Introduction. Loading Images
|
|
|
- Solomon Bates
- 5 years ago
- Views:
Transcription
1 Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: CellProfiler was developed by Anne Carpenter and Thouis Jones in the laboratory of David Sabatini at the Whitehead Institute for Biomedical Research and by Polina Golland at the CSAIL of the MIT. CellProfiler is designed to analyze large amounts of images automatically. Since the analysis of thousands of images is very computer intensive, a version is available to run on a Linux cluster allowing the parallelization of the analysis. The latest release of CellProfiler (January 2014, version 2.1) is multicore and the number of processors used can be adjusted in the preference settings. The software is written in the Python programming language and the source code can be downloaded and extended by the community. CellProfiler has an intuitive user interface with point and click actions that allow assembling modules in a pipeline for processing images. Several features in CellProfiler make it an easy-touse software. The website hosts many tutorials and examples of image processing pipelines together with the corresponding images. In the GUI of CellProfiler, every button or drop down box for adjusting parameters has a help button, allowing to quickly look up its functionality. CellProfiler provides many advanced image processing algorithms allowing developing image analysis solutions for a wide variety of assays in mammalian cells, tissues slices and small organisms. There are several modules for pre-processing images such as calculating illumination correction functions and applying them to images or measuring image quality. Other modules perform image processing functions such as morphological operations (opening, closing of the image etc.), enhancement of edges and many more. A small collection of nodes is dedicated to detect objects in images using many different algorithms for distinguishing background from foreground and drawing separation lines between objects. Lastly, features such as intensity, texture and morphology can be extracted on either a object-by-object or on an image-by-image basis and exported to either CSV files, a MySQLite or MySQL database. Visual control of the performance of the image analysis is essential in large scale screens to verify the segmentation quality of the algorithms. CellProfiler generates overlays of the detected object outlines on images, for instances on RGB color merges of the various channels created within CellProfiler or directly on the original images. Another useful feature of the software is the ability to annotate images and to extract metadata either from the path or the file name itself. The handling of the data in the statistical analysis is made much easier and allows to quickly perform quality control checks of the images and the assay. The following document sketches a very simple application to detect nuclei. The images were acquired with the Perkin Elmer Operetta. People interested in using more powerful features or solve more difficult challenges are strongly encouraged to take advantage of the online material or take a course. Loading Images
2 CellProfiler can read many image formats using the BIO-Formats library of LOCI. Images are read using the import modules found on the user interface. Alternatively the deprecated module LoadImages can be used. Drag the folder containing images on the field intended for this. Cellprofiler automatically searches for image files. If only a subset of images need to be analyzed, finer control over the list of images can be obtained by constructing logic filter with regular expression. In this case, only the Hoechst channel images will be loaded by selecting files with the regular expression:.*ch1.* Next metadata contained in the path to the images and their name is defined and extracted using again regular expressions. Metadata extraction from file name
3 Metadata extraction from image path A name variable has then to be associated with the loaded images. Lastly, the association of the images for multi tif stacks and movies can be specified, but is not applicable here.
4 Segmenting objects The most crucial and computer intensive step in image analysis is object segmentation. A common task in most image analysis routines is to detect nuclei as primary objects and cytoplasm of cells as secondary objects around the nuclei. We generally stain nuclei with Hoechst and the cytoplasm with HCS CellMask Blue (Life Technologies). Nuclei are detected using Otsu s three class thresholding and fused objects are separated by finding local maxima followed by watershed. These methods are available in the IdentifyPrimaryObjects module. The module is adjusted from top to bottom selecting the image to analyze, the name of the output objects, their size etc. For each customization step, help is available by clicking on the corresponding question mark. CellProfiler offers the option to automatically find the best segmentation method. While this is generally very efficient, we typically do not use it, since we wish to apply the same segmentation algorithm to all of our images and do not want signal variation due to different algorithms or settings. We typically use Otsu three level segmentation, set a lower threshold to define the segmentation levels and define smoothing filters and minimal distance between local maxima. We factorize all parameters to avoid different settings between images and to maintain consistency across batches. We also save the outlines of the identified objects in order to control for the correctness of the segmentation.
5 Result of the segmentation looks like this: Next, using the DetectSecondaryObjects module, the stained cytoplasms are detected.
6 We typically use the propagation algorithm and the nuclei as seeds. The local thresholds are again determined by using Otsu and all parameters are determined. The result is the following:
7 A further common task in many biological assays is to detect smaller objects such as organelles in the cytoplasm of cells. For small structures with high background intensities, a white top-hat filter is applied using the EnhanceOrSuppressFeatures module to increase the contrast. The tertiary objects are then detected, using the IdentifyPrimaryObject module with multi threshold Otsu and watershed seeded from local maxima. Finally, to obtain tertiary object data on a cellby-cell basis, the objects are linked to their parent cell with a RelateObjects module. Extracting parameters After segmenting all the objects of interest, features are extracted for the quantitative phenotypic description. Several measurement modules are available in CellProfiler for both image-based measurements and object-based measurements.
8 Using a MeasureObjectIntensity module, many aspects of the intensity of objects can be captured such as, their integral, average or median intensity. Other modules measure the morphology (MeasureObjectSizeShape), the texture (MeasureTexture) or the spatial distribution of objects (MeasureObjectRadialDistribution, MeasureObjectNeighbors). Extracting all these parameters can easily generate tables of several hundreds of columns. Control images and exporting data When analyzing large screens, it is impossible to visually control the quality of segmentation of all the images. A spot control needs to be carried out and control images showing the outlines of the segmented objects are very useful for this. Furthermore, it is paramount to visualize all the images of hit wells to ensure that no spurious segmentation artifact leads to the obtained result. To create control images the OverlayOutlines module is added.
9 The outlines of each segmented object can be overlaid on an image and saved with the SaveImages module.
10 The last step of the analysis exports the data and associated metadata as comma-separated values file (CSV files) using the ExportToSpreadsheet module or to a database using the ExportToDatabase module.
11 Conclusion This simple pipeline will allow segmenting nuclei and extract parameters about the segmented objects. For more complex pipelines, the tutorial on the CellProfiler website should be followed. Alternatively, the TDS of the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden offers one day courses on CellProfiler (please contact Marc Bickle:
Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data
 Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data Mark-Anthony Bray, Ph.D Imaging Platform, Broad Institute Cambridge, Massachusetts, USA mbray@broadinstitute.org
Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data Mark-Anthony Bray, Ph.D Imaging Platform, Broad Institute Cambridge, Massachusetts, USA mbray@broadinstitute.org
Advanced Cell Classifier (ACC) user manual. v3.0 - June 2017
 Advanced Cell Classifier (ACC) user manual www.cellclassifier.org v3.0 - June 2017 Prof. Peter Horvath, PhD Synthetic and Systems Biology Unit Biological Research Centre of the Hungarian Academy of Sciences
Advanced Cell Classifier (ACC) user manual www.cellclassifier.org v3.0 - June 2017 Prof. Peter Horvath, PhD Synthetic and Systems Biology Unit Biological Research Centre of the Hungarian Academy of Sciences
Multiple Usage of KNIME in a Screening Laboratory Environment
 Multiple Usage of KNIME in a Screening Laboratory Environment KNIME UGM Zürich, 02.02.2012 Marc Bickle HT-TDS, MPI-CBG Outline Presentation of TDS Our problem: large complex datasets KNIME as data mining
Multiple Usage of KNIME in a Screening Laboratory Environment KNIME UGM Zürich, 02.02.2012 Marc Bickle HT-TDS, MPI-CBG Outline Presentation of TDS Our problem: large complex datasets KNIME as data mining
Background information:
 Image- based screening using subcellular localization of FOXO1A in osteosarcoma cells: A computer exercise using CellProfiler & CellProfiler Analyst software Carolina Wählby, Martin Simonsson, Megan Rokop
Image- based screening using subcellular localization of FOXO1A in osteosarcoma cells: A computer exercise using CellProfiler & CellProfiler Analyst software Carolina Wählby, Martin Simonsson, Megan Rokop
Extracting Rich Information from Images
 Chapter 14 Extracting Rich Information from Images Anne E. Carpenter Summary Now that automated image-acquisition instruments (high-throughput microscopes) are commercially available and becoming more
Chapter 14 Extracting Rich Information from Images Anne E. Carpenter Summary Now that automated image-acquisition instruments (high-throughput microscopes) are commercially available and becoming more
CellProfiler Analyst data exploration software
 CellProfiler Analyst data exploration software TM Created in the laboratories of Anne E. Carpenter, David M. Sabatini and Polina Golland at: Computer Sciences/ Artificial Intelligence Laboratory at the
CellProfiler Analyst data exploration software TM Created in the laboratories of Anne E. Carpenter, David M. Sabatini and Polina Golland at: Computer Sciences/ Artificial Intelligence Laboratory at the
Voronoi-Based Segmentation of Cells on Image Manifolds
 Voronoi-Based Segmentation of Cells on Image Manifolds Thouis R. Jones 1, Anne Carpenter 2, and Polina Golland 1 1 MIT CSAIL, Cambridge, MA, USA 2 Whitehead Institute for Biomedical Research, Cambridge,
Voronoi-Based Segmentation of Cells on Image Manifolds Thouis R. Jones 1, Anne Carpenter 2, and Polina Golland 1 1 MIT CSAIL, Cambridge, MA, USA 2 Whitehead Institute for Biomedical Research, Cambridge,
Technology Development Studio (TDS) MPI-CBG, Dresden, Germany. Marc Bickle HT-TDS, MPI-CBG
 Technology Development Studio (TDS) MPI-CBG, Dresden, Germany Marc Bickle HT-TDS, MPI-CBG TDS: Core Screening Facility Core Screening facility of the MPI-CBG specialized in high content imaging (es 2003)
Technology Development Studio (TDS) MPI-CBG, Dresden, Germany Marc Bickle HT-TDS, MPI-CBG TDS: Core Screening Facility Core Screening facility of the MPI-CBG specialized in high content imaging (es 2003)
Detection of Sub-resolution Dots in Microscopy Images
 Detection of Sub-resolution Dots in Microscopy Images Karel Štěpka, 2012 Centre for Biomedical Image Analysis, FI MU supervisor: prof. RNDr. Michal Kozubek, Ph.D. Outline Introduction Existing approaches
Detection of Sub-resolution Dots in Microscopy Images Karel Štěpka, 2012 Centre for Biomedical Image Analysis, FI MU supervisor: prof. RNDr. Michal Kozubek, Ph.D. Outline Introduction Existing approaches
Definiens. Tissue Studio 4.2. Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection
 Definiens Tissue Studio 4.2 Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection Imprint and Version Copyright 2015
Definiens Tissue Studio 4.2 Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection Tutorial 3: Metadata Import, Manual ROI Selection and Vessel Detection Imprint and Version Copyright 2015
Documentation to image analysis
 Documentation to image analysis The documentation below provides a detailed explanation and pointers towards the use of the MS-ECS-2D update site. It is made available in conjunction with the publication
Documentation to image analysis The documentation below provides a detailed explanation and pointers towards the use of the MS-ECS-2D update site. It is made available in conjunction with the publication
Using References in NIS-Elements
 Using References in NIS-Elements Overview This technical note describes general usage of References in NIS-Elements for image processing and analysis. The Reference feature in NIS-Elements refers to a
Using References in NIS-Elements Overview This technical note describes general usage of References in NIS-Elements for image processing and analysis. The Reference feature in NIS-Elements refers to a
Colocalization tutorial
 Colocalization tutorial I. Introduction Fluorescence microscopy is helpful for determining when two proteins colocalize in the cell. At the most basic level, images can be inspected by eye. Quantitative
Colocalization tutorial I. Introduction Fluorescence microscopy is helpful for determining when two proteins colocalize in the cell. At the most basic level, images can be inspected by eye. Quantitative
Chemotaxis Data Processing: Creating an Analysis Job
 essenbioscience.com/incucyte TECH NOTE IncuCyte ZOOM Live-Cell Imaging System Chemotaxis Data Processing: Creating an Analysis Job Data processing analyzes and summarizes qualitative images into quantitative
essenbioscience.com/incucyte TECH NOTE IncuCyte ZOOM Live-Cell Imaging System Chemotaxis Data Processing: Creating an Analysis Job Data processing analyzes and summarizes qualitative images into quantitative
Definiens. Tissue Studio 4.4. Tutorial 4: Manual ROI Selection and Marker Area Detection
 Definiens Tissue Studio 4.4 Tutorial 4: Manual ROI Selection and Marker Area Detection Tutorial 4: Manual ROI Selection and Marker Area Detection Imprint and Version Copyright 2017 Definiens AG. All rights
Definiens Tissue Studio 4.4 Tutorial 4: Manual ROI Selection and Marker Area Detection Tutorial 4: Manual ROI Selection and Marker Area Detection Imprint and Version Copyright 2017 Definiens AG. All rights
IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training
 IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training Multi Target Analysis Neurite Outgrowth Membrane Translocation Analysis Object Intensity Dual Area Object Analysis Granularity Morphology
IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training Multi Target Analysis Neurite Outgrowth Membrane Translocation Analysis Object Intensity Dual Area Object Analysis Granularity Morphology
Lecture: Segmentation I FMAN30: Medical Image Analysis. Anders Heyden
 Lecture: Segmentation I FMAN30: Medical Image Analysis Anders Heyden 2017-11-13 Content What is segmentation? Motivation Segmentation methods Contour-based Voxel/pixel-based Discussion What is segmentation?
Lecture: Segmentation I FMAN30: Medical Image Analysis Anders Heyden 2017-11-13 Content What is segmentation? Motivation Segmentation methods Contour-based Voxel/pixel-based Discussion What is segmentation?
Use stage 16 embryos see Pereanu et al., 2007 for morphological staging criteria. First start Vaa3d (http://vaa3d.org/; Peng et al., 2010).
 Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
SETTING UP AN HCS DATA ANALYSIS SYSTEM
 A WHITE PAPER FROM GENEDATA JANUARY 2010 SETTING UP AN HCS DATA ANALYSIS SYSTEM WHY YOU NEED ONE HOW TO CREATE ONE HOW IT WILL HELP HCS MARKET AND DATA ANALYSIS CHALLENGES High Content Screening (HCS)
A WHITE PAPER FROM GENEDATA JANUARY 2010 SETTING UP AN HCS DATA ANALYSIS SYSTEM WHY YOU NEED ONE HOW TO CREATE ONE HOW IT WILL HELP HCS MARKET AND DATA ANALYSIS CHALLENGES High Content Screening (HCS)
Last updated 24 June 2017
 Published in: Braude JP, Vijayakumar S, Baumgarner K, Laurine R, Jones TA, Jones SM, Pyott SJ. (2015) Deletion of Shank1 has minimal effects on the molecular composition and function of glutamatergic afferent
Published in: Braude JP, Vijayakumar S, Baumgarner K, Laurine R, Jones TA, Jones SM, Pyott SJ. (2015) Deletion of Shank1 has minimal effects on the molecular composition and function of glutamatergic afferent
Dragonfly Pro. Visual Pathway to Quantitative Answers ORS. Exclusive to ZEISS OBJECT RESEARCH SYSTEMS
 Dragonfly Pro Exclusive to ZEISS Visual Pathway to Quantitative Answers ORS OBJECT RESEARCH SYSTEMS Visualization and analysis software without bounds Dragonfly Pro by Object Research Systems (ORS) is
Dragonfly Pro Exclusive to ZEISS Visual Pathway to Quantitative Answers ORS OBJECT RESEARCH SYSTEMS Visualization and analysis software without bounds Dragonfly Pro by Object Research Systems (ORS) is
Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays.
 Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays. http://www.nexus.ethz.ch/ -> Software -> TMARKER The plugin Intensity Clustering starts on step 7. Steps 1-6
Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays. http://www.nexus.ethz.ch/ -> Software -> TMARKER The plugin Intensity Clustering starts on step 7. Steps 1-6
The Arena View. Tutorial. November The Arena is an easy-to-use image management system for analysis of high volume data.
 The Arena View Tutorial November 2014 The Arena is an easy-to-use image management system for analysis of high volume data. The Arena is your working centre in Imaris and an interface for visualizing,
The Arena View Tutorial November 2014 The Arena is an easy-to-use image management system for analysis of high volume data. The Arena is your working centre in Imaris and an interface for visualizing,
Pathology Image Informatics Platform (PathIIP)
 Pathology Image Informatics Platform (PathIIP) PIs: Anant Madabhushi (CWRU), Metin Gurcan (OSU), Anne Martel (UToronto) Year 2 Update Develop a digital pathology platform to facilitate wider adoption of
Pathology Image Informatics Platform (PathIIP) PIs: Anant Madabhushi (CWRU), Metin Gurcan (OSU), Anne Martel (UToronto) Year 2 Update Develop a digital pathology platform to facilitate wider adoption of
SENSITIVITY DIFFERENTLY TO SEE THINGS THE SPEED AND. Opera Phenix High Content Screening System
 THE SPEED AND SENSITIVITY TO SEE THINGS DIFFERENTLY Opera Phenix High Content Screening System For research use only. Not for use in diagnostic procedures. HIGH CONTENT SCREENING WITHOUT THE COMPROMISE
THE SPEED AND SENSITIVITY TO SEE THINGS DIFFERENTLY Opera Phenix High Content Screening System For research use only. Not for use in diagnostic procedures. HIGH CONTENT SCREENING WITHOUT THE COMPROMISE
Michal Kuneš
 The Open Microscopy Environment A DataBase for the storage and manipulation of image data Michal Kuneš xkunes@utia.cas.cz ZOI UTIA, ASCR, Friday seminar 13.12.2013 OMERO http://www.openmicroscopy.org/site/support/omero4/users/index.html
The Open Microscopy Environment A DataBase for the storage and manipulation of image data Michal Kuneš xkunes@utia.cas.cz ZOI UTIA, ASCR, Friday seminar 13.12.2013 OMERO http://www.openmicroscopy.org/site/support/omero4/users/index.html
Supplementary Figures
 Supplementary Figures re co rd ed ch annels darkf ield MPM2 PI cell cycle phases G1/S/G2 prophase metaphase anaphase telophase bri ghtfield 55 pixel Supplementary Figure 1 Images of Jurkat cells captured
Supplementary Figures re co rd ed ch annels darkf ield MPM2 PI cell cycle phases G1/S/G2 prophase metaphase anaphase telophase bri ghtfield 55 pixel Supplementary Figure 1 Images of Jurkat cells captured
Algorithm User Guide:
 Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
Crop Counting and Metrics Tutorial
 Crop Counting and Metrics Tutorial The ENVI Crop Science platform contains remote sensing analytic tools for precision agriculture and agronomy. In this tutorial you will go through a typical workflow
Crop Counting and Metrics Tutorial The ENVI Crop Science platform contains remote sensing analytic tools for precision agriculture and agronomy. In this tutorial you will go through a typical workflow
For questions please contact: Lei-Ann Arceneaux, 4/13/2011
 TMALab TMALab is a web-based function in Spectrum Plus, working for Tissue Microarrays (TMAs). TMAs allow researchers to validate new biomarkers or discover and dissect molecular pathways in hundreds of
TMALab TMALab is a web-based function in Spectrum Plus, working for Tissue Microarrays (TMAs). TMAs allow researchers to validate new biomarkers or discover and dissect molecular pathways in hundreds of
IDL Tutorial. Working with Images. Copyright 2008 ITT Visual Information Solutions All Rights Reserved
 IDL Tutorial Working with Images Copyright 2008 ITT Visual Information Solutions All Rights Reserved http://www.ittvis.com/ IDL is a registered trademark of ITT Visual Information Solutions for the computer
IDL Tutorial Working with Images Copyright 2008 ITT Visual Information Solutions All Rights Reserved http://www.ittvis.com/ IDL is a registered trademark of ITT Visual Information Solutions for the computer
Appendix I. TACTICS Toolbox v3.x. Interactive MATLAB Platform For Bioimaging informatics. User Guide TRACKING MODULE
 TACTICS Toolbox v3.x Interactive MATLAB Platform For Bioimaging informatics User Guide TRACKING MODULE -17- Cell Tracking Module 1 (user interface) Once the images were successfully segmented, the next
TACTICS Toolbox v3.x Interactive MATLAB Platform For Bioimaging informatics User Guide TRACKING MODULE -17- Cell Tracking Module 1 (user interface) Once the images were successfully segmented, the next
Nuclei Segmentation of Whole Slide Images in Digital Pathology
 Nuclei Segmentation of Whole Slide Images in Digital Pathology Dennis Ai Department of Electrical Engineering Stanford University Stanford, CA dennisai@stanford.edu Abstract Pathology is the study of the
Nuclei Segmentation of Whole Slide Images in Digital Pathology Dennis Ai Department of Electrical Engineering Stanford University Stanford, CA dennisai@stanford.edu Abstract Pathology is the study of the
FlowJo Software Lecture Outline:
 FlowJo Software Lecture Outline: Workspace Basics: 3 major components 1) The Ribbons (toolbar) The availability of buttons here can be customized. *One of the best assets of FlowJo is the help feature*
FlowJo Software Lecture Outline: Workspace Basics: 3 major components 1) The Ribbons (toolbar) The availability of buttons here can be customized. *One of the best assets of FlowJo is the help feature*
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences
 Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris Imaris State of the Art Image Visualization and Analysis Over the last 25 years Imaris has continuously improved
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris Imaris State of the Art Image Visualization and Analysis Over the last 25 years Imaris has continuously improved
Detailed Program Image Processing Summer School 2010
 Detailed Program Image Processing Summer School 2010 Monday 08.30-09.00: Registration 09.00-10.00: Introduction to image processing (Peter Horvath) Basic definitions (digital image, bit depth, sampling,
Detailed Program Image Processing Summer School 2010 Monday 08.30-09.00: Registration 09.00-10.00: Introduction to image processing (Peter Horvath) Basic definitions (digital image, bit depth, sampling,
Virtual Frap User Guide
 Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Advanced Image Processing using Imaris ( ) Zentrum für Mikroskopie und Bildanalyse
 Advanced Image Processing using Imaris (25. 11. 2008) Urs Ziegler Zentrum für Mikroskopie und Bildanalyse ziegler@zmb.uzh.ch Imaris integrated modules Todays seminar Open and Save data Isosurface Contrast
Advanced Image Processing using Imaris (25. 11. 2008) Urs Ziegler Zentrum für Mikroskopie und Bildanalyse ziegler@zmb.uzh.ch Imaris integrated modules Todays seminar Open and Save data Isosurface Contrast
Quantitative Image Analysis and 3-D Digital Reconstruction of Aortic Valve Leaflet
 Quantitative Image Analysis and 3-D Digital Reconstruction of Aortic Valve Leaflet Chi Zheng 1 Mentors: John A. Stella 2 and Michael S. Sacks, Ph. D. 2 1 Bioengineering and Bioinformatics Summer Institute,
Quantitative Image Analysis and 3-D Digital Reconstruction of Aortic Valve Leaflet Chi Zheng 1 Mentors: John A. Stella 2 and Michael S. Sacks, Ph. D. 2 1 Bioengineering and Bioinformatics Summer Institute,
GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow.
![GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow. GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow.](/thumbs/75/71593469.jpg) Manual KNIME Image Processing :: First steps This manual aims to provide first insights in KNIME Image Processing. The image processing workflow Count nuclei is described in detail and can be downloaded
Manual KNIME Image Processing :: First steps This manual aims to provide first insights in KNIME Image Processing. The image processing workflow Count nuclei is described in detail and can be downloaded
BioImageXD Getting started
 BioImageXD Getting started Updated 2012-07-01 by Pasi Kankaanpää Introduction Welcome to BioImageXD, free open source software for biomedical image post-processing, visualization and analysis. This guide
BioImageXD Getting started Updated 2012-07-01 by Pasi Kankaanpää Introduction Welcome to BioImageXD, free open source software for biomedical image post-processing, visualization and analysis. This guide
Robust Detection for Red Blood Cells in Thin Blood Smear Microscopy Using Deep Learning
 Robust Detection for Red Blood Cells in Thin Blood Smear Microscopy Using Deep Learning By Yasmin Kassim PhD Candidate in University of Missouri-Columbia Supervised by Dr. Kannappan Palaniappan Mentored
Robust Detection for Red Blood Cells in Thin Blood Smear Microscopy Using Deep Learning By Yasmin Kassim PhD Candidate in University of Missouri-Columbia Supervised by Dr. Kannappan Palaniappan Mentored
Image Analysis Lecture Segmentation. Idar Dyrdal
 Image Analysis Lecture 9.1 - Segmentation Idar Dyrdal Segmentation Image segmentation is the process of partitioning a digital image into multiple parts The goal is to divide the image into meaningful
Image Analysis Lecture 9.1 - Segmentation Idar Dyrdal Segmentation Image segmentation is the process of partitioning a digital image into multiple parts The goal is to divide the image into meaningful
Fast and accurate automated cell boundary determination for fluorescence microscopy
 Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
A Quick Guide to Using Cerebral in InnateDB
 A Quick Guide to Using Cerebral in InnateDB Cerebral can be used to visualize interaction networks from a set of interactions from InnateDB. Cerebral uses subcellular localization annotations to provide
A Quick Guide to Using Cerebral in InnateDB Cerebral can be used to visualize interaction networks from a set of interactions from InnateDB. Cerebral uses subcellular localization annotations to provide
a measurable difference
 Thermo Scientific Cell Analysis Software and Informatics Products a measurable difference for everyone Thermo Scientific HCS Studio Cell Analysis Software Thermo Scientific Store Image and Database Management
Thermo Scientific Cell Analysis Software and Informatics Products a measurable difference for everyone Thermo Scientific HCS Studio Cell Analysis Software Thermo Scientific Store Image and Database Management
SuRVoS Workbench. Super-Region Volume Segmentation. Imanol Luengo
 SuRVoS Workbench Super-Region Volume Segmentation Imanol Luengo Index - The project - What is SuRVoS - SuRVoS Overview - What can it do - Overview of the internals - Current state & Limitations - Future
SuRVoS Workbench Super-Region Volume Segmentation Imanol Luengo Index - The project - What is SuRVoS - SuRVoS Overview - What can it do - Overview of the internals - Current state & Limitations - Future
Arena. Managing, grouping and searching images, controlling Batch runs, compiling results for Vantage
 1 Arena Managing, grouping and searching images, controlling Batch runs, compiling results for Vantage Imaris 8 Arena Search field (files, tags ) Arena tree provides direct access to all the components
1 Arena Managing, grouping and searching images, controlling Batch runs, compiling results for Vantage Imaris 8 Arena Search field (files, tags ) Arena tree provides direct access to all the components
Segmentation of Three Dimensional Cell Culture Models from a Single Focal Plane
 Segmentation of Three Dimensional Cell Culture Models from a Single Focal Plane Hang Chang 1,2 and Bahram Parvin 1 1 Lawrence Berkeley National Laboratory, Berkeley, CA 94720 2 Institute of Automation,
Segmentation of Three Dimensional Cell Culture Models from a Single Focal Plane Hang Chang 1,2 and Bahram Parvin 1 1 Lawrence Berkeley National Laboratory, Berkeley, CA 94720 2 Institute of Automation,
OBCOL. (Organelle Based CO-Localisation) Users Guide
 OBCOL (Organelle Based CO-Localisation) Users Guide INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual
OBCOL (Organelle Based CO-Localisation) Users Guide INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual
Segmentation and descriptors for cellular immunofluoresence images
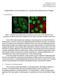 Christopher Probert cprobert@stanford.edu CS231A Project Report Segmentation and descriptors for cellular immunofluoresence images 1: Introduction Figure 1: Example immunoflouresence images of human cancer
Christopher Probert cprobert@stanford.edu CS231A Project Report Segmentation and descriptors for cellular immunofluoresence images 1: Introduction Figure 1: Example immunoflouresence images of human cancer
Babu Madhav Institute of Information Technology Years Integrated M.Sc.(IT)(Semester - 7)
 5 Years Integrated M.Sc.(IT)(Semester - 7) 060010707 Digital Image Processing UNIT 1 Introduction to Image Processing Q: 1 Answer in short. 1. What is digital image? 1. Define pixel or picture element?
5 Years Integrated M.Sc.(IT)(Semester - 7) 060010707 Digital Image Processing UNIT 1 Introduction to Image Processing Q: 1 Answer in short. 1. What is digital image? 1. Define pixel or picture element?
Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides
 1 Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides ways to flexibly merge your Mascot search and quantitation
1 Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides ways to flexibly merge your Mascot search and quantitation
A Visual Programming Environment for Machine Vision Engineers. Paul F Whelan
 A Visual Programming Environment for Machine Vision Engineers Paul F Whelan Vision Systems Group School of Electronic Engineering, Dublin City University, Dublin 9, Ireland. Ph: +353 1 700 5489 Fax: +353
A Visual Programming Environment for Machine Vision Engineers Paul F Whelan Vision Systems Group School of Electronic Engineering, Dublin City University, Dublin 9, Ireland. Ph: +353 1 700 5489 Fax: +353
UGviewer: a medical image viewer
 Appendix A UGviewer: a medical image viewer As a complement to this master s thesis, an own medical image viewer was programmed. This piece of software lets the user visualize and compare images. Designing
Appendix A UGviewer: a medical image viewer As a complement to this master s thesis, an own medical image viewer was programmed. This piece of software lets the user visualize and compare images. Designing
BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening. Alex Laude The BioImaging Unit
 BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening Alex Laude The BioImaging Unit Multi-dimensional, multi-modal imaging at the sub-cellular
BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening Alex Laude The BioImaging Unit Multi-dimensional, multi-modal imaging at the sub-cellular
Imaris 4.2 user information
 Imaris 4.2 user information There is also a manual to help you use Imaris. It is on the shelf above the Windows machine, to the left of the Adobe box. Please make sure you return it there when you are
Imaris 4.2 user information There is also a manual to help you use Imaris. It is on the shelf above the Windows machine, to the left of the Adobe box. Please make sure you return it there when you are
Zeiss Efficient Navigation (ZEN) Blue Edition Standard Operation Protocol
 Faculty Core Facility ZEN BLUE 2.3 SOP A-1 Zeiss Efficient Navigation (ZEN) Blue Edition Standard Operation Protocol Faculty Core Facility ZEN BLUE 2.3 SOP A-2 A. Content Overview. 3 Start up. 4 Display
Faculty Core Facility ZEN BLUE 2.3 SOP A-1 Zeiss Efficient Navigation (ZEN) Blue Edition Standard Operation Protocol Faculty Core Facility ZEN BLUE 2.3 SOP A-2 A. Content Overview. 3 Start up. 4 Display
Flicker Comparison of 2D Electrophoretic Gels
 Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute ++ SAIC-Frederick Frederick, MD, USA lemkin@ncifcrf.gov
Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute ++ SAIC-Frederick Frederick, MD, USA lemkin@ncifcrf.gov
Software Activation: inform is licensed software that must be activated for use of anything other than the viewer.
 inform Software Release Notes Software Version: 2.4.2 This file provides information about inform software, version 2.4.2 Please read this information carefully as it supersedes all printed material or
inform Software Release Notes Software Version: 2.4.2 This file provides information about inform software, version 2.4.2 Please read this information carefully as it supersedes all printed material or
Tour Guide for Windows and Macintosh
 Tour Guide for Windows and Macintosh 2011 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Suite 100A, Ann Arbor, MI 48108 USA phone 1.800.497.4939 or 1.734.769.7249 (fax) 1.734.769.7074
Tour Guide for Windows and Macintosh 2011 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Suite 100A, Ann Arbor, MI 48108 USA phone 1.800.497.4939 or 1.734.769.7249 (fax) 1.734.769.7074
TexRAD Research Version Client User Guide Version 3.9
 Imaging tools for medical decision makers Cambridge Computed Imaging Ltd Grange Park Broadway Bourn Cambridge CB23 2TA UK TexRAD Research Version Client User Guide Version 3.9 Release date 23/05/2016 Number
Imaging tools for medical decision makers Cambridge Computed Imaging Ltd Grange Park Broadway Bourn Cambridge CB23 2TA UK TexRAD Research Version Client User Guide Version 3.9 Release date 23/05/2016 Number
Flicker Comparison of 2D Electrophoretic Gels
 Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute ++ SAIC-Frederick Frederick, MD, USA lemkin@ncifcrf.gov
Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute ++ SAIC-Frederick Frederick, MD, USA lemkin@ncifcrf.gov
Software Activation: inform is licensed software that must be activated for use of anything other than the viewer.
 inform Software Release Notes Software Version: 2.4.1 This file provides information about inform software, version 2.4.1 Please read this information carefully as it supersedes all printed material or
inform Software Release Notes Software Version: 2.4.1 This file provides information about inform software, version 2.4.1 Please read this information carefully as it supersedes all printed material or
Morphology-form and structure. Who am I? structuring element (SE) Today s lecture. Morphological Transformation. Mathematical Morphology
 Mathematical Morphology Morphology-form and structure Sonka 13.1-13.6 Ida-Maria Sintorn Ida.sintorn@cb.uu.se mathematical framework used for: pre-processing - noise filtering, shape simplification,...
Mathematical Morphology Morphology-form and structure Sonka 13.1-13.6 Ida-Maria Sintorn Ida.sintorn@cb.uu.se mathematical framework used for: pre-processing - noise filtering, shape simplification,...
Cell Segmentation from Cellular Image
 Global Journal of Computer Science and Technology Vol. 10 Issue 13 (Ver. 1.0 ) October 2010 P a g e 55 Cell Segmentation from Cellular Image Mohammad Ibrahim Khan, Muhammad Kamal Hossen, Md.Sabbir Ali
Global Journal of Computer Science and Technology Vol. 10 Issue 13 (Ver. 1.0 ) October 2010 P a g e 55 Cell Segmentation from Cellular Image Mohammad Ibrahim Khan, Muhammad Kamal Hossen, Md.Sabbir Ali
Segmentation of Neuronal-Cell Images from Stained Fields and Monomodal Histograms
 Segmentation of Neuronal-Cell Images from Stained Fields and Monomodal Histograms Author D. Pham, Tuan, Crane, Denis Published 2005 Conference Title Proceedings of the 2005 IEEE Engineering in Medicine
Segmentation of Neuronal-Cell Images from Stained Fields and Monomodal Histograms Author D. Pham, Tuan, Crane, Denis Published 2005 Conference Title Proceedings of the 2005 IEEE Engineering in Medicine
9 Tumor Blood Vessels: 3-D Tubular Network Analysis
 Chap. 9 2015/8/20 14:02 page 1 1 9 Tumor Blood Vessels: 3-D Tubular Network Analysis Chap. 9 2015/8/20 14:02 page 2 2 Chap. 9 2015/8/20 14:02 page 3 3 Contents 9 Tumor Blood Vessels: 3-D Tubular Network
Chap. 9 2015/8/20 14:02 page 1 1 9 Tumor Blood Vessels: 3-D Tubular Network Analysis Chap. 9 2015/8/20 14:02 page 2 2 Chap. 9 2015/8/20 14:02 page 3 3 Contents 9 Tumor Blood Vessels: 3-D Tubular Network
Course 4 Advanced Image Analysis & Processing. April 2015
 Course 4 Advanced Image Analysis & Processing April 2015 1 From Survey Results Heterogenous audience Day 1 : ImageJ -Macro : language, batch, use of ActionBar. Day2 : ImageJ - Plugins : install, writing,
Course 4 Advanced Image Analysis & Processing April 2015 1 From Survey Results Heterogenous audience Day 1 : ImageJ -Macro : language, batch, use of ActionBar. Day2 : ImageJ - Plugins : install, writing,
Introduction to EBImage, an image processing and analysis toolkit for R
 Introduction to EBImage, an image processing and analysis toolkit for R Oleg Sklyar, Wolfgang Huber osklyar@ebi.ac.uk October 22, 2008 Contents 1 Introduction 1 2 Importing and exporting images 1 3 Displaying
Introduction to EBImage, an image processing and analysis toolkit for R Oleg Sklyar, Wolfgang Huber osklyar@ebi.ac.uk October 22, 2008 Contents 1 Introduction 1 2 Importing and exporting images 1 3 Displaying
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences
 Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris A Brief History Imaris was created 25 years ago to focus purely on the visualization of 3D fluorescence images
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris A Brief History Imaris was created 25 years ago to focus purely on the visualization of 3D fluorescence images
3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen
 3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen Dataset: Huh7.tif (original), obtained by Serial Block Face-SEM (Gatan 3View) Huh7_crop.tif
3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen Dataset: Huh7.tif (original), obtained by Serial Block Face-SEM (Gatan 3View) Huh7_crop.tif
Introduction to Celleste Imaging Analysis Software
 Introduction to Celleste Imaging Analysis Software USER GUIDE Publication Number MAN0018003 Revision A.0 For Research Use Only. Not for use in diagnostic procedures. Manufacturer: Life Technologies Corporation
Introduction to Celleste Imaging Analysis Software USER GUIDE Publication Number MAN0018003 Revision A.0 For Research Use Only. Not for use in diagnostic procedures. Manufacturer: Life Technologies Corporation
Image Analysis and Morphometry
 Image Analysis and Morphometry Lukas Schärer Evolutionary Biology Zoological Institute University of Basel 1 13. /15.3.2013 Zoology & Evolution Block Course Summary Quantifying morphology why do we need
Image Analysis and Morphometry Lukas Schärer Evolutionary Biology Zoological Institute University of Basel 1 13. /15.3.2013 Zoology & Evolution Block Course Summary Quantifying morphology why do we need
Rare Event Detection Algorithm. User s Guide
 Rare Event Detection Algorithm User s Guide Copyright 2008 Aperio Technologies, Inc. Part Number/Revision: MAN 0123, Revision A Date: September 2, 2008 This document applies to software versions Release
Rare Event Detection Algorithm User s Guide Copyright 2008 Aperio Technologies, Inc. Part Number/Revision: MAN 0123, Revision A Date: September 2, 2008 This document applies to software versions Release
SlideReader V12. Software Manual. System Requirements. Required Privileges
 SlideReader V12 Software Manual System Requirements - 1.8 GHz Processor or higher - Operating System: WIN XP professional or WIN 7 professional, 64-Bit - 1 Gb RAM or more - Minimum 2 Gb free disk space,
SlideReader V12 Software Manual System Requirements - 1.8 GHz Processor or higher - Operating System: WIN XP professional or WIN 7 professional, 64-Bit - 1 Gb RAM or more - Minimum 2 Gb free disk space,
Textbook de.nbi Bioimage Analysis Workshop
 BIOIMAGE ANALYSIS WORKSHOP WITHIN THE FRAMEWORK OF HD-HUB Textbook de.nbi Bioimage Analysis Workshop An introduction into KNIME image analysis for life scientists Jürgen Reymann ViroQuant-CellNetworks
BIOIMAGE ANALYSIS WORKSHOP WITHIN THE FRAMEWORK OF HD-HUB Textbook de.nbi Bioimage Analysis Workshop An introduction into KNIME image analysis for life scientists Jürgen Reymann ViroQuant-CellNetworks
TUTORIAL. HCS- Tools + Scripting Integrations
 TUTORIAL HCS- Tools + Scripting Integrations HCS- Tools... 3 Setup... 3 Task and Data... 4 1) Data Input Opera Reader... 7 2) Meta data integration Expand barcode... 8 3) Meta data integration Join Layout...
TUTORIAL HCS- Tools + Scripting Integrations HCS- Tools... 3 Setup... 3 Task and Data... 4 1) Data Input Opera Reader... 7 2) Meta data integration Expand barcode... 8 3) Meta data integration Join Layout...
An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data
 Supplementary material: User manual An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data Juan Prada, Manju
Supplementary material: User manual An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data Juan Prada, Manju
Histogram and watershed based segmentation of color images
 Histogram and watershed based segmentation of color images O. Lezoray H. Cardot LUSAC EA 2607 IUT Saint-Lô, 120 rue de l'exode, 50000 Saint-Lô, FRANCE Abstract A novel method for color image segmentation
Histogram and watershed based segmentation of color images O. Lezoray H. Cardot LUSAC EA 2607 IUT Saint-Lô, 120 rue de l'exode, 50000 Saint-Lô, FRANCE Abstract A novel method for color image segmentation
GPI, Exercise #1. Part 1
 GPI, Exercise #1 In this exercise you will gain some experience with GPI data and the basic reduction steps. Start by reading the three papers related to GPI s commissioning, first- light and observations
GPI, Exercise #1 In this exercise you will gain some experience with GPI data and the basic reduction steps. Start by reading the three papers related to GPI s commissioning, first- light and observations
MetaMorph Standard Operation Protocol Basic Application
 MetaMorph Standard Operation Protocol Basic Application Contents Basic Navigation and Image Handling... 2 Opening Images... 2 Separating Multichannel Images... 2 Cropping an Image... 3 Changing an 8 bit
MetaMorph Standard Operation Protocol Basic Application Contents Basic Navigation and Image Handling... 2 Opening Images... 2 Separating Multichannel Images... 2 Cropping an Image... 3 Changing an 8 bit
Analysis of multi-channel cell-based screens
 Analysis of multi-channel cell-based screens Lígia Brás, Michael Boutros and Wolfgang Huber August 6, 2006 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
Analysis of multi-channel cell-based screens Lígia Brás, Michael Boutros and Wolfgang Huber August 6, 2006 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
Image Processing: Final Exam November 10, :30 10:30
 Image Processing: Final Exam November 10, 2017-8:30 10:30 Student name: Student number: Put your name and student number on all of the papers you hand in (if you take out the staple). There are always
Image Processing: Final Exam November 10, 2017-8:30 10:30 Student name: Student number: Put your name and student number on all of the papers you hand in (if you take out the staple). There are always
A Tutorial Guide to Tribology Plug-in
 Supplementary Material A Tutorial Guide to Tribology Plug-in Tribology An ImageJ Plugin for surface topography analysis of laser textured surfaces. General Description This plugin presents an easy-to-use
Supplementary Material A Tutorial Guide to Tribology Plug-in Tribology An ImageJ Plugin for surface topography analysis of laser textured surfaces. General Description This plugin presents an easy-to-use
IMAGEQUANT. USER S GUIDE Version 5.0. for Microsoft Windows NT Apple Power Macintosh
 IMAGEQUANT USER S GUIDE Version 5.0 for Microsoft Windows NT Apple Power Macintosh Cover (Foreground): Image of a fluorescent 2-color TaqMan (assay in 96-well microplate scanned with a FluorImager 595.
IMAGEQUANT USER S GUIDE Version 5.0 for Microsoft Windows NT Apple Power Macintosh Cover (Foreground): Image of a fluorescent 2-color TaqMan (assay in 96-well microplate scanned with a FluorImager 595.
SIVIC GUI Overview. SIVIC GUI Layout Overview
 SIVIC GUI Overview SIVIC GUI Layout Overview At the top of the SIVIC GUI is a row of buttons called the Toolbar. It is a quick interface for loading datasets, controlling how the mouse manipulates the
SIVIC GUI Overview SIVIC GUI Layout Overview At the top of the SIVIC GUI is a row of buttons called the Toolbar. It is a quick interface for loading datasets, controlling how the mouse manipulates the
The walkthrough is available at /
 The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
TexRAD Research Version Client User Guide 1.1
 The Barn, Manor Road Church Lane Chilcompton Somerset BA3 4HP TexRAD Research Version Client User Guide 1.1 Release date 27/02/2013 Number of Pages 21 Replaces None Status Draft Comment Copyright 2013
The Barn, Manor Road Church Lane Chilcompton Somerset BA3 4HP TexRAD Research Version Client User Guide 1.1 Release date 27/02/2013 Number of Pages 21 Replaces None Status Draft Comment Copyright 2013
Software Activation: inform 2.3 is licensed software that must be activated for use of anything other than the viewer.
 inform Software Release Notes Software Version: 2.3 This file provides information about inform software, version 2.3 Please read this information carefully as it supersedes all printed material or online
inform Software Release Notes Software Version: 2.3 This file provides information about inform software, version 2.3 Please read this information carefully as it supersedes all printed material or online
Detection of Melanoma Skin Cancer using Segmentation and Classification Algorithm
 Detection of Melanoma Skin Cancer using Segmentation and Classification Algorithm Mrs. P. Jegadeeshwari Assistant professor/ece CK College of Engineering &Technology Abstract - Melanoma is the most dangerous
Detection of Melanoma Skin Cancer using Segmentation and Classification Algorithm Mrs. P. Jegadeeshwari Assistant professor/ece CK College of Engineering &Technology Abstract - Melanoma is the most dangerous
PhenoRipper Documentation
 PhenoRipper Documentation PhenoRipper Installation Windows We provide 32 bits and 64 bits installers. Choose the one that corresponds to your operating system, double click on it and follow the installer
PhenoRipper Documentation PhenoRipper Installation Windows We provide 32 bits and 64 bits installers. Choose the one that corresponds to your operating system, double click on it and follow the installer
n o r d i c B r a i n E x Tutorial DTI Module
 m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial DTI Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial DTI Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
Definiens. Tissue Studio 4.2. User Guide
 Definiens Tissue Studio 4.2 User Guide Definiens Documentation: Tissue Studio 4.2 User Guide Imprint 2015 Definiens AG. All rights reserved. This document may be copied and printed only in accordance with
Definiens Tissue Studio 4.2 User Guide Definiens Documentation: Tissue Studio 4.2 User Guide Imprint 2015 Definiens AG. All rights reserved. This document may be copied and printed only in accordance with
TOPIM-2018 schedule. Session 1: Introduction to OMERO & CellProfiler
 Presentations are available at https://downloads.openmicroscopy.org/presentations/2018/topim-crete/ TOPIM-2018 schedule Thursday 12th July: 19:40-20:15 (35 mins) OMERO workshop introduction Friday 13th
Presentations are available at https://downloads.openmicroscopy.org/presentations/2018/topim-crete/ TOPIM-2018 schedule Thursday 12th July: 19:40-20:15 (35 mins) OMERO workshop introduction Friday 13th
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences
 IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
IMAGEJ FINDFOCI Plugins
 IMAGEJ FINDFOCI Plugins Alex Herbert MRC Genome Damage and Stability Centre School of Life Sciences University of Sussex Science Road Falmer BN1 9RQ a.herbert@sussex.ac.uk Table of Contents Introduction...5
IMAGEJ FINDFOCI Plugins Alex Herbert MRC Genome Damage and Stability Centre School of Life Sciences University of Sussex Science Road Falmer BN1 9RQ a.herbert@sussex.ac.uk Table of Contents Introduction...5
Tutorial for VSDI. Table of Contents
 Tutorial for VSDI Table of Contents Before imaging... 2 Brain slices preparation... 2 Staining... 2 The EPHUS software... 2 Starting EPHUS... 3 Running a mapping experiment... 4 The BV software... 6 Setting
Tutorial for VSDI Table of Contents Before imaging... 2 Brain slices preparation... 2 Staining... 2 The EPHUS software... 2 Starting EPHUS... 3 Running a mapping experiment... 4 The BV software... 6 Setting
FiloQuant manual V1.0 Table of Contents
 FiloQuant manual V1.0 Table of Contents 1) FiloQuant aims and distribution license...2 2) Installation...3 3) FiloQuant, step-by-step instructions (single images)...4 1: Choose the region of interest to
FiloQuant manual V1.0 Table of Contents 1) FiloQuant aims and distribution license...2 2) Installation...3 3) FiloQuant, step-by-step instructions (single images)...4 1: Choose the region of interest to
Image Analysis & Cell Phenotyping
 Image Analysis & Cell Phenotyping Biological Image Processing Programs Feature ImageJ CellProfiler MetaMorph Definiens Matlab Neurolucida Price Free Free $$ $$$$ $ $$ Flexibility ++++ + ++ Optimized for
Image Analysis & Cell Phenotyping Biological Image Processing Programs Feature ImageJ CellProfiler MetaMorph Definiens Matlab Neurolucida Price Free Free $$ $$$$ $ $$ Flexibility ++++ + ++ Optimized for
