Tutorial using BEAST v2.4.7 Structured birth-death model Denise Kühnert and Jūlija Pečerska
|
|
|
- Myles Fisher
- 6 years ago
- Views:
Transcription
1 Tutorial using BEAST v2.4.7 Structured birth-death model Denise Kühnert and Jūlija Pečerska Population structure using the multi-type birth-death model 1 Introduction In this tutorial we will use the BEAST2 bdmm package to perform a Bayesian phylogenetic analysis of an influenza data set using the multi-type birth-death model (Kühnert et al. 2016). The data set used in this tutorial is a thinned 60 sequence subset of the 980 sequence H3N2 influenza data set used in the publication (Vaughan et al. 2014), which in turn was assembled from publicly-available data sets provided by various authors on GenBank. 1.1 Software Requirements BEAST2 - Bayesian Evolutionary Analysis Sampling Trees BEAST2 is a free software package for Bayesian evolutionary analysis of molecular sequences using MCMC and strictly oriented toward inference using rooted, time-measured phylogenetic trees. This tutorial is written for BEAST v2.4.7 (Bouckaert et al. 2014) BEAUti2 - Bayesian Evolutionary Analysis Utility BEAUti2 is a graphical user interface tool for generating BEAST2 XML configuration files. Both BEAST2 and BEAUti2 are Java programs, which means that the exact same code runs on all platforms. For us it simply means that the interface will be the same on all platforms. The screenshots used in this tutorial are taken on a Mac OS X computer; however, both programs will have the same layout and functionality on both Windows and Linux. BEAUti2 is provided as a part of the BEAST2 package so you do not need to install it separately TreeAnnotator TreeAnnotator is used to summarise the posterior sample of trees to produce a maximum clade credibility tree. It can also be used to summarise and visualise the posterior estimates of other tree parameters (e.g. node height). TreeAnnotator is provided as a part of the BEAST2 package so you do not need to install it separately Tracer Tracer ( is used to summarize the posterior estimates of the various parameters sampled by the Markov Chain. This program can be used for visual inspection and to assess convergence. It helps to quickly view median estimates and 95% highest posterior density intervals of the parameters, and calculates the effective sample sizes (ESS) of parameters. It can also be used to investigate potential parameter correlations. We will be using Tracer v
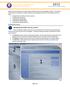
2 Figure 1: Install bdmm IcyTree IcyTree ( is a browser-based phylogenetic tree viewer. It is intended for rapid visualisation of phylogenetic tree files. It can also render phylogenetic networks provided in extended Newick format. IcyTree is compatible with current versions of Mozilla Firefox and Google Chrome. 1.2 Installing the bdmm package You can easily install the bdmm package via BEAUti s package manager. To do this, follow these steps: 1. Start BEAUti; 2. In the application menu, File > Manage packages. 3. Find bdmm in the list of packages shown, select it and then click Install/Upgrade. The BEAUTi window should look similar to what is shown in Figure 1. Note the actual version of bdmm may differ from the version shown in the figure, which is perfectly normal. Also note that bdmm depends on the MultiTypeTree package. BEAUTi will install the dependencies automatically once you select to install bdmm. Finally, restart BEAUti. The restart is necessary for the packages to be successfully installed. 2 Setting up the analysis using BEAUti 2.1 Loading the Template A BEAUTi template defines the basic structure and contents of your XML configuration file. By default BEAUTi will construct an XML file with standard uncoloured BEAST trees, however bdmm uses coloured trees which are defined in the MultiTypeTree package. To use the appropriate template for the configuration 2
3 Figure 2: Load the MultiTypeBirthDeath template. file, select File > Template > MultiTypeBirthDeath, as shown in Figure Loading the data Once the template is loaded, we can load in our example sequence data. In our case, this data is stored in a FASTA file, the first few lines of which look like this (the sequences have been truncated for better readability): > EU856841_HongKong_ GGGATAATTCTATTAACCATGAAGACTATCATTGCTTTGAGCTACATTT... > EU856989_HongKong_ CAAAAGCAGGGGATAATTCTATTAACCATGAAGACTATCATTGCTTTGAGCTACATTT... > CY039495_HongKong_ TTCTATTAACCATGAAGACTATCATTGCTTTGAGCTACATTC... > EU856853_HongKong_ TATTAACCATGAAGACTATCATTGCTTTGAGCTACATTC... > CY010084_NewZealand_ TATTAACCATGAAGACTATCATTGCTTTGAGCTACATTC... > CY007387_NewZealand_
4 Figure 3: Optional step: set the working directory to MultiTypeTree TATTAACCATGAAGACTATCATTGCTTTGAGCTACATTC... > CY012432_NewZealand_ CCATGAAGACTATCATTGCTTTGAGCTACATTT... The lines beginning with > are labels for the sequences immediately following. In general, these labels have no special format, but in this file each label is an underscore-delimited triple. The first element of each triple is the GenBank accession number of the sequence, the second is the geographical region from which it was sampled, and the third is the time at which it was sampled measured in calendar years or fractions thereof. In this tutorial we will be using the influenza sequence data which can be found in the examples folder of the MultiTypeTree package. To make it easier to find when loading the alignment, you can optionally set the working directory of BEAST2 to MultiTypeTree. This will make BEAUTi open the appropriate package folder when you look for the alignment. To set the working directory, select File > Set working dir > MultiTypeTree, as shown in Figure 3. 4
5 Figure 4: The alignment loaded into BEAUti. To load the file, select File > Add alignment. This will open a file selection dialog box. The example influenza sequence data file is named h3n2_2deme.fna. Assuming you have followed the previous step to set the working directory, this can be found in the examples/ directory shown when the file selection dialog box appears. In case you have not followed the previous step you will have to locate the folder containing the MultiTypeTree package and look for the examples/ folder there. Once the sequence file is loaded, your BEAUti screen should look similar to what is shown in Figure Setting up dates Once the data is loaded, the next step is to specify the times at which the sequences were sampled: 1. Select the Tip Dates panel. 2. Check the Use tip dates checkbox. 3. Click the Auto-configure button at the top-right of the panel. This opens a dialog that allows sample 5
6 Figure 5: Guessing the tip dates. times to be loaded from a file or inferred (guessed) from the sequence labels. 4. Because the times are included as the last element of the underscore-delimited sequence names, choose the use everything radio button and select after last from the drop-down menu. The default delimiter is already the underscore, so there is no need to change that. The date parsing setup will look as shown in Figure 5. After clicking OK you should find that the tip date table is filled with times that match those in the sequence headers, and that the last column of the table contains heights, i.e. times before most recent sample, calculated from the times. The BEAUTi panel should look as shown in Figure Setting up locations Now that we ve specified the sampling times, we move on to specifying the sampling locations. To do this, we follow a very similar set of steps to those we used to set the sample times: 1. Select the Tip Locations panel. You ll find that the locations are already filled with a single default value NOT_SET. 2. Click the Guess button at the top-right of the panel. This opens the same dialog that we saw in the 6
7 Figure 6: Sampling dates as seen in BEAUti. 7
8 Figure 7: Guessing the locations. previous section when setting up the dates. 3. The locations are included as the second element of the underscore-delimited sequence names. Therefore we choose the split on character radio button and select group 2 from the drop-down menu. Note again that the underscore character is already chosen as the delimiter. The location parsing setup will look as shown in Figure 7. After clicking OK you should find that the tip location table is filled with locations that match those in the sequence titles. The BEAUTi panel should look as shown in Figure Setting the substitution model For this analysis, we will use the HKY substitution model with 4 gamma categories and estimated base frequencies. To configure this in BEAUti, switch to the Site Model panel. First, we need to set up the rate category count. To approximate the continuous gamma rate distribution BEAST2 uses the discrete gamma distribution, where sites are divided into k equally probable rate categories. In general, 4-6 categories work well for most datasets, while having more categories involve a lot of computation at little precision gain, so we set the Gamma category count to 4. We would also like to estimate the Shape parameter, which describes the shape of the continuous gamma distribution we approximate. To do so, we need to set it to a non-zero value (e.g. the default 1.0) and tick the estimate checkbox. While the gamma categories account for rate variation, allowing some sites to have an evolutionary rate of 0 can improve fit to real data. To speed up the analysis we will fix this to the actual proportion of invariant sites we have in our alignment, which is
9 Figure 8: The locations in BEAUti. 9
10 Figure 9: Setup of the site model. Next, to set up the substitution model, select HKY from the drop-down menu (the default option is JC69). We would like to estimate the kappa parameter of HKY, so we leave the Kappa at the default value of 2.0 and leave the estimate checkbox checked. We would also like to estimate nucleotide frequencies, so we leave the Frequencies parameter at the default value (Estimated). The BEAUti panel should now look as shown in Figure 9. Note that the Substitution rate defined on this panel should not be estimated - we use the Clock rate defined in the Clock Model panel to determine the average per unit time rate of sequence evolution. This way, the Substitution rate is not actually a rate, but rather a rate multiplier that we fix to 1 to allow parameter identifiability. 2.6 Setting the clock model To speed up the analysis we will assume a strict clock for this small dataset. dataset. However, the selection of a clock model for a different, real analysis should not be taken lightly. Since our alignment contains sequences sampled at different times and those times are measured in years, we must use a clock rate expressed in units of expected substitutions per site per year. Usually the precise value is unknown 10
11 Figure 10: Fix the clock rate to speed up mixing. and so the default behaviour of BEAUti is to assume this rate has to be estimated. To speed up mixing we set the starting value of the Clock rate to 0.005, which we know from research to be much closer to the truth than the default value of 1. The Clock Model panel should now look as shown in Figure Adjusting Priors Setting up the bdmm tree prior bdmm defines a prior on the multi-type tree distribution. Thus it is particularly important for the analysis that we properly set up the priors. First, let s talk about the values that need to be set on the Priors panel. The first panel that you see at the top is the tree prior. bdmm is a model that can be used to explain data that is clearly divided into separate partitions, or demes. The demes can be geographical locations, as in our example, but the sequences can also be separated through other means than that, e.g. by a specific drug resistance mutation (strains can develop/lose drug resistance and thus move between demes, but can not transfer between demes otherwise), or location in the body (for example, for localised infections caused by the same agent). In this dataset we have strains from 11
12 2 different locations, New Zealand and Hong Kong, so the Number of demes should be set to 2, which also is the default value. Next, bdmm lets you estimate the Reproduction number per type and the BecomeUninfectiousRate per type. This will let us see the differences in reproduction fitness and speed of recovery between the two locations, so we leave the estimate checkboxes checked. We can leave the starting values at default as it will not influence the inference a lot. The next important thing one should take care of is setting the sampling proportions appropriately. In general, the trees that we build go back in time much further than the first sample that we have. If we set the same sampling proportion for the whole time period from our estimated tree origin to the time of the last sample, we will most likely run into trouble, as bdmm will try to produce a tree that has the same sampling proportion for the whole time, but no samples in the past and a lot of samples towards the present. In order to remove that bias from the trees, we need to make sure that we only have non-zero sampling starting from the first sample date (unless we know that there really weren t any related cases before the first sampled case). To do so, let s look at the SamplingProportion per type field. You will see that it has 4 values, which correspond to two values per type, lets call them [v1,v2,v3,v4]. v1 and v2 are the values for the first and second time interval for the first deme, and v3 and v4 are the values for the second deme. Thus, to do what we want we need to set the values v1 and v3 to zero. Because BEAST2 will use scalers to sample new values for the sampling proportions, the values which we set to 0 will remain so. Next, we also need to set the Sampling change time to the time slightly before the first sample. If we look back at the Tip dates panel, we can see that our oldest sample is the one labelled as EU856904_HongKong_ , for which the height, or the length of time from the first sample and the last, is We set the sampling change time in time units from the most recent sample and we need to make sure we include the first sample, thus we set the Sampling change time to 5.57, which is the height of the first sample rounded slightly up (and confirm the change with ENTER). The final setup of the tree prior can be seen in Figure 11. What if you have more demes? First things first, for an analysis with more demes you need to set the Number of demes to the appropriate value, e.g. N, that actually corresponds to the number of demes in the dataset. When you do that, the dimensions of the parameters Reproduction number, BecomeUninfectiousRate, SamplingProportion and Migration rates will change. The Reproduction number and the BecomeUninfectiousRate will have as many values as you have demes. The dimensionality of the SamplingProportion will be the number of demes times 2, so in case you have 4 demes your sampling proportion will need 8 values. You can view this parameter as a matrix of 2 x N values, which is flattened by row. The first column reflects the sampling rate before first sample and all of the values in it should be set to 0. The Sampling change time obviously does not change dimensionality, but has to be set to the appropriate time for your dataset. Finally, the Migration rates will have N * (N - 1) entries. As one can imagine, the matrix should have the dimensions of N * N, however since there is no migration from a deme to itself (values on the diagonal of the matrix), we subtract N values from the dimensionality, getting N * (N - 1) Setting up other priors By default, BEAST2 provides you with a prior distribution for each of the parameters of your model. This is done because otherwise BEAUTi will have a hard time displaying all of the parameters without any settings provided. Unfortunately, this means that some priors are very generic, and, moreover, some priors are in fact, improper the distribution does not integrate to one. This means that while the default setup might work and the runs will eventually mix, it can happen that the values are meaningless. So, let us go through the important parameters and set priors according to the information we have about 12
13 Figure 11: Set the change time for the sampling proportion so it is zero before the time of the first sample. 13
14 Figure 12: Set the prior for the R<sub>0</sub>. our dataset. The first important parameter is R0. In epidemiology, the basic reproduction number, R0, of an infection is the number of secondary cases one case generates on average over the course of its infectious period, in an otherwise uninfected population. Even though we do not have any information on R0 in the particular outbreak, infections rarely have R0 > 10, so we can set an upper limit on the sampled values. To do so, in the line denoted R0.t:h3n2_2deme click the button captioned initial = [2.0, 2.0] [0.0, Infinity] to get a pop up settings window (see Figure 12), where you can set the upper value. Other than that, the current prior sets ther median value of the distribution to e0 = 1, which will fit the endemic case of influenza. Next, we should adjust the prior for the rate of clearing the infection, which is labelled as becomeuninfectiousrate.t:h3n2_2deme. The value of the rate, say x, is the reciprocal of the average time a person with influenza is infectious, 1/x. From what we know about influenza we can say that an average infection lasts for about a week, however we would not want to impose too strong of a prior on this parameter. Let us change the distribution for this parameter to a LogNormal and tick the Mean in Real Space checkbox to make the setting easier. So, for a mean time of recovery of 7 days we need to set the mean of our distribution to 365/ (or to 52 for simplicity). Bear in mind that our time units are years, so we can not just set the rate to 1/7. This prior will ensure that we mainly sample realistic parameter values, but still gives BEAST2 quite a lot of freedom to go to extreme values if need be, as the 95% highest density interval for the prior is [4.44, 224], or [1.63, 82.21] infectious days. You can see the setup in Figure
15 Figure 13: Set the prior for the rate of recovery. 15
16 Figure 14: Set the prior for the clock rate. We will also set the prior for the clock rate to a distribution that is in accordance with what we know about RNA viruses, which is that in general their mean substitution rate is around We shall set the distribution for clockrate.c:h3n2_2deme to Log Normal with the mean of 0.001, with the Mean in Real Space checkbox checked. We will leave the S parameter (standard deviation) at the default value of 1.25 to allow BEAST2 a lot of freedom in case it is necessary. The appropriate prior setup can be seen in Figure 14. Lastly, we will set the sampling proportion prior to a more narrow distribution peaked around the very low values, as influenza spreads easily, but only few people actually get sampled. Taking into account that we are also using a thinned-down version of the dataset, we can use a diffuse prior with the mean around The default prior for the sampling proportion is a Beta distribution, which is only defined between 0 and 1, making it a natural choice for proportions. Here, however, we will use a Log Normal prior, with the mean M at 10-3 and the standard deviation S at 1.25 to allow a lot of variance. Once again we need to check that the Mean in Real Space checkbox is checked, and since the Log Normal distribution is defined outside the range of [0, 1] we also need to check that the Lower and Upper limits are set accordingly. You can see the sampling prior setup in Figure 15 For the purpose of this tutorial and given that we know little about the outbreak in question to set strict priors on the ratematrix, we will leave the other priors on the default values, but feel free to through them 16
17 Figure 15: Set the prior for sampling proportion. 17
18 yourself and verify their sensibility. 2.8 Saving the configuration Once you are done with setting all the appropriate parameters, you can save the configuration file. We will leave the MCMC panel parameters as they are by default. 3 Running the analysis using BEAST To run the analysis, simply start BEAST 2 in the manner appropriate for your platform, then select the configuration file you generated in the last section as the input. Unfortunately, this particular run will take quite some time to mix, e.g. on a 2.5 GHz i7 MacBook Pro it takes about 6 hours for samples. Feel free to run it and observe the results, but for the purpose of finishing the tutorial in a reasonable time, check out the provided log file to see the results. 4 Analyzing the results The results of the analysis primarily consist of two parts: 1. The parameter log, which is written to the file h3n2-bdmm.log. 2. The tree log, which is written to h3n2-bdmm.h3n2_2deme.trees. In addition, the file h3n2-bdmm.h3n2_2deme.map.trees contains the running estimate of the MAP tree as a function of MCMC step number, while the file h3n2-bdmm.h3n2_2deme.typednode.trees is the TreeAnnotator-compatible file we ll use to assemble a summary tree. 4.1 Parameter log file analysis We can use the program Tracer to view the parameter log file. To do this, start Tracer and then press the + button in the top-left hand corner of the window (under Trace files). Select the log file for this analysis (h3n2_2deme.log) from the file selection dialog box. You can also simply drag your log file from the file browser to the Tracer window. The Traces table will then be populated with parameters and summary statistics corresponding to our multitype birth-death analysis. Important traces are: R0.t:h3n2_2deme1 and R0.t:h3n2_2deme2: These give the effective reproduction numbers for deme 1 (Hong Kong) and 2 (New Zealand), respectively. becomeuninfectiousrate.t:h3n2_deme21 and becomeuninfectiousrate.t:h3n2_deme22: These are the rates of recovery for someone with flu in either of the locations. ratematrix.t:h3n2_2deme1 and ratematrix.t:h3n2_2deme2: These give the (per lineage per year) migration rates from deme 1 to 2 and vice versa. Tree.t:h3n2_2deme.count_HongKong_to_NewZealand: these give the actual number of ancestral migrations from Hong Kong to New Zealand. The tabs at the top-right of the window can be used to display one or more selected traces in various ways. We can look at the become uninfectious rate by selecting the becomeuninfectiousrate.t:h3n2_2deme1 trace (see Figure 16). The 95% HPD for the parameter is quite wide ([ , ]), which is most likely due to the fact that we have very little data, however the mean value is , which gives us an infectious period of 7.3 days. Next, selecting the two R0 traces (R0.t:h3n2_2deme1 and R0.t:h3n2_2deme2) and choosing the 18
19 Figure 16: Estimated become uninfectious rate marginal posterior. Marginal prob distribution panel results in useful comparison between the sampled population size marginal posterior distributions (see Figure 17). Looking at the posterior distributions we can not see any significant difference in R0 between the two demes. While the distributions are visibly different, they cover the same parameter range (deme 1 95% HPD interval [0.991, ], deme 2 95% HPD interval [0.9096, ]), so the values are indistinguishable through such analysis. In the case of our pre-cooked analysis all the ESS values are greater than 200 the arbitrary threshold for acceptability. However, it might happen that some values have not yet reached the appropriate ESS in the runs that you did on your own. If this analysis were part of a serious study you would want to run the chain for another few million iterations to improve the ESS values. In BEAST 2, analyses can be resumed the samples you already have will not be wasted. 4.2 Tree log visualization The popular phylogenetic tree visualizer FigTree can be used to visualize the sampled trees. However, Figtree can be quite slow with MultiTypeTree log files, so for this tutorial we suggest using IcyTree to view tree log files. IcyTree is a tree viewer that runs in a web browser, which runs best under recent versions of Google Chrome and Mozilla Firefox (in that order). To view MultiTypeTree log files using IcyTree, simply navigate to the IcyTree web page, select Load from 19
20 Figure 17: Estimated R<sub>0</sub> marginal posteriors. 20
21 Figure 18: An example of a sampled multi-type tree in IcyTree. file from the File menu, then select the h3n2-bdmm.h3n2_2deme.trees tree log file using the file selection dialog. Alternatively, you can simply drag the log file into your browser window. Once the file is loaded you will see the first tree it contains. In order to select a different tree, hover the mouse pointer over the box in the lower-left corner of the window. This box will expand to a small dialog containing buttons allowing you to navigate between trees. The < and > buttons move in steps of 1 tree, while << and >> move 10% of the tree file. You can also directly enter the index of a tree. Initially the tree edges will be uncoloured. To colour the edges according to the edge type (this is the strain location in our case), navigate to Style > Colour edges by and select type. A legend and axis can be added by choosing Display legend and Axis > Age from the same menu. You can browse the trees from your posterior sample (example of the trees you can see in Figure 18) to look at the traits they share, however in general we need some sort of a summary to be able to draw conclusions from our tree sample. One way of summarising is done by the special MultiTypeTree log, which logs the running estimates of the maximum a posteriori multi-type tree over the course of the analysis. In our case it is the h3n2-bdmm. h3n2_2deme.map.trees file. You can see the last tree from this file, which represents our sampled estimate of the MAP multi-type tree, in Figure Producing a summary tree using TreeAnnotator While it is tempting to view the MAP tree shown above as the primary result of the phylogenetic side of our analysis it is very important to remember that this is only a point estimate and says nothing about the uncertainty present in the result. This is an important drawback, as we have done a full Bayesian analysis 21
22 Figure 19: The final MAP multi-type tree in IcyTree. 22
23 Figure 20: Use TreeAnnotator to produce a summary tree. and have access to a large number of samples from the full posterior in the tree log files. The MAP tree discards almost all of this information. We can make better use of our raw analysis results by using the TreeAnnotator program which is distributed with BEAST2 to analyze the sample of trees which was produced by our MCMC run. To do this, simply start TreeAnnotator and select the h3n2-bdmm.h3n2_2deme.typednode.trees tree file as the input file and h3n2-bdmm. h3n2_2deme.summary.trees as the output file. We will set the Burnin percentage to 10, the Target tree type to the Maximum clade credibility tree (default) and for the Node heights we would like to have Mean heights. The setup can be seen in Figure 20. Pressing the Run button will produce an annotated summary tree. To visualize this tree, open IcyTree once more (maybe open it in a new browser tab), choose File > Open, then select the file h3n2_2deme.h3n2_2deme.summary.tree using the file selection dialog. Follow the instructions provided above to colour the tree by the type attribute and add the legend and time axis. In addition, open the Style menu and select Node height error bars > height_95%_hpd to add error bars to the internal node heights. Finally, open the Style menu and select Edge opacity > type.prob. This make cause the edges become increasingly transparent as the posterior probability for the displayed branch decreases. Once these style preferences have been set, you should see something similar to the tree shown in Figure 21. Here we have a full consensus tree annotated by the locations at coalescence nodes and showing node height uncertainty, with the width/transparency of the edges representing how certain we can be of the location 23
24 Figure 21: The summary tree in IcyTree. 24
25 estimate at each point on the tree. This is a much more comprehensive summary of the phylogenetic side of our analysis. One thing to pay attention to here is that the most probable root location in the summary tree is Hong Kong (under our model which assumes that only Hong Kong and New Zealand exist). Hovering the mouse cursor over the tiny edge above the root will bring up a table in which posterior probability of the displayed root location (type.prob) can be seen. In this analysis we see that it is about 88.8%. The analysis therefore strongly supports a Hong Kong origin over a New Zealand origin for this flu sample. 5 Acknowledgment The content of this tutorial is based on the Structured Coalescent tutorial by Tim Vaughan. 6 Useful Links Bayesian Evolutionary Analysis with BEAST 2 (Drummond and Bouckaert 2014) Multi-type birth-death process package (Kühnert et al. 2016) BEAST 2 website and documentation: This tutorial was written by Denise Kühnert and Jūlija Pečerska for Taming the BEAST and is licensed under a Creative Commons Attribution 4.0 International License. Version dated: February 8,
26 Relevant References Bouckaert, R, J Heled, D Kühnert, T Vaughan, C-H Wu, D Xie, MA Suchard, A Rambaut, and AJ Drummond Beast 2: a software platform for bayesian evolutionary analysis. PLoS computational biology 10: e Drummond, AJ and RR Bouckaert Bayesian evolutionary analysis with BEAST 2. Cambridge University Press, Kühnert, D, T Stadler, TG Vaughan, and AJ Drummond Phylodynamics with migration: a computational framework to quantify population structure from genomic data. Molecular Biology and Evolution Vaughan, TG, D Kühnert, A Popinga, D Welch, and AJ Drummond Efficient bayesian inference under the structured coalescent. Bioinformatics 30:
Tutorial using BEAST v2.4.7 MASCOT Tutorial Nicola F. Müller
 Tutorial using BEAST v2.4.7 MASCOT Tutorial Nicola F. Müller Parameter and State inference using the approximate structured coalescent 1 Background Phylogeographic methods can help reveal the movement
Tutorial using BEAST v2.4.7 MASCOT Tutorial Nicola F. Müller Parameter and State inference using the approximate structured coalescent 1 Background Phylogeographic methods can help reveal the movement
Tutorial using BEAST v2.4.1 Troubleshooting David A. Rasmussen
 Tutorial using BEAST v2.4.1 Troubleshooting David A. Rasmussen 1 Background The primary goal of most phylogenetic analyses in BEAST is to infer the posterior distribution of trees and associated model
Tutorial using BEAST v2.4.1 Troubleshooting David A. Rasmussen 1 Background The primary goal of most phylogenetic analyses in BEAST is to infer the posterior distribution of trees and associated model
Tutorial using BEAST v2.4.2 Introduction to BEAST2 Jūlija Pečerska and Veronika Bošková
 Tutorial using BEAST v2.4.2 Introduction to BEAST2 Jūlija Pečerska and Veronika Bošková This is a simple introductory tutorial to help you get started with using BEAST2 and its accomplices. 1 Background
Tutorial using BEAST v2.4.2 Introduction to BEAST2 Jūlija Pečerska and Veronika Bošková This is a simple introductory tutorial to help you get started with using BEAST2 and its accomplices. 1 Background
A STEP-BY-STEP TUTORIAL FOR DISCRETE STATE PHYLOGEOGRAPHY INFERENCE
 BEAST: Bayesian Evolutionary Analysis by Sampling Trees A STEP-BY-STEP TUTORIAL FOR DISCRETE STATE PHYLOGEOGRAPHY INFERENCE This step-by-step tutorial guides you through a discrete phylogeography analysis
BEAST: Bayesian Evolutionary Analysis by Sampling Trees A STEP-BY-STEP TUTORIAL FOR DISCRETE STATE PHYLOGEOGRAPHY INFERENCE This step-by-step tutorial guides you through a discrete phylogeography analysis
BEAST2 workflow. Jūlija Pečerska & Veronika Bošková
 BEAST2 workflow Jūlija Pečerska & Veronika Bošková Tools needed Tools needed BEAST2: Software implementing MCMC for model parameter and tree inference Tools needed BEAST2: Software implementing MCMC for
BEAST2 workflow Jūlija Pečerska & Veronika Bošková Tools needed Tools needed BEAST2: Software implementing MCMC for model parameter and tree inference Tools needed BEAST2: Software implementing MCMC for
Extended Bayesian Skyline Plot tutorial for BEAST 2
 Extended Bayesian Skyline Plot tutorial for BEAST 2 Joseph Heled (updated for BEAST 2 by Tim Vaughan) This short practical explains how to set up an Extended Bayesian Skyline Plot (EBSP) analysis in BEAST
Extended Bayesian Skyline Plot tutorial for BEAST 2 Joseph Heled (updated for BEAST 2 by Tim Vaughan) This short practical explains how to set up an Extended Bayesian Skyline Plot (EBSP) analysis in BEAST
Species Trees with Relaxed Molecular Clocks Estimating per-species substitution rates using StarBEAST2
 Species Trees with Relaxed Molecular Clocks Estimating per-species substitution rates using StarBEAST2 Joseph Heled, Remco Bouckaert, Walter Xie, Alexei J. Drummond and Huw A. Ogilvie 1 Background In this
Species Trees with Relaxed Molecular Clocks Estimating per-species substitution rates using StarBEAST2 Joseph Heled, Remco Bouckaert, Walter Xie, Alexei J. Drummond and Huw A. Ogilvie 1 Background In this
Estimating rates and dates from time-stamped sequences A hands-on practical
 Estimating rates and dates from time-stamped sequences A hands-on practical This chapter provides a step-by-step tutorial for analyzing a set of virus sequences which have been isolated at different points
Estimating rates and dates from time-stamped sequences A hands-on practical This chapter provides a step-by-step tutorial for analyzing a set of virus sequences which have been isolated at different points
Bayesian phylogenetic inference MrBayes (Practice)
 Bayesian phylogenetic inference MrBayes (Practice) The aim of this tutorial is to give a very short introduction to MrBayes. There is a website with most information about MrBayes: www.mrbayes.net (which
Bayesian phylogenetic inference MrBayes (Practice) The aim of this tutorial is to give a very short introduction to MrBayes. There is a website with most information about MrBayes: www.mrbayes.net (which
Bayesian Inference of Species Trees from Multilocus Data using *BEAST
 Bayesian Inference of Species Trees from Multilocus Data using *BEAST Alexei J Drummond, Walter Xie and Joseph Heled April 13, 2012 Introduction We describe a full Bayesian framework for species tree estimation.
Bayesian Inference of Species Trees from Multilocus Data using *BEAST Alexei J Drummond, Walter Xie and Joseph Heled April 13, 2012 Introduction We describe a full Bayesian framework for species tree estimation.
Phylogeographic inference in continuous space A hands-on practical
 Phylogeographic inference in continuous space A hands-on practical This chapter provides a step-by-step tutorial for reconstructing the spatial dynamics of the West Nile virus (WNV) invasion across North
Phylogeographic inference in continuous space A hands-on practical This chapter provides a step-by-step tutorial for reconstructing the spatial dynamics of the West Nile virus (WNV) invasion across North
*BEAST in BEAST 2.4.x Estimating Species Tree from Multilocus Data
 *BEAST in BEAST 2.4.x Estimating Species Tree from Multilocus Data Joseph Heled, Remco Bouckaert, Walter Xie and Alexei J Drummond modified by Chi Zhang 1 Introduction In this tutorial we describe a full
*BEAST in BEAST 2.4.x Estimating Species Tree from Multilocus Data Joseph Heled, Remco Bouckaert, Walter Xie and Alexei J Drummond modified by Chi Zhang 1 Introduction In this tutorial we describe a full
Phylogeographic inference in continuous space A hands-on practical
 Phylogeographic inference in continuous space A hands-on practical This chapter provides a step-by-step tutorial for reconstructing the spatial dynamics of the West Nile virus (WNV) invasion across North
Phylogeographic inference in continuous space A hands-on practical This chapter provides a step-by-step tutorial for reconstructing the spatial dynamics of the West Nile virus (WNV) invasion across North
Tutorial using BEAST v2.5.0 Introduction to BEAST2 Jūlija Pečerska, Veronika Bošková and Louis du Plessis
 Tutorial using BEAST v2.5.0 Introduction to BEAST2 Jūlija Pečerska, Veronika Bošková and Louis du Plessis This is a simple introductory tutorial to help you get started with using BEAST2 and its accomplices.
Tutorial using BEAST v2.5.0 Introduction to BEAST2 Jūlija Pečerska, Veronika Bošková and Louis du Plessis This is a simple introductory tutorial to help you get started with using BEAST2 and its accomplices.
Phylogeographic inference in discrete space A hands-on practical
 Phylogeographic inference in discrete space A hands-on practical This chapter provides a step-by-step tutorial on reconstructing the spatial dispersal and cross-species dynamics of rabies virus (RABV)
Phylogeographic inference in discrete space A hands-on practical This chapter provides a step-by-step tutorial on reconstructing the spatial dispersal and cross-species dynamics of rabies virus (RABV)
Performing whole genome SNP analysis with mapping performed locally
 BioNumerics Tutorial: Performing whole genome SNP analysis with mapping performed locally 1 Introduction 1.1 An introduction to whole genome SNP analysis A Single Nucleotide Polymorphism (SNP) is a variation
BioNumerics Tutorial: Performing whole genome SNP analysis with mapping performed locally 1 Introduction 1.1 An introduction to whole genome SNP analysis A Single Nucleotide Polymorphism (SNP) is a variation
10kTrees - Exercise #2. Viewing Trees Downloaded from 10kTrees: FigTree, R, and Mesquite
 10kTrees - Exercise #2 Viewing Trees Downloaded from 10kTrees: FigTree, R, and Mesquite The goal of this worked exercise is to view trees downloaded from 10kTrees, including tree blocks. You may wish to
10kTrees - Exercise #2 Viewing Trees Downloaded from 10kTrees: FigTree, R, and Mesquite The goal of this worked exercise is to view trees downloaded from 10kTrees, including tree blocks. You may wish to
Tips and Guidance for Analyzing Data. Executive Summary
 Tips and Guidance for Analyzing Data Executive Summary This document has information and suggestions about three things: 1) how to quickly do a preliminary analysis of time-series data; 2) key things to
Tips and Guidance for Analyzing Data Executive Summary This document has information and suggestions about three things: 1) how to quickly do a preliminary analysis of time-series data; 2) key things to
PRINCIPLES OF PHYLOGENETICS Spring 2008 Updated by Nick Matzke. Lab 11: MrBayes Lab
 Integrative Biology 200A University of California, Berkeley PRINCIPLES OF PHYLOGENETICS Spring 2008 Updated by Nick Matzke Lab 11: MrBayes Lab Note: try downloading and installing MrBayes on your own laptop,
Integrative Biology 200A University of California, Berkeley PRINCIPLES OF PHYLOGENETICS Spring 2008 Updated by Nick Matzke Lab 11: MrBayes Lab Note: try downloading and installing MrBayes on your own laptop,
B Y P A S S R D E G R Version 1.0.2
 B Y P A S S R D E G R Version 1.0.2 March, 2008 Contents Overview.................................. 2 Installation notes.............................. 2 Quick Start................................. 3 Examples
B Y P A S S R D E G R Version 1.0.2 March, 2008 Contents Overview.................................. 2 Installation notes.............................. 2 Quick Start................................. 3 Examples
Web-Friendly Sites. Planning & Design 1
 Planning & Design 1 This tutorial presents useful tips and tricks to help you achieve a more Web-friendly design and make your sites more efficient. The following topics are discussed: How Z-order and
Planning & Design 1 This tutorial presents useful tips and tricks to help you achieve a more Web-friendly design and make your sites more efficient. The following topics are discussed: How Z-order and
HybridCheck User Manual
 HybridCheck User Manual Ben J. Ward February 2015 HybridCheck is a software package to visualise the recombination signal in assembled next generation sequence data, and it can be used to detect recombination,
HybridCheck User Manual Ben J. Ward February 2015 HybridCheck is a software package to visualise the recombination signal in assembled next generation sequence data, and it can be used to detect recombination,
Geneious 5.6 Quickstart Manual. Biomatters Ltd
 Geneious 5.6 Quickstart Manual Biomatters Ltd October 15, 2012 2 Introduction This quickstart manual will guide you through the features of Geneious 5.6 s interface and help you orient yourself. You should
Geneious 5.6 Quickstart Manual Biomatters Ltd October 15, 2012 2 Introduction This quickstart manual will guide you through the features of Geneious 5.6 s interface and help you orient yourself. You should
Chapter 6. Building Maps with ArcGIS Online
 Chapter 6 Building Maps with ArcGIS Online Summary: ArcGIS Online is an on-line mapping software that allows you to upload tables with latitude and longitude geographic coordinates to create map layers
Chapter 6 Building Maps with ArcGIS Online Summary: ArcGIS Online is an on-line mapping software that allows you to upload tables with latitude and longitude geographic coordinates to create map layers
The NEXUS file includes a variety of node-date calibrations, including an offsetexp(min=45, mean=50) calibration for the root node:
 Appendix 1: Issues with the MrBayes dating analysis of Slater (2015). In setting up variant MrBayes analyses (Supplemental Table S2), a number of issues became apparent with the NEXUS file of the original
Appendix 1: Issues with the MrBayes dating analysis of Slater (2015). In setting up variant MrBayes analyses (Supplemental Table S2), a number of issues became apparent with the NEXUS file of the original
One of the fundamental kinds of websites that SharePoint 2010 allows
 Chapter 1 Getting to Know Your Team Site In This Chapter Requesting a new team site and opening it in the browser Participating in a team site Changing your team site s home page One of the fundamental
Chapter 1 Getting to Know Your Team Site In This Chapter Requesting a new team site and opening it in the browser Participating in a team site Changing your team site s home page One of the fundamental
SciGraphica. Tutorial Manual - Tutorials 1and 2 Version 0.8.0
 SciGraphica Tutorial Manual - Tutorials 1and 2 Version 0.8.0 Copyright (c) 2001 the SciGraphica documentation group Permission is granted to copy, distribute and/or modify this document under the terms
SciGraphica Tutorial Manual - Tutorials 1and 2 Version 0.8.0 Copyright (c) 2001 the SciGraphica documentation group Permission is granted to copy, distribute and/or modify this document under the terms
Tutorial: Resequencing Analysis using Tracks
 : Resequencing Analysis using Tracks September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : Resequencing
: Resequencing Analysis using Tracks September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : Resequencing
Gegenees genome format...7. Gegenees comparisons...8 Creating a fragmented all-all comparison...9 The alignment The analysis...
 User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
On the Web sun.com/aboutsun/comm_invest STAROFFICE 8 DRAW
 STAROFFICE 8 DRAW Graphics They say a picture is worth a thousand words. Pictures are often used along with our words for good reason. They help communicate our thoughts. They give extra information that
STAROFFICE 8 DRAW Graphics They say a picture is worth a thousand words. Pictures are often used along with our words for good reason. They help communicate our thoughts. They give extra information that
Tutorial for Windows and Macintosh SNP Hunting
 Tutorial for Windows and Macintosh SNP Hunting 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh SNP Hunting 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Introduction to the workbook and spreadsheet
 Excel Tutorial To make the most of this tutorial I suggest you follow through it while sitting in front of a computer with Microsoft Excel running. This will allow you to try things out as you follow along.
Excel Tutorial To make the most of this tutorial I suggest you follow through it while sitting in front of a computer with Microsoft Excel running. This will allow you to try things out as you follow along.
Euroopa Liit Euroopa Sotsiaalfond. Eesti tuleviku heaks. PC-Axis User Instructions
 Euroopa Liit Euroopa Sotsiaalfond Eesti tuleviku heaks PC-Axis User Instructions Table of Contents Introduction... 3 Installation... 3 Using the Program... 3 Support Functions in PC-Axis... 5 Opening a
Euroopa Liit Euroopa Sotsiaalfond Eesti tuleviku heaks PC-Axis User Instructions Table of Contents Introduction... 3 Installation... 3 Using the Program... 3 Support Functions in PC-Axis... 5 Opening a
3 Graphical Displays of Data
 3 Graphical Displays of Data Reading: SW Chapter 2, Sections 1-6 Summarizing and Displaying Qualitative Data The data below are from a study of thyroid cancer, using NMTR data. The investigators looked
3 Graphical Displays of Data Reading: SW Chapter 2, Sections 1-6 Summarizing and Displaying Qualitative Data The data below are from a study of thyroid cancer, using NMTR data. The investigators looked
Using the IMfig program.
 Using the IMfig program. IMfig is a program for generating a figure of an isolation-with-migration model from the output file generated by running the IMa2 program. IMfig generates an eps (encapsulated
Using the IMfig program. IMfig is a program for generating a figure of an isolation-with-migration model from the output file generated by running the IMa2 program. IMfig generates an eps (encapsulated
Week 5 Creating a Calendar. About Tables. Making a Calendar From a Table Template. Week 5 Word 2010
 Week 5 Creating a Calendar About Tables Tables are a good way to organize information. They can consist of only a few cells, or many cells that cover several pages. You can arrange boxes or cells vertically
Week 5 Creating a Calendar About Tables Tables are a good way to organize information. They can consist of only a few cells, or many cells that cover several pages. You can arrange boxes or cells vertically
Desktop Studio: Charts
 Desktop Studio: Charts Intellicus Enterprise Reporting and BI Platform Intellicus Technologies info@intellicus.com www.intellicus.com Working with Charts i Copyright 2011 Intellicus Technologies This document
Desktop Studio: Charts Intellicus Enterprise Reporting and BI Platform Intellicus Technologies info@intellicus.com www.intellicus.com Working with Charts i Copyright 2011 Intellicus Technologies This document
Tutorial for Windows and Macintosh SNP Hunting
 Tutorial for Windows and Macintosh SNP Hunting 2010 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh SNP Hunting 2010 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Your Name: Section: INTRODUCTION TO STATISTICAL REASONING Computer Lab #4 Scatterplots and Regression
 Your Name: Section: 36-201 INTRODUCTION TO STATISTICAL REASONING Computer Lab #4 Scatterplots and Regression Objectives: 1. To learn how to interpret scatterplots. Specifically you will investigate, using
Your Name: Section: 36-201 INTRODUCTION TO STATISTICAL REASONING Computer Lab #4 Scatterplots and Regression Objectives: 1. To learn how to interpret scatterplots. Specifically you will investigate, using
Desktop Studio: Charts. Version: 7.3
 Desktop Studio: Charts Version: 7.3 Copyright 2015 Intellicus Technologies This document and its content is copyrighted material of Intellicus Technologies. The content may not be copied or derived from,
Desktop Studio: Charts Version: 7.3 Copyright 2015 Intellicus Technologies This document and its content is copyrighted material of Intellicus Technologies. The content may not be copied or derived from,
Tutorial. Variant Detection. Sample to Insight. November 21, 2017
 Resequencing: Variant Detection November 21, 2017 Map Reads to Reference and Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Resequencing: Variant Detection November 21, 2017 Map Reads to Reference and Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Working with Charts Stratum.Viewer 6
 Working with Charts Stratum.Viewer 6 Getting Started Tasks Additional Information Access to Charts Introduction to Charts Overview of Chart Types Quick Start - Adding a Chart to a View Create a Chart with
Working with Charts Stratum.Viewer 6 Getting Started Tasks Additional Information Access to Charts Introduction to Charts Overview of Chart Types Quick Start - Adding a Chart to a View Create a Chart with
EXCEL BASICS: MICROSOFT OFFICE 2007
 EXCEL BASICS: MICROSOFT OFFICE 2007 GETTING STARTED PAGE 02 Prerequisites What You Will Learn USING MICROSOFT EXCEL PAGE 03 Opening Microsoft Excel Microsoft Excel Features Keyboard Review Pointer Shapes
EXCEL BASICS: MICROSOFT OFFICE 2007 GETTING STARTED PAGE 02 Prerequisites What You Will Learn USING MICROSOFT EXCEL PAGE 03 Opening Microsoft Excel Microsoft Excel Features Keyboard Review Pointer Shapes
Tutorial. Typing and Epidemiological Clustering of Common Pathogens (beta) Sample to Insight. November 21, 2017
 Typing and Epidemiological Clustering of Common Pathogens (beta) November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Typing and Epidemiological Clustering of Common Pathogens (beta) November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Getting Started With. A Step-by-Step Guide to Using WorldAPP Analytics to Analyze Survey Data, Create Charts, & Share Results Online
 Getting Started With A Step-by-Step Guide to Using WorldAPP Analytics to Analyze Survey, Create Charts, & Share Results Online Variables Crosstabs Charts PowerPoint Tables Introduction WorldAPP Analytics
Getting Started With A Step-by-Step Guide to Using WorldAPP Analytics to Analyze Survey, Create Charts, & Share Results Online Variables Crosstabs Charts PowerPoint Tables Introduction WorldAPP Analytics
3 Graphical Displays of Data
 3 Graphical Displays of Data Reading: SW Chapter 2, Sections 1-6 Summarizing and Displaying Qualitative Data The data below are from a study of thyroid cancer, using NMTR data. The investigators looked
3 Graphical Displays of Data Reading: SW Chapter 2, Sections 1-6 Summarizing and Displaying Qualitative Data The data below are from a study of thyroid cancer, using NMTR data. The investigators looked
Box-Cox Transformation for Simple Linear Regression
 Chapter 192 Box-Cox Transformation for Simple Linear Regression Introduction This procedure finds the appropriate Box-Cox power transformation (1964) for a dataset containing a pair of variables that are
Chapter 192 Box-Cox Transformation for Simple Linear Regression Introduction This procedure finds the appropriate Box-Cox power transformation (1964) for a dataset containing a pair of variables that are
Tutorial. Small RNA Analysis using Illumina Data. Sample to Insight. October 5, 2016
 Small RNA Analysis using Illumina Data October 5, 2016 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Small RNA Analysis using Illumina Data October 5, 2016 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
ArmCAD 6. reinforced concrete detailing program [updated for Build 2028]
![ArmCAD 6. reinforced concrete detailing program [updated for Build 2028] ArmCAD 6. reinforced concrete detailing program [updated for Build 2028]](/thumbs/89/97716434.jpg) ArmCAD 6 reinforced concrete detailing program [updated for Build 2028] This user manual explains only new program features and commands that have not been included in ArmCAD 2005, so it is thus primarily
ArmCAD 6 reinforced concrete detailing program [updated for Build 2028] This user manual explains only new program features and commands that have not been included in ArmCAD 2005, so it is thus primarily
GUARD1 PLUS Manual Version 2.8
 GUARD1 PLUS Manual Version 2.8 2002 TimeKeeping Systems, Inc. GUARD1 PLUS and THE PIPE are registered trademarks of TimeKeeping Systems, Inc. Table of Contents GUARD1 PLUS... 1 Introduction How to get
GUARD1 PLUS Manual Version 2.8 2002 TimeKeeping Systems, Inc. GUARD1 PLUS and THE PIPE are registered trademarks of TimeKeeping Systems, Inc. Table of Contents GUARD1 PLUS... 1 Introduction How to get
Small RNA Analysis using Illumina Data
 Small RNA Analysis using Illumina Data September 7, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Small RNA Analysis using Illumina Data September 7, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Step-by-Step Guide to Advanced Genetic Analysis
 Step-by-Step Guide to Advanced Genetic Analysis Page 1 Introduction In the previous document, 1 we covered the standard genetic analyses available in JMP Genomics. Here, we cover the more advanced options
Step-by-Step Guide to Advanced Genetic Analysis Page 1 Introduction In the previous document, 1 we covered the standard genetic analyses available in JMP Genomics. Here, we cover the more advanced options
Microsoft Excel 2007
 Microsoft Excel 2007 1 Excel is Microsoft s Spreadsheet program. Spreadsheets are often used as a method of displaying and manipulating groups of data in an effective manner. It was originally created
Microsoft Excel 2007 1 Excel is Microsoft s Spreadsheet program. Spreadsheets are often used as a method of displaying and manipulating groups of data in an effective manner. It was originally created
Comparative Sequencing
 Tutorial for Windows and Macintosh Comparative Sequencing 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh Comparative Sequencing 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
GETTING STARTED. A Step-by-Step Guide to Using MarketSight
 GETTING STARTED A Step-by-Step Guide to Using MarketSight Analyze any dataset Run crosstabs Test statistical significance Create charts and dashboards Share results online Introduction MarketSight is a
GETTING STARTED A Step-by-Step Guide to Using MarketSight Analyze any dataset Run crosstabs Test statistical significance Create charts and dashboards Share results online Introduction MarketSight is a
Tutorial: De Novo Assembly of Paired Data
 : De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
1 Introduction to Using Excel Spreadsheets
 Survey of Math: Excel Spreadsheet Guide (for Excel 2007) Page 1 of 6 1 Introduction to Using Excel Spreadsheets This section of the guide is based on the file (a faux grade sheet created for messing with)
Survey of Math: Excel Spreadsheet Guide (for Excel 2007) Page 1 of 6 1 Introduction to Using Excel Spreadsheets This section of the guide is based on the file (a faux grade sheet created for messing with)
Developer s Tip Print to Scale Feature in Slide
 Developer s Tip Print to Scale Feature in Slide The latest update to Slide 5.0 brings a number of improvements related to printing functionality, giving the user greater control over printed output. Users
Developer s Tip Print to Scale Feature in Slide The latest update to Slide 5.0 brings a number of improvements related to printing functionality, giving the user greater control over printed output. Users
CheckBook Pro 2 Help
 Get started with CheckBook Pro 9 Introduction 9 Create your Accounts document 10 Name your first Account 11 Your Starting Balance 12 Currency 13 We're not done yet! 14 AutoCompletion 15 Descriptions 16
Get started with CheckBook Pro 9 Introduction 9 Create your Accounts document 10 Name your first Account 11 Your Starting Balance 12 Currency 13 We're not done yet! 14 AutoCompletion 15 Descriptions 16
SAS Visual Analytics 8.2: Working with Report Content
 SAS Visual Analytics 8.2: Working with Report Content About Objects After selecting your data source and data items, add one or more objects to display the results. SAS Visual Analytics provides objects
SAS Visual Analytics 8.2: Working with Report Content About Objects After selecting your data source and data items, add one or more objects to display the results. SAS Visual Analytics provides objects
FLIR Tools+ and Report Studio
 Creating and Processing Word Templates http://www.infraredtraining.com 09-20-2017 2017, Infrared Training Center. 1 FLIR Report Studio Overview Report Studio is a Microsoft Word Reporting module that is
Creating and Processing Word Templates http://www.infraredtraining.com 09-20-2017 2017, Infrared Training Center. 1 FLIR Report Studio Overview Report Studio is a Microsoft Word Reporting module that is
G-PhoCS Generalized Phylogenetic Coalescent Sampler version 1.2.3
 G-PhoCS Generalized Phylogenetic Coalescent Sampler version 1.2.3 Contents 1. About G-PhoCS 2. Download and Install 3. Overview of G-PhoCS analysis: input and output 4. The sequence file 5. The control
G-PhoCS Generalized Phylogenetic Coalescent Sampler version 1.2.3 Contents 1. About G-PhoCS 2. Download and Install 3. Overview of G-PhoCS analysis: input and output 4. The sequence file 5. The control
A quick introduction to First Bayes
 A quick introduction to First Bayes Lawrence Joseph October 1, 2003 1 Introduction This document very briefly reviews the main features of the First Bayes statistical teaching package. For full details,
A quick introduction to First Bayes Lawrence Joseph October 1, 2003 1 Introduction This document very briefly reviews the main features of the First Bayes statistical teaching package. For full details,
Creating Page Layouts 25 min
 1 of 10 09/11/2011 19:08 Home > Design Tips > Creating Page Layouts Creating Page Layouts 25 min Effective document design depends on a clear visual structure that conveys and complements the main message.
1 of 10 09/11/2011 19:08 Home > Design Tips > Creating Page Layouts Creating Page Layouts 25 min Effective document design depends on a clear visual structure that conveys and complements the main message.
How to lay out a web page with CSS
 How to lay out a web page with CSS You can use table design features in Adobe Dreamweaver CS3 to create a simple page layout. However, a more powerful technique is to use Cascading Style Sheets (CSS).
How to lay out a web page with CSS You can use table design features in Adobe Dreamweaver CS3 to create a simple page layout. However, a more powerful technique is to use Cascading Style Sheets (CSS).
Using Adobe Contribute 4 A guide for new website authors
 Using Adobe Contribute 4 A guide for new website authors Adobe Contribute allows you to easily update websites without any knowledge of HTML. This handout will provide an introduction to Adobe Contribute
Using Adobe Contribute 4 A guide for new website authors Adobe Contribute allows you to easily update websites without any knowledge of HTML. This handout will provide an introduction to Adobe Contribute
Joint design of data analysis algorithms and user interface for video applications
 Joint design of data analysis algorithms and user interface for video applications Nebojsa Jojic Microsoft Research Sumit Basu Microsoft Research Nemanja Petrovic University of Illinois Brendan Frey University
Joint design of data analysis algorithms and user interface for video applications Nebojsa Jojic Microsoft Research Sumit Basu Microsoft Research Nemanja Petrovic University of Illinois Brendan Frey University
Introduction to MrBayes
 Introduction to MrBayes Fred(rik) Ronquist Dept. Bioinformatics and Genetics Swedish Museum of Natural History, Stockholm, Sweden Installing MrBayes! Two options:! Go to mrbayes.net, click Download and
Introduction to MrBayes Fred(rik) Ronquist Dept. Bioinformatics and Genetics Swedish Museum of Natural History, Stockholm, Sweden Installing MrBayes! Two options:! Go to mrbayes.net, click Download and
Tutorial. Phylogenetic Trees and Metadata. Sample to Insight. November 21, 2017
 Phylogenetic Trees and Metadata November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Phylogenetic Trees and Metadata November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Lastly, in case you don t already know this, and don t have Excel on your computers, you can get it for free through IT s website under software.
 Welcome to Basic Excel, presented by STEM Gateway as part of the Essential Academic Skills Enhancement, or EASE, workshop series. Before we begin, I want to make sure we are clear that this is by no means
Welcome to Basic Excel, presented by STEM Gateway as part of the Essential Academic Skills Enhancement, or EASE, workshop series. Before we begin, I want to make sure we are clear that this is by no means
Exercise 1: Introduction to MapInfo
 Geog 578 Exercise 1: Introduction to MapInfo Page: 1/22 Geog 578: GIS Applications Exercise 1: Introduction to MapInfo Assigned on January 25 th, 2006 Due on February 1 st, 2006 Total Points: 10 0. Convention
Geog 578 Exercise 1: Introduction to MapInfo Page: 1/22 Geog 578: GIS Applications Exercise 1: Introduction to MapInfo Assigned on January 25 th, 2006 Due on February 1 st, 2006 Total Points: 10 0. Convention
COMPUTER FOR BEGINNERS
 COMPUTER FOR BEGINNERS INTRODUCTION Class Objective: This class will familiarize you with using computers. By the end of the session you will be familiar with: Starting programs Quitting programs Saving
COMPUTER FOR BEGINNERS INTRODUCTION Class Objective: This class will familiarize you with using computers. By the end of the session you will be familiar with: Starting programs Quitting programs Saving
Tabular Building Template Manager (BTM)
 Tabular Building Template Manager (BTM) User Guide IES Vi rtual Environment Copyright 2015 Integrated Environmental Solutions Limited. All rights reserved. No part of the manual is to be copied or reproduced
Tabular Building Template Manager (BTM) User Guide IES Vi rtual Environment Copyright 2015 Integrated Environmental Solutions Limited. All rights reserved. No part of the manual is to be copied or reproduced
ArcGIS Online (AGOL) Quick Start Guide Fall 2018
 ArcGIS Online (AGOL) Quick Start Guide Fall 2018 ArcGIS Online (AGOL) is a web mapping tool available to UC Merced faculty, students and staff. The Spatial Analysis and Research Center (SpARC) provides
ArcGIS Online (AGOL) Quick Start Guide Fall 2018 ArcGIS Online (AGOL) is a web mapping tool available to UC Merced faculty, students and staff. The Spatial Analysis and Research Center (SpARC) provides
EXCEL BASICS: MICROSOFT OFFICE 2010
 EXCEL BASICS: MICROSOFT OFFICE 2010 GETTING STARTED PAGE 02 Prerequisites What You Will Learn USING MICROSOFT EXCEL PAGE 03 Opening Microsoft Excel Microsoft Excel Features Keyboard Review Pointer Shapes
EXCEL BASICS: MICROSOFT OFFICE 2010 GETTING STARTED PAGE 02 Prerequisites What You Will Learn USING MICROSOFT EXCEL PAGE 03 Opening Microsoft Excel Microsoft Excel Features Keyboard Review Pointer Shapes
Table Basics. The structure of an table
 TABLE -FRAMESET Table Basics A table is a grid of rows and columns that intersect to form cells. Two different types of cells exist: Table cell that contains data, is created with the A cell that
TABLE -FRAMESET Table Basics A table is a grid of rows and columns that intersect to form cells. Two different types of cells exist: Table cell that contains data, is created with the A cell that
Dreamweaver Basics Workshop
 Dreamweaver Basics Workshop Robert Rector idesign Lab - Fall 2013 What is Dreamweaver? o Dreamweaver is a web development tool o Dreamweaver is an HTML and CSS editor o Dreamweaver features a WYSIWIG (What
Dreamweaver Basics Workshop Robert Rector idesign Lab - Fall 2013 What is Dreamweaver? o Dreamweaver is a web development tool o Dreamweaver is an HTML and CSS editor o Dreamweaver features a WYSIWIG (What
Using Audacity A Tutorial
 Using Audacity A Tutorial Peter Graff Production Manager, KBCS FM These days, there are many digital audio editors out there that can do amazing things with sound. But, most of them cost money, and if
Using Audacity A Tutorial Peter Graff Production Manager, KBCS FM These days, there are many digital audio editors out there that can do amazing things with sound. But, most of them cost money, and if
Tracy Heath Workshop on Molecular Evolution, Woods Hole USA
 INTRODUCTION TO BAYESIAN PHYLOGENETIC SOFTWARE Tracy Heath Integrative Biology, University of California, Berkeley Ecology & Evolutionary Biology, University of Kansas 2013 Workshop on Molecular Evolution,
INTRODUCTION TO BAYESIAN PHYLOGENETIC SOFTWARE Tracy Heath Integrative Biology, University of California, Berkeley Ecology & Evolutionary Biology, University of Kansas 2013 Workshop on Molecular Evolution,
Phylogenetics on CUDA (Parallel) Architectures Bradly Alicea
 Descent w/modification Descent w/modification Descent w/modification Descent w/modification CPU Descent w/modification Descent w/modification Phylogenetics on CUDA (Parallel) Architectures Bradly Alicea
Descent w/modification Descent w/modification Descent w/modification Descent w/modification CPU Descent w/modification Descent w/modification Phylogenetics on CUDA (Parallel) Architectures Bradly Alicea
Computer vision: models, learning and inference. Chapter 10 Graphical Models
 Computer vision: models, learning and inference Chapter 10 Graphical Models Independence Two variables x 1 and x 2 are independent if their joint probability distribution factorizes as Pr(x 1, x 2 )=Pr(x
Computer vision: models, learning and inference Chapter 10 Graphical Models Independence Two variables x 1 and x 2 are independent if their joint probability distribution factorizes as Pr(x 1, x 2 )=Pr(x
Creating & Modifying Tables in Word 2003
 Creating & Modifying Tables in Word 2003 This bookl LaTonya Motley Trainer / Instructional Technology Specialist Staff Development 660-6452 Table of Contents Overview... 1 Inserting Tables... 1 Using Pre-Set
Creating & Modifying Tables in Word 2003 This bookl LaTonya Motley Trainer / Instructional Technology Specialist Staff Development 660-6452 Table of Contents Overview... 1 Inserting Tables... 1 Using Pre-Set
Box-Cox Transformation
 Chapter 190 Box-Cox Transformation Introduction This procedure finds the appropriate Box-Cox power transformation (1964) for a single batch of data. It is used to modify the distributional shape of a set
Chapter 190 Box-Cox Transformation Introduction This procedure finds the appropriate Box-Cox power transformation (1964) for a single batch of data. It is used to modify the distributional shape of a set
Lab 8: Using POY from your desktop and through CIPRES
 Integrative Biology 200A University of California, Berkeley PRINCIPLES OF PHYLOGENETICS Spring 2012 Updated by Michael Landis Lab 8: Using POY from your desktop and through CIPRES In this lab we re going
Integrative Biology 200A University of California, Berkeley PRINCIPLES OF PHYLOGENETICS Spring 2012 Updated by Michael Landis Lab 8: Using POY from your desktop and through CIPRES In this lab we re going
GGR 375 QGIS Tutorial
 GGR 375 QGIS Tutorial With text taken from: Sherman, Gary E. Shuffling Quantum GIS into the Open Source GIS Stack. Free and Open Source Software for Geospatial (FOSS4G) Conference. 2007. Available online
GGR 375 QGIS Tutorial With text taken from: Sherman, Gary E. Shuffling Quantum GIS into the Open Source GIS Stack. Free and Open Source Software for Geospatial (FOSS4G) Conference. 2007. Available online
Probabilistic Analysis Tutorial
 Probabilistic Analysis Tutorial 2-1 Probabilistic Analysis Tutorial This tutorial will familiarize the user with the Probabilistic Analysis features of Swedge. In a Probabilistic Analysis, you can define
Probabilistic Analysis Tutorial 2-1 Probabilistic Analysis Tutorial This tutorial will familiarize the user with the Probabilistic Analysis features of Swedge. In a Probabilistic Analysis, you can define
Working with Tables in Word 2010
 Working with Tables in Word 2010 Table of Contents INSERT OR CREATE A TABLE... 2 USE TABLE TEMPLATES (QUICK TABLES)... 2 USE THE TABLE MENU... 2 USE THE INSERT TABLE COMMAND... 2 KNOW YOUR AUTOFIT OPTIONS...
Working with Tables in Word 2010 Table of Contents INSERT OR CREATE A TABLE... 2 USE TABLE TEMPLATES (QUICK TABLES)... 2 USE THE TABLE MENU... 2 USE THE INSERT TABLE COMMAND... 2 KNOW YOUR AUTOFIT OPTIONS...
AGENT-BASED MODELING BOOTCAMP FOR HEALTH RESEARCHERS AUGUST 2012 A SIMPLE NETWORK-BASED INFECTION SPREAD AGENT-BASED MODEL
 Rating: Basic Prerequisites: Building a Minimalist Network-Based Model Framework Estimated Time Required: 1 hour This exercise rehearses basic steps that can be used to create a model of infection spread..
Rating: Basic Prerequisites: Building a Minimalist Network-Based Model Framework Estimated Time Required: 1 hour This exercise rehearses basic steps that can be used to create a model of infection spread..
Flames in Particle Flow
 Flames in Particle Flow In this tutorial we are going to take a look at creating some licking flames in Particle Flow. I warn you however, is that this method of fire creation is very processor intensive.
Flames in Particle Flow In this tutorial we are going to take a look at creating some licking flames in Particle Flow. I warn you however, is that this method of fire creation is very processor intensive.
ViTraM: VIsualization of TRAnscriptional Modules
 ViTraM: VIsualization of TRAnscriptional Modules Version 1.0 June 1st, 2009 Hong Sun, Karen Lemmens, Tim Van den Bulcke, Kristof Engelen, Bart De Moor and Kathleen Marchal KULeuven, Belgium 1 Contents
ViTraM: VIsualization of TRAnscriptional Modules Version 1.0 June 1st, 2009 Hong Sun, Karen Lemmens, Tim Van den Bulcke, Kristof Engelen, Bart De Moor and Kathleen Marchal KULeuven, Belgium 1 Contents
A Step-by-step guide to creating a Professional PowerPoint Presentation
 Quick introduction to Microsoft PowerPoint A Step-by-step guide to creating a Professional PowerPoint Presentation Created by Cruse Control creative services Tel +44 (0) 1923 842 295 training@crusecontrol.com
Quick introduction to Microsoft PowerPoint A Step-by-step guide to creating a Professional PowerPoint Presentation Created by Cruse Control creative services Tel +44 (0) 1923 842 295 training@crusecontrol.com
BusinessObjects Frequently Asked Questions
 BusinessObjects Frequently Asked Questions Contents Is there a quick way of printing together several reports from the same document?... 2 Is there a way of controlling the text wrap of a cell?... 2 How
BusinessObjects Frequently Asked Questions Contents Is there a quick way of printing together several reports from the same document?... 2 Is there a way of controlling the text wrap of a cell?... 2 How
Empty the Recycle Bin Right Click the Recycle Bin Select Empty Recycle Bin
 Taskbar Windows taskbar is that horizontal strip at the bottom of your desktop where your open files and programs appear. It s where the Start button lives. Below are improvements to the taskbar that will
Taskbar Windows taskbar is that horizontal strip at the bottom of your desktop where your open files and programs appear. It s where the Start button lives. Below are improvements to the taskbar that will
EXCEL BASICS: PROJECTS
 EXCEL BASICS: PROJECTS In this class, you will be practicing with three basic Excel worksheets to learn a variety of foundational skills necessary for more advanced projects. This class covers: Three Project
EXCEL BASICS: PROJECTS In this class, you will be practicing with three basic Excel worksheets to learn a variety of foundational skills necessary for more advanced projects. This class covers: Three Project
PhyloType User Manual V1.4
 PhyloType User Manual V1.4 francois.chevenet@ird.fr www.phylotype.org Screenshot of the PhyloType Web interface: www.phylotype.org (please contact the authors by e-mail for details or technical problems,
PhyloType User Manual V1.4 francois.chevenet@ird.fr www.phylotype.org Screenshot of the PhyloType Web interface: www.phylotype.org (please contact the authors by e-mail for details or technical problems,
BUCKy Bayesian Untangling of Concordance Knots (applied to yeast and other organisms)
 Introduction BUCKy Bayesian Untangling of Concordance Knots (applied to yeast and other organisms) Version 1.2, 17 January 2008 Copyright c 2008 by Bret Larget Last updated: 11 November 2008 Departments
Introduction BUCKy Bayesian Untangling of Concordance Knots (applied to yeast and other organisms) Version 1.2, 17 January 2008 Copyright c 2008 by Bret Larget Last updated: 11 November 2008 Departments
Heterotachy models in BayesPhylogenies
 Heterotachy models in is a general software package for inferring phylogenetic trees using Bayesian Markov Chain Monte Carlo (MCMC) methods. The program allows a range of models of gene sequence evolution,
Heterotachy models in is a general software package for inferring phylogenetic trees using Bayesian Markov Chain Monte Carlo (MCMC) methods. The program allows a range of models of gene sequence evolution,
MS Office Word Tabs & Tables Manual. Catraining.co.uk Tel:
 MS Office 2010 Word Tabs & Tables Manual Catraining.co.uk Tel: 020 7920 9500 Table of Contents TABS... 1 BASIC TABS WITH ALIGNMENT... 1 DEFAULT TAB STOP... 1 SET MANUAL TAB STOPS WITH RULER... 2 SET MANUAL
MS Office 2010 Word Tabs & Tables Manual Catraining.co.uk Tel: 020 7920 9500 Table of Contents TABS... 1 BASIC TABS WITH ALIGNMENT... 1 DEFAULT TAB STOP... 1 SET MANUAL TAB STOPS WITH RULER... 2 SET MANUAL
Site Owners: Cascade Basics. May 2017
 Site Owners: Cascade Basics May 2017 Page 2 Logging In & Your Site Logging In Open a browser and enter the following URL (or click this link): http://mordac.itcs.northwestern.edu/ OR http://www.northwestern.edu/cms/
Site Owners: Cascade Basics May 2017 Page 2 Logging In & Your Site Logging In Open a browser and enter the following URL (or click this link): http://mordac.itcs.northwestern.edu/ OR http://www.northwestern.edu/cms/
SharePoint List Booster Features
 SharePoint List Booster Features Contents Overview... 5 Supported Environment... 5 User Interface... 5 Disabling List Booster, Hiding List Booster Menu and Disabling Cross Page Queries for specific List
SharePoint List Booster Features Contents Overview... 5 Supported Environment... 5 User Interface... 5 Disabling List Booster, Hiding List Booster Menu and Disabling Cross Page Queries for specific List
