Workshop #7: Docking. Suggested Readings
|
|
|
- Rachel Porter
- 6 years ago
- Views:
Transcription
1 Workshop #7: Docking Protein protein docking is the prediction of a complex structure starting from its monomer components. The search space can be extremely large, so large amounts of computational resources are typically required. In this workshop, we will explore several of the techniques briefly; keep in mind that for real applications, many more decoys will need to be tested. Suggested Readings 1. J. J. Gray et al., Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations, J. Mol. Biol. 331, (2003). 2. S. Chaudhury & J. J. Gray, Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles, J. Mol. Biol. 381, (2008). Fast Fourier Transform Based Docking via ZDOCK There are several servers available based on fast Fourier transforms (FFTs). These servers are able to quickly carry out a global, grid-based matching searches. 1. Go to the ZDOCK server ( and upload trypsin (2PTN) and its inhibitor (1BA7 chain B) for docking. If completing this workshop for a class, do this in groups in order to not overload the server. When the jobs have finished (typically under an hour), download the output file. You will have to also download a script for creating complexes from the output file. Use the script to generate the top five models. Are these models similar or diverse? How so? 2. Are any of the models similar to the crystal structure of the bound complex (1AVW)? (Other servers include SmoothDock ( ClusPro ( Haddock ( and GRAMM-X ( Any of these provide global docking services to create models that might be useful for refinement by RosettaDock.) Docking Moves in Rosetta For the following exercises, download and clean the complex of colicin D and ImmD (1V74). Store three poses a full-atom starting pose and centroid and full-atom working poses. 45
2 46 Workshop #7: Docking The fundamental docking move is a rigid-body transformation consisting of a translation and rotation. Any rigid body move also needs to know which part moves and which part is fixed. In Rosetta, this division is known as a jump and the set of protein segments and jumps are stored in an object attached to a pose called a fold tree. print pose.fold_tree() In the fold tree printout, each three number sequence following the word EDGE is the beginning and ending residue number, then a code. The codes are -1 for stretches of protein and any positive integer for a jump, which represents the jump number. 3. View the fold tree of your full-atom pose. How many jumps are there in your pose? By default, there is a jump between the N-terminus of chain A and the N-terminus of chain B, but we can change this using the exposed method setup_foldtree(). from pyrosetta.teaching import * setup_foldtree(pose, "A_B", Vector1([1])) print pose.fold_tree() The argument "A_B" tells Rosetta to make chain A the rigid chain and allow chain B to move. If there were more chains in the pdb structure, supplying "AB_C" would hold chains A and B rigid together as a single unit and allow chain C to move. (The third argument Vector1([1]) is required. The function Vector1() creates a Rosetta vector object indexed from 1 from a Python list. In this case, we pass a list with one element that identifies the first jump in the fold tree for docking use.) 4. Set up a new fold tree for docking using the command above and output the new fold tree. What has changed? You can see the type of information in the jump by printing it from the pose: jump_num = 1 print pose.jump(jump_num).get_rotation() print pose.jump(jump_num).get_translation() 5. Write out the rotation matrix and the translation vector defined by the jump.!! # # $ $ (,, )
3 Workshop #7: Docking 47 The two basic manipulations are translations and rotations. For translation, the change in x, y, and z coordinates are needed as well as the jump number. A rotation requires a center and an axis about which to rotate. The rigid-body displacement can be altered directly with the RigidBodyTransMover for translations or the RigidBodySpinMover for rotations. However, for structure prediction calculations, we have a mover that is preconfigured to make random movements varying around set magnitudes (in this case, a mean of 8 rotation and 3 Å translation) located in the rosetta.protocols.rigid namespace, (which we will rename with an alias rigid_moves for our convenience) : import rosetta.protocols.rigid as rigid_moves pert_mover = rigid_moves.rigidbodyperturbmover(jump_num, 8, 3) 6. Apply the RigidBodyPerturbMover to a pose and use a PyMOLMover to confirm that the motions are what you expect. What are the new rotation matrix and translation vector in the jump? How many ångströms did the downstream protein move? Global perturbations are useful for making completely randomized starting structures. The following mover will rotate a protein about its geometric center. The final orientation is equally distributed over the globe. randomize1 = rigid_moves.rigidbodyrandomizemover(pose, jump_num, rigid_moves.partner_upstream) randomize2 = rigid_moves.rigidbodyrandomizemover(pose, jump_num, rigid_moves.partner_downstream) (partner_upstream and partner_downstream are predefined terms within the rosetta.protocols.rigid namespace, which in our case refer to chains A and B, respectively.) 7. Apply both movers to the starting structure, and view the structure in PyMOL. (You might view it along with the original pose.) Does the new conformation look like a candidate docked structure yet? Since proteins are not spherical, sometimes the random orientation creates severe clashes between the docking partners, and other times it places the partners so they are no longer
4 48 Workshop #7: Docking touching. The FaDockingSlideIntoContact mover will translate the downstream protein along the line of protein centers until the proteins are in contact. from rosetta.protocols.docking import * slide = DockingSlideIntoContact(jump_num) # for centroid mode slide = FaDockingSlideIntoContact(jump_num) # for fullatom mode slide.apply(pose) The MinMover, which we have previously used to change torsion angles to find the nearest minimum in the score function, can also operate on the jump translation and rotation. It suffices to set the jump variable as moveable in the MoveMap: movemap = MoveMap() movemap.set_jump(jump_num, True) min_mover = MinMover() min_mover.movemap(movemap) min_mover.score_function(scorefxn) # use any scorefxn scorefxn(pose) min_mover.apply(pose) 8. Apply the above MinMover. How much does the score change? What are the new rotation matrix and translation vector in the jump? How many Ångstroms did the downstream protein move? Low-Resolution Docking via RosettaDock RosettaDock can also perform global docking runs, but it can require significant time. Typically, 10 5 to 10 6 decoys are needed in a global run. For this workshop, we will create a much smaller number and learn the tools needed to handle large runs. Docking is available as a mover that completely encompasses the protocol. To use the mover, you will need a starting pose with both chains and a jump defined. The structure must be in low-resolution (centroid) mode, and you will need the low-resolution docking score function: scorefxn_low = create_score_function("interchain_cen")
5 Workshop #7: Docking 49 Randomize your centroid version of the complex. Then, create low-resolution docking structures as follows: dock_lowres = DockingLowRes(scorefxn_low, jump_num) dock_lowres.apply(pose) 9. You can compare structures by calculating the root-mean-squared deviation of all the C α atoms, using the function CA_rmsd(pose1, pose2). In docking, a more useful measure is the ligand RMSD, which is the deviation of the backbone C α atoms of the ligand after superposition of the receptor protein backbones. You can calculate ligand RMSD with calc_lrmsd(pose1, pose2, Vector1([1])). Using both measures, how far did your pose move from the low-resolution search? 10. Examine the created decoy in PyMOL. Does it look like a reasonable structure for a protein-protein complex? Explain. Job Distributor For exhaustive searches with Rosetta (docking, refinement, or folding), it is necessary to create a large number of candidate structures, termed decoys. This is often accomplished by spreading out the work over a large number of computers. Additionally, each decoy created needs to be individually labeled. The object that is responsible for managing the output is called a JobDistributor. Here, we will use a simple job distributor to create multiple structures. The following constructor sets the job distributor to create 10 decoys, with filenames like output_1.pdb, output_2.pdb, etc. The pdb files will also include scores according to the ScoreFunction provided. from pyrosetta.teaching import * jd = PyJobDistributor("output", 10, scorefxn_low) It is also useful to compare each decoy to the native structure (if it is known; otherwise any reference structure can be used). The job distributor will do the RMSD calculation and final scoring upon output. To set the native pose: native_pose = pose_from_pdb("your_favorite_protein.pdb") jd.native_pose = native_pose
6 50 Workshop #7: Docking 11. Create a randomized starting pose, working pose, fold tree, score function, job distributor, and low-resolution docking mover. Now, run the low-resolution docking protocol to create a structure, and output a decoy: pose.assign(starting_pose) dock_lowres.apply(pose) jd.output_decoy(pose) Do this twice and confirm that you have two output files. Whenever the output_decoy() method is called, the current_num variable of the JobDistributor is incremented, and it also outputs an updated score file: output.fasc. We can finish the set of 10 decoys by using the JobDistributor to set up a loop: while not jd.job_complete: pose.assign(starting_pose) dock_lowres.apply(pose) jd.output_decoy(pose) Note the jd.job_complete Boolean variable that indicates whether all 10 decoys have been created. 12. Run the loop to create 10 structures. The score file, output.fasc summarizes the energies and RMSDs of all structures created. Examine that file. What is the lowest score? What is the lowest energy? 13. Reset the JobDistributor to create 100 decoys (or more or less, as the speed of your processor allows) by reconstructing it. Rerun the loop above to make 100 decoys. Use your score file to plot score versus RMSD. (Two easy ways to do this are to import the score file into Excel or to use the Linux command gnuplot.) Do you see an energy funnel? Error! Reference source not found. includes more helpful information about using the PyJobDistributor to distribute jobs over multiple computers. High-Resolution Docking The high-resolution stage of RosettaDock is also available as a mover. This mover encompasses random rigid-body moves, side-chain packing, and gradient-based minimization in the rigid-body coordinates. High-resolution docking needs an all-atom score function. The optimized docking weights are available as a patch to the standard all-atom energy function. scorefxn_high = create_score_function_ws_patch( "talaris2013", "docking") dock_hires = DockMCMProtocol() dock_hires.set_scorefxn(scorefxn_high) dock_hires.set_partners("a_b")
7 Workshop #7: Docking 51 Note that unlike for DockingLowRes, we must supply the docking partners with "A_B" instead of jump_num. A high-resolution decoy needs side chains. One way to place the side chains is to call the PackMover, which will generate a conformation from rotamers. A second way is to copy the side chains from the original monomer structures. This is often helpful for docking calculations since the monomer crystal structures have good side chain positions. recover_sidechains = ReturnSidechainMover(starting_pose) recover_sidechains.apply(pose) 14. Load one of your low-resolution decoys, add the side chains from the starting pose, and refine the decoy using high-resolution docking. How far did the structure move during refinement? How much did the score improve? 15. Starting from your lowest-scoring low-resolution decoy, create three high-resolution decoys. (You might use the JobDistributor.) Do the same starting from the native structure. a. How do the refined-native scores compare to the refined-decoy scores? b. What is the RMSD of the refined native? Why is it not zero? c. How much variation do you see in the refined native scores? In the refined decoy scores? Is the difference between the refined natives and the refined decoys significant? Docking Funnel Using a job distributor and DockMCMProtocol, create 10 decoys starting with a RigidBodyRandomizeMover perturbation of partner_downstream, 10 decoys starting from different local random perturbations (8, 3 Å), 10 decoys starting from low-resolution decoys, and 10 starting from the native structure. Plot all of these points on a funnel plot. How is the sampling from each method? Does the scoring function discriminate good complexes?
8 52 Workshop #7: Docking Programming Exercises 1. Output a structure with a 10 Å translation and another with a 30 rotation (both starting from the same starting structure), and load them into PyMOL to confirm the motions are what you expect. 2. Diffusion. Make a series of random rigid body perturbations and record the RMSD after each. Plot RMSD versus the number of moves. Does this process emulate diffusion? If it did, how would you know? (Hint: there is a way to plot these data to make them linear.) 3. Starting from a low-resolution docking decoy, refine the structure in three separate ways: a. side-chain packing b. gradient-based minimization in the rigid-body coordinates c. gradient-based minimization in the torsional coordinates d. the docking high-resolution protocol For each, note the change in RMSD and the change in score. Which operations move the protein the most? Which make the most difference in the score? 4. Using the MonteCarlo object, the RigidBodyMover, PackRotamers, and the MinMover, create your own high-resolution docking protocol. Bonus: Can you tune it to beat the standard protocol? Beating the standard protocol could mean achieving lower energies, running in faster time, and/or being able to better predict complexes.
Workshop #8: Loop Modeling
 Workshop #8: Loop Modeling Loop modeling is an important step in building homology models, designing enzymes, or docking with flexible loops. Suggested Readings 1. A. A. Canutescu & R. L. Dunbrack, Cyclic
Workshop #8: Loop Modeling Loop modeling is an important step in building homology models, designing enzymes, or docking with flexible loops. Suggested Readings 1. A. A. Canutescu & R. L. Dunbrack, Cyclic
Mac OS X or Linux Python 2.7 Rosetta (https://www.rosettacommons.org/downloads/academic/2017/wk12) PyRosetta4 (http://www.pyrosetta.
 Hfq Rosetta FloppyTail Supplemental Protocol Materials and Equipment Mac OS X or Linux Python 2.7 Rosetta 2017.12 (https://www.rosettacommons.org/downloads/academic/2017/wk12) PyRosetta4 (http://www.pyrosetta.org/dow)
Hfq Rosetta FloppyTail Supplemental Protocol Materials and Equipment Mac OS X or Linux Python 2.7 Rosetta 2017.12 (https://www.rosettacommons.org/downloads/academic/2017/wk12) PyRosetta4 (http://www.pyrosetta.org/dow)
2) Model refinement via iterative local rebuilding
 2) Model refinement via iterative local rebuilding In this section, we introduce our cryoem refinement protocol, which uses an iterative local rebuilding procedure to escape local minima during refinement.
2) Model refinement via iterative local rebuilding In this section, we introduce our cryoem refinement protocol, which uses an iterative local rebuilding procedure to escape local minima during refinement.
Rosetta3.x as an library/api
 Rosetta3.x as an library/api With examples from an E2/E3 system Suggested reading for the tutorial: http://www.rosettacommons.org/manuals/rosetta3 _user_guide/adv_overview.html http://www.rosettacommons.org/manuals/rosetta3
Rosetta3.x as an library/api With examples from an E2/E3 system Suggested reading for the tutorial: http://www.rosettacommons.org/manuals/rosetta3 _user_guide/adv_overview.html http://www.rosettacommons.org/manuals/rosetta3
Computer Lab, Session 1
 Computer Lab, Session 1 1 Log in Please log in with username VSDD0xy where xy is your computer number ranging from 01, 02,, 20 2 Settings Open terminal In home directory (initial directory): cp /export/home/vsdd/vsdd001/.bashrc.
Computer Lab, Session 1 1 Log in Please log in with username VSDD0xy where xy is your computer number ranging from 01, 02,, 20 2 Settings Open terminal In home directory (initial directory): cp /export/home/vsdd/vsdd001/.bashrc.
A Parallel Implementation of a Higher-order Self Consistent Mean Field. Effectively solving the protein repacking problem is a key step to successful
 Karl Gutwin May 15, 2005 18.336 A Parallel Implementation of a Higher-order Self Consistent Mean Field Effectively solving the protein repacking problem is a key step to successful protein design. Put
Karl Gutwin May 15, 2005 18.336 A Parallel Implementation of a Higher-order Self Consistent Mean Field Effectively solving the protein repacking problem is a key step to successful protein design. Put
Molecular docking tutorial
 Molecular docking tutorial Sulfonamide-type D-Glu inhibitor docked into the MurD active site using ArgusLab In this tutorial [1] you will learn how to prepare and run molecular docking calculations using
Molecular docking tutorial Sulfonamide-type D-Glu inhibitor docked into the MurD active site using ArgusLab In this tutorial [1] you will learn how to prepare and run molecular docking calculations using
Workshop #11: Custom Movers & Energy Methods
 Workshop #11: Custom Movers & Energy Methods You may have noticed that every mover we have introduced in the preceding chapters has shared an apply() method, which takes a pose as its argument. One of
Workshop #11: Custom Movers & Energy Methods You may have noticed that every mover we have introduced in the preceding chapters has shared an apply() method, which takes a pose as its argument. One of
EMBO Practical Course on Image Processing for Cryo EM 4 14 September Practical 8: Fitting atomic structures into EM maps
 EMBO Practical Course on Image Processing for Cryo EM 4 14 September 2005 Practical 8: Fitting atomic structures into EM maps The best way to interpret the EM density map is to build a hybrid structure,
EMBO Practical Course on Image Processing for Cryo EM 4 14 September 2005 Practical 8: Fitting atomic structures into EM maps The best way to interpret the EM density map is to build a hybrid structure,
A FILTERING TECHNIQUE FOR FRAGMENT ASSEMBLY- BASED PROTEINS LOOP MODELING WITH CONSTRAINTS
 A FILTERING TECHNIQUE FOR FRAGMENT ASSEMBLY- BASED PROTEINS LOOP MODELING WITH CONSTRAINTS F. Campeotto 1,2 A. Dal Palù 3 A. Dovier 2 F. Fioretto 1 E. Pontelli 1 1. Dept. Computer Science, NMSU 2. Dept.
A FILTERING TECHNIQUE FOR FRAGMENT ASSEMBLY- BASED PROTEINS LOOP MODELING WITH CONSTRAINTS F. Campeotto 1,2 A. Dal Palù 3 A. Dovier 2 F. Fioretto 1 E. Pontelli 1 1. Dept. Computer Science, NMSU 2. Dept.
Workshop #2: PyRosetta
 Workshop #2: PyRosetta Rosetta is a suite of algorithms for biomolecular structure prediction and design. Rosetta is written in C++ and is available from www.rosettacommons.org. PyRosetta is a toolkit
Workshop #2: PyRosetta Rosetta is a suite of algorithms for biomolecular structure prediction and design. Rosetta is written in C++ and is available from www.rosettacommons.org. PyRosetta is a toolkit
Small-Molecule Ligand Docking into Comparative Models with Rosetta
 Supplementary Information Small-Molecule Ligand Docking into Comparative Models with Rosetta Steven A. Combs 1,2,*, Samuel L. DeLuca 1,3,*, Stephanie H. DeLuca 1,3,*, Gordon H. Lemmon 1,3,*, David P. Nannemann
Supplementary Information Small-Molecule Ligand Docking into Comparative Models with Rosetta Steven A. Combs 1,2,*, Samuel L. DeLuca 1,3,*, Stephanie H. DeLuca 1,3,*, Gordon H. Lemmon 1,3,*, David P. Nannemann
REDCAT LABORATORY EXERCISE BCMB/CHEM 8190, April 13, REDCAT may be downloaded from:
 REDCAT LABORATORY EXERCISE BCMB/CHEM 8190, April 13, 2012 Installation note for instructors: REDCAT may be downloaded from: http://ifestos.cse.sc.edu/software.php#redcat You will also find further instructions
REDCAT LABORATORY EXERCISE BCMB/CHEM 8190, April 13, 2012 Installation note for instructors: REDCAT may be downloaded from: http://ifestos.cse.sc.edu/software.php#redcat You will also find further instructions
HexServer: an FFT-based protein docking server powered by graphics processors
 Published online 5 May 2010 Nucleic Acids Research, 2010, Vol. 38, Web Server issue W445 W449 doi:10.1093/nar/gkq311 HexServer: an FFT-based protein docking server powered by graphics processors Gary Macindoe
Published online 5 May 2010 Nucleic Acids Research, 2010, Vol. 38, Web Server issue W445 W449 doi:10.1093/nar/gkq311 HexServer: an FFT-based protein docking server powered by graphics processors Gary Macindoe
REDCAT INSTALLATION. BCMB/CHEM 8190, April 7,
 REDCAT INSTALLATION BCMB/CHEM 8190, April 7, 2006 Installation note for instructors: Download REDCAT from: http://tesla.ccrc.uga.edu/software/redcat While REDCAT is developed on and for Linux systems,
REDCAT INSTALLATION BCMB/CHEM 8190, April 7, 2006 Installation note for instructors: Download REDCAT from: http://tesla.ccrc.uga.edu/software/redcat While REDCAT is developed on and for Linux systems,
AutoDockFR (ADFR) - Docking with flexible receptor sidechains
 AutoDockFR (ADFR) - Docking with flexible receptor sidechains Overview The input for ADFR includes a receptor and a ligand in PDBQT format, prepared affinity maps, and a set of 3D- fill points that are
AutoDockFR (ADFR) - Docking with flexible receptor sidechains Overview The input for ADFR includes a receptor and a ligand in PDBQT format, prepared affinity maps, and a set of 3D- fill points that are
Vrms v1.0 VeraChem LLC, 2006
 Vrms v1.0 VeraChem LLC, 2006 CONTENTS Overview Command-Line Options Example [The Vrms software, including executables, source code, this documentation, and other associated files, are protected by copyright.
Vrms v1.0 VeraChem LLC, 2006 CONTENTS Overview Command-Line Options Example [The Vrms software, including executables, source code, this documentation, and other associated files, are protected by copyright.
AutoDock Virtual Screening: Raccoon & Fox Tools
 AutoDock Virtual Screening: Raccoon & Fox Tools Stefano Forli Ruth Huey The Scripps Research Institute Molecular Graphics Laboratory 10550 N. Torrey Pines Rd. La Jolla, California 92037-1000 USA 3 December
AutoDock Virtual Screening: Raccoon & Fox Tools Stefano Forli Ruth Huey The Scripps Research Institute Molecular Graphics Laboratory 10550 N. Torrey Pines Rd. La Jolla, California 92037-1000 USA 3 December
4) De novo model-building guided by experimental density data
 4) De novo model-building guided by experimental density data In this scenario, we introduce a tool, denovo_density, aimed at automatically building backbone and placing sequence in 3-4.5 Å cryoem density
4) De novo model-building guided by experimental density data In this scenario, we introduce a tool, denovo_density, aimed at automatically building backbone and placing sequence in 3-4.5 Å cryoem density
Conformations of Proteins on Lattice Models. Jiangbo Miao Natalie Kantz
 Conformations of Proteins on Lattice Models Jiangbo Miao Natalie Kantz Lattice Model The lattice model offers a discrete space that limits the infinite number of protein conformations to the lattice space.
Conformations of Proteins on Lattice Models Jiangbo Miao Natalie Kantz Lattice Model The lattice model offers a discrete space that limits the infinite number of protein conformations to the lattice space.
static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, i
 MAESTRO static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
MAESTRO static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
Protein Flexibility Predictions
 Protein Flexibility Predictions Using Graph Theory by D.J. Jacobs, A.J. Rader et. al. from the Michigan State University Talk by Jan Christoph, 19th January 2007 Outline 1. Introduction / Motivation 2.
Protein Flexibility Predictions Using Graph Theory by D.J. Jacobs, A.J. Rader et. al. from the Michigan State University Talk by Jan Christoph, 19th January 2007 Outline 1. Introduction / Motivation 2.
static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, i
 GLIDE static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
GLIDE static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
7.36/7.91/20.390/20.490/6.802/6.874 PROBLEM SET 3. Gibbs Sampler, RNA secondary structure, Protein Structure with PyRosetta, Connections (25 Points)
 7.36/7.91/20.390/20.490/6.802/6.874 PROBLEM SET 3. Gibbs Sampler, RNA secondary structure, Protein Structure with PyRosetta, Connections (25 Points) Due: Thursday, April 3 th at noon. Python Scripts All
7.36/7.91/20.390/20.490/6.802/6.874 PROBLEM SET 3. Gibbs Sampler, RNA secondary structure, Protein Structure with PyRosetta, Connections (25 Points) Due: Thursday, April 3 th at noon. Python Scripts All
Practical 10: Protein dynamics and visualization Documentation
 Practical 10: Protein dynamics and visualization Documentation Release 1.0 Oliver Beckstein March 07, 2013 CONTENTS 1 Basics of protein structure 3 2 Analyzing protein structure and topology 5 2.1 Topology
Practical 10: Protein dynamics and visualization Documentation Release 1.0 Oliver Beckstein March 07, 2013 CONTENTS 1 Basics of protein structure 3 2 Analyzing protein structure and topology 5 2.1 Topology
The beginning of this guide offers a brief introduction to the Protein Data Bank, where users can download structure files.
 Structure Viewers Take a Class This guide supports the Galter Library class called Structure Viewers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
Structure Viewers Take a Class This guide supports the Galter Library class called Structure Viewers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
A First Introduction to Scientific Visualization Geoffrey Gray
 Visual Molecular Dynamics A First Introduction to Scientific Visualization Geoffrey Gray VMD on CIRCE: On the lower bottom left of your screen, click on the window start-up menu. In the search box type
Visual Molecular Dynamics A First Introduction to Scientific Visualization Geoffrey Gray VMD on CIRCE: On the lower bottom left of your screen, click on the window start-up menu. In the search box type
Graduate Topics in Biophysical Chemistry CH Assignment 0 (Programming Assignment) Due Monday, March 19
 Introduction and Goals Graduate Topics in Biophysical Chemistry CH 8990 03 Assignment 0 (Programming Assignment) Due Monday, March 19 It is virtually impossible to be a successful scientist today without
Introduction and Goals Graduate Topics in Biophysical Chemistry CH 8990 03 Assignment 0 (Programming Assignment) Due Monday, March 19 It is virtually impossible to be a successful scientist today without
Tutorial. Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Structural Parameters)
 Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Structural Parameters) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net
Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Structural Parameters) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net
Coot Tutorial. CSHL 2006 Workshop. October 20, Mousing 2
 Coot Tutorial CSHL 2006 Workshop October 20, 2006 Contents 1 Mousing 2 2 Introductory Tutorial 2 2.1 Get the files................................. 2 2.2 Start Coot................................... 2
Coot Tutorial CSHL 2006 Workshop October 20, 2006 Contents 1 Mousing 2 2 Introductory Tutorial 2 2.1 Get the files................................. 2 2.2 Start Coot................................... 2
Predicting Protein Folding Paths. S.Will, , Fall 2011
 Predicting Protein Folding Paths Protein Folding by Robotics Probabilistic Roadmap Planning (PRM): Thomas, Song, Amato. Protein folding by motion planning. Phys. Biol., 2005 Aims Find good quality folding
Predicting Protein Folding Paths Protein Folding by Robotics Probabilistic Roadmap Planning (PRM): Thomas, Song, Amato. Protein folding by motion planning. Phys. Biol., 2005 Aims Find good quality folding
Coot Tutorial. CCP4 Workshop. April 8, Mousing 2
 Coot Tutorial CCP4 Workshop April 8, 2005 Contents 1 Mousing 2 2 First Steps 2 2.1 Get the files................................. 2 2.2 Start Coot................................... 2 2.3 Display Coordinates............................
Coot Tutorial CCP4 Workshop April 8, 2005 Contents 1 Mousing 2 2 First Steps 2 2.1 Get the files................................. 2 2.2 Start Coot................................... 2 2.3 Display Coordinates............................
Assignment 4: Molecular Dynamics (50 points)
 Chemistry 380.37 Fall 2015 Dr. Jean M. Standard November 2, 2015 Assignment 4: Molecular Dynamics (50 points) In this assignment, you will use the Tinker molecular modeling software package to carry out
Chemistry 380.37 Fall 2015 Dr. Jean M. Standard November 2, 2015 Assignment 4: Molecular Dynamics (50 points) In this assignment, you will use the Tinker molecular modeling software package to carry out
PyMOL is a molecular visualization tool. There are many such tools available both commercially and publicly available:
 Workshop #1: PyMOL PyMOL is a molecular visualization tool. There are many such tools available both commercially and publicly available: PyMOL (www.pymol.org) Swiss-PdbViewer (spdbv.vital-it.ch) RasMol
Workshop #1: PyMOL PyMOL is a molecular visualization tool. There are many such tools available both commercially and publicly available: PyMOL (www.pymol.org) Swiss-PdbViewer (spdbv.vital-it.ch) RasMol
MD-TASK Documentation. Release 1.0.1
 MD-TASK Documentation Release 1.0.1 Feb 04, 2018 Getting started 1 MD-TASK 1 1.1 Contribute................................................ 1 1.2 Citing MD-TASK............................................
MD-TASK Documentation Release 1.0.1 Feb 04, 2018 Getting started 1 MD-TASK 1 1.1 Contribute................................................ 1 1.2 Citing MD-TASK............................................
Automatic MR Garib Murshudov MRC-LMB, Cambridge
 Automatic MR Garib Murshudov MRC-LMB, Cambridge These slides provided by Ronan Keegan MR pipelines in CCP4 MrBUMP - Ronan Keegan, Martin Winn BALBES Fei Long, Alexei Vagin, Garib Murshudov Ample - Ronan
Automatic MR Garib Murshudov MRC-LMB, Cambridge These slides provided by Ronan Keegan MR pipelines in CCP4 MrBUMP - Ronan Keegan, Martin Winn BALBES Fei Long, Alexei Vagin, Garib Murshudov Ample - Ronan
Geometry Sixth Grade
 Standard 6-4: The student will demonstrate through the mathematical processes an understanding of shape, location, and movement within a coordinate system; similarity, complementary, and supplementary
Standard 6-4: The student will demonstrate through the mathematical processes an understanding of shape, location, and movement within a coordinate system; similarity, complementary, and supplementary
ssrna Example SASSIE Workflow Data Interpolation
 NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/sassie2/docs/sample_work_flows/ssrna_example/ssrna_example.html.
NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/sassie2/docs/sample_work_flows/ssrna_example/ssrna_example.html.
Discovery Studio 1.5. Online tutorial. Standalone server documentation
 Discovery Studio 1.5 Online tutorial A tutorial helps you to increase your knowledge of Discovery Studio. The lessons are available for skill levels from beginning to advanced. This is a good place to
Discovery Studio 1.5 Online tutorial A tutorial helps you to increase your knowledge of Discovery Studio. The lessons are available for skill levels from beginning to advanced. This is a good place to
Optimization in Shape Matching in the Context of Molecular Docking
 Optimization in Shape Matching in the Context of Molecular Docking Vipin Kumar Tripathi Thesis Supervisors Dr. Bhaskar Dasgupta and Dr. Kalyanmoy Deb Department of Mechanical Engineering Indian Institute
Optimization in Shape Matching in the Context of Molecular Docking Vipin Kumar Tripathi Thesis Supervisors Dr. Bhaskar Dasgupta and Dr. Kalyanmoy Deb Department of Mechanical Engineering Indian Institute
Interactive Model Building with ModelMaker
 University of Illinois at Urbana-Champaign Beckman Institute for Advanced Science and Technology Theoretical and Computational Biophysics Group Computational Biophysics Workshop Interactive Model Building
University of Illinois at Urbana-Champaign Beckman Institute for Advanced Science and Technology Theoretical and Computational Biophysics Group Computational Biophysics Workshop Interactive Model Building
NMR structure calculation with ARIA
 NMR structure calculation with ARIA EMBO Practical Course, 2013, Basel B. Using CCPNmr Benjamin Bardiaux and Michael Nilges Unité de Bio-Informatique Structurale, Institut Pasteur, Paris, France bardiaux@pasteur.fr;
NMR structure calculation with ARIA EMBO Practical Course, 2013, Basel B. Using CCPNmr Benjamin Bardiaux and Michael Nilges Unité de Bio-Informatique Structurale, Institut Pasteur, Paris, France bardiaux@pasteur.fr;
Learning Log Title: CHAPTER 6: TRANSFORMATIONS AND SIMILARITY. Date: Lesson: Chapter 6: Transformations and Similarity
 Chapter 6: Transformations and Similarity CHAPTER 6: TRANSFORMATIONS AND SIMILARITY Date: Lesson: Learning Log Title: Date: Lesson: Learning Log Title: Chapter 6: Transformations and Similarity Date: Lesson:
Chapter 6: Transformations and Similarity CHAPTER 6: TRANSFORMATIONS AND SIMILARITY Date: Lesson: Learning Log Title: Date: Lesson: Learning Log Title: Chapter 6: Transformations and Similarity Date: Lesson:
Homology Modeling Professional for HyperChem Release Notes
 Homology Modeling Professional for HyperChem Release Notes This document lists additional information about Homology Modeling Professional for HyperChem. Current Revision Revision H1 (Version 8.1.1) Current
Homology Modeling Professional for HyperChem Release Notes This document lists additional information about Homology Modeling Professional for HyperChem. Current Revision Revision H1 (Version 8.1.1) Current
CS612 - Algorithms in Bioinformatics
 Spring 2016 Docking April 17, 2016 Docking Docking attempts to find the best matching between two molecules Docking A More Serious Definition Given two biological molecules determine: Whether the two molecules
Spring 2016 Docking April 17, 2016 Docking Docking attempts to find the best matching between two molecules Docking A More Serious Definition Given two biological molecules determine: Whether the two molecules
Sphero Lightning Lab Cheat Sheet
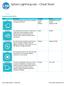 Actions Tool Description Variables Ranges Roll Combines heading, speed and time variables to make the robot roll. Duration Speed Heading (0 to 999999 seconds) (degrees 0-359) Set Speed Sets the speed of
Actions Tool Description Variables Ranges Roll Combines heading, speed and time variables to make the robot roll. Duration Speed Heading (0 to 999999 seconds) (degrees 0-359) Set Speed Sets the speed of
Tutorial. Basic operation of MolDesk. MolDesk Basic/Screening ver Biomodeling Research Co., Ltd. 2018/08/30 ...
 Biomodeling Research Co., Ltd. Tutorial Basic operation of MolDesk MolDesk Basic/Screening ver. 1.1.53 Biomodeling Research Co., Ltd. 2018/08/30... Table of contents 1. Introduction... 1 1.1. Overview...
Biomodeling Research Co., Ltd. Tutorial Basic operation of MolDesk MolDesk Basic/Screening ver. 1.1.53 Biomodeling Research Co., Ltd. 2018/08/30... Table of contents 1. Introduction... 1 1.1. Overview...
Model-Building of Proteins Using X-ray Data With Coot. Paul Emsley May 2013
 Model-Building of Proteins Using X-ray Data With Coot Paul Emsley May 2013 Which coot? Coot-0.7.1-pre revision 4600 (or so) Linux: http://www.ccp4.ac.uk/coot/nightlies/pre-release/ Mac OS X Windows Bill
Model-Building of Proteins Using X-ray Data With Coot Paul Emsley May 2013 Which coot? Coot-0.7.1-pre revision 4600 (or so) Linux: http://www.ccp4.ac.uk/coot/nightlies/pre-release/ Mac OS X Windows Bill
Protein Crystallography
 Protein Crystallography BBMB 334 Lab 4 Whitman College Program in Biochemistry, Biophysics & Molecular Biology (BBMB) Biophysics Laboratory Prof. Douglas Juers Part III. Structure Determination/Refinement
Protein Crystallography BBMB 334 Lab 4 Whitman College Program in Biochemistry, Biophysics & Molecular Biology (BBMB) Biophysics Laboratory Prof. Douglas Juers Part III. Structure Determination/Refinement
JET 2 User Manual 1 INSTALLATION 2 EXECUTION AND FUNCTIONALITIES. 1.1 Download. 1.2 System requirements. 1.3 How to install JET 2
 JET 2 User Manual 1 INSTALLATION 1.1 Download The JET 2 package is available at www.lcqb.upmc.fr/jet2. 1.2 System requirements JET 2 runs on Linux or Mac OS X. The program requires some external tools
JET 2 User Manual 1 INSTALLATION 1.1 Download The JET 2 package is available at www.lcqb.upmc.fr/jet2. 1.2 System requirements JET 2 runs on Linux or Mac OS X. The program requires some external tools
BIOC351: Proteins. PyMOL Laboratory #3. Scripting
 BIOC351: Proteins PyMOL Laboratory 3 Scripting Some information and figures for this handout were obtained from the following source: http://www.virology.wisc.edu/acp/tutorials/pymol-e-density.pdf In our
BIOC351: Proteins PyMOL Laboratory 3 Scripting Some information and figures for this handout were obtained from the following source: http://www.virology.wisc.edu/acp/tutorials/pymol-e-density.pdf In our
IMA Preprint Series # 2179
 PRTAD: A DATABASE FOR PROTEIN RESIDUE TORSION ANGLE DISTRIBUTIONS By Xiaoyong Sun Di Wu Robert Jernigan and Zhijun Wu IMA Preprint Series # 2179 ( November 2007 ) INSTITUTE FOR MATHEMATICS AND ITS APPLICATIONS
PRTAD: A DATABASE FOR PROTEIN RESIDUE TORSION ANGLE DISTRIBUTIONS By Xiaoyong Sun Di Wu Robert Jernigan and Zhijun Wu IMA Preprint Series # 2179 ( November 2007 ) INSTITUTE FOR MATHEMATICS AND ITS APPLICATIONS
Reducing the Search Space in Protein Structure Prediction
 FACULTY OF SCIENCE UNIVERSITY OF COPENHAGEN Ph.D. thesis Rasmus Fonseca Reducing the Search Space in Protein Structure Prediction Department of Computer Science Academic advisor: Pawel Winter Submitted:
FACULTY OF SCIENCE UNIVERSITY OF COPENHAGEN Ph.D. thesis Rasmus Fonseca Reducing the Search Space in Protein Structure Prediction Department of Computer Science Academic advisor: Pawel Winter Submitted:
Small Libraries of Protein Fragments through Clustering
 Small Libraries of Protein Fragments through Clustering Varun Ganapathi Department of Computer Science Stanford University June 8, 2005 Abstract When trying to extract information from the information
Small Libraries of Protein Fragments through Clustering Varun Ganapathi Department of Computer Science Stanford University June 8, 2005 Abstract When trying to extract information from the information
Tips and Tricks using Discovery Studio
 Tips and Tricks using Discovery Studio Allister J. Maynard, Ph.D. Senior Manager, R&D July 31 st, 2008 New Science and Customized Workflows for Drug Discovery Research Webinar Series June 12, 2008 - Advances
Tips and Tricks using Discovery Studio Allister J. Maynard, Ph.D. Senior Manager, R&D July 31 st, 2008 New Science and Customized Workflows for Drug Discovery Research Webinar Series June 12, 2008 - Advances
Energy Minimization on Manifolds for Docking Flexible Molecules
 pubs.acs.org/jctc Energy Minimization on Manifolds for Docking Flexible Molecules Hanieh Mirzaei, Shahrooz Zarbafian, Elizabeth Villar, Scott Mottarella, Dmitri Beglov, Sandor Vajda, Ioannis Ch. Paschalidis,
pubs.acs.org/jctc Energy Minimization on Manifolds for Docking Flexible Molecules Hanieh Mirzaei, Shahrooz Zarbafian, Elizabeth Villar, Scott Mottarella, Dmitri Beglov, Sandor Vajda, Ioannis Ch. Paschalidis,
CS612 - Algorithms in Bioinformatics
 Fall 2017 Structural Manipulation November 22, 2017 Rapid Structural Analysis Methods Emergence of large structural databases which do not allow manual (visual) analysis and require efficient 3-D search
Fall 2017 Structural Manipulation November 22, 2017 Rapid Structural Analysis Methods Emergence of large structural databases which do not allow manual (visual) analysis and require efficient 3-D search
Extra-Homework Problem Set
 Extra-Homework Problem Set => Will not be graded, but might be a good idea for self-study => Solutions are posted at the end of the problem set Your adviser asks you to find out about a so far unpublished
Extra-Homework Problem Set => Will not be graded, but might be a good idea for self-study => Solutions are posted at the end of the problem set Your adviser asks you to find out about a so far unpublished
A/D Converter. Sampling. Figure 1.1: Block Diagram of a DSP System
 CHAPTER 1 INTRODUCTION Digital signal processing (DSP) technology has expanded at a rapid rate to include such diverse applications as CDs, DVDs, MP3 players, ipods, digital cameras, digital light processing
CHAPTER 1 INTRODUCTION Digital signal processing (DSP) technology has expanded at a rapid rate to include such diverse applications as CDs, DVDs, MP3 players, ipods, digital cameras, digital light processing
Cecil Jones Academy Mathematics Fundamental Map
 Fundamentals Year 7 (Core) Knowledge Unit 1 Unit 2 Solve problems involving prime numbers Use highest common factors to solve problems Use lowest common multiples to solve problems Explore powers and roots
Fundamentals Year 7 (Core) Knowledge Unit 1 Unit 2 Solve problems involving prime numbers Use highest common factors to solve problems Use lowest common multiples to solve problems Explore powers and roots
Tutorial: Rosetta tools for structure determination in density Ray Y.-R. Wang, Frank DiMaio Keystone Conference on Hybrid Methods March 2015
 Tutorial: Rosetta tools for structure determination in density Ray Y.-R. Wang, Frank DiMaio Keystone Conference on Hybrid Methods March 2015 This tutorial is intended to introduce users to several different
Tutorial: Rosetta tools for structure determination in density Ray Y.-R. Wang, Frank DiMaio Keystone Conference on Hybrid Methods March 2015 This tutorial is intended to introduce users to several different
Viewing Molecular Structures
 Viewing Molecular Structures Proteins fulfill a wide range of biological functions which depend upon their three dimensional structures. Therefore, deciphering the structure of proteins has been the quest
Viewing Molecular Structures Proteins fulfill a wide range of biological functions which depend upon their three dimensional structures. Therefore, deciphering the structure of proteins has been the quest
Proteins, Particles, and Pseudo-Max- Marginals: A Submodular Approach Jason Pacheco Erik Sudderth
 Proteins, Particles, and Pseudo-Max- Marginals: A Submodular Approach Jason Pacheco Erik Sudderth Department of Computer Science Brown University, Providence RI Protein Side Chain Prediction Estimate side
Proteins, Particles, and Pseudo-Max- Marginals: A Submodular Approach Jason Pacheco Erik Sudderth Department of Computer Science Brown University, Providence RI Protein Side Chain Prediction Estimate side
Justin T. Douglas 1/21/09 A short guide to NMR structure calculations using Xplor-NIH.
 A short guide to NMR structure calculations using Xplor-NIH. The purpose of this guide is help people get started using Xplor-NIH for NMR structure calculations. It is not an exhaustive guide, nor is it
A short guide to NMR structure calculations using Xplor-NIH. The purpose of this guide is help people get started using Xplor-NIH for NMR structure calculations. It is not an exhaustive guide, nor is it
クートオオバン. Introduction to Coot
 大鷭大鳥類 クートオオバン Introduction to Coot Model-Building with Coot: An Introduction and low resolution tools Bernhard Lohkamp Karolinska Institutet Bernhard.Lohkamp@ki.se Tsukuba PF November 2014 Paul Emsley
大鷭大鳥類 クートオオバン Introduction to Coot Model-Building with Coot: An Introduction and low resolution tools Bernhard Lohkamp Karolinska Institutet Bernhard.Lohkamp@ki.se Tsukuba PF November 2014 Paul Emsley
Is there a different way to get the same result? Did we give enough information? How can we describe the position? CPM Materials modified by Mr.
 Common Core Standard: 8.G.3 Is there a different way to get the same result? Did we give enough information? How can we describe the position? CPM Materials modified by Mr. Deyo Title: IM8 Ch. 6.2.1 What
Common Core Standard: 8.G.3 Is there a different way to get the same result? Did we give enough information? How can we describe the position? CPM Materials modified by Mr. Deyo Title: IM8 Ch. 6.2.1 What
BIOC351: Proteins. PyMOL Laboratory #4. Movie Making
 BIOC351: Proteins PyMOL Laboratory #4 Movie Making Version 2 Some information and figures for this handout were obtained from the following sources: http://www.pymolwiki.org/index.php/movieschool http://www.chem.ucsb.edu/~kalju/csuperb/public/pymol_movies.html
BIOC351: Proteins PyMOL Laboratory #4 Movie Making Version 2 Some information and figures for this handout were obtained from the following sources: http://www.pymolwiki.org/index.php/movieschool http://www.chem.ucsb.edu/~kalju/csuperb/public/pymol_movies.html
SASSIE-web: Quick Start Introduction. SASSIE-web Interface
 NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/docs/sassie-web-quick-start/quick-start.html. Return to Main Documents
NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/docs/sassie-web-quick-start/quick-start.html. Return to Main Documents
Introduction to Hermes
 Introduction to Hermes Version 2.0 November 2017 Hermes v1.9 Table of Contents Introduction... 2 Visualising and Editing the MLL1 fusion protein... 2 Opening Files in Hermes... 3 Setting Style Preferences...
Introduction to Hermes Version 2.0 November 2017 Hermes v1.9 Table of Contents Introduction... 2 Visualising and Editing the MLL1 fusion protein... 2 Opening Files in Hermes... 3 Setting Style Preferences...
Re-dock of Roscovitine Against Human Cyclin-Dependent Kinase 2 with Molegro Virtual Docker
 Tutorial Re-dock of Roscovitine Against Human Cyclin-Dependent Kinase 2 with Molegro Virtual Docker Prof. Dr. Walter Filgueira de Azevedo Jr. walter@azevedolab.net azevedolab.net 1 Introduction In this
Tutorial Re-dock of Roscovitine Against Human Cyclin-Dependent Kinase 2 with Molegro Virtual Docker Prof. Dr. Walter Filgueira de Azevedo Jr. walter@azevedolab.net azevedolab.net 1 Introduction In this
Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex
 CORRECTION NOTICE Nat. Struct. Mol. Biol. advance online publication, doi:10.1038/nsmb.1618 (7 June 2009) Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex
CORRECTION NOTICE Nat. Struct. Mol. Biol. advance online publication, doi:10.1038/nsmb.1618 (7 June 2009) Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex
How Do We Measure Protein Shape? A Pattern Matching Example. A Simple Pattern Matching Algorithm. Comparing Protein Structures II
 How Do We Measure Protein Shape? omparing Protein Structures II Protein function is largely based on the proteins geometric shape Protein substructures with similar shapes are likely to share a common
How Do We Measure Protein Shape? omparing Protein Structures II Protein function is largely based on the proteins geometric shape Protein substructures with similar shapes are likely to share a common
ALGORITHMS EXPLOITING THE CHAIN STRUCTURE OF PROTEINS
 ALGORITHMS EXPLOITING THE CHAIN STRUCTURE OF PROTEINS a dissertation submitted to the department of computer science and the committee on graduate studies of stanford university in partial fulfillment
ALGORITHMS EXPLOITING THE CHAIN STRUCTURE OF PROTEINS a dissertation submitted to the department of computer science and the committee on graduate studies of stanford university in partial fulfillment
DNA Tutorial for Arcimboldo
 BORGES ARCIMBOLDO DNA Tutorial for Arcimboldo Aims of the tutorial This tutorial shows how to launch BORGES ARCIMBOLDO in the particular case of DNA binding protein libraries (Pröpper et al., 2014). In
BORGES ARCIMBOLDO DNA Tutorial for Arcimboldo Aims of the tutorial This tutorial shows how to launch BORGES ARCIMBOLDO in the particular case of DNA binding protein libraries (Pröpper et al., 2014). In
Molecular Distance Geometry and Atomic Orderings
 Molecular Distance Geometry and Atomic Orderings Antonio Mucherino IRISA, University of Rennes 1 Workshop Set Computation for Control ENSTA Bretagne, Brest, France December 5 th 2013 The Distance Geometry
Molecular Distance Geometry and Atomic Orderings Antonio Mucherino IRISA, University of Rennes 1 Workshop Set Computation for Control ENSTA Bretagne, Brest, France December 5 th 2013 The Distance Geometry
Chapter 2: Transformations. Chapter 2 Transformations Page 1
 Chapter 2: Transformations Chapter 2 Transformations Page 1 Unit 2: Vocabulary 1) transformation 2) pre-image 3) image 4) map(ping) 5) rigid motion (isometry) 6) orientation 7) line reflection 8) line
Chapter 2: Transformations Chapter 2 Transformations Page 1 Unit 2: Vocabulary 1) transformation 2) pre-image 3) image 4) map(ping) 5) rigid motion (isometry) 6) orientation 7) line reflection 8) line
Examination in Image Processing
 Umeå University, TFE Ulrik Söderström 203-03-27 Examination in Image Processing Time for examination: 4.00 20.00 Please try to extend the answers as much as possible. Do not answer in a single sentence.
Umeå University, TFE Ulrik Söderström 203-03-27 Examination in Image Processing Time for examination: 4.00 20.00 Please try to extend the answers as much as possible. Do not answer in a single sentence.
Refinement. DLS-CCP4 Data Collection and Structure Solution Workshop December Diamond Light Source, Oxfordshire, UK
 Refinement DLS-CCP4 Data Collection and Structure Solution Workshop December 13-20 2016 Diamond Light Source, Oxfordshire, UK Oleg Kovalevskiy Rob Nicholls okovalev@mrc-lmb.cam.ac.uk nicholls@mrc-lmb.cam.ac.uk
Refinement DLS-CCP4 Data Collection and Structure Solution Workshop December 13-20 2016 Diamond Light Source, Oxfordshire, UK Oleg Kovalevskiy Rob Nicholls okovalev@mrc-lmb.cam.ac.uk nicholls@mrc-lmb.cam.ac.uk
Tutorials for the User-Friendly EST Software
 Tutorials for the User-Friendly EST Software Installing the Program 1) To begin installation, run the setup.exe file. 2) Agree to install the Runtime Compiler for Matlab as it is essential to running the
Tutorials for the User-Friendly EST Software Installing the Program 1) To begin installation, run the setup.exe file. 2) Agree to install the Runtime Compiler for Matlab as it is essential to running the
Unit 1, Lesson 1: Moving in the Plane
 Unit 1, Lesson 1: Moving in the Plane Let s describe ways figures can move in the plane. 1.1: Which One Doesn t Belong: Diagrams Which one doesn t belong? 1.2: Triangle Square Dance m.openup.org/1/8-1-1-2
Unit 1, Lesson 1: Moving in the Plane Let s describe ways figures can move in the plane. 1.1: Which One Doesn t Belong: Diagrams Which one doesn t belong? 1.2: Triangle Square Dance m.openup.org/1/8-1-1-2
II: Single particle cryoem - an averaging technique
 II: Single particle cryoem - an averaging technique Radiation damage limits the total electron dose that can be used to image biological sample. Thus, images of frozen hydrated macromolecules are very
II: Single particle cryoem - an averaging technique Radiation damage limits the total electron dose that can be used to image biological sample. Thus, images of frozen hydrated macromolecules are very
DynOmics Portal and ENM server
 DynOmics Portal and ENM server 1. URLs of the DynOmics portal, ENM server, ANM server, and ignm database. DynOmics portal: http://dynomics.pitt.edu/ ENM server: ANM server: http://enm.pitt.edu/ http://anm.csb.pitt.edu/
DynOmics Portal and ENM server 1. URLs of the DynOmics portal, ENM server, ANM server, and ignm database. DynOmics portal: http://dynomics.pitt.edu/ ENM server: ANM server: http://enm.pitt.edu/ http://anm.csb.pitt.edu/
Optimization methods for side-chain positioning and macromolecular docking
 Boston University OpenBU Theses & Dissertations http://open.bu.edu Boston University Theses & Dissertations 2015 Optimization methods for side-chain positioning and macromolecular docking Moghadasi, Mohammad
Boston University OpenBU Theses & Dissertations http://open.bu.edu Boston University Theses & Dissertations 2015 Optimization methods for side-chain positioning and macromolecular docking Moghadasi, Mohammad
Table S1 Comparison of CypA observed Bragg data vs. calculated from Normal Modes.
 Table S1 Comparison of CypA observed Bragg data vs. calculated from Normal Modes. Resolution (Å) R-factor CC 44.560-11.070 0.5713 0.4144 11.070-8.698 0.3325 0.6957 8.697-6.834 0.2987 0.7324 6.831-5.369
Table S1 Comparison of CypA observed Bragg data vs. calculated from Normal Modes. Resolution (Å) R-factor CC 44.560-11.070 0.5713 0.4144 11.070-8.698 0.3325 0.6957 8.697-6.834 0.2987 0.7324 6.831-5.369
BIOC351: Proteins. PyMOL Laboratory #2. Objects, Distances & Images
 BIOC351: Proteins PyMOL Laboratory #2 Objects, Distances & Images Version 2.1 Valid commands in PyMOL are shown in green. Exercise A: Let s fetch 1fzc, a crystallographic dimer of a fragment of human fibrinogen
BIOC351: Proteins PyMOL Laboratory #2 Objects, Distances & Images Version 2.1 Valid commands in PyMOL are shown in green. Exercise A: Let s fetch 1fzc, a crystallographic dimer of a fragment of human fibrinogen
LibProtNMR: A reusable software library for manipulation of protein structures and analysis of NMR data
 LibProtNMR: A reusable software library for manipulation of protein structures and analysis of NMR data May 7, 2014 The Library for Protein NMR (LibProtNmr) is an open-source library of about 35,000 lines
LibProtNMR: A reusable software library for manipulation of protein structures and analysis of NMR data May 7, 2014 The Library for Protein NMR (LibProtNmr) is an open-source library of about 35,000 lines
Mobile Human Detection Systems based on Sliding Windows Approach-A Review
 Mobile Human Detection Systems based on Sliding Windows Approach-A Review Seminar: Mobile Human detection systems Njieutcheu Tassi cedrique Rovile Department of Computer Engineering University of Heidelberg
Mobile Human Detection Systems based on Sliding Windows Approach-A Review Seminar: Mobile Human detection systems Njieutcheu Tassi cedrique Rovile Department of Computer Engineering University of Heidelberg
Lab III: MD II & Analysis Linux & OS X. In this lab you will:
 Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
Approximation of Protein Structure for Fast Similarity Measures
 Approximation of Protein Structure for Fast Similarity Measures Itay Lotan 1 Department of Computer Science Stanford University Stanford, CA 94305 itayl@stanford.edu Fabian Schwarzer Department of Computer
Approximation of Protein Structure for Fast Similarity Measures Itay Lotan 1 Department of Computer Science Stanford University Stanford, CA 94305 itayl@stanford.edu Fabian Schwarzer Department of Computer
Multivariate Calibration Quick Guide
 Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Manual for RNA-As-Graphs Topology (RAGTOP) software suite
 Manual for RNA-As-Graphs Topology (RAGTOP) software suite Schlick lab Contents 1 General Information 1 1.1 Copyright statement....................................... 1 1.2 Citation requirements.......................................
Manual for RNA-As-Graphs Topology (RAGTOP) software suite Schlick lab Contents 1 General Information 1 1.1 Copyright statement....................................... 1 1.2 Citation requirements.......................................
11.1 Rigid Motions. Symmetry
 11.1 Rigid Motions Rigid Motions We will now take a closer look at the ideas behind the different types of symmetries that we have discussed by studying four different rigid motions. The act of taking
11.1 Rigid Motions Rigid Motions We will now take a closer look at the ideas behind the different types of symmetries that we have discussed by studying four different rigid motions. The act of taking
Version 1.0 November2016 Hermes V1.8.2
 Hermes in a Nutshell Version 1.0 November2016 Hermes V1.8.2 Table of Contents Hermes in a Nutshell... 1 Introduction... 2 Example 1. Visualizing and Editing the MLL1 fusion protein... 3 Setting Your Display...
Hermes in a Nutshell Version 1.0 November2016 Hermes V1.8.2 Table of Contents Hermes in a Nutshell... 1 Introduction... 2 Example 1. Visualizing and Editing the MLL1 fusion protein... 3 Setting Your Display...
BioLuminate 1.9. Quick Start Guide. Schrödinger Press
 BioLuminate Quick Start Guide BioLuminate 1.9 Quick Start Guide Schrödinger Press BioLuminate Quick Start Guide Copyright 2015 Schrödinger, LLC. All rights reserved. While care has been taken in the preparation
BioLuminate Quick Start Guide BioLuminate 1.9 Quick Start Guide Schrödinger Press BioLuminate Quick Start Guide Copyright 2015 Schrödinger, LLC. All rights reserved. While care has been taken in the preparation
Modeling solution conformation of a non-coding RNA molecule using PyRy3D web-server.
 Modeling solution conformation of a non-coding RNA molecule using PyRy3D web-server. During this exercise you will learn how to build a 3D model of an RNA molecule based on secondary structure restraints
Modeling solution conformation of a non-coding RNA molecule using PyRy3D web-server. During this exercise you will learn how to build a 3D model of an RNA molecule based on secondary structure restraints
Tutorial. Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Introduction)
 Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Introduction) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net
Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Introduction) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net
This week. CENG 732 Computer Animation. Warping an Object. Warping an Object. 2D Grid Deformation. Warping an Object.
 CENG 732 Computer Animation Spring 2006-2007 Week 4 Shape Deformation Animating Articulated Structures: Forward Kinematics/Inverse Kinematics This week Shape Deformation FFD: Free Form Deformation Hierarchical
CENG 732 Computer Animation Spring 2006-2007 Week 4 Shape Deformation Animating Articulated Structures: Forward Kinematics/Inverse Kinematics This week Shape Deformation FFD: Free Form Deformation Hierarchical
Nature Structural & Molecular Biology: doi: /nsmb.2467
 Supplementary Figure 1. Cryo-electron tomography and GRAFIX optimization of COPII cages. a) Tomogram of Sec13-31 cages viewed along the air/water interface. The tomogram shows a large amount of disordered
Supplementary Figure 1. Cryo-electron tomography and GRAFIX optimization of COPII cages. a) Tomogram of Sec13-31 cages viewed along the air/water interface. The tomogram shows a large amount of disordered
Jacobians. 6.1 Linearized Kinematics. Y: = k2( e6)
 Jacobians 6.1 Linearized Kinematics In previous chapters we have seen how kinematics relates the joint angles to the position and orientation of the robot's endeffector. This means that, for a serial robot,
Jacobians 6.1 Linearized Kinematics In previous chapters we have seen how kinematics relates the joint angles to the position and orientation of the robot's endeffector. This means that, for a serial robot,
Acceleration of a Production Rigid Molecule Docking Code Λy
 Acceleration of a Production Rigid Molecule Docking Code Λy Bharat Sukhwani Martin C. Herbordt Computer Architecture and Automated Design Laboratory Department of Electrical and Computer Engineering Boston
Acceleration of a Production Rigid Molecule Docking Code Λy Bharat Sukhwani Martin C. Herbordt Computer Architecture and Automated Design Laboratory Department of Electrical and Computer Engineering Boston
