CLAMP. Reference Manual. A Guide to the Extraction of Clinical Concepts
|
|
|
- Austin Henderson
- 5 years ago
- Views:
Transcription
1 CLAMP Reference Manual A Guide to the Extraction of Clinical Concepts
2 Table of Contents 1. Introduction System Requirements Installation How to run CLAMP Package Description Import existing projects into the new version Annotated documents used in CLAMP Contact us NLP Components NLP Components Sentence Detector Tokenizer Pos Tagger Chunker Named Entity Recognizer Assertion Identifier Ruta_ Rule_Engine Section Identifier UMLS Encoder User_Defined_Components Machine Learning components NER Feature Extractor DF_Brown_clustering_feature DF_Dictionary_lookup_feature DF_Ngram_feature DF_prefix_suffix_feature DF_Random_indexing_feature Clamp Documentation Page 1
3 9.1.6 DF_Section_feature DF_Sentence_pattern_feature DF_Word_embedding_feature DF_Word_shape_feature DF_Words_regular_expression_feature Build a Pipeline Create and Run a Pipeline Configure the pipeline Component dependency & Auto fix Import input files Run the pipeline Output visualization Built-in pipelines Export pipeline as a jar file Annotation Annotate corpus Create a project Import annotation files Define entity & relation types Start Annotation Visualization of entity & relation Pre-Annotation of entity and relation Machine learning model development Building machine learning models (NER model) Check output models & logs Use your own model in pipeline Visualization for error analysis Clamp Documentation Page 2
4 Clinical Language Annotation Modeling and Processing Toolkit System 1. Introduction Version The CLAMP System is a comprehensive clinical Natural Language Processing software that enables recognition and automatic encoding of clinical information in narrative patient reports. In addition to running a clinical concept extraction pipeline as well as an annotation pipeline, the individual components of the system can also be used as independent modules. The system lends itself for diverse applications in a broad range of clinical domains. The high performance language processing framework in CLAMP consists of the following key building blocks: NLP Pipelines CLAMP components builds on a set of high performance NLP components that were proven in several clinical NLP challenges such as i2b2, ShARe/CLEF, and SemEVAL. A pipeline can be created and customized by a simple drag and drop on the individual CLAMP components in the order that is desired. Upon creation of the pipeline, CLAMP checks for errors in sequence and directs the user to the appropriate logical order with insertion of the required components for a working pipeline. The CLAMP components are supported by knowledge resources consisting of medical abbreviations, dictionaries, section headers, and a corpus of 400 annotated clinical notes derived from MTsamples, a freely available resource of clinical narrative text. CLAMP also provides built-in pipelines ready for use out of the box for a series of common clinical applications. Clamp Documentation Page 3
5 Machine learning and hybrid approaches The CLAMP framework provides alternative components for some tasks, utilizing rule based methods and/or machine learning methods such as support vector machines, and conditional random fields. These components can be customized by re-training on an annotated corpus, or editing the rule sets within the CLAMP GUI to achieve a custom NLP task. The CLAMP GUI version also provides built-in functionality to test the model, using the annotated corpora or n-fold cross validation. Corpus management and annotation tool: The user interface also provides required tools to maintain and annotate text corpora. It hosts an improved version of the brat annotation tool (reference?) for textual annotations. 2. System Requirements CLAMP is a stand-alone Java application based on the Eclipse platform technologies. CLAMP uses the Apache UIMA (Unstructured Information Management Architecture) framework. The annotation module of CLAMP incorporates and enhances the brat rapid annotation tool. For the other individual constituents, Apache OpenNLP toolkit, Liblinear and CRF Suite are utilized in addition to in-house rule-based components. CLAMP also use the UIMA Ruta (Rule based Text Annotation) as a rule engine to help users specify rules. CLAMP is distributed as a ready-to-use binary package that can either be executed at the command line or carries the associated Graphic User Interface (GUI). Our distribution package includes components for jar files, CRFSuite, and a Lucene index of all levels of UMLS data. The only prerequisite necessary to compile CLAMP is JRE 1.8 (Java Runtime Environment). Please ensure that you have Java 8 or higher installed in your system. Run the following command in both Mac and Windows to check your version: java version Clamp Documentation Page 4
6 Here is an example of what you will see when running the command in Windows: If your java version is not 1.8, it is available for download from the Oracle website at html. An UMLS account is required in order to use Level 2 and higher data in the UMLS encoding component of the system. The account can be created at You will have to enter your UMLS username and password when prompted by CLAMP in order to utilise the UMLS encoding component. CLAMP also uses the computer s default browsers to visualize the clinical documents. Since all browsers do not completely support all the aspects of the technologies used to implement the visualization, limitations exist in term of running the CLAMP annotation module in the browsers. On the Windows OS, the Internet Explorer should be higher than IE9; On Macintosh computers, Safari (all versions) works well with CLAMP. Clamp Documentation Page 5
7 3. Installation Extract the contents of archive (Clamp_1.1.7_win.zip or Clamp_1.1.7_mac.zip) to your directory of choice. CLAMP application resides in the root folder of your archive. After extraction, double click on StartClamp(.exe) file to start CLAMP. For the CLAMP command line version please refer to the readme file. For further information and troubleshooting, please refer CLAMP website at 4. How to run CLAMP You can run the GUI version of CLAMP by double clicking on the startclamp icon. Once the software is completely loaded, you will notice a welcome tab. Close the tab to go to CLAMP working environment. Close the tab to go to CLAMP working environment Clamp Documentation Page 6
8 5. Package Description Since CLAMP is a stand-alone eclipse plugin, its folder structure is similar to other eclipse plugins. Configuration Folder: This folder contains CLAMP configuration files. StartCLAMP: This is the launching point for the CLAMP GUI. In Windows, this is an executable file while in Mac, this is an application. Workspace Folder: This folder contains seven sub-folders: 1. ComponentLibrary: contains the components used in machine learning feature extraction and NLP functions. 2. MyCorpus: contains the customized corpus built by the users. 3. MyPipeline: contains the customized pipeline created by users for clinical notes processing. 4. PipelineLibrary: contains the built-in pipelines ready to use for a series of common clinical applications. 5. Log: Includes CLAMP run-time log files 6. Metadata: The metadata used by CLAMP are included in this folder. 7. Resources: This folder includes third-party libraries. Currently it has two items: 7.1 CRFSuite: the CRF implementation for Name Entity Recognition tasks Clamp Documentation Page 7
9 7.2 Umls_index: the Lucene index built for CLAMP based on the UMLS thesaurus. If you want to use UMLS terminologies, then you will need to create an UMLS account. Please follow the following link to create an UMLS account if you do not have any The following table lists libraries included in CLAMP. groupid artifactid version org.cleartk cleartk-ml-liblinear org.ini4j ini4j org.apache.uima uimafit-core com.google.code.gson gson 2.3 org.apache.uima uimaj-core org.apache.uima uimaj-cpe org.apache.uima uimaj-document-annotation de.bwaldvogel liblinear 1.94 org.apache.lucene lucene-core org.apache.lucene lucene-analyzers-common org.apache.lucene lucene-queryparser org.apache.opennlp opennlp-tools incubating org.apache.ctakes ctakes-type-system org.cleartk cleartk-named-entity com.googlecode.clearnlp clearnlp commons-codec commons-codec dom4j dom4j org.apache.uima ruta-ep-engine javax.servlet servlet-api 3.0-alpha-1 com.sun.jersey jersey-client 1.19 junit junit 4.12 commons-cli commons-cli 1.3 net.jodah concurrentunit org.javatuples javatuples 1.2 Clamp Documentation Page 8
10 6. Import existing projects into the new version On Windows, simply copy contents of your previous work folder (i.e. from older Clamp_x.xx.xx_win\workspace\MyPipeline\ contents to new Clamp_x.xx.xx_win\workspace\MyPipeline\ contents) using Windows Explorer and restart CLAMP if it s already running. On startup, CLAMP will recognize these projects and import them into your workspace. On MacOSX, similarly copy contents of your previous work folder (i.e. from older Clamp_x.xx.xx_win/workspace/MyPipeline/ contents to new Clamp_x.xx.xx_win/workspace/MyPipeline/ contents) using Finder and restart CLAMP if it s already running. On startup, CLAMP will recognize these projects and import them into your new workspace. 6.1 Annotated documents used in CLAMP The pre-annotated notes are crawled from ' that has lots of publicly available de-identified notes. But only 'discharge summary' and 'general medicine' are included in CLAMP. We annotated all the 'problem', 'treatment' and 'test' mentions in the notes, based on the I2B NER guideline Contact us The CLAMP System was developed by Dr. Hua Xu s team from the School of Biomedical Informatics at the University of Texas Health Science Center in Houston. For technical issues, please contact: Jingqi.Wang@uth.tmc.edu For any other issues, please contact: Anupama.E.Gururaj@uth.tmc.edu Clamp Documentation Page 9
11 8. NLP Components 8.1 NLP Components NLP components are used for processing text. CLAMP offers multiple NLP components that are the building blocks for performing any of the NLP tasks. The individual components are provided as pre-built modules that are to be used in building pipelines for automatic text processing, as well as training customized machine learning models. Figure 8.1 displays the CLAMP NLP components, as well as its associated tools, algorithms and resources. In this section, we will provide details about each NLP component including their function as well as ways to customize them using your own dictionary/model. In Build a Pipeline section, we have tutorials that touch on use cases wherein the components are utilized in various applications. Figure 8.1 Schema of NLP Components Clamp Documentation Page 10
12 8.2 Sentence Detector A sentence is defined as the longest whitespace trimmed character sequence between two punctuation marks. A Sentence Detector utilizes different methods to detect a sentence. As shown in Figure 8.2, CLAMP provides three different models to detect a sentence: 1. DF_CLAMP_Sentence_Detector 2. DF_CLAMP_Sentence_by_newline, and 3. DF_CLAMP_OpenNLP_sentence_detector Each model is described in detail in the following sections. Figure 8.2 Three sentence detectors and their configuration files Clamp Documentation Page 11
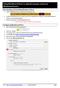
13 1. DF_CLAMP_Sentence_Detector: DF_CLAMP_Sentence_Detector is the default sentence detector in CLAMP. It is designed specifically for clinical notes and takes into account the distinctive characteristics observed in sentences found in clinical texts. To configure the DF_CLAMP_Sentence_Detector, please click on the config file. A pop-up window opens where you can modify two parameters: Medical Abbreviation, and Max Sentence Length. Medical Abbreviation: There are some medical abbreviations that have punctuation marks at their beginning (.NO2) while some of them have it at the end (spec.). Providing a list of such abbreviations would help the detector to identify sentences more accurately. By default, CLAMP has provided a comprehensive list of medical abbreviation which can be found in this file: defaultabbrs.txt A) To replace the abbreviation file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click the button with three dots to browse for your own file A) 3- Click on the Open button Clamp Documentation Page 12
14 B) To edit the current file: 1. Double click on the defaultabbrs.txt file to open it 2. Add the terms that you want to include in the abbreviation file 3. Click on the Save button on the toolbar B) 3- Click the Save button B) 2- Add the terms that you want to include in the file B) 1- Double click on defaultabbrs.txt file Max Sentence Length Checking the checkbox for Break long sentences or not? allows users to break long sentences into the number of words that they have specified in the input textbox. Please refer to Figure 8.3 for more information. Figure 8.3 Interface for config.conf of the DF_CLAMP_Sentence_Detector Clamp Documentation Page 13
15 2. DF_CLAMP_Sentence_by_newline This detector will identify new sentences using the line breaks in the file, i.e., each line in the file is treated as a single sentence. 3. DF_CLAMP_OpenNLP_sentence_detector. This is an OpenNLP sentence detector which advanced users can use its config.conf file to change its default model. A) To replace the default model: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your file A) 3- Click on the Open button Clamp Documentation Page 14
16 8.3 Tokenizer A Tokenizer segments the text into a sequence of tokens. As shown in Figure 8.4, CLAMP provides three different models of tokenizer: 1. DF_CLAMP_Tokenizer 2. DF_ OpenNLP_Tokenizer 3. DF_Tokenize_by_spaces Each model will be described in more details. Figure 8.4 Three tokenizers and their configuration files Clamp Documentation Page 15
17 1) DF_CLAMP_Tokenizer DF_CLAMP_Tokenizer is the default tokenizer designed specifically for clinical notes. Advanced users can use the config.conf file to change the default tokenization. A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your file A) 3- Click on the open button Clamp Documentation Page 16
18 2) DF_ OpenNLP_Tokenizer This is an OpenNLP tokenizer. Advanced users can use its config.conf file to change its default model, en-token.bin. A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your file A) 3- Click on the open button 3) DF_Tokenize_by_spaces. This tokenizer uses the spaces in a sentence to separate the tokens. Clamp Documentation Page 17
19 8.4 Pos Tagger A Pos tagger allows users to assign parts of speech to each token. As shown in Figure 8.5, CLAMP currently provides only one pos tagger, DF_OpenNLP_pos_tagger, designed specifically for clinical text. This tagger is built from re-training the OpenNLP pos tagger on a dataset of clinical notes, namely, the MiPACQ corpus. ( Advanced users can use the config.conf file to change the default pos tagger modelmipacq_pos.bin. Figure 8.5 DF_OpenNLP_pos_tagger and its configuration files Clamp Documentation Page 18
20 8.5 Chunker A chunker does a shallow parsing of a sentence and identifies the syntactic constituents such as noun phrases, verb phrases, and etc. As shown in Figure 8.6, CLAMP currently provides only one single chunker, DF_OpenNLP_chunker, which is a wrapper of the chunker in OpenNLP. Advanced users can use the config.conf file to change the default chunker model- en-chunker.bin. Figure 8.6 DF_OpenNLP_chunker and its configuration files Clamp Documentation Page 19
21 8.6 Named Entity Recognizer A named entity recognizer identifies named entities and their semantic types in text. Typically, named entities refer to clinical concepts in CLAMP. As shown in Figure 8.7, CLAMP provides two different models for named entity recognition: 1. DF_CRF_based_named_entity_recognizer,and 2. DF_Dictionary_lookup 3. DF_Regular_expression_NER Each model will be described in more details. Figure 8.7 Three named entity recognizers and their configuration files Clamp Documentation Page 20
22 1. DF_CRF_based_named_entity_recognizer DF_CRF_based_named_entity_recognizer is the default named entity recognizer used in CLAMP. The recognizer identifies three types of clinical concepts: Problems, treatments, and tests. It is built from training the CRF model on a dataset of clinical notes, namely, the i2b challenge corpus ( Advanced users can use the config.conf file to change the default recognizer model as in the file defaultmodel.jar. A) To replace the default file: 1. Double click on the config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your file A) 3- Click on the open button Clamp Documentation Page 21
23 2. DF_Dictionary_lookup DF_Dictionary_lookup uses terms in the dictionary to match them directly with the identified named entities. Currently the defaultdic.txt used in CLAMP consists of terms and their semantic types from UMLS ( The semantic type of the matched term in UMLS is assigned to the recognized named entity. To configure DF_Dictionary_lookup: First, click on the config file under the DF_Dictionary_matcher folder. This will open up a new window that takes the following three parameters: Case sensitive, Stemming and Dictionaries.( Figure 6.8) Case sensitive If you check the checkbox for Case sensitive, the matcher will differentiate between capital and lowercase letters when searching for a term in the dictionary. For example, Breast Cancer will not matched with breast cancer. Stemming If you check the checkbox for Stemming, the matcher will match the stemmed form of a candidate named entity with the terms in the dictionary. For example, breast cancers will be matched to breast cancer. Dictionaries You can also replace or edit the dictionary file suggested for this function. Clamp Documentation Page 22
24 A) To replace the default dictionary file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your file a)3- Click on the open button Clamp Documentation Page 23
25 B) To edit the current dictionary file: 1. Double click on the defaultdict.txt file to open it 2. Add the terms that you want to include in the dictionary file 3. Click the Save button at the top of the page B) 3- Click the save button B) 1- Doubel click on the defaultdict.txt file to open B) 2- Add the terms that you want to include in the dictionary file file 3. DF_Regular_expression_NER Using the defaultregexpr.txt file, this module can identify named entities. defaultregexpr.txt file can contain several regular expression. If a phrase matches a regular expression, it is recognized as a named entity. You can add your own regular expression to the existing file by double clicking the file and add the items that you want to include. Clamp Documentation Page 24
26 8.7 Assertion Identifier An Assertion identifier checks whether there is a negation related to a specific clinical concepts in the text. A negation means the absence or opposite of something positive. CLAMP Assertion Identifier provides a mechanism to examine the real-world implications of annotations in a given clinical text. The defaultnegexdict.txt file which contains common negation patterns is used by CLAMP to check for negation in a clinical text. You can either replace or edit this file by following the steps below (Figure 8.8). Figure 8.8 Assertion identifier and its configuration file Clamp Documentation Page 25
27 A) To replace the Negation list file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own file A) 3- Click on the Open B) To edit the current dictionary file: 1. Double click on the defaultnegexdict.txt file to open it 2. Add the terms that you want to include in the dictionary file 3. Click the Save button at the top of the page B) 3- Click the Save button B) 2- Add the terms that you want to include in the dictionary file B) 1- Double click on the defaultnegexdict.txt file to open it Clamp Documentation Page 26
28 8.8 Ruta_ Rule_Engine UIMA Ruta rules can be used to create or modify annotations as well as create features for annotations. Ruta rules in general can consist of a sequence of rule elements. A simple rule elements consist of four parts: A matching condition, an optional quantifier, an optional list of conditions and an optional list of actions. For more information please visit: A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own file A) 3- Click on the Open button Clamp Documentation Page 27
29 B) To edit the current dictionary file: 1. Double click on the default.ruta file to open it 2. Add the terms that you want to include in the dictionary file 3. Click the Save button at the top of the page B) 3- Click the Save button at the top of the page B) 2- Add the rules that you want to include in the dictionary file B) 1- Double click on the default.ruta file to open it 8.9 Section Identifier The section header identifier component identifies the section headers in a clinical note based on a predefined dictionary and categorizes them into general categories (Figure 8.9). E.g. the section header ICD 10 code will be assigned to the icd_code category. Figure 8.9 Section header identifier and its configuration file Clamp Documentation Page 28
30 You can replace or edit the default dictionary, section_map.txt, following the steps below: A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own file A) 3- Click on the Open button B) To add additional section headers to the current file: 1. Double click on the section_map.txt file to open it 2. Add the terms that you want to include in the file 3. Click the Save button at the top of the page B) 3- Click the Save button at the top of the page B) 1- Double click on the section_map.txt file to open it B) 2- Add the terms that you want to include in the dictionary file Clamp Documentation Page 29
31 8.10 UMLS Encoder A UMLS Encoder matches the terms of clinical concepts to its corresponding CUIs in UMLS. For example, the term breast cancer will be encoded into the CUI of C in UMLS. Currently CLAMP provides a default dictionary based on the UMLS encoder as shown in Figure Figure 8.10 A dictionary based UMLS encoder Clamp Documentation Page 30
32 8.11 User_Defined_Components DF_Drug_Attribute_Connector: This is a context free grammar parser which is extracted from Medex. It is used to connect medication to its possible attributes such as dose. DF_Relation_connector_after_ruta: While connecting two named entities using Ruta is relatively easy, it can not be used to provide a name for that relationship. Advanced users can generate their own file and replace it with the system s default file or edit the default file. A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own file A) 3- Click on the Open button Clamp Documentation Page 31
33 B) To edit the current dictionary file: 1. Double click on the relationconnection.txt file to open it 2. Add the terms that you want to include in the file 3. Click the Save button at the top of the page B) 3- Click the Save button at the top of the page B) 2- Add the rules that you want to include in the file B) 1- Double click on the relationconnection.txt file to open it Clamp Documentation Page 32
34 9. Machine Learning components 9.1 NER Feature Extractor This component consists of different feature extractors (Figure 9.1), which are used for extracting different types of features for named entity recognition, CLAMP users will use this component to build their own named entity recognizer in a corpus annotation project (Refer to Section 4.2). Similar to the previous components, we can customize these features by changing or replacing their default config files. Explanation of each extractor is as follows: Figure 9.1 List of NER feature extractors Clamp Documentation Page 33
35 9.1.1 DF_Brown_clustering_feature It is a type of word representation feature generated on the unlabeled data which is provided by the SemEval 2014 Challenge. Advanced users can eplace their own Brwon clustering file with the system s default file. A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own file A) 3- Click on the Open button For more information on how to create your own Brown Clustring file visit: Clamp Documentation Page 34
36 9.1.2 DF_Dictionary_lookup_feature This extractor uses a dictionary consisting of terms and their semantic types from UMLS to extract potential features. Advanced users can replace or edit the default file following the steps below: Note: The format of the content should be as the same as the default file: (phrase then tab then semantic type) A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own A) 3- Click on the Open button Clamp Documentation Page 35
37 B) To edit the default file: 1. Double click on the word_path.txt file to open it 2. Add the terms that you want to include in the file 3. Click the Save button at the top of the page B) 3- Click the save button B) 1- Doubel click on the defaultdict.txt file to open B) 2- Add the terms that you want to include in the dictionary file file Clamp Documentation Page 36
38 9.1.3 DF_Ngram_feature This module uses the words along with their part-of-speech (pos) tagging as NER features DF_prefix_suffix_feature This function extracts the prefix and suffix of words that may be a representative of a specific type of named entities DF_Random_indexing_feature Similar to the brown clustering, it is a type of word representation feature generated on unlabeled data using a 3 rd party package. For more information visit: A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own file A) 3- Click on the Open button Clamp Documentation Page 37
39 9.1.6 DF_Section_feature This function extracts the section in which a candidate named entity presents DF_Sentence_pattern_feature This function distinguishes the pattern of a sentence by CLAMP built in rules DF_Word_embedding_feature Similar to the brown clustering and random indexing, it is a type of distributed word representation feature generated on the unlabeled data (MIMIC II) provided by the SemEval 2014 Challenge using a neural network.advanced users can replace the default file with their own file. A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own file A) 3- Click on the Open button Clamp Documentation Page 38
40 9.1.9 DF_Word_shape_feature This function extracts the type of a word; it identifies whether or not it begins with an english letter, number, and etc DF_Words_regular_expression_feature This function extracts the regular expression patterns of words that may indicate a specific type of named entity. Advanced users can create their own regular expressions or edit the default file. A) To replace the default file: 1. Double click on config.conf file to open it 2. Click on the button with three dots to browse for your own file 3. Click on the open button A) 2- Click on the button with three dots to browse for your own file A) 3- Click on the Open button Clamp Documentation Page 39
41 B) To edit the default file: 1. Double click on the reglist.txt file to open it 2. Add the terms that you want to include in the file 3. Click the Save button at the top of the page B) 3- Click the save button B) 2- Add the terms that you want to include in the file file B) 1-Doubel click on the reglist.txt file to open it Clamp Documentation Page 40
42 10. Build a Pipeline 10.1 Create and Run a Pipeline Running a pipeline refers to the use of a set of NLP components to identify the specified information, including sentence segmentation, tokenization, part of speech tagging, abbreviations, etc. The NLP components are executed in a sequence based on the functional dependency amongst them. In order to recognize clinical concepts within clinical text: 1. You need to create a project 2. You need to configure the pipeline 3. You need to import the files that you want to be analyzed 4. You need to process the imported files by running them through the pipeline. Follow the steps below to build a pipeline: A) Create a new project 1. Click on the plus (+) sign at the top left corner of the screen as shown in Figure Figure 10.1 Create a new project 2. On the pop-up window (Figure 10.2), enter a name for your project, for example: Clinical_concept_recognition. 3. Select NLP Pipeline as the project type. 4. Click the Finish button. Clamp Documentation Page 41
43 A new project with the specified name is created and is placed under Mypipeline folder on the Pipeline panel at the lower left of the screen (Figure 10.3) A) 2- Enter a name for the project A) 3- Select NLP Pipeline as the project type. A) 4- Click the Finish button Figure 10.2 Creating a new NLP pipeline project Figure 10.3 A project with the specified name is created and is placed under Mypipeline folder Double click the pipeline name to view its content. As you can see, it contains two folders Components, and Data. The Components folder contains the pipeline configuration file. The Data folder includes two folders: Input, and Output. The Input folder holds the files that are processed by the pipeline. The results obtained by running the pipeline are saved in the output folder. Clamp Documentation Page 42
44 10.2 Configure the pipeline To configure a pipeline double click on the.pipeline file from the newly created pipeline project to open it in the middle window on the screen. (Figure 10.4). Here you can drag and drop the NLP components from the Component panel. Since we want to recognize clinical concepts using NLP components, we drag the DF_CRF_based_name_entity_recognizer from the NLP_components to the pipeline. Figure 10.4 Pipeline configuration window Clamp Documentation Page 43
45 10.3 Component dependency & Auto fix As shown in Figure 10.5, there is a red X sign in front of the newly added component, CRF based named entity recognizer. This sign indicates that the named entity recognizer component is dependant on other NLP components that are missing from the current pipeline. In our example, the clinical notes first need to be processed by the sentence detector, tokenizer, section identifier, and POS tagger components before processing by the named entity recognizer. To fix this issue, simply click on the Auto fix button at the top of the panel. This automatically adds the required components to the pipeline. The sequence of the individual components from top to bottom reflect the order in which they will run to process your input data. After the required components are added (Figure 10.6), the red X sign changes to the green circle sign indicating the accuracy of the order of the components. Once you see the green sign for each of the displayed NLP components, click on the Save button at the top of the screen to save your changes. Figure 10.5 A wrong pipeline for clinical concept recognition needs to be fixed with dependent NLP models Clamp Documentation Page 44
46 Figure A correct pipeline for clinical concept recognition with all necessary NLP models. Clamp Documentation Page 45
47 10.4 Import input files Once the pipeline is configured, you will need to import your desired files to the Input folder using the the following steps: 1. In the PipelineView, right click on the Input folder under the Data folder, then select the import (Figure 10.7). A pop-up menu appears which lets you select the files that you want to import. 2. Click on the small arrow next to the General folder to expand it, then select File System as the import source. 3. Click on the Next button (Figure 10.8) 1- right click on the Input folder under the Data folder, then select the import Figure 10.7 Drop-downContext menu for importing the input files Clamp Documentation Page 46
48 2- Click on the small arrow next to the General folder to expand it, then select File System Figure 10.8 Import resources from the local file system into an existing project 4. Next, as shown in Figure 10.9, click on the Browse button on the top of the window to choose the folder of your choice. The selected folder will be displayed on the left side of the window, and the files inside the folder will be displayed on the right side. 5. Click the checkbox for the files that you want to run the pipeline on; currently the CLAMP pipeline can only process files with the.txt extension. Also, CLAMP assists you in selecting your desired files in three different ways: Filter Types, Select All and Deselect All. Filter Types: Allows you to define the type of files that will be imported. For example, you may only want to import files with the.txt extension Select All: Allows you to choose all displayed files Deselect All: Allow you to deselect the files that have already been selected Clamp Documentation Page 47
49 6. Click on the Browse button next to the Into folder field to choose the folder that you want to import your files to. Here, we keep the default directory 7. Click on the Finish button. 4- choose the folder of your choice 5- Click the checkbox for the files that you want to run the pipeline on 6- choose the folder that you want to import your files to 7- Click on the Finish button Figure 10.9 Window of input files selection Now, you can double click on the Input folder to see the imported files (Figure 10.10). Similarly, you can also double click on each file to view its content (Figure 10.11). Figure Imported files under the Input folder Clamp Documentation Page 48
50 Figure View the content of input file sample_1054.txt directly in the interface Clamp Documentation Page 49
51 11. Run the pipeline After you have configured the pipeline and imported the input files, you can start running the pipeline. To run a pipeline, simply click on the run icon at the top of the screen as shown in Figure Once the pipeline starts running, you can check the progress of the input file processing from the Console window and the progress bar at the bottom of the screen (Figure 11.2). You can always stop the processing at anytime by clicking on the red stop button next to the progress bar. Figure 11.1 Running the pipeline Figure 11.2 check the progress of the input file processing from the Console window Clamp Documentation Page 50
52 12. Output visualization Once running the pipeline is completed, the generated files are displayed in the Output folder. These files can be viewed in two different formats (.xmi,.txt): Clicking on a file with the.xmi extension allows you to view its original content annotated with recognized clinical concepts. Different types of clinical concepts will be highlighted with different colors. (Figure 12.1) Figure 12.1 View of text annotated with recognized clinical concepts Clicking on a file with the.txt extension will display a view of tab delimited, detailed output information in a new window. As shown in Figure 12.2, each line in the file illustrates the detailed information of one recognized clinical concept. The following information is included in a tab delimited output: 1. Start Index: Starting position of the recognized concept. 2. End Index: Ending position of the recognized concept. 3. Semantic Type: Semantic type of the recognized concept. 4. CUI: The Concept Unique Identifier of the concept in Unified Medical Language System (UMLS). If the pipeline does not include the model of UMLS encoder, the value of this column will be null. Clamp Documentation Page 51
53 5. Assertion: If the pipeline does not include the model of Assertion identifier, the value of this column will be null. 6. Concept Mention: Referring to a concept, i.e., named entity in the text. 6- Concept Mention 1- Start Index 5- Assertion 2- End Index 4- CUI 3- Semantic Type Figure 12.2 Tab delimited format of output files Clamp Documentation Page 52
54 13. Built-in pipelines In order to facilitate a convenient utility of CLAMP, a series of pipelines that could be directly adopted in common clinical applications are pre_built and displayed in PipelineLibrary (Figure 13.1). Users can directly drag one of them (e.g., smoking_status, Figure 13.2) from the PipelineLibrary and drop it under My Pipeline. The required NLP components of these pipelines are already configured, as illustrated in Figure CLAMP allows you to customize each of these components to fit your needs. Now, you need to import your files; for more information go to Import input files section. Figure 13.1 Built-in pipeline library in CLAMP Clamp Documentation Page 53
55 Figure 13.2 Dragging smoking_status and drop it under MyPipeline Figure 13.3 Built-in pipeline library in CLAMP Clamp Documentation Page 54
56 Depending on what your use case is, the current built-in pipelines are divided into the following categories: 1. General: automatically annotates concepts and their attribute for general use, including: CLAMP-ner: annotates the disease, procedure and medication concepts CLAMP-ner-attribute: annotates the attributes of disease (e.g., body location of a disease), lab procedure (e.g., value of a lab test ) and medication (e.g., dosage of a medication) concepts Disease-attribute: annotate the attributes of diseases, including body locations (e.g., left atrium), severity degrees (e.g., mild, severe) and uncertainty (e.g., probably). Lab-attribute: annotates the attributes of lab procedures Medication-attribute: annotates the attributes of medications 2. Disease_symptom: automatically annotates symptoms of diseases, including: Bleeding_extraction: annotates bleeding symptoms Colorectal_cancer: annotates symptoms of colorectal cancer 3. Behavior: automatically annotates behaviors of patients, including: Smoking_status: annotates whether or not the patient is in a smoking status, and whether the patient has a smoking history. Figure 13.4 illustrates an example of using the disease-attribute pipeline in our pipeline library to annotate attributes and their relations with diseases. Clamp Documentation Page 55
57 Figure 13.4 An example of disease attribute annotation using the pipeline library in CLAMP Clamp Documentation Page 56
58 14. Export pipeline as a jar file In order to export a pipeline as a jar file and use it in the command line version, please follow the steps below (Figure 14.1): 1. Go to your desired pipeline folder 2. Click on the small arrow next to it to expand it 3. Right click on the Components folder and select Export as jar 1. Go to your desired pipeline folder 2. Click on the small arrow to expand the folder 3. Right click on the Components folder and select Export as jar Figure 14.1 Export a pipeline as a jar Clamp Documentation Page 57
59 15. Annotation 15.1 Annotate corpus The CLAMP annotation module enables you to annotate customized entities and specify relations between them in your desired corpus. These annotations enable you to assign additional clinical information to a selected text and develop an annotated corpus that s more suitable to the specific task that you have. Task-specific models can be developed and used in the machine-learning modules of CLAMP or any other system of your choice. Before using this function, you need to: 1. Create a project 2. Import the files that you want to annotate After completing these steps, you will be able to annotate the imported files based on some predefined structure. The following steps will guide you on how to perform the steps mentioned above Create a project A) To create a project: 1. Click on the plus (+) sign at the top left corner of the screen as shown in Figure15.1. A) 1- Click on the plus sign Index Figure 15.1 Step 1 to create a new project 2. On the pop-up window, enter a name for your project, e.g., Drug_name_annotation, (Figure 15.2). 3. Select Corpus Annotation as the project type. 4. Click the Finish button. Clamp Documentation Page 58
60 A) 2- enter a name for your project Index A) 3- Select Corpus Annotation as the project type A) 4- Click the Finish button Figure 15.2 Creating a new Corpus Annotation project A new project with the name that you have specified is created and placed in the Corpus panel. (Figure 15.3) Figure 15.3 Creating a new Corpus Annotation project Double click the project name to view its content. The created project contains two main folders: Corpus: Contains the files that will be annotated Models: Contains the machine learning models generated from the annotated files.in addition, the prediction results generated from the n-fold cross-validation process and gold standard annotations are included in this folder. Clamp Documentation Page 59
61 Import annotation files After you have created a project, follow the steps defined below to import the files that you want to annotate: (Please note that you can import the files to either train or test folders.) A) To import the files that you want to annotate: 1. Right click on the train folder under the corpus folder in the CorpusView panel 2. Select the import function from the context menu (Figure 15.4). A pop-up window will appear. A) 1- Right click on the train folder under the corpus folder in the CorpusView panel A) 2- Select the import function from the context menu Figure 15.4 Context menu for importing the input files 3. On the pop-up window, select the import source. Here, you need to select File System which is already selected by default. 4. Click on the Next button Clamp Documentation Page 60
62 A) 3- Select the import sourse A) 4- Click on the next button 5. Click on the Browse button on the top of the window to choose the folder of your choice. The selected folder will be displayed on the left side of the window, and the content of the folder will be displayed on the right side. To import you desired files, check the checkboxes next to the files of your choice. You also have three options to choose from: Filter Types/ Select All/ Deselect All Filter Types: Allows you to define the type of files that will be imported. The only extensions that you will work with in CLAMP are txt, and.xmi. For example, you may only want to import files with the.txt extension Select All: Allows you to choose all displayed files Deselect All: Allow you to deselect the files that have already been selected 6. Click on the Browse button next to the Into folder field to choose the folder that you want to import your files to. Here, we keep the default directory 7. Click on the Finish button. Clamp Documentation Page 61
63 A) 5- Chosse the folder of your choice A) 6- choose the folder that you want to import your files to A) 7- Click the Finish button Now that the selected files have been imported to your desired folder, you can start annotating them. Double click on the files to open theim in the middle window and annotate them. Upon double clicking each file, you will notice that another file with the same name but a different extension (.xmi) has been added to your folder and displayed on the screen. This is the file type used by CLAMP for display and interaction purposes (Figure 15.5). Upon double clicking each file, you will notice that another file with the same name but a different extension (.xmi) has been added to your folder Figure 15.5 The content of an annotation file Clamp Documentation Page 62
64 Define entity & relation types Before starting annotation, you need to define the semantic types that you will use for this purpose. Semantic types in CLAMP refer to entities(e.g, problem/treatment/test ) and the relations between them. A) To define a new entity type: 1. Double click on the typedef.xmi file under the models folder to open it. Using this file, you will be able to define a schema for entities and the relation types among them: 2. Right click on the Entities node 3. Go to Add Child 4. Click on New Element A) 2- Right click on the enteties node A) 3- Go to Add Child 5. In the pop up window, enter a name for the element 6. Click the OK button A) 4- Click on new elements A) 5- Enter a name for the element A) 6- Click the OK button Clamp Documentation Page 63
65 The created element will be added to the Entity node (Figure 15.6) Figure 15.6 The created element will be added to the Entity node The above steps should be repeated for every element that you want to add to the Entity node. B) To define a new relation type: 1. Right click on the Relations node 2. Go to Add Child 3. Click on New Element B) 1- Right click on the Relations node B) 2- Go to Add Child B) 3- Click on new elements 4. In the pop up window, enter a name for the relation 5. Click the OK button Clamp Documentation Page 64
66 B) 4- Enter a name for the element B) 5- Click the OK button Then, you need to decide which entities are involved in this relation. There are two roles of arguments an entity can hold in a relation: From, and To. From refers to an independent entity while To indicates the dependent entity. C) To select the entities that are involved in a relation: 1. Right click on the newly created relation 2. Go to Add Attribute 3. Click on New Attribute C) 1- Right click on the newly created relation C) 3- Click on new Attribute C) 2- Go to Add Attribute Clamp Documentation Page 65
67 4. In the pop up window, enter a name for the new attribute (you will use from for the independent entity, and to for the dependent entity) 5. Enter the actual name of the entities for the Value field 6. Click the OK button 7. Click on Save at the top of the window A) 4- Enter a name for the new attribute A) 5- Enter the actual name of the entities A) 6- Click the OK button Clamp Documentation Page 66
68 Start Annotation Now that you have set your desired schema, you are ready to start annotating your corpus. First, open your desired.xmi file, then: To assign entity: Place your mouse over a word/phrase to highlight it and assign an appropriate entity to the selected text. (Figure 15.6) Figure 15.6 Named entity annotation To assign relation: By dragging your mouse from an independent entity and dropping it to a dependent entity, the names of possible relations will occur. Choose the appropriate relation name by clicking on one of the displayed names. (Figure 15.7) Figure 15.7 Relation annotation Please remember that you can only assign a relation to the entities that have already been defined in that relation. Once you have completed annotating the corpus, save your changes by clicking on the save button at the top of the screen. Clamp Documentation Page 67
69 Visualization of entity & relation Although different colors are automatically assigned to the different items in the Display Option, (Figure 15.8), you are able to change them at any time. Figure 15.8 Change visualization of the annotated entity/relation A) To change the default colors for semantic types: 1. Double click on the default color for the entity of your choice 2. Pick a new color from the color picker window 3. Press the OK button A) 1- Double click the on the default color of your choice A) 2- Pick a new color A) 3- Press the OK button Clamp Documentation Page 68
70 Pre-Annotation of entity and relation As annotating a corpus from scratch may be a time-consuming and costly process, CLAMP offers an advanced feature called pre-annotation function which facilitates this process. The pre-annotation function relies on the existing models in CLAMP and is highly customizable. A) Using pre-annotation function 1. Choose your desired pipeline to annotate your files in a corpus project. For more information on how to run a pipeline, go to Run the pipeline section. 2. Select the.xmi files which contains the predicted named entities from the output folder. 3. Copy them into the train/test folder of your desired corpus annotation project. To copy, right click on the selected files and choose copy. A) 1- Choose your desired created pipeline A) 3- Copy them into the train/test folder of your desired corpus annotation project A) 2- Select the.xmi files which contains the predicted named entities from the output folder pipeline Double click on the files to view their contents in a new window. As you can see in Figure 15.9, the identified named entities in the file are already highlighted. Now you can start your own annotation. Clamp Documentation Page 69
71 Figure 15.9 A file with pre-annotated named entities Clamp Documentation Page 70
72 16. Machine learning model development Clamp enables you to build your own machine learning model based on a corpus that you have already annotated or a pre annotated one that you have imported into a corpus annotation project. The model can be used for predictions on new files. In the current version of Clamp, CRF (Conditional Random Field) is used to build machine-learning model for named entity recognition (NER). The first step to build a Machine Learning model is to configure its schema. After configuring the schema, you will be able to start running the training model and evaluation processes. Once these processes are completed, you can view the generated model, its associated log files, and named entities predicted by the model in the output folder. The following steps will guide you on how to perform the steps mentioned above Building machine learning models (NER model) 1. Select your desired train folder on the Corpus panel 2. Click on the Train Model button at the top of the window as shown in Figure 16.1 Figure On the pop up window as shown in Figure 16.2, enter a name for the model that you are building 4. Click the checkbox for the features that you want to include in your model Clamp Documentation Page 71
73 5. In the Evaluation box, choose if you want to test the built model against a test dataset and/or if you want to do a n-fold cross-validation during the training process. If you choose to test the model against a test set, make sure that you have your desired annotated xmi files in the folder of your choice. You can browse for the folder by clicking on the three dot button next to the checkboxes. With the n-fold cross validation, you are not required to do so as the training data will be used to test the model performance. 6. Click on the Finish button to start building the model. Once the building process starts, you can check the progress in the Console window, as well as the progress bar at the bottom of the screen. You can also stop the building process at anytime by clicking the red stop button in the Progress window. 3. Enter a name for the model that you are building 4. Click the checkbox for the features that you want to include in your model 5. Choose if you want to test the built model with a test dataset and/or if you want to do a n-fold cross-validation during the training process 6. Click on the Finish button to start building the model Figure 16.2 Configuration window for machine learning model building Clamp Documentation Page 72
74 Note: During the model building process, the training files can not be annotated. Clicking on the text of the training files pops up an alert window indicating that the user operation is waiting for a function to complete, (Figure 16.3). Figure 16.3 Annotations on the training file will be paused during the model building process Clamp Documentation Page 73
75 16.2 Check output models & logs By default the built models, their associated logs, and the named entities predicted by each model (in the output sub-folder) are stored in the models folder. As shown in Figure 16.4, both the model built during n-fold cross validation and the model trained on the whole training set are also stored in the directory. The content of the log files includes the output information of the training process and the evaluation performance of each specific folder for cross validation. (Figure 16.5) Figure 16.4 Annotations on the training file will be paused during the model building process Figure 16.5 Cross-validation performance in training.fold0.log Clamp Documentation Page 74
76 16.3 Use your own model in pipeline The steps below show how you should use your own model to recognize named entities: 1. Make sure that you have selected your desired project folder on the Corpus panel 2. Go to models> model_xxx> output, then, right click on the file labled model.jar and select copy (Figure 16.6) 3. Go to the pipeline panel and select the pipeline of your choice 4. Click on the small arrow next to it to expand it 5. Go to Components -> Named Entity Recognizer -> DF_CRF_based_named_entity_recognize 6. Paste the copied file into CRF based name entity recognizer folder by righ clicking on the folder and choosing past 7. Double click on the Config.conf file in the CRF based named entity recognizer folder to open it 8. Click on the three dots button to replace the default model for CRF based named entity recognizer with your own model (Figure 16.7) 9. Click on the Open button 10. Click the save button at the top of the page to save the changes (Figure 16.8) 2. Go to models> model_xxx> output, then, right click on the file labled model.jar and select copy 5.Go to Components > Named Entity Recognizer > DF_CRF_based_named_entity_recognizer Figure 16.6 How to use your own model to recognize name entities Clamp Documentation Page 75
77 8. Click on the three dots button to replace the default model for CRF based named entity recognizer with your own model 9. Click on the Open button Figure 16.7 Replace the default model with your own model Figure 16.8 Click the Save button at the top of the page Clamp Documentation Page 76
78 16.4 Visualization for error analysis You will be able to evaluate the performance of the NER model only if you have already checked the checkbox(es) for Use test set or/and CV, fold when creating the model. For more information on creating a new NER model, go to Building machine learning models section. Once the model is built, you can conduct an error analysis to compare the goldstandard annotations with the predicted ones (the annotations that are built based on the model that you have specified). To perform error analysis: Double click on one of the.xmi files listed in the output folder of your choice on the corpus panel. This will open a new window where you can see the original text along with both goldstandard and predicted annotations (Figure 16.9). Figure The.xmi file content which includes the original text along with both gold-standard and predicted annotations Please note that all named entities in both gold-standard and predicted annotations are listed on the "Display Options" panel. You can choose which named entities to be highlighted in the text file and assign different colors to them as described in Visualization of entity and relation types section. Clamp Documentation Page 77
CSC 5930/9010: Text Mining GATE Developer Overview
 1 CSC 5930/9010: Text Mining GATE Developer Overview Dr. Paula Matuszek Paula.Matuszek@villanova.edu Paula.Matuszek@gmail.com (610) 647-9789 GATE Components 2 We will deal primarily with GATE Developer:
1 CSC 5930/9010: Text Mining GATE Developer Overview Dr. Paula Matuszek Paula.Matuszek@villanova.edu Paula.Matuszek@gmail.com (610) 647-9789 GATE Components 2 We will deal primarily with GATE Developer:
Using Relations for Identification and Normalization of Disorders: Team CLEAR in the ShARe/CLEF 2013 ehealth Evaluation Lab
 Using Relations for Identification and Normalization of Disorders: Team CLEAR in the ShARe/CLEF 2013 ehealth Evaluation Lab James Gung University of Colorado, Department of Computer Science Boulder, CO
Using Relations for Identification and Normalization of Disorders: Team CLEAR in the ShARe/CLEF 2013 ehealth Evaluation Lab James Gung University of Colorado, Department of Computer Science Boulder, CO
An UIMA based Tool Suite for Semantic Text Processing
 An UIMA based Tool Suite for Semantic Text Processing Katrin Tomanek, Ekaterina Buyko, Udo Hahn Jena University Language & Information Engineering Lab StemNet Knowledge Management for Immunology in life
An UIMA based Tool Suite for Semantic Text Processing Katrin Tomanek, Ekaterina Buyko, Udo Hahn Jena University Language & Information Engineering Lab StemNet Knowledge Management for Immunology in life
In this tutorial, we will understand how to use the OpenNLP library to build an efficient text processing service.
 About the Tutorial Apache OpenNLP is an open source Java library which is used process Natural Language text. OpenNLP provides services such as tokenization, sentence segmentation, part-of-speech tagging,
About the Tutorial Apache OpenNLP is an open source Java library which is used process Natural Language text. OpenNLP provides services such as tokenization, sentence segmentation, part-of-speech tagging,
i2b2 Workbench Developer s Guide: Eclipse Neon & i2b2 Source Code
 i2b2 Workbench Developer s Guide: Eclipse Neon & i2b2 Source Code About this guide Informatics for Integrating Biology and the Bedside (i2b2) began as one of the sponsored initiatives of the NIH Roadmap
i2b2 Workbench Developer s Guide: Eclipse Neon & i2b2 Source Code About this guide Informatics for Integrating Biology and the Bedside (i2b2) began as one of the sponsored initiatives of the NIH Roadmap
Module 1: Information Extraction
 Module 1: Information Extraction Introduction to GATE Developer The University of Sheffield, 1995-2014 This work is licenced under the Creative Commons Attribution-NonCommercial-ShareAlike Licence About
Module 1: Information Extraction Introduction to GATE Developer The University of Sheffield, 1995-2014 This work is licenced under the Creative Commons Attribution-NonCommercial-ShareAlike Licence About
Tutorial. Identification of Variants Using GATK. Sample to Insight. November 21, 2017
 Identification of Variants Using GATK November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Identification of Variants Using GATK November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Downloading and editing signatures!
 Downloading and editing email signatures! The templates are customizable and can be edited and saved with any html editor like Dreamweaver. If this is not available we have two alternatives for editing/saving
Downloading and editing email signatures! The templates are customizable and can be edited and saved with any html editor like Dreamweaver. If this is not available we have two alternatives for editing/saving
Installation and Configuration Guide
 Sysgem SysMan Utilities (Professional Edition) Installation and Configuration Guide By Sysgem AG Sysgem is a trademark of Sysgem AG. Other brands and products are registered trademarks of their respective
Sysgem SysMan Utilities (Professional Edition) Installation and Configuration Guide By Sysgem AG Sysgem is a trademark of Sysgem AG. Other brands and products are registered trademarks of their respective
Implementing a Variety of Linguistic Annotations
 Implementing a Variety of Linguistic Annotations through a Common Web-Service Interface Adam Funk, Ian Roberts, Wim Peters University of Sheffield 18 May 2010 Adam Funk, Ian Roberts, Wim Peters Implementing
Implementing a Variety of Linguistic Annotations through a Common Web-Service Interface Adam Funk, Ian Roberts, Wim Peters University of Sheffield 18 May 2010 Adam Funk, Ian Roberts, Wim Peters Implementing
Natural Language Processing Pipelines to Annotate BioC Collections with an Application to the NCBI Disease Corpus
 Natural Language Processing Pipelines to Annotate BioC Collections with an Application to the NCBI Disease Corpus Donald C. Comeau *, Haibin Liu, Rezarta Islamaj Doğan and W. John Wilbur National Center
Natural Language Processing Pipelines to Annotate BioC Collections with an Application to the NCBI Disease Corpus Donald C. Comeau *, Haibin Liu, Rezarta Islamaj Doğan and W. John Wilbur National Center
The Goal of this Document. Where to Start?
 A QUICK INTRODUCTION TO THE SEMILAR APPLICATION Mihai Lintean, Rajendra Banjade, and Vasile Rus vrus@memphis.edu linteam@gmail.com rbanjade@memphis.edu The Goal of this Document This document introduce
A QUICK INTRODUCTION TO THE SEMILAR APPLICATION Mihai Lintean, Rajendra Banjade, and Vasile Rus vrus@memphis.edu linteam@gmail.com rbanjade@memphis.edu The Goal of this Document This document introduce
Eclipse Environment Setup
 Eclipse Environment Setup Adapted from a document from Jeffrey Miller and the CS201 team by Shiyuan Sheng. Introduction This lab document will go over the steps to install and set up Eclipse, which is
Eclipse Environment Setup Adapted from a document from Jeffrey Miller and the CS201 team by Shiyuan Sheng. Introduction This lab document will go over the steps to install and set up Eclipse, which is
This tutorial is designed for all Java enthusiasts who want to learn document type detection and content extraction using Apache Tika.
 About the Tutorial This tutorial provides a basic understanding of Apache Tika library, the file formats it supports, as well as content and metadata extraction using Apache Tika. Audience This tutorial
About the Tutorial This tutorial provides a basic understanding of Apache Tika library, the file formats it supports, as well as content and metadata extraction using Apache Tika. Audience This tutorial
Unstructured Information Management Architecture (UIMA) Graham Wilcock University of Helsinki
 Unstructured Information Management Architecture (UIMA) Graham Wilcock University of Helsinki Overview What is UIMA? A framework for NLP tasks and tools Part-of-Speech Tagging Full Parsing Shallow Parsing
Unstructured Information Management Architecture (UIMA) Graham Wilcock University of Helsinki Overview What is UIMA? A framework for NLP tasks and tools Part-of-Speech Tagging Full Parsing Shallow Parsing
Let s get parsing! Each component processes the Doc object, then passes it on. doc.is_parsed attribute checks whether a Doc object has been parsed
 Let s get parsing! SpaCy default model includes tagger, parser and entity recognizer nlp = spacy.load('en ) tells spacy to use "en" with ["tagger", "parser", "ner"] Each component processes the Doc object,
Let s get parsing! SpaCy default model includes tagger, parser and entity recognizer nlp = spacy.load('en ) tells spacy to use "en" with ["tagger", "parser", "ner"] Each component processes the Doc object,
EUSurvey OSS Installation Guide
 Prerequisites... 2 Tools... 2 Java 7 SDK... 2 MySQL 5.6 DB and Client (Workbench)... 4 Tomcat 7... 8 Spring Tool Suite... 11 Knowledge... 12 Control System Services... 12 Prepare the Database... 14 Create
Prerequisites... 2 Tools... 2 Java 7 SDK... 2 MySQL 5.6 DB and Client (Workbench)... 4 Tomcat 7... 8 Spring Tool Suite... 11 Knowledge... 12 Control System Services... 12 Prepare the Database... 14 Create
CSCI 201 Lab 1 Environment Setup
 CSCI 201 Lab 1 Environment Setup "The journey of a thousand miles begins with one step." - Lao Tzu Introduction This lab document will go over the steps to install and set up Eclipse, which is a Java integrated
CSCI 201 Lab 1 Environment Setup "The journey of a thousand miles begins with one step." - Lao Tzu Introduction This lab document will go over the steps to install and set up Eclipse, which is a Java integrated
Multi-Sponsor Environment. SAS Clinical Trial Data Transparency User Guide
 Multi-Sponsor Environment SAS Clinical Trial Data Transparency User Guide Version 6.0 01 December 2017 Contents Contents 1 Overview...1 2 Setting up Your Account...3 2.1 Completing the Initial Email and
Multi-Sponsor Environment SAS Clinical Trial Data Transparency User Guide Version 6.0 01 December 2017 Contents Contents 1 Overview...1 2 Setting up Your Account...3 2.1 Completing the Initial Email and
Vision Plan. For KDD- Service based Numerical Entity Searcher (KSNES) Version 2.0
 Vision Plan For KDD- Service based Numerical Entity Searcher (KSNES) Version 2.0 Submitted in partial fulfillment of the Masters of Software Engineering Degree. Naga Sowjanya Karumuri CIS 895 MSE Project
Vision Plan For KDD- Service based Numerical Entity Searcher (KSNES) Version 2.0 Submitted in partial fulfillment of the Masters of Software Engineering Degree. Naga Sowjanya Karumuri CIS 895 MSE Project
Apache UIMA and Mayo ctakes
 Apache and Mayo and how it is used in the clinical domain March 16, 2012 Apache and Mayo Outline 1 Apache and Mayo Outline 1 2 Introducing Pipeline Modules Apache and Mayo What is? (You - eee - muh) Unstructured
Apache and Mayo and how it is used in the clinical domain March 16, 2012 Apache and Mayo Outline 1 Apache and Mayo Outline 1 2 Introducing Pipeline Modules Apache and Mayo What is? (You - eee - muh) Unstructured
Install Guide. Version 1.0 Demonstration. Copyright 2007 MGH
 Install Guide Version 1.0 Demonstration Copyright 2007 MGH Table of Contents About this Guide iii Prerequisites 1 Downloads and Installation 1 Install & Configure 3 Setting up the cell and hive within
Install Guide Version 1.0 Demonstration Copyright 2007 MGH Table of Contents About this Guide iii Prerequisites 1 Downloads and Installation 1 Install & Configure 3 Setting up the cell and hive within
Beginner Workshop Activity Guide 2012 User Conference
 Beginner Workshop Activity Guide 2012 User Conference TUESDAY, MARCH 6 2:00PM 5:00 PM Beginner Training Workshop Attendees will learn the end user functions of OU Campus TM. They will learn how to log
Beginner Workshop Activity Guide 2012 User Conference TUESDAY, MARCH 6 2:00PM 5:00 PM Beginner Training Workshop Attendees will learn the end user functions of OU Campus TM. They will learn how to log
23 - Report & Export
 23 - Report & Export Contents 23 - REPORT & EXPORT... 1 SMART PUBLISHER... 1 Opening Smart Publisher... 1 Smart Publisher Settings... 2 The Finished Report... 5 Alias Names for Codes... 6 The Word Template
23 - Report & Export Contents 23 - REPORT & EXPORT... 1 SMART PUBLISHER... 1 Opening Smart Publisher... 1 Smart Publisher Settings... 2 The Finished Report... 5 Alias Names for Codes... 6 The Word Template
Story Workbench Quickstart Guide Version 1.2.0
 1 Basic Concepts Story Workbench Quickstart Guide Version 1.2.0 Mark A. Finlayson (markaf@mit.edu) Annotation An indivisible piece of data attached to a text is called an annotation. Annotations, also
1 Basic Concepts Story Workbench Quickstart Guide Version 1.2.0 Mark A. Finlayson (markaf@mit.edu) Annotation An indivisible piece of data attached to a text is called an annotation. Annotations, also
EUSurvey Installation Guide
 EUSurvey Installation Guide Guide to a successful installation of EUSurvey May 20 th, 2015 Version 1.2 (version family) 1 Content 1. Overview... 3 2. Prerequisites... 3 Tools... 4 Java SDK... 4 MySQL Database
EUSurvey Installation Guide Guide to a successful installation of EUSurvey May 20 th, 2015 Version 1.2 (version family) 1 Content 1. Overview... 3 2. Prerequisites... 3 Tools... 4 Java SDK... 4 MySQL Database
Privacy and Security in Online Social Networks Department of Computer Science and Engineering Indian Institute of Technology, Madras
 Privacy and Security in Online Social Networks Department of Computer Science and Engineering Indian Institute of Technology, Madras Lecture - 25 Tutorial 5: Analyzing text using Python NLTK Hi everyone,
Privacy and Security in Online Social Networks Department of Computer Science and Engineering Indian Institute of Technology, Madras Lecture - 25 Tutorial 5: Analyzing text using Python NLTK Hi everyone,
@Note2 tutorial. Hugo Costa Ruben Rodrigues Miguel Rocha
 @Note2 tutorial Hugo Costa (hcosta@silicolife.com) Ruben Rodrigues (pg25227@alunos.uminho.pt) Miguel Rocha (mrocha@di.uminho.pt) 23-01-2018 The document presents a typical workflow using @Note2 platform
@Note2 tutorial Hugo Costa (hcosta@silicolife.com) Ruben Rodrigues (pg25227@alunos.uminho.pt) Miguel Rocha (mrocha@di.uminho.pt) 23-01-2018 The document presents a typical workflow using @Note2 platform
Using Blackboard Drive to upload/manage content in Blackboard Learn
 Using Blackboard Drive to upload/manage content in Blackboard Learn Downloading and Installing Blackboard Drive 1. Login to Blackboard Learn, and click on the Utilities Tab. 2. Locate the Blackboard Drive
Using Blackboard Drive to upload/manage content in Blackboard Learn Downloading and Installing Blackboard Drive 1. Login to Blackboard Learn, and click on the Utilities Tab. 2. Locate the Blackboard Drive
User Guide. Copyright Wordfast, LLC All rights reserved.
 User Guide All rights reserved. Table of Contents About this Guide... 7 Conventions...7 Typographical... 7 Icons... 7 1 Release Notes Summary... 8 New Features and Improvements... 8 Fixed Issues... 8 Known
User Guide All rights reserved. Table of Contents About this Guide... 7 Conventions...7 Typographical... 7 Icons... 7 1 Release Notes Summary... 8 New Features and Improvements... 8 Fixed Issues... 8 Known
Using Microsoft Office 2003 Intermediate Word Handout INFORMATION TECHNOLOGY SERVICES California State University, Los Angeles Version 1.
 Using Microsoft Office 2003 Intermediate Word Handout INFORMATION TECHNOLOGY SERVICES California State University, Los Angeles Version 1.2 Summer 2010 Table of Contents Intermediate Microsoft Word 2003...
Using Microsoft Office 2003 Intermediate Word Handout INFORMATION TECHNOLOGY SERVICES California State University, Los Angeles Version 1.2 Summer 2010 Table of Contents Intermediate Microsoft Word 2003...
UIMA Tools Guide and Reference
 UIMA Tools Guide and Reference Written and maintained by the Apache UIMA Development Community Version 2.3.0-incubating Copyright 2004, 2006 International Business Machines Corporation Copyright 2006,
UIMA Tools Guide and Reference Written and maintained by the Apache UIMA Development Community Version 2.3.0-incubating Copyright 2004, 2006 International Business Machines Corporation Copyright 2006,
McMaster Service-Based ehealth Integration Environment (MACSeie) Installation Guide July 24, 2009
 McMaster Service-Based ehealth Integration Environment (MACSeie) Installation Guide July 24, 2009 Richard Lyn lynrf@mcmaster.ca Jianwei Yang yangj29@mcmaster.ca Document Revision History Rev. Level Date
McMaster Service-Based ehealth Integration Environment (MACSeie) Installation Guide July 24, 2009 Richard Lyn lynrf@mcmaster.ca Jianwei Yang yangj29@mcmaster.ca Document Revision History Rev. Level Date
PowerNotes. Physician. Created November 2011 by: Clinical Informaticist Team
 PowerNotes Physician Created November 2011 by: Clinical Informaticist Team Table of Contents Opening PowerNotes 1 Finding a PowerNote 1 Encounter Pathway 1 Existing 2 Precompleted 2 Catalog 3 Recent 3
PowerNotes Physician Created November 2011 by: Clinical Informaticist Team Table of Contents Opening PowerNotes 1 Finding a PowerNote 1 Encounter Pathway 1 Existing 2 Precompleted 2 Catalog 3 Recent 3
EMC Documentum Composer
 EMC Documentum Composer Version 6.0 SP1.5 User Guide P/N 300 005 253 A02 EMC Corporation Corporate Headquarters: Hopkinton, MA 01748 9103 1 508 435 1000 www.emc.com Copyright 2008 EMC Corporation. All
EMC Documentum Composer Version 6.0 SP1.5 User Guide P/N 300 005 253 A02 EMC Corporation Corporate Headquarters: Hopkinton, MA 01748 9103 1 508 435 1000 www.emc.com Copyright 2008 EMC Corporation. All
Machine Learning in GATE
 Machine Learning in GATE Angus Roberts, Horacio Saggion, Genevieve Gorrell Recap Previous two days looked at knowledge engineered IE This session looks at machine learned IE Supervised learning Effort
Machine Learning in GATE Angus Roberts, Horacio Saggion, Genevieve Gorrell Recap Previous two days looked at knowledge engineered IE This session looks at machine learned IE Supervised learning Effort
Table of Contents. Troubleshooting Guide for Home Users
 Table of Contents Introduction... 1 Chapter 1: System Requirements... 2 Recommended and Minimum Supported Requirements... 2 Additional Information... 3 Internet Connectivity... 3 Tablet Compatibility...
Table of Contents Introduction... 1 Chapter 1: System Requirements... 2 Recommended and Minimum Supported Requirements... 2 Additional Information... 3 Internet Connectivity... 3 Tablet Compatibility...
LAB 3: Text processing + Apache OpenNLP
 LAB 3: Text processing + Apache OpenNLP 1. Motivation: The text that was derived (e.g., crawling + using Apache Tika) must be processed before being used in an information retrieval system. Text processing
LAB 3: Text processing + Apache OpenNLP 1. Motivation: The text that was derived (e.g., crawling + using Apache Tika) must be processed before being used in an information retrieval system. Text processing
Mend for Eclipse quick start guide local analysis
 The Semmle Mend for Eclipse plugin allows users to view Semmle results in Eclipse. This document describes how to install and use the plugin for local analysis. You can install the plugin using a Semmle
The Semmle Mend for Eclipse plugin allows users to view Semmle results in Eclipse. This document describes how to install and use the plugin for local analysis. You can install the plugin using a Semmle
Halcyon Message Sender GUI. v6.0 User Guide
 GUI v6.0 User Guide Copyright Copyright HelpSystems, LLC. All rights reserved. www.helpsystems.com US: +1 952-933-0609 Outside the U.S.: +44 (0) 870 120 3148 IBM, AS/400, OS/400, System i, System i5, i5/os,
GUI v6.0 User Guide Copyright Copyright HelpSystems, LLC. All rights reserved. www.helpsystems.com US: +1 952-933-0609 Outside the U.S.: +44 (0) 870 120 3148 IBM, AS/400, OS/400, System i, System i5, i5/os,
Installation and Upgrade Guide Zend Studio 9.x
 Installation and Upgrade Guide Zend Studio 9.x By Zend Technologies, Inc. www.zend.com Disclaimer The information in this document is subject to change without notice and does not represent a commitment
Installation and Upgrade Guide Zend Studio 9.x By Zend Technologies, Inc. www.zend.com Disclaimer The information in this document is subject to change without notice and does not represent a commitment
NLP Chain. Giuseppe Castellucci Web Mining & Retrieval a.a. 2013/2014
 NLP Chain Giuseppe Castellucci castellucci@ing.uniroma2.it Web Mining & Retrieval a.a. 2013/2014 Outline NLP chains RevNLT Exercise NLP chain Automatic analysis of texts At different levels Token Morphological
NLP Chain Giuseppe Castellucci castellucci@ing.uniroma2.it Web Mining & Retrieval a.a. 2013/2014 Outline NLP chains RevNLT Exercise NLP chain Automatic analysis of texts At different levels Token Morphological
Talend Open Studio for Data Quality. User Guide 5.5.2
 Talend Open Studio for Data Quality User Guide 5.5.2 Talend Open Studio for Data Quality Adapted for v5.5. Supersedes previous releases. Publication date: January 29, 2015 Copyleft This documentation is
Talend Open Studio for Data Quality User Guide 5.5.2 Talend Open Studio for Data Quality Adapted for v5.5. Supersedes previous releases. Publication date: January 29, 2015 Copyleft This documentation is
Homework 2: Parsing and Machine Learning
 Homework 2: Parsing and Machine Learning COMS W4705_001: Natural Language Processing Prof. Kathleen McKeown, Fall 2017 Due: Saturday, October 14th, 2017, 2:00 PM This assignment will consist of tasks in
Homework 2: Parsing and Machine Learning COMS W4705_001: Natural Language Processing Prof. Kathleen McKeown, Fall 2017 Due: Saturday, October 14th, 2017, 2:00 PM This assignment will consist of tasks in
Enterprise Architect. User Guide Series. Maintenance. Author: Sparx Systems. Date: 30/06/2017. Version: 1.0 CREATED WITH
 Enterprise Architect User Guide Series Maintenance Author: Sparx Systems Date: 30/06/2017 Version: 1.0 CREATED WITH Table of Contents Maintenance 3 Working on Maintenance Items 5 Create Maintenance Items
Enterprise Architect User Guide Series Maintenance Author: Sparx Systems Date: 30/06/2017 Version: 1.0 CREATED WITH Table of Contents Maintenance 3 Working on Maintenance Items 5 Create Maintenance Items
UIMA-based Annotation Type System for a Text Mining Architecture
 UIMA-based Annotation Type System for a Text Mining Architecture Udo Hahn, Ekaterina Buyko, Katrin Tomanek, Scott Piao, Yoshimasa Tsuruoka, John McNaught, Sophia Ananiadou Jena University Language and
UIMA-based Annotation Type System for a Text Mining Architecture Udo Hahn, Ekaterina Buyko, Katrin Tomanek, Scott Piao, Yoshimasa Tsuruoka, John McNaught, Sophia Ananiadou Jena University Language and
Installation and Upgrade Guide Zend Studio 9.x
 Installation and Upgrade Guide Zend Studio 9.x By Zend Technologies, Inc. www.zend.com Disclaimer The information in this document is subject to change without notice and does not represent a commitment
Installation and Upgrade Guide Zend Studio 9.x By Zend Technologies, Inc. www.zend.com Disclaimer The information in this document is subject to change without notice and does not represent a commitment
EMC Documentum Composer
 EMC Documentum Composer Version 6 SP1 User Guide P/N 300 005 253 A01 EMC Corporation Corporate Headquarters: Hopkinton, MA 01748 9103 1 508 435 1000 www.emc.com Copyright 2008 EMC Corporation. All rights
EMC Documentum Composer Version 6 SP1 User Guide P/N 300 005 253 A01 EMC Corporation Corporate Headquarters: Hopkinton, MA 01748 9103 1 508 435 1000 www.emc.com Copyright 2008 EMC Corporation. All rights
UIMA Tools Guide and Reference
 UIMA Tools Guide and Reference Written and maintained by the Apache UIMA Development Community Version 3.0.0 Copyright 2006, 2018 The Apache Software Foundation License and Disclaimer. The ASF licenses
UIMA Tools Guide and Reference Written and maintained by the Apache UIMA Development Community Version 3.0.0 Copyright 2006, 2018 The Apache Software Foundation License and Disclaimer. The ASF licenses
IBM. IBM WebSphere Application Server Migration Toolkit. WebSphere Application Server. Version 9.0 Release
 WebSphere Application Server IBM IBM WebSphere Application Server Migration Toolkit Version 9.0 Release 18.0.0.3 Contents Chapter 1. Overview......... 1 Chapter 2. What's new........ 5 Chapter 3. Support..........
WebSphere Application Server IBM IBM WebSphere Application Server Migration Toolkit Version 9.0 Release 18.0.0.3 Contents Chapter 1. Overview......... 1 Chapter 2. What's new........ 5 Chapter 3. Support..........
Enterprise Architect. User Guide Series. Maintenance
 Enterprise Architect User Guide Series Maintenance In Sparx Systems Enterprise Architect, Maintenance items (such as defects, tasks and events) are managed as element properties. Change and Issue elements
Enterprise Architect User Guide Series Maintenance In Sparx Systems Enterprise Architect, Maintenance items (such as defects, tasks and events) are managed as element properties. Change and Issue elements
User Guide. Copyright Wordfast, LLC All rights reserved.
 User Guide All rights reserved. Table of Contents Summary... 7 New Features and Improvements... 7 Fixed Issues... 8 1 About Pro... 9 Key Advantages... 9 2 Get Started... 10 Requirements... 10 Install and
User Guide All rights reserved. Table of Contents Summary... 7 New Features and Improvements... 7 Fixed Issues... 8 1 About Pro... 9 Key Advantages... 9 2 Get Started... 10 Requirements... 10 Install and
Tutorial on Text Mining for the Going Digital initiative. Natural Language Processing (NLP), University of Essex
 Tutorial on Text Mining for the Going Digital initiative Natural Language Processing (NLP), University of Essex 6 February, 2013 Topics of This Tutorial o Information Extraction (IE) o Examples of IE systems
Tutorial on Text Mining for the Going Digital initiative Natural Language Processing (NLP), University of Essex 6 February, 2013 Topics of This Tutorial o Information Extraction (IE) o Examples of IE systems
Precise Medication Extraction using Agile Text Mining
 Precise Medication Extraction using Agile Text Mining Chaitanya Shivade *, James Cormack, David Milward * The Ohio State University, Columbus, Ohio, USA Linguamatics Ltd, Cambridge, UK shivade@cse.ohio-state.edu,
Precise Medication Extraction using Agile Text Mining Chaitanya Shivade *, James Cormack, David Milward * The Ohio State University, Columbus, Ohio, USA Linguamatics Ltd, Cambridge, UK shivade@cse.ohio-state.edu,
Tutorial to QuotationFinder_0.4.4
 Tutorial to QuotationFinder_0.4.4 What is Quotation Finder and for which purposes can it be used? Quotation Finder is a tool for the automatic comparison of fully digitized texts. It can detect quotations,
Tutorial to QuotationFinder_0.4.4 What is Quotation Finder and for which purposes can it be used? Quotation Finder is a tool for the automatic comparison of fully digitized texts. It can detect quotations,
NLP in practice, an example: Semantic Role Labeling
 NLP in practice, an example: Semantic Role Labeling Anders Björkelund Lund University, Dept. of Computer Science anders.bjorkelund@cs.lth.se October 15, 2010 Anders Björkelund NLP in practice, an example:
NLP in practice, an example: Semantic Role Labeling Anders Björkelund Lund University, Dept. of Computer Science anders.bjorkelund@cs.lth.se October 15, 2010 Anders Björkelund NLP in practice, an example:
EMC Documentum Composer
 EMC Documentum Composer Version 6.5 SP2 User Guide P/N 300-009-462 A01 EMC Corporation Corporate Headquarters: Hopkinton, MA 01748-9103 1-508-435-1000 www.emc.com Copyright 2008 2009 EMC Corporation. All
EMC Documentum Composer Version 6.5 SP2 User Guide P/N 300-009-462 A01 EMC Corporation Corporate Headquarters: Hopkinton, MA 01748-9103 1-508-435-1000 www.emc.com Copyright 2008 2009 EMC Corporation. All
WA2018 Programming REST Web Services with JAX-RS WebLogic 12c / Eclipse. Student Labs. Web Age Solutions Inc.
 WA2018 Programming REST Web Services with JAX-RS 1.1 - WebLogic 12c / Eclipse Student Labs Web Age Solutions Inc. Copyright 2012 Web Age Solutions Inc. 1 Table of Contents Lab 1 - Configure the Development
WA2018 Programming REST Web Services with JAX-RS 1.1 - WebLogic 12c / Eclipse Student Labs Web Age Solutions Inc. Copyright 2012 Web Age Solutions Inc. 1 Table of Contents Lab 1 - Configure the Development
User Guide. Copyright Wordfast, LLC All rights reserved.
 User Guide All rights reserved. Table of Contents About this Guide... 7 Conventions...7 Typographical... 7 Icons... 7 1 Release Notes Summary... 8 New Features and Improvements... 8 Fixed Issues... 8 2
User Guide All rights reserved. Table of Contents About this Guide... 7 Conventions...7 Typographical... 7 Icons... 7 1 Release Notes Summary... 8 New Features and Improvements... 8 Fixed Issues... 8 2
Read&Write 5 GOLD FOR MAC MANUAL
 Read&Write 5 GOLD FOR MAC MANUAL ABBYY FineReader Engine 8.0 ABBYY Software Ltd. 2005. ABBYY FineReader the keenest eye in OCR. ABBYY, FINEREADER and ABBYY FineReader are registered trademarks of ABBYY
Read&Write 5 GOLD FOR MAC MANUAL ABBYY FineReader Engine 8.0 ABBYY Software Ltd. 2005. ABBYY FineReader the keenest eye in OCR. ABBYY, FINEREADER and ABBYY FineReader are registered trademarks of ABBYY
Designing Reports. eivf Designing Reports Note Types 1
 Designing Reports Designing Reports...1 Note Types...3 Notes...3 Shorthands...3 Quick Note...3 Click N Build...3 Reports (Data Plates )...3 Most commonly use of the Note Types...4 Notes...5 To create a
Designing Reports Designing Reports...1 Note Types...3 Notes...3 Shorthands...3 Quick Note...3 Click N Build...3 Reports (Data Plates )...3 Most commonly use of the Note Types...4 Notes...5 To create a
POOSL IDE Installation Manual
 Embedded Systems Innovation by TNO POOSL IDE Installation Manual Tool version 4.1.0 7 th November 2017 1 POOSL IDE Installation Manual 1 Installation... 4 1.1 Minimal system requirements... 4 1.2 Installing
Embedded Systems Innovation by TNO POOSL IDE Installation Manual Tool version 4.1.0 7 th November 2017 1 POOSL IDE Installation Manual 1 Installation... 4 1.1 Minimal system requirements... 4 1.2 Installing
Welcome to InSite: A GUIDE TO PROOFING ONLINE. Getting Started Viewing Job Information Uploading Files Viewing Pages Requesting Corrections
 Welcome to InSite: A GUIDE TO PROOFING ONLINE Getting Started Viewing Job Information Uploading Files Viewing Pages Requesting Corrections PROOF.REGISTERGRAPHICS.COM *NOTE: There is no www in this address.
Welcome to InSite: A GUIDE TO PROOFING ONLINE Getting Started Viewing Job Information Uploading Files Viewing Pages Requesting Corrections PROOF.REGISTERGRAPHICS.COM *NOTE: There is no www in this address.
Nimsoft Monitor. websphere Guide. v1.5 series
 Nimsoft Monitor websphere Guide v1.5 series Legal Notices Copyright 2012, Nimsoft Corporation Warranty The material contained in this document is provided "as is," and is subject to being changed, without
Nimsoft Monitor websphere Guide v1.5 series Legal Notices Copyright 2012, Nimsoft Corporation Warranty The material contained in this document is provided "as is," and is subject to being changed, without
Administrator Guide. Oracle Health Sciences Central Designer 2.0. Part Number: E
 Administrator Guide Oracle Health Sciences Central Designer 2.0 Part Number: E37912-01 Copyright 2013, Oracle and/or its affiliates. All rights reserved. The Programs (which include both the software and
Administrator Guide Oracle Health Sciences Central Designer 2.0 Part Number: E37912-01 Copyright 2013, Oracle and/or its affiliates. All rights reserved. The Programs (which include both the software and
Instructions for using Borg and Ide CONNECT. Referring Provider Portal. Version 3.5.
 Instructions for using Borg and Ide CONNECT Referring Provider Portal www.borgideconnect.com Version 3.5 Topic Table of Name Contents Description Getting Started...1 Home Page and Finding Patient Exams...2
Instructions for using Borg and Ide CONNECT Referring Provider Portal www.borgideconnect.com Version 3.5 Topic Table of Name Contents Description Getting Started...1 Home Page and Finding Patient Exams...2
READ&WRITE 5 GOLD FOR MAC USER GUIDE
 READ&WRITE 5 GOLD FOR MAC USER GUIDE ABBYY FineReader Engine 8.0 ABBYY Software Ltd. 2005. ABBYY FineReader the keenest eye in OCR. ABBYY, FINEREADER and ABBYY FineReader are registered trademarks of ABBYY
READ&WRITE 5 GOLD FOR MAC USER GUIDE ABBYY FineReader Engine 8.0 ABBYY Software Ltd. 2005. ABBYY FineReader the keenest eye in OCR. ABBYY, FINEREADER and ABBYY FineReader are registered trademarks of ABBYY
CIM University: Track 3 Tools Session CIM User s Group June 13, 2017
 CIM University: Track 3 Tools Session CIM User s Group June 13, 2017 Margaret Goodrich Principal Consultant, Project Consultants, LLC Email: margaret@j-mgoodrich.com Phone: 903-477-7176 1 Using CIMTool
CIM University: Track 3 Tools Session CIM User s Group June 13, 2017 Margaret Goodrich Principal Consultant, Project Consultants, LLC Email: margaret@j-mgoodrich.com Phone: 903-477-7176 1 Using CIMTool
i2b2 User Guide Informatics for Integrating Biology & the Bedside Version 1.0 October 2012
 i2b2 (Informatics for Integrating Biology and the Bedside) is an informatics framework designed to simplify the process of using existing, de-identified, clinical data for preliminary research cohort discovery
i2b2 (Informatics for Integrating Biology and the Bedside) is an informatics framework designed to simplify the process of using existing, de-identified, clinical data for preliminary research cohort discovery
Tutorial to QuotationFinder_0.4.3
 Tutorial to QuotationFinder_0.4.3 What is Quotation Finder and for which purposes can it be used? Quotation Finder is a tool for the automatic comparison of fully digitized texts. It can either detect
Tutorial to QuotationFinder_0.4.3 What is Quotation Finder and for which purposes can it be used? Quotation Finder is a tool for the automatic comparison of fully digitized texts. It can either detect
CONSOLIDATED LABORATORY SERVICES
 TABLE OF CONTENTS 2 INTRODUCTION 3 LOGIN 4 DESKTOP 5 TEST RESULTS 6 Basic Features 12 Advanced Features 16 TEST ORDERS Coming Soon 17 ACTIVITY REPORTS 17 Trace Accession 18 Activity Report 19 ADMINISTRATOR
TABLE OF CONTENTS 2 INTRODUCTION 3 LOGIN 4 DESKTOP 5 TEST RESULTS 6 Basic Features 12 Advanced Features 16 TEST ORDERS Coming Soon 17 ACTIVITY REPORTS 17 Trace Accession 18 Activity Report 19 ADMINISTRATOR
Contents. Anaplan Connector for MuleSoft
 SW Version 1.1.2 Contents 1 Overview... 3 2 Mulesoft Prerequisites... 4 3 Anaplan Prerequisites for the Demos... 5 3.1 export demo mule-app.properties file...5 3.2 import demo mule-app.properties file...5
SW Version 1.1.2 Contents 1 Overview... 3 2 Mulesoft Prerequisites... 4 3 Anaplan Prerequisites for the Demos... 5 3.1 export demo mule-app.properties file...5 3.2 import demo mule-app.properties file...5
Alpha 1 i2b2 User Guide
 Alpha 1 i2b2 User Guide About i2b2 Accessing i2b2 Data Available in i2b2 Navigating the Workbench Workbench Screen Layout Additional Workbench Features Creating and Running a Query Creating a Query Excluding
Alpha 1 i2b2 User Guide About i2b2 Accessing i2b2 Data Available in i2b2 Navigating the Workbench Workbench Screen Layout Additional Workbench Features Creating and Running a Query Creating a Query Excluding
A tool for Cross-Language Pair Annotations: CLPA
 A tool for Cross-Language Pair Annotations: CLPA August 28, 2006 This document describes our tool called Cross-Language Pair Annotator (CLPA) that is capable to automatically annotate cognates and false
A tool for Cross-Language Pair Annotations: CLPA August 28, 2006 This document describes our tool called Cross-Language Pair Annotator (CLPA) that is capable to automatically annotate cognates and false
MyEclipse ER-Designer Quickstart
 MyEclipse ER-Designer Quickstart Last Revision: Outline 1. Preface 2. Requirements 3. Introduction 4. Creating an ER Diagram from a Database 5. Working with an Entity-Relationship Diagram 1. Notation and
MyEclipse ER-Designer Quickstart Last Revision: Outline 1. Preface 2. Requirements 3. Introduction 4. Creating an ER Diagram from a Database 5. Working with an Entity-Relationship Diagram 1. Notation and
CaliberRM 5.1 Integration for Describe Enterprise
 CaliberRM 5.1 Integration for Describe Enterprise Describe Enterprise integration is now available from within the Borland CaliberRM 5.1 software suite. This document describes how to set up and configure
CaliberRM 5.1 Integration for Describe Enterprise Describe Enterprise integration is now available from within the Borland CaliberRM 5.1 software suite. This document describes how to set up and configure
Hadoop Tutorial. General Instructions
 CS246H: Mining Massive Datasets Hadoop Lab Winter 2018 Hadoop Tutorial General Instructions The purpose of this tutorial is to get you started with Hadoop. Completing the tutorial is optional. Here you
CS246H: Mining Massive Datasets Hadoop Lab Winter 2018 Hadoop Tutorial General Instructions The purpose of this tutorial is to get you started with Hadoop. Completing the tutorial is optional. Here you
eiconsole for Healthcare Getting Started Tutorial
 eiconsole for Healthcare Getting Started Tutorial https://cms.pilotfishtechnology.com/eiconsole-for-healthcare-getting-started-tutorial Welcome to the eiconsole for Healthcare Getting Started Tutorial.
eiconsole for Healthcare Getting Started Tutorial https://cms.pilotfishtechnology.com/eiconsole-for-healthcare-getting-started-tutorial Welcome to the eiconsole for Healthcare Getting Started Tutorial.
eiconsole for Healthcare Getting Started Tutorial
 eiconsole for Healthcare Getting Started Tutorial http://cms.pilotfishtechnology.com/eiconsole-for-healthcare-getting-started-tutorial Welcome to the eiconsole for Healthcare Getting Started Tutorial.
eiconsole for Healthcare Getting Started Tutorial http://cms.pilotfishtechnology.com/eiconsole-for-healthcare-getting-started-tutorial Welcome to the eiconsole for Healthcare Getting Started Tutorial.
Oracle Enterprise Data Quality
 Oracle Enterprise Data Quality Hands-on-Lab 7653 Oracle Openworld 2017 Table of Contents Scenario... 3 Part 1 Launch the Director User Interface... 4 Part 2 Profiling the data using EDQ Product Data Services...
Oracle Enterprise Data Quality Hands-on-Lab 7653 Oracle Openworld 2017 Table of Contents Scenario... 3 Part 1 Launch the Director User Interface... 4 Part 2 Profiling the data using EDQ Product Data Services...
ECM-VNA Convergence Connector
 ECM-VNA Convergence Connector Installation and Setup Guide Version: 1.0.x Written by: Product Knowledge, R&D Date: September 2016 2016 Lexmark. All rights reserved. Lexmark is a trademark of Lexmark International
ECM-VNA Convergence Connector Installation and Setup Guide Version: 1.0.x Written by: Product Knowledge, R&D Date: September 2016 2016 Lexmark. All rights reserved. Lexmark is a trademark of Lexmark International
vsphere Web Client SDK Documentation VMware vsphere Web Client SDK VMware ESXi vcenter Server 6.5.1
 vsphere Web Client SDK Documentation VMware vsphere Web Client SDK 6.5.1 VMware ESXi 6.5.1 vcenter Server 6.5.1 You can find the most up-to-date technical documentation on the VMware website at: https://docs.vmware.com/
vsphere Web Client SDK Documentation VMware vsphere Web Client SDK 6.5.1 VMware ESXi 6.5.1 vcenter Server 6.5.1 You can find the most up-to-date technical documentation on the VMware website at: https://docs.vmware.com/
Word counts and match values
 integrated translation environment Word counts and match values 2004-2014 Kilgray Translation Technologies. All rights reserved. 2004-2014 ZAAC. All rights reserved. Contents Contents... 2 1 General...
integrated translation environment Word counts and match values 2004-2014 Kilgray Translation Technologies. All rights reserved. 2004-2014 ZAAC. All rights reserved. Contents Contents... 2 1 General...
Slide 1 CS 170 Java Programming 1 Duration: 00:00:49 Advance mode: Auto
 CS 170 Java Programming 1 Eclipse@Home Downloading, Installing and Customizing Eclipse at Home Slide 1 CS 170 Java Programming 1 Eclipse@Home Duration: 00:00:49 What is Eclipse? A full-featured professional
CS 170 Java Programming 1 Eclipse@Home Downloading, Installing and Customizing Eclipse at Home Slide 1 CS 170 Java Programming 1 Eclipse@Home Duration: 00:00:49 What is Eclipse? A full-featured professional
JSF Tools Reference Guide. Version: M5
 JSF Tools Reference Guide Version: 3.3.0.M5 1. Introduction... 1 1.1. Key Features of JSF Tools... 1 2. 3. 4. 5. 1.2. Other relevant resources on the topic... 2 JavaServer Faces Support... 3 2.1. Facelets
JSF Tools Reference Guide Version: 3.3.0.M5 1. Introduction... 1 1.1. Key Features of JSF Tools... 1 2. 3. 4. 5. 1.2. Other relevant resources on the topic... 2 JavaServer Faces Support... 3 2.1. Facelets
Lab #1: A Quick Introduction to the Eclipse IDE
 Lab #1: A Quick Introduction to the Eclipse IDE Eclipse is an integrated development environment (IDE) for Java programming. Actually, it is capable of much more than just compiling Java programs but that
Lab #1: A Quick Introduction to the Eclipse IDE Eclipse is an integrated development environment (IDE) for Java programming. Actually, it is capable of much more than just compiling Java programs but that
Searching Guide. September 16, Version 9.3
 Searching Guide September 16, 2016 - Version 9.3 - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Searching Guide September 16, 2016 - Version 9.3 - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Building TPS Web Pages with Dreamweaver
 Building TPS Web Pages with Dreamweaver Title Pages 1. Folder Management 7 2. Defining Your Site 8-11 3. Overview of Design Features 12-22 4. Working with Templates 23-25 5. Publishing Your Site to the
Building TPS Web Pages with Dreamweaver Title Pages 1. Folder Management 7 2. Defining Your Site 8-11 3. Overview of Design Features 12-22 4. Working with Templates 23-25 5. Publishing Your Site to the
Cisco Unified Serviceability
 Cisco Unified Serviceability Introduction, page 1 Installation, page 5 Introduction This document uses the following abbreviations to identify administration differences for these Cisco products: Unified
Cisco Unified Serviceability Introduction, page 1 Installation, page 5 Introduction This document uses the following abbreviations to identify administration differences for these Cisco products: Unified
Contents. About This Book...1 Audience... 1 Prerequisites... 1 Conventions... 2
 Contents About This Book...1 Audience... 1 Prerequisites... 1 Conventions... 2 1 About SAS Text Summarizer Studio...3 1.1 What is SAS Text Summarizer Studio?... 3 1.2 Benefits to Using SAS Text Summarizer
Contents About This Book...1 Audience... 1 Prerequisites... 1 Conventions... 2 1 About SAS Text Summarizer Studio...3 1.1 What is SAS Text Summarizer Studio?... 3 1.2 Benefits to Using SAS Text Summarizer
Product Documentation. ER/Studio Portal. User Guide. Version Published February 21, 2012
 Product Documentation ER/Studio Portal User Guide Version 1.6.3 Published February 21, 2012 2012 Embarcadero Technologies, Inc. Embarcadero, the Embarcadero Technologies logos, and all other Embarcadero
Product Documentation ER/Studio Portal User Guide Version 1.6.3 Published February 21, 2012 2012 Embarcadero Technologies, Inc. Embarcadero, the Embarcadero Technologies logos, and all other Embarcadero
5. Digital Library Creation and Management 5.2. Greenstone digital library software practical
 5. Digital Library Creation and Management 5.2. Greenstone digital library software practical Learning objectives To learn the procedure involved in the installation of GSDL To understand practical step
5. Digital Library Creation and Management 5.2. Greenstone digital library software practical Learning objectives To learn the procedure involved in the installation of GSDL To understand practical step
A Survey Of Different Text Mining Techniques Varsha C. Pande 1 and Dr. A.S. Khandelwal 2
 A Survey Of Different Text Mining Techniques Varsha C. Pande 1 and Dr. A.S. Khandelwal 2 1 Department of Electronics & Comp. Sc, RTMNU, Nagpur, India 2 Department of Computer Science, Hislop College, Nagpur,
A Survey Of Different Text Mining Techniques Varsha C. Pande 1 and Dr. A.S. Khandelwal 2 1 Department of Electronics & Comp. Sc, RTMNU, Nagpur, India 2 Department of Computer Science, Hislop College, Nagpur,
CHAPTER 6. Java Project Configuration
 CHAPTER 6 Java Project Configuration Eclipse includes features such as Content Assist and code templates that enhance rapid development and others that accelerate your navigation and learning of unfamiliar
CHAPTER 6 Java Project Configuration Eclipse includes features such as Content Assist and code templates that enhance rapid development and others that accelerate your navigation and learning of unfamiliar
Configuring GNS3 for CCNA Security Exam (for Windows) Software Requirements to Run GNS3
 Configuring GNS3 for CCNA Security Exam (for Windows) Software Requirements to Run GNS3 From Cisco s website, here are the minimum requirements for CCP 2.7 and CCP 2.8: The following info comes from many
Configuring GNS3 for CCNA Security Exam (for Windows) Software Requirements to Run GNS3 From Cisco s website, here are the minimum requirements for CCP 2.7 and CCP 2.8: The following info comes from many
Optical Character Recognition. SDK Reference. Issue 04 Date
 Issue 04 Date 2018-09-12 Contents Contents 1 SDK Environment Setup...1 1.1 Applying for a Service...1 1.2 Obtaining the SDK... 1 1.3 Preparing a Java Development Environment... 1 1.4 Installing Eclipse
Issue 04 Date 2018-09-12 Contents Contents 1 SDK Environment Setup...1 1.1 Applying for a Service...1 1.2 Obtaining the SDK... 1 1.3 Preparing a Java Development Environment... 1 1.4 Installing Eclipse
Kendo UI. Builder by Progress : Using Kendo UI Designer
 Kendo UI Builder by Progress : Using Kendo UI Designer Copyright 2017 Telerik AD. All rights reserved. December 2017 Last updated with new content: Version 2.1 Updated: 2017/12/22 3 Copyright 4 Contents
Kendo UI Builder by Progress : Using Kendo UI Designer Copyright 2017 Telerik AD. All rights reserved. December 2017 Last updated with new content: Version 2.1 Updated: 2017/12/22 3 Copyright 4 Contents
Using the Text Import Node
 Using the Text Import Node 1/4 Using the Text Import Node Contents You can use the Text Import node to import documents from a directory or the Web. See the following for examples of how to use the Text
Using the Text Import Node 1/4 Using the Text Import Node Contents You can use the Text Import node to import documents from a directory or the Web. See the following for examples of how to use the Text
ADOBE DRIVE 4.2 USER GUIDE
 ADOBE DRIVE 4.2 USER GUIDE 2 2013 Adobe Systems Incorporated. All rights reserved. Adobe Drive 4.2 User Guide Adobe, the Adobe logo, Creative Suite, Illustrator, InCopy, InDesign, and Photoshop are either
ADOBE DRIVE 4.2 USER GUIDE 2 2013 Adobe Systems Incorporated. All rights reserved. Adobe Drive 4.2 User Guide Adobe, the Adobe logo, Creative Suite, Illustrator, InCopy, InDesign, and Photoshop are either
Using Eclipse for Java. Using Eclipse for Java 1 / 1
 Using Eclipse for Java Using Eclipse for Java 1 / 1 Using Eclipse IDE for Java Development Download the latest version of Eclipse (Eclipse for Java Developers or the Standard version) from the website:
Using Eclipse for Java Using Eclipse for Java 1 / 1 Using Eclipse IDE for Java Development Download the latest version of Eclipse (Eclipse for Java Developers or the Standard version) from the website:
