Version 1.0 PROTEOMICA DEMYSTIFYING PROTEINS. Indian Institute of Technology Bombay, India Bhushan N Kharbikar
|
|
|
- Elijah Stanley
- 5 years ago
- Views:
Transcription
1 Version 1.0 PROTEOMICA DEMYSTIFYING PROTEINS Indian Institute of Technology Bombay, India Bhushan N Kharbikar
2 Software User s Manual Version 1.0 Copyright 2013 Bhushan N Kharbikar, Proteomics Lab Department of Biosciences and Bioengineering Indian Institute of Technology Bombay
3 CONTENT PROTEOMICA 3 INTRODUCTION 3 WHAT IS PROTEOMICA? 4 FIRST TIME USER EXPERIENCES (FTUE) 5 CONFIGURING ENVIRONMENT 5 LAUNCHING PROTEOMICA 5 PRINCIPAL GRAPHICAL USER INTERFACE 8 KNOW THE CONTROLS 9 IMAGE DOMAIN BASED APPROACH 9 DESCRIBING PROTEOMICA GUI 10 WORKFLOW 10 COMMAND BUTTONS 12 SAMPLE GEL ANALYSIS USING PROTEOMICA 15 SET UP AN EXPERIMENT 15 CREATE MATCHSET 15 PRE-PROCESSING OF IMAGES 17 GEL IMAGE MATCHING 20 MATCH AND ALIGN 20 GLOBAL GEL 24 SPOT DETECTION 26 SEGMENT 26 QUANTIFY 32 RESULT AND UTILITY TOOLS 34 STATISTICS 34 3D VIEW VISUALISATION TOOL 39 PROFILE PLOT 44 HELP 50 EXIT 50 REFERENCES 51
4 Proteomica Introduction The processing and analysis of 2 DE gel images is crucial step in proteomics workflow and have substantial impact on final qualitative and quantitative outcome. Due to large complexity in protein patterns in gel images, its analysis is very subtle and tedious hence computer assisted automated processing and analysis is inevitable. While 2 DE technique for study of proteome is matured over last 3-4 decades, computer assisted automated techniques are still evolving, even after their introduction between late 70s to early 80s. Majority of research publications and commercial packages emerged in due course of time offers varied approaches towards implementation of image processing workflows for 2 DE gel images. More robust algorithms comes up which effectively address the major underlying challenges in processing of 2DE gel images e.g., presence of multiple noise, horizontal and vertical streaking, spot segmentation, spot intensity saturation, overlapping of spots, geometrical variations, image registrations etc. Although plenty of commercial software packages are available for analysing 2DE gel images, their high cost, unavailability of standard image analysis workflow, not-so promising results and black box development led many research groups to open-source and freeware to perform analysis of 2DE gel images. It s unfortunate that the freeware and open-source resources available do not offers all the steps required to complete analysis of 2DE gel images. Hence till date not widely used and adopted and eventually not proved to be the alternative for costly commercial software.
5 What is Proteomica? Proteomica Demystifying Proteins developed by us at Proteomics Laboratory, Indian Institute of Technology Bombay is also motivated by the factors described in the previous section above. Proteomica is complete open source and freeware alternative solution to commercial software which provides handy and cost effective option for research community. Proteomica incorporates all the required operation for complete analysis of 2DE gel images. Proteomica implement the image domain based analysis workflow which is powered with established and widely accepted algorithms [][] for processing gel images in each steps. Development of Proteomica is inspired by []. Also It also incorporates plot and visualisation tool. Proteomica is a plugin developed for ImageJ and use complete ImageJ framework. Some of the ImageJ original codes are also modified for its operations hence distributed as bundled with ImageJ. For Proteomica we request you to use the Proteomica bundled ImageJ distribution provided on plugin download page. Bhushan N Kharbikar, Biomedical Engineering, Indian Institute of Technology Bombay April 2013
6 First Time User Experiences (FTUE) Configuring environment Please follow all the steps provided at link required for installation of ImageJ as per your operating system (OS) for installing Proteomica bundled ImageJ distributions. Setup the required Java environments by setting environmental variables. For details please refer Alternatively download the archive (.zip) from link. Create directory unzip the distribution. Proteomica is set to use. Launching Proteomica Open the directory created after unzipping the structure as follows to enter you will get directory
7 Double click on the highlighted above. It will launch the ImageJ. From all the available menu option in the IJ menu bar go to plugins Dropdown menu will pop-up. Proteomica distribution is available here and highlighted a
8 Click on the Proteomica. It launches Splash welcome screen projecting product and its tagline along with information for copyright. The programs then load and display the Proteomica s principal graphical user interface as below.
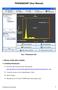
9 Principal Graphical User Interface Principal screen is divided in three area Product viz., label text area (marked as 1), Copyright text area (marked as 3) and command area (marked as 2). The command area is further divided subarea depending group of operations involved in workflow. Also the arrangement flows as per flow of steps in the image processing workflow for 2DE gel image analysis.
10 Know the controls Proteomica do the 2DE image analysis in image domain. Image domain based approach In image domain based approach the image alignment step precedes the spot detection. Reference image has been selected which has maximum no of spots patterns. Other gels images in match set then aligned to the referenced gel by warping. If alignment is not satisfactory, the process is then manually intervened and representative spot are selected over the gels which can be acts landmark points. Algorithm, then realign the images with the landmark in position and generated smooth interpolated warping for global alignment and with estimation of intermediary interpolation local alignment is achieved. Effective image also called fusion image or mean image is created by combining all the aligned images. Fusion image thus used for further analysis helps in reducing noise and to overcome variable features and artefacts. The fusion image is then subjected to background subtraction and intensity normalization. Use of fusion image for spot segmentation and quantification tends to increase sensitivity over single spots detection. Spot parameters are computed for spots on individual gel images. The resultant match sets have no missing spots and fidelity of analysis is high as compared with conventional method. Also the correctness of algorithm is limited by degree of correctness of gel alignments.
11 Describing Proteomica GUI Workflow Gel analysis starts with setting up an experiment by creating match set After match-set creation, all the gels added to the match-set are pre-processed to remove all the noise and background. Pre-procesing can be done repetitively till we get satisfactory results. After pre-processing, we have to determine best gel with minimum geometrical distortion and align all the gels with best one selected as target gel image. Match and alignment removes all the geometric distortions present in the gel images. Followed by the same Global gel image is created which would be the combination of all the gel images present in the match-set of an experiment for analysis of 2DE gel images.
12 Workflow further advances with the spot detections on the Global gel and after generating the spot list the spot s volume are quantified on each individual gels. Further, the results are generated with spot quantified on each and every gel image signifying their differential expressions if any. Result also includes few basics statistical parameters about detected protein s spots on the analysed 2DE gel images.
13 Command Buttons Several command buttons on panel marked as 2 above performs all the important functions in the image processing and analysis of 2DE gel images. Command buttons controls the calculations or help in accessing the other functions, as listed below. Match set button initiate the process and creates the match set by including gel images in order to setup an experiment Pre-process button fires the de-noising and background subtraction filters in order to clean the images and make it ready for the next step in workflow Match and Align button pops up the window in which you can select source and target gel images you want to align along with other options to set up or refines parameters and perform the matching and alignments and ask you to save the aligned image. Global gel button close all the open gel windows and launch the file selection popups to select all the aligned gel images to create global gel which is the combination of all the 2DE gel images which is pre-processed and aligned in the previous steps. Also it generates the stack of all the gel images. Segment button executes the mark and segment the protein spots on the Global gel image obtained in previous steps and add the list of detected protein spots in the spot manger / roi manager. Quantify button distributed all the protein spots detected in previous steps to all the gel image in the stack and quantify the corresponding volume of proteins for the same spot on every gel images.
14 Spot s Manager / ROI s manager button launches the spot / roi manages which displays and manages all the detected spots on the global gel also it has capability to save / open (load) / select / add and delete spots from the global gel which are auto-detected during segmentation of global gel image. Statistics button generate results for the entire analysis and also basic statistics describing all the protein spots detected on all the 2DE gel images included in the match-set for the experiment done as specified. 3D view button opens the JAVA3D interface to visualize the 3D surface profile of protein spots in gel images. Profile Plots button plots the 2D pots of the protein spot intensities. Also it can be operated at real-time or in the live mode. It also saves spot locations and the plot in the file. Help button displays the general FAQs about the Proteomica, Credits and Citations / References used for developing the current version of the Proteomica. Exit button closes all the windows and exit the Proteomica.
15 Hovering of the move pointer over the button provides you with the brief information about function of the said buttons.
16 Sample gel analysis using Proteomica Sample gels used in this demonstration are obtained from Proteomics Laboratory, Department of Biosciences and Bioengineering, IIT Bombay. Set up an experiment Create matchset We start with creating the match set by clicking on Matchset button and selecting all the gels which we intend to include in the experiments
17 After selecting all the images click open in file chooser dialog, it will create your matchset by opening all the selected images
18 Pre-processing of images We now proceeds towards the pre-processing steps Activate the image window frame by clicking on them and then click on pre-processing button on the Proteomica Input dialog window pops up asking for the various parameters to be put into it. The filters are already optimised for best performance based on the default values which appeared in the popup.
19 If you still fill to provide different values kindly fill in the same for improving the output of the step. Add the end of pre-processing the image is free from most of the noise and also the uneven background is subtracted from the image under processing. This step has been repeated for every image added to the image set if you feels that image requires pre-processing.
20 This figure below shows the output images before and after pre-processing step.
21 Gel Image Matching Match and Align Next step come is gel image matching. Click on the Match and align button. Popup dialog comes up asking for the source and target gel image to match. Select target image as which is best and which may be a reference image and select source image as the image which you want to align with reference image. And hit done.
22 Image alignment is started and you see real animated output when registration is in process.
23 After alignment you can see the image stack of which first image is output aligned image second image is the target (reference) image and other two in the stacks are distortion vectors and warping mask.
24 After successful alignment popup appears asking for the name of the images to save. It will only save the first image from the stack which the output aligned image in the match set directory.
25 Popup message appears displaying the successful alignment of source image with the target / reference image Global Gel Now we move towards creating the Global gel which is the effective combination of the all the aligned gel images. Click on Global gel button, file chooser window pops up.
26 Select only the aligned registered/aligned.jpeg file and click open button on UI. Popup appears in case if the all gel images are not of the same size. Select copy centre method from the method dropdown as it provide best results You select other methods to as which can be decided based on 2DE gel image. Click on OK button.
27 The process generates the effective Global gel as shown below. Spot Detection Segment Now we have to detect spot on the Global gel. Hit segment button in the spot detection sub panel.
28 Window pops up asking for the parameters. Provide necessary parameters and click on start watershed button. It initiates watershed segmentation on Global gel image.
29 Output of the watershed steps appears as follows. After the successful watershed click on segment button.
30 Clicking on the segment button opens up the advanced popup asking for the parameters. Default parameters which appear are already optimised for good performance. Still feel free to play around with these parameters. Click OK on the pop-up. It starts detecting the spots and add the detected protein spots on the Global gel to the ROI/ Spot Manager.
31 All the detected spots are appeared to be labelled on Global gel and added to ROI/ Spot Manager. Before clicking on the start watershed button or segment button, makes sure that Global gel window is active.
32 You are able to save the detected spot list in the match-set directory by clicking on the save button in the ROI/Spot Manager.
33 Quantify After detection of the spots on the global gel we have to determine the volume of the protein spots on the individual gel images. Quantify does the same for us. Activate the image stack window by click the image stack window and click on the Quantify button
34 It put the same spot pattern detected on the Global gel and quantify the volume of the protein on the individual gel images.
35 Result and Utility Tools Statistics Finally to view results click on statistics button. Popup message appears stating that generating statistics is under process.
36 Again popup appears asking how to arrange the results in the result tables. Provide suitable option and click ok. Result table starts appearing
37 Result tables displaying the results which include gel label, spot area, standard deviation, intden (Spot Volume), median, kurtosis and skewness.
38 We can save results file save as and then by giving std extensions (.txt or.csv or.xls)
39 We can even compute few more results and advanced parameters and display it in result table to configure the same go to the result window. Click on Results Set Measurements check all the required parameters and click ok.
40 3D view visualisation tool Click on the 3D view button. It launch Java3D viewer in which complete gel and/or even the part of the gel or protein spot are visualised in 3 Dimensions. For details please follow the below snapshots
41
42
43
44
45 Profile Plot It helps us to see the 2 dimensional intensity profile of proteins spot on the gel image. Also the profile of only spots on detected on the stack of the gel image can be visualised.
46 For visualising the profile of spots detected on the stack of the gel image. The spots must be loaded in the spot manager / roi manager.
47 After loading the save spot list in the Spot Manager / ROI manager use ImageJ selection tool to select the area of the gel for which profile needs to be visualized.
48 Below the profile plot of spot intensity are displayed.
49 Interface provides us with option to save the profile plots for 2DE gel images. We have option to see the live plotting by changing the selection and plot get updated for the same. This is achieved by clicking on the Live button on the UI.
50
51 Help Help button displays the general FAQs about the Proteomica, Credits and Citations / References used for developing the current version of the Proteomica. Exit Exit button closes all the windows and exit the Proteomica.
52 References Will be updated soon possible.
Flicker Comparison of 2D Electrophoretic Gels
 Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute - Frederick ++ SAIC - Frederick lemkin@ncifcrf.gov
Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute - Frederick ++ SAIC - Frederick lemkin@ncifcrf.gov
Matthew Baker. Visualisation of Electrophoresis Gels using TEX
 Visualisation of Electrophoresis Gels using TEX 1 Visualisation of Electrophoresis Gels using TEX Matthew Baker This paper describes a TEX system for creating interactive PDF files to visualize electrophoresis
Visualisation of Electrophoresis Gels using TEX 1 Visualisation of Electrophoresis Gels using TEX Matthew Baker This paper describes a TEX system for creating interactive PDF files to visualize electrophoresis
Guide for using the Photo Monitoring Plugins
 Guide for using the Photo Monitoring Plugins 24 June 2014 Overview This guide explains how to use the photo monitoring plugins written to work with ImageJ and a variant of ImageJ called Fiji. Since Fiji
Guide for using the Photo Monitoring Plugins 24 June 2014 Overview This guide explains how to use the photo monitoring plugins written to work with ImageJ and a variant of ImageJ called Fiji. Since Fiji
1 Download the latest version of ImageJ for your platform from the website:
 Using ImageJ with OMERO 4.4 This covers and version 4.4.x and the initial section assumes you have never installed ImageJ or Fiji before. For OMERO versions 5.1.x and 5.0.x see the Using ImageJ with OMERO
Using ImageJ with OMERO 4.4 This covers and version 4.4.x and the initial section assumes you have never installed ImageJ or Fiji before. For OMERO versions 5.1.x and 5.0.x see the Using ImageJ with OMERO
Image Processing Guideline for TMU 7T MRI
 Image Processing Guideline for TMU 7T MRI Chia Feng Lu Laboratory of NeuroImage Biomarker Analysis, Translational Imaging Research Center, TMU 08/18/2015, version 1.0 Section 1: Installation of ImageJ
Image Processing Guideline for TMU 7T MRI Chia Feng Lu Laboratory of NeuroImage Biomarker Analysis, Translational Imaging Research Center, TMU 08/18/2015, version 1.0 Section 1: Installation of ImageJ
The walkthrough is available at /
 The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
Tutorial 3D Max (for beginners) PART I
 Tutorial 3D Max (for beginners) PART I The Interface Introduction This tutorial gives a brief explanation of the MAX interface items commonly used and introduces you to the important areas of the interface.
Tutorial 3D Max (for beginners) PART I The Interface Introduction This tutorial gives a brief explanation of the MAX interface items commonly used and introduces you to the important areas of the interface.
BIOCAT User Guide Nov 2012 The main screen in BIOCAT allows the user to select three tasks:
 BIOCAT User Guide Nov 2012 The main screen in BIOCAT allows the user to select three tasks: 1. Model Selection for creating classification/annotation model 2. Training only without testing to create the
BIOCAT User Guide Nov 2012 The main screen in BIOCAT allows the user to select three tasks: 1. Model Selection for creating classification/annotation model 2. Training only without testing to create the
Workflow 1. Description
 Workflow 1 Description Determine protein staining intensities and distances in mitotic apparatus in z-stack intensity images, which were stained for the inner-centromere protein INCENP, and compare the
Workflow 1 Description Determine protein staining intensities and distances in mitotic apparatus in z-stack intensity images, which were stained for the inner-centromere protein INCENP, and compare the
OBCOL. (Organelle Based CO-Localisation) Users Guide
 OBCOL (Organelle Based CO-Localisation) Users Guide INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual
OBCOL (Organelle Based CO-Localisation) Users Guide INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual
Flint - Eclipse Based IDE User Manual
 1 E M B I E N T E C H N O L O G I E S Flint - Eclipse Based IDE User Manual Embien Technologies No 3, Sankarapandian Street, Madurai, India 625017 www.embien.com 2 3 Table of Contents 1 Introduction...
1 E M B I E N T E C H N O L O G I E S Flint - Eclipse Based IDE User Manual Embien Technologies No 3, Sankarapandian Street, Madurai, India 625017 www.embien.com 2 3 Table of Contents 1 Introduction...
English. Delta2D ANALYZING 2D GELS AS EASY AS POINT AND CLICK EXPLORING LIFE
 Getting started English 2D Western Blots Delta2D ANALYZING 2D GELS AS EASY AS POINT AND CLICK EXPLORING LIFE 2 Copyright DECODON GmbH. DECODON makes no representations, express or implied, with respect
Getting started English 2D Western Blots Delta2D ANALYZING 2D GELS AS EASY AS POINT AND CLICK EXPLORING LIFE 2 Copyright DECODON GmbH. DECODON makes no representations, express or implied, with respect
Lesson 1 Parametric Modeling Fundamentals
 1-1 Lesson 1 Parametric Modeling Fundamentals Create Simple Parametric Models. Understand the Basic Parametric Modeling Process. Create and Profile Rough Sketches. Understand the "Shape before size" approach.
1-1 Lesson 1 Parametric Modeling Fundamentals Create Simple Parametric Models. Understand the Basic Parametric Modeling Process. Create and Profile Rough Sketches. Understand the "Shape before size" approach.
Manual and tutorials for CurveAlign V4.0 Beta (August 31, 2017) UW-Madison
 Manual and tutorials for CurveAlign V4.0 Beta (August 31, 2017) LOCI @ UW-Madison http://loci.wisc.edu/software/curvealign 1 Introduction Overview: CurveAlign 4.0 framework is a curvelet transform based
Manual and tutorials for CurveAlign V4.0 Beta (August 31, 2017) LOCI @ UW-Madison http://loci.wisc.edu/software/curvealign 1 Introduction Overview: CurveAlign 4.0 framework is a curvelet transform based
ColorAnt - Quick Start Guide
 ColorAnt - Quick Start Guide ColorAnt is a software which, among other things, enables you to smooth and correct measurement value files (spectrophotometrically) and to average them intelligently. This
ColorAnt - Quick Start Guide ColorAnt is a software which, among other things, enables you to smooth and correct measurement value files (spectrophotometrically) and to average them intelligently. This
Flicker Comparison of 2D Electrophoretic Gels
 Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute ++ SAIC-Frederick Frederick, MD, USA lemkin@ncifcrf.gov
Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute ++ SAIC-Frederick Frederick, MD, USA lemkin@ncifcrf.gov
Align3_TP Manual. J. Anthony Parker, MD PhD Beth Israel Deaconess Medical Center Boston, MA Revised: 26 November 2004
 Align3_TP Manual J. Anthony Parker, MD PhD Beth Israel Deaconess Medical Center Boston, MA J.A.Parker@IEEE.org Revised: 26 November 2004 General ImageJ is a highly versatile image processing program written
Align3_TP Manual J. Anthony Parker, MD PhD Beth Israel Deaconess Medical Center Boston, MA J.A.Parker@IEEE.org Revised: 26 November 2004 General ImageJ is a highly versatile image processing program written
POOSL IDE Installation Manual
 Embedded Systems Innovation by TNO POOSL IDE Installation Manual Tool version 4.1.0 7 th November 2017 1 POOSL IDE Installation Manual 1 Installation... 4 1.1 Minimal system requirements... 4 1.2 Installing
Embedded Systems Innovation by TNO POOSL IDE Installation Manual Tool version 4.1.0 7 th November 2017 1 POOSL IDE Installation Manual 1 Installation... 4 1.1 Minimal system requirements... 4 1.2 Installing
3D Processing and Analysis with ImageJ
 3D Processing and Analysis with ImageJ Rapid development of technologies yielding multidimensional data LSCM / Video Electron tomography IRM / CT scan Thomas Boudier, Associate Professor, Cellular Imaging
3D Processing and Analysis with ImageJ Rapid development of technologies yielding multidimensional data LSCM / Video Electron tomography IRM / CT scan Thomas Boudier, Associate Professor, Cellular Imaging
Flicker Comparison of 2D Electrophoretic Gels
 Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute ++ SAIC-Frederick Frederick, MD, USA lemkin@ncifcrf.gov
Flicker Comparison of 2D Electrophoretic Gels Peter F. Lemkin +, Greg Thornwall ++ Lab. Experimental & Computational Biology + National Cancer Institute ++ SAIC-Frederick Frederick, MD, USA lemkin@ncifcrf.gov
S-PLUS INSTRUCTIONS FOR CASE STUDIES IN THE STATISTICAL SLEUTH
 S-PLUS INSTRUCTIONS FOR CASE STUDIES IN THE STATISTICAL SLEUTH Dan Schafer January, 2002 This guide contains brief instructions for accomplishing the graphical and numerical analyses of the case studies
S-PLUS INSTRUCTIONS FOR CASE STUDIES IN THE STATISTICAL SLEUTH Dan Schafer January, 2002 This guide contains brief instructions for accomplishing the graphical and numerical analyses of the case studies
Server Edition USER MANUAL. For Microsoft Windows
 Server Edition USER MANUAL For Microsoft Windows Copyright Notice & Proprietary Information Redstor Limited, 2016. All rights reserved. Trademarks - Microsoft, Windows, Microsoft Windows, Microsoft Windows
Server Edition USER MANUAL For Microsoft Windows Copyright Notice & Proprietary Information Redstor Limited, 2016. All rights reserved. Trademarks - Microsoft, Windows, Microsoft Windows, Microsoft Windows
Neuron Morpho plugin for ImageJ Version May 25, 2007
 Neuron Morpho plugin for ImageJ Version 1.1.6 - May 25, 2007 Giampaolo D Alessandro Department of Mathematics, University of Southampton, Southampton, SO17 1BJ, England, UK 1 Introduction The Neuron Morpho
Neuron Morpho plugin for ImageJ Version 1.1.6 - May 25, 2007 Giampaolo D Alessandro Department of Mathematics, University of Southampton, Southampton, SO17 1BJ, England, UK 1 Introduction The Neuron Morpho
Server Edition. V8 Peregrine User Manual. for Microsoft Windows
 Server Edition V8 Peregrine User Manual for Microsoft Windows Copyright Notice and Proprietary Information All rights reserved. Attix5, 2015 Trademarks - Microsoft, Windows, Microsoft Windows, Microsoft
Server Edition V8 Peregrine User Manual for Microsoft Windows Copyright Notice and Proprietary Information All rights reserved. Attix5, 2015 Trademarks - Microsoft, Windows, Microsoft Windows, Microsoft
'phred dist acd.tar.z'
 Phred is free for academic use but does require registration and for you to obtain a licence. Please visit http://www.phrap.org/consed/consed.html#howtoget and follow the instructions. A copy of the Phred
Phred is free for academic use but does require registration and for you to obtain a licence. Please visit http://www.phrap.org/consed/consed.html#howtoget and follow the instructions. A copy of the Phred
Image Analysis and Morphometry
 Image Analysis and Morphometry Lukas Schärer Evolutionary Biology Zoological Institute University of Basel 1 13. /15.3.2013 Zoology & Evolution Block Course Summary Quantifying morphology why do we need
Image Analysis and Morphometry Lukas Schärer Evolutionary Biology Zoological Institute University of Basel 1 13. /15.3.2013 Zoology & Evolution Block Course Summary Quantifying morphology why do we need
Design solutions worth your while. User Manual
 Design solutions worth your while Content 1. Product Search 04 2. Scope of the available information 06 3. Scope and form of presentation of the viewed files 07 4. Downloading drawing and models 09 5.
Design solutions worth your while Content 1. Product Search 04 2. Scope of the available information 06 3. Scope and form of presentation of the viewed files 07 4. Downloading drawing and models 09 5.
Gale Digital Scholar Lab Getting Started Walkthrough Guide
 Getting Started Logging In Your library or institution will provide you with your login link. You will have the option to sign in with a Google or Microsoft Account, this is so you have a personal account
Getting Started Logging In Your library or institution will provide you with your login link. You will have the option to sign in with a Google or Microsoft Account, this is so you have a personal account
<Partner Name> RSA ARCHER GRC Platform Implementation Guide. RiskLens <Partner Product>
 RSA ARCHER GRC Platform Implementation Guide 2.4.1 Wesley Loeffler, RSA Engineering Last Modified: April 25 th, 2018 2.4 Solution Summary The & Archer integration connects
RSA ARCHER GRC Platform Implementation Guide 2.4.1 Wesley Loeffler, RSA Engineering Last Modified: April 25 th, 2018 2.4 Solution Summary The & Archer integration connects
MasterPlex ReaderFit TUTORIAL
 TUTORIAL MasterPlex ReaderFit A H I T A C H I S O F T W A R E C O M P A N Y For Research Use Only 601 Gateway Blvd. Suite 100 South San Francisco, CA 94080 TELEPHONE 1.888.615.9600 (toll free) 1.650.615.7600
TUTORIAL MasterPlex ReaderFit A H I T A C H I S O F T W A R E C O M P A N Y For Research Use Only 601 Gateway Blvd. Suite 100 South San Francisco, CA 94080 TELEPHONE 1.888.615.9600 (toll free) 1.650.615.7600
You can download missing data from the course website, together with the codes R and python that we will run in this exercise.
 Exercise 4: Samples Characterization Aim: Sampling environmental conditions Principal Component Analysis of environmental conditions Hierarchical clustering of sampling spots Interpretation of the environmental
Exercise 4: Samples Characterization Aim: Sampling environmental conditions Principal Component Analysis of environmental conditions Hierarchical clustering of sampling spots Interpretation of the environmental
Panorama Sharing Skyline Documents
 Panorama Sharing Skyline Documents Panorama is a freely available, open-source web server database application for targeted proteomics assays that integrates into a Skyline proteomics workflow. It has
Panorama Sharing Skyline Documents Panorama is a freely available, open-source web server database application for targeted proteomics assays that integrates into a Skyline proteomics workflow. It has
QuickVol II Users Guide
 QuickVol II Users Guide Karl Schmidt (karl.schmidt@umassmed.edu) Working Draft 3/28/2006 This document is a working draft. If you find an error, please submit a bug on the Quickvol website. Thank you.
QuickVol II Users Guide Karl Schmidt (karl.schmidt@umassmed.edu) Working Draft 3/28/2006 This document is a working draft. If you find an error, please submit a bug on the Quickvol website. Thank you.
Guidelines for the MorphoLeaf application.
 Guidelines for the MorphoLeaf application. (Version 1.0, December 2015) Presentation of MorphoLeaf 2 Installation of MorphoLeaf 3 Some advice to start with optimal leaf images 4 1. Importing leaf images
Guidelines for the MorphoLeaf application. (Version 1.0, December 2015) Presentation of MorphoLeaf 2 Installation of MorphoLeaf 3 Some advice to start with optimal leaf images 4 1. Importing leaf images
IA L17 Auto Image Replication, a hands-on experience Hands-On Lab
 IA L17 Auto Image Replication, a hands-on experience Hands-On Lab Description Get hands on with Storage Lifecycle Policies Auto Image Replication in this session, which includes several different scenarios
IA L17 Auto Image Replication, a hands-on experience Hands-On Lab Description Get hands on with Storage Lifecycle Policies Auto Image Replication in this session, which includes several different scenarios
Tutorial 2: Analysis of DIA/SWATH data in Skyline
 Tutorial 2: Analysis of DIA/SWATH data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantification.
Tutorial 2: Analysis of DIA/SWATH data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantification.
Installation Guide - Windows
 Kony Visualizer Enterprise Installation Guide - Windows Release V8 SP3 Document Relevance and Accuracy This document is considered relevant to the Release stated on this title page and the document version
Kony Visualizer Enterprise Installation Guide - Windows Release V8 SP3 Document Relevance and Accuracy This document is considered relevant to the Release stated on this title page and the document version
e-bridge Color Profile Tool Quick Start Guide
 e-bridge Color Profile Tool Quick Start Guide 1 Contents 1. Installation... 3 1.1. Installing the e-bridge Color Profile Tool Software... 3 1.1. Removing the e-bridge Color Profile Tool... 4 1.2. Installing
e-bridge Color Profile Tool Quick Start Guide 1 Contents 1. Installation... 3 1.1. Installing the e-bridge Color Profile Tool Software... 3 1.1. Removing the e-bridge Color Profile Tool... 4 1.2. Installing
Tutorial files are available from the Exelis VIS website or on the ENVI Resource DVD in the image_reg directory.
 Image Registration Tutorial In this tutorial, you will use the Image Registration workflow in different scenarios to geometrically align two overlapping images with different viewing geometry and different
Image Registration Tutorial In this tutorial, you will use the Image Registration workflow in different scenarios to geometrically align two overlapping images with different viewing geometry and different
Mend for Eclipse quick start guide local analysis
 The Semmle Mend for Eclipse plugin allows users to view Semmle results in Eclipse. This document describes how to install and use the plugin for local analysis. You can install the plugin using a Semmle
The Semmle Mend for Eclipse plugin allows users to view Semmle results in Eclipse. This document describes how to install and use the plugin for local analysis. You can install the plugin using a Semmle
Vector Xpression 3. Speed Tutorial: III. Creating a Script for Automating Normalization of Data
 Vector Xpression 3 Speed Tutorial: III. Creating a Script for Automating Normalization of Data Table of Contents Table of Contents...1 Important: Please Read...1 Opening Data in Raw Data Viewer...2 Creating
Vector Xpression 3 Speed Tutorial: III. Creating a Script for Automating Normalization of Data Table of Contents Table of Contents...1 Important: Please Read...1 Opening Data in Raw Data Viewer...2 Creating
CF Vector User Manual V
 CF Vector User Manual V 1.0.0.0 1. The CF Vector application is a Windows Metro 8.1 application that can be used as an aid in learning about vectors. Two dimensional vectors [x,y] are used for simplicity.
CF Vector User Manual V 1.0.0.0 1. The CF Vector application is a Windows Metro 8.1 application that can be used as an aid in learning about vectors. Two dimensional vectors [x,y] are used for simplicity.
DeCyder 2D Software, Version 6.5
 GE Healthcare DeCyder 2D Software, Version 6.5 User Manual Contents Contents 1 Introduction 1.1 Introduction... 7 1.2 The DeCyder 2D Software User Manual... 7 1.3 Measuring differential protein abundance
GE Healthcare DeCyder 2D Software, Version 6.5 User Manual Contents Contents 1 Introduction 1.1 Introduction... 7 1.2 The DeCyder 2D Software User Manual... 7 1.3 Measuring differential protein abundance
Geographical Information Systems Institute. Center for Geographic Analysis, Harvard University. LAB EXERCISE 1: Basic Mapping in ArcMap
 Harvard University Introduction to ArcMap Geographical Information Systems Institute Center for Geographic Analysis, Harvard University LAB EXERCISE 1: Basic Mapping in ArcMap Individual files (lab instructions,
Harvard University Introduction to ArcMap Geographical Information Systems Institute Center for Geographic Analysis, Harvard University LAB EXERCISE 1: Basic Mapping in ArcMap Individual files (lab instructions,
TSM800 / TSM1330 Update Instructions
 TSM800 / TSM1330 Update Instructions Contents Contents... 1 Automatic Updates... 1 Manual Update to the Latest Version of N2KView... 2 Manual Update of the Complete Operating System... 3 Automatic Updates
TSM800 / TSM1330 Update Instructions Contents Contents... 1 Automatic Updates... 1 Manual Update to the Latest Version of N2KView... 2 Manual Update of the Complete Operating System... 3 Automatic Updates
PHASEQUANT (User Manual)
 PHASEQUANT (User Manual) ROI Pane Toolbar Threshold Bar Canvas Threshold Display Training Pane Buttons Fig 1. PhaseQuant GUI 1. INSTALLATION AND LOADING 1.1 Installing PhaseQuant Download the zip file
PHASEQUANT (User Manual) ROI Pane Toolbar Threshold Bar Canvas Threshold Display Training Pane Buttons Fig 1. PhaseQuant GUI 1. INSTALLATION AND LOADING 1.1 Installing PhaseQuant Download the zip file
Importing and processing a DGGE gel image
 BioNumerics Tutorial: Importing and processing a DGGE gel image 1 Aim Comprehensive tools for the processing of electrophoresis fingerprints, both from slab gels and capillary sequencers are incorporated
BioNumerics Tutorial: Importing and processing a DGGE gel image 1 Aim Comprehensive tools for the processing of electrophoresis fingerprints, both from slab gels and capillary sequencers are incorporated
Use stage 16 embryos see Pereanu et al., 2007 for morphological staging criteria. First start Vaa3d (http://vaa3d.org/; Peng et al., 2010).
 Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
Lauetools. A software package for Laue microdiffraction data analysis. https://sourceforge.net/projects/lauetools /
 Lauetools A software package for Laue microdiffraction data analysis https://sourceforge.net/projects/lauetools / Motivations Motivations ImageJ LAUE raw data XMAS fit2d Some codes Motivations LAUE raw
Lauetools A software package for Laue microdiffraction data analysis https://sourceforge.net/projects/lauetools / Motivations Motivations ImageJ LAUE raw data XMAS fit2d Some codes Motivations LAUE raw
CellTracer 1.0 Quick Start Guide
 CellTracer 1.0 Quick Start Guide Quanli Wang 1,3, Lingchong You 2,3 and Mike West 1,3 1 Department of Statistical Science, Duke University 2 Department of Biomedical Engineering, Duke University 3 Institute
CellTracer 1.0 Quick Start Guide Quanli Wang 1,3, Lingchong You 2,3 and Mike West 1,3 1 Department of Statistical Science, Duke University 2 Department of Biomedical Engineering, Duke University 3 Institute
PART A Three-Dimensional Measurement with iwitness
 PART A Three-Dimensional Measurement with iwitness A1. The Basic Process The iwitness software system enables a user to convert two-dimensional (2D) coordinate (x,y) information of feature points on an
PART A Three-Dimensional Measurement with iwitness A1. The Basic Process The iwitness software system enables a user to convert two-dimensional (2D) coordinate (x,y) information of feature points on an
DAME Web Application REsource Plugin Creator User Manual
 DAME Web Application REsource Plugin Creator User Manual DAMEWARE-MAN-NA-0016 Issue: 2.1 Date: March 20, 2014 Authors: S. Cavuoti, A. Nocella, S. Riccardi, M. Brescia Doc. : ModelPlugin_UserManual_DAMEWARE-MAN-NA-0016-Rel2.1
DAME Web Application REsource Plugin Creator User Manual DAMEWARE-MAN-NA-0016 Issue: 2.1 Date: March 20, 2014 Authors: S. Cavuoti, A. Nocella, S. Riccardi, M. Brescia Doc. : ModelPlugin_UserManual_DAMEWARE-MAN-NA-0016-Rel2.1
Mosaic Tutorial: Advanced Workflow
 Mosaic Tutorial: Advanced Workflow This tutorial demonstrates how to mosaic two scenes with different color variations. You will learn how to: Reorder the display of the input scenes Achieve a consistent
Mosaic Tutorial: Advanced Workflow This tutorial demonstrates how to mosaic two scenes with different color variations. You will learn how to: Reorder the display of the input scenes Achieve a consistent
The LViz Users Guide
 The LViz Users Guide Joshua Coyan Christopher Crosby Ramón Arrowsmith Jeff Conner* School of Earth and Space Exploration Arizona State University 24 October 2007 http://lidar.asu.edu Table of Contents
The LViz Users Guide Joshua Coyan Christopher Crosby Ramón Arrowsmith Jeff Conner* School of Earth and Space Exploration Arizona State University 24 October 2007 http://lidar.asu.edu Table of Contents
Exploring Data. This guide describes the facilities in SPM to gain initial insights about a dataset by viewing and generating descriptive statistics.
 This guide describes the facilities in SPM to gain initial insights about a dataset by viewing and generating descriptive statistics. 2018 by Minitab Inc. All rights reserved. Minitab, SPM, SPM Salford
This guide describes the facilities in SPM to gain initial insights about a dataset by viewing and generating descriptive statistics. 2018 by Minitab Inc. All rights reserved. Minitab, SPM, SPM Salford
MARS: Multiple Atlases Robust Segmentation
 Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
EPBscore user guide. Requirements:
 EPBscore user guide Requirements: Up to Matlab 2014a. Windows-only versions (both 32- and 64-bit) were tested. Toolboxes: Control, Images, Local, Matlab, Optim, Shared, Signal, Simulink, Stats, Wavelet
EPBscore user guide Requirements: Up to Matlab 2014a. Windows-only versions (both 32- and 64-bit) were tested. Toolboxes: Control, Images, Local, Matlab, Optim, Shared, Signal, Simulink, Stats, Wavelet
AN HIERARCHICAL APPROACH TO HULL FORM DESIGN
 AN HIERARCHICAL APPROACH TO HULL FORM DESIGN Marcus Bole and B S Lee Department of Naval Architecture and Marine Engineering, Universities of Glasgow and Strathclyde, Glasgow, UK 1 ABSTRACT As ship design
AN HIERARCHICAL APPROACH TO HULL FORM DESIGN Marcus Bole and B S Lee Department of Naval Architecture and Marine Engineering, Universities of Glasgow and Strathclyde, Glasgow, UK 1 ABSTRACT As ship design
Progenesis LC-MS Tutorial Including Data File Import, Alignment, Filtering, Progenesis Stats and Protein ID
 Progenesis LC-MS Tutorial Including Data File Import, Alignment, Filtering, Progenesis Stats and Protein ID 1 Introduction This tutorial takes you through a complete analysis of 9 LC-MS runs (3 replicate
Progenesis LC-MS Tutorial Including Data File Import, Alignment, Filtering, Progenesis Stats and Protein ID 1 Introduction This tutorial takes you through a complete analysis of 9 LC-MS runs (3 replicate
SYNTHETIC SCHLIEREN. Stuart B Dalziel, Graham O Hughes & Bruce R Sutherland. Keywords: schlieren, internal waves, image processing
 8TH INTERNATIONAL SYMPOSIUM ON FLOW VISUALIZATION (998) SYNTHETIC SCHLIEREN Keywords: schlieren, internal waves, image processing Abstract This paper outlines novel techniques for producing qualitative
8TH INTERNATIONAL SYMPOSIUM ON FLOW VISUALIZATION (998) SYNTHETIC SCHLIEREN Keywords: schlieren, internal waves, image processing Abstract This paper outlines novel techniques for producing qualitative
A³ Platform Quick Start
 A³ Platform Quick Start Last Update: Sep 07, 2011 Copyright 2002-2011 Visual Paradigm International Ltd. Table of Contents Table of Contents... 2 Getting Started... 3 Download A3 Platform... 3 Start-up
A³ Platform Quick Start Last Update: Sep 07, 2011 Copyright 2002-2011 Visual Paradigm International Ltd. Table of Contents Table of Contents... 2 Getting Started... 3 Download A3 Platform... 3 Start-up
Puma Energy elearning Hub. How To Guide for Staff Includes FAQs and Troubleshooting Guide
 Puma Energy elearning Hub How To Guide for Staff Includes FAQs and Troubleshooting Guide How To Guide for Staff Welcome to the elearning Hub. This How To Guide will highlight the steps you need to take
Puma Energy elearning Hub How To Guide for Staff Includes FAQs and Troubleshooting Guide How To Guide for Staff Welcome to the elearning Hub. This How To Guide will highlight the steps you need to take
There are two ways to launch Graphical User Interface (GUI). You can either
 How to get started? There are two ways to launch Graphical User Interface (GUI). You can either 1. Click on the Guide icon 2. Type guide at the prompt Just follow the instruction below: To start GUI we
How to get started? There are two ways to launch Graphical User Interface (GUI). You can either 1. Click on the Guide icon 2. Type guide at the prompt Just follow the instruction below: To start GUI we
MBB100 / DSM800 Update Instructions
 MBB100 / DSM800 Update Instructions Contents Contents... 1 Automatic Updates... 1 Manual Update to the Latest Version of N2KView... 2 Manual Update of the Complete Operating System... 3 Automatic Updates
MBB100 / DSM800 Update Instructions Contents Contents... 1 Automatic Updates... 1 Manual Update to the Latest Version of N2KView... 2 Manual Update of the Complete Operating System... 3 Automatic Updates
Documentation of Woocommerce Login / Sign up Premium. Installation of Woocommerce Login / Sign up Premium
 Documentation of Woocommerce Login / Sign up Premium Installation of Woocommerce Login / Sign up Premium Installation Install Word Press from http://codex.wordpress.org/installing_wordpress. Upload via
Documentation of Woocommerce Login / Sign up Premium Installation of Woocommerce Login / Sign up Premium Installation Install Word Press from http://codex.wordpress.org/installing_wordpress. Upload via
Software Requirements Specification. for WAVED. Version 3.0. Prepared By:
 Software Requirements Specification for WAVED Version 3.0 Prepared By: Sean Bluestein, Kristian Calhoun, Keith Horrocks, Steven Nguyen, Hannah Pinkos Advisor: Kurt Schmidt Stakeholder: Climate Central
Software Requirements Specification for WAVED Version 3.0 Prepared By: Sean Bluestein, Kristian Calhoun, Keith Horrocks, Steven Nguyen, Hannah Pinkos Advisor: Kurt Schmidt Stakeholder: Climate Central
Stratigraphy Modeling Horizon Coverages
 GMS TUTORIALS Stratigraphy Modeling Horizon Coverages This tutorial builds on the concepts taught in the tutorial entitled Stratigraphy Modeling Horizons and Solids. In that tutorial, you created solids
GMS TUTORIALS Stratigraphy Modeling Horizon Coverages This tutorial builds on the concepts taught in the tutorial entitled Stratigraphy Modeling Horizons and Solids. In that tutorial, you created solids
CIE L*a*b* color model
 CIE L*a*b* color model To further strengthen the correlation between the color model and human perception, we apply the following non-linear transformation: with where (X n,y n,z n ) are the tristimulus
CIE L*a*b* color model To further strengthen the correlation between the color model and human perception, we apply the following non-linear transformation: with where (X n,y n,z n ) are the tristimulus
Advanced Topics UNIT 2 PERFORMANCE EVALUATIONS
 Advanced Topics UNIT 2 PERFORMANCE EVALUATIONS Structure Page Nos. 2.0 Introduction 4 2. Objectives 5 2.2 Metrics for Performance Evaluation 5 2.2. Running Time 2.2.2 Speed Up 2.2.3 Efficiency 2.3 Factors
Advanced Topics UNIT 2 PERFORMANCE EVALUATIONS Structure Page Nos. 2.0 Introduction 4 2. Objectives 5 2.2 Metrics for Performance Evaluation 5 2.2. Running Time 2.2.2 Speed Up 2.2.3 Efficiency 2.3 Factors
TOPIM-2018 schedule. Session 1: Introduction to OMERO & CellProfiler
 Presentations are available at https://downloads.openmicroscopy.org/presentations/2018/topim-crete/ TOPIM-2018 schedule Thursday 12th July: 19:40-20:15 (35 mins) OMERO workshop introduction Friday 13th
Presentations are available at https://downloads.openmicroscopy.org/presentations/2018/topim-crete/ TOPIM-2018 schedule Thursday 12th July: 19:40-20:15 (35 mins) OMERO workshop introduction Friday 13th
Laboratory Assignment #4 Debugging in Eclipse CDT 1
 Lab 4 (10 points) November 20, 2013 CS-2301, System Programming for Non-majors, B-term 2013 Objective Laboratory Assignment #4 Debugging in Eclipse CDT 1 Due: at 11:59 pm on the day of your lab session
Lab 4 (10 points) November 20, 2013 CS-2301, System Programming for Non-majors, B-term 2013 Objective Laboratory Assignment #4 Debugging in Eclipse CDT 1 Due: at 11:59 pm on the day of your lab session
Vincent Mouchi, Quentin G. Crowley, Teresa Ubide, 2016
 AERYN: Walkthrough This short report will help you operate AERYN with all its capabilities. You can use the example files available with this report (also available at http://www.tcd.ie/geology/staff/crowleyq/aeryn/).
AERYN: Walkthrough This short report will help you operate AERYN with all its capabilities. You can use the example files available with this report (also available at http://www.tcd.ie/geology/staff/crowleyq/aeryn/).
TAX REPORTING SUITE MODULE IDES VERSION 1712
 TAX REPORTING SUITE MODULE IDES VERSION 1712 USERS S MANUAL Published: Jan 2018 For the latest information and to leave feedback, please visit Vogele IT-Services at http://www.section11.ch. 2 The information
TAX REPORTING SUITE MODULE IDES VERSION 1712 USERS S MANUAL Published: Jan 2018 For the latest information and to leave feedback, please visit Vogele IT-Services at http://www.section11.ch. 2 The information
The Regional Climate Model Viewer
 Introduction The Regional Climate Model viewer provides access to Regional Climate Model weather variables averaged by calendar month for a selection of time slices. The Regional Climate Model viewer has
Introduction The Regional Climate Model viewer provides access to Regional Climate Model weather variables averaged by calendar month for a selection of time slices. The Regional Climate Model viewer has
/5 Stacks. Displays the slice that follows the currently displayed slice. As a shortcut, press the > key.
 20-02-2018 1/5 Stacks Stacks This submenu contains commands that work with stacks. Add Slice Inserts a blank slice after the currently displayed slice. Hold down the Alt key to add the slice before the
20-02-2018 1/5 Stacks Stacks This submenu contains commands that work with stacks. Add Slice Inserts a blank slice after the currently displayed slice. Hold down the Alt key to add the slice before the
QuickStart Guide: Leica Cyclone REGISTER 360
 Leica Geosystems QuickStart Guide: Cyclone REGISTER 360 Product Leica Cyclone REGISTER 360 1.4 Date 17 October 2017 From HDS Software Product Management Contents Introduction... 2 Installation... 3 Licensing
Leica Geosystems QuickStart Guide: Cyclone REGISTER 360 Product Leica Cyclone REGISTER 360 1.4 Date 17 October 2017 From HDS Software Product Management Contents Introduction... 2 Installation... 3 Licensing
IMAGE STUDIO LITE. Tutorial Guide Featuring Image Studio Analysis Software Version 3.1
 IMAGE STUDIO LITE Tutorial Guide Featuring Image Studio Analysis Software Version 3.1 Notice The information contained in this document is subject to change without notice. LI-COR MAKES NO WARRANTY OF
IMAGE STUDIO LITE Tutorial Guide Featuring Image Studio Analysis Software Version 3.1 Notice The information contained in this document is subject to change without notice. LI-COR MAKES NO WARRANTY OF
Simpleware: Converting 3D Images into Models for Visualisation, Measurement and Computational Simulation
 converting 3d images into numerical models Simpleware: Converting 3D Images into Models for Visualisation, Measurement and Computational Simulation Dr Ross Cotton (Senior Application Engineer) r.cotton@simpleware.com
converting 3d images into numerical models Simpleware: Converting 3D Images into Models for Visualisation, Measurement and Computational Simulation Dr Ross Cotton (Senior Application Engineer) r.cotton@simpleware.com
Working with Standards and Duplicates in Geochemistry for ArcGIS
 Working with Standards and Duplicates in Geochemistry for ArcGIS After you have merged your locations and assay results, you may want to extract your control values (standards and duplicates). This process
Working with Standards and Duplicates in Geochemistry for ArcGIS After you have merged your locations and assay results, you may want to extract your control values (standards and duplicates). This process
Icy Training - Level 1 - Introduction
 Icy Training - Level 1 - Introduction Plan What is Icy? Installing Icy Graphical User Interface (GUI) Histograms & Colormap / Look up table Basic operations Overlays / Layers 3D view Icy Preferences Investigate
Icy Training - Level 1 - Introduction Plan What is Icy? Installing Icy Graphical User Interface (GUI) Histograms & Colormap / Look up table Basic operations Overlays / Layers 3D view Icy Preferences Investigate
Image Quant TL (PSC 563, 537, NSC 338, 438)
 Image Quant TL (PSC 563, 537, NSC 338, 438) Contact: Hyuk Kyu Seoh hseoh@gsu.edu Rm: PSC 537 Tel: (404) 413-5379 This software can analyze 1D gels, dot and slot blots, microplates, other basic arrays and
Image Quant TL (PSC 563, 537, NSC 338, 438) Contact: Hyuk Kyu Seoh hseoh@gsu.edu Rm: PSC 537 Tel: (404) 413-5379 This software can analyze 1D gels, dot and slot blots, microplates, other basic arrays and
A Tutorial Guide to Tribology Plug-in
 Supplementary Material A Tutorial Guide to Tribology Plug-in Tribology An ImageJ Plugin for surface topography analysis of laser textured surfaces. General Description This plugin presents an easy-to-use
Supplementary Material A Tutorial Guide to Tribology Plug-in Tribology An ImageJ Plugin for surface topography analysis of laser textured surfaces. General Description This plugin presents an easy-to-use
Expression Analysis with the Advanced RNA-Seq Plugin
 Expression Analysis with the Advanced RNA-Seq Plugin May 24, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Expression Analysis with the Advanced RNA-Seq Plugin May 24, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
A Statistical Consistency Check for the Space Carving Algorithm.
 A Statistical Consistency Check for the Space Carving Algorithm. A. Broadhurst and R. Cipolla Dept. of Engineering, Univ. of Cambridge, Cambridge, CB2 1PZ aeb29 cipolla @eng.cam.ac.uk Abstract This paper
A Statistical Consistency Check for the Space Carving Algorithm. A. Broadhurst and R. Cipolla Dept. of Engineering, Univ. of Cambridge, Cambridge, CB2 1PZ aeb29 cipolla @eng.cam.ac.uk Abstract This paper
Data Import and Quality Control in Geochemistry for ArcGIS
 Data Import and Quality Control in Geochemistry for ArcGIS This Data Import and Quality Control in Geochemistry for ArcGIS How-To Guide will demonstrate how to create a new geochemistry project, import
Data Import and Quality Control in Geochemistry for ArcGIS This Data Import and Quality Control in Geochemistry for ArcGIS How-To Guide will demonstrate how to create a new geochemistry project, import
Study Of Spatial Biological Systems Using a Graphical User Interface
 Study Of Spatial Biological Systems Using a Graphical User Interface Nigel J. Burroughs, George D. Tsibidis Mathematics Institute, University of Warwick, Coventry, CV47AL, UK {njb,tsibidis}@maths.warwick.ac.uk
Study Of Spatial Biological Systems Using a Graphical User Interface Nigel J. Burroughs, George D. Tsibidis Mathematics Institute, University of Warwick, Coventry, CV47AL, UK {njb,tsibidis}@maths.warwick.ac.uk
Bonita Workflow. Development Guide BONITA WORKFLOW
 Bonita Workflow Development Guide BONITA WORKFLOW Bonita Workflow Development Guide BSOA Workflow v3.0 Software January 2007 Copyright Bull SAS Table of Contents Chapter 1. Overview... 11 1.1 Role of
Bonita Workflow Development Guide BONITA WORKFLOW Bonita Workflow Development Guide BSOA Workflow v3.0 Software January 2007 Copyright Bull SAS Table of Contents Chapter 1. Overview... 11 1.1 Role of
The BERGEN Plug-in for EEGLAB
 The BERGEN Plug-in for EEGLAB July 2009, Version 1.0 What is the Bergen Plug-in for EEGLAB? The Bergen plug-in is a set of Matlab tools developed at the fmri group, University of Bergen, Norway, which
The BERGEN Plug-in for EEGLAB July 2009, Version 1.0 What is the Bergen Plug-in for EEGLAB? The Bergen plug-in is a set of Matlab tools developed at the fmri group, University of Bergen, Norway, which
CITO2 Installation & User Instructions
 CITO2 Installation & User Instructions DD 56107 Stoneridge Electronics Ltd 1. Installation...4 1.1. System Requirements...4 1.2. Installing CITO2...4 1.3. Uninstalling CITO2...4 2. Starting and closing
CITO2 Installation & User Instructions DD 56107 Stoneridge Electronics Ltd 1. Installation...4 1.1. System Requirements...4 1.2. Installing CITO2...4 1.3. Uninstalling CITO2...4 2. Starting and closing
GRADnet Videoconference Lectures
 GRADnet Videoconference Lectures Recording Presenting & Editing Guidelines Curated by Sean Cooper Contents Lecture Set- Up.. Page 3 Stopping a Recording.. Page 6 Advice for Lecturers. Page 7 Installing
GRADnet Videoconference Lectures Recording Presenting & Editing Guidelines Curated by Sean Cooper Contents Lecture Set- Up.. Page 3 Stopping a Recording.. Page 6 Advice for Lecturers. Page 7 Installing
3 FEBRUARY Guidelines for the VMSdatacall_2017_proposedWorkflow.r
 3 FEBRUARY 2017 Guidelines for the VMSdatacall_2017_proposedWorkflow.r Part 1 This document is designed to aid analysts streamline the process of extracting VMS data in accordance with requirements of
3 FEBRUARY 2017 Guidelines for the VMSdatacall_2017_proposedWorkflow.r Part 1 This document is designed to aid analysts streamline the process of extracting VMS data in accordance with requirements of
move object resize object create a sphere create light source camera left view camera view animation tracks
 Computer Graphics & Animation: CS Day @ SIUC This session explores computer graphics and animation using software that will let you create, display and animate 3D Objects. Basically we will create a 3
Computer Graphics & Animation: CS Day @ SIUC This session explores computer graphics and animation using software that will let you create, display and animate 3D Objects. Basically we will create a 3
Download and Installation Instructions. Java JDK Software for Windows
 Download and Installation Instructions for Java JDK Software for Windows Updated October, 2017 The CompuScholar Java Programming and Android Programming courses use the Java Development Kit (JDK) software.
Download and Installation Instructions for Java JDK Software for Windows Updated October, 2017 The CompuScholar Java Programming and Android Programming courses use the Java Development Kit (JDK) software.
Chart User Interface (v 2.0) Feature Specification
 Chart User Interface (v 2.0) Feature Specification User Interface changes for Charts in BIRT v 2.0 This document is intended to give an overview of the UI changes for Charts in BIRT v 2.0. Table of Contents
Chart User Interface (v 2.0) Feature Specification User Interface changes for Charts in BIRT v 2.0 This document is intended to give an overview of the UI changes for Charts in BIRT v 2.0. Table of Contents
Progenesis SameSpots Tutorial Including QC, Pre-filtering Stats and Picking
 Progenesis SameSpots Tutorial Including QC, Pre-filtering Stats and Picking 1 Introduction Progenesis SameSpots Tutorial This tutorial takes you through the complete analysis of a 4 gel 2D DIGE experiment*
Progenesis SameSpots Tutorial Including QC, Pre-filtering Stats and Picking 1 Introduction Progenesis SameSpots Tutorial This tutorial takes you through the complete analysis of a 4 gel 2D DIGE experiment*
Welcome to The Discovery Series Quantity One 1-D Analysis Software
 Bulletin 2908 US/EG Rev A Imaging Products Division Bio-Rad Laboratories Life Science Group Welcome to The Discovery Series Quantity One 1-D Analysis Software Version 4.4 Update Training Guide Bio-Rad
Bulletin 2908 US/EG Rev A Imaging Products Division Bio-Rad Laboratories Life Science Group Welcome to The Discovery Series Quantity One 1-D Analysis Software Version 4.4 Update Training Guide Bio-Rad
Transport Gateway Installation / Registration / Configuration
 CHAPTER 2 Transport Gateway Installation / Registration / Configuration This chapter covers the following areas: Transport Gateway requirements. Security Considerations When Using a Transport Gateway.
CHAPTER 2 Transport Gateway Installation / Registration / Configuration This chapter covers the following areas: Transport Gateway requirements. Security Considerations When Using a Transport Gateway.
Installing and Configuring Cisco Unified Real-Time Monitoring Tool
 CHAPTER 2 Installing and Configuring Cisco Unified Real-Time Monitoring Tool You can install Cisco Unified Real-Time Monitoring Tool (RTMT), which works for resolutions 800*600 and above, on a computer
CHAPTER 2 Installing and Configuring Cisco Unified Real-Time Monitoring Tool You can install Cisco Unified Real-Time Monitoring Tool (RTMT), which works for resolutions 800*600 and above, on a computer
Use of Shape Deformation to Seamlessly Stitch Historical Document Images
 Use of Shape Deformation to Seamlessly Stitch Historical Document Images Wei Liu Wei Fan Li Chen Jun Sun Satoshi Naoi In China, efforts are being made to preserve historical documents in the form of digital
Use of Shape Deformation to Seamlessly Stitch Historical Document Images Wei Liu Wei Fan Li Chen Jun Sun Satoshi Naoi In China, efforts are being made to preserve historical documents in the form of digital
TotalLab TL100 Quick Start
 TotalLab TL100 Quick Start Contents of thetl100 Quick Start Introduction to TL100 and Installation Instructions The Control Centre Getting Started The TL100 Interface 1D Gel Analysis Array Analysis Colony
TotalLab TL100 Quick Start Contents of thetl100 Quick Start Introduction to TL100 and Installation Instructions The Control Centre Getting Started The TL100 Interface 1D Gel Analysis Array Analysis Colony
