Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex
|
|
|
- Chloe Powers
- 5 years ago
- Views:
Transcription
1 CORRECTION NOTICE Nat. Struct. Mol. Biol. advance online publication, doi: /nsmb.1618 (7 June 2009) Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex Martin Kampmann & Günter Blobel In the version of the supplementary information originally posted online, the captions for the Supplementary Movies were in the incorrect order. This has been corrected as of 14 June nature structural & molecular biology
2 SUPPLEMENTARY INFORMATION Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex Martin Kampmann and Günter Blobel Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA blobel@rockefeller.edu
3 Supplementary Figure 1 Angles between particle segments, as defined in Figure 3a. All binary combinations of angles α, β, γ and δ are plotted; each marker represents two angles for a particle class. Marker area is proportional to class size. Particle-based correlation coefficients are indicated in purple.
4 Supplementary Figure 2 Principle component analysis (PCA) of the variation of angles α, β, γ and δ between particle segments (see also Fig. 3 and Supplementary Fig. 1). (a) Eigenvector 1 explains 73% of the variance; the remaining three eigenvectors explain the residual 27% of the variance. (b) Biplot: The black dots correspond to the original data projected onto the plane defined by the first two eigenvectors. The projections of the four original dimensions (angles) onto this plane are indicated by colored lines. Eigenvector 1 represents mainly the correlated variation in γ and δ. Eigenvector 2 represents mainly the anti-correlated variation in α and β.
5 Supplementary Figure 3 Micrograph tilt pair. A field of negatively stained particles was imaged at 50º and 0º tilt. Tilt pair images of the same particle were picked as illustrated by white ovals. Scale bar, 100 nm.
6 Supplementary Figure 4 Fourier Shell Correlation (FSC) for initial and final maps 1 and 2, as well as control maps that were reconstructed by projection matching, only using the best-matching 25% or 50% of particles, as shown in Supplementary Figures 5 and 6. The reproducible resolution, as defined by FSC=0.5, is ~58 Å for the initial maps and ~35 Å for the final maps.
7 Supplementary Figure 5 Cross-correlation coefficients for projection matching. Each grey dot represents one image of a particle from the tilted specimen. The non-normalized cross-correlation coefficient between the particle and the best-matching reference projection from initial map 2 is plotted versus the non-normalized cross-correlation coefficient between the particle and the best-matching reference projection from initial map 1. Red and blue dots represent particles from the classes that were used to construct initial maps 1 and 2, respectively. The diagonal (x=y) divides the particles into two sets of particles: those that have a higher score for alignment to a projection from initial volume 1 and were used for the reconstruction of final map 1 (dots below the diagonal), and those that have a higher score for alignment to a projection from initial volume 2 and were used for the reconstruction of final map 2 (dots above the diagonal). For control purposes, volumes were also reconstructed from subsets of particles that represented the 50% or 25% of the particles in each set with the highest cross-correlation coefficients for alignment to the best reference projection.
8 Supplementary Figure 6
9 Supplementary Figure 6 Three-dimensional structures of the heptameric complex (a) Initial maps obtained by random-conical tilt reconstruction from the classes shown in Figure 4a (b) Maps obtained by projection matching and simultaneous iterative reconstruction using for each map the 25% of particles which aligned to the initial maps with the highest cross-correlation coefficients (see text for details). 4a (c) Final maps obtained by projection matching and simultaneous iterative reconstruction using all particles. All structures are depicted to scale as isodensity contour surfaces that were lowpass filtered beyond the reproducible resolution. Scale bar, 100 Å.
10 Supplementary Figure 7
11 Supplementary Figure 7 Comparison of crystal structure docking into maps 1 and 2. Crystal structures were fitted computationally into maps 1 and 2 independently. Two views related by a 90º rotation around a vertical axis are shown. Color coding of crystal structures is the same as in Figure 6. Nup107 Nup133 was only docked into map 2, not map 1, since the stem hinge 2, which we showed to co-localize with the C-terminus of Nup133 (Fig. 5), is clearly visible in map 2 only. The crystal conformation of Nup107 Nup133 is likely to differ from the actual Nup84 Nup133 conformation in map 2, as evidenced by the poor fit, and the structure is included for illustrative purposes only.
12 SUPPLEMENTARY DISCUSSION Projection matching As described in the main text, initial maps were constructed from two selected particle classes using the random conical tilt method (Fig. 4a,b). We suspected that other particle images may represent slightly different views of the particle in a similar conformation. Therefore, the signal-to-noise ratio of the initial maps may be improved by incorporating additional particle images. We calculated reference projections from the two initial maps and aligned all particle images from tilted specimen to both reference projections. Supplementary Figure 5 shows the cross-correlation coefficients (CCC) for alignment of each particle to the best-matching reference projection from both initial maps (grey dots). Each particle was assigned to either map 1 or map 2 to maximize the CCC. Red and blue dots in Supplementary Figure 5 correspond to the particles that were used to construct initial maps 1 and 2, respectively. Of the particles used for initial map 1, 86% have a higher CCC for alignment to initial map 1 projections than to initial map 2 projections; they lie below the x=y diagonal in Supplementary Figure 5. Of the particles used for initial map 2, 91% lie above the diagonal. Therefore, the CCC is a suitable criterion to distinguish the two different particle conformations. Interestingly, the CCC distributions of particles used to reconstruct initial maps 1 and overlap with the CCC distribution of all other particles, supporting the idea that some of the other particles are likely to represent different views of a particle in the same (or a highly similar) conformation as initial maps 1 and 2. However, the CCC distributions are unimodal; therefore, it is not possible to find by inspection a correct threshold to distinguish between particles with a conformation corresponding to the reference map and particles with a different conformation. This finding is compatible with our conclusion that there is a continuous spectrum of particle conformations. We therefore independently reconstructed maps from three subsets for each map 1 and map 2. The subsets contained the 25% particles with the highest CCCs, the 50% particles with the highest CCCs, or all particles. The maps obtained from the
13 first and last of these subsets are displayed in Supplementary Figure 6. These maps are highly similar, as shown in Movies 1 and 2 (overlays of Final maps in orange and best 25% maps in purple for maps 1 and 2, respectively). The Fourier shell correlation (FSC) curves for all maps are compared in Supplementary Figure 4. The reproducible resolution of the maps, as defined by FSC=0.5, increases monotonically with the number of particles used for the reconstruction. We therefore decided to use the final maps 1 and 2 (reconstructed from all particle images) for further analysis. Localization of nups within the heptameric complex The localization of several nups or nup complexes to different segments of the Nup84 complex was reported by Hurt and colleagues 1, and further supported by crystallographic studies 2-5. Specifically, the following aspects of Nup84 complex architecture were previously established: (i) Nup145C Sec13 and Nup85 Seh1 are stable heterodimers 1 in which the seventh blade of the Sec13 and Seh1 β-propellers is contributed by Nup145C and Nup85, respectively 2-4. (ii) Nup120 can form trimers with Nup145C Sec13 and with Nup85 Seh1, and together, these five proteins can form a pentamer which appears in EM as a triskelion with three short arms, and can thus be concluded to correspond to the two arms and the vertex-proximal stem segment of the heptameric complex 1. (iii) Nup84 forms a dimer with Nup133 1 ; more specifically, these two nups interact via their C termini 5. (iv) Nup133 Nup84 forms a tetramer with Nup145C Sec13. Nup84 alone, as well as the Nup133 Nup84 dimer bind the Nup120 Nup145C Sec13 Nup85 Seh1 pentamer, thereby extending one of the three arms into the elongated stem 1. Taken together, these observations show that the two arms of the heptameric complex are formed by Nup120 and Nup85 Seh1, which are connected to Nup145C Sec13 at the vertex region. Nup145C Sec13 forms the upper part of the stem, Nup84 the mid-stem and Nup133 the distal stem and the foot.
14 This assignment does not establish the identity of the short and the long arm. We therefore mapped the position of Seh1 by EM and found it to localize to the short arm of the heptamer. We also mapped the position of the Nup133 C terminus, which localized to the stem hinge 2 region. Details of crystal structure docking within the EM maps The Nup85 Seh1 dimer assumed highly similar orientations in EM maps 1 and 2 (Supplementary Fig. 7). The crystal structure of Nup85 lacks 174 C-terminal amino acids. The C terminus of the crystallized Nup85 fragment points towards the end of the short arm (Fig. 6c, Supplementary Movie 5). Since the end of the short arm is occupied with the Seh1 β -propeller, additional C-terminal residues of Nup85 likely fold back towards the vertex region. The crystallized domain of Nup85 does not completely fill the short arm of the EM map, thus leaving space for the Nup85 C terminus. However, the current resolution of the EM map is not sufficient to discern the exact location of the additional residues. The Nup145C Sec13 dimer also had highly similar orientations in maps 1 and 2 (Supplementary Fig. 7). The crystal structure of Nup145C lacks 125 N-terminal amino acids and 156 C-terminal amino acids. Both the N and C terminus of the crystallized Nup145C fragment point towards the vertex of the heptamer (Fig. 6d, Supplementary Movie 5), suggesting that the region between the knob and the vertex is filled with the remaining portions of Nup145C. The N-terminal Nup133 β-propeller docked into the foot regions of maps 1 and 2 in similar orientations (Fig. 6e, Supplementary Figure 7). The C terminus of the crystallized Nup133 β -propeller domain points away from the stem; the following residues will have to fold back to connect to the remaining part of Nup133. Again, the resolution of the EM map is not high enough to predict the exact path taken by protein fragments absent from the crystal structure. The global fitting method failed for the Nup107 Nup133 structure: none of the computed positions coincided with stem hinge 2. Therefore, we positioned the crystal structure manually in this region and optimized the local fit computationally.
15 SUPPLEMENTARY REFERENCES 1. Lutzmann, M., Kunze, R., Buerer, A., Aebi, U. & Hurt, E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J 21, (2002). 2. Hsia, K.C., Stavropoulos, P., Blobel, G. & Hoelz, A. Architecture of a coat for the nuclear pore membrane. Cell 131, (2007). 3. Debler, E.W. et al. A fence-like coat for the nuclear pore membrane. Mol Cell 32, (2008). 4. Brohawn, S.G., Leksa, N.C., Spear, E.D., Rajashankar, K.R. & Schwartz, T.U. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322, (2008). 5. Boehmer, T., Jeudy, S., Berke, I.C. & Schwartz, T.U. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell 30, (2008).
Structural Basis for RNA Processing by the Human. RISC-Loading Complex
 Supplementary Information Structural Basis for RNA Processing by the Human RISC-Loading Complex Hong-Wei Wang, Cameron Noland, Bunpote Siridechadilok, David W. Taylor, Enbo Ma, Karin Felderer, Jennifer
Supplementary Information Structural Basis for RNA Processing by the Human RISC-Loading Complex Hong-Wei Wang, Cameron Noland, Bunpote Siridechadilok, David W. Taylor, Enbo Ma, Karin Felderer, Jennifer
II: Single particle cryoem - an averaging technique
 II: Single particle cryoem - an averaging technique Radiation damage limits the total electron dose that can be used to image biological sample. Thus, images of frozen hydrated macromolecules are very
II: Single particle cryoem - an averaging technique Radiation damage limits the total electron dose that can be used to image biological sample. Thus, images of frozen hydrated macromolecules are very
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S. Visualization of Biological Molecules in Their Native Stat e.
 THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
Structure of a Biologically Active Estrogen Receptor-Coactivator Complex on DNA
 Molecular Cell Supplemental Information Structure of a Biologically Active Estrogen Receptor-Coactivator Complex on DNA Ping Yi, Zhao Wang, Qin Feng, Grigore D. Pintilie, Charles E. Foulds, Rainer B. Lanz,
Molecular Cell Supplemental Information Structure of a Biologically Active Estrogen Receptor-Coactivator Complex on DNA Ping Yi, Zhao Wang, Qin Feng, Grigore D. Pintilie, Charles E. Foulds, Rainer B. Lanz,
Nature Structural & Molecular Biology: doi: /nsmb.2467
 Supplementary Figure 1. Cryo-electron tomography and GRAFIX optimization of COPII cages. a) Tomogram of Sec13-31 cages viewed along the air/water interface. The tomogram shows a large amount of disordered
Supplementary Figure 1. Cryo-electron tomography and GRAFIX optimization of COPII cages. a) Tomogram of Sec13-31 cages viewed along the air/water interface. The tomogram shows a large amount of disordered
Supplementary Information. Structure of a RSC nucleosome Complex and Insights into Chromatin Remodeling
 Supplementary Information Structure of a RSC nucleosome Complex and Insights into Chromatin Remodeling Yuriy Chaban 1, Chukwudi Ezeokonkwo 1, Wen-Hsiang Chung 1, Fan Zhang 1, Roger D. Kornberg 2, Barbara
Supplementary Information Structure of a RSC nucleosome Complex and Insights into Chromatin Remodeling Yuriy Chaban 1, Chukwudi Ezeokonkwo 1, Wen-Hsiang Chung 1, Fan Zhang 1, Roger D. Kornberg 2, Barbara
EMBO Practical Course on Image Processing for Cryo EM 4 14 September Practical 8: Fitting atomic structures into EM maps
 EMBO Practical Course on Image Processing for Cryo EM 4 14 September 2005 Practical 8: Fitting atomic structures into EM maps The best way to interpret the EM density map is to build a hybrid structure,
EMBO Practical Course on Image Processing for Cryo EM 4 14 September 2005 Practical 8: Fitting atomic structures into EM maps The best way to interpret the EM density map is to build a hybrid structure,
Supplementary Information
 Supplementary Information Supplementary Figure S1 The scheme of MtbHadAB/MtbHadBC dehydration reaction. The reaction is reversible. However, in the context of FAS-II elongation cycle, this reaction tends
Supplementary Information Supplementary Figure S1 The scheme of MtbHadAB/MtbHadBC dehydration reaction. The reaction is reversible. However, in the context of FAS-II elongation cycle, this reaction tends
II: Single particle cryoem - an averaging technique
 II: Single particle cryoem - an averaging technique Radiation damage limits the total electron dose that can be used to image biological sample. Thus, images of frozen hydrated macromolecules are very
II: Single particle cryoem - an averaging technique Radiation damage limits the total electron dose that can be used to image biological sample. Thus, images of frozen hydrated macromolecules are very
EE368 Project: Visual Code Marker Detection
 EE368 Project: Visual Code Marker Detection Kahye Song Group Number: 42 Email: kahye@stanford.edu Abstract A visual marker detection algorithm has been implemented and tested with twelve training images.
EE368 Project: Visual Code Marker Detection Kahye Song Group Number: 42 Email: kahye@stanford.edu Abstract A visual marker detection algorithm has been implemented and tested with twelve training images.
Structural Information obtained
 Structural Information obtained from Electron Microscopy Christiane Schaffitzel, 09.05.2013 Including slides from John Briggs, Bettina Boettcher, Nicolas Boisset, Andy Hoenger, Michael Schatz, and more
Structural Information obtained from Electron Microscopy Christiane Schaffitzel, 09.05.2013 Including slides from John Briggs, Bettina Boettcher, Nicolas Boisset, Andy Hoenger, Michael Schatz, and more
Single Particle Reconstruction Techniques
 T H E U N I V E R S I T Y of T E X A S S C H O O L O F H E A L T H I N F O R M A T I O N S C I E N C E S A T H O U S T O N Single Particle Reconstruction Techniques For students of HI 6001-125 Computational
T H E U N I V E R S I T Y of T E X A S S C H O O L O F H E A L T H I N F O R M A T I O N S C I E N C E S A T H O U S T O N Single Particle Reconstruction Techniques For students of HI 6001-125 Computational
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror
 Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
Image Alignment and Application of 2D Fast Rotational Matching in Single Particle cryo-electron Microscopy. Yao Cong. Baylor College of Medicine
 Image Alignment and Application of 2D Fast Rotational Matching in Single Particle cryo-electron Microscopy Yao Cong Baylor College of Medicine cryo-em & single particle analysis Single-particle electron
Image Alignment and Application of 2D Fast Rotational Matching in Single Particle cryo-electron Microscopy Yao Cong Baylor College of Medicine cryo-em & single particle analysis Single-particle electron
Tutorial 3. Correlated Random Hydraulic Conductivity Field
 Tutorial 3 Correlated Random Hydraulic Conductivity Field Table of Contents Objective. 1 Step-by-Step Procedure... 2 Section 1 Generation of Correlated Random Hydraulic Conductivity Field 2 Step 1: Open
Tutorial 3 Correlated Random Hydraulic Conductivity Field Table of Contents Objective. 1 Step-by-Step Procedure... 2 Section 1 Generation of Correlated Random Hydraulic Conductivity Field 2 Step 1: Open
3. Image formation, Fourier analysis and CTF theory. Paula da Fonseca
 3. Image formation, Fourier analysis and CTF theory Paula da Fonseca EM course 2017 - Agenda - Overview of: Introduction to Fourier analysis o o o o Sine waves Fourier transform (simple examples of 1D
3. Image formation, Fourier analysis and CTF theory Paula da Fonseca EM course 2017 - Agenda - Overview of: Introduction to Fourier analysis o o o o Sine waves Fourier transform (simple examples of 1D
Initial Model Generation
 Initial Model Generation Workshop on Advanced Topics in EM Structure Determination The Scripps Research Institute La Jolla, November 2007 The issue: Structures of the IP3 receptor as determined by single
Initial Model Generation Workshop on Advanced Topics in EM Structure Determination The Scripps Research Institute La Jolla, November 2007 The issue: Structures of the IP3 receptor as determined by single
Transmission Electron Microscopy 3D Construction by IPET Method TCBG GPU Workshop
 Transmission Electron Microscopy 3D Construction by IPET Method TCBG GPU Workshop Xing Zhang 8/4/2013 Molecular Foundry Lawrence Berkeley National Laboratory Department of Applied Physics Xi an Jiaotong
Transmission Electron Microscopy 3D Construction by IPET Method TCBG GPU Workshop Xing Zhang 8/4/2013 Molecular Foundry Lawrence Berkeley National Laboratory Department of Applied Physics Xi an Jiaotong
3D Reconstruction in EM
 3D Reconstruction in EM C.O.S. Sorzano Biocomputing Unit, CNB-CSIC Instruct Image Processing Center Contents Initial volume Angular assignment 3D Reconstruction 3D Classification Resolution Amplitude correction
3D Reconstruction in EM C.O.S. Sorzano Biocomputing Unit, CNB-CSIC Instruct Image Processing Center Contents Initial volume Angular assignment 3D Reconstruction 3D Classification Resolution Amplitude correction
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS.
 SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
Science Olympiad Protein Modeling Event Guide to Scoring Zinc Finger Motif
 Science Olympiad Protein Modeling Event Guide to Scoring Zinc Finger Motif Once you have folded the zinc finger motif (chain C, residues 4-31 of 1ZAA.pdb), use this guide in conjunction with the rubric
Science Olympiad Protein Modeling Event Guide to Scoring Zinc Finger Motif Once you have folded the zinc finger motif (chain C, residues 4-31 of 1ZAA.pdb), use this guide in conjunction with the rubric
Cover Page. The handle holds various files of this Leiden University dissertation
 Cover Page The handle http://hdl.handle.net/1887/48877 holds various files of this Leiden University dissertation Author: Li, Y. Title: A new method to reconstruct the structure from crystal images Issue
Cover Page The handle http://hdl.handle.net/1887/48877 holds various files of this Leiden University dissertation Author: Li, Y. Title: A new method to reconstruct the structure from crystal images Issue
III: Single particle cryoem - practical approaches
 III: Single particle cryoem - practical approaches Single particle EM analysis can be performed at both 2D and 3D. Single particle EM (both negative stain and cryo) is to extract structural information
III: Single particle cryoem - practical approaches Single particle EM analysis can be performed at both 2D and 3D. Single particle EM (both negative stain and cryo) is to extract structural information
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12009 Supplementary Figure 1. Experimental tilt series of 104 projections with a tilt range of ±72.6 and equal slope increments, acquired from a Pt nanoparticle using HAADF- STEM (energy:
doi:10.1038/nature12009 Supplementary Figure 1. Experimental tilt series of 104 projections with a tilt range of ±72.6 and equal slope increments, acquired from a Pt nanoparticle using HAADF- STEM (energy:
Revolve Vertices. Axis of revolution. Angle of revolution. Edge sense. Vertex to be revolved. Figure 2-47: Revolve Vertices operation
 Revolve Vertices The Revolve Vertices operation (edge create revolve command) creates circular arc edges or helixes by revolving existing real and/or non-real vertices about a specified axis. The command
Revolve Vertices The Revolve Vertices operation (edge create revolve command) creates circular arc edges or helixes by revolving existing real and/or non-real vertices about a specified axis. The command
Machine Learning : supervised versus unsupervised
 Machine Learning : supervised versus unsupervised Neural Networks: supervised learning makes use of a known property of the data: the digit as classified by a human the ground truth needs a training set,
Machine Learning : supervised versus unsupervised Neural Networks: supervised learning makes use of a known property of the data: the digit as classified by a human the ground truth needs a training set,
Assessing the homo- or heterogeneity of noisy experimental data. Kay Diederichs Konstanz, 01/06/2017
 Assessing the homo- or heterogeneity of noisy experimental data Kay Diederichs Konstanz, 01/06/2017 What is the problem? Why do an experiment? because we want to find out a property (or several) of an
Assessing the homo- or heterogeneity of noisy experimental data Kay Diederichs Konstanz, 01/06/2017 What is the problem? Why do an experiment? because we want to find out a property (or several) of an
Cryo-electron microscopy Cryo-EM. Garry Taylor
 Cryo-electron microscopy Cryo-EM Garry Taylor www.st-andrews.ac.uk/~glt2/bl3301 Electron has a wavelength de Broglie relationship: m v = h / λ or λ = h / mv Accelerate e - in a field of potential V, it
Cryo-electron microscopy Cryo-EM Garry Taylor www.st-andrews.ac.uk/~glt2/bl3301 Electron has a wavelength de Broglie relationship: m v = h / λ or λ = h / mv Accelerate e - in a field of potential V, it
Salvador Capella-Gutiérrez, Jose M. Silla-Martínez and Toni Gabaldón
 trimal: a tool for automated alignment trimming in large-scale phylogenetics analyses Salvador Capella-Gutiérrez, Jose M. Silla-Martínez and Toni Gabaldón Version 1.2b Index of contents 1. General features
trimal: a tool for automated alignment trimming in large-scale phylogenetics analyses Salvador Capella-Gutiérrez, Jose M. Silla-Martínez and Toni Gabaldón Version 1.2b Index of contents 1. General features
Locating ego-centers in depth for hippocampal place cells
 204 5th Joint Symposium on Neural Computation Proceedings UCSD (1998) Locating ego-centers in depth for hippocampal place cells Kechen Zhang,' Terrence J. Sejeowski112 & Bruce L. ~cnau~hton~ 'Howard Hughes
204 5th Joint Symposium on Neural Computation Proceedings UCSD (1998) Locating ego-centers in depth for hippocampal place cells Kechen Zhang,' Terrence J. Sejeowski112 & Bruce L. ~cnau~hton~ 'Howard Hughes
Supplementary Material
 Supplementary Material Figure 1S: Scree plot of the 400 dimensional data. The Figure shows the 20 largest eigenvalues of the (normalized) correlation matrix sorted in decreasing order; the insert shows
Supplementary Material Figure 1S: Scree plot of the 400 dimensional data. The Figure shows the 20 largest eigenvalues of the (normalized) correlation matrix sorted in decreasing order; the insert shows
Batch Processing of Tomograms with IMOD
 Batch Processing of Tomograms with IMOD Batch Processing Can Be Useful for a Wide Range of Tilt Series Routine plastic section tilt series can be fully reconstructed automatically with 80-95% success rate
Batch Processing of Tomograms with IMOD Batch Processing Can Be Useful for a Wide Range of Tilt Series Routine plastic section tilt series can be fully reconstructed automatically with 80-95% success rate
Combinatorial Problems on Strings with Applications to Protein Folding
 Combinatorial Problems on Strings with Applications to Protein Folding Alantha Newman 1 and Matthias Ruhl 2 1 MIT Laboratory for Computer Science Cambridge, MA 02139 alantha@theory.lcs.mit.edu 2 IBM Almaden
Combinatorial Problems on Strings with Applications to Protein Folding Alantha Newman 1 and Matthias Ruhl 2 1 MIT Laboratory for Computer Science Cambridge, MA 02139 alantha@theory.lcs.mit.edu 2 IBM Almaden
Alignment and Other Challenges in Reconstructing Cryotomograms with IMOD
 Alignment and Other Challenges in Reconstructing Cryotomograms with IMOD Challenges in Cryotomography Alignment, alignment, alignment It can be hard to get fiducials onto/in the sample The low SNR makes
Alignment and Other Challenges in Reconstructing Cryotomograms with IMOD Challenges in Cryotomography Alignment, alignment, alignment It can be hard to get fiducials onto/in the sample The low SNR makes
BMB/Bi/Ch 173 Winter 2017
 BMB/Bi/Ch 173 Winter 2017 Homework Set 3.2 Assigned 1/26/2017, Due 1/31/17 by 10:30am TA - Sara Weaver sjweaver [a] Caltech.edu Office hours Broad 3rd floor kitchen - Friday 1/27 1:30pm-2:30pm, Monday
BMB/Bi/Ch 173 Winter 2017 Homework Set 3.2 Assigned 1/26/2017, Due 1/31/17 by 10:30am TA - Sara Weaver sjweaver [a] Caltech.edu Office hours Broad 3rd floor kitchen - Friday 1/27 1:30pm-2:30pm, Monday
A First Introduction to Scientific Visualization Geoffrey Gray
 Visual Molecular Dynamics A First Introduction to Scientific Visualization Geoffrey Gray VMD on CIRCE: On the lower bottom left of your screen, click on the window start-up menu. In the search box type
Visual Molecular Dynamics A First Introduction to Scientific Visualization Geoffrey Gray VMD on CIRCE: On the lower bottom left of your screen, click on the window start-up menu. In the search box type
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10934 Supplementary Methods Mathematical implementation of the EST method. The EST method begins with padding each projection with zeros (that is, embedding
SUPPLEMENTARY INFORMATION doi:10.1038/nature10934 Supplementary Methods Mathematical implementation of the EST method. The EST method begins with padding each projection with zeros (that is, embedding
Computational Molecular Biology
 Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
Likelihood-based optimization of cryo-em data. Sjors Scheres National Center for Biotechnology CSIC Madrid, Spain
 Likelihood-based optimization of cryo-em data Sjors Scheres National Center for Biotechnology CSIC Madrid, Spain Life based on molecular machines DNA replication Protein synthesis Dynein motion Molecular
Likelihood-based optimization of cryo-em data Sjors Scheres National Center for Biotechnology CSIC Madrid, Spain Life based on molecular machines DNA replication Protein synthesis Dynein motion Molecular
Supporting information SI 0: TEM & STEM acquisition parameters, in 2D and 3D
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Supporting information SI 0: TEM & STEM acquisition parameters, in 2D and 3D Electron tomography
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Supporting information SI 0: TEM & STEM acquisition parameters, in 2D and 3D Electron tomography
Supporting Information: FIB-SEM Tomography Probes the Meso-Scale Pore Space of an Individual Catalytic Cracking Particle
 Supporting Information: FIB-SEM Tomography Probes the Meso-Scale Pore Space of an Individual Catalytic Cracking Particle D.A.Matthijs de Winter, Florian Meirer, Bert M. Weckhuysen*, Inorganic Chemistry
Supporting Information: FIB-SEM Tomography Probes the Meso-Scale Pore Space of an Individual Catalytic Cracking Particle D.A.Matthijs de Winter, Florian Meirer, Bert M. Weckhuysen*, Inorganic Chemistry
Chapter 4. Clustering Core Atoms by Location
 Chapter 4. Clustering Core Atoms by Location In this chapter, a process for sampling core atoms in space is developed, so that the analytic techniques in section 3C can be applied to local collections
Chapter 4. Clustering Core Atoms by Location In this chapter, a process for sampling core atoms in space is developed, so that the analytic techniques in section 3C can be applied to local collections
Resting state network estimation in individual subjects
 Resting state network estimation in individual subjects Data 3T NIL(21,17,10), Havard-MGH(692) Young adult fmri BOLD Method Machine learning algorithm MLP DR LDA Network image Correlation Spatial Temporal
Resting state network estimation in individual subjects Data 3T NIL(21,17,10), Havard-MGH(692) Young adult fmri BOLD Method Machine learning algorithm MLP DR LDA Network image Correlation Spatial Temporal
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Performance analysis under four different clustering scenarios. Performance analysis under four different clustering scenarios. i) Standard Conditions, ii) a sparse data set with
Supplementary Figure 1 Performance analysis under four different clustering scenarios. Performance analysis under four different clustering scenarios. i) Standard Conditions, ii) a sparse data set with
A Short Rasmol Tutorial: trna
 A Short Rasmol Tutorial: trna Note that this tutorial is due at the beginning of class on Wednesday, October 3. amino acid attaches here 3 end trna secondary structure. The sequence of yeast trna Phe is
A Short Rasmol Tutorial: trna Note that this tutorial is due at the beginning of class on Wednesday, October 3. amino acid attaches here 3 end trna secondary structure. The sequence of yeast trna Phe is
Version 1.0 November2016 Hermes V1.8.2
 Hermes in a Nutshell Version 1.0 November2016 Hermes V1.8.2 Table of Contents Hermes in a Nutshell... 1 Introduction... 2 Example 1. Visualizing and Editing the MLL1 fusion protein... 3 Setting Your Display...
Hermes in a Nutshell Version 1.0 November2016 Hermes V1.8.2 Table of Contents Hermes in a Nutshell... 1 Introduction... 2 Example 1. Visualizing and Editing the MLL1 fusion protein... 3 Setting Your Display...
Supporting Information
 Supporting Information Highly stable bimetallic AuIr/TiO 2 catalyst: physical origins of the intrinsic high stability against sintering Chang Wan Han 1, Paulami Majumdar 2, Ernesto E. Marinero 1, Antonio
Supporting Information Highly stable bimetallic AuIr/TiO 2 catalyst: physical origins of the intrinsic high stability against sintering Chang Wan Han 1, Paulami Majumdar 2, Ernesto E. Marinero 1, Antonio
Chimera EM Map Tutorial: RNA Polymerase II
 Chimera EM Map Tutorial: RNA Polymerase II May 1, 2007 This tutorial focuses on display of volume data from single particle EM reconstructions. We'll look at maps of two conformations of human RNA polymerase
Chimera EM Map Tutorial: RNA Polymerase II May 1, 2007 This tutorial focuses on display of volume data from single particle EM reconstructions. We'll look at maps of two conformations of human RNA polymerase
Expression pattern of nub-gal4. The expression pattern of nub-gal4 was. assayed by combining it with the reporter UAS-His2ADsRed as in Figure 1.
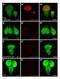 Supplementary Figure 1. Expression pattern of nub-gal4. The expression pattern of nub-gal4 was assayed by combining it with the reporter UAS-His2ADsRed as in Figure 1. His2ADsRed is localized to the nuclei
Supplementary Figure 1. Expression pattern of nub-gal4. The expression pattern of nub-gal4 was assayed by combining it with the reporter UAS-His2ADsRed as in Figure 1. His2ADsRed is localized to the nuclei
Image Enhancement in Spatial Domain. By Dr. Rajeev Srivastava
 Image Enhancement in Spatial Domain By Dr. Rajeev Srivastava CONTENTS Image Enhancement in Spatial Domain Spatial Domain Methods 1. Point Processing Functions A. Gray Level Transformation functions for
Image Enhancement in Spatial Domain By Dr. Rajeev Srivastava CONTENTS Image Enhancement in Spatial Domain Spatial Domain Methods 1. Point Processing Functions A. Gray Level Transformation functions for
3D Models and Matching
 3D Models and Matching representations for 3D object models particular matching techniques alignment-based systems appearance-based systems GC model of a screwdriver 1 3D Models Many different representations
3D Models and Matching representations for 3D object models particular matching techniques alignment-based systems appearance-based systems GC model of a screwdriver 1 3D Models Many different representations
Molecular Modeling Protocol
 Molecular Modeling of an unknown protein 1. Register for your own SWISS-MODEL Workspace at http://swissmodel.expasy.org/workspace/index. Follow the Login link in the upper right hand corner. Bring your
Molecular Modeling of an unknown protein 1. Register for your own SWISS-MODEL Workspace at http://swissmodel.expasy.org/workspace/index. Follow the Login link in the upper right hand corner. Bring your
X-RAY crystallography [1], [2] and nuclear magnetic. Computational Approaches for Automatic Structural Analysis of Large Bio-molecular Complexes
![X-RAY crystallography [1], [2] and nuclear magnetic. Computational Approaches for Automatic Structural Analysis of Large Bio-molecular Complexes X-RAY crystallography [1], [2] and nuclear magnetic. Computational Approaches for Automatic Structural Analysis of Large Bio-molecular Complexes](/thumbs/84/89164539.jpg) IEEE/ACM TRANSACTIONS ON COMPUTATIONAL BIOLOGY AND BIOINFORMATICS 1 Computational Approaches for Automatic Structural Analysis of Large Bio-molecular Complexes Zeyun Yu, Student Member, IEEE, Chandrajit
IEEE/ACM TRANSACTIONS ON COMPUTATIONAL BIOLOGY AND BIOINFORMATICS 1 Computational Approaches for Automatic Structural Analysis of Large Bio-molecular Complexes Zeyun Yu, Student Member, IEEE, Chandrajit
Chapter 3. Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C.
 Chapter 3 Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C. elegans Embryos Abstract Differential interference contrast (DIC) microscopy is
Chapter 3 Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C. elegans Embryos Abstract Differential interference contrast (DIC) microscopy is
Enhanced material contrast by dual-energy microct imaging
 Enhanced material contrast by dual-energy microct imaging Method note Page 1 of 12 2 Method note: Dual-energy microct analysis 1. Introduction 1.1. The basis for dual energy imaging Micro-computed tomography
Enhanced material contrast by dual-energy microct imaging Method note Page 1 of 12 2 Method note: Dual-energy microct analysis 1. Introduction 1.1. The basis for dual energy imaging Micro-computed tomography
5. Consider the tree solution for a minimum cost network flow problem shown in Figure 5.
 FIGURE 1. Data for a network flow problem. As usual the numbers above the nodes are supplies (negative values represent demands) and numbers shown above the arcs are unit shipping costs. The darkened arcs
FIGURE 1. Data for a network flow problem. As usual the numbers above the nodes are supplies (negative values represent demands) and numbers shown above the arcs are unit shipping costs. The darkened arcs
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Schematic demonstrating the preferred orientation sampling problem in single-particle cryo-em. Specimens sticking to the air-water interface or a substrate support of a cryo-em grid
Supplementary Figure 1 Schematic demonstrating the preferred orientation sampling problem in single-particle cryo-em. Specimens sticking to the air-water interface or a substrate support of a cryo-em grid
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror
 Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
2, the coefficient of variation R 2, and properties of the photon counts traces
 Supplementary Figure 1 Quality control of FCS traces. (a) Typical trace that passes the quality control (QC) according to the parameters shown in f. The QC is based on thresholds applied to fitting parameters
Supplementary Figure 1 Quality control of FCS traces. (a) Typical trace that passes the quality control (QC) according to the parameters shown in f. The QC is based on thresholds applied to fitting parameters
(Refer Slide Time 00:17) Welcome to the course on Digital Image Processing. (Refer Slide Time 00:22)
 Digital Image Processing Prof. P. K. Biswas Department of Electronics and Electrical Communications Engineering Indian Institute of Technology, Kharagpur Module Number 01 Lecture Number 02 Application
Digital Image Processing Prof. P. K. Biswas Department of Electronics and Electrical Communications Engineering Indian Institute of Technology, Kharagpur Module Number 01 Lecture Number 02 Application
Introduction to Hermes
 Introduction to Hermes Version 2.0 November 2017 Hermes v1.9 Table of Contents Introduction... 2 Visualising and Editing the MLL1 fusion protein... 2 Opening Files in Hermes... 3 Setting Style Preferences...
Introduction to Hermes Version 2.0 November 2017 Hermes v1.9 Table of Contents Introduction... 2 Visualising and Editing the MLL1 fusion protein... 2 Opening Files in Hermes... 3 Setting Style Preferences...
CS 231A Computer Vision (Winter 2014) Problem Set 3
 CS 231A Computer Vision (Winter 2014) Problem Set 3 Due: Feb. 18 th, 2015 (11:59pm) 1 Single Object Recognition Via SIFT (45 points) In his 2004 SIFT paper, David Lowe demonstrates impressive object recognition
CS 231A Computer Vision (Winter 2014) Problem Set 3 Due: Feb. 18 th, 2015 (11:59pm) 1 Single Object Recognition Via SIFT (45 points) In his 2004 SIFT paper, David Lowe demonstrates impressive object recognition
Tutorial 7: Automated Peak Picking in Skyline
 Tutorial 7: Automated Peak Picking in Skyline Skyline now supports the ability to create custom advanced peak picking and scoring models for both selected reaction monitoring (SRM) and data-independent
Tutorial 7: Automated Peak Picking in Skyline Skyline now supports the ability to create custom advanced peak picking and scoring models for both selected reaction monitoring (SRM) and data-independent
Classification of Hyperspectral Breast Images for Cancer Detection. Sander Parawira December 4, 2009
 1 Introduction Classification of Hyperspectral Breast Images for Cancer Detection Sander Parawira December 4, 2009 parawira@stanford.edu In 2009 approximately one out of eight women has breast cancer.
1 Introduction Classification of Hyperspectral Breast Images for Cancer Detection Sander Parawira December 4, 2009 parawira@stanford.edu In 2009 approximately one out of eight women has breast cancer.
SUPPORTING INFORMATION. Maltose Binding Protein Effectively Stabilizes the Partially. Closed Conformation of the ATP-binding Cassette Transporter
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2017 SUPPORTING INFORMATION Maltose Binding Protein Effectively Stabilizes the Partially
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2017 SUPPORTING INFORMATION Maltose Binding Protein Effectively Stabilizes the Partially
Network-based auto-probit modeling for protein function prediction
 Network-based auto-probit modeling for protein function prediction Supplementary material Xiaoyu Jiang, David Gold, Eric D. Kolaczyk Derivation of Markov Chain Monte Carlo algorithm with the GO annotation
Network-based auto-probit modeling for protein function prediction Supplementary material Xiaoyu Jiang, David Gold, Eric D. Kolaczyk Derivation of Markov Chain Monte Carlo algorithm with the GO annotation
TEAMS National Competition Middle School Version Photometry 25 Questions
 TEAMS National Competition Middle School Version Photometry 25 Questions Page 1 of 13 Telescopes and their Lenses Although telescopes provide us with the extraordinary power to see objects miles away,
TEAMS National Competition Middle School Version Photometry 25 Questions Page 1 of 13 Telescopes and their Lenses Although telescopes provide us with the extraordinary power to see objects miles away,
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
3DEM > GENERAL METHODS. SINGLE PARTICLE RECONSTRUCTION (SPA): Introduction
 3DEM > GENERAL METHODS SINGLE PARTICLE RECONSTRUCTION (SPA): Introduction SINGLE PARTICLE RECONSTRUCTION > INTRODUCTION General principle of single particle reconstruction o 2D projection images of a 3D
3DEM > GENERAL METHODS SINGLE PARTICLE RECONSTRUCTION (SPA): Introduction SINGLE PARTICLE RECONSTRUCTION > INTRODUCTION General principle of single particle reconstruction o 2D projection images of a 3D
Small Libraries of Protein Fragments through Clustering
 Small Libraries of Protein Fragments through Clustering Varun Ganapathi Department of Computer Science Stanford University June 8, 2005 Abstract When trying to extract information from the information
Small Libraries of Protein Fragments through Clustering Varun Ganapathi Department of Computer Science Stanford University June 8, 2005 Abstract When trying to extract information from the information
This research aims to present a new way of visualizing multi-dimensional data using generalized scatterplots by sensitivity coefficients to highlight
 This research aims to present a new way of visualizing multi-dimensional data using generalized scatterplots by sensitivity coefficients to highlight local variation of one variable with respect to another.
This research aims to present a new way of visualizing multi-dimensional data using generalized scatterplots by sensitivity coefficients to highlight local variation of one variable with respect to another.
Supplementary Materials
 Supplementary Materials Microtubule doublets isolated from sea urchin sperm were studied by cryoelectron tomography, producing a density map in which we can identify a number of nontubulin components,
Supplementary Materials Microtubule doublets isolated from sea urchin sperm were studied by cryoelectron tomography, producing a density map in which we can identify a number of nontubulin components,
Intrinsic Classification of Single Particle Images by Spectral Clustering
 Intrinsic Classification of Single Particle Images by Spectral Clustering Yutaka Ueno 1, Masaki Kawata 2 and Shinji Umeyama 1 1 Neuroscience Research Institute, 2 Grid Technology Research Center, National
Intrinsic Classification of Single Particle Images by Spectral Clustering Yutaka Ueno 1, Masaki Kawata 2 and Shinji Umeyama 1 1 Neuroscience Research Institute, 2 Grid Technology Research Center, National
PERFORMANCE CAPTURE FROM SPARSE MULTI-VIEW VIDEO
 Stefan Krauß, Juliane Hüttl SE, SoSe 2011, HU-Berlin PERFORMANCE CAPTURE FROM SPARSE MULTI-VIEW VIDEO 1 Uses of Motion/Performance Capture movies games, virtual environments biomechanics, sports science,
Stefan Krauß, Juliane Hüttl SE, SoSe 2011, HU-Berlin PERFORMANCE CAPTURE FROM SPARSE MULTI-VIEW VIDEO 1 Uses of Motion/Performance Capture movies games, virtual environments biomechanics, sports science,
think of the molecular atoms or ligands L
 2 nd year Crystal and olecular Architecture course The Octahedral Point Group As an inorganic or materials chemist the octahedral point group is one of the most important, Figure o a vast number of T or
2 nd year Crystal and olecular Architecture course The Octahedral Point Group As an inorganic or materials chemist the octahedral point group is one of the most important, Figure o a vast number of T or
Variations of images to increase their visibility
 Variations of images to increase their visibility Amelia Carolina Sparavigna Department of Applied Science and Technology Politecnico di Torino, Torino, Italy The calculus of variations applied to the
Variations of images to increase their visibility Amelia Carolina Sparavigna Department of Applied Science and Technology Politecnico di Torino, Torino, Italy The calculus of variations applied to the
Identification of Shielding Material Configurations Using NMIS Imaging
 Identification of Shielding Material Configurations Using NMIS Imaging B. R. Grogan, J. T. Mihalczo, S. M. McConchie, and J. A. Mullens Oak Ridge National Laboratory, P.O. Box 2008, MS-6010, Oak Ridge,
Identification of Shielding Material Configurations Using NMIS Imaging B. R. Grogan, J. T. Mihalczo, S. M. McConchie, and J. A. Mullens Oak Ridge National Laboratory, P.O. Box 2008, MS-6010, Oak Ridge,
doi: /
 Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
ERROR RECOGNITION and IMAGE ANALYSIS
 PREAMBLE TO ERROR RECOGNITION and IMAGE ANALYSIS 2 Why are these two topics in the same lecture? ERROR RECOGNITION and IMAGE ANALYSIS Ed Fomalont Error recognition is used to determine defects in the data
PREAMBLE TO ERROR RECOGNITION and IMAGE ANALYSIS 2 Why are these two topics in the same lecture? ERROR RECOGNITION and IMAGE ANALYSIS Ed Fomalont Error recognition is used to determine defects in the data
Sept. 9, An Introduction to Bioinformatics. Special Topics BSC5936:
 Special Topics BSC5936: An Introduction to Bioinformatics. Florida State University The Department of Biological Science www.bio.fsu.edu Sept. 9, 2003 The Dot Matrix Method Steven M. Thompson Florida State
Special Topics BSC5936: An Introduction to Bioinformatics. Florida State University The Department of Biological Science www.bio.fsu.edu Sept. 9, 2003 The Dot Matrix Method Steven M. Thompson Florida State
ICON User Guide
 ICON 1.7.2 User Guide (Copyright: Fei Sun s lab in Institute of Biophysics & Fa Zhang s lab in Institute of Computation Technology, Chinese Academy of Sciences) 1. Installation Go into the installation
ICON 1.7.2 User Guide (Copyright: Fei Sun s lab in Institute of Biophysics & Fa Zhang s lab in Institute of Computation Technology, Chinese Academy of Sciences) 1. Installation Go into the installation
Lecture 8. Conical tilt reconstruction
 Lecture 8 Conical tilt reconstruction Central section theorem Euler angles Conical tilt series Missing cone artefact Early D studies and negative staining problems Perspectives and new trends Tilt series
Lecture 8 Conical tilt reconstruction Central section theorem Euler angles Conical tilt series Missing cone artefact Early D studies and negative staining problems Perspectives and new trends Tilt series
Gradient-Free Boundary Tracking* Zhong Hu Faculty Advisor: Todd Wittman August 2007 UCLA Department of Mathematics
 Section I. Introduction Gradient-Free Boundary Tracking* Zhong Hu Faculty Advisor: Todd Wittman August 2007 UCLA Department of Mathematics In my paper, my main objective is to track objects in hyperspectral
Section I. Introduction Gradient-Free Boundary Tracking* Zhong Hu Faculty Advisor: Todd Wittman August 2007 UCLA Department of Mathematics In my paper, my main objective is to track objects in hyperspectral
Mu lt i s p e c t r a l
 Viewing Angle Analyser Revolutionary system for full spectral and polarization measurement in the entire viewing angle EZContrastMS80 & EZContrastMS88 ADVANCED LIGHT ANALYSIS by Field iris Fourier plane
Viewing Angle Analyser Revolutionary system for full spectral and polarization measurement in the entire viewing angle EZContrastMS80 & EZContrastMS88 ADVANCED LIGHT ANALYSIS by Field iris Fourier plane
The Dot Matrix Method
 Special Topics BS5936: An Introduction to Bioinformatics. Florida State niversity The Department of Biological Science www.bio.fsu.edu Sept. 9, 2003 The Dot Matrix Method Steven M. Thompson Florida State
Special Topics BS5936: An Introduction to Bioinformatics. Florida State niversity The Department of Biological Science www.bio.fsu.edu Sept. 9, 2003 The Dot Matrix Method Steven M. Thompson Florida State
Database Searching Using BLAST
 Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Basic relations between pixels (Chapter 2)
 Basic relations between pixels (Chapter 2) Lecture 3 Basic Relationships Between Pixels Definitions: f(x,y): digital image Pixels: q, p (p,q f) A subset of pixels of f(x,y): S A typology of relations:
Basic relations between pixels (Chapter 2) Lecture 3 Basic Relationships Between Pixels Definitions: f(x,y): digital image Pixels: q, p (p,q f) A subset of pixels of f(x,y): S A typology of relations:
Pedestrian Detection Using Correlated Lidar and Image Data EECS442 Final Project Fall 2016
 edestrian Detection Using Correlated Lidar and Image Data EECS442 Final roject Fall 2016 Samuel Rohrer University of Michigan rohrer@umich.edu Ian Lin University of Michigan tiannis@umich.edu Abstract
edestrian Detection Using Correlated Lidar and Image Data EECS442 Final roject Fall 2016 Samuel Rohrer University of Michigan rohrer@umich.edu Ian Lin University of Michigan tiannis@umich.edu Abstract
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014
 Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
Patch-Based Image Classification Using Image Epitomes
 Patch-Based Image Classification Using Image Epitomes David Andrzejewski CS 766 - Final Project December 19, 2005 Abstract Automatic image classification has many practical applications, including photo
Patch-Based Image Classification Using Image Epitomes David Andrzejewski CS 766 - Final Project December 19, 2005 Abstract Automatic image classification has many practical applications, including photo
CHAPTER 6 PERCEPTUAL ORGANIZATION BASED ON TEMPORAL DYNAMICS
 CHAPTER 6 PERCEPTUAL ORGANIZATION BASED ON TEMPORAL DYNAMICS This chapter presents a computational model for perceptual organization. A figure-ground segregation network is proposed based on a novel boundary
CHAPTER 6 PERCEPTUAL ORGANIZATION BASED ON TEMPORAL DYNAMICS This chapter presents a computational model for perceptual organization. A figure-ground segregation network is proposed based on a novel boundary
Practical 10: Protein dynamics and visualization Documentation
 Practical 10: Protein dynamics and visualization Documentation Release 1.0 Oliver Beckstein March 07, 2013 CONTENTS 1 Basics of protein structure 3 2 Analyzing protein structure and topology 5 2.1 Topology
Practical 10: Protein dynamics and visualization Documentation Release 1.0 Oliver Beckstein March 07, 2013 CONTENTS 1 Basics of protein structure 3 2 Analyzing protein structure and topology 5 2.1 Topology
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST
 An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
CSE/EE-576, Final Project
 1 CSE/EE-576, Final Project Torso tracking Ke-Yu Chen Introduction Human 3D modeling and reconstruction from 2D sequences has been researcher s interests for years. Torso is the main part of the human
1 CSE/EE-576, Final Project Torso tracking Ke-Yu Chen Introduction Human 3D modeling and reconstruction from 2D sequences has been researcher s interests for years. Torso is the main part of the human
Week 7 Picturing Network. Vahe and Bethany
 Week 7 Picturing Network Vahe and Bethany Freeman (2005) - Graphic Techniques for Exploring Social Network Data The two main goals of analyzing social network data are identification of cohesive groups
Week 7 Picturing Network Vahe and Bethany Freeman (2005) - Graphic Techniques for Exploring Social Network Data The two main goals of analyzing social network data are identification of cohesive groups
Lab Report: Optical Image Processing
 Lab Report: Optical Image Processing Kevin P. Chen * Advanced Labs for Special Topics in Photonics (ECE 1640H) University of Toronto March 5, 1999 Abstract This report describes the experimental principle,
Lab Report: Optical Image Processing Kevin P. Chen * Advanced Labs for Special Topics in Photonics (ECE 1640H) University of Toronto March 5, 1999 Abstract This report describes the experimental principle,
Assembly dynamics of microtubules at molecular resolution
 Supplementary Information with: Assembly dynamics of microtubules at molecular resolution Jacob W.J. Kerssemakers 1,2, E. Laura Munteanu 1, Liedewij Laan 1, Tim L. Noetzel 2, Marcel E. Janson 1,3, and
Supplementary Information with: Assembly dynamics of microtubules at molecular resolution Jacob W.J. Kerssemakers 1,2, E. Laura Munteanu 1, Liedewij Laan 1, Tim L. Noetzel 2, Marcel E. Janson 1,3, and
FLUENT Secondary flow in a teacup Author: John M. Cimbala, Penn State University Latest revision: 26 January 2016
 FLUENT Secondary flow in a teacup Author: John M. Cimbala, Penn State University Latest revision: 26 January 2016 Note: These instructions are based on an older version of FLUENT, and some of the instructions
FLUENT Secondary flow in a teacup Author: John M. Cimbala, Penn State University Latest revision: 26 January 2016 Note: These instructions are based on an older version of FLUENT, and some of the instructions
Spiraling close-packers of 3D space
 Spiraling close-packers of 3D space Frans Snik Abstract Utrecht University, the Netherlands f.snik@astro.uu.nl This paper presents a new class of three-dimensional mathematical objects that closepack to
Spiraling close-packers of 3D space Frans Snik Abstract Utrecht University, the Netherlands f.snik@astro.uu.nl This paper presents a new class of three-dimensional mathematical objects that closepack to
ACCURATE TEXTURE MEASUREMENTS ON THIN FILMS USING A POWDER X-RAY DIFFRACTOMETER
 ACCURATE TEXTURE MEASUREMENTS ON THIN FILMS USING A POWDER X-RAY DIFFRACTOMETER MARK D. VAUDIN NIST, Gaithersburg, MD, USA. Abstract A fast and accurate method that uses a conventional powder x-ray diffractometer
ACCURATE TEXTURE MEASUREMENTS ON THIN FILMS USING A POWDER X-RAY DIFFRACTOMETER MARK D. VAUDIN NIST, Gaithersburg, MD, USA. Abstract A fast and accurate method that uses a conventional powder x-ray diffractometer
