Joint Sulci Detection Using Graphical Models and Boosted Priors
|
|
|
- Dustin Nicholson
- 6 years ago
- Views:
Transcription
1 Joint Sulci Detection Using Graphical Models and Boosted Priors Yonggang Shi 1,ZhuowenTu 1, Allan L. Reiss 2, Rebecca A. Dutton 1, Agatha D. Lee 1, Albert M. Galaburda 3,IvoDinov 1, Paul M. Thompson 1, and Arthur W. Toga 1 1 Lab of Neuro Imaging, UCLA School of Medicine, Los Angeles, CA, USA {yshi,ztu,ivo.dinov,thompson,toga}@loni.ucla.edu 2 School of Medicine, Stanford University, Stanford, CA, USA 3 Harvard Medical School, Boston, MA, USA Abstract. In this paper we propose an automated approach for joint sulci detection on cortical surfaces by using graphical models and boosting techniques to incorporate shape priors of major sulci and their Markovian relations. For each sulcus, we represent it as a node in the graphical model and associate it with a sample space of candidate curves, which is generated automatically using the Hamilton-Jacobi skeleton of sulcal regions. To take into account individual as well as joint priors about the shape of major sulci, we learn the potential functions of the graphical model using AdaBoost algorithm to select and fuse information from a large set of features. This discriminative approach is especially powerful in capturing the neighboring relations between sulcal lines, which are otherwise hard to be captured by generative models. Using belief propagation, efficient inferencing is then performed on the graphical model to estimate each sulcus as the maximizer of its final belief. On a data set of 40 cortical surfaces, we demonstrate the advantage of joint detection on four major sulci: central, precentral, postcentral and the sylvian fissure. 1 Introduction Cortical sulci are important landmarks in human brain mapping because they encode rich information about the convolution patterns of human brains[1] and provide guidance for registration tasks[2], but the variability of the brain morphometry poses serious challenges for their automatic detection, thus manual annotation remains the golden standard in practice. In this paper, we propose a novel approach to incorporate prior knowledge from manual tracing by modeling the relation of major sulci with boosting techniques and detect them jointly via the solution of an inference problem on graphical models. Many algorithms were proposed for sulci detection in previous works. Curvature features were used in [3,4,5] to detect sulci semi-automatically with the need of manually inputing the starting and ending points. Depth features with respect to a shrink wrap surface were also used for sulci detection on surfaces[6,7]. The extraction of sulci from volume images were proposed in [8,9,10], but human interactions are still necessary to pick out specific sulci from the results. N. Karssemeijer and B. Lelieveldt (Eds.): IPMI 2007, LNCS 4584, pp , c Springer-Verlag Berlin Heidelberg 2007
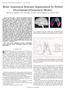
2 Joint Sulci Detection Using Graphical Models and Boosted Priors 99 Central(C 1 ) Precentral(C 2 ) Postcentral(C 3 ) Superior Frontal (C 7 ) Intraparietal(C 5 ) C 7 C 5 C 2 C 1 C 3 Inferior Frontal (C 6 ) Sylvian(C 4 ) C 6 C 4 (a) (b) Fig. 1. An illustration of joint sulci detection. (a) Seven major sulci on a cortical surface. (b) A graphical model representing the local dependency between neighboring sulci. To address this challenge, learning-based approaches were introduced into the detection process. Because of its simplicity, techniques based on the principal component analysis(pca) of point sets[11] were used in[12,13]. Graphical models and neural networks were used in [14] for automatic recognition of sulcal ribbons, which only identify subsets of major sulci. A learning-based technique based on probabilistic boosting trees[15] was proposed in [16] for the automatic detection of cortical sulci from volume images, but each sulcus was detected separately. We propose in this work to detect multiple sulci jointly with graphical models. From the experience of manual tracing, this seems a natural choice as the knowledge about the relative location of sulci is frequently utilized for the correct identification of these curves. Modeling the relation of multiple objects has also been shown useful in medical image segmentation[17]. As an example, seven major sulci are plotted on the lateral surface of a cortex in Fig. 1(a). Even though we can see that most parts of sulci follow furrows of high curvature, choices have to be made at intersections of multiple furrows because of the variability of the cortex. What makes this task more difficult is that the gyral regions have to be crossed sometimes to ensure a continuous curve is generated for each sulcus. To counter this kind of complications, protocols are defined in practice on how to use local dependency of sulcal lines for manual annotation. For the example in Fig. 1(a), the precentral sulcus has to cross a gyrus to meet the requirement that it should follow a path as parallel as possible to the central sulcus. In determining the inferior portion of postcentral sulcus that is highly variable, its relation with respect to the tail of the sylvian fissure also helps to provide critical information. There are two main challenges, however, to formulate a tractable inference problem over graphical models for joint sulci detection. (1) With each node of the graph representing a sulcus, the random variables of interest here live in high
3 100 Y. Shi et al. dimensional shape spaces and it is generally hard to do inferences directly over such spaces. (2) Every major sulcal curve observes flexible shape and it is hard to capture their individual as well as joint regularity by a good prior. In our joint detection framework, we tackle these challenges by first constructing a sample space containing a finite number of candidate curves at each node of the graph. These sample spaces greatly reduce the search range of inference algorithms and they are generated automatically based on the Hamilton-Jacobi skeleton of sulcal regions on triangulated cortical surfaces. We then use boosting techniques[18] to learn discriminative shape models of each sulcus and their neighboring relations. The advantages are twofold. (1) The algorithm is able to automatically select and fuse a set of informative shape features, characterizing unary as well as binary relationships of the sulcal curves, from a large set of candidate features. (2) The priors are learned directly from the training data and there is no parameter to tune for different sucal curves. Traditional generative model based algorithms, e.g. PCA, have difficulty in modeling such complicated priors due to its Gaussian assumptions. In making the inference, a belief propagation algorithm[19,20] is used to perform inferences efficiently on these sample spaces for joint sulci detection. In the rest of the paper, we first propose our joint detection framework with graphical models in section 2. The generation of sample spaces for each node of the graph is then described in section 3. After that, we develop a boosting approach in section 4 to learn potential functions in graphical models for sulci of interest. In section 5, experimental results on the joint detection of four major sulci: central, precentral, postcentral and the sylvian fissure on 40 cortical surfaces are presented. Finally, conclusions are made in section 6. 2 The Joint Detection Framework For the detection of a set of major sulci C 1,C 2,,C L on a cortical surface M, we assume an undirected graphical model G =(V,E) that represents the Markovian relation among sulci, where V = {C 1,C 2,,C L } are the set of nodes, and E is the set of edges in the graph. As an example, a graphical model for the seven sulci in Fig. 1(a) is shown in Fig. 1(b). Because the number of major sulci is small on the cortical surface, it is straightforward to construct the graph structure of such models and this only needs to be done once for the same detection task. To perform inferences on sulcal lines with graphical models, it is critical to first specify a proper sample space for each node as the general space of curves is infinite dimensional. One possible solution is to reduce the dimension of shape spaces with PCA, but there is no guarantee that these parameterized curves will live on the cortical surface and follow the bottom of sulcal regions. To overcome this problem, we generate a set of candidate curves S i automatically for each sulcus C i using a novel algorithm that will be developed in section 3. These candidate curves are weighted geodesics on the cortical surface and follow the skeleton of sulcal regions closely. By adopting the set of candidate curves S i as
4 Joint Sulci Detection Using Graphical Models and Boosted Priors 101 Table 1. The AdaBoost algorithm[18] Given training data: (x 1,y 1,w1), 1, (x n,y n,wn) 1 where x i are the sample data, y i { 1, 1} are the corresponding class labels, and wi 1 are the initial weights. For t =1,,T Train a weak classifier h t given the current weights. Compute the weighted error rate ɛ t of the classifier h t. Update the weights: w t+1 i = wt ie αty ih t(x i ) Z t where P α t = log((1 ɛ t)/ɛ t)/2 and Z t is a normalization constant such that n i=1 wt+1 i =1. Output the final classifier H = sign(f) with the decision function f = P T t=1 αtht. the sample space for each node C i, we are able to model each sulcus as a discrete random variable with values in a finite set and this makes the inference on the graphical model computationally tractable. Over the sample spaces of all the nodes in a graphical model, we define two types of potential functions to complete the construction of the graphical model: the local evidence function φ i : S i R at each node C i and the compatibility function ψ i,j : S i S j R for (C i,c j ) E. To incorporate shape priors about individual sulcus and their relations, we propose a discriminative approach using AdaBoost[18] in section 4 to learn both types of potential functions from manually annotated training data. Given input data x, the discriminative method learns the posterior probability p(y x) of a label y. With the discriminative approach, there is no need of specifying parametric forms for prior models of sulcal lines and their neighboring relations. Instead we use a large set of features derived from training data and selectively combine information from these features with boosting techniques. The central idea of AdaBoost, as listed in Table I, is the formation of a strong classifier through the combination of a series of weak classifiers. Using the decision function f generated from AdaBoost, an approximate posterior can then be defined and used as the potential function. With the graphical model defined, we can write down the joint distribution of all the sulci as: p(c 1,,C L )= 1 Z (C i,c j) E ψ i,j (C i,c j ) C i V φ i (C i ) (1) where Z is the partition function for normalization. For sulci detection, we use belief propagation to compute the marginal distribution of each sulcus from the joint distribution because it is applicable to graphs both with and without cycles. With belief propagation, each node in the graph receives and sends out messages at every iteration of the algorithm. For a node C i, the message it sends to its
5 102 Y. Shi et al. C 7 m 5,3 C 5 m 3,1 C 2 C 1 C 3 m 4,3 C 6 C 4 Fig. 2. An example of message passing in the graphical model of Fig. 1(b) neighbor C j is defined as: m i,j (C j )= C i S i ψ i,j (C i,c j )φ i (C i ) C k N (C i)\c j m k,i (C i ) (2) where N (C i )areneighborsofc i in the graph. This message takes into account not only the local evidence φ i and the compatibility function ψ i,j, but also the messages the node C i received from its neighbors except C j. As an illustration, we show in Fig. 2 the flow of messages from the node C 4 and C 5 to C 3,andthen to C 1 in the graphical model shown in Fig. 1(b). If we continue this message passing process until it converges, i.e., when the messages stop changing, we obtain the final belief at each node of the graph as: b i (C i )=φ i (C i ) m j,i (C i ) i =1, 2,,L. (3) C j N (C i) Using belief propagation, we collect information from all the nodes to form the final belief for every sulcus. For graphs without cycles[19], such as trees, the final belief function corresponds to the marginal distribution of each sulcus derived from the joint distribution in (1), thus we can detect a sulcus by maximizing the final belief: Ci =argmaxb i (C i ) i =1, 2,,L. (4) C i S i Even for graphs with cycles, belief propagation is known to frequently perform well and generate excellent results[20], most notably for its near Shannon limit performance in turbo decoding[21]. Thus the above equation is also applicable for joint sulci detection in the case that the graphical model has loops. 3 Generation of Sample Spaces Given a cortical surface, there are three main stages in our algorithm of sample space generation for each node of a graphical model: skeletonization of sulcal regions, a learning-based approach that picks out candidates for the starting and ending point of the sulcus, and the generation of member curves of the sample space as weighted geodesics on the cortical surface.
6 Joint Sulci Detection Using Graphical Models and Boosted Priors 103 (a) (b) (c) (d) Fig. 3. The process of generating sample spaces for sulci detection. (a) The original cortical surface. (b) Mesh segmentation results. Red: sulcal regions; blue: gyral regions. (c) The skeleton of sulcal regions plotted in black. (d) Curves from the sample space of the central sulcus plotted in red. In the first stage, we compute the skeleton of sulcal regions with the method we reported in[22]. For completeness we briefly review the main steps of this algorithm. We represent the cortical surface M as a triangular mesh and first compute the principle curvatures at each vertex. Using the curvature features, the cortical surface is then partitioned into sulcal and gyral regions using graph cuts. As an illustration, we show in Fig. 3(b) the result of the partition algorithm for the cortical surface in Fig. 3(a). Finally an extension of the method of Hamilton-Jacobi skeleton[23] is used to calculate the skeleton of sulcal regions as shown in Fig. 3(c). For all vertices of M in the skeleton, we classify them into three types using the neighboring system of the triangular mesh: End points: vertices with one neighbor in the skeleton. Knot points: vertices with three or more neighbors in the skeleton. Middle points: vertices with two neighbors in the skeleton. We learn a two-class classifier with AdaBoost in the second stage to pick a set of candidate points from the union of end and knot points in the skeleton for the starting and ending point of each sulcus. We use a set of K cortical surfaces with manually labeled sulci for the construction of our training data. These cortical surfaces are assumed to be registered to a common coordinate space such as ICBM. To learn the classifier for the starting point of a sulcus C i,weconstruct
7 104 Y. Shi et al. the training data as follows. The starting point of the manually labeled sulcus C i on the K cortical surfaces are assigned label 1 and weight 1/2K. The union of all the knot and end points from the skeletons of the K cortical surfaces are assigned label 1 andweight1/2ñ where Ñ is the total number of points in this set. At each iteration of AdaBoost, we train a perceptron as the weak classifier using the pocket algorithm with ratchet[24]. As shown in Table I, the final decision function generated from AdaBoost is of the following form: f = T α t h t (5) t=1 where h t : R 3 { 1, 1} is the weak classifier at the t-th iteration of AdaBoost and α t is the weight for this weak classifier. Applying the decision function f to all the end and knot points in the skeleton of a cortical surface, we pick the candidates for the starting point of C i as M points with the largest f values. Typically we choose M = 10 in our experiments and this generates a sufficiently large candidate set for the final detection of sulcal lines according to our experience. Similarly, a candidate set of M points can also be generated for the ending point of C i. Given the candidate set for the starting and ending points of the sulcal line C i, we connect them with weighted geodesics on M to generate the sample space S i. The weights are derived from the geodesic distance transform d of the skeleton on M generated in the first stage. We compute these geodesics with the fast marching algorithm on triangular meshes[25]. For a candidate of starting point X s and a candidate for the ending point X e, we connect them through a weighted geodesic with the weight defined as F = e d2 /2σ 2 (6) to encourage the weighted geodesic overlapping as much as possible with the skeleton of sulcal regions. To find this geodesic, we first compute a weighted distance transform d w on M by solving the Eikonal equation d w F =1 (7) intrinsically over the cortical surface and trace backward from X e to X s in gradient descent directions. This geodesic is then added to the sample space S i. We choose the parameter σ =1, 2, 3, 4, 5 to cover a wide range of possible routes. So overall the sample space S i of C i is composed of 5M 2 curves. As an illustration, all the candidate curves in the sample space of the central sulcus on the cortical surface in Fig. 1(a) are plotted in Fig. 1(d) and we can see that the true central sulcus is included in this sample space. 4 Learning Potential Functions Using AdaBoost In this section, we describe our learning-based approach for the construction of the potential functions in graphical models to take into account individual
8 Joint Sulci Detection Using Graphical Models and Boosted Priors 105 and joint shape priors of major sulci. Our approach is discriminative and learns both the local evidence functions φ i and the compatibility functions ψ i,j with AdaBoost. For each potential function, we construct a set of features and learn a strong two-class classifier by combining with AdaBoost a series of weak classifiers on these features. It is shown in [26] that AdaBoost approximates logistic regression and its result can be used to estimate the probability of a class label, which we then use to define the potential function. ForasulcusC i we learn its local evidence function φ i from a training data set of K cortical surfaces with the sulcus manually labeled on each surface. We also assume these cortical surfaces are registered to the same common coordinate system as in section 3. For the same sulcus, we generate a sample space on each of the cortical surface in the training data. All curves are parameterized with N (typically 100) uniformly sampled points such that one-to-one correspondences are established between curves. In the training data set, all manually labeled sulcus for C i on the K cortical surfaces are assigned label 1 and weight 1/2K and all the curves in the sample spaces are assigned a label 1 andweight 1/10KM 2. The same perceptron in section 3 is used as our weak classifier. At each iteration of AdaBoost, we train N perceptrons P n (1 n N) withthe n-th point from all the curves and pick the one with the best performance as the weak classifier h t. By combining all the weak classifiers, we obtain the final decision function f(c i ) = T t=1 α th t (C i ). Following [26], we define the local evidence function for C i as: φ i (C i )= e2f(ci) C 1+e 2f(Ci) i S i. (8) The local evidence function approaches the value 1 at curves in the sample space that are similar to the manually labeled sulcus in the training data, indicating intuitively that they exhibit strong evidence to be the sulcus we want to detect. To learn the compatibility function between two sulci C i and C j, we define the feature used for training as d i,j = C i C j, which is the difference between the two curves. For each cortical surface in the training data, the manually labeled sulci for C i and C j are used to construct the feature vector d i,j with label 1 and weight 1/2K. From the sample spaces of C i and C j on the K cortical surfaces, we randomly pick 5KM 2 pairs to construct the negative sample data with label 1 andweight1/10km 2. The same boosting process as above is applied to learn a decision function for the compatibility between the two sulci f(c i,c j )= T t=1 α th t (d i,j ) and we can define the compatibility function between C i and C j as: ψ i,j (C i,c j )= e2f(ci,cj) C 1+e 2f(Ci,Cj) i S i,c j S j. (9) With AdaBoost, we have developed a common solution for the learning of both local evidence functions and compatibility functions. Even though we used only features derived from coordinate information in our current work, the boosting method for learning priors is general and allows the inclusion of more features in future work. By inserting the potential functions into graphical models, the belief propagation process can then be used for the joint detection of major sulci.
9 106 Y. Shi et al. (a) Without graphical models. (b) With graphical models. (c) Without graphical models. (d) With graphical models. Fig. 4. Sulci detection results on two cortical surfaces. (a) and (b) show results from a surface in the training data. (c) and (d) show results from one of the surface in the testing data. For all the results, manually labeled results are plotted in black for comparison. The detected curves for the central, precentral, postcentral, and sylvian fissure are plotted in red, green, blue and cyan. 5 Experimental Results In this section we present preliminary experimental results for the joint detection of four major sulci: the central, precentral, postcentral sulcus and sylvian fissure. The graphical model used for the joint detection of these four sulci is the subgraph in Fig. 1(b) that includes the nodes C 1, C 2, C 3 and C 4. In our experiments, we have a dataset of 40 cortical surfaces with the four sulci manually labeled. We used 20 of them as the training data and the other 20 for testing. During the training stage, we first computed the skeleton of sulcal regions for the 20 training data. Decision functions were then learned for each sulcus such that a set of candidate points can be generated for both of its starting and ending point. After that, a sample space was generated for each sulcus with weighted geodesics as described in section 3. With these sample spaces, the local evidence functions and the compatibility functions between neighboring vertices in the graphical model were learned using the boosting method developed in section 4. For every cortical surface in the testing data, the sample space of each sulcus was generated with the same classifier learned from the training data. Once all the potential functions and sample spaces were constructed, we applied the joint detection algorithm with graphical models in section 2 to detect
10 Joint Sulci Detection Using Graphical Models and Boosted Priors 107 the four sulci on all 40 cortical surfaces. As a comparison, we also detected each sulcus independently without using graphical models. This is realized by simply picking the curve in the sample space that maximizes the local evidence function. To illustrate the advantage of the joint detection algorithm, we show in Fig. 4 the detection results on two cortical surfaces, one from the training data and the other from the testing data, using these two different methods. For the first surface, the results without using graphical models is shown in Fig. 4(a) and we can see the inferior part of the postcentral sulcus overlaps with the central sulcus and this is clearly undesirable. In the joint detection results shown in Fig. 4(b), it is easy to see the postcentral sulcus is correctly detected. Thanks to the compatibility functions learned with boosting, the tail of the sylvian fissure detected in Fig. 4(b) also agrees better with manually labeled results than the sylvian fissure detected in Fig. 4(a). In results of the second surface shown in Fig. 4(c) and (d), improved results were also obtained with graphical models for the central and precentral sulcus as compared to the results detected independently. We next perform a quantitative analysis of the performance of our sulci detection algorithm by comparing with manually labeled curves. For two curves C 1 and C 2 that are parameterized with N points, we define two distances: D a ( C 1, C 2 )= 1 N x n C 1 min x n y m (10) y m C 2 and D h ( C 1, C 2 )= max x n C 1 min x n y m. (11) y m C 2 The distance D a is an average of the distance from points on C 1 to C 2, while D h measures the Hausdorff distance from C 1 to C 2. For a detected sulcus C i and its manually labeled result C i, we use four distances to measure the difference between them: Da 1 = D a (C i, C i ), Da 2 = D a (C i,c i ), Dh 1 = D h(c i, C i ), and Dh 2 = D h(c i,c i ). Quantitative comparisons to manually labeled results were performed for both the sulci detected jointly with graphical models and independently without graphical models. For these two different groups of results, the average of the four distances over the training and testing data were computed and listed separately for each sulcus in Table 2. Overall we have 32 average distances for each group of results. The advantage of the jointly detected results are clear as they outperform results without graphical models in 28 of the 32 distances. A more detailed analysis shows that we achieve very good performance with our joint sulci detection algorithm in terms of Da 1 and Da 2 for all four sulci, and the results are especially encouraging for the central sulcus where an average distance of around 1mm is obtained to the manually annotated curves. The relatively large distances in terms of Dh 1 and D2 h are mostly due to the variability of the starting and ending part of sulci. Considering the lack of consensus among manual tracers on these parts, it might be interesting to use other metrics in our future research to measure how well the main body of each sulcal curve is detected.
11 108 Y. Shi et al. Table 2. Differences between sulci detected jointly with graphical models and independently as compared to manually annotated results (in millimeters) Sulci Central (training) Precentral (training) Postcentral(training) Sylvian (training) Central (testing) Precentral (testing) Postcentral (testing) Sylvian (testing) Without graphical models With graphical models Da 1 Da 2 Dh 1 Dh 2 Da 1 Da 2 Dh 1 Dh Conclusions A general framework for the joint detection of major sulci was proposed in this paper. Using boosting techniques, we integrated discriminative shape priors of each sulcus and their Markovian relations into graphical models and transformed sulci detection into a tractable inference problem over sample spaces of candidate curves. The boosting approach is flexible and allows the inclusion of new features to capture more detailed priors in future work. We are also testing the joint detection of more major sulci with our algorithm. Acknowledgment This work was funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 RR entitled Center for Computational Biology (CCB). References 1. Ono, M., Kubik, S., Abarnathey, C.: Atlas of the Cerebral Sulci. Thieme Medical Publishers (1990) 2. Thompson, P.M., Hayashi, K.M., Sowell, E.R., Gogtay, N., Giedd, J.N., Rapoport, J.L., de Zubicaray, G.I., Janke, A.L., Rose, S.E., Semple, J., Doddrell, D.M., Wang, Y., van Erp, T.G.M., Cannon, T.D., Toga, A.W.: Mapping cortical change in alzheimers disease, brain development, and schizophrenia. NeuroImage 23, S2 S18 (2004) 3. Khaneja, N., Miller, M., Grenander, U.: Dynamic programming generation of curves on brain surfaces. IEEE Trans. Pattern Anal. Machine Intell. 20(11), (1998) 4. Bartesaghi, A., Sapiro, G.: A system for the generation of curves on 3D brain images. Human Brain Mapping 14, 1 15 (2001) 5. Lui, L.M., Wang, Y., Chan, T.F., Thompson, P.M.: Automatic landmark and its application to the optimization of brain conformal mapping. In: Proc. CVPR. vol. 2, pp (2006) 6. Rettmann, M.E., Han, X., Xu, C., Prince, J.L.: Automated sulcal segmentation using watersheds on the cortical surface. NeuroImage 15(2), (2002)
12 Joint Sulci Detection Using Graphical Models and Boosted Priors Kao, C., Hofer, M., Sapiro, G., Stern, J., Rotternberg, D.: A geometric method for automatic extraction of sulcal fundi. In: Proc. ISBI 2006, pp (2006) 8. Lohmann, G.: Extracting line representations of sulcal and gyral patterns in MR images of the human brain. IEEE Trans. Med. Imag. 17(6), (1998) 9. Zhou, Y., Thompson, P.M., Toga, A.W.: Extracting and representing the cortical sulci. IEEE Computer Graphics and Applications 19(3), (1999) 10. Mangin, J.F., Frouin, V., Bloch, I., Régis, J., López-Krahe, J.: From 3d magnetic resonance images to structural representations of the cortex topography using topology preserving deformations. Journal of Mathematical Imaging and Vision 5(4), (1995) 11. Cootes, T., Taylor, C., Cooper, D., Graham, J.: Active shape models-their training and application. Computer Vision and Image Understanding 61(1), (1995) 12. Lohmann, G., Cramon, D.: Automatic labelling of the human cortical surface using sulcal basins. Medical Image Analysis 4, (2000) 13. Tao, X., Prince, J., Davatzikos, C.: Using a statistical shape model to extract sulcal curves on the outer cortex of the human brain. IEEE Trans. Med. Imag. 21(5), (2002) 14. Rivière, D., Mangin, J., Papadopoulos-Orfanos, D., Martinez, J., Frouin, V., Régis, J.: Automatic recognition of cortical sulci of the human brain using a congregation of neural networks. Medical Image Analysis 6, (2002) 15. Tu, Z.: Probabilistic boosting-tree: learning discriminative models for classification, recognition, and clustering. In: Proc. ICCV 2005, vol. 2, pp (2005) 16. Zheng, S., Tu, Z., Yuille, A., Reiss, A., Dutton, R., Lee, A., Galaburda, A., Thompson, P., Dinov, I., Toga, A.: A learning-based algorithm for automatic extraction of the cortical sulci. In: Larsen, R., Nielsen, M., Sporring, J. (eds.) MICCAI LNCS, vol. 4190, pp Springer, Heidelberg (2006) 17. Pizer, S., Jeong, J., Lu, C., Joshi, S.: Estimating the statistics of multi-object anatomic geometry using inter-object relationships. In: Proc. DSSCV 2005, pp (2005) 18. Freund, Y., Schapire, R.: A decision-theoretic generalization of on-line learning and an application to boosting. Journal of Computer and System Sciences 55(1), (1997) 19. Perl, J.: Probabilistic Reasoning in Intelligent Systems. Morgan Kaufman, San Mateo (1988) 20. Yedidia, J., Freeman, W., Weiss, Y.: Understanding Belief Propagation and Its Generalizations, pp Morgan Kaufmann Publishers Inc, San Francisco (2003) 21. Berrou, C., Glavieus, A., Thitimajshima, P.: Near Shannon limit error-correcting coding and decoding: Turbo-codes. In: Proc. IEEE Int. Conf. on Communications, pp (1993) 22. Shi, Y., Reiss, A., Lee, A., Dutton, R., Bellugi, U., Galaburda, A., Korenberg, J., Mills, D., Dinov, I., Thompson, P., Toga, A.: Hamilton-Jacobi skeletons on cortical surfaces with applications in characterizing the gyrification pattern in Williams syndrome. In: Proc. ISBI 2007 (2007) 23. Siddiqi, K., Bouix, S., Tannebaum, A., Zuker, S.: Hamilton-Jacobi skeletons. Int l Journal of Computer Vision 48(3), (2002) 24. Gallant, S.: Perceptron-based learning algorithms. IEEE Trans. Neural Networks 1(2), (1990) 25. Kimmel, R., Sethian, J.A.: Computing geodesic paths on manifolds. Proc. Natl. Acad. Sci. USA 95(15), (1998) 26. Friedman, J., Hastie, T., Tibshirani, R.: Additive logistic regression: a statistical view of boosting. Ann. Statist. 28(2), (2000)
A Learning Based Algorithm for Automatic Extraction of the Cortical Sulci
 A Learning Based Algorithm for Automatic Extraction of the Cortical Sulci Songfeng Zheng 1, Zhuowen Tu 2, Alan L. Yuille 1, Allan L. Reiss 3, Rebecca A. Dutton 2, Agatha D. Lee 2, Albert M. Galaburda 4,
A Learning Based Algorithm for Automatic Extraction of the Cortical Sulci Songfeng Zheng 1, Zhuowen Tu 2, Alan L. Yuille 1, Allan L. Reiss 3, Rebecca A. Dutton 2, Agatha D. Lee 2, Albert M. Galaburda 4,
CORTICAL sulci are important structures of the brain
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007 541 Automated Extraction of the Cortical Sulci Based on a Supervised Learning Approach Zhuowen Tu, Songfeng Zheng, Alan L. Yuille, Allan
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007 541 Automated Extraction of the Cortical Sulci Based on a Supervised Learning Approach Zhuowen Tu, Songfeng Zheng, Alan L. Yuille, Allan
IMA Preprint Series # 2078
 A GEOMETRIC METHOD FOR AUTOMATIC EXTRACTION OF SULCAL FUNDI By C.-Y. Kao M. Hofer G. Sapiro J. Stern and D. Rottenberg IMA Preprint Series # 2078 ( November 2005 ) INSTITUTE FOR MATHEMATICS AND ITS APPLICATIONS
A GEOMETRIC METHOD FOR AUTOMATIC EXTRACTION OF SULCAL FUNDI By C.-Y. Kao M. Hofer G. Sapiro J. Stern and D. Rottenberg IMA Preprint Series # 2078 ( November 2005 ) INSTITUTE FOR MATHEMATICS AND ITS APPLICATIONS
Watersheds on the Cortical Surface for Automated Sulcal Segmentation
 Watersheds on the Cortical Surface for Automated Sulcal Segmentation Maryam E. Rettmann,XiaoHan y, and Jerry L. Prince y Department of Biomedical Engineering y Department of Electrical and Computer Engineering
Watersheds on the Cortical Surface for Automated Sulcal Segmentation Maryam E. Rettmann,XiaoHan y, and Jerry L. Prince y Department of Biomedical Engineering y Department of Electrical and Computer Engineering
Brain Warping Via Landmark Points and Curves with a Level Set Representation
 Brain Warping Via Landmark Points and Curves with a Level Set Representation Andrew Y. Wang, Alex D. Leow, 2 Hillary D. Protas, Arthur W. Toga, Paul M. Thompson UCLA Laboratory of Neuro Imaging, Los Angeles,
Brain Warping Via Landmark Points and Curves with a Level Set Representation Andrew Y. Wang, Alex D. Leow, 2 Hillary D. Protas, Arthur W. Toga, Paul M. Thompson UCLA Laboratory of Neuro Imaging, Los Angeles,
Towards Learning-Based Holistic Brain Image Segmentation
 Towards Learning-Based Holistic Brain Image Segmentation Zhuowen Tu Lab of Neuro Imaging University of California, Los Angeles in collaboration with (A. Toga, P. Thompson et al.) Supported by (NIH CCB
Towards Learning-Based Holistic Brain Image Segmentation Zhuowen Tu Lab of Neuro Imaging University of California, Los Angeles in collaboration with (A. Toga, P. Thompson et al.) Supported by (NIH CCB
Using a Statistical Shape Model to Extract Sulcal Curves on the Outer Cortex of the Human Brain
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 21, NO. 5, MAY 2002 513 Using a Statistical Shape Model to Extract Sulcal Curves on the Outer Cortex of the Human Brain Xiaodong Tao, Jerry L. Prince, and Christos
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 21, NO. 5, MAY 2002 513 Using a Statistical Shape Model to Extract Sulcal Curves on the Outer Cortex of the Human Brain Xiaodong Tao, Jerry L. Prince, and Christos
COMBINATIONOF BRAIN CONFORMAL MAPPING AND LANDMARKS: A VARIATIONALAPPROACH
 COMBINATIONOF BRAIN CONFORMAL MAPPING AND LANDMARKS: A VARIATIONALAPPROACH Yalin Wang Mathematics Department, UCLA email: ylwang@math.ucla.edu Lok Ming Lui Mathematics Department, UCLA email: malmlui@math.ucla.edu
COMBINATIONOF BRAIN CONFORMAL MAPPING AND LANDMARKS: A VARIATIONALAPPROACH Yalin Wang Mathematics Department, UCLA email: ylwang@math.ucla.edu Lok Ming Lui Mathematics Department, UCLA email: malmlui@math.ucla.edu
Simultaneous Cortical Surface Labeling and Sulcal Curve Extraction
 Simultaneous Cortical Surface Labeling and Sulcal Curve Extraction Zhen Yang a, Aaron Carass a, Chen Chen a, Jerry L Prince a,b a Electrical and Computer Engineering, b Biomedical Engineering, Johns Hopkins
Simultaneous Cortical Surface Labeling and Sulcal Curve Extraction Zhen Yang a, Aaron Carass a, Chen Chen a, Jerry L Prince a,b a Electrical and Computer Engineering, b Biomedical Engineering, Johns Hopkins
Segmentation of a Human Brain Cortical Surface Mesh Using Watersheds
 Segmentation of a Human Brain Cortical Surface Mesh Using Watersheds 1.0 Abstract A 3D cortical surface mesh is a convoluted structure. The purpose of segmenting such a mesh is to impose a higher level
Segmentation of a Human Brain Cortical Surface Mesh Using Watersheds 1.0 Abstract A 3D cortical surface mesh is a convoluted structure. The purpose of segmenting such a mesh is to impose a higher level
Brain Surface Conformal Parameterization with Algebraic Functions
 Brain Surface Conformal Parameterization with Algebraic Functions Yalin Wang 1,2, Xianfeng Gu 3, Tony F. Chan 1, Paul M. Thompson 2, and Shing-Tung Yau 4 1 Mathematics Department, UCLA, Los Angeles, CA
Brain Surface Conformal Parameterization with Algebraic Functions Yalin Wang 1,2, Xianfeng Gu 3, Tony F. Chan 1, Paul M. Thompson 2, and Shing-Tung Yau 4 1 Mathematics Department, UCLA, Los Angeles, CA
Multivariate Tensor-based Brain Anatomical Surface Morphometry via Holomorphic One-Forms
 Multivariate Tensor-based Brain Anatomical Surface Morphometry via Holomorphic One-Forms Yalin Wang 1,2, Tony F. Chan 2, Arthur W. Toga 1, and Paul M. Thompson 1 1 Lab. of Neuro Imaging, UCLA School of
Multivariate Tensor-based Brain Anatomical Surface Morphometry via Holomorphic One-Forms Yalin Wang 1,2, Tony F. Chan 2, Arthur W. Toga 1, and Paul M. Thompson 1 1 Lab. of Neuro Imaging, UCLA School of
Statistical Study on Cortical Sulci of Human Brains
 Statistical Study on Cortical Sulci of Human Brains Xiaodong Tao 1,3,XiaoHan 1, Maryam E. Rettmann 2, Jerry L. Prince 1,2,3,and Christos Davatzikos 3 1 Electrical and Computer Engineering Johns Hopkins
Statistical Study on Cortical Sulci of Human Brains Xiaodong Tao 1,3,XiaoHan 1, Maryam E. Rettmann 2, Jerry L. Prince 1,2,3,and Christos Davatzikos 3 1 Electrical and Computer Engineering Johns Hopkins
A Fast and Accurate Method for Automated Cortical Surface Registration and Labeling
 A Fast and Accurate Method for Automated Cortical Surface Registration and Labeling Anand A Joshi 1, David W Shattuck 2 and Richard M Leahy 1 1 Signal and Image Processing Institute, University of Southern
A Fast and Accurate Method for Automated Cortical Surface Registration and Labeling Anand A Joshi 1, David W Shattuck 2 and Richard M Leahy 1 1 Signal and Image Processing Institute, University of Southern
Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity
 Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity Pahal Dalal 1, Feng Shi 2, Dinggang Shen 2, and Song Wang 1 1 Department of Computer Science and Engineering, University of South
Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity Pahal Dalal 1, Feng Shi 2, Dinggang Shen 2, and Song Wang 1 1 Department of Computer Science and Engineering, University of South
Automated Segmentation Using a Fast Implementation of the Chan-Vese Models
 Automated Segmentation Using a Fast Implementation of the Chan-Vese Models Huan Xu, and Xiao-Feng Wang,,3 Intelligent Computation Lab, Hefei Institute of Intelligent Machines, Chinese Academy of Science,
Automated Segmentation Using a Fast Implementation of the Chan-Vese Models Huan Xu, and Xiao-Feng Wang,,3 Intelligent Computation Lab, Hefei Institute of Intelligent Machines, Chinese Academy of Science,
Towards Whole Brain Segmentation by a Hybrid Model
 Towards Whole Brain Segmentation by a Hybrid Model Zhuowen Tu and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los Angeles, USA Abstract. Segmenting cortical and sub-cortical
Towards Whole Brain Segmentation by a Hybrid Model Zhuowen Tu and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los Angeles, USA Abstract. Segmenting cortical and sub-cortical
Parameterization of 3D Brain Structures for Statistical Shape Analysis
 Parameterization of 3D Brain Structures for Statistical Shape Analysis Litao Zhu and Tianzi Jiang * National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing
Parameterization of 3D Brain Structures for Statistical Shape Analysis Litao Zhu and Tianzi Jiang * National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing
Registration of Deformable Objects
 Registration of Deformable Objects Christopher DeCoro Includes content from: Consistent Mesh Parameterizations, Praun et. al, Siggraph 2001 The Space of Human Body Shapes, Allen et. al, Siggraph 2003 Shape-based
Registration of Deformable Objects Christopher DeCoro Includes content from: Consistent Mesh Parameterizations, Praun et. al, Siggraph 2001 The Space of Human Body Shapes, Allen et. al, Siggraph 2003 Shape-based
Automatic Subcortical Segmentation Using a Contextual Model
 Automatic Subcortical Segmentation Using a Contextual Model Jonathan H. Morra 1, Zhuowen Tu 1, Liana G. Apostolova 1,2, Amity E. Green 1,2, Arthur W. Toga 1, and Paul M. Thompson 1 1 Laboratory of Neuro
Automatic Subcortical Segmentation Using a Contextual Model Jonathan H. Morra 1, Zhuowen Tu 1, Liana G. Apostolova 1,2, Amity E. Green 1,2, Arthur W. Toga 1, and Paul M. Thompson 1 1 Laboratory of Neuro
BRAIN ANATOMICAL FEATURE DETECTION BY SOLVING PARTIAL DIFFERENTIAL EQUATIONS ON GENERAL MANIFOLDS. Los Angeles, CA , USA
 DISCRETE AND CONTINUOUS Website: http://aimsciences.org DYNAMICAL SYSTEMS SERIES B Volume 7, Number 3, May 2007 pp. 605 618 BRAIN ANATOMICAL FEATURE DETECTION BY SOLVING PARTIAL DIFFERENTIAL EQUATIONS
DISCRETE AND CONTINUOUS Website: http://aimsciences.org DYNAMICAL SYSTEMS SERIES B Volume 7, Number 3, May 2007 pp. 605 618 BRAIN ANATOMICAL FEATURE DETECTION BY SOLVING PARTIAL DIFFERENTIAL EQUATIONS
Wavelet-Based Representation of Biological Shapes
 Wavelet-Based Representation of Biological Shapes Bin Dong 1,YuMao 2, Ivo D. Dinov 3,ZhuowenTu 3, Yonggang Shi 3, Yalin Wang 3, and Arthur W. Toga 3 1 Department of Mathematics, University of California,
Wavelet-Based Representation of Biological Shapes Bin Dong 1,YuMao 2, Ivo D. Dinov 3,ZhuowenTu 3, Yonggang Shi 3, Yalin Wang 3, and Arthur W. Toga 3 1 Department of Mathematics, University of California,
Deformable Registration of Cortical Structures via Hybrid Volumetric and Surface Warping
 Deformable Registration of Cortical Structures via Hybrid Volumetric and Surface Warping Tianming Liu, Dinggang Shen, and Christos Davatzikos Section of Biomedical Image Analysis, Department of Radiology,
Deformable Registration of Cortical Structures via Hybrid Volumetric and Surface Warping Tianming Liu, Dinggang Shen, and Christos Davatzikos Section of Biomedical Image Analysis, Department of Radiology,
Labeling the Brain Surface Using a Deformable Multiresolution Mesh
 Labeling the Brain Surface Using a Deformable Multiresolution Mesh Sylvain Jaume 1, Benoît Macq 2, and Simon K. Warfield 3 1 Multiresolution Modeling Group, California Institute of Technology, 256-80,
Labeling the Brain Surface Using a Deformable Multiresolution Mesh Sylvain Jaume 1, Benoît Macq 2, and Simon K. Warfield 3 1 Multiresolution Modeling Group, California Institute of Technology, 256-80,
Anatomically Informed Convolution Kernels for the Projection of fmri Data on the Cortical Surface
 Anatomically Informed Convolution Kernels for the Projection of fmri Data on the Cortical Surface Grégory Operto 1,Rémy Bulot 1,Jean-LucAnton 2, and Olivier Coulon 1 1 Laboratoire LSIS, UMR 6168, CNRS,
Anatomically Informed Convolution Kernels for the Projection of fmri Data on the Cortical Surface Grégory Operto 1,Rémy Bulot 1,Jean-LucAnton 2, and Olivier Coulon 1 1 Laboratoire LSIS, UMR 6168, CNRS,
Optimization of Brain Conformal Mapping with Landmarks
 Optimization of Brain Conformal Mapping with Landmarks Yalin Wang 1,LokMingLui 1,TonyF.Chan 1, and Paul M. Thompson 2 Mathematics Department, UCLA, Los Angeles, CA 90095, USA Lab. of Neuro Imaging, UCLA
Optimization of Brain Conformal Mapping with Landmarks Yalin Wang 1,LokMingLui 1,TonyF.Chan 1, and Paul M. Thompson 2 Mathematics Department, UCLA, Los Angeles, CA 90095, USA Lab. of Neuro Imaging, UCLA
Automatic Landmark Tracking and its Application to the Optimization of Brain Conformal Mapping
 Automatic Landmark Tracking and its Application to the Optimization of Brain Conformal apping Lok ing Lui, Yalin Wang, Tony F. Chan Department of athematics University of California, Los Angeles {malmlui,
Automatic Landmark Tracking and its Application to the Optimization of Brain Conformal apping Lok ing Lui, Yalin Wang, Tony F. Chan Department of athematics University of California, Los Angeles {malmlui,
Level-set MCMC Curve Sampling and Geometric Conditional Simulation
 Level-set MCMC Curve Sampling and Geometric Conditional Simulation Ayres Fan John W. Fisher III Alan S. Willsky February 16, 2007 Outline 1. Overview 2. Curve evolution 3. Markov chain Monte Carlo 4. Curve
Level-set MCMC Curve Sampling and Geometric Conditional Simulation Ayres Fan John W. Fisher III Alan S. Willsky February 16, 2007 Outline 1. Overview 2. Curve evolution 3. Markov chain Monte Carlo 4. Curve
Population Modeling in Neuroscience Using Computer Vision and Machine Learning to learn from Brain Images. Overview. Image Registration
 Population Modeling in Neuroscience Using Computer Vision and Machine Learning to learn from Brain Images Overview 1. Part 1: Theory 1. 2. Learning 2. Part 2: Applications ernst.schwartz@meduniwien.ac.at
Population Modeling in Neuroscience Using Computer Vision and Machine Learning to learn from Brain Images Overview 1. Part 1: Theory 1. 2. Learning 2. Part 2: Applications ernst.schwartz@meduniwien.ac.at
NIH Public Access Author Manuscript Conf Proc IEEE Eng Med Biol Soc. Author manuscript; available in PMC 2009 December 11.
 NIH Public Access Author Manuscript Published in final edited form as: Conf Proc IEEE Eng Med Biol Soc. 2004 ; 2: 1521 1524. doi:10.1109/iembs.2004.1403466. An Elliptic PDE Approach for Shape Characterization
NIH Public Access Author Manuscript Published in final edited form as: Conf Proc IEEE Eng Med Biol Soc. 2004 ; 2: 1521 1524. doi:10.1109/iembs.2004.1403466. An Elliptic PDE Approach for Shape Characterization
Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow
 Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow Abstract. Finding meaningful 1-1 correspondences between hippocampal (HP) surfaces is an important but difficult
Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow Abstract. Finding meaningful 1-1 correspondences between hippocampal (HP) surfaces is an important but difficult
Wavelet-Based Representation of Biological Shapes
 Wavelet-Based Representation of Biological Shapes Bin Dong*, Yu Mao*, Ivo D. Dinov**, Zhuowen Tu**, Yonggang Shi**, Yalin Wang**, Arthur W. Toga** * Department of Mathematics, University of California,
Wavelet-Based Representation of Biological Shapes Bin Dong*, Yu Mao*, Ivo D. Dinov**, Zhuowen Tu**, Yonggang Shi**, Yalin Wang**, Arthur W. Toga** * Department of Mathematics, University of California,
Estimating Human Pose in Images. Navraj Singh December 11, 2009
 Estimating Human Pose in Images Navraj Singh December 11, 2009 Introduction This project attempts to improve the performance of an existing method of estimating the pose of humans in still images. Tasks
Estimating Human Pose in Images Navraj Singh December 11, 2009 Introduction This project attempts to improve the performance of an existing method of estimating the pose of humans in still images. Tasks
CRFs for Image Classification
 CRFs for Image Classification Devi Parikh and Dhruv Batra Carnegie Mellon University Pittsburgh, PA 15213 {dparikh,dbatra}@ece.cmu.edu Abstract We use Conditional Random Fields (CRFs) to classify regions
CRFs for Image Classification Devi Parikh and Dhruv Batra Carnegie Mellon University Pittsburgh, PA 15213 {dparikh,dbatra}@ece.cmu.edu Abstract We use Conditional Random Fields (CRFs) to classify regions
Face Alignment Under Various Poses and Expressions
 Face Alignment Under Various Poses and Expressions Shengjun Xin and Haizhou Ai Computer Science and Technology Department, Tsinghua University, Beijing 100084, China ahz@mail.tsinghua.edu.cn Abstract.
Face Alignment Under Various Poses and Expressions Shengjun Xin and Haizhou Ai Computer Science and Technology Department, Tsinghua University, Beijing 100084, China ahz@mail.tsinghua.edu.cn Abstract.
Research Proposal: Computational Geometry with Applications on Medical Images
 Research Proposal: Computational Geometry with Applications on Medical Images MEI-HENG YUEH yueh@nctu.edu.tw National Chiao Tung University 1 Introduction My research mainly focuses on the issues of computational
Research Proposal: Computational Geometry with Applications on Medical Images MEI-HENG YUEH yueh@nctu.edu.tw National Chiao Tung University 1 Introduction My research mainly focuses on the issues of computational
Learning-based Neuroimage Registration
 Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Retrospective Evaluation of Inter-subject Brain Registration
 Retrospective Evaluation of Inter-subject Brain Registration P. Hellier 1, C. Barillot 1, I. Corouge 1,B.Gibaud 2, G. Le Goualher 2,3, L. Collins 3, A. Evans 3, G. Malandain 4, and N. Ayache 4 1 Projet
Retrospective Evaluation of Inter-subject Brain Registration P. Hellier 1, C. Barillot 1, I. Corouge 1,B.Gibaud 2, G. Le Goualher 2,3, L. Collins 3, A. Evans 3, G. Malandain 4, and N. Ayache 4 1 Projet
Loopy Belief Propagation
 Loopy Belief Propagation Research Exam Kristin Branson September 29, 2003 Loopy Belief Propagation p.1/73 Problem Formalization Reasoning about any real-world problem requires assumptions about the structure
Loopy Belief Propagation Research Exam Kristin Branson September 29, 2003 Loopy Belief Propagation p.1/73 Problem Formalization Reasoning about any real-world problem requires assumptions about the structure
Correspondence Detection Using Wavelet-Based Attribute Vectors
 Correspondence Detection Using Wavelet-Based Attribute Vectors Zhong Xue, Dinggang Shen, and Christos Davatzikos Section of Biomedical Image Analysis, Department of Radiology University of Pennsylvania,
Correspondence Detection Using Wavelet-Based Attribute Vectors Zhong Xue, Dinggang Shen, and Christos Davatzikos Section of Biomedical Image Analysis, Department of Radiology University of Pennsylvania,
Brain Mapping with the Ricci Flow Conformal Parameterization and Multivariate Statistics on Deformation Tensors
 Brain Mapping with the Ricci Flow Conformal Parameterization and Multivariate Statistics on Deformation Tensors Yalin Wang 1,2, Xiaotian Yin 3, Jie Zhang 4, Xianfeng Gu 3, Tony F. Chan 2, Paul M. Thompson
Brain Mapping with the Ricci Flow Conformal Parameterization and Multivariate Statistics on Deformation Tensors Yalin Wang 1,2, Xiaotian Yin 3, Jie Zhang 4, Xianfeng Gu 3, Tony F. Chan 2, Paul M. Thompson
Learning Algorithms for Medical Image Analysis. Matteo Santoro slipguru
 Learning Algorithms for Medical Image Analysis Matteo Santoro slipguru santoro@disi.unige.it June 8, 2010 Outline 1. learning-based strategies for quantitative image analysis 2. automatic annotation of
Learning Algorithms for Medical Image Analysis Matteo Santoro slipguru santoro@disi.unige.it June 8, 2010 Outline 1. learning-based strategies for quantitative image analysis 2. automatic annotation of
3D Surface Matching and Registration through Shape Images
 3D Surface Matching and Registration through Shape Images Zhaoqiang Lai and Jing Hua Department of Computer Science, Wayne State University, Detroit, MI 48202, USA Abstract. In this paper, we present a
3D Surface Matching and Registration through Shape Images Zhaoqiang Lai and Jing Hua Department of Computer Science, Wayne State University, Detroit, MI 48202, USA Abstract. In this paper, we present a
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Semi-supervised Data Representation via Affinity Graph Learning
 1 Semi-supervised Data Representation via Affinity Graph Learning Weiya Ren 1 1 College of Information System and Management, National University of Defense Technology, Changsha, Hunan, P.R China, 410073
1 Semi-supervised Data Representation via Affinity Graph Learning Weiya Ren 1 1 College of Information System and Management, National University of Defense Technology, Changsha, Hunan, P.R China, 410073
Brain Surface Conformal Spherical Mapping
 Brain Surface Conformal Spherical Mapping Min Zhang Department of Industrial Engineering, Arizona State University mzhang33@asu.edu Abstract It is well known and proved that any genus zero surface can
Brain Surface Conformal Spherical Mapping Min Zhang Department of Industrial Engineering, Arizona State University mzhang33@asu.edu Abstract It is well known and proved that any genus zero surface can
A Hierarchical Compositional System for Rapid Object Detection
 A Hierarchical Compositional System for Rapid Object Detection Long Zhu and Alan Yuille Department of Statistics University of California at Los Angeles Los Angeles, CA 90095 {lzhu,yuille}@stat.ucla.edu
A Hierarchical Compositional System for Rapid Object Detection Long Zhu and Alan Yuille Department of Statistics University of California at Los Angeles Los Angeles, CA 90095 {lzhu,yuille}@stat.ucla.edu
Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images
 Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images MICCAI 2013: Workshop on Medical Computer Vision Authors: Quan Wang, Dijia Wu, Le Lu, Meizhu Liu, Kim L. Boyer,
Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images MICCAI 2013: Workshop on Medical Computer Vision Authors: Quan Wang, Dijia Wu, Le Lu, Meizhu Liu, Kim L. Boyer,
Multiple Motion and Occlusion Segmentation with a Multiphase Level Set Method
 Multiple Motion and Occlusion Segmentation with a Multiphase Level Set Method Yonggang Shi, Janusz Konrad, W. Clem Karl Department of Electrical and Computer Engineering Boston University, Boston, MA 02215
Multiple Motion and Occlusion Segmentation with a Multiphase Level Set Method Yonggang Shi, Janusz Konrad, W. Clem Karl Department of Electrical and Computer Engineering Boston University, Boston, MA 02215
Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm
 Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm Lin Chen and Gudrun Wagenknecht Central Institute for Electronics, Research Center Jülich, Jülich, Germany Email: l.chen@fz-juelich.de
Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm Lin Chen and Gudrun Wagenknecht Central Institute for Electronics, Research Center Jülich, Jülich, Germany Email: l.chen@fz-juelich.de
Medical Image Segmentation using Level Sets
 Medical Image Segmentation using Level Sets Technical Report #CS-8-1 Tenn Francis Chen Abstract Segmentation is a vital aspect of medical imaging. It aids in the visualization of medical data and diagnostics
Medical Image Segmentation using Level Sets Technical Report #CS-8-1 Tenn Francis Chen Abstract Segmentation is a vital aspect of medical imaging. It aids in the visualization of medical data and diagnostics
Global Minimization of the Active Contour Model with TV-Inpainting and Two-Phase Denoising
 Global Minimization of the Active Contour Model with TV-Inpainting and Two-Phase Denoising Shingyu Leung and Stanley Osher Department of Mathematics, UCLA, Los Angeles, CA 90095, USA {syleung, sjo}@math.ucla.edu
Global Minimization of the Active Contour Model with TV-Inpainting and Two-Phase Denoising Shingyu Leung and Stanley Osher Department of Mathematics, UCLA, Los Angeles, CA 90095, USA {syleung, sjo}@math.ucla.edu
City, University of London Institutional Repository
 City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
Selecting Models from Videos for Appearance-Based Face Recognition
 Selecting Models from Videos for Appearance-Based Face Recognition Abdenour Hadid and Matti Pietikäinen Machine Vision Group Infotech Oulu and Department of Electrical and Information Engineering P.O.
Selecting Models from Videos for Appearance-Based Face Recognition Abdenour Hadid and Matti Pietikäinen Machine Vision Group Infotech Oulu and Department of Electrical and Information Engineering P.O.
Topology-Invariant Similarity and Diffusion Geometry
 1 Topology-Invariant Similarity and Diffusion Geometry Lecture 7 Alexander & Michael Bronstein tosca.cs.technion.ac.il/book Numerical geometry of non-rigid shapes Stanford University, Winter 2009 Intrinsic
1 Topology-Invariant Similarity and Diffusion Geometry Lecture 7 Alexander & Michael Bronstein tosca.cs.technion.ac.il/book Numerical geometry of non-rigid shapes Stanford University, Winter 2009 Intrinsic
Active Geodesics: Region-based Active Contour Segmentation with a Global Edge-based Constraint
 Active Geodesics: Region-based Active Contour Segmentation with a Global Edge-based Constraint Vikram Appia Anthony Yezzi Georgia Institute of Technology, Atlanta, GA, USA. Abstract We present an active
Active Geodesics: Region-based Active Contour Segmentation with a Global Edge-based Constraint Vikram Appia Anthony Yezzi Georgia Institute of Technology, Atlanta, GA, USA. Abstract We present an active
3D Statistical Shape Model Building using Consistent Parameterization
 3D Statistical Shape Model Building using Consistent Parameterization Matthias Kirschner, Stefan Wesarg Graphisch Interaktive Systeme, TU Darmstadt matthias.kirschner@gris.tu-darmstadt.de Abstract. We
3D Statistical Shape Model Building using Consistent Parameterization Matthias Kirschner, Stefan Wesarg Graphisch Interaktive Systeme, TU Darmstadt matthias.kirschner@gris.tu-darmstadt.de Abstract. We
Random projection for non-gaussian mixture models
 Random projection for non-gaussian mixture models Győző Gidófalvi Department of Computer Science and Engineering University of California, San Diego La Jolla, CA 92037 gyozo@cs.ucsd.edu Abstract Recently,
Random projection for non-gaussian mixture models Győző Gidófalvi Department of Computer Science and Engineering University of California, San Diego La Jolla, CA 92037 gyozo@cs.ucsd.edu Abstract Recently,
Cortical surface thickness as a classifier: Boosting for autism classification
 Cortical surface thickness as a classifier: Boosting for autism classification Vikas Singh 1, Lopamudra Mukherjee 2, and Moo K. Chung 1 1 Biostatistics and Medical Informatics, University of Wisconsin-Madison,
Cortical surface thickness as a classifier: Boosting for autism classification Vikas Singh 1, Lopamudra Mukherjee 2, and Moo K. Chung 1 1 Biostatistics and Medical Informatics, University of Wisconsin-Madison,
SEGMENTING subcortical structures from 3-D brain images
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 27, NO. 4, APRIL 2008 495 Brain Anatomical Structure Segmentation by Hybrid Discriminative/Generative Models Zhuowen Tu, Katherine L. Narr, Piotr Dollár, Ivo
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 27, NO. 4, APRIL 2008 495 Brain Anatomical Structure Segmentation by Hybrid Discriminative/Generative Models Zhuowen Tu, Katherine L. Narr, Piotr Dollár, Ivo
D-Separation. b) the arrows meet head-to-head at the node, and neither the node, nor any of its descendants, are in the set C.
 D-Separation Say: A, B, and C are non-intersecting subsets of nodes in a directed graph. A path from A to B is blocked by C if it contains a node such that either a) the arrows on the path meet either
D-Separation Say: A, B, and C are non-intersecting subsets of nodes in a directed graph. A path from A to B is blocked by C if it contains a node such that either a) the arrows on the path meet either
Model-Driven Harmonic Parameterization of the Cortical Surface
 Model-Driven Harmonic Parameterization of the Cortical Surface Guillaume Auzias 1, Julien Lefèvre 1,2, Arnaud Le Troter 1,ClaraFischer 3, Matthieu Perrot 3,JeanRégis 4, and Olivier Coulon 1 1 LSIS Lab,
Model-Driven Harmonic Parameterization of the Cortical Surface Guillaume Auzias 1, Julien Lefèvre 1,2, Arnaud Le Troter 1,ClaraFischer 3, Matthieu Perrot 3,JeanRégis 4, and Olivier Coulon 1 1 LSIS Lab,
Robust Kernel Methods in Clustering and Dimensionality Reduction Problems
 Robust Kernel Methods in Clustering and Dimensionality Reduction Problems Jian Guo, Debadyuti Roy, Jing Wang University of Michigan, Department of Statistics Introduction In this report we propose robust
Robust Kernel Methods in Clustering and Dimensionality Reduction Problems Jian Guo, Debadyuti Roy, Jing Wang University of Michigan, Department of Statistics Introduction In this report we propose robust
Supervised Learning for Image Segmentation
 Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search
 Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
CRF Based Point Cloud Segmentation Jonathan Nation
 CRF Based Point Cloud Segmentation Jonathan Nation jsnation@stanford.edu 1. INTRODUCTION The goal of the project is to use the recently proposed fully connected conditional random field (CRF) model to
CRF Based Point Cloud Segmentation Jonathan Nation jsnation@stanford.edu 1. INTRODUCTION The goal of the project is to use the recently proposed fully connected conditional random field (CRF) model to
Intensity-Depth Face Alignment Using Cascade Shape Regression
 Intensity-Depth Face Alignment Using Cascade Shape Regression Yang Cao 1 and Bao-Liang Lu 1,2 1 Center for Brain-like Computing and Machine Intelligence Department of Computer Science and Engineering Shanghai
Intensity-Depth Face Alignment Using Cascade Shape Regression Yang Cao 1 and Bao-Liang Lu 1,2 1 Center for Brain-like Computing and Machine Intelligence Department of Computer Science and Engineering Shanghai
Tensor-based Brain Surface Modeling and Analysis
 Tensor-based Brain Surface Modeling and Analysis Moo K. Chung 1, Keith J. Worsley 2, Steve Robbins 2 and Alan C. Evans 2 1 Department of Statistics, Department of Biostatistics and Medical Informatics
Tensor-based Brain Surface Modeling and Analysis Moo K. Chung 1, Keith J. Worsley 2, Steve Robbins 2 and Alan C. Evans 2 1 Department of Statistics, Department of Biostatistics and Medical Informatics
Recovering Intrinsic Images from a Single Image
 Recovering Intrinsic Images from a Single Image Marshall F Tappen William T Freeman Edward H Adelson MIT Artificial Intelligence Laboratory Cambridge, MA 02139 mtappen@ai.mit.edu, wtf@ai.mit.edu, adelson@ai.mit.edu
Recovering Intrinsic Images from a Single Image Marshall F Tappen William T Freeman Edward H Adelson MIT Artificial Intelligence Laboratory Cambridge, MA 02139 mtappen@ai.mit.edu, wtf@ai.mit.edu, adelson@ai.mit.edu
STATISTICS AND ANALYSIS OF SHAPE
 Control and Cybernetics vol. 36 (2007) No. 2 Book review: STATISTICS AND ANALYSIS OF SHAPE by H. Krim, A. Yezzi, Jr., eds. There are numerous definitions of a notion of shape of an object. These definitions
Control and Cybernetics vol. 36 (2007) No. 2 Book review: STATISTICS AND ANALYSIS OF SHAPE by H. Krim, A. Yezzi, Jr., eds. There are numerous definitions of a notion of shape of an object. These definitions
Retrospective evaluation of inter-subject brain registration
 Retrospective evaluation of inter-subject brain registration P. Hellier 1, C. Barillot 1, I. Corouge 1,B.Gibaud 2, G. Le Goualher 2,3, L. Collins 3, A. Evans 3, G. Malandain 4, N. Ayache 4 1 Projet Vista,
Retrospective evaluation of inter-subject brain registration P. Hellier 1, C. Barillot 1, I. Corouge 1,B.Gibaud 2, G. Le Goualher 2,3, L. Collins 3, A. Evans 3, G. Malandain 4, N. Ayache 4 1 Projet Vista,
Matching 3D Shapes Using 2D Conformal Representations
 Matching 3D Shapes Using 2D Conformal Representations Xianfeng Gu 1 and Baba C. Vemuri 2 Computer and Information Science and Engineering, Gainesville, FL 32611-6120, USA {gu,vemuri}@cise.ufl.edu Abstract.
Matching 3D Shapes Using 2D Conformal Representations Xianfeng Gu 1 and Baba C. Vemuri 2 Computer and Information Science and Engineering, Gainesville, FL 32611-6120, USA {gu,vemuri}@cise.ufl.edu Abstract.
Shape Analysis with Conformal Invariants for Multiply Connected Domains and its Application to Analyzing Brain Morphology
 Shape Analysis with Conformal Invariants for Multiply Connected Domains and its Application to Analyzing Brain Morphology Yalin Wang Dept of Neurology/Math UCLA ylwang@math.ucla.edu Xianfeng Gu Dept of
Shape Analysis with Conformal Invariants for Multiply Connected Domains and its Application to Analyzing Brain Morphology Yalin Wang Dept of Neurology/Math UCLA ylwang@math.ucla.edu Xianfeng Gu Dept of
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs
 Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
Improving Recognition through Object Sub-categorization
 Improving Recognition through Object Sub-categorization Al Mansur and Yoshinori Kuno Graduate School of Science and Engineering, Saitama University, 255 Shimo-Okubo, Sakura-ku, Saitama-shi, Saitama 338-8570,
Improving Recognition through Object Sub-categorization Al Mansur and Yoshinori Kuno Graduate School of Science and Engineering, Saitama University, 255 Shimo-Okubo, Sakura-ku, Saitama-shi, Saitama 338-8570,
Multi-scale Shape Modeling by Markov Random Fields
 Multi-scale Shape Modeling by Markov Random Fields Conglin Lu, Stephen M. Pizer, and Sarang Joshi Medical Image Display and Analysis Group University of North Carolina, Chapel Hill, USA Abstract. The geometric
Multi-scale Shape Modeling by Markov Random Fields Conglin Lu, Stephen M. Pizer, and Sarang Joshi Medical Image Display and Analysis Group University of North Carolina, Chapel Hill, USA Abstract. The geometric
Comparison of Vessel Segmentations Using STAPLE
 Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Performance Issues in Shape Classification
 Performance Issues in Shape Classification Samson J. Timoner 1, Pollina Golland 1, Ron Kikinis 2, Martha E. Shenton 3, W. Eric L. Grimson 1, and William M. Wells III 1,2 1 MIT AI Laboratory, Cambridge
Performance Issues in Shape Classification Samson J. Timoner 1, Pollina Golland 1, Ron Kikinis 2, Martha E. Shenton 3, W. Eric L. Grimson 1, and William M. Wells III 1,2 1 MIT AI Laboratory, Cambridge
RECENT advances in medical imaging of the brain allow
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 18, NO. 6, JUNE 1999 467 Reconstruction of the Human Cerebral Cortex from Magnetic Resonance Images Chenyang Xu, Dzung L. Pham, Maryam E. Rettmann, Daphne N.
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 18, NO. 6, JUNE 1999 467 Reconstruction of the Human Cerebral Cortex from Magnetic Resonance Images Chenyang Xu, Dzung L. Pham, Maryam E. Rettmann, Daphne N.
RESEARCH STATEMENT Ronald Lok Ming Lui
 Ronald Lok Ming Lui 1/11 RESEARCH STATEMENT Ronald Lok Ming Lui (malmlui@math.harvard.edu) The main focus of my research has been on problems in computational conformal geometry and human brain mapping.
Ronald Lok Ming Lui 1/11 RESEARCH STATEMENT Ronald Lok Ming Lui (malmlui@math.harvard.edu) The main focus of my research has been on problems in computational conformal geometry and human brain mapping.
Multiple Constraint Satisfaction by Belief Propagation: An Example Using Sudoku
 Multiple Constraint Satisfaction by Belief Propagation: An Example Using Sudoku Todd K. Moon and Jacob H. Gunther Utah State University Abstract The popular Sudoku puzzle bears structural resemblance to
Multiple Constraint Satisfaction by Belief Propagation: An Example Using Sudoku Todd K. Moon and Jacob H. Gunther Utah State University Abstract The popular Sudoku puzzle bears structural resemblance to
Linear combinations of simple classifiers for the PASCAL challenge
 Linear combinations of simple classifiers for the PASCAL challenge Nik A. Melchior and David Lee 16 721 Advanced Perception The Robotics Institute Carnegie Mellon University Email: melchior@cmu.edu, dlee1@andrew.cmu.edu
Linear combinations of simple classifiers for the PASCAL challenge Nik A. Melchior and David Lee 16 721 Advanced Perception The Robotics Institute Carnegie Mellon University Email: melchior@cmu.edu, dlee1@andrew.cmu.edu
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Labeling Irregular Graphs with Belief Propagation
 Labeling Irregular Graphs with Belief Propagation Ifeoma Nwogu and Jason J. Corso State University of New York at Buffalo Department of Computer Science and Engineering 201 Bell Hall, Buffalo, NY 14260
Labeling Irregular Graphs with Belief Propagation Ifeoma Nwogu and Jason J. Corso State University of New York at Buffalo Department of Computer Science and Engineering 201 Bell Hall, Buffalo, NY 14260
Graph-Shifts Anatomic 3D Segmentation by Dynamic Hierarchical Minimization
 Graph-Shifts Anatomic 3D Segmentation by Dynamic Hierarchical Minimization Jason Corso Postdoctoral Fellow UCLA LONI/CCB jcorso@ucla.edu Motivation The work deals with the problem of automatically labeling
Graph-Shifts Anatomic 3D Segmentation by Dynamic Hierarchical Minimization Jason Corso Postdoctoral Fellow UCLA LONI/CCB jcorso@ucla.edu Motivation The work deals with the problem of automatically labeling
A Generic Probabilistic Active Shape Model for Organ Segmentation
 A Generic Probabilistic Active Shape Model for Organ Segmentation Andreas Wimmer 1,2, Grzegorz Soza 2, and Joachim Hornegger 1 1 Chair of Pattern Recognition, Department of Computer Science, Friedrich-Alexander
A Generic Probabilistic Active Shape Model for Organ Segmentation Andreas Wimmer 1,2, Grzegorz Soza 2, and Joachim Hornegger 1 1 Chair of Pattern Recognition, Department of Computer Science, Friedrich-Alexander
Anatomical landmark and region mapping based on a template surface deformation for foot bone morphology
 Anatomical landmark and region mapping based on a template surface deformation for foot bone morphology Jaeil Kim 1, Sang Gyo Seo 2, Dong Yeon Lee 2, Jinah Park 1 1 Department of Computer Science, KAIST,
Anatomical landmark and region mapping based on a template surface deformation for foot bone morphology Jaeil Kim 1, Sang Gyo Seo 2, Dong Yeon Lee 2, Jinah Park 1 1 Department of Computer Science, KAIST,
Snakes reparameterization for noisy images segmentation and targets tracking
 Snakes reparameterization for noisy images segmentation and targets tracking Idrissi Sidi Yassine, Samir Belfkih. Lycée Tawfik Elhakim Zawiya de Noaceur, route de Marrakech, Casablanca, maroc. Laboratoire
Snakes reparameterization for noisy images segmentation and targets tracking Idrissi Sidi Yassine, Samir Belfkih. Lycée Tawfik Elhakim Zawiya de Noaceur, route de Marrakech, Casablanca, maroc. Laboratoire
3D model classification using convolutional neural network
 3D model classification using convolutional neural network JunYoung Gwak Stanford jgwak@cs.stanford.edu Abstract Our goal is to classify 3D models directly using convolutional neural network. Most of existing
3D model classification using convolutional neural network JunYoung Gwak Stanford jgwak@cs.stanford.edu Abstract Our goal is to classify 3D models directly using convolutional neural network. Most of existing
STIC AmSud Project. Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach
 STIC AmSud Project Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach Caroline Petitjean A joint work with Damien Grosgeorge, Pr Su Ruan, Pr JN Dacher, MD October 22,
STIC AmSud Project Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach Caroline Petitjean A joint work with Damien Grosgeorge, Pr Su Ruan, Pr JN Dacher, MD October 22,
Structural Correspondence as A Contour Grouping Problem
 IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis, San Francisco, CA, June 14, 2010 Structural Correspondence as A Contour Grouping Problem Elena Bernardis University
IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis, San Francisco, CA, June 14, 2010 Structural Correspondence as A Contour Grouping Problem Elena Bernardis University
Probabilistic Graphical Models
 Overview of Part Two Probabilistic Graphical Models Part Two: Inference and Learning Christopher M. Bishop Exact inference and the junction tree MCMC Variational methods and EM Example General variational
Overview of Part Two Probabilistic Graphical Models Part Two: Inference and Learning Christopher M. Bishop Exact inference and the junction tree MCMC Variational methods and EM Example General variational
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 29, NO. 12, DECEMBER
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 29, NO. 12, DECEMBER 2010 2009 Robust Surface Reconstruction via Laplace-Beltrami Eigen-Projection and Boundary Deformation Yonggang Shi, Member, IEEE, Rongjie
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 29, NO. 12, DECEMBER 2010 2009 Robust Surface Reconstruction via Laplace-Beltrami Eigen-Projection and Boundary Deformation Yonggang Shi, Member, IEEE, Rongjie
Open-Curve Shape Correspondence Without Endpoint Correspondence
 Open-Curve Shape Correspondence Without Endpoint Correspondence Theodor Richardson and Song Wang Department of Computer Science and Engineering, University of South Carolina, Columbia, SC 29208, USA richa268@cse.sc.edu,
Open-Curve Shape Correspondence Without Endpoint Correspondence Theodor Richardson and Song Wang Department of Computer Science and Engineering, University of South Carolina, Columbia, SC 29208, USA richa268@cse.sc.edu,
Matching 3D Shapes Using 2D Conformal Representations
 Matching 3D Shapes Using 2D Conformal Representations Xianfeng Gu 1 and Baba C. Vemuri 2 Computer and Information Science and Engineering, Gainesville, FL 32611-6120, USA {gu,vemuri}@cise.ufl.edu Matching
Matching 3D Shapes Using 2D Conformal Representations Xianfeng Gu 1 and Baba C. Vemuri 2 Computer and Information Science and Engineering, Gainesville, FL 32611-6120, USA {gu,vemuri}@cise.ufl.edu Matching
Multi-modal Image Registration Using the Generalized Survival Exponential Entropy
 Multi-modal Image Registration Using the Generalized Survival Exponential Entropy Shu Liao and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Multi-modal Image Registration Using the Generalized Survival Exponential Entropy Shu Liao and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Effects of Registration Regularization and Atlas Sharpness on Segmentation Accuracy
 Effects of egistration egularization and Atlas Sharpness on Segmentation Accuracy B.T. Thomas Yeo 1, Mert. Sabuncu 1, ahul Desikan 3, Bruce Fischl 2, and Polina Golland 1 1 Computer Science and Artificial
Effects of egistration egularization and Atlas Sharpness on Segmentation Accuracy B.T. Thomas Yeo 1, Mert. Sabuncu 1, ahul Desikan 3, Bruce Fischl 2, and Polina Golland 1 1 Computer Science and Artificial
Sparse Shape Registration for Occluded Facial Feature Localization
 Shape Registration for Occluded Facial Feature Localization Fei Yang, Junzhou Huang and Dimitris Metaxas Abstract This paper proposes a sparsity driven shape registration method for occluded facial feature
Shape Registration for Occluded Facial Feature Localization Fei Yang, Junzhou Huang and Dimitris Metaxas Abstract This paper proposes a sparsity driven shape registration method for occluded facial feature
Using Decision Boundary to Analyze Classifiers
 Using Decision Boundary to Analyze Classifiers Zhiyong Yan Congfu Xu College of Computer Science, Zhejiang University, Hangzhou, China yanzhiyong@zju.edu.cn Abstract In this paper we propose to use decision
Using Decision Boundary to Analyze Classifiers Zhiyong Yan Congfu Xu College of Computer Science, Zhejiang University, Hangzhou, China yanzhiyong@zju.edu.cn Abstract In this paper we propose to use decision
Learning and Recognizing Visual Object Categories Without First Detecting Features
 Learning and Recognizing Visual Object Categories Without First Detecting Features Daniel Huttenlocher 2007 Joint work with D. Crandall and P. Felzenszwalb Object Category Recognition Generic classes rather
Learning and Recognizing Visual Object Categories Without First Detecting Features Daniel Huttenlocher 2007 Joint work with D. Crandall and P. Felzenszwalb Object Category Recognition Generic classes rather
