Refining Health Outcomes of Interest using Formal Concept Analysis and Semantic Query Expansion
|
|
|
- August Stevens
- 5 years ago
- Views:
Transcription
1 Refining Health Outcomes of Interest using Formal Concept Analysis and Semantic Query Expansion Olivier Curé 1, Henri Maurer 2, Nigma H. Shah 3, Paea Le Pendu 3 1 Université Paris-Est, LIGM - UMR CNRS 8049, France ocure@univ-mlv.fr 2 University of Edinburgh, Scotland, UK henri.maurer@gmail.com 3 Stanford University, Stanford, California, USA {nigam,plependu}@stanford.edu Abstract. Clinicians and researchers using Electronic Health Records (EHRs) often search for, extract, and analyze groups of patients by defining a health outcome of interest, which may include a set of diseases, conditions, signs, or symptoms. In our work on pharmacovigilance using clinical notes, for example, we use a method that operates over many (potentially hundreds) of ontologies at once, expands the input query, and increases the search space over clinical text as well as structured data. This method requires specifying an initial set of seed concepts, based on concept unique identifiers from the UMLS Metathesaurus. In some cases, such as for progressive multifocal leukoencephalopathy, the seed query is easy to specify, but in other cases this task can be more subtle, such as for chronic obstructive pulmonary disease. We have developed a method using formal concept analysis and semantic query expansion that assists the user in defining their seed query and in refining the expanded search space that it encompasses. These methods can be used for text-mining clinical notes from EHRs (this paper) and cohort building tools. 1 Introduction In applications that use Electronic Health Records (EHRs), such as in drug safety surveillance or observational studies, groups of patients are selected, extracted, compared, and analyzed based on definitions of certain health outcomes of interest (HOIs) [1,5]. Common examples of HOIs include myocardial infarction (MI), juvenile idiopathic arthritis, acute renal failure, chronic obstructive pulmonary disease (COPD), or peripheral artery disease. While the entry points for a search may at times be obvious, their full definitions are anything but easy to obtain. In work that also incorporates clinical text as data inputs, such as discharge summaries or nurses notes, these definitions should also capture variations of terms that are likely to appear in written notes so that the text-mining process has good recall [4]. For example, there are at least 47 distinct ways that we have seen so far for saying myocardial infarction that appears with frequency
2 in real clinical notes. The challenge in defining an HOI arises because medical and health terminologies are numerous and complex. There are currently over 160 terminologies in the Unified Medical Language System (UMLS) Metathesaurus with over 2-million distinct Concept Unique Identifiers (CUIs) and over 6-million distinct strings related to health and medicine. Some of these, like the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM), are well-known and often used to define the selection criteria for health outcomes of interest. We have found that most clinicians and researchers will say that choosing from lists of ICD-9 or SNOMED CT codes, is not a good way to perform this task. The problem is that the organization and presentation of concepts in any one of these terminologies, let alone 160 of them, might be intuitive for one person yet completely obtuse to another. This work addresses these challenges by enabling the user to specify their HOI using plain terms, much like an ordinary search query. We use Semantic Query Expansion (SQE) and Formal Concept Analysis (FCA) [3] to induce a lattice of concepts over hundreds of ontologies and terminologies at once and to find the best-matching concepts. Briefly, a lattice is a partially ordered set where every two elements have a least and greatest upper bounds. FCA is a machine learning technique that uses a lattice to reveal associations between elements of some pre-defined structure in this case, a set of ontologies with hierarchies and mappings. SQE leverages the hierarchies and mappings to expand concepts to include all subsumed ones. The system aims for broad coverage initially and asks for feedback on the furthest matches at the highest points of the lattice so that the search space can be rapidly refined with minimal input from the user. At the same time the user is issuing queries and confirming or denying concept matches, we display search results that highlight snippets from clinical notes matching their HOI, which provides instant feedback for the user as they refine their HOI. We have implemented these tools as REST service WEB APIs. Used together, FCA and SQE make it possible to order concepts semantically and search for matches that best reflect the intension of the user, i.e., the medical concepts she wants to express through a set of proposed terms. Intuitively, the system aims initially for greater coverage and relies on user feedback to improve relevance. The user s search query and feedback essentially helps to pinpoint and prune sections of the lattice until only those concepts that fit their intension remain. The amount of feedback can be minimized by measuring coverage of concepts at each major branch. The paper is organized as follows. In the next section, background and methods are presented. In Section 3, implementation details are provided. Section 4 presents preliminary results conducted on the i2b2 Obesity NLP reference set. A discussion and some future works are provided in section 5.
3 2 Background and Methods 2.1 A previous method for myocardial infarction HOI In [4], a query-driven approach called 2-hop was used to create a set of HOIs for detecting drug-adverse event associations and adverse events associated with drug-drug interactions. The algorithm takes a set of concepts C and derives all subconcepts C in each ontology O available in an ontology repository. This process is repeated one more time for all derived concepts C to obtain another set of concepts denoted C. Because concepts are mapped across ontologies, the process traverses simultaneously all ontologies that contain C (and C ), thereby hopping across ontologies twice. With this approach, C can capture more concepts from the adjacent ontologies that would have otherwise been missed with a single iteration. In principle, recursion with a least fixed-point semantics would apply; however, recursion does not work well in practice because of differing abstraction levels among ontologies, which induce cycles. We have found that two hops achieve an adequate balance between soundness and completeness for the current use case. This method was able to recognize events and exposures with enough accuracy for the drug safety use case. This accuracy was determined using a goldstandard corpus from the 2008 i2b2 Obesity Challenge. This corpus has been manually annotated by two annotators for 16 conditions and was designed to evaluate the ability of NLP systems to identify a condition present for a patient given a textual note. Moreover, this corpus was extended by manually annotating each of the events. Using the set of terms corresponding to the definition of the event of interest and the set of terms recognized by our annotation workflow in the i2b2 notes, we evaluate the sensitivity and specificity of identifying each of the events. Although efficient, this method requires a significant amount of manual effort, i.e., some medical experts have to provide concept identifiers, implying a deep knowledge of some ontologies. The goal of this work is to increase automation by only requiring from the medical experts to provide terms associated to their serach. Our goal is also to retain/improve previous results. 2.2 Semantic Query Expansion Theory One of the major assets of ontologies is the set of hierarchical relationships they often include. For every concept, a set of parent or super-concepts will usually induce a directed acyclic graph (DAG) structure for most biomedical ontologies. We can use standard graph traversal algorithms to compute the transitive closure and store the set of all ancestors and descendants of every concept. 4 SQE is the process of taking a set of concepts as a query and utilizing the transitive closure to expand that set to include all descendants or ancestors depending on whether the goal is to generalize or specialize the query. 4 We should note that ontologies in general can be more structurally complex than a DAG, in which case inference engines should be used to compute the subsumptions hierarchy in place of graph traversal algorithms.
4 2.3 Formal Concept Analysis Theory FCA is the process of abstracting conceptual descriptions from a set of objects described by some attributes and finds practical application in fields such as data and text mining, machine learning, and knowledge management. Given a set of ontologies, we represent all objects and their attributes including hierarchical relations as a binary matrix. Using the standard machinery of FCA, a concept lattice can be generated from this matrix. Because every lattice is a partial order, FCA will group similar objects according to their attributes in an ordered manner. We utilize these properties to identify improbable relations that are not explicitly stated in the ontology as a means of ranking questions to the user that will cover the greatest benefit in narrowing the SQE search space. More formally, FCA is based on the notion of a formal context, which is a triple K = (G, M, I), where G is a set of objects, M is a set of attributes and I is a binary relation between G and M, i.e., I G M. For an object g and an attribute m, (g, m) I is read as object g has attribute m. Given a formal context, we can define the notion of formal concepts, where, for A G, we define A = {m M g A : (g, m) I} and for B M, we define B = {g G m B : (g, m) I}. A formal concept of K is defined as a pair (A, B) with A G, B M, A = B and B = A. The hierarchy of formal concepts is formalized by (A 1, B 1 ) (A 2, B 2 ) A 1 A 2 and B 2 B 1. The concept lattice of K is the set of all its formal concepts with the partial order. This hierarchy of formal concepts obeys the mathematical axioms defining a lattice, and is called a concept lattice since the relation between the sets of objects and attributes is a Galois connection. A Galois connection plays an important role in lattice theory, universal algebras, and recently in computer science [2]. Let (P, ) and (Q, ) be two partially ordered sets (poset). A Galois connection between P and Q is a pair of mappings (Φ, Ψ) such that Φ : P Q, Ψ : Q P and: (i) x x implies Φ(x) Φ(x ), (ii) y y implies Ψ(y) Ψ(y ) and (iii) x Ψ(Φ(x)) and y Ψ(Φ(y)), for x,x P and y,y Q. 3 Implementation 3.1 Expansion using SQE The user inputs a free-text query representing their HOI, such as pituitary cancer. The query is tokenized and matched against all synonyms of every concept from our set of ontologies. For UMLS, this results in a set of matching CUIs. This step is purely lexical and does not use any semantics. We utilize the structure of the ontologies to expand the search and identify matches such as neoplasm or tumor that are closely related to the search. We perform this expansion by navigating to all super-concepts in each ontology incrementally until we achieve a minimal cover of lexical matches. Broadening the query using SQE in this way will identify many closely related terms (i.e., increase coverage), but it may also introduce many unrelated ones (i.e., decrease relevance). Thus, we have to
5 Fig. 1. Overview of ontology concept hierarchy prune the expanded set aggressively to improve relevance, which we facilitate using FCA (described in more detail below). In Figure 1, for example, the m concepts at the bottom represent the initial lexical matches, and the p concepts are the set of super-concepts that cover them. Based on the poor coverage of d 1 (perhaps because c 1 and c 2 seem too unrelated to m 1 and m 2 ) the user may be asked whether d 1 is a fit, and if not, it is pruned (which automatically eliminates c 1 and c 2 and everything else subsumed by d 1 ). 3.2 Pruning using FCA Table 3.2 provides an FCA lattice example using only hierarchical relations for a query on Hypertriglyceridemia. In the lattices we are producing with our approach, both objects (left value at each row) and attributes (top value of each column) are ontology concepts with the attributes corresponding to the super cocnepts of the objects. This lattice uses internal identifiers: stands for the Hyperpoproteinemia type IV concept and 0 correspond to the top concept of our set of ontologies. Hence is a sub-concept of the concepts 0, and Intuitively, FCA can identify rectangles such as the one made of columns {0,19118,740154} and rows {10365,12115} or the one made of columns {0,19118}. These rectangles are formal concepts as introduced in Section 2. The FCA lattice is defined over these rectangles. Figure 2(b) displays the extract of the lattice corresponding to the table in Figure 2(a).
6 (a) Hypertriglyceridemia FCA matrix extract (b) Lattice extract Fig. 2. Extract of Hypertriglyceridemia FCA matrix and lattice One advantage of the FCA lattice is a compact representation of the hierarchy that enables us to efficiently find the highest cut among concepts in our ontologies that provide maximal coverage of terms identified via lexical matching. It also enables us to identify concepts worth pruning in the following manner: Let F i = O i, A i be a formal concept traversed with O i and A i respectively the set of objects and attributes. For each F i traversed during our top-down navigation of the lattice, we create the two following lists: one denoted L A i, corresponding to the transitive closure of the sub-concepts for each element of A i and another one denoted L O, corresponding to the transitive closure of the sub-concepts of all O i. We compute the intersection of L O i with each L A i and we use the ratio L A i LO i / LA i. If this value is below a predefined threshold (denoted pruning threshold), e.g., 75%, then we consider that the considered A i concept is not relevant to the search, i.e., it has too many sub-concepts not corresponding to sub-concepts of the matched concepts. Otherwise it is relevant and we store it in a candidate list which will be proposed later on to the physician. Example (hypercholesterolemia): Using hypercholesterolemia as an example search query and a set of 18 ontologies, we identify 20 objects and 102 attributes initially, as in Figure 1. This induces a lattice of 67 formal concepts. Its top most formal concept contains all 67 objects with an empty attribute set. The lattice s second level has 2 formal concepts, one with 23 objects (and one attribute) and another one with 17 objects (and one attribute). Both of these concepts have too many sub-concepts not corresponding to the set of subconcepts of our 20 original objects, hence they are pruned. At the fourth level of the lattice, we discover a first potential concept contained in a formal concept containing 9 objects and 9 attributes one of which has a 75% ratio, i.e., satisfying our pruning threshold. It has 16 sub-concepts out of which only 4 are not covered by the sub-concepts of the 9 objects sub-concepts. Some of these 4 concepts could be unrelated, so we drill down futher, identify the specific area of the lattice with the smallest ratio, and ask the user whether this concept is a fit, if not, we prune the lattice above and work at this lower level instead until the user is satisfied. In the example, the labels of the 4 non-covered concepts are: hypercholesterolemia, cholesterolosis, secondary hypercholesterolemia and hyperlipidemia.
7 3.3 Web-based API Fig. 3. Example graphical interface based on API Figure 3 presents a web-based interface using the APIs for SEQ and FCA based search and refinement. The user provides and receives feedback in multiple ways (numbered from 1 to 4). First of all, the physician enters some terms associated to her search (area #1) and runs the query (area #2). The terms are sent to REST service API, which uses SEQ and FCA to identify those concepts that best fit the query, and those that require confirmation from the user. The concepts are also simultaneously matched against a pre-indexed set of clinical notes (area #3), which place those concepts within the context of their realworld use. The user can then confirm or deny the correctness of the match and update the search (area #4). The number of patients is also displayed in area #3, informing the euse on how many hits are being discovered. 4 Evaluation In [4], the online Supplementary Data S3 reports that the 2-hop method identifies HOIs with a sensitivity of 74% and a specificity of 96% for the i2b2 Obesity NLP reference set, henceforth i2b2 obesity. Our evaluation is performed on this same dataset in order to enable a comparison of our method. As such, we consider the set of terms associated to the 16 different conditions surveyed in i2b2 Obesity and retrieve from our database the corresponding concept identifiers.
8 Given this setting, we are able to compute the sensitivity and specificity of our approach. This is presented in Table 1. The experiments have been conducted on commodity hardware running a Java implementation using a MySQL database instance, we thus consider that there rooms for performance gains. The analysis of our method results is given in 3 dimensions: sensitivity, specificity and processing duration (not including end-user interactions). The obtained statistical measures are encouraging with averages of sensitivity and specificity of respectively 77.3 and 99.1%, hence improving on the results of the less automatized 2-hop solution. In the first hand, these values have to be considered in the context of a fast processing of potential terms, i.e., duration in seconds range from 0.6 to The 2 order of magnitude between the slowest and fastest executions are explained by the size of the matching concepts retrieved from the query patterns, the size of their subsumption relationship transitive closure and the structure of the associated FCA lattice. The slowest, i.e. Diabetes mellitus, involves the computation of matrix of more than 330 objects and 7000 attributes resulting in an FCA lattice of more than 3000 formal concepts out of which most candidates concepts are pruned. In the second hand, the matching and candidate concepts are proposed to the physician for acceptance and rejection. Hence, through an intuitive and user-friendly interface, she is able to easily improve the sensitivity measure. An evaluation on the efficiency and interactivity of the Web interface has yet to be conducted on with physicians on real case scenarios. Finally, we consider for some of the false negative concepts discovered by our method may end up being positive propositions. Moreover, these propositions come from both the matching (e.g., Sitosterolemia for hypercholesterolemia for hypercholesterolemia) and potential (e.g., h/o: raised blood, familial hyperlipoproteinemia, fh: raised blood lipids for hypercholesterolemia while the gold standard contains concepts such as hyperlipoproteinemia type ii ) concepts which confirms the relevance of using a semantic approach. Note that among our true positive, depending on the use case, a significant number of items have been retrieved from the potential concept set, i.e., using our FCA statistical approach. 5 Conclusion We have presented a novel, semi-automatic solution for defining health outcomes of interest. Our work is inspired by previous, manually-intensive work done for the purpose of text-mining clinical notes from EHRs. Our approach is composed of a cooperation between Semantic Query Expansion, to leverage the hierarchical structure of ontologies, and Formal Concept Analysis, to organize, reason, and prune discovered concepts. We implemented a RESTful API and a graphical Web-based interface to illustrate the process that users would follow to browse data and refine their HOI query rapidly. A preliminary evaluation of this work emphasizes positive results for sensitivity and specificity measures. The most promising aspect of this approach is the discovery of positive results not present
9 Table 1. i2b2 Obesity NLP reference evaluation Condition Se Sp D(s) Asthma 92.7% 99.1% 3.8 CAD 75.5% 99.4% 5.0 Congestive heart failure (CHF) 74.2% 99.4% 5.5 Depression 69.9% 100% 5.6 Diabetes mellitus 82.0% 99.2% 67.3 Gallstones / Cholcystectomy 81.2% 98.7% 4.9 GERD 56.2% 100% 4.8 Gout 94.4.% 100% 8.7 Hypercholesterolemia 82.4% 100% 4.9 Hypertension 82.6 % 98.6% 8.4 Hypertriglyceridemia 60.0% 98.8% 0.9 Obstructive sleep apnea (OSA) 100 % 100 % 1.5 Osteoarthritis 61.2% 98.9% 2.3 Peripheral vascular disease 66.3 % 99.4% 1.1 Venous insufficiency 77.8% 99.1% 2.8 Obesity 86.1% 99.2% 0.6 Average 77.3% 99.1% 8.0 Notes: Se (Sensitivity), Sp (Specificity), D (Duration), CAD (Atherosclerotic cardiovascular disease), GERD (Gastoresophageal reflux disease) our i2b2 Obesity NLP reference set. Thus, this approach provides better recall with high precision of the obtained results. Some of our future goals include (i) conducting user driven evaluation of the Web interface, (ii) analyzing the acceptance/rejection of physicians in several practical scenarios, (iii) using active learning over past query refinements to improve future queries, and (iv) studying our method impact s on mining EHRs clinical notes or cohort building tools. References 1. R. M. Carnahan and K. G. Moores. Mini-sentinel systematic reviews of validated methods for identifying health outcomes using administrative and claims data: methods and lessons learned. Pharmacoepidemiol Drug Saf, 21 Suppl 1:82 9, B. A. Davey and H. A. Priestley. Introduction to Lattices and Order (2. ed.). Cambridge University Press, B. Ganter and R. Wille. Formal concept analysis - mathematical foundations. Springer, P. Lependu, S. V. Iyer, A. Bauer-Mehren, R. Harpaz, J. M. Mortensen, T. Podchiyska, T. A. Ferris, and N. H. Shah. Pharmacovigilance using clinical notes. Clin Pharmacol Ther, P. Stang, P. Ryan, J. Racoosin, J. Overhage, A. Hartzema, C. Reich, E. Welebob, T. Scarnecchia, and J. Woodcock. Advancing the science for active surveillance: rationale and design for the observational medical outcomes partnership. Ann Intern Med, 153(9):600 6, 2010.
A Semantic Web-Based Approach for Harvesting Multilingual Textual. definitions from Wikipedia to support ICD-11 revision
 A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
warwick.ac.uk/lib-publications
 Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
Document Retrieval using Predication Similarity
 Document Retrieval using Predication Similarity Kalpa Gunaratna 1 Kno.e.sis Center, Wright State University, Dayton, OH 45435 USA kalpa@knoesis.org Abstract. Document retrieval has been an important research
Document Retrieval using Predication Similarity Kalpa Gunaratna 1 Kno.e.sis Center, Wright State University, Dayton, OH 45435 USA kalpa@knoesis.org Abstract. Document retrieval has been an important research
SNOMED Clinical Terms
 Representing clinical information using SNOMED Clinical Terms with different structural information models KR-MED 2008 - Phoenix David Markwell Laura Sato The Clinical Information Consultancy Ltd NHS Connecting
Representing clinical information using SNOMED Clinical Terms with different structural information models KR-MED 2008 - Phoenix David Markwell Laura Sato The Clinical Information Consultancy Ltd NHS Connecting
Languages and tools for building and using ontologies. Simon Jupp, James Malone
 An overview of ontology technology Languages and tools for building and using ontologies Simon Jupp, James Malone jupp@ebi.ac.uk, malone@ebi.ac.uk Outline Languages OWL and OBO classes, individuals, relations,
An overview of ontology technology Languages and tools for building and using ontologies Simon Jupp, James Malone jupp@ebi.ac.uk, malone@ebi.ac.uk Outline Languages OWL and OBO classes, individuals, relations,
CPRD Aurum Frequently asked questions (FAQs)
 CPRD Aurum Frequently asked questions (FAQs) Version 2.0 Date: 10 th April 2019 Authors: Helen Booth, Daniel Dedman, Achim Wolf (CPRD, UK) 1 Documentation Control Sheet During the course of the project
CPRD Aurum Frequently asked questions (FAQs) Version 2.0 Date: 10 th April 2019 Authors: Helen Booth, Daniel Dedman, Achim Wolf (CPRD, UK) 1 Documentation Control Sheet During the course of the project
Precise Medication Extraction using Agile Text Mining
 Precise Medication Extraction using Agile Text Mining Chaitanya Shivade *, James Cormack, David Milward * The Ohio State University, Columbus, Ohio, USA Linguamatics Ltd, Cambridge, UK shivade@cse.ohio-state.edu,
Precise Medication Extraction using Agile Text Mining Chaitanya Shivade *, James Cormack, David Milward * The Ohio State University, Columbus, Ohio, USA Linguamatics Ltd, Cambridge, UK shivade@cse.ohio-state.edu,
Graphical Methods for Reducing, Visualizing and Analyzing Large Data Sets Using Hierarchical Terminologies
 Graphical Methods for Reducing, Visualizing and Analyzing Large Data Sets Using Hierarchical Terminologies Xia Jing, MB, PhD 1, James J. Cimino, MD 1, 2 1 National Library of Medicine, 2 Clinical Center,
Graphical Methods for Reducing, Visualizing and Analyzing Large Data Sets Using Hierarchical Terminologies Xia Jing, MB, PhD 1, James J. Cimino, MD 1, 2 1 National Library of Medicine, 2 Clinical Center,
Open Self Medication on LOD
 Open Self Medication on LOD Olivier Curé Université Paris-Est, LIGM, CNRS UMR 8049, France ocure@univ-mlv.fr Abstract. Open Self Medication 1 is a Web application that better informs people when treating
Open Self Medication on LOD Olivier Curé Université Paris-Est, LIGM, CNRS UMR 8049, France ocure@univ-mlv.fr Abstract. Open Self Medication 1 is a Web application that better informs people when treating
Acquiring Experience with Ontology and Vocabularies
 Acquiring Experience with Ontology and Vocabularies Walt Melo Risa Mayan Jean Stanford The author's affiliation with The MITRE Corporation is provided for identification purposes only, and is not intended
Acquiring Experience with Ontology and Vocabularies Walt Melo Risa Mayan Jean Stanford The author's affiliation with The MITRE Corporation is provided for identification purposes only, and is not intended
Terminology Harmonization
 Terminology Harmonization Rob McClure, MD; Lisa Anderson, MSN, RN-BC; Angie Glotstein, BSN, RN November 14-15, 2018 Washington, DC Table of contents OVERVIEW OF CODE SYSTEMS AND TERMINOLOGY TOOLS USING
Terminology Harmonization Rob McClure, MD; Lisa Anderson, MSN, RN-BC; Angie Glotstein, BSN, RN November 14-15, 2018 Washington, DC Table of contents OVERVIEW OF CODE SYSTEMS AND TERMINOLOGY TOOLS USING
NIH Public Access Author Manuscript Methods Inf Med. Author manuscript; available in PMC 2014 October 27.
 NIH Public Access Author Manuscript Published in final edited form as: Methods Inf Med. 2014 ; 53(3): 173 185. doi:10.3414/me13-01-0075. A Complementary Graphical Method for Reducing and Analyzing Large
NIH Public Access Author Manuscript Published in final edited form as: Methods Inf Med. 2014 ; 53(3): 173 185. doi:10.3414/me13-01-0075. A Complementary Graphical Method for Reducing and Analyzing Large
A Semantic Multi-Field Clinical Search for Patient Medical Records
 BULGARIAN ACADEMY OF SCIENCES CYBERNETICS AND INFORMATION TECHNOLOGIES Volume 18, No 1 Sofia 2018 Print ISSN: 1311-9702; Online ISSN: 1314-4081 DOI: 10.2478/cait-2018-0014 A Semantic Multi-Field Clinical
BULGARIAN ACADEMY OF SCIENCES CYBERNETICS AND INFORMATION TECHNOLOGIES Volume 18, No 1 Sofia 2018 Print ISSN: 1311-9702; Online ISSN: 1314-4081 DOI: 10.2478/cait-2018-0014 A Semantic Multi-Field Clinical
CHAPTER 3 INFORMATION RETRIEVAL BASED ON QUERY EXPANSION AND LATENT SEMANTIC INDEXING
 43 CHAPTER 3 INFORMATION RETRIEVAL BASED ON QUERY EXPANSION AND LATENT SEMANTIC INDEXING 3.1 INTRODUCTION This chapter emphasizes the Information Retrieval based on Query Expansion (QE) and Latent Semantic
43 CHAPTER 3 INFORMATION RETRIEVAL BASED ON QUERY EXPANSION AND LATENT SEMANTIC INDEXING 3.1 INTRODUCTION This chapter emphasizes the Information Retrieval based on Query Expansion (QE) and Latent Semantic
Powering Knowledge Discovery. Insights from big data with Linguamatics I2E
 Powering Knowledge Discovery Insights from big data with Linguamatics I2E Gain actionable insights from unstructured data The world now generates an overwhelming amount of data, most of it written in natural
Powering Knowledge Discovery Insights from big data with Linguamatics I2E Gain actionable insights from unstructured data The world now generates an overwhelming amount of data, most of it written in natural
FCA-Map Results for OAEI 2016
 FCA-Map Results for OAEI 2016 Mengyi Zhao 1 and Songmao Zhang 2 1,2 Institute of Mathematics, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, P. R. China 1 myzhao@amss.ac.cn,
FCA-Map Results for OAEI 2016 Mengyi Zhao 1 and Songmao Zhang 2 1,2 Institute of Mathematics, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, P. R. China 1 myzhao@amss.ac.cn,
Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR
 Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR Guoqian Jiang 1, Eric Prud Hommeax 2, and Harold R. Solbrig 1 1 Mayo Clinic, Rochester, MN, 55905, USA 2
Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR Guoqian Jiang 1, Eric Prud Hommeax 2, and Harold R. Solbrig 1 1 Mayo Clinic, Rochester, MN, 55905, USA 2
Controlled Medical Vocabulary in the CPR Generations
 Tutorials, B. Hieb, M.D. Research Note 5 November 2003 Controlled Medical Vocabulary in the CPR Generations A CMV capability becomes progressively more important as computer-based patient record systems
Tutorials, B. Hieb, M.D. Research Note 5 November 2003 Controlled Medical Vocabulary in the CPR Generations A CMV capability becomes progressively more important as computer-based patient record systems
ENTITIES IN THE OBJECT-ORIENTED DESIGN PROCESS MODEL
 INTERNATIONAL DESIGN CONFERENCE - DESIGN 2000 Dubrovnik, May 23-26, 2000. ENTITIES IN THE OBJECT-ORIENTED DESIGN PROCESS MODEL N. Pavković, D. Marjanović Keywords: object oriented methodology, design process
INTERNATIONAL DESIGN CONFERENCE - DESIGN 2000 Dubrovnik, May 23-26, 2000. ENTITIES IN THE OBJECT-ORIENTED DESIGN PROCESS MODEL N. Pavković, D. Marjanović Keywords: object oriented methodology, design process
Open Research Online The Open University s repository of research publications and other research outputs
 Open Research Online The Open University s repository of research publications and other research outputs Bottom-Up Ontology Construction with Contento Conference or Workshop Item How to cite: Daga, Enrico;
Open Research Online The Open University s repository of research publications and other research outputs Bottom-Up Ontology Construction with Contento Conference or Workshop Item How to cite: Daga, Enrico;
CPRD Aurum Frequently asked questions (FAQs)
 CPRD Aurum Frequently asked questions (FAQs) Version 1.0 Date: 6 December 2017 Author: Helen Booth and Daniel Dedman, CPRD, UK Documentation Control Sheet During the course of the project it may be necessary
CPRD Aurum Frequently asked questions (FAQs) Version 1.0 Date: 6 December 2017 Author: Helen Booth and Daniel Dedman, CPRD, UK Documentation Control Sheet During the course of the project it may be necessary
IDECSE: A Semantic Integrated Development Environment for Composite Services Engineering
 IDECSE: A Semantic Integrated Development Environment for Composite Services Engineering Ahmed Abid 1, Nizar Messai 1, Mohsen Rouached 2, Thomas Devogele 1 and Mohamed Abid 3 1 LI, University Francois
IDECSE: A Semantic Integrated Development Environment for Composite Services Engineering Ahmed Abid 1, Nizar Messai 1, Mohsen Rouached 2, Thomas Devogele 1 and Mohamed Abid 3 1 LI, University Francois
A Session-based Ontology Alignment Approach for Aligning Large Ontologies
 Undefined 1 (2009) 1 5 1 IOS Press A Session-based Ontology Alignment Approach for Aligning Large Ontologies Editor(s): Name Surname, University, Country Solicited review(s): Name Surname, University,
Undefined 1 (2009) 1 5 1 IOS Press A Session-based Ontology Alignment Approach for Aligning Large Ontologies Editor(s): Name Surname, University, Country Solicited review(s): Name Surname, University,
Executive Summary for deliverable D6.1: Definition of the PFS services (requirements, initial design)
 Electronic Health Records for Clinical Research Executive Summary for deliverable D6.1: Definition of the PFS services (requirements, initial design) Project acronym: EHR4CR Project full title: Electronic
Electronic Health Records for Clinical Research Executive Summary for deliverable D6.1: Definition of the PFS services (requirements, initial design) Project acronym: EHR4CR Project full title: Electronic
Apache UIMA and Mayo ctakes
 Apache and Mayo and how it is used in the clinical domain March 16, 2012 Apache and Mayo Outline 1 Apache and Mayo Outline 1 2 Introducing Pipeline Modules Apache and Mayo What is? (You - eee - muh) Unstructured
Apache and Mayo and how it is used in the clinical domain March 16, 2012 Apache and Mayo Outline 1 Apache and Mayo Outline 1 2 Introducing Pipeline Modules Apache and Mayo What is? (You - eee - muh) Unstructured
AROMA results for OAEI 2009
 AROMA results for OAEI 2009 Jérôme David 1 Université Pierre-Mendès-France, Grenoble Laboratoire d Informatique de Grenoble INRIA Rhône-Alpes, Montbonnot Saint-Martin, France Jerome.David-at-inrialpes.fr
AROMA results for OAEI 2009 Jérôme David 1 Université Pierre-Mendès-France, Grenoble Laboratoire d Informatique de Grenoble INRIA Rhône-Alpes, Montbonnot Saint-Martin, France Jerome.David-at-inrialpes.fr
Intelligent flexible query answering Using Fuzzy Ontologies
 International Conference on Control, Engineering & Information Technology (CEIT 14) Proceedings - Copyright IPCO-2014, pp. 262-277 ISSN 2356-5608 Intelligent flexible query answering Using Fuzzy Ontologies
International Conference on Control, Engineering & Information Technology (CEIT 14) Proceedings - Copyright IPCO-2014, pp. 262-277 ISSN 2356-5608 Intelligent flexible query answering Using Fuzzy Ontologies
Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR
 Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR Guoqian Jiang 1, Eric Prud Hommeaux 2, Guohui Xiao 3, and Harold R. Solbrig 1 1 Mayo Clinic, Rochester,
Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR Guoqian Jiang 1, Eric Prud Hommeaux 2, Guohui Xiao 3, and Harold R. Solbrig 1 1 Mayo Clinic, Rochester,
A Model of Machine Learning Based on User Preference of Attributes
 1 A Model of Machine Learning Based on User Preference of Attributes Yiyu Yao 1, Yan Zhao 1, Jue Wang 2 and Suqing Han 2 1 Department of Computer Science, University of Regina, Regina, Saskatchewan, Canada
1 A Model of Machine Learning Based on User Preference of Attributes Yiyu Yao 1, Yan Zhao 1, Jue Wang 2 and Suqing Han 2 1 Department of Computer Science, University of Regina, Regina, Saskatchewan, Canada
Enumerating Pseudo-Intents in a Partial Order
 Enumerating Pseudo-Intents in a Partial Order Alexandre Bazin and Jean-Gabriel Ganascia Université Pierre et Marie Curie, Laboratoire d Informatique de Paris 6 Paris, France Alexandre.Bazin@lip6.fr Jean-Gabriel@Ganascia.name
Enumerating Pseudo-Intents in a Partial Order Alexandre Bazin and Jean-Gabriel Ganascia Université Pierre et Marie Curie, Laboratoire d Informatique de Paris 6 Paris, France Alexandre.Bazin@lip6.fr Jean-Gabriel@Ganascia.name
Profile-based Retrieval of Records in Medical Databases
 Profile-based Retrieval of Records in Medical Databases Journal: AMIA 2009 Annual Symposium Manuscript ID: AMIA-0314-A2009.R1 Manuscript Type: Paper Date Submitted by the Author: 29-Jul-2009 Complete List
Profile-based Retrieval of Records in Medical Databases Journal: AMIA 2009 Annual Symposium Manuscript ID: AMIA-0314-A2009.R1 Manuscript Type: Paper Date Submitted by the Author: 29-Jul-2009 Complete List
2017 Partners in Excellence Executive Overview, Targets, and Methodology
 2017 Partners in Excellence Executive Overview, s, and Methodology Overview The Partners in Excellence program forms the basis for HealthPartners financial and public recognition for medical or specialty
2017 Partners in Excellence Executive Overview, s, and Methodology Overview The Partners in Excellence program forms the basis for HealthPartners financial and public recognition for medical or specialty
Web Page Classification using FP Growth Algorithm Akansha Garg,Computer Science Department Swami Vivekanad Subharti University,Meerut, India
 Web Page Classification using FP Growth Algorithm Akansha Garg,Computer Science Department Swami Vivekanad Subharti University,Meerut, India Abstract - The primary goal of the web site is to provide the
Web Page Classification using FP Growth Algorithm Akansha Garg,Computer Science Department Swami Vivekanad Subharti University,Meerut, India Abstract - The primary goal of the web site is to provide the
An FCA classification of durations of time for textual databases
 An FCA classification of durations of time for textual databases Ulrik Sandborg-Petersen Department of Communication and Psychology Kroghstræde 3, DK 9220 Aalborg East, Denmark ulrikp@hum.aau.dk Abstract.
An FCA classification of durations of time for textual databases Ulrik Sandborg-Petersen Department of Communication and Psychology Kroghstræde 3, DK 9220 Aalborg East, Denmark ulrikp@hum.aau.dk Abstract.
Aggregating Educational Data for Patient Empowerment
 Nick Portokallidis, George Drosatos, Eleni Kaldoudi School of Medicine, Democritus University of Thrace 6 th Panhellenic Conference on Biomedical Technology Athens, Greece, 6-8 May 2015 This work was supported
Nick Portokallidis, George Drosatos, Eleni Kaldoudi School of Medicine, Democritus University of Thrace 6 th Panhellenic Conference on Biomedical Technology Athens, Greece, 6-8 May 2015 This work was supported
NEO: Systematic Non-Lattice Embedding of Ontologies for Comparing the Subsumption Relationship in SNOMED CT and in FMA Using MapReduce
 NEO: Systematic Non-Lattice Embedding of Ontologies for Comparing the Subsumption Relationship in SNOMED CT and in FMA Using MapReduce Wei Zhu, Guo-Qiang Zhang, PhD, Shiqiang Tao, MS, Mengmeng Sun, Licong
NEO: Systematic Non-Lattice Embedding of Ontologies for Comparing the Subsumption Relationship in SNOMED CT and in FMA Using MapReduce Wei Zhu, Guo-Qiang Zhang, PhD, Shiqiang Tao, MS, Mengmeng Sun, Licong
Posted on March 8, formally published on December 5, 2000 by Linkoping University Electronic Press Linkoping, Sweden Linkoping Electronic Artic
 Linkoping Electronic Articles in Computer and Information Science Vol. 5(2000): nr 5 A Knowledge Representation for Information Filtering Using Formal Concept Analysis Peter Eklund and Richard Cole Linkoping
Linkoping Electronic Articles in Computer and Information Science Vol. 5(2000): nr 5 A Knowledge Representation for Information Filtering Using Formal Concept Analysis Peter Eklund and Richard Cole Linkoping
Medical Records Clustering Based on the Text Fetched from Records
 Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology ISSN 2320 088X IMPACT FACTOR: 5.258 IJCSMC,
Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology ISSN 2320 088X IMPACT FACTOR: 5.258 IJCSMC,
2018 Partners in Excellence Executive Overview, Targets, and Methodology
 2018 Partners in Excellence Executive Overview, s, and Methodology Overview The Partners in Excellence program forms the basis for HealthPartners financial and public recognition for medical or specialty
2018 Partners in Excellence Executive Overview, s, and Methodology Overview The Partners in Excellence program forms the basis for HealthPartners financial and public recognition for medical or specialty
The SHARPn phenotyping funnel. Phenotype specific patient cohorts
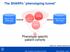 The SHARPn phenotyping funnel Mayo Clinic EHR data QDMs CEMs Intermountain EHR data DRLs Phenotype specific patient cohorts [Welch et al., JBI 2012; 45(4):763-71] 2012 MFMER slide-1 Algorithm Development
The SHARPn phenotyping funnel Mayo Clinic EHR data QDMs CEMs Intermountain EHR data DRLs Phenotype specific patient cohorts [Welch et al., JBI 2012; 45(4):763-71] 2012 MFMER slide-1 Algorithm Development
Theme Identification in RDF Graphs
 Theme Identification in RDF Graphs Hanane Ouksili PRiSM, Univ. Versailles St Quentin, UMR CNRS 8144, Versailles France hanane.ouksili@prism.uvsq.fr Abstract. An increasing number of RDF datasets is published
Theme Identification in RDF Graphs Hanane Ouksili PRiSM, Univ. Versailles St Quentin, UMR CNRS 8144, Versailles France hanane.ouksili@prism.uvsq.fr Abstract. An increasing number of RDF datasets is published
CIS UDEL Working Notes on ImageCLEF 2015: Compound figure detection task
 CIS UDEL Working Notes on ImageCLEF 2015: Compound figure detection task Xiaolong Wang, Xiangying Jiang, Abhishek Kolagunda, Hagit Shatkay and Chandra Kambhamettu Department of Computer and Information
CIS UDEL Working Notes on ImageCLEF 2015: Compound figure detection task Xiaolong Wang, Xiangying Jiang, Abhishek Kolagunda, Hagit Shatkay and Chandra Kambhamettu Department of Computer and Information
POMap results for OAEI 2017
 POMap results for OAEI 2017 Amir Laadhar 1, Faiza Ghozzi 2, Imen Megdiche 1, Franck Ravat 1, Olivier Teste 1, and Faiez Gargouri 2 1 Paul Sabatier University, IRIT (CNRS/UMR 5505) 118 Route de Narbonne
POMap results for OAEI 2017 Amir Laadhar 1, Faiza Ghozzi 2, Imen Megdiche 1, Franck Ravat 1, Olivier Teste 1, and Faiez Gargouri 2 1 Paul Sabatier University, IRIT (CNRS/UMR 5505) 118 Route de Narbonne
2015 Partners in Excellence Executive Overview, Targets, and Methodology
 2015 Partners in Excellence Executive Overview, s, and Methodology Overview The Partners in Excellence program forms the basis for HealthPartners financial and public recognition for medical or specialty
2015 Partners in Excellence Executive Overview, s, and Methodology Overview The Partners in Excellence program forms the basis for HealthPartners financial and public recognition for medical or specialty
The National Cancer Institute's Thésaurus and Ontology
 The National Cancer Institute's Thésaurus and Ontology Jennifer Golbeck 1, Gilberto Fragoso 2, Frank Hartel 2, Jim Hendler 1, Jim Oberthaler 2, Bijan Parsia 1 1 University of Maryland, College Park 2 National
The National Cancer Institute's Thésaurus and Ontology Jennifer Golbeck 1, Gilberto Fragoso 2, Frank Hartel 2, Jim Hendler 1, Jim Oberthaler 2, Bijan Parsia 1 1 University of Maryland, College Park 2 National
Prototyping a Biomedical Ontology Recommender Service
 Prototyping a Biomedical Ontology Recommender Service Clement Jonquet Nigam H. Shah Mark A. Musen jonquet@stanford.edu 1 Ontologies & data & annota@ons (1/2) Hard for biomedical researchers to find the
Prototyping a Biomedical Ontology Recommender Service Clement Jonquet Nigam H. Shah Mark A. Musen jonquet@stanford.edu 1 Ontologies & data & annota@ons (1/2) Hard for biomedical researchers to find the
New Approach to Graph Databases
 Paper PP05 New Approach to Graph Databases Anna Berg, Capish, Malmö, Sweden Henrik Drews, Capish, Malmö, Sweden Catharina Dahlbo, Capish, Malmö, Sweden ABSTRACT Graph databases have, during the past few
Paper PP05 New Approach to Graph Databases Anna Berg, Capish, Malmö, Sweden Henrik Drews, Capish, Malmö, Sweden Catharina Dahlbo, Capish, Malmö, Sweden ABSTRACT Graph databases have, during the past few
A Tagging Approach to Ontology Mapping
 A Tagging Approach to Ontology Mapping Colm Conroy 1, Declan O'Sullivan 1, Dave Lewis 1 1 Knowledge and Data Engineering Group, Trinity College Dublin {coconroy,declan.osullivan,dave.lewis}@cs.tcd.ie Abstract.
A Tagging Approach to Ontology Mapping Colm Conroy 1, Declan O'Sullivan 1, Dave Lewis 1 1 Knowledge and Data Engineering Group, Trinity College Dublin {coconroy,declan.osullivan,dave.lewis}@cs.tcd.ie Abstract.
Model-Solver Integration in Decision Support Systems: A Web Services Approach
 Model-Solver Integration in Decision Support Systems: A Web Services Approach Keun-Woo Lee a, *, Soon-Young Huh a a Graduate School of Management, Korea Advanced Institute of Science and Technology 207-43
Model-Solver Integration in Decision Support Systems: A Web Services Approach Keun-Woo Lee a, *, Soon-Young Huh a a Graduate School of Management, Korea Advanced Institute of Science and Technology 207-43
FCA-based Search for Duplicate objects in Ontologies
 FCA-based Search for Duplicate objects in Ontologies Dmitry A. Ilvovsky and Mikhail A. Klimushkin National Research University Higher School of Economics, School of Applied Mathematics and Informational
FCA-based Search for Duplicate objects in Ontologies Dmitry A. Ilvovsky and Mikhail A. Klimushkin National Research University Higher School of Economics, School of Applied Mathematics and Informational
High Accuracy Information Retrieval and Information Extraction System for Electronic Clinical Notes
 High Accuracy Information Retrieval and Information Extraction System for Electronic Clinical Notes Jon Patrick and Min Li Health Information Technology Research Laboratory, School of IT The University
High Accuracy Information Retrieval and Information Extraction System for Electronic Clinical Notes Jon Patrick and Min Li Health Information Technology Research Laboratory, School of IT The University
Knowledge Retrieval. Franz J. Kurfess. Computer Science Department California Polytechnic State University San Luis Obispo, CA, U.S.A.
 Knowledge Retrieval Franz J. Kurfess Computer Science Department California Polytechnic State University San Luis Obispo, CA, U.S.A. 1 Acknowledgements This lecture series has been sponsored by the European
Knowledge Retrieval Franz J. Kurfess Computer Science Department California Polytechnic State University San Luis Obispo, CA, U.S.A. 1 Acknowledgements This lecture series has been sponsored by the European
Ontology Matching with CIDER: Evaluation Report for the OAEI 2008
 Ontology Matching with CIDER: Evaluation Report for the OAEI 2008 Jorge Gracia, Eduardo Mena IIS Department, University of Zaragoza, Spain {jogracia,emena}@unizar.es Abstract. Ontology matching, the task
Ontology Matching with CIDER: Evaluation Report for the OAEI 2008 Jorge Gracia, Eduardo Mena IIS Department, University of Zaragoza, Spain {jogracia,emena}@unizar.es Abstract. Ontology matching, the task
On Demand Phenotype Ranking through Subspace Clustering
 On Demand Phenotype Ranking through Subspace Clustering Xiang Zhang, Wei Wang Department of Computer Science University of North Carolina at Chapel Hill Chapel Hill, NC 27599, USA {xiang, weiwang}@cs.unc.edu
On Demand Phenotype Ranking through Subspace Clustering Xiang Zhang, Wei Wang Department of Computer Science University of North Carolina at Chapel Hill Chapel Hill, NC 27599, USA {xiang, weiwang}@cs.unc.edu
LexGrid Philosophy, Model and Interfaces Harold R Solbrig Division of Biomedical Statistics and Informatics Mayo Clinic
 LexGrid Philosophy, Model and Interfaces Harold R Solbrig Division of Biomedical Statistics and Informatics Mayo Clinic Outline Why the LexGrid model was created LexGrid approach and principles Key aspects
LexGrid Philosophy, Model and Interfaces Harold R Solbrig Division of Biomedical Statistics and Informatics Mayo Clinic Outline Why the LexGrid model was created LexGrid approach and principles Key aspects
Joint Entity Resolution
 Joint Entity Resolution Steven Euijong Whang, Hector Garcia-Molina Computer Science Department, Stanford University 353 Serra Mall, Stanford, CA 94305, USA {swhang, hector}@cs.stanford.edu No Institute
Joint Entity Resolution Steven Euijong Whang, Hector Garcia-Molina Computer Science Department, Stanford University 353 Serra Mall, Stanford, CA 94305, USA {swhang, hector}@cs.stanford.edu No Institute
is easing the creation of new ontologies by promoting the reuse of existing ones and automating, as much as possible, the entire ontology
 Preface The idea of improving software quality through reuse is not new. After all, if software works and is needed, just reuse it. What is new and evolving is the idea of relative validation through testing
Preface The idea of improving software quality through reuse is not new. After all, if software works and is needed, just reuse it. What is new and evolving is the idea of relative validation through testing
From Lexicon To Mammographic Ontology: Experiences and Lessons
 From Lexicon To Mammographic : Experiences and Lessons Bo Hu, Srinandan Dasmahapatra and Nigel Shadbolt Department of Electronics and Computer Science University of Southampton United Kingdom Email: {bh,
From Lexicon To Mammographic : Experiences and Lessons Bo Hu, Srinandan Dasmahapatra and Nigel Shadbolt Department of Electronics and Computer Science University of Southampton United Kingdom Email: {bh,
Chapter 6 Evaluation Metrics and Evaluation
 Chapter 6 Evaluation Metrics and Evaluation The area of evaluation of information retrieval and natural language processing systems is complex. It will only be touched on in this chapter. First the scientific
Chapter 6 Evaluation Metrics and Evaluation The area of evaluation of information retrieval and natural language processing systems is complex. It will only be touched on in this chapter. First the scientific
An FCA Framework for Knowledge Discovery in SPARQL Query Answers
 An FCA Framework for Knowledge Discovery in SPARQL Query Answers Melisachew Wudage Chekol, Amedeo Napoli To cite this version: Melisachew Wudage Chekol, Amedeo Napoli. An FCA Framework for Knowledge Discovery
An FCA Framework for Knowledge Discovery in SPARQL Query Answers Melisachew Wudage Chekol, Amedeo Napoli To cite this version: Melisachew Wudage Chekol, Amedeo Napoli. An FCA Framework for Knowledge Discovery
Navigating and Exploring RDF Data using Formal Concept Analysis
 Navigating and Exploring RDF Data using Formal Concept Analysis Mehwish Alam and Amedeo Napoli LORIA (CNRS Inria Nancy Grand Est Université de Lorraine) BP 239, 54506 Vandoeuvre-les-Nancy, France {mehwish.alam,amedeo.napoli@loria.fr}
Navigating and Exploring RDF Data using Formal Concept Analysis Mehwish Alam and Amedeo Napoli LORIA (CNRS Inria Nancy Grand Est Université de Lorraine) BP 239, 54506 Vandoeuvre-les-Nancy, France {mehwish.alam,amedeo.napoli@loria.fr}
Text Retrival for Medical Discharge Summaries using. SNOMED and Formal Concept Analysis. Abstract
 Text Retrival for Medical Discharge Summaries using SNOMED and Formal Concept Analysis R.J. Cole Department of Computer Science The University of Adelaide Adelaide 55, Australia rjcole@cs.adelaide.edu.au
Text Retrival for Medical Discharge Summaries using SNOMED and Formal Concept Analysis R.J. Cole Department of Computer Science The University of Adelaide Adelaide 55, Australia rjcole@cs.adelaide.edu.au
A method for recommending ontology alignment strategies
 A method for recommending ontology alignment strategies He Tan and Patrick Lambrix Department of Computer and Information Science Linköpings universitet, Sweden This is a pre-print version of the article
A method for recommending ontology alignment strategies He Tan and Patrick Lambrix Department of Computer and Information Science Linköpings universitet, Sweden This is a pre-print version of the article
Mining XML Functional Dependencies through Formal Concept Analysis
 Mining XML Functional Dependencies through Formal Concept Analysis Viorica Varga May 6, 2010 Outline Definitions for XML Functional Dependencies Introduction to FCA FCA tool to detect XML FDs Finding XML
Mining XML Functional Dependencies through Formal Concept Analysis Viorica Varga May 6, 2010 Outline Definitions for XML Functional Dependencies Introduction to FCA FCA tool to detect XML FDs Finding XML
Weights Conditions ICD-8 codes ICD-10 codes 1 Myocardial 410 I21; I22; I23. Dementia ; F00-F03; F05.1; G30 Chronic pulmonary
 Supplementary tables Table A. Charlson comorbidity index Weights Conditions ICD-8 codes ICD-10 codes 1 Myocardial 410 I21; I22; I23 infarction Congestive heart 427.09; 427.10; 427.11; 427.19; I50; I11.0;
Supplementary tables Table A. Charlson comorbidity index Weights Conditions ICD-8 codes ICD-10 codes 1 Myocardial 410 I21; I22; I23 infarction Congestive heart 427.09; 427.10; 427.11; 427.19; I50; I11.0;
Quality Data Model (QDM)
 Quality Data Model (QDM) Overview Document National Quality Forum 4/20/2011 QUALITY DATA MODEL (QDM): OVERVIEW TABLE OF CONTENTS Quality Data Model (QDM): Overview... 2 National Quality Forum: Overview
Quality Data Model (QDM) Overview Document National Quality Forum 4/20/2011 QUALITY DATA MODEL (QDM): OVERVIEW TABLE OF CONTENTS Quality Data Model (QDM): Overview... 2 National Quality Forum: Overview
WHO ICD11 Wiki LexWiki, Semantic MediaWiki and the International Classification of Diseases
 WHO ICD11 Wiki LexWiki, Semantic MediaWiki and the International Classification of Diseases Guoqian Jiang, PhD Harold Solbrig Division of Biomedical Statistics and Informatics Mayo Clinic College of Medicine
WHO ICD11 Wiki LexWiki, Semantic MediaWiki and the International Classification of Diseases Guoqian Jiang, PhD Harold Solbrig Division of Biomedical Statistics and Informatics Mayo Clinic College of Medicine
HealthCyberMap: Mapping the Health Cyberspace Using Hypermedia GIS and Clinical Codes
 HealthCyberMap: Mapping the Health Cyberspace Using Hypermedia GIS and Clinical Codes PhD Research Project Maged Nabih Kamel Boulos MBBCh, MSc (Derm & Vener), MSc (Medical Informatics) 1 Summary The application
HealthCyberMap: Mapping the Health Cyberspace Using Hypermedia GIS and Clinical Codes PhD Research Project Maged Nabih Kamel Boulos MBBCh, MSc (Derm & Vener), MSc (Medical Informatics) 1 Summary The application
2 Which Methodology for Building Ontologies? 2.1 A Work Still in Progress Many approaches (for a complete survey, the reader can refer to the OntoWeb
 Semantic Commitment for Designing Ontologies: A Proposal Bruno Bachimont 1,Antoine Isaac 1;2, Raphaël Troncy 1;3 1 Institut National de l'audiovisuel, Direction de la Recherche 4, Av. de l'europe - 94366
Semantic Commitment for Designing Ontologies: A Proposal Bruno Bachimont 1,Antoine Isaac 1;2, Raphaël Troncy 1;3 1 Institut National de l'audiovisuel, Direction de la Recherche 4, Av. de l'europe - 94366
The NIH Collaboratory Distributed Research Network: A Privacy Protecting Method for Sharing Research Data Sets
 The NIH Collaboratory Distributed Research Network: A Privacy Protecting Method for Sharing Research Data Sets Jeffrey Brown, Lesley Curtis, and Rich Platt June 13, 2014 Previously The NIH Collaboratory:
The NIH Collaboratory Distributed Research Network: A Privacy Protecting Method for Sharing Research Data Sets Jeffrey Brown, Lesley Curtis, and Rich Platt June 13, 2014 Previously The NIH Collaboratory:
i2b2 User Guide University of Minnesota Clinical and Translational Science Institute
 Clinical and Translational Science Institute i2b2 User Guide i2b2 is a tool for discovering research cohorts using existing, de-identified, clinical data This guide is provided by the Office of Biomedical
Clinical and Translational Science Institute i2b2 User Guide i2b2 is a tool for discovering research cohorts using existing, de-identified, clinical data This guide is provided by the Office of Biomedical
Keywords: clustering algorithms, unsupervised learning, cluster validity
 Volume 6, Issue 1, January 2016 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com Clustering Based
Volume 6, Issue 1, January 2016 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com Clustering Based
Implementing a Document Standard: Perspectives on Adoption, Interoperability, and Utilization
 Implementing a Document Standard: Perspectives on Adoption, Interoperability, and Utilization Health Level 7 Clinical Document Architecture: What is it? Who is using it? Why in ehealth? Scottish Health
Implementing a Document Standard: Perspectives on Adoption, Interoperability, and Utilization Health Level 7 Clinical Document Architecture: What is it? Who is using it? Why in ehealth? Scottish Health
7. SNOMED CT Expressions
 7. SNOMED CT Expressions This section provides an overview of: Precoordinated Expressions Postcoordinated Expressions Why is this important? SNOMED CT provides a mechanism that enables clinical phrases
7. SNOMED CT Expressions This section provides an overview of: Precoordinated Expressions Postcoordinated Expressions Why is this important? SNOMED CT provides a mechanism that enables clinical phrases
Chapter S:II. II. Search Space Representation
 Chapter S:II II. Search Space Representation Systematic Search Encoding of Problems State-Space Representation Problem-Reduction Representation Choosing a Representation S:II-1 Search Space Representation
Chapter S:II II. Search Space Representation Systematic Search Encoding of Problems State-Space Representation Problem-Reduction Representation Choosing a Representation S:II-1 Search Space Representation
Core Technology Development Team Meeting
 Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
YAM++ Results for OAEI 2013
 YAM++ Results for OAEI 2013 DuyHoa Ngo, Zohra Bellahsene University Montpellier 2, LIRMM {duyhoa.ngo, bella}@lirmm.fr Abstract. In this paper, we briefly present the new YAM++ 2013 version and its results
YAM++ Results for OAEI 2013 DuyHoa Ngo, Zohra Bellahsene University Montpellier 2, LIRMM {duyhoa.ngo, bella}@lirmm.fr Abstract. In this paper, we briefly present the new YAM++ 2013 version and its results
Formal Concept Analysis and Hierarchical Classes Analysis
 Formal Concept Analysis and Hierarchical Classes Analysis Yaohua Chen, Yiyu Yao Department of Computer Science, University of Regina Regina, Saskatchewan, Canada S4S 0A2 E-mail: {chen115y, yyao}@cs.uregina.ca
Formal Concept Analysis and Hierarchical Classes Analysis Yaohua Chen, Yiyu Yao Department of Computer Science, University of Regina Regina, Saskatchewan, Canada S4S 0A2 E-mail: {chen115y, yyao}@cs.uregina.ca
Multimorbidity Cluster Analysis Toolkit January 2016
 Multimorbidity Cluster Analysis Toolkit January 2016 1 P a g e Contents Background... 3 Development Of This Toolkit... 3 Who Should Use This Toolkit?.... 4 What Does This Toolkit Contain?.. 4 How Should
Multimorbidity Cluster Analysis Toolkit January 2016 1 P a g e Contents Background... 3 Development Of This Toolkit... 3 Who Should Use This Toolkit?.... 4 What Does This Toolkit Contain?.. 4 How Should
PyMedTermino: an open-source generic API for advanced terminology services
 924 Digital Healthcare Empowering Europeans R. Cornet et al. (Eds.) 2015 European Federation for Medical Informatics (EFMI). This article is published online with Open Access by IOS Press and distributed
924 Digital Healthcare Empowering Europeans R. Cornet et al. (Eds.) 2015 European Federation for Medical Informatics (EFMI). This article is published online with Open Access by IOS Press and distributed
BioNav: An Ontology-Based Framework to Discover Semantic Links in the Cloud of Linked Data
 BioNav: An Ontology-Based Framework to Discover Semantic Links in the Cloud of Linked Data María-Esther Vidal 1, Louiqa Raschid 2, Natalia Márquez 1, Jean Carlo Rivera 1, and Edna Ruckhaus 1 1 Universidad
BioNav: An Ontology-Based Framework to Discover Semantic Links in the Cloud of Linked Data María-Esther Vidal 1, Louiqa Raschid 2, Natalia Márquez 1, Jean Carlo Rivera 1, and Edna Ruckhaus 1 1 Universidad
BiTeM / SIB Text Mining group Geneva, Switzerland. IHE World Summit Amsterdam, June 7
 BiTeM / SIB Text Mining group Geneva, Switzerland IHE World Summit Amsterdam, June 7 The group Patrick Ruch, Fund raising Arnaud Gaudinat, Web IT Julien Gobeill, Computer sciences Igor Mihlit, Information
BiTeM / SIB Text Mining group Geneva, Switzerland IHE World Summit Amsterdam, June 7 The group Patrick Ruch, Fund raising Arnaud Gaudinat, Web IT Julien Gobeill, Computer sciences Igor Mihlit, Information
A Formalization of Transition P Systems
 Fundamenta Informaticae 49 (2002) 261 272 261 IOS Press A Formalization of Transition P Systems Mario J. Pérez-Jiménez and Fernando Sancho-Caparrini Dpto. Ciencias de la Computación e Inteligencia Artificial
Fundamenta Informaticae 49 (2002) 261 272 261 IOS Press A Formalization of Transition P Systems Mario J. Pérez-Jiménez and Fernando Sancho-Caparrini Dpto. Ciencias de la Computación e Inteligencia Artificial
Specification of a Cardiovascular Metadata Model: A Consensus Standard
 Specification of a Cardiovascular Metadata Model: A Consensus Standard Rebecca Wilgus, RN MSN 1 ; Dana Pinchotti 2 ; Salvatore Mungal 3 ; David F Kong, MD AM 1 ; James E Tcheng, MD 1 ; Brian McCourt 1
Specification of a Cardiovascular Metadata Model: A Consensus Standard Rebecca Wilgus, RN MSN 1 ; Dana Pinchotti 2 ; Salvatore Mungal 3 ; David F Kong, MD AM 1 ; James E Tcheng, MD 1 ; Brian McCourt 1
Knowledge Representations. How else can we represent knowledge in addition to formal logic?
 Knowledge Representations How else can we represent knowledge in addition to formal logic? 1 Common Knowledge Representations Formal Logic Production Rules Semantic Nets Schemata and Frames 2 Production
Knowledge Representations How else can we represent knowledge in addition to formal logic? 1 Common Knowledge Representations Formal Logic Production Rules Semantic Nets Schemata and Frames 2 Production
PopMedNet TM Distributed Networking Technologies for Population Medicine
 PopMedNet TM Distributed Networking Technologies for Population Medicine ONC Summer Concert Series on Distributed Population Queries Jeffrey Brown Richard Platt August 3, 2011 Department of Population
PopMedNet TM Distributed Networking Technologies for Population Medicine ONC Summer Concert Series on Distributed Population Queries Jeffrey Brown Richard Platt August 3, 2011 Department of Population
Text Retrival for Medical Discharge Summaries using SNOMED and Formal. Concept Analysis. R.J. Cole and P.W Eklund. The University of Adelaide
 Text Retrival for Medical Discharge Summaries using SNOMED and Formal Concept Analysis R.J. Cole and P.W Eklund Department of Computer Science The University of Adelaide Adelaide 5005, Australia ABSTRACT
Text Retrival for Medical Discharge Summaries using SNOMED and Formal Concept Analysis R.J. Cole and P.W Eklund Department of Computer Science The University of Adelaide Adelaide 5005, Australia ABSTRACT
Designing a self-medication application on Semantic Web technologies. Olivier Curé UPEM LIGM, France
 Designing a self-medication application on Semantic Web technologies Olivier Curé UPEM LIGM, France Overview Self-medication applications Symptom & Drug DB Overview Self-medication applications Symptom
Designing a self-medication application on Semantic Web technologies Olivier Curé UPEM LIGM, France Overview Self-medication applications Symptom & Drug DB Overview Self-medication applications Symptom
Racer: An OWL Reasoning Agent for the Semantic Web
 Racer: An OWL Reasoning Agent for the Semantic Web Volker Haarslev and Ralf Möller Concordia University, Montreal, Canada (haarslev@cs.concordia.ca) University of Applied Sciences, Wedel, Germany (rmoeller@fh-wedel.de)
Racer: An OWL Reasoning Agent for the Semantic Web Volker Haarslev and Ralf Möller Concordia University, Montreal, Canada (haarslev@cs.concordia.ca) University of Applied Sciences, Wedel, Germany (rmoeller@fh-wedel.de)
Geosemantically-enhanced PubMed Queries Using the Geonames Ontology and Web Services
 Geosemantically-enhanced PubMed Queries Using the Geonames Ontology and Web Services Maged N. Kamel Boulos, PhD, MSc, MBBCh Plymouth University, UK mnkboulos@ieee.org Agenda About PubMed and MeSH The Problem
Geosemantically-enhanced PubMed Queries Using the Geonames Ontology and Web Services Maged N. Kamel Boulos, PhD, MSc, MBBCh Plymouth University, UK mnkboulos@ieee.org Agenda About PubMed and MeSH The Problem
Method for the Mapping between Health Terminologies aiming Systems Interoperability
 Method for the Mapping between Health Terminologies aiming Systems Interoperability Thiago Fernandes de Freitas Dias Inter Bioengineering Postgraduate Program, School of Engineering of São Carlos/Faculty
Method for the Mapping between Health Terminologies aiming Systems Interoperability Thiago Fernandes de Freitas Dias Inter Bioengineering Postgraduate Program, School of Engineering of São Carlos/Faculty
Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology (DDI)
 ICBO 2013 Workshop on Vaccine and Drug Ontology Studies Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology (DDI) Zhe He 1 Christopher Ochs 1 Larisa
ICBO 2013 Workshop on Vaccine and Drug Ontology Studies Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology (DDI) Zhe He 1 Christopher Ochs 1 Larisa
RS TechMedic BV the Netherlands T: vision.com
 Dyna Vision Remote Patient Monitoring: Demonstration of Real Time 12 lead ECG Acquisition and Waveform Analysis using Integrated Cellular Transmission Telemonitoring Session Med e Tel 2010 Definitions
Dyna Vision Remote Patient Monitoring: Demonstration of Real Time 12 lead ECG Acquisition and Waveform Analysis using Integrated Cellular Transmission Telemonitoring Session Med e Tel 2010 Definitions
MEDINFO Panel II: Do we need a Common Ontology between ICD 11 and SNOMED CT to ensure seamless re-use and semantic interoperability?
 MEDINFO Panel II: Do we need a Common Ontology between ICD 11 and SNOMED CT to ensure seamless re-use and semantic interoperability? Common ontology between the Founda2on Component (FC) of ICD 11 and SNOMED
MEDINFO Panel II: Do we need a Common Ontology between ICD 11 and SNOMED CT to ensure seamless re-use and semantic interoperability? Common ontology between the Founda2on Component (FC) of ICD 11 and SNOMED
Investigating Collaboration Dynamics in Different Ontology Development Environments
 Investigating Collaboration Dynamics in Different Ontology Development Environments Marco Rospocher DKM, Fondazione Bruno Kessler rospocher@fbk.eu Tania Tudorache and Mark Musen BMIR, Stanford University
Investigating Collaboration Dynamics in Different Ontology Development Environments Marco Rospocher DKM, Fondazione Bruno Kessler rospocher@fbk.eu Tania Tudorache and Mark Musen BMIR, Stanford University
MIRACLE at ImageCLEFmed 2008: Evaluating Strategies for Automatic Topic Expansion
 MIRACLE at ImageCLEFmed 2008: Evaluating Strategies for Automatic Topic Expansion Sara Lana-Serrano 1,3, Julio Villena-Román 2,3, José C. González-Cristóbal 1,3 1 Universidad Politécnica de Madrid 2 Universidad
MIRACLE at ImageCLEFmed 2008: Evaluating Strategies for Automatic Topic Expansion Sara Lana-Serrano 1,3, Julio Villena-Román 2,3, José C. González-Cristóbal 1,3 1 Universidad Politécnica de Madrid 2 Universidad
EXPLOITING SEMANTICS FOR IMPROVING CLINICAL INFORMATION RETRIEVAL
 T EXPLOTNG SEMANTCS FOR MPROVNG CLNCAL NFORMATON RETREVAL Atanaz Babashzadeh A THESS SUBMTTED TO THE FACULTY OF GRADUATE STUDES N PARTAL FULFLMENT OF THE REQUREMENTS FOR THE DEGREE OF MASTER OF ARTS GRADUATE
T EXPLOTNG SEMANTCS FOR MPROVNG CLNCAL NFORMATON RETREVAL Atanaz Babashzadeh A THESS SUBMTTED TO THE FACULTY OF GRADUATE STUDES N PARTAL FULFLMENT OF THE REQUREMENTS FOR THE DEGREE OF MASTER OF ARTS GRADUATE
Optimised Classification for Taxonomic Knowledge Bases
 Optimised Classification for Taxonomic Knowledge Bases Dmitry Tsarkov and Ian Horrocks University of Manchester, Manchester, UK {tsarkov horrocks}@cs.man.ac.uk Abstract Many legacy ontologies are now being
Optimised Classification for Taxonomic Knowledge Bases Dmitry Tsarkov and Ian Horrocks University of Manchester, Manchester, UK {tsarkov horrocks}@cs.man.ac.uk Abstract Many legacy ontologies are now being
Entailment-based Text Exploration with Application to the Health-care Domain
 Entailment-based Text Exploration with Application to the Health-care Domain Meni Adler Bar Ilan University Ramat Gan, Israel adlerm@cs.bgu.ac.il Jonathan Berant Tel Aviv University Tel Aviv, Israel jonatha6@post.tau.ac.il
Entailment-based Text Exploration with Application to the Health-care Domain Meni Adler Bar Ilan University Ramat Gan, Israel adlerm@cs.bgu.ac.il Jonathan Berant Tel Aviv University Tel Aviv, Israel jonatha6@post.tau.ac.il
i2b2 User Guide Informatics for Integrating Biology & the Bedside Version 1.0 October 2012
 i2b2 (Informatics for Integrating Biology and the Bedside) is an informatics framework designed to simplify the process of using existing, de-identified, clinical data for preliminary research cohort discovery
i2b2 (Informatics for Integrating Biology and the Bedside) is an informatics framework designed to simplify the process of using existing, de-identified, clinical data for preliminary research cohort discovery
