Atlas-Based Under-Segmentation
|
|
|
- Juniper Wells
- 6 years ago
- Views:
Transcription
1 Atlas-Based Under-Segmentation Christian Wachinger 1,2 and Polina Golland 1 1 Computer Science and Artificial Intelligence Lab, MIT 2 Massachusetts General Hospital, Harvard Medical School Abstract. We study the widespread, but rarely discussed, tendency of atlas-based segmentation to under-segment the organs of interest. Commonly used error measures do not distinguish between under- and oversegmentation, contributing to the problem. We explicitly quantify overand under-segmentation in several typical examples and present a new hypothesis for the cause. We provide evidence that segmenting only one organ of interest and merging all surrounding structures into one label creates bias towards background in the label estimates suggested by the atlas. We propose a generative model that corrects for this effect by learning the background structures from the data. Inference in the model separates the background into distinct structures and consequently improves the segmentation accuracy. Our experiments demonstrate a clear improvement in several applications. 1 Introduction Atlas-based segmentation exploits knowledge from previously labeled training images to segment the target image. In this paper, we focus on multi-atlas segmentation methods that map all labeled images onto the target image, which helps to reduce segmentation errors [6, 8, 11]. Label fusion combines the transferred labels into the final segmentation [9]. A common tendency of atlas-based segmentation to under-segment has largely been ignored in the field. We conjecture that one of the reasons that this phenomenon has not received more attention is that common error metrics do not capture the under-segmentation effect. For instance, the Dice volume overlap [3] and the Hausdorff distance [4] do not indicate if the segmentation is too large or too small. We are only aware of one recent article that addresses the spatial bias in atlas-based segmentation [12]. In that work, the bias is approximated by spatial convolution with an isotropic Gaussian kernel, modeling the distribution of residual registration errors. This model implies under-segmentation of convex shapes and over-segmentation of concave shapes. To reduce the spatial bias, a deconvolution is applied to the label maps. Results were reported for the segmentation of the hippocampus [12]. We present an alternative hypothesis for the bias in segmentation and propose a strategy to correct for such bias. First, we quantify the under-segmentation in atlas-based segmentation with new volume overlap measures. Our hypothesis ties the under-segmentation to the asymmetry of most segmentation setups where we seek to identify a single organ and merge all surrounding structures
2 2 C. Wachinger, P. Golland into one large background class. We show that this foreground-background segmentation strategy exhibits stronger bias than multi-organ segmentation. We propose a generative model of the background to correct under-segmentation even if the segmentation labels for multiple organs are not available. The posterior probability distribution of the Dirichlet process mixture model yields the splitting of the background into several components. Our experiments illustrate that this refined voting scheme improves the segmentation accuracy. 2 Under-segmentation in multi-atlas segmentation In multi-atlas segmentation, the training set includes images I = {I 1,..., I n } with the corresponding manual segmentations S = {S 1,..., S n } and S i (x) {1,..., η}, where η is the number of labels. The objective is to infer segmentation S for a new input image I. Probabilistic label maps L = {L 1,..., L η } specify the likelihood of each label l {1,..., η} at location x Ω in the new image L l (x) = n p(s(x) = l S i ) p(i(x) I i ). (1) i=1 The label maps satisfy l Ll (x) = 1 and L l (x) 1. For obtaining label likelihood, we register all training images I to the test image I, yielding deformation fields {φ 1,..., φ n }, and define p(s(x) = l S i ) = { 1 if Si (φ i (x)) = l, otherwise. (2) Alternatively, probabilistic segmentations S i can be included in the label likelihood, with the rest of the analysis unchanged. For majority voting (MV) [6, 8], the image likelihood is constant, p(i(x) I i ) 1. For intensity-weighted (IW) voting [9], also referred to as locally-weighted voting, the likelihood depends on image intensities p(i(x) I i ) exp ( (I(x) I i (φ i (x))) 2 /2σ 2), (3) where σ 2 is the variance of the image noise. We obtain the final segmentation Ŝ(x) by choosing the most likely label Ŝ(x) = arg max L l (x). (4) l For one structure (η=2), we directly compare foreground and background likelihoods to obtain the segmentation by identifying image locations x for which L f (x) > L b (x), or equivalently L f (x) > Quantifying under-segmentation Since the Dice volume overlap [3] and the Hausdorff distance [4] do not capture the type of segmentation error, we introduce two measures that explicitly quantify the over- and under-segmentation. Given the manual segmentation S and
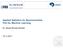
3 Atlas-Based Under-Segmentation Foreground Background Multi Organ WM GM HC CA PU PA AM AC VE WM GM HC CA PU PA AM AC VE Comparison of over and under segmentation 5 4 * Left Parotid Right Parotid Left Atrium Fig. 1: Statistical analysis of over-segmentation O (left bar) and under-segmentation U (right bar). Top: Segmentation of brain structures with foreground-background (left panel) and multi-organ (right panel) scheme. Left: Segmentation statistics for foreground-background segmentation of parotid glands and left atrium. Red line indicates the median, the boxes extend to the 25 th and 75 th percentiles, and the whiskers reach to the most extreme values not considered outliers (red crosses). *,, and indicate statistical significance levels at.5,.1, and.1, respectively. the automatic segmentation Ŝ, we define O(Ŝ, S) = Ŝ \ S S and U(Ŝ, S) = S \ Ŝ S (5) to quantify over- and under-segmentation, respectively. To examine the problem of under-segmentation, we compute and report statistics in three different segmentation applications using intensity-weighted voting in Fig. 1. The applications target the segmentation of (i) nine brain structures in magnetic resonance (MR) images, (ii) left and right parotid glands in CT images, and (iii) the left atrium of the heart in magnetic resonance angiography (MRA) images. For the brain, we perform foreground-background segmentation by segmenting each brain structure separately and merging all other structures into a background label. The structures we segment are white matter (WM), gray matter (GM), hippocampus (HC), caudate (CA), putamen (PU), pallidum (PA), amygdala (AM), accumbens (AC), ventricles (VE). Under-segmentation errors are significantly higher than over-segmentation errors in all three applications, suggesting a bias towards under-segmentation in atlas-based segmentation. 2.2 Foreground-background segmentation causes spatial bias Our hypothesis for the cause of under-segmentation is the asymmetry in how the foreground and background labels are treated by binary classification methods. Merging all surrounding structures into background causes this new meta-label to dominate in the voting process even if the evidence for the foreground label
4 VisualizeUnderseg.m 4 C. Wachinger, P. Golland 2 Multi Organ Voting 25 Frgrd Bckgrd Organ Bckgrd Voting Segmentation of Amygdala Votes 1 Votes 15 1 Dice WM GM HC AM Other Backgrd AM Merged Structures Fig. 2: Left: Manual segmentation of amygdala is shown in red, the outline of the automatic segmentation with foreground-background scheme is shown in yellow. Two middle panels: Distribution of votes for the location marked in black in the image on the left. Multi-organ segmentation correctly assigns the AM label. The foreground-background segmentation assigns the background label, which is an error. Right: Dice volume overlap as a function of the number of merged neighboring structures. is stronger than that for any of the surrounding structures. We illustrate this phenomenon on the example of the amygdala in Fig. 2. The atlas-based segmentation with the foreground-background scheme yields an under-segmentation (yellow outline). Investigating the votes for one location (black voxel in the left image), we observe that labels from several structures are present. Amygdala is assigned the highest number of votes and would win the voting in a multi-organ scheme. However, merging all other structures into a background label causes the background to win, leading to a segmentation error. To further illustrate the impact of merging neighboring structures, we examine the drop in the Dice volume overlap as we accumulate more and more structures into the background label (Fig. 2, right panel). To further quantify the difference between foreground-background and multiorgan segmentation, we report the under- and over-segmentation statistics for the brain segmentation in Fig. 1. In comparison to the foreground-background segmentation, the under-segmentation is reduced and most of the significant differences between over- and under-segmentation are reduced or eliminated when we use multiple labels. Interestingly, the gray matter segmentation changes from under- to over-segmentation, which may be attributed to its complex shape. While it is possible for the brain segmentation algorithms to use multi-organ schemes because many structures have been delineated in training data, it is not possible for many other applications (e.g., left atrium or parotid glands), where no multi-label training data exists. 3 Latent multi-label model of the background We introduce a generative model that estimates latent labels for the background structures from the available images. We emphasize that the method does not re-
5 Atlas-Based Under-Segmentation 5 quire multi-label training segmentations. We use image intensities of the training data to perform unsupervised separation of the background into K components while simultaneously estimating the number of components K. Estimated components serve as labels in the voting procedure. We assume that image patches P ix = I i (M(x)), with patch neighborhood M, are sampled from a Gaussian mixture model (GMM). Since we do not know the number of components a priori, we employ a Dirichlet process Gaussian mixture model (DP-GMM) [5, 1] to account for the potentially infinite number of components. In practice, the number of components is determined as part of the inference procedure. Formally, our generative model and the corresponding graphical model are as follows P ix z ix N (µ zix, Σ zix ), z ix Cat(π), π GEM(α), (µ k, Σ k ) H(λ), µ k, k 1 z ix P ix N where (µ k, Σ k ) are the mean and covariance of the normal distribution. We choose the conjugate Normal-Wishart distribution H with hyperparameter λ as a prior on the parameters (µ k, Σ k ) [5]. Mixture weights π follow a stickbreaking process GEM with parameter α [1]. Setting Σ u = σi, the asymptotic case of σ yields the DP-means algorithm [7], which is an extension of the k-means algorithm that assumes a variable number of clusters during the estimation procedure. We compare the performance of k-means, DP-means, GMM, and DP-GMM in our experiments. For k-means and DP-means, we use k-means++ seeding for initialization [1]. Once the inference yields a model with K components, the index z ix {1,..., K} specifies the component that generates the patch P ix. Since we only consider background patches, we replace the background label S i (x) = b with the component index S i (x) = z ix in the voting procedure. The labels for the foreground-background segmentation therefore change from {f, b} to {f, 1,..., K}. Voting on this updated label set as defined in Eq. (4) yields the segmentation. 3.1 Model Inference The increased model complexity of DP mixture models makes the posterior inference difficult. Variational inference algorithms that approximate the result lack convergence guarantees. Instead, we use a recently proposed inference scheme based on efficient Markov chain Monte Carlo sampling, which shows improved convergence properties [2]. The method combines non-ergodic, restricted Gibbs iteration with split-merge moves yielding an ergodic Markov chain. It is not necessary to perform the inference on the entire background region, as it will affect the voting only in voxels close to the organ boundary. We restrict the inference to the atlas-induced region Γ = {x Ω :.1 < L f (x) <.5},
6 6 C. Wachinger, P. Golland 8 Comparison of over and under segmentation, Latent Labels Improvement over foregr backgr for multi organ and latent label Dice difference * * WM GM HC CA PU PA AM AC VE WM GM HC CA PU PA AM AC VE Fig. 3: Segmentation statistics for brain data. Left: Over- and undersegmentation for each brain structure after inference of latent labels. Right: Improvement offered by multi-organ (left bar) and latent label estimation (right bar) over foreground-background segmentation. since our procedure does not change the vote of foreground locations. Within this region, we investigate a global and a local approach. The global approach considers background patches in the region Γ for all training images, P = {P ix : x Γ, S i (x) = b}. The local approach selects patches in a small region R around a current location P(x) = {P iy : y R(x), S i (y) = b}. Considering patches in a small region is necessary to have more relevant samples for learning the parameters. For the local approach, we perform separate inferences for each location on P(x), instead of one global inference on P. 4 Results We evaluate our approach on three datasets. The first set contains 39 brain MR T1 scans with 1mm isotropic resolution and dimensions that were used to construct the FreeSurfer atlas. The second dataset includes 18 CT scans from patients with head and neck cancer [11], containing between 8 and 2 axial slices with a slice thickness of 2.5mm. The in-plane resolution is.9mm, the slice size is pixels. The third dataset contains 16 heart MRA images that are electro-cardiogram gated to counteract considerable volume changes of the left atrium and contrast-enhanced (.2 mmol/kg, Gadolinium-DTPA, CIDA sequence, TR=4.3ms, TE=2.ms). The in-plane resolution varies from.51mm to.68mm and slice thickness varies from 1.2mm to 1.7mm with an image resolution of We use intensity-weighted voting for creating baseline label maps that serve as input to our algorithm (σ = 1 for brain, σ = 45 for head and neck, σ =.5 for heart). We compare to the deconvolution with a generalized Gaussian [12], where we sweep kernel parameters for each application to determine the best setting. We quantify the segmentation accuracy with the Dice volume overlap between manual and automatic segmentation. We set the patch size M to (3, 3, 3) for brain and (3, 3, 1) for the other two applications to account for anisotropy in the data. For the global approach, we evaluate k-means, DP-means, GMM, and DP-GMM. We set α =.1 for DP-
7 Atlas-Based Under-Segmentation 7 Left Parotid Gland Right Parotid Gland Dice.8 Dice IW Deconv g kmeans g DPmeans g GMM g DPGMM l kmeans l DPmeans IW Deconv g kmeans g DPmeans g GMM g DPGMM l kmeans l DPmeans Dice Left atrium IW Deconv g kmeans g DPmeans g GMM g DPGMM l kmeans l DPmeans Comparison of over and under segmentation Left Parotid Right Parotid Left Atrium Fig. 4: Segmentation accuracy of parotid glands (top) and left atrium (bottom left). We compare global (g-) and local (l-) approaches with intensity-weighted voting (IW) [9] and the deconvolution approach (Deconv) [12] as baseline methods. The plot in the bottom right shows the over- and under-segmentation statistics after latent label estimation for all three structures. GMM and create a new cluster in DP-means if the distance to a cluster center exceeds 1 times the average distance within a cluster. For GMM and k-means, we set the number of clusters to 5. For the local approach, we set the region R to (3, 3, 3) and only consider k-means and DP-means because other methods become computationally prohibitive. The number of clusters is set to 3 for the local approach, as we expect fewer structures to be present at one location. Fig. 3 reports segmentation results of brain structures with the inference of latent background labels using DP-GMM. The under-segmentation is reduced when compared to the foreground-background segmentation in Fig. 1. The latent label estimation offers improvements in accuracy that are comparable to those of the multi-organ scheme, without requiring the multiple organ segmentations for the training set. Fig. 4 illustrates segmentation results for the parotid glands and the left atrium, where we experiment with different inference methods and add the deconvolution approach to the comparison. We also quantify the undersegmentation for the proposed method. We observe that the differences between over- and under-segmentation are no longer significant. Our results demonstrate the advantage of estimating latent background labels over foreground-background segmentation. The non-parametric methods based on the Dirichlet process yield a slight additional gain compared to their parametric counterparts. This is a consequence of the simultaneous estimation of component membership and number of components, which enables dynamic adaptation to the data. The comparison of global and local approaches indicates that the performance is application dependent. While local approaches perform better for parotid glands, they are slightly worse for the left atrium. Our exper-
8 8 C. Wachinger, P. Golland iments with majority voting are not included in the article but they confirm the presented results. 5 Conclusion We demonstrated that a significant bias exists in atlas-based segmentation that leads to under-segmentation. We proposed the asymmetry in foreground-background segmentation as a new hypothesis for the cause of this phenomenon. To reduce the domination of the voting by the background, we introduced a generative model for the background based on the Dirichlet process mixture model. Inference of latent labels yielded partitioning of the background. Segmentation results for brain structures, parotid glands, and the left atrium illustrated clear improvement in the segmentation quality. Acknowledgements: We thank Jason Chang and Greg Sharp. This work was supported in part by the National Alliance for Medical Image Computing (U54- EB5149) and the NeuroImaging Analysis Center (P41-EB1592). References 1. Arthur, D., Vassilvitskii, S.: k-means++: the advantages of careful seeding. In: ACM-SIAM Symposium on Discrete Algorithms (SODA). pp (27) 2. Chang, J., Fisher III, J.W.: Parallel sampling of dp mixture models using subclusters splits. In: Neural Information Processing Systems. pp (213) 3. Dice, L.: Measures of the amount of ecologic association between species. Ecology 26(3), (1945) 4. Dubuisson, M., Jain, A.: A modified hausdorff distance for object matching. In: International Conference on Pattern Recognition. vol. 1, pp (1994) 5. Görür, D., Rasmussen, C.E.: Dirichlet process gaussian mixture models: Choice of the base distribution. Computer Science and Technology 25(4), (21) 6. Heckemann, R., Hajnal, J., Aljabar, P., Rueckert, D., Hammers, A.: Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage 33(1), (26) 7. Kulis, B., Jordan, M.I.: Revisiting k-means: New algorithms via bayesian nonparametrics. In: International Conference on Machine Learning. pp (212) 8. Rohlfing, T., Brandt, R., Menzel, R., Maurer, C., et al.: Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. NeuroImage 21(4), (24) 9. Sabuncu, M., Yeo, B., Van Leemput, K., Fischl, B., Golland, P.: A Generative Model for Image Segmentation Based on Label Fusion. IEEE Transactions on Medical Imaging 29 (21) 1. Sudderth, E.B.: Graphical Models for Visual Object Recognition and Tracking. Ph.D. thesis, Massachusetts Institute of Technology (26) 11. Wachinger, C., Sharp, G.C., Golland, P.: Contour-driven regression for label inference in atlas-based segmentation. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 213, Part III. LNCS, vol. 8151, pp Springer, Heidelberg (213) 12. Wang, H., Yushkevich, P.A.: Spatial bias in multi-atlas based segmentation. In: Computer Vision and Pattern Recognition (CVPR). pp (212)
Counter-Driven Regression for Label Inference in Atlas- Based Segmentation
 Counter-Driven Regression for Label Inference in Atlas- Based Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation
Counter-Driven Regression for Label Inference in Atlas- Based Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation
Contour-Driven Atlas-Based Segmentation
 Contour-Driven Atlas-Based Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Wachinger,
Contour-Driven Atlas-Based Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Wachinger,
A Generative Model for Image Segmentation Based on Label Fusion Mert R. Sabuncu*, B. T. Thomas Yeo, Koen Van Leemput, Bruce Fischl, and Polina Golland
 1714 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 29, NO. 10, OCTOBER 2010 A Generative Model for Image Segmentation Based on Label Fusion Mert R. Sabuncu*, B. T. Thomas Yeo, Koen Van Leemput, Bruce Fischl,
1714 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 29, NO. 10, OCTOBER 2010 A Generative Model for Image Segmentation Based on Label Fusion Mert R. Sabuncu*, B. T. Thomas Yeo, Koen Van Leemput, Bruce Fischl,
Norbert Schuff VA Medical Center and UCSF
 Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
3D Brain Segmentation Using Active Appearance Models and Local Regressors
 3D Brain Segmentation Using Active Appearance Models and Local Regressors K.O. Babalola, T.F. Cootes, C.J. Twining, V. Petrovic, and C.J. Taylor Division of Imaging Science and Biomedical Engineering,
3D Brain Segmentation Using Active Appearance Models and Local Regressors K.O. Babalola, T.F. Cootes, C.J. Twining, V. Petrovic, and C.J. Taylor Division of Imaging Science and Biomedical Engineering,
Multi-Atlas Segmentation of the Cardiac MR Right Ventricle
 Multi-Atlas Segmentation of the Cardiac MR Right Ventricle Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department of Radiology, University
Multi-Atlas Segmentation of the Cardiac MR Right Ventricle Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department of Radiology, University
An Introduction To Automatic Tissue Classification Of Brain MRI. Colm Elliott Mar 2014
 An Introduction To Automatic Tissue Classification Of Brain MRI Colm Elliott Mar 2014 Tissue Classification Tissue classification is part of many processing pipelines. We often want to classify each voxel
An Introduction To Automatic Tissue Classification Of Brain MRI Colm Elliott Mar 2014 Tissue Classification Tissue classification is part of many processing pipelines. We often want to classify each voxel
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation
 Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion
 Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department
Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department
Robust atlas-based segmentation of highly variable anatomy: left atrium segmentation
 Robust atlas-based segmentation of highly variable anatomy: left atrium segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters.
Robust atlas-based segmentation of highly variable anatomy: left atrium segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters.
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION
 ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
Atlas of Classifiers for Brain MRI Segmentation
 Atlas of Classifiers for Brain MRI Segmentation B. Kodner 1,2, S. H. Gordon 1,2, J. Goldberger 3 and T. Riklin Raviv 1,2 1 Department of Electrical and Computer Engineering, 2 The Zlotowski Center for
Atlas of Classifiers for Brain MRI Segmentation B. Kodner 1,2, S. H. Gordon 1,2, J. Goldberger 3 and T. Riklin Raviv 1,2 1 Department of Electrical and Computer Engineering, 2 The Zlotowski Center for
Nonparametric Mixture Models for Supervised Image Parcellation
 Nonparametric Mixture Models for Supervised Image Parcellation Mert R. Sabuncu 1,B.T.Thomas Yeo 1, Koen Van Leemput 1,2,3,Bruce Fischl 1,2, and Polina Golland 1 1 Computer Science and Artificial Intelligence
Nonparametric Mixture Models for Supervised Image Parcellation Mert R. Sabuncu 1,B.T.Thomas Yeo 1, Koen Van Leemput 1,2,3,Bruce Fischl 1,2, and Polina Golland 1 1 Computer Science and Artificial Intelligence
A Nonparametric Bayesian Approach to Detecting Spatial Activation Patterns in fmri Data
 A Nonparametric Bayesian Approach to Detecting Spatial Activation Patterns in fmri Data Seyoung Kim, Padhraic Smyth, and Hal Stern Bren School of Information and Computer Sciences University of California,
A Nonparametric Bayesian Approach to Detecting Spatial Activation Patterns in fmri Data Seyoung Kim, Padhraic Smyth, and Hal Stern Bren School of Information and Computer Sciences University of California,
Efficient Descriptor-Based Segmentation of Parotid Glands With Nonlocal Means
 1492 IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING, VOL. 64, NO. 7, JULY 2017 Efficient Descriptor-Based Segmentation of Parotid Glands With Nonlocal Means Christian Wachinger, Matthew Brennan, Greg C. Sharp,
1492 IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING, VOL. 64, NO. 7, JULY 2017 Efficient Descriptor-Based Segmentation of Parotid Glands With Nonlocal Means Christian Wachinger, Matthew Brennan, Greg C. Sharp,
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 10: Medical Image Segmentation as an Energy Minimization Problem
 SPRING 07 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
SPRING 07 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
Spatial Latent Dirichlet Allocation
 Spatial Latent Dirichlet Allocation Xiaogang Wang and Eric Grimson Computer Science and Computer Science and Artificial Intelligence Lab Massachusetts Tnstitute of Technology, Cambridge, MA, 02139, USA
Spatial Latent Dirichlet Allocation Xiaogang Wang and Eric Grimson Computer Science and Computer Science and Artificial Intelligence Lab Massachusetts Tnstitute of Technology, Cambridge, MA, 02139, USA
Optimal MAP Parameters Estimation in STAPLE - Learning from Performance Parameters versus Image Similarity Information
 Optimal MAP Parameters Estimation in STAPLE - Learning from Performance Parameters versus Image Similarity Information Subrahmanyam Gorthi 1, Alireza Akhondi-Asl 1, Jean-Philippe Thiran 2, and Simon K.
Optimal MAP Parameters Estimation in STAPLE - Learning from Performance Parameters versus Image Similarity Information Subrahmanyam Gorthi 1, Alireza Akhondi-Asl 1, Jean-Philippe Thiran 2, and Simon K.
Ensemble registration: Combining groupwise registration and segmentation
 PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
STIC AmSud Project. Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach
 STIC AmSud Project Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach Caroline Petitjean A joint work with Damien Grosgeorge, Pr Su Ruan, Pr JN Dacher, MD October 22,
STIC AmSud Project Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach Caroline Petitjean A joint work with Damien Grosgeorge, Pr Su Ruan, Pr JN Dacher, MD October 22,
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 10: Medical Image Segmentation as an Energy Minimization Problem
 SPRING 06 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
SPRING 06 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans
 Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans Eva M. van Rikxoort, Ivana Isgum, Marius Staring, Stefan Klein and Bram van Ginneken Image Sciences Institute,
Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans Eva M. van Rikxoort, Ivana Isgum, Marius Staring, Stefan Klein and Bram van Ginneken Image Sciences Institute,
Applied Bayesian Nonparametrics 5. Spatial Models via Gaussian Processes, not MRFs Tutorial at CVPR 2012 Erik Sudderth Brown University
 Applied Bayesian Nonparametrics 5. Spatial Models via Gaussian Processes, not MRFs Tutorial at CVPR 2012 Erik Sudderth Brown University NIPS 2008: E. Sudderth & M. Jordan, Shared Segmentation of Natural
Applied Bayesian Nonparametrics 5. Spatial Models via Gaussian Processes, not MRFs Tutorial at CVPR 2012 Erik Sudderth Brown University NIPS 2008: E. Sudderth & M. Jordan, Shared Segmentation of Natural
Subvoxel Segmentation and Representation of Brain Cortex Using Fuzzy Clustering and Gradient Vector Diffusion
 Subvoxel Segmentation and Representation of Brain Cortex Using Fuzzy Clustering and Gradient Vector Diffusion Ming-Ching Chang Xiaodong Tao GE Global Research Center {changm, taox} @ research.ge.com SPIE
Subvoxel Segmentation and Representation of Brain Cortex Using Fuzzy Clustering and Gradient Vector Diffusion Ming-Ching Chang Xiaodong Tao GE Global Research Center {changm, taox} @ research.ge.com SPIE
8/3/2017. Contour Assessment for Quality Assurance and Data Mining. Objective. Outline. Tom Purdie, PhD, MCCPM
 Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Auto-contouring the Prostate for Online Adaptive Radiotherapy
 Auto-contouring the Prostate for Online Adaptive Radiotherapy Yan Zhou 1 and Xiao Han 1 Elekta Inc., Maryland Heights, MO, USA yan.zhou@elekta.com, xiao.han@elekta.com, Abstract. Among all the organs under
Auto-contouring the Prostate for Online Adaptive Radiotherapy Yan Zhou 1 and Xiao Han 1 Elekta Inc., Maryland Heights, MO, USA yan.zhou@elekta.com, xiao.han@elekta.com, Abstract. Among all the organs under
Manifold Learning-based Data Sampling for Model Training
 Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
An Efficient Model Selection for Gaussian Mixture Model in a Bayesian Framework
 IEEE SIGNAL PROCESSING LETTERS, VOL. XX, NO. XX, XXX 23 An Efficient Model Selection for Gaussian Mixture Model in a Bayesian Framework Ji Won Yoon arxiv:37.99v [cs.lg] 3 Jul 23 Abstract In order to cluster
IEEE SIGNAL PROCESSING LETTERS, VOL. XX, NO. XX, XXX 23 An Efficient Model Selection for Gaussian Mixture Model in a Bayesian Framework Ji Won Yoon arxiv:37.99v [cs.lg] 3 Jul 23 Abstract In order to cluster
Modified Expectation Maximization Method for Automatic Segmentation of MR Brain Images
 Modified Expectation Maximization Method for Automatic Segmentation of MR Brain Images R.Meena Prakash, R.Shantha Selva Kumari 1 P.S.R.Engineering College, Sivakasi, Tamil Nadu, India 2 Mepco Schlenk Engineering
Modified Expectation Maximization Method for Automatic Segmentation of MR Brain Images R.Meena Prakash, R.Shantha Selva Kumari 1 P.S.R.Engineering College, Sivakasi, Tamil Nadu, India 2 Mepco Schlenk Engineering
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations
 Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
IMAGE SEGMENTATION. Václav Hlaváč
 IMAGE SEGMENTATION Václav Hlaváč Czech Technical University in Prague Faculty of Electrical Engineering, Department of Cybernetics Center for Machine Perception http://cmp.felk.cvut.cz/ hlavac, hlavac@fel.cvut.cz
IMAGE SEGMENTATION Václav Hlaváč Czech Technical University in Prague Faculty of Electrical Engineering, Department of Cybernetics Center for Machine Perception http://cmp.felk.cvut.cz/ hlavac, hlavac@fel.cvut.cz
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
Spatial Regularization of Functional Connectivity Using High-Dimensional Markov Random Fields
 Spatial Regularization of Functional Connectivity Using High-Dimensional Markov Random Fields Wei Liu 1, Peihong Zhu 1, Jeffrey S. Anderson 2, Deborah Yurgelun-Todd 3, and P. Thomas Fletcher 1 1 Scientific
Spatial Regularization of Functional Connectivity Using High-Dimensional Markov Random Fields Wei Liu 1, Peihong Zhu 1, Jeffrey S. Anderson 2, Deborah Yurgelun-Todd 3, and P. Thomas Fletcher 1 1 Scientific
Multi-atlas spectral PatchMatch: Application to cardiac image segmentation
 Multi-atlas spectral PatchMatch: Application to cardiac image segmentation W. Shi 1, H. Lombaert 3, W. Bai 1, C. Ledig 1, X. Zhuang 2, A. Marvao 1, T. Dawes 1, D. O Regan 1, and D. Rueckert 1 1 Biomedical
Multi-atlas spectral PatchMatch: Application to cardiac image segmentation W. Shi 1, H. Lombaert 3, W. Bai 1, C. Ledig 1, X. Zhuang 2, A. Marvao 1, T. Dawes 1, D. O Regan 1, and D. Rueckert 1 1 Biomedical
Machine Learning and Data Mining. Clustering (1): Basics. Kalev Kask
 Machine Learning and Data Mining Clustering (1): Basics Kalev Kask Unsupervised learning Supervised learning Predict target value ( y ) given features ( x ) Unsupervised learning Understand patterns of
Machine Learning and Data Mining Clustering (1): Basics Kalev Kask Unsupervised learning Supervised learning Predict target value ( y ) given features ( x ) Unsupervised learning Understand patterns of
PBSI: A symmetric probabilistic extension of the Boundary Shift Integral
 PBSI: A symmetric probabilistic extension of the Boundary Shift Integral Christian Ledig 1, Robin Wolz 1, Paul Aljabar 1,2, Jyrki Lötjönen 3, Daniel Rueckert 1 1 Department of Computing, Imperial College
PBSI: A symmetric probabilistic extension of the Boundary Shift Integral Christian Ledig 1, Robin Wolz 1, Paul Aljabar 1,2, Jyrki Lötjönen 3, Daniel Rueckert 1 1 Department of Computing, Imperial College
Norbert Schuff Professor of Radiology VA Medical Center and UCSF
 Norbert Schuff Professor of Radiology Medical Center and UCSF Norbert.schuff@ucsf.edu 2010, N.Schuff Slide 1/67 Overview Definitions Role of Segmentation Segmentation methods Intensity based Shape based
Norbert Schuff Professor of Radiology Medical Center and UCSF Norbert.schuff@ucsf.edu 2010, N.Schuff Slide 1/67 Overview Definitions Role of Segmentation Segmentation methods Intensity based Shape based
PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION
 PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION Ms. Vaibhavi Nandkumar Jagtap 1, Mr. Santosh D. Kale 2 1 PG Scholar, 2 Assistant Professor, Department of Electronics and Telecommunication,
PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION Ms. Vaibhavi Nandkumar Jagtap 1, Mr. Santosh D. Kale 2 1 PG Scholar, 2 Assistant Professor, Department of Electronics and Telecommunication,
NIH Public Access Author Manuscript Conf Comput Vis Pattern Recognit Workshops. Author manuscript; available in PMC 2012 May 01.
 NIH Public Access Author Manuscript Conf Comput Vis Pattern Recognit Workshops. Author manuscript; available in PMC 2012 May 01. Published in final edited form as: Conf Comput Vis Pattern Recognit Workshops.
NIH Public Access Author Manuscript Conf Comput Vis Pattern Recognit Workshops. Author manuscript; available in PMC 2012 May 01. Published in final edited form as: Conf Comput Vis Pattern Recognit Workshops.
Shape-Aware Multi-Atlas Segmentation
 Shape-Aware Multi-Atlas Segmentation Jennifer Alvén, Fredrik Kahl, Matilda Landgren, Viktor Larsson and Johannes Ulén Department of Signals and Systems, Chalmers University of Technology, Sweden Email:
Shape-Aware Multi-Atlas Segmentation Jennifer Alvén, Fredrik Kahl, Matilda Landgren, Viktor Larsson and Johannes Ulén Department of Signals and Systems, Chalmers University of Technology, Sweden Email:
Homework #4 Programming Assignment Due: 11:59 pm, November 4, 2018
 CSCI 567, Fall 18 Haipeng Luo Homework #4 Programming Assignment Due: 11:59 pm, ovember 4, 2018 General instructions Your repository will have now a directory P4/. Please do not change the name of this
CSCI 567, Fall 18 Haipeng Luo Homework #4 Programming Assignment Due: 11:59 pm, ovember 4, 2018 General instructions Your repository will have now a directory P4/. Please do not change the name of this
Performance Evaluation of the TINA Medical Image Segmentation Algorithm on Brainweb Simulated Images
 Tina Memo No. 2008-003 Internal Memo Performance Evaluation of the TINA Medical Image Segmentation Algorithm on Brainweb Simulated Images P. A. Bromiley Last updated 20 / 12 / 2007 Imaging Science and
Tina Memo No. 2008-003 Internal Memo Performance Evaluation of the TINA Medical Image Segmentation Algorithm on Brainweb Simulated Images P. A. Bromiley Last updated 20 / 12 / 2007 Imaging Science and
Markov Random Fields and Gibbs Sampling for Image Denoising
 Markov Random Fields and Gibbs Sampling for Image Denoising Chang Yue Electrical Engineering Stanford University changyue@stanfoed.edu Abstract This project applies Gibbs Sampling based on different Markov
Markov Random Fields and Gibbs Sampling for Image Denoising Chang Yue Electrical Engineering Stanford University changyue@stanfoed.edu Abstract This project applies Gibbs Sampling based on different Markov
Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation
 Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Ezequiel Geremia 2, Nicholas Ayache 2, and Gabor Szekely 1 1 Computer
Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Ezequiel Geremia 2, Nicholas Ayache 2, and Gabor Szekely 1 1 Computer
Ischemic Stroke Lesion Segmentation Proceedings 5th October 2015 Munich, Germany
 0111010001110001101000100101010111100111011100100011011101110101101012 Ischemic Stroke Lesion Segmentation www.isles-challenge.org Proceedings 5th October 2015 Munich, Germany Preface Stroke is the second
0111010001110001101000100101010111100111011100100011011101110101101012 Ischemic Stroke Lesion Segmentation www.isles-challenge.org Proceedings 5th October 2015 Munich, Germany Preface Stroke is the second
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Histograms. h(r k ) = n k. p(r k )= n k /NM. Histogram: number of times intensity level rk appears in the image
 Histograms h(r k ) = n k Histogram: number of times intensity level rk appears in the image p(r k )= n k /NM normalized histogram also a probability of occurence 1 Histogram of Image Intensities Create
Histograms h(r k ) = n k Histogram: number of times intensity level rk appears in the image p(r k )= n k /NM normalized histogram also a probability of occurence 1 Histogram of Image Intensities Create
Image Segmentation Using Iterated Graph Cuts BasedonMulti-scaleSmoothing
 Image Segmentation Using Iterated Graph Cuts BasedonMulti-scaleSmoothing Tomoyuki Nagahashi 1, Hironobu Fujiyoshi 1, and Takeo Kanade 2 1 Dept. of Computer Science, Chubu University. Matsumoto 1200, Kasugai,
Image Segmentation Using Iterated Graph Cuts BasedonMulti-scaleSmoothing Tomoyuki Nagahashi 1, Hironobu Fujiyoshi 1, and Takeo Kanade 2 1 Dept. of Computer Science, Chubu University. Matsumoto 1200, Kasugai,
Keypoint Transfer Segmentation
 Keypoint Transfer Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Wachinger, C.; Toews,
Keypoint Transfer Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Wachinger, C.; Toews,
Unsupervised Learning
 Networks for Pattern Recognition, 2014 Networks for Single Linkage K-Means Soft DBSCAN PCA Networks for Kohonen Maps Linear Vector Quantization Networks for Problems/Approaches in Machine Learning Supervised
Networks for Pattern Recognition, 2014 Networks for Single Linkage K-Means Soft DBSCAN PCA Networks for Kohonen Maps Linear Vector Quantization Networks for Problems/Approaches in Machine Learning Supervised
Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas
 Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas Tobias Gass, Gabor Szekely, and Orcun Goksel Computer Vision Lab, Dep. of Electrical Engineering, ETH Zurich, Switzerland.
Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas Tobias Gass, Gabor Szekely, and Orcun Goksel Computer Vision Lab, Dep. of Electrical Engineering, ETH Zurich, Switzerland.
Automatic Subcortical Segmentation Using a Contextual Model
 Automatic Subcortical Segmentation Using a Contextual Model Jonathan H. Morra 1, Zhuowen Tu 1, Liana G. Apostolova 1,2, Amity E. Green 1,2, Arthur W. Toga 1, and Paul M. Thompson 1 1 Laboratory of Neuro
Automatic Subcortical Segmentation Using a Contextual Model Jonathan H. Morra 1, Zhuowen Tu 1, Liana G. Apostolova 1,2, Amity E. Green 1,2, Arthur W. Toga 1, and Paul M. Thompson 1 1 Laboratory of Neuro
MRI Segmentation. MRI Bootcamp, 14 th of January J. Miguel Valverde
 MRI Segmentation MRI Bootcamp, 14 th of January 2019 Segmentation Segmentation Information Segmentation Algorithms Approach Types of Information Local 4 45 100 110 80 50 76 42 27 186 177 120 167 111 56
MRI Segmentation MRI Bootcamp, 14 th of January 2019 Segmentation Segmentation Information Segmentation Algorithms Approach Types of Information Local 4 45 100 110 80 50 76 42 27 186 177 120 167 111 56
PET Image Reconstruction using Anatomical Information through Mutual Information Based Priors
 2005 IEEE Nuclear Science Symposium Conference Record M11-354 PET Image Reconstruction using Anatomical Information through Mutual Information Based Priors Sangeetha Somayajula, Evren Asma, and Richard
2005 IEEE Nuclear Science Symposium Conference Record M11-354 PET Image Reconstruction using Anatomical Information through Mutual Information Based Priors Sangeetha Somayajula, Evren Asma, and Richard
ABSTRACT 1. INTRODUCTION 2. METHODS
 Finding Seeds for Segmentation Using Statistical Fusion Fangxu Xing *a, Andrew J. Asman b, Jerry L. Prince a,c, Bennett A. Landman b,c,d a Department of Electrical and Computer Engineering, Johns Hopkins
Finding Seeds for Segmentation Using Statistical Fusion Fangxu Xing *a, Andrew J. Asman b, Jerry L. Prince a,c, Bennett A. Landman b,c,d a Department of Electrical and Computer Engineering, Johns Hopkins
CS 229 Midterm Review
 CS 229 Midterm Review Course Staff Fall 2018 11/2/2018 Outline Today: SVMs Kernels Tree Ensembles EM Algorithm / Mixture Models [ Focus on building intuition, less so on solving specific problems. Ask
CS 229 Midterm Review Course Staff Fall 2018 11/2/2018 Outline Today: SVMs Kernels Tree Ensembles EM Algorithm / Mixture Models [ Focus on building intuition, less so on solving specific problems. Ask
Clustering. Chapter 10 in Introduction to statistical learning
 Clustering Chapter 10 in Introduction to statistical learning 16 14 12 10 8 6 4 2 0 2 4 6 8 10 12 14 1 Clustering ² Clustering is the art of finding groups in data (Kaufman and Rousseeuw, 1990). ² What
Clustering Chapter 10 in Introduction to statistical learning 16 14 12 10 8 6 4 2 0 2 4 6 8 10 12 14 1 Clustering ² Clustering is the art of finding groups in data (Kaufman and Rousseeuw, 1990). ² What
mritc: A Package for MRI Tissue Classification
 mritc: A Package for MRI Tissue Classification Dai Feng 1 Luke Tierney 2 1 Merck Research Labratories 2 University of Iowa July 2010 Feng & Tierney (Merck & U of Iowa) MRI Tissue Classification July 2010
mritc: A Package for MRI Tissue Classification Dai Feng 1 Luke Tierney 2 1 Merck Research Labratories 2 University of Iowa July 2010 Feng & Tierney (Merck & U of Iowa) MRI Tissue Classification July 2010
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion
 Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
Smooth Image Segmentation by Nonparametric Bayesian Inference
 Smooth Image Segmentation by Nonparametric Bayesian Inference Peter Orbanz and Joachim M. Buhmann Institute of Computational Science, ETH Zurich European Conference on Computer Vision (ECCV), 2006 The
Smooth Image Segmentation by Nonparametric Bayesian Inference Peter Orbanz and Joachim M. Buhmann Institute of Computational Science, ETH Zurich European Conference on Computer Vision (ECCV), 2006 The
Image Segmentation Using Iterated Graph Cuts Based on Multi-scale Smoothing
 Image Segmentation Using Iterated Graph Cuts Based on Multi-scale Smoothing Tomoyuki Nagahashi 1, Hironobu Fujiyoshi 1, and Takeo Kanade 2 1 Dept. of Computer Science, Chubu University. Matsumoto 1200,
Image Segmentation Using Iterated Graph Cuts Based on Multi-scale Smoothing Tomoyuki Nagahashi 1, Hironobu Fujiyoshi 1, and Takeo Kanade 2 1 Dept. of Computer Science, Chubu University. Matsumoto 1200,
Pictorial Multi-atlas Segmentation of Brain MRI
 Pictorial Multi-atlas Segmentation of Brain MRI Cheng-Yi Liu 1, Juan Eugenio Iglesias 1,2, Zhuowen Tu 1 1 University of California, Los Angeles, Laboratory of Neuro Imaging 2 Athinoula A. Martinos Center
Pictorial Multi-atlas Segmentation of Brain MRI Cheng-Yi Liu 1, Juan Eugenio Iglesias 1,2, Zhuowen Tu 1 1 University of California, Los Angeles, Laboratory of Neuro Imaging 2 Athinoula A. Martinos Center
Blind Image Deblurring Using Dark Channel Prior
 Blind Image Deblurring Using Dark Channel Prior Jinshan Pan 1,2,3, Deqing Sun 2,4, Hanspeter Pfister 2, and Ming-Hsuan Yang 3 1 Dalian University of Technology 2 Harvard University 3 UC Merced 4 NVIDIA
Blind Image Deblurring Using Dark Channel Prior Jinshan Pan 1,2,3, Deqing Sun 2,4, Hanspeter Pfister 2, and Ming-Hsuan Yang 3 1 Dalian University of Technology 2 Harvard University 3 UC Merced 4 NVIDIA
Clustering Relational Data using the Infinite Relational Model
 Clustering Relational Data using the Infinite Relational Model Ana Daglis Supervised by: Matthew Ludkin September 4, 2015 Ana Daglis Clustering Data using the Infinite Relational Model September 4, 2015
Clustering Relational Data using the Infinite Relational Model Ana Daglis Supervised by: Matthew Ludkin September 4, 2015 Ana Daglis Clustering Data using the Infinite Relational Model September 4, 2015
Applied Statistics for Neuroscientists Part IIa: Machine Learning
 Applied Statistics for Neuroscientists Part IIa: Machine Learning Dr. Seyed-Ahmad Ahmadi 04.04.2017 16.11.2017 Outline Machine Learning Difference between statistics and machine learning Modeling the problem
Applied Statistics for Neuroscientists Part IIa: Machine Learning Dr. Seyed-Ahmad Ahmadi 04.04.2017 16.11.2017 Outline Machine Learning Difference between statistics and machine learning Modeling the problem
Segmentation of the Pectoral Muscle in Breast MRI Using Atlas-Based Approaches
 Segmentation of the Pectoral Muscle in Breast MRI Using Atlas-Based Approaches Albert Gubern-Mérida 1, Michiel Kallenberg 2, Robert Martí 1, and Nico Karssemeijer 2 1 University of Girona, Spain {agubern,marly}@eia.udg.edu
Segmentation of the Pectoral Muscle in Breast MRI Using Atlas-Based Approaches Albert Gubern-Mérida 1, Michiel Kallenberg 2, Robert Martí 1, and Nico Karssemeijer 2 1 University of Girona, Spain {agubern,marly}@eia.udg.edu
MARS: Multiple Atlases Robust Segmentation
 Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Correspondence Detection Using Wavelet-Based Attribute Vectors
 Correspondence Detection Using Wavelet-Based Attribute Vectors Zhong Xue, Dinggang Shen, and Christos Davatzikos Section of Biomedical Image Analysis, Department of Radiology University of Pennsylvania,
Correspondence Detection Using Wavelet-Based Attribute Vectors Zhong Xue, Dinggang Shen, and Christos Davatzikos Section of Biomedical Image Analysis, Department of Radiology University of Pennsylvania,
arxiv: v1 [cs.cv] 27 Feb 2017
![arxiv: v1 [cs.cv] 27 Feb 2017 arxiv: v1 [cs.cv] 27 Feb 2017](/thumbs/79/78828884.jpg) DeepNAT: Deep Convolutional Neural Network for Segmenting Neuroanatomy Christian Wachinger a, Martin Reuter b,c, Tassilo Klein d a Department of Child and Adolescent Psychiatry, Psychosomatic and Psychotherapy,
DeepNAT: Deep Convolutional Neural Network for Segmenting Neuroanatomy Christian Wachinger a, Martin Reuter b,c, Tassilo Klein d a Department of Child and Adolescent Psychiatry, Psychosomatic and Psychotherapy,
PSU Student Research Symposium 2017 Bayesian Optimization for Refining Object Proposals, with an Application to Pedestrian Detection Anthony D.
 PSU Student Research Symposium 2017 Bayesian Optimization for Refining Object Proposals, with an Application to Pedestrian Detection Anthony D. Rhodes 5/10/17 What is Machine Learning? Machine learning
PSU Student Research Symposium 2017 Bayesian Optimization for Refining Object Proposals, with an Application to Pedestrian Detection Anthony D. Rhodes 5/10/17 What is Machine Learning? Machine learning
Scene-Based Segmentation of Multiple Muscles from MRI in MITK
 Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2014 February 03.
 NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2014 February 03. Published in final edited form as: Med Image Comput Comput Assist Interv.
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2014 February 03. Published in final edited form as: Med Image Comput Comput Assist Interv.
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2014 October 07.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
Regional Manifold Learning for Deformable Registration of Brain MR Images
 Regional Manifold Learning for Deformable Registration of Brain MR Images Dong Hye Ye, Jihun Hamm, Dongjin Kwon, Christos Davatzikos, and Kilian M. Pohl Department of Radiology, University of Pennsylvania,
Regional Manifold Learning for Deformable Registration of Brain MR Images Dong Hye Ye, Jihun Hamm, Dongjin Kwon, Christos Davatzikos, and Kilian M. Pohl Department of Radiology, University of Pennsylvania,
A Cautionary Analysis of STAPLE Using Direct Inference of Segmentation Truth
 A Cautionary Analysis of STAPLE Using Direct Inference of Segmentation Truth Koen Van Leemput 1,2 and Mert R. Sabuncu 1 1 A.A. Martinos Center for Biomedical Imaging, MGH, Harvard Medical School, USA 2
A Cautionary Analysis of STAPLE Using Direct Inference of Segmentation Truth Koen Van Leemput 1,2 and Mert R. Sabuncu 1 1 A.A. Martinos Center for Biomedical Imaging, MGH, Harvard Medical School, USA 2
Exploratory fmri analysis without spatial normalization
 Exploratory fmri analysis without spatial normalization The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher
Exploratory fmri analysis without spatial normalization The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher
Introduction to Pattern Recognition Part II. Selim Aksoy Bilkent University Department of Computer Engineering
 Introduction to Pattern Recognition Part II Selim Aksoy Bilkent University Department of Computer Engineering saksoy@cs.bilkent.edu.tr RETINA Pattern Recognition Tutorial, Summer 2005 Overview Statistical
Introduction to Pattern Recognition Part II Selim Aksoy Bilkent University Department of Computer Engineering saksoy@cs.bilkent.edu.tr RETINA Pattern Recognition Tutorial, Summer 2005 Overview Statistical
Learning-based Neuroimage Registration
 Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
A Model-Independent, Multi-Image Approach to MR Inhomogeneity Correction
 Tina Memo No. 2007-003 Published in Proc. MIUA 2007 A Model-Independent, Multi-Image Approach to MR Inhomogeneity Correction P. A. Bromiley and N.A. Thacker Last updated 13 / 4 / 2007 Imaging Science and
Tina Memo No. 2007-003 Published in Proc. MIUA 2007 A Model-Independent, Multi-Image Approach to MR Inhomogeneity Correction P. A. Bromiley and N.A. Thacker Last updated 13 / 4 / 2007 Imaging Science and
Colorado School of Mines. Computer Vision. Professor William Hoff Dept of Electrical Engineering &Computer Science.
 Professor William Hoff Dept of Electrical Engineering &Computer Science http://inside.mines.edu/~whoff/ 1 Image Segmentation Some material for these slides comes from https://www.csd.uwo.ca/courses/cs4487a/
Professor William Hoff Dept of Electrical Engineering &Computer Science http://inside.mines.edu/~whoff/ 1 Image Segmentation Some material for these slides comes from https://www.csd.uwo.ca/courses/cs4487a/
ECG782: Multidimensional Digital Signal Processing
 ECG782: Multidimensional Digital Signal Processing Object Recognition http://www.ee.unlv.edu/~b1morris/ecg782/ 2 Outline Knowledge Representation Statistical Pattern Recognition Neural Networks Boosting
ECG782: Multidimensional Digital Signal Processing Object Recognition http://www.ee.unlv.edu/~b1morris/ecg782/ 2 Outline Knowledge Representation Statistical Pattern Recognition Neural Networks Boosting
for Images A Bayesian Deformation Model
 Statistics in Imaging Workshop July 8, 2004 A Bayesian Deformation Model for Images Sining Chen Postdoctoral Fellow Biostatistics Division, Dept. of Oncology, School of Medicine Outline 1. Introducing
Statistics in Imaging Workshop July 8, 2004 A Bayesian Deformation Model for Images Sining Chen Postdoctoral Fellow Biostatistics Division, Dept. of Oncology, School of Medicine Outline 1. Introducing
EXPECTED LABEL VALUE COMPUTATION FOR ATLAS-BASED IMAGE SEGMENTATION. Iman Aganj and Bruce Fischl
 EXPECTED LABEL VALUE COMPUTATION FOR ATLAS-BASED IMAGE SEGMENTATION Iman Aganj and Bruce Fischl Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School Computer Science
EXPECTED LABEL VALUE COMPUTATION FOR ATLAS-BASED IMAGE SEGMENTATION Iman Aganj and Bruce Fischl Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School Computer Science
AN AUTOMATED SEGMENTATION FRAMEWORK FOR BRAIN MRI VOLUMES BASED ON ADAPTIVE MEAN-SHIFT CLUSTERING
 AN AUTOMATED SEGMENTATION FRAMEWORK FOR BRAIN MRI VOLUMES BASED ON ADAPTIVE MEAN-SHIFT CLUSTERING Sam Ponnachan 1, Paul Pandi 2,A.Winifred 3,Joselin Jose 4 UG Scholars 1,2,3,4 Department of EIE 1,2 Department
AN AUTOMATED SEGMENTATION FRAMEWORK FOR BRAIN MRI VOLUMES BASED ON ADAPTIVE MEAN-SHIFT CLUSTERING Sam Ponnachan 1, Paul Pandi 2,A.Winifred 3,Joselin Jose 4 UG Scholars 1,2,3,4 Department of EIE 1,2 Department
Data mining for neuroimaging data. John Ashburner
 Data mining for neuroimaging data John Ashburner MODELLING The Scientific Process MacKay, David JC. Bayesian interpolation. Neural computation 4, no. 3 (1992): 415-447. Model Selection Search for the best
Data mining for neuroimaging data John Ashburner MODELLING The Scientific Process MacKay, David JC. Bayesian interpolation. Neural computation 4, no. 3 (1992): 415-447. Model Selection Search for the best
Multi-atlas labeling with population-specific template and non-local patch-based label fusion
 Multi-atlas labeling with population-specific template and non-local patch-based label fusion Vladimir Fonov, Pierrick Coupé, Simon Eskildsen, Jose Manjon, Louis Collins To cite this version: Vladimir
Multi-atlas labeling with population-specific template and non-local patch-based label fusion Vladimir Fonov, Pierrick Coupé, Simon Eskildsen, Jose Manjon, Louis Collins To cite this version: Vladimir
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Graph-Shifts Anatomic 3D Segmentation by Dynamic Hierarchical Minimization
 Graph-Shifts Anatomic 3D Segmentation by Dynamic Hierarchical Minimization Jason Corso Postdoctoral Fellow UCLA LONI/CCB jcorso@ucla.edu Motivation The work deals with the problem of automatically labeling
Graph-Shifts Anatomic 3D Segmentation by Dynamic Hierarchical Minimization Jason Corso Postdoctoral Fellow UCLA LONI/CCB jcorso@ucla.edu Motivation The work deals with the problem of automatically labeling
Automatic Generation of Shape Models Using Nonrigid Registration with a Single Segmented Template Mesh
 Automatic Generation of Shape Models Using Nonrigid Registration with a Single Segmented Template Mesh Geremy Heitz, Torsten Rohlfing, and Calvin R. Maurer, Jr. Image Guidance Laboratories Department of
Automatic Generation of Shape Models Using Nonrigid Registration with a Single Segmented Template Mesh Geremy Heitz, Torsten Rohlfing, and Calvin R. Maurer, Jr. Image Guidance Laboratories Department of
Sparsity Based Spectral Embedding: Application to Multi-Atlas Echocardiography Segmentation!
 Sparsity Based Spectral Embedding: Application to Multi-Atlas Echocardiography Segmentation Ozan Oktay, Wenzhe Shi, Jose Caballero, Kevin Keraudren, and Daniel Rueckert Department of Compu.ng Imperial
Sparsity Based Spectral Embedding: Application to Multi-Atlas Echocardiography Segmentation Ozan Oktay, Wenzhe Shi, Jose Caballero, Kevin Keraudren, and Daniel Rueckert Department of Compu.ng Imperial
A Generative Model for Probabilistic Label Fusion of Multimodal Data
 A Generative Model for Probabilistic Label Fusion of Multimodal Data Juan Eugenio Iglesias 1, Mert Rory Sabuncu 1,, and Koen Van Leemput 1,2,3, 1 Martinos Center for Biomedical Imaging, MGH, Harvard Medical
A Generative Model for Probabilistic Label Fusion of Multimodal Data Juan Eugenio Iglesias 1, Mert Rory Sabuncu 1,, and Koen Van Leemput 1,2,3, 1 Martinos Center for Biomedical Imaging, MGH, Harvard Medical
Preprocessing II: Between Subjects John Ashburner
 Preprocessing II: Between Subjects John Ashburner Pre-processing Overview Statistics or whatever fmri time-series Anatomical MRI Template Smoothed Estimate Spatial Norm Motion Correct Smooth Coregister
Preprocessing II: Between Subjects John Ashburner Pre-processing Overview Statistics or whatever fmri time-series Anatomical MRI Template Smoothed Estimate Spatial Norm Motion Correct Smooth Coregister
Methods in Computer Vision: Mixture Models and their Applications
 Methods in Computer Vision: Mixture Models and their Applications Oren Freifeld Computer Science, Ben-Gurion University May 7, 2017 May 7, 2017 1 / 40 1 Background Modeling Digression: Mixture Models GMM
Methods in Computer Vision: Mixture Models and their Applications Oren Freifeld Computer Science, Ben-Gurion University May 7, 2017 May 7, 2017 1 / 40 1 Background Modeling Digression: Mixture Models GMM
Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques
 Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques Sea Chen Department of Biomedical Engineering Advisors: Dr. Charles A. Bouman and Dr. Mark J. Lowe S. Chen Final Exam October
Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques Sea Chen Department of Biomedical Engineering Advisors: Dr. Charles A. Bouman and Dr. Mark J. Lowe S. Chen Final Exam October
Coherent Motion Segmentation in Moving Camera Videos using Optical Flow Orientations - Supplementary material
 Coherent Motion Segmentation in Moving Camera Videos using Optical Flow Orientations - Supplementary material Manjunath Narayana narayana@cs.umass.edu Allen Hanson hanson@cs.umass.edu University of Massachusetts,
Coherent Motion Segmentation in Moving Camera Videos using Optical Flow Orientations - Supplementary material Manjunath Narayana narayana@cs.umass.edu Allen Hanson hanson@cs.umass.edu University of Massachusetts,
Robust 3D Organ Localization with Dual Learning Architectures and Fusion
 Robust 3D Organ Localization with Dual Learning Architectures and Fusion Xiaoguang Lu (B), Daguang Xu, and David Liu Medical Imaging Technologies, Siemens Medical Solutions, Inc., Princeton, NJ, USA xiaoguang.lu@siemens.com
Robust 3D Organ Localization with Dual Learning Architectures and Fusion Xiaoguang Lu (B), Daguang Xu, and David Liu Medical Imaging Technologies, Siemens Medical Solutions, Inc., Princeton, NJ, USA xiaoguang.lu@siemens.com
Counting People by Clustering Person Detector Outputs
 2014 11th IEEE International Conference on Advanced Video and Signal Based Surveillance (AVSS) Counting People by Clustering Person Detector Outputs Ibrahim Saygin Topkaya Sabanci University Alcatel-Lucent
2014 11th IEEE International Conference on Advanced Video and Signal Based Surveillance (AVSS) Counting People by Clustering Person Detector Outputs Ibrahim Saygin Topkaya Sabanci University Alcatel-Lucent
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS
 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
