Image-based Modelling of Organogenesis
|
|
|
- Rudolf Maxwell
- 6 years ago
- Views:
Transcription
1 Image-based Modelling of Organogenesis Dagmar Iber 1,2,*, Zahra Karimaddini 1,2, Erkan Ünal 1,2,3 Affiliations: 1 Department of Biosystems, Science and Engineering (D-BSSE), ETH Zurich, Switzerland 2 - Swiss Institute of Bioinformatics (SIB), Mattenstraße 26, 4058 Basel, Switzerland 3 Department of Biomedicine, University of Basel, Mattenstraße 28, 4058 Basel, Switzerland * Corresponding Author: Dagmar Iber (dagmar.iber@bsse.ethz.ch) Key Words: Image-based Modelling, Computational Biology, Developmental Biology Summary Statements: - Methods for image-based modelling to explore Organogenesis - A pipeline for the extraction of boundaries and displacement fields from 2D and 3D microscopy images - Strategies for meshing and the solution of partial differential equations (PDEs) on the extracted (growing) domains
2 ABSTRACT One of the major challenges in biology concerns the integration of data across length and time scales into a consistent framework: how do macroscopic properties and functionalities arise from the molecular regulatory networks - and how can they change as a result of mutations? Morphogenesis provides an excellent model system to study how simple molecular networks robustly control complex processes on the macroscopic scale in spite of molecular noise, and how important functional variants can emerge from small genetic changes. Recent advancements in 3D imaging technologies, computer algorithms, and computer power now allow us to develop and analyse increasingly realistic models of biological control. Here we present our pipeline for image-based modeling that includes the segmentation of images, the determination of displacement fields, and the solution of systems of partial differential equations (PDEs) on the growing, embryonic domains. The development of suitable mathematical models, the data-based inference of parameter sets, and the evaluation of competing models are still challenging, and current approaches are discussed.
3 INTRODUCTION The process by which embryos self-organize into mature organisms with their particular shape and organs has fascinated scientists for a long time. Decades of experiments in developmental biology have led to the identification of many of the regulatory factors that control organogenesis and tissue development [1, 2]. Although these factors and their main regulatory interactions have been defined, the regulatory logic is often still elusive. A deeper understanding of the regulatory networks that govern developmental processes is important to enable the design and engineering of new functionalities and the repair of organs and tissues in regenerative medicine. The size and morphology of organs and organisms are the result of the net effect of cell proliferation, differentiation, migration and apoptosis, and must be strictly controlled. Coordination of cell behavior within the tissue is achieved by cell-cell communication [3, 4] either between neighbouring cells via cell surface proteins (Notch-Delta, Cadherins etc) or gap junctions, or over longer distances via diffusible, secreted factors, called morphogens [5], or by cellular processes, referred to as cytonemes [6]. Mechanical feedbacks can further coordinate cell behavior within tissue [7-9]. Spatio-temporal gene expression and cellular events during morphogenesis are highly dynamic and involve complex interactions. Mathematical modeling has identified a number of candidate mechanisms for the control of such complex dynamic patterning processes [10, 11]. One of the oldest and most prominent ideas in developmental patterning is the Turing patterning mechanism. Alan Turing recognized that chemical substances, which he
4 termed morphogens, can lead to the emergence of spatial patterns from noisy initial conditions by a diffusion-driven instability [5]. This mechanism has been highly attractive to explain biological patterning phenomena because deterministic, reliable symmetry breaks can be obtained with noisy initial conditions. A large number of patterning phenomena have been suggested to be controlled by a Turing mechanism, including those in the following references [12-16]. While all these models provide a good match with experimental observations, an experimental confirmation of the proposed Turing mechanisms is still outstanding because the kinetic rate constants have so far proved impossible to measure in the tissue. Pattern likeness itself is not a proof for the applicability of a Turing mechanism, and subsequent experiments in the Drosophila embryo have for instance ruled out the applicability of a Turing mechanisms in spite of excellent pattern likeness [17]. More quantitative experimental data is therefore very much needed to test Turing models and alternative models, where proposed [13, 18]. The second important concept is the so-called French-flag model [19]. Assuming that a symmetry break has already taken place such that a particular morphogen is secreted only in some part of the tissue, this model stipulates that cells may determine their position relative to this source (and thus their cell fate) by reading out the concentration gradient that the morphogen will create across the tissue. Also here many open questions remain, in particular regarding the precision of the patterning mechanism [20-22], and its robustness to noise [23] and to variable environmental conditions, e.g. temperature, as well as to differences in the size of the embryonic domain [24-27].
5 Many further concepts have been analysed [10], and have been applied to various patterning phenomena, including the emergence of travelling waves [28], wave pinning [29], shuttling [30], chemotaxis [31, 32], and mechanical feedbacks [9, 33]. Further experimental testing and refinement of all these mathematical concepts and ideas will be important. Most data in developmental biology is based on images, and recent developments in microscopy further enhance the opportunities to visualize developmental processes [34, 35]. Imaging enables the spatio-temporal visualization of regulatory aspects such as gene expression domains and protein localization, cell-based phenomena such as cell migration, division, and apoptosis [36], and the quantification of mechanical effects during morphogenesis [37-41]. Moreover, high-quality 3D imaging now provides detailed information on the complex architecture of tissues and organs [42, 43]. Most of these aspects have so far been analysed in isolation. Imagebased modeling provides an opportunity to integrate the wide range of imagebased spatio-temporal data with other biological datasets, e.g. qpcr, RNASeq, and proteomic data, into a consistent framework to arrive at a mechanistic understanding of morphogenesis [12, 14, 44-47]. Here, the gene expression domains indicate where the different model components are produced, while spatio-temporal information on protein distribution provide information regarding the spatio-temporal distribution of the model components. Moreover, cell-based data provides information on tissue growth and thus on advection and dilution terms (see below). In the following, we will explain the current approaches and methods for image-based modeling.
6 SPATIO-TEMPORAL MODELS OF ORGANOGENESIS Spatio-temporal models of regulatory networks can be formulated as a set of reaction-diffusion equations of the form!!!!" =!!! +!(!! ). (1)! Here, c i denotes the concentration and D i the diffusion coefficient of component i. Δ represents the Laplace operator of the diffusion term, and R(c k ) represents the reactions that alter the concentration of c i. The spatiotemporal models must be solved on suitable domains, and boundary and initial conditions need to be specified. While initial values are typically not known, reasonable approximations can typically be made. Simulations may then need to be run for a certain time period to achieve a reasonable starting point for physiological simulations, e.g. a steady-state. Boundary conditions can be implemented either as a set number (Dirichlet boundary conditions), a flux (von-neumann boundary condition), or a combination of the two. Sometimes, the domain of interest needs to be embedded into a larger domain to obtain realistic boundary effects for the (smaller) domain of interest [48]. As domains, idealized approximations to reality are typically employed [47]. Advances in microscopy and computer algorithms now, however, also permit an image-based approach, where computational domains are obtained from microscopy. In the following, we will present a pipeline for image-based modeling (Figure 1). A PIPELINE FOR IMAGE-BASED MODELLING OF ORGANOGENESIS
7 Before the computational work can be started, imaging data needs to be obtained of the tissue of interest. If different sub-structures are of interest, the tissue needs to be labeled accordingly. The sample preparation and staining methods of choice depends on the tissue, the substructure of interest, and the image acquisition technique and have their own challenges, which we will not discuss here. A large range of imaging techniques are available and are still being developed, as reviewed elsewhere [49]. The choice of imaging software is as important as the choice of the hardware. Thus, specialised software is required to control the functions of the optical microscopy tools and to collect and store the images that are acquired over time. A list of commercial, opensource and microscope companies software packages have been reviewed elsewhere [50]. Once the imaging data has been obtained, the images have to be visualized and analyzed to extract meaningful quantitative data from the microscopy images. Algorithms have been developed to analyse and process images, and we refer the interested reader to the wide range of available textbooks and further texts on the topic, e.g. [51-53]. For the user, software packages are available, including 3-D Slicer, AMIRA, Arivis, BioImageXD, Icy, Drishti, Fijii, IMARIS, MorphoGraphX, Rhino, and Simpleware. For a review of the available software packages, bioimaging libraries, and toolkits with their applications we refer the reader to [50, 54]. In the following, we will provide an overview of the specific steps that need to be taken for image-based modeling, and we will describe the procedure in detail for the software package AMIRA (Box 1). To use images in simulations, the following steps need to be performed: 1) image alignment and averaging, 2) image cropping and segmentation, and, 3)
8 in case of growth processes, determination of the mapping between images at subsequent developmental stages. The domains can subsequently be meshed and used to solve mathematical models of the regulatory networks on the physiological domain. The finite element method (FEM) is the numerical method of choice to solve the PDE models on the physiological domains. A large number of FEM solvers are available, including Abaqus, ANSYS, and COMSOL Multiphysics as commercial packages and AMDiS, deal.ii, DUNE, FEniCS, FreeFEM, LifeV as open source academic packages. We will focus on the FEM solver COMSOL. In the following, we will discuss the different steps in detail. Alignment & Averaging If multiple image recordings of the organ or tissue are available at a given stage, then the 3D images can be aligned and averaged. The alignment procedure is a computationally non-trivial problem and requires the use of suitable optimization algorithms, as described in textbooks and reviews [51, 55] (Box 1). Averaging is subsequently performed by averaging pixel intensities of corresponding pixels in multiple datasets of the same size and resolution. This helps to assess the variability between embryos and identifies common features. It also reduces the variability due to experimental handling. However, averaging of badly aligned datasets can result in loss of biologically relevant spatial information. It is therefore important to run the alignment algorithm several times, starting with different initial positions of the objects, which are to be aligned. Image Segmentation
9 The next step is to crop out the region of interest and to perform image segmentation. During image segmentation the digital image is partitioned into multiple subdomains, usually corresponding to anatomic features such as particular gene expression regions or different tissue layers (Figure 2). A wide range of algorithms is available for image segmentation, including thresholding, region-based segmentation, edge-based segmentation, levelsets, atlas-guided approaches, mean shift segmentation, and we refer the interested reader to textbooks and reviews on the topic [51-53]. Most segmentation algorithms are based on differences in pixel intensity and thus require a high signal-to-noise ratio in the images (see Box 1). Further manual work may be needed to complete domains. 2D Virtual sections and isosurfaces of 3D images 3D simulations are expensive and simulations are therefore often better first run on 2D sections (Figure 2). Following the alignment of the specimens in 3-D, 2-D optical sections can be extracted (see Box 1). SIMULATION OF NETWORK MODELS To simulate network models on the realistic geometries, the following steps need be performed: 1) generation of meshes, 2) import of meshes into an FEM solver, and 3) implementation of the PDE models and simulation of the models. The three steps will be discussed in the following. Mesh Generation & Import
10 To numerically solve the partial derivative equations (PDEs) that describe the signaling networks of interest, surface and volume meshes of sufficient quality must be generated (Figure 2). A large variety of algorithms, including advancing front methods, grid, quadtree, and octree algorithms, as well as Delaunay refinement algorithms have been developed to create suitable meshes in 2D and 3D with different shapes, e.g. triangular, tetrahedral, quadrilateral and hexahedral, and we refer the interested reader to textbooks on the matter [56]. Image processing and FEM software packages (see above) as well as specialized meshing software, e.g. BioMesh3D, Gmsh, and MeshLab, can be used for mesh generation and subsequent improvement. The quality of the mesh depends on the mesh size and the ratio of the sides of the mesh elements. The linear size of the mesh should be much smaller than any feature of interest in the computational solution, i.e. if the gradient length scale in the model is 50 µm then the linear size of the mesh should be at least several times lower than 50 µm. Additionally, the ratio of the length of the shortest side to the longest side should be 0.1 or more. To confirm the convergence of the simulation, the model must be solved on a series of refined meshes. When the geometries are extracted from experimental images, then the resolution is often so high that the resulting meshes are very fine. Too dense meshes increase the simulation time. The resolution must therefore be decreased. If the original resolution is very high, this may have to be done in multiple steps. While doing this, it is important to avoid intersections and inverted mesh elements. PDE Solvers
11 To exchange meshes between the image processing software and the simulation software, suitable file formats need to be chosen (Box 1). Once the meshes have been imported to an FEM solver, the PDE models can be solved on the domains of interest. Solving computational models of organogenesis on physiological domains can be computationally expensive and computational complexity should therefore be minimized as much as possible. This can be achieved, in particular, by minimizing the number of mesh elements and by smoothing sharp transitions and domain boundaries as much as possible. Details on how to solve PDE models of organogenesis and some approaches to reduce simulation time in COMSOL have been described in [57-59]. SIMULATIONS ON GROWING DOMAINS During embryonic development, tissue morphogenesis and signaling are tightly coupled. It is therefore important to simulate both tissue morphogenesis and signaling simultaneously in in silico models of developmental processes. The development of mechanistic models of tissue growth is challenging and requires detailed knowledge of the gene regulatory network, mechanical properties of the tissue, and its response to physical and biochemical cues. If these are not available, 3D image data can be used to solve the models on realistically expanding domains. Confocal microscopy [14, 60] and light-sheet microscopy [36, 61] can be used for live imaging. If light-sheet microscopy is not available and the specimen is too large for confocal microscopy, a
12 developmental series based on fixed samples can be acquired with optical projection tomography (OPT) [62]. Determination of displacement fields To describe the growing domains, suitable displacement fields need to be determined from the data. In the ideal case, tissue development can be imaged continuously, and single cells can be tracked during live microscopy [63]. In that case, the displacement field can be determined as the displacement field of those cells. However, in most applications such data is not available. In the latter case, approximate displacement fields can be obtained by determining a mapping between the tissue boundaries of two stages that have been extracted at sequential time points during development. Ideally, the spacing between two time points should be rather small, at least small enough that the deformations are small. However, the minimal distance between two time points is often dictated by experimental limitations, in particular when biological processes cannot be imaged continuously and separate specimen at staged developmental time points have to be used. Even when cultured organs are imaged continuously, frame rates may be limiting. Accordingly, suitable algorithms need to be employed that can deal with the experimental time series. While there are, in principle, an infinite number of possible mappings between such consecutive shapes, there are a number of important constraints. First of all, mappings should generally not intersect because this would create numerical problems and would generally also not be realistic because cells (in epithelia) will normally not swap positions with cells at other positions.
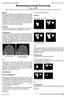
13 Moreover, given that the physiological displacement field is not known, it is sensible to create a displacement field that is both numerically favourable and biologically sensible. A range of mapping algorithms can be employed and which algorithm is most suitable depends on details of the geometries and their deformations (Algorithm 1) [64]. Algorithms for estimating displacement fields Before applying a mapping algorithm between the curve C 1 at time t and the curve C 2 at time t+δt it is important to carry out three pre-processing steps: If the structure has grown substantially in between the two measurements, it is often helpful to first stretch the curves uniformly such that the two curves are similar (Figure 3A-D). Next, the same number of points, N, should be interpolated on both boundary curves such that the Euclidean distance is identical between all points on each boundary curve. Finally, the direction, in which the points on C 1 and C 2 are sampled, should be the same. After applying the above pre-processing steps, a map between the interpolated points can be established. Mappings based on the minimal distance between the two curves are easy to implement, but frequently fail (Figure 3A) [60, 64]. If C 1 and C 2 are open boundary curves, then the uniform mapping algorithm is more suitable, where the mapping is established between an equal number of equidistant points on the two curves (Figure 3E, Algorithm 2). When the growth of the domain is not uniform, then the displacement field is, however, biased towards the direction of maximal growth (Figure 3F). In case of closed boundaries, we recommend the use of normal mapping, where the mapping is based on vectors that are normal to the original curve C 1 and end on the
14 target curve C 2 (Figure 3G, Algorithm 3). If the normal vectors intersect (Figure 3G), then either a reverse-normal mapping can be employed (Figure 3H), or the diffusion-mapping algorithm can be used instead (Figure 3I-L, Algorithm 4). In case of the diffusion mapping, the steady state diffusion equation is solved in between the curves C 1 and C 2 (Figure 3I), and the displacement field is based on the start and end points of streamlines (Figure 3J), which never cross. Depending on the shape of the domain the diffusion mapping is better calculated from C 1 to C 2 (Figure 3K) or in the reverse direction (Figure 3L). While the diffusion mapping prevents the crossing of the displacement vectors, we note that the diffusion mapping is computationally more expensive and the resulting displacement fields are more likely to result in mesh distortions. Diffusion mapping should therefore only be used when normal mapping or minimal distance mapping fail. If a domain contains subdomains, then the boundaries that divide the subdomains intersect (Figure 4A). Intersections need to be avoided and the intersected curve should therefore be split into two curves at the intersection point (Algorithm 5) [65]. This step has to be done for all intersected boundaries at time t and at time t+δt (Figure 4B,C). The displacement fields for all independent segments can then be determined as discussed above. We note that Algorithm 1 only works well as long as the shapes are relatively close. In case of larger differences, we used the landmark-based Bookstein algorithm [66], as illustrated in Figure 5 [14]. Here we obtained lung buds at different developmental stages (Figure 5A), and determined the displacement field (Figure 5B) and thereby the growth fields (Figure 5C) between those
15 stages. We then imported the mesh of the earlier stage lung bud into the FEM solver and solved signaling models on the imported shape (Figure 5D). In certain cases, the maxima of the predicted signaling fields coincided with the maxima of the embryonic growth fields (Figure 5E). We note that while the landmark-based Bookstein algorithm [66] provided a straightforward method to obtain the displacement fields for the shapes, the usage of such a manual approach is very work intensive and dependent on the user and thus to a certain extent arbitrary. Moreover, for complex structures such as branched organs, the identification of mapping points can be difficult even when stereoscopic visualization technologies are available. We therefore recommend to adapt (semi-) automatic mapping algorithms as described above to the particular situation as much as possible. Simulations on growing domains Once the displacement fields have been determined, they can be used to solve the PDE models on a realistically growing and deforming domain (Figure 6). To carry out the FEM-based simulations, the meshed geometries (Figure 6A) and displacement fields need to be imported into an FEM solver. On growing domains, advection and dilution effects need to be incorporated [67], such that Eq. (1) becomes on a growing domain!!!!" + (!!!) =!!! +!(!! ). (2)! Here! denotes the velocity field. To solve coupled PDE models on complex growing geometries, the Arbitrary Lagrangian Eulerian (ALE) method should be used. The ALE method is available in the commercial FEM solver
16 COMSOL, which we use to solve our PDE models of organogenesis on growing domains (Figure 6B) [15, 26, 48, 60, 68-70]. PARAMETER ESTIMATION AND MODEL SELECTION FOR IMAGE- BASED MODELLING In the final step, the image-based data can be used to infer parameter sets for the models and to evaluate models and hypotheses. To evaluate the match between simulations and data, first a suitable metric needs to be defined to quantify the difference between model prediction and experimental data. Which metric is most suitable depends on the type of experimental data and the model [14, 71, 72]. Thus, only very little data is truly quantitative, such as the strength of the growth fields discussed above. A lot of data is purely qualitative, e.g. in situ hybridization data, and some data is semi-quantitative, e.g. fluorescent data. Given that the choice of a metric is to a certain extent arbitrary, but can affect the ranking of parameter sets and models, different metrics should ideally be evaluated [14, 71, 72]. For most models, several data sets need to be combined, and appropriate weighting functions need to be defined. Once a metric and a weighting of the different datasets has been defined, an optimization algorithm has to be chosen to minimize the distance between the data and the simulation output. If a prior probability distribution for the parameter values is known, Bayesian optimization is the method of choice to incorporate this information into the optimization process. A wide range of global and local optimization algorithms have been developed, and we refer the interested reader to textbooks for details [73] and to previous
17 reviews on parameter estimation for biological models [74-77]. If the models are sufficiently simple, then it is computationally feasible to first globally screen the parameter sets over the entire physiological range of the parameter values, and to subsequently carry out a local optimization procedure for the best parameter sets to further improve the fit between simulation output and data [14]. Based on the parameter screens, the posterior probability distributions for the parameters can be reconstructed and the relative likelihood of different models can be evaluated [78]. In case of multiple objectives (because of distinct datasets, genotypes etc.) in can be sensible to evaluate the Pareto optimality [71]. In case of more complex signaling models, a dense sampling of the parameter space is no longer possible. Interpolation algorithms such as Gaussian Process Upper Confidence Bound (GC-UCB) and Kriging methods can be used to guide the sampling process and thus reach the optimal parameter set with less sampling steps [79, 80]. Nonetheless it is sensible to keep model complexity to a minimum, and the field of model reduction is currently very active as reviewed elsewhere [81, 82]. Many biological models are modular and modules may be important on different time scales. In that case, parameter values can be estimated sequentially, as done recently in a tree-based approach [83]. CONCLUSION
18 Recent advances in imaging techniques, algorithms and computer power now permit the use of spatio-temporal imaging data for the generation and testing of increasingly realistic models of developmental processes. We have reviewed a pipeline for image-based modeling that comprises image acquisition, processing (segmentation, alignment, averaging), the determination of displacement fields, meshing of domains, transfer to an appropriate FEM solver, parameter inference and model selection. The computational approach reviewed here should enable a wider adoption of realistic spatio-temporal computational models by both biologists and computational biologists. The field is rapidly moving forward and offers ample challenges and opportunities for computational scientists. Further developments are required, in particular in the areas of PDE solvers, cell-based models, and parameter inference. Thus, available PDE solvers were developed to tackle physical rather than biological problems and often cannot easily and efficiently be used to model organogenesis, especially on 3D growing domains. A number of software packages for cell-based tissue simulations already exist, but further developments are required to fully represent and analyse tissue mechanics and signaling in 3D [84]. Finally, current methods for parameter estimation cannot easily handle the complexity of biological models. We expect that further developments in these three areas will allow to fully exploit the 3D dynamic imaging data that can now be obtained with lightsheet microscopy for a mechanistic understanding of developmental processes.
19 Acknowledgements The authors thank past and present members of the CoBi group for discussions and acknowledge funding from a SystemsX iphd grant and the SystemsX NeurostemX RTD.
20 REFERENCE 1. Gilbert SF. Developmental Biology. Sinauer Associates, Wolpert L. Principles of Development. Oxford University Press (OUP), Restrepo S, Zartman JJ, Basler K. Coordination of Patterning and Growth by the Morphogen DPP., Curr Biol 2014;24:R245-R Müller P, Rogers KW, Yu SR et al., Development 2013;140: Turing AM. The chemical basis of morphogenesis, Phil. Trans. Roy. Soc. Lond 1952;B237: Kornberg TB, Roy S. Cytonemes as specialized signaling filopodia, Development 2014;141: Gjorevski N, Nelson CM. The mechanics of development: Models and methods for tissue morphogenesis, Birth Defects Res C Embryo Today 2010;90: Aegerter-Wilmsen T, Heimlicher MB, Smith AC et al. Integrating forcesensing and signaling pathways in a model for the regulation of wing imaginal disc size., Development 2012;139: Hufnagel L, Teleman AA, Rouault H et al. On the mechanism of wing size determination in fly development., Proceedings of the National Academy of Sciences of the United States of America 2007;104: Murray JD. Mathematical Biology. 3rd edition in 2 volumes: Mathematical Biology: II. Spatial Models and Biomedical Applications.: Springer, Umulis DM, Othmer HG. The role of mathematical models in understanding pattern formation in developmental biology, Bull Math Biol 2015;77: Salazar-Ciudad I, Jernvall J. A computational model of teeth and the developmental origins of morphological variation., Nature 2010;464: Sick S, Reinker S, Timmer J et al. WNT and DKK determine hair follicle spacing through a reaction-diffusion mechanism., Science 2006;314: Menshykau D, Blanc P, Unal E et al. An interplay of geometry and signaling enables robust lung branching morphogenesis, Development 2014;141: Badugu A, Kraemer C, Germann P et al. Digit patterning during limb development as a result of the BMP-receptor interaction., Sci Rep 2012;2: Kondo, Asai. A reaction diffusion wave on the skin of the marine angelfish Pomacanthus, Nature 1995;376: Akam M. Drosophila development: making stripes inelegantly. Nature. 1989, Cheng CW, Niu B, Warren M et al. Predicting the spatiotemporal dynamics of hair follicle patterns in the developing mouse, Proc Natl Acad Sci U S A 2014;111: Wolpert L. Positional information and the spatial pattern of cellular differentiation, J Theor Biol 1969;25:1-47.
21 20. Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo, Nature 2002;415: Gregor T, Tank DW, Wieschaus EF et al. Probing the Limits to Positional Information, Cell 2007;130: Bollenbach T, Pantazis P, Kicheva A et al. Precision of the Dpp gradient., Development 2008;135: Saunders TE, Howard M. Morphogen profiles can be optimized to buffer against noise., Phys Rev E Stat Nonlin Soft Matter Phys 2009;80: Gregor T, Bialek W, de Ruyter van Steveninck RR et al. Diffusion and scaling during early embryonic pattern formation., Proceedings of the National Academy of Sciences of the United States of America 2005;102: Wartlick O, Jülicher F, González-Gaitán M. Growth control by a moving morphogen gradient during Drosophila eye development, Development 2014;141: Fried P, Iber D. Dynamic scaling of morphogen gradients on growing domains, Nat Commun 2014;5: Umulis DM, Othmer HG. Mechanisms of scaling in pattern formation, Development 2013;140: Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis, J Theor Biol 1976;58: Mori Y, Jilkine A, Edelstein-Keshet L. Wave-pinning and cell polarity from a bistable reaction-diffusion system, Biophys J 2008;94: Eldar A, Dorfman R, Weiss D et al. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning, Nature 2002;419: Painter KJ, Maini PK, Othmer HG. A chemotactic model for the advance and retreat of the primitive streak in avian development, Bull Math Biol 2000;62: Painter KJ, Maini PK, Othmer HG. Stripe formation in juvenile Pomacanthus explained by a generalized turing mechanism with chemotaxis, Proc Natl Acad Sci U S A 1999;96: Aegerter-Wilmsen T, Aegerter CM, Hafen E. Model for the regulation of size in the wing imaginal disc of Drosophila, Mechanisms of Oates AC, Gorfinkiel N, Gonzalez-Gaitan M et al. Quantitative approaches in developmental biology., Nature reviews. Genetics 2009;10: Kicheva A, Pantazis P, Bollenbach T et al. Kinetics of Morphogen Gradient Formation, Science 2007;315: Keller PJ, Schmidt AD, Wittbrodt J et al. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy, Science 2008;322: Routier-Kierzkowska AL, Weber A, Kochova P et al. Cellular force microscopy for in vivo measurements of plant tissue mechanics, Plant Physiol 2012;158: Heisenberg CP, Bellaiche Y. Forces in tissue morphogenesis and patterning, Cell 2013;153:
22 39. Blanchard GB, Kabla AJ, Schultz NL et al. Tissue tectonics: morphogenetic strain rates, cell shape change and intercalation, Nat Methods 2009;6: Wyczalkowski MA, Chen Z, Filas BA et al. Computational models for mechanics of morphogenesis, Birth Defects Res C Embryo Today 2012;96: LeGoff L, Rouault H, Lecuit T. A global pattern of mechanical stress polarizes cell divisions and cell shape in the growing Drosophila wing disc, Development 2013;140: Short K, Hodson M, Smyth I. Spatial mapping and quantification of developmental branching morphogenesis., Development Combes AN, Short KM, Lefevre J et al. An integrated pipeline for the multidimensional analysis of branching morphogenesis, Nat Protoc 2014;9: Iber D, Zeller R. Curr Opin Genet Dev 2012;22: Iber D, Menshykau D. The control of branching morphogenesis., Open biology 2013;3: Iber D, Germann P. How do digits emerge? - mathematical models of limb development., Birth Defects Res C Embryo Today 2014;102: Probst S, Kraemer C, Demougin P et al. SHH propagates distal limb bud development by enhancing CYP26B1-mediated retinoic acid clearance via AER-FGF signalling., Development 2011;138: Menshykau D, Kraemer C, Iber D. Branch mode selection during early lung development, PLoS Comput Biol 2012;8:e Howard GC, Brown WE, Auer M. Imaging Life: Biological Systems from Atoms to Tissues. Oxford University Press, Eliceiri KW, Berthold MR, Goldberg IG et al. Biological imaging software tools, Nat Methods 2012;9: Szeliski R. Computer Vision. Springer, Russ JC. The Image Processing Handbook, Sixth Edition. CRC Press, Burger W, Burge MJ. Principles of Digital Image Processing: Core Algorithms London: Springer, Wiesmann V, Franz D, Held C et al. Review of free software tools for image analysis of fluorescence cell micrographs, J Microsc 2015;257: Goshtasby, Ardeshir A. Image Registration - Principles, Tools and Methods. Springer, Lo DSH. Finite Element Mesh Generation. CRC Press, Germann P, Menshykau D, Tanaka S et al. Simulating Organogenesis in Comsol. COMSOL conference. Stuttgart, Menshykau D, Iber D. Simulating Organogenesis with Comsol: Interacting and Deforming Domains. In: COMSOL Conference Milan, COMSOL Proceedings. 59. Vollmer J, Menshykau D, Iber D. Simulating Organogenesis in COMSOL: Cell-based Signaling Models, CORD Conference Proceedings Adivarahan S, Menshykau D, Michos O et al. Dynamic Image-Based Modelling of Kidney Branching Morphogenesis. Berlin, Heidelberg: Springer Berlin Heidelberg, 2013,
23 61. Stelzer EH. Light-sheet fluorescence microscopy for quantitative biology, Nat Methods 2015;12: Sharpe J, Ahlgren U, Perry P et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies., Science 2002;296: Gros J, Hu JK-H, Vinegoni C et al. WNT5A/JNK and FGF/MAPK pathways regulate the cellular events shaping the vertebrate limb bud., Curr Biol 2010;20: Schwaninger CA, Menshykau D, Iber D. Simulating Organogenesis: Algorithms for the Image-Based Determination of Displacement Fields, Acm Transactions on Modeling and Computer Simulation 2015; Karimaddini Z, Ünal E, Menshykau D et al. Simulating Organogenesis in COMSOL: Image-based Modeling. COMSOL conference. Cambridge: COMSOL Proceedings, Bookstein FL. Principal warps: Thin-plate splines and the decomposition of deformations, Pattern Analysis and Machine Intelligence Iber D, Tanaka S, Fried P et al. Simulating Tissue Morphogenesis and Signaling. In: Nelson C. M. (ed) Methods in Molecular Biology (Springer). New York, NY: Springer New York, 2015, Tanaka S, Iber D. Inter-dependent tissue growth and Turing patterning in a model for long bone development, Phys Biol 2013;10: Bachler M, Menshykau D, De Geyter C et al. Species-specific differences in follicular antral sizes result from diffusion-based limitations on the thickness of the granulosa cell layer, Mol Hum Reprod 2014;20: Celliere G, Menshykau D, Iber D. Simulations demonstrate a simple network to be sufficient to control branch point selection, smooth muscle and vasculature formation during lung branching morphogenesis, Biol Open 2012;1: Pargett M, Rundell AE, Buzzard GT et al. Model-based analysis for qualitative data: an application in Drosophila germline stem cell regulation, PLoS Comput Biol 2014;10:e Hengenius JB, Gribskov M, Rundell AE et al. Making models match measurements: model optimization for morphogen patterning networks, Semin Cell Dev Biol 2014;35: Chong EKP, Zak SH. An Introduction to Optimization. John Wiley & Sons, Jaqaman K, Danuser G. Linking data to models: data regression, Nat Rev Mol Cell Biol 2006;7: Ashyraliyev M, Fomekong-Nanfack Y, Kaandorp JA et al. Systems biology: parameter estimation for biochemical models., FEBS Journal 2009;276: Geier F, Fengos G, Felizzi F et al. Analyzing and constraining signaling networks: parameter estimation for the user., Methods in molecular biology (Clifton, N.J.) 2012;880: Raue A, Karlsson J, Saccomani MP et al. Comparison of approaches for parameter identifiability analysis of biological systems, Bioinformatics (Oxford, England) 2014;30: Burnham KP, Anderson DR. Model Selection and Multimodel Inference. New York, NY: Springer New York, 2004.
24 79. Srinivas N, Krause A, Kakade SM et al. Information-Theoretic Regret Bounds for Gaussian Process Optimization in the Bandit Setting, Ieee Transactions on Information Theory 2012;58: Rasmussen CE, Williams C. Gaussian Processes for Machine Learning, Model Selection and Adaptation of Hyperparameters, Chapter Manzoni A, Quarteroni A, Rozza G. Computational Reduction for Parametrized PDEs: Strategies and Applications, Milan Journal of Mathematics 2012;80: Leugering Gn. Trends in PDE constrained optimization. Cham Switzerland ; New York: Birkhäuser ; Springer, Velten B, Ünal E, Iber D. Image-based Parameter Inference for Spatiotemporal models of Organogenesis. International Symposium on Nonlinear Theory & its Applications (NOLTA 2014 ). Luzern, CH, Tanaka S, Sichau D, Iber D. LBIBCell: a cell-based simulation environment for morphogenetic problems., Bioinformatics (Oxford, England) Deserno TM. Biomedical Image Processing. Springer, 2011.
25 BOX 1: SOFTWARE DETAILS FOR IMAGE PROCESSING, MESH GENERATION & PARAMETER OPTIMIZATION In the following, we provide details on AMIRA-based image processing and mesh generation as well as mesh import, PDE model simulation, and parameter optimization in the FEM solver COMSOL. 1. Alignment & Averaging To align images a number of iterative hierarchical optimization algorithms, e.g. QuasiNewton, are available in the software package AMIRA, as well as similarity measures, e.g. Euclidean distance. Averaging is subsequently performed by averaging pixel intensities of corresponding pixels in multiple datasets of the same size and resolution. 2. Image Segmentation In AMIRA, a variety of algorithms are available for image segmentation, including a thresholding algorithm, which is suitable for most 3-D images, a watershed algorithm for the segmentation of more complex structures, and a top-hat algorithm for the detection of dark or white regions [52, 85]. Given the dependency of these algorithms on differences in pixel intensity and thus on a sufficient signal-to-noise ratio in the images, further manual work may be needed to complete domains. 3. 2D Virtual sections and isosurfaces of 3D images In AMIRA, the SurfaceCut module is available for this purpose. The boundaries of the 2-D sections can be extracted in AMIRA using the Intersect module. Once saved in ASCII format, the section boundaries can be further
26 processed in MATLAB to smooth and fine-tune the subdomains of the boundaries. 4. AMIRA Mesh Import into the FEM solver COMSOL AMIRA meshes can be saved in the I-DEAS universal data format, which can be read by the Gmsh software and saved as a Nastran Bulk data file. The file extension of the Nastran Bulk data file needs to be changed from UNV to DAT to make it compatible with the simulation software COMSOL Multiphysics. In the COMSOL mesh module, the mesh can then be imported. 5. Parameter Optimization in COMSOL COMSOL provides a range of optimization solvers, including gradient-based ones, e.g. Levenberg-Marquardt, Method of Moving Asymptotes (MMA), Sparse Nonlinear Optimizer (SNOP), and derivative free ones, e.g. constraint optimization by linear approximation (COBYLA), and bound optimization by quadratic approximation (BOBYQA).
27 BOX 2: MAPPING ALGORITHMS Uniform Mapping (Algorithm 2) This algorithm maps each point on C 1 to a point on C 2 such that the order of points on C 1 and C 2 is conserved, assuming that both boundary curves contain the same number of equidistant points. Normal Mapping (Algorithm 3) The normal vector at every point on the curve C 1 is computed. Then, each point on C 1 is mapped to the closest point on C2, where the normal vector and C2 intersect. Diffusion Mapping (Algorithm 4) This algorithm solves the steady state diffusion equation (Δc=0) with Dirichlet boundary conditions (c=1 on C 1 and c=0 on C 2 ) in the area between the two boundaries. The mapping vectors are calculated as the difference between the start and the end point of the streamlines. The streamlines can be estimated using the COMSOL Multiphsyics particle tracking module. Intersecting Curves (Algorithm 5) In case of two boundary curves B 1 ={P 11,, P n1 } and B 2 ={P,, P m2 } that intersect at point P, B 1 should be split into two segments B 1,seqment1 ={P 11,, P} and B 1,seqment2 ={P,, P n1 }, while B 2 remains the same. This step has to be done at time t and at time t+δt. To determine the displacement fields, uniform mapping (Algorithm 2) should subsequently be applied to each open segment.
28 FIGURES Figure 1: A Pipeline for Image-based Modelling of Organogenesis The figure summarizes the steps for image-based modeling. For details see the main text. Figure 2: Extraction of physiological domains for FEM modelling After acquisition of the 3D image of the organ of interest (here an embryonic lung), the structure of interest is identified (here the tip of a lung bud) and cropped out for further processing. Based on markers, sub-structures of interest can be segmented. Finally, surfaces can be extracted. In a separate step, 2-D surface cuts can be generated to extract 2D planes and their boundaries. Finally, the domain can be meshed. Figure 3: Determination of Displacement Fields (A-D) Transformation prior to the mapping can improve the resulting displacement field. (A) The displacement field (red) was computed between C 1 (green) and C 2 (blue) using the minimal distance algorithm. (B) C 1 is scaled to be more similar to C 2 ; the new curve (red, dashed line) is called C 1,s. (C) The displacement field (red) is computed from the transformed curve C 1,s (green) onto C 2. (D) The starting points of the vectors on C 1,s where finally transformed back onto the original curve C 1. Comparison of the displacement fields (red) in panels A and D shows that, as a result of the transformation, the
29 number of regions on C 2, onto which no points were mapped, has decreased and the displacement field quality has therefore increased. (E-F) Uniform mapping. (E) The displacement field of open curves is determined by interpolating both curves with equidistant points and by mapping the points consecutively onto each other. (F) When the growth of the domain is not uniform, the displacement field is biased towards the direction of maximal growth. (G-H) Normal mapping. (G) If C 1 encloses a non-convex domain and both curves are not very close to each other in the concave regions of C 1, the displacement field vectors generated by normal mapping cross each other. (H) The problem can be resolved by using reverse normal mapping, i.e. by mapping C 2 onto C 1. (I-L) Diffusion mapping. (I) The steady-state solution of the diffusion equation. (J) The streamlines, as obtained by particle tracking from C 1 to C 2. (K) The displacement field vectors are obtained by connecting the start and the end points of the streamlines. (L) Reverse diffusion mapping, i.e. C 2 is mapped onto C 1. This yields better displacement fields when C 2 is much larger than C 1. The figure reproduces panels from [64]. Figure 4: Displacement Fields for Domains with Internal Boundaries
30 (A) A domain with three subdomains. The boundary curves (black, red, green) intersect at different points. (B) C 1 (green) and C 2 (red) intersect at point P. (C) To map the intersection point P (blue point) at time t to the intersection point at t+1, the curves are divided into segments, such that the point P becomes the start/end point of the intersecting curves. The figure reproduces panels from [65]. Figure 5: Image-based Modelling with embryonic data (A) 3D lung bud shapes at two different stages. (B) A displacement field between the two stages shown in panel A. (C) The displacement vectors with out-ward pointing vectors correspond to the growth field. (D) The solution of a ligand-receptor based Turing model on the first stage of the segmented lung in panel A. (E) The predicted distribution of the ligand-receptor signaling pattern (solid colours) coincides with the embryonic growth field (arrows). The Figure reproduces panels from [14]. Figure 6: Simulations on growing domains (A) The meshed domain of an ureteric bud. (B) The solution of a computational model (solid colours) and the estimated growth fields (coloured arrows) of a growing ureteric bud. The figure reproduces panels from [64].
31
32
33 Figure 3
34
35 Figure 5
36
Lecture 10: Image-Based Modelling
 Computational Biology Group (CoBI), D-BSSE, ETHZ Lecture 10: Image-Based Modelling Prof Dagmar Iber, PhD DPhil MSc Computational Biology 2015 Contents 1 Image-based Domains for Simulations Staining & Imaging
Computational Biology Group (CoBI), D-BSSE, ETHZ Lecture 10: Image-Based Modelling Prof Dagmar Iber, PhD DPhil MSc Computational Biology 2015 Contents 1 Image-based Domains for Simulations Staining & Imaging
Modelling With Comsol: Gradients and Pattern Formation Direct questions and suggestions to Denis
 Modelling With Comsol: Gradients and Pattern Formation Direct questions and suggestions to Denis (dzianis.menshykau@bsse.ethz.ch) Problem 1 Solve simple diffusion equation (no reactions!) on a 1D domain.
Modelling With Comsol: Gradients and Pattern Formation Direct questions and suggestions to Denis (dzianis.menshykau@bsse.ethz.ch) Problem 1 Solve simple diffusion equation (no reactions!) on a 1D domain.
Simulating Organogenesis in COMSOL: Comparison Of Methods For Simulating Branching Morphogenesis
 Simulating Organogenesis in COMSOL: Comparison Of Methods For Simulating Branching Morphogenesis Lucas D. Wittwer 1, Michael Peters 1,2, Sebastian Aland 3, Dagmar Iber *1,2 1 D-BSSE, ETH Zürich, Zurich,
Simulating Organogenesis in COMSOL: Comparison Of Methods For Simulating Branching Morphogenesis Lucas D. Wittwer 1, Michael Peters 1,2, Sebastian Aland 3, Dagmar Iber *1,2 1 D-BSSE, ETH Zürich, Zurich,
Chapter 3. Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C.
 Chapter 3 Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C. elegans Embryos Abstract Differential interference contrast (DIC) microscopy is
Chapter 3 Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C. elegans Embryos Abstract Differential interference contrast (DIC) microscopy is
Partial Differential Equations
 Simulation in Computer Graphics Partial Differential Equations Matthias Teschner Computer Science Department University of Freiburg Motivation various dynamic effects and physical processes are described
Simulation in Computer Graphics Partial Differential Equations Matthias Teschner Computer Science Department University of Freiburg Motivation various dynamic effects and physical processes are described
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS.
 SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
Morphometric Analysis of Biomedical Images. Sara Rolfe 10/9/17
 Morphometric Analysis of Biomedical Images Sara Rolfe 10/9/17 Morphometric Analysis of Biomedical Images Object surface contours Image difference features Compact representation of feature differences
Morphometric Analysis of Biomedical Images Sara Rolfe 10/9/17 Morphometric Analysis of Biomedical Images Object surface contours Image difference features Compact representation of feature differences
B. Pattern Formation. Differentiation & Pattern Formation
 B. Pattern Formation 1/25/12 1 Differentiation & Pattern Formation A central problem in development: How do cells differentiate to fulfill different purposes? How do complex systems generate spatial &
B. Pattern Formation 1/25/12 1 Differentiation & Pattern Formation A central problem in development: How do cells differentiate to fulfill different purposes? How do complex systems generate spatial &
Introduction to C omputational F luid Dynamics. D. Murrin
 Introduction to C omputational F luid Dynamics D. Murrin Computational fluid dynamics (CFD) is the science of predicting fluid flow, heat transfer, mass transfer, chemical reactions, and related phenomena
Introduction to C omputational F luid Dynamics D. Murrin Computational fluid dynamics (CFD) is the science of predicting fluid flow, heat transfer, mass transfer, chemical reactions, and related phenomena
An explicit feature control approach in structural topology optimization
 th World Congress on Structural and Multidisciplinary Optimisation 07 th -2 th, June 205, Sydney Australia An explicit feature control approach in structural topology optimization Weisheng Zhang, Xu Guo
th World Congress on Structural and Multidisciplinary Optimisation 07 th -2 th, June 205, Sydney Australia An explicit feature control approach in structural topology optimization Weisheng Zhang, Xu Guo
Inclusion of Aleatory and Epistemic Uncertainty in Design Optimization
 10 th World Congress on Structural and Multidisciplinary Optimization May 19-24, 2013, Orlando, Florida, USA Inclusion of Aleatory and Epistemic Uncertainty in Design Optimization Sirisha Rangavajhala
10 th World Congress on Structural and Multidisciplinary Optimization May 19-24, 2013, Orlando, Florida, USA Inclusion of Aleatory and Epistemic Uncertainty in Design Optimization Sirisha Rangavajhala
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search
 Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
1.2 Numerical Solutions of Flow Problems
 1.2 Numerical Solutions of Flow Problems DIFFERENTIAL EQUATIONS OF MOTION FOR A SIMPLIFIED FLOW PROBLEM Continuity equation for incompressible flow: 0 Momentum (Navier-Stokes) equations for a Newtonian
1.2 Numerical Solutions of Flow Problems DIFFERENTIAL EQUATIONS OF MOTION FOR A SIMPLIFIED FLOW PROBLEM Continuity equation for incompressible flow: 0 Momentum (Navier-Stokes) equations for a Newtonian
CHAPTER 1. Introduction
 ME 475: Computer-Aided Design of Structures 1-1 CHAPTER 1 Introduction 1.1 Analysis versus Design 1.2 Basic Steps in Analysis 1.3 What is the Finite Element Method? 1.4 Geometrical Representation, Discretization
ME 475: Computer-Aided Design of Structures 1-1 CHAPTER 1 Introduction 1.1 Analysis versus Design 1.2 Basic Steps in Analysis 1.3 What is the Finite Element Method? 1.4 Geometrical Representation, Discretization
Image Segmentation and Registration
 Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S. Visualization of Biological Molecules in Their Native Stat e.
 THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
THREE-DIMENSIONA L ELECTRON MICROSCOP Y OF MACROMOLECULAR ASSEMBLIE S Visualization of Biological Molecules in Their Native Stat e Joachim Frank CHAPTER 1 Introduction 1 1 The Electron Microscope and
3D Finite Element Software for Cracks. Version 3.2. Benchmarks and Validation
 3D Finite Element Software for Cracks Version 3.2 Benchmarks and Validation October 217 1965 57 th Court North, Suite 1 Boulder, CO 831 Main: (33) 415-1475 www.questintegrity.com http://www.questintegrity.com/software-products/feacrack
3D Finite Element Software for Cracks Version 3.2 Benchmarks and Validation October 217 1965 57 th Court North, Suite 1 Boulder, CO 831 Main: (33) 415-1475 www.questintegrity.com http://www.questintegrity.com/software-products/feacrack
Finite Element Convergence for Time-Dependent PDEs with a Point Source in COMSOL 4.2
 Finite Element Convergence for Time-Dependent PDEs with a Point Source in COMSOL 4.2 David W. Trott and Matthias K. Gobbert Department of Mathematics and Statistics, University of Maryland, Baltimore County,
Finite Element Convergence for Time-Dependent PDEs with a Point Source in COMSOL 4.2 David W. Trott and Matthias K. Gobbert Department of Mathematics and Statistics, University of Maryland, Baltimore County,
Ultrasonic Multi-Skip Tomography for Pipe Inspection
 18 th World Conference on Non destructive Testing, 16-2 April 212, Durban, South Africa Ultrasonic Multi-Skip Tomography for Pipe Inspection Arno VOLKER 1, Rik VOS 1 Alan HUNTER 1 1 TNO, Stieltjesweg 1,
18 th World Conference on Non destructive Testing, 16-2 April 212, Durban, South Africa Ultrasonic Multi-Skip Tomography for Pipe Inspection Arno VOLKER 1, Rik VOS 1 Alan HUNTER 1 1 TNO, Stieltjesweg 1,
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes
 Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Probabilistic Tracking and Model-based Segmentation of 3D Tubular Structures
 Probabilistic Tracking and Model-based Segmentation of 3D Tubular Structures Stefan Wörz, William J. Godinez, Karl Rohr University of Heidelberg, BIOQUANT, IPMB, and DKFZ Heidelberg, Dept. Bioinformatics
Probabilistic Tracking and Model-based Segmentation of 3D Tubular Structures Stefan Wörz, William J. Godinez, Karl Rohr University of Heidelberg, BIOQUANT, IPMB, and DKFZ Heidelberg, Dept. Bioinformatics
"The real world is nonlinear"... 7 main Advantages using Abaqus
 "The real world is nonlinear"... 7 main Advantages using Abaqus FEA SERVICES LLC 6000 FAIRVIEW ROAD, SUITE 1200 CHARLOTTE, NC 28210 704.552.3841 WWW.FEASERVICES.NET AN OFFICIAL DASSAULT SYSTÈMES VALUE
"The real world is nonlinear"... 7 main Advantages using Abaqus FEA SERVICES LLC 6000 FAIRVIEW ROAD, SUITE 1200 CHARLOTTE, NC 28210 704.552.3841 WWW.FEASERVICES.NET AN OFFICIAL DASSAULT SYSTÈMES VALUE
Particle Velocimetry Data from COMSOL Model of Micro-channels
 Particle Velocimetry Data from COMSOL Model of Micro-channels P.Mahanti *,1, M.Keebaugh 1, N.Weiss 1, P.Jones 1, M.Hayes 1, T.Taylor 1 Arizona State University, Tempe, Arizona *Corresponding author: GWC
Particle Velocimetry Data from COMSOL Model of Micro-channels P.Mahanti *,1, M.Keebaugh 1, N.Weiss 1, P.Jones 1, M.Hayes 1, T.Taylor 1 Arizona State University, Tempe, Arizona *Corresponding author: GWC
Morphogenesis. Simulation Results
 Morphogenesis Simulation Results This document contains the results of the simulations designed to investigate the regeneration strength of the computational model of the planarium. Specific portions of
Morphogenesis Simulation Results This document contains the results of the simulations designed to investigate the regeneration strength of the computational model of the planarium. Specific portions of
Statistical Shape Analysis
 Statistical Shape Analysis I. L. Dryden and K. V. Mardia University ofleeds, UK JOHN WILEY& SONS Chichester New York Weinheim Brisbane Singapore Toronto Contents Preface Acknowledgements xv xix 1 Introduction
Statistical Shape Analysis I. L. Dryden and K. V. Mardia University ofleeds, UK JOHN WILEY& SONS Chichester New York Weinheim Brisbane Singapore Toronto Contents Preface Acknowledgements xv xix 1 Introduction
Finite Element Simulation of Moving Targets in Radio Therapy
 Finite Element Simulation of Moving Targets in Radio Therapy Pan Li, Gregor Remmert, Jürgen Biederer, Rolf Bendl Medical Physics, German Cancer Research Center, 69120 Heidelberg Email: pan.li@dkfz.de Abstract.
Finite Element Simulation of Moving Targets in Radio Therapy Pan Li, Gregor Remmert, Jürgen Biederer, Rolf Bendl Medical Physics, German Cancer Research Center, 69120 Heidelberg Email: pan.li@dkfz.de Abstract.
Step 1: Problem Type Specification. (1) Open COMSOL Multiphysics 4.1. (2) Under Select Space Dimension tab, select 2D Axisymmetric.
 Step 1: Problem Type Specification (1) Open COMSOL Multiphysics 4.1. (2) Under Select Space Dimension tab, select 2D Axisymmetric. (3) Click on blue arrow next to Select Space Dimension title. (4) Click
Step 1: Problem Type Specification (1) Open COMSOL Multiphysics 4.1. (2) Under Select Space Dimension tab, select 2D Axisymmetric. (3) Click on blue arrow next to Select Space Dimension title. (4) Click
Modeling with Uncertainty Interval Computations Using Fuzzy Sets
 Modeling with Uncertainty Interval Computations Using Fuzzy Sets J. Honda, R. Tankelevich Department of Mathematical and Computer Sciences, Colorado School of Mines, Golden, CO, U.S.A. Abstract A new method
Modeling with Uncertainty Interval Computations Using Fuzzy Sets J. Honda, R. Tankelevich Department of Mathematical and Computer Sciences, Colorado School of Mines, Golden, CO, U.S.A. Abstract A new method
MATHEMATICAL ANALYSIS, MODELING AND OPTIMIZATION OF COMPLEX HEAT TRANSFER PROCESSES
 MATHEMATICAL ANALYSIS, MODELING AND OPTIMIZATION OF COMPLEX HEAT TRANSFER PROCESSES Goals of research Dr. Uldis Raitums, Dr. Kārlis Birģelis To develop and investigate mathematical properties of algorithms
MATHEMATICAL ANALYSIS, MODELING AND OPTIMIZATION OF COMPLEX HEAT TRANSFER PROCESSES Goals of research Dr. Uldis Raitums, Dr. Kārlis Birģelis To develop and investigate mathematical properties of algorithms
Unstructured Mesh Generation for Implicit Moving Geometries and Level Set Applications
 Unstructured Mesh Generation for Implicit Moving Geometries and Level Set Applications Per-Olof Persson (persson@mit.edu) Department of Mathematics Massachusetts Institute of Technology http://www.mit.edu/
Unstructured Mesh Generation for Implicit Moving Geometries and Level Set Applications Per-Olof Persson (persson@mit.edu) Department of Mathematics Massachusetts Institute of Technology http://www.mit.edu/
Robust Design Methodology of Topologically optimized components under the effect of uncertainties
 Robust Design Methodology of Topologically optimized components under the effect of uncertainties Joshua Amrith Raj and Arshad Javed Department of Mechanical Engineering, BITS-Pilani Hyderabad Campus,
Robust Design Methodology of Topologically optimized components under the effect of uncertainties Joshua Amrith Raj and Arshad Javed Department of Mechanical Engineering, BITS-Pilani Hyderabad Campus,
Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences
 Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences W. J. Godinez 1, M. Lampe 2, S. Wörz 1, B. Müller 2, R. Eils 1 and K. Rohr 1 1 University of Heidelberg, IPMB, and DKFZ
Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences W. J. Godinez 1, M. Lampe 2, S. Wörz 1, B. Müller 2, R. Eils 1 and K. Rohr 1 1 University of Heidelberg, IPMB, and DKFZ
A study on fractality properties of nano particles scanning electron microscopy images
 Leonardo Journal of Sciences ISSN 1583-0233 Issue 25, July-December 2014 p. 111-116 A study on fractality properties of nano particles scanning electron microscopy images Shahrbano MALEKI, Shahriar GHAMMAMY
Leonardo Journal of Sciences ISSN 1583-0233 Issue 25, July-December 2014 p. 111-116 A study on fractality properties of nano particles scanning electron microscopy images Shahrbano MALEKI, Shahriar GHAMMAMY
Digital Volume Correlation for Materials Characterization
 19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
Visualization of errors of finite element solutions P. Beckers, H.G. Zhong, Ph. Andry Aerospace Department, University of Liege, B-4000 Liege, Belgium
 Visualization of errors of finite element solutions P. Beckers, H.G. Zhong, Ph. Andry Aerospace Department, University of Liege, B-4000 Liege, Belgium Abstract The aim of this paper is to show how to use
Visualization of errors of finite element solutions P. Beckers, H.G. Zhong, Ph. Andry Aerospace Department, University of Liege, B-4000 Liege, Belgium Abstract The aim of this paper is to show how to use
Three-dimensional nondestructive evaluation of cylindrical objects (pipe) using an infrared camera coupled to a 3D scanner
 Three-dimensional nondestructive evaluation of cylindrical objects (pipe) using an infrared camera coupled to a 3D scanner F. B. Djupkep Dizeu, S. Hesabi, D. Laurendeau, A. Bendada Computer Vision and
Three-dimensional nondestructive evaluation of cylindrical objects (pipe) using an infrared camera coupled to a 3D scanner F. B. Djupkep Dizeu, S. Hesabi, D. Laurendeau, A. Bendada Computer Vision and
Morphological Analysis of Brain Structures Using Spatial Normalization
 Morphological Analysis of Brain Structures Using Spatial Normalization C. Davatzikos 1, M. Vaillant 1, S. Resnick 2, J.L. Prince 3;1, S. Letovsky 1, and R.N. Bryan 1 1 Department of Radiology, Johns Hopkins
Morphological Analysis of Brain Structures Using Spatial Normalization C. Davatzikos 1, M. Vaillant 1, S. Resnick 2, J.L. Prince 3;1, S. Letovsky 1, and R.N. Bryan 1 1 Department of Radiology, Johns Hopkins
Module 1 Lecture Notes 2. Optimization Problem and Model Formulation
 Optimization Methods: Introduction and Basic concepts 1 Module 1 Lecture Notes 2 Optimization Problem and Model Formulation Introduction In the previous lecture we studied the evolution of optimization
Optimization Methods: Introduction and Basic concepts 1 Module 1 Lecture Notes 2 Optimization Problem and Model Formulation Introduction In the previous lecture we studied the evolution of optimization
CHAPTER 2. Morphometry on rodent brains. A.E.H. Scheenstra J. Dijkstra L. van der Weerd
 CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
Programmable Pattern-Formation and Scale-Independence
 Programmable Pattern-Formation and Scale-Independence Radhika Nagpal PostDoctoral Lecturer, MIT Artificial Intelligence Lab radhi@ai.mit.edu This paper presents a programming language for pattern-formation
Programmable Pattern-Formation and Scale-Independence Radhika Nagpal PostDoctoral Lecturer, MIT Artificial Intelligence Lab radhi@ai.mit.edu This paper presents a programming language for pattern-formation
Modeling Flexibility with Spline Approximations for Fast VR Visualizations
 Modeling Flexibility with Spline Approximations for Fast VR Visualizations Abstract: Kevin Tatur a and Renate Sitte a a Faculty of Engineering and Information Technology, Griffith University, Gold Coast,
Modeling Flexibility with Spline Approximations for Fast VR Visualizations Abstract: Kevin Tatur a and Renate Sitte a a Faculty of Engineering and Information Technology, Griffith University, Gold Coast,
A NURBS-BASED APPROACH FOR SHAPE AND TOPOLOGY OPTIMIZATION OF FLOW DOMAINS
 6th European Conference on Computational Mechanics (ECCM 6) 7th European Conference on Computational Fluid Dynamics (ECFD 7) 11 15 June 2018, Glasgow, UK A NURBS-BASED APPROACH FOR SHAPE AND TOPOLOGY OPTIMIZATION
6th European Conference on Computational Mechanics (ECCM 6) 7th European Conference on Computational Fluid Dynamics (ECFD 7) 11 15 June 2018, Glasgow, UK A NURBS-BASED APPROACH FOR SHAPE AND TOPOLOGY OPTIMIZATION
A New Smoothing Algorithm for Quadrilateral and Hexahedral Meshes
 A New Smoothing Algorithm for Quadrilateral and Hexahedral Meshes Sanjay Kumar Khattri Department of Mathematics, University of Bergen, Norway sanjay@mi.uib.no http://www.mi.uib.no/ sanjay Abstract. Mesh
A New Smoothing Algorithm for Quadrilateral and Hexahedral Meshes Sanjay Kumar Khattri Department of Mathematics, University of Bergen, Norway sanjay@mi.uib.no http://www.mi.uib.no/ sanjay Abstract. Mesh
Surgical Cutting on a Multimodal Object Representation
 Surgical Cutting on a Multimodal Object Representation Lenka Jeřábková and Torsten Kuhlen Virtual Reality Group, RWTH Aachen University, 52074 Aachen Email: jerabkova@rz.rwth-aachen.de Abstract. In this
Surgical Cutting on a Multimodal Object Representation Lenka Jeřábková and Torsten Kuhlen Virtual Reality Group, RWTH Aachen University, 52074 Aachen Email: jerabkova@rz.rwth-aachen.de Abstract. In this
RESTORING ARTIFACT-FREE MICROSCOPY IMAGE SEQUENCES. Robotics Institute Carnegie Mellon University 5000 Forbes Ave, Pittsburgh, PA 15213, USA
 RESTORING ARTIFACT-FREE MICROSCOPY IMAGE SEQUENCES Zhaozheng Yin Takeo Kanade Robotics Institute Carnegie Mellon University 5000 Forbes Ave, Pittsburgh, PA 15213, USA ABSTRACT Phase contrast and differential
RESTORING ARTIFACT-FREE MICROSCOPY IMAGE SEQUENCES Zhaozheng Yin Takeo Kanade Robotics Institute Carnegie Mellon University 5000 Forbes Ave, Pittsburgh, PA 15213, USA ABSTRACT Phase contrast and differential
New developments in LS-OPT
 7. LS-DYNA Anwenderforum, Bamberg 2008 Optimierung II New developments in LS-OPT Nielen Stander, Tushar Goel, Willem Roux Livermore Software Technology Corporation, Livermore, CA94551, USA Summary: This
7. LS-DYNA Anwenderforum, Bamberg 2008 Optimierung II New developments in LS-OPT Nielen Stander, Tushar Goel, Willem Roux Livermore Software Technology Corporation, Livermore, CA94551, USA Summary: This
MODELING OF A MICRO-GRIPPER COMPLIANT JOINT USING COMSOL MULTIPHYSICS SIMULATION
 MODELING OF A MICRO-GRIPPER COMPLIANT JOINT USING COMSOL MULTIPHYSICS SIMULATION Mihăiţă Nicolae ARDELEANU, Veronica DESPA, Ioan Alexandru IVAN Valahia University from Targoviste E-mail: mihai.ardeleanu@valahia.ro,
MODELING OF A MICRO-GRIPPER COMPLIANT JOINT USING COMSOL MULTIPHYSICS SIMULATION Mihăiţă Nicolae ARDELEANU, Veronica DESPA, Ioan Alexandru IVAN Valahia University from Targoviste E-mail: mihai.ardeleanu@valahia.ro,
CS443: Digital Imaging and Multimedia Binary Image Analysis. Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University
 CS443: Digital Imaging and Multimedia Binary Image Analysis Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University Outlines A Simple Machine Vision System Image segmentation by thresholding
CS443: Digital Imaging and Multimedia Binary Image Analysis Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University Outlines A Simple Machine Vision System Image segmentation by thresholding
Engineering 1000 Chapter 6: Abstraction and Modeling
 Engineering 1000 Chapter 6: Abstraction and Modeling Outline Why is abstraction useful? What are models? how are models different from theory and simulation? Examples from microelectronics Types of model
Engineering 1000 Chapter 6: Abstraction and Modeling Outline Why is abstraction useful? What are models? how are models different from theory and simulation? Examples from microelectronics Types of model
Geometric Representations. Stelian Coros
 Geometric Representations Stelian Coros Geometric Representations Languages for describing shape Boundary representations Polygonal meshes Subdivision surfaces Implicit surfaces Volumetric models Parametric
Geometric Representations Stelian Coros Geometric Representations Languages for describing shape Boundary representations Polygonal meshes Subdivision surfaces Implicit surfaces Volumetric models Parametric
Supporting Information. High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images
 Supporting Information High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images Christine R. Laramy, 1, Keith A. Brown, 2, Matthew N. O Brien, 2 and Chad. A.
Supporting Information High-Throughput, Algorithmic Determination of Nanoparticle Structure From Electron Microscopy Images Christine R. Laramy, 1, Keith A. Brown, 2, Matthew N. O Brien, 2 and Chad. A.
Chapter 8 Visualization and Optimization
 Chapter 8 Visualization and Optimization Recommended reference books: [1] Edited by R. S. Gallagher: Computer Visualization, Graphics Techniques for Scientific and Engineering Analysis by CRC, 1994 [2]
Chapter 8 Visualization and Optimization Recommended reference books: [1] Edited by R. S. Gallagher: Computer Visualization, Graphics Techniques for Scientific and Engineering Analysis by CRC, 1994 [2]
A Novel Image Super-resolution Reconstruction Algorithm based on Modified Sparse Representation
 , pp.162-167 http://dx.doi.org/10.14257/astl.2016.138.33 A Novel Image Super-resolution Reconstruction Algorithm based on Modified Sparse Representation Liqiang Hu, Chaofeng He Shijiazhuang Tiedao University,
, pp.162-167 http://dx.doi.org/10.14257/astl.2016.138.33 A Novel Image Super-resolution Reconstruction Algorithm based on Modified Sparse Representation Liqiang Hu, Chaofeng He Shijiazhuang Tiedao University,
Surgery Simulation and Planning
 Surgery Simulation and Planning S. H. Martin Roth Dr. Rolf M. Koch Daniel Bielser Prof. Dr. Markus Gross Facial surgery project in collaboration with Prof. Dr. Dr. H. Sailer, University Hospital Zurich,
Surgery Simulation and Planning S. H. Martin Roth Dr. Rolf M. Koch Daniel Bielser Prof. Dr. Markus Gross Facial surgery project in collaboration with Prof. Dr. Dr. H. Sailer, University Hospital Zurich,
STATISTICS AND ANALYSIS OF SHAPE
 Control and Cybernetics vol. 36 (2007) No. 2 Book review: STATISTICS AND ANALYSIS OF SHAPE by H. Krim, A. Yezzi, Jr., eds. There are numerous definitions of a notion of shape of an object. These definitions
Control and Cybernetics vol. 36 (2007) No. 2 Book review: STATISTICS AND ANALYSIS OF SHAPE by H. Krim, A. Yezzi, Jr., eds. There are numerous definitions of a notion of shape of an object. These definitions
Morphological Image Processing
 Morphological Image Processing Binary image processing In binary images, we conventionally take background as black (0) and foreground objects as white (1 or 255) Morphology Figure 4.1 objects on a conveyor
Morphological Image Processing Binary image processing In binary images, we conventionally take background as black (0) and foreground objects as white (1 or 255) Morphology Figure 4.1 objects on a conveyor
Example 24 Spring-back
 Example 24 Spring-back Summary The spring-back simulation of sheet metal bent into a hat-shape is studied. The problem is one of the famous tests from the Numisheet 93. As spring-back is generally a quasi-static
Example 24 Spring-back Summary The spring-back simulation of sheet metal bent into a hat-shape is studied. The problem is one of the famous tests from the Numisheet 93. As spring-back is generally a quasi-static
Modeling the Acoustic Scattering from Axially Symmetric Fluid, Elastic, and Poroelastic Objects due to Nonsymmetric Forcing Using COMSOL Multiphysics
 Modeling the Acoustic Scattering from Axially Symmetric Fluid, Elastic, and Poroelastic Objects due to Nonsymmetric Forcing Using COMSOL Multiphysics Anthony L. Bonomo *1 and Marcia J. Isakson 1 1 Applied
Modeling the Acoustic Scattering from Axially Symmetric Fluid, Elastic, and Poroelastic Objects due to Nonsymmetric Forcing Using COMSOL Multiphysics Anthony L. Bonomo *1 and Marcia J. Isakson 1 1 Applied
Morphological Image Processing
 Morphological Image Processing Megha Goyal Dept. of ECE, Doaba Institute of Engineering and Technology, Kharar, Mohali, Punjab, India Abstract The purpose of this paper is to provide readers with an in-depth
Morphological Image Processing Megha Goyal Dept. of ECE, Doaba Institute of Engineering and Technology, Kharar, Mohali, Punjab, India Abstract The purpose of this paper is to provide readers with an in-depth
Parametric. Practices. Patrick Cunningham. CAE Associates Inc. and ANSYS Inc. Proprietary 2012 CAE Associates Inc. and ANSYS Inc. All rights reserved.
 Parametric Modeling Best Practices Patrick Cunningham July, 2012 CAE Associates Inc. and ANSYS Inc. Proprietary 2012 CAE Associates Inc. and ANSYS Inc. All rights reserved. E-Learning Webinar Series This
Parametric Modeling Best Practices Patrick Cunningham July, 2012 CAE Associates Inc. and ANSYS Inc. Proprietary 2012 CAE Associates Inc. and ANSYS Inc. All rights reserved. E-Learning Webinar Series This
Isogeometric Analysis of Fluid-Structure Interaction
 Isogeometric Analysis of Fluid-Structure Interaction Y. Bazilevs, V.M. Calo, T.J.R. Hughes Institute for Computational Engineering and Sciences, The University of Texas at Austin, USA e-mail: {bazily,victor,hughes}@ices.utexas.edu
Isogeometric Analysis of Fluid-Structure Interaction Y. Bazilevs, V.M. Calo, T.J.R. Hughes Institute for Computational Engineering and Sciences, The University of Texas at Austin, USA e-mail: {bazily,victor,hughes}@ices.utexas.edu
A Survey of Light Source Detection Methods
 A Survey of Light Source Detection Methods Nathan Funk University of Alberta Mini-Project for CMPUT 603 November 30, 2003 Abstract This paper provides an overview of the most prominent techniques for light
A Survey of Light Source Detection Methods Nathan Funk University of Alberta Mini-Project for CMPUT 603 November 30, 2003 Abstract This paper provides an overview of the most prominent techniques for light
Adaptive-Mesh-Refinement Pattern
 Adaptive-Mesh-Refinement Pattern I. Problem Data-parallelism is exposed on a geometric mesh structure (either irregular or regular), where each point iteratively communicates with nearby neighboring points
Adaptive-Mesh-Refinement Pattern I. Problem Data-parallelism is exposed on a geometric mesh structure (either irregular or regular), where each point iteratively communicates with nearby neighboring points
Skåne University Hospital Lund, Lund, Sweden 2 Deparment of Numerical Analysis, Centre for Mathematical Sciences, Lund University, Lund, Sweden
 Volume Tracking: A New Method for Visualization of Intracardiac Blood Flow from Three-Dimensional, Time-Resolved, Three-Component Magnetic Resonance Velocity Mapping Appendix: Theory and Numerical Implementation
Volume Tracking: A New Method for Visualization of Intracardiac Blood Flow from Three-Dimensional, Time-Resolved, Three-Component Magnetic Resonance Velocity Mapping Appendix: Theory and Numerical Implementation
Journal of Engineering Research and Studies E-ISSN
 Journal of Engineering Research and Studies E-ISS 0976-79 Research Article SPECTRAL SOLUTIO OF STEADY STATE CODUCTIO I ARBITRARY QUADRILATERAL DOMAIS Alavani Chitra R 1*, Joshi Pallavi A 1, S Pavitran
Journal of Engineering Research and Studies E-ISS 0976-79 Research Article SPECTRAL SOLUTIO OF STEADY STATE CODUCTIO I ARBITRARY QUADRILATERAL DOMAIS Alavani Chitra R 1*, Joshi Pallavi A 1, S Pavitran
EE795: Computer Vision and Intelligent Systems
 EE795: Computer Vision and Intelligent Systems Spring 2012 TTh 17:30-18:45 FDH 204 Lecture 14 130307 http://www.ee.unlv.edu/~b1morris/ecg795/ 2 Outline Review Stereo Dense Motion Estimation Translational
EE795: Computer Vision and Intelligent Systems Spring 2012 TTh 17:30-18:45 FDH 204 Lecture 14 130307 http://www.ee.unlv.edu/~b1morris/ecg795/ 2 Outline Review Stereo Dense Motion Estimation Translational
Accurate 3D Face and Body Modeling from a Single Fixed Kinect
 Accurate 3D Face and Body Modeling from a Single Fixed Kinect Ruizhe Wang*, Matthias Hernandez*, Jongmoo Choi, Gérard Medioni Computer Vision Lab, IRIS University of Southern California Abstract In this
Accurate 3D Face and Body Modeling from a Single Fixed Kinect Ruizhe Wang*, Matthias Hernandez*, Jongmoo Choi, Gérard Medioni Computer Vision Lab, IRIS University of Southern California Abstract In this
Quantifying Three-Dimensional Deformations of Migrating Fibroblasts
 45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
Three-Dimensional Computer Vision
 \bshiaki Shirai Three-Dimensional Computer Vision With 313 Figures ' Springer-Verlag Berlin Heidelberg New York London Paris Tokyo Table of Contents 1 Introduction 1 1.1 Three-Dimensional Computer Vision
\bshiaki Shirai Three-Dimensional Computer Vision With 313 Figures ' Springer-Verlag Berlin Heidelberg New York London Paris Tokyo Table of Contents 1 Introduction 1 1.1 Three-Dimensional Computer Vision
3D Modeling of Objects Using Laser Scanning
 1 3D Modeling of Objects Using Laser Scanning D. Jaya Deepu, LPU University, Punjab, India Email: Jaideepudadi@gmail.com Abstract: In the last few decades, constructing accurate three-dimensional models
1 3D Modeling of Objects Using Laser Scanning D. Jaya Deepu, LPU University, Punjab, India Email: Jaideepudadi@gmail.com Abstract: In the last few decades, constructing accurate three-dimensional models
3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set
 3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set John M. Sullivan, Jr., Ziji Wu, and Anand Kulkarni Worcester Polytechnic Institute Worcester,
3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set John M. Sullivan, Jr., Ziji Wu, and Anand Kulkarni Worcester Polytechnic Institute Worcester,
It has been widely accepted that the finite element
 Advancements of Extrusion Simulation in DEFORM-3D By G. Li, J. Yang, J.Y. Oh, M. Foster, and W. Wu, Scientific Forming Tech. Corp.; and P. Tsai and W. Chang, MIRDC Introduction It has been widely accepted
Advancements of Extrusion Simulation in DEFORM-3D By G. Li, J. Yang, J.Y. Oh, M. Foster, and W. Wu, Scientific Forming Tech. Corp.; and P. Tsai and W. Chang, MIRDC Introduction It has been widely accepted
doi: /
 Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
CFD Project Workflow Guide
 CFD Project Workflow Guide Contents Select a problem with known results for proof-of-concept testing... 1 Set up and run a coarse test case... 2 Select and calibrate numerical methods... 3 Minimize & quantify
CFD Project Workflow Guide Contents Select a problem with known results for proof-of-concept testing... 1 Set up and run a coarse test case... 2 Select and calibrate numerical methods... 3 Minimize & quantify
MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS
 Mathematical and Computational Applications, Vol. 5, No. 2, pp. 240-247, 200. Association for Scientific Research MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS Volkan Uslan and Đhsan Ömür Bucak
Mathematical and Computational Applications, Vol. 5, No. 2, pp. 240-247, 200. Association for Scientific Research MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS Volkan Uslan and Đhsan Ömür Bucak
Outline. COMSOL Multyphysics: Overview of software package and capabilities
 COMSOL Multyphysics: Overview of software package and capabilities Lecture 5 Special Topics: Device Modeling Outline Basic concepts and modeling paradigm Overview of capabilities Steps in setting-up a
COMSOL Multyphysics: Overview of software package and capabilities Lecture 5 Special Topics: Device Modeling Outline Basic concepts and modeling paradigm Overview of capabilities Steps in setting-up a
Virtual Frap User Guide
 Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
PTC Newsletter January 14th, 2002
 PTC Email Newsletter January 14th, 2002 PTC Product Focus: Pro/MECHANICA (Structure) Tip of the Week: Creating and using Rigid Connections Upcoming Events and Training Class Schedules PTC Product Focus:
PTC Email Newsletter January 14th, 2002 PTC Product Focus: Pro/MECHANICA (Structure) Tip of the Week: Creating and using Rigid Connections Upcoming Events and Training Class Schedules PTC Product Focus:
David Wagner, Kaan Divringi, Can Ozcan Ozen Engineering
 Internal Forces of the Femur: An Automated Procedure for Applying Boundary Conditions Obtained From Inverse Dynamic Analysis to Finite Element Simulations David Wagner, Kaan Divringi, Can Ozcan Ozen Engineering
Internal Forces of the Femur: An Automated Procedure for Applying Boundary Conditions Obtained From Inverse Dynamic Analysis to Finite Element Simulations David Wagner, Kaan Divringi, Can Ozcan Ozen Engineering
Bioimage Informatics. Lecture 8, Spring Bioimage Data Analysis (II): Point Feature Detection
 Bioimage Informatics Lecture 8, Spring 2012 Bioimage Data Analysis (II): Point Feature Detection Lecture 8 February 13, 2012 1 Outline Project assignment 02 Comments on reading assignment 01 Review: pixel
Bioimage Informatics Lecture 8, Spring 2012 Bioimage Data Analysis (II): Point Feature Detection Lecture 8 February 13, 2012 1 Outline Project assignment 02 Comments on reading assignment 01 Review: pixel
Introduction to Computational Fluid Dynamics Mech 122 D. Fabris, K. Lynch, D. Rich
 Introduction to Computational Fluid Dynamics Mech 122 D. Fabris, K. Lynch, D. Rich 1 Computational Fluid dynamics Computational fluid dynamics (CFD) is the analysis of systems involving fluid flow, heat
Introduction to Computational Fluid Dynamics Mech 122 D. Fabris, K. Lynch, D. Rich 1 Computational Fluid dynamics Computational fluid dynamics (CFD) is the analysis of systems involving fluid flow, heat
A new 8-node quadrilateral spline finite element
 Journal of Computational and Applied Mathematics 195 (2006) 54 65 www.elsevier.com/locate/cam A new 8-node quadrilateral spline finite element Chong-Jun Li, Ren-Hong Wang Institute of Mathematical Sciences,
Journal of Computational and Applied Mathematics 195 (2006) 54 65 www.elsevier.com/locate/cam A new 8-node quadrilateral spline finite element Chong-Jun Li, Ren-Hong Wang Institute of Mathematical Sciences,
Controlling the spread of dynamic self-organising maps
 Neural Comput & Applic (2004) 13: 168 174 DOI 10.1007/s00521-004-0419-y ORIGINAL ARTICLE L. D. Alahakoon Controlling the spread of dynamic self-organising maps Received: 7 April 2004 / Accepted: 20 April
Neural Comput & Applic (2004) 13: 168 174 DOI 10.1007/s00521-004-0419-y ORIGINAL ARTICLE L. D. Alahakoon Controlling the spread of dynamic self-organising maps Received: 7 April 2004 / Accepted: 20 April
Lecture 2 Unstructured Mesh Generation
 Lecture 2 Unstructured Mesh Generation MIT 16.930 Advanced Topics in Numerical Methods for Partial Differential Equations Per-Olof Persson (persson@mit.edu) February 13, 2006 1 Mesh Generation Given a
Lecture 2 Unstructured Mesh Generation MIT 16.930 Advanced Topics in Numerical Methods for Partial Differential Equations Per-Olof Persson (persson@mit.edu) February 13, 2006 1 Mesh Generation Given a
CFD Post-Processing of Rampressor Rotor Compressor
 Gas Turbine Industrial Fellowship Program 2006 CFD Post-Processing of Rampressor Rotor Compressor Curtis Memory, Brigham Young niversity Ramgen Power Systems Mentor: Rob Steele I. Introduction Recent movements
Gas Turbine Industrial Fellowship Program 2006 CFD Post-Processing of Rampressor Rotor Compressor Curtis Memory, Brigham Young niversity Ramgen Power Systems Mentor: Rob Steele I. Introduction Recent movements
3/27/2012 WHY SPECT / CT? SPECT / CT Basic Principles. Advantages of SPECT. Advantages of CT. Dr John C. Dickson, Principal Physicist UCLH
 3/27/212 Advantages of SPECT SPECT / CT Basic Principles Dr John C. Dickson, Principal Physicist UCLH Institute of Nuclear Medicine, University College London Hospitals and University College London john.dickson@uclh.nhs.uk
3/27/212 Advantages of SPECT SPECT / CT Basic Principles Dr John C. Dickson, Principal Physicist UCLH Institute of Nuclear Medicine, University College London Hospitals and University College London john.dickson@uclh.nhs.uk
Invariant shape similarity. Invariant shape similarity. Invariant similarity. Equivalence. Equivalence. Equivalence. Equal SIMILARITY TRANSFORMATION
 1 Invariant shape similarity Alexer & Michael Bronstein, 2006-2009 Michael Bronstein, 2010 tosca.cs.technion.ac.il/book 2 Invariant shape similarity 048921 Advanced topics in vision Processing Analysis
1 Invariant shape similarity Alexer & Michael Bronstein, 2006-2009 Michael Bronstein, 2010 tosca.cs.technion.ac.il/book 2 Invariant shape similarity 048921 Advanced topics in vision Processing Analysis
Prof. Fanny Ficuciello Robotics for Bioengineering Visual Servoing
 Visual servoing vision allows a robotic system to obtain geometrical and qualitative information on the surrounding environment high level control motion planning (look-and-move visual grasping) low level
Visual servoing vision allows a robotic system to obtain geometrical and qualitative information on the surrounding environment high level control motion planning (look-and-move visual grasping) low level
Registration: Rigid vs. Deformable
 Lecture 20 Deformable / Non-Rigid Registration ch. 11 of Insight into Images edited by Terry Yoo, et al. Spring 2017 16-725 (CMU RI) : BioE 2630 (Pitt) Dr. John Galeotti The content of these slides by
Lecture 20 Deformable / Non-Rigid Registration ch. 11 of Insight into Images edited by Terry Yoo, et al. Spring 2017 16-725 (CMU RI) : BioE 2630 (Pitt) Dr. John Galeotti The content of these slides by
CHAPTER 4. Numerical Models. descriptions of the boundary conditions, element types, validation, and the force
 CHAPTER 4 Numerical Models This chapter presents the development of numerical models for sandwich beams/plates subjected to four-point bending and the hydromat test system. Detailed descriptions of the
CHAPTER 4 Numerical Models This chapter presents the development of numerical models for sandwich beams/plates subjected to four-point bending and the hydromat test system. Detailed descriptions of the
Non-Linear Finite Element Methods in Solid Mechanics Attilio Frangi, Politecnico di Milano, February 3, 2017, Lesson 1
 Non-Linear Finite Element Methods in Solid Mechanics Attilio Frangi, attilio.frangi@polimi.it Politecnico di Milano, February 3, 2017, Lesson 1 1 Politecnico di Milano, February 3, 2017, Lesson 1 2 Outline
Non-Linear Finite Element Methods in Solid Mechanics Attilio Frangi, attilio.frangi@polimi.it Politecnico di Milano, February 3, 2017, Lesson 1 1 Politecnico di Milano, February 3, 2017, Lesson 1 2 Outline
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION
 CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
Introduction to the Finite Element Method (3)
 Introduction to the Finite Element Method (3) Petr Kabele Czech Technical University in Prague Faculty of Civil Engineering Czech Republic petr.kabele@fsv.cvut.cz people.fsv.cvut.cz/~pkabele 1 Outline
Introduction to the Finite Element Method (3) Petr Kabele Czech Technical University in Prague Faculty of Civil Engineering Czech Republic petr.kabele@fsv.cvut.cz people.fsv.cvut.cz/~pkabele 1 Outline
Mathematical Morphology and Distance Transforms. Robin Strand
 Mathematical Morphology and Distance Transforms Robin Strand robin.strand@it.uu.se Morphology Form and structure Mathematical framework used for: Pre-processing Noise filtering, shape simplification,...
Mathematical Morphology and Distance Transforms Robin Strand robin.strand@it.uu.se Morphology Form and structure Mathematical framework used for: Pre-processing Noise filtering, shape simplification,...
Cellular Learning Automata-Based Color Image Segmentation using Adaptive Chains
 Cellular Learning Automata-Based Color Image Segmentation using Adaptive Chains Ahmad Ali Abin, Mehran Fotouhi, Shohreh Kasaei, Senior Member, IEEE Sharif University of Technology, Tehran, Iran abin@ce.sharif.edu,
Cellular Learning Automata-Based Color Image Segmentation using Adaptive Chains Ahmad Ali Abin, Mehran Fotouhi, Shohreh Kasaei, Senior Member, IEEE Sharif University of Technology, Tehran, Iran abin@ce.sharif.edu,
Generalized Principal Component Analysis CVPR 2007
 Generalized Principal Component Analysis Tutorial @ CVPR 2007 Yi Ma ECE Department University of Illinois Urbana Champaign René Vidal Center for Imaging Science Institute for Computational Medicine Johns
Generalized Principal Component Analysis Tutorial @ CVPR 2007 Yi Ma ECE Department University of Illinois Urbana Champaign René Vidal Center for Imaging Science Institute for Computational Medicine Johns
Data Mining Practical Machine Learning Tools and Techniques. Slides for Chapter 6 of Data Mining by I. H. Witten and E. Frank
 Data Mining Practical Machine Learning Tools and Techniques Slides for Chapter 6 of Data Mining by I. H. Witten and E. Frank Implementation: Real machine learning schemes Decision trees Classification
Data Mining Practical Machine Learning Tools and Techniques Slides for Chapter 6 of Data Mining by I. H. Witten and E. Frank Implementation: Real machine learning schemes Decision trees Classification
Tomographic Algorithm for Industrial Plasmas
 Tomographic Algorithm for Industrial Plasmas More info about this article: http://www.ndt.net/?id=22342 1 Sudhir K. Chaudhary, 1 Kavita Rathore, 2 Sudeep Bhattacharjee, 1 Prabhat Munshi 1 Nuclear Engineering
Tomographic Algorithm for Industrial Plasmas More info about this article: http://www.ndt.net/?id=22342 1 Sudhir K. Chaudhary, 1 Kavita Rathore, 2 Sudeep Bhattacharjee, 1 Prabhat Munshi 1 Nuclear Engineering
Medial Scaffolds for 3D data modelling: status and challenges. Frederic Fol Leymarie
 Medial Scaffolds for 3D data modelling: status and challenges Frederic Fol Leymarie Outline Background Method and some algorithmic details Applications Shape representation: From the Medial Axis to the
Medial Scaffolds for 3D data modelling: status and challenges Frederic Fol Leymarie Outline Background Method and some algorithmic details Applications Shape representation: From the Medial Axis to the
CS 231. Deformation simulation (and faces)
 CS 231 Deformation simulation (and faces) 1 Cloth Simulation deformable surface model Represent cloth model as a triangular or rectangular grid Points of finite mass as vertices Forces or energies of points
CS 231 Deformation simulation (and faces) 1 Cloth Simulation deformable surface model Represent cloth model as a triangular or rectangular grid Points of finite mass as vertices Forces or energies of points
