OBCOL. (Organelle Based CO-Localisation) Users Guide
|
|
|
- Clemence King
- 5 years ago
- Views:
Transcription
1 OBCOL (Organelle Based CO-Localisation) Users Guide
2 INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual objects. It is capable of determining object dimension, colour composition and overlap as well producing a variety of colocalisation statistics.
3 USING OBCOL 1. Image Preparation Format OBCOL works with any file format supported by ImageJ. Removal of Unwanted Content OBCOL analyses all visible data within an image. Any data not to be analysed should be removed. For example: - If there are other cells within the image these should be cropped out. - If only interested in internal objects, fluorescence at the plasma membrane should be cropped out. Filtering Background fluorescence and noise may effect OBCOL s ability to correctly detect the edges of objects within an image. If this is the case images should be filtered prior to analysis. For example 1 pixel Median filter has been applied to this image. Intensity Cutoff To remove background fluorescence and to ensure OBCOL only examines objects of interest the range of channel intensity should be set. By setting an intensity threshold any object/noise/artefact with an intensity below that threshold will be excluded from analysis.
4 To do this: 1. Split the image into separate channels (IMAGE>COLOR>RGB SPLIT). RED GREEN BLUE 2. Select the RED channel and use the THRESHOLD tool to determine the intensity range you are interested in analysing. (IMAGE>ADJUST>THRESHOLD ). Any signal covered by the red mask is above the minimum threshold (61 in this example). For 3D image stacks it is a good idea to examine all slices to ensure the mask is covering all objects of interest and removing all unwanted objects. 3. Click the Reset button in THRESHOLD and close this window. With the RED channel still open apply the threshold in OBCOL (PLUGINS> COLOCALISATION PIPELINE>CUTOFF ). OBCOL will the remove all fluorescence below this intensity.
5 4. Repeat the above steps each channel in the image (OBCOL can process up to 255 channels). 2. Running OBCOL Adding Image Channels to OBCOL After setting the intensity cutoff for each channel have all the image channels open in ImageJ. Make sure they are saved to the hard drive after filtering, intensity cutoffs etc. To add an image to OBCOL analysis open the OBCOL menu (PLUGINS>COLOCALISATION PIPELINE>PIPELINE) and click Add all. All the channels should appear in the dialogue box. Additional channels can be opened and then added separately using the Add single button. One, two, three or more images may be added. Note that the order of the images selected is important. A number of the statistics reported by OBCOL later in the pipeline refer to channels by number, not by name. Therefore adding images in the red, green, blue order will make things easier in the long run. Once all the channels have been added click Finished Selection. Start Pipeline. Analysis options Upon starting the pipeline a window will open (shown below) providing several options to adjust the analysis process.
6 - Watershed tolerance/threshold o Adjusts the watershed algorithm used to separate adjacent objects. Based on the ImageJ plugin Find Maxima see ImageJ documentation for more details. o Briefly, for a local maximum of intensity I, the contiguous region with intensities in the range I-tolerance to I will be selected. A minimum threshold for maxima may also be set here. o Default settings are: Tolerance = 5, Threshold =1. - Local Threshold o Gives the option of using ImageJ s automatic local thresholding (defaults to on). - Pixel Overlap options o These parameters dictate the number of pixels required to be overlapping before an object in another channel are considered joined. o Default to 1 pixel. - Slices in time point. o The image stack may represent multiple time points. Enter the number of slices in a time point, or 0 if the stack is a single time point. Once parameters have been set click OK OBCOL s processing time will vary depending upon image size. A stack of a single cell take on average 5-10 minutes, whilst a single frame will take only a few seconds. 3. OBCOL analysis The Object Viewer Menu Once OBCOL has processed the image data the Coalesced Object Viewer Menu will appear. From this menu you can select several image viewing options as well as the generation of statistical tables.
7 - Image Viewing Options o Before generating an output image from analysis OBCOL will ask wether you wish to apply a min pixel filter. o Selecting this option will allow you to exclude objects under a certain number of pixels (removing artefact objects or objects deemed to small for analysis) or over a certain number of pixels. o If you have not already decided on a minimum object size try various cut off using the visualisation options below. Once you have found parameters most representative for the objects you are interested use them for the statistical analysis options. o View All Objects Coloured by Object This option will generate an image/image stack with each object coloured according to its number. Note: If there are more objects present in the image than identification colours available some separate objects will share the same colour. o View All Objects Coloured by Stack This option will generate an image/image stack using the original channel colours (e.g. Red/Green/Blue). This view does not use intensity gradients, colour is either present (256) or absent (0). This allows an easier observation of channel overlap.
8 o View All Objects as Mask This option will generate an image/image stack using the original channel colours in addition to the intensity gradients present in the original input image. To generate this image it is necessary to open the original channels used for OBCOL analysis. A menu box will open to allow you to select the correct images file for each channel. - Viewing individual objects o An important and extremely useful aspect of OBCOL is the ability to select and examine an individual object at any time. o Following the generation of an image via the options above you may click on an object within that image and a new image window will be generated containing information relating only to that object in the coloured by stack viewing format. o A table of statistics for that object will also be generated. o This allows you to visually identify an object in an image and determine its OBJECT ID. This is useful if you wish to analyse a few specific objects. OR to identify and remove specific objects from the data output later. - Mask stats o This option will generate colocalisation statistics for each individual object based on the intensity values found in the original data. As with the View All Objects as Mask option it will be necessary to use the original channels used for OBCOL analysis. Open the pre-obcol analysis image (and split it into separate channels). Select the appropriate image files for each channel in the dialogue window that follows. The option for a min pixel filter to be applied is given. Parameters most representative of the real objects you wish to analyse should be chosen (if not already decided, use the visualisation options mentioned prior to determine these) o Once Min Pixel Filter has been applied OBCOL will generate a table of statistics through ImageJ (this can be selected, copied and pasted into excel) o Values in the table: Blank column ImageJ s reference number for each line in the table
9 ID The identification number assigned by OBCOL to an object in the image Intensity Average (whole object) 1/2/3/etc The average intensity of the channel across the whole object (i.e. including regions of the object where this channel is absent) Numbers denote channel number according to how they were added into OBCOL (typically 1 RED, 2 GREEN, 3 BLUE) Intensity Average (only structures in this Channel) 1/2/3/etc The average intensity of the channel within the object, ignoring regions where its value is zero. Pearson s Coefficient r (1&2/2&3/etc) The Pearson s correlation between the two channels mentioned within the object Calculated using the JACOP colocalisation plugin for ImageJ Other statistics Including Manders coefficients, cytofluorogram and Li s intensity correlation coefficient. Please refer to the JaCoP paper for full details on these statistics S. Bolte & F. P. Cordelières, A guided tour into subcellular colocalization analysis in light microscopy, Journal of Microscopy, Volume 224, Issue 3: View All Object Statistics o This option will generate statistics on the size and channel composition for each individual object. o Once Min Pixel Filter has been applied OBCOL will generate a table of statistics through ImageJ (this can be selected, copied and pasted into excel) o Values in the table: Blank column ImageJ s reference number for each line in the table Object ID The identification number assigned by OBCOL to an object in the image ROIs in the stack 1/2/3 etc The number of regions of this channel found in the object. Pixels in stack(s) 1/2/1&2/3/etc The number of pixels of this channel within the object In the case of a RGB image 7 columns will be created o 1, 2, 1&2, 3, 1&3, 2&3, 1&2&3 (R, G, RG, B, RB, GB, RGB) o For example: Column 1 = number of red pixels Column 1&2 = number of pixels containing both R and G.
10 Num Pixels The total number of pixels in the object Size of Bounding Box 1/2/3 The dimensions (in pixels) of a 3 dimensional box that would contain the object. 1 (x-axis), 2 (y-axis), 3 (z-axis) Middle of Bounding Box Axis 1/2/3 The location of the object within the image (x,y,z). Note that these statistics correspond to the numbers in the master ImageJ window, not the image window, as shown in the figure below. This means that if an object is in the first slice only, then the Middle of Bounding Box Axis 3 will be 0. Final Note: OBCOL may readily utilise segmentations produced by other software if a mask image has been created by that software for each channel. The OBCOL pipeline may then be run on these mask images to create the objects. Overlap statistics, positional information and so forth may then be calculated. By opening the original images, colocalisation statistics may then be calculated for the source images in the objects found in the mask images.
COLOCALISATION. Alexia Loynton-Ferrand. Imaging Core Facility Biozentrum Basel
 COLOCALISATION Alexia Loynton-Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to
COLOCALISATION Alexia Loynton-Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to
COLOCALISATION. Alexia Ferrand. Imaging Core Facility Biozentrum Basel
 COLOCALISATION Alexia Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to analyse
COLOCALISATION Alexia Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to analyse
WCIF COLOCALIS ATION PLUGINS
 WCIF COLOCALIS ATION PLUGINS Colocalisation Test Background When a coefficient is calculated for two images, it is often unclear quite what this means, in particular for intermediate values. This raises
WCIF COLOCALIS ATION PLUGINS Colocalisation Test Background When a coefficient is calculated for two images, it is often unclear quite what this means, in particular for intermediate values. This raises
Colocalisation/Correlation
 Colocalisation/Correlation The past: I see yellow - therefore there is colocalisation but published images look over exposed. No colocalisation definition + No stats = No Science. From Now On: 3D. Quantification.
Colocalisation/Correlation The past: I see yellow - therefore there is colocalisation but published images look over exposed. No colocalisation definition + No stats = No Science. From Now On: 3D. Quantification.
Lecture 19 - Applied Image Analysis
 Lecture 19 - Applied Image Analysis Graeme Ball Three Questions 1. Why should you be interested in image processing & analysis? 2. What are the key concepts & techniques? 3. How can you apply them most
Lecture 19 - Applied Image Analysis Graeme Ball Three Questions 1. Why should you be interested in image processing & analysis? 2. What are the key concepts & techniques? 3. How can you apply them most
Colocalization tutorial
 Colocalization tutorial I. Introduction Fluorescence microscopy is helpful for determining when two proteins colocalize in the cell. At the most basic level, images can be inspected by eye. Quantitative
Colocalization tutorial I. Introduction Fluorescence microscopy is helpful for determining when two proteins colocalize in the cell. At the most basic level, images can be inspected by eye. Quantitative
Colocalization in Confocal Microscopy. Quantitative Colocalization Analysis
 Quantitative Colocalization Analysis 1 Quantitative Colocalization Analysis Colocalization of fluorescently labeled molecules (immunostainings, fluorescent proteins, etc.) is often used as the first indicator
Quantitative Colocalization Analysis 1 Quantitative Colocalization Analysis Colocalization of fluorescently labeled molecules (immunostainings, fluorescent proteins, etc.) is often used as the first indicator
/5 Stacks. Displays the slice that follows the currently displayed slice. As a shortcut, press the > key.
 20-02-2018 1/5 Stacks Stacks This submenu contains commands that work with stacks. Add Slice Inserts a blank slice after the currently displayed slice. Hold down the Alt key to add the slice before the
20-02-2018 1/5 Stacks Stacks This submenu contains commands that work with stacks. Add Slice Inserts a blank slice after the currently displayed slice. Hold down the Alt key to add the slice before the
Presentation and analysis of multidimensional data sets
 Presentation and analysis of multidimensional data sets Overview 1. 3D data visualisation Multidimensional images Data pre-processing Visualisation methods Multidimensional images wavelength time 3D image
Presentation and analysis of multidimensional data sets Overview 1. 3D data visualisation Multidimensional images Data pre-processing Visualisation methods Multidimensional images wavelength time 3D image
Topic 01 - Installation, configuration, help and updates
 Topic 01 - Installation, configuration, help and updates Solution 1.1: Installation Go to the ImageJ website at http://imagej.nih.gov/ij/. Click on Download. Illustration 1: The ImageJ website http://imagej.nih.gov/ij/
Topic 01 - Installation, configuration, help and updates Solution 1.1: Installation Go to the ImageJ website at http://imagej.nih.gov/ij/. Click on Download. Illustration 1: The ImageJ website http://imagej.nih.gov/ij/
Image Processing: Final Exam November 10, :30 10:30
 Image Processing: Final Exam November 10, 2017-8:30 10:30 Student name: Student number: Put your name and student number on all of the papers you hand in (if you take out the staple). There are always
Image Processing: Final Exam November 10, 2017-8:30 10:30 Student name: Student number: Put your name and student number on all of the papers you hand in (if you take out the staple). There are always
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences
 IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
EPBscore user guide. Requirements:
 EPBscore user guide Requirements: Up to Matlab 2014a. Windows-only versions (both 32- and 64-bit) were tested. Toolboxes: Control, Images, Local, Matlab, Optim, Shared, Signal, Simulink, Stats, Wavelet
EPBscore user guide Requirements: Up to Matlab 2014a. Windows-only versions (both 32- and 64-bit) were tested. Toolboxes: Control, Images, Local, Matlab, Optim, Shared, Signal, Simulink, Stats, Wavelet
PHASEQUANT (User Manual)
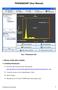 PHASEQUANT (User Manual) ROI Pane Toolbar Threshold Bar Canvas Threshold Display Training Pane Buttons Fig 1. PhaseQuant GUI 1. INSTALLATION AND LOADING 1.1 Installing PhaseQuant Download the zip file
PHASEQUANT (User Manual) ROI Pane Toolbar Threshold Bar Canvas Threshold Display Training Pane Buttons Fig 1. PhaseQuant GUI 1. INSTALLATION AND LOADING 1.1 Installing PhaseQuant Download the zip file
Neuron Morpho plugin for ImageJ Version May 25, 2007
 Neuron Morpho plugin for ImageJ Version 1.1.6 - May 25, 2007 Giampaolo D Alessandro Department of Mathematics, University of Southampton, Southampton, SO17 1BJ, England, UK 1 Introduction The Neuron Morpho
Neuron Morpho plugin for ImageJ Version 1.1.6 - May 25, 2007 Giampaolo D Alessandro Department of Mathematics, University of Southampton, Southampton, SO17 1BJ, England, UK 1 Introduction The Neuron Morpho
WHAT S NEW IN SLIDEBOOK 5.0
 WHAT S NEW IN SLIDEBOOK 5.0 We at 3i are pleased to announce the much anticipated release of SlideBook 5.0. In this latest installment of our cutting edge digital microscopy software, you will find innovative
WHAT S NEW IN SLIDEBOOK 5.0 We at 3i are pleased to announce the much anticipated release of SlideBook 5.0. In this latest installment of our cutting edge digital microscopy software, you will find innovative
Use stage 16 embryos see Pereanu et al., 2007 for morphological staging criteria. First start Vaa3d (http://vaa3d.org/; Peng et al., 2010).
 Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
Textbook de.nbi Bioimage Analysis Workshop
 BIOIMAGE ANALYSIS WORKSHOP WITHIN THE FRAMEWORK OF HD-HUB Textbook de.nbi Bioimage Analysis Workshop An introduction into KNIME image analysis for life scientists Jürgen Reymann ViroQuant-CellNetworks
BIOIMAGE ANALYSIS WORKSHOP WITHIN THE FRAMEWORK OF HD-HUB Textbook de.nbi Bioimage Analysis Workshop An introduction into KNIME image analysis for life scientists Jürgen Reymann ViroQuant-CellNetworks
FindSpots (OME v2.5.1) user guide
 FindSpots (OME v2.5.1) user guide User guide by David A. Schiffmann, Paul L. Appleton & Ilya G. Goldberg * * Email: igg@nih.gov 24th February 2006 1 The small print Whilst every effort has been taken to
FindSpots (OME v2.5.1) user guide User guide by David A. Schiffmann, Paul L. Appleton & Ilya G. Goldberg * * Email: igg@nih.gov 24th February 2006 1 The small print Whilst every effort has been taken to
Adaptive thresholding. in CTAn
 Adaptive thresholding in CTAn Method note Page 1 of 8 2 Bruker-MicroCT method note:adaptive thresholding in CTAn Introduction 2D or 3D morphometric analysis of micro-ct images always requires binarization
Adaptive thresholding in CTAn Method note Page 1 of 8 2 Bruker-MicroCT method note:adaptive thresholding in CTAn Introduction 2D or 3D morphometric analysis of micro-ct images always requires binarization
IMAGEJ FINDFOCI Plugins
 IMAGEJ FINDFOCI Plugins Alex Herbert MRC Genome Damage and Stability Centre School of Life Sciences University of Sussex Science Road Falmer BN1 9RQ a.herbert@sussex.ac.uk Table of Contents Introduction...5
IMAGEJ FINDFOCI Plugins Alex Herbert MRC Genome Damage and Stability Centre School of Life Sciences University of Sussex Science Road Falmer BN1 9RQ a.herbert@sussex.ac.uk Table of Contents Introduction...5
45 µm polystyrene bead embedded in scattering tissue phantom. (a,b) raw images under oblique
 Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
Virtual Frap User Guide
 Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Supporting Information. Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking
 Supporting Information Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking Daan van der Zwaag 1,2, Nane Vanparijs 3, Sjors Wijnands 1,4, Riet De Rycke 5, Bruno G. De Geest 2* and
Supporting Information Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking Daan van der Zwaag 1,2, Nane Vanparijs 3, Sjors Wijnands 1,4, Riet De Rycke 5, Bruno G. De Geest 2* and
Presentation of GcoPS Icy plugin
 Presentation of GcoPS Icy plugin GcoPS is a fully automatic and non parametric colocalization algorithm with four possible outputs: Statistical test: GcoPS computes a statistical test to evaluate the global
Presentation of GcoPS Icy plugin GcoPS is a fully automatic and non parametric colocalization algorithm with four possible outputs: Statistical test: GcoPS computes a statistical test to evaluate the global
Babu Madhav Institute of Information Technology Years Integrated M.Sc.(IT)(Semester - 7)
 5 Years Integrated M.Sc.(IT)(Semester - 7) 060010707 Digital Image Processing UNIT 1 Introduction to Image Processing Q: 1 Answer in short. 1. What is digital image? 1. Define pixel or picture element?
5 Years Integrated M.Sc.(IT)(Semester - 7) 060010707 Digital Image Processing UNIT 1 Introduction to Image Processing Q: 1 Answer in short. 1. What is digital image? 1. Define pixel or picture element?
Frequency tables Create a new Frequency Table
 Frequency tables Create a new Frequency Table Contents FREQUENCY TABLES CREATE A NEW FREQUENCY TABLE... 1 Results Table... 2 Calculate Descriptive Statistics for Frequency Tables... 6 Transfer Results
Frequency tables Create a new Frequency Table Contents FREQUENCY TABLES CREATE A NEW FREQUENCY TABLE... 1 Results Table... 2 Calculate Descriptive Statistics for Frequency Tables... 6 Transfer Results
Ulrik Söderström 16 Feb Image Processing. Segmentation
 Ulrik Söderström ulrik.soderstrom@tfe.umu.se 16 Feb 2011 Image Processing Segmentation What is Image Segmentation? To be able to extract information from an image it is common to subdivide it into background
Ulrik Söderström ulrik.soderstrom@tfe.umu.se 16 Feb 2011 Image Processing Segmentation What is Image Segmentation? To be able to extract information from an image it is common to subdivide it into background
Version 1.0 PROTEOMICA DEMYSTIFYING PROTEINS. Indian Institute of Technology Bombay, India Bhushan N Kharbikar
 Version 1.0 PROTEOMICA DEMYSTIFYING PROTEINS Indian Institute of Technology Bombay, India Bhushan N Kharbikar Software User s Manual Version 1.0 Copyright 2013 Bhushan N Kharbikar, Proteomics Lab Department
Version 1.0 PROTEOMICA DEMYSTIFYING PROTEINS Indian Institute of Technology Bombay, India Bhushan N Kharbikar Software User s Manual Version 1.0 Copyright 2013 Bhushan N Kharbikar, Proteomics Lab Department
Physical Color. Color Theory - Center for Graphics and Geometric Computing, Technion 2
 Color Theory Physical Color Visible energy - small portion of the electro-magnetic spectrum Pure monochromatic colors are found at wavelengths between 380nm (violet) and 780nm (red) 380 780 Color Theory
Color Theory Physical Color Visible energy - small portion of the electro-magnetic spectrum Pure monochromatic colors are found at wavelengths between 380nm (violet) and 780nm (red) 380 780 Color Theory
TexRAD Research Version Client User Guide Version 3.9
 Imaging tools for medical decision makers Cambridge Computed Imaging Ltd Grange Park Broadway Bourn Cambridge CB23 2TA UK TexRAD Research Version Client User Guide Version 3.9 Release date 23/05/2016 Number
Imaging tools for medical decision makers Cambridge Computed Imaging Ltd Grange Park Broadway Bourn Cambridge CB23 2TA UK TexRAD Research Version Client User Guide Version 3.9 Release date 23/05/2016 Number
BioImageXD Getting started
 BioImageXD Getting started Updated 2012-07-01 by Pasi Kankaanpää Introduction Welcome to BioImageXD, free open source software for biomedical image post-processing, visualization and analysis. This guide
BioImageXD Getting started Updated 2012-07-01 by Pasi Kankaanpää Introduction Welcome to BioImageXD, free open source software for biomedical image post-processing, visualization and analysis. This guide
Introduction to Medical Imaging (5XSA0) Module 5
 Introduction to Medical Imaging (5XSA0) Module 5 Segmentation Jungong Han, Dirk Farin, Sveta Zinger ( s.zinger@tue.nl ) 1 Outline Introduction Color Segmentation region-growing region-merging watershed
Introduction to Medical Imaging (5XSA0) Module 5 Segmentation Jungong Han, Dirk Farin, Sveta Zinger ( s.zinger@tue.nl ) 1 Outline Introduction Color Segmentation region-growing region-merging watershed
DIFFUSION TENSOR IMAGING ANALYSIS. Using Analyze
 DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
HitFilm Express - Editing
 HitFilm Express - Editing Table of Contents Getting Started 3 Create New Project 3 Workspaces 4 The Interface 5 Trimmer 5 Viewer 5 Panels 6 Timeline 7 Import Media 7 Editing 9 Preview 9 Trim 9 Add Clip
HitFilm Express - Editing Table of Contents Getting Started 3 Create New Project 3 Workspaces 4 The Interface 5 Trimmer 5 Viewer 5 Panels 6 Timeline 7 Import Media 7 Editing 9 Preview 9 Trim 9 Add Clip
Visible Color. 700 (red) 580 (yellow) 520 (green)
 Color Theory Physical Color Visible energy - small portion of the electro-magnetic spectrum Pure monochromatic colors are found at wavelengths between 380nm (violet) and 780nm (red) 380 780 Color Theory
Color Theory Physical Color Visible energy - small portion of the electro-magnetic spectrum Pure monochromatic colors are found at wavelengths between 380nm (violet) and 780nm (red) 380 780 Color Theory
Scale-adaptive Wavelet-based Particle Detection in Microscopy Images
 Scale-adaptive Wavelet-based Particle Detection in Microscopy Images Oliver Greß 1, Birgit Möller 1, Nadine Stöhr 2, Stefan Hüttelmaier 2, Stefan Posch 1 1 Institute of Computer Science, Martin Luther
Scale-adaptive Wavelet-based Particle Detection in Microscopy Images Oliver Greß 1, Birgit Möller 1, Nadine Stöhr 2, Stefan Hüttelmaier 2, Stefan Posch 1 1 Institute of Computer Science, Martin Luther
Algorithm User Guide:
 Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
1. Basic Steps for Data Analysis Data Editor. 2.4.To create a new SPSS file
 1 SPSS Guide 2009 Content 1. Basic Steps for Data Analysis. 3 2. Data Editor. 2.4.To create a new SPSS file 3 4 3. Data Analysis/ Frequencies. 5 4. Recoding the variable into classes.. 5 5. Data Analysis/
1 SPSS Guide 2009 Content 1. Basic Steps for Data Analysis. 3 2. Data Editor. 2.4.To create a new SPSS file 3 4 3. Data Analysis/ Frequencies. 5 4. Recoding the variable into classes.. 5 5. Data Analysis/
Colin Paul Updated 14 November Preparation of publication-quality videos using ImageJ
 Preparation of publication-quality videos using ImageJ Statements made in scientific papers are often much easier to understand if they are supplemented by representative videos. In the best case, these
Preparation of publication-quality videos using ImageJ Statements made in scientific papers are often much easier to understand if they are supplemented by representative videos. In the best case, these
SAS Visual Analytics 8.2: Working with Report Content
 SAS Visual Analytics 8.2: Working with Report Content About Objects After selecting your data source and data items, add one or more objects to display the results. SAS Visual Analytics provides objects
SAS Visual Analytics 8.2: Working with Report Content About Objects After selecting your data source and data items, add one or more objects to display the results. SAS Visual Analytics provides objects
MetaMorph Standard Operation Protocol Basic Application
 MetaMorph Standard Operation Protocol Basic Application Contents Basic Navigation and Image Handling... 2 Opening Images... 2 Separating Multichannel Images... 2 Cropping an Image... 3 Changing an 8 bit
MetaMorph Standard Operation Protocol Basic Application Contents Basic Navigation and Image Handling... 2 Opening Images... 2 Separating Multichannel Images... 2 Cropping an Image... 3 Changing an 8 bit
Image Processing Guideline for TMU 7T MRI
 Image Processing Guideline for TMU 7T MRI Chia Feng Lu Laboratory of NeuroImage Biomarker Analysis, Translational Imaging Research Center, TMU 08/18/2015, version 1.0 Section 1: Installation of ImageJ
Image Processing Guideline for TMU 7T MRI Chia Feng Lu Laboratory of NeuroImage Biomarker Analysis, Translational Imaging Research Center, TMU 08/18/2015, version 1.0 Section 1: Installation of ImageJ
Excel Core Certification
 Microsoft Office Specialist 2010 Microsoft Excel Core Certification 2010 Lesson 6: Working with Charts Lesson Objectives This lesson introduces you to working with charts. You will look at how to create
Microsoft Office Specialist 2010 Microsoft Excel Core Certification 2010 Lesson 6: Working with Charts Lesson Objectives This lesson introduces you to working with charts. You will look at how to create
Tracking Tool PRO v2.0. manual
 Tracking Tool PRO v2.0 User manual Introduction Save hours using the new Tracking Tool PRO software! automated Tracking Tool PRO, the new automated tracking tool software developed by Gradientech, provides
Tracking Tool PRO v2.0 User manual Introduction Save hours using the new Tracking Tool PRO software! automated Tracking Tool PRO, the new automated tracking tool software developed by Gradientech, provides
URBAN FOOTPRINT MAPPING WITH SENTINEL-1 DATA
 URBAN FOOTPRINT MAPPING WITH SENTINEL-1 DATA Data: Sentinel-1A IW SLC 1SSV: S1A_IW_SLC 1SSV_20160102T005143_20160102T005208_009308_00D72A_849D S1A_IW_SLC 1SSV_20160126T005142_20160126T005207_009658_00E14A_49C0
URBAN FOOTPRINT MAPPING WITH SENTINEL-1 DATA Data: Sentinel-1A IW SLC 1SSV: S1A_IW_SLC 1SSV_20160102T005143_20160102T005208_009308_00D72A_849D S1A_IW_SLC 1SSV_20160126T005142_20160126T005207_009658_00E14A_49C0
Agilent Seahorse XF Imaging & Cell Counting Software
 Tech Note Agilent Seahorse XF Imaging & Cell Counting Software Adjusting Analysis Parameters in BioTek Gen5 for Difficult to Count Cell Lines This document describes how to use BioTek s Gen5 software to
Tech Note Agilent Seahorse XF Imaging & Cell Counting Software Adjusting Analysis Parameters in BioTek Gen5 for Difficult to Count Cell Lines This document describes how to use BioTek s Gen5 software to
Example Workflow for the Analysis of the Provided Maize Dataset
 Image Analysis with IAP Example Workflow for the Analysis of the Provided Maize Dataset Image Analysis Group - Leibniz Institute of Plant Genetics and Crop Plant Research IPK, Corrensstr. 3, 06466 Gatersleben
Image Analysis with IAP Example Workflow for the Analysis of the Provided Maize Dataset Image Analysis Group - Leibniz Institute of Plant Genetics and Crop Plant Research IPK, Corrensstr. 3, 06466 Gatersleben
Year 10 General Mathematics Unit 2
 Year 11 General Maths Year 10 General Mathematics Unit 2 Bivariate Data Chapter 4 Chapter Four 1 st Edition 2 nd Edition 2013 4A 1, 2, 3, 4, 6, 7, 8, 9, 10, 11 1, 2, 3, 4, 6, 7, 8, 9, 10, 11 2F (FM) 1,
Year 11 General Maths Year 10 General Mathematics Unit 2 Bivariate Data Chapter 4 Chapter Four 1 st Edition 2 nd Edition 2013 4A 1, 2, 3, 4, 6, 7, 8, 9, 10, 11 1, 2, 3, 4, 6, 7, 8, 9, 10, 11 2F (FM) 1,
Creating a Basic Chart in Excel 2007
 Creating a Basic Chart in Excel 2007 A chart is a pictorial representation of the data you enter in a worksheet. Often, a chart can be a more descriptive way of representing your data. As a result, those
Creating a Basic Chart in Excel 2007 A chart is a pictorial representation of the data you enter in a worksheet. Often, a chart can be a more descriptive way of representing your data. As a result, those
Quaternion-based color difference measure for removing impulse noise in color images
 2014 International Conference on Informative and Cybernetics for Computational Social Systems (ICCSS) Quaternion-based color difference measure for removing impulse noise in color images Lunbo Chen, Yicong
2014 International Conference on Informative and Cybernetics for Computational Social Systems (ICCSS) Quaternion-based color difference measure for removing impulse noise in color images Lunbo Chen, Yicong
Icy Training - Level 1 - Introduction
 Icy Training - Level 1 - Introduction Plan What is Icy? Installing Icy Graphical User Interface (GUI) Histograms & Colormap / Look up table Basic operations Overlays / Layers 3D view Icy Preferences Investigate
Icy Training - Level 1 - Introduction Plan What is Icy? Installing Icy Graphical User Interface (GUI) Histograms & Colormap / Look up table Basic operations Overlays / Layers 3D view Icy Preferences Investigate
3D Processing and Analysis with ImageJ
 3D Processing and Analysis with ImageJ Rapid development of technologies yielding multidimensional data LSCM / Video Electron tomography IRM / CT scan Thomas Boudier, Associate Professor, Cellular Imaging
3D Processing and Analysis with ImageJ Rapid development of technologies yielding multidimensional data LSCM / Video Electron tomography IRM / CT scan Thomas Boudier, Associate Professor, Cellular Imaging
Last updated 24 June 2017
 Published in: Braude JP, Vijayakumar S, Baumgarner K, Laurine R, Jones TA, Jones SM, Pyott SJ. (2015) Deletion of Shank1 has minimal effects on the molecular composition and function of glutamatergic afferent
Published in: Braude JP, Vijayakumar S, Baumgarner K, Laurine R, Jones TA, Jones SM, Pyott SJ. (2015) Deletion of Shank1 has minimal effects on the molecular composition and function of glutamatergic afferent
TUTORIAL - [MITOCHONDRIAL MEMBRANE POTENTIAL MEASUREMENT] July 26, 2015
![TUTORIAL - [MITOCHONDRIAL MEMBRANE POTENTIAL MEASUREMENT] July 26, 2015 TUTORIAL - [MITOCHONDRIAL MEMBRANE POTENTIAL MEASUREMENT] July 26, 2015](/thumbs/84/89490383.jpg) Example data set: MitoMemPot1.zip Pipelines used: Mitochondrial membrane potential measurement (TMRM/PMPI) QUICK START GUIDE 1. Unzip contents of MitoMemPot1.zip onto your hard drive. 2. Open the recording
Example data set: MitoMemPot1.zip Pipelines used: Mitochondrial membrane potential measurement (TMRM/PMPI) QUICK START GUIDE 1. Unzip contents of MitoMemPot1.zip onto your hard drive. 2. Open the recording
Lauetools. A software package for Laue microdiffraction data analysis. https://sourceforge.net/projects/lauetools /
 Lauetools A software package for Laue microdiffraction data analysis https://sourceforge.net/projects/lauetools / Motivations Motivations ImageJ LAUE raw data XMAS fit2d Some codes Motivations LAUE raw
Lauetools A software package for Laue microdiffraction data analysis https://sourceforge.net/projects/lauetools / Motivations Motivations ImageJ LAUE raw data XMAS fit2d Some codes Motivations LAUE raw
Matthew Baker. Visualisation of Electrophoresis Gels using TEX
 Visualisation of Electrophoresis Gels using TEX 1 Visualisation of Electrophoresis Gels using TEX Matthew Baker This paper describes a TEX system for creating interactive PDF files to visualize electrophoresis
Visualisation of Electrophoresis Gels using TEX 1 Visualisation of Electrophoresis Gels using TEX Matthew Baker This paper describes a TEX system for creating interactive PDF files to visualize electrophoresis
Vincent Mouchi, Quentin G. Crowley, Teresa Ubide, 2016
 AERYN: Walkthrough This short report will help you operate AERYN with all its capabilities. You can use the example files available with this report (also available at http://www.tcd.ie/geology/staff/crowleyq/aeryn/).
AERYN: Walkthrough This short report will help you operate AERYN with all its capabilities. You can use the example files available with this report (also available at http://www.tcd.ie/geology/staff/crowleyq/aeryn/).
Data processing. Filters and normalisation. Mélanie Pétéra W4M Core Team 31/05/2017 v 1.0.0
 Data processing Filters and normalisation Mélanie Pétéra W4M Core Team 31/05/2017 v 1.0.0 Presentation map 1) Processing the data W4M table format for Galaxy 2) A generic tool to filter in Galaxy a) Generic
Data processing Filters and normalisation Mélanie Pétéra W4M Core Team 31/05/2017 v 1.0.0 Presentation map 1) Processing the data W4M table format for Galaxy 2) A generic tool to filter in Galaxy a) Generic
Data Visualization (CIS/DSC 468)
 Data Visualization (CIS/DSC 46) Volume Rendering Dr. David Koop Visualizing Volume (3D) Data 2D visualization slice images (or multi-planar reformating MPR) Indirect 3D visualization isosurfaces (or surface-shaded
Data Visualization (CIS/DSC 46) Volume Rendering Dr. David Koop Visualizing Volume (3D) Data 2D visualization slice images (or multi-planar reformating MPR) Indirect 3D visualization isosurfaces (or surface-shaded
AnalySIS Tutorial part 2
 AnalySIS Tutorial part 2 Sveinung Lillehaug Neural Systems and Graphics Computing Laboratory Department of Anatomy University of Oslo N-0317 Oslo Norway www.nesys.uio.no Using AnalySIS to automatically
AnalySIS Tutorial part 2 Sveinung Lillehaug Neural Systems and Graphics Computing Laboratory Department of Anatomy University of Oslo N-0317 Oslo Norway www.nesys.uio.no Using AnalySIS to automatically
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS.
 SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
Guidelines for the MorphoLeaf application.
 Guidelines for the MorphoLeaf application. (Version 1.0, December 2015) Presentation of MorphoLeaf 2 Installation of MorphoLeaf 3 Some advice to start with optimal leaf images 4 1. Importing leaf images
Guidelines for the MorphoLeaf application. (Version 1.0, December 2015) Presentation of MorphoLeaf 2 Installation of MorphoLeaf 3 Some advice to start with optimal leaf images 4 1. Importing leaf images
MS data processing. Filtering and correcting data. W4M Core Team. 22/09/2015 v 1.0.0
 MS data processing Filtering and correcting data W4M Core Team 22/09/2015 v 1.0.0 Presentation map 1) Processing the data W4M table format for Galaxy 2) Filters for mass spectrometry extracted data a)
MS data processing Filtering and correcting data W4M Core Team 22/09/2015 v 1.0.0 Presentation map 1) Processing the data W4M table format for Galaxy 2) Filters for mass spectrometry extracted data a)
Emerge Workflow CE8 SAMPLE IMAGE. Simon Voisey July 2008
 Emerge Workflow SAMPLE IMAGE CE8 Simon Voisey July 2008 Introduction The document provides a step-by-step guide for producing Emerge predicted petrophysical volumes based on log data of the same type.
Emerge Workflow SAMPLE IMAGE CE8 Simon Voisey July 2008 Introduction The document provides a step-by-step guide for producing Emerge predicted petrophysical volumes based on log data of the same type.
Display. Introduction page 67 2D Images page 68. All Orientations page 69 Single Image page 70 3D Images page 71
 Display Introduction page 67 2D Images page 68 All Orientations page 69 Single Image page 70 3D Images page 71 Intersecting Sections page 71 Cube Sections page 72 Render page 73 1. Tissue Maps page 77
Display Introduction page 67 2D Images page 68 All Orientations page 69 Single Image page 70 3D Images page 71 Intersecting Sections page 71 Cube Sections page 72 Render page 73 1. Tissue Maps page 77
VERY LARGE TELESCOPE 3D Visualization Tool Cookbook
 European Organisation for Astronomical Research in the Southern Hemisphere VERY LARGE TELESCOPE 3D Visualization Tool Cookbook VLT-SPE-ESO-19500-5652 Issue 1.0 10 July 2012 Prepared: Mark Westmoquette
European Organisation for Astronomical Research in the Southern Hemisphere VERY LARGE TELESCOPE 3D Visualization Tool Cookbook VLT-SPE-ESO-19500-5652 Issue 1.0 10 July 2012 Prepared: Mark Westmoquette
Graphics for VEs. Ruth Aylett
 Graphics for VEs Ruth Aylett Overview VE Software Graphics for VEs The graphics pipeline Projections Lighting Shading Runtime VR systems Two major parts: initialisation and update loop. Initialisation
Graphics for VEs Ruth Aylett Overview VE Software Graphics for VEs The graphics pipeline Projections Lighting Shading Runtime VR systems Two major parts: initialisation and update loop. Initialisation
Product information. Hi-Tech Electronics Pte Ltd
 Product information Introduction TEMA Motion is the world leading software for advanced motion analysis. Starting with digital image sequences the operator uses TEMA Motion to track objects in images,
Product information Introduction TEMA Motion is the world leading software for advanced motion analysis. Starting with digital image sequences the operator uses TEMA Motion to track objects in images,
RT_Image v0.2β User s Guide
 RT_Image v0.2β User s Guide RT_Image is a three-dimensional image display and analysis suite developed in IDL (ITT, Boulder, CO). It offers a range of flexible tools for the visualization and quantitation
RT_Image v0.2β User s Guide RT_Image is a three-dimensional image display and analysis suite developed in IDL (ITT, Boulder, CO). It offers a range of flexible tools for the visualization and quantitation
Crop Counting and Metrics Tutorial
 Crop Counting and Metrics Tutorial The ENVI Crop Science platform contains remote sensing analytic tools for precision agriculture and agronomy. In this tutorial you will go through a typical workflow
Crop Counting and Metrics Tutorial The ENVI Crop Science platform contains remote sensing analytic tools for precision agriculture and agronomy. In this tutorial you will go through a typical workflow
DTC-350. VISUALmpeg PRO MPEG Analyser Software.
 VISUALmpeg PRO MPEG Analyser Software 1. Introduction VISUALmpeg PRO is a powerful tool set intended for detailed off-line analysis of Video Elementary Streams in MPEG-1 or MPEG-2 video format. The analysis
VISUALmpeg PRO MPEG Analyser Software 1. Introduction VISUALmpeg PRO is a powerful tool set intended for detailed off-line analysis of Video Elementary Streams in MPEG-1 or MPEG-2 video format. The analysis
Text & Design 2015 Wojciech Piskor
 Text & Design 2015 Wojciech Piskor www.wojciechpiskor.wordpress.com wojciech.piskor@gmail.com All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means,
Text & Design 2015 Wojciech Piskor www.wojciechpiskor.wordpress.com wojciech.piskor@gmail.com All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means,
A Visual Programming Environment for Machine Vision Engineers. Paul F Whelan
 A Visual Programming Environment for Machine Vision Engineers Paul F Whelan Vision Systems Group School of Electronic Engineering, Dublin City University, Dublin 9, Ireland. Ph: +353 1 700 5489 Fax: +353
A Visual Programming Environment for Machine Vision Engineers Paul F Whelan Vision Systems Group School of Electronic Engineering, Dublin City University, Dublin 9, Ireland. Ph: +353 1 700 5489 Fax: +353
Tutorial 7: Automated Peak Picking in Skyline
 Tutorial 7: Automated Peak Picking in Skyline Skyline now supports the ability to create custom advanced peak picking and scoring models for both selected reaction monitoring (SRM) and data-independent
Tutorial 7: Automated Peak Picking in Skyline Skyline now supports the ability to create custom advanced peak picking and scoring models for both selected reaction monitoring (SRM) and data-independent
Appendix I. TACTICS Toolbox v3.x. Interactive MATLAB Platform For Bioimaging informatics. User Guide TRACKING MODULE
 TACTICS Toolbox v3.x Interactive MATLAB Platform For Bioimaging informatics User Guide TRACKING MODULE -17- Cell Tracking Module 1 (user interface) Once the images were successfully segmented, the next
TACTICS Toolbox v3.x Interactive MATLAB Platform For Bioimaging informatics User Guide TRACKING MODULE -17- Cell Tracking Module 1 (user interface) Once the images were successfully segmented, the next
SuRVoS Workbench. Super-Region Volume Segmentation. Imanol Luengo
 SuRVoS Workbench Super-Region Volume Segmentation Imanol Luengo Index - The project - What is SuRVoS - SuRVoS Overview - What can it do - Overview of the internals - Current state & Limitations - Future
SuRVoS Workbench Super-Region Volume Segmentation Imanol Luengo Index - The project - What is SuRVoS - SuRVoS Overview - What can it do - Overview of the internals - Current state & Limitations - Future
Handling Multi-Dimensional Data
 Handling Multi-Dimensional Data Advanced Technology Development Group Gray Cancer Institute Data, data, everybody wants more data. In our cell biology work investigating the mechanisms of cancer we find
Handling Multi-Dimensional Data Advanced Technology Development Group Gray Cancer Institute Data, data, everybody wants more data. In our cell biology work investigating the mechanisms of cancer we find
Digital Imaging I: 2nd Part. Martin Offterdinger
 Digital Imaging I: 2nd Part Martin Offterdinger Biooptics at MUI http://www.i-med.ac.at/neurobiochemistry/neurobiochemistry/biooptics/main.html What MAY we (not) do with digital images? The Journal of
Digital Imaging I: 2nd Part Martin Offterdinger Biooptics at MUI http://www.i-med.ac.at/neurobiochemistry/neurobiochemistry/biooptics/main.html What MAY we (not) do with digital images? The Journal of
Geneious Microsatellite Plugin. Biomatters Ltd
 Geneious Microsatellite Plugin Biomatters Ltd November 24, 2018 2 Introduction This plugin imports ABI fragment analysis files and allows you to visualize traces, fit ladders, call peaks, predict bins,
Geneious Microsatellite Plugin Biomatters Ltd November 24, 2018 2 Introduction This plugin imports ABI fragment analysis files and allows you to visualize traces, fit ladders, call peaks, predict bins,
STATA 13 INTRODUCTION
 STATA 13 INTRODUCTION Catherine McGowan & Elaine Williamson LONDON SCHOOL OF HYGIENE & TROPICAL MEDICINE DECEMBER 2013 0 CONTENTS INTRODUCTION... 1 Versions of STATA... 1 OPENING STATA... 1 THE STATA
STATA 13 INTRODUCTION Catherine McGowan & Elaine Williamson LONDON SCHOOL OF HYGIENE & TROPICAL MEDICINE DECEMBER 2013 0 CONTENTS INTRODUCTION... 1 Versions of STATA... 1 OPENING STATA... 1 THE STATA
Pedestrian Detection Using Correlated Lidar and Image Data EECS442 Final Project Fall 2016
 edestrian Detection Using Correlated Lidar and Image Data EECS442 Final roject Fall 2016 Samuel Rohrer University of Michigan rohrer@umich.edu Ian Lin University of Michigan tiannis@umich.edu Abstract
edestrian Detection Using Correlated Lidar and Image Data EECS442 Final roject Fall 2016 Samuel Rohrer University of Michigan rohrer@umich.edu Ian Lin University of Michigan tiannis@umich.edu Abstract
Methodology for spot quality evaluation
 Methodology for spot quality evaluation Semi-automatic pipeline in MAIA The general workflow of the semi-automatic pipeline analysis in MAIA is shown in Figure 1A, Manuscript. In Block 1 raw data, i.e..tif
Methodology for spot quality evaluation Semi-automatic pipeline in MAIA The general workflow of the semi-automatic pipeline analysis in MAIA is shown in Figure 1A, Manuscript. In Block 1 raw data, i.e..tif
Particle localization and tracking GUI: TrackingGUI_rp.m
 Particle localization and tracking GUI: TrackingGUI_rp.m Raghuveer Parthasarathy Department of Physics The University of Oregon raghu@uoregon.edu Begun April, 2012 (based on earlier work). Last modified
Particle localization and tracking GUI: TrackingGUI_rp.m Raghuveer Parthasarathy Department of Physics The University of Oregon raghu@uoregon.edu Begun April, 2012 (based on earlier work). Last modified
Digital Image Processing
 Digital Image Processing Jen-Hui Chuang Department of Computer Science National Chiao Tung University 2 3 Image Enhancement in the Spatial Domain 3.1 Background 3.4 Enhancement Using Arithmetic/Logic Operations
Digital Image Processing Jen-Hui Chuang Department of Computer Science National Chiao Tung University 2 3 Image Enhancement in the Spatial Domain 3.1 Background 3.4 Enhancement Using Arithmetic/Logic Operations
3-D. Here red spheres show the location of gold nanoparticles inside/around a cell nucleus.
 3-D The CytoViva 3-D System allows the user can locate objects of interest in a 3-D space. It does this by acquiring multiple Z planes and performing our custom software routines to locate and observe
3-D The CytoViva 3-D System allows the user can locate objects of interest in a 3-D space. It does this by acquiring multiple Z planes and performing our custom software routines to locate and observe
IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training
 IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training Multi Target Analysis Neurite Outgrowth Membrane Translocation Analysis Object Intensity Dual Area Object Analysis Granularity Morphology
IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training Multi Target Analysis Neurite Outgrowth Membrane Translocation Analysis Object Intensity Dual Area Object Analysis Granularity Morphology
Introduction. Loading Images
 Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: http://www.cellprofiler.org/. CellProfiler
Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: http://www.cellprofiler.org/. CellProfiler
Rare Event Detection Algorithm. User s Guide
 Rare Event Detection Algorithm User s Guide Copyright 2008 Aperio Technologies, Inc. Part Number/Revision: MAN 0123, Revision A Date: September 2, 2008 This document applies to software versions Release
Rare Event Detection Algorithm User s Guide Copyright 2008 Aperio Technologies, Inc. Part Number/Revision: MAN 0123, Revision A Date: September 2, 2008 This document applies to software versions Release
LAB 1 INSTRUCTIONS DESCRIBING AND DISPLAYING DATA
 LAB 1 INSTRUCTIONS DESCRIBING AND DISPLAYING DATA This lab will assist you in learning how to summarize and display categorical and quantitative data in StatCrunch. In particular, you will learn how to
LAB 1 INSTRUCTIONS DESCRIBING AND DISPLAYING DATA This lab will assist you in learning how to summarize and display categorical and quantitative data in StatCrunch. In particular, you will learn how to
Detecting Geometric Faults from Measured Data
 Detecting Geometric s from Measured Data A.L. Gower 1 1 School of Mathematics, Statistics and Applied Mathematics, National University of Ireland Galway, Ireland. May 4, 214 Abstract Manufactured artefacts
Detecting Geometric s from Measured Data A.L. Gower 1 1 School of Mathematics, Statistics and Applied Mathematics, National University of Ireland Galway, Ireland. May 4, 214 Abstract Manufactured artefacts
Importing and processing a DGGE gel image
 BioNumerics Tutorial: Importing and processing a DGGE gel image 1 Aim Comprehensive tools for the processing of electrophoresis fingerprints, both from slab gels and capillary sequencers are incorporated
BioNumerics Tutorial: Importing and processing a DGGE gel image 1 Aim Comprehensive tools for the processing of electrophoresis fingerprints, both from slab gels and capillary sequencers are incorporated
An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data
 Supplementary material: User manual An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data Juan Prada, Manju
Supplementary material: User manual An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data Juan Prada, Manju
Mineral Browser - Viewing Minerals and Assays
 Manual 86-1(1) Starting Mineral database The Mineral database can be accessed by clicking on the button on the GEO main toolbar on the left side of the screen. After clicking the Mineral Browser main window
Manual 86-1(1) Starting Mineral database The Mineral database can be accessed by clicking on the button on the GEO main toolbar on the left side of the screen. After clicking the Mineral Browser main window
Sample Sizes: up to 1 X1 X 1/4. Scanners: 50 X 50 X 17 microns and 15 X 15 X 7 microns
 R-AFM100 For Nanotechnology Researchers Wanting to do routine scanning of nano-structures Instrument Innovators Using AFM as a platform to create a new instrument Educators Teaching students about AFM
R-AFM100 For Nanotechnology Researchers Wanting to do routine scanning of nano-structures Instrument Innovators Using AFM as a platform to create a new instrument Educators Teaching students about AFM
Manual for Microfilament Analyzer
 Manual for Microfilament Analyzer Eveline Jacques & Jan Buytaert, Michał Lewandowski, Joris Dirckx, Jean-Pierre Verbelen, Kris Vissenberg University of Antwerp 1 Image data 2 image files are needed: a
Manual for Microfilament Analyzer Eveline Jacques & Jan Buytaert, Michał Lewandowski, Joris Dirckx, Jean-Pierre Verbelen, Kris Vissenberg University of Antwerp 1 Image data 2 image files are needed: a
SmartWorks MFP V4 Help File
 SmartWorks MFP V Help File. Overview. Navigation - Select Function. Start/Stop. Reset. Program Timeout Reset. Default Presets Document Types. COPY Presets. SCAN Presets. PRINT Presets.7 Color Modes & Fine
SmartWorks MFP V Help File. Overview. Navigation - Select Function. Start/Stop. Reset. Program Timeout Reset. Default Presets Document Types. COPY Presets. SCAN Presets. PRINT Presets.7 Color Modes & Fine
GPR Analyzer version 1.23 User s Manual
 GPR Analyzer version 1.23 User s Manual GPR Analyzer is a tool to quickly analyze multi- species microarrays, especially designed for use with the MIDTAL (Microarray Detection of Toxic ALgae) chip. It
GPR Analyzer version 1.23 User s Manual GPR Analyzer is a tool to quickly analyze multi- species microarrays, especially designed for use with the MIDTAL (Microarray Detection of Toxic ALgae) chip. It
Documentation to image analysis
 Documentation to image analysis The documentation below provides a detailed explanation and pointers towards the use of the MS-ECS-2D update site. It is made available in conjunction with the publication
Documentation to image analysis The documentation below provides a detailed explanation and pointers towards the use of the MS-ECS-2D update site. It is made available in conjunction with the publication
The walkthrough is available at /
 The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
