Introduction: microarray quality assessment with arrayqualitymetrics
|
|
|
- Alisha Sanders
- 6 years ago
- Views:
Transcription
1 : microarray quality assessment with arrayqualitymetrics Audrey Kauffmann, Wolfgang Huber October 30, 2017 Contents 1 Basic use Affymetrix data - before preprocessing Affymetrix data - after preprocessing ExpressionSet and ExpressionSetIllumina Two colour arrays, NChannelSet, RGList, MAList Loading data from ArrayExpress Making the report more informative by adding a factor of interest Extended use Spatial layout of the array Mapping of the reporters RNA quality Introduction The arrayqualitymetrics package produces, through a single function call, a comprehensive HTML report of quality metrics about a microarray dataset [1, 2, 3]. The quality metrics are mainly on the per array level, i. e. they can be used to assess the relative quality of different arrays within a dataset. Some of the metrics can also be used to diagnose batch effects, and thus the quality of the overall dataset. The report can be extended to contain further diagnostics through additional arguments, and we will see examples for this in Section 3.
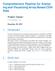
2 Figure 1: Left: An example plot from the report The plot shows the arrays (points) in a two-dimensional plot area spanned by the first two axes of a principal component analysis (PCA). By moving the mouse over the points, the corresponding array s metadata is displayed in the table to the right of the plot. By clicking on a point, it can be selected or deselected. Selected arrays are indicated by larger points or wider lines in the plots and by ticked checkboxes in the array table shown in the right panel. Arrays can also be (de)selected by clicking the checkboxes. Initially, when the report is loaded (or reloaded) by the browser, all arrays are selected that were called outliers by at least one criterion. The aim of the arrayqualitymetrics package is to produce information that is relevant for your decision making - not, to make the decision. It will often be applied to two, somewhat distinct, use cases: (i) assessing quality of a raw dataset, in order to get feedback on the experimental procedures that produced the data; (ii) assessing quality of a normalised dataset, in order to decide whether and how to use the dataset (or subsets of arrays in it) for subsequent data analysis. Different types of microarray data (one colour, two colour, Affymetrix, Illumina) are represented by different object classes in Bioconductor. The function arrayquality Metrics will work in the same way for all of them. Further information about its arguments can be found in its manual page. When the function arrayqualitymetrics is finished, a report is produced in the directory specified by the function s outdir argument. By default, a directory with a suitable name is created in the current working directory. This directory contains an HTML page index.html that can be opened by a browser. The report contains a series of plots explained by text. Some of the plots are interactive (see Figure 1). Technically, this is achieved by the use of SVG (scalable vector graphics) and JavaScript, and it requires that you use a recent (HTML5 capable) web browser 1. Other plots, where interactivity is less relevant, are provided as bitmaps (PNG format) and are also linked to PDF files that provide high resolution versions e. g. for publication. Plus (+) or minus (-) symbols at the begin of different section headings of the report (as in the left panel of Figure 1) indicate that you can show or hide these sections by clicking on the heading. After (re)loading, all sections are shown except for the Outlier detection barplots, which are hidden and can be expanded by clicking on them. 1 If in doubt, please see the notes about browser compatibility at the top of the report; or contact the Bioconductor mailing list. 2
3 Metadata about the arrays is shown at the top of the report as a table (see Figure 1). It is extracted from the phenodata slot of the data object supplied to arrayquali tymetrics. It can be useful to adjust the contents this slot before producing the report, and to make sure it contains the right quantity of information to make an informative report - not too much, not too little. In the case of AffyBatch input, some Affymetrix specific sections are added to the standard report. Also for other types of arrays, sections can be added to the standard report if certain metadata are present in the input object (see Section 3). The function arrayqualitymetrics also produces an R object (essentially, a big list) with all the information contained in the report, and this object can be used by downstream tools for programmatic analysis of the report. This is discussed in the vignette Advanced topics: Customizing arrayqualitymetrics reports and programmatic processing of the output 1 Basic use 1.1 Affymetrix data - before preprocessing If you are working with Affymetrix GeneChips, an AffyBatch object is the most appropriate way to import your raw data into Bioconductor. Starting from CEL files, this is typically done using the function ReadAffy from the affy package 2. Here, we use the dataset MLL.A, an object of class AffyBatch provided in the data package ALLMLL. library("allmll") data("mll.a") Now that the data are loaded, we can call arrayqualitymetrics 3. library("arrayqualitymetrics") arrayqualitymetrics(expressionset = MLL.A[, 1:5], outdir = "Report_for_MLL_A", force = TRUE, do.logtransform = TRUE) 2 For more information on how to produce an AffyBatch from your data, please see the documentation of the affy package. 3 For this vignette, in order to save computation time, we only call the function on the first 5 arrays; in your own application, you can call it on the complete data object. This is the simplest way of calling the function. We give a name to the directory (outdir) and we overwrite the possibly existing files of this directory (force). Finally, we set do.logtransform to logarithm transform the intensities. You can then view the report by directing your browser to the file index.html in the directory whose name is indicated by outdir. 3
4 1.2 Affymetrix data - after preprocessing We can call the RMA algorithm on MLL.A to obtain a preprocessed dataset. The preprocessing includes background correction, between array intensity adjustment (normalisation) and probeset summarisation. The resulting object nmll is of class ExpressionSet and contains one value (expression estimate) for each gene for each array. nmll = rma(mll.a) We can then call again the function arrayqualitymetrics. arrayqualitymetrics(expressionset = nmll, outdir = "Report_for_nMLL", force = TRUE) We do not need to set do.logtransform as after rma the data are already logarithm transformed. 1.3 ExpressionSet and ExpressionSetIllumina If you are working on one colour arrays other than Affymetrix genechips, you can load your data into Bioconductor as an ExpressionSet object 4, or if you work with Illumina data and the beadarray package, as an ExpressionSetIllumina object. You can then proceed exactly as above. 4 See the documentation of the Biobase package. 1.4 Two colour arrays, NChannelSet, RGList, MAList The package limma imports a wide range of data formats used for two colour arrays and produces objects of class RGList or MAList. When presented with an object of these classes, arrayqualitymetrics tries to convert them into an NChannelSet and then proceeds with calling its NChannelSet method. Alternatively, you can create an NChannelSet to contain your data from scratch. The documentation of the Biobase package gives instructions on how to do so. The arrayqualitymetrics function expects the assaydata slot of the NChannelSet object to contain the elements R and G, for the red and the green intensities. Optionally, it can contain elements Rb and Gb for associated background intensities. As an alternative to all that, the arrayqualitymetrics function also accepts NChannelSet objects with a single slot exprs, and will then simply behave like it does for (single-colour) ExpressionSet objects. As an example, we consider the dataset CCl4 from the data package CCl4 and normalize it using the variance stabilization method available in the package vsn. 4
5 library("vsn") library("ccl4") data("ccl4") nccl4 = justvsn(ccl4, subsample = 15000) arrayqualitymetrics(expressionset = nccl4, outdir = "Report_for_nCCl4", force = TRUE) 1.5 Loading data from ArrayExpress You can use the ArrayExpress package [4] to download datasets from the EBI s ArrayExpress database. The resulting ExpressionSet, AffyBatch or NChannelSet objects can be directly fed into arrayqualitymetrics. 2 Making the report more informative by adding a factor of interest A useful feature of arrayqualitymetrics is the possibility to show the results in the context of an experimental factor of interest, i. e. a categorical variable associated with the arrays such as hybridisation date, treatment level or replicate number. Specifying a factor does not change how the quality metrics are computed. By setting the argument intgroup to contain the names of one or multiple columns of the data object s phenodata slot 5, a bar on the side of the heatmap with colours representing the respective factors is added. Similarly, the colours of the boxplots and density plots reflect the levels of the first of the factors named by intgroup. 5 This is where Bioconductor objects store array annotation We use the nmll example again, and create artificial array metadata factors condi tion and batch (see Section 3.3 for a more realistic example). pdata(nmll)$condition = rep(letters[1:4], times = 5) pdata(nmll)$batch = rep(paste(1:4), each = 5) arrayqualitymetrics(expressionset = nmll, outdir = "Report_for_nMLL_with_factors", force = TRUE, intgroup = c("condition", "batch")) 5
6 3 Extended use Some of the quality metrics that the package can compute require specific information about the features on the arrays. To use these, you need to make sure that this information is provided in your input object. We use the nccl4 example again. 3.1 Spatial layout of the array To plot the spatial distributions of the intensities of the arrays, arrayqualitymet rics needs the spatial coordinates of the features on the chip. For AffyBatch or BeadLevelList, this information is automatically available without further user input. For the other types of objects, two columns corresponding to X and Y coordinates of the features are required in the featuredata slot of the object. These columns should be named "X" and "Y". If the arrays are split into blocks, rows and columns, then the function addxyfromgal (please check its manual page for details) can used to convert the row, column and blocks indices into absolute "X" and "Y" coordinates on the array. In the example of the dataset CCl4, the coordinates of the spots are in the columns named "Row" and "Column" of the featuredata (the slot of the object containing the annotation of the probes). We copy this information into columns named "X" and "Y" respectively featuredata(nccl4)$x = featuredata(nccl4)$row featuredata(nccl4)$y = featuredata(nccl4)$column The next call to arrayqualitymetrics with this refined version of nccl4 (see Section 3.3) will now include this information in the report, and the spatial distribution of the intensities will be shown. 3.2 Mapping of the reporters The report can also include an assessment of the effect of the target mapping of the reporters. You can define a featuredata column named hastarget that indicates, by logical TRUE, if the reporter matches a known transcript, and by FALSE, if not. In the CCl4 example, many of the reporter names are RefSeq identifiers, while others are not. Thus, we let hastarget indicate whether the name begins with "NM". featuredata(nccl4)$hastarget = (regexpr("^nm", featuredata(nccl4)$name) > 0) table(featuredata(nccl4)$hastarget) ## ## FALSE TRUE ##
7 The next call to arrayqualitymetrics with this refined version of nccl4 (see Section 3.3) will now include this information in the report, and the spatial distribution of the intensities will be shown. 3.3 RNA quality The RNA hybridized to the arrays in the CCl4 dataset was intentionally made to good, medium or poor quality, and this is recorded by a so-called RIN value (see CCl4 vignette). pd = pdata(ccl4) rownames(pd) = NULL pd ## Cy3 Cy5 RIN.Cy3 RIN.Cy5 ## 1 DMSO CCl ## 2 DMSO CCl ## 3 DMSO CCl ## 4 DMSO CCl ## 5 CCl4 DMSO ## 6 CCl4 DMSO ## 7 CCl4 DMSO ## 8 DMSO CCl ## 9 CCl4 DMSO ## 10 CCl4 DMSO ## 11 CCl4 DMSO ## 12 DMSO CCl ## 13 DMSO CCl ## 14 DMSO CCl ## 15 DMSO CCl ## 16 CCl4 DMSO ## 17 CCl4 DMSO ## 18 CCl4 DMSO The RIN is always 9 for the reference (DMSO), the relevant value is that for the test sample (CCl4). RIN = with(pd, ifelse( Cy3=="CCl4", RIN.Cy3, RIN.Cy5)) frin = factor(rin) levels(frin) = c("poor", "medium", "good") pdata(nccl4)$"rna-integrity" = frin Now we can use this to set the argument intgroup when calling the function ar rayqualitymetrics. 7
8 arrayqualitymetrics(expressionset = nccl4, outdir = "Report_for_nCCl4_with_RIN", force = TRUE, intgroup = "RNA-integrity") Boxplots, PCA plot and heatmap in the report will now indicate the values of the factor RNA-integrity for each array. Session Info R Under development (unstable) ( r73567), x86_64-pc-linux-gnu Locale: LC_CTYPE=en_US.UTF-8, LC_NUMERIC=C, LC_TIME=en_US.UTF-8, LC_COLLATE=C, LC_MONETARY=en_US.UTF-8, LC_MESSAGES=en_US.UTF-8, LC_PAPER=en_US.UTF-8, LC_NAME=C, LC_ADDRESS=C, LC_TELEPHONE=C, LC_MEASUREMENT=en_US.UTF-8, LC_IDENTIFICATION=C Running under: Ubuntu LTS Matrix products: default BLAS: /home/biocbuild/bbs-3.7-bioc/r/lib/librblas.so LAPACK: /home/biocbuild/bbs-3.7-bioc/r/lib/librlapack.so Base packages: base, datasets, grdevices, graphics, methods, parallel, stats, utils Other packages: ALLMLL , Biobase , BiocGenerics , CCl , affy , arrayqualitymetrics , hexbin , hgu133acdf , limma , vsn Loaded via a namespace (and not attached): AnnotationDbi , BeadDataPackR , BiocInstaller , BiocStyle 2.7.0, Biostrings , Cairo 1.5-9, DBI 0.7, Formula 1.2-2, GenomeInfoDb , GenomeInfoDbData , GenomicRanges , Hmisc 4.0-3, IRanges , KernSmooth , Matrix , RColorBrewer 1.1-2, RCurl , RSQLite 2.0, Rcpp , S4Vectors , XML , XVector , acepack 1.4.1, affyplm , affyio , annotate , backports 1.1.1, base64 2.0, base64enc 0.1-3, beadarray , bit , bit , bitops 1.0-6, blob 1.1.0, checkmate 1.8.5, cluster 2.0.6, codetools , colorspace 1.3-2, compiler 3.5.0, data.table , digest , evaluate , foreign , gcrma , genefilter , ggplot , grid 3.5.0, gridextra 2.3, gridsvg 1.6-0, gtable 0.2.0, highr 0.6, htmltable 1.9, htmltools 0.3.6, htmlwidgets 0.9, hwriter 1.3.2, illuminaio , jsonlite 1.5, knitr 1.17, labeling 0.3, lattice , latticeextra , lazyeval 0.2.1, magrittr 1.5, memoise 1.1.0, munsell 0.4.3, nnet , openssl 0.9.7, 8
9 plyr 1.8.4, preprocesscore , reshape , rlang 0.1.2, rmarkdown 1.6, rpart , rprojroot 1.2, scales 0.5.0, setrng , splines 3.5.0, stats , stringi 1.1.5, stringr 1.2.0, survival , tibble 1.3.4, tools 3.5.0, xtable 1.8-2, yaml , zlibbioc References [1] Audrey Kauffmann, Robert Gentleman, and Wolfgang Huber. arrayqualitymetrics - a Bioconductor package for quality assessment of microarray data. Bioinformatics, 25: , [2] Audrey Kauffmann and Wolfgang Huber. Microarray data quality control improves the detection of differentially expressed genes. Genomics, 95: , [3] Matthew N. McCall, Peter N. Murakami, and Rafael A. Irizarry. Assessing microarray quality. Technical report, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, [4] Audrey Kauffmann, Tim F. Rayner, Helen Parkinson, Misha Kapushesky, Margus Lukk, Alivs Brazma, and Wolfgang Huber. Importing ArrayExpress datasets into R/Bioconductor. Bioinformatics, 25: ,
Introduction: microarray quality assessment with arrayqualitymetrics
 Introduction: microarray quality assessment with arrayqualitymetrics Audrey Kauffmann, Wolfgang Huber April 4, 2013 Contents 1 Basic use 3 1.1 Affymetrix data - before preprocessing.......................
Introduction: microarray quality assessment with arrayqualitymetrics Audrey Kauffmann, Wolfgang Huber April 4, 2013 Contents 1 Basic use 3 1.1 Affymetrix data - before preprocessing.......................
Image Analysis with beadarray
 Mike Smith October 30, 2017 Introduction From version 2.0 beadarray provides more flexibility in the processing of array images and the extraction of bead intensities than its predecessor. In the past
Mike Smith October 30, 2017 Introduction From version 2.0 beadarray provides more flexibility in the processing of array images and the extraction of bead intensities than its predecessor. In the past
RMassBank for XCMS. Erik Müller. January 4, Introduction 2. 2 Input files LC/MS data Additional Workflow-Methods 2
 RMassBank for XCMS Erik Müller January 4, 2019 Contents 1 Introduction 2 2 Input files 2 2.1 LC/MS data........................... 2 3 Additional Workflow-Methods 2 3.1 Options..............................
RMassBank for XCMS Erik Müller January 4, 2019 Contents 1 Introduction 2 2 Input files 2 2.1 LC/MS data........................... 2 3 Additional Workflow-Methods 2 3.1 Options..............................
Visualisation, transformations and arithmetic operations for grouped genomic intervals
 ## Warning: replacing previous import ggplot2::position by BiocGenerics::Position when loading soggi Visualisation, transformations and arithmetic operations for grouped genomic intervals Thomas Carroll
## Warning: replacing previous import ggplot2::position by BiocGenerics::Position when loading soggi Visualisation, transformations and arithmetic operations for grouped genomic intervals Thomas Carroll
A complete analysis of peptide microarray binding data using the pepstat framework
 A complete analysis of peptide microarray binding data using the pepstat framework Greg Imholte, Renan Sauteraud, Mike Jiang and Raphael Gottardo April 30, 2018 This document present a full analysis, from
A complete analysis of peptide microarray binding data using the pepstat framework Greg Imholte, Renan Sauteraud, Mike Jiang and Raphael Gottardo April 30, 2018 This document present a full analysis, from
Package arrayqualitymetrics
 Package arrayqualitymetrics January 17, 2018 Title Quality metrics report for microarray data sets Version 3.34.0 Author Maintainer Mike Smith BugReports https://github.com/grimbough/arrayqualitymetrics/issues
Package arrayqualitymetrics January 17, 2018 Title Quality metrics report for microarray data sets Version 3.34.0 Author Maintainer Mike Smith BugReports https://github.com/grimbough/arrayqualitymetrics/issues
SimBindProfiles: Similar Binding Profiles, identifies common and unique regions in array genome tiling array data
 SimBindProfiles: Similar Binding Profiles, identifies common and unique regions in array genome tiling array data Bettina Fischer October 30, 2018 Contents 1 Introduction 1 2 Reading data and normalisation
SimBindProfiles: Similar Binding Profiles, identifies common and unique regions in array genome tiling array data Bettina Fischer October 30, 2018 Contents 1 Introduction 1 2 Reading data and normalisation
NacoStringQCPro. Dorothee Nickles, Thomas Sandmann, Robert Ziman, Richard Bourgon. Modified: April 10, Compiled: April 24, 2017.
 NacoStringQCPro Dorothee Nickles, Thomas Sandmann, Robert Ziman, Richard Bourgon Modified: April 10, 2017. Compiled: April 24, 2017 Contents 1 Scope 1 2 Quick start 1 3 RccSet loaded with NanoString data
NacoStringQCPro Dorothee Nickles, Thomas Sandmann, Robert Ziman, Richard Bourgon Modified: April 10, 2017. Compiled: April 24, 2017 Contents 1 Scope 1 2 Quick start 1 3 RccSet loaded with NanoString data
Advanced analysis using bayseq; generic distribution definitions
 Advanced analysis using bayseq; generic distribution definitions Thomas J Hardcastle October 30, 2017 1 Generic Prior Distributions bayseq now offers complete user-specification of underlying distributions
Advanced analysis using bayseq; generic distribution definitions Thomas J Hardcastle October 30, 2017 1 Generic Prior Distributions bayseq now offers complete user-specification of underlying distributions
segmentseq: methods for detecting methylation loci and differential methylation
 segmentseq: methods for detecting methylation loci and differential methylation Thomas J. Hardcastle October 30, 2018 1 Introduction This vignette introduces analysis methods for data from high-throughput
segmentseq: methods for detecting methylation loci and differential methylation Thomas J. Hardcastle October 30, 2018 1 Introduction This vignette introduces analysis methods for data from high-throughput
Analysis of two-way cell-based assays
 Analysis of two-way cell-based assays Lígia Brás, Michael Boutros and Wolfgang Huber April 16, 2015 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
Analysis of two-way cell-based assays Lígia Brás, Michael Boutros and Wolfgang Huber April 16, 2015 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
MIGSA: Getting pbcmc datasets
 MIGSA: Getting pbcmc datasets Juan C Rodriguez Universidad Católica de Córdoba Universidad Nacional de Córdoba Cristóbal Fresno Instituto Nacional de Medicina Genómica Andrea S Llera Fundación Instituto
MIGSA: Getting pbcmc datasets Juan C Rodriguez Universidad Católica de Córdoba Universidad Nacional de Córdoba Cristóbal Fresno Instituto Nacional de Medicina Genómica Andrea S Llera Fundación Instituto
riboseqr Introduction Getting Data Workflow Example Thomas J. Hardcastle, Betty Y.W. Chung October 30, 2017
 riboseqr Thomas J. Hardcastle, Betty Y.W. Chung October 30, 2017 Introduction Ribosome profiling extracts those parts of a coding sequence currently bound by a ribosome (and thus, are likely to be undergoing
riboseqr Thomas J. Hardcastle, Betty Y.W. Chung October 30, 2017 Introduction Ribosome profiling extracts those parts of a coding sequence currently bound by a ribosome (and thus, are likely to be undergoing
Introduction to the Codelink package
 Introduction to the Codelink package Diego Diez October 30, 2018 1 Introduction This package implements methods to facilitate the preprocessing and analysis of Codelink microarrays. Codelink is a microarray
Introduction to the Codelink package Diego Diez October 30, 2018 1 Introduction This package implements methods to facilitate the preprocessing and analysis of Codelink microarrays. Codelink is a microarray
How to Use pkgdeptools
 How to Use pkgdeptools Seth Falcon February 10, 2018 1 Introduction The pkgdeptools package provides tools for computing and analyzing dependency relationships among R packages. With it, you can build
How to Use pkgdeptools Seth Falcon February 10, 2018 1 Introduction The pkgdeptools package provides tools for computing and analyzing dependency relationships among R packages. With it, you can build
Introduction to the TSSi package: Identification of Transcription Start Sites
 Introduction to the TSSi package: Identification of Transcription Start Sites Julian Gehring, Clemens Kreutz October 30, 2017 Along with the advances in high-throughput sequencing, the detection of transcription
Introduction to the TSSi package: Identification of Transcription Start Sites Julian Gehring, Clemens Kreutz October 30, 2017 Along with the advances in high-throughput sequencing, the detection of transcription
Creating a New Annotation Package using SQLForge
 Creating a New Annotation Package using SQLForge Marc Carlson, HervÃľ PagÃĺs, Nianhua Li April 30, 2018 1 Introduction The AnnotationForge package provides a series of functions that can be used to build
Creating a New Annotation Package using SQLForge Marc Carlson, HervÃľ PagÃĺs, Nianhua Li April 30, 2018 1 Introduction The AnnotationForge package provides a series of functions that can be used to build
Working with aligned nucleotides (WORK- IN-PROGRESS!)
 Working with aligned nucleotides (WORK- IN-PROGRESS!) Hervé Pagès Last modified: January 2014; Compiled: November 17, 2017 Contents 1 Introduction.............................. 1 2 Load the aligned reads
Working with aligned nucleotides (WORK- IN-PROGRESS!) Hervé Pagès Last modified: January 2014; Compiled: November 17, 2017 Contents 1 Introduction.............................. 1 2 Load the aligned reads
INSPEcT - INference of Synthesis, Processing and degradation rates in Time-course analysis
 INSPEcT - INference of Synthesis, Processing and degradation rates in Time-course analysis Stefano de Pretis October 30, 2017 Contents 1 Introduction.............................. 1 2 Quantification of
INSPEcT - INference of Synthesis, Processing and degradation rates in Time-course analysis Stefano de Pretis October 30, 2017 Contents 1 Introduction.............................. 1 2 Quantification of
An Overview of the S4Vectors package
 Patrick Aboyoun, Michael Lawrence, Hervé Pagès Edited: February 2018; Compiled: June 7, 2018 Contents 1 Introduction.............................. 1 2 Vector-like and list-like objects...................
Patrick Aboyoun, Michael Lawrence, Hervé Pagès Edited: February 2018; Compiled: June 7, 2018 Contents 1 Introduction.............................. 1 2 Vector-like and list-like objects...................
An Introduction to the methylumi package
 An Introduction to the methylumi package Sean Davis and Sven Bilke April 30, 2018 1 Introduction Gene expression patterns are very important in understanding any biologic system. The regulation of gene
An Introduction to the methylumi package Sean Davis and Sven Bilke April 30, 2018 1 Introduction Gene expression patterns are very important in understanding any biologic system. The regulation of gene
Analysis of screens with enhancer and suppressor controls
 Analysis of screens with enhancer and suppressor controls Lígia Brás, Michael Boutros and Wolfgang Huber April 16, 2015 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
Analysis of screens with enhancer and suppressor controls Lígia Brás, Michael Boutros and Wolfgang Huber April 16, 2015 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
MMDiff2: Statistical Testing for ChIP-Seq Data Sets
 MMDiff2: Statistical Testing for ChIP-Seq Data Sets Gabriele Schwikert 1 and David Kuo 2 [1em] 1 The Informatics Forum, University of Edinburgh and The Wellcome
MMDiff2: Statistical Testing for ChIP-Seq Data Sets Gabriele Schwikert 1 and David Kuo 2 [1em] 1 The Informatics Forum, University of Edinburgh and The Wellcome
RNAseq Differential Expression Analysis Jana Schor and Jörg Hackermüller November, 2017
 RNAseq Differential Expression Analysis Jana Schor and Jörg Hackermüller November, 2017 RNA-Seq: Example dataset and download} Dataset taken from Somel et al. 2012: (check publication for details) 3 human
RNAseq Differential Expression Analysis Jana Schor and Jörg Hackermüller November, 2017 RNA-Seq: Example dataset and download} Dataset taken from Somel et al. 2012: (check publication for details) 3 human
Creating IGV HTML reports with tracktables
 Thomas Carroll 1 [1em] 1 Bioinformatics Core, MRC Clinical Sciences Centre; thomas.carroll (at)imperial.ac.uk June 13, 2018 Contents 1 The tracktables package.................... 1 2 Creating IGV sessions
Thomas Carroll 1 [1em] 1 Bioinformatics Core, MRC Clinical Sciences Centre; thomas.carroll (at)imperial.ac.uk June 13, 2018 Contents 1 The tracktables package.................... 1 2 Creating IGV sessions
An Introduction to the methylumi package
 An Introduction to the methylumi package Sean Davis and Sven Bilke October 13, 2014 1 Introduction Gene expression patterns are very important in understanding any biologic system. The regulation of gene
An Introduction to the methylumi package Sean Davis and Sven Bilke October 13, 2014 1 Introduction Gene expression patterns are very important in understanding any biologic system. The regulation of gene
HowTo: Querying online Data
 HowTo: Querying online Data Jeff Gentry and Robert Gentleman November 12, 2017 1 Overview This article demonstrates how you can make use of the tools that have been provided for on-line querying of data
HowTo: Querying online Data Jeff Gentry and Robert Gentleman November 12, 2017 1 Overview This article demonstrates how you can make use of the tools that have been provided for on-line querying of data
segmentseq: methods for detecting methylation loci and differential methylation
 segmentseq: methods for detecting methylation loci and differential methylation Thomas J. Hardcastle October 13, 2015 1 Introduction This vignette introduces analysis methods for data from high-throughput
segmentseq: methods for detecting methylation loci and differential methylation Thomas J. Hardcastle October 13, 2015 1 Introduction This vignette introduces analysis methods for data from high-throughput
How to use cghmcr. October 30, 2017
 How to use cghmcr Jianhua Zhang Bin Feng October 30, 2017 1 Overview Copy number data (arraycgh or SNP) can be used to identify genomic regions (Regions Of Interest or ROI) showing gains or losses that
How to use cghmcr Jianhua Zhang Bin Feng October 30, 2017 1 Overview Copy number data (arraycgh or SNP) can be used to identify genomic regions (Regions Of Interest or ROI) showing gains or losses that
INSPEcT - INference of Synthesis, Processing and degradation rates in Time-course analysis
 INSPEcT - INference of Synthesis, Processing and degradation rates in Time-course analysis Stefano de Pretis April 7, 20 Contents 1 Introduction 1 2 Quantification of Exon and Intron features 3 3 Time-course
INSPEcT - INference of Synthesis, Processing and degradation rates in Time-course analysis Stefano de Pretis April 7, 20 Contents 1 Introduction 1 2 Quantification of Exon and Intron features 3 3 Time-course
How To Use GOstats and Category to do Hypergeometric testing with unsupported model organisms
 How To Use GOstats and Category to do Hypergeometric testing with unsupported model organisms M. Carlson October 30, 2017 1 Introduction This vignette is meant as an extension of what already exists in
How To Use GOstats and Category to do Hypergeometric testing with unsupported model organisms M. Carlson October 30, 2017 1 Introduction This vignette is meant as an extension of what already exists in
The bumphunter user s guide
 The bumphunter user s guide Kasper Daniel Hansen khansen@jhsph.edu Martin Aryee aryee.martin@mgh.harvard.edu Rafael A. Irizarry rafa@jhu.edu Modified: November 23, 2012. Compiled: April 30, 2018 Introduction
The bumphunter user s guide Kasper Daniel Hansen khansen@jhsph.edu Martin Aryee aryee.martin@mgh.harvard.edu Rafael A. Irizarry rafa@jhu.edu Modified: November 23, 2012. Compiled: April 30, 2018 Introduction
Analysis of Bead-level Data using beadarray
 Mark Dunning October 30, 2017 Introduction beadarray is a package for the pre-processing and analysis of Illumina BeadArray. The main advantage is being able to read raw data output by Illumina s scanning
Mark Dunning October 30, 2017 Introduction beadarray is a package for the pre-processing and analysis of Illumina BeadArray. The main advantage is being able to read raw data output by Illumina s scanning
An Introduction to ShortRead
 Martin Morgan Modified: 21 October, 2013. Compiled: April 30, 2018 > library("shortread") The ShortRead package provides functionality for working with FASTQ files from high throughput sequence analysis.
Martin Morgan Modified: 21 October, 2013. Compiled: April 30, 2018 > library("shortread") The ShortRead package provides functionality for working with FASTQ files from high throughput sequence analysis.
Creating a New Annotation Package using SQLForge
 Creating a New Annotation Package using SQLForge Marc Carlson, Herve Pages, Nianhua Li November 19, 2013 1 Introduction The AnnotationForge package provides a series of functions that can be used to build
Creating a New Annotation Package using SQLForge Marc Carlson, Herve Pages, Nianhua Li November 19, 2013 1 Introduction The AnnotationForge package provides a series of functions that can be used to build
Bioconductor: Annotation Package Overview
 Bioconductor: Annotation Package Overview April 30, 2018 1 Overview In its current state the basic purpose of annotate is to supply interface routines that support user actions that rely on the different
Bioconductor: Annotation Package Overview April 30, 2018 1 Overview In its current state the basic purpose of annotate is to supply interface routines that support user actions that rely on the different
GenoGAM: Genome-wide generalized additive
 GenoGAM: Genome-wide generalized additive models Georg Stricker 1, Julien Gagneur 1 [1em] 1 Technische Universität München, Department of Informatics, Garching, Germany Abstract April 30, 2018 Many genomic
GenoGAM: Genome-wide generalized additive models Georg Stricker 1, Julien Gagneur 1 [1em] 1 Technische Universität München, Department of Informatics, Garching, Germany Abstract April 30, 2018 Many genomic
Count outlier detection using Cook s distance
 Count outlier detection using Cook s distance Michael Love August 9, 2014 1 Run DE analysis with and without outlier removal The following vignette produces the Supplemental Figure of the effect of replacing
Count outlier detection using Cook s distance Michael Love August 9, 2014 1 Run DE analysis with and without outlier removal The following vignette produces the Supplemental Figure of the effect of replacing
Cardinal design and development
 Kylie A. Bemis October 30, 2017 Contents 1 Introduction.............................. 2 2 Design overview........................... 2 3 iset: high-throughput imaging experiments............ 3 3.1 SImageSet:
Kylie A. Bemis October 30, 2017 Contents 1 Introduction.............................. 2 2 Design overview........................... 2 3 iset: high-throughput imaging experiments............ 3 3.1 SImageSet:
SCnorm: robust normalization of single-cell RNA-seq data
 SCnorm: robust normalization of single-cell RNA-seq data Rhonda Bacher and Christina Kendziorski February 18, 2019 Contents 1 Introduction.............................. 1 2 Run SCnorm.............................
SCnorm: robust normalization of single-cell RNA-seq data Rhonda Bacher and Christina Kendziorski February 18, 2019 Contents 1 Introduction.............................. 1 2 Run SCnorm.............................
DupChecker: a bioconductor package for checking high-throughput genomic data redundancy in meta-analysis
 DupChecker: a bioconductor package for checking high-throughput genomic data redundancy in meta-analysis Quanhu Sheng, Yu Shyr, Xi Chen Center for Quantitative Sciences, Vanderbilt University, Nashville,
DupChecker: a bioconductor package for checking high-throughput genomic data redundancy in meta-analysis Quanhu Sheng, Yu Shyr, Xi Chen Center for Quantitative Sciences, Vanderbilt University, Nashville,
Getting started with flowstats
 Getting started with flowstats F. Hahne, N. Gopalakrishnan October, 7 Abstract flowstats is a collection of algorithms for the statistical analysis of flow cytometry data. So far, the focus is on automated
Getting started with flowstats F. Hahne, N. Gopalakrishnan October, 7 Abstract flowstats is a collection of algorithms for the statistical analysis of flow cytometry data. So far, the focus is on automated
PROPER: PROspective Power Evaluation for RNAseq
 PROPER: PROspective Power Evaluation for RNAseq Hao Wu [1em]Department of Biostatistics and Bioinformatics Emory University Atlanta, GA 303022 [1em] hao.wu@emory.edu October 30, 2017 Contents 1 Introduction..............................
PROPER: PROspective Power Evaluation for RNAseq Hao Wu [1em]Department of Biostatistics and Bioinformatics Emory University Atlanta, GA 303022 [1em] hao.wu@emory.edu October 30, 2017 Contents 1 Introduction..............................
mocluster: Integrative clustering using multiple
 mocluster: Integrative clustering using multiple omics data Chen Meng Modified: June 03, 2015. Compiled: October 30, 2018. Contents 1 mocluster overview......................... 1 2 Run mocluster............................
mocluster: Integrative clustering using multiple omics data Chen Meng Modified: June 03, 2015. Compiled: October 30, 2018. Contents 1 mocluster overview......................... 1 2 Run mocluster............................
survsnp: Power and Sample Size Calculations for SNP Association Studies with Censored Time to Event Outcomes
 survsnp: Power and Sample Size Calculations for SNP Association Studies with Censored Time to Event Outcomes Kouros Owzar Zhiguo Li Nancy Cox Sin-Ho Jung Chanhee Yi June 29, 2016 1 Introduction This vignette
survsnp: Power and Sample Size Calculations for SNP Association Studies with Censored Time to Event Outcomes Kouros Owzar Zhiguo Li Nancy Cox Sin-Ho Jung Chanhee Yi June 29, 2016 1 Introduction This vignette
Analysis of Pairwise Interaction Screens
 Analysis of Pairwise Interaction Screens Bernd Fischer April 11, 2014 Contents 1 Introduction 1 2 Installation of the RNAinteract package 1 3 Creating an RNAinteract object 2 4 Single Perturbation effects
Analysis of Pairwise Interaction Screens Bernd Fischer April 11, 2014 Contents 1 Introduction 1 2 Installation of the RNAinteract package 1 3 Creating an RNAinteract object 2 4 Single Perturbation effects
bayseq: Empirical Bayesian analysis of patterns of differential expression in count data
 bayseq: Empirical Bayesian analysis of patterns of differential expression in count data Thomas J. Hardcastle October 30, 2017 1 Introduction This vignette is intended to give a rapid introduction to the
bayseq: Empirical Bayesian analysis of patterns of differential expression in count data Thomas J. Hardcastle October 30, 2017 1 Introduction This vignette is intended to give a rapid introduction to the
The rtracklayer package
 The rtracklayer package Michael Lawrence January 22, 2018 Contents 1 Introduction 2 2 Gene expression and microrna target sites 2 2.1 Creating a target site track..................... 2 2.1.1 Constructing
The rtracklayer package Michael Lawrence January 22, 2018 Contents 1 Introduction 2 2 Gene expression and microrna target sites 2 2.1 Creating a target site track..................... 2 2.1.1 Constructing
Shrinkage of logarithmic fold changes
 Shrinkage of logarithmic fold changes Michael Love August 9, 2014 1 Comparing the posterior distribution for two genes First, we run a DE analysis on the Bottomly et al. dataset, once with shrunken LFCs
Shrinkage of logarithmic fold changes Michael Love August 9, 2014 1 Comparing the posterior distribution for two genes First, we run a DE analysis on the Bottomly et al. dataset, once with shrunken LFCs
How to use CNTools. Overview. Algorithms. Jianhua Zhang. April 14, 2011
 How to use CNTools Jianhua Zhang April 14, 2011 Overview Studies have shown that genomic alterations measured as DNA copy number variations invariably occur across chromosomal regions that span over several
How to use CNTools Jianhua Zhang April 14, 2011 Overview Studies have shown that genomic alterations measured as DNA copy number variations invariably occur across chromosomal regions that span over several
qplexanalyzer Matthew Eldridge, Kamal Kishore, Ashley Sawle January 4, Overview 1 2 Import quantitative dataset 2 3 Quality control 2
 qplexanalyzer Matthew Eldridge, Kamal Kishore, Ashley Sawle January 4, 2019 Contents 1 Overview 1 2 Import quantitative dataset 2 3 Quality control 2 4 Data normalization 8 5 Aggregation of peptide intensities
qplexanalyzer Matthew Eldridge, Kamal Kishore, Ashley Sawle January 4, 2019 Contents 1 Overview 1 2 Import quantitative dataset 2 3 Quality control 2 4 Data normalization 8 5 Aggregation of peptide intensities
Evaluation of VST algorithm in lumi package
 Evaluation of VST algorithm in lumi package Pan Du 1, Simon Lin 1, Wolfgang Huber 2, Warrren A. Kibbe 1 December 22, 2010 1 Robert H. Lurie Comprehensive Cancer Center Northwestern University, Chicago,
Evaluation of VST algorithm in lumi package Pan Du 1, Simon Lin 1, Wolfgang Huber 2, Warrren A. Kibbe 1 December 22, 2010 1 Robert H. Lurie Comprehensive Cancer Center Northwestern University, Chicago,
The analysis of rtpcr data
 The analysis of rtpcr data Jitao David Zhang, Markus Ruschhaupt October 30, 2017 With the help of this document, an analysis of rtpcr data can be performed. For this, the user has to specify several parameters
The analysis of rtpcr data Jitao David Zhang, Markus Ruschhaupt October 30, 2017 With the help of this document, an analysis of rtpcr data can be performed. For this, the user has to specify several parameters
MethylAid: Visual and Interactive quality control of Illumina Human DNA Methylation array data
 MethylAid: Visual and Interactive quality control of Illumina Human DNA Methylation array Maarten van Iterson, Elmar Tobi, Roderick Slieker, Wouter den Hollander, Rene Luijk, Eline Slagboom and Bas Heijmans
MethylAid: Visual and Interactive quality control of Illumina Human DNA Methylation array Maarten van Iterson, Elmar Tobi, Roderick Slieker, Wouter den Hollander, Rene Luijk, Eline Slagboom and Bas Heijmans
SCnorm: robust normalization of single-cell RNA-seq data
 SCnorm: robust normalization of single-cell RNA-seq data Rhonda Bacher and Christina Kendziorski September 24, 207 Contents Introduction.............................. 2 Run SCnorm.............................
SCnorm: robust normalization of single-cell RNA-seq data Rhonda Bacher and Christina Kendziorski September 24, 207 Contents Introduction.............................. 2 Run SCnorm.............................
An Introduction to GenomeInfoDb
 Martin Morgan, Hervé Pagès, Marc Carlson, Sonali Arora Modified: 17 January, 2014. Compiled: January 21, 2018 Contents 1 Introduction.............................. 2 2 Functionality for all existing organisms...............
Martin Morgan, Hervé Pagès, Marc Carlson, Sonali Arora Modified: 17 January, 2014. Compiled: January 21, 2018 Contents 1 Introduction.............................. 2 2 Functionality for all existing organisms...............
CQN (Conditional Quantile Normalization)
 CQN (Conditional Quantile Normalization) Kasper Daniel Hansen khansen@jhsph.edu Zhijin Wu zhijin_wu@brown.edu Modified: August 8, 2012. Compiled: April 30, 2018 Introduction This package contains the CQN
CQN (Conditional Quantile Normalization) Kasper Daniel Hansen khansen@jhsph.edu Zhijin Wu zhijin_wu@brown.edu Modified: August 8, 2012. Compiled: April 30, 2018 Introduction This package contains the CQN
Using branchpointer for annotation of intronic human splicing branchpoints
 Using branchpointer for annotation of intronic human splicing branchpoints Beth Signal October 30, 2018 Contents 1 Introduction.............................. 1 2 Preparation..............................
Using branchpointer for annotation of intronic human splicing branchpoints Beth Signal October 30, 2018 Contents 1 Introduction.............................. 1 2 Preparation..............................
RDAVIDWebService: a versatile R interface to DAVID
 RDAVIDWebService: a versatile R interface to DAVID Cristóbal Fresno 1,2 and Elmer A. Fernández 1,2 1 Bio-science Data Mining Group, Catholic University of Córdoba, Córdoba, Argentina. 2 CONICET, Argentina
RDAVIDWebService: a versatile R interface to DAVID Cristóbal Fresno 1,2 and Elmer A. Fernández 1,2 1 Bio-science Data Mining Group, Catholic University of Córdoba, Córdoba, Argentina. 2 CONICET, Argentina
Using the qrqc package to gather information about sequence qualities
 Using the qrqc package to gather information about sequence qualities Vince Buffalo Bioinformatics Core UC Davis Genome Center vsbuffalo@ucdavis.edu 2012-02-19 Abstract Many projects in bioinformatics
Using the qrqc package to gather information about sequence qualities Vince Buffalo Bioinformatics Core UC Davis Genome Center vsbuffalo@ucdavis.edu 2012-02-19 Abstract Many projects in bioinformatics
Preprocessing and Genotyping Illumina Arrays for Copy Number Analysis
 Preprocessing and Genotyping Illumina Arrays for Copy Number Analysis Rob Scharpf September 18, 2012 Abstract This vignette illustrates the steps required prior to copy number analysis for Infinium platforms.
Preprocessing and Genotyping Illumina Arrays for Copy Number Analysis Rob Scharpf September 18, 2012 Abstract This vignette illustrates the steps required prior to copy number analysis for Infinium platforms.
Building R objects from ArrayExpress datasets
 Building R objects from ArrayExpress datasets Audrey Kauffmann October 30, 2017 1 ArrayExpress database ArrayExpress is a public repository for transcriptomics and related data, which is aimed at storing
Building R objects from ArrayExpress datasets Audrey Kauffmann October 30, 2017 1 ArrayExpress database ArrayExpress is a public repository for transcriptomics and related data, which is aimed at storing
ssviz: A small RNA-seq visualizer and analysis toolkit
 ssviz: A small RNA-seq visualizer and analysis toolkit Diana HP Low Institute of Molecular and Cell Biology Agency for Science, Technology and Research (A*STAR), Singapore dlow@imcb.a-star.edu.sg August
ssviz: A small RNA-seq visualizer and analysis toolkit Diana HP Low Institute of Molecular and Cell Biology Agency for Science, Technology and Research (A*STAR), Singapore dlow@imcb.a-star.edu.sg August
puma User Guide R. D. Pearson, X. Liu, M. Rattray, M. Milo, N. D. Lawrence G. Sanguinetti, Li Zhang October 30, Abstract 2 2 Citing puma 2
 puma User Guide R. D. Pearson, X. Liu, M. Rattray, M. Milo, N. D. Lawrence G. Sanguinetti, Li Zhang October 30, 2017 Contents 1 Abstract 2 2 Citing puma 2 3 Introduction 3 4 Introductory example analysis
puma User Guide R. D. Pearson, X. Liu, M. Rattray, M. Milo, N. D. Lawrence G. Sanguinetti, Li Zhang October 30, 2017 Contents 1 Abstract 2 2 Citing puma 2 3 Introduction 3 4 Introductory example analysis
An Introduction to Bioconductor s ExpressionSet Class
 An Introduction to Bioconductor s ExpressionSet Class Seth Falcon, Martin Morgan, and Robert Gentleman 6 October, 2006; revised 9 February, 2007 1 Introduction Biobase is part of the Bioconductor project,
An Introduction to Bioconductor s ExpressionSet Class Seth Falcon, Martin Morgan, and Robert Gentleman 6 October, 2006; revised 9 February, 2007 1 Introduction Biobase is part of the Bioconductor project,
The REDseq user s guide
 The REDseq user s guide Lihua Julie Zhu * October 30, 2017 Contents 1 Introduction 1 2 Examples of using REDseq 2 2.1 Task 1: Build a RE map for a genome..................... 2 2.2 Task 2: Assign mapped
The REDseq user s guide Lihua Julie Zhu * October 30, 2017 Contents 1 Introduction 1 2 Examples of using REDseq 2 2.1 Task 1: Build a RE map for a genome..................... 2 2.2 Task 2: Assign mapped
Analyze Illumina Infinium methylation microarray data
 Analyze Illumina Infinium methylation microarray data Pan Du, Gang Feng, Spencer Huang, Warren A. Kibbe, Simon Lin May 3, 2016 Genentech Inc., South San Francisco, CA, 94080, USA Biomedical Informatics
Analyze Illumina Infinium methylation microarray data Pan Du, Gang Feng, Spencer Huang, Warren A. Kibbe, Simon Lin May 3, 2016 Genentech Inc., South San Francisco, CA, 94080, USA Biomedical Informatics
Package frma. R topics documented: March 8, Version Date Title Frozen RMA and Barcode
 Version 1.34.0 Date 2017-11-08 Title Frozen RMA and Barcode Package frma March 8, 2019 Preprocessing and analysis for single microarrays and microarray batches. Author Matthew N. McCall ,
Version 1.34.0 Date 2017-11-08 Title Frozen RMA and Barcode Package frma March 8, 2019 Preprocessing and analysis for single microarrays and microarray batches. Author Matthew N. McCall ,
Introduction to GenomicFiles
 Valerie Obenchain, Michael Love, Martin Morgan Last modified: October 2014; Compiled: October 30, 2018 Contents 1 Introduction.............................. 1 2 Quick Start..............................
Valerie Obenchain, Michael Love, Martin Morgan Last modified: October 2014; Compiled: October 30, 2018 Contents 1 Introduction.............................. 1 2 Quick Start..............................
Comprehensive Pipeline for Analyzing and Visualizing Array-Based CGH Data
 Comprehensive Pipeline for Analyzing and Visualizing Array-Based CGH Data Frederic Commo fredcommo@gmail.com November 30, 2017 1 Introduction Genomic profiling using array-based comparative genomic hybridization
Comprehensive Pipeline for Analyzing and Visualizing Array-Based CGH Data Frederic Commo fredcommo@gmail.com November 30, 2017 1 Introduction Genomic profiling using array-based comparative genomic hybridization
The DMRcate package user s guide
 The DMRcate package user s guide Peters TJ, Buckley MJ, Statham A, Pidsley R, Clark SJ, Molloy PL March 31, 2017 Summary DMRcate extracts the most differentially methylated regions (DMRs) and variably
The DMRcate package user s guide Peters TJ, Buckley MJ, Statham A, Pidsley R, Clark SJ, Molloy PL March 31, 2017 Summary DMRcate extracts the most differentially methylated regions (DMRs) and variably
Bioconductor tutorial
 Bioconductor tutorial Adapted by Alex Sanchez from tutorials by (1) Steffen Durinck, Robert Gentleman and Sandrine Dudoit (2) Laurent Gautier (3) Matt Ritchie (4) Jean Yang Outline The Bioconductor Project
Bioconductor tutorial Adapted by Alex Sanchez from tutorials by (1) Steffen Durinck, Robert Gentleman and Sandrine Dudoit (2) Laurent Gautier (3) Matt Ritchie (4) Jean Yang Outline The Bioconductor Project
Overview of ensemblvep
 Overview of ensemblvep Lori Shepherd and Valerie Obenchain October 30, 2017 Contents 1 Introduction 1 2 Results as R objects 1 3 Write results to a file 4 4 Configuring runtime options 5 5 sessioninfo()
Overview of ensemblvep Lori Shepherd and Valerie Obenchain October 30, 2017 Contents 1 Introduction 1 2 Results as R objects 1 3 Write results to a file 4 4 Configuring runtime options 5 5 sessioninfo()
Comparing methods for differential expression analysis of RNAseq data with the compcoder
 Comparing methods for differential expression analysis of RNAseq data with the compcoder package Charlotte Soneson October 30, 2017 Contents 1 Introduction.............................. 2 2 The compdata
Comparing methods for differential expression analysis of RNAseq data with the compcoder package Charlotte Soneson October 30, 2017 Contents 1 Introduction.............................. 2 2 The compdata
Triform: peak finding in ChIP-Seq enrichment profiles for transcription factors
 Triform: peak finding in ChIP-Seq enrichment profiles for transcription factors Karl Kornacker * and Tony Håndstad October 30, 2018 A guide for using the Triform algorithm to predict transcription factor
Triform: peak finding in ChIP-Seq enrichment profiles for transcription factors Karl Kornacker * and Tony Håndstad October 30, 2018 A guide for using the Triform algorithm to predict transcription factor
Introduction to QDNAseq
 Ilari Scheinin October 30, 2017 Contents 1 Running QDNAseq.......................... 1 1.1 Bin annotations.......................... 1 1.2 Processing bam files....................... 2 1.3 Downstream analyses......................
Ilari Scheinin October 30, 2017 Contents 1 Running QDNAseq.......................... 1 1.1 Bin annotations.......................... 1 1.2 Processing bam files....................... 2 1.3 Downstream analyses......................
MLSeq package: Machine Learning Interface to RNA-Seq Data
 MLSeq package: Machine Learning Interface to RNA-Seq Data Gokmen Zararsiz 1, Dincer Goksuluk 2, Selcuk Korkmaz 2, Vahap Eldem 3, Izzet Parug Duru 4, Turgay Unver 5, Ahmet Ozturk 5 1 Erciyes University,
MLSeq package: Machine Learning Interface to RNA-Seq Data Gokmen Zararsiz 1, Dincer Goksuluk 2, Selcuk Korkmaz 2, Vahap Eldem 3, Izzet Parug Duru 4, Turgay Unver 5, Ahmet Ozturk 5 1 Erciyes University,
Fitting Baselines to Spectra
 Fitting Baselines to Spectra Claudia Beleites DIA Raman Spectroscopy Group, University of Trieste/Italy (2005 2008) Spectroscopy Imaging, IPHT, Jena/Germany (2008 2016) ÖPV, JKI,
Fitting Baselines to Spectra Claudia Beleites DIA Raman Spectroscopy Group, University of Trieste/Italy (2005 2008) Spectroscopy Imaging, IPHT, Jena/Germany (2008 2016) ÖPV, JKI,
MultipleAlignment Objects
 MultipleAlignment Objects Marc Carlson Bioconductor Core Team Fred Hutchinson Cancer Research Center Seattle, WA April 30, 2018 Contents 1 Introduction 1 2 Creation and masking 1 3 Analytic utilities 7
MultipleAlignment Objects Marc Carlson Bioconductor Core Team Fred Hutchinson Cancer Research Center Seattle, WA April 30, 2018 Contents 1 Introduction 1 2 Creation and masking 1 3 Analytic utilities 7
The fastliquidassociation Package
 The fastliquidassociation Package Tina Gunderson October 30, 2018 1 Introduction The fastliquidassociation package is intended to provide more efficient tools for genome-wide application of liquid association.
The fastliquidassociation Package Tina Gunderson October 30, 2018 1 Introduction The fastliquidassociation package is intended to provide more efficient tools for genome-wide application of liquid association.
crlmm to downstream data analysis
 crlmm to downstream data analysis VJ Carey, B Carvalho March, 2012 1 Running CRLMM on a nontrivial set of CEL files To use the crlmm algorithm, the user must load the crlmm package, as described below:
crlmm to downstream data analysis VJ Carey, B Carvalho March, 2012 1 Running CRLMM on a nontrivial set of CEL files To use the crlmm algorithm, the user must load the crlmm package, as described below:
Introduction to BatchtoolsParam
 Nitesh Turaga 1, Martin Morgan 2 Edited: March 22, 2018; Compiled: January 4, 2019 1 Nitesh.Turaga@ RoswellPark.org 2 Martin.Morgan@ RoswellPark.org Contents 1 Introduction..............................
Nitesh Turaga 1, Martin Morgan 2 Edited: March 22, 2018; Compiled: January 4, 2019 1 Nitesh.Turaga@ RoswellPark.org 2 Martin.Morgan@ RoswellPark.org Contents 1 Introduction..............................
Frozen Robust Multi-Array Analysis and the Gene Expression Barcode
 Frozen Robust Multi-Array Analysis and the Gene Expression Barcode Matthew N. McCall November 9, 2017 Contents 1 Frozen Robust Multiarray Analysis (frma) 2 1.1 From CEL files to expression estimates...................
Frozen Robust Multi-Array Analysis and the Gene Expression Barcode Matthew N. McCall November 9, 2017 Contents 1 Frozen Robust Multiarray Analysis (frma) 2 1.1 From CEL files to expression estimates...................
Starr: Simple Tiling ARRay analysis for Affymetrix ChIP-chip data
 Starr: Simple Tiling ARRay analysis for Affymetrix ChIP-chip data Benedikt Zacher, Achim Tresch October 30, 2017 1 Introduction Starr is an extension of the Ringo [6] package for the analysis of ChIP-chip
Starr: Simple Tiling ARRay analysis for Affymetrix ChIP-chip data Benedikt Zacher, Achim Tresch October 30, 2017 1 Introduction Starr is an extension of the Ringo [6] package for the analysis of ChIP-chip
rhdf5 - HDF5 interface for R
 Bernd Fischer October 30, 2017 Contents 1 Introduction 1 2 Installation of the HDF5 package 2 3 High level R -HDF5 functions 2 31 Creating an HDF5 file and group hierarchy 2 32 Writing and reading objects
Bernd Fischer October 30, 2017 Contents 1 Introduction 1 2 Installation of the HDF5 package 2 3 High level R -HDF5 functions 2 31 Creating an HDF5 file and group hierarchy 2 32 Writing and reading objects
Overview of ensemblvep
 Overview of ensemblvep Lori Shepherd and Valerie Obenchain October 30, 2018 Contents 1 Introduction 1 2 Results as R objects 1 3 Write results to a file 4 4 Configuring runtime options 5 5 sessioninfo()
Overview of ensemblvep Lori Shepherd and Valerie Obenchain October 30, 2018 Contents 1 Introduction 1 2 Results as R objects 1 3 Write results to a file 4 4 Configuring runtime options 5 5 sessioninfo()
RAPIDR. Kitty Lo. November 20, Intended use of RAPIDR 1. 2 Create binned counts file from BAMs Masking... 1
 RAPIDR Kitty Lo November 20, 2014 Contents 1 Intended use of RAPIDR 1 2 Create binned counts file from BAMs 1 2.1 Masking.................................................... 1 3 Build the reference 2 3.1
RAPIDR Kitty Lo November 20, 2014 Contents 1 Intended use of RAPIDR 1 2 Create binned counts file from BAMs 1 2.1 Masking.................................................... 1 3 Build the reference 2 3.1
M 3 D: Statistical Testing for RRBS Data Sets
 Tom Mayo Modified: 5 September, 2016. Compiled: October 30, 2018 Contents 1 Introduction 1 2 Analysis Pipeline 2 2.1 Reading in Data. 2 2.2 Computing the MMD, and coverage-mmd. 5 2.3 Generating p-values..
Tom Mayo Modified: 5 September, 2016. Compiled: October 30, 2018 Contents 1 Introduction 1 2 Analysis Pipeline 2 2.1 Reading in Data. 2 2.2 Computing the MMD, and coverage-mmd. 5 2.3 Generating p-values..
Making and Utilizing TxDb Objects
 Marc Carlson, Patrick Aboyoun, HervÃľ PagÃĺs, Seth Falcon, Martin Morgan October 30, 2017 1 Introduction The GenomicFeatures package retrieves and manages transcript-related features from the UCSC Genome
Marc Carlson, Patrick Aboyoun, HervÃľ PagÃĺs, Seth Falcon, Martin Morgan October 30, 2017 1 Introduction The GenomicFeatures package retrieves and manages transcript-related features from the UCSC Genome
Reproducible Homerange Analysis
 Reproducible Homerange Analysis (Sat Aug 09 15:28:43 2014) based on the rhr package This is an automatically generated file with all parameters and settings, in order to enable later replication of the
Reproducible Homerange Analysis (Sat Aug 09 15:28:43 2014) based on the rhr package This is an automatically generated file with all parameters and settings, in order to enable later replication of the
Exploring cdna Data. Achim Tresch, Andreas Buness, Wolfgang Huber, Tim Beißbarth
 Exploring cdna Data Achim Tresch, Andreas Buness, Wolfgang Huber, Tim Beißbarth Practical DNA Microarray Analysis http://compdiag.molgen.mpg.de/ngfn/pma0nov.shtml The following exercise will guide you
Exploring cdna Data Achim Tresch, Andreas Buness, Wolfgang Huber, Tim Beißbarth Practical DNA Microarray Analysis http://compdiag.molgen.mpg.de/ngfn/pma0nov.shtml The following exercise will guide you
Raman Spectra of Chondrocytes in Cartilage: hyperspec s chondro data set
 Raman Spectra of Chondrocytes in Cartilage: hyperspec s chondro data set Claudia Beleites CENMAT and DI3, University of Trieste Spectroscopy Imaging, IPHT Jena e.v. February 13,
Raman Spectra of Chondrocytes in Cartilage: hyperspec s chondro data set Claudia Beleites CENMAT and DI3, University of Trieste Spectroscopy Imaging, IPHT Jena e.v. February 13,
Biobase development and the new eset
 Biobase development and the new eset Martin T. Morgan * 7 August, 2006 Revised 4 September, 2006 featuredata slot. Revised 20 April 2007 minor wording changes; verbose and other arguments passed through
Biobase development and the new eset Martin T. Morgan * 7 August, 2006 Revised 4 September, 2006 featuredata slot. Revised 20 April 2007 minor wording changes; verbose and other arguments passed through
Wave correction for arrays
 Wave correction for arrays Eitan Halper-Stromberg and Robert B. Scharpf April 30, 2018 Abstract ArrayTV is a GC correction algorithm for microarray data designed to mitigate the appearance of waves. This
Wave correction for arrays Eitan Halper-Stromberg and Robert B. Scharpf April 30, 2018 Abstract ArrayTV is a GC correction algorithm for microarray data designed to mitigate the appearance of waves. This
Analyse RT PCR data with ddct
 Analyse RT PCR data with ddct Jitao David Zhang, Rudolf Biczok and Markus Ruschhaupt October 30, 2017 Abstract Quantitative real time PCR (qrt PCR or RT PCR for short) is a laboratory technique based on
Analyse RT PCR data with ddct Jitao David Zhang, Rudolf Biczok and Markus Ruschhaupt October 30, 2017 Abstract Quantitative real time PCR (qrt PCR or RT PCR for short) is a laboratory technique based on
Simulation of molecular regulatory networks with graphical models
 Simulation of molecular regulatory networks with graphical models Inma Tur 1 inma.tur@upf.edu Alberto Roverato 2 alberto.roverato@unibo.it Robert Castelo 1 robert.castelo@upf.edu 1 Universitat Pompeu Fabra,
Simulation of molecular regulatory networks with graphical models Inma Tur 1 inma.tur@upf.edu Alberto Roverato 2 alberto.roverato@unibo.it Robert Castelo 1 robert.castelo@upf.edu 1 Universitat Pompeu Fabra,
Using metama for differential gene expression analysis from multiple studies
 Using metama for differential gene expression analysis from multiple studies Guillemette Marot and Rémi Bruyère Modified: January 28, 2015. Compiled: January 28, 2015 Abstract This vignette illustrates
Using metama for differential gene expression analysis from multiple studies Guillemette Marot and Rémi Bruyère Modified: January 28, 2015. Compiled: January 28, 2015 Abstract This vignette illustrates
Preprocessing Affymetrix Data. Educational Materials 2005 R. Irizarry and R. Gentleman Modified 21 November, 2009, M. Morgan
 Preprocessing Affymetrix Data Educational Materials 2005 R. Irizarry and R. Gentleman Modified 21 November, 2009, M. Morgan 1 Input, Quality Assessment, and Pre-processing Fast Track Identify CEL files.
Preprocessing Affymetrix Data Educational Materials 2005 R. Irizarry and R. Gentleman Modified 21 November, 2009, M. Morgan 1 Input, Quality Assessment, and Pre-processing Fast Track Identify CEL files.
VariantFiltering: filtering of coding and noncoding genetic variants
 VariantFiltering: filtering of coding and noncoding genetic variants Dei M. Elurbe 1,2 and Robert Castelo 3,4 June 26, 2018 1 CIBER de Enfermedades Raras (CIBERER), Barcelona, Spain 2 Present address:
VariantFiltering: filtering of coding and noncoding genetic variants Dei M. Elurbe 1,2 and Robert Castelo 3,4 June 26, 2018 1 CIBER de Enfermedades Raras (CIBERER), Barcelona, Spain 2 Present address:
Exploring cdna Data. Achim Tresch, Andreas Buness, Tim Beißbarth, Wolfgang Huber
 Exploring cdna Data Achim Tresch, Andreas Buness, Tim Beißbarth, Wolfgang Huber Practical DNA Microarray Analysis http://compdiag.molgen.mpg.de/ngfn/pma0nov.shtml The following exercise will guide you
Exploring cdna Data Achim Tresch, Andreas Buness, Tim Beißbarth, Wolfgang Huber Practical DNA Microarray Analysis http://compdiag.molgen.mpg.de/ngfn/pma0nov.shtml The following exercise will guide you
