Stacked Multiscale Feature Learning for Domain Independent Medical Image Segmentation
|
|
|
- Wilfrid Nichols
- 6 years ago
- Views:
Transcription
1 Stacked Multiscale Feature Learning for Domain Independent Medical Image Segmentation Ryan Kiros, Karteek Popuri, Dana Cobzas, and Martin Jagersand University of Alberta, Canada Abstract. In this work we propose a feature-based segmentation approach that is domain independent. While most existing approaches are based on application-specific hand-crafted features, we propose a framework for learning features from data itself at multiple scales and depth. Our features can be easily integrated into classifiers or energy-based segmentation algorithms. We test the performance of our proposed method on two MICCAI grand challenges, obtaining the top score on VESSEL12 and competitive performance on BRATS Introduction The choice of image representation plays a crucial role in the success of medical image segmentation algorithms. Most existing methods utilize hand-crafted features incorporated into an energy-based segmentation method or into a machine learning classifier. Commonly, energy-based methods utilize engineered features such as Gabor filters for texture-based segmentation [1], while machine learning approaches use many more simple features like Haar or steerable filters leaving the classification method to disambiguate the ones that are significant for the segmentation task. Popular examples of machine learning methods are ones based on decision trees [2] or random forests [3]. Some methods use very specialized filters designed for a particular task, such as extracting linear structures based on eigenvalues of the image Hessian matrix [4]. Recently there has been much interest within the machine learning and computer vision communities to automatically learn feature representations from scratch. Feature learning methods are general, while hand-crafted features require a certain insight and understanding of the given image data to be analyzed, thus they are often not optimal when applied to a new dataset. Moreover, feature learning algorithms can benefit from many unlabeled examples, even those that may come from a different distribution than the target data [5]. Features can be learned either in an unsupervised setting or in a joint end-to-end system trained with supervision. Successful applications have included object recognition [6] [7], scene parsing and segmentation [8], annotation and retrieval [9], multimodal applications [10] and large-scale learning [11]. What these methods have in common is the emphasis on learning hierarchical representations as opposed to single-layer algorithms such as sparse coding. Unfortunately, most of the above methods are not directly applicable to medical imaging tasks as they often assume the use of natural images and require a
2 2 Ryan Kiros, Karteek Popuri, Dana Cobzas, and Martin Jagersand Table 1: A comparison of different feature learning architectures for application to medical image segmentation: Y is yes, N is no and S is sometimes. Multi-scale and multi-depth methods can often improve performance while patch-based and stagewise learning improve speed. Here sparse coding eefers to any method that aims to learn a filter bank with a sparsity cost. Method Patch-based Multi-scale Multi-depth Stagewise Sparse coding Y N N Y Convolutional sparse coding N N N Y Convolutional networks N S Y N Proposed approach Y Y Y Y large number of labeled examples to be effective. There exist few feature learning methods applied to medical data like the segmentation of linear [12] and curvilinear [13] structures; segmentation of electron microscopy (EM) images [14] and a recent work on MS lesions segmentation [15]. In this paper we propose a framework that is domain independent and utilizes features learned from multiple scales and depth. The key features that make our method fast and thus suitable for medical data are detailed below and can be summarized as: (1) patch-based, (2) stage-based system and a (3) fast dictionary learning method. Table 1 summarizes and distinguishes four types of feature learning architectures. Simpler single layer sparse coding methods like [15] also use patches but with no scales or depth. Convolutional sparse coding algorithms, such as those used by [12],[16] and [13], differ from standard sparse coding methods as convolution is incorporated into the optimization procedure. The third architecture describes convolutional networks, used by [14] for EM segmentation, which are learned jointly with supervision. While convolutional networks are often very effective, jointly training the whole model can be time consuming. Furthermore, convolutional networks require many labeled examples in order to avoid overfitting. The last architectures illustrates our proposed framework. Features are learned one stage at a time using patch-based learning at multiple scales. Since the model does not require joint learning, features can be learned efficiently and quickly. Our framework is the first to utilize the encoding versus training principle of [17] in the context of image segmentation. The emphasis of this work is the importance of the feature encoding as opposed to the filter learning algorithm itself. Due to this, we suggest that more expensive convolutional filter learning is unnecessary, so long as a proper encoding is performed after learning. Experimentally we demonstrate that the same algorithm can be used to obtain strong performance on two completely different medical segmentation tasks. We report superior results on the vessel segmentation of the lung (VESSEL12) challenge data and competitive performance on multimodal brain tumor segmentation (BRATS2012) data. Furthermore, our system is able to learn features in under ten minutes on both challenges. Code for our approach will be released upon publication.
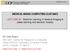
3 Stacked Multiscale Feature Learning 3 2 Method We assume we are given m volumes with s modalities {{V (j) } s j=1 }m i=1, where each V (j) i R n V n H n. n V n H is the spatial dimension of a slice and n is the number of slices. For simplicity, we assume that each volume V (j) i has dimensionality n V n H n although this is not needed. As a specific example, brain tumor segmentation tasks can use s = 4 modalites consisting of FLAIR, T1, T2 and post-gadolinium T1. The general outline of our feature learning framework is as follows: Extract multimodal patches at multiple scales using a Gaussian pyramid. Learn a filter bank using orthogonal matching pursuit. Convolutionally extract feature maps using the learned filters as kernels. Repeat the above steps, using the computed features maps as input to another layer. The number of feature maps (next layer modalities) corresponds to the number of filters. In each of the following subsections, we describe the above operations in detail. Fig. 1: Visualization of our feature learning approach. Each volume slice is scaled using a Gaussian pyramid. Patches are extracted at each scale to learn a dictionary D using OMP. Convolution is performed over all scales with the dictionary filters, resulting in Γ k feature maps. After training the first layer, the feature maps can then be used as input to a second layer. 2.1 Pre-processing and dictionary learning Given a volume V, a Gaussian pyramid with Γ scales is applied to each modality of each slice. Let {p (1),..., p (m P ) } denote a set of m P patches randomly extracted from the scaled volumes. Each patch p (l) is of spatial dimension r c s where r c is the receptive field size. These patches are then flattened into column
4 4 Ryan Kiros, Karteek Popuri, Dana Cobzas, and Martin Jagersand vectors. Per patch contrast normalization and patch-wise mean subtraction is performed. For dictionary learning we use orthogonal matching pursuit (OMP). OMP aims to solve the following optimization problem: minimize D,x (i) subject to m P Dx (i) p (i) 2 2 i=1 D (l) 2 2 = 1, l x (i) 0 q, i (1) where D R n P k and D (l) is the l-th column of D. Optimization is done using alternation over the dictionary D and codes x. For all our experiments we set q = 1, which reduces to a form of gain-shape vector quantization. In particular, given a dictionary D, an index κ is chosen as κ = argmax D (l)t p (i) (2) l for which the κ-th index of x (i) is set as x κ (i) = D (κ)t p (i) with all other indices left as zero in order to satisfy the constraint x (i) 0 1 for all i. Given the onehot codes X, the dictionary is easily updated by first solving the unconstrained problem, followed by re-normalization to satisfy the constraint D (l) 2 2 = 1 for all l. 2.2 Convolutional feature extraction Let T γ j denote a volume slice of modality j and scale γ. Each r c s patch in T γ j is pre-processed by contrast normalization and mean subtraction. Let D (l) j R r c denote the l-th basis for modality j of D. We will define the feature encoding for basis l as: s f γ l = T γ j D(l) j (3) j=1 where * denotes convolution. The resulting feature maps {f γ l }k l=1 are of the same spatial dimensions as T γ j. The feature maps are finally upsampled to the original n V n H spatial dimension. Figure 1 illustrates our approach. 2.3 Stacking multiple layers Our described setup for feature learning has involved scaling, dictionary learning and convolutional extraction. Just as the volumes slices were inputs to a first layer with s modalities, the upsampled output feature maps {{f γ l }Γ γ=1} k l=1 may be seen as inputs to a second layer but with Γ k modalities. The same described operations are applied a second time resulting in additional second layer output feature maps. These groups of feature maps can be concatenated together resulting in a total number of Γ 1 k 1 + Γ 2 k 2 feature maps, where Γ 1, k 1 are the number
5 Stacked Multiscale Feature Learning 5 of first layer scales and filters while Γ 2, k 2 are the number of second layer scales and filters. Thus each pixel in a volume slice can be represented as a Γ 1 k 1 +Γ 2 k 2 dimensional feature vector. Fig. 2: Visualizing the importance of scale and depth for vessel segmentation. 3 Experiments We perform experimental evaluation using data from two MICCAI grad challenges: vessel segmentation of the lung 1 and multimodal brain tumor segmentation Vessel segmentation The vessel segmentation challenge consists of 20 volumes of CT scans to segment with 3 additional volumes that include 882 labeled pixels based on the agreement of at least 3 experts. Each slice is of size with each volume containing a few hundred slices. We performed feature learning with 2 depths, 6 scales, a receptive field size of 5 5, 32 first layer filters and 64 second layer filters. The final feature vector is thus of size 6 (32+64) = 576. In order to perform segmentation, we extracted features for the existing labeled pixels and trained a L2-regularized logistic regression classifier, using 10-fold cross validation in order to tune the L2 hyperparameter. Each pixel of a new slice is then classified, resulting in a probability of whether or not the pixel is a vessel. For our submission to the challenge, the probabilities are scaled and rounded to unsigned 8-bit integers as requested. Figure 2 illustrates the importance of adding depth and scale to segmentation. The first image is the original CT scan. The second image shows segmentation when neither depth nor scale is added while the third image shows segmentation with added depth and scale. Without scale, larger vessels are less likely to be segmented while without depth, segmentation is much more scattered and less contiguous. For visualization purposes, a pixel is labeled as being a vessel if the probability of a vessel given the pixel features is greater than
6 6 Ryan Kiros, Karteek Popuri, Dana Cobzas, and Martin Jagersand Table 2: The top 5 results from the VESSEL12 challenge leaderboard. Team Method type score our method feature learning + classification LKEBChina Krissian-inspired vesselness FME LungVessels Frangi vesselness + region growing LKEBChina Krissian-inspired vesselness with bi-gaussian kernel FME LungVessels Frangi vesselness + region growing (raw) Fig. 3: Sample vessel segmentation results. Table 2 shows the top 5 performing methods on the VESSEL12 challenge. Our proposed method tops all existing approaches. The top performing methods in the competition are largely based on the use of Frangi [4] and Krissian vesselness [18] all of which derive structural properties from the eigenvalues of the Hessian. 3.2 Brain tumor segmentation To emphasize that the proposed method is domain independent, we evaluated it on the BRATS2012 multimodal brain tumor segmentation challenge, a dataset that has totally different properties and segmentation task than the vessel data. Due to BRATS2012 site maintenance, the test volume labels were unavailable at the time we did our experiments. Instead we perform evaluation using leave-oneout cross validation on the training set. Two types of tumour data are evaluated: high-grade and low-grade. Each volume voxel is labeled as being one of three classes: tumor, edema and other. We utilized our approach with one scale and two depths, with 16 bases in each depth for a total of 32 features. A 2 hidden layer network with dropout [19] is used to make predictions. Within each training fold, 10-fold cross validation is used to select the dropout parameters. Table 3 shows our results in comparison to the top 2 methods in the competition. We note again that out comparison is not on the same held-out data. None the less, our results are competitive with the top performing methods.
7 Stacked Multiscale Feature Learning 7 Flair T1 T1C T2 Manual Automatic Fig. 4: Sample brain tumor segmentation results. Table 3: Comparison against the top two performers in the BRATS2012 competition. HG and LG stand for high-grade and low-grade, respectively. Team region mean dice coeff. region mean dice coeff. our method HG edema LG edema Bauer et al. HG edema LG edema Zikic et al. HG edema LG edema our method HG tumor LG tumor Bauer et al. HG tumor LG tumor Zikic et al. HG tumor LG tumor our method HG GTV LG GTV Conclusion In this paper we proposed a domain independent approach for segmenting medical images. Our approaches involves learning feature representations at multiple scales and depths which are compatible with existing classification and energybased segmentation methods. We obtain the best performing result on the VES- SEL12 challenge and competitive results on the BRATS2012 multimodal brain tumor segmentation challenge. For future work we intend to further evaluate our approach on additional grand challenge problems. We also intend to study various transfer learning scenarios between domains and modalities. References 1. Paragios, N., Deriche, R.: Geodesic active regions and level set methods for supervised texture segmentation. IJCV 50(3) (2002)
8 8 Ryan Kiros, Karteek Popuri, Dana Cobzas, and Martin Jagersand 2. Zheng, Y., Barbu, A., Georgescu, B., Scheuering, M., Comaniciu, D.: Fourchamber heart modeling and automatic segmentation for 3d cardiac ct volumes using marginal space learning and steerable features. IEEE Trans. Medical Imaging 27(11) (2008) Criminisi, A., Robertson, D., Konukoglu, E., Shotton, J., Pathak, S., White, S., Siddiqui, K.: Regression forests for efficient anatomy detection and localization in computed tomography scans. Medical Image Analysis (2013) 4. Frangi, A., Niessen, W.J., Vincken, K.L., Viergever, M.A.: Multiscale vessel enhancement filtering. In: MICCAI. (1998) Raina, R., Battle, A., Lee, H., Packer, B., Ng, A.Y.: Self-taught learning: transfer learning from unlabeled data. In: ICML. (2007) Bo, L., Ren, X., Fox, D.: Unsupervised Feature Learning for RGB-D Based Object Recognition. In: ISER. (June 2012) 7. Krizhevsky, A., Sutskever, I., Hinton, G.: Imagenet classification with deep convolutional neural networks. In: NIPS. (2012) Farabet, C., Couprie, C., Najman, L., LeCun, Y.: Scene parsing with multiscale feature learning, purity trees, and optimal covers. In: ICML. (2012) 9. Kiros, R., Szepesvari, C.: Deep representations and codes for image autoannotation. In: NIPS. (2012) Srivastava, N., Salakhutdinov, R.: Multimodal learning with deep boltzmann machines. In: NIPS. (2012) Dean, J., Corrado, G., Monga, R., Chen, K., Devin, M., Le, Q., Mao, M., Ranzato, M., Senior, A., Tucker, P., Yang, K., Ng, A.: Large scale distributed deep networks. In: NIPS. (2012) Rigamonti, R., Lepetit, V.: Accurate and efficient linear structure. segmentation by leveraging ad hoc features with learned filters. In: MICCAI. (2012) Becker, C., Rigamonti, R., Lepetit, V., Fua, P.: Supervised feature learning for curvilinear structure segmentation. In: MICCAI. (2013) Ciresan, D., Giusti, A., Schmidhuber, J., et al.: Deep neural networks segment neuronal membranes in electron microscopy images. In: NIPS. (2012) Weiss, N., Rueckert, D., Rao, A.: Multiple sclerosis lesion segmentation using dictionary learning and sparse coding. In: MICCAI. (2013) Rigamonti, R., Türetken, E., González, G., Fua, P., Lepetit, V.: Filter learning for linear structure segmentation. Technical report, Technical report, EPFL (2011) 17. Coates, A., Ng, A.Y.: The importance of encoding versus training with sparse coding and vector quantization. In: ICML. Volume 8. (2011) Krissian, K., Malandain, G., Ayache, N., Vaillant, R., Trousset, Y.: Model-based detection of tubular structures in 3d images. Computer vision and image understanding 80(2) (2000) Hinton, G.E., Srivastava, N., Krizhevsky, A., Sutskever, I., Salakhutdinov, R.R.: Improving neural networks by preventing co-adaptation of feature detectors. arxiv preprint arxiv: (2012)
University of Alberta. Ryan Kiros. Master of Science in Statistical Machine Learning. Department of Computing Science
 University of Alberta LEARNING DEEP REPRESENTATIONS, EMBEDDINGS AND CODES FROM THE PIXEL LEVEL OF NATURAL AND MEDICAL IMAGES by Ryan Kiros A thesis submitted to the Faculty of Graduate Studies and Research
University of Alberta LEARNING DEEP REPRESENTATIONS, EMBEDDINGS AND CODES FROM THE PIXEL LEVEL OF NATURAL AND MEDICAL IMAGES by Ryan Kiros A thesis submitted to the Faculty of Graduate Studies and Research
ImageNet Classification with Deep Convolutional Neural Networks
 ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky Ilya Sutskever Geoffrey Hinton University of Toronto Canada Paper with same name to appear in NIPS 2012 Main idea Architecture
ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky Ilya Sutskever Geoffrey Hinton University of Toronto Canada Paper with same name to appear in NIPS 2012 Main idea Architecture
Reconstructive Sparse Code Transfer for Contour Detection and Semantic Labeling
 Reconstructive Sparse Code Transfer for Contour Detection and Semantic Labeling Michael Maire 1,2 Stella X. Yu 3 Pietro Perona 2 1 TTI Chicago 2 California Institute of Technology 3 University of California
Reconstructive Sparse Code Transfer for Contour Detection and Semantic Labeling Michael Maire 1,2 Stella X. Yu 3 Pietro Perona 2 1 TTI Chicago 2 California Institute of Technology 3 University of California
3D Densely Convolutional Networks for Volumetric Segmentation. Toan Duc Bui, Jitae Shin, and Taesup Moon
 3D Densely Convolutional Networks for Volumetric Segmentation Toan Duc Bui, Jitae Shin, and Taesup Moon School of Electronic and Electrical Engineering, Sungkyunkwan University, Republic of Korea arxiv:1709.03199v2
3D Densely Convolutional Networks for Volumetric Segmentation Toan Duc Bui, Jitae Shin, and Taesup Moon School of Electronic and Electrical Engineering, Sungkyunkwan University, Republic of Korea arxiv:1709.03199v2
Facial Expression Classification with Random Filters Feature Extraction
 Facial Expression Classification with Random Filters Feature Extraction Mengye Ren Facial Monkey mren@cs.toronto.edu Zhi Hao Luo It s Me lzh@cs.toronto.edu I. ABSTRACT In our work, we attempted to tackle
Facial Expression Classification with Random Filters Feature Extraction Mengye Ren Facial Monkey mren@cs.toronto.edu Zhi Hao Luo It s Me lzh@cs.toronto.edu I. ABSTRACT In our work, we attempted to tackle
Boundary-aware Fully Convolutional Network for Brain Tumor Segmentation
 Boundary-aware Fully Convolutional Network for Brain Tumor Segmentation Haocheng Shen, Ruixuan Wang, Jianguo Zhang, and Stephen J. McKenna Computing, School of Science and Engineering, University of Dundee,
Boundary-aware Fully Convolutional Network for Brain Tumor Segmentation Haocheng Shen, Ruixuan Wang, Jianguo Zhang, and Stephen J. McKenna Computing, School of Science and Engineering, University of Dundee,
Convolutional-Recursive Deep Learning for 3D Object Classification
 Convolutional-Recursive Deep Learning for 3D Object Classification Richard Socher, Brody Huval, Bharath Bhat, Christopher D. Manning, Andrew Y. Ng NIPS 2012 Iro Armeni, Manik Dhar Motivation Hand-designed
Convolutional-Recursive Deep Learning for 3D Object Classification Richard Socher, Brody Huval, Bharath Bhat, Christopher D. Manning, Andrew Y. Ng NIPS 2012 Iro Armeni, Manik Dhar Motivation Hand-designed
LEARNING A SPARSE DICTIONARY OF VIDEO STRUCTURE FOR ACTIVITY MODELING. Nandita M. Nayak, Amit K. Roy-Chowdhury. University of California, Riverside
 LEARNING A SPARSE DICTIONARY OF VIDEO STRUCTURE FOR ACTIVITY MODELING Nandita M. Nayak, Amit K. Roy-Chowdhury University of California, Riverside ABSTRACT We present an approach which incorporates spatiotemporal
LEARNING A SPARSE DICTIONARY OF VIDEO STRUCTURE FOR ACTIVITY MODELING Nandita M. Nayak, Amit K. Roy-Chowdhury University of California, Riverside ABSTRACT We present an approach which incorporates spatiotemporal
NIH Public Access Author Manuscript Proc IEEE Int Symp Biomed Imaging. Author manuscript; available in PMC 2014 November 15.
 NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging. 2013 April ; 2013: 748 751. doi:10.1109/isbi.2013.6556583. BRAIN TUMOR SEGMENTATION WITH SYMMETRIC
NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging. 2013 April ; 2013: 748 751. doi:10.1109/isbi.2013.6556583. BRAIN TUMOR SEGMENTATION WITH SYMMETRIC
Robust 3D Organ Localization with Dual Learning Architectures and Fusion
 Robust 3D Organ Localization with Dual Learning Architectures and Fusion Xiaoguang Lu (B), Daguang Xu, and David Liu Medical Imaging Technologies, Siemens Medical Solutions, Inc., Princeton, NJ, USA xiaoguang.lu@siemens.com
Robust 3D Organ Localization with Dual Learning Architectures and Fusion Xiaoguang Lu (B), Daguang Xu, and David Liu Medical Imaging Technologies, Siemens Medical Solutions, Inc., Princeton, NJ, USA xiaoguang.lu@siemens.com
Deep Learning With Noise
 Deep Learning With Noise Yixin Luo Computer Science Department Carnegie Mellon University yixinluo@cs.cmu.edu Fan Yang Department of Mathematical Sciences Carnegie Mellon University fanyang1@andrew.cmu.edu
Deep Learning With Noise Yixin Luo Computer Science Department Carnegie Mellon University yixinluo@cs.cmu.edu Fan Yang Department of Mathematical Sciences Carnegie Mellon University fanyang1@andrew.cmu.edu
arxiv: v1 [cs.mm] 12 Jan 2016
![arxiv: v1 [cs.mm] 12 Jan 2016 arxiv: v1 [cs.mm] 12 Jan 2016](/thumbs/73/69424856.jpg) Learning Subclass Representations for Visually-varied Image Classification Xinchao Li, Peng Xu, Yue Shi, Martha Larson, Alan Hanjalic Multimedia Information Retrieval Lab, Delft University of Technology
Learning Subclass Representations for Visually-varied Image Classification Xinchao Li, Peng Xu, Yue Shi, Martha Larson, Alan Hanjalic Multimedia Information Retrieval Lab, Delft University of Technology
Supervised Learning for Image Segmentation
 Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
COMP 551 Applied Machine Learning Lecture 16: Deep Learning
 COMP 551 Applied Machine Learning Lecture 16: Deep Learning Instructor: Ryan Lowe (ryan.lowe@cs.mcgill.ca) Slides mostly by: Class web page: www.cs.mcgill.ca/~hvanho2/comp551 Unless otherwise noted, all
COMP 551 Applied Machine Learning Lecture 16: Deep Learning Instructor: Ryan Lowe (ryan.lowe@cs.mcgill.ca) Slides mostly by: Class web page: www.cs.mcgill.ca/~hvanho2/comp551 Unless otherwise noted, all
Deep Learning & Neural Networks
 Deep Learning & Neural Networks Machine Learning CSE4546 Sham Kakade University of Washington November 29, 2016 Sham Kakade 1 Announcements: HW4 posted Poster Session Thurs, Dec 8 Today: Review: EM Neural
Deep Learning & Neural Networks Machine Learning CSE4546 Sham Kakade University of Washington November 29, 2016 Sham Kakade 1 Announcements: HW4 posted Poster Session Thurs, Dec 8 Today: Review: EM Neural
Deep Tracking: Biologically Inspired Tracking with Deep Convolutional Networks
 Deep Tracking: Biologically Inspired Tracking with Deep Convolutional Networks Si Chen The George Washington University sichen@gwmail.gwu.edu Meera Hahn Emory University mhahn7@emory.edu Mentor: Afshin
Deep Tracking: Biologically Inspired Tracking with Deep Convolutional Networks Si Chen The George Washington University sichen@gwmail.gwu.edu Meera Hahn Emory University mhahn7@emory.edu Mentor: Afshin
Deep Similarity Learning for Multimodal Medical Images
 Deep Similarity Learning for Multimodal Medical Images Xi Cheng, Li Zhang, and Yefeng Zheng Siemens Corporation, Corporate Technology, Princeton, NJ, USA Abstract. An effective similarity measure for multi-modal
Deep Similarity Learning for Multimodal Medical Images Xi Cheng, Li Zhang, and Yefeng Zheng Siemens Corporation, Corporate Technology, Princeton, NJ, USA Abstract. An effective similarity measure for multi-modal
Lung Nodule Detection using 3D Convolutional Neural Networks Trained on Weakly Labeled Data
 Lung Nodule Detection using 3D Convolutional Neural Networks Trained on Weakly Labeled Data Rushil Anirudh 1, Jayaraman J. Thiagarajan 2, Timo Bremer 2, and Hyojin Kim 2 1 School of Electrical, Computer
Lung Nodule Detection using 3D Convolutional Neural Networks Trained on Weakly Labeled Data Rushil Anirudh 1, Jayaraman J. Thiagarajan 2, Timo Bremer 2, and Hyojin Kim 2 1 School of Electrical, Computer
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues
 Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
arxiv: v1 [cs.lg] 20 Dec 2013
![arxiv: v1 [cs.lg] 20 Dec 2013 arxiv: v1 [cs.lg] 20 Dec 2013](/thumbs/89/99594013.jpg) Unsupervised Feature Learning by Deep Sparse Coding Yunlong He Koray Kavukcuoglu Yun Wang Arthur Szlam Yanjun Qi arxiv:1312.5783v1 [cs.lg] 20 Dec 2013 Abstract In this paper, we propose a new unsupervised
Unsupervised Feature Learning by Deep Sparse Coding Yunlong He Koray Kavukcuoglu Yun Wang Arthur Szlam Yanjun Qi arxiv:1312.5783v1 [cs.lg] 20 Dec 2013 Abstract In this paper, we propose a new unsupervised
Machine Learning. Deep Learning. Eric Xing (and Pengtao Xie) , Fall Lecture 8, October 6, Eric CMU,
 Machine Learning 10-701, Fall 2015 Deep Learning Eric Xing (and Pengtao Xie) Lecture 8, October 6, 2015 Eric Xing @ CMU, 2015 1 A perennial challenge in computer vision: feature engineering SIFT Spin image
Machine Learning 10-701, Fall 2015 Deep Learning Eric Xing (and Pengtao Xie) Lecture 8, October 6, 2015 Eric Xing @ CMU, 2015 1 A perennial challenge in computer vision: feature engineering SIFT Spin image
Apparel Classifier and Recommender using Deep Learning
 Apparel Classifier and Recommender using Deep Learning Live Demo at: http://saurabhg.me/projects/tag-that-apparel Saurabh Gupta sag043@ucsd.edu Siddhartha Agarwal siagarwa@ucsd.edu Apoorve Dave a1dave@ucsd.edu
Apparel Classifier and Recommender using Deep Learning Live Demo at: http://saurabhg.me/projects/tag-that-apparel Saurabh Gupta sag043@ucsd.edu Siddhartha Agarwal siagarwa@ucsd.edu Apoorve Dave a1dave@ucsd.edu
Learning Visual Semantics: Models, Massive Computation, and Innovative Applications
 Learning Visual Semantics: Models, Massive Computation, and Innovative Applications Part II: Visual Features and Representations Liangliang Cao, IBM Watson Research Center Evolvement of Visual Features
Learning Visual Semantics: Models, Massive Computation, and Innovative Applications Part II: Visual Features and Representations Liangliang Cao, IBM Watson Research Center Evolvement of Visual Features
arxiv: v1 [cs.cv] 16 Oct 2014
![arxiv: v1 [cs.cv] 16 Oct 2014 arxiv: v1 [cs.cv] 16 Oct 2014](/thumbs/95/122492349.jpg) Reconstructive Sparse Code Transfer for Contour Detection and Semantic Labeling Michael Maire 1,2, Stella X. Yu 3, and Pietro Perona 2 arxiv:1410.4521v1 [cs.cv] 16 Oct 2014 1 TTI Chicago 2 California Institute
Reconstructive Sparse Code Transfer for Contour Detection and Semantic Labeling Michael Maire 1,2, Stella X. Yu 3, and Pietro Perona 2 arxiv:1410.4521v1 [cs.cv] 16 Oct 2014 1 TTI Chicago 2 California Institute
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Fast Semantic Segmentation of RGB-D Scenes with GPU-Accelerated Deep Neural Networks
 Fast Semantic Segmentation of RGB-D Scenes with GPU-Accelerated Deep Neural Networks Nico Höft, Hannes Schulz, and Sven Behnke Rheinische Friedrich-Wilhelms-Universität Bonn Institut für Informatik VI,
Fast Semantic Segmentation of RGB-D Scenes with GPU-Accelerated Deep Neural Networks Nico Höft, Hannes Schulz, and Sven Behnke Rheinische Friedrich-Wilhelms-Universität Bonn Institut für Informatik VI,
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests)
 SPRING 2016 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
SPRING 2016 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
Deep Learning for Computer Vision II
 IIIT Hyderabad Deep Learning for Computer Vision II C. V. Jawahar Paradigm Shift Feature Extraction (SIFT, HoG, ) Part Models / Encoding Classifier Sparrow Feature Learning Classifier Sparrow L 1 L 2 L
IIIT Hyderabad Deep Learning for Computer Vision II C. V. Jawahar Paradigm Shift Feature Extraction (SIFT, HoG, ) Part Models / Encoding Classifier Sparrow Feature Learning Classifier Sparrow L 1 L 2 L
Deep Representations and Codes for Image Auto-Annotation
 Deep Representations and Codes for Image Auto-Annotation Ryan Kiros Department of Computing Science University of Alberta Edmonton, AB, Canada rkiros@ualberta.ca Csaba Szepesvári Department of Computing
Deep Representations and Codes for Image Auto-Annotation Ryan Kiros Department of Computing Science University of Alberta Edmonton, AB, Canada rkiros@ualberta.ca Csaba Szepesvári Department of Computing
Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation
 Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Ezequiel Geremia 2, Nicholas Ayache 2, and Gabor Szekely 1 1 Computer
Segmenting Glioma in Multi-Modal Images using a Generative-Discriminative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Ezequiel Geremia 2, Nicholas Ayache 2, and Gabor Szekely 1 1 Computer
Deep Learning with Tensorflow AlexNet
 Machine Learning and Computer Vision Group Deep Learning with Tensorflow http://cvml.ist.ac.at/courses/dlwt_w17/ AlexNet Krizhevsky, Alex, Ilya Sutskever, and Geoffrey E. Hinton, "Imagenet classification
Machine Learning and Computer Vision Group Deep Learning with Tensorflow http://cvml.ist.ac.at/courses/dlwt_w17/ AlexNet Krizhevsky, Alex, Ilya Sutskever, and Geoffrey E. Hinton, "Imagenet classification
Deep Neural Networks:
 Deep Neural Networks: Part II Convolutional Neural Network (CNN) Yuan-Kai Wang, 2016 Web site of this course: http://pattern-recognition.weebly.com source: CNN for ImageClassification, by S. Lazebnik,
Deep Neural Networks: Part II Convolutional Neural Network (CNN) Yuan-Kai Wang, 2016 Web site of this course: http://pattern-recognition.weebly.com source: CNN for ImageClassification, by S. Lazebnik,
Gated Boltzmann Machine in Texture Modeling
 Gated Boltzmann Machine in Texture Modeling Tele Hao, Tapani Rao, Alexander Ilin, and Juha Karhunen Department of Information and Computer Science Aalto University, Espoo, Finland firstname.lastname@aalto.fi
Gated Boltzmann Machine in Texture Modeling Tele Hao, Tapani Rao, Alexander Ilin, and Juha Karhunen Department of Information and Computer Science Aalto University, Espoo, Finland firstname.lastname@aalto.fi
From Maxout to Channel-Out: Encoding Information on Sparse Pathways
 From Maxout to Channel-Out: Encoding Information on Sparse Pathways Qi Wang and Joseph JaJa Department of Electrical and Computer Engineering and, University of Maryland Institute of Advanced Computer
From Maxout to Channel-Out: Encoding Information on Sparse Pathways Qi Wang and Joseph JaJa Department of Electrical and Computer Engineering and, University of Maryland Institute of Advanced Computer
MRI Tumor Segmentation with Densely Connected 3D CNN. Lele Chen, Yue Wu, Adora M. DSouze, Anas Z. Abidin, Axel Wismüller, and Chenliang Xu
 1 MRI Tumor Segmentation with Densely Connected 3D CNN Lele Chen, Yue Wu, Adora M. DSouze, Anas Z. Abidin, Axel Wismüller, and MRI Brain Tumor Segmentation 2 Image source: https://github.com/naldeborgh7575/brain_segmentation
1 MRI Tumor Segmentation with Densely Connected 3D CNN Lele Chen, Yue Wu, Adora M. DSouze, Anas Z. Abidin, Axel Wismüller, and MRI Brain Tumor Segmentation 2 Image source: https://github.com/naldeborgh7575/brain_segmentation
Computer Vision Lecture 16
 Announcements Computer Vision Lecture 16 Deep Learning Applications 11.01.2017 Seminar registration period starts on Friday We will offer a lab course in the summer semester Deep Robot Learning Topic:
Announcements Computer Vision Lecture 16 Deep Learning Applications 11.01.2017 Seminar registration period starts on Friday We will offer a lab course in the summer semester Deep Robot Learning Topic:
Cost-alleviative Learning for Deep Convolutional Neural Network-based Facial Part Labeling
 [DOI: 10.2197/ipsjtcva.7.99] Express Paper Cost-alleviative Learning for Deep Convolutional Neural Network-based Facial Part Labeling Takayoshi Yamashita 1,a) Takaya Nakamura 1 Hiroshi Fukui 1,b) Yuji
[DOI: 10.2197/ipsjtcva.7.99] Express Paper Cost-alleviative Learning for Deep Convolutional Neural Network-based Facial Part Labeling Takayoshi Yamashita 1,a) Takaya Nakamura 1 Hiroshi Fukui 1,b) Yuji
Machine Learning. MGS Lecture 3: Deep Learning
 Dr Michel F. Valstar http://cs.nott.ac.uk/~mfv/ Machine Learning MGS Lecture 3: Deep Learning Dr Michel F. Valstar http://cs.nott.ac.uk/~mfv/ WHAT IS DEEP LEARNING? Shallow network: Only one hidden layer
Dr Michel F. Valstar http://cs.nott.ac.uk/~mfv/ Machine Learning MGS Lecture 3: Deep Learning Dr Michel F. Valstar http://cs.nott.ac.uk/~mfv/ WHAT IS DEEP LEARNING? Shallow network: Only one hidden layer
DEEP LEARNING TO DIVERSIFY BELIEF NETWORKS FOR REMOTE SENSING IMAGE CLASSIFICATION
 DEEP LEARNING TO DIVERSIFY BELIEF NETWORKS FOR REMOTE SENSING IMAGE CLASSIFICATION S.Dhanalakshmi #1 #PG Scholar, Department of Computer Science, Dr.Sivanthi Aditanar college of Engineering, Tiruchendur
DEEP LEARNING TO DIVERSIFY BELIEF NETWORKS FOR REMOTE SENSING IMAGE CLASSIFICATION S.Dhanalakshmi #1 #PG Scholar, Department of Computer Science, Dr.Sivanthi Aditanar college of Engineering, Tiruchendur
Hierarchical Matching Pursuit for Image Classification: Architecture and Fast Algorithms
 Hierarchical Matching Pursuit for Image Classification: Architecture and Fast Algorithms Liefeng Bo University of Washington Seattle WA 98195, USA Xiaofeng Ren ISTC-Pervasive Computing Intel Labs Seattle
Hierarchical Matching Pursuit for Image Classification: Architecture and Fast Algorithms Liefeng Bo University of Washington Seattle WA 98195, USA Xiaofeng Ren ISTC-Pervasive Computing Intel Labs Seattle
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation
 Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Advanced Machine Learning
 Advanced Machine Learning Convolutional Neural Networks for Handwritten Digit Recognition Andreas Georgopoulos CID: 01281486 Abstract Abstract At this project three different Convolutional Neural Netwroks
Advanced Machine Learning Convolutional Neural Networks for Handwritten Digit Recognition Andreas Georgopoulos CID: 01281486 Abstract Abstract At this project three different Convolutional Neural Netwroks
Advanced Introduction to Machine Learning, CMU-10715
 Advanced Introduction to Machine Learning, CMU-10715 Deep Learning Barnabás Póczos, Sept 17 Credits Many of the pictures, results, and other materials are taken from: Ruslan Salakhutdinov Joshua Bengio
Advanced Introduction to Machine Learning, CMU-10715 Deep Learning Barnabás Póczos, Sept 17 Credits Many of the pictures, results, and other materials are taken from: Ruslan Salakhutdinov Joshua Bengio
Comparing Dropout Nets to Sum-Product Networks for Predicting Molecular Activity
 000 001 002 003 004 005 006 007 008 009 010 011 012 013 014 015 016 017 018 019 020 021 022 023 024 025 026 027 028 029 030 031 032 033 034 035 036 037 038 039 040 041 042 043 044 045 046 047 048 049 050
000 001 002 003 004 005 006 007 008 009 010 011 012 013 014 015 016 017 018 019 020 021 022 023 024 025 026 027 028 029 030 031 032 033 034 035 036 037 038 039 040 041 042 043 044 045 046 047 048 049 050
Isointense infant brain MRI segmentation with a dilated convolutional neural network Moeskops, P.; Pluim, J.P.W.
 Isointense infant brain MRI segmentation with a dilated convolutional neural network Moeskops, P.; Pluim, J.P.W. Published: 09/08/2017 Document Version Author s version before peer-review Please check
Isointense infant brain MRI segmentation with a dilated convolutional neural network Moeskops, P.; Pluim, J.P.W. Published: 09/08/2017 Document Version Author s version before peer-review Please check
Computer Vision Lecture 16
 Computer Vision Lecture 16 Deep Learning Applications 11.01.2017 Bastian Leibe RWTH Aachen http://www.vision.rwth-aachen.de leibe@vision.rwth-aachen.de Announcements Seminar registration period starts
Computer Vision Lecture 16 Deep Learning Applications 11.01.2017 Bastian Leibe RWTH Aachen http://www.vision.rwth-aachen.de leibe@vision.rwth-aachen.de Announcements Seminar registration period starts
Learning to Segment 3D Linear Structures Using Only 2D Annotations
 Learning to Segment 3D Linear Structures Using Only 2D Annotations Mateusz Koziński, Agata Mosinska, Mathieu Salzmann, and Pascal Fua {name.surname}@epfl.ch Computer Vision Laboratory, École Polytechnique
Learning to Segment 3D Linear Structures Using Only 2D Annotations Mateusz Koziński, Agata Mosinska, Mathieu Salzmann, and Pascal Fua {name.surname}@epfl.ch Computer Vision Laboratory, École Polytechnique
Computer Vision Lecture 16
 Computer Vision Lecture 16 Deep Learning for Object Categorization 14.01.2016 Bastian Leibe RWTH Aachen http://www.vision.rwth-aachen.de leibe@vision.rwth-aachen.de Announcements Seminar registration period
Computer Vision Lecture 16 Deep Learning for Object Categorization 14.01.2016 Bastian Leibe RWTH Aachen http://www.vision.rwth-aachen.de leibe@vision.rwth-aachen.de Announcements Seminar registration period
A Deep Learning Framework for Authorship Classification of Paintings
 A Deep Learning Framework for Authorship Classification of Paintings Kai-Lung Hua ( 花凱龍 ) Dept. of Computer Science and Information Engineering National Taiwan University of Science and Technology Taipei,
A Deep Learning Framework for Authorship Classification of Paintings Kai-Lung Hua ( 花凱龍 ) Dept. of Computer Science and Information Engineering National Taiwan University of Science and Technology Taipei,
Microscopy Cell Counting with Fully Convolutional Regression Networks
 Microscopy Cell Counting with Fully Convolutional Regression Networks Weidi Xie, J. Alison Noble, Andrew Zisserman Department of Engineering Science, University of Oxford,UK Abstract. This paper concerns
Microscopy Cell Counting with Fully Convolutional Regression Networks Weidi Xie, J. Alison Noble, Andrew Zisserman Department of Engineering Science, University of Oxford,UK Abstract. This paper concerns
Convolutional Neural Networks
 Lecturer: Barnabas Poczos Introduction to Machine Learning (Lecture Notes) Convolutional Neural Networks Disclaimer: These notes have not been subjected to the usual scrutiny reserved for formal publications.
Lecturer: Barnabas Poczos Introduction to Machine Learning (Lecture Notes) Convolutional Neural Networks Disclaimer: These notes have not been subjected to the usual scrutiny reserved for formal publications.
arxiv: v2 [cs.ne] 2 Dec 2015
![arxiv: v2 [cs.ne] 2 Dec 2015 arxiv: v2 [cs.ne] 2 Dec 2015](/thumbs/75/72075077.jpg) An Introduction to Convolutional Neural Networks Keiron O Shea 1 and Ryan Nash 2 1 Department of Computer Science, Aberystwyth University, Ceredigion, SY23 3DB keo7@aber.ac.uk 2 School of Computing and
An Introduction to Convolutional Neural Networks Keiron O Shea 1 and Ryan Nash 2 1 Department of Computer Science, Aberystwyth University, Ceredigion, SY23 3DB keo7@aber.ac.uk 2 School of Computing and
A Sparse and Locally Shift Invariant Feature Extractor Applied to Document Images
 A Sparse and Locally Shift Invariant Feature Extractor Applied to Document Images Marc Aurelio Ranzato Yann LeCun Courant Institute of Mathematical Sciences New York University - New York, NY 10003 Abstract
A Sparse and Locally Shift Invariant Feature Extractor Applied to Document Images Marc Aurelio Ranzato Yann LeCun Courant Institute of Mathematical Sciences New York University - New York, NY 10003 Abstract
Locally Scale-Invariant Convolutional Neural Networks
 Locally Scale-Invariant Convolutional Neural Networks Angjoo Kanazawa Department of Computer Science University of Maryland, College Park, MD 20740 kanazawa@umiacs.umd.edu Abhishek Sharma Department of
Locally Scale-Invariant Convolutional Neural Networks Angjoo Kanazawa Department of Computer Science University of Maryland, College Park, MD 20740 kanazawa@umiacs.umd.edu Abhishek Sharma Department of
On Linear Embeddings and Unsupervised Feature Learning
 Ryan Kiros Csaba Szepesvári University of Alberta, Canada rkiros@ualberta.ca szepesva@ualberta.ca Abstract The ability to train deep architectures has led to many developments in parametric, non-linear
Ryan Kiros Csaba Szepesvári University of Alberta, Canada rkiros@ualberta.ca szepesva@ualberta.ca Abstract The ability to train deep architectures has led to many developments in parametric, non-linear
Histograms of Sparse Codes for Object Detection
 Histograms of Sparse Codes for Object Detection Xiaofeng Ren (Amazon), Deva Ramanan (UC Irvine) Presented by Hossein Azizpour What does the paper do? (learning) a new representation local histograms of
Histograms of Sparse Codes for Object Detection Xiaofeng Ren (Amazon), Deva Ramanan (UC Irvine) Presented by Hossein Azizpour What does the paper do? (learning) a new representation local histograms of
Text Detection and Character Recognition in Scene Images with Unsupervised Feature Learning
 Text Detection and Character Recognition in Scene Images with Unsupervised Feature Learning Adam Coates, Blake Carpenter, Carl Case, Sanjeev Satheesh, Bipin Suresh, Tao Wang, Andrew Y. Ng Computer Science
Text Detection and Character Recognition in Scene Images with Unsupervised Feature Learning Adam Coates, Blake Carpenter, Carl Case, Sanjeev Satheesh, Bipin Suresh, Tao Wang, Andrew Y. Ng Computer Science
A Sparse and Locally Shift Invariant Feature Extractor Applied to Document Images
 A Sparse and Locally Shift Invariant Feature Extractor Applied to Document Images Marc Aurelio Ranzato Yann LeCun Courant Institute of Mathematical Sciences New York University - New York, NY 10003 Abstract
A Sparse and Locally Shift Invariant Feature Extractor Applied to Document Images Marc Aurelio Ranzato Yann LeCun Courant Institute of Mathematical Sciences New York University - New York, NY 10003 Abstract
Scale and Curvature Invariant Ridge Detector for Tortuous and Fragmented Structures
 Scale and Curvature Invariant Ridge Detector for Tortuous and Fragmented Structures Roberto Annunziata, Ahmad Kheirkhah, Pedram Hamrah, and Emanuele Trucco School of Computing, University of Dundee, Dundee,
Scale and Curvature Invariant Ridge Detector for Tortuous and Fragmented Structures Roberto Annunziata, Ahmad Kheirkhah, Pedram Hamrah, and Emanuele Trucco School of Computing, University of Dundee, Dundee,
Edge Detection Using Convolutional Neural Network
 Edge Detection Using Convolutional Neural Network Ruohui Wang (B) Department of Information Engineering, The Chinese University of Hong Kong, Hong Kong, China wr013@ie.cuhk.edu.hk Abstract. In this work,
Edge Detection Using Convolutional Neural Network Ruohui Wang (B) Department of Information Engineering, The Chinese University of Hong Kong, Hong Kong, China wr013@ie.cuhk.edu.hk Abstract. In this work,
SIIM 2017 Scientific Session Analytics & Deep Learning Part 2 Friday, June 2 8:00 am 9:30 am
 SIIM 2017 Scientific Session Analytics & Deep Learning Part 2 Friday, June 2 8:00 am 9:30 am Performance of Deep Convolutional Neural Networks for Classification of Acute Territorial Infarct on Brain MRI:
SIIM 2017 Scientific Session Analytics & Deep Learning Part 2 Friday, June 2 8:00 am 9:30 am Performance of Deep Convolutional Neural Networks for Classification of Acute Territorial Infarct on Brain MRI:
Analysis of Learned Features for Remote Sensing Image Classification
 Analysis of Learned Features for Remote Sensing Image Classification Vladimir Risojević, Member, IEEE Abstract Convolutional neural networks (convnets) have shown excellent results in various image classification
Analysis of Learned Features for Remote Sensing Image Classification Vladimir Risojević, Member, IEEE Abstract Convolutional neural networks (convnets) have shown excellent results in various image classification
End-To-End Spam Classification With Neural Networks
 End-To-End Spam Classification With Neural Networks Christopher Lennan, Bastian Naber, Jan Reher, Leon Weber 1 Introduction A few years ago, the majority of the internet s network traffic was due to spam
End-To-End Spam Classification With Neural Networks Christopher Lennan, Bastian Naber, Jan Reher, Leon Weber 1 Introduction A few years ago, the majority of the internet s network traffic was due to spam
Preface to the Second Edition. Preface to the First Edition. 1 Introduction 1
 Preface to the Second Edition Preface to the First Edition vii xi 1 Introduction 1 2 Overview of Supervised Learning 9 2.1 Introduction... 9 2.2 Variable Types and Terminology... 9 2.3 Two Simple Approaches
Preface to the Second Edition Preface to the First Edition vii xi 1 Introduction 1 2 Overview of Supervised Learning 9 2.1 Introduction... 9 2.2 Variable Types and Terminology... 9 2.3 Two Simple Approaches
IMPROVING THE ROBUSTNESS OF CONVOLUTIONAL NETWORKS TO APPEARANCE VARIABILITY IN BIOMEDICAL IMAGES
 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) April 4-7, 2018, Washington, D.C., USA IMPROVING THE ROBUSTNESS OF CONVOLUTIONAL NETWORKS TO APPEARANCE VARIABILITY IN BIOMEDICAL
2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) April 4-7, 2018, Washington, D.C., USA IMPROVING THE ROBUSTNESS OF CONVOLUTIONAL NETWORKS TO APPEARANCE VARIABILITY IN BIOMEDICAL
Accurate and Efficient Linear Structure Segmentation by Leveraging Ad Hoc Features with Learned Filters
 Accurate and Efficient Linear Structure Segmentation by Leveraging Ad Hoc Features with Learned Filters CVLab, Roberto Rigamonti and Vincent Lepetit École Polytechnique Fédérale de Lausanne, Lausanne,
Accurate and Efficient Linear Structure Segmentation by Leveraging Ad Hoc Features with Learned Filters CVLab, Roberto Rigamonti and Vincent Lepetit École Polytechnique Fédérale de Lausanne, Lausanne,
3 Object Detection. BVM 2018 Tutorial: Advanced Deep Learning Methods. Paul F. Jaeger, Division of Medical Image Computing
 3 Object Detection BVM 2018 Tutorial: Advanced Deep Learning Methods Paul F. Jaeger, of Medical Image Computing What is object detection? classification segmentation obj. detection (1 label per pixel)
3 Object Detection BVM 2018 Tutorial: Advanced Deep Learning Methods Paul F. Jaeger, of Medical Image Computing What is object detection? classification segmentation obj. detection (1 label per pixel)
Single Image Depth Estimation via Deep Learning
 Single Image Depth Estimation via Deep Learning Wei Song Stanford University Stanford, CA Abstract The goal of the project is to apply direct supervised deep learning to the problem of monocular depth
Single Image Depth Estimation via Deep Learning Wei Song Stanford University Stanford, CA Abstract The goal of the project is to apply direct supervised deep learning to the problem of monocular depth
CPSC340. State-of-the-art Neural Networks. Nando de Freitas November, 2012 University of British Columbia
 CPSC340 State-of-the-art Neural Networks Nando de Freitas November, 2012 University of British Columbia Outline of the lecture This lecture provides an overview of two state-of-the-art neural networks:
CPSC340 State-of-the-art Neural Networks Nando de Freitas November, 2012 University of British Columbia Outline of the lecture This lecture provides an overview of two state-of-the-art neural networks:
Tiny ImageNet Visual Recognition Challenge
 Tiny ImageNet Visual Recognition Challenge Ya Le Department of Statistics Stanford University yle@stanford.edu Xuan Yang Department of Electrical Engineering Stanford University xuany@stanford.edu Abstract
Tiny ImageNet Visual Recognition Challenge Ya Le Department of Statistics Stanford University yle@stanford.edu Xuan Yang Department of Electrical Engineering Stanford University xuany@stanford.edu Abstract
An Analysis of Single-Layer Networks in Unsupervised Feature Learning
 An Analysis of Single-Layer Networks in Unsupervised Feature Learning Adam Coates Honglak Lee Andrew Y. Ng Stanford University Computer Science Dept. 353 Serra Mall Stanford, CA 94305 University of Michigan
An Analysis of Single-Layer Networks in Unsupervised Feature Learning Adam Coates Honglak Lee Andrew Y. Ng Stanford University Computer Science Dept. 353 Serra Mall Stanford, CA 94305 University of Michigan
Rotation Invariance Neural Network
 Rotation Invariance Neural Network Shiyuan Li Abstract Rotation invariance and translate invariance have great values in image recognition. In this paper, we bring a new architecture in convolutional neural
Rotation Invariance Neural Network Shiyuan Li Abstract Rotation invariance and translate invariance have great values in image recognition. In this paper, we bring a new architecture in convolutional neural
Deep Learning in Visual Recognition. Thanks Da Zhang for the slides
 Deep Learning in Visual Recognition Thanks Da Zhang for the slides Deep Learning is Everywhere 2 Roadmap Introduction Convolutional Neural Network Application Image Classification Object Detection Object
Deep Learning in Visual Recognition Thanks Da Zhang for the slides Deep Learning is Everywhere 2 Roadmap Introduction Convolutional Neural Network Application Image Classification Object Detection Object
Multipath Sparse Coding Using Hierarchical Matching Pursuit
 Multipath Sparse Coding Using Hierarchical Matching Pursuit Liefeng Bo, Xiaofeng Ren ISTC Pervasive Computing, Intel Labs Seattle WA 98195, USA {liefeng.bo,xiaofeng.ren}@intel.com Dieter Fox University
Multipath Sparse Coding Using Hierarchical Matching Pursuit Liefeng Bo, Xiaofeng Ren ISTC Pervasive Computing, Intel Labs Seattle WA 98195, USA {liefeng.bo,xiaofeng.ren}@intel.com Dieter Fox University
arxiv: v1 [cs.cv] 6 Jul 2016
![arxiv: v1 [cs.cv] 6 Jul 2016 arxiv: v1 [cs.cv] 6 Jul 2016](/thumbs/80/81416563.jpg) arxiv:607.079v [cs.cv] 6 Jul 206 Deep CORAL: Correlation Alignment for Deep Domain Adaptation Baochen Sun and Kate Saenko University of Massachusetts Lowell, Boston University Abstract. Deep neural networks
arxiv:607.079v [cs.cv] 6 Jul 206 Deep CORAL: Correlation Alignment for Deep Domain Adaptation Baochen Sun and Kate Saenko University of Massachusetts Lowell, Boston University Abstract. Deep neural networks
CS231A Course Project Final Report Sign Language Recognition with Unsupervised Feature Learning
 CS231A Course Project Final Report Sign Language Recognition with Unsupervised Feature Learning Justin Chen Stanford University justinkchen@stanford.edu Abstract This paper focuses on experimenting with
CS231A Course Project Final Report Sign Language Recognition with Unsupervised Feature Learning Justin Chen Stanford University justinkchen@stanford.edu Abstract This paper focuses on experimenting with
A FRAMEWORK OF EXTRACTING MULTI-SCALE FEATURES USING MULTIPLE CONVOLUTIONAL NEURAL NETWORKS. Kuan-Chuan Peng and Tsuhan Chen
 A FRAMEWORK OF EXTRACTING MULTI-SCALE FEATURES USING MULTIPLE CONVOLUTIONAL NEURAL NETWORKS Kuan-Chuan Peng and Tsuhan Chen School of Electrical and Computer Engineering, Cornell University, Ithaca, NY
A FRAMEWORK OF EXTRACTING MULTI-SCALE FEATURES USING MULTIPLE CONVOLUTIONAL NEURAL NETWORKS Kuan-Chuan Peng and Tsuhan Chen School of Electrical and Computer Engineering, Cornell University, Ithaca, NY
Extended Dictionary Learning : Convolutional and Multiple Feature Spaces
 Extended Dictionary Learning : Convolutional and Multiple Feature Spaces Konstantina Fotiadou, Greg Tsagkatakis & Panagiotis Tsakalides kfot@ics.forth.gr, greg@ics.forth.gr, tsakalid@ics.forth.gr ICS-
Extended Dictionary Learning : Convolutional and Multiple Feature Spaces Konstantina Fotiadou, Greg Tsagkatakis & Panagiotis Tsakalides kfot@ics.forth.gr, greg@ics.forth.gr, tsakalid@ics.forth.gr ICS-
arxiv: v1 [cs.lg] 16 Jan 2013
![arxiv: v1 [cs.lg] 16 Jan 2013 arxiv: v1 [cs.lg] 16 Jan 2013](/thumbs/80/80526613.jpg) Stochastic Pooling for Regularization of Deep Convolutional Neural Networks arxiv:131.3557v1 [cs.lg] 16 Jan 213 Matthew D. Zeiler Department of Computer Science Courant Institute, New York University zeiler@cs.nyu.edu
Stochastic Pooling for Regularization of Deep Convolutional Neural Networks arxiv:131.3557v1 [cs.lg] 16 Jan 213 Matthew D. Zeiler Department of Computer Science Courant Institute, New York University zeiler@cs.nyu.edu
Introduction to Deep Learning
 ENEE698A : Machine Learning Seminar Introduction to Deep Learning Raviteja Vemulapalli Image credit: [LeCun 1998] Resources Unsupervised feature learning and deep learning (UFLDL) tutorial (http://ufldl.stanford.edu/wiki/index.php/ufldl_tutorial)
ENEE698A : Machine Learning Seminar Introduction to Deep Learning Raviteja Vemulapalli Image credit: [LeCun 1998] Resources Unsupervised feature learning and deep learning (UFLDL) tutorial (http://ufldl.stanford.edu/wiki/index.php/ufldl_tutorial)
JOINT INTENT DETECTION AND SLOT FILLING USING CONVOLUTIONAL NEURAL NETWORKS. Puyang Xu, Ruhi Sarikaya. Microsoft Corporation
 JOINT INTENT DETECTION AND SLOT FILLING USING CONVOLUTIONAL NEURAL NETWORKS Puyang Xu, Ruhi Sarikaya Microsoft Corporation ABSTRACT We describe a joint model for intent detection and slot filling based
JOINT INTENT DETECTION AND SLOT FILLING USING CONVOLUTIONAL NEURAL NETWORKS Puyang Xu, Ruhi Sarikaya Microsoft Corporation ABSTRACT We describe a joint model for intent detection and slot filling based
Deep Learning for Vision: Tricks of the Trade
 Deep Learning for Vision: Tricks of the Trade Marc'Aurelio Ranzato Facebook, AI Group www.cs.toronto.edu/~ranzato BAVM Friday, 4 October 2013 Ideal Features Ideal Feature Extractor - window, right - chair,
Deep Learning for Vision: Tricks of the Trade Marc'Aurelio Ranzato Facebook, AI Group www.cs.toronto.edu/~ranzato BAVM Friday, 4 October 2013 Ideal Features Ideal Feature Extractor - window, right - chair,
Image Question Answering using Convolutional Neural Network with Dynamic Parameter Prediction
 Image Question Answering using Convolutional Neural Network with Dynamic Parameter Prediction by Noh, Hyeonwoo, Paul Hongsuck Seo, and Bohyung Han.[1] Presented : Badri Patro 1 1 Computer Vision Reading
Image Question Answering using Convolutional Neural Network with Dynamic Parameter Prediction by Noh, Hyeonwoo, Paul Hongsuck Seo, and Bohyung Han.[1] Presented : Badri Patro 1 1 Computer Vision Reading
A HMAX with LLC for Visual Recognition
 A HMAX with LLC for Visual Recognition Kean Hong Lau, Yong Haur Tay, Fook Loong Lo Centre for Computing and Intelligent System Universiti Tunku Abdul Rahman, Kuala Lumpur, Malaysia {laukh,tayyh,lofl}@utar.edu.my
A HMAX with LLC for Visual Recognition Kean Hong Lau, Yong Haur Tay, Fook Loong Lo Centre for Computing and Intelligent System Universiti Tunku Abdul Rahman, Kuala Lumpur, Malaysia {laukh,tayyh,lofl}@utar.edu.my
Multi-modal Unsupervised Feature Learning for RGB-D Scene Labeling
 Multi-modal Unsupervised Feature Learning for RGB-D Scene Labeling Anran Wang 1, Jiwen Lu 2, Gang Wang 1,2, Jianfei Cai 1, and Tat-Jen Cham 1 1 Nanyang Technological University, Singapore 2 Advanced Digital
Multi-modal Unsupervised Feature Learning for RGB-D Scene Labeling Anran Wang 1, Jiwen Lu 2, Gang Wang 1,2, Jianfei Cai 1, and Tat-Jen Cham 1 1 Nanyang Technological University, Singapore 2 Advanced Digital
SELF SUPERVISED DEEP REPRESENTATION LEARNING FOR FINE-GRAINED BODY PART RECOGNITION
 SELF SUPERVISED DEEP REPRESENTATION LEARNING FOR FINE-GRAINED BODY PART RECOGNITION Pengyue Zhang Fusheng Wang Yefeng Zheng Medical Imaging Technologies, Siemens Medical Solutions USA Inc., Princeton,
SELF SUPERVISED DEEP REPRESENTATION LEARNING FOR FINE-GRAINED BODY PART RECOGNITION Pengyue Zhang Fusheng Wang Yefeng Zheng Medical Imaging Technologies, Siemens Medical Solutions USA Inc., Princeton,
Multipath Sparse Coding Using Hierarchical Matching Pursuit
 2013 IEEE Conference on Computer Vision and Pattern Recognition Multipath Sparse Coding Using Hierarchical Matching Pursuit Liefeng Bo ISTC-PC Intel Labs liefeng.bo@intel.com Xiaofeng Ren ISTC-PC Intel
2013 IEEE Conference on Computer Vision and Pattern Recognition Multipath Sparse Coding Using Hierarchical Matching Pursuit Liefeng Bo ISTC-PC Intel Labs liefeng.bo@intel.com Xiaofeng Ren ISTC-PC Intel
Study of Residual Networks for Image Recognition
 Study of Residual Networks for Image Recognition Mohammad Sadegh Ebrahimi Stanford University sadegh@stanford.edu Hossein Karkeh Abadi Stanford University hosseink@stanford.edu Abstract Deep neural networks
Study of Residual Networks for Image Recognition Mohammad Sadegh Ebrahimi Stanford University sadegh@stanford.edu Hossein Karkeh Abadi Stanford University hosseink@stanford.edu Abstract Deep neural networks
Channel Locality Block: A Variant of Squeeze-and-Excitation
 Channel Locality Block: A Variant of Squeeze-and-Excitation 1 st Huayu Li Northern Arizona University Flagstaff, United State Northern Arizona University hl459@nau.edu arxiv:1901.01493v1 [cs.lg] 6 Jan
Channel Locality Block: A Variant of Squeeze-and-Excitation 1 st Huayu Li Northern Arizona University Flagstaff, United State Northern Arizona University hl459@nau.edu arxiv:1901.01493v1 [cs.lg] 6 Jan
Yiqi Yan. May 10, 2017
 Yiqi Yan May 10, 2017 P a r t I F u n d a m e n t a l B a c k g r o u n d s Convolution Single Filter Multiple Filters 3 Convolution: case study, 2 filters 4 Convolution: receptive field receptive field
Yiqi Yan May 10, 2017 P a r t I F u n d a m e n t a l B a c k g r o u n d s Convolution Single Filter Multiple Filters 3 Convolution: case study, 2 filters 4 Convolution: receptive field receptive field
arxiv: v1 [cs.cv] 20 Dec 2016
![arxiv: v1 [cs.cv] 20 Dec 2016 arxiv: v1 [cs.cv] 20 Dec 2016](/thumbs/73/68905842.jpg) End-to-End Pedestrian Collision Warning System based on a Convolutional Neural Network with Semantic Segmentation arxiv:1612.06558v1 [cs.cv] 20 Dec 2016 Heechul Jung heechul@dgist.ac.kr Min-Kook Choi mkchoi@dgist.ac.kr
End-to-End Pedestrian Collision Warning System based on a Convolutional Neural Network with Semantic Segmentation arxiv:1612.06558v1 [cs.cv] 20 Dec 2016 Heechul Jung heechul@dgist.ac.kr Min-Kook Choi mkchoi@dgist.ac.kr
Compressing Deep Neural Networks using a Rank-Constrained Topology
 Compressing Deep Neural Networks using a Rank-Constrained Topology Preetum Nakkiran Department of EECS University of California, Berkeley, USA preetum@berkeley.edu Raziel Alvarez, Rohit Prabhavalkar, Carolina
Compressing Deep Neural Networks using a Rank-Constrained Topology Preetum Nakkiran Department of EECS University of California, Berkeley, USA preetum@berkeley.edu Raziel Alvarez, Rohit Prabhavalkar, Carolina
Convolutional Neural Networks: Applications and a short timeline. 7th Deep Learning Meetup Kornel Kis Vienna,
 Convolutional Neural Networks: Applications and a short timeline 7th Deep Learning Meetup Kornel Kis Vienna, 1.12.2016. Introduction Currently a master student Master thesis at BME SmartLab Started deep
Convolutional Neural Networks: Applications and a short timeline 7th Deep Learning Meetup Kornel Kis Vienna, 1.12.2016. Introduction Currently a master student Master thesis at BME SmartLab Started deep
Fast 3D Mean Shift Filter for CT Images
 Fast 3D Mean Shift Filter for CT Images Gustavo Fernández Domínguez, Horst Bischof, and Reinhard Beichel Institute for Computer Graphics and Vision, Graz University of Technology Inffeldgasse 16/2, A-8010,
Fast 3D Mean Shift Filter for CT Images Gustavo Fernández Domínguez, Horst Bischof, and Reinhard Beichel Institute for Computer Graphics and Vision, Graz University of Technology Inffeldgasse 16/2, A-8010,
The Automated Learning of Deep Features for Breast Mass Classification from Mammograms
 The Automated Learning of Deep Features for Breast Mass Classification from Mammograms Neeraj Dhungel Gustavo Carneiro Andrew P. Bradley ACVT, School of Computer Science, The University of Adelaide School
The Automated Learning of Deep Features for Breast Mass Classification from Mammograms Neeraj Dhungel Gustavo Carneiro Andrew P. Bradley ACVT, School of Computer Science, The University of Adelaide School
Machine Learning 13. week
 Machine Learning 13. week Deep Learning Convolutional Neural Network Recurrent Neural Network 1 Why Deep Learning is so Popular? 1. Increase in the amount of data Thanks to the Internet, huge amount of
Machine Learning 13. week Deep Learning Convolutional Neural Network Recurrent Neural Network 1 Why Deep Learning is so Popular? 1. Increase in the amount of data Thanks to the Internet, huge amount of
Tiled convolutional neural networks
 Tiled convolutional neural networks Quoc V. Le, Jiquan Ngiam, Zhenghao Chen, Daniel Chia, Pang Wei Koh, Andrew Y. Ng Computer Science Department, Stanford University {quocle,jngiam,zhenghao,danchia,pangwei,ang}@cs.stanford.edu
Tiled convolutional neural networks Quoc V. Le, Jiquan Ngiam, Zhenghao Chen, Daniel Chia, Pang Wei Koh, Andrew Y. Ng Computer Science Department, Stanford University {quocle,jngiam,zhenghao,danchia,pangwei,ang}@cs.stanford.edu
Multipath Sparse Coding Using Hierarchical Matching Pursuit
 Multipath Sparse Coding Using Hierarchical Matching Pursuit Liefeng Bo ISTC-PC Intel Labs liefeng.bo@intel.com Xiaofeng Ren ISTC-PC Intel Labs xren@cs.washington.edu Dieter Fox University of Washington
Multipath Sparse Coding Using Hierarchical Matching Pursuit Liefeng Bo ISTC-PC Intel Labs liefeng.bo@intel.com Xiaofeng Ren ISTC-PC Intel Labs xren@cs.washington.edu Dieter Fox University of Washington
An Exploration of Computer Vision Techniques for Bird Species Classification
 An Exploration of Computer Vision Techniques for Bird Species Classification Anne L. Alter, Karen M. Wang December 15, 2017 Abstract Bird classification, a fine-grained categorization task, is a complex
An Exploration of Computer Vision Techniques for Bird Species Classification Anne L. Alter, Karen M. Wang December 15, 2017 Abstract Bird classification, a fine-grained categorization task, is a complex
Beyond Spatial Pyramids: Receptive Field Learning for Pooled Image Features
 Beyond Spatial Pyramids: Receptive Field Learning for Pooled Image Features Yangqing Jia UC Berkeley EECS jiayq@berkeley.edu Chang Huang NEC Labs America chuang@sv.nec-labs.com Abstract We examine the
Beyond Spatial Pyramids: Receptive Field Learning for Pooled Image Features Yangqing Jia UC Berkeley EECS jiayq@berkeley.edu Chang Huang NEC Labs America chuang@sv.nec-labs.com Abstract We examine the
