CSSS 510: Lab 2. Introduction to Maximum Likelihood Estimation
|
|
|
- Lily Baker
- 5 years ago
- Views:
Transcription
1 CSSS 510: Lab 2 Introduction to Maximum Likelihood Estimation
2 0. Agenda 1. Housekeeping: simcf, tile 2. Questions about Homework 1 or lecture 3. Simulating heteroskedastic normal data 4. Fitting a least squares regression model using the simulated data 5. Calculating predicted values 6. Fitting the heteroskedastic normal model using ML 7. Simulating predicted values and confidence intervals
3 1. Housekeeping 1. Please make sure that you have R or R Studio installed on your computers with Chris s simcf and tile packages installed You can find these packages on the course website and follow the instructions to install (do not unzip the.tgz files)
4 2. Questions
5 3. Simulating heteroskedastic normal data y w2
6 3. Simulating heteroskedastic normal data Stochastic component: y i N(µ i, σ 2 i ) Systematic components: µ i = x i β y σ 2 i = exp(z i γ) w2
7 3. Simulating heteroskedastic normal data 1. Set the number of observations to 1500 (n) 2. Set a parameter vector for the mean (assume 2 covariates plus the constant) (β) 3. Set a parameter vector for the variance (assume heteroskedasticity) (γ) 4. Generate the constant and the covariates, length 1500 for each (draw from a uniform distribution) (x i, z i ) 5. Create the systematic component for the mean (x i β) 6. Create the systematic component for the variance (the same covariates affect mu and sigma) exp(z i γ) 7. Generate the response variable (y i ) 8. Save the data to a data frame 9. Plot the data
8 3. Simulating heteroskedastic normal data rm(list=ls()) # Clear memory set.seed(123456) # To reproduce random numbers library(mass) # Load packages library(simcf) n < # Generate 1500 observations beta <- c(0, 5, 15) # Set a parameter vector for the mean # One for constant, one for covariate 1, one for covariate 2. gamma <- c(1, 0, 3) # Set a parameter vector for the variance # Gamma estimate for covariate 2 is set to be 3, creating heteroskedasticity w0 <- rep(1, n) # Create the constant and covariates w1 <- runif(n) # Length of each vector is 1500 w2 <- runif(n)
9 3. Simulating heteroskedastic normal data x <- cbind(w0, w1, w2) # Create a matrix of the covariates mu <- x%*%beta # Create the systemtic component for the mean z <- x # i.e., same covariates affect mu and sigma sigma2 <- exp(x%*%gamma) # Create the systematic component for the variance # z is 1500 by 3 matrix, gamma is 3 by 1 matrix #ith row of sigma 2 thus equals exp(1+0+w2_i*3). i.e., it is a function of w2 y <- mu + rnorm(n)*sqrt(sigma2) # Create the response variable #y <-rnorm(obs, mean=mu, sd=sqrt(sigma2)) data <- cbind(y,w1,w2) # Save the data to a data frame data <- as.data.frame(data) names(data) <- c("y","w1","w2")
10 3. Plot the data par(mfrow=c(1,3)) #Plot the data plot(y=y,x=w1) plot(y=y,x=w2) plot(y=w1,x=w2) w y y w w w2
11 4. Fitting a model using the simulated data 1. Assume we don t know the true value of the parameters and fit a model using least squares (use the lm() function and regress the response variable on the two covariates) 2. Calculate and print the AIC
12 4. Fitting a model using the simulated data ls.result <- lm(y ~ w1 + w2) #Fit a linear model using the simulated data print(summary(ls.result)) ## ## Call: ## lm(formula = y ~ w1 + w2) ## ## Residuals: ## Min 1Q Median 3Q Max ## ## ## Coefficients: ## Estimate Std. Error t value Pr(> t ) ## (Intercept) ## w <2e-16 *** ## w <2e-16 *** ## --- ## Signif. codes: 0 '***' '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 4.17 on 1497 degrees of freedom ## Multiple R-squared: , Adjusted R-squared: ## F-statistic: on 2 and 1497 DF, p-value: < 2.2e-16
13 4. Fitting a model using the simulated data ls.aic <- AIC(ls.result) # Calculate and print the AIC print(ls.aic) ## [1]
14 Lecture Recap: Key Terms Take a minute and try to define 1. Systematic and stochastic component 2. Expected value and predicted value (and fitted value) 3. Confidence interval and prediction interval 4. Bayes Theorem 5. Bayesian inference and likelihood inference 6. Maximum likelihood estimation
15 Questions
16 5. Calculating predicted values Scenario 1: Vary covariate 1 1. Create a data frame with a set of hypothetical scenarios for covariate 1 while keeping covariate 2 at its mean 2. Calculate the predicted values using the predict() function 3. Plot the predicted values
17 5. Calculating predicted values - Scenario 1 # Calculate predicted values using predict() # Start by calculating P(Y w1) for different w1 values w1range <- seq(0:20)/20 # Set as necessary # Set up a dataframe with the hypothetical scenarios # (varied w1, all else equal) baseline <- c(mean(w1), mean(w2)) # Set as necessary xhypo <- matrix(baseline, nrow=length(w1range), ncol=2, byrow= TRUE) # Set ncol to # of x's # same as: xhypo <- # matrix(rep(baseline,21), nrow=length(w1range), ncol=2, byrow= TRUE) xhypo <- as.data.frame(xhypo) names(xhypo) <- c("w1","w2") xhypo[,1] <- w1range # Scenarios: Changing values in the first column # Keeping second column values at the mean of w2. head(xhypo) ## w1 w2 ## ## ## ## ## ##
18 5. Calculating predicted values - Scenario 1 # Calculate Predicted Y using predict() simls.w1<-predict(ls.result,newdata = xhypo,interval="prediction",level=0.95) head(simls.w1) ## fit lwr upr ## ## ## ## ## ## # Plot them yplot <- simls.w1 xplot <- cbind(w1range,w1range,w1range) # need to have the same dimension [21,3] #pdf("homoyvsw1.pdf")
19 5. Calculating predicted values - Scenario 1 matplot(y=yplot, x=xplot, type="l", lty=c("solid","dashed","dashed"), col=c("black"), xlab = "w1", ylab = "Predicted Y") Predicted Y #dev.off() w1
20 5. Calculating predicted values - Scenario 1 # Calculate Predicted Y using predict() # Compare prediction intervals with confidence intervals simls.w1<-predict(ls.result,newdata = xhypo,interval="confidence",level=0.95) head(simls.w1) ## fit lwr upr ## ## ## ## ## ## # Plot them yplot <- simls.w1 xplot <- cbind(w1range,w1range,w1range) # need to have the same dimension [21,3] #pdf("homoyvsw1.pdf")
21 5. Calculating predicted values - Scenario 1 matplot(y=yplot, x=xplot, type="l", lty=c("solid","dashed","dashed"), col=c("black"), xlab = "w1", ylab = "Predicted Y") Predicted Y #dev.off() w1
22 5. Calculating predicted values Scenario 2: Vary covariate 2 1. Create a data frame with a set of hypothetical scenarios for covariate 2 while keeping covariate 1 at its mean 2. Calculate the predicted values using the predict() function 3. Plot the predicted values
23 5. Calculating predicted values - Scenario 2 # Calculate predicted values using predict() # Start by calculating P(Y w1) for different w1 values w2range <- seq(0:20)/20 # Set as necessary # Set up a dataframe with the hypothetical scenarios # (varied w1, all else equal) baseline <- c(mean(w1), mean(w2)) # Set as necessary xhypo <- matrix(baseline, nrow=length(w2range), ncol=2, byrow= TRUE) xhypo <- as.data.frame(xhypo) # Set ncol to # of x's names(xhypo) <- c("w1","w2") # Set by user xhypo[,2] <- w1range # Change as necessary # Calculate Predicted Y using predict() simls.w2 <- predict(ls.result,newdata=xhypo,interval="prediction",level=0.95) # Plot them yplot <- simls.w2 xplot <- cbind(w2range,w2range,w2range) #pdf("homoyvsw2.pdf")
24 5. Calculating predicted values - Scenario 2 matplot(y=yplot, x=xplot, type="l", lty=c("solid","dashed","dashed"), col=c("black"), xlab = "w1", ylab = "Predicted Y") Predicted Y #dev.off() w1
25 6. Fitting the heteroskedastic normal model using ML Create the input matrices (the two covariates) Write a likelihood function for the heteroskedastic normal model Find the MLEs using the optim() function Extract the point estimates Compute the standard errors Compare with the least squares estimates Find the log likelihood at its maximum Compute the AIC Simulate the results by drawing from the model s predictive distribution Separate the simulated betas from the simulated gammas
26 Agenda Clarification of lecture or Homework 2 Finish example using optim from last week Review numerical optimization (gradient descent/ascent) Review MLE standard errors Discuss logistic regression Take questions about Homework 2
27 Clarification of lecture or Homework 2
28 6. Fitting the heteroskedastic normal model using ML 2. Write a likelihood function for the heteroskedastic normal model Recall from lecture: L(µ, σ 2 y) P(y µ, σ 2 ) L(µ, σ 2 y) = k(y)p(y µ, σ 2 ) n ( L(µ, σ 2 y) = k(y) (2πσ 2 ) 1/2 (yi µ i ) 2 ) exp 2σ i= L(β, σ 2 y) = 1 n logσ 2 1 n (y i x i β) σ i=1 i=1 2 L(β, γ y) = 1 n z i γ 1 n (y i x i β) exp(z i=1 i=1 i γ)
29 6. Fitting the heteroskedastic normal model using ML # A likelihood function for ML heteroskedastic Normal llk.hetnormlin <- function(param,y,x,z) { x <- as.matrix(x) #x as a matrix z <- as.matrix(z) #z as a matrix os <- rep(1,nrow(x)) #1 for the intercept x <- cbind(os,x) #combine z <- cbind(os,z) b <- param[ 1 : ncol(x) ] # i.e., the first three spaces in the param vector g <- param[ (ncol(x)+1) : (ncol(x) + ncol(z)) ] # i.e., the three remaining spaces xb <- x%*%b # systematic components for the mean s2 <- exp(z%*%g) # systematic components for the variance sum(0.5*(log(s2)+(y-xb)^2/s2)) # "optim" command minimizes a function by default. # Minimalization of -lnl is the same as maximization of lnl # so we will put -lnl(param y) here #-sum(0.5*(log(s2)+(y-xb)^2/s2)) # Alternativly, you can use lnl(param y) and set optim to be a maximizer }
30 6. Fitting the heteroskedastic normal model using ML # Create input matrices xcovariates <- cbind(w1,w2) zcovariates <- cbind(w1,w2) # initial guesses of beta0, beta1,..., gamma0, gamma1,... # we need one entry per parameter, in order! stval <- c(0,0,0,0,0,0) # also include beta and gamma estiamtes for constants help(optim) # Run ML, get the output we need hetnorm.result <- optim(stval,llk.hetnormlin,method="bfgs", hessian=t,y=y,x=xcovariates,z=zcovariates) # by default, calls minimizer procedure. # you can make optim a maximizer by adding control=list(fnscale=-1)
31 6. Fitting the heteroskedastic normal model using ML pe <- hetnorm.result$par pe # point estimates ## [1] vc <- solve(hetnorm.result$hessian) # 6x6 var-cov matrix (allows to compute standard errors) round(vc,5) ## [,1] [,2] [,3] [,4] [,5] [,6] ## [1,] ## [2,] ## [3,] ## [4,] ## [5,] ## [6,] se <- sqrt(diag(vc)) # standard errors # the ML standard errors are the square roots of the diagonal of the Hessian # or inverse of the matrix of second derivaties se ## [1]
32 6. Fitting the heteroskedastic normal model using ML mle.result<-round(cbind(pe[1:3], se[1:3]),2) # see pe and se colnames(mle.result)<-c("estimate", "Std.Error") rownames(mle.result)<-c("(intercept)", "w1","w2" ) mle.result ## Estimate Std.Error ## (Intercept) ## w ## w round(summary(ls.result)$coefficients[,c(1,2)],2) #compare with the ls result ## Estimate Std. Error ## (Intercept) ## w ## w ll <- -hetnorm.result$value # likelihood at maximum, no need to # have a negative sign if you set optime to be a maximizer. ll ## [1]
33 6. Fitting the heteroskedastic normal model using ML # The AIC is the deviance or -2*ll at its max plus 2*number of # parameters or the dimension hetnorm.aic <- 2*length(stval) - 2*ll # first component to penalizing the number of parameters # (i.e., the loss of degree of freedom). Lower aic is better print(hetnorm.aic) ## [1] # remember AIC from LS fit? print(ls.aic) ## [1]
34 6. Numerical optimization Q: What is numerical optimization and why do we use it? optim uses gradient descent as a method of numerical optimization constructs a quadratic approximation of the likelihood function (using Taylor Series) starts with an initial set of parameter values uses the first derivative to check if the function is minimized increases (or decreases) the initial values based on the first derivatives decides on the magnitude of the increase (or decrease) based on the second derivatives continues until the first derivatives reaches some tolerance level near zero
35 6. MLE Standard Errors Recall that the MLE standard errors are computed using the matrix of second derivatives where the likelihood is at its max Intuitively, when these are large, the likelihood function is steeper, and when these are small, it is flatter MLEs are more precise in the former and less precise in the latter The variance-covariance matrix is the inverse of the matrix of second derivatives (Hessian matrix) The MLE standard errors are therefore computed as the square root of the diagonal of the inverse of the Hessian or matrix of second derivatives
36 7. Simulating predicted values and confidence intervals Scenario 1: Vary covariate 1 1. Create a data frame with a set of hypothetical scenarios for covariate 1 while keeping covariate 2 at its mean 2. Simulate the predicted values and the confidence intervals using simcf 3. Plot the results Recall from lecture:
37 7. Simulating predicted values and confidence intervals - Scenario 1 # Simulate results by drawing from the model predictive distribution sims < simparam <- mvrnorm(sims,pe,vc) # draw parameters store them in 10000x6 matrix. # We assume that parameter estimates are distributed # according to a multivariate normal distribution with population mean pe # and population variance-covariance matrix vc. # Separate into the simulated betas and simulated gammas simbetas <- simparam[,1:(ncol(xcovariates)+1)] # first three columns store simulated beta coefficients simgammas <- simparam[,(ncol(simbetas)+1):ncol(simparam)] # then simulated gamma coefficients # Put our models in "formula" form model <- (y ~ w1 + w2) varmodel <- (y ~ w1 + w2) # Scenario 1: Vary w1 # Start by calculating P(Y w1) for different w1 values w1range <- seq(0:20)/20
38 7. Simulating predicted values and confidence intervals - Scenario 1 # Set up a matrix with the hypothetical scenarios (varied w1, all else equal) xhypo <- cfmake(model, data, nscen = length(w1range)) # creating a set of scenarios for (i in 1:length(w1range)) { xhypo <- cfchange(xhypo, "w1", x=w1range[i], scen=i) # change the values of the variables of your interest, set others at the mean } zhypo <- cfmake(varmodel, data, nscen = length(w1range)) for (i in 1:length(w1range)) { zhypo <- cfchange(zhypo, "w1", x=w1range[i], scen=i) } # Simulate the predicted Y's and CI's simres.w1 <- hetnormsimpv(xhypo,simbetas, zhypo,simgammas, ci=0.95, constant=1,varconstant=1) #simy<-rnorm(sims)*sqrt(simsigma2)+ simmu # Plot them yplot <- cbind(simres.w1$pe, simres.w1$lower, simres.w1$upper) xplot <- cbind(w1range,w1range,w1range)
39 7. Simulating predicted values and confidence intervals - Scenario 1 #pdf("heteroyvsw1.pdf") matplot(y=yplot, x=xplot, type="l", lty=c("solid","dashed","dashed"), col=c("black"), xlab = "w1", ylab = "Predicted Y") Predicted Y #dev.off() w1
40 7. Simulating predicted values and confidence intervals - Scenario 2 1. Create a data frame with a set of hypothetical scenarios for covariate 2 while keeping covariate 1 at its mean 2. Simulate the predicted values and the confidence intervals using simcf 3. Plot the results
41 7. Simulating predicted values and confidence intervals - Scenario 2 # Start by calculating P(Y w2) for different w1 values w2range <- seq(0:20)/20 # Set up a matrix with the hypothetical scenarios (varied w1, all else equal) xhypo <- cfmake(model, data, nscen = length(w2range)) for (i in 1:length(w2range)) { xhypo <- cfchange(xhypo, "w2", x=w2range[i], scen=i) } zhypo <- cfmake(varmodel, data, nscen = length(w2range)) for (i in 1:length(w2range)) { zhypo <- cfchange(zhypo, "w2", x=w2range[i], scen=i) } # Simulate the predicted Y's and CI's simres.w2 <- hetnormsimpv(xhypo,simbetas, zhypo,simgammas, ci=0.95, constant=1,varconstant=1) # Plot them yplot <- cbind(simres.w2$pe, simres.w2$lower, simres.w2$upper) xplot <- cbind(w1range,w1range,w1range) #pdf("heteroyvsw2.pdf")
42 7. Simulating predicted values and confidence intervals - Scenario 2 matplot(y=yplot, x=xplot, type="l", lty=c("solid","dashed","dashed"), col=c("black"), xlab = "w2", ylab = "Predicted Y") Predicted Y #dev.off() w2
Exercise 2.23 Villanova MAT 8406 September 7, 2015
 Exercise 2.23 Villanova MAT 8406 September 7, 2015 Step 1: Understand the Question Consider the simple linear regression model y = 50 + 10x + ε where ε is NID(0, 16). Suppose that n = 20 pairs of observations
Exercise 2.23 Villanova MAT 8406 September 7, 2015 Step 1: Understand the Question Consider the simple linear regression model y = 50 + 10x + ε where ε is NID(0, 16). Suppose that n = 20 pairs of observations
Section 2.3: Simple Linear Regression: Predictions and Inference
 Section 2.3: Simple Linear Regression: Predictions and Inference Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.4 1 Simple
Section 2.3: Simple Linear Regression: Predictions and Inference Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.4 1 Simple
Estimating R 0 : Solutions
 Estimating R 0 : Solutions John M. Drake and Pejman Rohani Exercise 1. Show how this result could have been obtained graphically without the rearranged equation. Here we use the influenza data discussed
Estimating R 0 : Solutions John M. Drake and Pejman Rohani Exercise 1. Show how this result could have been obtained graphically without the rearranged equation. Here we use the influenza data discussed
9.1 Random coefficients models Constructed data Consumer preference mapping of carrots... 10
 St@tmaster 02429/MIXED LINEAR MODELS PREPARED BY THE STATISTICS GROUPS AT IMM, DTU AND KU-LIFE Module 9: R 9.1 Random coefficients models...................... 1 9.1.1 Constructed data........................
St@tmaster 02429/MIXED LINEAR MODELS PREPARED BY THE STATISTICS GROUPS AT IMM, DTU AND KU-LIFE Module 9: R 9.1 Random coefficients models...................... 1 9.1.1 Constructed data........................
Poisson Regression and Model Checking
 Poisson Regression and Model Checking Readings GH Chapter 6-8 September 27, 2017 HIV & Risk Behaviour Study The variables couples and women_alone code the intervention: control - no counselling (both 0)
Poisson Regression and Model Checking Readings GH Chapter 6-8 September 27, 2017 HIV & Risk Behaviour Study The variables couples and women_alone code the intervention: control - no counselling (both 0)
Regression on the trees data with R
 > trees Girth Height Volume 1 8.3 70 10.3 2 8.6 65 10.3 3 8.8 63 10.2 4 10.5 72 16.4 5 10.7 81 18.8 6 10.8 83 19.7 7 11.0 66 15.6 8 11.0 75 18.2 9 11.1 80 22.6 10 11.2 75 19.9 11 11.3 79 24.2 12 11.4 76
> trees Girth Height Volume 1 8.3 70 10.3 2 8.6 65 10.3 3 8.8 63 10.2 4 10.5 72 16.4 5 10.7 81 18.8 6 10.8 83 19.7 7 11.0 66 15.6 8 11.0 75 18.2 9 11.1 80 22.6 10 11.2 75 19.9 11 11.3 79 24.2 12 11.4 76
1 Lab 1. Graphics and Checking Residuals
 R is an object oriented language. We will use R for statistical analysis in FIN 504/ORF 504. To download R, go to CRAN (the Comprehensive R Archive Network) at http://cran.r-project.org Versions for Windows
R is an object oriented language. We will use R for statistical analysis in FIN 504/ORF 504. To download R, go to CRAN (the Comprehensive R Archive Network) at http://cran.r-project.org Versions for Windows
Applied Statistics and Econometrics Lecture 6
 Applied Statistics and Econometrics Lecture 6 Giuseppe Ragusa Luiss University gragusa@luiss.it http://gragusa.org/ March 6, 2017 Luiss University Empirical application. Data Italian Labour Force Survey,
Applied Statistics and Econometrics Lecture 6 Giuseppe Ragusa Luiss University gragusa@luiss.it http://gragusa.org/ March 6, 2017 Luiss University Empirical application. Data Italian Labour Force Survey,
Risk Management Using R, SoSe 2013
 1. Problem (vectors and factors) a) Create a vector containing the numbers 1 to 10. In this vector, replace all numbers greater than 4 with 5. b) Create a sequence of length 5 starting at 0 with an increment
1. Problem (vectors and factors) a) Create a vector containing the numbers 1 to 10. In this vector, replace all numbers greater than 4 with 5. b) Create a sequence of length 5 starting at 0 with an increment
Statistics Lab #7 ANOVA Part 2 & ANCOVA
 Statistics Lab #7 ANOVA Part 2 & ANCOVA PSYCH 710 7 Initialize R Initialize R by entering the following commands at the prompt. You must type the commands exactly as shown. options(contrasts=c("contr.sum","contr.poly")
Statistics Lab #7 ANOVA Part 2 & ANCOVA PSYCH 710 7 Initialize R Initialize R by entering the following commands at the prompt. You must type the commands exactly as shown. options(contrasts=c("contr.sum","contr.poly")
Standard Errors in OLS Luke Sonnet
 Standard Errors in OLS Luke Sonnet Contents Variance-Covariance of ˆβ 1 Standard Estimation (Spherical Errors) 2 Robust Estimation (Heteroskedasticity Constistent Errors) 4 Cluster Robust Estimation 7
Standard Errors in OLS Luke Sonnet Contents Variance-Covariance of ˆβ 1 Standard Estimation (Spherical Errors) 2 Robust Estimation (Heteroskedasticity Constistent Errors) 4 Cluster Robust Estimation 7
Bayes Estimators & Ridge Regression
 Bayes Estimators & Ridge Regression Readings ISLR 6 STA 521 Duke University Merlise Clyde October 27, 2017 Model Assume that we have centered (as before) and rescaled X o (original X) so that X j = X o
Bayes Estimators & Ridge Regression Readings ISLR 6 STA 521 Duke University Merlise Clyde October 27, 2017 Model Assume that we have centered (as before) and rescaled X o (original X) so that X j = X o
Canopy Light: Synthesizing multiple data sources
 Canopy Light: Synthesizing multiple data sources Tree growth depends upon light (previous example, lab 7) Hard to measure how much light an ADULT tree receives Multiple sources of proxy data Exposed Canopy
Canopy Light: Synthesizing multiple data sources Tree growth depends upon light (previous example, lab 7) Hard to measure how much light an ADULT tree receives Multiple sources of proxy data Exposed Canopy
Predictive Checking. Readings GH Chapter 6-8. February 8, 2017
 Predictive Checking Readings GH Chapter 6-8 February 8, 2017 Model Choice and Model Checking 2 Questions: 1. Is my Model good enough? (no alternative models in mind) 2. Which Model is best? (comparison
Predictive Checking Readings GH Chapter 6-8 February 8, 2017 Model Choice and Model Checking 2 Questions: 1. Is my Model good enough? (no alternative models in mind) 2. Which Model is best? (comparison
Bayesian model selection and diagnostics
 Bayesian model selection and diagnostics A typical Bayesian analysis compares a handful of models. Example 1: Consider the spline model for the motorcycle data, how many basis functions? Example 2: Consider
Bayesian model selection and diagnostics A typical Bayesian analysis compares a handful of models. Example 1: Consider the spline model for the motorcycle data, how many basis functions? Example 2: Consider
Package RegressionFactory
 Type Package Package RegressionFactory September 8, 2016 Title Expander Functions for Generating Full Gradient and Hessian from Single-Slot and Multi-Slot Base Distributions Version 0.7.2 Date 2016-09-07
Type Package Package RegressionFactory September 8, 2016 Title Expander Functions for Generating Full Gradient and Hessian from Single-Slot and Multi-Slot Base Distributions Version 0.7.2 Date 2016-09-07
Section 3.4: Diagnostics and Transformations. Jared S. Murray The University of Texas at Austin McCombs School of Business
 Section 3.4: Diagnostics and Transformations Jared S. Murray The University of Texas at Austin McCombs School of Business 1 Regression Model Assumptions Y i = β 0 + β 1 X i + ɛ Recall the key assumptions
Section 3.4: Diagnostics and Transformations Jared S. Murray The University of Texas at Austin McCombs School of Business 1 Regression Model Assumptions Y i = β 0 + β 1 X i + ɛ Recall the key assumptions
Introduction to R, Github and Gitlab
 Introduction to R, Github and Gitlab 27/11/2018 Pierpaolo Maisano Delser mail: maisanop@tcd.ie ; pm604@cam.ac.uk Outline: Why R? What can R do? Basic commands and operations Data analysis in R Github and
Introduction to R, Github and Gitlab 27/11/2018 Pierpaolo Maisano Delser mail: maisanop@tcd.ie ; pm604@cam.ac.uk Outline: Why R? What can R do? Basic commands and operations Data analysis in R Github and
FMA901F: Machine Learning Lecture 3: Linear Models for Regression. Cristian Sminchisescu
 FMA901F: Machine Learning Lecture 3: Linear Models for Regression Cristian Sminchisescu Machine Learning: Frequentist vs. Bayesian In the frequentist setting, we seek a fixed parameter (vector), with value(s)
FMA901F: Machine Learning Lecture 3: Linear Models for Regression Cristian Sminchisescu Machine Learning: Frequentist vs. Bayesian In the frequentist setting, we seek a fixed parameter (vector), with value(s)
Model Selection and Inference
 Model Selection and Inference Merlise Clyde January 29, 2017 Last Class Model for brain weight as a function of body weight In the model with both response and predictor log transformed, are dinosaurs
Model Selection and Inference Merlise Clyde January 29, 2017 Last Class Model for brain weight as a function of body weight In the model with both response and predictor log transformed, are dinosaurs
The grplasso Package
 The grplasso Package June 27, 2007 Type Package Title Fitting user specified models with Group Lasso penalty Version 0.2-1 Date 2007-06-27 Author Lukas Meier Maintainer Lukas Meier
The grplasso Package June 27, 2007 Type Package Title Fitting user specified models with Group Lasso penalty Version 0.2-1 Date 2007-06-27 Author Lukas Meier Maintainer Lukas Meier
Model selection. Peter Hoff. 560 Hierarchical modeling. Statistics, University of Washington 1/41
 1/41 Model selection 560 Hierarchical modeling Peter Hoff Statistics, University of Washington /41 Modeling choices Model: A statistical model is a set of probability distributions for your data. In HLM,
1/41 Model selection 560 Hierarchical modeling Peter Hoff Statistics, University of Washington /41 Modeling choices Model: A statistical model is a set of probability distributions for your data. In HLM,
GAMs semi-parametric GLMs. Simon Wood Mathematical Sciences, University of Bath, U.K.
 GAMs semi-parametric GLMs Simon Wood Mathematical Sciences, University of Bath, U.K. Generalized linear models, GLM 1. A GLM models a univariate response, y i as g{e(y i )} = X i β where y i Exponential
GAMs semi-parametric GLMs Simon Wood Mathematical Sciences, University of Bath, U.K. Generalized linear models, GLM 1. A GLM models a univariate response, y i as g{e(y i )} = X i β where y i Exponential
Solution to Bonus Questions
 Solution to Bonus Questions Q2: (a) The histogram of 1000 sample means and sample variances are plotted below. Both histogram are symmetrically centered around the true lambda value 20. But the sample
Solution to Bonus Questions Q2: (a) The histogram of 1000 sample means and sample variances are plotted below. Both histogram are symmetrically centered around the true lambda value 20. But the sample
Getting started with simulating data in R: some helpful functions and how to use them Ariel Muldoon August 28, 2018
 Getting started with simulating data in R: some helpful functions and how to use them Ariel Muldoon August 28, 2018 Contents Overview 2 Generating random numbers 2 rnorm() to generate random numbers from
Getting started with simulating data in R: some helpful functions and how to use them Ariel Muldoon August 28, 2018 Contents Overview 2 Generating random numbers 2 rnorm() to generate random numbers from
Stat 4510/7510 Homework 4
 Stat 45/75 1/7. Stat 45/75 Homework 4 Instructions: Please list your name and student number clearly. In order to receive credit for a problem, your solution must show sufficient details so that the grader
Stat 45/75 1/7. Stat 45/75 Homework 4 Instructions: Please list your name and student number clearly. In order to receive credit for a problem, your solution must show sufficient details so that the grader
Package StVAR. February 11, 2017
 Type Package Title Student's t Vector Autoregression (StVAR) Version 1.1 Date 2017-02-10 Author Niraj Poudyal Maintainer Niraj Poudyal Package StVAR February 11, 2017 Description Estimation
Type Package Title Student's t Vector Autoregression (StVAR) Version 1.1 Date 2017-02-10 Author Niraj Poudyal Maintainer Niraj Poudyal Package StVAR February 11, 2017 Description Estimation
Organizing data in R. Fitting Mixed-Effects Models Using the lme4 Package in R. R packages. Accessing documentation. The Dyestuff data set
 Fitting Mixed-Effects Models Using the lme4 Package in R Deepayan Sarkar Fred Hutchinson Cancer Research Center 18 September 2008 Organizing data in R Standard rectangular data sets (columns are variables,
Fitting Mixed-Effects Models Using the lme4 Package in R Deepayan Sarkar Fred Hutchinson Cancer Research Center 18 September 2008 Organizing data in R Standard rectangular data sets (columns are variables,
36-402/608 HW #1 Solutions 1/21/2010
 36-402/608 HW #1 Solutions 1/21/2010 1. t-test (20 points) Use fullbumpus.r to set up the data from fullbumpus.txt (both at Blackboard/Assignments). For this problem, analyze the full dataset together
36-402/608 HW #1 Solutions 1/21/2010 1. t-test (20 points) Use fullbumpus.r to set up the data from fullbumpus.txt (both at Blackboard/Assignments). For this problem, analyze the full dataset together
Regression Analysis and Linear Regression Models
 Regression Analysis and Linear Regression Models University of Trento - FBK 2 March, 2015 (UNITN-FBK) Regression Analysis and Linear Regression Models 2 March, 2015 1 / 33 Relationship between numerical
Regression Analysis and Linear Regression Models University of Trento - FBK 2 March, 2015 (UNITN-FBK) Regression Analysis and Linear Regression Models 2 March, 2015 1 / 33 Relationship between numerical
Gradient Descent. Wed Sept 20th, James McInenrey Adapted from slides by Francisco J. R. Ruiz
 Gradient Descent Wed Sept 20th, 2017 James McInenrey Adapted from slides by Francisco J. R. Ruiz Housekeeping A few clarifications of and adjustments to the course schedule: No more breaks at the midpoint
Gradient Descent Wed Sept 20th, 2017 James McInenrey Adapted from slides by Francisco J. R. Ruiz Housekeeping A few clarifications of and adjustments to the course schedule: No more breaks at the midpoint
Week 5: Multiple Linear Regression II
 Week 5: Multiple Linear Regression II Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Adjusted R
Week 5: Multiple Linear Regression II Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Adjusted R
Practice in R. 1 Sivan s practice. 2 Hetroskadasticity. January 28, (pdf version)
 Practice in R January 28, 2010 (pdf version) 1 Sivan s practice Her practice file should be (here), or check the web for a more useful pointer. 2 Hetroskadasticity ˆ Let s make some hetroskadastic data:
Practice in R January 28, 2010 (pdf version) 1 Sivan s practice Her practice file should be (here), or check the web for a more useful pointer. 2 Hetroskadasticity ˆ Let s make some hetroskadastic data:
Logistic Regression. (Dichotomous predicted variable) Tim Frasier
 Logistic Regression (Dichotomous predicted variable) Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information.
Logistic Regression (Dichotomous predicted variable) Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information.
Exponential Random Graph Models for Social Networks
 Exponential Random Graph Models for Social Networks ERGM Introduction Martina Morris Departments of Sociology, Statistics University of Washington Departments of Sociology, Statistics, and EECS, and Institute
Exponential Random Graph Models for Social Networks ERGM Introduction Martina Morris Departments of Sociology, Statistics University of Washington Departments of Sociology, Statistics, and EECS, and Institute
Beta-Regression with SPSS Michael Smithson School of Psychology, The Australian National University
 9/1/2005 Beta-Regression with SPSS 1 Beta-Regression with SPSS Michael Smithson School of Psychology, The Australian National University (email: Michael.Smithson@anu.edu.au) SPSS Nonlinear Regression syntax
9/1/2005 Beta-Regression with SPSS 1 Beta-Regression with SPSS Michael Smithson School of Psychology, The Australian National University (email: Michael.Smithson@anu.edu.au) SPSS Nonlinear Regression syntax
STENO Introductory R-Workshop: Loading a Data Set Tommi Suvitaival, Steno Diabetes Center June 11, 2015
 STENO Introductory R-Workshop: Loading a Data Set Tommi Suvitaival, tsvv@steno.dk, Steno Diabetes Center June 11, 2015 Contents 1 Introduction 1 2 Recap: Variables 2 3 Data Containers 2 3.1 Vectors................................................
STENO Introductory R-Workshop: Loading a Data Set Tommi Suvitaival, tsvv@steno.dk, Steno Diabetes Center June 11, 2015 Contents 1 Introduction 1 2 Recap: Variables 2 3 Data Containers 2 3.1 Vectors................................................
NEURAL NETWORKS. Cement. Blast Furnace Slag. Fly Ash. Water. Superplasticizer. Coarse Aggregate. Fine Aggregate. Age
 NEURAL NETWORKS As an introduction, we ll tackle a prediction task with a continuous variable. We ll reproduce research from the field of cement and concrete manufacturing that seeks to model the compressive
NEURAL NETWORKS As an introduction, we ll tackle a prediction task with a continuous variable. We ll reproduce research from the field of cement and concrete manufacturing that seeks to model the compressive
CIS 520, Machine Learning, Fall 2015: Assignment 7 Due: Mon, Nov 16, :59pm, PDF to Canvas [100 points]
![CIS 520, Machine Learning, Fall 2015: Assignment 7 Due: Mon, Nov 16, :59pm, PDF to Canvas [100 points] CIS 520, Machine Learning, Fall 2015: Assignment 7 Due: Mon, Nov 16, :59pm, PDF to Canvas [100 points]](/thumbs/89/100746783.jpg) CIS 520, Machine Learning, Fall 2015: Assignment 7 Due: Mon, Nov 16, 2015. 11:59pm, PDF to Canvas [100 points] Instructions. Please write up your responses to the following problems clearly and concisely.
CIS 520, Machine Learning, Fall 2015: Assignment 7 Due: Mon, Nov 16, 2015. 11:59pm, PDF to Canvas [100 points] Instructions. Please write up your responses to the following problems clearly and concisely.
A Knitr Demo. Charles J. Geyer. February 8, 2017
 A Knitr Demo Charles J. Geyer February 8, 2017 1 Licence This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License http://creativecommons.org/licenses/by-sa/4.0/.
A Knitr Demo Charles J. Geyer February 8, 2017 1 Licence This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License http://creativecommons.org/licenses/by-sa/4.0/.
Multiple Linear Regression
 Multiple Linear Regression Rebecca C. Steorts, Duke University STA 325, Chapter 3 ISL 1 / 49 Agenda How to extend beyond a SLR Multiple Linear Regression (MLR) Relationship Between the Response and Predictors
Multiple Linear Regression Rebecca C. Steorts, Duke University STA 325, Chapter 3 ISL 1 / 49 Agenda How to extend beyond a SLR Multiple Linear Regression (MLR) Relationship Between the Response and Predictors
Correlation and Regression
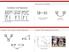 How are X and Y Related Correlation and Regression Causation (regression) p(y X=) Correlation p(y=, X=) http://kcd.com/552/ Correlation Can be Induced b Man Mechanisms # of Pups 1 2 3 4 5 6 7 8 Inbreeding
How are X and Y Related Correlation and Regression Causation (regression) p(y X=) Correlation p(y=, X=) http://kcd.com/552/ Correlation Can be Induced b Man Mechanisms # of Pups 1 2 3 4 5 6 7 8 Inbreeding
Comparing Fitted Models with the fit.models Package
 Comparing Fitted Models with the fit.models Package Kjell Konis Acting Assistant Professor Computational Finance and Risk Management Dept. Applied Mathematics, University of Washington History of fit.models
Comparing Fitted Models with the fit.models Package Kjell Konis Acting Assistant Professor Computational Finance and Risk Management Dept. Applied Mathematics, University of Washington History of fit.models
Missing Data Analysis for the Employee Dataset
 Missing Data Analysis for the Employee Dataset 67% of the observations have missing values! Modeling Setup For our analysis goals we would like to do: Y X N (X, 2 I) and then interpret the coefficients
Missing Data Analysis for the Employee Dataset 67% of the observations have missing values! Modeling Setup For our analysis goals we would like to do: Y X N (X, 2 I) and then interpret the coefficients
Section 2.2: Covariance, Correlation, and Least Squares
 Section 2.2: Covariance, Correlation, and Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 A Deeper
Section 2.2: Covariance, Correlation, and Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 A Deeper
More advanced use of mgcv. Simon Wood Mathematical Sciences, University of Bath, U.K.
 More advanced use of mgcv Simon Wood Mathematical Sciences, University of Bath, U.K. Fine control of smoothness: gamma Suppose that we fit a model but a component is too wiggly. For GCV/AIC we can increase
More advanced use of mgcv Simon Wood Mathematical Sciences, University of Bath, U.K. Fine control of smoothness: gamma Suppose that we fit a model but a component is too wiggly. For GCV/AIC we can increase
Lab #13 - Resampling Methods Econ 224 October 23rd, 2018
 Lab #13 - Resampling Methods Econ 224 October 23rd, 2018 Introduction In this lab you will work through Section 5.3 of ISL and record your code and results in an RMarkdown document. I have added section
Lab #13 - Resampling Methods Econ 224 October 23rd, 2018 Introduction In this lab you will work through Section 5.3 of ISL and record your code and results in an RMarkdown document. I have added section
Section 3.2: Multiple Linear Regression II. Jared S. Murray The University of Texas at Austin McCombs School of Business
 Section 3.2: Multiple Linear Regression II Jared S. Murray The University of Texas at Austin McCombs School of Business 1 Multiple Linear Regression: Inference and Understanding We can answer new questions
Section 3.2: Multiple Linear Regression II Jared S. Murray The University of Texas at Austin McCombs School of Business 1 Multiple Linear Regression: Inference and Understanding We can answer new questions
Analysis of variance - ANOVA
 Analysis of variance - ANOVA Based on a book by Julian J. Faraway University of Iceland (UI) Estimation 1 / 50 Anova In ANOVAs all predictors are categorical/qualitative. The original thinking was to try
Analysis of variance - ANOVA Based on a book by Julian J. Faraway University of Iceland (UI) Estimation 1 / 50 Anova In ANOVAs all predictors are categorical/qualitative. The original thinking was to try
Simulating power in practice
 Simulating power in practice Author: Nicholas G Reich This material is part of the statsteachr project Made available under the Creative Commons Attribution-ShareAlike 3.0 Unported License: http://creativecommons.org/licenses/by-sa/3.0/deed.en
Simulating power in practice Author: Nicholas G Reich This material is part of the statsteachr project Made available under the Creative Commons Attribution-ShareAlike 3.0 Unported License: http://creativecommons.org/licenses/by-sa/3.0/deed.en
This is called a linear basis expansion, and h m is the mth basis function For example if X is one-dimensional: f (X) = β 0 + β 1 X + β 2 X 2, or
 STA 450/4000 S: February 2 2005 Flexible modelling using basis expansions (Chapter 5) Linear regression: y = Xβ + ɛ, ɛ (0, σ 2 ) Smooth regression: y = f (X) + ɛ: f (X) = E(Y X) to be specified Flexible
STA 450/4000 S: February 2 2005 Flexible modelling using basis expansions (Chapter 5) Linear regression: y = Xβ + ɛ, ɛ (0, σ 2 ) Smooth regression: y = f (X) + ɛ: f (X) = E(Y X) to be specified Flexible
Bootstrapping Methods
 Bootstrapping Methods example of a Monte Carlo method these are one Monte Carlo statistical method some Bayesian statistical methods are Monte Carlo we can also simulate models using Monte Carlo methods
Bootstrapping Methods example of a Monte Carlo method these are one Monte Carlo statistical method some Bayesian statistical methods are Monte Carlo we can also simulate models using Monte Carlo methods
Inference for Generalized Linear Mixed Models
 Inference for Generalized Linear Mixed Models Christina Knudson, Ph.D. University of St. Thomas October 18, 2018 Reviewing the Linear Model The usual linear model assumptions: responses normally distributed
Inference for Generalized Linear Mixed Models Christina Knudson, Ph.D. University of St. Thomas October 18, 2018 Reviewing the Linear Model The usual linear model assumptions: responses normally distributed
Markov Chain Monte Carlo (part 1)
 Markov Chain Monte Carlo (part 1) Edps 590BAY Carolyn J. Anderson Department of Educational Psychology c Board of Trustees, University of Illinois Spring 2018 Depending on the book that you select for
Markov Chain Monte Carlo (part 1) Edps 590BAY Carolyn J. Anderson Department of Educational Psychology c Board of Trustees, University of Illinois Spring 2018 Depending on the book that you select for
An Introductory Guide to R
 An Introductory Guide to R By Claudia Mahler 1 Contents Installing and Operating R 2 Basics 4 Importing Data 5 Types of Data 6 Basic Operations 8 Selecting and Specifying Data 9 Matrices 11 Simple Statistics
An Introductory Guide to R By Claudia Mahler 1 Contents Installing and Operating R 2 Basics 4 Importing Data 5 Types of Data 6 Basic Operations 8 Selecting and Specifying Data 9 Matrices 11 Simple Statistics
Section 4: FWL and model fit
 Section 4: FWL and model fit Ed Rubin Contents 1 Admin 1 1.1 What you will need........................................ 1 1.2 Last week............................................. 2 1.3 This week.............................................
Section 4: FWL and model fit Ed Rubin Contents 1 Admin 1 1.1 What you will need........................................ 1 1.2 Last week............................................. 2 1.3 This week.............................................
The linear mixed model: modeling hierarchical and longitudinal data
 The linear mixed model: modeling hierarchical and longitudinal data Analysis of Experimental Data AED The linear mixed model: modeling hierarchical and longitudinal data 1 of 44 Contents 1 Modeling Hierarchical
The linear mixed model: modeling hierarchical and longitudinal data Analysis of Experimental Data AED The linear mixed model: modeling hierarchical and longitudinal data 1 of 44 Contents 1 Modeling Hierarchical
Conditional and Unconditional Regression with No Measurement Error
 Conditional and with No Measurement Error /* reg2ways.sas */ %include 'readsenic.sas'; title2 ''; proc reg; title3 'Conditional Regression'; model infrisk = stay census; proc calis cov; /* Analyze the
Conditional and with No Measurement Error /* reg2ways.sas */ %include 'readsenic.sas'; title2 ''; proc reg; title3 'Conditional Regression'; model infrisk = stay census; proc calis cov; /* Analyze the
OVERVIEW OF ESTIMATION FRAMEWORKS AND ESTIMATORS
 OVERVIEW OF ESTIMATION FRAMEWORKS AND ESTIMATORS Set basic R-options upfront and load all required R packages: > options(prompt = " ", digits = 4) setwd('c:/klaus/aaec5126/test/')#r Sweaves to the default
OVERVIEW OF ESTIMATION FRAMEWORKS AND ESTIMATORS Set basic R-options upfront and load all required R packages: > options(prompt = " ", digits = 4) setwd('c:/klaus/aaec5126/test/')#r Sweaves to the default
Package bayescl. April 14, 2017
 Package bayescl April 14, 2017 Version 0.0.1 Date 2017-04-10 Title Bayesian Inference on a GPU using OpenCL Author Rok Cesnovar, Erik Strumbelj Maintainer Rok Cesnovar Description
Package bayescl April 14, 2017 Version 0.0.1 Date 2017-04-10 Title Bayesian Inference on a GPU using OpenCL Author Rok Cesnovar, Erik Strumbelj Maintainer Rok Cesnovar Description
Missing Data Analysis for the Employee Dataset
 Missing Data Analysis for the Employee Dataset 67% of the observations have missing values! Modeling Setup Random Variables: Y i =(Y i1,...,y ip ) 0 =(Y i,obs, Y i,miss ) 0 R i =(R i1,...,r ip ) 0 ( 1
Missing Data Analysis for the Employee Dataset 67% of the observations have missing values! Modeling Setup Random Variables: Y i =(Y i1,...,y ip ) 0 =(Y i,obs, Y i,miss ) 0 R i =(R i1,...,r ip ) 0 ( 1
DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R
 DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R Lee, Rönnegård & Noh LRN@du.se Lee, Rönnegård & Noh HGLM book 1 / 25 Overview 1 Background to the book 2 A motivating example from my own
DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R Lee, Rönnegård & Noh LRN@du.se Lee, Rönnegård & Noh HGLM book 1 / 25 Overview 1 Background to the book 2 A motivating example from my own
EXST 7014, Lab 1: Review of R Programming Basics and Simple Linear Regression
 EXST 7014, Lab 1: Review of R Programming Basics and Simple Linear Regression OBJECTIVES 1. Prepare a scatter plot of the dependent variable on the independent variable 2. Do a simple linear regression
EXST 7014, Lab 1: Review of R Programming Basics and Simple Linear Regression OBJECTIVES 1. Prepare a scatter plot of the dependent variable on the independent variable 2. Do a simple linear regression
DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R
 DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R Lee, Rönnegård & Noh LRN@du.se Lee, Rönnegård & Noh HGLM book 1 / 24 Overview 1 Background to the book 2 Crack growth example 3 Contents
DATA ANALYSIS USING HIERARCHICAL GENERALIZED LINEAR MODELS WITH R Lee, Rönnegård & Noh LRN@du.se Lee, Rönnegård & Noh HGLM book 1 / 24 Overview 1 Background to the book 2 Crack growth example 3 Contents
5.5 Regression Estimation
 5.5 Regression Estimation Assume a SRS of n pairs (x, y ),..., (x n, y n ) is selected from a population of N pairs of (x, y) data. The goal of regression estimation is to take advantage of a linear relationship
5.5 Regression Estimation Assume a SRS of n pairs (x, y ),..., (x n, y n ) is selected from a population of N pairs of (x, y) data. The goal of regression estimation is to take advantage of a linear relationship
An Introduction to Model-Fitting with the R package glmm
 An Introduction to Model-Fitting with the R package glmm Christina Knudson November 7, 2017 Contents 1 Introduction 2 2 Formatting the Data 2 3 Fitting the Model 4 4 Reading the Model Summary 6 5 Isolating
An Introduction to Model-Fitting with the R package glmm Christina Knudson November 7, 2017 Contents 1 Introduction 2 2 Formatting the Data 2 3 Fitting the Model 4 4 Reading the Model Summary 6 5 Isolating
Theoretical Concepts of Machine Learning
 Theoretical Concepts of Machine Learning Part 2 Institute of Bioinformatics Johannes Kepler University, Linz, Austria Outline 1 Introduction 2 Generalization Error 3 Maximum Likelihood 4 Noise Models 5
Theoretical Concepts of Machine Learning Part 2 Institute of Bioinformatics Johannes Kepler University, Linz, Austria Outline 1 Introduction 2 Generalization Error 3 Maximum Likelihood 4 Noise Models 5
Multivariate Analysis Multivariate Calibration part 2
 Multivariate Analysis Multivariate Calibration part 2 Prof. Dr. Anselmo E de Oliveira anselmo.quimica.ufg.br anselmo.disciplinas@gmail.com Linear Latent Variables An essential concept in multivariate data
Multivariate Analysis Multivariate Calibration part 2 Prof. Dr. Anselmo E de Oliveira anselmo.quimica.ufg.br anselmo.disciplinas@gmail.com Linear Latent Variables An essential concept in multivariate data
Getting to the Bottom of Regression with Gradient Descent
 Getting to the Bottom of Regression with Gradient Descent Jocelyn T. Chi and Eric C. Chi January 10, 2014 This article on gradient descent was written as part of the Getting to the Bottom learning series
Getting to the Bottom of Regression with Gradient Descent Jocelyn T. Chi and Eric C. Chi January 10, 2014 This article on gradient descent was written as part of the Getting to the Bottom learning series
Cross-Validation Alan Arnholt 3/22/2016
 Cross-Validation Alan Arnholt 3/22/2016 Note: Working definitions and graphs are taken from Ugarte, Militino, and Arnholt (2016) The Validation Set Approach The basic idea behind the validation set approach
Cross-Validation Alan Arnholt 3/22/2016 Note: Working definitions and graphs are taken from Ugarte, Militino, and Arnholt (2016) The Validation Set Approach The basic idea behind the validation set approach
LAB #2: SAMPLING, SAMPLING DISTRIBUTIONS, AND THE CLT
 NAVAL POSTGRADUATE SCHOOL LAB #2: SAMPLING, SAMPLING DISTRIBUTIONS, AND THE CLT Statistics (OA3102) Lab #2: Sampling, Sampling Distributions, and the Central Limit Theorem Goal: Use R to demonstrate sampling
NAVAL POSTGRADUATE SCHOOL LAB #2: SAMPLING, SAMPLING DISTRIBUTIONS, AND THE CLT Statistics (OA3102) Lab #2: Sampling, Sampling Distributions, and the Central Limit Theorem Goal: Use R to demonstrate sampling
Calibration of Quinine Fluorescence Emission Vignette for the Data Set flu of the R package hyperspec
 Calibration of Quinine Fluorescence Emission Vignette for the Data Set flu of the R package hyperspec Claudia Beleites CENMAT and DI3, University of Trieste Spectroscopy Imaging,
Calibration of Quinine Fluorescence Emission Vignette for the Data Set flu of the R package hyperspec Claudia Beleites CENMAT and DI3, University of Trieste Spectroscopy Imaging,
Gov Troubleshooting the Linear Model II: Heteroskedasticity
 Gov 2000-10. Troubleshooting the Linear Model II: Heteroskedasticity Matthew Blackwell December 4, 2015 1 / 64 1. Heteroskedasticity 2. Clustering 3. Serial Correlation 4. What s next for you? 2 / 64 Where
Gov 2000-10. Troubleshooting the Linear Model II: Heteroskedasticity Matthew Blackwell December 4, 2015 1 / 64 1. Heteroskedasticity 2. Clustering 3. Serial Correlation 4. What s next for you? 2 / 64 Where
Introduction to Mixed Models: Multivariate Regression
 Introduction to Mixed Models: Multivariate Regression EPSY 905: Multivariate Analysis Spring 2016 Lecture #9 March 30, 2016 EPSY 905: Multivariate Regression via Path Analysis Today s Lecture Multivariate
Introduction to Mixed Models: Multivariate Regression EPSY 905: Multivariate Analysis Spring 2016 Lecture #9 March 30, 2016 EPSY 905: Multivariate Regression via Path Analysis Today s Lecture Multivariate
Machine Learning / Jan 27, 2010
 Revisiting Logistic Regression & Naïve Bayes Aarti Singh Machine Learning 10-701/15-781 Jan 27, 2010 Generative and Discriminative Classifiers Training classifiers involves learning a mapping f: X -> Y,
Revisiting Logistic Regression & Naïve Bayes Aarti Singh Machine Learning 10-701/15-781 Jan 27, 2010 Generative and Discriminative Classifiers Training classifiers involves learning a mapping f: X -> Y,
MS&E 226: Small Data
 MS&E 226: Small Data Lecture 13: The bootstrap (v3) Ramesh Johari ramesh.johari@stanford.edu 1 / 30 Resampling 2 / 30 Sampling distribution of a statistic For this lecture: There is a population model
MS&E 226: Small Data Lecture 13: The bootstrap (v3) Ramesh Johari ramesh.johari@stanford.edu 1 / 30 Resampling 2 / 30 Sampling distribution of a statistic For this lecture: There is a population model
Stat 579: More Preliminaries, Reading from Files
 Stat 579: More Preliminaries, Reading from Files Ranjan Maitra 2220 Snedecor Hall Department of Statistics Iowa State University. Phone: 515-294-7757 maitra@iastate.edu September 1, 2011, 1/10 Some more
Stat 579: More Preliminaries, Reading from Files Ranjan Maitra 2220 Snedecor Hall Department of Statistics Iowa State University. Phone: 515-294-7757 maitra@iastate.edu September 1, 2011, 1/10 Some more
Chapters 5-6: Statistical Inference Methods
 Chapters 5-6: Statistical Inference Methods Chapter 5: Estimation (of population parameters) Ex. Based on GSS data, we re 95% confident that the population mean of the variable LONELY (no. of days in past
Chapters 5-6: Statistical Inference Methods Chapter 5: Estimation (of population parameters) Ex. Based on GSS data, we re 95% confident that the population mean of the variable LONELY (no. of days in past
Outline. Bayesian Data Analysis Hierarchical models. Rat tumor data. Errandum: exercise GCSR 3.11
 Outline Bayesian Data Analysis Hierarchical models Helle Sørensen May 15, 2009 Today: More about the rat tumor data: model, derivation of posteriors, the actual computations in R. : a hierarchical normal
Outline Bayesian Data Analysis Hierarchical models Helle Sørensen May 15, 2009 Today: More about the rat tumor data: model, derivation of posteriors, the actual computations in R. : a hierarchical normal
PSY 9556B (Feb 5) Latent Growth Modeling
 PSY 9556B (Feb 5) Latent Growth Modeling Fixed and random word confusion Simplest LGM knowing how to calculate dfs How many time points needed? Power, sample size Nonlinear growth quadratic Nonlinear growth
PSY 9556B (Feb 5) Latent Growth Modeling Fixed and random word confusion Simplest LGM knowing how to calculate dfs How many time points needed? Power, sample size Nonlinear growth quadratic Nonlinear growth
Bivariate (Simple) Regression Analysis
 Revised July 2018 Bivariate (Simple) Regression Analysis This set of notes shows how to use Stata to estimate a simple (two-variable) regression equation. It assumes that you have set Stata up on your
Revised July 2018 Bivariate (Simple) Regression Analysis This set of notes shows how to use Stata to estimate a simple (two-variable) regression equation. It assumes that you have set Stata up on your
Monte Carlo Analysis
 Monte Carlo Analysis Andrew Q. Philips* February 7, 27 *Ph.D Candidate, Department of Political Science, Texas A&M University, 2 Allen Building, 4348 TAMU, College Station, TX 77843-4348. aphilips@pols.tamu.edu.
Monte Carlo Analysis Andrew Q. Philips* February 7, 27 *Ph.D Candidate, Department of Political Science, Texas A&M University, 2 Allen Building, 4348 TAMU, College Station, TX 77843-4348. aphilips@pols.tamu.edu.
Confirmatory Factor Analysis on the Twin Data: Try One
 Confirmatory Factor Analysis on the Twin Data: Try One /************ twinfac2.sas ********************/ TITLE2 'Confirmatory Factor Analysis'; %include 'twinread.sas'; proc calis corr; /* Analyze the correlation
Confirmatory Factor Analysis on the Twin Data: Try One /************ twinfac2.sas ********************/ TITLE2 'Confirmatory Factor Analysis'; %include 'twinread.sas'; proc calis corr; /* Analyze the correlation
Lecture 6 - Multivariate numerical optimization
 Lecture 6 - Multivariate numerical optimization Björn Andersson (w/ Jianxin Wei) Department of Statistics, Uppsala University February 13, 2014 1 / 36 Table of Contents 1 Plotting functions of two variables
Lecture 6 - Multivariate numerical optimization Björn Andersson (w/ Jianxin Wei) Department of Statistics, Uppsala University February 13, 2014 1 / 36 Table of Contents 1 Plotting functions of two variables
NCSS Statistical Software
 Chapter 327 Geometric Regression Introduction Geometric regression is a special case of negative binomial regression in which the dispersion parameter is set to one. It is similar to regular multiple regression
Chapter 327 Geometric Regression Introduction Geometric regression is a special case of negative binomial regression in which the dispersion parameter is set to one. It is similar to regular multiple regression
Week 10: Heteroskedasticity II
 Week 10: Heteroskedasticity II Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Dealing with heteroskedasticy
Week 10: Heteroskedasticity II Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Dealing with heteroskedasticy
Linear Modeling with Bayesian Statistics
 Linear Modeling with Bayesian Statistics Bayesian Approach I I I I I Estimate probability of a parameter State degree of believe in specific parameter values Evaluate probability of hypothesis given the
Linear Modeling with Bayesian Statistics Bayesian Approach I I I I I Estimate probability of a parameter State degree of believe in specific parameter values Evaluate probability of hypothesis given the
References R's single biggest strenght is it online community. There are tons of free tutorials on R.
 Introduction to R Syllabus Instructor Grant Cavanaugh Department of Agricultural Economics University of Kentucky E-mail: gcavanugh@uky.edu Course description Introduction to R is a short course intended
Introduction to R Syllabus Instructor Grant Cavanaugh Department of Agricultural Economics University of Kentucky E-mail: gcavanugh@uky.edu Course description Introduction to R is a short course intended
BART STAT8810, Fall 2017
 BART STAT8810, Fall 2017 M.T. Pratola November 1, 2017 Today BART: Bayesian Additive Regression Trees BART: Bayesian Additive Regression Trees Additive model generalizes the single-tree regression model:
BART STAT8810, Fall 2017 M.T. Pratola November 1, 2017 Today BART: Bayesian Additive Regression Trees BART: Bayesian Additive Regression Trees Additive model generalizes the single-tree regression model:
Linear Model Selection and Regularization. especially usefull in high dimensions p>>100.
 Linear Model Selection and Regularization especially usefull in high dimensions p>>100. 1 Why Linear Model Regularization? Linear models are simple, BUT consider p>>n, we have more features than data records
Linear Model Selection and Regularization especially usefull in high dimensions p>>100. 1 Why Linear Model Regularization? Linear models are simple, BUT consider p>>n, we have more features than data records
Section 2.1: Intro to Simple Linear Regression & Least Squares
 Section 2.1: Intro to Simple Linear Regression & Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 Regression:
Section 2.1: Intro to Simple Linear Regression & Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 Regression:
Random coefficients models
 enote 9 1 enote 9 Random coefficients models enote 9 INDHOLD 2 Indhold 9 Random coefficients models 1 9.1 Introduction.................................... 2 9.2 Example: Constructed data...........................
enote 9 1 enote 9 Random coefficients models enote 9 INDHOLD 2 Indhold 9 Random coefficients models 1 9.1 Introduction.................................... 2 9.2 Example: Constructed data...........................
Week 4: Simple Linear Regression III
 Week 4: Simple Linear Regression III Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Goodness of
Week 4: Simple Linear Regression III Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Goodness of
Generalized Least Squares (GLS) and Estimated Generalized Least Squares (EGLS)
 Generalized Least Squares (GLS) and Estimated Generalized Least Squares (EGLS) Linear Model in matrix notation for the population Y = Xβ + Var ( ) = In GLS, the error covariance matrix is known In EGLS
Generalized Least Squares (GLS) and Estimated Generalized Least Squares (EGLS) Linear Model in matrix notation for the population Y = Xβ + Var ( ) = In GLS, the error covariance matrix is known In EGLS
Linear discriminant analysis and logistic
 Practical 6: classifiers Linear discriminant analysis and logistic This practical looks at two different methods of fitting linear classifiers. The linear discriminant analysis is implemented in the MASS
Practical 6: classifiers Linear discriminant analysis and logistic This practical looks at two different methods of fitting linear classifiers. The linear discriminant analysis is implemented in the MASS
Section 2.1: Intro to Simple Linear Regression & Least Squares
 Section 2.1: Intro to Simple Linear Regression & Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 Regression:
Section 2.1: Intro to Simple Linear Regression & Least Squares Jared S. Murray The University of Texas at Austin McCombs School of Business Suggested reading: OpenIntro Statistics, Chapter 7.1, 7.2 1 Regression:
Week 4: Simple Linear Regression II
 Week 4: Simple Linear Regression II Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Algebraic properties
Week 4: Simple Linear Regression II Marcelo Coca Perraillon University of Colorado Anschutz Medical Campus Health Services Research Methods I HSMP 7607 2017 c 2017 PERRAILLON ARR 1 Outline Algebraic properties
Logical operators: R provides an extensive list of logical operators. These include
 meat.r: Explanation of code Goals of code: Analyzing a subset of data Creating data frames with specified X values Calculating confidence and prediction intervals Lists and matrices Only printing a few
meat.r: Explanation of code Goals of code: Analyzing a subset of data Creating data frames with specified X values Calculating confidence and prediction intervals Lists and matrices Only printing a few
K-Means and Gaussian Mixture Models
 K-Means and Gaussian Mixture Models David Rosenberg New York University June 15, 2015 David Rosenberg (New York University) DS-GA 1003 June 15, 2015 1 / 43 K-Means Clustering Example: Old Faithful Geyser
K-Means and Gaussian Mixture Models David Rosenberg New York University June 15, 2015 David Rosenberg (New York University) DS-GA 1003 June 15, 2015 1 / 43 K-Means Clustering Example: Old Faithful Geyser
STAT 5200 Handout #25. R-Square & Design Matrix in Mixed Models
 STAT 5200 Handout #25 R-Square & Design Matrix in Mixed Models I. R-Square in Mixed Models (with Example from Handout #20): For mixed models, the concept of R 2 is a little complicated (and neither PROC
STAT 5200 Handout #25 R-Square & Design Matrix in Mixed Models I. R-Square in Mixed Models (with Example from Handout #20): For mixed models, the concept of R 2 is a little complicated (and neither PROC
