Tutorial. Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Structural Parameters)
|
|
|
- Candice Hart
- 6 years ago
- Views:
Transcription
1 Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Structural Parameters) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net azevedolab.net walter@azevedolab.net 1
2 2
3 We can analyze the correlation of docking (Ensemble-Docking) results against structural parameters (derived from the crystallographic information). This task reads docking RMSD and the values for scoring functions stored in a CSV file as the one shown below (for the first twenty lines). Ensemble-Docking results means that one docking protocol was applied to all structures in the dataset. 3
4 The first column brings ligand identification, and we need to have a column for RMSD results. This column has to have RMSD as header. We also need a column for the number of torsion angles, this column should have Torsions as header. The rest of columns is for the scoring functions. We have 100 structural parameters, the list can be accessed here. 4
5 To use SAnDReS to investigate the correlation between a structural parameter and docking results we proceed in a similar way as described for the Ensemble Docking. The major difference is that the results are stored in stdock.csv files. 5
6 Firstly we have to check if all ligands in the chklig.in file are in the Ensemble- Docking file (ensembledock.csv). To do that, click on Structural Parameters->Check Ligands in ensembledock.csv File, as shown here. 6
7 If you gets the message No missing ligands!, everything is fine and you can go ahead and carry out the statistical analysis. Otherwise, you have to edit the CSV file and add missing structure information. 7
8 To proceed, click on Structural Parameters- >Generate Input for Statistical Analysis of Docking Results. 8
9 On the new pop-up window click on the Generate button. Click on the Close button. SAnDReS stdock1.in and files. generates stdock2.in 9
10 Below we have the first two lines of stdock1.in file. The first line indicates that a cutoff criteria will be used, we have only ligands with number of torsion angles below 40. This avoids very flexible ligands. We also have the option to use RMSD as cutoff criteria. The second line brings the keyword STDOCK, followed by PDB access code, ligand information (code, chain, and number), RMSD in angstrom, number of torsion angles, and binding affinity. The rest of the lines follow the same overall structure of the second line. stdock1.in CUTOFF,NTOR,40 STDOCK,1E1V,CMG,A, 401,2.8349,3,
11 Below we have the first five lines of stdock2.in file. We used the keyword SPHERE followed by the radius in angstrom, separated by comma. The first three lines indicate that we will carry out analysis of structural parameters centered at active ligand, with radii of 15.0, 20.0 and 25.0 angstroms. This allows us to investigate the influence of structural parameters such as B-factors and occupancy around the active ligand. The rest of the lines follow the same overall structure described for stdock1.in file. stdock2.in SPHERE, 15.0 SPHERE, 20.0 SPHERE, 25.0 CUTOFF,NTOR,40 STDOCK,1E1V,CMG,A, 401,2.8349,3,
12 We may carry out a short analysis, with few structural parameters (stdock1.in) or a long analysis (stdock2.in), we all structural parameters. Click on Structural Parameters->Statistical Analysis of Docking Results (Long). 12
13 Click on the Yes button. 13
14 It may take few seconds 14
15 Once finished, the message Done! SAnDReS finished running stdock2.in is shown in the information strip, and the log file is shown in the text and GUI windows, as shown here. Besides the log file, SAnDReS also generated CSV files for each structural parameter. To generate plots, we proceed as we have previously described. 15
16 Here we have a typical scatter plot generated by SAnDReS. 16
Tutorial. Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Scoring Function Analysis)
 Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Scoring Function Analysis) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology
Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Scoring Function Analysis) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology
Tutorial. Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Docking Hub)
 Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Docking Hub) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net
Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Docking Hub) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net
Tutorial. Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Introduction)
 Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Introduction) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net
Tutorial Docking School SAnDReS Tutorial Cyclin-Dependent Kinases with K i Information (Introduction) Prof. Dr. Walter Filgueira de Azevedo Jr. Laboratory of Computational Systems Biology azevedolab.net
Re-dock of Roscovitine Against Human Cyclin-Dependent Kinase 2 with Molegro Virtual Docker
 Tutorial Re-dock of Roscovitine Against Human Cyclin-Dependent Kinase 2 with Molegro Virtual Docker Prof. Dr. Walter Filgueira de Azevedo Jr. walter@azevedolab.net azevedolab.net 1 Introduction In this
Tutorial Re-dock of Roscovitine Against Human Cyclin-Dependent Kinase 2 with Molegro Virtual Docker Prof. Dr. Walter Filgueira de Azevedo Jr. walter@azevedolab.net azevedolab.net 1 Introduction In this
Annotating a single sequence
 BioNumerics Tutorial: Annotating a single sequence 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn how
BioNumerics Tutorial: Annotating a single sequence 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn how
static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, i
 GLIDE static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
GLIDE static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
AutoDockFR (ADFR) - Docking with flexible receptor sidechains
 AutoDockFR (ADFR) - Docking with flexible receptor sidechains Overview The input for ADFR includes a receptor and a ligand in PDBQT format, prepared affinity maps, and a set of 3D- fill points that are
AutoDockFR (ADFR) - Docking with flexible receptor sidechains Overview The input for ADFR includes a receptor and a ligand in PDBQT format, prepared affinity maps, and a set of 3D- fill points that are
AutoDock Virtual Screening: Raccoon & Fox Tools
 AutoDock Virtual Screening: Raccoon & Fox Tools Stefano Forli Ruth Huey The Scripps Research Institute Molecular Graphics Laboratory 10550 N. Torrey Pines Rd. La Jolla, California 92037-1000 USA 3 December
AutoDock Virtual Screening: Raccoon & Fox Tools Stefano Forli Ruth Huey The Scripps Research Institute Molecular Graphics Laboratory 10550 N. Torrey Pines Rd. La Jolla, California 92037-1000 USA 3 December
Introduction to Hermes
 Introduction to Hermes Version 2.0 November 2017 Hermes v1.9 Table of Contents Introduction... 2 Visualising and Editing the MLL1 fusion protein... 2 Opening Files in Hermes... 3 Setting Style Preferences...
Introduction to Hermes Version 2.0 November 2017 Hermes v1.9 Table of Contents Introduction... 2 Visualising and Editing the MLL1 fusion protein... 2 Opening Files in Hermes... 3 Setting Style Preferences...
Tips and Tricks using Discovery Studio
 Tips and Tricks using Discovery Studio Allister J. Maynard, Ph.D. Senior Manager, R&D July 31 st, 2008 New Science and Customized Workflows for Drug Discovery Research Webinar Series June 12, 2008 - Advances
Tips and Tricks using Discovery Studio Allister J. Maynard, Ph.D. Senior Manager, R&D July 31 st, 2008 New Science and Customized Workflows for Drug Discovery Research Webinar Series June 12, 2008 - Advances
PFstats User Guide. Aspartate/ornithine carbamoyltransferase Case Study. Neli Fonseca
 PFstats User Guide Aspartate/ornithine carbamoyltransferase Case Study 1 Contents Overview 3 Obtaining An Alignment 3 Methods 4 Alignment Filtering............................................ 4 Reference
PFstats User Guide Aspartate/ornithine carbamoyltransferase Case Study 1 Contents Overview 3 Obtaining An Alignment 3 Methods 4 Alignment Filtering............................................ 4 Reference
RClimTool USER MANUAL
 RClimTool USER MANUAL By Lizeth Llanos Herrera, student Statistics This tool is designed to support, process automation and analysis of climatic series within the agreement of CIAT-MADR. It is not intended
RClimTool USER MANUAL By Lizeth Llanos Herrera, student Statistics This tool is designed to support, process automation and analysis of climatic series within the agreement of CIAT-MADR. It is not intended
Molecular docking tutorial
 Molecular docking tutorial Sulfonamide-type D-Glu inhibitor docked into the MurD active site using ArgusLab In this tutorial [1] you will learn how to prepare and run molecular docking calculations using
Molecular docking tutorial Sulfonamide-type D-Glu inhibitor docked into the MurD active site using ArgusLab In this tutorial [1] you will learn how to prepare and run molecular docking calculations using
Small-Molecule Ligand Docking into Comparative Models with Rosetta
 Supplementary Information Small-Molecule Ligand Docking into Comparative Models with Rosetta Steven A. Combs 1,2,*, Samuel L. DeLuca 1,3,*, Stephanie H. DeLuca 1,3,*, Gordon H. Lemmon 1,3,*, David P. Nannemann
Supplementary Information Small-Molecule Ligand Docking into Comparative Models with Rosetta Steven A. Combs 1,2,*, Samuel L. DeLuca 1,3,*, Stephanie H. DeLuca 1,3,*, Gordon H. Lemmon 1,3,*, David P. Nannemann
Protein structure. Enter the name of the protein: rhamnogalacturonan acetylesterase in the search field and click Go.
 In this practical you will learn how to Protein structure search the Protein Structure Databank for information critically choose the best structure, when more than one is available visualize a protein
In this practical you will learn how to Protein structure search the Protein Structure Databank for information critically choose the best structure, when more than one is available visualize a protein
Introduction to the Protein Data Bank Master Chimie Info Roland Stote Page #
 Introduction to the Protein Data Bank Master Chimie Info - 2009 Roland Stote The purpose of the Protein Data Bank is to collect and organize 3D structures of proteins, nucleic acids, protein-nucleic acid
Introduction to the Protein Data Bank Master Chimie Info - 2009 Roland Stote The purpose of the Protein Data Bank is to collect and organize 3D structures of proteins, nucleic acids, protein-nucleic acid
Supplementary Material. EASYMIFs and SITEHOUND: a toolkit for the identification of ligand-binding sites in protein structures
 Supplementary Material EASYMIFs and SITEHOUND: a toolkit for the identification of ligand-binding sites in protein structures Dario Ghersi and Roberto Sanchez 1 Running EASYMIFs or SITEHOUND separately
Supplementary Material EASYMIFs and SITEHOUND: a toolkit for the identification of ligand-binding sites in protein structures Dario Ghersi and Roberto Sanchez 1 Running EASYMIFs or SITEHOUND separately
Simple REFMAC tutorials
 Simple REFMAC tutorials Prerequisites: To use this tutorial you need to have ccp4. For jelly body, automatic and local ncs restraints, occupancy refinement you need to have the latest version of ccp4-6.2
Simple REFMAC tutorials Prerequisites: To use this tutorial you need to have ccp4. For jelly body, automatic and local ncs restraints, occupancy refinement you need to have the latest version of ccp4-6.2
Version 1.0 November2016 Hermes V1.8.2
 Hermes in a Nutshell Version 1.0 November2016 Hermes V1.8.2 Table of Contents Hermes in a Nutshell... 1 Introduction... 2 Example 1. Visualizing and Editing the MLL1 fusion protein... 3 Setting Your Display...
Hermes in a Nutshell Version 1.0 November2016 Hermes V1.8.2 Table of Contents Hermes in a Nutshell... 1 Introduction... 2 Example 1. Visualizing and Editing the MLL1 fusion protein... 3 Setting Your Display...
Protein Structure and Visualization
 Protein Structure and Visualization In this practical you will learn how to By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
Protein Structure and Visualization In this practical you will learn how to By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
Depositing small-angle scattering data and models to the Small-Angle Scattering Biological Data Bank (SASBDB).
 Depositing small-angle scattering data and models to the Small-Angle Scattering Biological Data Bank (SASBDB). Introduction. The following guide provides a basic outline of the minimum requirements necessary
Depositing small-angle scattering data and models to the Small-Angle Scattering Biological Data Bank (SASBDB). Introduction. The following guide provides a basic outline of the minimum requirements necessary
PSF Project MMS Virtual LAB
 PSF Project 2016-2017 MMS Virtual LAB Molecular Modeling Section (MMS) Dip. Scienze del Farmaco Target SMILE Nc1nc(N)c2nc(c(N)nc2n1)-c1cccc(Cl)c1 Aim of the Project 1. Characterize the reference 6-(3-chlorophenyl)pteridine-2,4,7-triamine
PSF Project 2016-2017 MMS Virtual LAB Molecular Modeling Section (MMS) Dip. Scienze del Farmaco Target SMILE Nc1nc(N)c2nc(c(N)nc2n1)-c1cccc(Cl)c1 Aim of the Project 1. Characterize the reference 6-(3-chlorophenyl)pteridine-2,4,7-triamine
Workshop #7: Docking. Suggested Readings
 Workshop #7: Docking Protein protein docking is the prediction of a complex structure starting from its monomer components. The search space can be extremely large, so large amounts of computational resources
Workshop #7: Docking Protein protein docking is the prediction of a complex structure starting from its monomer components. The search space can be extremely large, so large amounts of computational resources
Docking Study with HyperChem Release Notes
 Docking Study with HyperChem Release Notes This document lists additional information about Docking Study with HyperChem family, Essential, Premium Essential, Professional, Advanced, Ultimat, and Cluster.
Docking Study with HyperChem Release Notes This document lists additional information about Docking Study with HyperChem family, Essential, Premium Essential, Professional, Advanced, Ultimat, and Cluster.
This manual describes step-by-step instructions to perform basic operations for data analysis.
 HDXanalyzer User Manual The program HDXanalyzer is available for the analysis of the deuterium exchange mass spectrometry data obtained on high-resolution mass spectrometers. Currently, the program is
HDXanalyzer User Manual The program HDXanalyzer is available for the analysis of the deuterium exchange mass spectrometry data obtained on high-resolution mass spectrometers. Currently, the program is
Tutorial. Basic operation of MolDesk. MolDesk Basic/Screening ver Biomodeling Research Co., Ltd. 2018/08/30 ...
 Biomodeling Research Co., Ltd. Tutorial Basic operation of MolDesk MolDesk Basic/Screening ver. 1.1.53 Biomodeling Research Co., Ltd. 2018/08/30... Table of contents 1. Introduction... 1 1.1. Overview...
Biomodeling Research Co., Ltd. Tutorial Basic operation of MolDesk MolDesk Basic/Screening ver. 1.1.53 Biomodeling Research Co., Ltd. 2018/08/30... Table of contents 1. Introduction... 1 1.1. Overview...
Model-Building of Proteins Using X-ray Data With Coot. Paul Emsley May 2013
 Model-Building of Proteins Using X-ray Data With Coot Paul Emsley May 2013 Which coot? Coot-0.7.1-pre revision 4600 (or so) Linux: http://www.ccp4.ac.uk/coot/nightlies/pre-release/ Mac OS X Windows Bill
Model-Building of Proteins Using X-ray Data With Coot Paul Emsley May 2013 Which coot? Coot-0.7.1-pre revision 4600 (or so) Linux: http://www.ccp4.ac.uk/coot/nightlies/pre-release/ Mac OS X Windows Bill
Using CSD-CrossMiner to Create a Feature Database
 Table of Contents Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Features and Pharmacophore Representation... 4 Building your Own Feature Database... 5 Using CSD-CrossMiner
Table of Contents Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Features and Pharmacophore Representation... 4 Building your Own Feature Database... 5 Using CSD-CrossMiner
Protein Structure and Visualization
 In this practical you will learn how to Protein Structure and Visualization By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
In this practical you will learn how to Protein Structure and Visualization By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
Quick Start Guide for Teachers
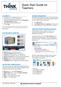 Quick Start Guide for Teachers LOGGING IN 1. Navigate to www-k6.thinkcentral.com. 2. Select your state, district, and school. 3. To make subsequent logins easier, check Remember My Organization (optional).
Quick Start Guide for Teachers LOGGING IN 1. Navigate to www-k6.thinkcentral.com. 2. Select your state, district, and school. 3. To make subsequent logins easier, check Remember My Organization (optional).
Quick Tutorial. Running CellDesigner TM Simulation with ControlPanel
 Running CellDesigner TM Simulation with ControlPanel Quick Tutorial 2005/09/01 The Systems Biology Institute http://www.systems-biology.org/ Mitsui Knowledge Industry Co. Ltd. http://bio.mki.co.jp/en/
Running CellDesigner TM Simulation with ControlPanel Quick Tutorial 2005/09/01 The Systems Biology Institute http://www.systems-biology.org/ Mitsui Knowledge Industry Co. Ltd. http://bio.mki.co.jp/en/
ECO375 Tutorial 1 Introduction to Stata
 ECO375 Tutorial 1 Introduction to Stata Matt Tudball University of Toronto Mississauga September 14, 2017 Matt Tudball (University of Toronto) ECO375H5 September 14, 2017 1 / 25 What Is Stata? Stata is
ECO375 Tutorial 1 Introduction to Stata Matt Tudball University of Toronto Mississauga September 14, 2017 Matt Tudball (University of Toronto) ECO375H5 September 14, 2017 1 / 25 What Is Stata? Stata is
Practical 10: Protein dynamics and visualization Documentation
 Practical 10: Protein dynamics and visualization Documentation Release 1.0 Oliver Beckstein March 07, 2013 CONTENTS 1 Basics of protein structure 3 2 Analyzing protein structure and topology 5 2.1 Topology
Practical 10: Protein dynamics and visualization Documentation Release 1.0 Oliver Beckstein March 07, 2013 CONTENTS 1 Basics of protein structure 3 2 Analyzing protein structure and topology 5 2.1 Topology
Vrms v1.0 VeraChem LLC, 2006
 Vrms v1.0 VeraChem LLC, 2006 CONTENTS Overview Command-Line Options Example [The Vrms software, including executables, source code, this documentation, and other associated files, are protected by copyright.
Vrms v1.0 VeraChem LLC, 2006 CONTENTS Overview Command-Line Options Example [The Vrms software, including executables, source code, this documentation, and other associated files, are protected by copyright.
Easy manual for SIRD interface
 Easy manual for SIRD interface What is SIRD interface (1) SIRDi is the web-based interface for SIRD (Structure-Interaction Relational Database) (2) You can search for protein structure models with sequence,
Easy manual for SIRD interface What is SIRD interface (1) SIRDi is the web-based interface for SIRD (Structure-Interaction Relational Database) (2) You can search for protein structure models with sequence,
QUICK GUIDE TO SLIDE VERSION 3.4
 QUICK GUIDE TO SLIDE VERSION 3.4 Protein Structural Analysis & Design Lab MSU 1 Preparing to use SLIDE (User Guide Chapter 3) Setting SLIDE environment variables. Add the following lines to your.cshrc
QUICK GUIDE TO SLIDE VERSION 3.4 Protein Structural Analysis & Design Lab MSU 1 Preparing to use SLIDE (User Guide Chapter 3) Setting SLIDE environment variables. Add the following lines to your.cshrc
Investigating the Sine and Cosine Functions Part 1
 Investigating the Sine and Cosine Functions Part 1 Name: Period: Date: Set-Up Press. Move down to 5: Cabri Jr and press. Press for the F1 menu and select New. Press for F5 and select Hide/Show > Axes.
Investigating the Sine and Cosine Functions Part 1 Name: Period: Date: Set-Up Press. Move down to 5: Cabri Jr and press. Press for the F1 menu and select New. Press for F5 and select Hide/Show > Axes.
DynOmics Portal and ENM server
 DynOmics Portal and ENM server 1. URLs of the DynOmics portal, ENM server, ANM server, and ignm database. DynOmics portal: http://dynomics.pitt.edu/ ENM server: ANM server: http://enm.pitt.edu/ http://anm.csb.pitt.edu/
DynOmics Portal and ENM server 1. URLs of the DynOmics portal, ENM server, ANM server, and ignm database. DynOmics portal: http://dynomics.pitt.edu/ ENM server: ANM server: http://enm.pitt.edu/ http://anm.csb.pitt.edu/
ssrna Example SASSIE Workflow Data Interpolation
 NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/sassie2/docs/sample_work_flows/ssrna_example/ssrna_example.html.
NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/sassie2/docs/sample_work_flows/ssrna_example/ssrna_example.html.
Computer Lab, Session 1
 Computer Lab, Session 1 1 Log in Please log in with username VSDD0xy where xy is your computer number ranging from 01, 02,, 20 2 Settings Open terminal In home directory (initial directory): cp /export/home/vsdd/vsdd001/.bashrc.
Computer Lab, Session 1 1 Log in Please log in with username VSDD0xy where xy is your computer number ranging from 01, 02,, 20 2 Settings Open terminal In home directory (initial directory): cp /export/home/vsdd/vsdd001/.bashrc.
About This User Guide
 About This User Guide This user guide is a practical guide to using the Relibase and Relibase+ tools for searching protein/ ligand structures. It includes instructions on using the graphical user interface,
About This User Guide This user guide is a practical guide to using the Relibase and Relibase+ tools for searching protein/ ligand structures. It includes instructions on using the graphical user interface,
static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, i
 MAESTRO static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
MAESTRO static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
A: if no run time errors or exceptions B: (-1 point) If you got one or more run time error or exception but the application is still running.
 Computer Programming Project Evaluation - Summer 2018 Criteria for grading and Grading 14 points in total - Minimum to pass 8, according to 5 dimensions: 1) Functionality (Coverage of the requirements)
Computer Programming Project Evaluation - Summer 2018 Criteria for grading and Grading 14 points in total - Minimum to pass 8, according to 5 dimensions: 1) Functionality (Coverage of the requirements)
Houghton Mifflin Harcourt and its logo are trademarks of Houghton Mifflin Harcourt Publishing Company.
 Guide for Teachers Updated September 2013 Houghton Mifflin Harcourt Publishing Company. All rights reserved. Houghton Mifflin Harcourt and its logo are trademarks of Houghton Mifflin Harcourt Publishing
Guide for Teachers Updated September 2013 Houghton Mifflin Harcourt Publishing Company. All rights reserved. Houghton Mifflin Harcourt and its logo are trademarks of Houghton Mifflin Harcourt Publishing
Glide 6.7. Quick Start Guide. Schrödinger Press
 Glide Quick Start Guide Glide 6.7 Quick Start Guide Schrödinger Press Glide Quick Start Guide Copyright 2015 Schrödinger, LLC. All rights reserved. While care has been taken in the preparation of this
Glide Quick Start Guide Glide 6.7 Quick Start Guide Schrödinger Press Glide Quick Start Guide Copyright 2015 Schrödinger, LLC. All rights reserved. While care has been taken in the preparation of this
Trajectory Analysis. Release Ahmet Bakan
 Trajectory Analysis Release 1.5.1 Ahmet Bakan December 24, 2013 CONTENTS 1 Introduction 2 1.1 Required Programs........................................... 2 1.2 Recommended Programs.......................................
Trajectory Analysis Release 1.5.1 Ahmet Bakan December 24, 2013 CONTENTS 1 Introduction 2 1.1 Required Programs........................................... 2 1.2 Recommended Programs.......................................
AMass Tutorial: Introduction to the Java AMass applet
 AMass Tutorial: Introduction to the 13.021 Java AMass applet The purpose of this document is to familiarize you with the 13.021 Java AMass applet. This applet can be found at http://web.mit.edu/13.021/www/software.html.
AMass Tutorial: Introduction to the 13.021 Java AMass applet The purpose of this document is to familiarize you with the 13.021 Java AMass applet. This applet can be found at http://web.mit.edu/13.021/www/software.html.
AMass Tutorial. Introduction to the 2.20 Java AMass applet
 AMass Tutorial Introduction to the 2.20 Java AMass applet The purpose of this document is to familiarize you with the 2.20 Java AMass applet. This applet can be found at http://ocw.mit.edu/ocwweb/mechanical-engineering/2-20spring-
AMass Tutorial Introduction to the 2.20 Java AMass applet The purpose of this document is to familiarize you with the 2.20 Java AMass applet. This applet can be found at http://ocw.mit.edu/ocwweb/mechanical-engineering/2-20spring-
Contents. Protus Messaging Services User Guide Web Fax Merge
 Contents Protus Messaging Services User Guide Getting Started... 1 Setting up an account... 1 Requirements... 1 Logging In... 1 Sending a New... 2 Who are you sending your fax to?... 2 Sample Merge List...
Contents Protus Messaging Services User Guide Getting Started... 1 Setting up an account... 1 Requirements... 1 Logging In... 1 Sending a New... 2 Who are you sending your fax to?... 2 Sample Merge List...
CCP4 Tutorial: Molecular Replacement and MrBUMP
 CCP4 Tutorial: Molecular Replacement and MrBUMP This tutorial demonstrates how to use the MrBUMP program to carry out much of the work involved in the molecular replacement (MR) process automatically.
CCP4 Tutorial: Molecular Replacement and MrBUMP This tutorial demonstrates how to use the MrBUMP program to carry out much of the work involved in the molecular replacement (MR) process automatically.
BIOC351: Proteins. PyMOL Laboratory #2. Objects, Distances & Images
 BIOC351: Proteins PyMOL Laboratory #2 Objects, Distances & Images Version 2.1 Valid commands in PyMOL are shown in green. Exercise A: Let s fetch 1fzc, a crystallographic dimer of a fragment of human fibrinogen
BIOC351: Proteins PyMOL Laboratory #2 Objects, Distances & Images Version 2.1 Valid commands in PyMOL are shown in green. Exercise A: Let s fetch 1fzc, a crystallographic dimer of a fragment of human fibrinogen
Statistics with a Hemacytometer
 Statistics with a Hemacytometer Overview This exercise incorporates several different statistical analyses. Data gathered from cell counts with a hemacytometer is used to explore frequency distributions
Statistics with a Hemacytometer Overview This exercise incorporates several different statistical analyses. Data gathered from cell counts with a hemacytometer is used to explore frequency distributions
Project Number: MQP-GTH-MQIS CLUSTER ANALYSIS USING QUANTILE PLOTS WITH APPLICATIONS IN COMPUTATIONAL CHEMISTRY. A Major Qualifying Project Report
 Project Number: MQP-GTH-MQIS CLUSTER ANALYSIS USING QUANTILE PLOTS WITH APPLICATIONS IN COMPUTATIONAL CHEMISTRY A Major Qualifying Project Report Submitted to the Faculty of the WORCESTER POLYTECHNIC INSTITUTE
Project Number: MQP-GTH-MQIS CLUSTER ANALYSIS USING QUANTILE PLOTS WITH APPLICATIONS IN COMPUTATIONAL CHEMISTRY A Major Qualifying Project Report Submitted to the Faculty of the WORCESTER POLYTECHNIC INSTITUTE
/4 Directions: Graph the functions, then answer the following question.
 1.) Graph y = x. Label the graph. Standard: F-BF.3 Identify the effect on the graph of replacing f(x) by f(x) +k, k f(x), f(kx), and f(x+k), for specific values of k; find the value of k given the graphs.
1.) Graph y = x. Label the graph. Standard: F-BF.3 Identify the effect on the graph of replacing f(x) by f(x) +k, k f(x), f(kx), and f(x+k), for specific values of k; find the value of k given the graphs.
Lab 5, part b: Scatterplots and Correlation
 Lab 5, part b: Scatterplots and Correlation Toews, Math 160, Fall 2014 November 21, 2014 Objectives: 1. Get more practice working with data frames 2. Start looking at relationships between two variables
Lab 5, part b: Scatterplots and Correlation Toews, Math 160, Fall 2014 November 21, 2014 Objectives: 1. Get more practice working with data frames 2. Start looking at relationships between two variables
Annotating sequences in batch
 BioNumerics Tutorial: Annotating sequences in batch 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn
BioNumerics Tutorial: Annotating sequences in batch 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn
MOLECULAR VISUALIZATION LAB USING PYMOL a supplement to Chapter 11. Please complete this tutorial before coming to your lab section
 MOLECULAR VISUALIZATION LAB USING PYMOL a supplement to Chapter 11 Please complete this tutorial before coming to your lab section (Adapted from Dr. Vardar-Ulu Fall 2015) Before coming to your lab section
MOLECULAR VISUALIZATION LAB USING PYMOL a supplement to Chapter 11 Please complete this tutorial before coming to your lab section (Adapted from Dr. Vardar-Ulu Fall 2015) Before coming to your lab section
Before we begin, there are a few important points that we wish to highlight.
 We are very excited to present our new Web Access Portal. We hope you enjoy using it. If you have any questions, please call Patt at (650) 875 3800 or Client Services at (650) 875 3600. Before we begin,
We are very excited to present our new Web Access Portal. We hope you enjoy using it. If you have any questions, please call Patt at (650) 875 3800 or Client Services at (650) 875 3600. Before we begin,
Protein Flexibility Predictions
 Protein Flexibility Predictions Using Graph Theory by D.J. Jacobs, A.J. Rader et. al. from the Michigan State University Talk by Jan Christoph, 19th January 2007 Outline 1. Introduction / Motivation 2.
Protein Flexibility Predictions Using Graph Theory by D.J. Jacobs, A.J. Rader et. al. from the Michigan State University Talk by Jan Christoph, 19th January 2007 Outline 1. Introduction / Motivation 2.
Discovery Studio 1.5. Online tutorial. Standalone server documentation
 Discovery Studio 1.5 Online tutorial A tutorial helps you to increase your knowledge of Discovery Studio. The lessons are available for skill levels from beginning to advanced. This is a good place to
Discovery Studio 1.5 Online tutorial A tutorial helps you to increase your knowledge of Discovery Studio. The lessons are available for skill levels from beginning to advanced. This is a good place to
Modelling With Comsol: Gradients and Pattern Formation Direct questions and suggestions to Denis
 Modelling With Comsol: Gradients and Pattern Formation Direct questions and suggestions to Denis (dzianis.menshykau@bsse.ethz.ch) Problem 1 Solve simple diffusion equation (no reactions!) on a 1D domain.
Modelling With Comsol: Gradients and Pattern Formation Direct questions and suggestions to Denis (dzianis.menshykau@bsse.ethz.ch) Problem 1 Solve simple diffusion equation (no reactions!) on a 1D domain.
Enabling Scalable Data Analysis for Large Computa9onal Structural Biology Datasets on Distributed Memory Systems
 Enabling Scalable Data Analysis for Large Computa9onal Structural Biology Datasets on Distributed Memory Systems Michela Taufer Global Compu9ng Laboratory Computer and Informa9on Sciences University of
Enabling Scalable Data Analysis for Large Computa9onal Structural Biology Datasets on Distributed Memory Systems Michela Taufer Global Compu9ng Laboratory Computer and Informa9on Sciences University of
LONI IMAGE & DATA ARCHIVE USER MANUAL
 LONI IMAGE & DATA ARCHIVE USER MANUAL Laboratory of Neuro Imaging Dr. Arthur W. Toga, Director April, 2017 LONI Image & Data Archive INTRODUCTION The LONI Image & Data Archive (IDA) is a user-friendly
LONI IMAGE & DATA ARCHIVE USER MANUAL Laboratory of Neuro Imaging Dr. Arthur W. Toga, Director April, 2017 LONI Image & Data Archive INTRODUCTION The LONI Image & Data Archive (IDA) is a user-friendly
Package press. April 9, 2011
 Type Package Title Protein REsidue-level Structural Statistics Version 1.0 Package press April 9, 2011 Depends pressdata, gwidgets, RGtk2, gwidgetsrgtk2, Rcmdr, rgl, car,outliers, MASS Date 2010-07-20
Type Package Title Protein REsidue-level Structural Statistics Version 1.0 Package press April 9, 2011 Depends pressdata, gwidgets, RGtk2, gwidgetsrgtk2, Rcmdr, rgl, car,outliers, MASS Date 2010-07-20
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S. Visualization of Biological Molecules in Their Native Stat e.
 THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
Data Mining With Weka A Short Tutorial
 Data Mining With Weka A Short Tutorial Dr. Wenjia Wang School of Computing Sciences University of East Anglia (UEA), Norwich, UK Content 1. Introduction to Weka 2. Data Mining Functions and Tools 3. Data
Data Mining With Weka A Short Tutorial Dr. Wenjia Wang School of Computing Sciences University of East Anglia (UEA), Norwich, UK Content 1. Introduction to Weka 2. Data Mining Functions and Tools 3. Data
Working with Lists: Adding and Importing Addresses
 JangoMail Tutorial Working with Lists: Adding and Importing Email Addresses Overview: You store your email lists in JangoMail s database using the Lists tab. Here, you create your own Lists, each with
JangoMail Tutorial Working with Lists: Adding and Importing Email Addresses Overview: You store your email lists in JangoMail s database using the Lists tab. Here, you create your own Lists, each with
v Introduction to WMS Become familiar with the WMS interface WMS Tutorials Time minutes Prerequisite Tutorials None
 s v. 10.0 WMS 10.0 Tutorial Become familiar with the WMS interface Objectives Read files into WMS and change modules and display options to become familiar with the WMS interface. Prerequisite Tutorials
s v. 10.0 WMS 10.0 Tutorial Become familiar with the WMS interface Objectives Read files into WMS and change modules and display options to become familiar with the WMS interface. Prerequisite Tutorials
Tutorial. Step 1. Step 2. Figure 1
 Tutorial Welcome to the MISTIC Tutorial! In the next pages we will use an example case study to help you load data, submit the job and then analyze and visualize the results. Step 1 We will be using the
Tutorial Welcome to the MISTIC Tutorial! In the next pages we will use an example case study to help you load data, submit the job and then analyze and visualize the results. Step 1 We will be using the
Workshop. Import Workshop
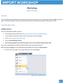 Import Overview This workshop will help participants understand the tools and techniques used in importing a variety of different types of data. It will also showcase a couple of the new import features
Import Overview This workshop will help participants understand the tools and techniques used in importing a variety of different types of data. It will also showcase a couple of the new import features
WMS 9.1 Tutorial Hydraulics and Floodplain Modeling Floodplain Delineation Learn how to us the WMS floodplain delineation tools
 v. 9.1 WMS 9.1 Tutorial Hydraulics and Floodplain Modeling Floodplain Delineation Learn how to us the WMS floodplain delineation tools Objectives Experiment with the various floodplain delineation options
v. 9.1 WMS 9.1 Tutorial Hydraulics and Floodplain Modeling Floodplain Delineation Learn how to us the WMS floodplain delineation tools Objectives Experiment with the various floodplain delineation options
REFRESHER TRAINING OF FOCAL POINTS ON THE
 CountrySTAT / FENIX Overview of the DSD editor in the new CountrySTAT platform REFRESHER TRAINING OF FOCAL POINTS ON THE CountrySTAT/FENIX SYSTEM 24 28 July 2016 ECO Secretariat, Teheran, Iran Stefania
CountrySTAT / FENIX Overview of the DSD editor in the new CountrySTAT platform REFRESHER TRAINING OF FOCAL POINTS ON THE CountrySTAT/FENIX SYSTEM 24 28 July 2016 ECO Secretariat, Teheran, Iran Stefania
CHEM /12/01 INSTRUCTIONS FOR AB-INITIO CALCULATIONS Prepared by: Dr. Cynthia HARWOOD, Ahu AKIN, Prof. Tim KEIDERLING
 CHEM 542 10/12/01 INSTRUCTIONS FOR AB-INITIO CALCULATIONS Prepared by: Dr. Cynthia HARWOOD, Ahu AKIN, Prof. Tim KEIDERLING SETTLING IN: - Type your login name : Chem542 and press Enter - Type your password
CHEM 542 10/12/01 INSTRUCTIONS FOR AB-INITIO CALCULATIONS Prepared by: Dr. Cynthia HARWOOD, Ahu AKIN, Prof. Tim KEIDERLING SETTLING IN: - Type your login name : Chem542 and press Enter - Type your password
JOB AID Searching for Information in KC
 There are a variety of search options available to users. This Job Aid will guide faculty and administrators through several common searches as well as provide information for organizing and exporting
There are a variety of search options available to users. This Job Aid will guide faculty and administrators through several common searches as well as provide information for organizing and exporting
Goals and Objectives
 Goals and Objectives This example lists NJ Core Content Standards, with the strands for each standard shown as links in outline form. The Special Education Management Module also has choices for non core
Goals and Objectives This example lists NJ Core Content Standards, with the strands for each standard shown as links in outline form. The Special Education Management Module also has choices for non core
File Importing - Text Files
 File Importing - Text Files With this tutorial we are going to go through the basic elements of importing a text file that contains several records (lines) each containing several fields. Sample Data -
File Importing - Text Files With this tutorial we are going to go through the basic elements of importing a text file that contains several records (lines) each containing several fields. Sample Data -
.txt - Exporting and Importing. Table of Contents
 .txt - Exporting and Importing Table of Contents Export... 2 Using Add Skip... 3 Delimiter... 3 Other Options... 4 Saving Templates of Options Chosen... 4 Editing Information in the lower Grid... 5 Import...
.txt - Exporting and Importing Table of Contents Export... 2 Using Add Skip... 3 Delimiter... 3 Other Options... 4 Saving Templates of Options Chosen... 4 Editing Information in the lower Grid... 5 Import...
TimeStudio process manual Static AOIs analysis
 TimeStudio process manual Static AOIs analysis Author: Pär Nyström Last revision: 2014-02-11 (Peter Johnsson) Innehåll TimeStudio process manual Static AOIs analysis... 1 1. Purpose... 2 2. Definitions...
TimeStudio process manual Static AOIs analysis Author: Pär Nyström Last revision: 2014-02-11 (Peter Johnsson) Innehåll TimeStudio process manual Static AOIs analysis... 1 1. Purpose... 2 2. Definitions...
A First Introduction to Scientific Visualization Geoffrey Gray
 Visual Molecular Dynamics A First Introduction to Scientific Visualization Geoffrey Gray VMD on CIRCE: On the lower bottom left of your screen, click on the window start-up menu. In the search box type
Visual Molecular Dynamics A First Introduction to Scientific Visualization Geoffrey Gray VMD on CIRCE: On the lower bottom left of your screen, click on the window start-up menu. In the search box type
Using McKesson Specialty Care Solutions US Oncology Order Center
 Using Specialty Care Solutions US Oncology Order The, mscs.mckesson.com, is an online destination that provides easy access to everything you need to manage your purchasing relationship with Specialty
Using Specialty Care Solutions US Oncology Order The, mscs.mckesson.com, is an online destination that provides easy access to everything you need to manage your purchasing relationship with Specialty
SASSIE-web: Quick Start Introduction. SASSIE-web Interface
 NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/docs/sassie-web-quick-start/quick-start.html. Return to Main Documents
NOTE: This PDF file is for reference purposes only. This lab should be accessed directly from the web at https://sassieweb.chem.utk.edu/docs/sassie-web-quick-start/quick-start.html. Return to Main Documents
The R and R-commander software
 The R and R-commander software This course uses the statistical package 'R' and the 'R-commander' graphical user interface (Rcmdr). Full details about these packages and the advantages associated with
The R and R-commander software This course uses the statistical package 'R' and the 'R-commander' graphical user interface (Rcmdr). Full details about these packages and the advantages associated with
Introduction to PyMOL
 Introduction to PyMOL Introduction to PyMOL 1.0...1 General Introduction...2 Layout and Features...2 Basic Mouse Controls...4 Visualization Presets...7 Making Measurements...9 Saving Images...10 Saving
Introduction to PyMOL Introduction to PyMOL 1.0...1 General Introduction...2 Layout and Features...2 Basic Mouse Controls...4 Visualization Presets...7 Making Measurements...9 Saving Images...10 Saving
DOCK 5.2 User Manual
 DOCK 5.2 User Manual Irwin D. Kuntz Demetri T. Moustakas Paula Therese Lang Copyright University of California 2005 Last updated March 2005 Introduction to DOCK 5.2 We are proud to announce the release
DOCK 5.2 User Manual Irwin D. Kuntz Demetri T. Moustakas Paula Therese Lang Copyright University of California 2005 Last updated March 2005 Introduction to DOCK 5.2 We are proud to announce the release
Lecture 3: Geometric and Signal 3D Processing (and some Visualization)
 Lecture 3: Geometric and Signal 3D Processing (and some Visualization) Chandrajit Bajaj Algorithms & Tools Structure elucidation: filtering, contrast enhancement, segmentation, skeletonization, subunit
Lecture 3: Geometric and Signal 3D Processing (and some Visualization) Chandrajit Bajaj Algorithms & Tools Structure elucidation: filtering, contrast enhancement, segmentation, skeletonization, subunit
Graduate Topics in Biophysical Chemistry CH Assignment 0 (Programming Assignment) Due Monday, March 19
 Introduction and Goals Graduate Topics in Biophysical Chemistry CH 8990 03 Assignment 0 (Programming Assignment) Due Monday, March 19 It is virtually impossible to be a successful scientist today without
Introduction and Goals Graduate Topics in Biophysical Chemistry CH 8990 03 Assignment 0 (Programming Assignment) Due Monday, March 19 It is virtually impossible to be a successful scientist today without
LOOMIS EXPRESS HOW TO IMPORT THE E-BILL LOOMIS ( ) Technical Support Hotline
 LOOMIS EXPRESS HOW TO IMPORT THE E-BILL www.loomis-express.com 1.855.2LOOMIS (1.855.256.6647) Technical Support Hotline 1.877.549.3638 HOW TO IMPORT THE E-BILL INTO EXCEL Thank you for choosing Loomis
LOOMIS EXPRESS HOW TO IMPORT THE E-BILL www.loomis-express.com 1.855.2LOOMIS (1.855.256.6647) Technical Support Hotline 1.877.549.3638 HOW TO IMPORT THE E-BILL INTO EXCEL Thank you for choosing Loomis
Docking with GOLD. Novembre 2009 CCDC. G. Marcou, E. Kellenberger. G. Marcou, E. Kellenberger () Docking with GOLD Novembre / 25
 Docking with GOLD CCDC G. Marcou, E. Kellenberger Novembre 2009 G. Marcou, E. Kellenberger () Docking with GOLD Novembre 2009 1 / 25 Sommaire 1 GOLD Suite organization 2 GOLD run setting - Wizard 3 GOLD
Docking with GOLD CCDC G. Marcou, E. Kellenberger Novembre 2009 G. Marcou, E. Kellenberger () Docking with GOLD Novembre 2009 1 / 25 Sommaire 1 GOLD Suite organization 2 GOLD run setting - Wizard 3 GOLD
Lab III: MD II & Analysis Linux & OS X. In this lab you will:
 Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
Bayesian Spam Detection System Using Hybrid Feature Selection Method
 2016 International Conference on Manufacturing Science and Information Engineering (ICMSIE 2016) ISBN: 978-1-60595-325-0 Bayesian Spam Detection System Using Hybrid Feature Selection Method JUNYING CHEN,
2016 International Conference on Manufacturing Science and Information Engineering (ICMSIE 2016) ISBN: 978-1-60595-325-0 Bayesian Spam Detection System Using Hybrid Feature Selection Method JUNYING CHEN,
Docking Tutorial Documentation
 Docking Tutorial Documentation Release 1.0 Noel O Boyle Nov 03, 2017 Contents 1 Overview of the AutoDock world 3 2 Configure PyRx to find AutoDock 5 3 Tutorial - Docking a HIV Protease Inhibitor 7 3.1
Docking Tutorial Documentation Release 1.0 Noel O Boyle Nov 03, 2017 Contents 1 Overview of the AutoDock world 3 2 Configure PyRx to find AutoDock 5 3 Tutorial - Docking a HIV Protease Inhibitor 7 3.1
Quick Reference: Returned Funds Report. Returned Funds Report Overview. Running a Returned Funds Report
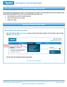 Returned Funds Report Overview The Returned Funds reporting process allows you to generate and view reports on returned funds activities tied to transactions and batches that meet specified criteria. You
Returned Funds Report Overview The Returned Funds reporting process allows you to generate and view reports on returned funds activities tied to transactions and batches that meet specified criteria. You
VEGETATION DESCRIPTION AND ANALYSIS
 VEGETATION DESCRIPTION AND ANALYSIS LABORATORY 5 AND 6 ORDINATIONS USING PC-ORD AND INSTRUCTIONS FOR LAB AND WRITTEN REPORT Introduction LABORATORY 5 (OCT 4, 2017) PC-ORD 1 BRAY & CURTIS ORDINATION AND
VEGETATION DESCRIPTION AND ANALYSIS LABORATORY 5 AND 6 ORDINATIONS USING PC-ORD AND INSTRUCTIONS FOR LAB AND WRITTEN REPORT Introduction LABORATORY 5 (OCT 4, 2017) PC-ORD 1 BRAY & CURTIS ORDINATION AND
Frequency Distributions and Descriptive Statistics in SPS
 230 Combs Building 859.622.3050 studentcomputing.eku.edu studentcomputing@eku.edu Frequency Distributions and Descriptive Statistics in SPSS In this tutorial, we re going to work through a sample problem
230 Combs Building 859.622.3050 studentcomputing.eku.edu studentcomputing@eku.edu Frequency Distributions and Descriptive Statistics in SPSS In this tutorial, we re going to work through a sample problem
PCGENESIS PAYROLL SYSTEM OPERATIONS GUIDE
 PCGENESIS PAYROLL SYSTEM OPERATIONS GUIDE 6/30/2010 Section C: Payroll Check and Direct Deposit Statement Processing, V2.0 [Topic 7: Import PCGenesis Paystub Leave Data for the Leave Management System,
PCGENESIS PAYROLL SYSTEM OPERATIONS GUIDE 6/30/2010 Section C: Payroll Check and Direct Deposit Statement Processing, V2.0 [Topic 7: Import PCGenesis Paystub Leave Data for the Leave Management System,
ANOMALY DETECTION ON MACHINE LOG
 ANOMALY DETECTION ON MACHINE LOG Data Mining Prof. Sunnie S Chung Ankur Pandit 2619650 Raw Data: NASA HTTP access logs It contain two month's of all HTTP requests to the NASA Kennedy Space Center WWW server
ANOMALY DETECTION ON MACHINE LOG Data Mining Prof. Sunnie S Chung Ankur Pandit 2619650 Raw Data: NASA HTTP access logs It contain two month's of all HTTP requests to the NASA Kennedy Space Center WWW server
Lab III: MD II & Analysis Linux & OS X. In this lab you will:
 Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
Lab III: MD II & Analysis Linux & OS X In this lab you will: 1) Add water, ions, and set up and minimize a solvated protein system 2) Learn how to create fixed atom files and systematically minimize a
Advanced Graphics: NOMAD Summer. Interactive analysis and visualization of complex datasets
 NOMAD Summer A hands-on course on tools for novel-materials discovery September 25-29, 2017, Berlin Advanced Graphics: Interactive analysis and visualization of complex datasets Michele Compostella Markus
NOMAD Summer A hands-on course on tools for novel-materials discovery September 25-29, 2017, Berlin Advanced Graphics: Interactive analysis and visualization of complex datasets Michele Compostella Markus
User s Guide. Using the R-Peridot Graphical User Interface (GUI) on Windows and GNU/Linux Systems
 User s Guide Using the R-Peridot Graphical User Interface (GUI) on Windows and GNU/Linux Systems Pitágoras Alves 01/06/2018 Natal-RN, Brazil Index 1. The R Environment Manager...
User s Guide Using the R-Peridot Graphical User Interface (GUI) on Windows and GNU/Linux Systems Pitágoras Alves 01/06/2018 Natal-RN, Brazil Index 1. The R Environment Manager...
User Guide to. NovaFold. Version 12.3
 User Guide to NovaFold Version 12.3 DNASTAR, Inc. 2015 NovaFold Overview NovaFold, DNASTAR s 3D protein structure prediction program, can use a sequence file to predict ligand binding sites and protein
User Guide to NovaFold Version 12.3 DNASTAR, Inc. 2015 NovaFold Overview NovaFold, DNASTAR s 3D protein structure prediction program, can use a sequence file to predict ligand binding sites and protein
