StrAuto. Automation and Parallelization of STRUCTURE Analysis. User Manual Version 1.0.
|
|
|
- Marjorie Reynolds
- 6 years ago
- Views:
Transcription
1 StrAuto Automation and Parallelization of STRUCTURE Analysis User Manual Version Vikram E. Chhatre 1,2 & Kevin J. Emerson 3 1 Dept. of Plant Biology, University of Vermont 2 INBRE Bioinformatics Core, University of Wyoming 3 Dept. of Biology, St. Mary s College of Maryland crypticlineage@gmail.com kjemerson@smcm.edu February 18, 2018
2 Contents 1 For the Impatient 3 2 How to cite 3 3 Package Contents 4 4 Requirements 4 5 What StrAuto Does The input.py Template Example Data File sim.str Running StrAuto StrAuto Output Files Begin Structure analysis Output Files Behind the Scenes: runstructure script A Note on Random Seed Number Using High Performance Computing Cluster Caution Getting Started The structurecommands Script The runstructure Script Submit the job Exploratory Analysis using Random Subsets Input File Usage Output File Example Bibliography Author Contributions & Acknowledgements 11 2
3 1 For the Impatient If you are a returning user and/or otherwise do not have time to read through the manual, just read the following quickstart instructions. 1. Make sure the following are installed on your computer/server: Python 2.7.x or 3.0.x (see folder labeled py3 for that version) STRUCTURE backend version (Pritchard et al., 2000) structureharvester (Earl and vonholdt, 2012) (Optional) GNU Parallel (Tange, 2011) (Optional) 2. Open the input.py file in a text editor (we recommend vim) and provide information about your data and the planned analysis. 3. An example data file sim.str based on Hubisz et al. (2009) has been provided for testing purposes. 4. Save the input.py file. Do not make any changes to the name of this file (input.py) including to the.py extension. 5. Launch the StrAuto program by typing./strauto.1.py in the terminal window. 6. StrAuto writes several output files: runstructure, structurecommands, mainparams, and extraparams. If needed, make any changes to extraparams, which only contains default options. 7. Launch Structure: screen -S mysession -d -m./runstructure. This launches a detached screen session. 8. You can reattach that session by typing screen -r 9. The session can be detached again with [Ctrl-a + d], that is, type [control a] and [d] keys in quick succession. 10. Once Structure analysis is finished, a folder named harvester will contain a zip archive of the results. 11. The script will then collate the results using locally installed structureharvester. The rest of this document provides these instructions in more detail. 2 How to cite If you found StrAuto useful in your research, please cite our article. Chhatre, VE & Emerson KJ. StrAuto: Automation and parallelization of STRUCTURE analysis. BMC Bioinformatics (2017) 18:192. doi:
4 3 Package Contents Unpacking of the source archive will populate the current folder with following files. After unpacking, verify the MD5sum of the main script against the one provided in strauto 1.md5. Both scripts listed below work with Python version 2.7.x. For Python 3.0 functionality, see the py3 folder. strauto 1.py - The main script strauto doc.pdf - This guide input.py - Template file sim.str - Example data file from Hubisz et al (2009) samplestructurefile.py - A program for subsetting your data for testing purposes. 4 Requirements Python or 3.0.x (see folder named py3) Structure version ( (Pritchard et al., 2000) Structure Harvester (Earl and vonholdt, 2012) ( dearl) (Optional) GNU Parallel (Tange, 2011) ( This is an optional install. If you are working with very small data sets or do not have access to at least two processor cores, you can skip installing GNU Parallel. On Mac OS, this package is available through Fink and Homebrew. With Homebrew, you can install it by typing brew install parallel in the terminal. Verify the installation with which parallel. 5 What StrAuto Does StrAuto and the resulting scripts will work on Mac OS and Linux. On Windows, same functionality can be obtained and on Windows via Cygwin. But the resulting shell script will only work with Mac OS and Linux. The StrAuto script: 1. Collects information about your data anlysis project from the template (input.py) 2. Checks whether Structure, structureharvester and GNU Parallel are in system path 3. Writes Structure parameter files: mainparams and extraparams 4. Prepares a shell script (runstructure) and a companion commands file (structurecommands) when parallel implementation is sought. 5. When executed, the runstructure script will run Structure analysis, and store results and log files to respective folders. 6. A zip archive for Structure Harvester input is created. 7. If Harvester is available locally, it is run on the output files from above. 4
5 5.1 The input.py Template This file is provided with the program as a template for entering information about your data analysis project. It contains 24 variables that need to be defined. The template refers to example data file sim.str based on Hubisz et al. (2009). Last four questions in the template concern parallelization and processing the results through Structure Harvester. If you would like to disable these options, make appropriate changes to the template. 5.2 Example Data File sim.str We highly recommend that you set the burn-in and MCMC to 100 each and set up an example run with this data file. Once you verify that the trial run worked as intended, proceed with your own data. 5.3 Running StrAuto Once you have the input.py template filled out completely, execute StrAuto as follows. $./strauto_1.py If you get an error message, your $PATH variable is not set correctly to execute the python binary. In that case, do the following (replace /path/to/ with the actual path to the Python binary). $ /path/to/python strauto_1.py The program will present an introductory screen and wait for you to proceed. A carriage return allows the program to read the template input.py file. If the reading is successful, all parameter values will be displayed on screen for verification. If you find any errors, type (q)uit to exit the program and make corrections to your input file before proceeding. Once verified, type (a)ccept to begin writing the output files. 5.4 StrAuto Output Files A successful run of StrAuto will generate following files: $ ls -lh -rw-r--r--@ 1 user group 1.0K Apr 2 11:42 extraparams -rw-r--r--@ 1 user group 2.8K Apr 2 08:59 input.py -rw-r--r--@ 1 user group 567B Apr 2 11:42 mainparams -rwxr-xr-x@ 1 user group 634B Apr 2 11:42 runstructure -rw-r--r--@ 1 user group 6.6K Apr 2 11:42 structurecommands The program also checks the system $PATH and current directory for STRUCTURE, structureharvester and GNU Parallel binaries. If one or more are not found, a message is printed to the screen. Out- 5
6 put files are still generated, if these binaries are not found, but then we assume that you will populate the current directory with them. 6 Begin Structure analysis Now you have everything you need to begin your structure run. If you ran StrAuto on your local workstation, but want to perform the analysis on a remote server, copy this folder in its entirety to that server. Then proceed with the analysis as follows. $ screen -S my_session -d -m./runstructure Where: screen is a GNU ( utility to run processes in the background -S switch names the screen session using assigned variable e.g. my session -d switch creates a detached session -m invokes a new session (which is a detached session due to the -d switch we used)./runstructure executes the shell script You will notice that the shell prompt has returned and is not showing any activity. This is because the Structure analysis is running in the background in a detached screen session. In order to see the progress, you will need to re-attach the session. Here is how to resume (notice the -r switch below) a session: $ screen -r To disconnect from the session (i.e. to detach it): $ Ctrl-a + d If you are wondering why we used screen, it is because screen allows the analysis to run in the background, as opposed to in an open-ended terminal session which, if you accidentally close or disconnect from the server, will disrupt your analysis. Structure analysis may, depending upon the size of your data and the number of markers used, be computationally intensive and will likely need hours, if not days to complete. The screen session can be detached and reattached any number of times without interrupting the process to monitor the progress. Even after the analysis is complete, the session remains in place so you could log back in to check progress. For more information, type man screen in the terminal window to read the user manual for this neat Unix utility. Another Unix utility to check the status of the run is top. Try it as follows: 6
7 $ top -c 6.1 Output Files If all went well, you should now have results. Check the following against your output. We are showing only relevant files/folders here for simplicity. Folder results f contains individual results files for K clusters tested and the companion folder, log contains runtime screen output. Results from structureharvester such as K (Evanno et al., 2005) and relevant plots are are stored in the folder harvester. If you chose not to run harvester locally, only the zip archive ready for upload to its website will be stored in this folder. $ ls -lh drwxr-xr-x 3 user group 102B Apr 2 12:44 harvester drwxr-xr-x 6 user group 204B Apr 2 12:44 log drwxr-xr-x 6 user group 204B Apr 2 12:44 results_f -rw-r--r-- 1 user group 88B Apr 2 12:44 seed.txt 7 Behind the Scenes: runstructure script If you plan on using this program, it is probably a good idea to familiarize yourself with how the runstructure script works, so that you can spot any errors or if the script is not doing what it should. The script follows these steps: 1. Create folders: results f, log, and harvester 2. Create subfolders K1 through Kn under results f and log folders 3. Read structurecommands and execute STRUCTURE iteratively for all K clusters 4. Store the results f and the.log files for each run in their respective folders 5. Once all runs are finished, move results folders K1 through Kn inside results f 6. Cull individual results files in one place and compress them into a zip archive 7. Move the zip archive to the harvester folder 8. If structureharvester option was set to 1, run the script on results and store the output in harvester folder. StrAuto expects that the harvester script is named structureharvester.py and that it is available in the current directory. 9. Operation is complete. 7.1 A Note on Random Seed Number To ensure assignment of unique seeds for the random number generator in each instance of structure, by default, we disable the RANDOMIZE function and initiate every new instance of structure using a unique random seed in the command line. This assures that replicate runs of structure are independent and that replicate runs of the same runstructure scripts are exactly the same. 7
8 8 Using High Performance Computing Cluster This section describes protocol for deploying StrAuto based STRUCTURE analysis on high performance computing clusters (HPCC) to make use of the multi-node environment. The HPCCs almost always run on some flavor of the Linux OS. The jobs are queued using a scheduler. The examples below are specific to the SLURM Workload Manager. If your cluster uses PBS, your system administrator can provide further help. 8.1 Caution Please consult your HPCC system administrator before launching these scripts. Engaging large number of nodes at once can overload the system. You have been warned. 8.2 Getting Started Following example assumes that you have access to 3 compute nodes on a HPC cluster, each with 16 cores. We will also assume that you have run StrAuto and generated these files: runstructure, structurecommands, mainparams, and extraparams. The first two files (scripts) will need to be modified The structurecommands Script Currently, your script looks something like this./structure -K 1 -m mainparams -o k1/sim_k1_run1 2>&1 tee log/k1/sim_k1_run1.log./structure -K 1 -m mainparams -o k1/sim_k1_run2 2>&1 tee log/k1/sim_k1_run2.log./structure -K 1 -m mainparams -o k1/sim_k1_run3 2>&1 tee log/k1/sim_k1_run3.log./structure -K 1 -m mainparams -o k1/sim_k1_run4 2>&1 tee log/k1/sim_k1_run4.log Every line of this code needs to be prefixed with a srun statement which ensures one analysis process per physical core. This avoids hyperthreading and helps maximize performance scaling. The file should look like this after change (lines truncated due to space limitation). srun -N1 -n1 --exclusive structure -K 1 -m mainparams -o... srun -N1 -n1 --exclusive structure -K 1 -m mainparams -o... srun -N1 -n1 --exclusive structure -K 1 -m mainparams -o... srun -N1 -n1 --exclusive structure -K 1 -m mainparams -o... In vim text editor, you can accomplish this with two commands. First command inserts the srun argument on each line. Second command is removing reference to local binary of structure (since we are using it as a module). :%s/^/srun \-N1 \-n1 \-\-exclusive /g :%s/\.\/structure/structure/g 8
9 8.2.2 The runstructure Script We assume that 3 nodes (16x3=48 cores) will be deployed. #!/bin/bash #SBATCH --nodes=3 #SBATCH --tasks-per-node=16 #SBATCH --account=my_account_name #SBATCH --time=00:05:00 module load intel/16.3 module load structure/2.3.4 module load perl module load parallel mkdir results_f log harvester mkdir k1 mkdir k2 mkdir k3 mkdir k4 mkdir log/k1 mkdir log/k2 mkdir log/k3 mkdir log/k4 cat structurecommands parallel -j $SLURM_NTASKS --joblog parallel.log "{}"... rest of the script follows Submit the job Once both files are edited as above and you have consulted with the sysadmins and ensured that it is okay to use whatever number of nodes you want, go ahead and submit the job. You can monitor the current status of the job at any time by using the squeue function. $ sbatch runstructure $ squeue -u username 9
10 9 Exploratory Analysis using Random Subsets In order to do experimental tests in a reasonable amount of time with a large dataset, we recommend trial runs with a representative sample of your datatset. Our usage suggests that qualitative results details of the MCMC process are similar with datasets ranging from ,000 loci. Therefore, to determine the length of MCMC chain for structure and other parameters, or to test different underlying models, we recommend that tests are done with 500 loci sampled from larger datasets. Once the user develops some intuition about the dataset and feels comfortable with the parameters to be used, they may then set up the process with the complete dataset. To this end, we supply a python script samplestructurefile.py that randomly samples a given number of loci from a structure-formatted input file. 9.1 Input File Input for this program is a STRUCTURE formatted file. Currently the script only supports files where data for each individual is on two rows (i.e. ONEROWPERIND=0). If your data is formatted to indicate ONEROWPERIND=1, you will need to switch the format before using this script. Additionally it is assumed that (1) each locus column has a label (MARKERNAMES=1) and (2) all fields in the data file are tab separated (space separated genotype data will not work). 9.2 Usage There are four required arguments for the script: -f the number of fixed columns in the structure file (i.e. the number of columns that are not genotype data these are assumed to be the left-most columns such as individual labels and location identifiers). -n the number of loci that you would like to sample from the dataset -i input file name -o output file name 9.3 Output File The output file is a structure-formatted file that contains the same fixed columns as the input file, and an additional n columns of genotype data (as determined by the -n parameter above). Additionally, a file called samplestructure.log is created that lists the names of all of the loci that are retained in the sampled datafile. 9.4 Example As an example, consider a structure-formatted file test.str that has one column with individual names and one with population names and 13,000 columns of genotype data. From this we wish to sample 700 loci. We would run python samplestructurefile.py -i test.str -o sampled_test.str -f 2 -n
11 This would generate two files sampled test.str that contained a column of individual names, a column of population names, and 700 columns of genotype data, and samplestructure.log that gives information about the sampling process, including the names of all of the loci sampled. 10 Bibliography D.A. Earl and B.M. vonholdt. Structure harvester: A website and program for visualizing structure output and implementing the Evanno method. Conservation Genetics Resources, 4(2): , G. Evanno, S. Regnaut, and J. Goudet. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology, 14: , M.J. Hubisz, D. Falush, M. Stephens, and J.K. Pritchard. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources, 9(5): , J.K. Pritchard, M. Stephens, and P. Donnelly. Inference of population structure using multilocus genotype data. Genetics, 155: , O. Tange. Gnu parallel - the command-line power tool. The USENIX Magazine, 36(1):42 47, Feb URL 11 Author Contributions & Acknowledgements Original StrAuto was developed by VEC during his graduate training at Texas A&M University. It benefited from valuable feedback from Tomasz Koralewski and Tiffani Williams of Texas A&M University. VEC acknowledges indirect financial support from Texas A&M University s Genetics Graduate Program, Texas Forest Service and University of Vermont. KJE added parallelization, harvester analysis functionality and the random subset script. KJE acknowledges funding from St. Mary s College of Maryland. We thank Jared Baker of University of Wyoming Advanced Research Computing Center for help with testing StrAuto on HPC platform. 11
Contents. Note: pay attention to where you are. Note: Plaintext version. Note: pay attention to where you are... 1 Note: Plaintext version...
 Contents Note: pay attention to where you are........................................... 1 Note: Plaintext version................................................... 1 Hello World of the Bash shell 2 Accessing
Contents Note: pay attention to where you are........................................... 1 Note: Plaintext version................................................... 1 Hello World of the Bash shell 2 Accessing
HPC Course Session 3 Running Applications
 HPC Course Session 3 Running Applications Checkpointing long jobs on Iceberg 1.1 Checkpointing long jobs to safeguard intermediate results For long running jobs we recommend using checkpointing this allows
HPC Course Session 3 Running Applications Checkpointing long jobs on Iceberg 1.1 Checkpointing long jobs to safeguard intermediate results For long running jobs we recommend using checkpointing this allows
Introduction to Joker Cyber Infrastructure Architecture Team CIA.NMSU.EDU
 Introduction to Joker Cyber Infrastructure Architecture Team CIA.NMSU.EDU What is Joker? NMSU s supercomputer. 238 core computer cluster. Intel E-5 Xeon CPUs and Nvidia K-40 GPUs. InfiniBand innerconnect.
Introduction to Joker Cyber Infrastructure Architecture Team CIA.NMSU.EDU What is Joker? NMSU s supercomputer. 238 core computer cluster. Intel E-5 Xeon CPUs and Nvidia K-40 GPUs. InfiniBand innerconnect.
Introduction: What is Unix?
 Introduction Introduction: What is Unix? An operating system Developed at AT&T Bell Labs in the 1960 s Command Line Interpreter GUIs (Window systems) are now available Introduction: Unix vs. Linux Unix
Introduction Introduction: What is Unix? An operating system Developed at AT&T Bell Labs in the 1960 s Command Line Interpreter GUIs (Window systems) are now available Introduction: Unix vs. Linux Unix
USER S MANUAL FOR THE AMaCAID PROGRAM
 USER S MANUAL FOR THE AMaCAID PROGRAM TABLE OF CONTENTS Introduction How to download and install R Folder Data The three AMaCAID models - Model 1 - Model 2 - Model 3 - Processing times Changing directory
USER S MANUAL FOR THE AMaCAID PROGRAM TABLE OF CONTENTS Introduction How to download and install R Folder Data The three AMaCAID models - Model 1 - Model 2 - Model 3 - Processing times Changing directory
CSC BioWeek 2018: Using Taito cluster for high throughput data analysis
 CSC BioWeek 2018: Using Taito cluster for high throughput data analysis 7. 2. 2018 Running Jobs in CSC Servers Exercise 1: Running a simple batch job in Taito We will run a small alignment using BWA: https://research.csc.fi/-/bwa
CSC BioWeek 2018: Using Taito cluster for high throughput data analysis 7. 2. 2018 Running Jobs in CSC Servers Exercise 1: Running a simple batch job in Taito We will run a small alignment using BWA: https://research.csc.fi/-/bwa
Using a Linux System 6
 Canaan User Guide Connecting to the Cluster 1 SSH (Secure Shell) 1 Starting an ssh session from a Mac or Linux system 1 Starting an ssh session from a Windows PC 1 Once you're connected... 1 Ending an
Canaan User Guide Connecting to the Cluster 1 SSH (Secure Shell) 1 Starting an ssh session from a Mac or Linux system 1 Starting an ssh session from a Windows PC 1 Once you're connected... 1 Ending an
Working with Shell Scripting. Daniel Balagué
 Working with Shell Scripting Daniel Balagué Editing Text Files We offer many text editors in the HPC cluster. Command-Line Interface (CLI) editors: vi / vim nano (very intuitive and easy to use if you
Working with Shell Scripting Daniel Balagué Editing Text Files We offer many text editors in the HPC cluster. Command-Line Interface (CLI) editors: vi / vim nano (very intuitive and easy to use if you
UoW HPC Quick Start. Information Technology Services University of Wollongong. ( Last updated on October 10, 2011)
 UoW HPC Quick Start Information Technology Services University of Wollongong ( Last updated on October 10, 2011) 1 Contents 1 Logging into the HPC Cluster 3 1.1 From within the UoW campus.......................
UoW HPC Quick Start Information Technology Services University of Wollongong ( Last updated on October 10, 2011) 1 Contents 1 Logging into the HPC Cluster 3 1.1 From within the UoW campus.......................
Lab 1 Introduction to UNIX and C
 Name: Lab 1 Introduction to UNIX and C This first lab is meant to be an introduction to computer environments we will be using this term. You must have a Pitt username to complete this lab. NOTE: Text
Name: Lab 1 Introduction to UNIX and C This first lab is meant to be an introduction to computer environments we will be using this term. You must have a Pitt username to complete this lab. NOTE: Text
Working with Basic Linux. Daniel Balagué
 Working with Basic Linux Daniel Balagué How Linux Works? Everything in Linux is either a file or a process. A process is an executing program identified with a PID number. It runs in short or long duration
Working with Basic Linux Daniel Balagué How Linux Works? Everything in Linux is either a file or a process. A process is an executing program identified with a PID number. It runs in short or long duration
CSC BioWeek 2016: Using Taito cluster for high throughput data analysis
 CSC BioWeek 2016: Using Taito cluster for high throughput data analysis 4. 2. 2016 Running Jobs in CSC Servers A note on typography: Some command lines are too long to fit a line in printed form. These
CSC BioWeek 2016: Using Taito cluster for high throughput data analysis 4. 2. 2016 Running Jobs in CSC Servers A note on typography: Some command lines are too long to fit a line in printed form. These
Image Sharpening. Practical Introduction to HPC Exercise. Instructions for Cirrus Tier-2 System
 Image Sharpening Practical Introduction to HPC Exercise Instructions for Cirrus Tier-2 System 2 1. Aims The aim of this exercise is to get you used to logging into an HPC resource, using the command line
Image Sharpening Practical Introduction to HPC Exercise Instructions for Cirrus Tier-2 System 2 1. Aims The aim of this exercise is to get you used to logging into an HPC resource, using the command line
HPC Introductory Course - Exercises
 HPC Introductory Course - Exercises The exercises in the following sections will guide you understand and become more familiar with how to use the Balena HPC service. Lines which start with $ are commands
HPC Introductory Course - Exercises The exercises in the following sections will guide you understand and become more familiar with how to use the Balena HPC service. Lines which start with $ are commands
CS Fundamentals of Programming II Fall Very Basic UNIX
 CS 215 - Fundamentals of Programming II Fall 2012 - Very Basic UNIX This handout very briefly describes how to use Unix and how to use the Linux server and client machines in the CS (Project) Lab (KC-265)
CS 215 - Fundamentals of Programming II Fall 2012 - Very Basic UNIX This handout very briefly describes how to use Unix and how to use the Linux server and client machines in the CS (Project) Lab (KC-265)
Introduction to the Linux Command Line
 Introduction to the Linux Command Line May, 2015 How to Connect (securely) ssh sftp scp Basic Unix or Linux Commands Files & directories Environment variables Not necessarily in this order.? Getting Connected
Introduction to the Linux Command Line May, 2015 How to Connect (securely) ssh sftp scp Basic Unix or Linux Commands Files & directories Environment variables Not necessarily in this order.? Getting Connected
ClaNC: The Manual (v1.1)
 ClaNC: The Manual (v1.1) Alan R. Dabney June 23, 2008 Contents 1 Installation 3 1.1 The R programming language............................... 3 1.2 X11 with Mac OS X....................................
ClaNC: The Manual (v1.1) Alan R. Dabney June 23, 2008 Contents 1 Installation 3 1.1 The R programming language............................... 3 1.2 X11 with Mac OS X....................................
Moving Materials from Blackboard to Moodle
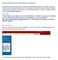 Moving Materials from Blackboard to Moodle Blackboard and Moodle organize course material somewhat differently and the conversion process can be a little messy (but worth it). Because of this, we ve gathered
Moving Materials from Blackboard to Moodle Blackboard and Moodle organize course material somewhat differently and the conversion process can be a little messy (but worth it). Because of this, we ve gathered
Introduction. File System. Note. Achtung!
 3 Unix Shell 1: Introduction Lab Objective: Explore the basics of the Unix Shell. Understand how to navigate and manipulate file directories. Introduce the Vim text editor for easy writing and editing
3 Unix Shell 1: Introduction Lab Objective: Explore the basics of the Unix Shell. Understand how to navigate and manipulate file directories. Introduce the Vim text editor for easy writing and editing
Introduction to Linux Basics
 Introduction to Linux Basics Part-I Georgia Advanced Computing Resource Center University of Georgia Zhuofei Hou, HPC Trainer zhuofei@uga.edu Outline What is GACRC? What is Linux? Linux Command, Shell
Introduction to Linux Basics Part-I Georgia Advanced Computing Resource Center University of Georgia Zhuofei Hou, HPC Trainer zhuofei@uga.edu Outline What is GACRC? What is Linux? Linux Command, Shell
Exercise Sheet 2. (Classifications of Operating Systems)
 Exercise Sheet 2 Exercise 1 (Classifications of Operating Systems) 1. At any given moment, only a single program can be executed. What is the technical term for this operation mode? 2. What are half multi-user
Exercise Sheet 2 Exercise 1 (Classifications of Operating Systems) 1. At any given moment, only a single program can be executed. What is the technical term for this operation mode? 2. What are half multi-user
An Introduction to Gauss. Paul D. Baines University of California, Davis November 20 th 2012
 An Introduction to Gauss Paul D. Baines University of California, Davis November 20 th 2012 What is Gauss? * http://wiki.cse.ucdavis.edu/support:systems:gauss * 12 node compute cluster (2 x 16 cores per
An Introduction to Gauss Paul D. Baines University of California, Davis November 20 th 2012 What is Gauss? * http://wiki.cse.ucdavis.edu/support:systems:gauss * 12 node compute cluster (2 x 16 cores per
SOLOMON: Parentage Analysis 1. Corresponding author: Mark Christie
 SOLOMON: Parentage Analysis 1 Corresponding author: Mark Christie christim@science.oregonstate.edu SOLOMON: Parentage Analysis 2 Table of Contents: Installing SOLOMON on Windows/Linux Pg. 3 Installing
SOLOMON: Parentage Analysis 1 Corresponding author: Mark Christie christim@science.oregonstate.edu SOLOMON: Parentage Analysis 2 Table of Contents: Installing SOLOMON on Windows/Linux Pg. 3 Installing
Quick Start Guide. by Burak Himmetoglu. Supercomputing Consultant. Enterprise Technology Services & Center for Scientific Computing
 Quick Start Guide by Burak Himmetoglu Supercomputing Consultant Enterprise Technology Services & Center for Scientific Computing E-mail: bhimmetoglu@ucsb.edu Linux/Unix basic commands Basic command structure:
Quick Start Guide by Burak Himmetoglu Supercomputing Consultant Enterprise Technology Services & Center for Scientific Computing E-mail: bhimmetoglu@ucsb.edu Linux/Unix basic commands Basic command structure:
Backup App V7. Quick Start Guide for Windows
 Backup App V7 Quick Start Guide for Windows Revision History Date Descriptions Type of modification 30 Jun 2016 First Draft New 25 Nov 2016 Added Restore Options to Ch 8 Restoring Data; Combined Technical
Backup App V7 Quick Start Guide for Windows Revision History Date Descriptions Type of modification 30 Jun 2016 First Draft New 25 Nov 2016 Added Restore Options to Ch 8 Restoring Data; Combined Technical
Operating System Interaction via bash
 Operating System Interaction via bash bash, or the Bourne-Again Shell, is a popular operating system shell that is used by many platforms bash uses the command line interaction style generally accepted
Operating System Interaction via bash bash, or the Bourne-Again Shell, is a popular operating system shell that is used by many platforms bash uses the command line interaction style generally accepted
CMSC 201 Spring 2017 Lab 01 Hello World
 CMSC 201 Spring 2017 Lab 01 Hello World Assignment: Lab 01 Hello World Due Date: Sunday, February 5th by 8:59:59 PM Value: 10 points At UMBC, our General Lab (GL) system is designed to grant students the
CMSC 201 Spring 2017 Lab 01 Hello World Assignment: Lab 01 Hello World Due Date: Sunday, February 5th by 8:59:59 PM Value: 10 points At UMBC, our General Lab (GL) system is designed to grant students the
Xton Access Manager GETTING STARTED GUIDE
 Xton Access Manager GETTING STARTED GUIDE XTON TECHNOLOGIES, LLC PHILADELPHIA Copyright 2017. Xton Technologies LLC. Contents Introduction... 2 Technical Support... 2 What is Xton Access Manager?... 3
Xton Access Manager GETTING STARTED GUIDE XTON TECHNOLOGIES, LLC PHILADELPHIA Copyright 2017. Xton Technologies LLC. Contents Introduction... 2 Technical Support... 2 What is Xton Access Manager?... 3
CSC209. Software Tools and Systems Programming. https://mcs.utm.utoronto.ca/~209
 CSC209 Software Tools and Systems Programming https://mcs.utm.utoronto.ca/~209 What is this Course About? Software Tools Using them Building them Systems Programming Quirks of C The file system System
CSC209 Software Tools and Systems Programming https://mcs.utm.utoronto.ca/~209 What is this Course About? Software Tools Using them Building them Systems Programming Quirks of C The file system System
FEPS. SSH Access with Two-Factor Authentication. RSA Key-pairs
 FEPS SSH Access with Two-Factor Authentication RSA Key-pairs access.eps.surrey.ac.uk Contents: Introduction - 3 RSA Key-pairs - 3 Where can I use my RSA Key-Pair? - 3 Step 1 Prepare to generate your RSA
FEPS SSH Access with Two-Factor Authentication RSA Key-pairs access.eps.surrey.ac.uk Contents: Introduction - 3 RSA Key-pairs - 3 Where can I use my RSA Key-Pair? - 3 Step 1 Prepare to generate your RSA
Introduction to Linux for BlueBEAR. January
 Introduction to Linux for BlueBEAR January 2019 http://intranet.birmingham.ac.uk/bear Overview Understanding of the BlueBEAR workflow Logging in to BlueBEAR Introduction to basic Linux commands Basic file
Introduction to Linux for BlueBEAR January 2019 http://intranet.birmingham.ac.uk/bear Overview Understanding of the BlueBEAR workflow Logging in to BlueBEAR Introduction to basic Linux commands Basic file
CSE 101 Introduction to Computers Development / Tutorial / Lab Environment Setup
 CSE 101 Introduction to Computers Development / Tutorial / Lab Environment Setup Purpose: The purpose of this lab is to setup software that you will be using throughout the term for learning about Python
CSE 101 Introduction to Computers Development / Tutorial / Lab Environment Setup Purpose: The purpose of this lab is to setup software that you will be using throughout the term for learning about Python
New User Tutorial. OSU High Performance Computing Center
 New User Tutorial OSU High Performance Computing Center TABLE OF CONTENTS Logging In... 3-5 Windows... 3-4 Linux... 4 Mac... 4-5 Changing Password... 5 Using Linux Commands... 6 File Systems... 7 File
New User Tutorial OSU High Performance Computing Center TABLE OF CONTENTS Logging In... 3-5 Windows... 3-4 Linux... 4 Mac... 4-5 Changing Password... 5 Using Linux Commands... 6 File Systems... 7 File
SCALABLE HYBRID PROTOTYPE
 SCALABLE HYBRID PROTOTYPE Scalable Hybrid Prototype Part of the PRACE Technology Evaluation Objectives Enabling key applications on new architectures Familiarizing users and providing a research platform
SCALABLE HYBRID PROTOTYPE Scalable Hybrid Prototype Part of the PRACE Technology Evaluation Objectives Enabling key applications on new architectures Familiarizing users and providing a research platform
Semester 2, 2018: Lab 1
 Semester 2, 2018: Lab 1 S2 2018 Lab 1 This lab has two parts. Part A is intended to help you familiarise yourself with the computing environment found on the CSIT lab computers which you will be using
Semester 2, 2018: Lab 1 S2 2018 Lab 1 This lab has two parts. Part A is intended to help you familiarise yourself with the computing environment found on the CSIT lab computers which you will be using
9.2 Linux Essentials Exam Objectives
 9.2 Linux Essentials Exam Objectives This chapter will cover the topics for the following Linux Essentials exam objectives: Topic 3: The Power of the Command Line (weight: 10) 3.3: Turning Commands into
9.2 Linux Essentials Exam Objectives This chapter will cover the topics for the following Linux Essentials exam objectives: Topic 3: The Power of the Command Line (weight: 10) 3.3: Turning Commands into
Lab 1 1 Due Wed., 2 Sept. 2015
 Lab 1 1 Due Wed., 2 Sept. 2015 CMPSC 112 Introduction to Computer Science II (Fall 2015) Prof. John Wenskovitch http://cs.allegheny.edu/~jwenskovitch/teaching/cmpsc112 Lab 1 - Version Control with Git
Lab 1 1 Due Wed., 2 Sept. 2015 CMPSC 112 Introduction to Computer Science II (Fall 2015) Prof. John Wenskovitch http://cs.allegheny.edu/~jwenskovitch/teaching/cmpsc112 Lab 1 - Version Control with Git
Introduction to UNIX. SURF Research Boot Camp April Jeroen Engelberts Consultant Supercomputing
 Introduction to UNIX SURF Research Boot Camp April 2018 Jeroen Engelberts jeroen.engelberts@surfsara.nl Consultant Supercomputing Outline Introduction to UNIX What is UNIX? (Short) history of UNIX Cartesius
Introduction to UNIX SURF Research Boot Camp April 2018 Jeroen Engelberts jeroen.engelberts@surfsara.nl Consultant Supercomputing Outline Introduction to UNIX What is UNIX? (Short) history of UNIX Cartesius
Duke Compute Cluster Workshop. 3/28/2018 Tom Milledge rc.duke.edu
 Duke Compute Cluster Workshop 3/28/2018 Tom Milledge rc.duke.edu rescomputing@duke.edu Outline of talk Overview of Research Computing resources Duke Compute Cluster overview Running interactive and batch
Duke Compute Cluster Workshop 3/28/2018 Tom Milledge rc.duke.edu rescomputing@duke.edu Outline of talk Overview of Research Computing resources Duke Compute Cluster overview Running interactive and batch
Module 4: Working with MPI
 Module 4: Working with MPI Objective Learn how to develop, build and launch a parallel (MPI) program on a remote parallel machine Contents Remote project setup Building with Makefiles MPI assistance features
Module 4: Working with MPI Objective Learn how to develop, build and launch a parallel (MPI) program on a remote parallel machine Contents Remote project setup Building with Makefiles MPI assistance features
IBM z Systems Development and Test Environment Tools User's Guide IBM
 IBM z Systems Development and Test Environment Tools User's Guide IBM ii IBM z Systems Development and Test Environment Tools User's Guide Contents Chapter 1. Overview......... 1 Introduction..............
IBM z Systems Development and Test Environment Tools User's Guide IBM ii IBM z Systems Development and Test Environment Tools User's Guide Contents Chapter 1. Overview......... 1 Introduction..............
CS 215 Fundamentals of Programming II Spring 2019 Very Basic UNIX
 CS 215 Fundamentals of Programming II Spring 2019 Very Basic UNIX This handout very briefly describes how to use Unix and how to use the Linux server and client machines in the EECS labs that dual boot
CS 215 Fundamentals of Programming II Spring 2019 Very Basic UNIX This handout very briefly describes how to use Unix and how to use the Linux server and client machines in the EECS labs that dual boot
CENG 334 Computer Networks. Laboratory I Linux Tutorial
 CENG 334 Computer Networks Laboratory I Linux Tutorial Contents 1. Logging In and Starting Session 2. Using Commands 1. Basic Commands 2. Working With Files and Directories 3. Permission Bits 3. Introduction
CENG 334 Computer Networks Laboratory I Linux Tutorial Contents 1. Logging In and Starting Session 2. Using Commands 1. Basic Commands 2. Working With Files and Directories 3. Permission Bits 3. Introduction
Emile R. Chimusa Division of Human Genetics Department of Pathology University of Cape Town
 Advanced Genomic data manipulation and Quality Control with plink Emile R. Chimusa (emile.chimusa@uct.ac.za) Division of Human Genetics Department of Pathology University of Cape Town Outlines: 1.Introduction
Advanced Genomic data manipulation and Quality Control with plink Emile R. Chimusa (emile.chimusa@uct.ac.za) Division of Human Genetics Department of Pathology University of Cape Town Outlines: 1.Introduction
Introduction to Linux
 Introduction to Linux The command-line interface A command-line interface (CLI) is a type of interface, that is, a way to interact with a computer. Window systems, punched cards or a bunch of dials, buttons
Introduction to Linux The command-line interface A command-line interface (CLI) is a type of interface, that is, a way to interact with a computer. Window systems, punched cards or a bunch of dials, buttons
CMSC 201 Spring 2018 Lab 01 Hello World
 CMSC 201 Spring 2018 Lab 01 Hello World Assignment: Lab 01 Hello World Due Date: Sunday, February 4th by 8:59:59 PM Value: 10 points At UMBC, the GL system is designed to grant students the privileges
CMSC 201 Spring 2018 Lab 01 Hello World Assignment: Lab 01 Hello World Due Date: Sunday, February 4th by 8:59:59 PM Value: 10 points At UMBC, the GL system is designed to grant students the privileges
FileCatalyst HotFolder Quickstart
 FileCatalyst HotFolder Quickstart Contents 1 Installation... 2 1.1 Verify Java Version... 2 1.2 Perform Installation... 3 1.2.1 Windows... 3 1.2.2 Mac OSX... 3 1.2.3 Linux, Solaris, *nix... 3 1.3 Enable
FileCatalyst HotFolder Quickstart Contents 1 Installation... 2 1.1 Verify Java Version... 2 1.2 Perform Installation... 3 1.2.1 Windows... 3 1.2.2 Mac OSX... 3 1.2.3 Linux, Solaris, *nix... 3 1.3 Enable
Introduction to Molecular Dynamics on ARCHER: Instructions for running parallel jobs on ARCHER
 Introduction to Molecular Dynamics on ARCHER: Instructions for running parallel jobs on ARCHER 1 Introduction This handout contains basic instructions for how to login in to ARCHER and submit jobs to the
Introduction to Molecular Dynamics on ARCHER: Instructions for running parallel jobs on ARCHER 1 Introduction This handout contains basic instructions for how to login in to ARCHER and submit jobs to the
CSC209. Software Tools and Systems Programming. https://mcs.utm.utoronto.ca/~209
 CSC209 Software Tools and Systems Programming https://mcs.utm.utoronto.ca/~209 What is this Course About? Software Tools Using them Building them Systems Programming Quirks of C The file system System
CSC209 Software Tools and Systems Programming https://mcs.utm.utoronto.ca/~209 What is this Course About? Software Tools Using them Building them Systems Programming Quirks of C The file system System
Unit: Making a move (using FTP)
 Data Introduction to Unix and HPC (HPC for Wimps) Unit: Making a move (using FTP) Goals: Can login via Secure FTP and see home directory. Can transfer a file from local machine via FTP to home directory.
Data Introduction to Unix and HPC (HPC for Wimps) Unit: Making a move (using FTP) Goals: Can login via Secure FTP and see home directory. Can transfer a file from local machine via FTP to home directory.
Mills HPC Tutorial Series. Linux Basics I
 Mills HPC Tutorial Series Linux Basics I Objectives Command Line Window Anatomy Command Structure Command Examples Help Files and Directories Permissions Wildcards and Home (~) Redirection and Pipe Create
Mills HPC Tutorial Series Linux Basics I Objectives Command Line Window Anatomy Command Structure Command Examples Help Files and Directories Permissions Wildcards and Home (~) Redirection and Pipe Create
Week 2 Lecture 3. Unix
 Lecture 3 Unix Terminal and Shell 2 Terminal Prompt Command Argument Result 3 Shell Intro A system program that allows a user to execute: shell functions (e.g., ls -la) other programs (e.g., eclipse) shell
Lecture 3 Unix Terminal and Shell 2 Terminal Prompt Command Argument Result 3 Shell Intro A system program that allows a user to execute: shell functions (e.g., ls -la) other programs (e.g., eclipse) shell
Introduction to Unix: Fundamental Commands
 Introduction to Unix: Fundamental Commands Ricky Patterson UVA Library Based on slides from Turgut Yilmaz Istanbul Teknik University 1 What We Will Learn The fundamental commands of the Unix operating
Introduction to Unix: Fundamental Commands Ricky Patterson UVA Library Based on slides from Turgut Yilmaz Istanbul Teknik University 1 What We Will Learn The fundamental commands of the Unix operating
Unix and C Program Development SEEM
 Unix and C Program Development SEEM 3460 1 Operating Systems A computer system cannot function without an operating system (OS). There are many different operating systems available for PCs, minicomputers,
Unix and C Program Development SEEM 3460 1 Operating Systems A computer system cannot function without an operating system (OS). There are many different operating systems available for PCs, minicomputers,
CISC 220 fall 2011, set 1: Linux basics
 CISC 220: System-Level Programming instructor: Margaret Lamb e-mail: malamb@cs.queensu.ca office: Goodwin 554 office phone: 533-6059 (internal extension 36059) office hours: Tues/Wed/Thurs 2-3 (this week
CISC 220: System-Level Programming instructor: Margaret Lamb e-mail: malamb@cs.queensu.ca office: Goodwin 554 office phone: 533-6059 (internal extension 36059) office hours: Tues/Wed/Thurs 2-3 (this week
Part I. Introduction to Linux
 Part I Introduction to Linux 7 Chapter 1 Linux operating system Goal-of-the-Day Familiarisation with basic Linux commands and creation of data plots. 1.1 What is Linux? All astronomical data processing
Part I Introduction to Linux 7 Chapter 1 Linux operating system Goal-of-the-Day Familiarisation with basic Linux commands and creation of data plots. 1.1 What is Linux? All astronomical data processing
Ion Client User Manual
 Ion Client User Manual Table of Contents About Ion Protocol...3 System Requirements... 4 Hardware (Client)... 4 Hardware (Server Connecting to)... 4 Software (Ion Client)... 4 Software (Server Connecting
Ion Client User Manual Table of Contents About Ion Protocol...3 System Requirements... 4 Hardware (Client)... 4 Hardware (Server Connecting to)... 4 Software (Ion Client)... 4 Software (Server Connecting
USING NGC WITH GOOGLE CLOUD PLATFORM
 USING NGC WITH GOOGLE CLOUD PLATFORM DU-08962-001 _v02 April 2018 Setup Guide TABLE OF CONTENTS Chapter 1. Introduction to... 1 Chapter 2. Deploying an NVIDIA GPU Cloud Image from the GCP Console...3 2.1.
USING NGC WITH GOOGLE CLOUD PLATFORM DU-08962-001 _v02 April 2018 Setup Guide TABLE OF CONTENTS Chapter 1. Introduction to... 1 Chapter 2. Deploying an NVIDIA GPU Cloud Image from the GCP Console...3 2.1.
CSE 560M Computer Systems Architecture I
 CSE 560M Computer Systems Architecture I Assignment 1, due Friday, Oct. 19, 2018 This goal of this lab assignment is to help familiarize you with simulating a system using gem5 by simulating two programs
CSE 560M Computer Systems Architecture I Assignment 1, due Friday, Oct. 19, 2018 This goal of this lab assignment is to help familiarize you with simulating a system using gem5 by simulating two programs
Archivists Toolkit Internal Database
 Archivists Toolkit Internal Database The Archivists Toolkit now includes (AT 2.0, update 9 and later), support for an internal database based on HyperSQL 2.0 (HSQLDB). HyperSQL is a small, reliable, high
Archivists Toolkit Internal Database The Archivists Toolkit now includes (AT 2.0, update 9 and later), support for an internal database based on HyperSQL 2.0 (HSQLDB). HyperSQL is a small, reliable, high
Quick Start Guide. by Burak Himmetoglu. Supercomputing Consultant. Enterprise Technology Services & Center for Scientific Computing
 Quick Start Guide by Burak Himmetoglu Supercomputing Consultant Enterprise Technology Services & Center for Scientific Computing E-mail: bhimmetoglu@ucsb.edu Contents User access, logging in Linux/Unix
Quick Start Guide by Burak Himmetoglu Supercomputing Consultant Enterprise Technology Services & Center for Scientific Computing E-mail: bhimmetoglu@ucsb.edu Contents User access, logging in Linux/Unix
Cloud Computing and Unix: An Introduction. Dr. Sophie Shaw University of Aberdeen, UK
 Cloud Computing and Unix: An Introduction Dr. Sophie Shaw University of Aberdeen, UK s.shaw@abdn.ac.uk Aberdeen London Exeter What We re Going To Do Why Unix? Cloud Computing Connecting to AWS Introduction
Cloud Computing and Unix: An Introduction Dr. Sophie Shaw University of Aberdeen, UK s.shaw@abdn.ac.uk Aberdeen London Exeter What We re Going To Do Why Unix? Cloud Computing Connecting to AWS Introduction
Processing Big Data with Hadoop in Azure HDInsight
 Processing Big Data with Hadoop in Azure HDInsight Lab 1 - Getting Started with HDInsight Overview In this lab, you will provision an HDInsight cluster. You will then run a sample MapReduce job on the
Processing Big Data with Hadoop in Azure HDInsight Lab 1 - Getting Started with HDInsight Overview In this lab, you will provision an HDInsight cluster. You will then run a sample MapReduce job on the
Temple University Computer Science Programming Under the Linux Operating System January 2017
 Temple University Computer Science Programming Under the Linux Operating System January 2017 Here are the Linux commands you need to know to get started with Lab 1, and all subsequent labs as well. These
Temple University Computer Science Programming Under the Linux Operating System January 2017 Here are the Linux commands you need to know to get started with Lab 1, and all subsequent labs as well. These
When talking about how to launch commands and other things that is to be typed into the terminal, the following syntax is used:
 Linux Tutorial How to read the examples When talking about how to launch commands and other things that is to be typed into the terminal, the following syntax is used: $ application file.txt
Linux Tutorial How to read the examples When talking about how to launch commands and other things that is to be typed into the terminal, the following syntax is used: $ application file.txt
DAVE LIDDAMENT INTRODUCTION TO BASH
 DAVE LIDDAMENT INTRODUCTION TO BASH @daveliddament FORMAT Short lectures Practical exercises (help each other) Write scripts LEARNING OBJECTIVES What is Bash When should you use Bash Basic concepts of
DAVE LIDDAMENT INTRODUCTION TO BASH @daveliddament FORMAT Short lectures Practical exercises (help each other) Write scripts LEARNING OBJECTIVES What is Bash When should you use Bash Basic concepts of
Exercise sheet 1 To be corrected in tutorials in the week from 23/10/2017 to 27/10/2017
 Einführung in die Programmierung für Physiker WS 207/208 Marc Wagner Francesca Cuteri: cuteri@th.physik.uni-frankfurt.de Alessandro Sciarra: sciarra@th.physik.uni-frankfurt.de Exercise sheet To be corrected
Einführung in die Programmierung für Physiker WS 207/208 Marc Wagner Francesca Cuteri: cuteri@th.physik.uni-frankfurt.de Alessandro Sciarra: sciarra@th.physik.uni-frankfurt.de Exercise sheet To be corrected
Cloud Computing and Unix: An Introduction. Dr. Sophie Shaw University of Aberdeen, UK
 Cloud Computing and Unix: An Introduction Dr. Sophie Shaw University of Aberdeen, UK s.shaw@abdn.ac.uk Aberdeen London Exeter What We re Going To Do Why Unix? Cloud Computing Connecting to AWS Introduction
Cloud Computing and Unix: An Introduction Dr. Sophie Shaw University of Aberdeen, UK s.shaw@abdn.ac.uk Aberdeen London Exeter What We re Going To Do Why Unix? Cloud Computing Connecting to AWS Introduction
Backpacking with Code: Software Portability for DHTC Wednesday morning, 9:00 am Christina Koch Research Computing Facilitator
 Backpacking with Code: Software Portability for DHTC Wednesday morning, 9:00 am Christina Koch (ckoch5@wisc.edu) Research Computing Facilitator University of Wisconsin - Madison Understand the basics of...
Backpacking with Code: Software Portability for DHTC Wednesday morning, 9:00 am Christina Koch (ckoch5@wisc.edu) Research Computing Facilitator University of Wisconsin - Madison Understand the basics of...
Week - 01 Lecture - 04 Downloading and installing Python
 Programming, Data Structures and Algorithms in Python Prof. Madhavan Mukund Department of Computer Science and Engineering Indian Institute of Technology, Madras Week - 01 Lecture - 04 Downloading and
Programming, Data Structures and Algorithms in Python Prof. Madhavan Mukund Department of Computer Science and Engineering Indian Institute of Technology, Madras Week - 01 Lecture - 04 Downloading and
Hands-on Exercise Hadoop
 Department of Economics and Business Administration Chair of Business Information Systems I Prof. Dr. Barbara Dinter Big Data Management Hands-on Exercise Hadoop Building and Testing a Hadoop Cluster by
Department of Economics and Business Administration Chair of Business Information Systems I Prof. Dr. Barbara Dinter Big Data Management Hands-on Exercise Hadoop Building and Testing a Hadoop Cluster by
For Dr Landau s PHYS8602 course
 For Dr Landau s PHYS8602 course Shan-Ho Tsai (shtsai@uga.edu) Georgia Advanced Computing Resource Center - GACRC January 7, 2019 You will be given a student account on the GACRC s Teaching cluster. Your
For Dr Landau s PHYS8602 course Shan-Ho Tsai (shtsai@uga.edu) Georgia Advanced Computing Resource Center - GACRC January 7, 2019 You will be given a student account on the GACRC s Teaching cluster. Your
Pre-Workshop Training materials to move you from Data to Discovery. Get Science Done. Reproducibly.
 Pre-Workshop Packet Training materials to move you from Data to Discovery Get Science Done Reproducibly Productively @CyVerseOrg Introduction to CyVerse... 3 What is Cyberinfrastructure?... 3 What to do
Pre-Workshop Packet Training materials to move you from Data to Discovery Get Science Done Reproducibly Productively @CyVerseOrg Introduction to CyVerse... 3 What is Cyberinfrastructure?... 3 What to do
Introduction to BioHPC
 Introduction to BioHPC New User Training [web] [email] portal.biohpc.swmed.edu biohpc-help@utsouthwestern.edu 1 Updated for 2015-06-03 Overview Today we re going to cover: What is BioHPC? How do I access
Introduction to BioHPC New User Training [web] [email] portal.biohpc.swmed.edu biohpc-help@utsouthwestern.edu 1 Updated for 2015-06-03 Overview Today we re going to cover: What is BioHPC? How do I access
GUT. GUT Installation Guide
 Date : 17 Mar 2011 1/6 GUT Contents 1 Introduction...2 2 Installing GUT...2 2.1 Optional Extensions...2 2.2 Installation using the Binary package...2 2.2.1 Linux or Mac OS X...2 2.2.2 Windows...4 2.3 Installing
Date : 17 Mar 2011 1/6 GUT Contents 1 Introduction...2 2 Installing GUT...2 2.1 Optional Extensions...2 2.2 Installation using the Binary package...2 2.2.1 Linux or Mac OS X...2 2.2.2 Windows...4 2.3 Installing
Working With Unix. Scott A. Handley* September 15, *Adapted from UNIX introduction material created by Dr. Julian Catchen
 Working With Unix Scott A. Handley* September 15, 2014 *Adapted from UNIX introduction material created by Dr. Julian Catchen What is UNIX? An operating system (OS) Designed to be multiuser and multitasking
Working With Unix Scott A. Handley* September 15, 2014 *Adapted from UNIX introduction material created by Dr. Julian Catchen What is UNIX? An operating system (OS) Designed to be multiuser and multitasking
Lab 3. On-Premises Deployments (Optional)
 Lab 3 On-Premises Deployments (Optional) Overview This Lab is considered optional to the completion of the API-Led Connectivity Workshop. Using Runtime Manager, you can register and set up the properties
Lab 3 On-Premises Deployments (Optional) Overview This Lab is considered optional to the completion of the API-Led Connectivity Workshop. Using Runtime Manager, you can register and set up the properties
Using CSC Environment Efficiently,
 Using CSC Environment Efficiently, 13.2.2017 1 Exercises a) Log in to Taito either with your training or CSC user account, either from a terminal (with X11 forwarding) or using NX client b) Go to working
Using CSC Environment Efficiently, 13.2.2017 1 Exercises a) Log in to Taito either with your training or CSC user account, either from a terminal (with X11 forwarding) or using NX client b) Go to working
CS197U: A Hands on Introduction to Unix
 CS197U: A Hands on Introduction to Unix Lecture 11: WWW and Wrap up Tian Guo University of Massachusetts Amherst CICS 1 Reminders Assignment 4 was graded and scores on Moodle Assignment 5 was due and you
CS197U: A Hands on Introduction to Unix Lecture 11: WWW and Wrap up Tian Guo University of Massachusetts Amherst CICS 1 Reminders Assignment 4 was graded and scores on Moodle Assignment 5 was due and you
Running Sentaurus on the DOE Network
 Running Sentaurus on the DOE Network Department of Electronics Carleton University April 2016 Warning: This primer is intended to cover the DOE.CARLETON.CA domain. Other domains are not covered in this
Running Sentaurus on the DOE Network Department of Electronics Carleton University April 2016 Warning: This primer is intended to cover the DOE.CARLETON.CA domain. Other domains are not covered in this
ENCM 339 Fall 2017: Editing and Running Programs in the Lab
 page 1 of 8 ENCM 339 Fall 2017: Editing and Running Programs in the Lab Steve Norman Department of Electrical & Computer Engineering University of Calgary September 2017 Introduction This document is a
page 1 of 8 ENCM 339 Fall 2017: Editing and Running Programs in the Lab Steve Norman Department of Electrical & Computer Engineering University of Calgary September 2017 Introduction This document is a
TEMPO INSTALLATION I O A. Platform Independent Notes 1. Installing Tempo 3. Installing Tools for the Plugins 5. v0.2.
 TEMPO INSTALLATION v0.2.2 (BETA) 2/7/2008 Platform Independent Notes 1 On Windows: 2 On Linux: 2 On OS X (Tiger 10.4.7 and later) 2 I O A Installing Tempo 3 Installing on Windows (Vista/XP/W2K) 3 Installing
TEMPO INSTALLATION v0.2.2 (BETA) 2/7/2008 Platform Independent Notes 1 On Windows: 2 On Linux: 2 On OS X (Tiger 10.4.7 and later) 2 I O A Installing Tempo 3 Installing on Windows (Vista/XP/W2K) 3 Installing
Linux Training. for New Users of Cluster. Georgia Advanced Computing Resource Center University of Georgia Suchitra Pakala
 Linux Training for New Users of Cluster Georgia Advanced Computing Resource Center University of Georgia Suchitra Pakala pakala@uga.edu 1 Overview GACRC Linux Operating System Shell, Filesystem, and Common
Linux Training for New Users of Cluster Georgia Advanced Computing Resource Center University of Georgia Suchitra Pakala pakala@uga.edu 1 Overview GACRC Linux Operating System Shell, Filesystem, and Common
Linux Command Line Interface. December 27, 2017
 Linux Command Line Interface December 27, 2017 Foreword It is supposed to be a refresher (?!) If you are familiar with UNIX/Linux/MacOS X CLI, this is going to be boring... I will not talk about editors
Linux Command Line Interface December 27, 2017 Foreword It is supposed to be a refresher (?!) If you are familiar with UNIX/Linux/MacOS X CLI, this is going to be boring... I will not talk about editors
Backup App v7. Quick Start Guide for Windows
 Backup App v7 Quick Start Guide for Windows Revision History Date Descriptions Type of modification 30 Jun 2016 First Draft New 25 Nov 2016 Added Restore Options to Ch 8 Restore Data; Combined Technical
Backup App v7 Quick Start Guide for Windows Revision History Date Descriptions Type of modification 30 Jun 2016 First Draft New 25 Nov 2016 Added Restore Options to Ch 8 Restore Data; Combined Technical
CS 2400 Laboratory Assignment #1: Exercises in Compilation and the UNIX Programming Environment (100 pts.)
 1 Introduction 1 CS 2400 Laboratory Assignment #1: Exercises in Compilation and the UNIX Programming Environment (100 pts.) This laboratory is intended to give you some brief experience using the editing/compiling/file
1 Introduction 1 CS 2400 Laboratory Assignment #1: Exercises in Compilation and the UNIX Programming Environment (100 pts.) This laboratory is intended to give you some brief experience using the editing/compiling/file
What is CMS? How do I access my CMS site? How do I log into the CMS site? www1
 What is CMS? CMS stands for Content Management System and it is a generic name for a web application used for managing websites and web content. Web content is documents, news, events, and images. MSN.com
What is CMS? CMS stands for Content Management System and it is a generic name for a web application used for managing websites and web content. Web content is documents, news, events, and images. MSN.com
C++ Programming on Linux
 C++ Programming on Linux What is Linux? CS 2308 Spring 2017 Jill Seaman Slides 14-end are for your information only, you will not be tested over that material. 1 l an operating system l Unix-like l Open
C++ Programming on Linux What is Linux? CS 2308 Spring 2017 Jill Seaman Slides 14-end are for your information only, you will not be tested over that material. 1 l an operating system l Unix-like l Open
Introduction in Unix. Linus Torvalds Ken Thompson & Dennis Ritchie
 Introduction in Unix Linus Torvalds Ken Thompson & Dennis Ritchie My name: John Donners John.Donners@surfsara.nl Consultant at SURFsara And Cedric Nugteren Cedric.Nugteren@surfsara.nl Consultant at SURFsara
Introduction in Unix Linus Torvalds Ken Thompson & Dennis Ritchie My name: John Donners John.Donners@surfsara.nl Consultant at SURFsara And Cedric Nugteren Cedric.Nugteren@surfsara.nl Consultant at SURFsara
InstallShield AdminStudio Evaluator s Guide
 InstallShield AdminStudio Evaluator s Guide Published: April 2003 Abstract This guide helps system administrators and other reviewers evaluate the key functionality of InstallShield AdminStudio. It provides
InstallShield AdminStudio Evaluator s Guide Published: April 2003 Abstract This guide helps system administrators and other reviewers evaluate the key functionality of InstallShield AdminStudio. It provides
ITNPBD7 Cluster Computing Spring Using Condor
 The aim of this practical is to work through the Condor examples demonstrated in the lectures and adapt them to alternative tasks. Before we start, you will need to map a network drive to \\wsv.cs.stir.ac.uk\datasets
The aim of this practical is to work through the Condor examples demonstrated in the lectures and adapt them to alternative tasks. Before we start, you will need to map a network drive to \\wsv.cs.stir.ac.uk\datasets
IBM WebSphere Java Batch Lab
 IBM WebSphere Java Batch Lab What are we going to do? First we are going to set up a development environment on your workstation. Download and install Eclipse IBM WebSphere Developer Tools IBM Liberty
IBM WebSphere Java Batch Lab What are we going to do? First we are going to set up a development environment on your workstation. Download and install Eclipse IBM WebSphere Developer Tools IBM Liberty
Introduction to Unix. University of Massachusetts Medical School. October, 2014
 .. Introduction to Unix University of Massachusetts Medical School October, 2014 . DISCLAIMER For the sake of clarity, the concepts mentioned in these slides have been simplified significantly. Most of
.. Introduction to Unix University of Massachusetts Medical School October, 2014 . DISCLAIMER For the sake of clarity, the concepts mentioned in these slides have been simplified significantly. Most of
Linux Operating System Environment Computadors Grau en Ciència i Enginyeria de Dades Q2
 Linux Operating System Environment Computadors Grau en Ciència i Enginyeria de Dades 2017-2018 Q2 Facultat d Informàtica de Barcelona This first lab session is focused on getting experience in working
Linux Operating System Environment Computadors Grau en Ciència i Enginyeria de Dades 2017-2018 Q2 Facultat d Informàtica de Barcelona This first lab session is focused on getting experience in working
An Introductory Tutorial on UNIX
 An Introductory Tutorial on UNIX Kevin Keay February 6 2009 Introduction The purpose of this document is to guide you through the sequence of: 1. Describing a quick method of connecting to a remote UNIX
An Introductory Tutorial on UNIX Kevin Keay February 6 2009 Introduction The purpose of this document is to guide you through the sequence of: 1. Describing a quick method of connecting to a remote UNIX
Lab Assignment #1. University of Pittsburgh Department of Electrical and Computer Engineering
 Fall 2017 ECE1192/2192 Lab Assignment #1 University of Pittsburgh Department of Electrical and Computer Engineering 1 Objective The objective of this handout is to help you get familiar with the UNIX/Linux
Fall 2017 ECE1192/2192 Lab Assignment #1 University of Pittsburgh Department of Electrical and Computer Engineering 1 Objective The objective of this handout is to help you get familiar with the UNIX/Linux
CSC105, Introduction to Computer Science I. Introduction. Perl Directions NOTE : It is also a good idea to
 CSC105, Introduction to Computer Science Lab03: Introducing Perl I. Introduction. [NOTE: This material assumes that you have reviewed Chapters 1, First Steps in Perl and 2, Working With Simple Values in
CSC105, Introduction to Computer Science Lab03: Introducing Perl I. Introduction. [NOTE: This material assumes that you have reviewed Chapters 1, First Steps in Perl and 2, Working With Simple Values in
AASPI Software Structure
 AASPI Software Structure Introduction The AASPI software comprises a rich collection of seismic attribute generation, data conditioning, and multiattribute machine-learning analysis tools constructed by
AASPI Software Structure Introduction The AASPI software comprises a rich collection of seismic attribute generation, data conditioning, and multiattribute machine-learning analysis tools constructed by
Unix L555. Dept. of Linguistics, Indiana University Fall Unix. Unix. Directories. Files. Useful Commands. Permissions. tar.
 L555 Dept. of Linguistics, Indiana University Fall 2010 1 / 21 What is? is an operating system, like DOS or Windows developed in 1969 by Bell Labs works well for single computers as well as for servers
L555 Dept. of Linguistics, Indiana University Fall 2010 1 / 21 What is? is an operating system, like DOS or Windows developed in 1969 by Bell Labs works well for single computers as well as for servers
CS CS Tutorial 2 2 Winter 2018
 CS CS 230 - Tutorial 2 2 Winter 2018 Sections 1. Unix Basics and connecting to CS environment 2. MIPS Introduction & CS230 Interface 3. Connecting Remotely If you haven t set up a CS environment password,
CS CS 230 - Tutorial 2 2 Winter 2018 Sections 1. Unix Basics and connecting to CS environment 2. MIPS Introduction & CS230 Interface 3. Connecting Remotely If you haven t set up a CS environment password,
