Lezione 7. Bioinformatica. Mauro Ceccanti e Alberto Paoluzzi
|
|
|
- Ophelia Fisher
- 5 years ago
- Views:
Transcription
1 Lezione 7 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza
2 BioPython Installing and exploration Tutorial First Course Project First Start First Start with Biopython
3 Contents BioPython Installing and exploration Tutorial First Course Project First Start First Start with Biopython
4 BioPython Biopython is a set of freely available tools for biological computation written in Python by an international team of developers. [<+->] The web site provides an online resource for modules, scripts, and web links for developers of Python-based software for life science BioPython makes it as easy as possible to use Python for bioinformatics by creating high-quality, reusable modules and scripts
5 BioPython Biopython is a set of freely available tools for biological computation written in Python by an international team of developers. [<+->] The web site provides an online resource for modules, scripts, and web links for developers of Python-based software for life science BioPython makes it as easy as possible to use Python for bioinformatics by creating high-quality, reusable modules and scripts
6 BioPython Biopython is a set of freely available tools for biological computation written in Python by an international team of developers. This wiki will help you download and install Biopython, and start using the libraries and tools
7 Download Current Release
8 Biopython installation Short version [<+->] installer for windows: download Python msi standard install on MacOSX, Linux and Unix: download the source from command line in a terminal: 1 > python setup.py build 2 > python setup.py test 3 > sudo python setup.py install
9 Biopython installation Short version [<+->] installer for windows: download Python msi standard install on MacOSX, Linux and Unix: download the source from command line in a terminal: 1 > python setup.py build 2 > python setup.py test 3 > sudo python setup.py install
10 Biopython installation Short version [<+->] installer for windows: download Python msi standard install on MacOSX, Linux and Unix: download the source from command line in a terminal: 1 > python setup.py build 2 > python setup.py test 3 > sudo python setup.py install
11 Biopython installation Short version [<+->] installer for windows: download Python msi standard install on MacOSX, Linux and Unix: download the source from command line in a terminal: 1 > python setup.py build 2 > python setup.py test 3 > sudo python setup.py install
12 Biopython installation Long version
13 Biopython installation Best version ;-) from a terminal, with easy_install package already installed: 1 > easy_install -f
14 Contents BioPython Installing and exploration Tutorial First Course Project First Start First Start with Biopython
15 Biopython Tutorial and cookbook Biopython Tutorial and Cookbook by Jeff Chang, Brad Chapman, Iddo Friedberg, Thomas Hamelryck, Michiel de Hoon, Peter Cock, and Tiago Antão
16 Biopython Tutorial and Cookbook Chapter 1 Introduction Chapter 2 Quick Start What can you do with Biopython? Chapter 3 Sequence objects Chapter 4 Sequence Record objects Chapter 5 Sequence Input/Output Chapter 6 Sequence Alignment Input/Output, and Alignment Tools Chapter 7 BLAST Chapter 8 Accessing NCBI s Entrez databases Chapter 9 Swiss-Prot and ExPASy Chapter 10 Going 3D: The PDB module Chapter 11 Bio.PopGen: Population genetics Chapter 12 Supervised learning methods Chapter 13 Graphics including GenomeDiagram Chapter 14 Cookbook Cool things to do with it Chapter 15 The Biopython testing framework Chapter 16 Advanced Chapter 17 Where to go from here contributing to Biopython Chapter 18 Appendix: Useful stuff about Python
17 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
18 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
19 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
20 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
21 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
22 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
23 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
24 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
25 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
26 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. The ability to parse bioinformatics files into Python utilizable data structures, including support for the following formats: Blast output both from standalone and WWW Blast Clustalw FASTA GenBank PubMed and Medline ExPASy files, like Enzyme and Prosite SCOP, including dom and lin files UniGene SwissProt
27 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
28 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
29 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
30 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
31 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
32 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
33 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
34 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
35 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
36 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Files in the supported formats can be iterated over record by record or indexed and accessed via a Dictionary interface. 2. Code to deal with popular on-line bioinformatics destinations such as: NCBI Blast, Entrez and PubMed services ExPASy Swiss-Prot and Prosite entries, as well as Prosite searches 3. Interfaces to common bioinformatics programs such as: Standalone Blast from NCBI Clustalw alignment program EMBOSS command line tools 4. A standard sequence class that deals with sequences, ids on sequences, and sequence features. 5. Tools for performing common operations on sequences, such as translation, transcription and weight calculations.
37 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Code to perform classification of data using k Nearest Neighbors, Naive Bayes or Support Vector Machines. 2. Code for dealing with alignments, including a standard way to create and deal with substitution matrices. 3. Code making it easy to split up parallelizable tasks into separate processes. 4. GUI-based programs to do basic sequence manipulations, translations, BLASTing, etc. 5. Extensive documentation and help with using the modules, including this file, on-line wiki documentation, 6. the web site, and the mailing list. 7. Integration with BioSQL, a sequence database schema also supported by the BioPerl and BioJava projects.
38 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Code to perform classification of data using k Nearest Neighbors, Naive Bayes or Support Vector Machines. 2. Code for dealing with alignments, including a standard way to create and deal with substitution matrices. 3. Code making it easy to split up parallelizable tasks into separate processes. 4. GUI-based programs to do basic sequence manipulations, translations, BLASTing, etc. 5. Extensive documentation and help with using the modules, including this file, on-line wiki documentation, 6. the web site, and the mailing list. 7. Integration with BioSQL, a sequence database schema also supported by the BioPerl and BioJava projects.
39 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Code to perform classification of data using k Nearest Neighbors, Naive Bayes or Support Vector Machines. 2. Code for dealing with alignments, including a standard way to create and deal with substitution matrices. 3. Code making it easy to split up parallelizable tasks into separate processes. 4. GUI-based programs to do basic sequence manipulations, translations, BLASTing, etc. 5. Extensive documentation and help with using the modules, including this file, on-line wiki documentation, 6. the web site, and the mailing list. 7. Integration with BioSQL, a sequence database schema also supported by the BioPerl and BioJava projects.
40 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Code to perform classification of data using k Nearest Neighbors, Naive Bayes or Support Vector Machines. 2. Code for dealing with alignments, including a standard way to create and deal with substitution matrices. 3. Code making it easy to split up parallelizable tasks into separate processes. 4. GUI-based programs to do basic sequence manipulations, translations, BLASTing, etc. 5. Extensive documentation and help with using the modules, including this file, on-line wiki documentation, 6. the web site, and the mailing list. 7. Integration with BioSQL, a sequence database schema also supported by the BioPerl and BioJava projects.
41 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Code to perform classification of data using k Nearest Neighbors, Naive Bayes or Support Vector Machines. 2. Code for dealing with alignments, including a standard way to create and deal with substitution matrices. 3. Code making it easy to split up parallelizable tasks into separate processes. 4. GUI-based programs to do basic sequence manipulations, translations, BLASTing, etc. 5. Extensive documentation and help with using the modules, including this file, on-line wiki documentation, 6. the web site, and the mailing list. 7. Integration with BioSQL, a sequence database schema also supported by the BioPerl and BioJava projects.
42 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Code to perform classification of data using k Nearest Neighbors, Naive Bayes or Support Vector Machines. 2. Code for dealing with alignments, including a standard way to create and deal with substitution matrices. 3. Code making it easy to split up parallelizable tasks into separate processes. 4. GUI-based programs to do basic sequence manipulations, translations, BLASTing, etc. 5. Extensive documentation and help with using the modules, including this file, on-line wiki documentation, 6. the web site, and the mailing list. 7. Integration with BioSQL, a sequence database schema also supported by the BioPerl and BioJava projects.
43 Biopython Package contents The main Biopython releases have lots of functionality, including: 1. Code to perform classification of data using k Nearest Neighbors, Naive Bayes or Support Vector Machines. 2. Code for dealing with alignments, including a standard way to create and deal with substitution matrices. 3. Code making it easy to split up parallelizable tasks into separate processes. 4. GUI-based programs to do basic sequence manipulations, translations, BLASTing, etc. 5. Extensive documentation and help with using the modules, including this file, on-line wiki documentation, 6. the web site, and the mailing list. 7. Integration with BioSQL, a sequence database schema also supported by the BioPerl and BioJava projects.
44 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
45 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
46 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
47 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
48 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
49 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
50 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
51 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
52 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
53 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
54 PDB: Atomic Coordinate Entry Format Description Learn to parse PDB files, locally and on the web Protein Data Bank Contents Guide Introduction Title Section Primary Structure Section Heterogen Section Secondary Structure Section Connectivity Annotation Section Miscellaneous Features Section Crystallographic and Coordinate Transformation Section Coordinate Section Connectivity Section Bookkeeping Section
55 Contents BioPython Installing and exploration Tutorial First Course Project First Start First Start with Biopython
56 First project Curation of records of PDB files of aminoacids Start from: Amino Acids web page and Library of 3-D Molecular Structures, in particular from Amino Acids Section
57 Contents BioPython Installing and exploration Tutorial First Course Project First Start First Start with Biopython
58 Start using Biopython Importing the package 1 Python (r263:75184, Oct , 07:56:03) 2 [GCC (Apple Inc. build 5493)] on darwin 3 Type "copyright", "credits" or "license()" for more information. 4 5 IDLE >>> import Bio 7 >>> print Bio. version >>>
59 Start using Biopython help() on the package 1 >>> help(bio) 2 Help on package Bio: 3 4 NAME 5 Bio - Collection of modules for dealing with biological data in Python. 6 7 FILE 8 /Library/Frameworks/Python.framework/Versions/2.6/ lib/python2.6/site-packages/biopython-1.51-py2.6- macosx-10.3-fat.egg/bio/ init.py 9 10 DESCRIPTION 11 The Biopython Project is an international association of developers 12 of freely available Python tools for computational molecular biology
60 Start using Biopython import the PDB package 1 >>> from Bio.PDB import * 2 >>> dir() 3 [ AbstractPropertyMap, Atom, Bio, CaPPBuilder, Chain, DSSP, Dice, Entity, ExposureCN, FragmentMapper, HSExposure, HSExposureCA, HSExposureCB, Model, NeighborSearch, PDBExceptions, PDBIO, PDBList, PDBParser, PPBuilder, Polypeptide, Residue, ResidueDepth, Select, Selection, Structure, StructureAlignment, StructureBuilder, Superimposer, Vector, builtins, doc, name, package, calc_angle, calc_dihedral, extract, get_surface, is_aa, m2rotaxis, make_dssp_dict, mmcif, parse_pdb_header, refmat, rotaxis, rotaxis2m, rotmat, standard_aa_names, to_one_letter_code, vector_to_axis ]
61 Start using Biopython help() on the package 1 >>> help(bio.pdb) 2 Help on package Bio.PDB in Bio: 3 4 NAME 5 Bio.PDB 6 7 FILE 8 /Library/Frameworks/Python.framework/Versions/2.6/ lib/python2.6/site-packages/biopython-1.51-py2.6- macosx-10.3-fat.egg/bio/pdb/ init.py 9 10 DESCRIPTION 11 Classes that deal with macromolecular crystal structures. (eg. 12 PDB and mmcif parsers, a Structure class, a module to keep 13 a local copy of the PDB up-to-date, selective IO of PDB files, 14 etc.). Author: Thomas Hamelryck. Additional code by
62 Start using Biopython help() on the module Atom 1 >>> Atom 2 <module Bio.PDB.Atom from /Library/Frameworks/Python. framework/versions/2.6/lib/python2.6/site-packages/ biopython-1.51-py2.6-macosx-10.3-fat.egg/bio/pdb/atom.pyc > 3 >>> from Bio.PDB.Atom import * 4 >>> help(bio.pdb.atom) 5 Help on module Bio.PDB.Atom in Bio.PDB: 6 7 NAME 8 Bio.PDB.Atom - Atom class, used in Structure objects FILE 11 /Library/Frameworks/Python.framework/Versions/2.6/ lib/python2.6/site-packages/biopython-1.51-py2.6- macosx-10.3-fat.egg/bio/pdb/atom.py CLASSES
63 Start using Biopython Look the atom.py file... 1 # Copyright (C) 2002, Thomas Hamelryck (thamelry@binf.ku.dk) 2 # This code is part of the Biopython distribution and governed by its 3 # license. Please see the LICENSE file that should have been included 4 # as part of this package. 5 6 # Python stuff 7 import numpy 8 9 # My stuff 10 from Entity import DisorderedEntityWrapper 11 from Vector import Vector doc ="Atom class, used in Structure objects." class Atom: 16 def init (self, name, coord, bfactor, occupancy,
Lezione 7. BioPython. Contents. BioPython Installing and exploration Tutorial. Bioinformatica. Mauro Ceccanti e Alberto Paoluzzi
 Lezione 7 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza with Biopython Biopython is a set of freely available
Lezione 7 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza with Biopython Biopython is a set of freely available
Lezione 7. BioPython. Contents. BioPython Installing and exploration Tutorial First Course Project First Start First Start with Biopython
 Lezione 7 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza with Biopython Biopython is a set of freely available
Lezione 7 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza with Biopython Biopython is a set of freely available
Using Biopython for Laboratory Analysis Pipelines
 Using Biopython for Laboratory Analysis Pipelines Brad Chapman 27 June 2003 What is Biopython? Official blurb The Biopython Project is an international association of developers of freely available Python
Using Biopython for Laboratory Analysis Pipelines Brad Chapman 27 June 2003 What is Biopython? Official blurb The Biopython Project is an international association of developers of freely available Python
Biopython: Python tools for computation biology
 Biopython: Python tools for computation biology Brad Chapman and Jeff Chang August 2000 Contents 1 Abstract 1 2 Introduction 2 3 Parsers for Biological Data 2 3.1 Design Goals.............................................
Biopython: Python tools for computation biology Brad Chapman and Jeff Chang August 2000 Contents 1 Abstract 1 2 Introduction 2 3 Parsers for Biological Data 2 3.1 Design Goals.............................................
Scientific Programming Practical 10
 Scientific Programming Practical 10 Introduction Luca Bianco - Academic Year 2017-18 luca.bianco@fmach.it Biopython FROM Biopython s website: The Biopython Project is an international association of developers
Scientific Programming Practical 10 Introduction Luca Bianco - Academic Year 2017-18 luca.bianco@fmach.it Biopython FROM Biopython s website: The Biopython Project is an international association of developers
Lezione 13. Bioinformatica. Mauro Ceccanti e Alberto Paoluzzi
 Lezione 13 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza Lecture 13: Alignment of sequences Sequence alignment
Lezione 13 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza Lecture 13: Alignment of sequences Sequence alignment
Managing Your Biological Data with Python
 Chapman & Hall/CRC Mathematical and Computational Biology Series Managing Your Biological Data with Python Ailegra Via Kristian Rother Anna Tramontano CRC Press Taylor & Francis Group Boca Raton London
Chapman & Hall/CRC Mathematical and Computational Biology Series Managing Your Biological Data with Python Ailegra Via Kristian Rother Anna Tramontano CRC Press Taylor & Francis Group Boca Raton London
Geneious 5.6 Quickstart Manual. Biomatters Ltd
 Geneious 5.6 Quickstart Manual Biomatters Ltd October 15, 2012 2 Introduction This quickstart manual will guide you through the features of Geneious 5.6 s interface and help you orient yourself. You should
Geneious 5.6 Quickstart Manual Biomatters Ltd October 15, 2012 2 Introduction This quickstart manual will guide you through the features of Geneious 5.6 s interface and help you orient yourself. You should
Biopython Project Update
 Biopython Project Update Peter Cock, Plant Pathology, SCRI, Dundee, UK 10 th Annual Bioinformatics Open Source Conference (BOSC) Stockholm, Sweden, 28 June 2009 Contents Brief introduction to Biopython
Biopython Project Update Peter Cock, Plant Pathology, SCRI, Dundee, UK 10 th Annual Bioinformatics Open Source Conference (BOSC) Stockholm, Sweden, 28 June 2009 Contents Brief introduction to Biopython
Viewing Molecular Structures
 Viewing Molecular Structures Proteins fulfill a wide range of biological functions which depend upon their three dimensional structures. Therefore, deciphering the structure of proteins has been the quest
Viewing Molecular Structures Proteins fulfill a wide range of biological functions which depend upon their three dimensional structures. Therefore, deciphering the structure of proteins has been the quest
Contents. Lezione 3. Reference sources. Contents
 Contents Lezione 3 Introduzione alla programmazione con Python Mauro Ceccanti and Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza, escape
Contents Lezione 3 Introduzione alla programmazione con Python Mauro Ceccanti and Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza, escape
Lecture 5 Advanced BLAST
 Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
Introduction to the Protein Data Bank Master Chimie Info Roland Stote Page #
 Introduction to the Protein Data Bank Master Chimie Info - 2009 Roland Stote The purpose of the Protein Data Bank is to collect and organize 3D structures of proteins, nucleic acids, protein-nucleic acid
Introduction to the Protein Data Bank Master Chimie Info - 2009 Roland Stote The purpose of the Protein Data Bank is to collect and organize 3D structures of proteins, nucleic acids, protein-nucleic acid
from scratch A primer for scientists working with Next-Generation- Sequencing data CHAPTER 8 biopython
 from scratch A primer for scientists working with Next-Generation- Sequencing data CHAPTER 8 biopython Chapter 8: Biopython Biopython is a collection of modules that implement common bioinformatical tasks
from scratch A primer for scientists working with Next-Generation- Sequencing data CHAPTER 8 biopython Chapter 8: Biopython Biopython is a collection of modules that implement common bioinformatical tasks
Structural Bioinformatics
 Structural Bioinformatics Elucidation of the 3D structures of biomolecules. Analysis and comparison of biomolecular structures. Prediction of biomolecular recognition. Handles three-dimensional (3-D) structures.
Structural Bioinformatics Elucidation of the 3D structures of biomolecules. Analysis and comparison of biomolecular structures. Prediction of biomolecular recognition. Handles three-dimensional (3-D) structures.
The beginning of this guide offers a brief introduction to the Protein Data Bank, where users can download structure files.
 Structure Viewers Take a Class This guide supports the Galter Library class called Structure Viewers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
Structure Viewers Take a Class This guide supports the Galter Library class called Structure Viewers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
Lezione 6. Installing NumPy. Contents
 Lezione 6 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza Lab 01: Contents As with a lot of open-source
Lezione 6 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza Lab 01: Contents As with a lot of open-source
PyMod 2. User s Guide. PyMod 2 Documention (Last updated: 7/11/2016)
 PyMod 2 User s Guide PyMod 2 Documention (Last updated: 7/11/2016) http://schubert.bio.uniroma1.it/pymod/index.html Department of Biochemical Sciences A. Rossi Fanelli, Sapienza University of Rome, Italy
PyMod 2 User s Guide PyMod 2 Documention (Last updated: 7/11/2016) http://schubert.bio.uniroma1.it/pymod/index.html Department of Biochemical Sciences A. Rossi Fanelli, Sapienza University of Rome, Italy
Biopython Project Update Peter Cock & the Biopython Developers, BOSC 2013, Berlin, Germany
 Biopython Project Update 2013 Peter Cock & the Biopython Developers, BOSC 2013, Berlin, Germany Twitter: @pjacock & @biopython Introduction 2 My Employer After PhD joined Scottish Crop Research Institute
Biopython Project Update 2013 Peter Cock & the Biopython Developers, BOSC 2013, Berlin, Germany Twitter: @pjacock & @biopython Introduction 2 My Employer After PhD joined Scottish Crop Research Institute
Introduction to Biopython
 Introduction to Biopython Python libraries for computational molecular biology http://www.biopython.org Biopython functionality and tools Tools to parse bioinformatics files into Python data structures
Introduction to Biopython Python libraries for computational molecular biology http://www.biopython.org Biopython functionality and tools Tools to parse bioinformatics files into Python data structures
BLAST. NCBI BLAST Basic Local Alignment Search Tool
 BLAST NCBI BLAST Basic Local Alignment Search Tool http://www.ncbi.nlm.nih.gov/blast/ Global versus local alignments Global alignments: Attempt to align every residue in every sequence, Most useful when
BLAST NCBI BLAST Basic Local Alignment Search Tool http://www.ncbi.nlm.nih.gov/blast/ Global versus local alignments Global alignments: Attempt to align every residue in every sequence, Most useful when
Fastest Protein Databank parser is written in Haskell
 Fastest Protein Databank parser is written in Haskell Michał J. Gajda Fast parsing intro Tips and tricks Benchmarks Library interface Compilers Code analyzers Text analyzers Parser are used in many applications
Fastest Protein Databank parser is written in Haskell Michał J. Gajda Fast parsing intro Tips and tricks Benchmarks Library interface Compilers Code analyzers Text analyzers Parser are used in many applications
mpmorfsdb: A database of Molecular Recognition Features (MoRFs) in membrane proteins. Introduction
 mpmorfsdb: A database of Molecular Recognition Features (MoRFs) in membrane proteins. Introduction Molecular Recognition Features (MoRFs) are short, intrinsically disordered regions in proteins that undergo
mpmorfsdb: A database of Molecular Recognition Features (MoRFs) in membrane proteins. Introduction Molecular Recognition Features (MoRFs) are short, intrinsically disordered regions in proteins that undergo
Genome 559 Intro to Statistical and Computational Genomics. Lecture 17b: Biopython Larry Ruzzo
 Genome 559 Intro to Statistical and Computational Genomics Lecture 17b: Biopython Larry Ruzzo Biopython What is Biopython? How do I get it to run on my computer? What can it do? Biopython Biopython is
Genome 559 Intro to Statistical and Computational Genomics Lecture 17b: Biopython Larry Ruzzo Biopython What is Biopython? How do I get it to run on my computer? What can it do? Biopython Biopython is
PyMod Documentation (Version 2.1, September 2011)
 PyMod User s Guide PyMod Documentation (Version 2.1, September 2011) http://schubert.bio.uniroma1.it/pymod/ Emanuele Bramucci & Alessandro Paiardini, Francesco Bossa, Stefano Pascarella, Department of
PyMod User s Guide PyMod Documentation (Version 2.1, September 2011) http://schubert.bio.uniroma1.it/pymod/ Emanuele Bramucci & Alessandro Paiardini, Francesco Bossa, Stefano Pascarella, Department of
Homology Modeling FABP
 Homology Modeling FABP Homology modeling is a technique used to approximate the 3D structure of a protein when no experimentally determined structure exists. It operates under the principle that protein
Homology Modeling FABP Homology modeling is a technique used to approximate the 3D structure of a protein when no experimentally determined structure exists. It operates under the principle that protein
Data Mining Technologies for Bioinformatics Sequences
 Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Proceedings of the Postgraduate Annual Research Seminar
 Proceedings of the Postgraduate Annual Research Seminar 2006 202 Database Integration Approaches for Heterogeneous Biological Data Sources: An overview Iskandar Ishak, Naomie Salim Faculty of Computer
Proceedings of the Postgraduate Annual Research Seminar 2006 202 Database Integration Approaches for Heterogeneous Biological Data Sources: An overview Iskandar Ishak, Naomie Salim Faculty of Computer
Massive Automatic Functional Annotation MAFA
 Massive Automatic Functional Annotation MAFA José Nelson Perez-Castillo 1, Cristian Alejandro Rojas-Quintero 2, Nelson Enrique Vera-Parra 3 1 GICOGE Research Group - Director Center for Scientific Research
Massive Automatic Functional Annotation MAFA José Nelson Perez-Castillo 1, Cristian Alejandro Rojas-Quintero 2, Nelson Enrique Vera-Parra 3 1 GICOGE Research Group - Director Center for Scientific Research
Lecture 8: Introduction to Python and Biopython April 3, 2017
 ICQB Introduction to Computational & Quantitative Biology (G4120) Spring 2017 Oliver Jovanovic, Ph.D. Columbia University Department of Microbiology & Immunology Python The Python programming language
ICQB Introduction to Computational & Quantitative Biology (G4120) Spring 2017 Oliver Jovanovic, Ph.D. Columbia University Department of Microbiology & Immunology Python The Python programming language
BLAST, Profile, and PSI-BLAST
 BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
BMMB 597D - Practical Data Analysis for Life Scientists. Week 12 -Lecture 23. István Albert Huck Institutes for the Life Sciences
 BMMB 597D - Practical Data Analysis for Life Scientists Week 12 -Lecture 23 István Albert Huck Institutes for the Life Sciences Tapping into data sources Entrez: Cross-Database Search System EntrezGlobal
BMMB 597D - Practical Data Analysis for Life Scientists Week 12 -Lecture 23 István Albert Huck Institutes for the Life Sciences Tapping into data sources Entrez: Cross-Database Search System EntrezGlobal
EasyModeller 4.0 : A new GUI to MODELLER
 EasyModeller 4.0 : A new GUI to MODELLER Background Homology modeling has become a key component in structural bioinformatics for prediction of the three-dimensional structure of proteins from their sequences
EasyModeller 4.0 : A new GUI to MODELLER Background Homology modeling has become a key component in structural bioinformatics for prediction of the three-dimensional structure of proteins from their sequences
Protein Structure and Visualization
 In this practical you will learn how to Protein Structure and Visualization By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
In this practical you will learn how to Protein Structure and Visualization By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
Protein Structure and Visualization
 Protein Structure and Visualization In this practical you will learn how to By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
Protein Structure and Visualization In this practical you will learn how to By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
2) NCBI BLAST tutorial This is a users guide written by the education department at NCBI.
 Web resources -- Tour. page 1 of 8 This is a guided tour. Any homework is separate. In fact, this exercise is used for multiple classes and is publicly available to everyone. The entire tour will take
Web resources -- Tour. page 1 of 8 This is a guided tour. Any homework is separate. In fact, this exercise is used for multiple classes and is publicly available to everyone. The entire tour will take
Tutorial 4 BLAST Searching the CHO Genome
 Tutorial 4 BLAST Searching the CHO Genome Accessing the CHO Genome BLAST Tool The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting BLAST from the menu bar
Tutorial 4 BLAST Searching the CHO Genome Accessing the CHO Genome BLAST Tool The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting BLAST from the menu bar
Scientific Computing Using. Atriya Sen
 Scientific Computing Using Atriya Sen Broad Outline Part I, in which I discuss several aspects of the Python programming language Part II, in which I talk about some Python modules for scientific computing
Scientific Computing Using Atriya Sen Broad Outline Part I, in which I discuss several aspects of the Python programming language Part II, in which I talk about some Python modules for scientific computing
Basic Local Alignment Search Tool (BLAST)
 BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
INTRODUCTION TO BIOINFORMATICS
 Molecular Biology-2019 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Molecular Biology-2019 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
BIOINFORMATICS A PRACTICAL GUIDE TO THE ANALYSIS OF GENES AND PROTEINS
 BIOINFORMATICS A PRACTICAL GUIDE TO THE ANALYSIS OF GENES AND PROTEINS EDITED BY Genome Technology Branch National Human Genome Research Institute National Institutes of Health Bethesda, Maryland B. F.
BIOINFORMATICS A PRACTICAL GUIDE TO THE ANALYSIS OF GENES AND PROTEINS EDITED BY Genome Technology Branch National Human Genome Research Institute National Institutes of Health Bethesda, Maryland B. F.
Bioinforma)cs Resources
 Bioinforma)cs Resources Lecture & Exercises Prof. B. Rost, Dr. L. Richter, J. Reeb Ins)tut für Informa)k I12 Bioinforma)cs Resources Organiza)on Schedule Overview Organiza)on Lecture: Friday 9-12, i.e.
Bioinforma)cs Resources Lecture & Exercises Prof. B. Rost, Dr. L. Richter, J. Reeb Ins)tut für Informa)k I12 Bioinforma)cs Resources Organiza)on Schedule Overview Organiza)on Lecture: Friday 9-12, i.e.
Similarity searches in biological sequence databases
 Similarity searches in biological sequence databases Volker Flegel september 2004 Page 1 Outline Keyword search in databases General concept Examples SRS Entrez Expasy Similarity searches in databases
Similarity searches in biological sequence databases Volker Flegel september 2004 Page 1 Outline Keyword search in databases General concept Examples SRS Entrez Expasy Similarity searches in databases
Bio3D: Interactive Tools for Structural Bioinformatics.
 Bio3D: Interactive Tools for Structural Bioinformatics http://thegrantlab.org/bio3d/ What is Bio3D A freely distributed and widely used R package for structural bioinformatics. Provides a large number
Bio3D: Interactive Tools for Structural Bioinformatics http://thegrantlab.org/bio3d/ What is Bio3D A freely distributed and widely used R package for structural bioinformatics. Provides a large number
Wilson Leung 01/03/2018 An Introduction to NCBI BLAST. Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment
 An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
Bioinformatics Database Worksheet
 Bioinformatics Database Worksheet (based on http://www.usm.maine.edu/~rhodes/goodies/matics.html) Where are the opsin genes in the human genome? Point your browser to the NCBI Map Viewer at http://www.ncbi.nlm.nih.gov/mapview/.
Bioinformatics Database Worksheet (based on http://www.usm.maine.edu/~rhodes/goodies/matics.html) Where are the opsin genes in the human genome? Point your browser to the NCBI Map Viewer at http://www.ncbi.nlm.nih.gov/mapview/.
Biostatistics and Bioinformatics Molecular Sequence Databases
 . 1 Description of Module Subject Name Paper Name Module Name/Title 13 03 Dr. Vijaya Khader Dr. MC Varadaraj 2 1. Objectives: In the present module, the students will learn about 1. Encoding linear sequences
. 1 Description of Module Subject Name Paper Name Module Name/Title 13 03 Dr. Vijaya Khader Dr. MC Varadaraj 2 1. Objectives: In the present module, the students will learn about 1. Encoding linear sequences
Bioinformatics explained: Smith-Waterman
 Bioinformatics Explained Bioinformatics explained: Smith-Waterman May 1, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com
Bioinformatics Explained Bioinformatics explained: Smith-Waterman May 1, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com
Sequence Alignment. GBIO0002 Archana Bhardwaj University of Liege
 Sequence Alignment GBIO0002 Archana Bhardwaj University of Liege 1 What is Sequence Alignment? A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity.
Sequence Alignment GBIO0002 Archana Bhardwaj University of Liege 1 What is Sequence Alignment? A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity.
What is Internet COMPUTER NETWORKS AND NETWORK-BASED BIOINFORMATICS RESOURCES
 What is Internet COMPUTER NETWORKS AND NETWORK-BASED BIOINFORMATICS RESOURCES Global Internet DNS Internet IP Internet Domain Name System Domain Name System The Domain Name System (DNS) is a hierarchical,
What is Internet COMPUTER NETWORKS AND NETWORK-BASED BIOINFORMATICS RESOURCES Global Internet DNS Internet IP Internet Domain Name System Domain Name System The Domain Name System (DNS) is a hierarchical,
LinkDB: A Database of Cross Links between Molecular Biology Databases
 LinkDB: A Database of Cross Links between Molecular Biology Databases Susumu Goto, Yutaka Akiyama, Minoru Kanehisa Institute for Chemical Research, Kyoto University Introduction We have developed a molecular
LinkDB: A Database of Cross Links between Molecular Biology Databases Susumu Goto, Yutaka Akiyama, Minoru Kanehisa Institute for Chemical Research, Kyoto University Introduction We have developed a molecular
Protein structure. Enter the name of the protein: rhamnogalacturonan acetylesterase in the search field and click Go.
 In this practical you will learn how to Protein structure search the Protein Structure Databank for information critically choose the best structure, when more than one is available visualize a protein
In this practical you will learn how to Protein structure search the Protein Structure Databank for information critically choose the best structure, when more than one is available visualize a protein
Geneious 2.0. Biomatters Ltd
 Geneious 2.0 Biomatters Ltd August 2, 2006 2 Contents 1 Getting Started 5 1.1 Downloading & Installing Geneious.......................... 5 1.2 Using Geneious for the first time............................
Geneious 2.0 Biomatters Ltd August 2, 2006 2 Contents 1 Getting Started 5 1.1 Downloading & Installing Geneious.......................... 5 1.2 Using Geneious for the first time............................
Rules of Thumb. 1/25/05 CAP5510/CGS5166 (Lec 5) 1
 Rules of Thumb Most sequences with significant similarity over their entire lengths are homologous. Matches that are > 50% identical in a 20-40 aa region occur frequently by chance. Distantly related homologs
Rules of Thumb Most sequences with significant similarity over their entire lengths are homologous. Matches that are > 50% identical in a 20-40 aa region occur frequently by chance. Distantly related homologs
An I/O device driver for bioinformatics tools: the case for BLAST
 An I/O device driver for bioinformatics tools 563 An I/O device driver for bioinformatics tools: the case for BLAST Renato Campos Mauro and Sérgio Lifschitz Departamento de Informática PUC-RIO, Pontifícia
An I/O device driver for bioinformatics tools 563 An I/O device driver for bioinformatics tools: the case for BLAST Renato Campos Mauro and Sérgio Lifschitz Departamento de Informática PUC-RIO, Pontifícia
org.hs.ipi.db November 7, 2017 annotation data package
 org.hs.ipi.db November 7, 2017 org.hs.ipi.db annotation data package Welcome to the org.hs.ipi.db annotation Package. The annotation package was built using a downloadable R package - PAnnBuilder (download
org.hs.ipi.db November 7, 2017 org.hs.ipi.db annotation data package Welcome to the org.hs.ipi.db annotation Package. The annotation package was built using a downloadable R package - PAnnBuilder (download
Software review. Biomolecular Interaction Network Database
 Biomolecular Interaction Network Database Keywords: protein interactions, visualisation, biology data integration, web access Abstract This software review looks at the utility of the Biomolecular Interaction
Biomolecular Interaction Network Database Keywords: protein interactions, visualisation, biology data integration, web access Abstract This software review looks at the utility of the Biomolecular Interaction
Lezione 8. Shell command language Introduction. Sommario. Bioinformatica. Mauro Ceccanti e Alberto Paoluzzi
 Lezione 8 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza Sommario Shell command language Introduction A
Lezione 8 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza Sommario Shell command language Introduction A
BioRuby and the KEGG API. Toshiaki Katayama Bioinformatics center, Kyoto U., Japan
 BioRuby and the KEGG API Toshiaki Katayama k@bioruby.org Bioinformatics center, Kyoto U., Japan Use the source! What is BioRuby? Yet another BioPerl written in Ruby since Nov 2000 Developed in Japan includes
BioRuby and the KEGG API Toshiaki Katayama k@bioruby.org Bioinformatics center, Kyoto U., Japan Use the source! What is BioRuby? Yet another BioPerl written in Ruby since Nov 2000 Developed in Japan includes
An Introduction to Taverna Workflows Katy Wolstencroft University of Manchester
 An Introduction to Taverna Workflows Katy Wolstencroft University of Manchester Download Taverna from http://taverna.sourceforge.net Windows or linux If you are using either a modern version of Windows
An Introduction to Taverna Workflows Katy Wolstencroft University of Manchester Download Taverna from http://taverna.sourceforge.net Windows or linux If you are using either a modern version of Windows
I 2 LOR Tutorial: Applications for the Semantic Web (II)
 I 2 LOR Tutorial: Applications for the Semantic Web (II) René Witte Faculty of Informatics Institute for Program Structures and Data Organization (IPD) Universität Karlsruhe (TH), Germany http://rene-witte.net
I 2 LOR Tutorial: Applications for the Semantic Web (II) René Witte Faculty of Informatics Institute for Program Structures and Data Organization (IPD) Universität Karlsruhe (TH), Germany http://rene-witte.net
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics. ECS 254; Phone: x3748
 CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 2/12/07 CAP5510 1 Perl: Practical Extraction & Report Language
CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 2/12/07 CAP5510 1 Perl: Practical Extraction & Report Language
Bioinformatics resources for data management. Etienne de Villiers KEMRI-Wellcome Trust, Kilifi
 Bioinformatics resources for data management Etienne de Villiers KEMRI-Wellcome Trust, Kilifi Typical Bioinformatic Project Pose Hypothesis Store data in local database Read Relevant Papers Retrieve data
Bioinformatics resources for data management Etienne de Villiers KEMRI-Wellcome Trust, Kilifi Typical Bioinformatic Project Pose Hypothesis Store data in local database Read Relevant Papers Retrieve data
Using many concepts related to bioinformatics, an application was created to
 Patrick Graves Bioinformatics Thursday, April 26, 2007 1 - ABSTRACT Using many concepts related to bioinformatics, an application was created to visually display EST s. Each EST was displayed in the correct
Patrick Graves Bioinformatics Thursday, April 26, 2007 1 - ABSTRACT Using many concepts related to bioinformatics, an application was created to visually display EST s. Each EST was displayed in the correct
Lezione 8. Shell command language Introduction. Sommario. Bioinformatica. Esercitazione Introduzione al linguaggio di shell
 Lezione 8 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Esercitazione Introduzione al linguaggio di shell Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza
Lezione 8 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Esercitazione Introduzione al linguaggio di shell Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza
Ontology-Based Mediation in the. Pisa June 2007
 http://asp.uma.es Ontology-Based Mediation in the Amine System Project Pisa June 2007 Prof. Dr. José F. Aldana Montes (jfam@lcc.uma.es) Prof. Dr. Francisca Sánchez-Jiménez Ismael Navas Delgado Raúl Montañez
http://asp.uma.es Ontology-Based Mediation in the Amine System Project Pisa June 2007 Prof. Dr. José F. Aldana Montes (jfam@lcc.uma.es) Prof. Dr. Francisca Sánchez-Jiménez Ismael Navas Delgado Raúl Montañez
Extra-Homework Problem Set
 Extra-Homework Problem Set => Will not be graded, but might be a good idea for self-study => Solutions are posted at the end of the problem set Your adviser asks you to find out about a so far unpublished
Extra-Homework Problem Set => Will not be graded, but might be a good idea for self-study => Solutions are posted at the end of the problem set Your adviser asks you to find out about a so far unpublished
Bioinformatics explained: BLAST. March 8, 2007
 Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Activities of PDBj and wwpdb: A new PDB format, Data Deposition, Validation, and Data Integration
 Activities of PDBj and wwpdb: A new PDB format, Data Deposition, Validation, and Data Integration Biophysical Society of Japan, Kyoto Luncheon Seminar on 28 October 2013 Institute for Protein Research
Activities of PDBj and wwpdb: A new PDB format, Data Deposition, Validation, and Data Integration Biophysical Society of Japan, Kyoto Luncheon Seminar on 28 October 2013 Institute for Protein Research
Easy manual for SIRD interface
 Easy manual for SIRD interface What is SIRD interface (1) SIRDi is the web-based interface for SIRD (Structure-Interaction Relational Database) (2) You can search for protein structure models with sequence,
Easy manual for SIRD interface What is SIRD interface (1) SIRDi is the web-based interface for SIRD (Structure-Interaction Relational Database) (2) You can search for protein structure models with sequence,
Bioinformatics Data Distribution and Integration via Web Services and XML
 Letter Bioinformatics Data Distribution and Integration via Web Services and XML Xiao Li and Yizheng Zhang* College of Life Science, Sichuan University/Sichuan Key Laboratory of Molecular Biology and Biotechnology,
Letter Bioinformatics Data Distribution and Integration via Web Services and XML Xiao Li and Yizheng Zhang* College of Life Science, Sichuan University/Sichuan Key Laboratory of Molecular Biology and Biotechnology,
Integrated Access to Biological Data. A use case
 Integrated Access to Biological Data. A use case Marta González Fundación ROBOTIKER, Parque Tecnológico Edif 202 48970 Zamudio, Vizcaya Spain marta@robotiker.es Abstract. This use case reflects the research
Integrated Access to Biological Data. A use case Marta González Fundación ROBOTIKER, Parque Tecnológico Edif 202 48970 Zamudio, Vizcaya Spain marta@robotiker.es Abstract. This use case reflects the research
INTRODUCTION TO BIOINFORMATICS
 Molecular Biology-2017 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Molecular Biology-2017 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics. ECS 254; Phone: x3748
 CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 1/18/07 CAP5510 1 Molecular Biology Background 1/18/07 CAP5510
CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 1/18/07 CAP5510 1 Molecular Biology Background 1/18/07 CAP5510
Biopython. Karin Lagesen.
 Biopython Karin Lagesen karin.lagesen@bio.uio.no Object oriented programming Biopython is object-oriented Some knowledge helps understand how biopython works OOP is a way of organizing data and methods
Biopython Karin Lagesen karin.lagesen@bio.uio.no Object oriented programming Biopython is object-oriented Some knowledge helps understand how biopython works OOP is a way of organizing data and methods
Bioinformatics Hubs on the Web
 Bioinformatics Hubs on the Web Take a class The Galter Library teaches a related class called Bioinformatics Hubs on the Web. See our Classes schedule for the next available offering. If this class is
Bioinformatics Hubs on the Web Take a class The Galter Library teaches a related class called Bioinformatics Hubs on the Web. See our Classes schedule for the next available offering. If this class is
BioExtract Server User Manual
 BioExtract Server User Manual University of South Dakota About Us The BioExtract Server harnesses the power of online informatics tools for creating and customizing workflows. Users can query online sequence
BioExtract Server User Manual University of South Dakota About Us The BioExtract Server harnesses the power of online informatics tools for creating and customizing workflows. Users can query online sequence
Integrating Data with Publications: Greater Interactivity and Challenges for Long-Term Preservation of the Scientific Record
 Integrating Data with Publications: Greater Interactivity and Challenges for Long-Term Preservation of the Scientific Record Brian McMahon International Union of Crystallography 5 Abbey Square Chester
Integrating Data with Publications: Greater Interactivity and Challenges for Long-Term Preservation of the Scientific Record Brian McMahon International Union of Crystallography 5 Abbey Square Chester
Supplementary Material
 Software Requirements: Python Version 2.7 Biopython Version 1.60 Python Modules: httplib, urllib2, ete2 (An Environment for Tree Exploration (ETE), Optional) ARB Version 5.1 Goal: Importing alignment,
Software Requirements: Python Version 2.7 Biopython Version 1.60 Python Modules: httplib, urllib2, ete2 (An Environment for Tree Exploration (ETE), Optional) ARB Version 5.1 Goal: Importing alignment,
CAP BIOINFORMATICS Su-Shing Chen CISE. 8/19/2005 Su-Shing Chen, CISE 1
 CAP 5510-2 BIOINFORMATICS Su-Shing Chen CISE 8/19/2005 Su-Shing Chen, CISE 1 Building Local Genomic Databases Genomic research integrates sequence data with gene function knowledge. Gene ontology to represent
CAP 5510-2 BIOINFORMATICS Su-Shing Chen CISE 8/19/2005 Su-Shing Chen, CISE 1 Building Local Genomic Databases Genomic research integrates sequence data with gene function knowledge. Gene ontology to represent
BioJava. What is BioJava? An Open-Source Java Library for Bioinformatics
 BioJava An Open-Source Java Library for Bioinformatics (taken from M. Pocock, BioJava Consulting LTD, presentation) What is BioJava? Java code (Java2 required 1.2 and higher) Open-Source Bioinformatics
BioJava An Open-Source Java Library for Bioinformatics (taken from M. Pocock, BioJava Consulting LTD, presentation) What is BioJava? Java code (Java2 required 1.2 and higher) Open-Source Bioinformatics
Tips and Tricks using Discovery Studio
 Tips and Tricks using Discovery Studio Allister J. Maynard, Ph.D. Senior Manager, R&D July 31 st, 2008 New Science and Customized Workflows for Drug Discovery Research Webinar Series June 12, 2008 - Advances
Tips and Tricks using Discovery Studio Allister J. Maynard, Ph.D. Senior Manager, R&D July 31 st, 2008 New Science and Customized Workflows for Drug Discovery Research Webinar Series June 12, 2008 - Advances
Multiple Sequence Alignments
 Multiple Sequence Alignments Pair-wise Alignments Blast and FASTA first find small high-scoring alignments to build words which are used as a starting points for alignments Blast words default size is
Multiple Sequence Alignments Pair-wise Alignments Blast and FASTA first find small high-scoring alignments to build words which are used as a starting points for alignments Blast words default size is
The GenAlg Project: Developing a New Integrating Data Model, Language, and Tool for Managing and Querying Genomic Information
 The GenAlg Project: Developing a New Integrating Data Model, Language, and Tool for Managing and Querying Genomic Information Joachim Hammer and Markus Schneider Department of Computer and Information
The GenAlg Project: Developing a New Integrating Data Model, Language, and Tool for Managing and Querying Genomic Information Joachim Hammer and Markus Schneider Department of Computer and Information
Web-based tools for Bioinformatics; A (free) introduction to (freely available) NCBI, MUSC and World-wide Bioinformatics Resources.
 1 of 12 9/10/2003 11:15 AM Web-based tools for Bioinformatics; A (free) introduction to (freely available) NCBI, MUSC and World-wide Bioinformatics Resources. When and Where---Wednesdays at 1pm Room 438
1 of 12 9/10/2003 11:15 AM Web-based tools for Bioinformatics; A (free) introduction to (freely available) NCBI, MUSC and World-wide Bioinformatics Resources. When and Where---Wednesdays at 1pm Room 438
Public Repositories Tutorial: Bulk Downloads
 Public Repositories Tutorial: Bulk Downloads Almost all of the public databases, genome browsers, and other tools you have explored so far offer some form of access to rapidly download all or large chunks
Public Repositories Tutorial: Bulk Downloads Almost all of the public databases, genome browsers, and other tools you have explored so far offer some form of access to rapidly download all or large chunks
Geneious Biomatters Ltd
 Geneious 2.5.4 Biomatters Ltd February 26, 2007 2 Contents 1 Getting Started 5 1.1 Downloading & Installing Geneious.......................... 5 1.2 Using Geneious for the first time............................
Geneious 2.5.4 Biomatters Ltd February 26, 2007 2 Contents 1 Getting Started 5 1.1 Downloading & Installing Geneious.......................... 5 1.2 Using Geneious for the first time............................
Unix, Perl and BioPerl
 Unix, Perl and BioPerl III: Sequence Analysis with Perl - Modules and BioPerl George Bell, Ph.D. WIBR Bioinformatics and Research Computing Sequence analysis with Perl Modules and BioPerl Regular expressions
Unix, Perl and BioPerl III: Sequence Analysis with Perl - Modules and BioPerl George Bell, Ph.D. WIBR Bioinformatics and Research Computing Sequence analysis with Perl Modules and BioPerl Regular expressions
Similarity Searches on Sequence Databases
 Similarity Searches on Sequence Databases Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Zürich, October 2004 Swiss Institute of Bioinformatics Swiss EMBnet node Outline Importance of
Similarity Searches on Sequence Databases Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Zürich, October 2004 Swiss Institute of Bioinformatics Swiss EMBnet node Outline Importance of
SARDAR PATEL UNIVERSITY S.Y.B.Sc. Bioinformatics (To be effective from June 2003)
 Course Structure fro B.Sc. Bioinformatics SARDAR PATEL UNIVERSITY S.Y.B.Sc. Bioinformatics (To be effective from June 2003) Principle Subjects Bioinformatics Bioinformatics Practicals Allied Subjects Molecular
Course Structure fro B.Sc. Bioinformatics SARDAR PATEL UNIVERSITY S.Y.B.Sc. Bioinformatics (To be effective from June 2003) Principle Subjects Bioinformatics Bioinformatics Practicals Allied Subjects Molecular
Patterns / Regular expressions
 Sequence bioinformatics http://bio.lundberg.gu.se/courses/ht07/bio2/ Perl programming (GK) Hidden Markov Models (MO) Methods and applications - Algorithms of sequence alignment, BLAST, multiple alignments
Sequence bioinformatics http://bio.lundberg.gu.se/courses/ht07/bio2/ Perl programming (GK) Hidden Markov Models (MO) Methods and applications - Algorithms of sequence alignment, BLAST, multiple alignments
Biological Interactions and Network Analysis Using BIANA Cytoscape Plugin. Emre Güney & Javier García García
 Biological Interactions and Network Analysis Using BIANA Cytoscape Plugin Emre Güney & Javier García García November 13, 2017 CONTENTS 1 Contents 1 Introduction 3 1.1 BIANA Overview.........................
Biological Interactions and Network Analysis Using BIANA Cytoscape Plugin Emre Güney & Javier García García November 13, 2017 CONTENTS 1 Contents 1 Introduction 3 1.1 BIANA Overview.........................
EBI patent related services
 EBI patent related services 4 th Annual Forum for SMEs October 18-19 th 2010 Jennifer McDowall Senior Scientist, EMBL-EBI EBI is an Outstation of the European Molecular Biology Laboratory. Overview Patent
EBI patent related services 4 th Annual Forum for SMEs October 18-19 th 2010 Jennifer McDowall Senior Scientist, EMBL-EBI EBI is an Outstation of the European Molecular Biology Laboratory. Overview Patent
NCBI News, November 2009
 Peter Cooper, Ph.D. NCBI cooper@ncbi.nlm.nh.gov Dawn Lipshultz, M.S. NCBI lipshult@ncbi.nlm.nih.gov Featured Resource: New Discovery-oriented PubMed and NCBI Homepage The NCBI Site Guide A new and improved
Peter Cooper, Ph.D. NCBI cooper@ncbi.nlm.nh.gov Dawn Lipshultz, M.S. NCBI lipshult@ncbi.nlm.nih.gov Featured Resource: New Discovery-oriented PubMed and NCBI Homepage The NCBI Site Guide A new and improved
Bioinforma)cs Resources
 Bioinforma)cs Resources Lecture & Exercises Prof. B. Rost, Dr. L. Richter, J. Reeb Ins)tut für Informa)k I12 Bioinforma)cs Resources Organiza)on Schedule Overview Organiza)on Lecture: Friday 9-12, i.e.
Bioinforma)cs Resources Lecture & Exercises Prof. B. Rost, Dr. L. Richter, J. Reeb Ins)tut für Informa)k I12 Bioinforma)cs Resources Organiza)on Schedule Overview Organiza)on Lecture: Friday 9-12, i.e.
Bioinformatics. Sequence alignment BLAST Significance. Next time Protein Structure
 Bioinformatics Sequence alignment BLAST Significance Next time Protein Structure 1 Experimental origins of sequence data The Sanger dideoxynucleotide method F Each color is one lane of an electrophoresis
Bioinformatics Sequence alignment BLAST Significance Next time Protein Structure 1 Experimental origins of sequence data The Sanger dideoxynucleotide method F Each color is one lane of an electrophoresis
Overview.
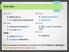 Overview day one 0. getting set up 1. text output and manipulation day two 2. reading and writing files 3. lists and loops today 4. writing functions 5. conditional statements day four day five day six
Overview day one 0. getting set up 1. text output and manipulation day two 2. reading and writing files 3. lists and loops today 4. writing functions 5. conditional statements day four day five day six
Bioinforma)cs Resources
 Bioinforma)cs Resources Lecture & Exercises Prof. B. Rost, Dr. L. Richter, J. Reeb Ins)tut für Informa)k I12 Bioinforma)cs Resources Organiza)on Schedule Overview Organiza)on Lecture: Friday 9-12, i.e.
Bioinforma)cs Resources Lecture & Exercises Prof. B. Rost, Dr. L. Richter, J. Reeb Ins)tut für Informa)k I12 Bioinforma)cs Resources Organiza)on Schedule Overview Organiza)on Lecture: Friday 9-12, i.e.
Overview.
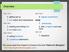 Overview day one 0. getting set up 1. text output and manipulation day two 2. reading and writing files 3. lists and loops day three 4. writing functions 5. conditional statements day four today day six
Overview day one 0. getting set up 1. text output and manipulation day two 2. reading and writing files 3. lists and loops day three 4. writing functions 5. conditional statements day four today day six
Integration of the Comprehensive Microbial Resource into the BioSPICE Data Warehouse
 Integration of the Comprehensive Microbial Resource into the BioSPICE Data Warehouse Department of Biomedical Informatics Stanford University BioMedIn 231: Computational Molecular Biology Final Project
Integration of the Comprehensive Microbial Resource into the BioSPICE Data Warehouse Department of Biomedical Informatics Stanford University BioMedIn 231: Computational Molecular Biology Final Project
