Heriot-Watt University
|
|
|
- Patience Wood
- 5 years ago
- Views:
Transcription
1 Heriot-Watt University Heriot-Watt University Research Gateway Adebayo, Solomon; McLeod, Kenneth; Tudose, Ilinca; Osumi-Sutherland, David; Baldock, Richard; Burger, Albert Georg; Parkinson, Helen Publication date: 2015 Document Version Publisher's PDF, also known as Version of record Link to publication in Heriot-Watt University Research Portal Citation for published version (APA): Adebayo, S., McLeod, K., Tudose, I., Osumi-Sutherland, D., Baldock, R., Burger, A. G., & Parkinson, H. (2015) Paper presented at Phenotype Day 2015 at 23rd Annual International Conference on Intelligent Systems for Molecular Biology, Dublin, Ireland. General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim.
2 Solomon Adebayo 1, Kenneth M c Leod 2, Ilinca Tudose 3, David Osumi-Sutherland 3, Richard Baldock 1, Albert Burger 2 and Helen Parkinson 3* 1 MRC Human Genetics Unit, IGMM, University of Edinburgh, Crewe Road, Edinburgh, UK, 2 Department of Computer Science, Heriot-Watt University, Edinburgh, UK, 3 European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK ABSTRACT Motivation: High throughput imaging is now available to many groups and it is possible to generate a large quantity of images quickly. PhenoImageShare provides a lightweight image data query and annotation tool, and a single point of access backed by a Solr server for programmatic access to a federated image collection and related image PhIS enabling improved community access. 1 INTRODUCTION As reference genomes and large-scale programs to generate model organism mutants and knock-outs are completed, there has been matching effort from projects such as the International Mouse Phenotyping Consortium (IMPC) to establish and codify phenotype with genomic coverage (Koscielny et al., 2013). Current phenotyping effort will deliver annotations held in independent databases associated with the primary data, which may be searched individually, but there is no mechanism for integration, cross-query and analysis, especially with respect to human abnormality and disease phenotypes for image derived data. Therefore we have developed PhenoImageShare (PhIS). PhIS is a cross browser, cross repository platform enabling semantic discovery, phenotypic browsing and annotation of federate phenotypic images. Resources such as OMERO (Allan et al., 2012) allow users to store their images, but do not provide ontology enabled user facing tools to share and query image PhIS across repositories using standard query engines such as Solr. Further, images are often siloed by imaging methodology or domain and access to multiple image repositories requires bespoke development against multiple modes of programmatic access each of which is evolving as each resource changes. PhenoImageShare is species and imaging technology neutral and currently provides access to 94,751 images federated from different data resources with 10,000 regions of interest (ROI) associated to anatomy or phenotype annotations via ontologies. These can be accessed via the GUI beta.phenoimageshare.org or via web services and URL. To date PhIS is populated with images from Drosophila and three different mouse projects. Code * To whom correspondence should be addressed. is available from and 2 METHODS The PhIS platform consists of three major components. The Image Discovery Infrastructure, consisting of a schema for image related PhIS representation, a Solr index offering queries on the data, and a web service hosted at the EBI which is accessible to the project and also as a service to external users wishing to access image PhIS. The IQS component, run at Heriot-Watt University, provides an integrated query service (IQS) supporting access to spatial reasoning queries integrated with the web services for the Solr index. Finally the user interface components hosted at Edinburgh University provide an intuitive GUI to query cross platform image meta-data and will in future host the annotation service to allow image owners, and third parties to annotate images using ontologies. 2.1 Image Discovery Infrastructure The query API is available from It offers a simple REST interface with a single response type (JSON). In future the annotation submission API will be exposed to programmatic access and will require authentication, however, at present we do not expose the image submission functionality to outside access. We currently perform batch submissions from XML export files. Data sources provide us with up-to-date XML exports of their data, following the PhIS schema (XSD). We then perform XML and semantic validation (checking URIs and that term labels match), followed by data enrichment before indexing. This includes addition of synonyms and ancestor relationships to the Solr index allowing an improved semantic search. The implementation is in Java and the query functionality is primarily via Solr. The database, XSD and Solr representations share a common high-level schema. The meta data is split into three types: image annotation, channel annotation and ROI annotation (described in detail below). The image entity is core and stores generic, immutable metadata. These are roughly encompassed by imaging procedure and details, data source, 1
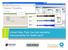
3 S. Adebayo et al. credits and sample information, including genotype or sample preparation techniques. The channel entity encapsulates visualization information, such as tags or labels and genomic information when needed, such as for expression images. The ROI entity holds both the coordinates, within the given image, of the ROI(s) as well as the annotation values. The metadata stored does not overlap so all associated entity types should be queried. For example an ROI entity associating an expression value in liver to a spatial region on the image should be analyzed with the associated image document containing genetic information and with channel entity providing information about the reporter used. The user interface makes the browsing seamless for users across these three categories. 2.2 Spatial Reasoning Within PhenoImageShare, spatial reasoning is used to find phenotypes via a spatial description e.g. associated with a location or with respect to some other entity from which location can be inferred. For example, given the phenotype cataract the anatomical structure eye can be inferred. Spatial reasoning can also be used to search for particular combinations or patterns in the data that are spatially similar. A range of possible definitions exists for similar spatial regions including overlapping and adjacent areas. In order to facilitate this reasoning the images must include direct or inferred from phenotype spatial annotation, e.g. from pulmonary fibrosis the anatomical term lung is inferred; bridge ontologies provided by the Monarch initiative (Köhler et al., 2013) support this inference. When images have appropriate spatial annotations spatial reasoning can identify spatially similar ROIs. Two anatomical terms that have an is-a or part-of relationship are considered to be close. Reasoning is pre-computed with the results stored in Solr for query. More sophisticated spatial reasoning can be undertaken by mapping the spatial information within the images onto an appropriate atlas that contains a representation of space including direction. Through the spatial model of the atlas a spatial relationship between two ROIs is obtained. For example, ROI A is distance Y further along the cranial axis than ROI B. Measurements of distance and direction present the user with the opportunity to select their own definition of similar by simply changing a threshold. The spatial reasoning API will be integrated into future releases via the Integrated Query Service (IQS). The IQS will turn the GUI s query into distinct queries for the spatial reasoning service and Solr. The two responses will be combined in a single result set and sent to the GUI. 2.3 User interface PhIS delivers its functionality through a highly responsive and configurable web-based interface that allows users to register, search, query and provide phenotypic annotations for image data using standard ontologies. It provides discover, query and subsequently annotate image data held in independent databases. These annotations and associated metadata can be imported at registration and also exported in standard formats such as CSV, RDF, etc. The first release of the PhIS GUI provides annotation discovery and query, multi-word free-text and ontology-based search with autosuggest, facet-based browsing and visual analytics providing a unified view of the distributed image databases. Feature implementation is requirements-driven and is prioritized based on user test sessions. The Model-View-Controller (MVC) design technique has been adopted due to the nature of PhIS data. Portability, re-usability and responsiveness are at the core of Graphical User Interface (GUI) design considerations. The core logic of the web interfaces is developed using Python, Django, JavaScript and AJAX. Bootstrap and Flat-UI technologies are used for the interface theme and styling (Views), and are integrated with the core to support changing of theme dynamically, while allowing application components to be developed as widgets. Figure 1. Users are presented with options for taking a tour of the website (a) or performing a quick free-text search (b) or navigating to the search interface (c). Clicking through points on the interactive charts launches the search page with imaging method filter (d) applied. Data labels can be used to toggle (e) the contributions of the data points and charts can be downloaded in various formats (f). Users can explore PhIS from the index page through options ranging from taking a tour of the web application, through performing a quick search or navigating search interface via the application menu, to performing quick filtering/browsing by clicking through points on the interactive charts, which present visual summaries of the data in the repository. Search text can be an anatomy or phenotype term, gene or allele symbol from standard ontologies, or user entered free-text annotation term. Points or slices on the pie charts represent canned queries, which map to data filters on the search interface. Clicking through points on the charts launched the search page with the corresponding filters selected, and the matching image data displayed. Users can also interact with the charts by toggling the data 2
4 labels in order to visualize the contributions of each data point. This interactivity affords users the facility to quickly drill down to images of interest by filtering out dominating data within a category of interest. The charts can be exported in various formats such as JPG, PNG, and SVG for local use. These features of the index page are shown in Figure 1. The search interface is the main entry point for users to browse through PhIS repository, through the use of facets or quick search feature. In addition to support for autosuggest, the search interface is responsiveness and search results are displayed in real-time. Text-based search and faceting Users can perform quick search for a gene or allele (using their symbols), anatomical or phenotypic terms from ontologies or free-text using the generic search box and autosuggest provides users with a drop-down list of terms predicted from their text entries. Search results are displayed in realtime as users enter their search text. Users can further narrow down their search by applying filters for taxon, anatomy, imaging method etc. Browsing and data filtering with facets Facets and sub-facets allow users to browse through PhIS dataset without having to type in text in the generic search box. This is useful for users with limited prior knowledge of what to search for. They can expand facets and check filters (checkboxes) - e.g. Mutant-Expression applicable to their search. Applied checkbox filters are displayed as tags above the facets. Users can click on the tag(s) to unselect the corresponding facet(s), and the updated records. Users can also perform ontology-based facetted search for gene or allele symbol, anatomy or phenotype term or a combination of these. Figure 2 shows the search functionality of PhIS web application Detail & Annotation Interface When a user finds an image of interest, clicking on it takes them to the detail. Genotype information and image metadata are presented on this page along with ROI and annotations associated with the selected image. Annotations associated with the image or ROI within the image are displayed. These can be downloaded from the interface. At present genotype information is captured at image registration, whereas phenotypic annotations are captured either at registration or through the user facing annotation submission interface to be released in the next version of the application. 3 DATA AND ONTOLOGIES The current PhIS release contains data from two sources, which cover a variety of image types (X-ray, macro, histopathology slides, and lacz expression). 1. The TRACER database (Chen et al., 2013) contains mouse data embryos carrying a transgenic insertion generated by the Sleeping Beauty transposon-based system. Embryo expression images are recorded and annotated with a controlled vocabulary for anatomy parts and are therefore suitable for inclusion in PhenoImageShare. 2. Images from the Wellcome Trust Sanger Institute represent single gene knock-outs and are of types: X-ray, histopathology or gene expression images using lacz reporter. Genotype, anatomy and phenotype annotations are provided for these data. Further data sources will be included in the next release, and will include Drosophila data from Virtual Fly Brain and mouse data from EMAGE. Figure 2. Query interface with ontology-based search and faceting. Results of auto-suggest for abnormal epidermis stratum basal morphology (a) filtered by development stage (postnatal) (b) and confocal imaging method (c). Example query and help on using facets (d) are also provided on the interface. 3
5 S. Adebayo et al. To add semantic value and make the integration possible we support the use different ontologies e.g. by species and application area and bridge between ontologies. For anatomy annotations we use the MA, EMAP, EMAPA and ontologies and for cross species integration UBERON (Mungall, Torniai, Gkoutos, Lewis, & Haendel, 2012) and the Monarch Initiative s bridge files. We also have phenotype currently from the MP (Smith & Eppig, 2012) and we expect to extend these to pathology, and other species in future releases. The species specific developmental stage ontologies and the Fbbi ontology for biological imaging methods, originally developed for the Cell Image Library (Orloff, Iwasa, Martone, Ellisman, & Kane, 2013) are critical for the development of PhenoImageShare. In order to accommodate data from different resources free text is also permitted, although the annotation interface will encourage the use of ontology terms through tailored auto-suggest and tools such as Zooma ( which can suggest ontology terms based on the free text description entered. 4 CONCLUSION The PhIS platform provides underlying infrastructure for informatics users and user facing tools for biologists enabling the query and annotation of federated images. It supports the use of ontologies and builds on these to deliver spatial reasoning. The releases of an ontology enabled annotation tool for image generators and third parties will allow projects such as IMPC (Koscielny et al., 2013) to federate image annotation tasks and will provide a collaboration platform for annotators. Features to be supported in future releases include query sharing and bookmarking, grid-style image gallery view, image discovery by similarities, complex query builder, drawings and annotations of ROIs on images, cross-species search, integration and mappings, spatial reasoning, links to reference atlas frameworks such as EMAP (Baldock et al., 2003), support for 3D image annotation, browsing and annotation of image collections, and query storage and analytics. The next data release includes EMAGE database and the Virtual Fly Brain (Milyaev et al., 2012) (VFB). The VFB dataset offers an interesting use case for cross species integration, but also adds value to our anatomy coverage with its focus on neuroanatomy and gene expression in the adult Drosophila melanogaster brain. Future infrastructural plans include offering the PhIS services as portable widgets such as those offered by BioJS (Gómez et al., 2013) for inclusion in third party tools. We encourage owners and generators of image datasets to expose their data via PhIS and view it as sustainable platform for the sharing of image data which requires minimal investment in disk, and supports a federated model of image data sharing. ACKNOWLEDGEMENTS PhenoImageShare is funded by BBSRC BBR grant BB/K020153/1 and EMBL Core funds to HP. Thanks to Jason Swedlow for useful discussions; Yiya Yang for export of Mouse Atlas data; Tony Burdett, Jeremy Mason and Gautier Koscielny for technical guidance; and Matt Pierce for assistance with Solr. REFERENCES Allan, C., Burel, J.-M., Moore, J., Blackburn, C., Linkert, M., Loynton, S., Swedlow, J. R. (2012). OMERO: flexible, modeldriven data management for experimental biology. Nature Methods, 9(3), doi: /nmeth.1896 Baldock, R. A., Bard, J. B. L., Burger, A., Burton, N., Christiansen, J., Feng, G., Davidson, D. R. (2003). EMAP and EMAGE: a framework for understanding spatially organized data. Neuroinformatics, 1(4), doi: /ni:1:4:309 Chen, C.-K., Symmons, O., Uslu, V. V., Tsujimura, T., Ruf, S., Smedley, D., & Spitz, F. (2013). TRACER: a resource to study the regulatory architecture of the mouse genome. BMC Genomics, 14(1), 215. doi: / Gómez, J., García, L. J., Salazar, G. A., Villaveces, J., Gore, S., García, A., Jiménez, R. C. (2013). BioJS: an open source JavaScript framework for biological data visualization. Bioinformatics (Oxford, England), 29(8), doi: /bioinformatics/btt100 Köhler, S., Doelken, S. C., Ruef, B. J., Bauer, S., Washington, N., Westerfield, M., Mungall, C. J. (2013). Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research, 2, 30. doi: /f1000research.2-30.v2 Koscielny, G., Yaikhom, G., Iyer, V., Meehan, T. F., Morgan, H., Atienza-Herrero, J., Parkinson, H. (2013). The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Research. doi: /nar/gkt977 Milyaev, N., Osumi-Sutherland, D., Reeve, S., Burton, N., Baldock, R. A., & Armstrong, J. D. (2012). The Virtual Fly Brain browser and query interface. Bioinformatics (Oxford, England), 28(3), doi: /bioinformatics/btr677 Mungall, C. J., Torniai, C., Gkoutos, G. V, Lewis, S. E., & Haendel, M. A. (2012). Uberon, an integrative multi-species anatomy ontology. Genome Biology, 13(1), R5. doi: /gb r5 Orloff, D. N., Iwasa, J. H., Martone, M. E., Ellisman, M. H., & Kane, C. M. (2013). The cell: an image library-ccdb: a curated repository of microscopy data. Nucleic Acids Research, 41(Database issue), D doi: /nar/gks1257 Smith, C. L., & Eppig, J. T. (2012). The Mammalian Phenotype Ontology as a unifying standard for experimental and highthroughput phenotyping data. Mammalian Genome, 23(9-10), doi: /s
PhenoImageShare: an image annotation and query infrastructure
 Adebayo et al. Journal of Biomedical Semantics (2016) 7:35 DOI 10.1186/s13326-016-0072-2 SOFTWARE Open Access PhenoImageShare: an image annotation and query infrastructure Solomon Adebayo 1, Kenneth McLeod
Adebayo et al. Journal of Biomedical Semantics (2016) 7:35 DOI 10.1186/s13326-016-0072-2 SOFTWARE Open Access PhenoImageShare: an image annotation and query infrastructure Solomon Adebayo 1, Kenneth McLeod
Facilitating Semantic Alignment of EBI Resources
 Facilitating Semantic Alignment of EBI Resources 17 th March, 2017 Tony Burdett Technical Co-ordinator Samples, Phenotypes and Ontologies Team www.ebi.ac.uk What is EMBL-EBI? Europe s home for biological
Facilitating Semantic Alignment of EBI Resources 17 th March, 2017 Tony Burdett Technical Co-ordinator Samples, Phenotypes and Ontologies Team www.ebi.ac.uk What is EMBL-EBI? Europe s home for biological
Human Disease Models Tutorial
 Mouse Genome Informatics www.informatics.jax.org The fundamental mission of the Mouse Genome Informatics resource is to facilitate the use of mouse as a model system for understanding human biology and
Mouse Genome Informatics www.informatics.jax.org The fundamental mission of the Mouse Genome Informatics resource is to facilitate the use of mouse as a model system for understanding human biology and
Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform
 Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform 1. Exploring the IDR This current IDR web user interface (WUI) is based on the open source
Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform 1. Exploring the IDR This current IDR web user interface (WUI) is based on the open source
Taking a view on bio-ontologies. Simon Jupp Functional Genomics Production Team ICBO, 2012 Graz, Austria
 Taking a view on bio-ontologies Simon Jupp Functional Genomics Production Team ICBO, 2012 Graz, Austria Who we are European Bioinformatics Institute one of world s largest bio data and service providers
Taking a view on bio-ontologies Simon Jupp Functional Genomics Production Team ICBO, 2012 Graz, Austria Who we are European Bioinformatics Institute one of world s largest bio data and service providers
University of Bath. Publication date: Document Version Publisher's PDF, also known as Version of record. Link to publication
 Citation for published version: Patel, M & Duke, M 2004, 'Knowledge Discovery in an Agents Environment' Paper presented at European Semantic Web Symposium 2004, Heraklion, Crete, UK United Kingdom, 9/05/04-11/05/04,.
Citation for published version: Patel, M & Duke, M 2004, 'Knowledge Discovery in an Agents Environment' Paper presented at European Semantic Web Symposium 2004, Heraklion, Crete, UK United Kingdom, 9/05/04-11/05/04,.
Enabling Open Science: Data Discoverability, Access and Use. Jo McEntyre Head of Literature Services
 Enabling Open Science: Data Discoverability, Access and Use Jo McEntyre Head of Literature Services www.ebi.ac.uk About EMBL-EBI Part of the European Molecular Biology Laboratory International, non-profit
Enabling Open Science: Data Discoverability, Access and Use Jo McEntyre Head of Literature Services www.ebi.ac.uk About EMBL-EBI Part of the European Molecular Biology Laboratory International, non-profit
The ELIXIR of Linked Data
 The ELIXIR of Linked Data Professor Carole Goble (UK node) Barend Mons (NL node), Helen Parkinson (EMBL-EBI node) The Interoperability Services Backbone Team European Life Sciences Infrastructure for Biological
The ELIXIR of Linked Data Professor Carole Goble (UK node) Barend Mons (NL node), Helen Parkinson (EMBL-EBI node) The Interoperability Services Backbone Team European Life Sciences Infrastructure for Biological
Bioinformatics Hubs on the Web
 Bioinformatics Hubs on the Web Take a class The Galter Library teaches a related class called Bioinformatics Hubs on the Web. See our Classes schedule for the next available offering. If this class is
Bioinformatics Hubs on the Web Take a class The Galter Library teaches a related class called Bioinformatics Hubs on the Web. See our Classes schedule for the next available offering. If this class is
Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment
 Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment Harry Hochheiser and Ilya G. Goldberg Image Informatics and Computational Biology Unit, Laboratory
Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment Harry Hochheiser and Ilya G. Goldberg Image Informatics and Computational Biology Unit, Laboratory
Ontology-based annotation of multiscale imaging data: Utilizing and building the Neuroscience Information Framework. Maryann E.
 Ontology-based annotation of multiscale imaging data: Utilizing and building the Neuroscience Information Framework Maryann E. Martone University of California, San Diego What does this mean? 3D Volumes
Ontology-based annotation of multiscale imaging data: Utilizing and building the Neuroscience Information Framework Maryann E. Martone University of California, San Diego What does this mean? 3D Volumes
Biobtree: A tool to search, map and visualize bioinformatics identifiers and special keywords [version 1; referees: awaiting peer review]
![Biobtree: A tool to search, map and visualize bioinformatics identifiers and special keywords [version 1; referees: awaiting peer review] Biobtree: A tool to search, map and visualize bioinformatics identifiers and special keywords [version 1; referees: awaiting peer review]](/thumbs/88/117074068.jpg) SOFTWARE TOOL ARTICLE Biobtree: A tool to search, map and visualize bioinformatics identifiers and special keywords [version 1; referees: awaiting peer review] Tamer Gur European Bioinformatics Institute,
SOFTWARE TOOL ARTICLE Biobtree: A tool to search, map and visualize bioinformatics identifiers and special keywords [version 1; referees: awaiting peer review] Tamer Gur European Bioinformatics Institute,
The MEG Metadata Schemas Registry Schemas and Ontologies: building a Semantic Infrastructure for GRIDs and digital libraries Edinburgh, 16 May 2003
 The MEG Metadata Schemas Registry Schemas and Ontologies: building a Semantic Infrastructure for GRIDs and digital libraries Edinburgh, 16 May 2003 Pete Johnston UKOLN, University of Bath Bath, BA2 7AY
The MEG Metadata Schemas Registry Schemas and Ontologies: building a Semantic Infrastructure for GRIDs and digital libraries Edinburgh, 16 May 2003 Pete Johnston UKOLN, University of Bath Bath, BA2 7AY
Data publication and discovery with Globus
 Data publication and discovery with Globus Questions and comments to outreach@globus.org The Globus data publication and discovery services make it easy for institutions and projects to establish collections,
Data publication and discovery with Globus Questions and comments to outreach@globus.org The Globus data publication and discovery services make it easy for institutions and projects to establish collections,
Mouse BIRN Data Integration. Maryann Martone Mouse All Hands Meeting
 Mouse BIRN Data Integration Maryann Martone 2005 Mouse All Hands Meeting Specific Aims Specific Aim 1: Data Access and Management Continue development of multi-scale databases along existing lines extending
Mouse BIRN Data Integration Maryann Martone 2005 Mouse All Hands Meeting Specific Aims Specific Aim 1: Data Access and Management Continue development of multi-scale databases along existing lines extending
Using Bio-ontologies as Data Annotation, Integration & Analytical Tools at Mouse Genome Informatics
 CASIMIR Workshop Database Interoperability & Ontologies Using Bio-ontologies as Data Annotation, Integration & Analytical Tools at Mouse Genome Informatics www.informatics.jax.org Anna V. Anagnostopoulos,
CASIMIR Workshop Database Interoperability & Ontologies Using Bio-ontologies as Data Annotation, Integration & Analytical Tools at Mouse Genome Informatics www.informatics.jax.org Anna V. Anagnostopoulos,
Software review. Biomolecular Interaction Network Database
 Biomolecular Interaction Network Database Keywords: protein interactions, visualisation, biology data integration, web access Abstract This software review looks at the utility of the Biomolecular Interaction
Biomolecular Interaction Network Database Keywords: protein interactions, visualisation, biology data integration, web access Abstract This software review looks at the utility of the Biomolecular Interaction
The Human PAX6 Mutation Database
 1998 Oxford University Press Nucleic Acids Research, 1998, Vol. 26, No. 1 259 264 The Human PAX6 Mutation Database Alastair Brown*, Mark McKie, Veronica van Heyningen and Jane Prosser Medical Research
1998 Oxford University Press Nucleic Acids Research, 1998, Vol. 26, No. 1 259 264 The Human PAX6 Mutation Database Alastair Brown*, Mark McKie, Veronica van Heyningen and Jane Prosser Medical Research
SEXTANT 1. Purpose of the Application
 SEXTANT 1. Purpose of the Application Sextant has been used in the domains of Earth Observation and Environment by presenting its browsing and visualization capabilities using a number of link geospatial
SEXTANT 1. Purpose of the Application Sextant has been used in the domains of Earth Observation and Environment by presenting its browsing and visualization capabilities using a number of link geospatial
Exercises. Biological Data Analysis Using InterMine workshop exercises with answers
 Exercises Biological Data Analysis Using InterMine workshop exercises with answers Exercise1: Faceted Search Use HumanMine for this exercise 1. Search for one or more of the following using the keyword
Exercises Biological Data Analysis Using InterMine workshop exercises with answers Exercise1: Faceted Search Use HumanMine for this exercise 1. Search for one or more of the following using the keyword
Brain, a library for the OWL2 EL profile
 Brain, a library for the OWL2 EL profile Samuel Croset 1, John Overington 1, and Dietrich Rebholz-Schuhmann 1 EMBL-EBI, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD UK croset@ebi.ac.uk Abstract.
Brain, a library for the OWL2 EL profile Samuel Croset 1, John Overington 1, and Dietrich Rebholz-Schuhmann 1 EMBL-EBI, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD UK croset@ebi.ac.uk Abstract.
A Semantic Web-Based Approach for Harvesting Multilingual Textual. definitions from Wikipedia to support ICD-11 revision
 A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
Developing Seamless Discovery of Scholarly and Trade Journal Resources Via OAI and RSS Chumbe, Santiago Segundo; MacLeod, Roddy
 Heriot-Watt University Heriot-Watt University Research Gateway Developing Seamless Discovery of Scholarly and Trade Journal Resources Via OAI and RSS Chumbe, Santiago Segundo; MacLeod, Roddy Publication
Heriot-Watt University Heriot-Watt University Research Gateway Developing Seamless Discovery of Scholarly and Trade Journal Resources Via OAI and RSS Chumbe, Santiago Segundo; MacLeod, Roddy Publication
Putting OWL into production at the European Bioinformatics Institute
 Putting OWL into production at the European Bioinformatics Institute James Malone, Tony Burdett, Jon Ison, Simon Jupp, Drashtti Vasant, Dani Welter, and Helen Parkinson European Bioinformatics Institute,
Putting OWL into production at the European Bioinformatics Institute James Malone, Tony Burdett, Jon Ison, Simon Jupp, Drashtti Vasant, Dani Welter, and Helen Parkinson European Bioinformatics Institute,
Reducing Consumer Uncertainty
 Spatial Analytics Reducing Consumer Uncertainty Towards an Ontology for Geospatial User-centric Metadata Introduction Cooperative Research Centre for Spatial Information (CRCSI) in Australia Communicate
Spatial Analytics Reducing Consumer Uncertainty Towards an Ontology for Geospatial User-centric Metadata Introduction Cooperative Research Centre for Spatial Information (CRCSI) in Australia Communicate
ELIXIR Human Data Use Case
 ELIXIR Human Data Use Case Mikael Borg, ELIXIR Sweden ELIXIR-EXCELERATE is funded by the European Commission within the Research Infrastructures programme of Horizon 2020, grant agreement number 676559.
ELIXIR Human Data Use Case Mikael Borg, ELIXIR Sweden ELIXIR-EXCELERATE is funded by the European Commission within the Research Infrastructures programme of Horizon 2020, grant agreement number 676559.
The Euro-BioImaging Image Data Repository: Update & Future Directions
 The Euro-BioImaging Image Data Repository: Update & Future Directions Jason Swedlow University of Dundee Open Microscopy Environment openmicroscopy.org eurobioimaging.eu @jrswedlow Mission & Scope Europe
The Euro-BioImaging Image Data Repository: Update & Future Directions Jason Swedlow University of Dundee Open Microscopy Environment openmicroscopy.org eurobioimaging.eu @jrswedlow Mission & Scope Europe
Ontology Servers and Metadata Vocabulary Repositories
 Ontology Servers and Metadata Vocabulary Repositories Dr. Manjula Patel Technical Research and Development m.patel@ukoln.ac.uk http://www.ukoln.ac.uk/ Overview agentcities.net deployment grant Background
Ontology Servers and Metadata Vocabulary Repositories Dr. Manjula Patel Technical Research and Development m.patel@ukoln.ac.uk http://www.ukoln.ac.uk/ Overview agentcities.net deployment grant Background
Development of an Ontology-Based Portal for Digital Archive Services
 Development of an Ontology-Based Portal for Digital Archive Services Ching-Long Yeh Department of Computer Science and Engineering Tatung University 40 Chungshan N. Rd. 3rd Sec. Taipei, 104, Taiwan chingyeh@cse.ttu.edu.tw
Development of an Ontology-Based Portal for Digital Archive Services Ching-Long Yeh Department of Computer Science and Engineering Tatung University 40 Chungshan N. Rd. 3rd Sec. Taipei, 104, Taiwan chingyeh@cse.ttu.edu.tw
Euro- OMERO Tanja Ninkovic, EMBL
 Euro- BioImaging @ OMERO 2015 02.06.2015 Tanja Ninkovic, EMBL Aim & Development Euro-BioImaging will become a pan-european research infrastructure, which provides: Open access and services to imaging technologies
Euro- BioImaging @ OMERO 2015 02.06.2015 Tanja Ninkovic, EMBL Aim & Development Euro-BioImaging will become a pan-european research infrastructure, which provides: Open access and services to imaging technologies
Enterprise Data Catalog for Microsoft Azure Tutorial
 Enterprise Data Catalog for Microsoft Azure Tutorial VERSION 10.2 JANUARY 2018 Page 1 of 45 Contents Tutorial Objectives... 4 Enterprise Data Catalog Overview... 5 Overview... 5 Objectives... 5 Enterprise
Enterprise Data Catalog for Microsoft Azure Tutorial VERSION 10.2 JANUARY 2018 Page 1 of 45 Contents Tutorial Objectives... 4 Enterprise Data Catalog Overview... 5 Overview... 5 Objectives... 5 Enterprise
Biosphere: the interoperation of web services in microarray cluster analysis
 Biosphere: the interoperation of web services in microarray cluster analysis Kei-Hoi Cheung 1,2,*, Remko de Knikker 1, Youjun Guo 1, Guoneng Zhong 1, Janet Hager 3,4, Kevin Y. Yip 5, Albert K.H. Kwan 5,
Biosphere: the interoperation of web services in microarray cluster analysis Kei-Hoi Cheung 1,2,*, Remko de Knikker 1, Youjun Guo 1, Guoneng Zhong 1, Janet Hager 3,4, Kevin Y. Yip 5, Albert K.H. Kwan 5,
Simile Tools Workshop Summary MacKenzie Smith, MIT Libraries
 Simile Tools Workshop Summary MacKenzie Smith, MIT Libraries Intro On June 10 th and 11 th, 2010 a group of Simile Exhibit users, software developers and architects met in Washington D.C. to discuss the
Simile Tools Workshop Summary MacKenzie Smith, MIT Libraries Intro On June 10 th and 11 th, 2010 a group of Simile Exhibit users, software developers and architects met in Washington D.C. to discuss the
The walkthrough is available at /
 The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
XML in the bipharmaceutical
 XML in the bipharmaceutical sector XML holds out the opportunity to integrate data across both the enterprise and the network of biopharmaceutical alliances - with little technological dislocation and
XML in the bipharmaceutical sector XML holds out the opportunity to integrate data across both the enterprise and the network of biopharmaceutical alliances - with little technological dislocation and
POMap results for OAEI 2017
 POMap results for OAEI 2017 Amir Laadhar 1, Faiza Ghozzi 2, Imen Megdiche 1, Franck Ravat 1, Olivier Teste 1, and Faiez Gargouri 2 1 Paul Sabatier University, IRIT (CNRS/UMR 5505) 118 Route de Narbonne
POMap results for OAEI 2017 Amir Laadhar 1, Faiza Ghozzi 2, Imen Megdiche 1, Franck Ravat 1, Olivier Teste 1, and Faiez Gargouri 2 1 Paul Sabatier University, IRIT (CNRS/UMR 5505) 118 Route de Narbonne
Spatial Data on the Web
 Spatial Data on the Web Tools and guidance for data providers The European Commission s science and knowledge service W3C Data on the Web Best Practices 35 W3C/OGC Spatial Data on the Web Best Practices
Spatial Data on the Web Tools and guidance for data providers The European Commission s science and knowledge service W3C Data on the Web Best Practices 35 W3C/OGC Spatial Data on the Web Best Practices
RIKEN MetaDatabase: A Database Platform for Health Care and Life Sciences as a Microcosm of Linked Open Data Cloud ABSTRACT
 RIKEN MetaDatabase: A Database Platform for Health Care and Life Sciences as a Microcosm of Linked Open Data Cloud Norio Kobayashi, Advanced Center for Computing and Communication (ACCC), BioResource Center,
RIKEN MetaDatabase: A Database Platform for Health Care and Life Sciences as a Microcosm of Linked Open Data Cloud Norio Kobayashi, Advanced Center for Computing and Communication (ACCC), BioResource Center,
Development of an Environment for Data Annotation in Bioengineering
 Development of an Environment for Data Annotation in Bioengineering Renato Galina Barbosa Azambuja Correia Instituto Superior Técnico, Universidade Técnica de Lisboa, Portugal renato_correia1@hotmail.com
Development of an Environment for Data Annotation in Bioengineering Renato Galina Barbosa Azambuja Correia Instituto Superior Técnico, Universidade Técnica de Lisboa, Portugal renato_correia1@hotmail.com
Minimal Metadata Standards and MIIDI Reports
 Dryad-UK Workshop Wolfson College, Oxford 12 September 2011 Minimal Metadata Standards and MIIDI Reports David Shotton, Silvio Peroni and Tanya Gray Image BioInformatics Research Group Department of Zoology
Dryad-UK Workshop Wolfson College, Oxford 12 September 2011 Minimal Metadata Standards and MIIDI Reports David Shotton, Silvio Peroni and Tanya Gray Image BioInformatics Research Group Department of Zoology
Ontology and Application for Reusable Search Interface Design
 Ontology and Application for Reusable Search Interface Design Plans for Advanced Semantic Technologies Final Project Eric Rozell, Tetherless World Constellation Outline Project Overview Paper Contents
Ontology and Application for Reusable Search Interface Design Plans for Advanced Semantic Technologies Final Project Eric Rozell, Tetherless World Constellation Outline Project Overview Paper Contents
Michal Kuneš
 The Open Microscopy Environment A DataBase for the storage and manipulation of image data Michal Kuneš xkunes@utia.cas.cz ZOI UTIA, ASCR, Friday seminar 13.12.2013 OMERO http://www.openmicroscopy.org/site/support/omero4/users/index.html
The Open Microscopy Environment A DataBase for the storage and manipulation of image data Michal Kuneš xkunes@utia.cas.cz ZOI UTIA, ASCR, Friday seminar 13.12.2013 OMERO http://www.openmicroscopy.org/site/support/omero4/users/index.html
Logi Info v12.5 WHAT S NEW
 Logi Info v12.5 WHAT S NEW Introduction Logi empowers companies to embed analytics into the fabric of their organizations and products enabling anyone to analyze data, share insights, and make informed
Logi Info v12.5 WHAT S NEW Introduction Logi empowers companies to embed analytics into the fabric of their organizations and products enabling anyone to analyze data, share insights, and make informed
HymenopteraMine Documentation
 HymenopteraMine Documentation Release 1.0 Aditi Tayal, Deepak Unni, Colin Diesh, Chris Elsik, Darren Hagen Apr 06, 2017 Contents 1 Welcome to HymenopteraMine 3 1.1 Overview of HymenopteraMine.....................................
HymenopteraMine Documentation Release 1.0 Aditi Tayal, Deepak Unni, Colin Diesh, Chris Elsik, Darren Hagen Apr 06, 2017 Contents 1 Welcome to HymenopteraMine 3 1.1 Overview of HymenopteraMine.....................................
Long-term preservation for INSPIRE: a metadata framework and geo-portal implementation
 Long-term preservation for INSPIRE: a metadata framework and geo-portal implementation INSPIRE 2010, KRAKOW Dr. Arif Shaon, Dr. Andrew Woolf (e-science, Science and Technology Facilities Council, UK) 3
Long-term preservation for INSPIRE: a metadata framework and geo-portal implementation INSPIRE 2010, KRAKOW Dr. Arif Shaon, Dr. Andrew Woolf (e-science, Science and Technology Facilities Council, UK) 3
National Centre for Text Mining NaCTeM. e-science and data mining workshop
 National Centre for Text Mining NaCTeM e-science and data mining workshop John Keane Co-Director, NaCTeM john.keane@manchester.ac.uk School of Informatics, University of Manchester What is text mining?
National Centre for Text Mining NaCTeM e-science and data mining workshop John Keane Co-Director, NaCTeM john.keane@manchester.ac.uk School of Informatics, University of Manchester What is text mining?
Syddansk Universitet. Værktøj til videndeling og interaktion. Dai, Zheng. Published in: inform. Publication date: 2009
 Syddansk Universitet Værktøj til videndeling og interaktion Dai, Zheng Published in: inform Publication date: 2009 Document version Peer-review version Citation for pulished version (APA): Dai, Z. (2009).
Syddansk Universitet Værktøj til videndeling og interaktion Dai, Zheng Published in: inform Publication date: 2009 Document version Peer-review version Citation for pulished version (APA): Dai, Z. (2009).
Update: MIRIAM Registry and SBO
 Update: MIRIAM Registry and SBO Nick Juty, EMBL-EBI 3rd Sept, 2011 Overview MIRIAM Registry MIRIAM Guidelines.. MIRIAM Registry content URIs (URN form), example Summary/current developments SBO Purpose
Update: MIRIAM Registry and SBO Nick Juty, EMBL-EBI 3rd Sept, 2011 Overview MIRIAM Registry MIRIAM Guidelines.. MIRIAM Registry content URIs (URN form), example Summary/current developments SBO Purpose
AGDASHBOARDS USER MANUAL AGDASHBOARDS USER MANUAL. Fostering stronger data driven planning and decision making in a more visually engaging format.
 AGDASHBOARDS USER MANUAL AGDASHBOARDS USER MANUAL Fostering stronger data driven planning and decision making in a more visually engaging format." Table of Contents General Instructions... 3 Logging Into
AGDASHBOARDS USER MANUAL AGDASHBOARDS USER MANUAL Fostering stronger data driven planning and decision making in a more visually engaging format." Table of Contents General Instructions... 3 Logging Into
enanomapper database, search tools and templates Nina Jeliazkova, Nikolay Kochev IdeaConsult Ltd. Sofia, Bulgaria
 enanomapper database, search tools and templates Nina Jeliazkova, Nikolay Kochev IdeaConsult Ltd. Sofia, Bulgaria www.ideaconsult.net Ø enanomapper database: data model, technology; NANoREG data transfer
enanomapper database, search tools and templates Nina Jeliazkova, Nikolay Kochev IdeaConsult Ltd. Sofia, Bulgaria www.ideaconsult.net Ø enanomapper database: data model, technology; NANoREG data transfer
User Manual. Ver. 3.0 March 19, 2012
 User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
The following topics describe how to work with reports in the Firepower System:
 The following topics describe how to work with reports in the Firepower System: Introduction to Reports Introduction to Reports, on page 1 Risk Reports, on page 1 Standard Reports, on page 2 About Working
The following topics describe how to work with reports in the Firepower System: Introduction to Reports Introduction to Reports, on page 1 Risk Reports, on page 1 Standard Reports, on page 2 About Working
Browsing the Semantic Web
 Proceedings of the 7 th International Conference on Applied Informatics Eger, Hungary, January 28 31, 2007. Vol. 2. pp. 237 245. Browsing the Semantic Web Peter Jeszenszky Faculty of Informatics, University
Proceedings of the 7 th International Conference on Applied Informatics Eger, Hungary, January 28 31, 2007. Vol. 2. pp. 237 245. Browsing the Semantic Web Peter Jeszenszky Faculty of Informatics, University
warwick.ac.uk/lib-publications
 Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
[ PARADIGM SCIENTIFIC SEARCH ] A POWERFUL SOLUTION for Enterprise-Wide Scientific Information Access
![[ PARADIGM SCIENTIFIC SEARCH ] A POWERFUL SOLUTION for Enterprise-Wide Scientific Information Access [ PARADIGM SCIENTIFIC SEARCH ] A POWERFUL SOLUTION for Enterprise-Wide Scientific Information Access](/thumbs/88/116124260.jpg) A POWERFUL SOLUTION for Enterprise-Wide Scientific Information Access ENABLING EASY ACCESS TO Enterprise-Wide Scientific Information Waters Paradigm Scientific Search Software enables fast, easy, high
A POWERFUL SOLUTION for Enterprise-Wide Scientific Information Access ENABLING EASY ACCESS TO Enterprise-Wide Scientific Information Waters Paradigm Scientific Search Software enables fast, easy, high
Web Resources. iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases
 Web Resources iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases Ethan W. Hollingsworth 1, 2, Jacob E. Vaughn 1, 2, Josh C. Orack 1,2, Chelsea Skinner
Web Resources iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases Ethan W. Hollingsworth 1, 2, Jacob E. Vaughn 1, 2, Josh C. Orack 1,2, Chelsea Skinner
Introduction to Cisco UCS Central
 Introducing Cisco UCS Central, page 1 Introducing Cisco UCS Central Cisco UCS Central provides scalable management solution for growing Cisco UCS environment. Cisco UCS Central simplifies the management
Introducing Cisco UCS Central, page 1 Introducing Cisco UCS Central Cisco UCS Central provides scalable management solution for growing Cisco UCS environment. Cisco UCS Central simplifies the management
Jazz for Service Management Version 1.1. Offering Guide
 Jazz for Service Management Version 1.1 Offering Guide Jazz for Service Management Version 1.1 Offering Guide Note Before using this information and the offering it supports, read the information in Notices
Jazz for Service Management Version 1.1 Offering Guide Jazz for Service Management Version 1.1 Offering Guide Note Before using this information and the offering it supports, read the information in Notices
Etanova Enterprise Solutions
 Etanova Enterprise Solutions Front End Development» 2018-09-23 http://www.etanova.com/technologies/front-end-development Contents HTML 5... 6 Rich Internet Applications... 6 Web Browser Hardware Acceleration...
Etanova Enterprise Solutions Front End Development» 2018-09-23 http://www.etanova.com/technologies/front-end-development Contents HTML 5... 6 Rich Internet Applications... 6 Web Browser Hardware Acceleration...
Core Technology Development Team Meeting
 Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
National Data Sharing and Accessibility Policy-2012 (NDSAP-2012)
 National Data Sharing and Accessibility Policy-2012 (NDSAP-2012) Department of Science & Technology Ministry of science & Technology Government of India Government of India Ministry of Science & Technology
National Data Sharing and Accessibility Policy-2012 (NDSAP-2012) Department of Science & Technology Ministry of science & Technology Government of India Government of India Ministry of Science & Technology
PROJECT PERIODIC REPORT
 PROJECT PERIODIC REPORT Grant Agreement number: 257403 Project acronym: CUBIST Project title: Combining and Uniting Business Intelligence and Semantic Technologies Funding Scheme: STREP Date of latest
PROJECT PERIODIC REPORT Grant Agreement number: 257403 Project acronym: CUBIST Project title: Combining and Uniting Business Intelligence and Semantic Technologies Funding Scheme: STREP Date of latest
PYRAMID Headline Features. April 2018 Release
 PYRAMID 2018.03 April 2018 Release The April release of Pyramid brings a big list of over 40 new features and functional upgrades, designed to make Pyramid s OS the leading solution for customers wishing
PYRAMID 2018.03 April 2018 Release The April release of Pyramid brings a big list of over 40 new features and functional upgrades, designed to make Pyramid s OS the leading solution for customers wishing
Core Technology Development Team Meeting
 Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Ensembl Core API. EMBL European Bioinformatics Institute Wellcome Trust Genome Campus Hinxton, Cambridge, CB10 1SD, UK
 Ensembl Core API EMBL European Bioinformatics Institute Wellcome Trust Genome Campus Hinxton, Cambridge, CB10 1SD, UK EBI is an Outstation of the European Molecular Biology Laboratory. Outline a. b. c.
Ensembl Core API EMBL European Bioinformatics Institute Wellcome Trust Genome Campus Hinxton, Cambridge, CB10 1SD, UK EBI is an Outstation of the European Molecular Biology Laboratory. Outline a. b. c.
Ontology-Driven Metadata Enrichment for Genomic Datasets
 Ontology-Driven Metadata Enrichment for Genomic Datasets Anna Bernasconi [0000 0001 8016 5750], Arif Canakoglu [0000 0003 4528 6586], Andrea Colombo, and Stefano Ceri [0000 0003 0671 2415] Politecnico
Ontology-Driven Metadata Enrichment for Genomic Datasets Anna Bernasconi [0000 0001 8016 5750], Arif Canakoglu [0000 0003 4528 6586], Andrea Colombo, and Stefano Ceri [0000 0003 0671 2415] Politecnico
Linked Data: Fast, low cost semantic interoperability for health care?
 Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
A Grid Middleware for Ontology Access
 Available online at http://www.ges2007.de This document is under the terms of the CC-BY-NC-ND Creative Commons Attribution A Grid Middleware for Access Michael Hartung 1 and Erhard Rahm 2 1 Interdisciplinary
Available online at http://www.ges2007.de This document is under the terms of the CC-BY-NC-ND Creative Commons Attribution A Grid Middleware for Access Michael Hartung 1 and Erhard Rahm 2 1 Interdisciplinary
A Semantic Model for Federated Queries Over a Normalized Corpus
 A Semantic Model for Federated Queries Over a Normalized Corpus Samuel Croset, Christoph Grabmüller, Dietrich Rebholz-Schuhmann 17 th March 2010, Hinxton EBI is an Outstation of the European Molecular
A Semantic Model for Federated Queries Over a Normalized Corpus Samuel Croset, Christoph Grabmüller, Dietrich Rebholz-Schuhmann 17 th March 2010, Hinxton EBI is an Outstation of the European Molecular
Information Resources in Molecular Biology Marcela Davila-Lopez How many and where
 Information Resources in Molecular Biology Marcela Davila-Lopez (marcela.davila@medkem.gu.se) How many and where Data growth DB: What and Why A Database is a shared collection of logically related data,
Information Resources in Molecular Biology Marcela Davila-Lopez (marcela.davila@medkem.gu.se) How many and where Data growth DB: What and Why A Database is a shared collection of logically related data,
Information Workbench
 Information Workbench The Optique Technical Solution Christoph Pinkel, fluid Operations AG Optique: What is it, really? 3 Optique: End-user Access to Big Data 4 Optique: Scalable Access to Big Data 5 The
Information Workbench The Optique Technical Solution Christoph Pinkel, fluid Operations AG Optique: What is it, really? 3 Optique: End-user Access to Big Data 4 Optique: Scalable Access to Big Data 5 The
SECTION 10 EXCHANGE PROTOCOL
 SECTION 10 EXCHANGE PROTOCOL The ADMS specification will facilitate the creation of a federation of disparate semantic asset repositories at the EU level. This federation will consist of Joinup setting
SECTION 10 EXCHANGE PROTOCOL The ADMS specification will facilitate the creation of a federation of disparate semantic asset repositories at the EU level. This federation will consist of Joinup setting
Seamless Dynamic Web (and Smart Device!) Reporting with SAS D.J. Penix, Pinnacle Solutions, Indianapolis, IN
 Paper RIV05 Seamless Dynamic Web (and Smart Device!) Reporting with SAS D.J. Penix, Pinnacle Solutions, Indianapolis, IN ABSTRACT The SAS Business Intelligence platform provides a wide variety of reporting
Paper RIV05 Seamless Dynamic Web (and Smart Device!) Reporting with SAS D.J. Penix, Pinnacle Solutions, Indianapolis, IN ABSTRACT The SAS Business Intelligence platform provides a wide variety of reporting
Working with Reports
 The following topics describe how to work with reports in the Firepower System: Introduction to Reports, page 1 Risk Reports, page 1 Standard Reports, page 2 About Working with Generated Reports, page
The following topics describe how to work with reports in the Firepower System: Introduction to Reports, page 1 Risk Reports, page 1 Standard Reports, page 2 About Working with Generated Reports, page
The European Variation Archive
 The European Variation Archive Webinar: A database of all types of genomic variation data from all species Hannah McLaren www.ebi.ac.uk/eva eva-helpdesk@ebi.ac.uk Learning objectives Establish the key
The European Variation Archive Webinar: A database of all types of genomic variation data from all species Hannah McLaren www.ebi.ac.uk/eva eva-helpdesk@ebi.ac.uk Learning objectives Establish the key
No Programming Required Create web apps rapidly with Web AppBuilder for ArcGIS
 No Programming Required Create web apps rapidly with Web AppBuilder for ArcGIS By Derek Law, Esri Product Manager, ArcGIS for Server Do you want to build web mapping applications you can run on desktop,
No Programming Required Create web apps rapidly with Web AppBuilder for ArcGIS By Derek Law, Esri Product Manager, ArcGIS for Server Do you want to build web mapping applications you can run on desktop,
Combining Business Intelligence with Semantic Technologies: The CUBIST Project
 Combining Business Intelligence with Semantic Technologies: The CUBIST Project DAU, Frithjof and ANDREWS, Simon Available from Sheffield Hallam University Research
Combining Business Intelligence with Semantic Technologies: The CUBIST Project DAU, Frithjof and ANDREWS, Simon Available from Sheffield Hallam University Research
Bioinformatics Data Distribution and Integration via Web Services and XML
 Letter Bioinformatics Data Distribution and Integration via Web Services and XML Xiao Li and Yizheng Zhang* College of Life Science, Sichuan University/Sichuan Key Laboratory of Molecular Biology and Biotechnology,
Letter Bioinformatics Data Distribution and Integration via Web Services and XML Xiao Li and Yizheng Zhang* College of Life Science, Sichuan University/Sichuan Key Laboratory of Molecular Biology and Biotechnology,
You need to use the URL provided by your institute s OMERO administrator to access the OMERO.web client.
 1 OMERO.web Client Using OMERO.web to view and work with image data via a web browser. You need to use the URL provided by your institute s OMERO administrator to access the OMERO.web client. Logging in,
1 OMERO.web Client Using OMERO.web to view and work with image data via a web browser. You need to use the URL provided by your institute s OMERO administrator to access the OMERO.web client. Logging in,
The Final Updates. Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University of Oxford, UK
 The Final Updates Supported by the NIH grant 1U24 AI117966-01 to UCSD PI, Co-Investigators at: Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University
The Final Updates Supported by the NIH grant 1U24 AI117966-01 to UCSD PI, Co-Investigators at: Philippe Rocca-Serra Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone, Oxford e-research Centre, University
ESG: Extended Similarity Group Job Submission
 ESG: Extended Similarity Group Job Submission Cite: Meghana Chitale, Troy Hawkins, Changsoon Park, & Daisuke Kihara ESG: Extended similarity group method for automated protein function prediction, Bioinformatics,
ESG: Extended Similarity Group Job Submission Cite: Meghana Chitale, Troy Hawkins, Changsoon Park, & Daisuke Kihara ESG: Extended similarity group method for automated protein function prediction, Bioinformatics,
LORE: A Compound Object Authoring and Publishing Tool for Literary Scholars based on the FRBR. Anna Gerber, Jane Hunter
 LORE: A Compound Object Authoring and Publishing Tool for Literary Scholars based on the FRBR Anna Gerber, Jane Hunter Open Repositories 2009 Overview LORE: Literature Object Reuse and Exchange Background
LORE: A Compound Object Authoring and Publishing Tool for Literary Scholars based on the FRBR Anna Gerber, Jane Hunter Open Repositories 2009 Overview LORE: Literature Object Reuse and Exchange Background
Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides
 1 Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides ways to flexibly merge your Mascot search and quantitation
1 Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides ways to flexibly merge your Mascot search and quantitation
Executive Summary for deliverable D6.1: Definition of the PFS services (requirements, initial design)
 Electronic Health Records for Clinical Research Executive Summary for deliverable D6.1: Definition of the PFS services (requirements, initial design) Project acronym: EHR4CR Project full title: Electronic
Electronic Health Records for Clinical Research Executive Summary for deliverable D6.1: Definition of the PFS services (requirements, initial design) Project acronym: EHR4CR Project full title: Electronic
SHARING YOUR RESEARCH DATA VIA
 SHARING YOUR RESEARCH DATA VIA SCHOLARBANK@NUS MEET OUR TEAM Gerrie Kow Head, Scholarly Communication NUS Libraries gerrie@nus.edu.sg Estella Ye Research Data Management Librarian NUS Libraries estella.ye@nus.edu.sg
SHARING YOUR RESEARCH DATA VIA SCHOLARBANK@NUS MEET OUR TEAM Gerrie Kow Head, Scholarly Communication NUS Libraries gerrie@nus.edu.sg Estella Ye Research Data Management Librarian NUS Libraries estella.ye@nus.edu.sg
SELF-SERVICE SEMANTIC DATA FEDERATION
 SELF-SERVICE SEMANTIC DATA FEDERATION WE LL MAKE YOU A DATA SCIENTIST Contact: IPSNP Computing Inc. Chris Baker, CEO Chris.Baker@ipsnp.com (506) 721 8241 BIG VISION: SELF-SERVICE DATA FEDERATION Biomedical
SELF-SERVICE SEMANTIC DATA FEDERATION WE LL MAKE YOU A DATA SCIENTIST Contact: IPSNP Computing Inc. Chris Baker, CEO Chris.Baker@ipsnp.com (506) 721 8241 BIG VISION: SELF-SERVICE DATA FEDERATION Biomedical
Ontology-based Architecture Documentation Approach
 4 Ontology-based Architecture Documentation Approach In this chapter we investigate how an ontology can be used for retrieving AK from SA documentation (RQ2). We first give background information on the
4 Ontology-based Architecture Documentation Approach In this chapter we investigate how an ontology can be used for retrieving AK from SA documentation (RQ2). We first give background information on the
Core Technology Development Team Meeting
 Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Semantic Web Company. PoolParty - Server. PoolParty - Technical White Paper.
 Semantic Web Company PoolParty - Server PoolParty - Technical White Paper http://www.poolparty.biz Table of Contents Introduction... 3 PoolParty Technical Overview... 3 PoolParty Components Overview...
Semantic Web Company PoolParty - Server PoolParty - Technical White Paper http://www.poolparty.biz Table of Contents Introduction... 3 PoolParty Technical Overview... 3 PoolParty Components Overview...
SAS Web Infrastructure Kit 1.0. Overview, Second Edition
 SAS Web Infrastructure Kit 1.0 Overview, Second Edition The correct bibliographic citation for this manual is as follows: SAS Institute Inc. 2006. SAS Web Infrastructure Kit 1.0: Overview, Second Edition.
SAS Web Infrastructure Kit 1.0 Overview, Second Edition The correct bibliographic citation for this manual is as follows: SAS Institute Inc. 2006. SAS Web Infrastructure Kit 1.0: Overview, Second Edition.
Knowledge Representations. How else can we represent knowledge in addition to formal logic?
 Knowledge Representations How else can we represent knowledge in addition to formal logic? 1 Common Knowledge Representations Formal Logic Production Rules Semantic Nets Schemata and Frames 2 Production
Knowledge Representations How else can we represent knowledge in addition to formal logic? 1 Common Knowledge Representations Formal Logic Production Rules Semantic Nets Schemata and Frames 2 Production
Powering Knowledge Discovery. Insights from big data with Linguamatics I2E
 Powering Knowledge Discovery Insights from big data with Linguamatics I2E Gain actionable insights from unstructured data The world now generates an overwhelming amount of data, most of it written in natural
Powering Knowledge Discovery Insights from big data with Linguamatics I2E Gain actionable insights from unstructured data The world now generates an overwhelming amount of data, most of it written in natural
Brown University Libraries Technology Plan
 Brown University Libraries Technology Plan 2009-2011 Technology Vision Brown University Library creates, develops, promotes, and uses technology to further the Library s mission and strategic directions
Brown University Libraries Technology Plan 2009-2011 Technology Vision Brown University Library creates, develops, promotes, and uses technology to further the Library s mission and strategic directions
The Now Platform Reference Guide
 The Now Platform Reference Guide A tour of key features and functionality START Introducing the Now Platform Digitize your business with intelligent apps The Now Platform is an application Platform-as-a-Service
The Now Platform Reference Guide A tour of key features and functionality START Introducing the Now Platform Digitize your business with intelligent apps The Now Platform is an application Platform-as-a-Service
D WSMO Data Grounding Component
 Project Number: 215219 Project Acronym: SOA4All Project Title: Instrument: Thematic Priority: Service Oriented Architectures for All Integrated Project Information and Communication Technologies Activity
Project Number: 215219 Project Acronym: SOA4All Project Title: Instrument: Thematic Priority: Service Oriented Architectures for All Integrated Project Information and Communication Technologies Activity
The Materials Data Facility
 The Materials Data Facility Ben Blaiszik (blaiszik@uchicago.edu), Kyle Chard (chard@uchicago.edu) Ian Foster (foster@uchicago.edu) materialsdatafacility.org What is MDF? We aim to make it simple for materials
The Materials Data Facility Ben Blaiszik (blaiszik@uchicago.edu), Kyle Chard (chard@uchicago.edu) Ian Foster (foster@uchicago.edu) materialsdatafacility.org What is MDF? We aim to make it simple for materials
Building a missing item in INSPIRE: The Re3gistry
 Building a missing item in INSPIRE: The Re3gistry www.jrc.ec.europa.eu Serving society Stimulating innovation Supporting legislation Key pillars of data interoperability Conceptual data models Encoding
Building a missing item in INSPIRE: The Re3gistry www.jrc.ec.europa.eu Serving society Stimulating innovation Supporting legislation Key pillars of data interoperability Conceptual data models Encoding
Blast2GO User Manual. Blast2GO Ortholog Group Annotation May, BioBam Bioinformatics S.L. Valencia, Spain
 Blast2GO User Manual Blast2GO Ortholog Group Annotation May, 2016 BioBam Bioinformatics S.L. Valencia, Spain Contents 1 Clusters of Orthologs 2 2 Orthologous Group Annotation Tool 2 3 Statistics for NOG
Blast2GO User Manual Blast2GO Ortholog Group Annotation May, 2016 BioBam Bioinformatics S.L. Valencia, Spain Contents 1 Clusters of Orthologs 2 2 Orthologous Group Annotation Tool 2 3 Statistics for NOG
Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR
 Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR Guoqian Jiang 1, Eric Prud Hommeaux 2, Guohui Xiao 3, and Harold R. Solbrig 1 1 Mayo Clinic, Rochester,
Developing A Semantic Web-based Framework for Executing the Clinical Quality Language Using FHIR Guoqian Jiang 1, Eric Prud Hommeaux 2, Guohui Xiao 3, and Harold R. Solbrig 1 1 Mayo Clinic, Rochester,
Core Technology Development Team Meeting
 Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
Core Technology Development Team Meeting To hear the meeting, you must call in Toll-free phone number: 1-866-740-1260 Access Code: 2201876 For international call in numbers, please visit: https://www.readytalk.com/account-administration/international-numbers
