3D RECONSTRUCTION OF PHASE CONTRAST IMAGES USING FOCUS MEASURES.
|
|
|
- Corey Farmer
- 5 years ago
- Views:
Transcription
1 3D RECONSTRUCTION OF PHASE CONTRAST IMAGES USING FOCUS MEASURES Vincent On 1, Atena Zahedi 2 and, Bir Bhanu 1,2 1 Dept. of Electrical and Computer Engineering, University of California, Riverside, CA 92521, USA 2 Depart. of Bioengineering, University of California, Riverside, CA 92521, USA von001@ucr.edu, azahe001@ucr.edu, bhanu@cris.ucr.edu ABSTRACT In this paper, we present an approach for 3D phase-contrast microscopy using focus measure features. By using fluorescence data from the same location as the phase contrast data, we can train supervised regression algorithms to compute a depth map indicating the height of objects in imaged volume. From these depth maps, a 3D reconstruction of phase contrast images can be generated. This paper has shown the ability to 3D reconstruct phase contrast images using a variance metric inspired by all-in-focus methods. The proposed method has been used on A549 lung epithelial cells. Index Terms 3D Reconstruction, Phase Contrast Images, Focus Measures, Fluorescence, Regression 1. INTRODUCTION Phase contrast microscopy is a contrast-enhancing optical imaging modality. It is commonly used to produce highcontrast images of specimens, such as cells, microorganisms, tissue, and other transparent objects. Phase contrast imaging has many distinct advantages over other imaging methods. Objects collected with phase contrast are more visible than traditional bright field microscopy, even displaying internal structures that were previously invisible. Additionally, living specimens do not have to be killed, fixed, or stained and can be observed during ongoing biological processes. Lastly, phase contrast is not orientation dependent like differential interference contrast (DIC) microscopy and can be imaged at angle without producing artifacts. Traditionally, phase contrast has been constrained to 2D images. However, as samples become thicker the microscope s depth-of-field is not sufficient to image the entire object [1]. Because of this, 3D image is important to fully analyze the object of interest. 3D construction can be useful for visualizing the internal compartments of cells and cell shape. Additionally, 3D phase contrast reconstruction can be used to identify individual cells in tightly packed colonies such as pluripotent stem cells, and cells which grow as tightly-packed monolayers such as lung cells. A single cell culture may contain multiple types of objects such as cell substructures, debris, media and differentiated cells. Each of these may appear differently in an image and can greatly benefit from height and shape information provided by 3D imaging. In this paper, we propose a novel machine learning based approach for three-dimensional rendering of cells in culture. Unlike previous methods, our method can be used with any microscopy system that has phase contrast and fluorescence. After training the machine learning algorithms, the fluorescence imaging is no longer necessary and can be used to generate 3D phase contrast volumes using only a z- stack of phase contrast images. 2. RELATED WORK AND CONTRIBUTION Traditionally, 3D microscopy has been acquired using confocal microscopy or super-resolution microscopy because of their ability to remove out of focus light rays [2]. However, there are very few systems that provide both phase contrast and confocal/super-resolution images at the same time. Because of this we have decided to work on a system that has both phase contrast and fluorescent imaging. In fluorescent imaging, fluorescent stains are introduced to the imaging sample. By marking the contour of an object with stains, we can obtain an accurate depth ground truth. The proposed method assumes that different parts of an object will be most in focus at different heights. Because of this, it is useful to relate our method to all-in-focus algorithms. Many of these algorithms begin by acquiring image focal stacks that are focused at different distances [3]. These methods will either use defocus or in-focus parameter to select the focal distance in which a pixel is most in focus [4]. By using an in-focus parameter, a depth map may be generated and can be used to 3D render the object of interest. In addition, these methods may be used to produce all-infocus microscopy images. Other potential applications can include finding and maintaining autofocus during time-lapse imaging. To the best of our knowledge 3D phase contrast has not been achieved without specialized equipment. Chen et al. [5] have produced a 3D reconstruction of polystyrene beads, however they constructed a custom LED array and axial motion stage to image the sample at different angles. The
2 Fig. 1: System overview for phase contrast 3D Reconstruction proposed method does not require the addition of specialized hardware to existing imaging systems. 3. THE PROPOSED METHOD The proposed method is shown in Figure 1. The system extracts variance features from phase contrast image volumes and estimates a depth value using supervised regression. Fluorescent images are used to generate a ground truth and to provide labels to the phase contrast data. The regressed values are then reshaped into a depth map which is used for 3D reconstruction Ground Truth Generation During data collection, both phase contrast and fluorescent images of size r x c were taken for various cell cultures. For each region of interest, an expert biologist focused the microscope, so that the center plane is the most in focus. From this plane, h planes above and below are collected across the z-axis of the microscope. Every plane is separated by a step size of s micrometers. In total, H = 2h+1 planes are collected for each cell in each imaging modality. Each image represents a different focal distance. Phase contrast and fluorescent images collected at various heights are shown in Figure 2. To generate a ground truth for the phase contrast 3D reconstruction, a fluorescent 3D reconstruction is generated on the same volume. Usually, these images are collected with a microscope that removes out of focused light such as confocal or structured illumination microscopy (SIM). Since there are few systems that can collect these types of images with phase, we must compute a depth map while taking blur into account. Our method for generating ground truth from a volume of fluorescence images is to generate a depth map representing the height of each pixel. Various in-focus measures can be used to estimate the height. Yao et al. [6] have shown that variance is a strong metric to compute an allin-focus image. Our method begins by generating a variance volume V by convolving a variance filter through each voxel. As we are only interested in the most in-focused image, the filter is an m x m 2D filter. We have found m=5 to work the best empirically. A depth map D is generated from the variance volume by the following algorithm. 1. Initialize D as a zeros matrix with the same size as the input images. 2. Loop across i and j for all pixels of D. 3. Compute the maximum value of V at (i,j) across the z-axis. 4. The value of D(i,j) is the index of the maximum value across the z-axis. As there may be some errors in the depth map, a median filter is applied to smooth the values.
3 data as we are analyzing in vitro cells on a dish. Once these cells attach to the substrate, the cell lay flat on the surface. Using this constraint, a labelled volume L is created from the depth map as follows: L(i, j, k) = { 1 if k D p(i, j) 0 otherwise (1) To reduce noise, a 2D median filter is convolved with the depth map before being used to create the labelled volume Datasets 4. EXPERIMENTAL RESULTS Fig. 2: Phase contrast and fluorescence images at different focal points 3.2. Input Features Our proposed method for phase contrast 3D reconstruction is to generate a depth map based on supervised regression. To generate the features, we begin by computing a variance volume V p using the same method to generate our ground truth. However, instead of using the fluorescence images, the phase contrast images are used. To reduce the effect of background information in our regression, we remove the background from the image before computing V p. This is done by using the edge based segmentation method used in [7] on every image in the stack. Once objects are segmented anything below a specific size is considered a part of the background and set to zero. After acquiring V p, data samples are extracted by examining variance values along the z-axis. For a volume of size r x c x H, there will be r*c samples of size H x 1. A label L(i,j) is also assigned to each voxel based on corresponding value on the depth map D(i,j). 3.3 Supervised Learning Once extracted, the samples will be used to train a regression model. We have tested three different regression models: regression trees [8], linear support vector regression [9], least squares boosting regression [10]. Each model will take a vector of H x 1 inputs and estimate a height value. After estimating a height value for the r*c samples of an image volume, these values are reshaped into a depth map D p of size r x c D Reconstruction from Depth Map Because the system is imaging only the top of cells, the method assumes that every voxel below the top voxel of the object is a part of the object. This is a safe assumption for our All images have a resolution of 800x600 pixels. To reduce processing time, all images were resized to 400x300 by subsampling the pixels in the image. Phase contrast and fluorescence 3D images were acquired using the BioStation IM (Nikon Instruments, Melville NY) which is a compact cell incubator and monitoring system. 99 slices were collected at 0.3 um spacing. A 20x objective was used to gather individual cells and small cell clusters. All images were of A549 lung epithelial cells. A549 lung epithelial cells (human type II pulmonary alveolar adenocarcinoma cells) were obtained from (ATCC CCL-185, Manassas, VA, USA) and cultured in Ham s F-12 media supplemented with 10% fetal bovine serum (ATCC, Rockville, MD). Cells were grown until 80% confluency, at which point they were detached using 0.25% trypsin EDTA/DPBS. A549 cells were passaged every 2-3 days and medium was replenished every other day. Both cells types were grown in a 37 C incubator with 90% humidity and 5% CO2. In the A549s, Cyclin D1 (Red) is a marker important in cell-cycle and is localized to the nucleus. CD44 is a cancer stem cell marker, and was localized to the plasma membrane. There was an increase in the Cyclin D1 and CD44 expression on the treated A549 lung cancer cells. Acetylated α -tubulin (Green) localized to the cytoplasm. 5.2 Results Our dataset consisted of 5 cell volumes with 400x300 data points each. A 5-fold cross-validation was performed for each regression model by training on 4 cell volumes and testing on the last volume. Figure 3 displays the depth maps of ground truth and the 3 regression models. The figure also shows the 3D reconstruction generated by the depth maps. All sub images are of the same cell. From figure 3, the thickness of the samples is overexaggerated. Since the center plane (z=50) is the most in-focus plane, z=1 is not bottom of the cell. The z=1 plane is likely to be below the bottom, which explains the extra
4 Fig. 3: Row 1: Ground truth and regression depth maps. Row 2: 3D reconstructions of volumes from depth maps thickness seen in the 3D reconstruction. There are also errors around borders of the cell due to segmentation. As the segmentation sets the background to zero, these boundaries form an edge. These artificial edges cause an increase in the variance metric. LSVM is especially susceptible to this as seen in the extreme boundaries in the depth map. Because the stains in the fluorescence images only target specific structures, other objects such as debris are not detected. However, they are still visible in phase contrast. One such object can be seen in the bottom of the trees depth map. These objects are then detected in the phase 3D reconstructions but not the fluorescence. Table 1 shows the precision and recall results for each cell. Trees and LSBoost performed well on all cells except for cell 3. Cell 3 was an image stack with large dead cells which likely affected the precision results. LSVM was very sensitive to boundary conditions, causing it to perform very poorly for all cells. Figure 4 displays the z=50 slice of cells 2 to 5. fluorescence data as the ground truth, the system was able to train various regression models to compute a depth map. From this depth map, a 3D reconstruction can be generated which can detect objects not shown in fluorescence. Unlike other 3D phase contrast methods, the proposed system does not require custom hardware. Also unlike other methods, our system only requires one angle to be imaged and can be used by any research group with a microscopy system capable of imaging in both phase and fluorescence. 5. ACKNOWLEDGEMENTS The authors would to thank Dr. Prue Talbot for providing the cells and materials used in this paper. The support for this work was provided in part by NSF IGERT: Video Bioinformatics grant number DGE CONCLUSIONS In this paper, we have developed a 3D reconstruction method that can be used on phase contrast images. By using Trees LSVM LSBoost Precision Recall Precision Recall Precision Recall cell % 89.12% 5.95% 50.52% 85.51% 69.02% cell % 56.09% 10.02% 52.09% 88.30% 47.85% cell % 86.65% 3.75% 95.76% 25.57% 88.46% cell % 86.00% 10.52% 45.50% 87.12% 72.15% cell % 75.87% 8.75% 48.75% 75.15% 53.90% Table 1: Depth map regression results Fig. 4: Center phase contrast images of cells 2 to 5
5 6. REFERENCES [1] E. Maalouf, Contribution to Fluorescence Microscopy, 3d Thick Samples Deconvolution and Depth-variant Psf Maalouf [2] A. Ray and A. K. Mitra, Nanotechnology in Intracellular Trafficking, Imaging, and Delivery of Therapeutic Agents, in Emerging Nanotechnologies for Diagnostics, Drug Delivery and Medical Devices, Elsevier, 2017, pp [3] D. Vaquero, N. Gelfand, M. Tico, K. Pulli, and M. Turk, Generalized autofocus, in 2011 IEEE Workshop on Applications of Computer Vision (WACV), 2011, pp [4] J. Koseki et al., The development of an all-in-focus algorithm for endoscopy, in MHS2013, 2013, no. 1, pp [5] M. Chen, L. Tian, and L. Waller, 3D differential phase contrast microscopy, Biomed. Opt. Express, vol. 7, no. 10, p. 3940, [6] Y. Yao, B. Abidi, N. Doggaz, and M. Abidi, Evaluation of sharpness measures and search algorithms for the auto focusing of high-magnification images, 2006, vol. 6246, p G. [7] A. Zahedi, V. On, S. C. Lin, B. C. Bays, and E. Omaiye, Evaluating Cell Processes, Quality, and Biomarkers in Pluripotent Stem Cells Using Video Bioinformatics, PLOS ONE, pp. 1 22, [8] W. Loh, Regression Trees with. Unbiased Variable Selection, Korean J. Appl. Stat., vol. 17, no. 3, pp , Nov [9] C.-H. Ho and C.-J. Lin, Large-scale Linear Support Vector Regression, Jmlr, vol. 13, pp , [10] J. H. Friedman, Greedy Function Approximation: A Gradient Boosting Machine, Ann. Stat., vol. 29, no. 5, pp , Oct
Automated Human Embryonic Stem Cell Detection
 212 IEEE Second Conference on Healthcare Informatics, Imaging and Systems Biology Automated Human Embryonic Stem Cell Detection Benjamin X. Guan, Bir Bhanu Center for Research in Intelligent Systems University
212 IEEE Second Conference on Healthcare Informatics, Imaging and Systems Biology Automated Human Embryonic Stem Cell Detection Benjamin X. Guan, Bir Bhanu Center for Research in Intelligent Systems University
doi: /
 Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Presentation and analysis of multidimensional data sets
 Presentation and analysis of multidimensional data sets Overview 1. 3D data visualisation Multidimensional images Data pre-processing Visualisation methods Multidimensional images wavelength time 3D image
Presentation and analysis of multidimensional data sets Overview 1. 3D data visualisation Multidimensional images Data pre-processing Visualisation methods Multidimensional images wavelength time 3D image
RESTORING ARTIFACT-FREE MICROSCOPY IMAGE SEQUENCES. Robotics Institute Carnegie Mellon University 5000 Forbes Ave, Pittsburgh, PA 15213, USA
 RESTORING ARTIFACT-FREE MICROSCOPY IMAGE SEQUENCES Zhaozheng Yin Takeo Kanade Robotics Institute Carnegie Mellon University 5000 Forbes Ave, Pittsburgh, PA 15213, USA ABSTRACT Phase contrast and differential
RESTORING ARTIFACT-FREE MICROSCOPY IMAGE SEQUENCES Zhaozheng Yin Takeo Kanade Robotics Institute Carnegie Mellon University 5000 Forbes Ave, Pittsburgh, PA 15213, USA ABSTRACT Phase contrast and differential
Detection of Sub-resolution Dots in Microscopy Images
 Detection of Sub-resolution Dots in Microscopy Images Karel Štěpka, 2012 Centre for Biomedical Image Analysis, FI MU supervisor: prof. RNDr. Michal Kozubek, Ph.D. Outline Introduction Existing approaches
Detection of Sub-resolution Dots in Microscopy Images Karel Štěpka, 2012 Centre for Biomedical Image Analysis, FI MU supervisor: prof. RNDr. Michal Kozubek, Ph.D. Outline Introduction Existing approaches
Restoring the Invisible Details in Differential Interference Contrast Microscopy Images
 Restoring the Invisible Details in Differential Interference Contrast Microscopy Images Wenchao Jiang and Zhaozheng Yin Department of Computer Science, Missouri University of Science and Technology, wjm84@mst.edu,
Restoring the Invisible Details in Differential Interference Contrast Microscopy Images Wenchao Jiang and Zhaozheng Yin Department of Computer Science, Missouri University of Science and Technology, wjm84@mst.edu,
Supporting Information. Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking
 Supporting Information Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking Daan van der Zwaag 1,2, Nane Vanparijs 3, Sjors Wijnands 1,4, Riet De Rycke 5, Bruno G. De Geest 2* and
Supporting Information Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking Daan van der Zwaag 1,2, Nane Vanparijs 3, Sjors Wijnands 1,4, Riet De Rycke 5, Bruno G. De Geest 2* and
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS.
 SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening. Alex Laude The BioImaging Unit
 BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening Alex Laude The BioImaging Unit Multi-dimensional, multi-modal imaging at the sub-cellular
BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening Alex Laude The BioImaging Unit Multi-dimensional, multi-modal imaging at the sub-cellular
Real-time quantitative differential interference contrast (DIC) microscopy implemented via novel liquid crystal prisms
 Real-time quantitative differential interference contrast (DIC) microscopy implemented via novel liquid crystal prisms Ramzi N. Zahreddine a, Robert H. Cormack a, Hugh Masterson b, Sharon V. King b, Carol
Real-time quantitative differential interference contrast (DIC) microscopy implemented via novel liquid crystal prisms Ramzi N. Zahreddine a, Robert H. Cormack a, Hugh Masterson b, Sharon V. King b, Carol
SPATIAL DENSITY ESTIMATION BASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES
 SPATIAL DENSITY ESTIMATION ASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES Kuan-Chieh Jackie Chen 1,2,3, Ge Yang 1,2, and Jelena Kovačević 3,1,2 1 Dept. of iomedical Eng., 2 Center
SPATIAL DENSITY ESTIMATION ASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES Kuan-Chieh Jackie Chen 1,2,3, Ge Yang 1,2, and Jelena Kovačević 3,1,2 1 Dept. of iomedical Eng., 2 Center
Contents. Supplementary Information. Detection and Segmentation of Cell Nuclei in Virtual Microscopy Images: A Minimum-Model Approach
 Supplementary Information Detection and Segmentation of Cell Nuclei in Virtual Microscopy Images: A Minimum-Model Approach Stephan Wienert 1,2, Daniel Heim 2, Kai Saeger 2, Albrecht Stenzinger 3, Michael
Supplementary Information Detection and Segmentation of Cell Nuclei in Virtual Microscopy Images: A Minimum-Model Approach Stephan Wienert 1,2, Daniel Heim 2, Kai Saeger 2, Albrecht Stenzinger 3, Michael
Fast Z-stacking 3D Microscopy Extended Depth of Field Autofocus Z Depth Measurement 3D Surface Analysis
 MICROSCOPE 3D ADD-ON FAST PRECISE AFFORDABLE 3D ADD-ON FOR MICROSCOPY Fast Z-stacking 3D Microscopy Extended Depth of Field Autofocus Z Depth Measurement 3D Surface Analysis Compatible With Transmitted
MICROSCOPE 3D ADD-ON FAST PRECISE AFFORDABLE 3D ADD-ON FOR MICROSCOPY Fast Z-stacking 3D Microscopy Extended Depth of Field Autofocus Z Depth Measurement 3D Surface Analysis Compatible With Transmitted
Independent Resolution Test of
 Independent Resolution Test of as conducted and published by Dr. Adam Puche, PhD University of Maryland June 2005 as presented by (formerly Thales Optem Inc.) www.qioptiqimaging.com Independent Resolution
Independent Resolution Test of as conducted and published by Dr. Adam Puche, PhD University of Maryland June 2005 as presented by (formerly Thales Optem Inc.) www.qioptiqimaging.com Independent Resolution
Introductory Guide to Light Microscopy - Biomedical Confocal Microscopy
 Introductory Guide to Light Microscopy - Biomedical Confocal Microscopy 7 May 2007 Michael Hooker Microscopy Facility Michael Chua microscopy@unc.edu 843-3268 6007 Thurston Bowles Wendy Salmon wendy_salmon@med.unc.edu
Introductory Guide to Light Microscopy - Biomedical Confocal Microscopy 7 May 2007 Michael Hooker Microscopy Facility Michael Chua microscopy@unc.edu 843-3268 6007 Thurston Bowles Wendy Salmon wendy_salmon@med.unc.edu
45 µm polystyrene bead embedded in scattering tissue phantom. (a,b) raw images under oblique
 Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
Colin Paul Updated 14 November Preparation of publication-quality videos using ImageJ
 Preparation of publication-quality videos using ImageJ Statements made in scientific papers are often much easier to understand if they are supplemented by representative videos. In the best case, these
Preparation of publication-quality videos using ImageJ Statements made in scientific papers are often much easier to understand if they are supplemented by representative videos. In the best case, these
Principles of Light Microscopy
 Monday 8 August 2011 Principles of Light Microscopy 09:00 09:30 Introduction 09:30 10:15 The story of the microscope / 10:15 Coffee 10:30 12:45 Limitations of the eye. Resolution, contrast, magnification.
Monday 8 August 2011 Principles of Light Microscopy 09:00 09:30 Introduction 09:30 10:15 The story of the microscope / 10:15 Coffee 10:30 12:45 Limitations of the eye. Resolution, contrast, magnification.
Lab 4 - The Microscope
 Lab 4 - The Microscope Part A: The parts of the compound microscope. Obtain a microscope as indicated by your instructor. Always carry the microscope with one hand under the base and the other hand holding
Lab 4 - The Microscope Part A: The parts of the compound microscope. Obtain a microscope as indicated by your instructor. Always carry the microscope with one hand under the base and the other hand holding
4D IMAGING AT YOUR FINGERTIPS Real-time, Portable, High-Resolution Solutions for your Quality Control Needs
 STDO Dynamic 3D 4D Imaging Microscopy Instrument Systems 4D IMAGING AT YOUR FINGERTIPS Real-time, Portable, High-Resolution Solutions for your Quality Control Needs STDO-HOLO Overview: STDO-HOLO enables
STDO Dynamic 3D 4D Imaging Microscopy Instrument Systems 4D IMAGING AT YOUR FINGERTIPS Real-time, Portable, High-Resolution Solutions for your Quality Control Needs STDO-HOLO Overview: STDO-HOLO enables
Look Ahead to Get Ahead
 Microscopy from Carl Zeiss Archive 3D Deconvolution Z-stack 3D Deconvolution Look Ahead to Get Ahead Mark & Find Multichannel Timelapse Greater brilliance and resolution More convenient and precise than
Microscopy from Carl Zeiss Archive 3D Deconvolution Z-stack 3D Deconvolution Look Ahead to Get Ahead Mark & Find Multichannel Timelapse Greater brilliance and resolution More convenient and precise than
Fully Automatic Methodology for Human Action Recognition Incorporating Dynamic Information
 Fully Automatic Methodology for Human Action Recognition Incorporating Dynamic Information Ana González, Marcos Ortega Hortas, and Manuel G. Penedo University of A Coruña, VARPA group, A Coruña 15071,
Fully Automatic Methodology for Human Action Recognition Incorporating Dynamic Information Ana González, Marcos Ortega Hortas, and Manuel G. Penedo University of A Coruña, VARPA group, A Coruña 15071,
Structured Light II. Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov
 Structured Light II Johannes Köhler Johannes.koehler@dfki.de Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov Introduction Previous lecture: Structured Light I Active Scanning Camera/emitter
Structured Light II Johannes Köhler Johannes.koehler@dfki.de Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov Introduction Previous lecture: Structured Light I Active Scanning Camera/emitter
Cell Segmentation and Tracking in Phase Contrast Images using Graph Cut with Asymmetric Boundary Costs
 Cell Segmentation and Tracking in Phase Contrast Images using Graph Cut with Asymmetric Boundary Costs Robert Bensch and Olaf Ronneberger Computer Science Department and BIOSS Centre for Biological Signalling
Cell Segmentation and Tracking in Phase Contrast Images using Graph Cut with Asymmetric Boundary Costs Robert Bensch and Olaf Ronneberger Computer Science Department and BIOSS Centre for Biological Signalling
New Olympus FV3000RS Laser Scanning Confocal System. Quotation Number V2
 New Olympus FV3000RS Laser Scanning Confocal System Quotation Number 00056013 V2 Quotation Date: 08/08/2018 Revised Date: 07/09/2018 Quotation number: 00056013 Quotation Expiry: 07/11/2018 Macquarie University
New Olympus FV3000RS Laser Scanning Confocal System Quotation Number 00056013 V2 Quotation Date: 08/08/2018 Revised Date: 07/09/2018 Quotation number: 00056013 Quotation Expiry: 07/11/2018 Macquarie University
Quantifying Three-Dimensional Deformations of Migrating Fibroblasts
 45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
APPROACHES IN QUANTITATIVE, MULTI-DIMENSIONAL MICROSCOPY
 APPROACHES IN QUANTITATIVE, MULTI-DIMENSIONAL MICROSCOPY Profs. Zvi Kam and Benjamin Geiger Department of Molecular Cell Biology The Weizmann Institute of Science Rehovot, Israel In this presentation we
APPROACHES IN QUANTITATIVE, MULTI-DIMENSIONAL MICROSCOPY Profs. Zvi Kam and Benjamin Geiger Department of Molecular Cell Biology The Weizmann Institute of Science Rehovot, Israel In this presentation we
Spinning Disk Protocol
 Spinning Disk Protocol 1) System Startup F Please note our sign-up policy You must inform the facility at least 24 hours beforehand if you can t come; otherwise, you will receive a charge for unused time
Spinning Disk Protocol 1) System Startup F Please note our sign-up policy You must inform the facility at least 24 hours beforehand if you can t come; otherwise, you will receive a charge for unused time
SuRVoS Workbench. Super-Region Volume Segmentation. Imanol Luengo
 SuRVoS Workbench Super-Region Volume Segmentation Imanol Luengo Index - The project - What is SuRVoS - SuRVoS Overview - What can it do - Overview of the internals - Current state & Limitations - Future
SuRVoS Workbench Super-Region Volume Segmentation Imanol Luengo Index - The project - What is SuRVoS - SuRVoS Overview - What can it do - Overview of the internals - Current state & Limitations - Future
CFIM MICROSCOPY COURSE TIMETABLE PRINCIPLES OF MICROSCOPY MONDAY 6 TH OF JANUARY 2014 FRIDAY 10 TH OF JANUARY 2014
 MICROSCOPY COURSE TIMETABLE PRINCIPLES OF MICROSCOPY MONDAY 6 TH OF JANUARY 2014 FRIDAY 10 TH OF JANUARY 2014 CONFOCAL AND FLUORESCENCE MICROSCOPY MONDAY 20 TH OF JANUARY 2014 FRIDAY 24 TH OF JANUARY 2014
MICROSCOPY COURSE TIMETABLE PRINCIPLES OF MICROSCOPY MONDAY 6 TH OF JANUARY 2014 FRIDAY 10 TH OF JANUARY 2014 CONFOCAL AND FLUORESCENCE MICROSCOPY MONDAY 20 TH OF JANUARY 2014 FRIDAY 24 TH OF JANUARY 2014
Lab 5 Microscopy. Introduction. Carrying the Microscope. Depth of Focus. Parts of the Compound Microscope. Magnification
 Lab 5 Microscopy Introduction The microscope is an instrument that contains one or more lenses and is used to view objects that are too small be seen with the unaided eye. A magnifying glass is a simple
Lab 5 Microscopy Introduction The microscope is an instrument that contains one or more lenses and is used to view objects that are too small be seen with the unaided eye. A magnifying glass is a simple
Elucidating the Mechanism Governing Particle Alignment and Movement by DEP Yu Zhao 1, Jozef Brcka 2, Jacques Faguet 2, Guigen Zhang *,1,3,4 1
 Elucidating the Mechanism Governing Particle Alignment and Movement by DEP Yu Zhao 1, Jozef Brcka 2, Jacques Faguet 2, Guigen Zhang *,1,3,4 1 Department of Bioengineering, Clemson University, Clemson,
Elucidating the Mechanism Governing Particle Alignment and Movement by DEP Yu Zhao 1, Jozef Brcka 2, Jacques Faguet 2, Guigen Zhang *,1,3,4 1 Department of Bioengineering, Clemson University, Clemson,
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror
 Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
Leica Microsystems Intelligent Structured Illumination Microscopy
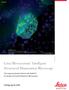 Widefield Mouse kidney section. Maximum projection of a stack containing 65 planes. Green: glomeruli and convoluted tubules (wheat germ agglutinin Alexa Fluor 488) Blue: Nuclei (DAPI). Structured Illumination
Widefield Mouse kidney section. Maximum projection of a stack containing 65 planes. Green: glomeruli and convoluted tubules (wheat germ agglutinin Alexa Fluor 488) Blue: Nuclei (DAPI). Structured Illumination
EPFL SV PTBIOP BIOP COURSE 2015 OPTICAL SLICING METHODS
 BIOP COURSE 2015 OPTICAL SLICING METHODS OPTICAL SLICING METHODS Scanning Methods Wide field Methods Point Scanning Deconvolution Line Scanning Multiple Beam Scanning Single Photon Multiple Photon Total
BIOP COURSE 2015 OPTICAL SLICING METHODS OPTICAL SLICING METHODS Scanning Methods Wide field Methods Point Scanning Deconvolution Line Scanning Multiple Beam Scanning Single Photon Multiple Photon Total
3D Computer Vision. Structured Light II. Prof. Didier Stricker. Kaiserlautern University.
 3D Computer Vision Structured Light II Prof. Didier Stricker Kaiserlautern University http://ags.cs.uni-kl.de/ DFKI Deutsches Forschungszentrum für Künstliche Intelligenz http://av.dfki.de 1 Introduction
3D Computer Vision Structured Light II Prof. Didier Stricker Kaiserlautern University http://ags.cs.uni-kl.de/ DFKI Deutsches Forschungszentrum für Künstliche Intelligenz http://av.dfki.de 1 Introduction
IDENTIFYING OPTICAL TRAP
 IDENTIFYING OPTICAL TRAP Yulwon Cho, Yuxin Zheng 12/16/2011 1. BACKGROUND AND MOTIVATION Optical trapping (also called optical tweezer) is widely used in studying a variety of biological systems in recent
IDENTIFYING OPTICAL TRAP Yulwon Cho, Yuxin Zheng 12/16/2011 1. BACKGROUND AND MOTIVATION Optical trapping (also called optical tweezer) is widely used in studying a variety of biological systems in recent
High-Resolution 3D Reconstruction of the Surface of Live Early-Stage Toad Embryo
 BIOL 507 PROJECT REPORT High-Resolution 3D Reconstruction of the Surface of Live Early-Stage Toad Embryo PLEDGE As my honor, I pledge that I have neither received nor given help in this homework. SIGNATURE
BIOL 507 PROJECT REPORT High-Resolution 3D Reconstruction of the Surface of Live Early-Stage Toad Embryo PLEDGE As my honor, I pledge that I have neither received nor given help in this homework. SIGNATURE
Minimizing Noise and Bias in 3D DIC. Correlated Solutions, Inc.
 Minimizing Noise and Bias in 3D DIC Correlated Solutions, Inc. Overview Overview of Noise and Bias Digital Image Correlation Background/Tracking Function Minimizing Noise Focus Contrast/Lighting Glare
Minimizing Noise and Bias in 3D DIC Correlated Solutions, Inc. Overview Overview of Noise and Bias Digital Image Correlation Background/Tracking Function Minimizing Noise Focus Contrast/Lighting Glare
ONBI Practical 7: Comparison of techniques
 ONBI Practical 7: Comparison of techniques RM Parton 2014 Aims of practical 7: One of the most common issues confronting people new to microscopy is the confusing array of different techniques available.
ONBI Practical 7: Comparison of techniques RM Parton 2014 Aims of practical 7: One of the most common issues confronting people new to microscopy is the confusing array of different techniques available.
Digital Volume Correlation for Materials Characterization
 19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
Neuron Crawler: An Automatic Tracing Algorithm for Very Large Neuron Images
 Neuron Crawler: An Automatic Tracing Algorithm for Very Large Neuron Images Zhi Zhou, Staci A. Sorensen, and Hanchuan Peng* Allen Institute for Brain Science, Seattle, WA 98103. * Corresponding author.
Neuron Crawler: An Automatic Tracing Algorithm for Very Large Neuron Images Zhi Zhou, Staci A. Sorensen, and Hanchuan Peng* Allen Institute for Brain Science, Seattle, WA 98103. * Corresponding author.
Segmentation and Tracking of Partial Planar Templates
 Segmentation and Tracking of Partial Planar Templates Abdelsalam Masoud William Hoff Colorado School of Mines Colorado School of Mines Golden, CO 800 Golden, CO 800 amasoud@mines.edu whoff@mines.edu Abstract
Segmentation and Tracking of Partial Planar Templates Abdelsalam Masoud William Hoff Colorado School of Mines Colorado School of Mines Golden, CO 800 Golden, CO 800 amasoud@mines.edu whoff@mines.edu Abstract
Nikon T1-E microscope with A1 confocal and STORM super-resolution Zeiss LSM510/Meta confocal microscope.
 Nikon T1-E microscope with A1 confocal and STORM super-resolution. This system allows for the imaging of cells and tissues labeled with fluorescent probes using live or fixed specimens to obtain 3D images
Nikon T1-E microscope with A1 confocal and STORM super-resolution. This system allows for the imaging of cells and tissues labeled with fluorescent probes using live or fixed specimens to obtain 3D images
What should I know about the 3D Restoration module? Improvision Technical Note No. 142 Last updated: 28 June, 2000
 What should I know about the 3D Restoration module? Improvision Technical Note No. 142 Last updated: 28 June, 2000 Topic This technical note discusses some aspects of iterative deconvolution and how to
What should I know about the 3D Restoration module? Improvision Technical Note No. 142 Last updated: 28 June, 2000 Topic This technical note discusses some aspects of iterative deconvolution and how to
Using Machine Learning for Classification of Cancer Cells
 Using Machine Learning for Classification of Cancer Cells Camille Biscarrat University of California, Berkeley I Introduction Cell screening is a commonly used technique in the development of new drugs.
Using Machine Learning for Classification of Cancer Cells Camille Biscarrat University of California, Berkeley I Introduction Cell screening is a commonly used technique in the development of new drugs.
Distributed Ray Tracing
 CT5510: Computer Graphics Distributed Ray Tracing BOCHANG MOON Distributed Ray Tracing Motivation The classical ray tracing produces very clean images (look fake) Perfect focus Perfect reflections Sharp
CT5510: Computer Graphics Distributed Ray Tracing BOCHANG MOON Distributed Ray Tracing Motivation The classical ray tracing produces very clean images (look fake) Perfect focus Perfect reflections Sharp
Bioimage Informatics
 Bioimage Informatics Lecture 10, Spring 2012 Bioimage Data Analysis (II): Applications of Point Feature Detection Techniques: Super Resolution Fluorescence Microscopy Bioimage Data Analysis (III): Edge
Bioimage Informatics Lecture 10, Spring 2012 Bioimage Data Analysis (II): Applications of Point Feature Detection Techniques: Super Resolution Fluorescence Microscopy Bioimage Data Analysis (III): Edge
A web interface for the quantification of microtubule dynamics
 A web interface for the quantification of microtubule dynamics Koon Yin Kong, Georgia Institute of Technology Adam Marcus, Emory University Paraskevi Giaanakakou, Emory University May D. Wang, Georgia
A web interface for the quantification of microtubule dynamics Koon Yin Kong, Georgia Institute of Technology Adam Marcus, Emory University Paraskevi Giaanakakou, Emory University May D. Wang, Georgia
Effect of OTF attenuation on single-slice SR-SIM reconstruction
 Effect of OTF attenuation on single-slice SR-SIM reconstruction Supplementary Figure 1: Actin filaments in U2OS cells, labelled with Phalloidin-Atto488, measured on a DeltaVision OMX, excited at 488 nm
Effect of OTF attenuation on single-slice SR-SIM reconstruction Supplementary Figure 1: Actin filaments in U2OS cells, labelled with Phalloidin-Atto488, measured on a DeltaVision OMX, excited at 488 nm
Display. Introduction page 67 2D Images page 68. All Orientations page 69 Single Image page 70 3D Images page 71
 Display Introduction page 67 2D Images page 68 All Orientations page 69 Single Image page 70 3D Images page 71 Intersecting Sections page 71 Cube Sections page 72 Render page 73 1. Tissue Maps page 77
Display Introduction page 67 2D Images page 68 All Orientations page 69 Single Image page 70 3D Images page 71 Intersecting Sections page 71 Cube Sections page 72 Render page 73 1. Tissue Maps page 77
Supplementary Figures
 Supplementary Figures re co rd ed ch annels darkf ield MPM2 PI cell cycle phases G1/S/G2 prophase metaphase anaphase telophase bri ghtfield 55 pixel Supplementary Figure 1 Images of Jurkat cells captured
Supplementary Figures re co rd ed ch annels darkf ield MPM2 PI cell cycle phases G1/S/G2 prophase metaphase anaphase telophase bri ghtfield 55 pixel Supplementary Figure 1 Images of Jurkat cells captured
3-D. Here red spheres show the location of gold nanoparticles inside/around a cell nucleus.
 3-D The CytoViva 3-D System allows the user can locate objects of interest in a 3-D space. It does this by acquiring multiple Z planes and performing our custom software routines to locate and observe
3-D The CytoViva 3-D System allows the user can locate objects of interest in a 3-D space. It does this by acquiring multiple Z planes and performing our custom software routines to locate and observe
Tomographic Modeling of Casted Blood Vascular Systems
 Tomographic Modeling of Casted Blood Vascular Systems Roger C. Wagner & Fred E. Hossler The three dimensional architecture of blood vascular systems is intimately associated with their function of irrigating
Tomographic Modeling of Casted Blood Vascular Systems Roger C. Wagner & Fred E. Hossler The three dimensional architecture of blood vascular systems is intimately associated with their function of irrigating
DeltaVision OMX SR super-resolution microscope. Super-resolution doesn t need to be complicated
 DeltaVision OMX SR super-resolution microscope Super-resolution doesn t need to be complicated Live-cell super-resolution microscopy DeltaVision OMX SR is a compact super-resolution microscope system optimized
DeltaVision OMX SR super-resolution microscope Super-resolution doesn t need to be complicated Live-cell super-resolution microscopy DeltaVision OMX SR is a compact super-resolution microscope system optimized
Multi-Plane Tomographic Phase Retrieval for 4D Cell Microscopy. Emrah Bostan Computational Imaging Lab UC Berkeley
 Multi-Plane Tomographic Phase Retrieval for 4D Cell Microscopy Emrah Bostan Computational Imaging Lab UC Berkeley IMA Workshop Series: Phaseless Imaging in Theory and Practice University of Minnesota,
Multi-Plane Tomographic Phase Retrieval for 4D Cell Microscopy Emrah Bostan Computational Imaging Lab UC Berkeley IMA Workshop Series: Phaseless Imaging in Theory and Practice University of Minnesota,
QUANTITATIVE PHASE IMAGING OF BIOLOGICAL CELLS USING OFF-AXIS METHOD OF WIDE FIELD DIGITAL INTERFERENCE MICROSCOPY (WFDIM)
 http:// QUANTITATIVE PHASE IMAGING OF BIOLOGICAL CELLS USING OFF-AXIS METHOD OF WIDE FIELD DIGITAL INTERFERENCE MICROSCOPY (WFDIM) Pradeep Kumar Behera 1, Dalip Singh Mehta 2 1,2 Physics,Indian Institute
http:// QUANTITATIVE PHASE IMAGING OF BIOLOGICAL CELLS USING OFF-AXIS METHOD OF WIDE FIELD DIGITAL INTERFERENCE MICROSCOPY (WFDIM) Pradeep Kumar Behera 1, Dalip Singh Mehta 2 1,2 Physics,Indian Institute
Determination of Volume Characteristics of Cells from Dynamical Microscopic Image
 JOURNAL OF APPLIED COMPUTER SCIENCE Vol. 23 No. 1 (2015), pp. 63-76 Determination of Volume Characteristics of Cells from Dynamical Microscopic Image Olga Nedzved 1,2, Sergey Ablameyko 2,3 1 Belarusian
JOURNAL OF APPLIED COMPUTER SCIENCE Vol. 23 No. 1 (2015), pp. 63-76 Determination of Volume Characteristics of Cells from Dynamical Microscopic Image Olga Nedzved 1,2, Sergey Ablameyko 2,3 1 Belarusian
Voxel-based Registration Methods in in vivo Imaging
 Vladimír Ulman Laboratory of Optical Microscopy, FI MU 14th April, 2005 Informatics seminar, FI MU Outline Introduction to bioinformatics Introduction to in vivo imaging Point-based registration Optical
Vladimír Ulman Laboratory of Optical Microscopy, FI MU 14th April, 2005 Informatics seminar, FI MU Outline Introduction to bioinformatics Introduction to in vivo imaging Point-based registration Optical
IN VITRO cell displacement has been shown to be a useful parameter
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 6, JUNE 2005 697 Tracking of Migrating Cells Under Phase-Contrast Video Microscopy With Combined Mean-Shift Processes O. Debeir*, P. Van Ham, R. Kiss,
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 6, JUNE 2005 697 Tracking of Migrating Cells Under Phase-Contrast Video Microscopy With Combined Mean-Shift Processes O. Debeir*, P. Van Ham, R. Kiss,
Learning to Count Cells: Applications to Lens-free Imaging of Large Fields
 1 Learning to Count Cells: Applications to Lens-free Imaging of Large Fields G. Flaccavento 1,2, V. Lempitsky 3, I. Pope 1,4, P.R. Barber 1, A. Zisserman 3, J.A. Noble 2, B. Vojnovic 1 1 Gray Institute
1 Learning to Count Cells: Applications to Lens-free Imaging of Large Fields G. Flaccavento 1,2, V. Lempitsky 3, I. Pope 1,4, P.R. Barber 1, A. Zisserman 3, J.A. Noble 2, B. Vojnovic 1 1 Gray Institute
ND Processing Tools in NIS-Elements
 ND Processing Tools in NIS-Elements Overview This technical note describes basic uses of the ND processing tools available in NIS-Elements. These tools are specifically designed for arithmetic functions
ND Processing Tools in NIS-Elements Overview This technical note describes basic uses of the ND processing tools available in NIS-Elements. These tools are specifically designed for arithmetic functions
The Paper Project Guide to Confocal Imaging at Home & in the Classroom
 The Paper Project Guide to Confocal Imaging at Home & in the Classroom http://lifesciences.asu.edu/paperproject The Paper Project Guide to Confocal Imaging at Home & in the Classroom CJ Kazilek & Dennis
The Paper Project Guide to Confocal Imaging at Home & in the Classroom http://lifesciences.asu.edu/paperproject The Paper Project Guide to Confocal Imaging at Home & in the Classroom CJ Kazilek & Dennis
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10934 Supplementary Methods Mathematical implementation of the EST method. The EST method begins with padding each projection with zeros (that is, embedding
SUPPLEMENTARY INFORMATION doi:10.1038/nature10934 Supplementary Methods Mathematical implementation of the EST method. The EST method begins with padding each projection with zeros (that is, embedding
Blob-Representation of Multidimensional Objects and Surfaces
 Blob-Representation of Multidimensional Objects and Surfaces Edgar Garduño and Gabor T. Herman Department of Computer Science The Graduate Center City University of New York Presentation Outline Reconstruction
Blob-Representation of Multidimensional Objects and Surfaces Edgar Garduño and Gabor T. Herman Department of Computer Science The Graduate Center City University of New York Presentation Outline Reconstruction
EXAM SOLUTIONS. Image Processing and Computer Vision Course 2D1421 Monday, 13 th of March 2006,
 School of Computer Science and Communication, KTH Danica Kragic EXAM SOLUTIONS Image Processing and Computer Vision Course 2D1421 Monday, 13 th of March 2006, 14.00 19.00 Grade table 0-25 U 26-35 3 36-45
School of Computer Science and Communication, KTH Danica Kragic EXAM SOLUTIONS Image Processing and Computer Vision Course 2D1421 Monday, 13 th of March 2006, 14.00 19.00 Grade table 0-25 U 26-35 3 36-45
Coherent Gradient Sensing Microscopy: Microinterferometric Technique. for Quantitative Cell Detection
 Coherent Gradient Sensing Microscopy: Microinterferometric Technique for Quantitative Cell Detection Proceedings of the SEM Annual Conference June 7-10, 010 Indianapolis, Indiana USA 010 Society for Experimental
Coherent Gradient Sensing Microscopy: Microinterferometric Technique for Quantitative Cell Detection Proceedings of the SEM Annual Conference June 7-10, 010 Indianapolis, Indiana USA 010 Society for Experimental
Chapter 36. Image Formation
 Chapter 36 Image Formation Apr 22, 2012 Light from distant things We learn about a distant thing from the light it generates or redirects. The lenses in our eyes create images of objects our brains can
Chapter 36 Image Formation Apr 22, 2012 Light from distant things We learn about a distant thing from the light it generates or redirects. The lenses in our eyes create images of objects our brains can
Fast and accurate automated cell boundary determination for fluorescence microscopy
 Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Rectification of distorted elemental image array using four markers in three-dimensional integral imaging
 Rectification of distorted elemental image array using four markers in three-dimensional integral imaging Hyeonah Jeong 1 and Hoon Yoo 2 * 1 Department of Computer Science, SangMyung University, Korea.
Rectification of distorted elemental image array using four markers in three-dimensional integral imaging Hyeonah Jeong 1 and Hoon Yoo 2 * 1 Department of Computer Science, SangMyung University, Korea.
Band-Pass Filter Design by Segmentation in Frequency Domain for Detection of Epithelial Cells in Endomicroscope Images
 Band-Pass Filter Design by Segmentation in Frequency Domain for Detection of Epithelial Cells in Endomicroscope Images Bastian Bier 1, Firas Mualla 1, Stefan Steidl 1, Christopher Bohr 2, Helmut Neumann
Band-Pass Filter Design by Segmentation in Frequency Domain for Detection of Epithelial Cells in Endomicroscope Images Bastian Bier 1, Firas Mualla 1, Stefan Steidl 1, Christopher Bohr 2, Helmut Neumann
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12009 Supplementary Figure 1. Experimental tilt series of 104 projections with a tilt range of ±72.6 and equal slope increments, acquired from a Pt nanoparticle using HAADF- STEM (energy:
doi:10.1038/nature12009 Supplementary Figure 1. Experimental tilt series of 104 projections with a tilt range of ±72.6 and equal slope increments, acquired from a Pt nanoparticle using HAADF- STEM (energy:
A Survey of Light Source Detection Methods
 A Survey of Light Source Detection Methods Nathan Funk University of Alberta Mini-Project for CMPUT 603 November 30, 2003 Abstract This paper provides an overview of the most prominent techniques for light
A Survey of Light Source Detection Methods Nathan Funk University of Alberta Mini-Project for CMPUT 603 November 30, 2003 Abstract This paper provides an overview of the most prominent techniques for light
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model
 Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 12,HannanAbdillahi 3, Lala Ceklic 3,Ute Wolf-Schnurrbusch 23,JensKowal 12 1 ARTORG Center
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 12,HannanAbdillahi 3, Lala Ceklic 3,Ute Wolf-Schnurrbusch 23,JensKowal 12 1 ARTORG Center
Supplementary Figure 1
 Supplementary Figure 1 Experimental unmodified 2D and astigmatic 3D PSFs. (a) The averaged experimental unmodified 2D PSF used in this study. PSFs at axial positions from -800 nm to 800 nm are shown. The
Supplementary Figure 1 Experimental unmodified 2D and astigmatic 3D PSFs. (a) The averaged experimental unmodified 2D PSF used in this study. PSFs at axial positions from -800 nm to 800 nm are shown. The
Image Acquisition Systems
 Image Acquisition Systems Goals and Terminology Conventional Radiography Axial Tomography Computer Axial Tomography (CAT) Magnetic Resonance Imaging (MRI) PET, SPECT Ultrasound Microscopy Imaging ITCS
Image Acquisition Systems Goals and Terminology Conventional Radiography Axial Tomography Computer Axial Tomography (CAT) Magnetic Resonance Imaging (MRI) PET, SPECT Ultrasound Microscopy Imaging ITCS
Introduction to Medical Image Processing
 Introduction to Medical Image Processing Δ Essential environments of a medical imaging system Subject Image Analysis Energy Imaging System Images Image Processing Feature Images Image processing may be
Introduction to Medical Image Processing Δ Essential environments of a medical imaging system Subject Image Analysis Energy Imaging System Images Image Processing Feature Images Image processing may be
Neuroscience Solutions. by leica microsystems
 Neuroscience Solutions by leica microsystems neuroscience solutions from leica microsystems Working with Acute Slice Preparations The nervous system works as a tightly regulated, highly complex cellular
Neuroscience Solutions by leica microsystems neuroscience solutions from leica microsystems Working with Acute Slice Preparations The nervous system works as a tightly regulated, highly complex cellular
Integrating Shape from Shading and Shape from Stereo for Variable Reflectance Surface Reconstruction from SEM Images
 Integrating Shape from Shading and Shape from Stereo for Variable Reflectance Surface Reconstruction from SEM Images Reinhard Danzl 1 and Stefan Scherer 2 1 Institute for Computer Graphics and Vision,
Integrating Shape from Shading and Shape from Stereo for Variable Reflectance Surface Reconstruction from SEM Images Reinhard Danzl 1 and Stefan Scherer 2 1 Institute for Computer Graphics and Vision,
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S. Visualization of Biological Molecules in Their Native Stat e.
 THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
Classification of Hyperspectral Breast Images for Cancer Detection. Sander Parawira December 4, 2009
 1 Introduction Classification of Hyperspectral Breast Images for Cancer Detection Sander Parawira December 4, 2009 parawira@stanford.edu In 2009 approximately one out of eight women has breast cancer.
1 Introduction Classification of Hyperspectral Breast Images for Cancer Detection Sander Parawira December 4, 2009 parawira@stanford.edu In 2009 approximately one out of eight women has breast cancer.
METRIC PLANE RECTIFICATION USING SYMMETRIC VANISHING POINTS
 METRIC PLANE RECTIFICATION USING SYMMETRIC VANISHING POINTS M. Lefler, H. Hel-Or Dept. of CS, University of Haifa, Israel Y. Hel-Or School of CS, IDC, Herzliya, Israel ABSTRACT Video analysis often requires
METRIC PLANE RECTIFICATION USING SYMMETRIC VANISHING POINTS M. Lefler, H. Hel-Or Dept. of CS, University of Haifa, Israel Y. Hel-Or School of CS, IDC, Herzliya, Israel ABSTRACT Video analysis often requires
Lecture 02 Microscopy, Staining, Classification (Ch3)
 Lecture 02 Microscopy, Staining, Classification (Ch3) Topics Methods of Culturing Microorganisms Microscope (History, Types, Definitions) Staining (Gram s) Dichotomous keys Micro for Health Sciences 1
Lecture 02 Microscopy, Staining, Classification (Ch3) Topics Methods of Culturing Microorganisms Microscope (History, Types, Definitions) Staining (Gram s) Dichotomous keys Micro for Health Sciences 1
Improvement of the correlative AFM and ToF-SIMS approach using an empirical sputter model for 3D chemical characterization
 Improvement of the correlative AFM and ToF-SIMS approach using an empirical sputter model for 3D chemical characterization T. Terlier 1, J. Lee 1, K. Lee 2, and Y. Lee 1 * 1 Advanced Analysis Center, Korea
Improvement of the correlative AFM and ToF-SIMS approach using an empirical sputter model for 3D chemical characterization T. Terlier 1, J. Lee 1, K. Lee 2, and Y. Lee 1 * 1 Advanced Analysis Center, Korea
Comparison of Vessel Segmentations using STAPLE
 Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
Chapter 7: Geometrical Optics. The branch of physics which studies the properties of light using the ray model of light.
 Chapter 7: Geometrical Optics The branch of physics which studies the properties of light using the ray model of light. Overview Geometrical Optics Spherical Mirror Refraction Thin Lens f u v r and f 2
Chapter 7: Geometrical Optics The branch of physics which studies the properties of light using the ray model of light. Overview Geometrical Optics Spherical Mirror Refraction Thin Lens f u v r and f 2
James Boedicker, Hernan Garcia, Manuel Razo Mejia, Griffin Chure, Elizabeth Eck, Meghan Turner
 Matlab Tutorial: Measuring cell growth rate James Boedicker, Hernan Garcia, Manuel Razo Mejia, Griffin Chure, Elizabeth Eck, Meghan Turner Introduction Reproduction is an essential process of life. In
Matlab Tutorial: Measuring cell growth rate James Boedicker, Hernan Garcia, Manuel Razo Mejia, Griffin Chure, Elizabeth Eck, Meghan Turner Introduction Reproduction is an essential process of life. In
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Courtois et al., http://www.jcb.org/cgi/content/full/jcb.201202135/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Measurement of distribution and size of MTOCs in the
Supplemental material Courtois et al., http://www.jcb.org/cgi/content/full/jcb.201202135/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Measurement of distribution and size of MTOCs in the
Nonlinear optics and two photon microscopy. Table of contents. Sam Whiteley and Seth Parker PHYS 173/BGGN 266 July 13, 2014
 Nonlinear optics and two photon microscopy Sam Whiteley and Seth Parker PHYS 173/BGGN 266 July 13, 2014 Table of contents 1. Introduction 2. Optical setup 3. Initial images and troubleshooting 4. Determining
Nonlinear optics and two photon microscopy Sam Whiteley and Seth Parker PHYS 173/BGGN 266 July 13, 2014 Table of contents 1. Introduction 2. Optical setup 3. Initial images and troubleshooting 4. Determining
Chapter 3. Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C.
 Chapter 3 Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C. elegans Embryos Abstract Differential interference contrast (DIC) microscopy is
Chapter 3 Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C. elegans Embryos Abstract Differential interference contrast (DIC) microscopy is
Exam Microscopic Measurement Techniques 4T th of April, 2008
 Exam Microscopic Measurement Techniques 4T300 29 th of April, 2008 Name / Initials: Ident. #: Education: This exam consists of 5 questions. Questions and sub questions will be rewarded with the amount
Exam Microscopic Measurement Techniques 4T300 29 th of April, 2008 Name / Initials: Ident. #: Education: This exam consists of 5 questions. Questions and sub questions will be rewarded with the amount
Other Reconstruction Techniques
 Other Reconstruction Techniques Ruigang Yang CS 684 CS 684 Spring 2004 1 Taxonomy of Range Sensing From Brain Curless, SIGGRAPH 00 Lecture notes CS 684 Spring 2004 2 Taxonomy of Range Scanning (cont.)
Other Reconstruction Techniques Ruigang Yang CS 684 CS 684 Spring 2004 1 Taxonomy of Range Sensing From Brain Curless, SIGGRAPH 00 Lecture notes CS 684 Spring 2004 2 Taxonomy of Range Scanning (cont.)
Vutara 350. Innovation with Integrity. The Fastest, Super-Resolution Microscope Deep 3D Imaging on Live Cells, Quickly and Easily
 Vutara 350 The Fastest, Super-Resolution Microscope Deep 3D Imaging on Live Cells, Quickly and Easily Innovation with Integrity Fluorescence Microscopy Vutara 350 Don t Get Left Behind Bruker s Vutara
Vutara 350 The Fastest, Super-Resolution Microscope Deep 3D Imaging on Live Cells, Quickly and Easily Innovation with Integrity Fluorescence Microscopy Vutara 350 Don t Get Left Behind Bruker s Vutara
Submitted to the Faculty. Drexel University. Surya Sreeram Vissapragada. In partial fulfillment of the. Requirements for the degree.
 Submitted to the Faculty Of Drexel University By Surya Sreeram Vissapragada In partial fulfillment of the Requirements for the degree Of Master of Science September 2008 1 Acknowledgement I would like
Submitted to the Faculty Of Drexel University By Surya Sreeram Vissapragada In partial fulfillment of the Requirements for the degree Of Master of Science September 2008 1 Acknowledgement I would like
Bosch Institute Advanced Microscopy Facility Workshops Summer/Autumn 2016
 Bosch Institute Advanced Microscopy Facility Workshops Summer/Autumn 2016 Presented by Dr Louise Cole and Dr Cathy Payne, Advanced Microscopy Facility, Bosch Institute, School of Medical Sciences, The
Bosch Institute Advanced Microscopy Facility Workshops Summer/Autumn 2016 Presented by Dr Louise Cole and Dr Cathy Payne, Advanced Microscopy Facility, Bosch Institute, School of Medical Sciences, The
Guided Image Super-Resolution: A New Technique for Photogeometric Super-Resolution in Hybrid 3-D Range Imaging
 Guided Image Super-Resolution: A New Technique for Photogeometric Super-Resolution in Hybrid 3-D Range Imaging Florin C. Ghesu 1, Thomas Köhler 1,2, Sven Haase 1, Joachim Hornegger 1,2 04.09.2014 1 Pattern
Guided Image Super-Resolution: A New Technique for Photogeometric Super-Resolution in Hybrid 3-D Range Imaging Florin C. Ghesu 1, Thomas Köhler 1,2, Sven Haase 1, Joachim Hornegger 1,2 04.09.2014 1 Pattern
Removing Atmospheric Turbulence
 Removing Atmospheric Turbulence Xiang Zhu, Peyman Milanfar EE Department University of California, Santa Cruz SIAM Imaging Science, May 20 th, 2012 1 What is the Problem? time 2 Atmospheric Turbulence
Removing Atmospheric Turbulence Xiang Zhu, Peyman Milanfar EE Department University of California, Santa Cruz SIAM Imaging Science, May 20 th, 2012 1 What is the Problem? time 2 Atmospheric Turbulence
Measuring surface geometry of adherent cells using oblique transillumination
 Advances in Bioscience and Biotechnology, 2011, 2, 359-363 doi:10.4236/abb.2011.25053 Published Online October 2011 (http://www.scirp.org/journal/abb/). Measuring surface geometry of adherent cells using
Advances in Bioscience and Biotechnology, 2011, 2, 359-363 doi:10.4236/abb.2011.25053 Published Online October 2011 (http://www.scirp.org/journal/abb/). Measuring surface geometry of adherent cells using
Quantitative Phase Imaging in Microscopy
 Invited Paper Quantitative Phase Imaging in Microscopy Colin JR Sheppard, Shan S Kou and Shalin Mehta Division of Bioengineering National University of Singapore Singapore 117574 1 Introduction There are
Invited Paper Quantitative Phase Imaging in Microscopy Colin JR Sheppard, Shan S Kou and Shalin Mehta Division of Bioengineering National University of Singapore Singapore 117574 1 Introduction There are
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences
 IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
