi-quantitative technique. J Bone Miner Res 8(9), (993). Glocker, B., Feulner, J., Criminisi, A., Haynor, D.R., Konukolu, E.: Automatic localization and identification of vertebrae in arbitrary field-of-view CT scans. In: Proc. MICCAI 22. LNCS, vol. 752, pp Springer (22). Glocker, B., Zikic, D., Konukoglu, E., Haynor, D.R., Criminisi, A.: Vertebrae Localization in Pathological Spine CT via Dense Classification from Sparse Annotations. In: Proc. MICCAI 23, LNCS, vol. 85, pp Springer (23) 2. Klinder, T., Ostermann, J., Ehm, M., Franz, A., Kneser, R., Lorenz, C.: Automated modelbased vertebra detection, identification, and segmentation in CT images. Med Image Anal 3(3), (29) 3. Leidig-Bruckner, G., Minne, H.W.: The Spine Deformity Index (SDI); A New Approach to Quantifying Vertebral Crush Fractures in Patients with Osteoporosis. In: Vertebral Fracture in Osteoporosis, pp Osteoporosis Research Group, University of California (995) 4. Lindner, C., Bromiley, P.A., Ionita, M., Cootes, T.F.: Robust and Accurate Shape Model Matching using Random Forest Regression-Voting. IEEE TPAMI p. In press (25) 5. McCloskey, E., Selby, P., de Takats, D., Bernard, J., Davies, M., Robinson, J., Francis, R., Adams, J., Pande, K., Beneton, M., Jalava, T., Loyttyniemi, E., Kanis, J.A.: Effects of clodronate on vertebral fracture risk in osteoporosis: a -year interim analysis. Bone 28(3), 3 35 (2) 6. Rachner, T.D., Khosla, S., Hofbauer, L.C.: Osteoporosis: now and the future. The Lancet 377(9773), (2) 7. Roberts, M.G., Cootes, T.F., Adams, J.E.: Automatic Location of Vertebrae on DXA Images Using Random Forest Regression. In: Proc. MICCAI 22. LNCS, vol. 752, pp Springer-Verlag, Berlin (22) 8. Viola, P., Jones, M.: Rapid object detection using a boosted cascade of simple features. In: Proc. CVPR. pp IEEE Computer Society (2)
Localisation of Vertebrae on DXA Images using Constrained Local Models with Random Forest Regression Voting
 Localisation of Vertebrae on DXA Images using Constrained Local Models with Random Forest Regression Voting P.A. Bromiley, J.E. Adams and T.F. Cootes Imaging Sciences Research Group, University of Manchester,
Localisation of Vertebrae on DXA Images using Constrained Local Models with Random Forest Regression Voting P.A. Bromiley, J.E. Adams and T.F. Cootes Imaging Sciences Research Group, University of Manchester,
Fully Automatic Localisation of Vertebrae in CT images using Random Forest Regression Voting
 Fully Automatic Localisation of Vertebrae in CT images using Random Forest Regression Voting Paul A. Bromiley 1, Eleni P. Kariki 2, Judith E. Adams 2, and Timothy F. Cootes 1 1 Imaging Sciences Research
Fully Automatic Localisation of Vertebrae in CT images using Random Forest Regression Voting Paul A. Bromiley 1, Eleni P. Kariki 2, Judith E. Adams 2, and Timothy F. Cootes 1 1 Imaging Sciences Research
Accurate Bone Segmentation in 2D Radiographs Using Fully Automatic Shape Model Matching Based On Regression-Voting
 Accurate Bone Segmentation in 2D Radiographs Using Fully Automatic Shape Model Matching Based On Regression-Voting C. Lindner, S. Thiagarajah 2, J.M. Wilkinson 2, arcogen Consortium, G.A. Wallis 3, and
Accurate Bone Segmentation in 2D Radiographs Using Fully Automatic Shape Model Matching Based On Regression-Voting C. Lindner, S. Thiagarajah 2, J.M. Wilkinson 2, arcogen Consortium, G.A. Wallis 3, and
THE ability to accurately detect feature points of
 IEEE TRANSACTIONS ON PATTERN ANALYSIS AND MACHINE INTELLIGENCE Robust and Accurate Shape Model Matching using Random Forest Regression-Voting Claudia Lindner*, Paul A. Bromiley, Mircea C. Ionita and Tim
IEEE TRANSACTIONS ON PATTERN ANALYSIS AND MACHINE INTELLIGENCE Robust and Accurate Shape Model Matching using Random Forest Regression-Voting Claudia Lindner*, Paul A. Bromiley, Mircea C. Ionita and Tim
Available online at ScienceDirect. Procedia Computer Science 90 (2016 ) 48 53
 Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 9 (26 ) 48 53 International Conference On Medical Imaging Understanding and Analysis 26, MIUA 26, 6-8 July 26, Loughborough,
Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 9 (26 ) 48 53 International Conference On Medical Imaging Understanding and Analysis 26, MIUA 26, 6-8 July 26, Loughborough,
Semi-Automatic Detection of Cervical Vertebrae in X-ray Images Using Generalized Hough Transform
 Semi-Automatic Detection of Cervical Vertebrae in X-ray Images Using Generalized Hough Transform Mohamed Amine LARHMAM, Saïd MAHMOUDI and Mohammed BENJELLOUN Faculty of Engineering, University of Mons,
Semi-Automatic Detection of Cervical Vertebrae in X-ray Images Using Generalized Hough Transform Mohamed Amine LARHMAM, Saïd MAHMOUDI and Mohammed BENJELLOUN Faculty of Engineering, University of Mons,
Fully automatic cephalometric evaluation using Random Forest regression-voting
 Fully automatic cephalometric evaluation using Random Forest regression-voting Claudia Lindner and Tim F. Cootes Centre for Imaging Sciences, University of Manchester, UK Abstract. Cephalometric analysis
Fully automatic cephalometric evaluation using Random Forest regression-voting Claudia Lindner and Tim F. Cootes Centre for Imaging Sciences, University of Manchester, UK Abstract. Cephalometric analysis
Application of the Active Shape Model in a commercial medical device for bone densitometry
 Application of the Active Shape Model in a commercial medical device for bone densitometry H. H. Thodberg and A. Rosholm, Pronosco, Kohavevej 5, 2950 Vedbaek, Denmark hht@pronosco.com Abstract Osteoporosis
Application of the Active Shape Model in a commercial medical device for bone densitometry H. H. Thodberg and A. Rosholm, Pronosco, Kohavevej 5, 2950 Vedbaek, Denmark hht@pronosco.com Abstract Osteoporosis
Improving an Active Shape Model with Random Classification Forest for Segmentation of Cervical Vertebrae
 Improving an Active Shape Model with Random Classification Forest for Segmentation of Cervical Vertebrae S M Masudur Rahman Al Arif 1, Michael Gundry 2, Karen Knapp 2 and Greg Slabaugh 1 1 Department of
Improving an Active Shape Model with Random Classification Forest for Segmentation of Cervical Vertebrae S M Masudur Rahman Al Arif 1, Michael Gundry 2, Karen Knapp 2 and Greg Slabaugh 1 1 Department of
Vertebrae Segmentation in 3D CT Images based on a Variational Framework
 Vertebrae Segmentation in 3D CT Images based on a Variational Framework Kerstin Hammernik, Thomas Ebner, Darko Stern, Martin Urschler, and Thomas Pock Abstract Automatic segmentation of 3D vertebrae is
Vertebrae Segmentation in 3D CT Images based on a Variational Framework Kerstin Hammernik, Thomas Ebner, Darko Stern, Martin Urschler, and Thomas Pock Abstract Automatic segmentation of 3D vertebrae is
Automatic Vertebra Detection in X-Ray Images
 Automatic Vertebra Detection in X-Ray Images Daniel C. Moura INEB - Instituto de Engenharia Biomédica, Laboratório de Sinal e Imagem, Porto, Portugal Instituto Politécnico de Viana do Castelo, Escola Superior
Automatic Vertebra Detection in X-Ray Images Daniel C. Moura INEB - Instituto de Engenharia Biomédica, Laboratório de Sinal e Imagem, Porto, Portugal Instituto Politécnico de Viana do Castelo, Escola Superior
Vertebrae Localization in Pathological Spine CT via Dense Classification from Sparse Annotations
 Vertebrae Localization in Pathological Spine CT via Dense Classification from Sparse Annotations B. Glocker 1, D. Zikic 1, E. Konukoglu 2, D. R. Haynor 3, A. Criminisi 1 1 Microsoft Research, Cambridge,
Vertebrae Localization in Pathological Spine CT via Dense Classification from Sparse Annotations B. Glocker 1, D. Zikic 1, E. Konukoglu 2, D. R. Haynor 3, A. Criminisi 1 1 Microsoft Research, Cambridge,
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests)
 SPRING 2016 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
SPRING 2016 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
Supplementary methods
 Supplementary methods This section provides additional technical details on the sample, the applied imaging and analysis steps and methods. Structural imaging Trained radiographers placed all participants
Supplementary methods This section provides additional technical details on the sample, the applied imaging and analysis steps and methods. Structural imaging Trained radiographers placed all participants
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues
 Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
Summarising Data. Mark Lunt 09/10/2018. Arthritis Research UK Epidemiology Unit University of Manchester
 Summarising Data Mark Lunt Arthritis Research UK Epidemiology Unit University of Manchester 09/10/2018 Summarising Data Today we will consider Different types of data Appropriate ways to summarise these
Summarising Data Mark Lunt Arthritis Research UK Epidemiology Unit University of Manchester 09/10/2018 Summarising Data Today we will consider Different types of data Appropriate ways to summarise these
Automated Model-Based Rib Cage Segmentation and Labeling in CT Images
 Automated Model-Based Rib Cage Segmentation and Labeling in CT Images Tobias Klinder 1,2,CristianLorenz 2,JensvonBerg 2, Sebastian P.M. Dries 2, Thomas Bülow 2,andJörn Ostermann 1 1 Institut für Informationsverarbeitung,
Automated Model-Based Rib Cage Segmentation and Labeling in CT Images Tobias Klinder 1,2,CristianLorenz 2,JensvonBerg 2, Sebastian P.M. Dries 2, Thomas Bülow 2,andJörn Ostermann 1 1 Institut für Informationsverarbeitung,
A Model-Independent, Multi-Image Approach to MR Inhomogeneity Correction
 Tina Memo No. 2007-003 Published in Proc. MIUA 2007 A Model-Independent, Multi-Image Approach to MR Inhomogeneity Correction P. A. Bromiley and N.A. Thacker Last updated 13 / 4 / 2007 Imaging Science and
Tina Memo No. 2007-003 Published in Proc. MIUA 2007 A Model-Independent, Multi-Image Approach to MR Inhomogeneity Correction P. A. Bromiley and N.A. Thacker Last updated 13 / 4 / 2007 Imaging Science and
Semiautomatic Segmentation of Vertebrae in Lateral X-rays Using a Conditional Shape Model 1
 MICCAI Joint Disease Workshop Semiautomatic Segmentation of Vertebrae in Lateral X-rays Using a Conditional Shape Model 1 J. Eugenio Iglesias, MSc, Marleen de Bruijne, PhD Rationale and Objectives. Manual
MICCAI Joint Disease Workshop Semiautomatic Segmentation of Vertebrae in Lateral X-rays Using a Conditional Shape Model 1 J. Eugenio Iglesias, MSc, Marleen de Bruijne, PhD Rationale and Objectives. Manual
Conference Biomedical Engineering
 Automatic Medical Image Analysis for Measuring Bone Thickness and Density M. Kovalovs *, A. Glazs Image Processing and Computer Graphics Department, Riga Technical University, Latvia * E-mail: mihails.kovalovs@rtu.lv
Automatic Medical Image Analysis for Measuring Bone Thickness and Density M. Kovalovs *, A. Glazs Image Processing and Computer Graphics Department, Riga Technical University, Latvia * E-mail: mihails.kovalovs@rtu.lv
Initialising Groupwise Non-rigid Registration Using Multiple Parts+Geometry Models
 Initialising Groupwise Non-rigid Registration Using Multiple Parts+Geometry Models Pei Zhang 1,Pew-ThianYap 1, Dinggang Shen 1, and Timothy F. Cootes 2 1 Department of Radiology and Biomedical Research
Initialising Groupwise Non-rigid Registration Using Multiple Parts+Geometry Models Pei Zhang 1,Pew-ThianYap 1, Dinggang Shen 1, and Timothy F. Cootes 2 1 Department of Radiology and Biomedical Research
Locating Salient Object Features
 Locating Salient Object Features K.N.Walker, T.F.Cootes and C.J.Taylor Dept. Medical Biophysics, Manchester University, UK knw@sv1.smb.man.ac.uk Abstract We present a method for locating salient object
Locating Salient Object Features K.N.Walker, T.F.Cootes and C.J.Taylor Dept. Medical Biophysics, Manchester University, UK knw@sv1.smb.man.ac.uk Abstract We present a method for locating salient object
Semi-Automatic Delineation of the Spino-Laminar Junction Curve on Lateral X-ray Radiographs of the Cervical Spine
 Semi-Automatic Delineation of the Spino-Laminar Junction Curve on Lateral X-ray Radiographs of the Cervical Spine Benjamin Narang a, Michael Phillips b, Karen Knapp c, Andy Appelboam d, Adam Reuben d,
Semi-Automatic Delineation of the Spino-Laminar Junction Curve on Lateral X-ray Radiographs of the Cervical Spine Benjamin Narang a, Michael Phillips b, Karen Knapp c, Andy Appelboam d, Adam Reuben d,
Generic Face Alignment Using an Improved Active Shape Model
 Generic Face Alignment Using an Improved Active Shape Model Liting Wang, Xiaoqing Ding, Chi Fang Electronic Engineering Department, Tsinghua University, Beijing, China {wanglt, dxq, fangchi} @ocrserv.ee.tsinghua.edu.cn
Generic Face Alignment Using an Improved Active Shape Model Liting Wang, Xiaoqing Ding, Chi Fang Electronic Engineering Department, Tsinghua University, Beijing, China {wanglt, dxq, fangchi} @ocrserv.ee.tsinghua.edu.cn
Ensemble registration: Combining groupwise registration and segmentation
 PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
Deep Learning for Automatic Localization, Identification, and Segmentation of Vertebral Bodies in Volumetric MR Images
 Deep Learning for Automatic Localization, Identification, and Segmentation of Vertebral Bodies in Volumetric MR Images Amin Suzani a, Abtin Rasoulian a, Alexander Seitel a,sidneyfels a, Robert N. Rohling
Deep Learning for Automatic Localization, Identification, and Segmentation of Vertebral Bodies in Volumetric MR Images Amin Suzani a, Abtin Rasoulian a, Alexander Seitel a,sidneyfels a, Robert N. Rohling
Accurate Intervertebral Disc Localisation and Segmentation in MRI using Vantage Point Hough Forests and Multi-Atlas Fusion
 Accurate Intervertebral Disc Localisation and Segmentation in MRI using Vantage Point Hough Forests and Multi-Atlas Fusion Mattias P. Heinrich 1 and Ozan Oktay 2 1 Institute of Medical Informatics, University
Accurate Intervertebral Disc Localisation and Segmentation in MRI using Vantage Point Hough Forests and Multi-Atlas Fusion Mattias P. Heinrich 1 and Ozan Oktay 2 1 Institute of Medical Informatics, University
Automatic Vertebrae Localization in Pathological Spine CT using Decision Forests
 Automatic Vertebrae Localization in Pathological Spine CT using Decision Forests Ana Jiménez-Pastor 1, Esther Tomás-González 1, Ángel Alberich-Bayarri 1,2, Fabio García-Castro 1, David García-Juan 1, Luis
Automatic Vertebrae Localization in Pathological Spine CT using Decision Forests Ana Jiménez-Pastor 1, Esther Tomás-González 1, Ángel Alberich-Bayarri 1,2, Fabio García-Castro 1, David García-Juan 1, Luis
Object Identification in Ultrasound Scans
 Object Identification in Ultrasound Scans Wits University Dec 05, 2012 Roadmap Introduction to the problem Motivation Related Work Our approach Expected Results Introduction Nowadays, imaging devices like
Object Identification in Ultrasound Scans Wits University Dec 05, 2012 Roadmap Introduction to the problem Motivation Related Work Our approach Expected Results Introduction Nowadays, imaging devices like
Face Alignment Under Various Poses and Expressions
 Face Alignment Under Various Poses and Expressions Shengjun Xin and Haizhou Ai Computer Science and Technology Department, Tsinghua University, Beijing 100084, China ahz@mail.tsinghua.edu.cn Abstract.
Face Alignment Under Various Poses and Expressions Shengjun Xin and Haizhou Ai Computer Science and Technology Department, Tsinghua University, Beijing 100084, China ahz@mail.tsinghua.edu.cn Abstract.
Bone Age Classification Using the Discriminative Generalized Hough Transform
 Bone Age Classification Using the Discriminative Generalized Hough Transform Markus Brunk 1, Heike Ruppertshofen 1,2, Sarah Schmidt 2,3, Peter Beyerlein 3, Hauke Schramm 1 1 Institute of Applied Computer
Bone Age Classification Using the Discriminative Generalized Hough Transform Markus Brunk 1, Heike Ruppertshofen 1,2, Sarah Schmidt 2,3, Peter Beyerlein 3, Hauke Schramm 1 1 Institute of Applied Computer
Robust Active Appearance Models with Iteratively Rescaled Kernels
 Robust Active Appearance Models with Iteratively Rescaled Kernels M.G. Roberts, T.F. Cootes, J.E. Adams Department of Imaging Science and Biomedical Engineering University of Manchester Manchester, M13
Robust Active Appearance Models with Iteratively Rescaled Kernels M.G. Roberts, T.F. Cootes, J.E. Adams Department of Imaging Science and Biomedical Engineering University of Manchester Manchester, M13
Automatic Labeling and Segmentation of Vertebrae in CT Images
 000 001 002 003 004 005 006 007 008 009 010 011 012 013 014 015 016 017 018 019 020 021 022 023 024 025 026 027 028 029 030 031 032 033 034 035 036 037 038 039 040 041 042 043 044 045 046 047 048 049 050
000 001 002 003 004 005 006 007 008 009 010 011 012 013 014 015 016 017 018 019 020 021 022 023 024 025 026 027 028 029 030 031 032 033 034 035 036 037 038 039 040 041 042 043 044 045 046 047 048 049 050
8/3/2017. Contour Assessment for Quality Assurance and Data Mining. Objective. Outline. Tom Purdie, PhD, MCCPM
 Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Vertebrae Localization in Pathological Spine CT via Dense Classification from Sparse Annotations
 Vertebrae Localization in Pathological Spine CT via Dense Classification from Sparse Annotations Ben Glocker 1,DarkoZikic 1, Ender Konukoglu 2, David R. Haynor 3, and Antonio Criminisi 1 1 Microsoft Research,
Vertebrae Localization in Pathological Spine CT via Dense Classification from Sparse Annotations Ben Glocker 1,DarkoZikic 1, Ender Konukoglu 2, David R. Haynor 3, and Antonio Criminisi 1 1 Microsoft Research,
A Robust Segmentation Framework for Spine Trauma Diagnosis
 A Robust Segmentation Framework for Spine Trauma Diagnosis Poay Hoon Lim 1, Ulas Bagci 2 and Li Bai 1 1 School of Computer Science, University of Nottingham, UK 2 Radiology and Imaging Sciences, National
A Robust Segmentation Framework for Spine Trauma Diagnosis Poay Hoon Lim 1, Ulas Bagci 2 and Li Bai 1 1 School of Computer Science, University of Nottingham, UK 2 Radiology and Imaging Sciences, National
Face detection and recognition. Detection Recognition Sally
 Face detection and recognition Detection Recognition Sally Face detection & recognition Viola & Jones detector Available in open CV Face recognition Eigenfaces for face recognition Metric learning identification
Face detection and recognition Detection Recognition Sally Face detection & recognition Viola & Jones detector Available in open CV Face recognition Eigenfaces for face recognition Metric learning identification
Image Segmentation and Registration
 Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model
 Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 1,2, Hannan Abdillahi 3, Lala Ceklic 3, Ute Wolf-Schnurrbusch 2,3, and Jens Kowal 1,2 1 ARTORG
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 1,2, Hannan Abdillahi 3, Lala Ceklic 3, Ute Wolf-Schnurrbusch 2,3, and Jens Kowal 1,2 1 ARTORG
Fully Automatic Vertebra Detection in X-Ray Images Based on Multi-Class SVM
 Fully Automatic Vertebra Detection in X-Ray Images Based on Multi-Class SVM Fabian Lecron, Mohammed Benjelloun, Saïd Mahmoudi University of Mons, Faculty of Engineering, Computer Science Department 20,
Fully Automatic Vertebra Detection in X-Ray Images Based on Multi-Class SVM Fabian Lecron, Mohammed Benjelloun, Saïd Mahmoudi University of Mons, Faculty of Engineering, Computer Science Department 20,
Ultrasonic Multi-Skip Tomography for Pipe Inspection
 18 th World Conference on Non destructive Testing, 16-2 April 212, Durban, South Africa Ultrasonic Multi-Skip Tomography for Pipe Inspection Arno VOLKER 1, Rik VOS 1 Alan HUNTER 1 1 TNO, Stieltjesweg 1,
18 th World Conference on Non destructive Testing, 16-2 April 212, Durban, South Africa Ultrasonic Multi-Skip Tomography for Pipe Inspection Arno VOLKER 1, Rik VOS 1 Alan HUNTER 1 1 TNO, Stieltjesweg 1,
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs
 Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
[PDR03] RECOMMENDED CT-SCAN PROTOCOLS
![[PDR03] RECOMMENDED CT-SCAN PROTOCOLS [PDR03] RECOMMENDED CT-SCAN PROTOCOLS](/thumbs/72/66454100.jpg) SURGICAL & PROSTHETIC DESIGN [PDR03] RECOMMENDED CT-SCAN PROTOCOLS WORK-INSTRUCTIONS DOCUMENT (CUSTOMER) RECOMMENDED CT-SCAN PROTOCOLS [PDR03_V1]: LIVE 1 PRESCRIBING SURGEONS Patient-specific implants,
SURGICAL & PROSTHETIC DESIGN [PDR03] RECOMMENDED CT-SCAN PROTOCOLS WORK-INSTRUCTIONS DOCUMENT (CUSTOMER) RECOMMENDED CT-SCAN PROTOCOLS [PDR03_V1]: LIVE 1 PRESCRIBING SURGEONS Patient-specific implants,
An Adaptive Eigenshape Model
 An Adaptive Eigenshape Model Adam Baumberg and David Hogg School of Computer Studies University of Leeds, Leeds LS2 9JT, U.K. amb@scs.leeds.ac.uk Abstract There has been a great deal of recent interest
An Adaptive Eigenshape Model Adam Baumberg and David Hogg School of Computer Studies University of Leeds, Leeds LS2 9JT, U.K. amb@scs.leeds.ac.uk Abstract There has been a great deal of recent interest
Model-Based Lower Limb Segmentation using Weighted Multiple Candidates
 Model-Based Lower Limb Segmentation using Weighted Multiple Candidates André Gooßen 1, Eugen Hermann 1, Thorsten Gernoth 1 Thomas Pralow 2, Rolf-Rainer Grigat 1 1 Vision Systems, Hamburg University of
Model-Based Lower Limb Segmentation using Weighted Multiple Candidates André Gooßen 1, Eugen Hermann 1, Thorsten Gernoth 1 Thomas Pralow 2, Rolf-Rainer Grigat 1 1 Vision Systems, Hamburg University of
Classifier Case Study: Viola-Jones Face Detector
 Classifier Case Study: Viola-Jones Face Detector P. Viola and M. Jones. Rapid object detection using a boosted cascade of simple features. CVPR 2001. P. Viola and M. Jones. Robust real-time face detection.
Classifier Case Study: Viola-Jones Face Detector P. Viola and M. Jones. Rapid object detection using a boosted cascade of simple features. CVPR 2001. P. Viola and M. Jones. Robust real-time face detection.
Iterative fully convolutional neural networks for automatic vertebra segmentation
 Iterative fully convolutional neural networks for automatic vertebra segmentation Nikolas Lessmann Image Sciences Institute University Medical Center Utrecht Pim A. de Jong Department of Radiology University
Iterative fully convolutional neural networks for automatic vertebra segmentation Nikolas Lessmann Image Sciences Institute University Medical Center Utrecht Pim A. de Jong Department of Radiology University
IOSR Journal of Electronics and communication Engineering (IOSR-JCE) ISSN: , ISBN: , PP:
 IOSR Journal of Electronics and communication Engineering (IOSR-JCE) ISSN: 2278-2834, ISBN: 2278-8735, PP: 48-53 www.iosrjournals.org SEGMENTATION OF VERTEBRAE FROM DIGITIZED X-RAY IMAGES Ashwini Shivdas
IOSR Journal of Electronics and communication Engineering (IOSR-JCE) ISSN: 2278-2834, ISBN: 2278-8735, PP: 48-53 www.iosrjournals.org SEGMENTATION OF VERTEBRAE FROM DIGITIZED X-RAY IMAGES Ashwini Shivdas
Face detection and recognition. Many slides adapted from K. Grauman and D. Lowe
 Face detection and recognition Many slides adapted from K. Grauman and D. Lowe Face detection and recognition Detection Recognition Sally History Early face recognition systems: based on features and distances
Face detection and recognition Many slides adapted from K. Grauman and D. Lowe Face detection and recognition Detection Recognition Sally History Early face recognition systems: based on features and distances
Modern Medical Image Analysis 8DC00 Exam
 Parts of answers are inside square brackets [... ]. These parts are optional. Answers can be written in Dutch or in English, as you prefer. You can use drawings and diagrams to support your textual answers.
Parts of answers are inside square brackets [... ]. These parts are optional. Answers can be written in Dutch or in English, as you prefer. You can use drawings and diagrams to support your textual answers.
An Automatic Face Identification System Using Flexible Appearance Models
 An Automatic Face Identification System Using Flexible Appearance Models A. Lanitis, C.J.Taylor and T.F.Cootes Dpt. of Medical Biophysics, University of Manchester email: lan@wiau.mb.man.ac.uk We describe
An Automatic Face Identification System Using Flexible Appearance Models A. Lanitis, C.J.Taylor and T.F.Cootes Dpt. of Medical Biophysics, University of Manchester email: lan@wiau.mb.man.ac.uk We describe
Ulas Bagci
 CAP5415-Computer Vision Lecture 14-Decision Forests for Computer Vision Ulas Bagci bagci@ucf.edu 1 Readings Slide Credits: Criminisi and Shotton Z. Tu R.Cipolla 2 Common Terminologies Randomized Decision
CAP5415-Computer Vision Lecture 14-Decision Forests for Computer Vision Ulas Bagci bagci@ucf.edu 1 Readings Slide Credits: Criminisi and Shotton Z. Tu R.Cipolla 2 Common Terminologies Randomized Decision
Automated Pivot Location for the Cartesian-Polar Hybrid Point Distribution Model
 Automated Pivot Location for the Cartesian-Polar Hybrid Point Distribution Model Tony Heap and David Hogg School of Computer Studies, University of Leeds, Leeds LS2 9JT, UK email: ajh@scs.leeds.ac.uk Abstract
Automated Pivot Location for the Cartesian-Polar Hybrid Point Distribution Model Tony Heap and David Hogg School of Computer Studies, University of Leeds, Leeds LS2 9JT, UK email: ajh@scs.leeds.ac.uk Abstract
Performance Evaluation of the TINA Medical Image Segmentation Algorithm on Brainweb Simulated Images
 Tina Memo No. 2008-003 Internal Memo Performance Evaluation of the TINA Medical Image Segmentation Algorithm on Brainweb Simulated Images P. A. Bromiley Last updated 20 / 12 / 2007 Imaging Science and
Tina Memo No. 2008-003 Internal Memo Performance Evaluation of the TINA Medical Image Segmentation Algorithm on Brainweb Simulated Images P. A. Bromiley Last updated 20 / 12 / 2007 Imaging Science and
Automatic Rapid Segmentation of Human Lung from 2D Chest X-Ray Images
 Automatic Rapid Segmentation of Human Lung from 2D Chest X-Ray Images Abstract. In this paper, we propose a complete framework that segments lungs from 2D Chest X-Ray (CXR) images automatically and rapidly.
Automatic Rapid Segmentation of Human Lung from 2D Chest X-Ray Images Abstract. In this paper, we propose a complete framework that segments lungs from 2D Chest X-Ray (CXR) images automatically and rapidly.
Knowledge-Based Organ Identification from CT Images. Masahara Kobashi and Linda Shapiro Best-Paper Prize in Pattern Recognition Vol. 28, No.
 Knowledge-Based Organ Identification from CT Images Masahara Kobashi and Linda Shapiro Best-Paper Prize in Pattern Recognition Vol. 28, No. 4 1995 1 Motivation The extraction of structure from CT volumes
Knowledge-Based Organ Identification from CT Images Masahara Kobashi and Linda Shapiro Best-Paper Prize in Pattern Recognition Vol. 28, No. 4 1995 1 Motivation The extraction of structure from CT volumes
Shape-aware Deep Convolutional Neural Network for Vertebrae Segmentation
 Shape-aware Deep Convolutional Neural Network for Vertebrae Segmentation S M Masudur Rahman Al Arif 1, Karen Knapp 2 and Greg Slabaugh 1 1 City, University of London 2 University of Exeter Abstract. Shape
Shape-aware Deep Convolutional Neural Network for Vertebrae Segmentation S M Masudur Rahman Al Arif 1, Karen Knapp 2 and Greg Slabaugh 1 1 City, University of London 2 University of Exeter Abstract. Shape
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model
 Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 12,HannanAbdillahi 3, Lala Ceklic 3,Ute Wolf-Schnurrbusch 23,JensKowal 12 1 ARTORG Center
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 12,HannanAbdillahi 3, Lala Ceklic 3,Ute Wolf-Schnurrbusch 23,JensKowal 12 1 ARTORG Center
The Quantification of Volumetric Asymmetry by Dynamic Surface Topography. Thomas Shannon Oxford Brookes University Oxford, U.K.
 The Quantification of Volumetric Asymmetry by Dynamic Surface Topography Thomas Shannon Oxford Brookes University Oxford, U.K. The psychosocial impact of the cosmetic defect on Adolescent Idiopathic Scoliosis
The Quantification of Volumetric Asymmetry by Dynamic Surface Topography Thomas Shannon Oxford Brookes University Oxford, U.K. The psychosocial impact of the cosmetic defect on Adolescent Idiopathic Scoliosis
Prostate Detection Using Principal Component Analysis
 Prostate Detection Using Principal Component Analysis Aamir Virani (avirani@stanford.edu) CS 229 Machine Learning Stanford University 16 December 2005 Introduction During the past two decades, computed
Prostate Detection Using Principal Component Analysis Aamir Virani (avirani@stanford.edu) CS 229 Machine Learning Stanford University 16 December 2005 Introduction During the past two decades, computed
2 Michael E. Leventon and Sarah F. F. Gibson a b c d Fig. 1. (a, b) Two MR scans of a person's knee. Both images have high resolution in-plane, but ha
 Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
arxiv: v1 [cs.cv] 13 Mar 2017
![arxiv: v1 [cs.cv] 13 Mar 2017 arxiv: v1 [cs.cv] 13 Mar 2017](/thumbs/79/79066286.jpg) A Localisation-Segmentation Approach for Multi-label Annotation of Lumbar Vertebrae using Deep Nets Anjany Sekuboyina 1,2,, Alexander Valentinitsch 2, Jan S. Kirschke 2, and Bjoern H. Menze 1 arxiv:1703.04347v1
A Localisation-Segmentation Approach for Multi-label Annotation of Lumbar Vertebrae using Deep Nets Anjany Sekuboyina 1,2,, Alexander Valentinitsch 2, Jan S. Kirschke 2, and Bjoern H. Menze 1 arxiv:1703.04347v1
Texture Analysis in Quantitative Osteoporosis Assessment
 Texture Analysis in Quantitative Osteoporosis Assessment Characterizing Micro-architecture in High Resolution Peripheral Quantitative Computed Tomography Alexander Valentinitsch 1, Janina Patsch 1, Dirk
Texture Analysis in Quantitative Osteoporosis Assessment Characterizing Micro-architecture in High Resolution Peripheral Quantitative Computed Tomography Alexander Valentinitsch 1, Janina Patsch 1, Dirk
AAM Based Facial Feature Tracking with Kinect
 BULGARIAN ACADEMY OF SCIENCES CYBERNETICS AND INFORMATION TECHNOLOGIES Volume 15, No 3 Sofia 2015 Print ISSN: 1311-9702; Online ISSN: 1314-4081 DOI: 10.1515/cait-2015-0046 AAM Based Facial Feature Tracking
BULGARIAN ACADEMY OF SCIENCES CYBERNETICS AND INFORMATION TECHNOLOGIES Volume 15, No 3 Sofia 2015 Print ISSN: 1311-9702; Online ISSN: 1314-4081 DOI: 10.1515/cait-2015-0046 AAM Based Facial Feature Tracking
Face Tracking in Video
 Face Tracking in Video Hamidreza Khazaei and Pegah Tootoonchi Afshar Stanford University 350 Serra Mall Stanford, CA 94305, USA I. INTRODUCTION Object tracking is a hot area of research, and has many practical
Face Tracking in Video Hamidreza Khazaei and Pegah Tootoonchi Afshar Stanford University 350 Serra Mall Stanford, CA 94305, USA I. INTRODUCTION Object tracking is a hot area of research, and has many practical
Motion Estimation for Video Coding Standards
 Motion Estimation for Video Coding Standards Prof. Ja-Ling Wu Department of Computer Science and Information Engineering National Taiwan University Introduction of Motion Estimation The goal of video compression
Motion Estimation for Video Coding Standards Prof. Ja-Ling Wu Department of Computer Science and Information Engineering National Taiwan University Introduction of Motion Estimation The goal of video compression
1. Estimation equations for strip transect sampling, using notation consistent with that used to
 Web-based Supplementary Materials for Line Transect Methods for Plant Surveys by S.T. Buckland, D.L. Borchers, A. Johnston, P.A. Henrys and T.A. Marques Web Appendix A. Introduction In this on-line appendix,
Web-based Supplementary Materials for Line Transect Methods for Plant Surveys by S.T. Buckland, D.L. Borchers, A. Johnston, P.A. Henrys and T.A. Marques Web Appendix A. Introduction In this on-line appendix,
Automated Lesion Detection Methods for 2D and 3D Chest X-Ray Images
 Automated Lesion Detection Methods for 2D and 3D Chest X-Ray Images Takeshi Hara, Hiroshi Fujita,Yongbum Lee, Hitoshi Yoshimura* and Shoji Kido** Department of Information Science, Gifu University Yanagido
Automated Lesion Detection Methods for 2D and 3D Chest X-Ray Images Takeshi Hara, Hiroshi Fujita,Yongbum Lee, Hitoshi Yoshimura* and Shoji Kido** Department of Information Science, Gifu University Yanagido
Performance Evaluation Metrics and Statistics for Positional Tracker Evaluation
 Performance Evaluation Metrics and Statistics for Positional Tracker Evaluation Chris J. Needham and Roger D. Boyle School of Computing, The University of Leeds, Leeds, LS2 9JT, UK {chrisn,roger}@comp.leeds.ac.uk
Performance Evaluation Metrics and Statistics for Positional Tracker Evaluation Chris J. Needham and Roger D. Boyle School of Computing, The University of Leeds, Leeds, LS2 9JT, UK {chrisn,roger}@comp.leeds.ac.uk
Chapter 6 Normal Probability Distributions
 Chapter 6 Normal Probability Distributions 6-1 Review and Preview 6-2 The Standard Normal Distribution 6-3 Applications of Normal Distributions 6-4 Sampling Distributions and Estimators 6-5 The Central
Chapter 6 Normal Probability Distributions 6-1 Review and Preview 6-2 The Standard Normal Distribution 6-3 Applications of Normal Distributions 6-4 Sampling Distributions and Estimators 6-5 The Central
Support Vector Machine Density Estimator as a Generalized Parzen Windows Estimator for Mutual Information Based Image Registration
 Support Vector Machine Density Estimator as a Generalized Parzen Windows Estimator for Mutual Information Based Image Registration Sudhakar Chelikani 1, Kailasnath Purushothaman 1, and James S. Duncan
Support Vector Machine Density Estimator as a Generalized Parzen Windows Estimator for Mutual Information Based Image Registration Sudhakar Chelikani 1, Kailasnath Purushothaman 1, and James S. Duncan
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information
 Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Out-of-Plane Rotated Object Detection using Patch Feature based Classifier
 Available online at www.sciencedirect.com Procedia Engineering 41 (2012 ) 170 174 International Symposium on Robotics and Intelligent Sensors 2012 (IRIS 2012) Out-of-Plane Rotated Object Detection using
Available online at www.sciencedirect.com Procedia Engineering 41 (2012 ) 170 174 International Symposium on Robotics and Intelligent Sensors 2012 (IRIS 2012) Out-of-Plane Rotated Object Detection using
Supervoxel Classification Forests for Estimating Pairwise Image Correspondences
 Supervoxel Classification Forests for Estimating Pairwise Image Correspondences Fahdi Kanavati 1, Tong Tong 1, Kazunari Misawa 2, Michitaka Fujiwara 3, Kensaku Mori 4, Daniel Rueckert 1, and Ben Glocker
Supervoxel Classification Forests for Estimating Pairwise Image Correspondences Fahdi Kanavati 1, Tong Tong 1, Kazunari Misawa 2, Michitaka Fujiwara 3, Kensaku Mori 4, Daniel Rueckert 1, and Ben Glocker
Component-based Face Recognition with 3D Morphable Models
 Component-based Face Recognition with 3D Morphable Models B. Weyrauch J. Huang benjamin.weyrauch@vitronic.com jenniferhuang@alum.mit.edu Center for Biological and Center for Biological and Computational
Component-based Face Recognition with 3D Morphable Models B. Weyrauch J. Huang benjamin.weyrauch@vitronic.com jenniferhuang@alum.mit.edu Center for Biological and Center for Biological and Computational
Automatic Vascular Tree Formation Using the Mahalanobis Distance
 Automatic Vascular Tree Formation Using the Mahalanobis Distance Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, Department of Radiology The University
Automatic Vascular Tree Formation Using the Mahalanobis Distance Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, Department of Radiology The University
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images
 A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
Computer Vision Group Prof. Daniel Cremers. 8. Boosting and Bagging
 Prof. Daniel Cremers 8. Boosting and Bagging Repetition: Regression We start with a set of basis functions (x) =( 0 (x), 1(x),..., M 1(x)) x 2 í d The goal is to fit a model into the data y(x, w) =w T
Prof. Daniel Cremers 8. Boosting and Bagging Repetition: Regression We start with a set of basis functions (x) =( 0 (x), 1(x),..., M 1(x)) x 2 í d The goal is to fit a model into the data y(x, w) =w T
A novel 3D torso image reconstruction procedure using a pair of digital stereo back images
 Modelling in Medicine and Biology VIII 257 A novel 3D torso image reconstruction procedure using a pair of digital stereo back images A. Kumar & N. Durdle Department of Electrical & Computer Engineering,
Modelling in Medicine and Biology VIII 257 A novel 3D torso image reconstruction procedure using a pair of digital stereo back images A. Kumar & N. Durdle Department of Electrical & Computer Engineering,
ICA vs. PCA Active Appearance Models: Application to Cardiac MR Segmentation
 ICA vs. PCA Active Appearance Models: Application to Cardiac MR Segmentation M. Üzümcü 1, A.F. Frangi 2, M. Sonka 3, J.H.C. Reiber 1, B.P.F. Lelieveldt 1 1 Div. of Image Processing, Dept. of Radiology
ICA vs. PCA Active Appearance Models: Application to Cardiac MR Segmentation M. Üzümcü 1, A.F. Frangi 2, M. Sonka 3, J.H.C. Reiber 1, B.P.F. Lelieveldt 1 1 Div. of Image Processing, Dept. of Radiology
Intensity-Depth Face Alignment Using Cascade Shape Regression
 Intensity-Depth Face Alignment Using Cascade Shape Regression Yang Cao 1 and Bao-Liang Lu 1,2 1 Center for Brain-like Computing and Machine Intelligence Department of Computer Science and Engineering Shanghai
Intensity-Depth Face Alignment Using Cascade Shape Regression Yang Cao 1 and Bao-Liang Lu 1,2 1 Center for Brain-like Computing and Machine Intelligence Department of Computer Science and Engineering Shanghai
Multimedia Computing: Algorithms, Systems, and Applications: Edge Detection
 Multimedia Computing: Algorithms, Systems, and Applications: Edge Detection By Dr. Yu Cao Department of Computer Science The University of Massachusetts Lowell Lowell, MA 01854, USA Part of the slides
Multimedia Computing: Algorithms, Systems, and Applications: Edge Detection By Dr. Yu Cao Department of Computer Science The University of Massachusetts Lowell Lowell, MA 01854, USA Part of the slides
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION
 CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation
 A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
3D registration of micro-ct images for the identification of trabecular fracture region.
 3D registration of micro-ct images for the identification of trabecular fracture region. S. Tassani 1,2, P.A. Asvestas 3, G.K. Matsopoulos 1, and F. Baruffaldi 2 1 Institute of Communication and Computer
3D registration of micro-ct images for the identification of trabecular fracture region. S. Tassani 1,2, P.A. Asvestas 3, G.K. Matsopoulos 1, and F. Baruffaldi 2 1 Institute of Communication and Computer
EPSRC Centre for Doctoral Training in Industrially Focused Mathematical Modelling
 EPSRC Centre for Doctoral Training in Industrially Focused Mathematical Modelling More Accurate Optical Measurements of the Cornea Raquel González Fariña Contents 1. Introduction... 2 Background... 2 2.
EPSRC Centre for Doctoral Training in Industrially Focused Mathematical Modelling More Accurate Optical Measurements of the Cornea Raquel González Fariña Contents 1. Introduction... 2 Background... 2 2.
An Automated Image-based Method for Multi-Leaf Collimator Positioning Verification in Intensity Modulated Radiation Therapy
 An Automated Image-based Method for Multi-Leaf Collimator Positioning Verification in Intensity Modulated Radiation Therapy Chenyang Xu 1, Siemens Corporate Research, Inc., Princeton, NJ, USA Xiaolei Huang,
An Automated Image-based Method for Multi-Leaf Collimator Positioning Verification in Intensity Modulated Radiation Therapy Chenyang Xu 1, Siemens Corporate Research, Inc., Princeton, NJ, USA Xiaolei Huang,
Supervised vs. Unsupervised Learning
 Clustering Supervised vs. Unsupervised Learning So far we have assumed that the training samples used to design the classifier were labeled by their class membership (supervised learning) We assume now
Clustering Supervised vs. Unsupervised Learning So far we have assumed that the training samples used to design the classifier were labeled by their class membership (supervised learning) We assume now
Segmentation and Tracking of Partial Planar Templates
 Segmentation and Tracking of Partial Planar Templates Abdelsalam Masoud William Hoff Colorado School of Mines Colorado School of Mines Golden, CO 800 Golden, CO 800 amasoud@mines.edu whoff@mines.edu Abstract
Segmentation and Tracking of Partial Planar Templates Abdelsalam Masoud William Hoff Colorado School of Mines Colorado School of Mines Golden, CO 800 Golden, CO 800 amasoud@mines.edu whoff@mines.edu Abstract
A fast breast nonlinear elastography reconstruction technique using the Veronda-Westman model
 A fast breast nonlinear elastography reconstruction technique using the Veronda-Westman model Mohammadhosein Amooshahi a and Abbas Samani abc a Department of Electrical & Computer Engineering, University
A fast breast nonlinear elastography reconstruction technique using the Veronda-Westman model Mohammadhosein Amooshahi a and Abbas Samani abc a Department of Electrical & Computer Engineering, University
Joint Classification-Regression Forests for Spatially Structured Multi-Object Segmentation
 Joint Classification-Regression Forests for Spatially Structured Multi-Object Segmentation Ben Glocker 1, Olivier Pauly 2,3, Ender Konukoglu 1, Antonio Criminisi 1 1 Microsoft Research, Cambridge, UK 2
Joint Classification-Regression Forests for Spatially Structured Multi-Object Segmentation Ben Glocker 1, Olivier Pauly 2,3, Ender Konukoglu 1, Antonio Criminisi 1 1 Microsoft Research, Cambridge, UK 2
Object Recognition Using Pictorial Structures. Daniel Huttenlocher Computer Science Department. In This Talk. Object recognition in computer vision
 Object Recognition Using Pictorial Structures Daniel Huttenlocher Computer Science Department Joint work with Pedro Felzenszwalb, MIT AI Lab In This Talk Object recognition in computer vision Brief definition
Object Recognition Using Pictorial Structures Daniel Huttenlocher Computer Science Department Joint work with Pedro Felzenszwalb, MIT AI Lab In This Talk Object recognition in computer vision Brief definition
A Method of Automated Landmark Generation for Automated 3D PDM Construction
 A Method of Automated Landmark Generation for Automated 3D PDM Construction A. D. Brett and C. J. Taylor Department of Medical Biophysics University of Manchester Manchester M13 9PT, Uk adb@sv1.smb.man.ac.uk
A Method of Automated Landmark Generation for Automated 3D PDM Construction A. D. Brett and C. J. Taylor Department of Medical Biophysics University of Manchester Manchester M13 9PT, Uk adb@sv1.smb.man.ac.uk
Assessing the Skeletal Age From a Hand Radiograph: Automating the Tanner-Whitehouse Method
 Assessing the Skeletal Age From a Hand Radiograph: Automating the Tanner-Whitehouse Method M. Niemeijer a, B. van Ginneken a, C.A. Maas a, F.J.A. Beek b and M.A. Viergever a a Image Sciences Institute,
Assessing the Skeletal Age From a Hand Radiograph: Automating the Tanner-Whitehouse Method M. Niemeijer a, B. van Ginneken a, C.A. Maas a, F.J.A. Beek b and M.A. Viergever a a Image Sciences Institute,
Features: representation, normalization, selection. Chapter e-9
 Features: representation, normalization, selection Chapter e-9 1 Features Distinguish between instances (e.g. an image that you need to classify), and the features you create for an instance. Features
Features: representation, normalization, selection Chapter e-9 1 Features Distinguish between instances (e.g. an image that you need to classify), and the features you create for an instance. Features
Assessing the Local Credibility of a Medical Image Segmentation
 Assessing the Local Credibility of a Medical Image Segmentation Joshua H. Levy, Robert E. Broadhurst, Surajit Ray, Edward L. Chaney, and Stephen M. Pizer Medical Image Display and Analysis Group (MIDAG),
Assessing the Local Credibility of a Medical Image Segmentation Joshua H. Levy, Robert E. Broadhurst, Surajit Ray, Edward L. Chaney, and Stephen M. Pizer Medical Image Display and Analysis Group (MIDAG),
Supervised Learning for Image Segmentation
 Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
