Documentation to image analysis
|
|
|
- Leon Hawkins
- 6 years ago
- Views:
Transcription
1 Documentation to image analysis The documentation below provides a detailed explanation and pointers towards the use of the MS-ECS-2D update site. It is made available in conjunction with the publication of the paper: Stapel, L. C., Lombardot, B., Broaddus, C., Kainmueller, D., Jug, F., Myers, E. W., & Vastenhouw, N. L. (2015). Automated detection and quantification of single RNAs at cellular resolution in zebrafish embryos. Development, dev The documentation details: - The PathFinder Fiji plugin - The Cell annotation plugin in Fiji - The Transcript analysis plugin in Fiji
2 PathFinder Documentation to PathFinder Fiji plugin This document describes the setup and usage of the PathFinder Fiji plugin for cell segmentation of tissue sections with a single bright membrane channel. The input to the PathFinder plugin are membrane and vertex classified images as obtained by the MS-ECS-2D KNIME workflow. 1. Installation Open Fiji, go to Help Update Manage update sites and make sure the following URL is added: and under Host add: webdav:ms-ecs-2d This adds a collection of files to the ImageJ Updater site that you need for the automated detection of single RNAs at subcellular resolution. In case you are only interested in using the PathFinder plugin, you can choose to install only those files with the prefix Path_Finder/. You can search for Path_Finder after pressing Advanced mode. Select these files and click Apply changes, which installs them to the standard location, resulting in paths: [Fiji home]/plugins/path_finder/* Go to Help Refresh Menus. You should see Plugins Path Finder main, if not try reopening Fiji. NOTE: of course you can also choose to install all files of the update site. The usage of the Cell annotation and Transcript analysis plugins to analyze single RNAs at subcellular resolution is explained in their respective documentations. 2. Overview The PathFinder plugin allows you to perform membrane segmentation across several images simultaneously. Before you begin, you should make sure that the storage of your images adheres to the following directory structure: [original image folder] [original image 1].tif [original image 2].tif [original image n].tif [memgrane classifier output folder] [classifier output 1].tif
3 PathFinder [classifier output 2].tif [classifier output n].tif [vertex classifier output folder] [classifier output 1].tif [classifier output 2].tif [classifier output n].tif The original image folder contains the same images that were used as input to the membrane and vertex classification procedures. You are free to use whichever filenaming convention is convenient, provided that the images in these different directories match up correctly when listed in alphabetical order. By default, the classification procedure names its output files in a way that maintains this order. The plugin operates on each image in sequence, and will save the segmented version of each image to the location: [original image folder]/results/[original image n]_scaled.tif Each of these resulting images has three layers (Figure 1): 1. segmented cells : Each set of pixels belonging to the same cell has the same pixel value. 2. original : The original image from [original image folder]. 3. paths : A binary image where all pixels belonging to the final set of paths have value 255. The width and height of the result is equal to that of the original image that was provided, even when the output of the membrane and vertex classifiers may be downsized by a small factor for computational efficiency.
4 PathFinder A A B B C C Figure 1. Example of output images from the pathfinder plugin. (A) Segmented cells. (B) Original membrane image. (C) Paths. 3. Usage Details
5 PathFinder Run the plugin with Plugins Path Finder main. The following screen will appear: The vertex threshold parameter determines the threshold probability above which pixels from the vertex probability map image are classified as vertices. If the membrane path threshold is smaller than the average probability across all pixels in a path, then the path is removed. The small-cell-removal scale factor is used after the initial path construction in the small-cell-removal phase where we remove bad edges that cause over segmentation. Increasing this value will cause the small-cell-removal step to keep higher (worse) scoring edges, and lowering it will cause more of them to be removed. Setting this parameter to zero will skip the small cell removal phase entirely. The membrane pixel threshold parameter determines the threshold probability above which pixels form the membrane probability map image are classified as membrane. It is only within membrane regions that paths between vertices can be constructed. After the parameters have been selected you will be asked to select the original image folder, membrane image folder and vertex image folder via a series of three folder-selection dialogs:
6 PathFinder And you re done! The plugin will briefly open new image windows in Fiji as it progresses through different stages of the algorithm, but no windows remain open when the plugin is finished running. Thus, it is currently not possible to continue to working on a separate image in Fiji simultaneously while the plugin is running, though this may change in future versions of the plugin. Finally, if there are any errors they will be written to the Fiji Console. ***
7 Cell annotation Documentation to the Cell annotation plugin in Fiji The Cell annotation tool is an interface to manually correct a (2D) cell segmentation and to annotate cells once segmented. The tool is composed of a control window that allows choosing the annotation modes (label inspection, cell segmentation correction and cell type annotation) and a display window that shows an overlay of i. the masks to annotate and ii. the raw data that is used for the annotation. Once an annotation is finished, the annotation can be exported as a 2-channel image (cell labelling and cell annotation) that can be used as input to the Transcript analysis plugin (see corresponding documentation). 1. Installation To use the plugin (assuming Fiji is installed): Install the plugin: o Start Fiji and go to the menu Help>Update o When the imagej updater window is open click the manage update sites button and then check the MS-ECS-2D update site in the Manage update sites window. o Close the Manage update sites window, press Apply changes in the ImageJ Updater window and when prompted to, Close Fiji. o The plugin will be accessible in the menu Plugins>Cell_Transcript>Cell annotation. Note: these steps are not necessary if you have already activated the complete MS-ECS-2D update site to obtain the Transcript analysis or Pathfinder plugins. 2. Plugin usage Before starting the plugin, load the cell segmented image (see Figure 1.A, output cell segmentation pipeline) you wish to annotate and the original raw data (see Figure 1.B). Both images should have the same number of pixels in the X and Y dimensions. Also, each cell region in the labelled image should be separated by a boundary of zeros (in white in Figure 1.A). Such a labelled image can be obtained as an output of the cell segmentation process. You can then start the plugin by going to the menu: Plugins>Cell_Transcript>Cell_annotation A window will open requesting the following information (see Figure 1.C): Original Image: raw data that will guide the annotation Cell Masks Image: Masks to annotate. It can be a one-channel image showing cell segmentation or a two-channel image showing the cell segmentation in channel 1 and a previously generated cell tissue annotation in the second channel.
8 Cell annotation Nuclei channel: channel showing the nuclei in the raw data (first channel has value 1). Membrane channel: channel showing the nuclei in the raw data. Note that these channels will only be used for display and in practice any channel can be chosen. It is for example possible to select a channel that contains an antibody staining to facilitates cell type annotation. When pressing OK an interface named annotation control opens (see Figure 1.D) as well as an image, the annotation display. The annotation display shows a maximum projection of the selected membrane and nuclei channels (see Figure 1.E), superimposed with a transparent overlay of the cell label. The name of the annotation display always starts with [Annotation tool]. Annotation Modes and control description: The Annotation control window allows selecting the interaction mode and the parameters for the annotation. By changing the selection in the Mode drop down menu, one can switch between three types of interactions with the annotation display. Label inspection (see Figure 2.B): This mode enables visual inspection of the cell segmentation output. In this mode the original image is overlaid with the image of labelled cells regions. When switching to this mode the image is automatically relabelled to account for recent adjustments of the cell segmentation. Besides the usual Fiji zooming (by pressing + or - keys) and panning (space bar + mouse drag) options no interaction is possible with the annotation display in that mode. Correction (see Figure 2.C and Figure 3): In this mode the cell segmentation can be corrected. The cell borders are shown in red as an overlay to the projection of the original image. On top of zooming and panning this mode allows modifying the cell regions by drawing and erasing boundaries between them. To add boundaries move the mouse while pressing the mouse left button. The same action while pressing the Alt key allows erasing boundaries. The size of the brush to draw or erase boundaries can be set in the field Brush size. Annotation (see Figure 2.D): In this mode, cells can be assigned to a specific cell or tissue type to create a cell type mask. An overlay showing how each region is associated to a cell type is shown. If no cell annotation was present with the cell labelling a default annotation where all regions are of type DEL is created. Cells can be annotated with the currently selected cell type when a cell is clicked on. For the purpose of our analysis we defined 4 types or regions EVL, YSL, DEL, outside. EVL and DEL correspond to cell regions while YSL is for the zebrafish yolk syncytial layer and outside literally is used to indicate regions outside of the embryo. The code can be adjusted to facilitate the annotation of user-selected cell types.
9 Cell annotation In all modes the transparency of the overlay can be adjusted using the Overlay alpha slide bar. Every time the mode is changed the masks are updated to ensure consistency between the overlays. This behaviour can also be triggered by clicking the Relabel button. When one is satisfied with the cell labelling and the cell type annotations, the corresponding mask can be exported to the Fiji interface by pressing the get Masks button. This image can be used as input to the Transcript analysis plugin to obtain cell (type) specific data on gene expression. Figure 1: Typical input data and annotation tool interfaces. (A) Cell label output from the PathFinder tool. (B) Raw data. (C) Opening window of the Annotation tool. (D) Annotation control window. (E) Maximum projection of the raw data with an overlay of the cell labels.
10 Cell annotation Figure 2: Illustration of the tool display overlays for the different modes of interaction. For the raw data (A) the Annotation tool overlays cell labels in Label inspection mode (B), red cell contours in correction mode (C) and cell type labels in annotation mode (D). Figure 3: The correction mode allows to fuse ( or split) regions by removing ( or adding) boundaries. Setting the mode to Correction overlays regions boundaries (in red) to the raw data (A). Dragging the mouse while pressing the mouse left button and alt key draws a yellow trace (B). When releasing the mouse the piece of boundary below the trace is erased (C). When clicking the relabel button those boundaries that are not separating 2 objects are removed (D) 3. Methods description The annotation tool relies on 3 images that are overlaid on top of the raw data and can be updated by user interaction: an image of the cell labels, an image of the cell boundaries, an image of the cell type (where images are coloured according to the type of the region). In this section we describe how these images are initialized and updated when the user interacts with the annotation tool. Initialization of the overlay images: The labelled image provided as input is used as provided while the cell boundary overlay is obtained by thresholding the labelled image such that only the zero valued pixel are selected in the mask. A cell type annotation overlay can be provided in the second channel of the input mask. If not provided, the overlay for cell type annotation is created by setting cell boundaries to zeros while the rest of the image is set to a default value corresponding to the DEL cell type (the most common cell type in our images). Finally, images are cleaned to remove non-separating boundaries (see details of the cleaning process in the next section).
11 Cell annotation Cleaning of overlay images: Label image cleaning refers to an operation removing non-separating boundaries in the cell segmentation images (i.e. boundaries that are surrounded by a unique label). These non-separating boundaries can erroneously be generated by the PathFinder plugin. The process used removes all non-separating boundaries less than or equal to 2 pixels width. Boundaries thicker than 2 pixels will remain. The filter works as follows: a dilated and a closed version (with radius set to 1 pixel) of the labelled regions are subtracted. In the resulting image the non-zero values match with the cleaned boundaries of the regions (i.e. non separating boundaries do not appear anymore). From this image we create an image that is zeros in the cleaned boundaries and 1 inside the regions. This image is multiplied with the dilated version of the original labelled image. The resulting image is a cleaned version of the original labelled image. Update of overlay images after boundary modification: To create an updated label image after modification of the cell boundaries, the cell boundaries image is inverted and a connected component labelling is applied to it. The resulting image is then cleaned (see previous bullet point) to give the new cell labels image. A new boundary image is deduced from the new cell labels image by selecting the zerosvalued pixels. Regarding the cell type mask, a map from region label to region type is built and used to recolor a copy of the new cell labels according to their regions type. If a region is split, the daughter regions will have the same region type as the initial region. If regions are merged the new region type will be according to the type of most bottom right region (i.e. in the group of regions to be merged). In this case, region type might have to be adjusted manually afterwards. Region type annotation: A flood-fill algorithm with 8-connected neighbourhood is initiated in the cell annotation overlay at the position of the mouse click. The value for the flood filling corresponds to the region type selected (EVL=1, YSL=2, DEL = 3, outside regions=4). The flood fill will update the cell type value of the cell that was clicked. ***
12 Documentation to the Transcript analysis plugin in Fiji (1.0.7) The Transcript analysis plugin is an image analysis pipeline for the detection of transcripts and transcription foci in 3D sections of tissue. In particular it was developed to support the analysis of transcript abundance during zebrafish early development. The pipeline is initiated with z-stack images of smfish data that include nuclear detection, and optionally the output of the Cell annotation tool (see corresponding documentation). The plugin outputs a multichannel image illustrating the detections (transcripts, nuclei, cells), and text files indicating plugin parameters, cell and transcript descriptors and a few plots illustrating the detections. The pipeline can be started with a user interface and from a script (for batch processing for instance). These usages are described in the following sections. 1. Installation To use the plugin, assuming Fiji is installed: Install the 3D imagej Suite (the plugin relies on this library for some of the 3D image analysis): o Start Fiji and go to the menu Help>Update o When the imagej updater window is open click the manage update sites button and then check the 3D imagej Suite update site in the Manage update sites window. o Close the Manage update sites window, press Apply changes in the ImageJ Updater window and when prompted to, Close Fiji. Install the plugin: o Start Fiji and go to the menu Help>Update o When the imagej updater window is open click the manage update sites button and then check the MS-ECS-2D update site in the Manage update sites window. o Close the Manage update sites window, press Apply changes in the ImageJ Updater window and when prompted to, Close Fiji. o The plugin will be accessible in the menu Plugins>Cell_Transcript>Cell transcript analysis. Note: these steps are not necessary if you have already activated the complete MS-ECS-2D update site to obtain the Cell annotation or PathFinder plugins.
13 2. Plugin usage Before starting the plugin, load the raw data and optionally the cell segmentation and annotation image (as outputted by the annotation tool for instance, see section 3). Both images should have the same X and Y dimensions. You can then start the plugin by going to the menu: Plugins>Cell_Transcripts>Cell transcript analysis v3. A user interface will open requesting information necessary for the analysis (see Figure 1.F). The parameters are separated in 5 sections: Raw data and Masks o Input image (Figure 1.A, B): here the raw data to analyse is selected. A drop down menu lets the user choose from all images that are open in Fiji. It allows selecting the raw data to analyse. The selected image is expected to have a nuclei channel and at least one transcripts (smfish) channel. o Cell/Tissue masks (Figure 1.C): this drop down menu can be used to select a cell segmentation/annotation image from the images that are open in Fiji. In case one does not want to use a cell/tissue mask for the analysis None can be selected. The selected image is expected to be a cell segmentation (labelled regions separated by zeros valued boundaries) or a 2-channel image with cell segmentation in the first channel and cell annotation in the second channel. The latter format corresponds to the output of the annotation tool presented in section 3.If no cell segmentation is provided a simple a simple one will be processed by dilating nuclei segmentation. Nuclei Segmentation parameters: these parameters can be adjusted to obtain optimal nuclear segmentation. The parameter notation (between brackets) corresponds to that used in the method description. o Nuclei channel: An integer indicating the channel with the nuclei signal (1 correspond to the first channel). o Nuclei size (radius in pixel) / (Rnuc): An integer value indicating the radius of the largest nuclei to be expected in the sample. This value will be used for nuclei background subtraction and other filtering related to nuclei segmentation. o Nuclei threshold adjustment / (αnuc): Nuclei are differentiated from their background with to an automated threshold. This threshold will be multiplied αnuc. For instance if the threshold is systematically too high, some nuclei with low intensity values are ignored and reducing αnuc to a value smaller than 1 can aid the detection. If no systematic bias is observed the value should be set to 1 (i.e. the default value) Transcripts segmentation parameters: these parameters are used for transcript detection.
14 o Transcripts channel(s): an integer indicating the channel with the transcript signal (1 correspond to the first channel). If multiple transcript channel have to be analysed they can be indicated separated by a comma (1, 2, 3, ). o Transcripts typical radius / (Rtx): An integer value indicating the typical transcript radius (i.e. the pixel distance between a transcript maximum of intensity and the neighbouring background). This value is used to remove the background around the transcripts. A radius of 2 or 3 pixels should be used for images in which the sub-resolution transcripts are imaged with Nyquist sampling. o Spot minimum intensity / (Ttx): A value corresponding to a fraction of the typical transcript dynamics (i.e. the difference between typical transcript maxima of intensity and the neighbouring background). If this value is too low there will be false positive detections corresponding to peaks in the background noise. If it is too high transcripts with low intensities will be missed. We propose the user to first run the analysis with a coarsely chosen value here and then to use the histogram of detected maxima outputted by the script to determine an adequate threshold. In that histogram one can see a very sharp peak close to zero corresponding to the noise outside of the tissue and a second peak corresponding to the noise in the tissue. The threshold should be placed in the tail of that second peak to accurately detect the transcripts. If multiple transcripts channels are analysed multiple values separated by a comma can be set. Foci segmentation parameters: these parameters are used for the detection of transcription foci. Transcription foci are characterized by their large size (area) and intensity compared to individual transcripts. This information is used for the automated detection of transcription foci. In addition, transcription foci must fit the criterion that they are located in a nucleus. o Foci intensity threshold adjustment / (αimax): αimax, is used to adjust an automatically determined threshold, TImax. TImax is based on the maximum intensity, Imax, of detected transcripts. TImax=median_Imax +3. αimax.mad_imax, where median_imax and MAD_Imax are the median and the median absolute deviation of Imax on transcripts. to help the user to select the appropriate TImax for an image set a scatter plot of transcripts maximum intensity and volume is outputted. A value between 1.5 and 3 usually allows good focus detection in our images. If multiple transcripts channels are analysed, multiple values separated by a comma can be set. o Foci volume threshold adjustment / (αvol): See previous parameter description where maximum intensity of transcript can be replaced by volume of transcripts.
15 o Enlarge nuclei to detect foci / (RExtNuc): transcription foci can be located at the border of nuclei and then end up just outside the nuclear segmentation. Since localization of a focus candidate in a nucleus is a prerequisite for foci detection these foci fail to be detected. To decrease the rate of false negatives the nuclear area can be dilated by a fraction of the nuclear radius. This value is expressed in pixel and can be chosen as a fraction of the parameter Nuclei size. o Correct transcript over-segmentation: foci can be bigger than transcripts and have a tendency to get split during transcript detection when foci candidate are first identified. When checked, Correct transcript oversegmentation triggers a filtering step that prevents larger object from being split (see methods section below for more details). o Foci Maximum radius / (Rfoci): This value is used only if Correct transcript over-segmentation is checked. It is an integer value indicating the typical foci radius. This radius can be estimated manually from the original smfish image. Miscellaneous parameters: o Display Graph: when checked, a maximum spot intensity histogram and a scatter plot of the area vs intensity of all detected transcripts and foci are displayed. o Save Graph to disk: when checked, the abovementioned plots are saved to a user-defined directory. o Display results image: when checked a multichannel image illustrating the results of cell, nuclear and transcript detections is displayed. o Save result image: when checked the result image is saved in tiff format to a user-defined directory o Display transcripts images: when checked an image of transcript segmentation (colored after each transcript id) is displayed o Save directory: Path where results files will be saved after the processing. When the processing is finished the script will output a file with measures for each individual cell (_cell) and a file with extensive information on each of the segmented transcripts (_spot_ch<n>): <Img name>_cell.txt: this file provides measures for each individual cell. <Img name> is the name of the analysed raw data without the file extension. The name of each measure in the file, and their meaning are listed below. o CellId: Label of the cell in the mask used o X and Y: average position in image units (i.e. as found in the image properties) of the cell along the X and Y axis. The Unit is consigned in the parameter file.
16 o AreaCell: surface area in image units of the cell region corresponding to CellId. o AreaNuc: surface area in image units of the nuclear region corresponding to the CellId. This value is set to 0 if no nucleus is found in that particular cell. o MaskLabel: value corresponding to the cell type annotation associated with the cell (EVL=1, YSL=2, DEL = 3, outside = 4). If no region type is provided at plugin start this value is set to 0. o nuclear fraction: given that nuclei and cell segmentations originate from different sources a step was required to attribute nuclei to cells in the analysis. The value nuclear fraction denotes an assessment of this attribution and indicates the overlap of a nucleus with its parent cell. 1 corresponds to a complete overlap and zeros would correspond to a null overlap. If no nucleus is attributed to a cell the value is set to 0. o NspotNuc_ch<N>: the number of transcripts detected in the nucleus of the cell. The foci contribution is not included. Additionally, if the nucleus does no fully overlap with a cell only the transcripts in the overlapping region are counted. <N> is the channel number from which the analysis originates. If multiple transcript channels are analysed, the file will show multiple columns corresponding to the different channel. The transcripts that overlap with the extended nuclear region are not taken into account here. o NspotCell_ch<N>: the number of transcripts in the entire cell, nuclei included. Transcripts that are located in transcription foci are not included. o Nfoci_ch<N>: the number of foci detected in the nucleus of the cell. In the case of false positive detections this value can be larger than 2. o TxDensity_ch<N>: the ratio of NspotCell_ch<N> to the cell area in pixel. o NspotFoci1_ch<N>: the number of transcripts in the transcription focus with the largest number of transcripts. The number of transcripts is measured as the ratio of transcription focus integrated intensity to median integrated intensity of all transcripts in the image. Object integrated intensity is the sum of the pixels intensity in that object. o NspotFoci2_ch<N>: the number of transcripts in the transcription focus with the second largest number of transcripts. o NtxFoci_Other_ch<N>: the number of transcripts in remaining object that were (incorrectly) detected as foci. <Img name>_spot_ch<n>.txt: this file provides measures for each individual transcript. <Img name> is name of the raw data analysed without the extension. <N> is the number of the transcript channel analysed in the raw data. There will be
17 as many files as there are transcript channels analysed. The name of each measure in the file and their meaning are listed below. o X, Y and Z: position of transcripts in dataset Unit o volume: volume of the transcripts in dataset units o Iavg: average intensity of the segmented transcript o maxi: maximum intensity in the region of the segmented transcript o sumi: sum of the voxel intensities in the segmented transcript o isfoci: this value is 1 if the detection is considered as a focus (it belongs to a nuclei and its volume and intensity are above the defined thresholds) otherwise the value is 0. o cellparent: the id of the cell, CellId, that the transcript belongs to. The value is 0 if the transcript does not belong to any cell. o nucparent: the id of the nucleus that the transcript belongs to. The id of a nucleus is given after its cell parent id. nucparent is 0 if the transcript is not located in a nucleus. o extnucparent: the id of the extended nucleus the transcript belongs to. Extended nucleus id is given after the cell it belongs to. This value is only relevant for transcription foci. <Img name>_parameters.txt: this file gathers information on the analysis parameters as well as the date of the processing and image resolution <Img name>_maxdistrib_ch<n>.png and Maxima distribution_ch<n> window: It is possible to create a histogram plot of transcript candidate maximum intensities (Figure 1G). This plot can be used to determine an appropriate threshold for transcript detection. The histogram always has 500 bins between 0 and the candidate maximum intensity. If the Display Graph checkbox is checked in the input dialog then a window with the graph will open at the end of the processing. If Save Graph to disk is checked an 800 time 600 pixels png image will be saved to the save directory. <Img name>_scatterplot_ch<n>.png and Transcript_ch<N> window: It is possible to create a scatter plot of transcript maximum intensity versus transcript area (Figure 1H). This plot can aid the determination of appropriate thresholds for the detection of transcription foci. In the plot, cytoplasmic transcripts are in green, nuclear transcript in blue and foci in red. The thresholds that distinguish foci from nuclear transcripts are represented as red lines. If the Display Graph checkbox is checked in the input dialog then a window with the graph will be open. If Save Graph to disk is checked an 800 time 600 pixels png image will be saved to the save directory.
18 <Img name>_resultimg.tif file and window: It is possible to display a result image showing the cell, nuclear and transcript detections (Figure D,E). For each transcript channel analysed, a transcript detection channel is created where transcripts and foci have value 1 and 2, respectively. If more than 2 foci are detected in a nucleus all foci from that nucleus have a value of 3. An additional channel shows nuclei and cell segmentation. The 2D contour of nuclei detection are shown with value 510 and the contour of cells segmentation are set to 255. The new channels created are appended to a copy of the raw image data.if Display results image checkbox is checked in the input dialog then an image window will open at the end of the processing. If Save results image to disk is checked the image is saved to the save directory in tiff format. 3. Script and Batch Processing 3 scripts are provided on the update site to facilitate the use of the transcript analysis plugin. They can be accessed via the Fiji menu Plugins>Cell Transcript: get_transcripts_maxima_distribution.py: It creates a list of maxima intensity in the filtered transcript image. This information data are useful to build a distribution of maxima intensity and determine an adequate threshold for transcript detection. When selecting this menu entry an GUI will pop up requesting the location of the file to analyze, the location where to save the list of maxima intensity, the typical transcript radius (see description in section 2) and the channel to analyze (see description in section 2) get_transcripts_maxima_distribution_batch.py: this is a batch version of the previous script. It will measure the maxima intensity of multiple files in a folder. Once started the script will request a Source directory, the name of the image subfolder (i.e. not the full path) which is contained into the source directory, the name of the directory where result will be written and finally the analysis parameters (transcript radius and channel to analyze). cell_transcript_analysis.py : this is a batch version of the transcript analysis plugin. It creates a complete analysis of the transcript for the image in a folder. The script expects that the data are organized in 2 folders belonging to the same source folder. The first subfolder contains the raw images and the second the corresponding cell mask. If the cell mask is missing the process will run without the mask. The result folder will also be created in that source folder. Once the script is started a user interface pops up requesting the analysis parameters: the source folder, the name of the raw image subfolder, the name of the mask subfolder, the name of the result
19 subfolder. All the other parameters are the same as those requested in the transcript analysis plugin. One can refer to section 2 for more details. Figure 4. Illustration of input and output of the Transcript analysis plugin. (A) smfish transcript signal. (B) DAPI signal for the nuclei. (C) Cell and tissue masks. (D) Detected transcripts. (E) Cell and nuclear segmentation, merged with the original DAPI signal. (F) GUI for the Transcript analysis plugin. (G) Intensity distribution plot of all detected maxima. The vertical red line indicates the threshold used for transcript detection. (H) Scatter plot of Area vs Maximum intensity of all detected transcripts and transcription foci. Green: cytoplasmic transcripts, blue: nuclear transcripts, red: transcription foci. Red lines depict thresholds used for the detection of transcription foci.
20 Figure 5. input interfaces of to the scripts. (A)GUI for the batch analysis of the transcript analysis. (B) Close up on the dashed square in A showing the details data parameters. (C) GUI for the transcript maxima distribution script. (D) GUI for the batch version of the transcript maxima distribution script. 4. Methods description 4.1 Analysis Pipeline overview The analysis pipeline is composed of two parts. First, labels are created to describe the nuclei, cells and transcripts regions (image analysis). In a second step, several descriptors are measured in each of these nuclei, cells and transcript regions to characterize them (data analysis). Image analysis: In: nuclei raw image, transcript raw image, (cell labels). Out: nuclei labelling, extended nuclei labelling, cell labelling, transcript labelling, filtered transcripts. The successive steps of the image processing are as follows: Segment nuclei from nuclei raw image (i.e. create nuclei labelling).
21 If a cell labelling is provided, assign nuclei to cells and relabel nuclei and cell labelling to fit the assignment. Otherwise create a cell labelling by dilating the nuclei labelling. Dilate the nuclei labelling to create an extended nuclei labelling for later foci detection. Segment the transcripts (i.e. perform transcripts labelling). If Correct transcript over-segmentation is checked: update the transcript label to prevent over-segmentation of transcription foci. Create a filtered transcript image for further transcript intensity measures by removing the background. Data analysis: In: nuclei labelling, extended nuclei labelling, cells labelling, transcript labelling, filtered transcripts, (cell annotation). Out: cell measures, transcript measures. The successive steps of the data analysis are as follows: Get cell measures. Get transcript measures. Update transcript measures with cell, nuclei and extended nuclei parenthood. Update cell measures with transcripts counts in nuclei and cells. Update transcript measures and cell measures with transcription foci information Image analysis methods details Segment nuclei In: nuclei raw image; Out: nuclei labelling. Method description: The segmentation of nuclei can be solved in 2D by making a maximum projection of the original nuclei image. This strategy is satisfying since the tissue sections are less than one cell layer thick, which makes it very unlikely that two nuclei overlap. A background subtraction is performed on the projection to remove slowly varying intensity in the image background such as out of focus signal. The filter radius is chosen to be similar or larger than the largest nuclear radius to make sure the nuclear signal is not affected by the filtering. For this, the input parameter Rnuc (i.e. nuclei size ) is used. Nuclei are then detected with a difference of Gaussian filter (DOG filter) followed by maxima detection. The DOG filter enhances roundish structures, such as nuclei, at the chosen scale 1 σ. That scale should not be too large or too small to avoid respectively fusion of close objects or splitting of large objects. We chose σ equal to Rnuc/4. The nuclei are then detected as positive maxima of intensity in the DOG filtered image. To avoid over-detection only the maxima with dynamics 2 superior to ε are kept. ε is not too sensitive. It should be superior to one and smaller than the dynamics of 1 The DOG filter smooths the image with a Gaussian kernel of variance σ and subtract from it a copy of the image smoothed with a kernel of variance 2σ. 2 The dynamic of a local maximum is the difference of intensity between its own intensity and the intensity of the saddle separating it from another local maximum. If many such saddles exist, the intensity of the highest saddle is used to calculate the difference.
22 objects. We chose a value of 15 but any value between 5 and 100 would give only marginally different detections. Finally this detection is used to initiate a watershed 3 on the nuclei image. The nuclei image is smoothed with a Gaussian kernel of standard deviation σsmooth (σsmooth = Rnuc/10). The watershed algorithms used invades the image from the highest to the lowest intensities and stops at a lower threshold Tnuc. This threshold is obtained by multiplying the results of the Otsu method, TOtsu, with a user defined adjustment, αnuc (Tnuc = αnuc.totsu). The output of the watershed is a 2D labelled image representing the segmentation of the nuclei. Cell labelling and nuclei-to-cells assignments In: nuclei labelling, cell labelling (optional); Out: nuclei labelling, cell labelling. Method description: If a cell labelling is provided at the plugin inputs, the cell regions it defines have to be matched with the nuclear regions. Each cell region should contain either one, or no nucleus (if the nucleus of the cell has not been captured in the image section). However, in exceptional cases a cell region may overlap with multiple nuclei. In order to assign nuclei to cells and to resolve possible ambiguities we proceed as follows: each nucleus is assigned to the cell with which it has the largest overlap assuming that this overlap is at least 50% of the nuclear surface. This last point ensures that a nucleus can be paired with only one cell. If multiple nuclei overlap (partially) with a cell then the nucleus with the largest overlap (and in case of equality the one with the largest volume) is assigned to the cell. Following this assignment, nuclear and cell labels are updated such that nuclei get the same label as the cell they are assigned to. Region of nuclei overlapping with other cells are set to the label 1 and will not be taken into account in further analysis. If no cell labelling is provided as input to the plugin, a naïve cell segmentation is generated by dilating individual nucleus regions. Each region is dilated by a distance of Rnuc. If two regions overlap after dilation, pixels are attributed to the closest nucleus. In the resulting image one cell region is associated with exactly one nuclear region and these regions have the same label. Create an extended nuclei labelling In: nuclei labelling, RExtNuc; Out: extended nuclei labelling. Method description: An extended nuclei labelling is created by dilating the nuclei labelling by a distance of RExtNuc pixels. If two regions overlap after dilation, pixels are 3 The watershed algorithm can be seen as the flooding of a landscape of hills and valleys. The landscape altitude is determined by image signal intensity. The region to segment can be distinguished by different colors of liquid during the flooding. Sources of distinct colors are set at each seed points and fill the landscape starting first with the lower altitudes and gradually invading higher altitudes of the landscape. When liquids from two distinct sources meet, a dam is build (a watershed line) to keep the liquids separated. Additionally a threshold, T, can be set to stop the flooding when it reaches altitude T. This algorithm describes an invasion of the landscape from the lower altitudes to the highest one but in practice the principle can also be reversed. In that case the process start on hills and gradually visit lower altitudes till the bottom of the landscape or a threshold is reached.
23 attributed to the closest nuclei. Finally, parts of extended nuclei that overlap other cell region are set to 0. Transcripts segmentation In: transcript raw image, Rtx, Ttx; Out: transcript labelling. Method description: the 3D transcripts image is first filtered to remove large-scale variation with a tophat filtering (i.e. a background subtraction) of parameter Rtx. Then the image is smoothed with a Gaussian kernel of standard deviation σtx to remove the noise. This value should not be too large to avoid smoothing out of the transcripts and is fixed to 1 pixel. The transcripts are then detected as local maxima of that image. To avoid multiple maxima detections within a single transcript a minimum distance between local maxima is imposed. Its value is set to Rtx. All maxima superior to the userdefined threshold for transcript intensity, Ttx are kept as transcripts detection. Finally these detections are used to initiate a watershed segmentation of the transcripts. The watershed process is stop at 2/3 of Ttx. As a result a 3D labelled image is obtained where each transcript region has a distinct integer value. Additionally a list of all maxima and their intensities is generated and used -if requested- to display a distribution of maxima intensity, which can be used to choose Ttx. Correct over-segmentation of large spots In: transcript labelling,rtx,rfoci,ttx; Out: corrected transcript labelling. Method description: The spots corresponding to foci are first detected during transcript detection. However, foci that are much larger than transcripts can be split during that process. This method is meant to correct over-segmentation of transcription foci. This method is not a detection of transcription foci but rather a filtering of the transcript segmentation. For that matter a background subtraction adapted to the radius of the large foci is then performed with a tophat filter of radius Rfoci. The maxima of the image are then detected. A threshold equal to the median plus 3 times the median absolute deviation of maxima intensity is then applied to the filtered image. All connected component smaller than a sphere of radius Rtx are then discarded. The remaining regions are then copied onto the image of the segmented transcript regions with unused labels value. Create a filtered transcript image In: transcript raw image, Rfoci; Out: filtered transcripts intensities. Method description: The raw image of transcripts is processed with a tophat filter of radius Rfoci to remove image background. That image will be used to do intensity-based characterisation of the transcripts and foci regions. Removing the background is important to prevent a systematic bias of the measures Data analysis methods details Get cell measures. In: cell labelling, nuclei labelling, cell annotation (optional); Out: cell measures;
24 Method description: cell labelling and image labels are processed to extract per cell: Id, surface area, surface area of the corresponding nucleus, average position along the image axis (i.e. X and Y) and the region type as indicated by the cell annotation masks. Get transcript measures. In: transcript labelling, filtered transcripts intensities; Out: transcripts measures; Method description: Transcript labelling and the filtered transcript image are read to obtain for each transcript region: Id, surface area, average intensity, maximum intensity, integrated intensity and position along x, y and z axis. Update transcript measures with cell-, nuclei- and extended nuclei label they belong to. In: transcript measures, transcript labelling, cell labelling, nuclei labelling, extended nuclei labelling; Out: localized transcript measures; Method description: For each transcript the nucleus (or extended nucleus, cell) it belongs to is determined by looking at the nuclear (or extended nuclear, cell) label at the position of the transcript. The z-position is not considered since cell and nuclear labelling are 2D. Update cell measures with transcripts counts in nuclei and cells. In: cell measures, localized transcripts measures, cell labelling, transcript labelling; Out: cell transcript measures; Method description: For each cell, the number of transcripts is counted in the whole cell and in the nuclei and a density of transcripts per pixel is measured. The transcript density per μm 3 can be calculated based on the pixel size. Update transcript measures and cell measures with foci information In: Cell transcript measures, localized transcript measures, foci threshold adjustments; Out: cell measures, transcripts measures; Method description: Foci are detected among the transcripts based on their maximum intensity, volume and position (extended nuclear region). Those transcripts which features significantly deviate from the median transcripts intensity and area are selected as foci. On both features we use the following threshold Tfeat: Tfeat = Medianfeat + 3.αfeat.MADfeat where Medianfeat and MADfeat are the median and the median absolute deviation of the feature among all transcripts detected, respectively. αfeat is a user-defined value that can be used to adjust the value of the threshold for the feature. In each detected transcription focus the number of transcripts is counted by dividing the integrated intensity of the focus with the median integrated intensity of the transcripts. The estimated number of transcripts per foci is then saved for each cell. If one extended nucleus contains more than 2 foci, the number of transcripts of the supernumerary (smallest) foci are summed and added to the cell information in a separate column. ***
EECS490: Digital Image Processing. Lecture #22
 Lecture #22 Gold Standard project images Otsu thresholding Local thresholding Region segmentation Watershed segmentation Frequency-domain techniques Project Images 1 Project Images 2 Project Images 3 Project
Lecture #22 Gold Standard project images Otsu thresholding Local thresholding Region segmentation Watershed segmentation Frequency-domain techniques Project Images 1 Project Images 2 Project Images 3 Project
Neural Circuit Tracer
 2014 Neural Circuit Tracer Software for automated tracing of neurites from light microscopy stacks of images User Guide Version 4.0 Contents 1. Introduction... 2 2. System requirements and installation...
2014 Neural Circuit Tracer Software for automated tracing of neurites from light microscopy stacks of images User Guide Version 4.0 Contents 1. Introduction... 2 2. System requirements and installation...
GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow.
![GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow. GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow.](/thumbs/75/71593469.jpg) Manual KNIME Image Processing :: First steps This manual aims to provide first insights in KNIME Image Processing. The image processing workflow Count nuclei is described in detail and can be downloaded
Manual KNIME Image Processing :: First steps This manual aims to provide first insights in KNIME Image Processing. The image processing workflow Count nuclei is described in detail and can be downloaded
Definiens. Tissue Studio 4.4. Tutorial 4: Manual ROI Selection and Marker Area Detection
 Definiens Tissue Studio 4.4 Tutorial 4: Manual ROI Selection and Marker Area Detection Tutorial 4: Manual ROI Selection and Marker Area Detection Imprint and Version Copyright 2017 Definiens AG. All rights
Definiens Tissue Studio 4.4 Tutorial 4: Manual ROI Selection and Marker Area Detection Tutorial 4: Manual ROI Selection and Marker Area Detection Imprint and Version Copyright 2017 Definiens AG. All rights
IMAGEJ FINDFOCI Plugins
 IMAGEJ FINDFOCI Plugins Alex Herbert MRC Genome Damage and Stability Centre School of Life Sciences University of Sussex Science Road Falmer BN1 9RQ a.herbert@sussex.ac.uk Table of Contents Introduction...5
IMAGEJ FINDFOCI Plugins Alex Herbert MRC Genome Damage and Stability Centre School of Life Sciences University of Sussex Science Road Falmer BN1 9RQ a.herbert@sussex.ac.uk Table of Contents Introduction...5
Vector Xpression 3. Speed Tutorial: III. Creating a Script for Automating Normalization of Data
 Vector Xpression 3 Speed Tutorial: III. Creating a Script for Automating Normalization of Data Table of Contents Table of Contents...1 Important: Please Read...1 Opening Data in Raw Data Viewer...2 Creating
Vector Xpression 3 Speed Tutorial: III. Creating a Script for Automating Normalization of Data Table of Contents Table of Contents...1 Important: Please Read...1 Opening Data in Raw Data Viewer...2 Creating
Segmentation
 Lecture 6: Segmentation 24--4 Robin Strand Centre for Image Analysis Dept. of IT Uppsala University Today What is image segmentation? A smörgåsbord of methods for image segmentation: Thresholding Edge-based
Lecture 6: Segmentation 24--4 Robin Strand Centre for Image Analysis Dept. of IT Uppsala University Today What is image segmentation? A smörgåsbord of methods for image segmentation: Thresholding Edge-based
Segmentation
 Lecture 6: Segmentation 215-13-11 Filip Malmberg Centre for Image Analysis Uppsala University 2 Today What is image segmentation? A smörgåsbord of methods for image segmentation: Thresholding Edge-based
Lecture 6: Segmentation 215-13-11 Filip Malmberg Centre for Image Analysis Uppsala University 2 Today What is image segmentation? A smörgåsbord of methods for image segmentation: Thresholding Edge-based
Introduction. Loading Images
 Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: http://www.cellprofiler.org/. CellProfiler
Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: http://www.cellprofiler.org/. CellProfiler
1. ABOUT INSTALLATION COMPATIBILITY SURESIM WORKFLOWS a. Workflow b. Workflow SURESIM TUTORIAL...
 SuReSim manual 1. ABOUT... 2 2. INSTALLATION... 2 3. COMPATIBILITY... 2 4. SURESIM WORKFLOWS... 2 a. Workflow 1... 3 b. Workflow 2... 4 5. SURESIM TUTORIAL... 5 a. Import Data... 5 b. Parameter Selection...
SuReSim manual 1. ABOUT... 2 2. INSTALLATION... 2 3. COMPATIBILITY... 2 4. SURESIM WORKFLOWS... 2 a. Workflow 1... 3 b. Workflow 2... 4 5. SURESIM TUTORIAL... 5 a. Import Data... 5 b. Parameter Selection...
TexRAD Research Version Client User Guide Version 3.9
 Imaging tools for medical decision makers Cambridge Computed Imaging Ltd Grange Park Broadway Bourn Cambridge CB23 2TA UK TexRAD Research Version Client User Guide Version 3.9 Release date 23/05/2016 Number
Imaging tools for medical decision makers Cambridge Computed Imaging Ltd Grange Park Broadway Bourn Cambridge CB23 2TA UK TexRAD Research Version Client User Guide Version 3.9 Release date 23/05/2016 Number
BRAT Manual V1.1. May 6th, Christian Göschl & Wolfgang Busch. for correspondence:
 BRAT Manual V1.1 May 6th, 2014 Christian Göschl & Wolfgang Busch Email for correspondence: wolfgang.busch@gmi.oeaw.ac.at Gregor Mendel Institute Dr. Bohr-Gasse 3 1030 Vienna Austria 1 2 Brat Segmentation
BRAT Manual V1.1 May 6th, 2014 Christian Göschl & Wolfgang Busch Email for correspondence: wolfgang.busch@gmi.oeaw.ac.at Gregor Mendel Institute Dr. Bohr-Gasse 3 1030 Vienna Austria 1 2 Brat Segmentation
C E N T E R A T H O U S T O N S C H O O L of H E A L T H I N F O R M A T I O N S C I E N C E S. Image Operations II
 T H E U N I V E R S I T Y of T E X A S H E A L T H S C I E N C E C E N T E R A T H O U S T O N S C H O O L of H E A L T H I N F O R M A T I O N S C I E N C E S Image Operations II For students of HI 5323
T H E U N I V E R S I T Y of T E X A S H E A L T H S C I E N C E C E N T E R A T H O U S T O N S C H O O L of H E A L T H I N F O R M A T I O N S C I E N C E S Image Operations II For students of HI 5323
Image Analysis Image Segmentation (Basic Methods)
 Image Analysis Image Segmentation (Basic Methods) Christophoros Nikou cnikou@cs.uoi.gr Images taken from: R. Gonzalez and R. Woods. Digital Image Processing, Prentice Hall, 2008. Computer Vision course
Image Analysis Image Segmentation (Basic Methods) Christophoros Nikou cnikou@cs.uoi.gr Images taken from: R. Gonzalez and R. Woods. Digital Image Processing, Prentice Hall, 2008. Computer Vision course
Definiens. Tissue Studio 4.2. Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection
 Definiens Tissue Studio 4.2 Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection Imprint and Version Copyright 2015
Definiens Tissue Studio 4.2 Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection Imprint and Version Copyright 2015
cief Data Analysis Chapter Overview Chapter 12:
 page 285 Chapter 12: cief Data Analysis Chapter Overview Analysis Screen Overview Opening Run Files How Run Data is Displayed Viewing Run Data Data Notifications and Warnings Checking Your Results Group
page 285 Chapter 12: cief Data Analysis Chapter Overview Analysis Screen Overview Opening Run Files How Run Data is Displayed Viewing Run Data Data Notifications and Warnings Checking Your Results Group
SuRVoS Workbench. Super-Region Volume Segmentation. Imanol Luengo
 SuRVoS Workbench Super-Region Volume Segmentation Imanol Luengo Index - The project - What is SuRVoS - SuRVoS Overview - What can it do - Overview of the internals - Current state & Limitations - Future
SuRVoS Workbench Super-Region Volume Segmentation Imanol Luengo Index - The project - What is SuRVoS - SuRVoS Overview - What can it do - Overview of the internals - Current state & Limitations - Future
Manual for Microfilament Analyzer
 Manual for Microfilament Analyzer Eveline Jacques & Jan Buytaert, Michał Lewandowski, Joris Dirckx, Jean-Pierre Verbelen, Kris Vissenberg University of Antwerp 1 Image data 2 image files are needed: a
Manual for Microfilament Analyzer Eveline Jacques & Jan Buytaert, Michał Lewandowski, Joris Dirckx, Jean-Pierre Verbelen, Kris Vissenberg University of Antwerp 1 Image data 2 image files are needed: a
Virtual Frap User Guide
 Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
9 Tumor Blood Vessels: 3-D Tubular Network Analysis
 Chap. 9 2015/8/20 14:02 page 1 1 9 Tumor Blood Vessels: 3-D Tubular Network Analysis Chap. 9 2015/8/20 14:02 page 2 2 Chap. 9 2015/8/20 14:02 page 3 3 Contents 9 Tumor Blood Vessels: 3-D Tubular Network
Chap. 9 2015/8/20 14:02 page 1 1 9 Tumor Blood Vessels: 3-D Tubular Network Analysis Chap. 9 2015/8/20 14:02 page 2 2 Chap. 9 2015/8/20 14:02 page 3 3 Contents 9 Tumor Blood Vessels: 3-D Tubular Network
LEGENDplex Data Analysis Software Version 8 User Guide
 LEGENDplex Data Analysis Software Version 8 User Guide Introduction Welcome to the user s guide for Version 8 of the LEGENDplex data analysis software for Windows based computers 1. This tutorial will
LEGENDplex Data Analysis Software Version 8 User Guide Introduction Welcome to the user s guide for Version 8 of the LEGENDplex data analysis software for Windows based computers 1. This tutorial will
Frequency Distributions
 Displaying Data Frequency Distributions After collecting data, the first task for a researcher is to organize and summarize the data so that it is possible to get a general overview of the results. Remember,
Displaying Data Frequency Distributions After collecting data, the first task for a researcher is to organize and summarize the data so that it is possible to get a general overview of the results. Remember,
Desktop Studio: Charts
 Desktop Studio: Charts Intellicus Enterprise Reporting and BI Platform Intellicus Technologies info@intellicus.com www.intellicus.com Working with Charts i Copyright 2011 Intellicus Technologies This document
Desktop Studio: Charts Intellicus Enterprise Reporting and BI Platform Intellicus Technologies info@intellicus.com www.intellicus.com Working with Charts i Copyright 2011 Intellicus Technologies This document
This version is available from Sussex Research Online:
 : a focus detection algorithm with automated parameter training that closely matches human assignments, reduces human inconsistencies and increases speed of analysis Article (Published Version) Herbert,
: a focus detection algorithm with automated parameter training that closely matches human assignments, reduces human inconsistencies and increases speed of analysis Article (Published Version) Herbert,
Desktop Studio: Charts. Version: 7.3
 Desktop Studio: Charts Version: 7.3 Copyright 2015 Intellicus Technologies This document and its content is copyrighted material of Intellicus Technologies. The content may not be copied or derived from,
Desktop Studio: Charts Version: 7.3 Copyright 2015 Intellicus Technologies This document and its content is copyrighted material of Intellicus Technologies. The content may not be copied or derived from,
SOP.8. Automated Analysis
 Circulating Tumor Cells TheRapeutic APheresis: a novel biotechnology enabling personalized therapy for all cancer patients SOP.8. Automated Analysis Scoring CTC with ICY Introduction This Standard Operating
Circulating Tumor Cells TheRapeutic APheresis: a novel biotechnology enabling personalized therapy for all cancer patients SOP.8. Automated Analysis Scoring CTC with ICY Introduction This Standard Operating
Part 3: Image Processing
 Part 3: Image Processing Image Filtering and Segmentation Georgy Gimel farb COMPSCI 373 Computer Graphics and Image Processing 1 / 60 1 Image filtering 2 Median filtering 3 Mean filtering 4 Image segmentation
Part 3: Image Processing Image Filtering and Segmentation Georgy Gimel farb COMPSCI 373 Computer Graphics and Image Processing 1 / 60 1 Image filtering 2 Median filtering 3 Mean filtering 4 Image segmentation
Lecture: Segmentation I FMAN30: Medical Image Analysis. Anders Heyden
 Lecture: Segmentation I FMAN30: Medical Image Analysis Anders Heyden 2017-11-13 Content What is segmentation? Motivation Segmentation methods Contour-based Voxel/pixel-based Discussion What is segmentation?
Lecture: Segmentation I FMAN30: Medical Image Analysis Anders Heyden 2017-11-13 Content What is segmentation? Motivation Segmentation methods Contour-based Voxel/pixel-based Discussion What is segmentation?
Matthew Baker. Visualisation of Electrophoresis Gels using TEX
 Visualisation of Electrophoresis Gels using TEX 1 Visualisation of Electrophoresis Gels using TEX Matthew Baker This paper describes a TEX system for creating interactive PDF files to visualize electrophoresis
Visualisation of Electrophoresis Gels using TEX 1 Visualisation of Electrophoresis Gels using TEX Matthew Baker This paper describes a TEX system for creating interactive PDF files to visualize electrophoresis
Introduction to Celleste Imaging Analysis Software
 Introduction to Celleste Imaging Analysis Software USER GUIDE Publication Number MAN0018003 Revision A.0 For Research Use Only. Not for use in diagnostic procedures. Manufacturer: Life Technologies Corporation
Introduction to Celleste Imaging Analysis Software USER GUIDE Publication Number MAN0018003 Revision A.0 For Research Use Only. Not for use in diagnostic procedures. Manufacturer: Life Technologies Corporation
A Tutorial Guide to Tribology Plug-in
 Supplementary Material A Tutorial Guide to Tribology Plug-in Tribology An ImageJ Plugin for surface topography analysis of laser textured surfaces. General Description This plugin presents an easy-to-use
Supplementary Material A Tutorial Guide to Tribology Plug-in Tribology An ImageJ Plugin for surface topography analysis of laser textured surfaces. General Description This plugin presents an easy-to-use
Algorithm User Guide:
 Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
CS443: Digital Imaging and Multimedia Binary Image Analysis. Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University
 CS443: Digital Imaging and Multimedia Binary Image Analysis Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University Outlines A Simple Machine Vision System Image segmentation by thresholding
CS443: Digital Imaging and Multimedia Binary Image Analysis Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University Outlines A Simple Machine Vision System Image segmentation by thresholding
DETAILED INSTRUCTIONS FOR RUNNING BINARY_TRAVERSER
 DETAILED INSTRUCTIONS FOR RUNNING BINARY_TRAVERSER These instructions are also available at http://www.geo.umass.edu/climate/lewis/analysis/ Install ImageJ from http://rsb.info.nih.gov/ij/download.html.
DETAILED INSTRUCTIONS FOR RUNNING BINARY_TRAVERSER These instructions are also available at http://www.geo.umass.edu/climate/lewis/analysis/ Install ImageJ from http://rsb.info.nih.gov/ij/download.html.
Insight: Measurement Tool. User Guide
 OMERO Beta v2.2: Measurement Tool User Guide - 1 - October 2007 Insight: Measurement Tool User Guide Open Microscopy Environment: http://www.openmicroscopy.org OMERO Beta v2.2: Measurement Tool User Guide
OMERO Beta v2.2: Measurement Tool User Guide - 1 - October 2007 Insight: Measurement Tool User Guide Open Microscopy Environment: http://www.openmicroscopy.org OMERO Beta v2.2: Measurement Tool User Guide
3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen
 3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen Dataset: Huh7.tif (original), obtained by Serial Block Face-SEM (Gatan 3View) Huh7_crop.tif
3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen Dataset: Huh7.tif (original), obtained by Serial Block Face-SEM (Gatan 3View) Huh7_crop.tif
QDA Miner. Addendum v2.0
 QDA Miner Addendum v2.0 QDA Miner is an easy-to-use qualitative analysis software for coding, annotating, retrieving and reviewing coded data and documents such as open-ended responses, customer comments,
QDA Miner Addendum v2.0 QDA Miner is an easy-to-use qualitative analysis software for coding, annotating, retrieving and reviewing coded data and documents such as open-ended responses, customer comments,
OpenForms360 Validation User Guide Notable Solutions Inc.
 OpenForms360 Validation User Guide 2011 Notable Solutions Inc. 1 T A B L E O F C O N T EN T S Introduction...5 What is OpenForms360 Validation?... 5 Using OpenForms360 Validation... 5 Features at a glance...
OpenForms360 Validation User Guide 2011 Notable Solutions Inc. 1 T A B L E O F C O N T EN T S Introduction...5 What is OpenForms360 Validation?... 5 Using OpenForms360 Validation... 5 Features at a glance...
Lab 12: Sampling and Interpolation
 Lab 12: Sampling and Interpolation What You ll Learn: -Systematic and random sampling -Majority filtering -Stratified sampling -A few basic interpolation methods Data for the exercise are in the L12 subdirectory.
Lab 12: Sampling and Interpolation What You ll Learn: -Systematic and random sampling -Majority filtering -Stratified sampling -A few basic interpolation methods Data for the exercise are in the L12 subdirectory.
Use stage 16 embryos see Pereanu et al., 2007 for morphological staging criteria. First start Vaa3d (http://vaa3d.org/; Peng et al., 2010).
 Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
TexRAD Research Version Client User Guide 1.1
 The Barn, Manor Road Church Lane Chilcompton Somerset BA3 4HP TexRAD Research Version Client User Guide 1.1 Release date 27/02/2013 Number of Pages 21 Replaces None Status Draft Comment Copyright 2013
The Barn, Manor Road Church Lane Chilcompton Somerset BA3 4HP TexRAD Research Version Client User Guide 1.1 Release date 27/02/2013 Number of Pages 21 Replaces None Status Draft Comment Copyright 2013
Lab Practical - Limit Equilibrium Analysis of Engineered Slopes
 Lab Practical - Limit Equilibrium Analysis of Engineered Slopes Part 1: Planar Analysis A Deterministic Analysis This exercise will demonstrate the basics of a deterministic limit equilibrium planar analysis
Lab Practical - Limit Equilibrium Analysis of Engineered Slopes Part 1: Planar Analysis A Deterministic Analysis This exercise will demonstrate the basics of a deterministic limit equilibrium planar analysis
Topic 4 Image Segmentation
 Topic 4 Image Segmentation What is Segmentation? Why? Segmentation important contributing factor to the success of an automated image analysis process What is Image Analysis: Processing images to derive
Topic 4 Image Segmentation What is Segmentation? Why? Segmentation important contributing factor to the success of an automated image analysis process What is Image Analysis: Processing images to derive
Time Stamp Detection and Recognition in Video Frames
 Time Stamp Detection and Recognition in Video Frames Nongluk Covavisaruch and Chetsada Saengpanit Department of Computer Engineering, Chulalongkorn University, Bangkok 10330, Thailand E-mail: nongluk.c@chula.ac.th
Time Stamp Detection and Recognition in Video Frames Nongluk Covavisaruch and Chetsada Saengpanit Department of Computer Engineering, Chulalongkorn University, Bangkok 10330, Thailand E-mail: nongluk.c@chula.ac.th
Image segmentation. Stefano Ferrari. Università degli Studi di Milano Methods for Image Processing. academic year
 Image segmentation Stefano Ferrari Università degli Studi di Milano stefano.ferrari@unimi.it Methods for Image Processing academic year 2017 2018 Segmentation by thresholding Thresholding is the simplest
Image segmentation Stefano Ferrari Università degli Studi di Milano stefano.ferrari@unimi.it Methods for Image Processing academic year 2017 2018 Segmentation by thresholding Thresholding is the simplest
CRITERION Vantage 3 Admin Training Manual Contents Introduction 5
 CRITERION Vantage 3 Admin Training Manual Contents Introduction 5 Running Admin 6 Understanding the Admin Display 7 Using the System Viewer 11 Variables Characteristic Setup Window 19 Using the List Viewer
CRITERION Vantage 3 Admin Training Manual Contents Introduction 5 Running Admin 6 Understanding the Admin Display 7 Using the System Viewer 11 Variables Characteristic Setup Window 19 Using the List Viewer
Image Analysis Lecture Segmentation. Idar Dyrdal
 Image Analysis Lecture 9.1 - Segmentation Idar Dyrdal Segmentation Image segmentation is the process of partitioning a digital image into multiple parts The goal is to divide the image into meaningful
Image Analysis Lecture 9.1 - Segmentation Idar Dyrdal Segmentation Image segmentation is the process of partitioning a digital image into multiple parts The goal is to divide the image into meaningful
PHASEQUANT (User Manual)
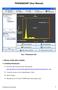 PHASEQUANT (User Manual) ROI Pane Toolbar Threshold Bar Canvas Threshold Display Training Pane Buttons Fig 1. PhaseQuant GUI 1. INSTALLATION AND LOADING 1.1 Installing PhaseQuant Download the zip file
PHASEQUANT (User Manual) ROI Pane Toolbar Threshold Bar Canvas Threshold Display Training Pane Buttons Fig 1. PhaseQuant GUI 1. INSTALLATION AND LOADING 1.1 Installing PhaseQuant Download the zip file
Elixir Ad-hoc Report. Release Elixir Technology Pte Ltd
 Elixir Ad-hoc Report Release 4.0.0 Elixir Technology Pte Ltd Elixir Ad-hoc Report: Release 4.0.0 Elixir Technology Pte Ltd Published 2015 Copyright 2015 Elixir Technology Pte Ltd All rights reserved. Java
Elixir Ad-hoc Report Release 4.0.0 Elixir Technology Pte Ltd Elixir Ad-hoc Report: Release 4.0.0 Elixir Technology Pte Ltd Published 2015 Copyright 2015 Elixir Technology Pte Ltd All rights reserved. Java
Multivariate Calibration Quick Guide
 Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
doi: /
 Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Edge and local feature detection - 2. Importance of edge detection in computer vision
 Edge and local feature detection Gradient based edge detection Edge detection by function fitting Second derivative edge detectors Edge linking and the construction of the chain graph Edge and local feature
Edge and local feature detection Gradient based edge detection Edge detection by function fitting Second derivative edge detectors Edge linking and the construction of the chain graph Edge and local feature
The walkthrough is available at /
 The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
Algorithm User Guide:
 Algorithm User Guide: Microvessel Analysis Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to use
Algorithm User Guide: Microvessel Analysis Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to use
Appendix 1: Manual for Fovea Software
 1 Appendix 1: Manual for Fovea Software Fovea is a software to calculate foveal width and depth by detecting local maxima and minima from fovea images in order to estimate foveal depth and width. This
1 Appendix 1: Manual for Fovea Software Fovea is a software to calculate foveal width and depth by detecting local maxima and minima from fovea images in order to estimate foveal depth and width. This
Crop Counting and Metrics Tutorial
 Crop Counting and Metrics Tutorial The ENVI Crop Science platform contains remote sensing analytic tools for precision agriculture and agronomy. In this tutorial you will go through a typical workflow
Crop Counting and Metrics Tutorial The ENVI Crop Science platform contains remote sensing analytic tools for precision agriculture and agronomy. In this tutorial you will go through a typical workflow
QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL
 QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL User manual for QIAseq Targeted RNAscan Panel Analysis 0.5.2 beta 1 Windows, Mac OS X and Linux February 5, 2018 This software is for research
QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL User manual for QIAseq Targeted RNAscan Panel Analysis 0.5.2 beta 1 Windows, Mac OS X and Linux February 5, 2018 This software is for research
Background information:
 Image- based screening using subcellular localization of FOXO1A in osteosarcoma cells: A computer exercise using CellProfiler & CellProfiler Analyst software Carolina Wählby, Martin Simonsson, Megan Rokop
Image- based screening using subcellular localization of FOXO1A in osteosarcoma cells: A computer exercise using CellProfiler & CellProfiler Analyst software Carolina Wählby, Martin Simonsson, Megan Rokop
Epithelial rosette detection in microscopic images
 Epithelial rosette detection in microscopic images Kun Liu,3, Sandra Ernst 2,3, Virginie Lecaudey 2,3 and Olaf Ronneberger,3 Department of Computer Science 2 Department of Developmental Biology 3 BIOSS
Epithelial rosette detection in microscopic images Kun Liu,3, Sandra Ernst 2,3, Virginie Lecaudey 2,3 and Olaf Ronneberger,3 Department of Computer Science 2 Department of Developmental Biology 3 BIOSS
4. TROUBLESHOOTING PREVIOUS VERSIONS RUN LOLITRACK ALWAYS AS ADMIN WIBU SOFTWARE PROTECTION... 30
 Version 4.2.0 CONTENTS 1. GETTING STARTED... 2 2. TYPICAL APPLICATIONS... 4 3. USER GUIDE... 5 3.1 SINGLE OBJECT MODE... 7 3.2 SINGLE ARENA MODE... 12 3.3 EVENT RECORDER... 19 3.4 BATCH TRACKING... 21
Version 4.2.0 CONTENTS 1. GETTING STARTED... 2 2. TYPICAL APPLICATIONS... 4 3. USER GUIDE... 5 3.1 SINGLE OBJECT MODE... 7 3.2 SINGLE ARENA MODE... 12 3.3 EVENT RECORDER... 19 3.4 BATCH TRACKING... 21
Digital Image Processing. Prof. P.K. Biswas. Department of Electronics & Electrical Communication Engineering
 Digital Image Processing Prof. P.K. Biswas Department of Electronics & Electrical Communication Engineering Indian Institute of Technology, Kharagpur Image Segmentation - III Lecture - 31 Hello, welcome
Digital Image Processing Prof. P.K. Biswas Department of Electronics & Electrical Communication Engineering Indian Institute of Technology, Kharagpur Image Segmentation - III Lecture - 31 Hello, welcome
Modify Panel. Flatten Tab
 AFM Image Processing Most images will need some post acquisition processing. A typical procedure is to: i) modify the image by flattening, using a planefit, and possibly also a mask, ii) analyzing the
AFM Image Processing Most images will need some post acquisition processing. A typical procedure is to: i) modify the image by flattening, using a planefit, and possibly also a mask, ii) analyzing the
Elixir Ad-hoc Report. Release Elixir Technology Pte Ltd
 Elixir Ad-hoc Report Release 3.5.0 Elixir Technology Pte Ltd Elixir Ad-hoc Report: Release 3.5.0 Elixir Technology Pte Ltd Published 2014 Copyright 2014 Elixir Technology Pte Ltd All rights reserved. Java
Elixir Ad-hoc Report Release 3.5.0 Elixir Technology Pte Ltd Elixir Ad-hoc Report: Release 3.5.0 Elixir Technology Pte Ltd Published 2014 Copyright 2014 Elixir Technology Pte Ltd All rights reserved. Java
Importing and processing a DGGE gel image
 BioNumerics Tutorial: Importing and processing a DGGE gel image 1 Aim Comprehensive tools for the processing of electrophoresis fingerprints, both from slab gels and capillary sequencers are incorporated
BioNumerics Tutorial: Importing and processing a DGGE gel image 1 Aim Comprehensive tools for the processing of electrophoresis fingerprints, both from slab gels and capillary sequencers are incorporated
Getting Started with NEAT-DATA: SOM Editing and Imputation Software Implementation
 Laboratory of Data Analysis University of Jyväskylä EUREDIT - WP6 reports Getting Started with NEAT-DATA: SOM Editing and Imputation Software Implementation Pasi P. Koikkalainen and Ismo Horppu University
Laboratory of Data Analysis University of Jyväskylä EUREDIT - WP6 reports Getting Started with NEAT-DATA: SOM Editing and Imputation Software Implementation Pasi P. Koikkalainen and Ismo Horppu University
Image Processing
 Image Processing 159.731 Canny Edge Detection Report Syed Irfanullah, Azeezullah 00297844 Danh Anh Huynh 02136047 1 Canny Edge Detection INTRODUCTION Edges Edges characterize boundaries and are therefore
Image Processing 159.731 Canny Edge Detection Report Syed Irfanullah, Azeezullah 00297844 Danh Anh Huynh 02136047 1 Canny Edge Detection INTRODUCTION Edges Edges characterize boundaries and are therefore
FIFI-LS: Basic Cube Analysis using SOSPEX
 FIFI-LS: Basic Cube Analysis using SOSPEX Date: 1 Oct 2018 Revision: - CONTENTS 1 INTRODUCTION... 1 2 INGREDIENTS... 1 3 INSPECTING THE CUBE... 3 4 COMPARING TO A REFERENCE IMAGE... 5 5 REFERENCE VELOCITY
FIFI-LS: Basic Cube Analysis using SOSPEX Date: 1 Oct 2018 Revision: - CONTENTS 1 INTRODUCTION... 1 2 INGREDIENTS... 1 3 INSPECTING THE CUBE... 3 4 COMPARING TO A REFERENCE IMAGE... 5 5 REFERENCE VELOCITY
OBCOL. (Organelle Based CO-Localisation) Users Guide
 OBCOL (Organelle Based CO-Localisation) Users Guide INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual
OBCOL (Organelle Based CO-Localisation) Users Guide INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual
EPBscore user guide. Requirements:
 EPBscore user guide Requirements: Up to Matlab 2014a. Windows-only versions (both 32- and 64-bit) were tested. Toolboxes: Control, Images, Local, Matlab, Optim, Shared, Signal, Simulink, Stats, Wavelet
EPBscore user guide Requirements: Up to Matlab 2014a. Windows-only versions (both 32- and 64-bit) were tested. Toolboxes: Control, Images, Local, Matlab, Optim, Shared, Signal, Simulink, Stats, Wavelet
Supporting Information. High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images
 Supporting Information High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images Christine R. Laramy, 1, Keith A. Brown, 2, Matthew N. O Brien, 2 and Chad. A.
Supporting Information High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images Christine R. Laramy, 1, Keith A. Brown, 2, Matthew N. O Brien, 2 and Chad. A.
Textbook de.nbi Bioimage Analysis Workshop
 BIOIMAGE ANALYSIS WORKSHOP WITHIN THE FRAMEWORK OF HD-HUB Textbook de.nbi Bioimage Analysis Workshop An introduction into KNIME image analysis for life scientists Jürgen Reymann ViroQuant-CellNetworks
BIOIMAGE ANALYSIS WORKSHOP WITHIN THE FRAMEWORK OF HD-HUB Textbook de.nbi Bioimage Analysis Workshop An introduction into KNIME image analysis for life scientists Jürgen Reymann ViroQuant-CellNetworks
CITS 4402 Computer Vision
 CITS 4402 Computer Vision A/Prof Ajmal Mian Adj/A/Prof Mehdi Ravanbakhsh, CEO at Mapizy (www.mapizy.com) and InFarm (www.infarm.io) Lecture 02 Binary Image Analysis Objectives Revision of image formation
CITS 4402 Computer Vision A/Prof Ajmal Mian Adj/A/Prof Mehdi Ravanbakhsh, CEO at Mapizy (www.mapizy.com) and InFarm (www.infarm.io) Lecture 02 Binary Image Analysis Objectives Revision of image formation
Fusion Detection Using QIAseq RNAscan Panels
 Fusion Detection Using QIAseq RNAscan Panels June 11, 2018 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com ts-bioinformatics@qiagen.com
Fusion Detection Using QIAseq RNAscan Panels June 11, 2018 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com ts-bioinformatics@qiagen.com
FiloQuant manual V1.0 Table of Contents
 FiloQuant manual V1.0 Table of Contents 1) FiloQuant aims and distribution license...2 2) Installation...3 3) FiloQuant, step-by-step instructions (single images)...4 1: Choose the region of interest to
FiloQuant manual V1.0 Table of Contents 1) FiloQuant aims and distribution license...2 2) Installation...3 3) FiloQuant, step-by-step instructions (single images)...4 1: Choose the region of interest to
Introduction to version Instruction date
 Introduction to version 1.1.0 Instruction date 16.5.2008 Windows and Files Start by creating the window Open FCS data file By right-clicking the axis the list of available parameters appear. Right-click
Introduction to version 1.1.0 Instruction date 16.5.2008 Windows and Files Start by creating the window Open FCS data file By right-clicking the axis the list of available parameters appear. Right-click
Automatic Partiicle Tracking Software USE ER MANUAL Update: May 2015
 Automatic Particle Tracking Software USER MANUAL Update: May 2015 File Menu The micrograph below shows the panel displayed when a movie is opened, including a playback menu where most of the parameters
Automatic Particle Tracking Software USER MANUAL Update: May 2015 File Menu The micrograph below shows the panel displayed when a movie is opened, including a playback menu where most of the parameters
Supplementary Figures
 Supplementary Figures re co rd ed ch annels darkf ield MPM2 PI cell cycle phases G1/S/G2 prophase metaphase anaphase telophase bri ghtfield 55 pixel Supplementary Figure 1 Images of Jurkat cells captured
Supplementary Figures re co rd ed ch annels darkf ield MPM2 PI cell cycle phases G1/S/G2 prophase metaphase anaphase telophase bri ghtfield 55 pixel Supplementary Figure 1 Images of Jurkat cells captured
Points Lines Connected points X-Y Scatter. X-Y Matrix Star Plot Histogram Box Plot. Bar Group Bar Stacked H-Bar Grouped H-Bar Stacked
 Plotting Menu: QCExpert Plotting Module graphs offers various tools for visualization of uni- and multivariate data. Settings and options in different types of graphs allow for modifications and customizations
Plotting Menu: QCExpert Plotting Module graphs offers various tools for visualization of uni- and multivariate data. Settings and options in different types of graphs allow for modifications and customizations
English. Delta2D ANALYZING 2D GELS AS EASY AS POINT AND CLICK EXPLORING LIFE
 Getting started English 2D Western Blots Delta2D ANALYZING 2D GELS AS EASY AS POINT AND CLICK EXPLORING LIFE 2 Copyright DECODON GmbH. DECODON makes no representations, express or implied, with respect
Getting started English 2D Western Blots Delta2D ANALYZING 2D GELS AS EASY AS POINT AND CLICK EXPLORING LIFE 2 Copyright DECODON GmbH. DECODON makes no representations, express or implied, with respect
Ulrik Söderström 16 Feb Image Processing. Segmentation
 Ulrik Söderström ulrik.soderstrom@tfe.umu.se 16 Feb 2011 Image Processing Segmentation What is Image Segmentation? To be able to extract information from an image it is common to subdivide it into background
Ulrik Söderström ulrik.soderstrom@tfe.umu.se 16 Feb 2011 Image Processing Segmentation What is Image Segmentation? To be able to extract information from an image it is common to subdivide it into background
Tutorial: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and Expression measures
 : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
Automated AFM Image Processing User Manual
 Automated AFM Image Processing User Manual Starting The Program Open and run the GUI_run_me.m script in Matlab to start the program. The first thing to do is to select the folder that contains the images
Automated AFM Image Processing User Manual Starting The Program Open and run the GUI_run_me.m script in Matlab to start the program. The first thing to do is to select the folder that contains the images
3DReshaper Help DReshaper Beginner's Guide. Surveying
 3DReshaper Beginner's Guide Surveying 1 of 29 Cross sections Exercise: Tunnel analysis Surface analysis Exercise: Complete analysis of a concrete floor Surveying extraction Exercise: Automatic extraction
3DReshaper Beginner's Guide Surveying 1 of 29 Cross sections Exercise: Tunnel analysis Surface analysis Exercise: Complete analysis of a concrete floor Surveying extraction Exercise: Automatic extraction
Tutorial 2: Analysis of DIA/SWATH data in Skyline
 Tutorial 2: Analysis of DIA/SWATH data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantification.
Tutorial 2: Analysis of DIA/SWATH data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantification.
Methodology for spot quality evaluation
 Methodology for spot quality evaluation Semi-automatic pipeline in MAIA The general workflow of the semi-automatic pipeline analysis in MAIA is shown in Figure 1A, Manuscript. In Block 1 raw data, i.e..tif
Methodology for spot quality evaluation Semi-automatic pipeline in MAIA The general workflow of the semi-automatic pipeline analysis in MAIA is shown in Figure 1A, Manuscript. In Block 1 raw data, i.e..tif
Cognalysis TM Reserving System User Manual
 Cognalysis TM Reserving System User Manual Return to Table of Contents 1 Table of Contents 1.0 Starting an Analysis 3 1.1 Opening a Data File....3 1.2 Open an Analysis File.9 1.3 Create Triangles.10 2.0
Cognalysis TM Reserving System User Manual Return to Table of Contents 1 Table of Contents 1.0 Starting an Analysis 3 1.1 Opening a Data File....3 1.2 Open an Analysis File.9 1.3 Create Triangles.10 2.0
Plotting: Customizing the Page Display
 Plotting: Customizing the Page Display Setting the Page Orientation Graphs can be viewed in landscape or portrait page orientation. To change the page orientation of the active graph window, select File:Page
Plotting: Customizing the Page Display Setting the Page Orientation Graphs can be viewed in landscape or portrait page orientation. To change the page orientation of the active graph window, select File:Page
CLC Server. End User USER MANUAL
 CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
Chapter Eight: Contents
 Volume Three Modules 01 March 2002 i Chapter Eight: Contents (Output Visualizer 04 March 2002 LA-UR-00-1725 TRANSIMS 3.0) 1. INTRODUCTION...1 1.1 OVERVIEW... 1 1.2 REQUIREMENTS... 1 2. USING THE OUTPUT
Volume Three Modules 01 March 2002 i Chapter Eight: Contents (Output Visualizer 04 March 2002 LA-UR-00-1725 TRANSIMS 3.0) 1. INTRODUCTION...1 1.1 OVERVIEW... 1 1.2 REQUIREMENTS... 1 2. USING THE OUTPUT
TotalLab TL100 Quick Start
 TotalLab TL100 Quick Start Contents of thetl100 Quick Start Introduction to TL100 and Installation Instructions The Control Centre Getting Started The TL100 Interface 1D Gel Analysis Array Analysis Colony
TotalLab TL100 Quick Start Contents of thetl100 Quick Start Introduction to TL100 and Installation Instructions The Control Centre Getting Started The TL100 Interface 1D Gel Analysis Array Analysis Colony
Vision MET/METCAD. 2D measurement system
 Vision MET/METCAD 2D measurement system September 2012 ~ Contents ~ 1 GENERAL INFORMATION:... 3 1.1 PRECISION AND RESOLUTION... 3 2 GETTING STARTED:... 5 2.1 USER IDENTIFICATION... 5 2.2 MAIN WINDOW OF
Vision MET/METCAD 2D measurement system September 2012 ~ Contents ~ 1 GENERAL INFORMATION:... 3 1.1 PRECISION AND RESOLUTION... 3 2 GETTING STARTED:... 5 2.1 USER IDENTIFICATION... 5 2.2 MAIN WINDOW OF
Local Features: Detection, Description & Matching
 Local Features: Detection, Description & Matching Lecture 08 Computer Vision Material Citations Dr George Stockman Professor Emeritus, Michigan State University Dr David Lowe Professor, University of British
Local Features: Detection, Description & Matching Lecture 08 Computer Vision Material Citations Dr George Stockman Professor Emeritus, Michigan State University Dr David Lowe Professor, University of British
m6aviewer Version Documentation
 m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
for the Analysis of Flow Cytometry Listmode Data Files
 Purdue University Cytometry Laboratories Introduction to WinMDI 2.8 (J. Trotter 1993-1998) for the Analysis of Flow Cytometry Listmode Data Files Objectives of this tutorial: - Open a listmode data file
Purdue University Cytometry Laboratories Introduction to WinMDI 2.8 (J. Trotter 1993-1998) for the Analysis of Flow Cytometry Listmode Data Files Objectives of this tutorial: - Open a listmode data file
Chapter 3 Image Registration. Chapter 3 Image Registration
 Chapter 3 Image Registration Distributed Algorithms for Introduction (1) Definition: Image Registration Input: 2 images of the same scene but taken from different perspectives Goal: Identify transformation
Chapter 3 Image Registration Distributed Algorithms for Introduction (1) Definition: Image Registration Input: 2 images of the same scene but taken from different perspectives Goal: Identify transformation
Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays.
 Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays. http://www.nexus.ethz.ch/ -> Software -> TMARKER The plugin Intensity Clustering starts on step 7. Steps 1-6
Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays. http://www.nexus.ethz.ch/ -> Software -> TMARKER The plugin Intensity Clustering starts on step 7. Steps 1-6
Fast and accurate automated cell boundary determination for fluorescence microscopy
 Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Solo 4.6 Release Notes
 June9, 2017 (Updated to include Solo 4.6.4 changes) Solo 4.6 Release Notes This release contains a number of new features, as well as enhancements to the user interface and overall performance. Together
June9, 2017 (Updated to include Solo 4.6.4 changes) Solo 4.6 Release Notes This release contains a number of new features, as well as enhancements to the user interface and overall performance. Together
SynPAnal User Manual. Eric Danielson and Sang H. Lee Medical College of Wisconsin
 SynPAnal User Manual Eric Danielson and Sang H. Lee Medical College of Wisconsin Installation Instructions Make sure the most recent version of JAVA is installed and is functioning. Unzip the SynPAnal_Installation.zip
SynPAnal User Manual Eric Danielson and Sang H. Lee Medical College of Wisconsin Installation Instructions Make sure the most recent version of JAVA is installed and is functioning. Unzip the SynPAnal_Installation.zip
Data Analysis and Solver Plugins for KSpread USER S MANUAL. Tomasz Maliszewski
 Data Analysis and Solver Plugins for KSpread USER S MANUAL Tomasz Maliszewski tmaliszewski@wp.pl Table of Content CHAPTER 1: INTRODUCTION... 3 1.1. ABOUT DATA ANALYSIS PLUGIN... 3 1.3. ABOUT SOLVER PLUGIN...
Data Analysis and Solver Plugins for KSpread USER S MANUAL Tomasz Maliszewski tmaliszewski@wp.pl Table of Content CHAPTER 1: INTRODUCTION... 3 1.1. ABOUT DATA ANALYSIS PLUGIN... 3 1.3. ABOUT SOLVER PLUGIN...
