Introduction to Systems Biology II: Lab
|
|
|
- Olivia Summers
- 5 years ago
- Views:
Transcription
1 Introduction to Systems Biology II: Lab Amin Emad NIH BD2K KnowEnG Center of Excellence in Big Data Computing Carl R. Woese Institute for Genomic Biology Department of Computer Science University of Illinois at Urbana-Champaign June, 2018 National Center for Supercomputing Applications University of Illinois at Urbana-Champaign
2 Summary Our goal in this lab is to use several pipelines of the KnowEnG platform to analyze omic and phenotypic spreadsheets We will focus on the Spreadsheet Visualization, Clustering, and Gene Prioritization pipelines implemented in KnowEnG We will try both network-guided and standard modes of operation for the pipelines (if applicable) NIH Big Data Center of Excellence 2
3 Data First download the data which we will use from the link below: 08_Systems_Biology_II.zip After the download is complete, Right Click and Extract the contents of the archive to your course directory. We will use the files found in: [course_directory]/08_systems_biology_ii/ NIH Big Data Center of Excellence 3
4 STEP1: Sign In Follow the link to the HubZero login screen: Enter your username and password for the course into the credentials boxes Click Sign In NIH Big Data Center of Excellence 4
5 Visualization and simple analysis of genomic spreadsheets: NIH Big Data Center of Excellence 5
6 STEP2: Spreadsheet Visualization We will use KnowEnG s Spreadsheet Visualization pipeline to explore various properties of a transcriptomic spreadsheet and the relationship between transcriptomic features and different clinical phenotypes We will use data corresponding to breast tumor samples from the METABRIC study NIH Big Data Center of Excellence 6
7 STEP2: Spreadsheet Visualization Dataset characteristics: Name Expression_METABRIC_Demo1 Description A matrix of (gene x samples) containing the expression (microarray) of 233 genes in 1058 samples. The expression profiles are normalized in advance. Phenotype_METABRIC_Demo1 A matrix of (samples x clinical phenotypes) including PAM50 subtype, treatment, stage, survival years, etc. NIH Big Data Center of Excellence 7
8 Upload the data: STEP2: Spreadsheet Visualization Select Data at the top of the page Click on Upload New Data Click BROWSE and find the file to upload: NIH Big Data Center of Excellence 8
9 Select the pipeline: STEP2: Spreadsheet Visualization Select Analysis Pipelines at the top of the page Select Spreadsheet Visualization and Click on Start Pipeline NIH Big Data Center of Excellence 9
10 STEP2: Spreadsheet Visualization Configure the pipeline: Select the files: - Expression_METABRIC_Demo1.txt - Phenotype_METABRIC_Demo1.txt Select Next at the right bottom corner of the page You can change the name of the results Then press Submit Job NIH Big Data Center of Excellence 10
11 The results: STEP2: Spreadsheet Visualization Select Go to Data Page Select the job you just ran Then View Results NIH Big Data Center of Excellence 11
12 STEP2: Spreadsheet Visualization Allows grouping/ sorting of columns using another spreadsheet samples gene names NIH Big Data Center of Excellence 12
13 STEP2: Spreadsheet Visualization Click the dropdown Group Columns By menu and select the phenotype spreadsheet (Phenotype_METABRIC_Demo1.txt) NIH Big Data Center of Excellence 13
14 STEP2: Spreadsheet Visualization Click the dropdown Group Columns By menu and select the phenotype spreadsheet (Phenotype_METABRIC_Demo1.txt) Select PAM50 Class : the columns of the heatmap will automatically reorganize accordingly. Then press Done. PAM50 Class represents different subtypes of Breast Cancer NIH Big Data Center of Excellence 14
15 STEP2: Spreadsheet Visualization Click the dropdown Sort Columns By menu and select the phenotype spreadsheet (Phenotype_METABRIC_Demo1.txt) again NIH Big Data Center of Excellence 15
16 STEP2: Spreadsheet Visualization Click the dropdown Sort Columns By menu and select the phenotype spreadsheet (Phenotype_METABRIC_Demo1.txt) again Select Treatment : the columns of the heatmap will automatically reorganize accordingly. Then press Done. NIH Big Data Center of Excellence 16
17 STEP2: Spreadsheet Visualization Bars show the status of each sample NIH Big Data Center of Excellence 17
18 STEP2: Spreadsheet Visualization Bars show the status of each sample More details can be seen by clicking on the bars NIH Big Data Center of Excellence 18
19 STEP2: Spreadsheet Visualization Bars show the status of each sample More details can be seen by clicking on the bars Bar charts show the histogram of each category NIH Big Data Center of Excellence 19
20 STEP2: Spreadsheet Visualization Click the dropdown Filter Rows By menu and select Correlation to Group. Click the dropdown Sort Rows By menu and select Correlation to Group. NIH Big Data Center of Excellence 20
21 STEP2: Spreadsheet Visualization Hover over G1-Basal and click on it NIH Big Data Center of Excellence 21
22 STEP2: Spreadsheet Visualization Hover over G1-Basal and click on it Click on the arrows to expand the group and observe the expressions NIH Big Data Center of Excellence 22
23 STEP2: Spreadsheet Visualization Click on the clock sign to perform Kaplan Meier survival analysis using a set of categories Use this table to configure Kaplan Meier analysis by selecting the events and time to events NIH Big Data Center of Excellence 23
24 STEP2: Spreadsheet Visualization Select the options below for Kaplan Meier analysis and press Done. NIH Big Data Center of Excellence 24
25 STEP2: Spreadsheet Visualization NIH Big Data Center of Excellence 25
26 Network-guided clustering of somatic mutations in different cancer types NIH Big Data Center of Excellence 26
27 STEP3: Sample Clustering We will use KnowEnG s clustering pipeline to perform both networkguided as well as standard clustering of samples The network-guided clustering implemented in KnowEnG is inspired by the network-based stratification approach: We will use some of the samples from the TCGA pancan12 dataset NIH Big Data Center of Excellence 27
28 STEP3: Sample Clustering Outline of Network-based Stratification: NIH Big Data Center of Excellence 28
29 STEP3: Sample Clustering Dataset characteristics: Name Description Demo2_Mutation_pancan12_30 A matrix of (gene x samples) containing the somatic mutation status of ~15k protein coding genes in 360 tumor samples. Demo2_Clinical_pancan12_30 A matrix of (samples x clinical phenotypes) including primary disease, PANCAN consensus cluster, survival years, etc. NIH Big Data Center of Excellence 29
30 STEP3: Sample Clustering (network-guided) Select the pipeline: Select Analysis Pipelines at the top of the page Select Sample Clustering and Click on Start Pipeline NIH Big Data Center of Excellence 30
31 STEP3: Sample Clustering (network-guided) Upload the data: Click on Upload New Data Click BROWSE and find the files to upload: - Demo2_Clinical_pancan12_30 - Demo2_Mutation_pancan12_30 NIH Big Data Center of Excellence 31
32 STEP3: Sample Clustering (network-guided) Configure the pipeline: For the omics file select: - Demo2_Mutation_pancan12_30 Click Next at the bottom right corner For the phenotype file select: - Demo2_Clinical_pancan12_30 Click Next at the bottom right corner NIH Big Data Center of Excellence 32
33 STEP3: Sample Clustering (network-guided) Select Yes in response to using the knowledge network: This allows us to perform networkguided clustering Keep the species as Human Select HumanNet Integrated Network as the network Keep network smoothing at 50% and click Next: This controls how much importance is put on network connections instead of the somatic mutations NIH Big Data Center of Excellence 33
34 STEP3: Sample Clustering (network-guided) Choose 8 as number of clusters and click Next Select Yes in response to using bootstrap sampling: This allows us to obtain a more robust final clustering Choose 5 as number of bootstraps We will use the default 80% rate to sample the data in each bootstrap NIH Big Data Center of Excellence 34
35 STEP3: Sample Clustering (network-guided) Review the summary of the job and change the default Job Name to easily recognize later Press Submit Job NIH Big Data Center of Excellence 35
36 STEP3: Sample Clustering (standard) Select the pipeline: Select Analysis Pipelines at the top of the page Select Sample Clustering and Click on Start Pipeline NIH Big Data Center of Excellence 36
37 STEP3: Sample Clustering (standard) Configure the pipeline: For the omics file select: - Demo2_Mutation_pancan12_30 Click Next at the bottom right corner For the phenotype file select: - Demo2_Clinical_pancan12_30 Click Next at the bottom right corner NIH Big Data Center of Excellence 37
38 STEP3: Sample Clustering (standard) Select No in response to using the knowledge network: This allows us to perform standard clustering on the data Choose 8 as number of clusters We will use the default K-Means clustering algorithm Click on Next at the bottom right corner NIH Big Data Center of Excellence 38
39 STEP3: Sample Clustering (standard) Select Yes in response to using bootstrap sampling: This allows us to obtain a more robust final clustering Choose 5 as number of bootstraps We will use the default 80% rate to sample the data in each bootstrap Click on Next at the bottom right corner NIH Big Data Center of Excellence 39
40 STEP3: Sample Clustering (standard) Review the summary of the job and change the default Job Name to easily recognize later Submit the job NIH Big Data Center of Excellence 40
41 STEP3: Sample Clustering (standard vs. network) Go to the Data page: Select SC_nonet_clust8 (or any other name you chose) Select View Results at the top right corner NIH Big Data Center of Excellence 41
42 STEP3: Sample Clustering (standard vs. network) Heatmap shows samples x samples or features x samples You can select the dropdown menu to choose between the two options The color of each cell indicates how often a pair of patients fell within the same cluster across all samplings NIH Big Data Center of Excellence 42
43 STEP3: Sample Clustering (standard vs. network) High degree of clustering bias You can add a phenotype to compare with NIH Big Data Center of Excellence 43
44 STEP3: Sample Clustering (standard vs. network) Go to the Data page: Select SC_HumanNet_clust8 (or any other name you chose) Select View Results at the top right corner NIH Big Data Center of Excellence 44
45 STEP3: Sample Clustering (standard vs. network) A more balanced clustering The color of each cell indicates how often a pair of patients fell within the same cluster across all samplings NIH Big Data Center of Excellence 45
46 STEP3: Sample Clustering (standard vs. network) Go to the Data page Click on triangle by SC_HumanNet_clust8 Select sample_labels_by_cluster Click on the name at the right top corner to edit and add _HumanNet to the end Repeat the same for SC_nonet_clust8 and add _nonet to the end NIH Big Data Center of Excellence 46
47 STEP3: Sample Clustering (standard vs. network) Let s evaluate the results in SSV Select Analysis Pipelines Select Spreadsheet Visualization and Click on Start Pipeline NIH Big Data Center of Excellence 47
48 STEP3: Sample Clustering (standard vs. network) Select these four files to evaluate simultaneously and press Next: Check the summary and change the job name if you like. Press Submit Job. NIH Big Data Center of Excellence 48
49 STEP3: Sample Clustering (standard vs. network) The results: Select Go to Data Page Select the job you just ran Then View Results NIH Big Data Center of Excellence 49
50 STEP3: Sample Clustering (standard vs. network) In Group Columns By select cluster_assignment from the sample_labels_by_cluster_humannet.txt By clicking on Show Rows add _primary_disease and _PANCAN_Cluster_Cluster_PANCAN from Demo2_Clinical_pancan12_30.txt NIH Big Data Center of Excellence 50
51 STEP3: Sample Clustering (standard vs. network) You can explore top genes, draw Kaplan Meier curves, etc. NIH Big Data Center of Excellence 51
52 STEP3: Sample Clustering (standard vs. network) Click on the clock sign to perform Kaplan Meier survival analysis using any of the categories Use this table to configure Kaplan Meier analysis by selecting the events and time to events NIH Big Data Center of Excellence 52
53 STEP3: Sample Clustering (standard vs. network) Select the parameters below and press Done to see Kaplan Meier curves of clusters identified using HumanNet network NIH Big Data Center of Excellence 53
54 Network-guided gene prioritization NIH Big Data Center of Excellence 54
55 STEP4: Gene Prioritization We will use KnowEnG s gene prioritization pipeline to perform networkguided gene prioritization The network-guided gene prioritization implemented in KnowEnG is a method called ProGENI: We will use samples from the CCLE dataset NIH Big Data Center of Excellence 55
56 STEP4: Gene Prioritization Outline of ProGENI: a) Genes Drug response (e.g. IC50) Cell lines Gene expressions Normalize w.r.t. global network distribu8on Perform Network transforma8on of gene expressions Iden8fy response correlated genes (RCG) and use them as the restart set for a RWR Obtain equilibrium probability distribu8on for the nodes Rank genes according to normalized probability scores Network NIH Big Data Center of Excellence 56
57 STEP4: Gene Prioritization Dataset characteristics: Name demo_fp.genomic Description A matrix of (gene x samples) containing the expression of ~17k genes in ~500 cell lines. The expression profiles are normalized in advance. demo_fp.phenotypic A matrix of (samples x drugs) containing IC50 values for 24 cytotoxic treatments. NIH Big Data Center of Excellence 57
58 STEP4: Gene Prioritization (network-guided) Select the pipeline: Select Analysis Pipelines at the top of the page Select Feature Prioritization and Click on Start Pipeline NIH Big Data Center of Excellence 58
59 STEP4: Gene Prioritization (network-guided) Configure the pipeline: For the omics file select Use Demo Data Click Next at the bottom right corner For the response file select Use Demo Data Click Next at the bottom right corner NIH Big Data Center of Excellence 59
60 STEP4: Gene Prioritization (network-guided) Select Yes in response to using the knowledge network: This allows us to perform networkguided prioritization (ProGENI) Keep the species as Human Select STRING Experimental PPI as the network Keep network smoothing at 50%: This controls how much importance is put on network connections instead of the somatic mutations NIH Big Data Center of Excellence 60
61 STEP4: Gene Prioritization (network-guided) Keep the default parameters on this page Used for continuousvalued response Size of RCG set Choose No for bootstrapping NIH Big Data Center of Excellence 61
62 STEP4: Gene Prioritization (network-guided) Review the summary of the job and change its name if you like Submit the job NIH Big Data Center of Excellence 62
63 STEP4: Gene Prioritization (network-guided) Go to the Data page Select View Results when the job is done Heatmap shows the top genes identified for each drug NIH Big Data Center of Excellence 63
64 STEP4: Gene Prioritization (network-guided) You can right-click on a drug to sort rows it and see its top genes You can also sort columns by a gene to see drugs for which the gene was among the top list NIH Big Data Center of Excellence 64
65 STEP4: Gene Prioritization (network-guided) Let s see the enrichment of the top genes in different GO terms Go to Analysis Pipelines page Select Gene Set Characterization pipeline NIH Big Data Center of Excellence 65
66 STEP4: Gene Prioritization (network-guided) Select the green triangle by the gene prioritization job you ran Select top_features_per_phenotype_matrix Press Next NIH Big Data Center of Excellence 66
67 STEP4: Gene Prioritization (network-guided) For gene sets, select your gene sets of interest (e.g. GO) and press Next Say No to using the knowledge network and press Next. Then press Submit Job. NIH Big Data Center of Excellence 67
68 STEP4: Gene Prioritization (network-guided) The results: Select Go to Data Page Select the job you just ran Then View Results NIH Big Data Center of Excellence 68
69 STEP4: Gene Prioritization (network-guided) This page shows the enriched gene sets for each drug You can change the filter (scores represent log10 (p-value) of enrichment) to see fewer or more enriched gene sets NIH Big Data Center of Excellence 69
70 Resources Tutorials: Quickstarts: YouTube: Resources: Data Preparation Guide: master/pipeline_readmes/readme-dataprep.md Knowledge Network Contents: Future Pipelines: Source Code: Docker Images: Github Repos: Other Cloud Platforms Contact Us with Questions and Feedback: NIH Big Data Center of Excellence 70
Drug/Cell-line Browser (DBC) v1.0 - User Manual
 Drug/Cell-line Browser (DBC) v1.0 - User Manual Written by Qiaonan Duan and Avi Ma ayan Last Updated: 6/26/2014 Table of Contents I. Abstract II. Installation and Requirements III. Instructions 1. Cell-line
Drug/Cell-line Browser (DBC) v1.0 - User Manual Written by Qiaonan Duan and Avi Ma ayan Last Updated: 6/26/2014 Table of Contents I. Abstract II. Installation and Requirements III. Instructions 1. Cell-line
Differential Expression Analysis at PATRIC
 Differential Expression Analysis at PATRIC The following step- by- step workflow is intended to help users learn how to upload their differential gene expression data to their private workspace using Expression
Differential Expression Analysis at PATRIC The following step- by- step workflow is intended to help users learn how to upload their differential gene expression data to their private workspace using Expression
PROMO 2017a - Tutorial
 PROMO 2017a - Tutorial Introduction... 2 Installing PROMO... 2 Step 1 - Importing data... 2 Step 2 - Preprocessing... 6 Step 3 Data Exploration... 9 Step 4 Clustering... 13 Step 5 Analysis of sample clusters...
PROMO 2017a - Tutorial Introduction... 2 Installing PROMO... 2 Step 1 - Importing data... 2 Step 2 - Preprocessing... 6 Step 3 Data Exploration... 9 Step 4 Clustering... 13 Step 5 Analysis of sample clusters...
ChIP-Seq Tutorial on Galaxy
 1 Introduction ChIP-Seq Tutorial on Galaxy 2 December 2010 (modified April 6, 2017) Rory Stark The aim of this practical is to give you some experience handling ChIP-Seq data. We will be working with data
1 Introduction ChIP-Seq Tutorial on Galaxy 2 December 2010 (modified April 6, 2017) Rory Stark The aim of this practical is to give you some experience handling ChIP-Seq data. We will be working with data
User Guide. v Released June Advaita Corporation 2016
 User Guide v. 0.9 Released June 2016 Copyright Advaita Corporation 2016 Page 2 Table of Contents Table of Contents... 2 Background and Introduction... 4 Variant Calling Pipeline... 4 Annotation Information
User Guide v. 0.9 Released June 2016 Copyright Advaita Corporation 2016 Page 2 Table of Contents Table of Contents... 2 Background and Introduction... 4 Variant Calling Pipeline... 4 Annotation Information
Tutorial. Introduction. Starting GenePattern. Prerequisites. Scientific Scenario
 Tutorial The GenePattern Tutorial introduces you to GenePattern by providing step-by-step instructions for analyzing gene expression. It takes approximately 40 minutes to complete. All of the information
Tutorial The GenePattern Tutorial introduces you to GenePattern by providing step-by-step instructions for analyzing gene expression. It takes approximately 40 minutes to complete. All of the information
User Manual. Ver. 3.0 March 19, 2012
 User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
mirnet Tutorial Starting with expression data
 mirnet Tutorial Starting with expression data Computer and Browser Requirements A modern web browser with Java Script enabled Chrome, Safari, Firefox, and Internet Explorer 9+ For best performance and
mirnet Tutorial Starting with expression data Computer and Browser Requirements A modern web browser with Java Script enabled Chrome, Safari, Firefox, and Internet Explorer 9+ For best performance and
SEEK User Manual. Introduction
 SEEK User Manual Introduction SEEK is a computational gene co-expression search engine. It utilizes a vast human gene expression compendium to deliver fast, integrative, cross-platform co-expression analyses.
SEEK User Manual Introduction SEEK is a computational gene co-expression search engine. It utilizes a vast human gene expression compendium to deliver fast, integrative, cross-platform co-expression analyses.
ROTS: Reproducibility Optimized Test Statistic
 ROTS: Reproducibility Optimized Test Statistic Fatemeh Seyednasrollah, Tomi Suomi, Laura L. Elo fatsey (at) utu.fi March 3, 2016 Contents 1 Introduction 2 2 Algorithm overview 3 3 Input data 3 4 Preprocessing
ROTS: Reproducibility Optimized Test Statistic Fatemeh Seyednasrollah, Tomi Suomi, Laura L. Elo fatsey (at) utu.fi March 3, 2016 Contents 1 Introduction 2 2 Algorithm overview 3 3 Input data 3 4 Preprocessing
Gene Survey: FAQ. Gene Survey: FAQ Tod Casasent DRAFT
 Gene Survey: FAQ Tod Casasent 2016-02-22-1245 DRAFT 1 What is this document? This document is intended for use by internal and external users of the Gene Survey package, results, and output. This document
Gene Survey: FAQ Tod Casasent 2016-02-22-1245 DRAFT 1 What is this document? This document is intended for use by internal and external users of the Gene Survey package, results, and output. This document
SVM Classification in -Arrays
 SVM Classification in -Arrays SVM classification and validation of cancer tissue samples using microarray expression data Furey et al, 2000 Special Topics in Bioinformatics, SS10 A. Regl, 7055213 What
SVM Classification in -Arrays SVM classification and validation of cancer tissue samples using microarray expression data Furey et al, 2000 Special Topics in Bioinformatics, SS10 A. Regl, 7055213 What
How to store and visualize RNA-seq data
 How to store and visualize RNA-seq data Gabriella Rustici Functional Genomics Group gabry@ebi.ac.uk EBI is an Outstation of the European Molecular Biology Laboratory. Talk summary How do we archive RNA-seq
How to store and visualize RNA-seq data Gabriella Rustici Functional Genomics Group gabry@ebi.ac.uk EBI is an Outstation of the European Molecular Biology Laboratory. Talk summary How do we archive RNA-seq
Predicting Disease-related Genes using Integrated Biomedical Networks
 Predicting Disease-related Genes using Integrated Biomedical Networks Jiajie Peng (jiajiepeng@nwpu.edu.cn) HanshengXue(xhs1892@gmail.com) Jin Chen* (chen.jin@uky.edu) Yadong Wang* (ydwang@hit.edu.cn) 1
Predicting Disease-related Genes using Integrated Biomedical Networks Jiajie Peng (jiajiepeng@nwpu.edu.cn) HanshengXue(xhs1892@gmail.com) Jin Chen* (chen.jin@uky.edu) Yadong Wang* (ydwang@hit.edu.cn) 1
2. This is a cell; this cell is designated as A1.
 Queen s Learning Commons: Microsoft Excel Basics 1. These are the columns. 2. This is a cell; this cell is designated as A1. 3. Let s make a table. Click on the box you want to put text in and simply begin
Queen s Learning Commons: Microsoft Excel Basics 1. These are the columns. 2. This is a cell; this cell is designated as A1. 3. Let s make a table. Click on the box you want to put text in and simply begin
cbioportal https://www.ncbi.nlm.nih.gov/pubmed/ /5/401
 cbioportal http://www.cbioportal.org/ https://www.ncbi.nlm.nih.gov/pubmed/23550210 http://cancerdiscovery.aacrjournals.org/content/ 2/5/401 Tutorials http://www.cbioportal.org/tutorial.jsp http://www.cbioportal.org/faq.jsp
cbioportal http://www.cbioportal.org/ https://www.ncbi.nlm.nih.gov/pubmed/23550210 http://cancerdiscovery.aacrjournals.org/content/ 2/5/401 Tutorials http://www.cbioportal.org/tutorial.jsp http://www.cbioportal.org/faq.jsp
Tutorial. RNA-Seq Analysis of Breast Cancer Data. Sample to Insight. November 21, 2017
 RNA-Seq Analysis of Breast Cancer Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
RNA-Seq Analysis of Breast Cancer Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
SALES REPORTING GUIDE TO MANUALLY ENTER SALES DATA IN TRADEMARX:
 SALES REPORTING GUIDE TO MANUALLY ENTER SALES DATA IN TRADEMARX: 1. Go to http://licensing.trademarxonline.com 2. Log in using your email address and password. If you don t know your password, please click
SALES REPORTING GUIDE TO MANUALLY ENTER SALES DATA IN TRADEMARX: 1. Go to http://licensing.trademarxonline.com 2. Log in using your email address and password. If you don t know your password, please click
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 15: Microarray clustering http://compbio.pbworks.com/f/wood2.gif Some slides were adapted from Dr. Shaojie Zhang (University of Central Florida) Microarray
EECS730: Introduction to Bioinformatics Lecture 15: Microarray clustering http://compbio.pbworks.com/f/wood2.gif Some slides were adapted from Dr. Shaojie Zhang (University of Central Florida) Microarray
This tutorial will guide a curator/user to create the files to upload phenotype experiment annotation and data onto T3.
 This tutorial will guide a curator/user to create the files to upload phenotype experiment annotation and data onto T3. Updated December 2012. Any feedback is welcome! Sections 1. Download the Template
This tutorial will guide a curator/user to create the files to upload phenotype experiment annotation and data onto T3. Updated December 2012. Any feedback is welcome! Sections 1. Download the Template
Analyzing Genomic Data with NOJAH
 Analyzing Genomic Data with NOJAH TAB A) GENOME WIDE ANALYSIS Step 1: Select the example dataset or upload your own. Two example datasets are available. Genome-Wide TCGA-BRCA Expression datasets and CoMMpass
Analyzing Genomic Data with NOJAH TAB A) GENOME WIDE ANALYSIS Step 1: Select the example dataset or upload your own. Two example datasets are available. Genome-Wide TCGA-BRCA Expression datasets and CoMMpass
ACCESSING TIDE. New Hampshire Statewide Assessment System RESETTING YOUR TIDE ACCOUNT FROM A PREVIOUS SCHOOL YEAR
 ACCESSING TIDE New Hampshire Statewide School administrators use the Test Information Distribution Engine (TIDE) to add and manage user accounts and to manage information about students who participate
ACCESSING TIDE New Hampshire Statewide School administrators use the Test Information Distribution Engine (TIDE) to add and manage user accounts and to manage information about students who participate
GARAGE SALE PERMIT APPLICATION ON LINE
 GARAGE SALE PERMIT APPLICATION ON LINE The Village of Pinecrest makes it easy for you to apply for certain types of permits online using etrakit.net. This also includes garage sale permits. To access etrakit.net,
GARAGE SALE PERMIT APPLICATION ON LINE The Village of Pinecrest makes it easy for you to apply for certain types of permits online using etrakit.net. This also includes garage sale permits. To access etrakit.net,
HMIS Guide to Running & Retrieving the CSV APR 2017 Export in ClientTrack An HMIS End User Training Resource
 2018 HMIS Guide to Running & Retrieving the CSV APR 2017 Export in ClientTrack An HMIS End User Training Resource This guide will demonstrate how to run the CSV APR 2017 Export and prepare files for submission
2018 HMIS Guide to Running & Retrieving the CSV APR 2017 Export in ClientTrack An HMIS End User Training Resource This guide will demonstrate how to run the CSV APR 2017 Export and prepare files for submission
1. Introduction. 2. System requirements
 Table of Contents 1. Introduction...2 2. System requirements...2 3. Installation...3 4. Running on exemplary datasets...3 5. Preparing your own data to run Oncodrive-fm...4 1. Introduction Oncodrive-fm
Table of Contents 1. Introduction...2 2. System requirements...2 3. Installation...3 4. Running on exemplary datasets...3 5. Preparing your own data to run Oncodrive-fm...4 1. Introduction Oncodrive-fm
EGAN Tutorial: A Basic Use-case
 EGAN Tutorial: A Basic Use-case July 2010 Jesse Paquette Biostatistics and Computational Biology Core Helen Diller Family Comprehensive Cancer Center University of California, San Francisco (AKA BCBC HDFCCC
EGAN Tutorial: A Basic Use-case July 2010 Jesse Paquette Biostatistics and Computational Biology Core Helen Diller Family Comprehensive Cancer Center University of California, San Francisco (AKA BCBC HDFCCC
Core Technology Development Team Meeting
 Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Employee User s Guide
 User Guide 1 12612 Challenger Parkway Suite 300 Orlando, FL 32826 www.ivisitor.com Employee User s Guide INTRODUCTION The instructions and information contained in this document outline the steps necessary
User Guide 1 12612 Challenger Parkway Suite 300 Orlando, FL 32826 www.ivisitor.com Employee User s Guide INTRODUCTION The instructions and information contained in this document outline the steps necessary
8:15 Introduction/Overview Michelle Giglio. 8:45 CloVR background W. Florian Fricke. 9:15 Hands-on: Start CloVR W. Florian Fricke
 Hands-On Exercises 2016 1 Agenda 8:15 Introduction/Overview Michelle Giglio 8:45 CloVR background W. Florian Fricke 9:15 Hands-on: Start CloVR W. Florian Fricke 9:45 Break 9:55 Hands-on: Start CloVR-Microbe
Hands-On Exercises 2016 1 Agenda 8:15 Introduction/Overview Michelle Giglio 8:45 CloVR background W. Florian Fricke 9:15 Hands-on: Start CloVR W. Florian Fricke 9:45 Break 9:55 Hands-on: Start CloVR-Microbe
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Schematic representation of the Workflow window in Perseus All data matrices uploaded in the running session of Perseus and all processing steps are displayed in the order of execution.
Supplementary Figure 1 Schematic representation of the Workflow window in Perseus All data matrices uploaded in the running session of Perseus and all processing steps are displayed in the order of execution.
File Storage. This manual contains pertinent information about your File Storage space at SLC.
 File Storage This manual contains pertinent information about your File Storage space at SLC. June, 2017 About All students at SLC are provided with an allotment of secure storage space to save and store
File Storage This manual contains pertinent information about your File Storage space at SLC. June, 2017 About All students at SLC are provided with an allotment of secure storage space to save and store
Tutorial 3 - Performing a Change-Point Analysis in Excel
 Tutorial 3 - Performing a Change-Point Analysis in Excel Introduction This tutorial teaches you how to perform a change-point analysis while using Microsoft Excel. The Change-Point Analyzer Add-In allows
Tutorial 3 - Performing a Change-Point Analysis in Excel Introduction This tutorial teaches you how to perform a change-point analysis while using Microsoft Excel. The Change-Point Analyzer Add-In allows
TUTORIAL for using the web interface
 Progression Analysis of Disease PAD web tool for the paper *: Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival M.Nicolau, A.Levine,
Progression Analysis of Disease PAD web tool for the paper *: Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival M.Nicolau, A.Levine,
SEARCH COMMITTEE/SEARCH CHAIR GUIDE
 SEARCH COMMITTEE/SEARCH CHAIR GUIDE Role of Search Committee Member in ATS: View the Posting Summary this is the vacancy announcement. View Applications and Move Applicants in the Workflow to show status
SEARCH COMMITTEE/SEARCH CHAIR GUIDE Role of Search Committee Member in ATS: View the Posting Summary this is the vacancy announcement. View Applications and Move Applicants in the Workflow to show status
CONTRIBUTION GATEWAY GUIDE
 CONTRIBUTION GATEWAY GUIDE USER-FRIENDLY INSTRUCTIONS TO SUBMIT PLAN CONTRIBUTIONS ALERUS RETIREMENT AND BENEFITS TABLE OF CONTENTS GETTING STARTED... PAGE 1 CONTRIBUTION GATEWAY UPLOAD INSTRUCTIONS...
CONTRIBUTION GATEWAY GUIDE USER-FRIENDLY INSTRUCTIONS TO SUBMIT PLAN CONTRIBUTIONS ALERUS RETIREMENT AND BENEFITS TABLE OF CONTENTS GETTING STARTED... PAGE 1 CONTRIBUTION GATEWAY UPLOAD INSTRUCTIONS...
CLC Server. End User USER MANUAL
 CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
SUPER USER GUIDE. For Practice Performance Registry. Rachel Bryan
 SUPER USER GUIDE For Practice Performance Registry Rachel Bryan Rachel.bryan@dartnet.info Super User Guide Welcome, Super User. You are a Super User because you have a special user account used for system
SUPER USER GUIDE For Practice Performance Registry Rachel Bryan Rachel.bryan@dartnet.info Super User Guide Welcome, Super User. You are a Super User because you have a special user account used for system
PlicElements Quick Start Guide
 PlicElements is a high-speed-low-drag web application used by Professional Photographers and/or Studios to upload and prepare class/school composites to participating labs for processing. Simply upload
PlicElements is a high-speed-low-drag web application used by Professional Photographers and/or Studios to upload and prepare class/school composites to participating labs for processing. Simply upload
Introduction to Galaxy
 Introduction to Galaxy Dr Jason Wong Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW Day 1 Thurs 28 th January 2016 Overview What is Galaxy? Description of
Introduction to Galaxy Dr Jason Wong Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW Day 1 Thurs 28 th January 2016 Overview What is Galaxy? Description of
Version 9 Client Workflow Interface (Dashboard) Quick Start Guide
 Version 9 Client Workflow Interface (Dashboard) Quick Start Guide Client Workflow Interface (Dashboard) Quick Start Guide The Quick Start Guide will walk you through the initial steps of using the Progeny
Version 9 Client Workflow Interface (Dashboard) Quick Start Guide Client Workflow Interface (Dashboard) Quick Start Guide The Quick Start Guide will walk you through the initial steps of using the Progeny
What is clustering. Organizing data into clusters such that there is high intra- cluster similarity low inter- cluster similarity
 Clustering What is clustering Organizing data into clusters such that there is high intra- cluster similarity low inter- cluster similarity Informally, finding natural groupings among objects. High dimensional
Clustering What is clustering Organizing data into clusters such that there is high intra- cluster similarity low inter- cluster similarity Informally, finding natural groupings among objects. High dimensional
Package DriverNet. R topics documented: October 4, Type Package
 Package DriverNet October 4, 2013 Type Package Title Drivernet: uncovering somatic driver mutations modulating transcriptional networks in cancer Version 1.0.0 Date 2012-03-21 Author Maintainer Jiarui
Package DriverNet October 4, 2013 Type Package Title Drivernet: uncovering somatic driver mutations modulating transcriptional networks in cancer Version 1.0.0 Date 2012-03-21 Author Maintainer Jiarui
How to register and enter scores for a NASP virtual tournament. Basic Steps:
 How to register and enter scores for a NASP virtual tournament. Basic Steps: 1. Register the school and the tournament roster. 2. Add group information, coaches, and other registration info. 3. Come back
How to register and enter scores for a NASP virtual tournament. Basic Steps: 1. Register the school and the tournament roster. 2. Add group information, coaches, and other registration info. 3. Come back
FunRich Tool Documentation
 FunRich Tool Documentation Version 2.1.2 Shivakumar Keerthikumar, Mohashin Pathan, Johnson Agbinya and Suresh Mathivanan Mathivanan Lab http://www.mathivananlab.org La Trobe University LIMS1 Department
FunRich Tool Documentation Version 2.1.2 Shivakumar Keerthikumar, Mohashin Pathan, Johnson Agbinya and Suresh Mathivanan Mathivanan Lab http://www.mathivananlab.org La Trobe University LIMS1 Department
Contents. Introduction 2. PerturbationClustering 2. SubtypingOmicsData 5. References 8
 PINSPlus: Clustering Algorithm for Data Integration and Disease Subtyping Hung Nguyen, Sangam Shrestha, and Tin Nguyen Department of Computer Science and Engineering University of Nevada, Reno, NV 89557
PINSPlus: Clustering Algorithm for Data Integration and Disease Subtyping Hung Nguyen, Sangam Shrestha, and Tin Nguyen Department of Computer Science and Engineering University of Nevada, Reno, NV 89557
Breeding Guide. Customer Services PHENOME-NETWORKS 4Ben Gurion Street, 74032, Nes-Ziona, Israel
 Breeding Guide Customer Services PHENOME-NETWORKS 4Ben Gurion Street, 74032, Nes-Ziona, Israel www.phenome-netwoks.com Contents PHENOME ONE - INTRODUCTION... 3 THE PHENOME ONE LAYOUT... 4 THE JOBS ICON...
Breeding Guide Customer Services PHENOME-NETWORKS 4Ben Gurion Street, 74032, Nes-Ziona, Israel www.phenome-netwoks.com Contents PHENOME ONE - INTRODUCTION... 3 THE PHENOME ONE LAYOUT... 4 THE JOBS ICON...
How to Create Student Accounts and Assignments
 How to Create Student Accounts and Assignments From the top navigation, select My Classes and click My Students Carolina Science Online will allow you to either create a single student account, one at
How to Create Student Accounts and Assignments From the top navigation, select My Classes and click My Students Carolina Science Online will allow you to either create a single student account, one at
Download Instructions
 Download Instructions The download page provides several options for importing data back into your applications. The Excel template will automatically format the data within the commadelimited file. The
Download Instructions The download page provides several options for importing data back into your applications. The Excel template will automatically format the data within the commadelimited file. The
Introduction to Cancer Genomics
 Introduction to Cancer Genomics Gene expression data analysis part I David Gfeller Computational Cancer Biology Ludwig Center for Cancer research david.gfeller@unil.ch 1 Overview 1. Basic understanding
Introduction to Cancer Genomics Gene expression data analysis part I David Gfeller Computational Cancer Biology Ludwig Center for Cancer research david.gfeller@unil.ch 1 Overview 1. Basic understanding
DRAGEN Bio-IT Platform Enabling the Global Genomic Infrastructure
 TM DRAGEN Bio-IT Platform Enabling the Global Genomic Infrastructure About DRAGEN Edico Genome s DRAGEN TM (Dynamic Read Analysis for GENomics) Bio-IT Platform provides ultra-rapid secondary analysis of
TM DRAGEN Bio-IT Platform Enabling the Global Genomic Infrastructure About DRAGEN Edico Genome s DRAGEN TM (Dynamic Read Analysis for GENomics) Bio-IT Platform provides ultra-rapid secondary analysis of
An Introduction to BigQuery
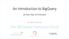 An Introduction to BigQuery (in less than 10 minutes) brought to you by The ISB Cancer Genomics Cloud This is what you should see the first time you go to the BigQuery Web UI at bigquery.cloud.google.com
An Introduction to BigQuery (in less than 10 minutes) brought to you by The ISB Cancer Genomics Cloud This is what you should see the first time you go to the BigQuery Web UI at bigquery.cloud.google.com
TexRAD Research Version Client User Guide Version 3.9
 Imaging tools for medical decision makers Cambridge Computed Imaging Ltd Grange Park Broadway Bourn Cambridge CB23 2TA UK TexRAD Research Version Client User Guide Version 3.9 Release date 23/05/2016 Number
Imaging tools for medical decision makers Cambridge Computed Imaging Ltd Grange Park Broadway Bourn Cambridge CB23 2TA UK TexRAD Research Version Client User Guide Version 3.9 Release date 23/05/2016 Number
Introduction to SPSS Faiez Mossa 2 nd Class
 Introduction to SPSS 16.0 Faiez Mossa 2 nd Class 1 Outline Review of Concepts (stats and scales) Data entry (the workspace and labels) By hand Import Excel Running an analysis- frequency, central tendency,
Introduction to SPSS 16.0 Faiez Mossa 2 nd Class 1 Outline Review of Concepts (stats and scales) Data entry (the workspace and labels) By hand Import Excel Running an analysis- frequency, central tendency,
DOWNLOADING YOUR BENEFICIARY SAMPLE Last Updated: 11/16/18. CMS Web Interface Excel Instructions
 DOWNLOADING YOUR BENEFICIARY SAMPLE Last Updated: 11/16/18 CMS Web Interface Excel Instructions Last updated: 11/16/2018 1 Smarter reporting. Smarter care. CMS Web Interface file upload. Using the Excel
DOWNLOADING YOUR BENEFICIARY SAMPLE Last Updated: 11/16/18 CMS Web Interface Excel Instructions Last updated: 11/16/2018 1 Smarter reporting. Smarter care. CMS Web Interface file upload. Using the Excel
How to Complete the CITI Online Tutorial
 How to Complete the CITI Online Tutorial Guidance for New Users Please note: Our previous online tutorials were NOT part of the CITI program. If you have never registered on the CITI website before, you
How to Complete the CITI Online Tutorial Guidance for New Users Please note: Our previous online tutorials were NOT part of the CITI program. If you have never registered on the CITI website before, you
How to Filter and Sort Excel Spreadsheets (Patient-Level Detail Report)
 How to Filter and Sort Excel Spreadsheets (Patient-Level Detail Report) When you use the filter and sort option on an excel spreadsheet, it allows you to narrow down a large spreadsheet to show just the
How to Filter and Sort Excel Spreadsheets (Patient-Level Detail Report) When you use the filter and sort option on an excel spreadsheet, it allows you to narrow down a large spreadsheet to show just the
A web app for guided and interactive generation of multimarker panels (www.combiroc.eu)
 COMBIROC S TUTORIAL. A web app for guided and interactive generation of multimarker panels (www.combiroc.eu) Overview of the CombiROC workflow. CombiROC delivers a simple workflow to help researchers in
COMBIROC S TUTORIAL. A web app for guided and interactive generation of multimarker panels (www.combiroc.eu) Overview of the CombiROC workflow. CombiROC delivers a simple workflow to help researchers in
Lab 19: Excel Formatting, Using Conditional Formatting and Sorting Records
 Lab 19: Excel Formatting, Using Conditional Formatting and Sorting Records () CONTENTS 1 Lab Topic... 2 1.1 In-Lab... 2 1.1.1 In-Lab Materials... 2 1.1.2 In-Lab Instructions... 2 1.2 Out-Lab... 9 1.2.1
Lab 19: Excel Formatting, Using Conditional Formatting and Sorting Records () CONTENTS 1 Lab Topic... 2 1.1 In-Lab... 2 1.1.1 In-Lab Materials... 2 1.1.2 In-Lab Instructions... 2 1.2 Out-Lab... 9 1.2.1
IHS Connect CONSTRUCTION 2015 IHS. ALL RIGHTS RESERVED.
 IHS Connect CONSTRUCTION IHS Connect is an online market and business intelligence platform enabling faster, smarter, and more efficient access to world renowned information and insight from IHS. A single
IHS Connect CONSTRUCTION IHS Connect is an online market and business intelligence platform enabling faster, smarter, and more efficient access to world renowned information and insight from IHS. A single
Open World 2008 Hands-On Lab. JD Edwards EnterpriseOne Embedded BI Publisher. OOW 2008 Hands On Labs JD Edwards EnterpriseOne
 Open World 2008 Hands-On Lab JD Edwards EnterpriseOne Embedded BI Publisher OOW 2008 Hands On Labs JD Edwards EnterpriseOne Table of Contents 1 TUTORIAL OVERVIEW...3 2 GENERATE XML...4 2.1 Sign-on...4
Open World 2008 Hands-On Lab JD Edwards EnterpriseOne Embedded BI Publisher OOW 2008 Hands On Labs JD Edwards EnterpriseOne Table of Contents 1 TUTORIAL OVERVIEW...3 2 GENERATE XML...4 2.1 Sign-on...4
DOLLAR GENERAL CAREER SITE CANDIDATE ONLINE APPLICATION REFERENCE GUIDE
 DOLLAR GENERAL CAREER SITE CANDIDATE ONLINE APPLICATION REFERENCE GUIDE In June 2016, Dollar General launched a new online application system. This Reference Guide is for the new system and includes the
DOLLAR GENERAL CAREER SITE CANDIDATE ONLINE APPLICATION REFERENCE GUIDE In June 2016, Dollar General launched a new online application system. This Reference Guide is for the new system and includes the
Network analysis. Martina Kutmon Department of Bioinformatics Maastricht University
 Network analysis Martina Kutmon Department of Bioinformatics Maastricht University What's gonna happen today? Network Analysis Introduction Quiz Hands-on session ConsensusPathDB interaction database Outline
Network analysis Martina Kutmon Department of Bioinformatics Maastricht University What's gonna happen today? Network Analysis Introduction Quiz Hands-on session ConsensusPathDB interaction database Outline
DAVID hands-on. by Ester Feldmesser, June 2017
 DAVID hands-on by Ester Feldmesser, June 2017 1. Go to the DAVID website (http://david.abcc.ncifcrf.gov/) 2. Press on Start Analysis: 3. Choose the Upload tab in the left panel: 4. Download the k-means5_arabidopsis.txt
DAVID hands-on by Ester Feldmesser, June 2017 1. Go to the DAVID website (http://david.abcc.ncifcrf.gov/) 2. Press on Start Analysis: 3. Choose the Upload tab in the left panel: 4. Download the k-means5_arabidopsis.txt
miscript mirna PCR Array Data Analysis v1.1 revision date November 2014
 miscript mirna PCR Array Data Analysis v1.1 revision date November 2014 Overview The miscript mirna PCR Array Data Analysis Quick Reference Card contains instructions for analyzing the data returned from
miscript mirna PCR Array Data Analysis v1.1 revision date November 2014 Overview The miscript mirna PCR Array Data Analysis Quick Reference Card contains instructions for analyzing the data returned from
Importing from Blackboard Learn Grade Center Data to Banner 9 User Learning Scenarios
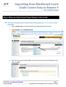 Importing from Blackboard Learn Grade Center Data to Banner 9 User Learning Scenarios Step 1: Make sure Final Grade Column Displays Letter Grade Ensure your final grade column in Grade Center has letter
Importing from Blackboard Learn Grade Center Data to Banner 9 User Learning Scenarios Step 1: Make sure Final Grade Column Displays Letter Grade Ensure your final grade column in Grade Center has letter
Web Resources. iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases
 Web Resources iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases Ethan W. Hollingsworth 1, 2, Jacob E. Vaughn 1, 2, Josh C. Orack 1,2, Chelsea Skinner
Web Resources iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases Ethan W. Hollingsworth 1, 2, Jacob E. Vaughn 1, 2, Josh C. Orack 1,2, Chelsea Skinner
OVERVIEW. User Dashboard
 OVERVIEW User Dashboard File & Serve Illinois USER DASHBOARD TABLE OF CONTENTS File & Serve Illinois Resources 3 File & Serve Illinois User Dashboard Overview 4 Submit a New Filing Tab 5 Completed Filings
OVERVIEW User Dashboard File & Serve Illinois USER DASHBOARD TABLE OF CONTENTS File & Serve Illinois Resources 3 File & Serve Illinois User Dashboard Overview 4 Submit a New Filing Tab 5 Completed Filings
Colorado State University Bioinformatics Algorithms Assignment 6: Analysis of High- Throughput Biological Data Hamidreza Chitsaz, Ali Sharifi- Zarchi
 Colorado State University Bioinformatics Algorithms Assignment 6: Analysis of High- Throughput Biological Data Hamidreza Chitsaz, Ali Sharifi- Zarchi Although a little- bit long, this is an easy exercise
Colorado State University Bioinformatics Algorithms Assignment 6: Analysis of High- Throughput Biological Data Hamidreza Chitsaz, Ali Sharifi- Zarchi Although a little- bit long, this is an easy exercise
Creating Classes and Issuing Licenses TUTORIAL
 Creating Classes and Issuing Licenses TUTORIAL 1 Contents CREATING STUDENT ACCOUNTS... 2 CREATING CLASSES... 4 IMPORTING A CLASS ROSTER... 8 ISSUING LICENSES... 14 CREATING STUDENT ACCOUNTS From your LabSim
Creating Classes and Issuing Licenses TUTORIAL 1 Contents CREATING STUDENT ACCOUNTS... 2 CREATING CLASSES... 4 IMPORTING A CLASS ROSTER... 8 ISSUING LICENSES... 14 CREATING STUDENT ACCOUNTS From your LabSim
Physics 211 E&M and Modern Physics Spring Lab #1 (to be done at home) Plotting with Excel. Good laboratory practices
 NORTHERN ILLINOIS UNIVERSITY PHYSICS DEPARTMENT Physics 211 E&M and Modern Physics Spring 2018 Lab #1 (to be done at home) Lab Writeup Due: Mon/Wed/Thu/Fri, Jan. 22/24/25/26, 2018 Read Serway & Vuille:
NORTHERN ILLINOIS UNIVERSITY PHYSICS DEPARTMENT Physics 211 E&M and Modern Physics Spring 2018 Lab #1 (to be done at home) Lab Writeup Due: Mon/Wed/Thu/Fri, Jan. 22/24/25/26, 2018 Read Serway & Vuille:
GlobalVetLINK s HealthLINK
 GlobalVetLINK s HealthLINK 1. To begin writing a health certificate (CVI), click on HealthLINK. This will take you directly to the create page (shown below). The first step is to select your Consignor.
GlobalVetLINK s HealthLINK 1. To begin writing a health certificate (CVI), click on HealthLINK. This will take you directly to the create page (shown below). The first step is to select your Consignor.
CITI ACCESS AND DIRECTIONS FOR EXTRAMURAL PERFORMERS NEW USERS
 CITI ACCESS AND DIRECTIONS FOR EXTRAMURAL PERFORMERS NEW USERS To access the CITI training program go to: http://www.citiprogram.org. The following directions will assist you as you work through the required
CITI ACCESS AND DIRECTIONS FOR EXTRAMURAL PERFORMERS NEW USERS To access the CITI training program go to: http://www.citiprogram.org. The following directions will assist you as you work through the required
The Allen Human Brain Atlas offers three types of searches to allow a user to: (1) obtain gene expression data for specific genes (or probes) of
 Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Tutorial:OverRepresentation - OpenTutorials
 Tutorial:OverRepresentation From OpenTutorials Slideshow OverRepresentation (about 12 minutes) (http://opentutorials.rbvi.ucsf.edu/index.php?title=tutorial:overrepresentation& ce_slide=true&ce_style=cytoscape)
Tutorial:OverRepresentation From OpenTutorials Slideshow OverRepresentation (about 12 minutes) (http://opentutorials.rbvi.ucsf.edu/index.php?title=tutorial:overrepresentation& ce_slide=true&ce_style=cytoscape)
Automating the Collection and Processing of Cancer Mutation Data
 Automating the Collection and Processing of Cancer Mutation Data Master s Project Report Jeremy Watson Advisor: Ben Raphael 1. Introduction 1 The Raphael lab has developed multiple software packages, such
Automating the Collection and Processing of Cancer Mutation Data Master s Project Report Jeremy Watson Advisor: Ben Raphael 1. Introduction 1 The Raphael lab has developed multiple software packages, such
GenePilot : The Next Step in MicroArray Analysis. Why Choose GenePilot? Addressing Your Specific Needs!
 GenePilot : The Next Step in MicroArray Analysis GenePilot is the new tool in the field of MicroArray Analysis, offering an integrated and sophisticated analysis suite which is more intuitive to use, uses
GenePilot : The Next Step in MicroArray Analysis GenePilot is the new tool in the field of MicroArray Analysis, offering an integrated and sophisticated analysis suite which is more intuitive to use, uses
CMN310A Advanced Office: Word and Excel or concurrent enrollment
 CLASS NUMBER AND NAME: TOTAL CLOCK HOURS/UNITS: PREREQUISITE: TEXT AND MATERIALS: CMN310C POST-ADVANCED OFFICE: Word And Excel 24 HOURS/1 UNIT CMN310A Advanced Office: Word and Excel or concurrent enrollment
CLASS NUMBER AND NAME: TOTAL CLOCK HOURS/UNITS: PREREQUISITE: TEXT AND MATERIALS: CMN310C POST-ADVANCED OFFICE: Word And Excel 24 HOURS/1 UNIT CMN310A Advanced Office: Word and Excel or concurrent enrollment
/ Computational Genomics. Normalization
 10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
Introduction Google Forms is used to plan events, provide quizzes, survey, or collect needed information quickly.
 Table of Contents Introduction Creating a Form from Drive Question Types Editing the Form Form Layout Reorder Questions Confirmation Page Sharing your Form Add Collaborators Choose a Form Response Destination
Table of Contents Introduction Creating a Form from Drive Question Types Editing the Form Form Layout Reorder Questions Confirmation Page Sharing your Form Add Collaborators Choose a Form Response Destination
Exploring gene expression datasets
 Exploring gene expression datasets Alexey Sergushichev Dec 4-5, St. Louis About the workshop We will cover the basic analysis of gene expression matrices No working with raw data The focus is on being
Exploring gene expression datasets Alexey Sergushichev Dec 4-5, St. Louis About the workshop We will cover the basic analysis of gene expression matrices No working with raw data The focus is on being
Tenant Coordination Website User Guide For Mall Management
 Tenant Coordination Website User Guide For Mall Management Website s Testing address: http://www.ninthdegree.com/westfield/ Contents View Specific Deal 1 Upload the Complete Design Criteria Package 5 View
Tenant Coordination Website User Guide For Mall Management Website s Testing address: http://www.ninthdegree.com/westfield/ Contents View Specific Deal 1 Upload the Complete Design Criteria Package 5 View
Microsoft Excel Using Excel in the Science Classroom
 Microsoft Excel Using Excel in the Science Classroom OBJECTIVE Students will take data and use an Excel spreadsheet to manipulate the information. This will include creating graphs, manipulating data,
Microsoft Excel Using Excel in the Science Classroom OBJECTIVE Students will take data and use an Excel spreadsheet to manipulate the information. This will include creating graphs, manipulating data,
User Guide. Last Updated June 201 6
 User Guide Last Updated June 201 6 1 TABLE OF CONTENTS 1 Basics 4 1.1Logging In 4 1.2Dashboard 5 1.2.1 Tabs 5 1.2.2 Action Items 6 1.2.3 Coordinator s Dashboard 6 2 Forms 6 2.1Common Form Operations 6
User Guide Last Updated June 201 6 1 TABLE OF CONTENTS 1 Basics 4 1.1Logging In 4 1.2Dashboard 5 1.2.1 Tabs 5 1.2.2 Action Items 6 1.2.3 Coordinator s Dashboard 6 2 Forms 6 2.1Common Form Operations 6
User guide for GEM-TREND
 User guide for GEM-TREND 1. Requirements for Using GEM-TREND GEM-TREND is implemented as a java applet which can be run in most common browsers and has been test with Internet Explorer 7.0, Internet Explorer
User guide for GEM-TREND 1. Requirements for Using GEM-TREND GEM-TREND is implemented as a java applet which can be run in most common browsers and has been test with Internet Explorer 7.0, Internet Explorer
Drug versus Disease (DrugVsDisease) package
 1 Introduction Drug versus Disease (DrugVsDisease) package The Drug versus Disease (DrugVsDisease) package provides a pipeline for the comparison of drug and disease gene expression profiles where negatively
1 Introduction Drug versus Disease (DrugVsDisease) package The Drug versus Disease (DrugVsDisease) package provides a pipeline for the comparison of drug and disease gene expression profiles where negatively
NMC Surveillance System (Android App) NMC Surveillance System (Android App) Page 01
 Page 01 Table of Contents Content Page Glossary 03 Register New User 04 Sign In to App for 1st time 05 Navigation 06-07 Inbox 08 New Case 09-10 View Case 11-12 Edit Case 13-14 Profile 15 Profile Link Facilities
Page 01 Table of Contents Content Page Glossary 03 Register New User 04 Sign In to App for 1st time 05 Navigation 06-07 Inbox 08 New Case 09-10 View Case 11-12 Edit Case 13-14 Profile 15 Profile Link Facilities
The walkthrough is available at /
 The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
Introduction to GE Microarray data analysis Practical Course MolBio 2012
 Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
Teacher User Easy Setup Guide. Teacher User Setup Guide - wushka.com.au -
 Teacher User Easy Setup Guide - - 5 Easy Steps to Set Up Your Students and Reading Groups Input your students details individually Step 1: Manage Class List Page OR Use the upload tool to set up all students
Teacher User Easy Setup Guide - - 5 Easy Steps to Set Up Your Students and Reading Groups Input your students details individually Step 1: Manage Class List Page OR Use the upload tool to set up all students
CITI ACCESS AND DIRECTIONS FOR SENIOR NAVY LEADERSHIP NEW USERS -CITI LOGIN AND REGISTRATION-
 CITI ACCESS AND DIRECTIONS FOR SENIOR NAVY LEADERSHIP NEW USERS To access the CITI training program, go to: http://www.citiprogram.org. The following directions will assist you as you work through the
CITI ACCESS AND DIRECTIONS FOR SENIOR NAVY LEADERSHIP NEW USERS To access the CITI training program, go to: http://www.citiprogram.org. The following directions will assist you as you work through the
Data Processing and Analysis in Systems Medicine. Milena Kraus Data Management for Digital Health Summer 2017
 Milena Kraus Digital Health Summer Agenda Real-world Use Cases Oncology Nephrology Heart Insufficiency Additional Topics Data Management & Foundations Biology Recap Data Sources Data Formats Business Processes
Milena Kraus Digital Health Summer Agenda Real-world Use Cases Oncology Nephrology Heart Insufficiency Additional Topics Data Management & Foundations Biology Recap Data Sources Data Formats Business Processes
Quick-Start Tutorial. Airavata Reference Gateway
 Quick-Start Tutorial Airavata Reference Gateway Test/Demo Environment Details Tutorial I - Gateway User Account Create Account Login to Account Password Recovery Tutorial II - Using Projects Create Project
Quick-Start Tutorial Airavata Reference Gateway Test/Demo Environment Details Tutorial I - Gateway User Account Create Account Login to Account Password Recovery Tutorial II - Using Projects Create Project
HEDIS Profile. The HEDIS Profile allows providers and groups to:
 HEDIS Profile *Please note that all screenshots in this document are mock-ups used for training purposes and do not display any real member information. These screenshots are subject to change. The HEDIS
HEDIS Profile *Please note that all screenshots in this document are mock-ups used for training purposes and do not display any real member information. These screenshots are subject to change. The HEDIS
HOW TO SUBMIT A CE WORKSHOP OR TEST-FOCUSED WORKSHOP FOR THE NAN ANNUAL CONFERENCE
 HOW TO SUBMIT A CE WORKSHOP OR TEST-FOCUSED WORKSHOP FOR THE NAN ANNUAL CONFERENCE Submission Deadline: March 29, 2019 1. Click here to access the submission site. 2. NAN Account Holders: please click
HOW TO SUBMIT A CE WORKSHOP OR TEST-FOCUSED WORKSHOP FOR THE NAN ANNUAL CONFERENCE Submission Deadline: March 29, 2019 1. Click here to access the submission site. 2. NAN Account Holders: please click
Import GEO Experiment into Partek Genomics Suite
 Import GEO Experiment into Partek Genomics Suite This tutorial will illustrate how to: Import a gene expression experiment from GEO SOFT files Specify annotations Import RAW data from GEO for gene expression
Import GEO Experiment into Partek Genomics Suite This tutorial will illustrate how to: Import a gene expression experiment from GEO SOFT files Specify annotations Import RAW data from GEO for gene expression
2. Click File and then select Import from the menu above the toolbar. 3. From the Import window click the Create File to Import button:
 Totality 4 Import How to Import data into Totality 4. Totality 4 will allow you to import data from an Excel spreadsheet or CSV (comma separated values). You must have Microsoft Excel installed in order
Totality 4 Import How to Import data into Totality 4. Totality 4 will allow you to import data from an Excel spreadsheet or CSV (comma separated values). You must have Microsoft Excel installed in order
How-To Efficiently Create Section Groups in a Merged Bb Course Page In the merged Parent course, click into the Full Grade Center.
 How-To Efficiently Create Section Groups in a Merged Bb Course Page 1 1. In the merged Parent course, click into the Full Grade Center. 2. From there, download a spreadsheet by clicking Work Offline >
How-To Efficiently Create Section Groups in a Merged Bb Course Page 1 1. In the merged Parent course, click into the Full Grade Center. 2. From there, download a spreadsheet by clicking Work Offline >
SciVerse ScienceDirect. User Guide. October SciVerse ScienceDirect. Open to accelerate science
 SciVerse ScienceDirect User Guide October 2010 SciVerse ScienceDirect Open to accelerate science Welcome to SciVerse ScienceDirect: How to get the most from your subscription SciVerse ScienceDirect is
SciVerse ScienceDirect User Guide October 2010 SciVerse ScienceDirect Open to accelerate science Welcome to SciVerse ScienceDirect: How to get the most from your subscription SciVerse ScienceDirect is
Scenario Implementation as the R SubtypeDiscovery Package
 Chapter 5 Scenario Implementation as the R SubtypeDiscovery Package To enable reproducibility of our analyses and to abstract from the application domains, we implemented in the R SubtypeDiscovery package
Chapter 5 Scenario Implementation as the R SubtypeDiscovery Package To enable reproducibility of our analyses and to abstract from the application domains, we implemented in the R SubtypeDiscovery package
Mitochondrial DNA Typing
 Tutorial for Windows and Macintosh Mitochondrial DNA Typing 2007 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere)
Tutorial for Windows and Macintosh Mitochondrial DNA Typing 2007 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere)
