Document Navigation: Ontologies or Knowledge Organisation Systems?
|
|
|
- Jonah Shelton
- 6 years ago
- Views:
Transcription
1 Document Navigation: Ontologies or Knowledge Organisation Systems? Simon Jupp *1, Sean Bechhofer 1, Patty Kostkova 2, Robert Stevens 1, Yeliz Yesilada 1 1 University of Manchester, Oxford Road, UK 2 City ehealth Research Centre, City University, London, UK addresses: Manchester: first.last [at] manchester.ac.uk London: patty [at] soi.city.ac.uk Abstract Bioinformatics relies heavily on web resources for information gathering. Ontologies are being developed to fill the background knowledge needed to drive Semantic Web applications. This paper discusses how formal ontologies are not always suited for document navigation on the web. Converting ontologies into a model with looser semantics, allows cheap and rapid generation of useful knowledge systems. The message is that ontologies are not the only knowledge artefact needed; vocabularies and other classification schemes with weaker semantics have their role and are the best solution in certain circumstances. Introduction Navigation via hypertext is a mainstay of the World Wide Web (WWW). The author owned and unary links of standard HTML often neither offer the link sources nor targets needed by a particular group. Conceptual hypermedia provides navigation between web resources, supported by a conceptual model. The content of the model is used to dynamically identify link sources in web documents, and also supply the link targets to relevant web-services. The field of bioinformatics relies heavily on web resources and the community is now rich in bio-medical ontologies that can be used to populate this conceptual model. The ability to browse documents on the web via hyperlinks embedded in text is still a fundamental part of the information gathering process used by bioinformaticians. As successful as hypertext is, it is not without its limitations; Hard Coding: Links are hard coded into the HTML source of a document. Ownership: Ownership of the page is required to place links in pages. Legacy: Link target can be deprecated leaving invalid links on pages. Unary targets: The current web links are restricted to point-to-point linking; there is only one target. Conceptual Open Hypermedia supports the construction of hypertext link structures built using information encoded in ontologies. Dynamic linking, supported by ontologies, offer a mechanism to help overcome some of these restrictions. The - 1 -
2 Conceptual Open Hypermedia Service (COHSE) 1 (Carr 2001) system enhances document resources through the addition of hypertext links (see Figure 1). These links are generated based on a mapping between concepts found in the document and lexicons available from the ontology. Links can have multiple targets based on the type of concept identified and in addition the structure of the ontology facilitates navigation to further targets based on sub/super concepts asserted in the ontology. The COHSE architecture has been demonstrated in several fields, the GOHSE (Bechhofer 2005) system was applied to bioinformatics using the Gene Ontology (GO) (Ashburner, Ball et al. 2000)as an ontology and GO associations 2 as link targets. The Sealife project 3 is now looking to extend this work and provide an ontology that integrates many of the ontologies being developed in biomedicine, to aid query by navigation to both scientists and health care professionals in the study of infectious diseases. One of the major obstacles at this stage is how to integrate all the necessary ontologies into a single model with appropriate semantics that suit navigation. We argue that the strict relationships held between concepts in ontologies are not well suited for navigational purposes. A thesaurus like artefact is better suited for this task, it allows us to capture relationships that are not formal or universal nor part of the integral definition of the term. Our goal is to benefit from the work being done in the bio-ontology community i.e. capturing specific domain knowledge, and bring this knowledge into a model that suits the application s needs. The proposed solution is to convert relevant bio-ontologies, medical vocabularies, thesauri, taxonomies and other concept schemes into one large Knowledge Organisation System (KOS). The Simple Knowledge Organisation System (SKOS) 4 is chosen as a model to hold this information. A use case from the Sealife project is used to demonstrate the application in the study of infectious disease. Sealife Use Case The Sealife project seeks to develop a series of browsers in the context of the Semantic Web and Semantic Grid. The grid offers an infrastructure for large scale in silico science via a large number of computational services. The Grid setting needs to be combined with the continuing presence and use of numbers of Web documents describing knowledge about biology. Ontologies and controlled vocabularies provide great benefits for describing and using their data. The Sealife browser aims to use these vocabularies and ontologies as description of knowledge in the life sciences to flexibly manage the inter-linking of these documents and services. One example application is to provide dynamic hyper-linking of resources from the National electronic Library of Infection (NeLI) 5 (Kostkova 2003) portal to other related resources on the web. NeLI is a digital library bringing together the best available on-line evidence-based, quality tagged resources on the investigation, treatment, prevention and control of infectious disease. Many documents on the NeLI site contain few, if any, hyperlinks to other resources on the web. It would take a large 1 See
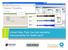
3 curational effort and cost to manually mark up these pages with links to other web resources. In addition to this problem, NeLI has a range of users; we want different link targets based on the kind of user browsing the NeLI site. COHSE can help to solve some of these problems. Terms from the ontologies can be used to identify concepts in web pages and create the link sources. Each link source can have multiple targets; the targets selected are tailored to suit the needs of each user group. The ability to identify user groups is important. Users can range from members of the public, molecular biologists to clinicians and GPs. Each group has a different view of the bio-medical domain, and is therefore interested in different kinds of information. By providing alternative vocabularies for different users, the system can identify link sources relevant to that user and also provide multiple targets to relevant web resources. Table 1 shows four different user groups, some questions they might want answering and the different kinds of target sites a Sealife browser would offer them based on the type of user (Madle 2006). The system is demonstrated with a simple use case involving a news site linking to NeLI. News sites are often the first to report on disease outbreaks via news feeds. Consider the scenario where a traveller is planning a trip to Namibia, only to find an article on the BBC website about a recent outbreak of Polio. COHSE can provide links to relevant resources that had not been included by the original author. Such resources could include information about the polio virus, its effect on humans, vaccination information and also geographical information about the local area. A family doctor, in contrast, might use a vocabulary skewed to the their interests to link through to sites on drugs, details of symptoms and clinical presentations, treatment and local hospital facilities etc. User Group Question Targets Family Doctor Tuberculosis drugs and side British National Formulary (BNF) (GP) effects? Clinicians Tuberculosis treatments guidelines? Public Health Observatories (PHO) Molecular Drug resistant tuberculosis PubMed Biologists species? General Public What is tuberculosis? Health Protection Agency (HPA) or the NHS direct online website. Table 1. NeLI users and example targets from the UK. Figure 1 shows the system in action. The first image shows the original BBC article, the second shows dynamic links that have been added based on concepts held in the vocabulary. It also shows a link box that is dynamically generated when a link is clicked. The link box contains a textual description of the term and targets to multiple web resources. In addition to targets for the selected link the system can provide targets for broader, narrower and related resources. For example, NeLI has a web-service which takes terms from a NeLI vocabulary as inputs, this service is invoked when the polio link is selected and targets are returned which link to relevant documents from the NeLI portal. This simple demonstration shows how the addition of a navigational layer based on the semantic content of documents can be added to the existing web
4 Fig. 1. Dynamic linking in action. Gathering the background knowledge For the Sealife browser to be useful across such a diverse subject as biology we, need a system to rapidly collect the current available resources together, and place them into a single representation that will facilitate navigation. The biomedical domain already has a rich collection of vocabularies and ontologies such as MesH6, UMLS7, GALEN8 and the OBO9 ontologies. There are also classification systems relating to genes, protein, drug and other terminological resources that would be useful to Sealife. The languages used to represent ontologies vary considerably, and can range from simple taxonomy languages through to rich, formal logic based languages such as OWL. Increasingly strict semantics can remove ambiguity in the representation and facilitate the use of machine processing. Similarly, these languages can be used with varying degrees of ontological formality, not all OWL ontologies, for example, make rigorous ontological distinctions. Experience with COHSE has suggested that formal ontological distinctions and strict semantics are not always best suited to the task of navigating a collection of resources. Strict sub/super class relationships are not necessarily appropriate for navigation rather, the looser notions of broader/narrower as found in vocabularies or thesauri provide the user with more appropriate linking. SKOS is a model for representing classification systems, thesauri, taxonomies and other concept schemes. SKOS is currently undergoing stanardisation by the W3C
5 and has a RDF/XML representation that makes it well suited for semantic web applications. By representing the biological knowledge in SKOS we have a simple model that provides a lexical resource for identifying concepts in our documents, as well as a framework for asserting semantic relationships between concepts. SKOS has a set of properties that are well suited for supporting navigation. These include preferred labels, alternate labels (synonyms) and textual definitions for describing concepts as well as broader, narrower and related, for representing the relationships between concepts. Converting ontologies to SKOS The semantics of some biomedical terminologies are already relatively weak. A good example for such a terminology that is commonly used in medicine is the Medical Subject Headings (MeSH). The semantics of A narrower B simply means that users interested in B might also be interested in A. The MeSH terms found under accident include kinds of accidents as expected (e.g. Traffic accidents), but also Accident prevention. This is not a good ontological distinction, but a valid one in the context of navigation and retrieval. In contrast in the Open Biomedical Ontologies (OBO), A is-a B, a common type of relationship in OBO ontologies implies that all A s are also instances of B. This contrast in semantics means that conversions from MeSH into OBO are not possible without misinterpreting the intended semantics. Despite this, we see that many of the OBO ontologies share concepts with MeSH, especially the Disease Ontology 11. From a navigation point of view we would like to combine these resources to gain maximum benefit from efforts in MeSH and OBO development. By converting them both into SKOS we can use a single representation and use the lightweight semantics to build a larger and richer vocabulary. The release of the 10 OBO relations (Smith 2005) gives OBO developers another level of expressivity in their ontologies. These relations have strong definitions with precise semantics; which are used to define relationships between terms in OBO ontologies. When converting OBO ontologies into SKOS we can use these relationships to assert broader, narrower and related relationships between SKOS concepts. Here is an example of the conversion one might make when mapping ontological properties to SKOS properties. rel:part_of -> skos:broader (e.g. finger part_of hand) rel:contains -> skos:narrower (e.g. skull contains brain) rel:has_name -> skos:related (e.g. Person has_name PersonName) Another advantage when converting properties from ontologies to SKOS is the ability to assert the inverse. Consider an ontology where Nucleus partof cell, from an ontological point of view this implies that every Nucleus is partof some Cell. However, the inverse is not true, every Cell does not havepart Nucleus. When converting to a SKOS model we can assert the inverse using the broader property to say that Nucleus has a broader term called Cell, which is quite reasonable. When navigating around documents about cells, the system could then also provide links to documents about nuclei users interested in cells are often also interested in nuclei. If we use the polio use case example we can show that a great deal of information can be acquired about polio from the various vocabularies available. When the
6 semantics are strict we have to be very careful how we bring all this related information together. With all this information in SKOS, Sealife can benefit from many different knowledge resources. Table 2 outlines the results from searching polio against a varying set of ontologies and vocabularies alongside the SKOS property used to relate them. Source Terms found SKOS relation to Poliovirus MeSH Brunhilde Virus skos:altterm Disease Ontology Spinal cord disease skos:broaderthan Postpoliomyelitis skos:narrowerthan Syndrome SNOMED Microorganism skos:broaderthan Enterovirus skos:broaderthan Table 2. Searching polio virus against different resources and converting intended semantics into SKOS semantics for navigation. (NLM 1960, Cote et al 1993) There is, however, likely to be some trade off associated when bringing multiple resources together, it is possible that a lot of unwanted terms are returned, especially when using formal ontologies. Ontologies can benefit from an upper-ontology (Rector 2003) that contains abstract categories; these can be used to build formal definitions for classes. An ontological definition which states whether the concept is a physical or non-physical entity may be crucial to the design of a robust ontology, but is largely irrelevant from a navigational point of view. To overcome this we must remove these properties at the stage of conversion into SKOS, how we do the conversion from ontologies to SKOS is something for future work. CONCLUSION A large community of ontology developers and knowledge engineers is forming in the life sciences. It is hoped that they will deliver the infrastructure needed to realise a real semantic-web, where computers can begin to interpret and interoperate biological data automatically. If applications like Sealife are to demonstrate the early potential of Semantic Web technologies, then the trade off associated with relaxing the semantics in the background knowledge has to be acceptable. The nature of formal ontologies can sometimes make it difficult to express relationships between concepts that experts from the domain would expect to find under some circumstances. Thesauri are much more suited to represent the way words and language are used in the field. The Sealife project will demonstrate how the effort and cost associated with building rich formal ontologies can also be used to feed into other knowledge artefacts, like thesaurae, vocabularies, classification schemes etc. in SKOS, which can then be used in different application scenarios. ACKNOWLEDGEMENTS Funding by the Sealife project (IST ) for Simon Jupp is kindly acknowledged
7 REFERENCES Ashburner, M., C. A. Ball, et al. (2000). "Gene ontology: tool for the unification of biology. The Gene Ontology Consortium." Nat Genet 25(1): Bechhofer, S. Stevens, R. Lord, P. (2005) Ontology Driven Dynamix Linking of Biology Resources. Pacific Symposium on Biocomputing, Hawaii. Carr, L. Bechhofer, S. Goble, C. Hall, W. (2001) Conceptual Linking; Ontologybased Open Hypermedia. WWW10, Tenth World Wide Web Conference, Hong Kong. Cote RA et a;., eds. SNOME International: the systematized nomenclature of human and veterinary medicine. Vols I-IV. Northfeild, IL, College of American Pathologists G. Madle, P. Kostkova, J. Mani-Saada, A. Roy. (2006) Lessons learned from Evaluation of the Use of the National electronic Library of Infection, Health Informatics Journal, Special Issue, Healthcare Digital Libraries, 12: National Library of Medicine. Medical subject headings: main heading, subheadings, and cross references used in the Index Medicus and the Natinal Library of Medcine Catalog. 1 st ed. Washinton, DC:U.S. Department of Health, Education, and Welfare.1960 P. Kostkova et al. (2003) Agent-Based Up-to-date Data Management in National electronic Library for Communicable Disease. In SI: "Applications of intelligent agents in health care", J. Nealon, T. Moreno Ed, in Whitestein Series in Software Agent Technologies, pages Rector, A. (2003) Modularisation of Domain Ontologies Implemented in Description Logics and related formalisms including OWL. Knowledge Capture, ACM, Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, Mungall CJ, Neuhaus F, Rector A, Rosse C Relations in Biomedical Ontologies. Genome Biology, 2005, 6:R46-7 -
Knowledge Representation for Web Navigation
 Knowledge Representation for Web Navigation Simon Jupp 1, Robert Stevens 1, Sean Bechhofer 1, Yeliz Yesilada 1, and Patty Kostkova 2 1 School of Computer Science, University of Manchester, Oxford Road,
Knowledge Representation for Web Navigation Simon Jupp 1, Robert Stevens 1, Sean Bechhofer 1, Yeliz Yesilada 1, and Patty Kostkova 2 1 School of Computer Science, University of Manchester, Oxford Road,
User Profiling for Semantic Browsing in Medical Digital Libraries
 User Profiling for Semantic Browsing in Medical Digital Libraries Patty Kostkova 1,, Gayo Diallo 1 and Gawesh Jawaheer 1 1 City ehalth Research Centre, City University, Northampton Square, London, EC1V
User Profiling for Semantic Browsing in Medical Digital Libraries Patty Kostkova 1,, Gayo Diallo 1 and Gawesh Jawaheer 1 1 City ehalth Research Centre, City University, Northampton Square, London, EC1V
Ontologies SKOS. COMP62342 Sean Bechhofer
 Ontologies SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
Ontologies SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
SKOS. COMP62342 Sean Bechhofer
 SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Ontologies Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
SKOS COMP62342 Sean Bechhofer sean.bechhofer@manchester.ac.uk Ontologies Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
A Semantic Web-Based Approach for Harvesting Multilingual Textual. definitions from Wikipedia to support ICD-11 revision
 A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
A Semantic Web-Based Approach for Harvesting Multilingual Textual Definitions from Wikipedia to Support ICD-11 Revision Guoqian Jiang 1,* Harold R. Solbrig 1 and Christopher G. Chute 1 1 Department of
Taking a view on bio-ontologies. Simon Jupp Functional Genomics Production Team ICBO, 2012 Graz, Austria
 Taking a view on bio-ontologies Simon Jupp Functional Genomics Production Team ICBO, 2012 Graz, Austria Who we are European Bioinformatics Institute one of world s largest bio data and service providers
Taking a view on bio-ontologies Simon Jupp Functional Genomics Production Team ICBO, 2012 Graz, Austria Who we are European Bioinformatics Institute one of world s largest bio data and service providers
0.1 Knowledge Organization Systems for Semantic Web
 0.1 Knowledge Organization Systems for Semantic Web 0.1 Knowledge Organization Systems for Semantic Web 0.1.1 Knowledge Organization Systems Why do we need to organize knowledge? Indexing Retrieval Organization
0.1 Knowledge Organization Systems for Semantic Web 0.1 Knowledge Organization Systems for Semantic Web 0.1.1 Knowledge Organization Systems Why do we need to organize knowledge? Indexing Retrieval Organization
Languages and tools for building and using ontologies. Simon Jupp, James Malone
 An overview of ontology technology Languages and tools for building and using ontologies Simon Jupp, James Malone jupp@ebi.ac.uk, malone@ebi.ac.uk Outline Languages OWL and OBO classes, individuals, relations,
An overview of ontology technology Languages and tools for building and using ontologies Simon Jupp, James Malone jupp@ebi.ac.uk, malone@ebi.ac.uk Outline Languages OWL and OBO classes, individuals, relations,
warwick.ac.uk/lib-publications
 Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
Original citation: Zhao, Lei, Lim Choi Keung, Sarah Niukyun and Arvanitis, Theodoros N. (2016) A BioPortalbased terminology service for health data interoperability. In: Unifying the Applications and Foundations
Integrated Access to Biological Data. A use case
 Integrated Access to Biological Data. A use case Marta González Fundación ROBOTIKER, Parque Tecnológico Edif 202 48970 Zamudio, Vizcaya Spain marta@robotiker.es Abstract. This use case reflects the research
Integrated Access to Biological Data. A use case Marta González Fundación ROBOTIKER, Parque Tecnológico Edif 202 48970 Zamudio, Vizcaya Spain marta@robotiker.es Abstract. This use case reflects the research
Annotating, linking and browsing provenance logs for e-science
 Annotating, linking and browsing provenance logs for e-science Jun Zhao, Carole Goble, Mark Greenwood, Chris Wroe, Robert Stevens Department of Computer Science, University of Manchester, Oxford Road,
Annotating, linking and browsing provenance logs for e-science Jun Zhao, Carole Goble, Mark Greenwood, Chris Wroe, Robert Stevens Department of Computer Science, University of Manchester, Oxford Road,
Adaptable and Adaptive Web Information Systems. Lecture 1: Introduction
 Adaptable and Adaptive Web Information Systems School of Computer Science and Information Systems Birkbeck College University of London Lecture 1: Introduction George Magoulas gmagoulas@dcs.bbk.ac.uk October
Adaptable and Adaptive Web Information Systems School of Computer Science and Information Systems Birkbeck College University of London Lecture 1: Introduction George Magoulas gmagoulas@dcs.bbk.ac.uk October
An Online Ontology: WiktionaryZ
 KR-MED 2006 "Biomedical Ontology in Action" November 8, 2006, Baltimore, Maryland, USA An Online Ontology: WiktionaryZ Erik M. van Mulligen, Ph.D. 1,2, Erik Möller, Peter-Jan Roes 3, Marc Weeber, Ph.D.
KR-MED 2006 "Biomedical Ontology in Action" November 8, 2006, Baltimore, Maryland, USA An Online Ontology: WiktionaryZ Erik M. van Mulligen, Ph.D. 1,2, Erik Möller, Peter-Jan Roes 3, Marc Weeber, Ph.D.
The National Cancer Institute's Thésaurus and Ontology
 The National Cancer Institute's Thésaurus and Ontology Jennifer Golbeck 1, Gilberto Fragoso 2, Frank Hartel 2, Jim Hendler 1, Jim Oberthaler 2, Bijan Parsia 1 1 University of Maryland, College Park 2 National
The National Cancer Institute's Thésaurus and Ontology Jennifer Golbeck 1, Gilberto Fragoso 2, Frank Hartel 2, Jim Hendler 1, Jim Oberthaler 2, Bijan Parsia 1 1 University of Maryland, College Park 2 National
EBP. Accessing the Biomedical Literature for the Best Evidence
 Accessing the Biomedical Literature for the Best Evidence Structuring the search for information and evidence Basic search resources Starting the search EBP Lab / Practice: Simple searches Using PubMed
Accessing the Biomedical Literature for the Best Evidence Structuring the search for information and evidence Basic search resources Starting the search EBP Lab / Practice: Simple searches Using PubMed
Copyright 2012 Taxonomy Strategies. All rights reserved. Semantic Metadata. A Tale of Two Types of Vocabularies
 Taxonomy Strategies July 17, 2012 Copyright 2012 Taxonomy Strategies. All rights reserved. Semantic Metadata A Tale of Two Types of Vocabularies What is semantic metadata? Semantic relationships in the
Taxonomy Strategies July 17, 2012 Copyright 2012 Taxonomy Strategies. All rights reserved. Semantic Metadata A Tale of Two Types of Vocabularies What is semantic metadata? Semantic relationships in the
Terminologies, Knowledge Organization Systems, Ontologies
 Terminologies, Knowledge Organization Systems, Ontologies Gerhard Budin University of Vienna TSS July 2012, Vienna Motivation and Purpose Knowledge Organization Systems In this unit of TSS 12, we focus
Terminologies, Knowledge Organization Systems, Ontologies Gerhard Budin University of Vienna TSS July 2012, Vienna Motivation and Purpose Knowledge Organization Systems In this unit of TSS 12, we focus
Brain, a library for the OWL2 EL profile
 Brain, a library for the OWL2 EL profile Samuel Croset 1, John Overington 1, and Dietrich Rebholz-Schuhmann 1 EMBL-EBI, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD UK croset@ebi.ac.uk Abstract.
Brain, a library for the OWL2 EL profile Samuel Croset 1, John Overington 1, and Dietrich Rebholz-Schuhmann 1 EMBL-EBI, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD UK croset@ebi.ac.uk Abstract.
Complex Query Formulation Over Diverse Information Sources Using an Ontology
 Complex Query Formulation Over Diverse Information Sources Using an Ontology Robert Stevens, Carole Goble, Norman Paton, Sean Bechhofer, Gary Ng, Patricia Baker and Andy Brass Department of Computer Science,
Complex Query Formulation Over Diverse Information Sources Using an Ontology Robert Stevens, Carole Goble, Norman Paton, Sean Bechhofer, Gary Ng, Patricia Baker and Andy Brass Department of Computer Science,
Semantics. Matthew J. Graham CACR. Methods of Computational Science Caltech, 2011 May 10. matthew graham
 Semantics Matthew J. Graham CACR Methods of Computational Science Caltech, 2011 May 10 semantic web The future of the Internet (Web 3.0) Decentralized platform for distributed knowledge A web of databases
Semantics Matthew J. Graham CACR Methods of Computational Science Caltech, 2011 May 10 semantic web The future of the Internet (Web 3.0) Decentralized platform for distributed knowledge A web of databases
SNOMED Clinical Terms
 Representing clinical information using SNOMED Clinical Terms with different structural information models KR-MED 2008 - Phoenix David Markwell Laura Sato The Clinical Information Consultancy Ltd NHS Connecting
Representing clinical information using SNOMED Clinical Terms with different structural information models KR-MED 2008 - Phoenix David Markwell Laura Sato The Clinical Information Consultancy Ltd NHS Connecting
The Model-Driven Semantic Web Emerging Standards & Technologies
 The Model-Driven Semantic Web Emerging Standards & Technologies Elisa Kendall Sandpiper Software March 24, 2005 1 Model Driven Architecture (MDA ) Insulates business applications from technology evolution,
The Model-Driven Semantic Web Emerging Standards & Technologies Elisa Kendall Sandpiper Software March 24, 2005 1 Model Driven Architecture (MDA ) Insulates business applications from technology evolution,
Ontology Languages. Frank Wolter. Department of Computer Science. University of Liverpool
 Ontology Languages Frank Wolter Department of Computer Science University of Liverpool About The Module These slides and other material for this module are available at the module site http://cgi.csc.liv.ac.uk/~frank/teaching/comp08/comp321.html
Ontology Languages Frank Wolter Department of Computer Science University of Liverpool About The Module These slides and other material for this module are available at the module site http://cgi.csc.liv.ac.uk/~frank/teaching/comp08/comp321.html
XML in the bipharmaceutical
 XML in the bipharmaceutical sector XML holds out the opportunity to integrate data across both the enterprise and the network of biopharmaceutical alliances - with little technological dislocation and
XML in the bipharmaceutical sector XML holds out the opportunity to integrate data across both the enterprise and the network of biopharmaceutical alliances - with little technological dislocation and
A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES
 A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES 1 Yuxin Mao 1 School of Computer and Information Engineering, Zhejiang Gongshang University, Hangzhou 310018, P.R. China E-mail: 1 maoyuxin@zjgsu.edu.cn ABSTRACT
A WEB-BASED TOOLKIT FOR LARGE-SCALE ONTOLOGIES 1 Yuxin Mao 1 School of Computer and Information Engineering, Zhejiang Gongshang University, Hangzhou 310018, P.R. China E-mail: 1 maoyuxin@zjgsu.edu.cn ABSTRACT
Acquiring Experience with Ontology and Vocabularies
 Acquiring Experience with Ontology and Vocabularies Walt Melo Risa Mayan Jean Stanford The author's affiliation with The MITRE Corporation is provided for identification purposes only, and is not intended
Acquiring Experience with Ontology and Vocabularies Walt Melo Risa Mayan Jean Stanford The author's affiliation with The MITRE Corporation is provided for identification purposes only, and is not intended
SEMANTIC SUPPORT FOR MEDICAL IMAGE SEARCH AND RETRIEVAL
 SEMANTIC SUPPORT FOR MEDICAL IMAGE SEARCH AND RETRIEVAL Wang Wei, Payam M. Barnaghi School of Computer Science and Information Technology The University of Nottingham Malaysia Campus {Kcy3ww, payam.barnaghi}@nottingham.edu.my
SEMANTIC SUPPORT FOR MEDICAL IMAGE SEARCH AND RETRIEVAL Wang Wei, Payam M. Barnaghi School of Computer Science and Information Technology The University of Nottingham Malaysia Campus {Kcy3ww, payam.barnaghi}@nottingham.edu.my
National Centre for Text Mining NaCTeM. e-science and data mining workshop
 National Centre for Text Mining NaCTeM e-science and data mining workshop John Keane Co-Director, NaCTeM john.keane@manchester.ac.uk School of Informatics, University of Manchester What is text mining?
National Centre for Text Mining NaCTeM e-science and data mining workshop John Keane Co-Director, NaCTeM john.keane@manchester.ac.uk School of Informatics, University of Manchester What is text mining?
Linked Data: Fast, low cost semantic interoperability for health care?
 Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
Linked Data: Fast, low cost semantic interoperability for health care? About the presentation Part I: Motivation Why we need semantic operability in health care Why enhancing existing systems to increase
Collecting Public Health Data at a Global Scale. D. Cenk Erdil, PhD Marist College
 Collecting Public Health Data at a Global Scale D. Cenk Erdil, PhD Marist College DataCloud 2015 November 15, 2015 Outline Background A Public Health CPS Data Collection Marist MAgIC Summary Q&A 2 Background
Collecting Public Health Data at a Global Scale D. Cenk Erdil, PhD Marist College DataCloud 2015 November 15, 2015 Outline Background A Public Health CPS Data Collection Marist MAgIC Summary Q&A 2 Background
Learning from the Masters: Understanding Ontologies found on the Web
 Learning from the Masters: Understanding Ontologies found on the Web Bernardo Cuenca Grau 1, Ian Horrocks 1, Bijan Parsia 1, Peter Patel-Schneider 2, and Ulrike Sattler 1 1 School of Computer Science,
Learning from the Masters: Understanding Ontologies found on the Web Bernardo Cuenca Grau 1, Ian Horrocks 1, Bijan Parsia 1, Peter Patel-Schneider 2, and Ulrike Sattler 1 1 School of Computer Science,
BIO-ONTOLOGIES: A KNOWLEDGE REPRESENTATION RESOURCE IN BIOINFORMATICS
 BIO-ONTOLOGIES: A KNOWLEDGE REPRESENTATION RESOURCE IN BIOINFORMATICS Carmen Galvez University of Granada Granada, Spain cgalvez@ugr.es Abstract Bioinformatics manages the information that has been gathered
BIO-ONTOLOGIES: A KNOWLEDGE REPRESENTATION RESOURCE IN BIOINFORMATICS Carmen Galvez University of Granada Granada, Spain cgalvez@ugr.es Abstract Bioinformatics manages the information that has been gathered
MeSH: A Thesaurus for PubMed
 Resources and tools for bibliographic research MeSH: A Thesaurus for PubMed October 24, 2012 What is MeSH? Who uses MeSH? Why use MeSH? Searching by using the MeSH Database What is MeSH? Acronym for Medical
Resources and tools for bibliographic research MeSH: A Thesaurus for PubMed October 24, 2012 What is MeSH? Who uses MeSH? Why use MeSH? Searching by using the MeSH Database What is MeSH? Acronym for Medical
Introduction to The Cochrane Library
 Introduction to The Cochrane Library What is The Cochrane Library? A collection of databases that include high quality, independent, reliable evidence from Cochrane and other systematic reviews, clinical
Introduction to The Cochrane Library What is The Cochrane Library? A collection of databases that include high quality, independent, reliable evidence from Cochrane and other systematic reviews, clinical
Converting a thesaurus into an ontology: the use case of URBISOC
 Advanced Information Systems Laboratory Cost Action C2 Converting a thesaurus into an ontology: the use case of URBISOC J. Nogueras-Iso, J. Lacasta Alcalá de Henares, 4-5 May 2007 http://iaaa.cps.unizar.es
Advanced Information Systems Laboratory Cost Action C2 Converting a thesaurus into an ontology: the use case of URBISOC J. Nogueras-Iso, J. Lacasta Alcalá de Henares, 4-5 May 2007 http://iaaa.cps.unizar.es
SKOS. COMP60421 Sean Bechhofer
 SKOS COMP60421 Sean Bechhofer sean.bechhofer@manchester.ac.uk Ontologies Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
SKOS COMP60421 Sean Bechhofer sean.bechhofer@manchester.ac.uk Ontologies Metadata Resources marked-up with descriptions of their content. No good unless everyone speaks the same language; Terminologies
Powering Knowledge Discovery. Insights from big data with Linguamatics I2E
 Powering Knowledge Discovery Insights from big data with Linguamatics I2E Gain actionable insights from unstructured data The world now generates an overwhelming amount of data, most of it written in natural
Powering Knowledge Discovery Insights from big data with Linguamatics I2E Gain actionable insights from unstructured data The world now generates an overwhelming amount of data, most of it written in natural
Disease Information and Semantic Web
 Rheinische Friedrich-Wilhelms-Universität Bonn Institute of Computer Science III Disease Information and Semantic Web Master s Thesis Supervisor: Prof. Sören Auer, Heiner OberKampf Turan Gojayev München,
Rheinische Friedrich-Wilhelms-Universität Bonn Institute of Computer Science III Disease Information and Semantic Web Master s Thesis Supervisor: Prof. Sören Auer, Heiner OberKampf Turan Gojayev München,
Ontology Summit2007 Survey Response Analysis. Ken Baclawski Northeastern University
 Ontology Summit2007 Survey Response Analysis Ken Baclawski Northeastern University Outline Communities Ontology value, issues, problems, solutions Ontology languages Terms for ontology Ontologies April
Ontology Summit2007 Survey Response Analysis Ken Baclawski Northeastern University Outline Communities Ontology value, issues, problems, solutions Ontology languages Terms for ontology Ontologies April
A Knowledge Model Driven Solution for Web-Based Telemedicine Applications
 Medical Informatics in a United and Healthy Europe K.-P. Adlassnig et al. (Eds.) IOS Press, 2009 2009 European Federation for Medical Informatics. All rights reserved. doi:10.3233/978-1-60750-044-5-443
Medical Informatics in a United and Healthy Europe K.-P. Adlassnig et al. (Eds.) IOS Press, 2009 2009 European Federation for Medical Informatics. All rights reserved. doi:10.3233/978-1-60750-044-5-443
SKOS Standards and Best Practises for USING Knowledge Organisation Systems ON THE Semantic Web
 NKOS workshop ECDL Bath 2004-09-16 SKOS Standards and Best Practises for USING Knowledge Organisation Systems ON THE Semantic Web Rutherford Appleton Laboratory Overview Intro SKOS Core SKOS API SKOS Mapping
NKOS workshop ECDL Bath 2004-09-16 SKOS Standards and Best Practises for USING Knowledge Organisation Systems ON THE Semantic Web Rutherford Appleton Laboratory Overview Intro SKOS Core SKOS API SKOS Mapping
Renae Barger, Executive Director NN/LM Middle Atlantic Region
 Renae Barger, Executive Director NN/LM Middle Atlantic Region rbarger@pitt.edu http://nnlm.gov/mar/ DANJ Meeting, November 4, 2011 Advanced PubMed (20 min) General Information PubMed Citation Types Automatic
Renae Barger, Executive Director NN/LM Middle Atlantic Region rbarger@pitt.edu http://nnlm.gov/mar/ DANJ Meeting, November 4, 2011 Advanced PubMed (20 min) General Information PubMed Citation Types Automatic
Semantic interoperability, e-health and Australian health statistics
 Semantic interoperability, e-health and Australian health statistics Sally Goodenough Abstract E-health implementation in Australia will depend upon interoperable computer systems to share information
Semantic interoperability, e-health and Australian health statistics Sally Goodenough Abstract E-health implementation in Australia will depend upon interoperable computer systems to share information
Interoperability and Semantics in Use- Application of UML, XMI and MDA to Precision Medicine and Cancer Research
 Interoperability and Semantics in Use- Application of UML, XMI and MDA to Precision Medicine and Cancer Research Ian Fore, D.Phil. Associate Director, Biorepository and Pathology Informatics Senior Program
Interoperability and Semantics in Use- Application of UML, XMI and MDA to Precision Medicine and Cancer Research Ian Fore, D.Phil. Associate Director, Biorepository and Pathology Informatics Senior Program
Google indexed 3,3 billion of pages. Google s index contains 8,1 billion of websites
 Access IT Training 2003 Google indexed 3,3 billion of pages http://searchenginewatch.com/3071371 2005 Google s index contains 8,1 billion of websites http://blog.searchenginewatch.com/050517-075657 Estimated
Access IT Training 2003 Google indexed 3,3 billion of pages http://searchenginewatch.com/3071371 2005 Google s index contains 8,1 billion of websites http://blog.searchenginewatch.com/050517-075657 Estimated
Semantic Technology. Opportunities
 Semantic Technology Opportunities Avinash Punekar Scientific Publishing Services April 2011 2 Semantic Technology April 2011 3 What is Semantic Technology? ² Semantic Web ² Web 3.0 ² Linked Open Data /
Semantic Technology Opportunities Avinash Punekar Scientific Publishing Services April 2011 2 Semantic Technology April 2011 3 What is Semantic Technology? ² Semantic Web ² Web 3.0 ² Linked Open Data /
Mapping between Digital Identity Ontologies through SISM
 Mapping between Digital Identity Ontologies through SISM Matthew Rowe The OAK Group, Department of Computer Science, University of Sheffield, Regent Court, 211 Portobello Street, Sheffield S1 4DP, UK m.rowe@dcs.shef.ac.uk
Mapping between Digital Identity Ontologies through SISM Matthew Rowe The OAK Group, Department of Computer Science, University of Sheffield, Regent Court, 211 Portobello Street, Sheffield S1 4DP, UK m.rowe@dcs.shef.ac.uk
Taxonomies and controlled vocabularies best practices for metadata
 Original Article Taxonomies and controlled vocabularies best practices for metadata Heather Hedden is the taxonomy manager at First Wind Energy LLC. Previously, she was a taxonomy consultant with Earley
Original Article Taxonomies and controlled vocabularies best practices for metadata Heather Hedden is the taxonomy manager at First Wind Energy LLC. Previously, she was a taxonomy consultant with Earley
ICT for Health Care and Life Sciences
 School of Information Engineering Laurea Magistrale in Information Engineering Dipartimento di Elettronica e Informazione ICT for Health Care and Life Sciences 7 th November 2012 Davide Chicco davide.chicco@gmail.com
School of Information Engineering Laurea Magistrale in Information Engineering Dipartimento di Elettronica e Informazione ICT for Health Care and Life Sciences 7 th November 2012 Davide Chicco davide.chicco@gmail.com
Information retrieval (IR)
 Information Retrieval (Search) What is Biomedical & Health Informatics? William Hersh, MD Copyright 2018 Oregon Health & Science University Information retrieval (IR) Field concerned with organization
Information Retrieval (Search) What is Biomedical & Health Informatics? William Hersh, MD Copyright 2018 Oregon Health & Science University Information retrieval (IR) Field concerned with organization
Maximizing the Value of STM Content through Semantic Enrichment. Frank Stumpf December 1, 2009
 Maximizing the Value of STM Content through Semantic Enrichment Frank Stumpf December 1, 2009 What is Semantics and Semantic Processing? Content Knowledge Framework Technology Framework Search Text Images
Maximizing the Value of STM Content through Semantic Enrichment Frank Stumpf December 1, 2009 What is Semantics and Semantic Processing? Content Knowledge Framework Technology Framework Search Text Images
Adaptive Personal Information Environment based on the Semantic Web
 Adaptive Personal Information Environment based on the Semantic Web Thanyalak Maneewatthana, Gary Wills, Wendy Hall Intelligence, Agents, Multimedia Group School of Electronics and Computer Science University
Adaptive Personal Information Environment based on the Semantic Web Thanyalak Maneewatthana, Gary Wills, Wendy Hall Intelligence, Agents, Multimedia Group School of Electronics and Computer Science University
The Semantic Planetary Data System
 The Semantic Planetary Data System J. Steven Hughes 1, Daniel J. Crichton 1, Sean Kelly 1, and Chris Mattmann 1 1 Jet Propulsion Laboratory 4800 Oak Grove Drive Pasadena, CA 91109 USA {steve.hughes, dan.crichton,
The Semantic Planetary Data System J. Steven Hughes 1, Daniel J. Crichton 1, Sean Kelly 1, and Chris Mattmann 1 1 Jet Propulsion Laboratory 4800 Oak Grove Drive Pasadena, CA 91109 USA {steve.hughes, dan.crichton,
MeSH : A Thesaurus for PubMed
 Scuola di dottorato di ricerca in Scienze Molecolari Resources and tools for bibliographic research MeSH : A Thesaurus for PubMed What is MeSH? Who uses MeSH? Why use MeSH? Searching by using the MeSH
Scuola di dottorato di ricerca in Scienze Molecolari Resources and tools for bibliographic research MeSH : A Thesaurus for PubMed What is MeSH? Who uses MeSH? Why use MeSH? Searching by using the MeSH
Literature Databases
 Literature Databases Introduction to Bioinformatics Dortmund, 16.-20.07.2007 Lectures: Sven Rahmann Exercises: Udo Feldkamp, Michael Wurst 1 Overview 1. Databases 2. Publications in Science 3. PubMed and
Literature Databases Introduction to Bioinformatics Dortmund, 16.-20.07.2007 Lectures: Sven Rahmann Exercises: Udo Feldkamp, Michael Wurst 1 Overview 1. Databases 2. Publications in Science 3. PubMed and
A Grid Middleware for Ontology Access
 Available online at http://www.ges2007.de This document is under the terms of the CC-BY-NC-ND Creative Commons Attribution A Grid Middleware for Access Michael Hartung 1 and Erhard Rahm 2 1 Interdisciplinary
Available online at http://www.ges2007.de This document is under the terms of the CC-BY-NC-ND Creative Commons Attribution A Grid Middleware for Access Michael Hartung 1 and Erhard Rahm 2 1 Interdisciplinary
On practical aspects of enhancing semantic interoperability using SKOS and KOS alignment
 On practical aspects of enhancing semantic interoperability using SKOS and KOS alignment Antoine ISAAC KRR lab, Vrije Universiteit Amsterdam National Library of the Netherlands ISKO UK Meeting, July 21,
On practical aspects of enhancing semantic interoperability using SKOS and KOS alignment Antoine ISAAC KRR lab, Vrije Universiteit Amsterdam National Library of the Netherlands ISKO UK Meeting, July 21,
Measuring inter-annotator agreement in GO annotations
 Measuring inter-annotator agreement in GO annotations Camon EB, Barrell DG, Dimmer EC, Lee V, Magrane M, Maslen J, Binns ns D, Apweiler R. An evaluation of GO annotation retrieval for BioCreAtIvE and GOA.
Measuring inter-annotator agreement in GO annotations Camon EB, Barrell DG, Dimmer EC, Lee V, Magrane M, Maslen J, Binns ns D, Apweiler R. An evaluation of GO annotation retrieval for BioCreAtIvE and GOA.
Semantic Web. Ontology Pattern. Gerd Gröner, Matthias Thimm. Institute for Web Science and Technologies (WeST) University of Koblenz-Landau
 Semantic Web Ontology Pattern Gerd Gröner, Matthias Thimm {groener,thimm}@uni-koblenz.de Institute for Web Science and Technologies (WeST) University of Koblenz-Landau July 18, 2013 Gerd Gröner, Matthias
Semantic Web Ontology Pattern Gerd Gröner, Matthias Thimm {groener,thimm}@uni-koblenz.de Institute for Web Science and Technologies (WeST) University of Koblenz-Landau July 18, 2013 Gerd Gröner, Matthias
From Lexicon To Mammographic Ontology: Experiences and Lessons
 From Lexicon To Mammographic : Experiences and Lessons Bo Hu, Srinandan Dasmahapatra and Nigel Shadbolt Department of Electronics and Computer Science University of Southampton United Kingdom Email: {bh,
From Lexicon To Mammographic : Experiences and Lessons Bo Hu, Srinandan Dasmahapatra and Nigel Shadbolt Department of Electronics and Computer Science University of Southampton United Kingdom Email: {bh,
Searching Healthcare Databases NHS Athens
 CAMBRIDGE UNIVERSITY LIBRARY MEDICAL LIBRARY Searching the Evidence Searching Healthcare Databases NHS Athens https://hdas.nice.org.uk July 2018 Searching the Evidence Searching Healthcare Databases NHS
CAMBRIDGE UNIVERSITY LIBRARY MEDICAL LIBRARY Searching the Evidence Searching Healthcare Databases NHS Athens https://hdas.nice.org.uk July 2018 Searching the Evidence Searching Healthcare Databases NHS
CD 485 Computer Applications in Communication Disorders and Sciences MODULE 3
 CD 485 Computer Applications in Communication Disorders and Sciences MODULE 3 SECTION VII IDENTIFYING THE APPROPRIATE DATABASES JOURNAL ARTICLES THROUGH PUBMED, MEDLINE AND COMMUNICATION DISORDERS MULTISEARCH
CD 485 Computer Applications in Communication Disorders and Sciences MODULE 3 SECTION VII IDENTIFYING THE APPROPRIATE DATABASES JOURNAL ARTICLES THROUGH PUBMED, MEDLINE AND COMMUNICATION DISORDERS MULTISEARCH
Development of an Ontology-Based Portal for Digital Archive Services
 Development of an Ontology-Based Portal for Digital Archive Services Ching-Long Yeh Department of Computer Science and Engineering Tatung University 40 Chungshan N. Rd. 3rd Sec. Taipei, 104, Taiwan chingyeh@cse.ttu.edu.tw
Development of an Ontology-Based Portal for Digital Archive Services Ching-Long Yeh Department of Computer Science and Engineering Tatung University 40 Chungshan N. Rd. 3rd Sec. Taipei, 104, Taiwan chingyeh@cse.ttu.edu.tw
Collaborative Ontology Development on the (Semantic) Web
 Collaborative Ontology Development on the (Semantic) Web Natalya F. Noy and Tania Tudorache Stanford Center for Biomedical Informatics Research Stanford University Stanford, CA 94305 {noy,tudorache}@stanford.edu
Collaborative Ontology Development on the (Semantic) Web Natalya F. Noy and Tania Tudorache Stanford Center for Biomedical Informatics Research Stanford University Stanford, CA 94305 {noy,tudorache}@stanford.edu
Modeling Relationships
 Modeling Relationships Welcome to Lecture on Modeling Relationships in the course on Healthcare Databases. In this lecture we are going to cover two types of relationships, namely, the subtype and the
Modeling Relationships Welcome to Lecture on Modeling Relationships in the course on Healthcare Databases. In this lecture we are going to cover two types of relationships, namely, the subtype and the
ISO INTERNATIONAL STANDARD. Health informatics Genomic Sequence Variation Markup Language (GSVML)
 INTERNATIONAL STANDARD ISO 25720 First edition 2009-08-15 Health informatics Genomic Sequence Variation Markup Language (GSVML) Informatique de santé Langage de balisage de la variation de séquence génomique
INTERNATIONAL STANDARD ISO 25720 First edition 2009-08-15 Health informatics Genomic Sequence Variation Markup Language (GSVML) Informatique de santé Langage de balisage de la variation de séquence génomique
Publishing Vocabularies on the Web. Guus Schreiber Antoine Isaac Vrije Universiteit Amsterdam
 Publishing Vocabularies on the Web Guus Schreiber Antoine Isaac Vrije Universiteit Amsterdam Acknowledgements Alistair Miles, Dan Brickley, Mark van Assem, Jan Wielemaker, Bob Wielinga Participants of
Publishing Vocabularies on the Web Guus Schreiber Antoine Isaac Vrije Universiteit Amsterdam Acknowledgements Alistair Miles, Dan Brickley, Mark van Assem, Jan Wielemaker, Bob Wielinga Participants of
Knowledge Representations. How else can we represent knowledge in addition to formal logic?
 Knowledge Representations How else can we represent knowledge in addition to formal logic? 1 Common Knowledge Representations Formal Logic Production Rules Semantic Nets Schemata and Frames 2 Production
Knowledge Representations How else can we represent knowledge in addition to formal logic? 1 Common Knowledge Representations Formal Logic Production Rules Semantic Nets Schemata and Frames 2 Production
SELF-SERVICE SEMANTIC DATA FEDERATION
 SELF-SERVICE SEMANTIC DATA FEDERATION WE LL MAKE YOU A DATA SCIENTIST Contact: IPSNP Computing Inc. Chris Baker, CEO Chris.Baker@ipsnp.com (506) 721 8241 BIG VISION: SELF-SERVICE DATA FEDERATION Biomedical
SELF-SERVICE SEMANTIC DATA FEDERATION WE LL MAKE YOU A DATA SCIENTIST Contact: IPSNP Computing Inc. Chris Baker, CEO Chris.Baker@ipsnp.com (506) 721 8241 BIG VISION: SELF-SERVICE DATA FEDERATION Biomedical
Korea Institute of Oriental Medicine, South Korea 2 Biomedical Knowledge Engineering Laboratory,
 A Medical Treatment System based on Traditional Korean Medicine Ontology Sang-Kyun Kim 1, SeJin Nam 2, Dong-Hun Park 1, Yong-Taek Oh 1, Hyunchul Jang 1 1 Literature & Informatics Research Division, Korea
A Medical Treatment System based on Traditional Korean Medicine Ontology Sang-Kyun Kim 1, SeJin Nam 2, Dong-Hun Park 1, Yong-Taek Oh 1, Hyunchul Jang 1 1 Literature & Informatics Research Division, Korea
What is Text Mining? Sophia Ananiadou National Centre for Text Mining University of Manchester
 National Centre for Text Mining www.nactem.ac.uk University of Manchester Outline Aims of text mining Text Mining steps Text Mining uses Applications 2 Aims Extract and discover knowledge hidden in text
National Centre for Text Mining www.nactem.ac.uk University of Manchester Outline Aims of text mining Text Mining steps Text Mining uses Applications 2 Aims Extract and discover knowledge hidden in text
Using Ontologies for Data and Semantic Integration
 Using Ontologies for Data and Semantic Integration Monica Crubézy Stanford Medical Informatics, Stanford University ~~ November 4, 2003 Ontologies Conceptualize a domain of discourse, an area of expertise
Using Ontologies for Data and Semantic Integration Monica Crubézy Stanford Medical Informatics, Stanford University ~~ November 4, 2003 Ontologies Conceptualize a domain of discourse, an area of expertise
Humboldt-University of Berlin
 Humboldt-University of Berlin Exploiting Link Structure to Discover Meaningful Associations between Controlled Vocabulary Terms exposé of diploma thesis of Andrej Masula 13th October 2008 supervisor: Louiqa
Humboldt-University of Berlin Exploiting Link Structure to Discover Meaningful Associations between Controlled Vocabulary Terms exposé of diploma thesis of Andrej Masula 13th October 2008 supervisor: Louiqa
Optimization of the PubMed Automatic Term Mapping
 238 Medical Informatics in a United and Healthy Europe K.-P. Adlassnig et al. (Eds.) IOS Press, 2009 2009 European Federation for Medical Informatics. All rights reserved. doi:10.3233/978-1-60750-044-5-238
238 Medical Informatics in a United and Healthy Europe K.-P. Adlassnig et al. (Eds.) IOS Press, 2009 2009 European Federation for Medical Informatics. All rights reserved. doi:10.3233/978-1-60750-044-5-238
The Semantic Web & Ontologies
 The Semantic Web & Ontologies Kwenton Bellette The semantic web is an extension of the current web that will allow users to find, share and combine information more easily (Berners-Lee, 2001, p.34) This
The Semantic Web & Ontologies Kwenton Bellette The semantic web is an extension of the current web that will allow users to find, share and combine information more easily (Berners-Lee, 2001, p.34) This
Metadata Standards and Applications. 6. Vocabularies: Attributes and Values
 Metadata Standards and Applications 6. Vocabularies: Attributes and Values Goals of Session Understand how different vocabularies are used in metadata Learn about relationships in vocabularies Understand
Metadata Standards and Applications 6. Vocabularies: Attributes and Values Goals of Session Understand how different vocabularies are used in metadata Learn about relationships in vocabularies Understand
Introduction to Ovid. As a Clinical Librarían tool! Masoud Mohammadi Golestan University of Medical Sciences
 Introduction to Ovid As a Clinical Librarían tool! Masoud Mohammadi Golestan University of Medical Sciences Overview Ovid helps researchers, librarians, clinicians, and other healthcare professionals find
Introduction to Ovid As a Clinical Librarían tool! Masoud Mohammadi Golestan University of Medical Sciences Overview Ovid helps researchers, librarians, clinicians, and other healthcare professionals find
A method for recommending ontology alignment strategies
 A method for recommending ontology alignment strategies He Tan and Patrick Lambrix Department of Computer and Information Science Linköpings universitet, Sweden This is a pre-print version of the article
A method for recommending ontology alignment strategies He Tan and Patrick Lambrix Department of Computer and Information Science Linköpings universitet, Sweden This is a pre-print version of the article
Wondering about either OWL ontologies or SKOS vocabularies? You need both!
 Making sense of content Wondering about either OWL ontologies or SKOS vocabularies? You need both! ISKO UK SKOS Event London, 21st July 2008 bernard.vatant@mondeca.com A few words about Mondeca Founded
Making sense of content Wondering about either OWL ontologies or SKOS vocabularies? You need both! ISKO UK SKOS Event London, 21st July 2008 bernard.vatant@mondeca.com A few words about Mondeca Founded
MeSH : A Thesaurus for PubMed
 Resources and tools for bibliographic research MeSH : A Thesaurus for PubMed What is MeSH? Who uses MeSH? Why use MeSH? Searching by using the MeSH Database What is MeSH? http://www.ncbi.nlm.nih.gov/mesh
Resources and tools for bibliographic research MeSH : A Thesaurus for PubMed What is MeSH? Who uses MeSH? Why use MeSH? Searching by using the MeSH Database What is MeSH? http://www.ncbi.nlm.nih.gov/mesh
Using DAML format for representation and integration of complex gene networks: implications in novel drug discovery
 Using DAML format for representation and integration of complex gene networks: implications in novel drug discovery K. Baclawski Northeastern University E. Neumann Beyond Genomics T. Niu Harvard School
Using DAML format for representation and integration of complex gene networks: implications in novel drug discovery K. Baclawski Northeastern University E. Neumann Beyond Genomics T. Niu Harvard School
Semantic Technologies for Nuclear Knowledge Modelling and Applications
 Semantic Technologies for Nuclear Knowledge Modelling and Applications D. Beraha 3 rd International Conference on Nuclear Knowledge Management 7.-11.11.2016, Vienna, Austria Why Semantics? Machines understanding
Semantic Technologies for Nuclear Knowledge Modelling and Applications D. Beraha 3 rd International Conference on Nuclear Knowledge Management 7.-11.11.2016, Vienna, Austria Why Semantics? Machines understanding
Searching the Evidence in PubMed
 CAMBRIDGE UNIVERSITY LIBRARY MEDICAL LIBRARY Supporting Literature Searching Searching the Evidence in PubMed July 2017 Supporting Literature Searching Searching the Evidence in PubMed How to access PubMed
CAMBRIDGE UNIVERSITY LIBRARY MEDICAL LIBRARY Supporting Literature Searching Searching the Evidence in PubMed July 2017 Supporting Literature Searching Searching the Evidence in PubMed How to access PubMed
ARKive-ERA Project Lessons and Thoughts
 ARKive-ERA Project Lessons and Thoughts Semantic Web for Scientific and Cultural Organisations Convitto della Calza 17 th June 2003 Paul Shabajee (ILRT, University of Bristol) 1 Contents Context Digitisation
ARKive-ERA Project Lessons and Thoughts Semantic Web for Scientific and Cultural Organisations Convitto della Calza 17 th June 2003 Paul Shabajee (ILRT, University of Bristol) 1 Contents Context Digitisation
Text mining tools for semantically enriching the scientific literature
 Text mining tools for semantically enriching the scientific literature Sophia Ananiadou Director National Centre for Text Mining School of Computer Science University of Manchester Need for enriching the
Text mining tools for semantically enriching the scientific literature Sophia Ananiadou Director National Centre for Text Mining School of Computer Science University of Manchester Need for enriching the
Enhancing information services using machine to machine terminology services
 Enhancing information services using machine to machine terminology services Gordon Dunsire Presented to the IFLA 2009 Satellite Conference Looking at the past and preparing for the future 20-21 Aug 2009,
Enhancing information services using machine to machine terminology services Gordon Dunsire Presented to the IFLA 2009 Satellite Conference Looking at the past and preparing for the future 20-21 Aug 2009,
SKOS and the Ontogenesis of Vocabularies
 SKOS and the Ontogenesis of Vocabularies Joseph T. Tennis The University of British Columbia Tel: +01 604 822 24321 Fax:+01 604 822 6006 jtennis@interchange.ubc.ca Abstract: The paper suggests extensions
SKOS and the Ontogenesis of Vocabularies Joseph T. Tennis The University of British Columbia Tel: +01 604 822 24321 Fax:+01 604 822 6006 jtennis@interchange.ubc.ca Abstract: The paper suggests extensions
Ontology driven voice-based interaction in mobile environment
 Ontology driven voice-based interaction in mobile environment Jiri Kopsa 1, Zdenek Mikovec 1, Pavel Slavik 1 1 Czech Technical University in Prague Karlovo namesti 13, Prague 2, Czech Republic j.kopsa@fee.ctup.cz,
Ontology driven voice-based interaction in mobile environment Jiri Kopsa 1, Zdenek Mikovec 1, Pavel Slavik 1 1 Czech Technical University in Prague Karlovo namesti 13, Prague 2, Czech Republic j.kopsa@fee.ctup.cz,
A Semantic Multi-Field Clinical Search for Patient Medical Records
 BULGARIAN ACADEMY OF SCIENCES CYBERNETICS AND INFORMATION TECHNOLOGIES Volume 18, No 1 Sofia 2018 Print ISSN: 1311-9702; Online ISSN: 1314-4081 DOI: 10.2478/cait-2018-0014 A Semantic Multi-Field Clinical
BULGARIAN ACADEMY OF SCIENCES CYBERNETICS AND INFORMATION TECHNOLOGIES Volume 18, No 1 Sofia 2018 Print ISSN: 1311-9702; Online ISSN: 1314-4081 DOI: 10.2478/cait-2018-0014 A Semantic Multi-Field Clinical
DBpedia-An Advancement Towards Content Extraction From Wikipedia
 DBpedia-An Advancement Towards Content Extraction From Wikipedia Neha Jain Government Degree College R.S Pura, Jammu, J&K Abstract: DBpedia is the research product of the efforts made towards extracting
DBpedia-An Advancement Towards Content Extraction From Wikipedia Neha Jain Government Degree College R.S Pura, Jammu, J&K Abstract: DBpedia is the research product of the efforts made towards extracting
BUILDING A CONCEPTUAL MODEL OF THE WORLD WIDE WEB FOR VISUALLY IMPAIRED USERS
 1 of 7 17/01/2007 10:39 BUILDING A CONCEPTUAL MODEL OF THE WORLD WIDE WEB FOR VISUALLY IMPAIRED USERS Mary Zajicek and Chris Powell School of Computing and Mathematical Sciences Oxford Brookes University,
1 of 7 17/01/2007 10:39 BUILDING A CONCEPTUAL MODEL OF THE WORLD WIDE WEB FOR VISUALLY IMPAIRED USERS Mary Zajicek and Chris Powell School of Computing and Mathematical Sciences Oxford Brookes University,
HealthCyberMap: Mapping the Health Cyberspace Using Hypermedia GIS and Clinical Codes
 HealthCyberMap: Mapping the Health Cyberspace Using Hypermedia GIS and Clinical Codes PhD Research Project Maged Nabih Kamel Boulos MBBCh, MSc (Derm & Vener), MSc (Medical Informatics) 1 Summary The application
HealthCyberMap: Mapping the Health Cyberspace Using Hypermedia GIS and Clinical Codes PhD Research Project Maged Nabih Kamel Boulos MBBCh, MSc (Derm & Vener), MSc (Medical Informatics) 1 Summary The application
An Introduction to PubMed Searching: A Reference Guide
 An Introduction to PubMed Searching: A Reference Guide Created by the Ontario Public Health Libraries Association (OPHLA) ACCESSING PubMed PubMed, the National Library of Medicine s free version of MEDLINE,
An Introduction to PubMed Searching: A Reference Guide Created by the Ontario Public Health Libraries Association (OPHLA) ACCESSING PubMed PubMed, the National Library of Medicine s free version of MEDLINE,
BioNav: An Ontology-Based Framework to Discover Semantic Links in the Cloud of Linked Data
 BioNav: An Ontology-Based Framework to Discover Semantic Links in the Cloud of Linked Data María-Esther Vidal 1, Louiqa Raschid 2, Natalia Márquez 1, Jean Carlo Rivera 1, and Edna Ruckhaus 1 1 Universidad
BioNav: An Ontology-Based Framework to Discover Semantic Links in the Cloud of Linked Data María-Esther Vidal 1, Louiqa Raschid 2, Natalia Márquez 1, Jean Carlo Rivera 1, and Edna Ruckhaus 1 1 Universidad
Retrieval of Highly Related Documents Containing Gene-Disease Association
 Retrieval of Highly Related Documents Containing Gene-Disease Association K. Santhosh kumar 1, P. Sudhakar 2 Department of Computer Science & Engineering Annamalai University Annamalai Nagar, India. santhosh09539@gmail.com,
Retrieval of Highly Related Documents Containing Gene-Disease Association K. Santhosh kumar 1, P. Sudhakar 2 Department of Computer Science & Engineering Annamalai University Annamalai Nagar, India. santhosh09539@gmail.com,
Opus: University of Bath Online Publication Store
 Patel, M. (2004) Semantic Interoperability in Digital Library Systems. In: WP5 Forum Workshop: Semantic Interoperability in Digital Library Systems, DELOS Network of Excellence in Digital Libraries, 2004-09-16-2004-09-16,
Patel, M. (2004) Semantic Interoperability in Digital Library Systems. In: WP5 Forum Workshop: Semantic Interoperability in Digital Library Systems, DELOS Network of Excellence in Digital Libraries, 2004-09-16-2004-09-16,
Searching Pubmed Database استخدام قاعدة المعلومات Pubmed
 Searching Pubmed Database استخدام قاعدة المعلومات Pubmed برنامج مهارات البحث العلمي مركز البحىث بأقسام العلىم والدراسات الطبية للطالبات األحد 1433/11/14 ه الموافق 30 2012 /9/ م د. سيناء عبد المحسن العقيل
Searching Pubmed Database استخدام قاعدة المعلومات Pubmed برنامج مهارات البحث العلمي مركز البحىث بأقسام العلىم والدراسات الطبية للطالبات األحد 1433/11/14 ه الموافق 30 2012 /9/ م د. سيناء عبد المحسن العقيل
Utilizing Semantic Web Technologies in Healthcare
 Utilizing Semantic Web Technologies in Healthcare Vassileios D. Kolias, John Stoitsis, Spyretta Golemati and Konstantina S. Nikita Abstract The technological breakthrough in biomedical engineering and
Utilizing Semantic Web Technologies in Healthcare Vassileios D. Kolias, John Stoitsis, Spyretta Golemati and Konstantina S. Nikita Abstract The technological breakthrough in biomedical engineering and
Adaptive Medical Information Delivery Combining User, Task and Situation Models
 Adaptive Medical Information Delivery Combining User, Task and Situation s Luis Francisco-Revilla and Frank M. Shipman III Department of Computer Science Texas A&M University College Station, TX 77843-3112,
Adaptive Medical Information Delivery Combining User, Task and Situation s Luis Francisco-Revilla and Frank M. Shipman III Department of Computer Science Texas A&M University College Station, TX 77843-3112,
NCI Thesaurus, managing towards an ontology
 NCI Thesaurus, managing towards an ontology CENDI/NKOS Workshop October 22, 2009 Gilberto Fragoso Outline Background on EVS The NCI Thesaurus BiomedGT Editing Plug-in for Protege Semantic Media Wiki supports
NCI Thesaurus, managing towards an ontology CENDI/NKOS Workshop October 22, 2009 Gilberto Fragoso Outline Background on EVS The NCI Thesaurus BiomedGT Editing Plug-in for Protege Semantic Media Wiki supports
