Deliverable A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data. 1st September 2015
|
|
|
- Brent Casey
- 5 years ago
- Views:
Transcription
1 Deliverable Project ID Project Title A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data. Project Acronym Start Date of the Project PhenoMeNal 1st September 2015 Duration of the Project 36 Months Work Number Package 9 Work Title Package WP9 Tools, Workflows, Audit and Data Management Deliverable Title D9.5.1 Updated Preprocess Virtual Machine Image Delivery Date M32 Work leader Package IPB Contributing Partners IPB, ICL, EMBL-EBI, SIB Authors Evangelos Chandakas, Tim Ebbels, Pablo Moreno, Steffen Neumann, Kristian Peters, Rico Rueedi, Daniel Schober Abstract This deliverable reports on the development and use of container images to enable data producers to (pre-)process raw data into standard community-supported formats, locally
2 or in the cloud. This deliverable is an update to D9.2.1 PhenoMeNal-Preprocess VM. Table of Contents 1. Executive Summary Contribution towards the project objectives Detailed report on the deliverable Background MS Data Preprocessing NMR Data Preprocessing nmrmlconv Container User documentation and training BATMAN Delivery and Schedule Conclusion
3 1.Executive Summary The PhenoMeNal project supports several of the most common workflows in metabolomics covering, amongst others, NMR and Mass Spectrometry data processing. The diversity of instrument vendor-specific data formats requires preprocessing tools to convert data files from proprietary formats into standardized open formats for instance, converting raw MS and NMR data files into the mzml or nmrml formats, respectively. This deliverable reports on the development and use of container images to enable data producers to (pre-)process raw data into standard community-supported formats, locally or in the cloud. This deliverable is an update to D9.2.1 PhenoMeNal-Preprocess VM. 2. Contribution towards the project objectives The deliverable has contributed towards the following project objectives for WP9: Specify and integrate software pipelines and tools utilised in the PhenoMeNal e- Infrastructure into VMIs, adhering to data standards developed in WP8 and supporting the interoperability and federation middleware developed in WP5. We will develop new applications only to complete missing links in pipelines. We will use public repositories and continuous integration to always provide development snapshots of the infrastructure VMIs. Develop methods to scale-up software pipelines for high-throughput analysis, supporting distributed execution on e.g. local clusters, private clouds, federated clouds, or GRIDs. 3. Detailed report on the deliverable 3.1 Background The PhenoMeNal project supports several of the most common workflows in metabolomics, covering NMR, Mass Spectrometry and downstream statistical analysis. The diversity of instrument vendor-specific data formats creates incompatibilities between processing tools and strategies that can be avoided by using community-accepted open standard data formats starting from the instrument level. Hence, PhenoMeNal supports preprocessing tools to convert data files from proprietary formats into standardized open formats for instance, converting raw MS and NMR data files into the mzml or nmrml formats, respectively. 3
4 This deliverable is an update to D9.2.1 PhenoMeNal-Preprocess VM: Virtual Machine Images to enable data producers to locally process raw data into standard formats supported in PhenoMeNal MS Data Preprocessing For the conversion vendor formats to mzml we are using the open source msconvert, developed by the ProteoWizard team ( 2, which is one of the reference implementations for mzml. It can convert to mzml from Sciex, Bruker, Thermo, Agilent, Shimadzu, Waters and also the earlier file formats like mzdata or mzxml and is consequently widely used. MSconvert and the integration into the continuous integration Jenkins and as Galaxy tool have already been described in D Since then, we have especially worked on the container image. The container was rebased from the seemingly unmaintained container suchja/wine:dev with pre-installed WINE (v1.8.0) windows layer, to the i386/debian:stretch-backports image that is part of the docker-library/official-images collection with regular updates. This also allowed to move to the WINE version 3.6.0, supporting a wider range of windows applications and better stability. The build file was also cleaned up, where several workarounds related to the earlier WINE version could be removed. The Proteowizard version was updated from version to The Dockerfile was also updated to conform to the latest PhenoMeNal guidelines, i.e. has updated LABELs that are used in the Build process, on the Jenkins continuous integration 3 and in the app library 4. A new data testing strategy was added, where we have started to collect a range of different MS files in vendor formats, which are then converted. We then check that the converted mzml matched a known-good output as Jenkins job Chambers, Matthew C et al. "A cross-platform toolkit for mass spectrometry and proteomics." Nature biotechnology (2012):
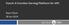
5 Figure 1: Screenshot of MSconvertGUI converting Thermo RAW data in a docker container on Ubuntu/Linux. It is now possible to also convert Thermo RAW files in this container using the GUI under Linux/X11, which is a long-standing wishlist item in the metabolomics community (see screenshot in Figure 1). However, it is not yet possible to perform this conversion also with the command line msconvert.exe tool. We are in contact with the Proteowizard team and identified the underlying reason, which lies in the complex interaction between VisualStudio 2012 C++ and the windows CLR runtime used in the msconvert.exe tool and vendor libraries. At the HUPO PSI 2018 meeting ( ) in Heidelberg (Germany), we were in contact with Jim Shofstahl from Thermo Scientific. We have received a version of the Thermo RawFileReader that is based on.net, and avoids the C++ runtime issues under WINE/Linux. We have created a local container- RawFileReader successfully running a command line program that extracts required information from the Thermo RAW file under linux, and informed the Proteowizard team. We can hence expect that an upcoming version of proteowizard will be able to successfully convert also Thermo data on the command line, and henceforth also in Galaxy.. We also worked on the license compliance. Due to the nature of the container registries and the kubernetes cloud setup it is not possible to request acknowledgement of the license terms during a download step, as is the case for the Proteowizard binaries 6. Instead, we are using a mechanism that is also used in the pwiz build process, where adding --i-agree-to-the-vendor-licenses while building will acknowledge the vendor licenses. With this analogy, the resulting container is called phnmnl/pwiz--i-agree-to-thevendor-licenses. All licensing information was added to the container repository and the container itself
6 Several tools in the PhenoMeNal workflows can process metabolomics data in mzml, including XCMS, CAMERA and MetFrag (via MSnbase), and the OpenMS tools. 3.3 NMR Data Preprocessing The nmrml standard is an open XML-based exchange and storage format for NMR spectral data. The nmrml format is intended to be fully compatible with existing NMR data for chemical, biochemical, and metabolomics experiments. nmrml can capture raw NMR data, spectral data acquisition parameters, and where available spectral metadata, such as chemical structures associated with spectral assignments, see Figure 2 for an overview of the role of nmrml. The nmrml format is compatible with pure-compound NMR data for reference spectral libraries as well as NMR data from complex biomixtures, i.e., metabolomics experiments. The manuscript nmrml: A Community Supported Open Data Standard for the Description, Storage, and Exchange of NMR Data has been published recently 7. Figure 2: workflow of nmrml data flow. The main route to nmrml-formatted data is using our open source converter nmrmlconv, which is part of the nmrml package. The converter is also packaged as a container image, together with the required Java runtime environment, and is available as a Galaxy tool
7 Since the Deliverable D9.2.1, we have updated the container image to the latest version of nmrmlconv and we have created a Galaxy module and a Galaxy workflow that uses nmrmlconv to process NMR RAW data, see Figure 3. The module can be used with both singular and many NMR RAW acquisitions that are bundled in a dataset collection. When processing the nmrmlconv module, the Galaxy workflow engine starts the nmrmlconvcontainer 8, which is launched within the PhenoMeNal e-infrastructure and is further described below. In the process, we have raised several github issues 9 to further improve the nmrml converter. Figure 3: Screenshot of the NMR workflow running in Galaxy. The module nmrmlconv has been emphasized and is now integral part of the NMR RAW processing workflow nmrmlconv Container The nmrmlconv container itself integrates the official nmrml converter sources 10 and packages them into a docker container. This container is launched within the PhenoMeNal e-infrastructure. In order to ensure that NMR RAW data is processed reproducibly, our Continuous Integration Framework automatically tests the container for
8 launching correctly 11 and with actual data 12. The nmrmlconv currently can process NMR RAW data from any of Bruker, Varian and Jeol vendor formats. The vendor to nmrml converter itself was annotated via the EDAM ontology and published under bio.tools repository (see Bio.tools now enables to find this tool via appropriate controlled vocabularies describing a tools function and I/O formats. We are also currently investigating how we can add/use nmrml as vendor independent NMR raw data standard within the newly emerging NMReData Record.zip NMR assignment data standard (see This standard could gain momentum at the late stages of NMR processing workflows, as it is supported by many molecule to spectrum feature assignment tools (e.g. mnova, TopSpin, ) and small molecule databases (e.g. NMRShiftDB, C6H6.org,...). The NMReData standard currently stores the vendor native formats in the Record.zip file, which impairs cross-vendor data access, e.g. as desired in molecule analytics databases. Repository providers (e.g. the drug data repository) have indicated interest to switch to nmrml due to that drawback and would therefore welcome nmrml inclusion User documentation and training The nmrml conversion is part of the NMR Workflow MTBLS1 Tutorial 13, was featured in a YouTube tutorial 14, and covered in the workshops CloudMET 15 and at the MetaboMeeting
9 Figure 4: User tutorial for the NMR workflow including the nmrml conversion BATMAN Bayesian AuTtomated Metabolite ANalyzer (BATMAN) for NMR is a protocol for automated metabolite deconvolution and quantification from complex NMR spectra. BATMAN deconvolves resonances from 1-dimensional NMR spectra and assigns them to specific metabolites from a target list and obtains concentration estimates. It applies a Markov Chain Monte Carlo (MCMC) algorithm to sample from a joint posterior distribution of the model parameters and obtains concentration estimates with reduced error compared with conventional numerical integration and comparable to manual deconvolution by experienced spectroscopists. BATMAN is available for PhenoMeNal galaxy users. The BATMAN workflow performs a 1D NMR spectra analysis using NMR raw data, coming from e.g. the MetaboLights database. The user can connect to this database via PhenoMeNal galaxy and can import data using the tool Metabolights downloader. Then, the raw NMR data is converted to multiple nmrml files using the tool nmrmlconv. As a next step, using the tool ZIP nmrml Collection a zip archive of a nmrml collection is created, which is imported via the nmrml2batman Converter, followed by the 9
10 remaining BATMAN data processing workflow, which will be covered in the upcoming D9.5.2 Updated Data Processing. Figure 5: NMR workflow that includes the nmrml2batman conversion and the BATMAN tool. 4. Delivery and Schedule The delivery is delayed: No 5. Conclusion In PhenoMeNal, we have worked to improve the preprocessing of metabolomics data, which starts with the conversion of raw vendor file formats to open formats supported in PhenoMeNal. We are now supporting the conversion to the mass spectrometry data format mzml and to nmrml on non-linux systems. Our efforts have been acknowledged by other developer in the mass spectrometry and workflow community. 10
A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data. 1st September 2015
 Deliverable 8.4.1 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype
Deliverable 8.4.1 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype
Deliverable 8.2. Project ID Project Title. Project Acronym. Start Date of the Project. Duration of the Project. Work Package Number 8
 Deliverable 8.2 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype
Deliverable 8.2 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype
Real converters, parsers & validators for NMR-ML. Standards Development. WP leader: Steffen Neumann IPB
 Deliverable D2.5 Project Title: Developing an efficient e-infrastructure, standards and dataflow for metabolomics and its interface to biomedical and life science e-infrastructures in Europe and world-wide
Deliverable D2.5 Project Title: Developing an efficient e-infrastructure, standards and dataflow for metabolomics and its interface to biomedical and life science e-infrastructures in Europe and world-wide
Deliverable A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data.
 Project ID 654241 Deliverable 9.2.3 Project Title Project Acronym A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data. PhenoMeNal Start Date of the Project
Project ID 654241 Deliverable 9.2.3 Project Title Project Acronym A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data. PhenoMeNal Start Date of the Project
Deliverable 6.3. A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data. 1st September 2015
 Deliverable 6.3 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project Work Package Number Work Package Title Deliverable Title Delivery Date Work Package leader
Deliverable 6.3 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project Work Package Number Work Package Title Deliverable Title Delivery Date Work Package leader
Deliverable 5.2. A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data. 1st September 2015
 Deliverable 5.2 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project Work Package Number Work Package Title Deliverable Title Delivery Date Work Package leader
Deliverable 5.2 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project Work Package Number Work Package Title Deliverable Title Delivery Date Work Package leader
Data Processing for Small Molecules
 Data Processing for Small Molecules Basic Metabolomics Workflows Metabolomics: the apogee of the omics trilogy Gary J. Patti, Oscar Yanes and Gary Siuzdak Molecular Cell Biology, 2012, 13, 263-269 MATTHEW
Data Processing for Small Molecules Basic Metabolomics Workflows Metabolomics: the apogee of the omics trilogy Gary J. Patti, Oscar Yanes and Gary Siuzdak Molecular Cell Biology, 2012, 13, 263-269 MATTHEW
Deliverable 8.3. A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data. 1st September 2015
 Deliverable 8.3 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project Work Package Number Work Package Title Deliverable Title Delivery Date Work Package leader
Deliverable 8.3 Project ID 654241 Project Title Project Acronym Start Date of the Project Duration of the Project Work Package Number Work Package Title Deliverable Title Delivery Date Work Package leader
MRMPROBS tutorial. Hiroshi Tsugawa RIKEN Center for Sustainable Resource Science
 MRMPROBS tutorial Edited in 2014/10/6 Introduction MRMPROBS is a tool for the analysis of data from multiple reaction monitoring (MRM)- or selected reaction monitoring (SRM)-based metabolomics studies.
MRMPROBS tutorial Edited in 2014/10/6 Introduction MRMPROBS is a tool for the analysis of data from multiple reaction monitoring (MRM)- or selected reaction monitoring (SRM)-based metabolomics studies.
MRMPROBS tutorial. Hiroshi Tsugawa RIKEN Center for Sustainable Resource Science MRMPROBS screenshot
 MRMPROBS tutorial Edited in 2016/11/16 Introduction MRMPROBS is launched as a universal program for targeted metabolomics using not only multiple reaction monitoring (MRM)- or selected reaction monitoring
MRMPROBS tutorial Edited in 2016/11/16 Introduction MRMPROBS is launched as a universal program for targeted metabolomics using not only multiple reaction monitoring (MRM)- or selected reaction monitoring
Package msdata. October 2, 2018
 Version 0.21.0 Package msdata October 2, 2018 Title Various Mass Spectrometry raw data example files Author Steffen Neumann , Laurent Gatto with contriutions from
Version 0.21.0 Package msdata October 2, 2018 Title Various Mass Spectrometry raw data example files Author Steffen Neumann , Laurent Gatto with contriutions from
Windocks Technical Backgrounder
 Windocks Technical Backgrounder Windocks is a port of Docker s open source to Windows used to modernize SQL Server workflows. Windocks is also an open, modern, data delivery solution that sources data
Windocks Technical Backgrounder Windocks is a port of Docker s open source to Windows used to modernize SQL Server workflows. Windocks is also an open, modern, data delivery solution that sources data
Technical Computing with MATLAB
 Technical Computing with MATLAB University Of Bath Seminar th 19 th November 2010 Adrienne James (Application Engineering) 1 Agenda Introduction to MATLAB Importing, visualising and analysing data from
Technical Computing with MATLAB University Of Bath Seminar th 19 th November 2010 Adrienne James (Application Engineering) 1 Agenda Introduction to MATLAB Importing, visualising and analysing data from
Metadata Models for Experimental Science Data Management
 Metadata Models for Experimental Science Data Management Brian Matthews Facilities Programme Manager Scientific Computing Department, STFC Co-Chair RDA Photon and Neutron Science Interest Group Task lead,
Metadata Models for Experimental Science Data Management Brian Matthews Facilities Programme Manager Scientific Computing Department, STFC Co-Chair RDA Photon and Neutron Science Interest Group Task lead,
Skyline High Resolution Metabolomics (Draft)
 Skyline High Resolution Metabolomics (Draft) The Skyline Targeted Proteomics Environment provides informative visual displays of the raw mass spectrometer data you import into your Skyline documents. Originally
Skyline High Resolution Metabolomics (Draft) The Skyline Targeted Proteomics Environment provides informative visual displays of the raw mass spectrometer data you import into your Skyline documents. Originally
Default output format. Conversi on software. Conversion output formats
 Machine Vendor Manufact urer software Default output format Conversi on software Conversion output formats UPLC Waters Masslynx.RAW directory Wolf *.mzxml, *.cdf.raw QTOF Waters Masslynx directory Wolf
Machine Vendor Manufact urer software Default output format Conversi on software Conversion output formats UPLC Waters Masslynx.RAW directory Wolf *.mzxml, *.cdf.raw QTOF Waters Masslynx directory Wolf
The OpenMS Developers
 User Tutorial The OpenMS Developers Creative Commons Attribution 4.0 International (CC BY 4.0) Contents 1 General remarks 6 2 Getting started 7 2.1 Installation.................................... 7 2.1.1
User Tutorial The OpenMS Developers Creative Commons Attribution 4.0 International (CC BY 4.0) Contents 1 General remarks 6 2 Getting started 7 2.1 Installation.................................... 7 2.1.1
TEXT MINING: THE NEXT DATA FRONTIER
 TEXT MINING: THE NEXT DATA FRONTIER An Infrastructural Approach Dr. Petr Knoth CORE (core.ac.uk) Knowledge Media institute, The Open University United Kingdom 2 OpenMinTeD Establish an open and sustainable
TEXT MINING: THE NEXT DATA FRONTIER An Infrastructural Approach Dr. Petr Knoth CORE (core.ac.uk) Knowledge Media institute, The Open University United Kingdom 2 OpenMinTeD Establish an open and sustainable
CompassXport CompassXport
 1 Overview... 2 1.1 Introduction... 2 1.2 License Agreement... 2 2 Manuals... 2 3 Site Specifications... 2 3.1 Operating Systems... 2 4 Installation... 3 4.1 First time Installation... 3 4.2 Upgrade from
1 Overview... 2 1.1 Introduction... 2 1.2 License Agreement... 2 2 Manuals... 2 3 Site Specifications... 2 3.1 Operating Systems... 2 4 Installation... 3 4.1 First time Installation... 3 4.2 Upgrade from
Containerizing GPU Applications with Docker for Scaling to the Cloud
 Containerizing GPU Applications with Docker for Scaling to the Cloud SUBBU RAMA FUTURE OF PACKAGING APPLICATIONS Turns Discrete Computing Resources into a Virtual Supercomputer GPU Mem Mem GPU GPU Mem
Containerizing GPU Applications with Docker for Scaling to the Cloud SUBBU RAMA FUTURE OF PACKAGING APPLICATIONS Turns Discrete Computing Resources into a Virtual Supercomputer GPU Mem Mem GPU GPU Mem
Part 3: Setup of Client PC
 Version 2.2, January 16, 2013 Contents 1. Introduction... 3 1.1 Requirements of Client PC... 3 1.2 Flow Chart of Client PC Setup... 4 2. Setting of Windows OS... 5 2.1 Setting to Display File Type Extension
Version 2.2, January 16, 2013 Contents 1. Introduction... 3 1.1 Requirements of Client PC... 3 1.2 Flow Chart of Client PC Setup... 4 2. Setting of Windows OS... 5 2.1 Setting to Display File Type Extension
NMRProcFlow Macro-command Reference Guide
 NMRProcFlow Macro-command Reference Guide This document is the reference guide of the macro-commands Daniel Jacob UMR 1332 BFP, Metabolomics Facility CGFB Bordeaux, MetaboHUB - 2018 1 NMRProcFlow - Macro-command
NMRProcFlow Macro-command Reference Guide This document is the reference guide of the macro-commands Daniel Jacob UMR 1332 BFP, Metabolomics Facility CGFB Bordeaux, MetaboHUB - 2018 1 NMRProcFlow - Macro-command
MassBank for NORMAN. Achievements in 2011 and further proposed steps for identification of unknowns
 MassBank for NRMAN Achievements in 2011 and further proposed steps for identification of unknowns Tobias Schulze NRMAN GA 2011, Stockholm 22-23 November 2011 tobias.schulze@ufz.de Background and objective
MassBank for NRMAN Achievements in 2011 and further proposed steps for identification of unknowns Tobias Schulze NRMAN GA 2011, Stockholm 22-23 November 2011 tobias.schulze@ufz.de Background and objective
OpenShift Roadmap Enterprise Kubernetes for Developers. Clayton Coleman, Architect, OpenShift
 OpenShift Roadmap Enterprise Kubernetes for Developers Clayton Coleman, Architect, OpenShift What Is OpenShift? Application-centric Platform INFRASTRUCTURE APPLICATIONS Use containers for efficiency Hide
OpenShift Roadmap Enterprise Kubernetes for Developers Clayton Coleman, Architect, OpenShift What Is OpenShift? Application-centric Platform INFRASTRUCTURE APPLICATIONS Use containers for efficiency Hide
Package Risa. November 28, 2017
 Version 1.20.0 Date 2013-08-15 Package R November 28, 2017 Title Converting experimental metadata from ISA-tab into Bioconductor data structures Author Alejandra Gonzalez-Beltran, Audrey Kauffmann, Steffen
Version 1.20.0 Date 2013-08-15 Package R November 28, 2017 Title Converting experimental metadata from ISA-tab into Bioconductor data structures Author Alejandra Gonzalez-Beltran, Audrey Kauffmann, Steffen
The OpenVX Computer Vision and Neural Network Inference
 The OpenVX Computer and Neural Network Inference Standard for Portable, Efficient Code Radhakrishna Giduthuri Editor, OpenVX Khronos Group radha.giduthuri@amd.com @RadhaGiduthuri Copyright 2018 Khronos
The OpenVX Computer and Neural Network Inference Standard for Portable, Efficient Code Radhakrishna Giduthuri Editor, OpenVX Khronos Group radha.giduthuri@amd.com @RadhaGiduthuri Copyright 2018 Khronos
Pathway Analysis of Untargeted Metabolomics Data using the MS Peaks to Pathways Module
 Pathway Analysis of Untargeted Metabolomics Data using the MS Peaks to Pathways Module By: Jasmine Chong, Jeff Xia Date: 14/02/2018 The aim of this tutorial is to demonstrate how the MS Peaks to Pathways
Pathway Analysis of Untargeted Metabolomics Data using the MS Peaks to Pathways Module By: Jasmine Chong, Jeff Xia Date: 14/02/2018 The aim of this tutorial is to demonstrate how the MS Peaks to Pathways
RMassBank: Run-through the Principles and Workflow in R
 RMassBank: Run-through the Principles and Workflow in R Emma Schymanski Michael Stravs, Heinz Singer & Juliane Hollender Eawag, Dübendorf, Switzerland Steffen Neumann, Erik Müller: IPB Halle, Germany Tobias
RMassBank: Run-through the Principles and Workflow in R Emma Schymanski Michael Stravs, Heinz Singer & Juliane Hollender Eawag, Dübendorf, Switzerland Steffen Neumann, Erik Müller: IPB Halle, Germany Tobias
A Parser for mzxml, mzdata and mzml files
 A Parser for mzxml, mzdata and mzml files Bernd Fischer Steffen Neumann Laurent Gatto December 23, 2012 Contents 1 Introduction 1 2 Mass spectrometry raw data 2 2.1 Spectral data access.........................
A Parser for mzxml, mzdata and mzml files Bernd Fischer Steffen Neumann Laurent Gatto December 23, 2012 Contents 1 Introduction 1 2 Mass spectrometry raw data 2 2.1 Spectral data access.........................
Orchestrating the Continuous Delivery Process
 Orchestrating the Continuous Delivery Process steven.g.harris@cloudbees.com @stevengharris SVP Products, CloudBees 1 Continuous Delivery Overview Feedback Loop App Lifecycle BUILD TEST STAGE Deploy Run
Orchestrating the Continuous Delivery Process steven.g.harris@cloudbees.com @stevengharris SVP Products, CloudBees 1 Continuous Delivery Overview Feedback Loop App Lifecycle BUILD TEST STAGE Deploy Run
Azure DevOps. Randy Pagels Intelligent Cloud Technical Specialist Great Lakes Region
 Azure DevOps Randy Pagels Intelligent Cloud Technical Specialist Great Lakes Region What is DevOps? People. Process. Products. Build & Test Deploy DevOps is the union of people, process, and products to
Azure DevOps Randy Pagels Intelligent Cloud Technical Specialist Great Lakes Region What is DevOps? People. Process. Products. Build & Test Deploy DevOps is the union of people, process, and products to
NORMAN MassBank and beyond Status and future of activities regarding mass spectral databases
 NORMAN MassBank and beyond Status and future of activities regarding mass spectral databases NORMAN Annual General Assembly Meeting Vienna, 30 November - 01 December 2016 commons.wikimedia.org Tobias Schulze,
NORMAN MassBank and beyond Status and future of activities regarding mass spectral databases NORMAN Annual General Assembly Meeting Vienna, 30 November - 01 December 2016 commons.wikimedia.org Tobias Schulze,
MilkyWay Proteomics Documentation
 MilkyWay Proteomics Documentation Release alpha1 William D. Barshop, Hee Jong Kim, James A. Wohlschlegel Mar 25, 2018 Contents: 1 Uploading files to MilkyWay via the R/Shiny interface 1 1.1 Accessing
MilkyWay Proteomics Documentation Release alpha1 William D. Barshop, Hee Jong Kim, James A. Wohlschlegel Mar 25, 2018 Contents: 1 Uploading files to MilkyWay via the R/Shiny interface 1 1.1 Accessing
H2020 RIA COMANOID H2020-RIA
 H2020 RIA COMANOID H2020-RIA-645097 Deliverable D5.1: Setup of the communication infrastructure M2 D5.1 H2020-RIA-645097 COMANOID M2 Project acronym: Project full title: COMANOID Multi-Contact Collaborative
H2020 RIA COMANOID H2020-RIA-645097 Deliverable D5.1: Setup of the communication infrastructure M2 D5.1 H2020-RIA-645097 COMANOID M2 Project acronym: Project full title: COMANOID Multi-Contact Collaborative
Continuous Delivery of Micro Applications with Jenkins, Docker & Kubernetes at Apollo
 Continuous Delivery of Micro Applications with Jenkins, Docker & Kubernetes at Apollo Ulrich Häberlein Team Manager Backend Systems Apollo-Optik Holding GmbH & Co KG Michael Steinfurth Linux / Unix Consultant
Continuous Delivery of Micro Applications with Jenkins, Docker & Kubernetes at Apollo Ulrich Häberlein Team Manager Backend Systems Apollo-Optik Holding GmbH & Co KG Michael Steinfurth Linux / Unix Consultant
Deliverable D5.5. D5.5 VRE-integrated PDBe Search and Query API. World-wide E-infrastructure for structural biology. Grant agreement no.
 Deliverable D5.5 Project Title: World-wide E-infrastructure for structural biology Project Acronym: West-Life Grant agreement no.: 675858 Deliverable title: D5.5 VRE-integrated PDBe Search and Query API
Deliverable D5.5 Project Title: World-wide E-infrastructure for structural biology Project Acronym: West-Life Grant agreement no.: 675858 Deliverable title: D5.5 VRE-integrated PDBe Search and Query API
Red Hat Roadmap for Containers and DevOps
 Red Hat Roadmap for Containers and DevOps Brian Gracely, Director of Strategy Diogenes Rettori, Principal Product Manager Red Hat September, 2016 Digital Transformation Requires an evolution in... 2 APPLICATIONS
Red Hat Roadmap for Containers and DevOps Brian Gracely, Director of Strategy Diogenes Rettori, Principal Product Manager Red Hat September, 2016 Digital Transformation Requires an evolution in... 2 APPLICATIONS
Package batman. June 19, 2016
 Version 1.2.1.08 Author Jie Hao, William Astle, Maria De Iorio, Timothy Ebbels Maintainer Jie Hao Package batman June 19, 2016 Title Bayesian AuTomated Metabolite Analyser for NMR spectra
Version 1.2.1.08 Author Jie Hao, William Astle, Maria De Iorio, Timothy Ebbels Maintainer Jie Hao Package batman June 19, 2016 Title Bayesian AuTomated Metabolite Analyser for NMR spectra
Preprocessing, Management, and Analysis of Mass Spectrometry Proteomics Data
 Preprocessing, Management, and Analysis of Mass Spectrometry Proteomics Data * Mario Cannataro University Magna Græcia of Catanzaro, Italy cannataro@unicz.it * Joint work with P. H. Guzzi, T. Mazza, P.
Preprocessing, Management, and Analysis of Mass Spectrometry Proteomics Data * Mario Cannataro University Magna Græcia of Catanzaro, Italy cannataro@unicz.it * Joint work with P. H. Guzzi, T. Mazza, P.
Chromatography Data System
 Chromatography Data System Single Platform - Simply Powerful CompassCDS is SCION Instruments universal Chromatography Data System designed for instrument control, data acquisition, processing and reporting.
Chromatography Data System Single Platform - Simply Powerful CompassCDS is SCION Instruments universal Chromatography Data System designed for instrument control, data acquisition, processing and reporting.
FuncX: A Function Serving Platform for HPC. Ryan Chard 28 Jan 2019
 FuncX: A Function Serving Platform for HPC Ryan Chard 28 Jan 2019 Outline - Motivation FuncX: FaaS for HPC Implementation status Preliminary applications - Machine learning inference Automating analysis
FuncX: A Function Serving Platform for HPC Ryan Chard 28 Jan 2019 Outline - Motivation FuncX: FaaS for HPC Implementation status Preliminary applications - Machine learning inference Automating analysis
Skyline MS1 Full Scan Filtering
 Skyline MS1 Full Scan Filtering The Skyline Targeted Proteomics Environment provides informative visual displays of the raw mass spectrometer data you import into your Skyline project. These displays allow
Skyline MS1 Full Scan Filtering The Skyline Targeted Proteomics Environment provides informative visual displays of the raw mass spectrometer data you import into your Skyline project. These displays allow
CONSIDERATIONS FOR YOUR NEXT CLOUD PROJECT CLOUDFORMS & OPENSTACK DO S AND DON TS
 CONSIDERATIONS FOR YOUR NEXT CLOUD PROJECT CLOUDFORMS & OPENSTACK DO S AND DON TS FREDERIK BIJLSMA Cloud Business Unit Manager, EMEA 6th December 2013 VIRTUALIZATION TO CLOUD CONTINUUM Server Virtualization
CONSIDERATIONS FOR YOUR NEXT CLOUD PROJECT CLOUDFORMS & OPENSTACK DO S AND DON TS FREDERIK BIJLSMA Cloud Business Unit Manager, EMEA 6th December 2013 VIRTUALIZATION TO CLOUD CONTINUUM Server Virtualization
Welcome to Docker Birthday # Docker Birthday events (list available at Docker.Party) RSVPs 600 mentors Big thanks to our global partners:
 Docker Birthday #3 Welcome to Docker Birthday #3 2 120 Docker Birthday events (list available at Docker.Party) 7000+ RSVPs 600 mentors Big thanks to our global partners: Travel Planet 24 e-food.gr The
Docker Birthday #3 Welcome to Docker Birthday #3 2 120 Docker Birthday events (list available at Docker.Party) 7000+ RSVPs 600 mentors Big thanks to our global partners: Travel Planet 24 e-food.gr The
Dataset-XML - A New CDISC Standard
 Dataset-XML - A New CDISC Standard Lex Jansen Principal Software Developer @ SAS CDISC XML Technologies Team Single Day Event CDISC Tools and Optimization September 29, 2014, Cary, NC Agenda Dataset-XML
Dataset-XML - A New CDISC Standard Lex Jansen Principal Software Developer @ SAS CDISC XML Technologies Team Single Day Event CDISC Tools and Optimization September 29, 2014, Cary, NC Agenda Dataset-XML
Near real-time processing of proteomics data using Hadoop. Hillman, Chris; Ahmad, Yasmeen; Whitehorn, Mark; Cobley, Andrew
 University of Dundee Near real-time processing of proteomics data using Hadoop. Hillman, Chris; Ahmad, Yasmeen; Whitehorn, Mark; Cobley, Andrew Published in: Big Data DOI: 10.1089/big.2013.0036 Publication
University of Dundee Near real-time processing of proteomics data using Hadoop. Hillman, Chris; Ahmad, Yasmeen; Whitehorn, Mark; Cobley, Andrew Published in: Big Data DOI: 10.1089/big.2013.0036 Publication
Medici for Digital Cultural Heritage Libraries. George Tsouloupas, PhD The LinkSCEEM Project
 Medici for Digital Cultural Heritage Libraries George Tsouloupas, PhD The LinkSCEEM Project Overview of Digital Libraries A Digital Library: "An informal definition of a digital library is a managed collection
Medici for Digital Cultural Heritage Libraries George Tsouloupas, PhD The LinkSCEEM Project Overview of Digital Libraries A Digital Library: "An informal definition of a digital library is a managed collection
MSFragger Manual. (build )
 MSFragger Manual (build 20170103.0) Introduction MSFragger is an ultrafast database search tool for peptide identifications in mass spectrometry-based proteomics. It differs from conventional search engines
MSFragger Manual (build 20170103.0) Introduction MSFragger is an ultrafast database search tool for peptide identifications in mass spectrometry-based proteomics. It differs from conventional search engines
DevOps and Continuous Delivery USE CASE
 DevOps and Continuous Delivery USE CASE CliQr DevOps and Continuous Delivery Page 2 DevOps and Continuous Delivery In the digital economy, increasing application velocity is key to success. In order to
DevOps and Continuous Delivery USE CASE CliQr DevOps and Continuous Delivery Page 2 DevOps and Continuous Delivery In the digital economy, increasing application velocity is key to success. In order to
Comply with Data Integrity Regulations with Chromeleon CDS Software
 Comply with Data Integrity Regulations with Chromeleon CDS Software Anna Severoni Sales Support Specialist for Chromatography Thermo Fisher Scientific, Rodano (MI) The world leader in serving science Introduction
Comply with Data Integrity Regulations with Chromeleon CDS Software Anna Severoni Sales Support Specialist for Chromatography Thermo Fisher Scientific, Rodano (MI) The world leader in serving science Introduction
Cloud interoperability and elasticity with COMPSs
 www.bsc.es Cloud interoperability and elasticity with COMPSs Interoperability Demo Days Dec 12-2014, London Daniele Lezzi Barcelona Supercomputing Center Outline COMPSs programming model COMPSs tools COMPSs
www.bsc.es Cloud interoperability and elasticity with COMPSs Interoperability Demo Days Dec 12-2014, London Daniele Lezzi Barcelona Supercomputing Center Outline COMPSs programming model COMPSs tools COMPSs
ProteomeXchange Submission Summary File Format Version (3 September 2014)
 ProteomeXchange Submission Summary File Format Version 2.2.0 (3 September 2014) 1 Overview... 1 2 Format specification... 1 2.1 Sections... 1 2.2 Use of Controlled Vocabulary (CV) terms... 2 2.3 Project
ProteomeXchange Submission Summary File Format Version 2.2.0 (3 September 2014) 1 Overview... 1 2 Format specification... 1 2.1 Sections... 1 2.2 Use of Controlled Vocabulary (CV) terms... 2 2.3 Project
TEN LAYERS OF CONTAINER SECURITY
 TEN LAYERS OF CONTAINER SECURITY Tim Hunt Kirsten Newcomer May 2017 ABOUT YOU Are you using containers? What s your role? Security professionals Developers / Architects Infrastructure / Ops Who considers
TEN LAYERS OF CONTAINER SECURITY Tim Hunt Kirsten Newcomer May 2017 ABOUT YOU Are you using containers? What s your role? Security professionals Developers / Architects Infrastructure / Ops Who considers
FCC An automated rule-based processing tool for life science data
 Barkow-Oesterreicher et al. Source Code for Biology and Medicine 2013, 8:3 SOFTWARE REVIEW Open Access FCC An automated rule-based processing tool for life science data Simon Barkow-Oesterreicher, Can
Barkow-Oesterreicher et al. Source Code for Biology and Medicine 2013, 8:3 SOFTWARE REVIEW Open Access FCC An automated rule-based processing tool for life science data Simon Barkow-Oesterreicher, Can
Red Hat Atomic Details Dockah, Dockah, Dockah! Containerization as a shift of paradigm for the GNU/Linux OS
 Red Hat Atomic Details Dockah, Dockah, Dockah! Containerization as a shift of paradigm for the GNU/Linux OS Daniel Riek Sr. Director Systems Design & Engineering In the beginning there was Stow... and
Red Hat Atomic Details Dockah, Dockah, Dockah! Containerization as a shift of paradigm for the GNU/Linux OS Daniel Riek Sr. Director Systems Design & Engineering In the beginning there was Stow... and
Rok: Data Management for Data Science
 Whitepaper Rok: Data Management for Data Science At Arrikto, we are building software to empower faster and easier collaboration for data scientists working in the same or different clouds, in the same
Whitepaper Rok: Data Management for Data Science At Arrikto, we are building software to empower faster and easier collaboration for data scientists working in the same or different clouds, in the same
Go Faster: Containers, Platforms and the Path to Better Software Development (Including Live Demo)
 RED HAT DAYS VANCOUVER Go Faster: Containers, Platforms and the Path to Better Software Development (Including Live Demo) Paul Armstrong Principal Solutions Architect Gerald Nunn Senior Middleware Solutions
RED HAT DAYS VANCOUVER Go Faster: Containers, Platforms and the Path to Better Software Development (Including Live Demo) Paul Armstrong Principal Solutions Architect Gerald Nunn Senior Middleware Solutions
PHP Composer 9 Benefits of Using a Binary Repository Manager
 PHP Composer 9 Benefits of Using a Binary Repository Manager White Paper Copyright 2017 JFrog Ltd. March 2017 www.jfrog.com Executive Summary PHP development has become one of the most popular platforms
PHP Composer 9 Benefits of Using a Binary Repository Manager White Paper Copyright 2017 JFrog Ltd. March 2017 www.jfrog.com Executive Summary PHP development has become one of the most popular platforms
ATAQS v1.0 User s Guide
 ATAQS v1.0 User s Guide Mi-Youn Brusniak Page 1 ATAQS is an open source software Licensed under the Apache License, Version 2.0 and it s source code, demo data and this guide can be downloaded at the http://tools.proteomecenter.org/ataqs/ataqs.html.
ATAQS v1.0 User s Guide Mi-Youn Brusniak Page 1 ATAQS is an open source software Licensed under the Apache License, Version 2.0 and it s source code, demo data and this guide can be downloaded at the http://tools.proteomecenter.org/ataqs/ataqs.html.
Mandi Walls. Technical Community #habitatsh
 Mandi Walls Technical Community Manager @lnxchk mandi@chef.io https://habitat.sh #habitatsh http://slack.habitat.sh/ Chef and Automation Infrastructure Automation Cloud early adopters Digital Transformation
Mandi Walls Technical Community Manager @lnxchk mandi@chef.io https://habitat.sh #habitatsh http://slack.habitat.sh/ Chef and Automation Infrastructure Automation Cloud early adopters Digital Transformation
Containerization Dockers / Mesospere. Arno Keller HPE
 Containerization Dockers / Mesospere Arno Keller HPE What is the Container technology Hypervisor vs. Containers (Huis vs artement) A container doesn't "boot" an OS instead it loads the application and
Containerization Dockers / Mesospere Arno Keller HPE What is the Container technology Hypervisor vs. Containers (Huis vs artement) A container doesn't "boot" an OS instead it loads the application and
An introduction to Baitmet package
 An introduction to Baitmet package Version 1.0.1 Xavier Domingo-Almenara (Maintainer) xdomingo@scripps.edu May 17, 2017 This vignette presents Baitmet, an R package for library driven compound profiling
An introduction to Baitmet package Version 1.0.1 Xavier Domingo-Almenara (Maintainer) xdomingo@scripps.edu May 17, 2017 This vignette presents Baitmet, an R package for library driven compound profiling
Building a government cloud Concepts and Solutions
 Building a government cloud Concepts and Solutions Dr. Gabor Szentivanyi, ULX Open Source Consulting & Distribution Background Over 18 years of experience in enterprise grade open source Based in Budapest,
Building a government cloud Concepts and Solutions Dr. Gabor Szentivanyi, ULX Open Source Consulting & Distribution Background Over 18 years of experience in enterprise grade open source Based in Budapest,
White Paper(Draft) Continuous Integration/Delivery/Deployment in Next Generation Data Integration
 Continuous Integration/Delivery/Deployment in Next Generation Data Integration 1 Contents Introduction...3 Challenges...3 Continuous Methodology Steps...3 Continuous Integration... 4 Code Build... 4 Code
Continuous Integration/Delivery/Deployment in Next Generation Data Integration 1 Contents Introduction...3 Challenges...3 Continuous Methodology Steps...3 Continuous Integration... 4 Code Build... 4 Code
Horizon 2020 Open Research Data Pilot: What is required? Sarah Jones Digital Curation Centre
 Horizon 2020 Open Research Data Pilot: What is required? Sarah Jones Digital Curation Centre sarah.jones@glasgow.ac.uk Twitter: @sjdcc Why open access and open data? The European Commission's vision is
Horizon 2020 Open Research Data Pilot: What is required? Sarah Jones Digital Curation Centre sarah.jones@glasgow.ac.uk Twitter: @sjdcc Why open access and open data? The European Commission's vision is
Preparing for Jenkins Certification
 Preparing for Jenkins Certification Agenda Jenkins Certification Prerequisites What exam do you plan to take? How to schedule and register for the test? Structure of the exams Plugins Organization of the
Preparing for Jenkins Certification Agenda Jenkins Certification Prerequisites What exam do you plan to take? How to schedule and register for the test? Structure of the exams Plugins Organization of the
YOUR APPLICATION S JOURNEY TO THE CLOUD. What s the best way to get cloud native capabilities for your existing applications?
 YOUR APPLICATION S JOURNEY TO THE CLOUD What s the best way to get cloud native capabilities for your existing applications? Introduction Moving applications to cloud is a priority for many IT organizations.
YOUR APPLICATION S JOURNEY TO THE CLOUD What s the best way to get cloud native capabilities for your existing applications? Introduction Moving applications to cloud is a priority for many IT organizations.
Bioshadock. O. Sallou - IRISA Nettab 2016 CC BY-CA 3.0
 Bioshadock O. Sallou - IRISA Nettab 2016 CC BY-CA 3.0 Containers 2 Docker, LXC, Rkt and Co Docker is the current leader in container ecosystem but not alone in ecosystem Rkt compatible with Docker images
Bioshadock O. Sallou - IRISA Nettab 2016 CC BY-CA 3.0 Containers 2 Docker, LXC, Rkt and Co Docker is the current leader in container ecosystem but not alone in ecosystem Rkt compatible with Docker images
Deliverable Project ID A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data
 Deliverable 1.4.4 Project ID 654241 Project Title A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data Project Acronym PhenoMeNal Start Date of the Project 1st
Deliverable 1.4.4 Project ID 654241 Project Title A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data Project Acronym PhenoMeNal Start Date of the Project 1st
AGILE DEVELOPMENT AND PAAS USING THE MESOSPHERE DCOS
 Sunil Shah AGILE DEVELOPMENT AND PAAS USING THE MESOSPHERE DCOS 1 THE DATACENTER OPERATING SYSTEM (DCOS) 2 DCOS INTRODUCTION The Mesosphere Datacenter Operating System (DCOS) is a distributed operating
Sunil Shah AGILE DEVELOPMENT AND PAAS USING THE MESOSPHERE DCOS 1 THE DATACENTER OPERATING SYSTEM (DCOS) 2 DCOS INTRODUCTION The Mesosphere Datacenter Operating System (DCOS) is a distributed operating
Continuous Integration and Delivery with Spinnaker
 White Paper Continuous Integration and Delivery with Spinnaker The field of software functional testing is undergoing a major transformation. What used to be an onerous manual process took a big step forward
White Paper Continuous Integration and Delivery with Spinnaker The field of software functional testing is undergoing a major transformation. What used to be an onerous manual process took a big step forward
SETTING UP AN HCS DATA ANALYSIS SYSTEM
 A WHITE PAPER FROM GENEDATA JANUARY 2010 SETTING UP AN HCS DATA ANALYSIS SYSTEM WHY YOU NEED ONE HOW TO CREATE ONE HOW IT WILL HELP HCS MARKET AND DATA ANALYSIS CHALLENGES High Content Screening (HCS)
A WHITE PAPER FROM GENEDATA JANUARY 2010 SETTING UP AN HCS DATA ANALYSIS SYSTEM WHY YOU NEED ONE HOW TO CREATE ONE HOW IT WILL HELP HCS MARKET AND DATA ANALYSIS CHALLENGES High Content Screening (HCS)
Transitioning to Symyx
 Whitepaper Transitioning to Symyx Notebook by Accelrys from Third-Party Electronic Lab Notebooks Ordinarily in a market with strong growth, vendors do not focus on competitive displacement of competitor
Whitepaper Transitioning to Symyx Notebook by Accelrys from Third-Party Electronic Lab Notebooks Ordinarily in a market with strong growth, vendors do not focus on competitive displacement of competitor
9 Reasons To Use a Binary Repository for Front-End Development with Bower
 9 Reasons To Use a Binary Repository for Front-End Development with Bower White Paper Introduction The availability of packages for front-end web development has somewhat lagged behind back-end systems.
9 Reasons To Use a Binary Repository for Front-End Development with Bower White Paper Introduction The availability of packages for front-end web development has somewhat lagged behind back-end systems.
Continuous integration & continuous delivery. COSC345 Software Engineering
 Continuous integration & continuous delivery COSC345 Software Engineering Outline Integrating different teams work, e.g., using git Defining continuous integration / continuous delivery We use continuous
Continuous integration & continuous delivery COSC345 Software Engineering Outline Integrating different teams work, e.g., using git Defining continuous integration / continuous delivery We use continuous
Reproducible & Transparent Computational Science with Galaxy. Jeremy Goecks The Galaxy Team
 Reproducible & Transparent Computational Science with Galaxy Jeremy Goecks The Galaxy Team 1 Doing Good Science Previous talks: performing an analysis setting up and scaling Galaxy adding tools libraries
Reproducible & Transparent Computational Science with Galaxy Jeremy Goecks The Galaxy Team 1 Doing Good Science Previous talks: performing an analysis setting up and scaling Galaxy adding tools libraries
BUILDING A GPU-FOCUSED CI SOLUTION
 BUILDING A GPU-FOCUSED CI SOLUTION Mike Wendt @mike_wendt github.com/nvidia github.com/mike-wendt Need for CPU CI Challenges of GPU CI Methods to Implement GPU CI AGENDA Improving GPU CI Today Demo Lessons
BUILDING A GPU-FOCUSED CI SOLUTION Mike Wendt @mike_wendt github.com/nvidia github.com/mike-wendt Need for CPU CI Challenges of GPU CI Methods to Implement GPU CI AGENDA Improving GPU CI Today Demo Lessons
Data processing. Filters and normalisation. Mélanie Pétéra W4M Core Team 31/05/2017 v 1.0.0
 Data processing Filters and normalisation Mélanie Pétéra W4M Core Team 31/05/2017 v 1.0.0 Presentation map 1) Processing the data W4M table format for Galaxy 2) A generic tool to filter in Galaxy a) Generic
Data processing Filters and normalisation Mélanie Pétéra W4M Core Team 31/05/2017 v 1.0.0 Presentation map 1) Processing the data W4M table format for Galaxy 2) A generic tool to filter in Galaxy a) Generic
Requirements for data catalogues within facilities
 Requirements for data catalogues within facilities Milan Prica 1, George Kourousias 1, Alistair Mills 2, Brian Matthews 2 1 Sincrotrone Trieste S.C.p.A, Trieste, Italy 2 Scientific Computing Department,
Requirements for data catalogues within facilities Milan Prica 1, George Kourousias 1, Alistair Mills 2, Brian Matthews 2 1 Sincrotrone Trieste S.C.p.A, Trieste, Italy 2 Scientific Computing Department,
Package batman. Installation and Testing
 Package batman Installation and Testing Table of Contents 1. INSTALLATION INSTRUCTIONS... 1 2. TESTING... 3 Test 1: Single spectrum from designed mixture data... 3 Test 2: Multiple spectra from designed
Package batman Installation and Testing Table of Contents 1. INSTALLATION INSTRUCTIONS... 1 2. TESTING... 3 Test 1: Single spectrum from designed mixture data... 3 Test 2: Multiple spectra from designed
Galaxy. Data intensive biology for everyone. / #usegalaxy
 Galaxy Data intensive biology for everyone. www.galaxyproject.org @jxtx / #usegalaxy Engineering Dannon Baker Dan Blankenberg Dave Bouvier Nate Coraor Carl Eberhard Jeremy Goecks Sam Guerler Greg von Kuster
Galaxy Data intensive biology for everyone. www.galaxyproject.org @jxtx / #usegalaxy Engineering Dannon Baker Dan Blankenberg Dave Bouvier Nate Coraor Carl Eberhard Jeremy Goecks Sam Guerler Greg von Kuster
BUDSS: A Software Shell for automated MS Data Processing
 BUDSS: A Software Shell for automated MS Data Processing Yang Su; Sequin Huang; Hua Huang; David H. Perlman; Catherine E. Costello; Mark E. McComb Cardiovascular Proteomics Center, Boston University School
BUDSS: A Software Shell for automated MS Data Processing Yang Su; Sequin Huang; Hua Huang; David H. Perlman; Catherine E. Costello; Mark E. McComb Cardiovascular Proteomics Center, Boston University School
Achieving Digital Transformation: FOUR MUST-HAVES FOR A MODERN VIRTUALIZATION PLATFORM WHITE PAPER
 Achieving Digital Transformation: FOUR MUST-HAVES FOR A MODERN VIRTUALIZATION PLATFORM WHITE PAPER Table of Contents The Digital Transformation 3 Four Must-Haves for a Modern Virtualization Platform 3
Achieving Digital Transformation: FOUR MUST-HAVES FOR A MODERN VIRTUALIZATION PLATFORM WHITE PAPER Table of Contents The Digital Transformation 3 Four Must-Haves for a Modern Virtualization Platform 3
D2.5 Data mediation. Project: ROADIDEA
 D2.5 Data mediation Project: ROADIDEA 215455 Document Number and Title: D2.5 Data mediation How to convert data with different formats Work-Package: WP2 Deliverable Type: Report Contractual Date of Delivery:
D2.5 Data mediation Project: ROADIDEA 215455 Document Number and Title: D2.5 Data mediation How to convert data with different formats Work-Package: WP2 Deliverable Type: Report Contractual Date of Delivery:
Java in the world of Software AG JCP EC May 2018
 Java in the world of Software AG JCP EC May 2018 Georgi Stanev Architect Software AG 2017 Software AG. All rights reserved. History of the Software AG 1969 The concept for an adaptable and extremely versatile
Java in the world of Software AG JCP EC May 2018 Georgi Stanev Architect Software AG 2017 Software AG. All rights reserved. History of the Software AG 1969 The concept for an adaptable and extremely versatile
Curatr: a web application for creating, curating, and sharing a mass spectral library
 Curatr: a web application for creating, curating, and sharing a mass spectral library Andrew Palmer (1), Prasad Phapale (1), Dominik Fay (1), Theodore Alexandrov (1,2) (1) European Molecular Biology Laboratory,
Curatr: a web application for creating, curating, and sharing a mass spectral library Andrew Palmer (1), Prasad Phapale (1), Dominik Fay (1), Theodore Alexandrov (1,2) (1) European Molecular Biology Laboratory,
Sunil Shah SECURE, FLEXIBLE CONTINUOUS DELIVERY PIPELINES WITH GITLAB AND DC/OS Mesosphere, Inc. All Rights Reserved.
 Sunil Shah SECURE, FLEXIBLE CONTINUOUS DELIVERY PIPELINES WITH GITLAB AND DC/OS 1 Introduction MOBILE, SOCIAL & CLOUD ARE RAISING CUSTOMER EXPECTATIONS We need a way to deliver software so fast that our
Sunil Shah SECURE, FLEXIBLE CONTINUOUS DELIVERY PIPELINES WITH GITLAB AND DC/OS 1 Introduction MOBILE, SOCIAL & CLOUD ARE RAISING CUSTOMER EXPECTATIONS We need a way to deliver software so fast that our
RED HAT OPENSHIFT A FOUNDATION FOR SUCCESSFUL DIGITAL TRANSFORMATION
 RED HAT OPENSHIFT A FOUNDATION FOR SUCCESSFUL DIGITAL TRANSFORMATION Stephanos D Bacon Product Portfolio Strategy, Application Platforms Stockholm, 13 September 2017 1 THE PATH TO DIGITAL LEADERSHIP IT
RED HAT OPENSHIFT A FOUNDATION FOR SUCCESSFUL DIGITAL TRANSFORMATION Stephanos D Bacon Product Portfolio Strategy, Application Platforms Stockholm, 13 September 2017 1 THE PATH TO DIGITAL LEADERSHIP IT
Tutorial 2: Analysis of DIA/SWATH data in Skyline
 Tutorial 2: Analysis of DIA/SWATH data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantification.
Tutorial 2: Analysis of DIA/SWATH data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantification.
Extracting reproducible simulation studies from model repositories using the CombineArchive Toolkit
 Extracting reproducible simulation studies from model repositories using the CombineArchive Toolkit Martin Scharm, Dagmar Waltemath Department of Systems Biology and Bioinformatics University of Rostock
Extracting reproducible simulation studies from model repositories using the CombineArchive Toolkit Martin Scharm, Dagmar Waltemath Department of Systems Biology and Bioinformatics University of Rostock
Copyright 2008 Casa Software Ltd.
 Quantification of Thermo Scientific K- Alpha Data The last ten years have seen the XPS technique develop from a technique used predominantly by experts of XPS to a tool used by application oriented scientists.
Quantification of Thermo Scientific K- Alpha Data The last ten years have seen the XPS technique develop from a technique used predominantly by experts of XPS to a tool used by application oriented scientists.
CoE CENTRE of EXCELLENCE ON DATA WAREHOUSING
 in partnership with Overall handbook to set up a S-DWH CoE: Deliverable: 4.6 Version: 3.1 Date: 3 November 2017 CoE CENTRE of EXCELLENCE ON DATA WAREHOUSING Handbook to set up a S-DWH 1 version 2.1 / 4
in partnership with Overall handbook to set up a S-DWH CoE: Deliverable: 4.6 Version: 3.1 Date: 3 November 2017 CoE CENTRE of EXCELLENCE ON DATA WAREHOUSING Handbook to set up a S-DWH 1 version 2.1 / 4
Data Management Plan
 Data Management Plan Mark Sanders, Martina Chýlková Document Identifier D1.9 Data Management Plan Version 1.0 Date Due M6 Submission date 30 November, 2015 WorkPackage WP1 Management and coordination Lead
Data Management Plan Mark Sanders, Martina Chýlková Document Identifier D1.9 Data Management Plan Version 1.0 Date Due M6 Submission date 30 November, 2015 WorkPackage WP1 Management and coordination Lead
The Go4IT project. Toward a TTCN-3 open environment for IPv6 protocols testing. Project identity card
 The Go4IT project Toward a TTCN-3 open environment for IPv6 protocols testing TTCN-3 User Conference 2006 - Berlin Project identity card Integrated Infrastructure Initiative Started in Nov 2005 30 month
The Go4IT project Toward a TTCN-3 open environment for IPv6 protocols testing TTCN-3 User Conference 2006 - Berlin Project identity card Integrated Infrastructure Initiative Started in Nov 2005 30 month
The ELIXIR of Linked Data
 The ELIXIR of Linked Data Professor Carole Goble (UK node) Barend Mons (NL node), Helen Parkinson (EMBL-EBI node) The Interoperability Services Backbone Team European Life Sciences Infrastructure for Biological
The ELIXIR of Linked Data Professor Carole Goble (UK node) Barend Mons (NL node), Helen Parkinson (EMBL-EBI node) The Interoperability Services Backbone Team European Life Sciences Infrastructure for Biological
A Greybeard's Worst Nightmare
 A Greybeard's Worst Nightmare How Kubernetes and Containers are re-defining the Linux OS Daniel Riek, Red Hat April 2017 Greybeard Greybeards fight Balrogs. They hate systemd. They fork distributions.
A Greybeard's Worst Nightmare How Kubernetes and Containers are re-defining the Linux OS Daniel Riek, Red Hat April 2017 Greybeard Greybeards fight Balrogs. They hate systemd. They fork distributions.
What is Cloud Computing? What are the Private and Public Clouds? What are IaaS, PaaS, and SaaS? What is the Amazon Web Services (AWS)?
 What is Cloud Computing? What are the Private and Public Clouds? What are IaaS, PaaS, and SaaS? What is the Amazon Web Services (AWS)? What is Amazon Machine Image (AMI)? Amazon Elastic Compute Cloud (EC2)?
What is Cloud Computing? What are the Private and Public Clouds? What are IaaS, PaaS, and SaaS? What is the Amazon Web Services (AWS)? What is Amazon Machine Image (AMI)? Amazon Elastic Compute Cloud (EC2)?
PoS(ISGC 2011 & OGF 31)119
 Enabling JChem application on grid Miklos Kozlovszky MTA SZTAKI H-1111 Kende str. 13-17, Budapest, Hungary E-mail: m.kozlovszky@sztaki.hu Akos Balasko MTA SZTAKI H-1111 Kende str. 13-17, Budapest, Hungary
Enabling JChem application on grid Miklos Kozlovszky MTA SZTAKI H-1111 Kende str. 13-17, Budapest, Hungary E-mail: m.kozlovszky@sztaki.hu Akos Balasko MTA SZTAKI H-1111 Kende str. 13-17, Budapest, Hungary
Managing Research Data for Diverse Scientific Experiments
 Managing Research Data for Diverse Scientific Experiments Erica Yang erica.yang@stfc.ac.uk Scientific Computing Department STFC Rutherford Appleton Laboratory Crystallographic Information and Data Management
Managing Research Data for Diverse Scientific Experiments Erica Yang erica.yang@stfc.ac.uk Scientific Computing Department STFC Rutherford Appleton Laboratory Crystallographic Information and Data Management
Deliverable Project ID A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data
 Deliverable 1.4.3 Project ID 654241 Project Title A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data Project Acronym PhenoMeNal Start Date of the Project 1st
Deliverable 1.4.3 Project ID 654241 Project Title A comprehensive and standardised e-infrastructure for analysing medical metabolic phenotype data Project Acronym PhenoMeNal Start Date of the Project 1st
