ssrna Example SASSIE Workflow Data Interpolation
|
|
|
- Elisabeth Barker
- 6 years ago
- Views:
Transcription
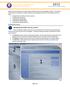
1 NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at Return to Main Documents Page ssrna Example SASSIE Workflow In this workflow example we will model the conformation of a portion of an intron from the HIV-1 viral single-stranded RNA. Retroviral RNAs are often exported from the cell nucleus with some or all of their introns. These RNAs eventually serve as genomes (encoding for Gag and other viral proteins) that are encapsulated in virions that leave the cell to infect other cells. The RNA portion to be examined consists of 80 nucleotides and is called trunc2a. A starting structure was built using the RNAComposer tool. The energyminimized structure, which is divided into 5 different regions, is shown in the figure above. The regions are listed by residues in the table alongside the structure. The 2 regions that were identified as flexible (colored red and green) are indicated in the table. It is not known whether the base pairings in the other 3 regions remain intact in solution. Therefore, SAXS data were obtained on the trunc2a sample at concentrations of 1.0 and 1.42 mg/ml. The data were consistent at these two concentrations, so the higher concentration will be used here for modeling. The purpose of this exercise is to test whether good fits to the SAXS data can be obtained by allowing only the green and red regions in the figure to be flexible. If structures with large enough Rg values can't be obtained, it will be necessary to allow some of the residues in the blue region to be flexible also in order to "open" up the structure and allow structures with larger Rg values to be sampled. To begin, log into your SASSIE-web account and choose an existing project or create a new project for your work. In this example, the project name is test. Download the trunc2a SAXS data and the minimized starting trunc2a structure. Data Interpolation Here we interpolate the SAXS data from trunc2a ssrna at a concentration of 1.42 mg/ml. More information on the Data Interpolation module is available in the Data Interpolation documentation. Select the 'Data Interpolation' button from this 'Tools' menu. You should now see a page like the one below. This page is used to enter all of the information needed to do the data interpolation. The figure shows the values for each field as required for data interpolation. Edit the values on your screen to match the screenshot. An explanation of the field and how to edit it can be found below.
2 run name: user defined name of folder that will contain the results. experimental data file: Name of input file with experimental data with at least three columns: q, I(q), and error in I(q). Here we use the trunc2a_saxs.sub file. output file name: Name of file that will contain the interpolated data. Here we choose the name trunc.dat. I(0): Experimentally determined value of scattering intensity at q = 0. Here we used the value of that was derived from a Guinier fit to the data. I(0) error: Experimentally determined value of the error of the scattering intensity at q = 0. Here we use the value of that was obtained from the Guinier fit to the data. new delta q: Desired spacing of q-values (1/Angstrom). This should be chosen so that your first interpolated data point falls within the q-range of the experimental data. For this tutorial, the value has been set to since the first data point occurs at a value of ~ number of new q-values: Integer number of desired q-values. For this tutorial, the value has been set to 72 to that the maximum q value is ~0.5. Click on the 'Submit' button to start simulation. Once complete the output should look similar to the figure below. The output will show a plot of the original and interpolated data, the name of the input file and the name of the interpolated data file as well as the directory in which it is located. Note that roll-over help will indicate options to resize, zoom and reset the view of the plot. What have we generated: test/run_0/data_interpolation trunc.dat: text file containing the interpolated SANS data
3 stn_trunc.dat: text file containing a the interpolated SANS data truncated at the q-value where the signal-to-noise drops below a value of 2 PDB Scan and Structure Minimization PDB Scan only supports PDB files for proteins at this time. Therefore, the ssrna PDB file had to be checked and corrected to conform to the CHARMM naming convention by hand. As mentioned above, the starting structure was minimized prior to the simulation. This can be done in SASSIE using the Energy Minimization module. More information can be found in the Energy Minimization documentation. Structure Variation - Monte Carlo (Monomer) Here we use the Monomer Monte Carlo module to vary the ssrna structure. More information can be found in the Monomer Monte Carlo documentation. Select the 'Simulate' button from the Main Menu of SASSIE-web and then click on the 'Monomer Monte Carlo' button. You should now see a page like the one below. This page is used to enter all of the information needed to run a Monte Carlo simulation. The figure shows the values for each field as required for our simulation. An explanation of some of the fields can be found below. reference pdb: The starting structure for the simulation. Here we use the trunc2a_min.pdb file. number of trial attempts: Number of times the simulation will try to vary the structure (some structures will be discarded by the Monte Carlo algorithm) For this tutorial set the value to For real studies tens of thousands of structures are needed. return to previous structure: Number of discarded structures in a row that are considered before returning to a randomly-selected structure that was previously accepted number of flexible regions to vary: single number residue range for each flexible region: comma-separated list of the range of residues to vary for each flexible region maximum angle(s): comma-separated list of the maximum angle sampled in a single Monte Carlo step for each flexible region structure alignment region: a single range of residues for structural alignment of all the flexible segments. This makes it easy to make visual comparisons of each frame in the output trajectory. overlap basis: Select either heavy atoms, all, backbone or enter atom name. The atom name option will spawn futher inputs: overlap basis: Enter an atom name to check for overlap. overlap cutoff (angsgtroms): Overlap basis atoms closer than this distance defines an overlap condition. Once you have understood the input fields and made sure that your values agree with the figure click on the 'Submit' button to start simulation. As the run continues the progress bar beneath the submit button should update. When the run is finished, some statistics are output in the white output box. Notice that 484 out of 1000 structures were accepted in this case. A graph beneath this should will show the variation of the radius of gyration over the steps of the Monte Carlo simulation. Once complete the output should look similar to the figure below.
4 What have we generated: test/run_0/monomer_monte_carlo run_0.dcd: DCD file containing all of the structures accepted by out Monte Carlo simulation run_0.dcd.accepted_rg_results_data.txt: text file containing radius of gyration for all structures that made it into the DCD file run_0.dcd.all_rg_results_data.txt: text file containing radius of gyration for all structures generated run_0.dcd.stats: text file containing statistics for our Monte Carlo run Visualization You should now download the output trajectory using the file browser. Click on the filing cabinet icon in the Session Management area. Note: the 'Configurations and statistics saved in' line in the output gives a relative path under the project directory. Click on the triangle next to the 'test' directory name to reveal the 'run_0' directory created by our simulation. Check the box next to 'run_0' to select it for download. Beneath the file tree is an option labelled 'Compression type'. Select an option suitable for your operating system from the list (for Windows select 'zipped' for Linux 'bzip2 tarball'). Click the 'Download' button. A progress bar will appear monitoring the upload of your files to the server. Once complete a link will appear beneath the download button.
5 Click on this to download your data. Once the download is complete uncompress the file in a location of your choice. You should now load the PDB (run_0/monomer_monte_carlo/trunc2a_min.pdb) and DCD (run_0/monomer_monte_carlo/run_0.dcd) into VMD to observe the variation produced even in our very short Monte Carlo simulation. Note that both of these files can be found in the run_0 directory you just downloaded. Remember the DCD file contains coordinates alone, you need to load the PDB first so that the visualization software knows about the atoms they represent and how they are connected. Initial SAS Curve Calculation - Single Structure We will use SasCalc to calculate the SAXS curves from the generated structures. More information can be found in the SasCalc documentation. The starting structure must be a complete structure without missing residues or atoms (including hydrogen atoms) in order to obtain accurate scattering profiles. Atom and residue naming must be compatiable with those defined in the CHARMM force field. The SasCalc module is first run using the "converged number of golden vectors" option on just one structure. The SASSIE workflow operates by calculating the scattering intensities at evenly spaced Q values and matching these against interpolated experimental values. The file trunc.dat contains our previously interpolated experimental data. In order to create the correct data points in our theoretical curves we need three pieces of information: To begin: Intensity and Q=0, I(0): Maximum value of Q: (units are inverse Angstroms) Number of points in the curve: 72 Select 'Beta' from the Main Menu. Click the 'SasCalc' button. Now you need to enter the information to run the scattering calculator. SasCalc can be used to calculate the scattering for SAXS and SANS and/or for several SANS contrasts at the same time. Other than the values listed above you can keep the default values for this tutorial, except for the Advanced Input section. Since this is the first time we are running SasCalc on this structure, we are using the "converged number of golden vectors" option. Choose this option from the SasCalc method menu in the Advanced Input section of the page (see figure below). The single structure that we used to start the simulation is used as both the reference pdb and the trajectory file filename (PDB in this case) so it is already uploaded to the SASSIE-web server. Thus, you can either upload it again from your local computer or locate it on the server and read it from there. To read the file from the server:
6 Click on the 'Browse server' button next to the appropriate field. Navigate to the file test/run_0/monomer_monte_carlo/trunc2a_min.pdb Click 'OK' Click on the 'Submit' button to start the calculation. A scattering curve will be calculated for the starting structure (the progress bar should reach 100% and a message stating the run finished appear in the window beneath when the job has completed). Note that the files are written to a the directory sascalc/xray. What have we generated: test/run_0/sascalc/xray run_0_00001.iq: files containing theoretical scattering data run_0_00001.log: log files containing information about the structure and calculation inputs Second SAS Curve Calculation - Entire Ensemble Next we calculate a theoretical scattering curve for each of the trial structures we have generated. The run_0_00001.log file from the inital SAS Curve calculation indicates that 43 golden vectors were required for convergence to the desired tolerance (0.01 in this case). We now use this information to calculate the scattering curves for all of the generated structures using the "fixed number of golden vectors" option from the SasCalc method menu as shown below. Since 484 structures are not sufficient to perform a complete analysis of conformation space, we will use a previously generated DCD file run_0_large.dcd that has almost 10,000 structures for further analysis. Download this file and use it as the trajectory filename (see figure below).
7 reference pdb: the starting structure that is already on the SASSIE-web server trajectory file filename: the run_0_large.dcd file that you just downloaded When all input fields are complete: Click 'Submit'. NOTE: This job takes ~16 min using GPUs, including setup time. Total time can be checked in the Jobs Manager after the job is finished. A scattering curve will be calculated for all of the structures generated by the Monte Carlo simulation (the progress bar should reach 100% and a message stating the run finished appear in the window beneath when the job has completed). Note that the files written during the initial SAS curve calculation will be overwritten since we chose the same run name (run_0) in both cases. If you wish to save the files from the initial calculation, use a different run name. What have we generated: test/run_0/sascalc/xray *.iq: files containing theoretical scattering data for all frames in the DCD file (9659 files in this case). *.log: log files containing information about each structure and calculation inputs (9659 files in this case).
8 Initial SAS Curve Comparison Now we compare our theoretical curves to the experimental data to see which of our structures are plausible models of the real protein using Chi-Square Filter. More information can be found in the Chi-Square Filter documention. Select 'Analyze' from the Main Menu. Click the 'Chi-Square Filter' button. We now need to select the path containing the theoretical scattering curves and the file containing the experimental data. In addition we need to input the value of I(0) to enable comparison of the two curves (see the picture below). To set the path to the scattering curves generated in the previus step: Click on the 'Browse server for a path' button Navigate to the test project folder and select the run_0/sascalc/xray folder (see figure below) Click 'OK' interpolated data file: I(0): Click on the 'Choose File' button Navigate to and select the trunc.dat file on your local computer or on the server. Click 'OK' Enter the value We eventually may want to create 'weight files' that record which frames meet criteria that make them successful models of our data. This means those with low chi square values. However, we don't know the range of chi square values we have at this stage. So, we set the 'number of weight files' to 0 at this time. Sas type: Choose SasCalc from the menu number of weight files: Set this to 0 using the down arrow associated with the input box. If you type '0' in the box, press the TAB button to make sure this value is
9 accepted. Note: There are list boxes that allow the selection of the format of the input theoretical curves and the metric used to compare the curves. Here we wish to use the defaults of 'SasCalc' and 'reduced chi-square'. Click 'Submit'. Once complete you should see outputs similar to those below.
10 In the text output you will see the minimum chi square (X2) values is given. The top plot shows the variation of chi squared (y-axis) with the radius of gyration (x-axis). Chi squared is a measure of the quality of fit of the theoretical curve to the experimental one. It is a percentage and the lower the value the better. The bottom plot shows a direct comparison of the best, worst and average theoretical curves with experiment (goal). Examine the x2 vs Rg plot using the mouse commands: drag to pan, double-click to zoom, to reset zoom and pan: click on title, axis labels or live coordinates box. What have we generated: test/run_0/chi_square_filter/xray /spectra averagefile.txt: average scattering curve for all structures bestworstfile.txt: best and worst scattering curves selected from all structures sas_spectra_plot.txt: goal, best, worst and average scattering curves x2_vs_rg_plot.txt: chi squared against radius of gyration for all structures x2file.txt: chi squared for all structures spec_*.ciq*: scattering curves scaled to correct I(0) for each structure Second SAS Curve Comparison
11 Now that we know the range of chi square values that we have, we can compare the theoretical curves to the data a second time and create two weight files that flag all structures with chi square values below a certain number. Now, we set the 'number of weight files' to 2. run name: Since we already have a chi_square_filter folder in the run_0 directory, set the run name to run_1. number of weight files: Set this to 2 using the down arrow associated with the input box. Weight files contain information on which frames in our simulation meet specific criteria provided in the expression box. enter expression[1]: Enter the following expression: x2 < 1 This selects all frames with a chi square less than 1. weight file name[1]: Enter x2_lt_1.txt low Rg cutoff[1]: Enter a value if you wish to also restrict the Rg range to be above this value. The default value is 0 so that all Rg values are acceptable. enter expression[2]: Now enter the following expression for the second weight file: x2 < 0.5 This selects all frames with a chi square less than 3.0. Adjust this value if necessary to suit the results from your simulation. weight file name[2]:
12 Enter x2_lt_0.5.txt low Rg cutoff[2]: Enter a value if you wish to also restrict the Rg range to be above this value. The default value is 0 so that all Rg values are acceptable. Click 'Submit'. Once complete the outputs should be exactly the same as for the previous run. The only difference is that the two weight files can now be found in the chi_square_filter/xray folder. What have we generated: test/run_1/chi_square_filter/xray /spectra averagefile.txt: average scattering curve for all structures bestworstfile.txt: best and worst scattering curves selected from all structures sas_spectra_plot.txt: goal, best, worst and average scattering curves x2_vs_rg_plot.txt: chi squared against radius of gyration for all structures x2file.txt: chi squared for all structures x2_lt_1.txt: weights file selecting only frames with chi squared < 1. x2_lt_0.5.txt: weights file selecting only frames with chi squared < 0.5 spec_*.ciq: scattering curves scaled to correct I(0) for each structure Trajectory Filtering Now we can filter out the best fit structures and vizualize them using the Extract Utilities. More information can be found in the Extract Utilities documentation. We will extract the frames from run_0_large.dcd that fit the data with x2 values less than 0.5. Select 'Tools' from the Main Menu. Click the 'Extract Utilities' button. In this module we can select structures from run_0_large.dcd using the weight files generated in the Chi-Square Filter module. In this case, we chose to select the weight file from the server. Check the tick box labelled 'extract trajectory' (this will reveal the options shown in the screenshot) Select the trunc2a_min.pdb 'reference pdb' and the run_0_large.dcd file from the server (or from your local computer). Input 'run_1_0.5wt.dcd' as the 'output filename'
13 Choose 'weight file' from the 'select option' listbox. Where it says 'input name of weight file' select the 'x2_lt_0.5.txt' file generated in the last step that selects only the frames which had a chi squared value of less 0.5. Click 'Submit' When the process is finished your output should look like the one below. What have we generated: test/run_1/extract_utilities run_1_0.5wt.dcd: DCD containing only the frames for which the theoretical scattering curve is a good match to experiment. Visualization Download the 'run_1_0.5wt.dcd' file as you did the unfiltered DCD and then vizualize both the 'run_0_large.dcd' and 'run_1_0.5wt.dcd' trajectories in VMD. (You will need to load a suitable PDB in both cases as before). Can you tell if the filtered structures are more compact than many of those in the unfiltered DCD? This is often difficult to do by examining the trajectories directly. Density Plot Another way to visualize the structures sampled in the 'run_0_large.dcd' and 'run_1_0.5wt.dcd' files for comparison is to use the Density Plot module. The purpose of the Density Plot module is to generate files with volumetric data to visualize results. Often, this is used to visualize sub-ensembles of structures that are in agreement with experimental data. More information can be found in the Density Plot documentation. The density plot below shows the envelope sampled by all of the accepted structures as well as that sampled by only the best fit structures with x2 < 0.5. The blue region is the envelope represented by residues 31-46, which is essentially the fixed alignment region that we defined in the Monomer Monte Carlo module. The red regions represent the envelope sampled by residues 1-30 for all accepted structures (mesh) and for the best fit structures (solid). The green regions represent the envelope sampled by residues for all accepted structures (mesh) and the best fit structures (solid). This representation makes it easier to see that the envelope represented by the best fit structures is significantly smaller than that represented by all of the accepted structures.
14 Instructions on how to make the above density plot can be found here. Final SAS Curve Comparison Now, we can compare the structures with x2 < 0.5 to the SAXS data by calculating their theoretical SAXS curves and comparing them to the SAXS data again. First, calculate the theoretical SAXS curves using SasCalc. run name: Set the run name to run_2. Once the inputs have been entered, click 'Submit'.
15 Once the run is complete, you should see outputs like those below. What have we generated: test/run_2/sascalc/xray *.iq: files containing theoretical scattering data for all frames in the DCD file (692 files in this case). *.log: log files containing information about each structure and calculation inputs (692 files in this case). Then, compare the theoretical SAXS curves to the SAXS data using Chi-Square Filter. run name: Since we already have a chi_square_filter folder in the run_1 directory, set the run name to run_2. number of weight files Set the 'number of weight files' to 0 since we are already dealing with the best fit structures. Click 'Submit'. Once the run is complete, you should see outputs like those below.
16 Notice that the best, worst and ensemble average curves now all fit the data well. This means that any single structure in the ensemble of structures with x2 < 0.5 and any combination of structures in the ensemble, including the entire ensemble itself, represents a good fit to the SAXS data. What have we generated:
17 test/run_2/chi_square_filter /spectra averagefile.txt: average scattering curve for all structures bestworstfile.txt: best and worst scattering curves selected from all structures sas_spectra_plot.txt: goal, best, worst and average scattering curves x2_vs_rg_plot.txt: chi squared against radius of gyration for all structures x2file.txt: chi squared for all structures spec_*.ciq: scattering curves scalled to correct I(0) for each structure Visualization Download the contents of the chi_square_filter directory and reproduce the output plots. This can be accomplished by plotting x2 vs Rg from the x2_vs_rg_plot.txt file and by plotting the goal, best, worst and average SAXS curves vs q from the sas_spectra_plot.txt file using your favorite plotting program. Minimization of Structures The structures in trajectories such as run_0_large.dcd or run_1_0.5wt.dcd should be minimized for at least 1000 steps if the trajectories will undergo further processing. More information can be found in the Energy Minimization documentation. References 1. SASSIE: A program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints J. E. Curtis, S. Raghunandan, H. Nanda, S. Krueger, Comp. Phys. Comm. 183, (2012). BIBTeX, EndNote, Plain Text Return to Main Documents Page Go to top Supported via CCP-SAS a joint EPSRC (EP/K039121/1) and NSF (CHE ) grant
SASSIE-web: Quick Start Introduction. SASSIE-web Interface
 NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/docs/sassie-web-quick-start/quick-start.html. Return to Main Documents
NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/docs/sassie-web-quick-start/quick-start.html. Return to Main Documents
REDCAT LABORATORY EXERCISE BCMB/CHEM 8190, April 13, REDCAT may be downloaded from:
 REDCAT LABORATORY EXERCISE BCMB/CHEM 8190, April 13, 2012 Installation note for instructors: REDCAT may be downloaded from: http://ifestos.cse.sc.edu/software.php#redcat You will also find further instructions
REDCAT LABORATORY EXERCISE BCMB/CHEM 8190, April 13, 2012 Installation note for instructors: REDCAT may be downloaded from: http://ifestos.cse.sc.edu/software.php#redcat You will also find further instructions
Chimera EM Map Tutorial: RNA Polymerase II
 Chimera EM Map Tutorial: RNA Polymerase II May 1, 2007 This tutorial focuses on display of volume data from single particle EM reconstructions. We'll look at maps of two conformations of human RNA polymerase
Chimera EM Map Tutorial: RNA Polymerase II May 1, 2007 This tutorial focuses on display of volume data from single particle EM reconstructions. We'll look at maps of two conformations of human RNA polymerase
static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, i
 GLIDE static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
GLIDE static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
The scan tools are used to convert and edit bitmap images so that they can be cut or routed by the Torchmate.
 Scan Tools The scan tools are used to convert and edit bitmap images so that they can be cut or routed by the Torchmate. Upon clicking the Scan tool, the flyout opens exposing the two scanning options.
Scan Tools The scan tools are used to convert and edit bitmap images so that they can be cut or routed by the Torchmate. Upon clicking the Scan tool, the flyout opens exposing the two scanning options.
fitted solution envelope of native cyt. c
 10-1 I(q) (cm -1 ) 10-2 10-3 fitted solution envelope of native cyt. c 10-4 0.1 q (Å -1 ) Acknowledge EMBL Hamburg BioSAXS team memebers effort in ATSAS software package. The data analysis software, ATSAS
10-1 I(q) (cm -1 ) 10-2 10-3 fitted solution envelope of native cyt. c 10-4 0.1 q (Å -1 ) Acknowledge EMBL Hamburg BioSAXS team memebers effort in ATSAS software package. The data analysis software, ATSAS
Tutorial. De Novo Assembly of Paired Data. Sample to Insight. November 21, 2017
 De Novo Assembly of Paired Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
De Novo Assembly of Paired Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Tutorial: De Novo Assembly of Paired Data
 : De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
REDCAT INSTALLATION. BCMB/CHEM 8190, April 7,
 REDCAT INSTALLATION BCMB/CHEM 8190, April 7, 2006 Installation note for instructors: Download REDCAT from: http://tesla.ccrc.uga.edu/software/redcat While REDCAT is developed on and for Linux systems,
REDCAT INSTALLATION BCMB/CHEM 8190, April 7, 2006 Installation note for instructors: Download REDCAT from: http://tesla.ccrc.uga.edu/software/redcat While REDCAT is developed on and for Linux systems,
RUNNING MOLECULAR DYNAMICS SIMULATIONS WITH CHARMM: A BRIEF TUTORIAL
 RUNNING MOLECULAR DYNAMICS SIMULATIONS WITH CHARMM: A BRIEF TUTORIAL While you can probably write a reasonable program that carries out molecular dynamics (MD) simulations, it s sometimes more efficient
RUNNING MOLECULAR DYNAMICS SIMULATIONS WITH CHARMM: A BRIEF TUTORIAL While you can probably write a reasonable program that carries out molecular dynamics (MD) simulations, it s sometimes more efficient
Lab III: MD II & Analysis Linux & OS X. In this lab you will:
 Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
1. Open the SPDBV_4.04_OSX folder on the desktop and double click DeepView to open.
 Molecular of inhibitor-bound Lysozyme This lab will not require a lab report. Rather each student will follow this tutorial, answer the italicized questions (worth 2 points each) directly on this protocol/worksheet,
Molecular of inhibitor-bound Lysozyme This lab will not require a lab report. Rather each student will follow this tutorial, answer the italicized questions (worth 2 points each) directly on this protocol/worksheet,
Lab III: MD II & Analysis Linux & OS X. In this lab you will:
 Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
JobSite OnLine User s Guide Table of Contents
 JobSite OnLine User s Guide Table of Contents For the Beginner: 2 Preparing and Logging In to Run JobSite OnLine...2 How Drawings and Specs are Organized - Ease of Use...3 Searching for Files or Containers...4
JobSite OnLine User s Guide Table of Contents For the Beginner: 2 Preparing and Logging In to Run JobSite OnLine...2 How Drawings and Specs are Organized - Ease of Use...3 Searching for Files or Containers...4
Genome Browsers - The UCSC Genome Browser
 Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Using OPUS to Process Evolved Gas Data (8/12/15 edits highlighted)
 Using OPUS to Process Evolved Gas Data (8/12/15 edits highlighted) Once FTIR data has been acquired for the gases evolved during your DSC/TGA run, you will process using the OPUS software package. Select
Using OPUS to Process Evolved Gas Data (8/12/15 edits highlighted) Once FTIR data has been acquired for the gases evolved during your DSC/TGA run, you will process using the OPUS software package. Select
APBS electrostatics in VMD
 APBS electrostatics in VMD The VMD molecular graphics software package provides support for both the execution of APBS and the visualization of the resulting electrostatic potentials. Documentation on
APBS electrostatics in VMD The VMD molecular graphics software package provides support for both the execution of APBS and the visualization of the resulting electrostatic potentials. Documentation on
Visual Physics - Introductory Lab Lab 0
 Your Introductory Lab will guide you through the steps necessary to utilize state-of-the-art technology to acquire and graph data of mechanics experiments. Throughout Visual Physics, you will be using
Your Introductory Lab will guide you through the steps necessary to utilize state-of-the-art technology to acquire and graph data of mechanics experiments. Throughout Visual Physics, you will be using
40. Sim Module - Common Tools
 HSC Sim Common Tools 15021-ORC-J 1 (33) 40. Sim Module - Common Tools Table of Contents 40.1. Drawing flowsheets and adding tables to flowsheets... 2 40.1.1. Drawing units... 2 40.1.2. Drawing streams...
HSC Sim Common Tools 15021-ORC-J 1 (33) 40. Sim Module - Common Tools Table of Contents 40.1. Drawing flowsheets and adding tables to flowsheets... 2 40.1.1. Drawing units... 2 40.1.2. Drawing streams...
Chapter 2 Surfer Tutorial
 Chapter 2 Surfer Tutorial Overview This tutorial introduces you to some of Surfer s features and shows you the steps to take to produce maps. In addition, the tutorial will help previous Surfer users learn
Chapter 2 Surfer Tutorial Overview This tutorial introduces you to some of Surfer s features and shows you the steps to take to produce maps. In addition, the tutorial will help previous Surfer users learn
Overview. Experiment Specifications. This tutorial will enable you to
 Defining a protocol in BioAssay Overview BioAssay provides an interface to store, manipulate, and retrieve biological assay data. The application allows users to define customized protocol tables representing
Defining a protocol in BioAssay Overview BioAssay provides an interface to store, manipulate, and retrieve biological assay data. The application allows users to define customized protocol tables representing
USER GUIDE PowerPhoto CRM
 USER GUIDE PowerPhoto CRM 2013 2015 Contents CONFIGURING POWERPHOTO Add, Remove, Set Default, Set Record Image, Thumbnail & View Text Display Original File Save Original File Resize Photo Resize Height
USER GUIDE PowerPhoto CRM 2013 2015 Contents CONFIGURING POWERPHOTO Add, Remove, Set Default, Set Record Image, Thumbnail & View Text Display Original File Save Original File Resize Photo Resize Height
Getting Started with DADiSP
 Section 1: Welcome to DADiSP Getting Started with DADiSP This guide is designed to introduce you to the DADiSP environment. It gives you the opportunity to build and manipulate your own sample Worksheets
Section 1: Welcome to DADiSP Getting Started with DADiSP This guide is designed to introduce you to the DADiSP environment. It gives you the opportunity to build and manipulate your own sample Worksheets
ACAS Tutorial. Simple Experiment Loader. Contents:
 Simple Experiment Loader Contents: Overall Workflow Loading a Generic Data File Loading a Concentration Data File Adding a Report File Adding Images Validation Errors Editing a Protocol Adding Files to
Simple Experiment Loader Contents: Overall Workflow Loading a Generic Data File Loading a Concentration Data File Adding a Report File Adding Images Validation Errors Editing a Protocol Adding Files to
MassHunter File Reader
 MassHunter File Reader vers 1.0.0 2015 Quadtech Associates, Inc. All Rights Reserved Release date: November 18, 2015 www.quadtechassociates.com MassHunter File Reader Welcome to MassHunter File Reader.
MassHunter File Reader vers 1.0.0 2015 Quadtech Associates, Inc. All Rights Reserved Release date: November 18, 2015 www.quadtechassociates.com MassHunter File Reader Welcome to MassHunter File Reader.
Tutorial: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and Expression measures
 : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
PSA-Manager Version 3 User Manual
 PSA-Manager Version 3 User Manual CONTENTS 1 Introduction...2 2 File Types and Locations...3 3 PSA-Manager Windows...4 4 Viewing Trace Files...7 4.1.1 The Trace Memo sub-window...8 4.1.2 Printing Trace
PSA-Manager Version 3 User Manual CONTENTS 1 Introduction...2 2 File Types and Locations...3 3 PSA-Manager Windows...4 4 Viewing Trace Files...7 4.1.1 The Trace Memo sub-window...8 4.1.2 Printing Trace
General Guidelines: SAS Analyst
 General Guidelines: SAS Analyst The Analyst application is a data analysis tool in SAS for Windows (version 7 and later) that provides easy access to basic statistical analyses using a point-and-click
General Guidelines: SAS Analyst The Analyst application is a data analysis tool in SAS for Windows (version 7 and later) that provides easy access to basic statistical analyses using a point-and-click
Tutorial (Beginner level): Orthomosaic and DEM Generation with Agisoft PhotoScan Pro 1.3 (with Ground Control Points)
 Tutorial (Beginner level): Orthomosaic and DEM Generation with Agisoft PhotoScan Pro 1.3 (with Ground Control Points) Overview Agisoft PhotoScan Professional allows to generate georeferenced dense point
Tutorial (Beginner level): Orthomosaic and DEM Generation with Agisoft PhotoScan Pro 1.3 (with Ground Control Points) Overview Agisoft PhotoScan Professional allows to generate georeferenced dense point
STIPlotDigitizer. User s Manual
 STIPlotDigitizer User s Manual Table of Contents What is STIPlotDigitizer?... 3 Installation Guide... 3 Initializing STIPlotDigitizer... 4 Project GroupBox... 4 Import Image GroupBox... 5 Exit Button...
STIPlotDigitizer User s Manual Table of Contents What is STIPlotDigitizer?... 3 Installation Guide... 3 Initializing STIPlotDigitizer... 4 Project GroupBox... 4 Import Image GroupBox... 5 Exit Button...
Client Care Desktop V4
 Client Care Desktop V4 V4.1 Quay Document Manager V4.1 Contents 1. LOCATIONS...3 1.1. Client... 3 1.2. Holdings... 4 1.3. Providers... 4 1.4. Contacts/Introducers... 5 1.5. Adviser... 5 2. ADDING EXISTING
Client Care Desktop V4 V4.1 Quay Document Manager V4.1 Contents 1. LOCATIONS...3 1.1. Client... 3 1.2. Holdings... 4 1.3. Providers... 4 1.4. Contacts/Introducers... 5 1.5. Adviser... 5 2. ADDING EXISTING
Instructions for automatically masking a black background in Agisoft PhotoScan
 A Simple Photogrammetry Rig for the Reliable Creation of 3D Artifact Models in the Field: Lithic Examples from the Early Upper Paleolithic Sequence of Les Cottés (France) Instructions for automatically
A Simple Photogrammetry Rig for the Reliable Creation of 3D Artifact Models in the Field: Lithic Examples from the Early Upper Paleolithic Sequence of Les Cottés (France) Instructions for automatically
Chemical Shift Perturbation Tutorial
 CcpNmr Analysis Version 3 Chemical Shift Perturbation Tutorial Written by: Luca Mureddu CCPN application developer, PhD Student MCB, University of Leicester, Lancaster Road, Leicester, LE1 9HN, UK 1 t:
CcpNmr Analysis Version 3 Chemical Shift Perturbation Tutorial Written by: Luca Mureddu CCPN application developer, PhD Student MCB, University of Leicester, Lancaster Road, Leicester, LE1 9HN, UK 1 t:
Introduction to the workbook and spreadsheet
 Excel Tutorial To make the most of this tutorial I suggest you follow through it while sitting in front of a computer with Microsoft Excel running. This will allow you to try things out as you follow along.
Excel Tutorial To make the most of this tutorial I suggest you follow through it while sitting in front of a computer with Microsoft Excel running. This will allow you to try things out as you follow along.
MetaMorph Standard Operation Protocol Basic Application
 MetaMorph Standard Operation Protocol Basic Application Contents Basic Navigation and Image Handling... 2 Opening Images... 2 Separating Multichannel Images... 2 Cropping an Image... 3 Changing an 8 bit
MetaMorph Standard Operation Protocol Basic Application Contents Basic Navigation and Image Handling... 2 Opening Images... 2 Separating Multichannel Images... 2 Cropping an Image... 3 Changing an 8 bit
Uploading Files to CMS
 Uploading Files to CMS Li Gardiner Room 1111 East Building 212-772- 4855 DesignSS@hunter.cuny.edu 1 Uploading a file (such as a PDF or an image) to your CMS Web site for the first time Log into the Hunter
Uploading Files to CMS Li Gardiner Room 1111 East Building 212-772- 4855 DesignSS@hunter.cuny.edu 1 Uploading a file (such as a PDF or an image) to your CMS Web site for the first time Log into the Hunter
DOCUWARE REFERENCE MANUAL
 DOCUWARE REFERENCE MANUAL Sierra Lands, Inc 2013 LOGGING INTO DOCUWARE Open a web browser and navigate to: https://docs.cariberoyale.com/dwwebclient. DocuWare will automatically log you in based on your
DOCUWARE REFERENCE MANUAL Sierra Lands, Inc 2013 LOGGING INTO DOCUWARE Open a web browser and navigate to: https://docs.cariberoyale.com/dwwebclient. DocuWare will automatically log you in based on your
v Importing Rasters SMS 11.2 Tutorial Requirements Raster Module Map Module Mesh Module Time minutes Prerequisites Overview Tutorial
 v. 11.2 SMS 11.2 Tutorial Objectives This tutorial teaches how to import a Raster, view elevations at individual points, change display options for multiple views of the data, show the 2D profile plots,
v. 11.2 SMS 11.2 Tutorial Objectives This tutorial teaches how to import a Raster, view elevations at individual points, change display options for multiple views of the data, show the 2D profile plots,
LEGENDplex Data Analysis Software Version 8 User Guide
 LEGENDplex Data Analysis Software Version 8 User Guide Introduction Welcome to the user s guide for Version 8 of the LEGENDplex data analysis software for Windows based computers 1. This tutorial will
LEGENDplex Data Analysis Software Version 8 User Guide Introduction Welcome to the user s guide for Version 8 of the LEGENDplex data analysis software for Windows based computers 1. This tutorial will
Client Care Desktop v4.3. Document Manager V4.3
 Client Care Desktop v4.3 Document Manager V4.3 Contents 1. LOCATIONS... 3 1.1. Client... 3 1.2. Enquiries... 4 1.3. Holdings... 4 1.4. Providers... 5 1.5. Contacts/Introducers... 6 1.6. Adviser... 6 2.
Client Care Desktop v4.3 Document Manager V4.3 Contents 1. LOCATIONS... 3 1.1. Client... 3 1.2. Enquiries... 4 1.3. Holdings... 4 1.4. Providers... 5 1.5. Contacts/Introducers... 6 1.6. Adviser... 6 2.
ROES EVENTS SYSTEM TUTORIAL
 table of contents 3 Introduction to ROES Events 4 Preparing your Data File 6 Recommended Order of Operations 6 Build Packages 9 Import Data & Images 10 Match Images & Identify Packages 11 Review Order
table of contents 3 Introduction to ROES Events 4 Preparing your Data File 6 Recommended Order of Operations 6 Build Packages 9 Import Data & Images 10 Match Images & Identify Packages 11 Review Order
A Guide to Processing Photos into 3D Models Using Agisoft PhotoScan
 A Guide to Processing Photos into 3D Models Using Agisoft PhotoScan Samantha T. Porter University of Minnesota, Twin Cities Fall 2015 Index 1) Automatically masking a black background / Importing Images.
A Guide to Processing Photos into 3D Models Using Agisoft PhotoScan Samantha T. Porter University of Minnesota, Twin Cities Fall 2015 Index 1) Automatically masking a black background / Importing Images.
IMPORTANT DATE: THE SUBMISSION DEADLINE FOR ALL ABSTRACTS IS EXTENDED TO Friday, March 9, 5:00 PM PT
 Thank you for your interest in submitting an Abstract for the 2018 Florida Society of Anesthesiologists Annual Meeting. This document is intended to be your guide in using the online submission software
Thank you for your interest in submitting an Abstract for the 2018 Florida Society of Anesthesiologists Annual Meeting. This document is intended to be your guide in using the online submission software
Sherlock Tutorial Project Overview
 Sherlock Tutorial Project Overview Background Sherlock organizes design files, inputs and analysis results as project folders that can be managed inside of the Sherlock application and shared between Sherlock
Sherlock Tutorial Project Overview Background Sherlock organizes design files, inputs and analysis results as project folders that can be managed inside of the Sherlock application and shared between Sherlock
Graphing on Excel. Open Excel (2013). The first screen you will see looks like this (it varies slightly, depending on the version):
 Graphing on Excel Open Excel (2013). The first screen you will see looks like this (it varies slightly, depending on the version): The first step is to organize your data in columns. Suppose you obtain
Graphing on Excel Open Excel (2013). The first screen you will see looks like this (it varies slightly, depending on the version): The first step is to organize your data in columns. Suppose you obtain
Using Microsoft Word. Working With Objects
 Using Microsoft Word Many Word documents will require elements that were created in programs other than Word, such as the picture to the right. Nontext elements in a document are referred to as Objects
Using Microsoft Word Many Word documents will require elements that were created in programs other than Word, such as the picture to the right. Nontext elements in a document are referred to as Objects
static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, i
 MAESTRO static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
MAESTRO static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
Protein Deconvolution Quick Start Guide
 Protein Deconvolution Quick Start Guide The electrospray ionization (ESI) of intact peptides and proteins produces mass spectra containing a series of multiply charged ions with associated mass-to-charge
Protein Deconvolution Quick Start Guide The electrospray ionization (ESI) of intact peptides and proteins produces mass spectra containing a series of multiply charged ions with associated mass-to-charge
A First Introduction to Scientific Visualization Geoffrey Gray
 Visual Molecular Dynamics A First Introduction to Scientific Visualization Geoffrey Gray VMD on CIRCE: On the lower bottom left of your screen, click on the window start-up menu. In the search box type
Visual Molecular Dynamics A First Introduction to Scientific Visualization Geoffrey Gray VMD on CIRCE: On the lower bottom left of your screen, click on the window start-up menu. In the search box type
THE SUBMISSION DEADLINE FOR ALL ABSTRACTS IS FRIDAY, SEPTEMBER 7, 2018, 5:00 PM PDT
 Thank you for your interest in submitting an abstract for the upcoming North American Skull Based Society Annual Meeting. This document is intended to be your guide in using the online submission software
Thank you for your interest in submitting an abstract for the upcoming North American Skull Based Society Annual Meeting. This document is intended to be your guide in using the online submission software
Creating a Basic Chart in Excel 2007
 Creating a Basic Chart in Excel 2007 A chart is a pictorial representation of the data you enter in a worksheet. Often, a chart can be a more descriptive way of representing your data. As a result, those
Creating a Basic Chart in Excel 2007 A chart is a pictorial representation of the data you enter in a worksheet. Often, a chart can be a more descriptive way of representing your data. As a result, those
Importing and processing a DGGE gel image
 BioNumerics Tutorial: Importing and processing a DGGE gel image 1 Aim Comprehensive tools for the processing of electrophoresis fingerprints, both from slab gels and capillary sequencers are incorporated
BioNumerics Tutorial: Importing and processing a DGGE gel image 1 Aim Comprehensive tools for the processing of electrophoresis fingerprints, both from slab gels and capillary sequencers are incorporated
IMPORTANT DATE: THE SUBMISSION DEADLINE FOR ALL ABSTRACTS IS JANUARY 5, 11:59 PM PST
 Thank you for your interest in submitting an abstract for the upcoming Emerging Technology Session. This document is intended to be your guide in using the online submission software and we strongly suggest
Thank you for your interest in submitting an abstract for the upcoming Emerging Technology Session. This document is intended to be your guide in using the online submission software and we strongly suggest
For the Beginner: c) Click the settings tab to set screen resolution d) Set resolution with slider bar.
 For the Beginner: Preparing and Logging In to Run JobSite OnLine 1) Jobsite works best with a screen resolution of at least 800 x 600 preferably 1024 x 768. To set this, follow the instructions below.
For the Beginner: Preparing and Logging In to Run JobSite OnLine 1) Jobsite works best with a screen resolution of at least 800 x 600 preferably 1024 x 768. To set this, follow the instructions below.
Moving Materials from Blackboard to Moodle
 Moving Materials from Blackboard to Moodle Blackboard and Moodle organize course material somewhat differently and the conversion process can be a little messy (but worth it). Because of this, we ve gathered
Moving Materials from Blackboard to Moodle Blackboard and Moodle organize course material somewhat differently and the conversion process can be a little messy (but worth it). Because of this, we ve gathered
INTViewer Tutorial Cube Tutorial
 INTViewer Tutorial Cube Tutorial This tutorial shows how to use INTViewer to display a seismic cube stored in a Seismic file. Windows created will include INLINE, XLINE, Time Slice and an arbitrary traverse,
INTViewer Tutorial Cube Tutorial This tutorial shows how to use INTViewer to display a seismic cube stored in a Seismic file. Windows created will include INLINE, XLINE, Time Slice and an arbitrary traverse,
IMPORTANT DATE: THE SUBMISSION DEADLINE FOR ALL ABSTRACTS AND VIDEOS IS SEPTEMBER 21, :59 PM PDT
 Thank you for your interest in submitting an abstract for the upcoming Scientific Session. This document is intended to be your guide in using the online submission software and we strongly suggest you
Thank you for your interest in submitting an abstract for the upcoming Scientific Session. This document is intended to be your guide in using the online submission software and we strongly suggest you
ProShake 2.0. The first step in this tutorial exercise is to start the ProShake 2.0 program. Click on the ProShake 2.0 icon to start the program.
 PROSHAKE 2.0 TUTORIAL The easiest way to learn the basics of ProShake s organization and operation is to complete the tutorial exercise detailed in this section. This tutorial will take you through nearly
PROSHAKE 2.0 TUTORIAL The easiest way to learn the basics of ProShake s organization and operation is to complete the tutorial exercise detailed in this section. This tutorial will take you through nearly
AutoCollage 2008 makes it easy to create an AutoCollage from a folder of Images. To create an AutoCollage:
 Page 1 of 18 Using AutoCollage 2008 AutoCollage 2008 makes it easy to create an AutoCollage from a folder of Images. To create an AutoCollage: 1. Click on a folder name in the Image Browser. 2. Once at
Page 1 of 18 Using AutoCollage 2008 AutoCollage 2008 makes it easy to create an AutoCollage from a folder of Images. To create an AutoCollage: 1. Click on a folder name in the Image Browser. 2. Once at
Creating Interactive PDF Forms
 Creating Interactive PDF Forms Using Adobe Acrobat X Pro for the Mac University Information Technology Services Training, Outreach, Learning Technologies and Video Production Copyright 2012 KSU Department
Creating Interactive PDF Forms Using Adobe Acrobat X Pro for the Mac University Information Technology Services Training, Outreach, Learning Technologies and Video Production Copyright 2012 KSU Department
Slope Stability Problem Session
 Slope Stability Problem Session Stability Analysis of a Proposed Soil Slope Using Slide 5.0 Tuesday, February 28, 2006 10:00 am - 12:00 pm GeoCongress 2006 Atlanta, GA software tools for rock and soil
Slope Stability Problem Session Stability Analysis of a Proposed Soil Slope Using Slide 5.0 Tuesday, February 28, 2006 10:00 am - 12:00 pm GeoCongress 2006 Atlanta, GA software tools for rock and soil
ABI PRISM GeneMapper Software Version 3.0 SNP Genotyping
 ABI PRISM GeneMapper Software Version 3.0 SNP Genotyping Tutorial ABI PRISM GeneMapper Software Version 3.0 SNP Genotyping Tutorial September 25, 2002 1:20 pm, 7x9_Title.fm Copyright 2002, Applied Biosystems.
ABI PRISM GeneMapper Software Version 3.0 SNP Genotyping Tutorial ABI PRISM GeneMapper Software Version 3.0 SNP Genotyping Tutorial September 25, 2002 1:20 pm, 7x9_Title.fm Copyright 2002, Applied Biosystems.
FLUENT Secondary flow in a teacup Author: John M. Cimbala, Penn State University Latest revision: 26 January 2016
 FLUENT Secondary flow in a teacup Author: John M. Cimbala, Penn State University Latest revision: 26 January 2016 Note: These instructions are based on an older version of FLUENT, and some of the instructions
FLUENT Secondary flow in a teacup Author: John M. Cimbala, Penn State University Latest revision: 26 January 2016 Note: These instructions are based on an older version of FLUENT, and some of the instructions
SIVIC GUI Overview. SIVIC GUI Layout Overview
 SIVIC GUI Overview SIVIC GUI Layout Overview At the top of the SIVIC GUI is a row of buttons called the Toolbar. It is a quick interface for loading datasets, controlling how the mouse manipulates the
SIVIC GUI Overview SIVIC GUI Layout Overview At the top of the SIVIC GUI is a row of buttons called the Toolbar. It is a quick interface for loading datasets, controlling how the mouse manipulates the
Multivariate Calibration Quick Guide
 Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Using Excel for Graphical Analysis of Data
 Using Excel for Graphical Analysis of Data Introduction In several upcoming labs, a primary goal will be to determine the mathematical relationship between two variable physical parameters. Graphs are
Using Excel for Graphical Analysis of Data Introduction In several upcoming labs, a primary goal will be to determine the mathematical relationship between two variable physical parameters. Graphs are
Steady Flow: Lid-Driven Cavity Flow
 STAR-CCM+ User Guide Steady Flow: Lid-Driven Cavity Flow 2 Steady Flow: Lid-Driven Cavity Flow This tutorial demonstrates the performance of STAR-CCM+ in solving a traditional square lid-driven cavity
STAR-CCM+ User Guide Steady Flow: Lid-Driven Cavity Flow 2 Steady Flow: Lid-Driven Cavity Flow This tutorial demonstrates the performance of STAR-CCM+ in solving a traditional square lid-driven cavity
A TUTORIAL ON WORD. Katie Gregory
 A TUTORIAL ON WORD Katie Gregory First, CLICK HERE Then, find Microsoft Word under programs and the Microsoft Office 2013 Folder This is what the document should look like when opened. SAVING A WORD DOCUMENT
A TUTORIAL ON WORD Katie Gregory First, CLICK HERE Then, find Microsoft Word under programs and the Microsoft Office 2013 Folder This is what the document should look like when opened. SAVING A WORD DOCUMENT
Tutorial files are available from the Exelis VIS website or on the ENVI Resource DVD in the image_reg directory.
 Image Registration Tutorial In this tutorial, you will use the Image Registration workflow in different scenarios to geometrically align two overlapping images with different viewing geometry and different
Image Registration Tutorial In this tutorial, you will use the Image Registration workflow in different scenarios to geometrically align two overlapping images with different viewing geometry and different
proteindiffraction.org Select
 This tutorial will walk you through the steps of processing the data from an X-ray diffraction experiment using HKL-2000. If you need to install HKL-2000, please see the instructions at the HKL Research
This tutorial will walk you through the steps of processing the data from an X-ray diffraction experiment using HKL-2000. If you need to install HKL-2000, please see the instructions at the HKL Research
HOW TO USE THE EXPORT FEATURE IN LCL
 HOW TO USE THE EXPORT FEATURE IN LCL In LCL go to the Go To menu and select Export. Select the items that you would like to have exported to the file. To select them you will click the item in the left
HOW TO USE THE EXPORT FEATURE IN LCL In LCL go to the Go To menu and select Export. Select the items that you would like to have exported to the file. To select them you will click the item in the left
Investigator Site OC RDC PDF User Guide
 Investigator Site OC RDC PDF User Guide Version 1.0 Page 1 of 40 TABLE OF CONTENTS Accessing OC RDC Steps for Access 3 Logging On 4 Change Password 4 Laptop and System Security 5 Change Study 5 Navigating
Investigator Site OC RDC PDF User Guide Version 1.0 Page 1 of 40 TABLE OF CONTENTS Accessing OC RDC Steps for Access 3 Logging On 4 Change Password 4 Laptop and System Security 5 Change Study 5 Navigating
Physics 211 E&M and Modern Physics Spring Lab #1 (to be done at home) Plotting with Excel. Good laboratory practices
 NORTHERN ILLINOIS UNIVERSITY PHYSICS DEPARTMENT Physics 211 E&M and Modern Physics Spring 2018 Lab #1 (to be done at home) Lab Writeup Due: Mon/Wed/Thu/Fri, Jan. 22/24/25/26, 2018 Read Serway & Vuille:
NORTHERN ILLINOIS UNIVERSITY PHYSICS DEPARTMENT Physics 211 E&M and Modern Physics Spring 2018 Lab #1 (to be done at home) Lab Writeup Due: Mon/Wed/Thu/Fri, Jan. 22/24/25/26, 2018 Read Serway & Vuille:
0 Graphical Analysis Use of Excel
 Lab 0 Graphical Analysis Use of Excel What You Need To Know: This lab is to familiarize you with the graphing ability of excels. You will be plotting data set, curve fitting and using error bars on the
Lab 0 Graphical Analysis Use of Excel What You Need To Know: This lab is to familiarize you with the graphing ability of excels. You will be plotting data set, curve fitting and using error bars on the
Visual Physics Introductory Lab [Lab 0]
![Visual Physics Introductory Lab [Lab 0] Visual Physics Introductory Lab [Lab 0]](/thumbs/74/69956143.jpg) Your Introductory Lab will guide you through the steps necessary to utilize state-of-the-art technology to acquire and graph data of mechanics experiments. Throughout Visual Physics, you will be using
Your Introductory Lab will guide you through the steps necessary to utilize state-of-the-art technology to acquire and graph data of mechanics experiments. Throughout Visual Physics, you will be using
SAS Studio: A New Way to Program in SAS
 SAS Studio: A New Way to Program in SAS Lora D Delwiche, Winters, CA Susan J Slaughter, Avocet Solutions, Davis, CA ABSTRACT SAS Studio is an important new interface for SAS, designed for both traditional
SAS Studio: A New Way to Program in SAS Lora D Delwiche, Winters, CA Susan J Slaughter, Avocet Solutions, Davis, CA ABSTRACT SAS Studio is an important new interface for SAS, designed for both traditional
The Kindred Directory allows you to search for employees and locations across all of our lines of business.
 Release 2.0 July 2017 1 GETTING STARTED The Kindred Directory allows you to search for employees and locations across all of our lines of business. The Kindred Directory can be accessed in a variety of
Release 2.0 July 2017 1 GETTING STARTED The Kindred Directory allows you to search for employees and locations across all of our lines of business. The Kindred Directory can be accessed in a variety of
The beginning of this guide offers a brief introduction to the Protein Data Bank, where users can download structure files.
 Structure Viewers Take a Class This guide supports the Galter Library class called Structure Viewers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
Structure Viewers Take a Class This guide supports the Galter Library class called Structure Viewers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
AIS Student Guide for submitting a Turnitin Assignment in Moodle
 AIS Student Guide for submitting a Turnitin Assignment in Moodle Before you start Turnitin currently accepts the following file types for upload into an assignment: Microsoft Word (.doc and.docx) Plain
AIS Student Guide for submitting a Turnitin Assignment in Moodle Before you start Turnitin currently accepts the following file types for upload into an assignment: Microsoft Word (.doc and.docx) Plain
Optimax USER S MANUAL v0.6.3
 Optimax USER S MANUAL v0.6.3 1 Introduction Matej Borovinšek January, 2017 This is a description of the Optimax software. The Optimax software allows the user to structure the design process, explore the
Optimax USER S MANUAL v0.6.3 1 Introduction Matej Borovinšek January, 2017 This is a description of the Optimax software. The Optimax software allows the user to structure the design process, explore the
Lab Practical - Limit Equilibrium Analysis of Engineered Slopes
 Lab Practical - Limit Equilibrium Analysis of Engineered Slopes Part 1: Planar Analysis A Deterministic Analysis This exercise will demonstrate the basics of a deterministic limit equilibrium planar analysis
Lab Practical - Limit Equilibrium Analysis of Engineered Slopes Part 1: Planar Analysis A Deterministic Analysis This exercise will demonstrate the basics of a deterministic limit equilibrium planar analysis
Docking Study with HyperChem Release Notes
 Docking Study with HyperChem Release Notes This document lists additional information about Docking Study with HyperChem family, Essential, Premium Essential, Professional, Advanced, Ultimat, and Cluster.
Docking Study with HyperChem Release Notes This document lists additional information about Docking Study with HyperChem family, Essential, Premium Essential, Professional, Advanced, Ultimat, and Cluster.
1. Open up PRO-DESKTOP from your programmes menu. Then click on the file menu > new> design.
 Radio Tutorial Draw your spatula shape by:- 1. Open up PRO-DESKTOP from your programmes menu. Then click on the file menu > new> design. 2. The new design window will now open. Double click on design 1
Radio Tutorial Draw your spatula shape by:- 1. Open up PRO-DESKTOP from your programmes menu. Then click on the file menu > new> design. 2. The new design window will now open. Double click on design 1
Depositing small-angle scattering data and models to the Small-Angle Scattering Biological Data Bank (SASBDB).
 Depositing small-angle scattering data and models to the Small-Angle Scattering Biological Data Bank (SASBDB). Introduction. The following guide provides a basic outline of the minimum requirements necessary
Depositing small-angle scattering data and models to the Small-Angle Scattering Biological Data Bank (SASBDB). Introduction. The following guide provides a basic outline of the minimum requirements necessary
MODFLOW Automated Parameter Estimation
 GMS 7.0 TUTORIALS MODFLOW Automated Parameter Estimation 1 Introduction The MODFLOW-Model Calibration tutorial describes the basic calibration tools provided in GMS. It illustrates how head levels from
GMS 7.0 TUTORIALS MODFLOW Automated Parameter Estimation 1 Introduction The MODFLOW-Model Calibration tutorial describes the basic calibration tools provided in GMS. It illustrates how head levels from
IGSS 13 Configuration Workshop - Exercises
 IGSS 13 Configuration Workshop - Exercises Contents IGSS 13 Configuration Workshop - Exercises... 1 Exercise 1: Working as an Operator in IGSS... 2 Exercise 2: Creating a new IGSS Project... 28 Exercise
IGSS 13 Configuration Workshop - Exercises Contents IGSS 13 Configuration Workshop - Exercises... 1 Exercise 1: Working as an Operator in IGSS... 2 Exercise 2: Creating a new IGSS Project... 28 Exercise
Using Adobe Contribute 4 A guide for new website authors
 Using Adobe Contribute 4 A guide for new website authors Adobe Contribute allows you to easily update websites without any knowledge of HTML. This handout will provide an introduction to Adobe Contribute
Using Adobe Contribute 4 A guide for new website authors Adobe Contribute allows you to easily update websites without any knowledge of HTML. This handout will provide an introduction to Adobe Contribute
Chemistry Excel. Microsoft 2007
 Chemistry Excel Microsoft 2007 This workshop is designed to show you several functionalities of Microsoft Excel 2007 and particularly how it applies to your chemistry course. In this workshop, you will
Chemistry Excel Microsoft 2007 This workshop is designed to show you several functionalities of Microsoft Excel 2007 and particularly how it applies to your chemistry course. In this workshop, you will
GIS DATA SUBMISSION USER GUIDE. Innovation and Networks Executive Agency
 Innovation and Networks Executive Agency GIS DATA SUBMISSION USER GUIDE Innovation and Networks Executive Agency (INEA) W910 Chaussée de Wavre 910 B-1049 Brussels, Belgium Tel: +32 (0)2 29 95252 Fax: +32
Innovation and Networks Executive Agency GIS DATA SUBMISSION USER GUIDE Innovation and Networks Executive Agency (INEA) W910 Chaussée de Wavre 910 B-1049 Brussels, Belgium Tel: +32 (0)2 29 95252 Fax: +32
Part A: Monitoring the Rotational Sensors of the Motor
 LEGO MINDSTORMS NXT Lab 1 This lab session is an introduction to the use of motors and rotational sensors for the Lego Mindstorm NXT. The first few parts of this exercise will introduce the use of the
LEGO MINDSTORMS NXT Lab 1 This lab session is an introduction to the use of motors and rotational sensors for the Lego Mindstorm NXT. The first few parts of this exercise will introduce the use of the
Tutorial: chloroplast genomes
 Tutorial: chloroplast genomes Stacia Wyman Department of Computer Sciences Williams College Williamstown, MA 01267 March 10, 2005 ASSUMPTIONS: You are using Internet Explorer under OS X on the Mac. You
Tutorial: chloroplast genomes Stacia Wyman Department of Computer Sciences Williams College Williamstown, MA 01267 March 10, 2005 ASSUMPTIONS: You are using Internet Explorer under OS X on the Mac. You
GIS DATA SUBMISSION USER GUIDE. Innovation and Networks Executive Agency
 Innovation and Networks Executive Agency GIS DATA SUBMISSION USER GUIDE Innovation and Networks Executive Agency (INEA) W910 Chaussée de Wavre 910 B-1049 Brussels, Belgium Tel: +32 (0)2 29 95252 Fax: +32
Innovation and Networks Executive Agency GIS DATA SUBMISSION USER GUIDE Innovation and Networks Executive Agency (INEA) W910 Chaussée de Wavre 910 B-1049 Brussels, Belgium Tel: +32 (0)2 29 95252 Fax: +32
Using Excel for Graphical Analysis of Data
 EXERCISE Using Excel for Graphical Analysis of Data Introduction In several upcoming experiments, a primary goal will be to determine the mathematical relationship between two variable physical parameters.
EXERCISE Using Excel for Graphical Analysis of Data Introduction In several upcoming experiments, a primary goal will be to determine the mathematical relationship between two variable physical parameters.
Poet Image Description Tool: Step-by-step Guide
 Poet Image Description Tool: Step-by-step Guide Introduction This guide is designed to help you use the Poet image description tool to add image descriptions to DAISY books. The tool assumes you have access
Poet Image Description Tool: Step-by-step Guide Introduction This guide is designed to help you use the Poet image description tool to add image descriptions to DAISY books. The tool assumes you have access
Adobe Dreamweaver CS5 Tutorial
 Adobe Dreamweaver CS5 Tutorial GETTING STARTED This tutorial focuses on the basic steps involved in creating an attractive, functional website. In using this tutorial you will learn to design a site layout,
Adobe Dreamweaver CS5 Tutorial GETTING STARTED This tutorial focuses on the basic steps involved in creating an attractive, functional website. In using this tutorial you will learn to design a site layout,
IMAGE STUDIO LITE. Tutorial Guide Featuring Image Studio Analysis Software Version 3.1
 IMAGE STUDIO LITE Tutorial Guide Featuring Image Studio Analysis Software Version 3.1 Notice The information contained in this document is subject to change without notice. LI-COR MAKES NO WARRANTY OF
IMAGE STUDIO LITE Tutorial Guide Featuring Image Studio Analysis Software Version 3.1 Notice The information contained in this document is subject to change without notice. LI-COR MAKES NO WARRANTY OF
24 - TEAMWORK... 1 HOW DOES MAXQDA SUPPORT TEAMWORK?... 1 TRANSFER A MAXQDA PROJECT TO OTHER TEAM MEMBERS... 2
 24 - Teamwork Contents 24 - TEAMWORK... 1 HOW DOES MAXQDA SUPPORT TEAMWORK?... 1 TRANSFER A MAXQDA PROJECT TO OTHER TEAM MEMBERS... 2 Sharing projects that include external files... 3 TRANSFER CODED SEGMENTS,
24 - Teamwork Contents 24 - TEAMWORK... 1 HOW DOES MAXQDA SUPPORT TEAMWORK?... 1 TRANSFER A MAXQDA PROJECT TO OTHER TEAM MEMBERS... 2 Sharing projects that include external files... 3 TRANSFER CODED SEGMENTS,
Autodesk Moldflow Insight AMI Getting Started Tutorial
 Autodesk Moldflow Insight 2012 AMI Getting Started Tutorial Revision 1, 30 March 2012. This document contains Autodesk and third-party software license agreements/notices and/or additional terms and conditions
Autodesk Moldflow Insight 2012 AMI Getting Started Tutorial Revision 1, 30 March 2012. This document contains Autodesk and third-party software license agreements/notices and/or additional terms and conditions
Supplemental Online Materials
 1 Supplemental Online Materials 1. Quick Guide to FORCulator 1.1 System Requirements FORCulator is a self-contained package written using Igor Pro by WaveMetrics (www.wavemetrics.com). Although Igor Pro
1 Supplemental Online Materials 1. Quick Guide to FORCulator 1.1 System Requirements FORCulator is a self-contained package written using Igor Pro by WaveMetrics (www.wavemetrics.com). Although Igor Pro
Intermediate Lab PHYS 3870
 Intermediate Lab PHYS 3870 CONVEYIMG INFORMATION Gathering Information Installing and Using DataThief References: PHYS 3870 Web Site Introduction Section 0 Lecture 1 Slide 1 USU Library Class Web Site
Intermediate Lab PHYS 3870 CONVEYIMG INFORMATION Gathering Information Installing and Using DataThief References: PHYS 3870 Web Site Introduction Section 0 Lecture 1 Slide 1 USU Library Class Web Site
