Supporting Information. Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking
|
|
|
- Oswin Conley
- 5 years ago
- Views:
Transcription
1 Supporting Information Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking Daan van der Zwaag 1,2, Nane Vanparijs 3, Sjors Wijnands 1,4, Riet De Rycke 5, Bruno G. De Geest 2* and Lorenzo Albertazzi 1,6* 1 Institute for Complex Molecular Systems, Eindhoven University of Technology, Eindhoven, Netherlands 2 Department of Chemical Engineering and Chemistry, Eindhoven University of Technology, Eindhoven, The Netherlands 3 Department of Pharmaceutics, Ghent University, Ghent, Belgium 4 Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands 5 VIB Department for Molecular Biomedical Research, Technologiepark 927, 9052 Ghent, Belgium 6 Institute for Bioengineering of Catalonia (IBEC), Barcelona, Spain these authors equally contributed as first authors * Prof.Dr. Bruno De Geest Department of Pharmaceutics, Ghent University, Ghent Belgium br.degeest@ugent.be * Dr. Lorenzo Albertazzi Institute for Bioengineering of Catalonia (IBEC), Barcelona, Spain lalbertazzi@ibecbarcelona.eu S1
2 Single Molecule Data Analysis Software 2D Bead Analysis Localization datasets corresponding to two-color, 2D-STORM images acquired as outlined before were imported into Matlab as.txt files. The datasets contain membrane localizations and bead localizations in different channels (see Chart 1 for an example), and were plotted to find regions of interest (ROI) containing a high density of beads inside a cell. These ROIs were automatically processed to efficiently obtain significant amounts of nanoparticles images. Chart 1 Raw data of a 2-color STORM image of nanoparticles (red) internalized in HeLa cells (membrane stained in green). A density-based clustering algorithm (dbscan.m) was applied to the NP channel to remove background localizations and identify individual objects in an image. In this algorithm with parameters k and ε, localizations which have at least k neighboring localization within ε nanometers are clustered together with these neighbors. Iteratively, neighboring points for these newly added neighbors are also detected, until no points are added anymore and a cluster is completely identified. In this way, background localizations due to detector noise, background fluorescence or other causes were eliminated and the individual clusters saved separately (see Chart 2) S2
3 Chart 2 Background removal from the nanoparticle channel (left) and isolation of localization clusters Subsequently, the clusters were fitted as circles to determine the exact location and size of each NP. Depending on the quality of reconstruction, size of the NP, microscope resolution and other experimental factors, this could be done using a fit to either an empty circle (using a closed expression in circfit.m) or a filled circle (optimization using a custom script). The procedure yields the midpoint and radius of every individual bead, which can be used for single-bead analysis. Each cluster was subjected to three quality checks to ensure only well-reconstructed single NP were taken into account in the analysis (see chart 3): - Sphericity. The sphericity was assessed by calculating the aspect ratio (AR) of the localization points cloud, defined as the ratio of the standard deviations in orthogonal directions: AR i = σ i σ, i Here, σ i is the standard deviation of the points cloud in direction i, and σ,i is the standard deviation of the points cloud in the orthogonal direction. If any of the AR i (or AR -1 i if σ,i > σ i ) exceeds a threshold value (1.5 in the 2D overlay), the cluster is rejected. - Size. The NP size was checked by comparing the found radius to the median value of the beads in an entire image, or to the radius given by the supplier. NP that are much smaller (R > 0.5*R ref ) or larger (R > 1.5*R ref ) than this reference radius are rejected. - Number of localizations. NP were rejected if the corresponding cluster contained too few localizations for accurate reconstruction, or too many to be due to normal dye behavior. S3
4 Image overlay was performed by aligning all beads according to their found midpoint and adding all localizations in a finely pixelated grid. For visualization, the Matlab function imagesc was used. Chart 3 2D quality check of localization clusters S4
5 3D Bead Analysis Analysis of 3D-STORM images consisted of largely similar steps to the 2D overlay, adapted for three-dimensional data. After plotting the localizations and selecting an ROI, densitybased clustering was applied (dbscan.m) to yield separated beads. Chart 4 Raw data (left), background removal (middle) and cluster identification (right) of 3D data for nanoparticles. Beads were fitted using hollow (spherefit.m) or filled (custom script) spheres, and identifying appropriate individual beads involved the three quality controls: - Sphericity. For the three-dimensional data, the aspect ratio (AR) was defined as: AR i maxσ j= x, y, z = min σ j= x, y, z If the AR i in any direction i exceeds a threshold value (1.8 in the 3D overlay), the cluster is rejected. Directions i are picked homogeneously in three dimensions using the script psphere.m. - Size. Same as 2D - Number of localizations. Same as 2D i, j i, j S5
6 Chart 5 3D quality check of localization clusters Image overlay was again performed by aligning all beads and adding up localization in a 3Dvoxel grid. Visualization of 2D-planes used the Matlab function slice, and the 3D-cutouts were created using the patch and isonormals commands. External Matlab scripts (via MathWorks) -dbscan.m (M. Daszykowski, University of Silesia) -circfit.m (I. Bucher) -psphere.m (J. Bowman) -spherefit.m (A. Jennings, University of Dayton) S6
7 Figure S1 - Confocal imaging of internalized PS nanoparticles. 80nm NPs are imaged with a confocal microscope to compare with the STORM image reported in Fig.1B. Subdiffraction NPs appears broadened due to the lack of resolution of confocal microscopy. Figure S2 3D reconstruction of internalized PS nanoparticles. 3D STORM imaging of PS nanoparticles and plasma membrane (top panel). Side view of PS nanoparticles (red) and cell membrane (green) highlighting membrane curvature. S7
8 Figure S3 Z sectioning of 330nm nanoparticles in HeLa cells. The 3D imaged reported in Fig. 1E and here in the left panel has been z-sectioned (z-sections of 50nm) to better solve the cluster of NPs internalized. On the right panels two sections (distance about 300nm) are reported highlighting the precise number, a cluster of three in this case, and positioning of the beads in 3D. Figure S4 Confocal z-stacking of internalized PS nanoparticles. 330nm particles internalized in HeLa cells are imaged by confocal microscopy. Z-stacking 3D rendering shows the reconstruction of the particles that due to the lack of z-resolution appears axially elongated. S8
9 Figure S5 STORM colocalization. HeLa cells that internalized 80nm PS nanoparticles (red) are co-stained for specific organelles. The colocalization of the two super resolution channels allows to determine with high accuracy the localization of NPs during intracellular trafficking. Figure S6 Imaging Macropynocytosis events. Conventional (left) and STORM (right) imaging of 220nm NPs interactions with the plasma membrane of HeLa cells. Macropynocytosis membrane ruffles engulfing the NP are imaged. S9
10 Figure S7 Dynamic light scattering and zeta potential. Dynamic light scattering data for the unlabeled and labeled nanoparticles used in this work. For all the particles labeling does not significantly affect NP size. Average size and zeta potentials for all studied NPs are reported in the table. Figure S8 In vitro NP STORM. Super resolution imaging of the NPs presented in this work after adsorption on a glass coverslip. Image size is 4µm. S10
11 Figure S9 Histogram of localizations. Histogram of number of localization per OVAparticle internalized in DC cells. Figure S10 TEM imaging in DC cells. TEM images of individual nanoparticles type internalized in DC cells. S11
Swammerdam Institute for Life Sciences (Universiteit van Amsterdam), 1098 XH Amsterdam, The Netherland
 Sparse deconvolution of high-density super-resolution images (SPIDER) Siewert Hugelier 1, Johan J. de Rooi 2,4, Romain Bernex 1, Sam Duwé 3, Olivier Devos 1, Michel Sliwa 1, Peter Dedecker 3, Paul H. C.
Sparse deconvolution of high-density super-resolution images (SPIDER) Siewert Hugelier 1, Johan J. de Rooi 2,4, Romain Bernex 1, Sam Duwé 3, Olivier Devos 1, Michel Sliwa 1, Peter Dedecker 3, Paul H. C.
Independent Resolution Test of
 Independent Resolution Test of as conducted and published by Dr. Adam Puche, PhD University of Maryland June 2005 as presented by (formerly Thales Optem Inc.) www.qioptiqimaging.com Independent Resolution
Independent Resolution Test of as conducted and published by Dr. Adam Puche, PhD University of Maryland June 2005 as presented by (formerly Thales Optem Inc.) www.qioptiqimaging.com Independent Resolution
TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy
 TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy Introduction The OptiGrid converts the illumination system of a conventional wide
TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy Introduction The OptiGrid converts the illumination system of a conventional wide
Supporting Information. High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images
 Supporting Information High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images Christine R. Laramy, 1, Keith A. Brown, 2, Matthew N. O Brien, 2 and Chad. A.
Supporting Information High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images Christine R. Laramy, 1, Keith A. Brown, 2, Matthew N. O Brien, 2 and Chad. A.
ClusterViSu, a method for clustering of protein complexes by Voronoi. tessellation in super-resolution microscopy
 ClusterViSu, a method for clustering of protein complexes by Voronoi tessellation in super-resolution microscopy Leonid Andronov 1,2,3,4, Igor Orlov 1,2,3,4, Yves Lutz 1,2,3,4, Jean-Luc Vonesch 1,2,3,4,
ClusterViSu, a method for clustering of protein complexes by Voronoi tessellation in super-resolution microscopy Leonid Andronov 1,2,3,4, Igor Orlov 1,2,3,4, Yves Lutz 1,2,3,4, Jean-Luc Vonesch 1,2,3,4,
COLOCALISATION. Alexia Ferrand. Imaging Core Facility Biozentrum Basel
 COLOCALISATION Alexia Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to analyse
COLOCALISATION Alexia Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to analyse
Virtual Frap User Guide
 Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
COLOCALISATION. Alexia Loynton-Ferrand. Imaging Core Facility Biozentrum Basel
 COLOCALISATION Alexia Loynton-Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to
COLOCALISATION Alexia Loynton-Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to
45 µm polystyrene bead embedded in scattering tissue phantom. (a,b) raw images under oblique
 Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
MAT 17C - DISCUSSION #4, Counting Proteins
 MAT 17C - DISCUSSION #4, Counting Proteins Visualization of molecules inside a living cell is one of the most important tools of molecular biology in the 21 st century. In fact, it was the subject of the
MAT 17C - DISCUSSION #4, Counting Proteins Visualization of molecules inside a living cell is one of the most important tools of molecular biology in the 21 st century. In fact, it was the subject of the
Optical Sectioning. Bo Huang. Pharmaceutical Chemistry
 Optical Sectioning Bo Huang Pharmaceutical Chemistry Approaches to 3D imaging Physical cutting Technical difficulty Highest resolution Highest sensitivity Optical sectioning Simple sample prep. No physical
Optical Sectioning Bo Huang Pharmaceutical Chemistry Approaches to 3D imaging Physical cutting Technical difficulty Highest resolution Highest sensitivity Optical sectioning Simple sample prep. No physical
Supporting information SI 0: TEM & STEM acquisition parameters, in 2D and 3D
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Supporting information SI 0: TEM & STEM acquisition parameters, in 2D and 3D Electron tomography
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Supporting information SI 0: TEM & STEM acquisition parameters, in 2D and 3D Electron tomography
MetaMorph Super-Resolution Software. Localization-based super-resolution microscopy
 MetaMorph Super-Resolution Software Localization-based super-resolution microscopy KEY BENEFITS Lateral Resolution up to 20 nm & Axial Resolution up to 40nm Real-time and offline superresolution image
MetaMorph Super-Resolution Software Localization-based super-resolution microscopy KEY BENEFITS Lateral Resolution up to 20 nm & Axial Resolution up to 40nm Real-time and offline superresolution image
FLUOVIEW FV1000/FV1200
 FLUOVIEW FV1000/FV1200 UPGRADE TO 3D NANOIMAGING AND SINGLE MOLECULE TRACKING FOR OLYMPUS FLUOVIEW FV1000/FV1200 Within the past few years, several methods have been devised in order to obtain images with
FLUOVIEW FV1000/FV1200 UPGRADE TO 3D NANOIMAGING AND SINGLE MOLECULE TRACKING FOR OLYMPUS FLUOVIEW FV1000/FV1200 Within the past few years, several methods have been devised in order to obtain images with
Supplementary Figure 1
 Supplementary Figure 1 Experimental unmodified 2D and astigmatic 3D PSFs. (a) The averaged experimental unmodified 2D PSF used in this study. PSFs at axial positions from -800 nm to 800 nm are shown. The
Supplementary Figure 1 Experimental unmodified 2D and astigmatic 3D PSFs. (a) The averaged experimental unmodified 2D PSF used in this study. PSFs at axial positions from -800 nm to 800 nm are shown. The
SPATIAL DENSITY ESTIMATION BASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES
 SPATIAL DENSITY ESTIMATION ASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES Kuan-Chieh Jackie Chen 1,2,3, Ge Yang 1,2, and Jelena Kovačević 3,1,2 1 Dept. of iomedical Eng., 2 Center
SPATIAL DENSITY ESTIMATION ASED SEGMENTATION OF SUPER-RESOLUTION LOCALIZATION MICROSCOPY IMAGES Kuan-Chieh Jackie Chen 1,2,3, Ge Yang 1,2, and Jelena Kovačević 3,1,2 1 Dept. of iomedical Eng., 2 Center
Faster STORM using compressed sensing. Supplementary Software. Nature Methods. Nature Methods: doi: /nmeth.1978
 Nature Methods Faster STORM using compressed sensing Lei Zhu, Wei Zhang, Daniel Elnatan & Bo Huang Supplementary File Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3 Supplementary
Nature Methods Faster STORM using compressed sensing Lei Zhu, Wei Zhang, Daniel Elnatan & Bo Huang Supplementary File Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3 Supplementary
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS.
 SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S. Visualization of Biological Molecules in Their Native Stat e.
 THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
OBCOL. (Organelle Based CO-Localisation) Users Guide
 OBCOL (Organelle Based CO-Localisation) Users Guide INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual
OBCOL (Organelle Based CO-Localisation) Users Guide INTRODUCTION OBCOL is an ImageJ plugin designed to autonomously detect objects within an image (or image stack) and analyse them separately as individual
II: Single particle cryoem - an averaging technique
 II: Single particle cryoem - an averaging technique Radiation damage limits the total electron dose that can be used to image biological sample. Thus, images of frozen hydrated macromolecules are very
II: Single particle cryoem - an averaging technique Radiation damage limits the total electron dose that can be used to image biological sample. Thus, images of frozen hydrated macromolecules are very
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror
 Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
Single-particle electron microscopy (cryo-electron microscopy) CS/CME/BioE/Biophys/BMI 279 Nov. 16 and 28, 2017 Ron Dror 1 Last month s Nobel Prize in Chemistry Awarded to Jacques Dubochet, Joachim Frank
Introductory Guide to Light Microscopy - Biomedical Confocal Microscopy
 Introductory Guide to Light Microscopy - Biomedical Confocal Microscopy 7 May 2007 Michael Hooker Microscopy Facility Michael Chua microscopy@unc.edu 843-3268 6007 Thurston Bowles Wendy Salmon wendy_salmon@med.unc.edu
Introductory Guide to Light Microscopy - Biomedical Confocal Microscopy 7 May 2007 Michael Hooker Microscopy Facility Michael Chua microscopy@unc.edu 843-3268 6007 Thurston Bowles Wendy Salmon wendy_salmon@med.unc.edu
Quantifying Three-Dimensional Deformations of Migrating Fibroblasts
 45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
Quantifying Caveolin-1 Molecular Domains via Machine Learning and Computational Network Analysis of Super-resolution Microscopy Data
 Quantifying Caveolin-1 Molecular Domains via Machine Learning and Computational Network Analysis of Super-resolution Microscopy Data Ismail M. Khater1, Fanrui Meng2, Ivan Robert Nabi2, Ghassan Hamarneh1
Quantifying Caveolin-1 Molecular Domains via Machine Learning and Computational Network Analysis of Super-resolution Microscopy Data Ismail M. Khater1, Fanrui Meng2, Ivan Robert Nabi2, Ghassan Hamarneh1
EMBO Practical Course on Image Processing for Cryo EM 1-11 September 2015
 EMBO Practical Course on Image Processing for Cryo EM 1-11 September 2015 Practical 4: Optional part for experienced IMOD users - Reconstructing a cryo tomogram and sub-tomogram averaging of GroEL IMOD
EMBO Practical Course on Image Processing for Cryo EM 1-11 September 2015 Practical 4: Optional part for experienced IMOD users - Reconstructing a cryo tomogram and sub-tomogram averaging of GroEL IMOD
2, the coefficient of variation R 2, and properties of the photon counts traces
 Supplementary Figure 1 Quality control of FCS traces. (a) Typical trace that passes the quality control (QC) according to the parameters shown in f. The QC is based on thresholds applied to fitting parameters
Supplementary Figure 1 Quality control of FCS traces. (a) Typical trace that passes the quality control (QC) according to the parameters shown in f. The QC is based on thresholds applied to fitting parameters
Colocalization in Confocal Microscopy. Quantitative Colocalization Analysis
 Quantitative Colocalization Analysis 1 Quantitative Colocalization Analysis Colocalization of fluorescently labeled molecules (immunostainings, fluorescent proteins, etc.) is often used as the first indicator
Quantitative Colocalization Analysis 1 Quantitative Colocalization Analysis Colocalization of fluorescently labeled molecules (immunostainings, fluorescent proteins, etc.) is often used as the first indicator
A SUPER-RESOLUTION MICROSCOPY WITH STANDING EVANESCENT LIGHT AND IMAGE RECONSTRUCTION METHOD
 A SUPER-RESOLUTION MICROSCOPY WITH STANDING EVANESCENT LIGHT AND IMAGE RECONSTRUCTION METHOD Hiroaki Nishioka, Satoru Takahashi Kiyoshi Takamasu Department of Precision Engineering, The University of Tokyo,
A SUPER-RESOLUTION MICROSCOPY WITH STANDING EVANESCENT LIGHT AND IMAGE RECONSTRUCTION METHOD Hiroaki Nishioka, Satoru Takahashi Kiyoshi Takamasu Department of Precision Engineering, The University of Tokyo,
Dragonfly Pro. Visual Pathway to Quantitative Answers ORS. Exclusive to ZEISS OBJECT RESEARCH SYSTEMS
 Dragonfly Pro Exclusive to ZEISS Visual Pathway to Quantitative Answers ORS OBJECT RESEARCH SYSTEMS Visualization and analysis software without bounds Dragonfly Pro by Object Research Systems (ORS) is
Dragonfly Pro Exclusive to ZEISS Visual Pathway to Quantitative Answers ORS OBJECT RESEARCH SYSTEMS Visualization and analysis software without bounds Dragonfly Pro by Object Research Systems (ORS) is
Supporting Information: FIB-SEM Tomography Probes the Meso-Scale Pore Space of an Individual Catalytic Cracking Particle
 Supporting Information: FIB-SEM Tomography Probes the Meso-Scale Pore Space of an Individual Catalytic Cracking Particle D.A.Matthijs de Winter, Florian Meirer, Bert M. Weckhuysen*, Inorganic Chemistry
Supporting Information: FIB-SEM Tomography Probes the Meso-Scale Pore Space of an Individual Catalytic Cracking Particle D.A.Matthijs de Winter, Florian Meirer, Bert M. Weckhuysen*, Inorganic Chemistry
Batch Processing of Tomograms with IMOD
 Batch Processing of Tomograms with IMOD Batch Processing Can Be Useful for a Wide Range of Tilt Series Routine plastic section tilt series can be fully reconstructed automatically with 80-95% success rate
Batch Processing of Tomograms with IMOD Batch Processing Can Be Useful for a Wide Range of Tilt Series Routine plastic section tilt series can be fully reconstructed automatically with 80-95% success rate
IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training
 IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training Multi Target Analysis Neurite Outgrowth Membrane Translocation Analysis Object Intensity Dual Area Object Analysis Granularity Morphology
IN Cell Analyzer 1000 Analysis Modules Multi Target Analysis Training Multi Target Analysis Neurite Outgrowth Membrane Translocation Analysis Object Intensity Dual Area Object Analysis Granularity Morphology
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12009 Supplementary Figure 1. Experimental tilt series of 104 projections with a tilt range of ±72.6 and equal slope increments, acquired from a Pt nanoparticle using HAADF- STEM (energy:
doi:10.1038/nature12009 Supplementary Figure 1. Experimental tilt series of 104 projections with a tilt range of ±72.6 and equal slope increments, acquired from a Pt nanoparticle using HAADF- STEM (energy:
DETECTION AND ROBUST ESTIMATION OF CYLINDER FEATURES IN POINT CLOUDS INTRODUCTION
 DETECTION AND ROBUST ESTIMATION OF CYLINDER FEATURES IN POINT CLOUDS Yun-Ting Su James Bethel Geomatics Engineering School of Civil Engineering Purdue University 550 Stadium Mall Drive, West Lafayette,
DETECTION AND ROBUST ESTIMATION OF CYLINDER FEATURES IN POINT CLOUDS Yun-Ting Su James Bethel Geomatics Engineering School of Civil Engineering Purdue University 550 Stadium Mall Drive, West Lafayette,
EPFL SV PTBIOP BIOP COURSE 2015 OPTICAL SLICING METHODS
 BIOP COURSE 2015 OPTICAL SLICING METHODS OPTICAL SLICING METHODS Scanning Methods Wide field Methods Point Scanning Deconvolution Line Scanning Multiple Beam Scanning Single Photon Multiple Photon Total
BIOP COURSE 2015 OPTICAL SLICING METHODS OPTICAL SLICING METHODS Scanning Methods Wide field Methods Point Scanning Deconvolution Line Scanning Multiple Beam Scanning Single Photon Multiple Photon Total
NDD FLIM Systems for Leica SP2 MP and SP5 MP Multiphoton Microscopes
 NDD FLIM Systems for Leica SP2 MP and SP5 MP Multiphoton Microscopes bh FLIM systems for the confocal and the multiphoton versions of the Leica SP2 and SP5 microscopes are available since 2002 [4]. These
NDD FLIM Systems for Leica SP2 MP and SP5 MP Multiphoton Microscopes bh FLIM systems for the confocal and the multiphoton versions of the Leica SP2 and SP5 microscopes are available since 2002 [4]. These
Calibrate the Confocal Volume for FCS Using the FCS Calibration Script
 Tutorial Calibrate the Confocal Volume for FCS Using the FCS Calibration Script Summary This tutorial shows step-by-step, how to calibrate the confocal volume for FCS measurements with a calibration dye.
Tutorial Calibrate the Confocal Volume for FCS Using the FCS Calibration Script Summary This tutorial shows step-by-step, how to calibrate the confocal volume for FCS measurements with a calibration dye.
MICROTUBULE FILAMENT TRACING AND ESTIMATION. Rohan Chabukswar. Department of Electrical and Computer Engineering Carnegie Mellon University
 MICROTUBULE FILAMENT TRACING AND ESTIMATION Rohan Chabukswar Department of Electrical and Computer Engineering Carnegie Mellon University ABSTRACT The project explores preprocessing of images to facilitate
MICROTUBULE FILAMENT TRACING AND ESTIMATION Rohan Chabukswar Department of Electrical and Computer Engineering Carnegie Mellon University ABSTRACT The project explores preprocessing of images to facilitate
Find the Correspondences
 Advanced Topics in BioImage Analysis - 3. Find the Correspondences from Registration to Motion Estimation CellNetworks Math-Clinic Qi Gao 03.02.2017 Math-Clinic core facility Provide services on bioinformatics
Advanced Topics in BioImage Analysis - 3. Find the Correspondences from Registration to Motion Estimation CellNetworks Math-Clinic Qi Gao 03.02.2017 Math-Clinic core facility Provide services on bioinformatics
Effect of OTF attenuation on single-slice SR-SIM reconstruction
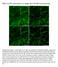 Effect of OTF attenuation on single-slice SR-SIM reconstruction Supplementary Figure 1: Actin filaments in U2OS cells, labelled with Phalloidin-Atto488, measured on a DeltaVision OMX, excited at 488 nm
Effect of OTF attenuation on single-slice SR-SIM reconstruction Supplementary Figure 1: Actin filaments in U2OS cells, labelled with Phalloidin-Atto488, measured on a DeltaVision OMX, excited at 488 nm
Gpufit: An open-source toolkit for GPU-accelerated curve fitting
 Gpufit: An open-source toolkit for GPU-accelerated curve fitting Adrian Przybylski, Björn Thiel, Jan Keller-Findeisen, Bernd Stock, and Mark Bates Supplementary Information Table of Contents Calculating
Gpufit: An open-source toolkit for GPU-accelerated curve fitting Adrian Przybylski, Björn Thiel, Jan Keller-Findeisen, Bernd Stock, and Mark Bates Supplementary Information Table of Contents Calculating
DeltaVision OMX SR super-resolution microscope. Super-resolution doesn t need to be complicated
 DeltaVision OMX SR super-resolution microscope Super-resolution doesn t need to be complicated Live-cell super-resolution microscopy DeltaVision OMX SR is a compact super-resolution microscope system optimized
DeltaVision OMX SR super-resolution microscope Super-resolution doesn t need to be complicated Live-cell super-resolution microscopy DeltaVision OMX SR is a compact super-resolution microscope system optimized
3D RECONSTRUCTION OF PHASE CONTRAST IMAGES USING FOCUS MEASURES.
 3D RECONSTRUCTION OF PHASE CONTRAST IMAGES USING FOCUS MEASURES Vincent On 1, Atena Zahedi 2 and, Bir Bhanu 1,2 1 Dept. of Electrical and Computer Engineering, University of California, Riverside, CA 92521,
3D RECONSTRUCTION OF PHASE CONTRAST IMAGES USING FOCUS MEASURES Vincent On 1, Atena Zahedi 2 and, Bir Bhanu 1,2 1 Dept. of Electrical and Computer Engineering, University of California, Riverside, CA 92521,
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences
 IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
WCIF COLOCALIS ATION PLUGINS
 WCIF COLOCALIS ATION PLUGINS Colocalisation Test Background When a coefficient is calculated for two images, it is often unclear quite what this means, in particular for intermediate values. This raises
WCIF COLOCALIS ATION PLUGINS Colocalisation Test Background When a coefficient is calculated for two images, it is often unclear quite what this means, in particular for intermediate values. This raises
Nonlinear optics and two photon microscopy. Table of contents. Sam Whiteley and Seth Parker PHYS 173/BGGN 266 July 13, 2014
 Nonlinear optics and two photon microscopy Sam Whiteley and Seth Parker PHYS 173/BGGN 266 July 13, 2014 Table of contents 1. Introduction 2. Optical setup 3. Initial images and troubleshooting 4. Determining
Nonlinear optics and two photon microscopy Sam Whiteley and Seth Parker PHYS 173/BGGN 266 July 13, 2014 Table of contents 1. Introduction 2. Optical setup 3. Initial images and troubleshooting 4. Determining
Advanced materials research using the Real-Time 3D Analytical FIB-SEM 'NX9000'
 SCIENTIFIC INSTRUMENT NEWS 2017 Vol. 9 SEPTEMBER Technical magazine of Electron Microscope and Analytical Instruments. Technical Explanation Advanced materials research using the Real-Time 3D Analytical
SCIENTIFIC INSTRUMENT NEWS 2017 Vol. 9 SEPTEMBER Technical magazine of Electron Microscope and Analytical Instruments. Technical Explanation Advanced materials research using the Real-Time 3D Analytical
Single particle tracking: principles and applications
 Single particle tracking: principles and applications Valeria Levi Laboratorio de Dinámica Intracelular Facultad de Ciencias Exactas y Naturales Universidad de Buenos Aires vlevi1@gmail.com Why single
Single particle tracking: principles and applications Valeria Levi Laboratorio de Dinámica Intracelular Facultad de Ciencias Exactas y Naturales Universidad de Buenos Aires vlevi1@gmail.com Why single
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Courtois et al., http://www.jcb.org/cgi/content/full/jcb.201202135/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Measurement of distribution and size of MTOCs in the
Supplemental material Courtois et al., http://www.jcb.org/cgi/content/full/jcb.201202135/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Measurement of distribution and size of MTOCs in the
3D-Analysis of Microstructures with Confocal Laser Scanning Microscopy
 3D-Analysis of Microstructures with Confocal Laser Scanning Microscopy Eckart Uhlmann, Dirk Oberschmidt Institute for Machine Tools and Factory Management (IWF), Technical University Berlin Gerald Kunath-Fandrei
3D-Analysis of Microstructures with Confocal Laser Scanning Microscopy Eckart Uhlmann, Dirk Oberschmidt Institute for Machine Tools and Factory Management (IWF), Technical University Berlin Gerald Kunath-Fandrei
Automatic Partiicle Tracking Software USE ER MANUAL Update: May 2015
 Automatic Particle Tracking Software USER MANUAL Update: May 2015 File Menu The micrograph below shows the panel displayed when a movie is opened, including a playback menu where most of the parameters
Automatic Particle Tracking Software USER MANUAL Update: May 2015 File Menu The micrograph below shows the panel displayed when a movie is opened, including a playback menu where most of the parameters
UNWRAP User Document. Figure 1. Workflow of the UNWRAP application.
 UNWRAP User Document This MATLAB application takes a set of confocal microscope z-stack images of fluorescently labeled cells in a 3D cylindrical geometry and creates an unwrapped 2D image from the tube
UNWRAP User Document This MATLAB application takes a set of confocal microscope z-stack images of fluorescently labeled cells in a 3D cylindrical geometry and creates an unwrapped 2D image from the tube
Digital Laminography and Computed Tomography with 600 kv for Aerospace Applications
 4th International Symposium on NDT in Aerospace 2012 - Tu.3.A.1 Digital Laminography and Computed Tomography with 600 kv for Aerospace Applications Malte KURFISS 1, Gerd STRECKENBACH 2 1 YXLON International
4th International Symposium on NDT in Aerospace 2012 - Tu.3.A.1 Digital Laminography and Computed Tomography with 600 kv for Aerospace Applications Malte KURFISS 1, Gerd STRECKENBACH 2 1 YXLON International
IDENTIFYING OPTICAL TRAP
 IDENTIFYING OPTICAL TRAP Yulwon Cho, Yuxin Zheng 12/16/2011 1. BACKGROUND AND MOTIVATION Optical trapping (also called optical tweezer) is widely used in studying a variety of biological systems in recent
IDENTIFYING OPTICAL TRAP Yulwon Cho, Yuxin Zheng 12/16/2011 1. BACKGROUND AND MOTIVATION Optical trapping (also called optical tweezer) is widely used in studying a variety of biological systems in recent
Fast and accurate automated cell boundary determination for fluorescence microscopy
 Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Improvement of the correlative AFM and ToF-SIMS approach using an empirical sputter model for 3D chemical characterization
 Improvement of the correlative AFM and ToF-SIMS approach using an empirical sputter model for 3D chemical characterization T. Terlier 1, J. Lee 1, K. Lee 2, and Y. Lee 1 * 1 Advanced Analysis Center, Korea
Improvement of the correlative AFM and ToF-SIMS approach using an empirical sputter model for 3D chemical characterization T. Terlier 1, J. Lee 1, K. Lee 2, and Y. Lee 1 * 1 Advanced Analysis Center, Korea
Look Ahead to Get Ahead
 Microscopy from Carl Zeiss Archive 3D Deconvolution Z-stack 3D Deconvolution Look Ahead to Get Ahead Mark & Find Multichannel Timelapse Greater brilliance and resolution More convenient and precise than
Microscopy from Carl Zeiss Archive 3D Deconvolution Z-stack 3D Deconvolution Look Ahead to Get Ahead Mark & Find Multichannel Timelapse Greater brilliance and resolution More convenient and precise than
Bioimage Informatics. Lecture 8, Spring Bioimage Data Analysis (II): Point Feature Detection
 Bioimage Informatics Lecture 8, Spring 2012 Bioimage Data Analysis (II): Point Feature Detection Lecture 8 February 13, 2012 1 Outline Project assignment 02 Comments on reading assignment 01 Review: pixel
Bioimage Informatics Lecture 8, Spring 2012 Bioimage Data Analysis (II): Point Feature Detection Lecture 8 February 13, 2012 1 Outline Project assignment 02 Comments on reading assignment 01 Review: pixel
3D Energy Dispersive Spectroscopy Elemental Tomography in the Scanning Transmission Electron Microscope
 3D Energy Dispersive Spectroscopy Elemental Tomography in the Scanning Transmission Electron Microscope Brian Van Devener Topics 1.Introduction to EDS in the STEM 2.Extending EDS into three dimensions
3D Energy Dispersive Spectroscopy Elemental Tomography in the Scanning Transmission Electron Microscope Brian Van Devener Topics 1.Introduction to EDS in the STEM 2.Extending EDS into three dimensions
Digital Volume Correlation for Materials Characterization
 19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
BME I5000: Biomedical Imaging
 BME I5000: Biomedical Imaging Lecture 1 Introduction Lucas C. Parra, parra@ccny.cuny.edu 1 Content Topics: Physics of medial imaging modalities (blue) Digital Image Processing (black) Schedule: 1. Introduction,
BME I5000: Biomedical Imaging Lecture 1 Introduction Lucas C. Parra, parra@ccny.cuny.edu 1 Content Topics: Physics of medial imaging modalities (blue) Digital Image Processing (black) Schedule: 1. Introduction,
Supporting Information. Characterization of Porous Materials by. Fluorescence Correlation Spectroscopy Superresolution. Optical Fluctuation Imaging
 Supporting Information Characterization of Porous Materials by Fluorescence Correlation Spectroscopy Superresolution Optical Fluctuation Imaging Lydia Kisley, 1 Rachel Brunetti, 2 Lawrence J. Tauzin, 1
Supporting Information Characterization of Porous Materials by Fluorescence Correlation Spectroscopy Superresolution Optical Fluctuation Imaging Lydia Kisley, 1 Rachel Brunetti, 2 Lawrence J. Tauzin, 1
Colocalisation/Correlation
 Colocalisation/Correlation The past: I see yellow - therefore there is colocalisation but published images look over exposed. No colocalisation definition + No stats = No Science. From Now On: 3D. Quantification.
Colocalisation/Correlation The past: I see yellow - therefore there is colocalisation but published images look over exposed. No colocalisation definition + No stats = No Science. From Now On: 3D. Quantification.
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences
 Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris A Brief History Imaris was created 25 years ago to focus purely on the visualization of 3D fluorescence images
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris A Brief History Imaris was created 25 years ago to focus purely on the visualization of 3D fluorescence images
EN1610 Image Understanding Lab # 3: Edges
 EN1610 Image Understanding Lab # 3: Edges The goal of this fourth lab is to ˆ Understanding what are edges, and different ways to detect them ˆ Understand different types of edge detectors - intensity,
EN1610 Image Understanding Lab # 3: Edges The goal of this fourth lab is to ˆ Understanding what are edges, and different ways to detect them ˆ Understand different types of edge detectors - intensity,
Live-cell 3D super-resolution imaging in thick biological samples
 Nature Methods Live-cell 3D super-resolution imaging in thick biological samples Francesca Cella Zanacchi, Zeno Lavagnino, Michela Perrone Donnorso, Alessio Del Bue, Lauria Furia, Mario Faretta & Alberto
Nature Methods Live-cell 3D super-resolution imaging in thick biological samples Francesca Cella Zanacchi, Zeno Lavagnino, Michela Perrone Donnorso, Alessio Del Bue, Lauria Furia, Mario Faretta & Alberto
Supplementary document to Programming 2D/3D shape-shifting with hobbyist 3D printers
 Electronic Supplementary Material (ESI) for Materials Horizons. This journal is The Royal Society of Chemistry 2017 Supplementary document to Programming 2D/3D shape-shifting with hobbyist 3D printers
Electronic Supplementary Material (ESI) for Materials Horizons. This journal is The Royal Society of Chemistry 2017 Supplementary document to Programming 2D/3D shape-shifting with hobbyist 3D printers
Raster Image Correlation Spectroscopy and Number and Brightness Analysis
 Raster Image Correlation Spectroscopy and Number and Brightness Analysis 15 Principles of Fluorescence Course Paolo Annibale, PhD On behalf of Dr. Enrico Gratton lfd Raster Image Correlation Spectroscopy
Raster Image Correlation Spectroscopy and Number and Brightness Analysis 15 Principles of Fluorescence Course Paolo Annibale, PhD On behalf of Dr. Enrico Gratton lfd Raster Image Correlation Spectroscopy
ND Processing Tools in NIS-Elements
 ND Processing Tools in NIS-Elements Overview This technical note describes basic uses of the ND processing tools available in NIS-Elements. These tools are specifically designed for arithmetic functions
ND Processing Tools in NIS-Elements Overview This technical note describes basic uses of the ND processing tools available in NIS-Elements. These tools are specifically designed for arithmetic functions
Supporting Information
 Supporting Information Gelbart et al. 10.1073/pnas.1205258109 SI Text Description of Embryo Development Geometry Explorer. This section describes in detail the five main steps of the Embryo Development
Supporting Information Gelbart et al. 10.1073/pnas.1205258109 SI Text Description of Embryo Development Geometry Explorer. This section describes in detail the five main steps of the Embryo Development
Resting state network estimation in individual subjects
 Resting state network estimation in individual subjects Data 3T NIL(21,17,10), Havard-MGH(692) Young adult fmri BOLD Method Machine learning algorithm MLP DR LDA Network image Correlation Spatial Temporal
Resting state network estimation in individual subjects Data 3T NIL(21,17,10), Havard-MGH(692) Young adult fmri BOLD Method Machine learning algorithm MLP DR LDA Network image Correlation Spatial Temporal
Epithelial rosette detection in microscopic images
 Epithelial rosette detection in microscopic images Kun Liu,3, Sandra Ernst 2,3, Virginie Lecaudey 2,3 and Olaf Ronneberger,3 Department of Computer Science 2 Department of Developmental Biology 3 BIOSS
Epithelial rosette detection in microscopic images Kun Liu,3, Sandra Ernst 2,3, Virginie Lecaudey 2,3 and Olaf Ronneberger,3 Department of Computer Science 2 Department of Developmental Biology 3 BIOSS
Alignment and Other Challenges in Reconstructing Cryotomograms with IMOD
 Alignment and Other Challenges in Reconstructing Cryotomograms with IMOD Challenges in Cryotomography Alignment, alignment, alignment It can be hard to get fiducials onto/in the sample The low SNR makes
Alignment and Other Challenges in Reconstructing Cryotomograms with IMOD Challenges in Cryotomography Alignment, alignment, alignment It can be hard to get fiducials onto/in the sample The low SNR makes
Assembly dynamics of microtubules at molecular resolution
 Supplementary Information with: Assembly dynamics of microtubules at molecular resolution Jacob W.J. Kerssemakers 1,2, E. Laura Munteanu 1, Liedewij Laan 1, Tim L. Noetzel 2, Marcel E. Janson 1,3, and
Supplementary Information with: Assembly dynamics of microtubules at molecular resolution Jacob W.J. Kerssemakers 1,2, E. Laura Munteanu 1, Liedewij Laan 1, Tim L. Noetzel 2, Marcel E. Janson 1,3, and
Note: FLIM-FRET is a robust method to determine the FRET efficiency of a suited donor acceptor pair.
 Tutorial FLIM-FRET-Calculation for Multi-Exponential Donors Summary This tutorial shows step-by-step, how the "Lifetime FRET Image" script of SymPhoTime 64 can be used to calculate pixel-by-pixel the average
Tutorial FLIM-FRET-Calculation for Multi-Exponential Donors Summary This tutorial shows step-by-step, how the "Lifetime FRET Image" script of SymPhoTime 64 can be used to calculate pixel-by-pixel the average
WETTING PROPERTIES OF STRUCTURED INTERFACES COMPOSED OF SURFACE-ATTACHED SPHERICAL NANOPARTICLES
 November 20, 2018 WETTING PROPERTIES OF STRUCTURED INTERFACES COMPOSED OF SURFACE-ATTACHED SPHERICAL NANOPARTICLES Bishal Bhattarai and Nikolai V. Priezjev Department of Mechanical and Materials Engineering
November 20, 2018 WETTING PROPERTIES OF STRUCTURED INTERFACES COMPOSED OF SURFACE-ATTACHED SPHERICAL NANOPARTICLES Bishal Bhattarai and Nikolai V. Priezjev Department of Mechanical and Materials Engineering
BSI scmos. High Quantum Efficiency. Cooled Scientific CMOS Camera
 95 BSI scmos High Quantum Efficiency Cooled Scientific CMOS Camera Backside-illuminated scmos technology Opening a new era of high sensitivity imaging applications! The Dhyana 95 is a highly sensitive
95 BSI scmos High Quantum Efficiency Cooled Scientific CMOS Camera Backside-illuminated scmos technology Opening a new era of high sensitivity imaging applications! The Dhyana 95 is a highly sensitive
BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening. Alex Laude The BioImaging Unit
 BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening Alex Laude The BioImaging Unit Multi-dimensional, multi-modal imaging at the sub-cellular
BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening Alex Laude The BioImaging Unit Multi-dimensional, multi-modal imaging at the sub-cellular
Advanced Image Reconstruction Methods for Photoacoustic Tomography
 Advanced Image Reconstruction Methods for Photoacoustic Tomography Mark A. Anastasio, Kun Wang, and Robert Schoonover Department of Biomedical Engineering Washington University in St. Louis 1 Outline Photoacoustic/thermoacoustic
Advanced Image Reconstruction Methods for Photoacoustic Tomography Mark A. Anastasio, Kun Wang, and Robert Schoonover Department of Biomedical Engineering Washington University in St. Louis 1 Outline Photoacoustic/thermoacoustic
Coherent Gradient Sensing Microscopy: Microinterferometric Technique. for Quantitative Cell Detection
 Coherent Gradient Sensing Microscopy: Microinterferometric Technique for Quantitative Cell Detection Proceedings of the SEM Annual Conference June 7-10, 010 Indianapolis, Indiana USA 010 Society for Experimental
Coherent Gradient Sensing Microscopy: Microinterferometric Technique for Quantitative Cell Detection Proceedings of the SEM Annual Conference June 7-10, 010 Indianapolis, Indiana USA 010 Society for Experimental
A Constrained Delaunay Triangle Mesh Method for Three-Dimensional Unstructured Boundary Point Cloud
 International Journal of Computer Systems (ISSN: 2394-1065), Volume 03 Issue 02, February, 2016 Available at http://www.ijcsonline.com/ A Constrained Delaunay Triangle Mesh Method for Three-Dimensional
International Journal of Computer Systems (ISSN: 2394-1065), Volume 03 Issue 02, February, 2016 Available at http://www.ijcsonline.com/ A Constrained Delaunay Triangle Mesh Method for Three-Dimensional
CS 231A Computer Vision (Winter 2014) Problem Set 3
 CS 231A Computer Vision (Winter 2014) Problem Set 3 Due: Feb. 18 th, 2015 (11:59pm) 1 Single Object Recognition Via SIFT (45 points) In his 2004 SIFT paper, David Lowe demonstrates impressive object recognition
CS 231A Computer Vision (Winter 2014) Problem Set 3 Due: Feb. 18 th, 2015 (11:59pm) 1 Single Object Recognition Via SIFT (45 points) In his 2004 SIFT paper, David Lowe demonstrates impressive object recognition
Workflow 1. Description
 Workflow 1 Description Determine protein staining intensities and distances in mitotic apparatus in z-stack intensity images, which were stained for the inner-centromere protein INCENP, and compare the
Workflow 1 Description Determine protein staining intensities and distances in mitotic apparatus in z-stack intensity images, which were stained for the inner-centromere protein INCENP, and compare the
Developments in Dimensional Metrology in X-ray Computed Tomography at NPL
 Developments in Dimensional Metrology in X-ray Computed Tomography at NPL Wenjuan Sun and Stephen Brown 10 th May 2016 1 Possible factors influencing XCT measurements Components Influencing variables Possible
Developments in Dimensional Metrology in X-ray Computed Tomography at NPL Wenjuan Sun and Stephen Brown 10 th May 2016 1 Possible factors influencing XCT measurements Components Influencing variables Possible
3-D. Here red spheres show the location of gold nanoparticles inside/around a cell nucleus.
 3-D The CytoViva 3-D System allows the user can locate objects of interest in a 3-D space. It does this by acquiring multiple Z planes and performing our custom software routines to locate and observe
3-D The CytoViva 3-D System allows the user can locate objects of interest in a 3-D space. It does this by acquiring multiple Z planes and performing our custom software routines to locate and observe
3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen
 3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen Dataset: Huh7.tif (original), obtained by Serial Block Face-SEM (Gatan 3View) Huh7_crop.tif
3D-Modeling with Microscopy Image Browser (im_browser) (pdf version) Ilya Belevich, Darshan Kumar, Helena Vihinen Dataset: Huh7.tif (original), obtained by Serial Block Face-SEM (Gatan 3View) Huh7_crop.tif
Comparison of Cost Function Against Positional Analysis for Automated Tracking of Three-Cell Interactions
 Syracuse University SURFACE Dissertations - ALL SURFACE January 2015 Comparison of Cost Function Against Positional Analysis for Automated Tracking of Three-Cell Interactions Ye He Syracuse University
Syracuse University SURFACE Dissertations - ALL SURFACE January 2015 Comparison of Cost Function Against Positional Analysis for Automated Tracking of Three-Cell Interactions Ye He Syracuse University
Digital Image Processing
 Digital Image Processing SPECIAL TOPICS CT IMAGES Hamid R. Rabiee Fall 2015 What is an image? 2 Are images only about visual concepts? We ve already seen that there are other kinds of image. In this lecture
Digital Image Processing SPECIAL TOPICS CT IMAGES Hamid R. Rabiee Fall 2015 What is an image? 2 Are images only about visual concepts? We ve already seen that there are other kinds of image. In this lecture
Algorithm User Guide:
 Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
CS229: Machine Learning
 Project Title: Recognition and Classification of human Embryonic Stem (hes) Cells Differentiation Level Team members Dennis Won (jwon2014), Jeong Soo Sim (digivice) Project Purpose 1. Given an image of
Project Title: Recognition and Classification of human Embryonic Stem (hes) Cells Differentiation Level Team members Dennis Won (jwon2014), Jeong Soo Sim (digivice) Project Purpose 1. Given an image of
Technology and equipment at the Bioimaging Center
 Technology and equipment at the Bioimaging Center Light microscopy / Widefields Name Type Applications and equipment LEICA AF6000LX Nikon BioStation Timelaps imaging, CO2 and temperature controlled (14
Technology and equipment at the Bioimaging Center Light microscopy / Widefields Name Type Applications and equipment LEICA AF6000LX Nikon BioStation Timelaps imaging, CO2 and temperature controlled (14
Correlative Light/Electron Microscopy
 Correlative Light/Electron Microscopy Dr. Guenter Resch Leica Beijing HPF Workshop, October 2014 Contents Overview Leica EM PACT2 CLEM Workflow Leica EM HPM100 CLEM Workflow Leica Cryo-LM Stage Section
Correlative Light/Electron Microscopy Dr. Guenter Resch Leica Beijing HPF Workshop, October 2014 Contents Overview Leica EM PACT2 CLEM Workflow Leica EM HPM100 CLEM Workflow Leica Cryo-LM Stage Section
TUTORIAL - [MITOCHONDRIAL MEMBRANE POTENTIAL MEASUREMENT] July 26, 2015
![TUTORIAL - [MITOCHONDRIAL MEMBRANE POTENTIAL MEASUREMENT] July 26, 2015 TUTORIAL - [MITOCHONDRIAL MEMBRANE POTENTIAL MEASUREMENT] July 26, 2015](/thumbs/84/89490383.jpg) Example data set: MitoMemPot1.zip Pipelines used: Mitochondrial membrane potential measurement (TMRM/PMPI) QUICK START GUIDE 1. Unzip contents of MitoMemPot1.zip onto your hard drive. 2. Open the recording
Example data set: MitoMemPot1.zip Pipelines used: Mitochondrial membrane potential measurement (TMRM/PMPI) QUICK START GUIDE 1. Unzip contents of MitoMemPot1.zip onto your hard drive. 2. Open the recording
Quantitative imaging of human red blood cells infected with Plasmodium falciparum
 Biophysical Journal, Volume 99 Supporting Material Quantitative imaging of human red blood cells infected with Plasmodium falciparum Alessandro Esposito, Jean-Baptiste Choimet, Jeremy N. Skepper, Jakob
Biophysical Journal, Volume 99 Supporting Material Quantitative imaging of human red blood cells infected with Plasmodium falciparum Alessandro Esposito, Jean-Baptiste Choimet, Jeremy N. Skepper, Jakob
Summary of Publicly Released CIPS Data Versions.
 Summary of Publicly Released CIPS Data Versions. Last Updated 13 May 2012 V3.11 - Baseline data version, available before July 2008 All CIPS V3.X data versions followed the data processing flow and data
Summary of Publicly Released CIPS Data Versions. Last Updated 13 May 2012 V3.11 - Baseline data version, available before July 2008 All CIPS V3.X data versions followed the data processing flow and data
1. ABOUT INSTALLATION COMPATIBILITY SURESIM WORKFLOWS a. Workflow b. Workflow SURESIM TUTORIAL...
 SuReSim manual 1. ABOUT... 2 2. INSTALLATION... 2 3. COMPATIBILITY... 2 4. SURESIM WORKFLOWS... 2 a. Workflow 1... 3 b. Workflow 2... 4 5. SURESIM TUTORIAL... 5 a. Import Data... 5 b. Parameter Selection...
SuReSim manual 1. ABOUT... 2 2. INSTALLATION... 2 3. COMPATIBILITY... 2 4. SURESIM WORKFLOWS... 2 a. Workflow 1... 3 b. Workflow 2... 4 5. SURESIM TUTORIAL... 5 a. Import Data... 5 b. Parameter Selection...
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences
 Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris Imaris State of the Art Image Visualization and Analysis Over the last 25 years Imaris has continuously improved
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris Imaris State of the Art Image Visualization and Analysis Over the last 25 years Imaris has continuously improved
Supporting Information for Critical Factors of the 3D Microstructural Formation. in Hybrid Conductive Adhesive Materials by X-ray Nano-tomography
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 Supporting Information for Critical Factors of the 3D Microstructural Formation in Hybrid Conductive
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 Supporting Information for Critical Factors of the 3D Microstructural Formation in Hybrid Conductive
The latest trend of hybrid instrumentation
 Multivariate Data Processing of Spectral Images: The Ugly, the Bad, and the True The results of various multivariate data-processing methods of Raman maps recorded with a dispersive Raman microscope are
Multivariate Data Processing of Spectral Images: The Ugly, the Bad, and the True The results of various multivariate data-processing methods of Raman maps recorded with a dispersive Raman microscope are
