Data Walkthrough: Background
|
|
|
- Lorin Ferguson
- 6 years ago
- Views:
Transcription
1 Data Walkthrough: Background File Types FASTA Files FASTA files are text-based representations of genetic information. They can contain nucleotide or amino acid sequences. For this activity, students will be working with a FASTA file which contains the nucleotide sequence of the mitochondrial hypervariable region II (mthvrii). Every FASTA sequence contains two main parts: a header line, beginning with > and containing the name and details of the sequence, and the sequence information. A FASTA file can contain multiple sequences so long as every header line begins with > and has a unique identifier. Most programs which process FASTA files allow files to be uploaded directly, but FASTA files can be opened in a text editor (such as Notepad) to be viewed and modified. Figure 1 FASTA format for the mthvrii sequence Variant Call Format (VCF) Files VCF files are text-based, tab-delimited, files containing information about where and how an observed sequence differed from the reference. These files contain a large amount of information about both the quality and frequency of reads, although the specific types of information will vary depending on what equipment was used to generate the VCF. Between the variety of information, and the condensed manner in which it is presented, VCF files can be overwhelming; therefore this data walkthrough uses a trimmed down VCF containing only the information necessary to complete our analysis process. The general format of a VCF file consists of two sections: a header section of rows beginning with ##, and a content section containing a table of information for the different variants observed. The header section contains information about the data used to generate the file (alignment tool, reference sequence, etc), as well as specifics about what types of information are presented in the table below. The tables always contain the same column headers: CHROM the chromosome where the variant is located POS the position of variant within that chromosome (each type of mutation observed has its own line in the VCF, so if a both single nucleotide change and an insert were observed at the same point, that position will have multiple rows assigned to it) ID if the reference is annotated with known mutation, they are indicated here REF what base/bases appear in the reference
2 ALT the alternate base/bases observed, multiple entries are separated by commas QUAL quality metric; higher numbers indicate more certainty in the accuracy of the read FILTER a combined measure of quality and frequency, a variant must pass both to be considered accurate and will indicate if either failed (LowGQ or LowVarFrequency) INFO general information about the variants observed FORMAT the order in which sample specific information will be presented, types of information are separated by colons, : [SAMPLE NAME] sample-specific information presented in the order described in format. There may be multiple samples represented in a single VCF file. Figure 2 Full VCF format, opened in Excel For this demo set of data, we have trimmed the VCFs down to the information vital to our analysis. We have kept the chromosome, position, reference, alternate, and filter information unchanged. In addition we kept the Allelic Depth (AD) information, but renamed it Read Frequency (RF) to avoid confusion over the terms allelic and depth (especially since the hypervariable regions are non-coding and allele is usually used with genes), to function as a tiebreaker when a position has multiple valid variants. Our abridged VCF is shown below, opened in both Excel (which formats the table to columns) and in Word (which keeps the tab delimiters, but will not always align columns neatly).
3 Figure 3 Trimmed VCF file opened in Excel (left) and Word (right). Clustal Omega Clustal Omega is a multiple alignment tool which compares FASTA sequences (nucleotide or protein) to determine how similar they are. It is a free, online tool provided at Figure 4 Clustal Omega input page. In this activity, students will import the mthvrii sequences they ve generated using the reference and VCF files into Clustal Omega and use the resultant similarity matrix to determine which are most genetically similar.
4 Data Walkthrough: Workflow References and Variants The first step of analysis is to generate the complete sequence for each individual. Since entire genomes can be extremely long, the sequencer doesn t write out the entire sequence for every sample. Instead, it generates a file listing only the places where the sample differed from the reference, this file is in a variant call format and referred to as a VCF. We re not looking at an entire genome though, and the hypervariable region (HVR) we re using is only about 430 base pairs (bp) long, so we can generate the full sequence fairly easily using the reference as a template and making the changes indicated in the VCF. Generate a sample specific sequence 1. Open Reference FASTA in Notepad (or text-editing program of your choice) 2. Copy/Save As to make a new, sample specific FASTA a. Be sure to change the header row to indicate which sample it represents 3. Open the appropriate VCF in Excel or Word (Notepad doesn t always format correctly) 4. Using the VCF, make the appropriate changes to the sample FASTA a. Only variants which pass filter should be considered. If the filter column has LowVariantFreq (did not pass the frequency filter) or LowGQ (did not pass the quality filter), the variant shouldn t be used. b. Use read frequencies (RF) to pick the most frequent of multiple, valid mutations at a single point. 5. Once all changes are made, save the file. 6. Repeat as necessary until all samples have FASTA sequences saved. Additional Advice for Editing FASTAs In notepad, when View>status bar is checked, you can use the col in the lower right hand corner to know which position you re at (number is for the position to the RIGHT of the cursor) It s often easier to add indels (inserts or deletions) last, since they ll change positon numbers for the rest of the FASTAs
5 Multiple Sequence Alignment Once all the sequences have been made, it s necessary to align and compare them. Since we ve used the same reference as the basis for inputting variants, alignment isn t going to be difficult, but the alignment program will highlight which nucleotides differ between samples and allows us to quantify the differences. 1. Go to Clustal Omega ( an online multiple sequence alignment tool 2. Input all the FASTAs you want to align 3. Make sure it s set to align DNA and click submit 4. Once it s finished aligning: a. Automatically brings up the alignment page i. Nucleotides with perfect matches are marked by * ii. Gaps which have been added are marked as 5. Go to the Results Summary tab and click the Percent Identity Matrix a. Matrix indicates that the row and column headers are going to be the same. b. Take a moment to review the matrix. Be sure to note any obvious patterns, and their possible explanations, before continuing. c. The matrix can used directly in the browser, or copied into Excel i. Do not copy header rows (which begin with #) ii. Paste into Excel, each row will now be in a single cell iii. Highlight the cells, navigate to the data tab and select Text to Columns in the Data Tools box. iv. Follow the prompts using Fixed Width as the data type 6. Use the percent identity matrix to identify most and least similar matches for each example sample Other useful tools on Clustal Omega Clustal Omega uses the presence and absence of mutations to predict how closely samples are related, since samples containing the same mutation are likely to have inherited it through a common ancestor. The Phylogenetic Tree tab displays this prediction similarly to how you may represent your immediate relatives on a family tree.
Intro to NGS Tutorial
 Intro to NGS Tutorial Release 8.6.0 Golden Helix, Inc. October 31, 2016 Contents 1. Overview 2 2. Import Variants and Quality Fields 3 3. Quality Filters 10 Generate Alternate Read Ratio.........................................
Intro to NGS Tutorial Release 8.6.0 Golden Helix, Inc. October 31, 2016 Contents 1. Overview 2 2. Import Variants and Quality Fields 3 3. Quality Filters 10 Generate Alternate Read Ratio.........................................
NGS Data Analysis. Roberto Preste
 NGS Data Analysis Roberto Preste 1 Useful info http://bit.ly/2r1y2dr Contacts: roberto.preste@gmail.com Slides: http://bit.ly/ngs-data 2 NGS data analysis Overview 3 NGS Data Analysis: the basic idea http://bit.ly/2r1y2dr
NGS Data Analysis Roberto Preste 1 Useful info http://bit.ly/2r1y2dr Contacts: roberto.preste@gmail.com Slides: http://bit.ly/ngs-data 2 NGS data analysis Overview 3 NGS Data Analysis: the basic idea http://bit.ly/2r1y2dr
Performing whole genome SNP analysis with mapping performed locally
 BioNumerics Tutorial: Performing whole genome SNP analysis with mapping performed locally 1 Introduction 1.1 An introduction to whole genome SNP analysis A Single Nucleotide Polymorphism (SNP) is a variation
BioNumerics Tutorial: Performing whole genome SNP analysis with mapping performed locally 1 Introduction 1.1 An introduction to whole genome SNP analysis A Single Nucleotide Polymorphism (SNP) is a variation
Analyzing Variant Call results using EuPathDB Galaxy, Part II
 Analyzing Variant Call results using EuPathDB Galaxy, Part II In this exercise, we will work in groups to examine the results from the SNP analysis workflow that we started yesterday. The first step is
Analyzing Variant Call results using EuPathDB Galaxy, Part II In this exercise, we will work in groups to examine the results from the SNP analysis workflow that we started yesterday. The first step is
Finding Selection in All the Right Places TA Notes and Key Lab 9
 Objectives: Finding Selection in All the Right Places TA Notes and Key Lab 9 1. Use published genome data to look for evidence of selection in individual genes. 2. Understand the need for DNA sequence
Objectives: Finding Selection in All the Right Places TA Notes and Key Lab 9 1. Use published genome data to look for evidence of selection in individual genes. 2. Understand the need for DNA sequence
User Guide. v Released June Advaita Corporation 2016
 User Guide v. 0.9 Released June 2016 Copyright Advaita Corporation 2016 Page 2 Table of Contents Table of Contents... 2 Background and Introduction... 4 Variant Calling Pipeline... 4 Annotation Information
User Guide v. 0.9 Released June 2016 Copyright Advaita Corporation 2016 Page 2 Table of Contents Table of Contents... 2 Background and Introduction... 4 Variant Calling Pipeline... 4 Annotation Information
MIRING: Minimum Information for Reporting Immunogenomic NGS Genotyping. Data Standards Hackathon for NGS HACKATHON 1.0 Bethesda, MD September
 MIRING: Minimum Information for Reporting Immunogenomic NGS Genotyping Data Standards Hackathon for NGS HACKATHON 1.0 Bethesda, MD September 27 2014 Static Dynamic Static Minimum Information for Reporting
MIRING: Minimum Information for Reporting Immunogenomic NGS Genotyping Data Standards Hackathon for NGS HACKATHON 1.0 Bethesda, MD September 27 2014 Static Dynamic Static Minimum Information for Reporting
University of North Dakota PeopleSoft Finance Tip Sheets. Utilizing the Query Download Feature
 There is a custom feature available in Query Viewer that allows files to be created from queries and copied to a user s PC. This feature doesn t have the same size limitations as running a query to HTML
There is a custom feature available in Query Viewer that allows files to be created from queries and copied to a user s PC. This feature doesn t have the same size limitations as running a query to HTML
Differential Expression Analysis at PATRIC
 Differential Expression Analysis at PATRIC The following step- by- step workflow is intended to help users learn how to upload their differential gene expression data to their private workspace using Expression
Differential Expression Analysis at PATRIC The following step- by- step workflow is intended to help users learn how to upload their differential gene expression data to their private workspace using Expression
BaseSpace Variant Interpreter Release Notes
 Document ID: EHAD_RN_010220118_0 Release Notes External v.2.4.1 (KN:v1.2.24) Release Date: Page 1 of 7 BaseSpace Variant Interpreter Release Notes BaseSpace Variant Interpreter v2.4.1 FOR RESEARCH USE
Document ID: EHAD_RN_010220118_0 Release Notes External v.2.4.1 (KN:v1.2.24) Release Date: Page 1 of 7 BaseSpace Variant Interpreter Release Notes BaseSpace Variant Interpreter v2.4.1 FOR RESEARCH USE
Lession #5: Adding a New Salary Using Batch Uploads
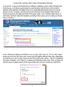 Lession #5: Adding a New Salary Using Batch Uploads In Lesson #4, we discussed the function of adding or updating a salary online through IWAS. In this lesson, we ll discuss performing the same function,
Lession #5: Adding a New Salary Using Batch Uploads In Lesson #4, we discussed the function of adding or updating a salary online through IWAS. In this lesson, we ll discuss performing the same function,
User Manual. Ver. 3.0 March 19, 2012
 User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
Tutorial: chloroplast genomes
 Tutorial: chloroplast genomes Stacia Wyman Department of Computer Sciences Williams College Williamstown, MA 01267 March 10, 2005 ASSUMPTIONS: You are using Internet Explorer under OS X on the Mac. You
Tutorial: chloroplast genomes Stacia Wyman Department of Computer Sciences Williams College Williamstown, MA 01267 March 10, 2005 ASSUMPTIONS: You are using Internet Explorer under OS X on the Mac. You
Tutorial 4 BLAST Searching the CHO Genome
 Tutorial 4 BLAST Searching the CHO Genome Accessing the CHO Genome BLAST Tool The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting BLAST from the menu bar
Tutorial 4 BLAST Searching the CHO Genome Accessing the CHO Genome BLAST Tool The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting BLAST from the menu bar
Instructions: DRDP Online Child Upload
 Instructions: DRDP Online Child Upload Version 1.00 10/29/2018 Table of Contents Introduction... 2 Upload Roles... 2 Upload Template... 2 Entering Data into the Template... 3 Uploading to DRDP Online...
Instructions: DRDP Online Child Upload Version 1.00 10/29/2018 Table of Contents Introduction... 2 Upload Roles... 2 Upload Template... 2 Entering Data into the Template... 3 Uploading to DRDP Online...
MiSeq Reporter TruSight Tumor 15 Workflow Guide
 MiSeq Reporter TruSight Tumor 15 Workflow Guide For Research Use Only. Not for use in diagnostic procedures. Introduction 3 TruSight Tumor 15 Workflow Overview 4 Reports 8 Analysis Output Files 9 Manifest
MiSeq Reporter TruSight Tumor 15 Workflow Guide For Research Use Only. Not for use in diagnostic procedures. Introduction 3 TruSight Tumor 15 Workflow Overview 4 Reports 8 Analysis Output Files 9 Manifest
CircosVCF workshop, TAU, 9/11/2017
 CircosVCF exercise In this exercise, we will create and design circos plots using CircosVCF. We will use vcf files of a published case "X-linked elliptocytosis with impaired growth is related to mutated
CircosVCF exercise In this exercise, we will create and design circos plots using CircosVCF. We will use vcf files of a published case "X-linked elliptocytosis with impaired growth is related to mutated
mtdna Variant Processor v1.0 BaseSpace App Guide
 mtdna Variant Processor v1.0 BaseSpace App Guide For Research Use Only. Not for use in diagnostic procedures. Introduction 3 Workflow Diagram 4 Workflow 5 Log In to BaseSpace 6 Set Analysis Parameters
mtdna Variant Processor v1.0 BaseSpace App Guide For Research Use Only. Not for use in diagnostic procedures. Introduction 3 Workflow Diagram 4 Workflow 5 Log In to BaseSpace 6 Set Analysis Parameters
Minimum Information for Reporting Immunogenomic NGS Genotyping (MIRING)
 Minimum Information for Reporting Immunogenomic NGS Genotyping (MIRING) Reporting guideline statement for HLA and KIR genotyping data generated via Next Generation Sequencing (NGS) technologies and analysis
Minimum Information for Reporting Immunogenomic NGS Genotyping (MIRING) Reporting guideline statement for HLA and KIR genotyping data generated via Next Generation Sequencing (NGS) technologies and analysis
INTRODUCTION AUX FORMATS DE FICHIERS
 INTRODUCTION AUX FORMATS DE FICHIERS Plan. Formats de séquences brutes.. Format fasta.2. Format fastq 2. Formats d alignements 2.. Format SAM 2.2. Format BAM 4. Format «Variant Calling» 4.. Format Varscan
INTRODUCTION AUX FORMATS DE FICHIERS Plan. Formats de séquences brutes.. Format fasta.2. Format fastq 2. Formats d alignements 2.. Format SAM 2.2. Format BAM 4. Format «Variant Calling» 4.. Format Varscan
Click on "+" button Select your VCF data files (see #Input Formats->1 above) Remove file from files list:
 CircosVCF: CircosVCF is a web based visualization tool of genome-wide variant data described in VCF files using circos plots. The provided visualization capabilities, gives a broad overview of the genomic
CircosVCF: CircosVCF is a web based visualization tool of genome-wide variant data described in VCF files using circos plots. The provided visualization capabilities, gives a broad overview of the genomic
Advanced UCSC Browser Functions
 Advanced UCSC Browser Functions Dr. Thomas Randall tarandal@email.unc.edu bioinformatics.unc.edu UCSC Browser: genome.ucsc.edu Overview Custom Tracks adding your own datasets Utilities custom tools for
Advanced UCSC Browser Functions Dr. Thomas Randall tarandal@email.unc.edu bioinformatics.unc.edu UCSC Browser: genome.ucsc.edu Overview Custom Tracks adding your own datasets Utilities custom tools for
Simulation of Molecular Evolution with Bioinformatics Analysis
 Simulation of Molecular Evolution with Bioinformatics Analysis Barbara N. Beck, Rochester Community and Technical College, Rochester, MN Project created by: Barbara N. Beck, Ph.D., Rochester Community
Simulation of Molecular Evolution with Bioinformatics Analysis Barbara N. Beck, Rochester Community and Technical College, Rochester, MN Project created by: Barbara N. Beck, Ph.D., Rochester Community
Importing Career Standards Benchmark Scores
 Importing Career Standards Benchmark Scores The Career Standards Benchmark assessments that are reported on the PIMS Student Fact Template for Career Standards Benchmarks can be imported en masse using
Importing Career Standards Benchmark Scores The Career Standards Benchmark assessments that are reported on the PIMS Student Fact Template for Career Standards Benchmarks can be imported en masse using
Helpful Galaxy screencasts are available at:
 This user guide serves as a simplified, graphic version of the CloudMap paper for applicationoriented end-users. For more details, please see the CloudMap paper. Video versions of these user guides and
This user guide serves as a simplified, graphic version of the CloudMap paper for applicationoriented end-users. For more details, please see the CloudMap paper. Video versions of these user guides and
INTRODUCTION TO BIOINFORMATICS
 Molecular Biology-2019 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Molecular Biology-2019 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Tutorial. Find Very Low Frequency Variants With QIAGEN GeneRead Panels. Sample to Insight. November 21, 2017
 Find Very Low Frequency Variants With QIAGEN GeneRead Panels November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Find Very Low Frequency Variants With QIAGEN GeneRead Panels November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Biostatistics and Bioinformatics Molecular Sequence Databases
 . 1 Description of Module Subject Name Paper Name Module Name/Title 13 03 Dr. Vijaya Khader Dr. MC Varadaraj 2 1. Objectives: In the present module, the students will learn about 1. Encoding linear sequences
. 1 Description of Module Subject Name Paper Name Module Name/Title 13 03 Dr. Vijaya Khader Dr. MC Varadaraj 2 1. Objectives: In the present module, the students will learn about 1. Encoding linear sequences
BIR pipeline steps and subsequent output files description STEP 1: BLAST search
 Lifeportal (Brief description) The Lifeportal at University of Oslo (https://lifeportal.uio.no) is a Galaxy based life sciences portal lifeportal.uio.no under the UiO tools section for phylogenomic analysis,
Lifeportal (Brief description) The Lifeportal at University of Oslo (https://lifeportal.uio.no) is a Galaxy based life sciences portal lifeportal.uio.no under the UiO tools section for phylogenomic analysis,
RNAseq analysis: SNP calling. BTI bioinformatics course, spring 2013
 RNAseq analysis: SNP calling BTI bioinformatics course, spring 2013 RNAseq overview RNAseq overview Choose technology 454 Illumina SOLiD 3 rd generation (Ion Torrent, PacBio) Library types Single reads
RNAseq analysis: SNP calling BTI bioinformatics course, spring 2013 RNAseq overview RNAseq overview Choose technology 454 Illumina SOLiD 3 rd generation (Ion Torrent, PacBio) Library types Single reads
Tutorial. Batching of Multi-Input Workflows. Sample to Insight. November 21, 2017
 Batching of Multi-Input Workflows November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Batching of Multi-Input Workflows November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
Introduction to Bioinformatics Online Course: IBT
 Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec2 Choosing the Right Sequences Choosing the Right Sequences Before you build your alignment,
Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec2 Choosing the Right Sequences Choosing the Right Sequences Before you build your alignment,
How to Edit Your Website
 How to Edit Your Website A guide to using your Content Management System Overview 2 Accessing the CMS 2 Choosing Your Language 2 Resetting Your Password 3 Sites 4 Favorites 4 Pages 5 Creating Pages 5 Managing
How to Edit Your Website A guide to using your Content Management System Overview 2 Accessing the CMS 2 Choosing Your Language 2 Resetting Your Password 3 Sites 4 Favorites 4 Pages 5 Creating Pages 5 Managing
Proteome Comparison: A fine-grained tool for comparative genomics
 Proteome Comparison: A fine-grained tool for comparative genomics In addition to the Protein Family Sorter that allows researchers to examine up to the protein families from up to 500 genomes at a time,
Proteome Comparison: A fine-grained tool for comparative genomics In addition to the Protein Family Sorter that allows researchers to examine up to the protein families from up to 500 genomes at a time,
2. Take a few minutes to look around the site. The goal is to familiarize yourself with a few key components of the NCBI.
 2 Navigating the NCBI Instructions Aim: To become familiar with the resources available at the National Center for Bioinformatics (NCBI) and the search engine Entrez. Instructions: Write the answers to
2 Navigating the NCBI Instructions Aim: To become familiar with the resources available at the National Center for Bioinformatics (NCBI) and the search engine Entrez. Instructions: Write the answers to
How to Edit Your Website
 How to Edit Your Website A guide to using your Content Management System Overview 2 Accessing the CMS 2 Choosing Your Language 2 Resetting Your Password 3 Sites 4 Favorites 4 Pages 5 Creating Pages 5 Managing
How to Edit Your Website A guide to using your Content Management System Overview 2 Accessing the CMS 2 Choosing Your Language 2 Resetting Your Password 3 Sites 4 Favorites 4 Pages 5 Creating Pages 5 Managing
Browser Exercises - I. Alignments and Comparative genomics
 Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Briefly: Bioinformatics File Formats. J Fass September 2018
 Briefly: Bioinformatics File Formats J Fass September 2018 Overview ASCII Text Sequence Fasta, Fastq ~Annotation TSV, CSV, BED, GFF, GTF, VCF, SAM Binary (Data, Compressed, Executable) Data HDF5 BAM /
Briefly: Bioinformatics File Formats J Fass September 2018 Overview ASCII Text Sequence Fasta, Fastq ~Annotation TSV, CSV, BED, GFF, GTF, VCF, SAM Binary (Data, Compressed, Executable) Data HDF5 BAM /
Excel Basics Rice Digital Media Commons Guide Written for Microsoft Excel 2010 Windows Edition by Eric Miller
 Excel Basics Rice Digital Media Commons Guide Written for Microsoft Excel 2010 Windows Edition by Eric Miller Table of Contents Introduction!... 1 Part 1: Entering Data!... 2 1.a: Typing!... 2 1.b: Editing
Excel Basics Rice Digital Media Commons Guide Written for Microsoft Excel 2010 Windows Edition by Eric Miller Table of Contents Introduction!... 1 Part 1: Entering Data!... 2 1.a: Typing!... 2 1.b: Editing
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014
 Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
CLC Server. End User USER MANUAL
 CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
Business Online TM. Positive Pay - Adding Issued Items. Quick Reference Guide
 Business Online TM Positive Pay - Adding Issued Items Quick Reference Guide Positive Pay Adding Issued Items Manually or Using Templates Positive Pay is a risk management solution that provides the ability
Business Online TM Positive Pay - Adding Issued Items Quick Reference Guide Positive Pay Adding Issued Items Manually or Using Templates Positive Pay is a risk management solution that provides the ability
Importing and Merging Data Tutorial
 Importing and Merging Data Tutorial Release 1.0 Golden Helix, Inc. February 17, 2012 Contents 1. Overview 2 2. Import Pedigree Data 4 3. Import Phenotypic Data 6 4. Import Genetic Data 8 5. Import and
Importing and Merging Data Tutorial Release 1.0 Golden Helix, Inc. February 17, 2012 Contents 1. Overview 2 2. Import Pedigree Data 4 3. Import Phenotypic Data 6 4. Import Genetic Data 8 5. Import and
Business Process Procedures
 Business Process Procedures 14.40 MICROSOFT EXCEL TIPS Overview These procedures document some helpful hints and tricks while using Microsoft Excel. Key Points This document will explore the following:
Business Process Procedures 14.40 MICROSOFT EXCEL TIPS Overview These procedures document some helpful hints and tricks while using Microsoft Excel. Key Points This document will explore the following:
Creating and Using Genome Assemblies Tutorial
 Creating and Using Genome Assemblies Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Create a Genome Assembly for Danio rerio 2 2. Building Annotation Sources 5 A. Creating a Reference
Creating and Using Genome Assemblies Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Create a Genome Assembly for Danio rerio 2 2. Building Annotation Sources 5 A. Creating a Reference
Sequence Genotyper Reference Guide
 Sequence Genotyper Reference Guide For Research Use Only. Not for use in diagnostic procedures. Introduction 3 Installation 4 Dashboard Overview 5 Projects 6 Targets 7 Samples 9 Reports 12 Revision History
Sequence Genotyper Reference Guide For Research Use Only. Not for use in diagnostic procedures. Introduction 3 Installation 4 Dashboard Overview 5 Projects 6 Targets 7 Samples 9 Reports 12 Revision History
AgroMarker Finder manual (1.1)
 AgroMarker Finder manual (1.1) 1. Introduction 2. Installation 3. How to run? 4. How to use? 5. Java program for calculating of restriction enzyme sites (TaqαI). 1. Introduction AgroMarker Finder (AMF)is
AgroMarker Finder manual (1.1) 1. Introduction 2. Installation 3. How to run? 4. How to use? 5. Java program for calculating of restriction enzyme sites (TaqαI). 1. Introduction AgroMarker Finder (AMF)is
QIAseq DNA V3 Panel Analysis Plugin USER MANUAL
 QIAseq DNA V3 Panel Analysis Plugin USER MANUAL User manual for QIAseq DNA V3 Panel Analysis 1.0.1 Windows, Mac OS X and Linux January 25, 2018 This software is for research purposes only. QIAGEN Aarhus
QIAseq DNA V3 Panel Analysis Plugin USER MANUAL User manual for QIAseq DNA V3 Panel Analysis 1.0.1 Windows, Mac OS X and Linux January 25, 2018 This software is for research purposes only. QIAGEN Aarhus
Welcome to MAPHiTS (Mapping Analysis Pipeline for High-Throughput Sequences) tutorial page.
 Welcome to MAPHiTS (Mapping Analysis Pipeline for High-Throughput Sequences) tutorial page. In this page you will learn to use the tools of the MAPHiTS suite. A little advice before starting : rename your
Welcome to MAPHiTS (Mapping Analysis Pipeline for High-Throughput Sequences) tutorial page. In this page you will learn to use the tools of the MAPHiTS suite. A little advice before starting : rename your
CEU Online System, The Friday Center for Continuing Education, UNC-Chapel Hill How to Obtain Participant IDs for Awarding of CEUs
 The Friday Center for Continuing Education has the responsibility of approving continuing education activities for which CEUs are recorded and maintained as a permanent record for individual participants.
The Friday Center for Continuing Education has the responsibility of approving continuing education activities for which CEUs are recorded and maintained as a permanent record for individual participants.
Tutorial for Windows and Macintosh SNP Hunting
 Tutorial for Windows and Macintosh SNP Hunting 2010 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh SNP Hunting 2010 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Microsoft Office 365 includes the entire Office Suite (Word, Excel, PowerPoint, Access, Publisher, Lync, Outlook, etc ) and an OneDrive account.
 Microsoft Office 365 Contents What is Office 365?... 2 What is OneDrive?... 2 What if you already have a Microsoft Account?... 2 Download Office for FREE... 3 How to Access OneDrive... 4 Office Online...
Microsoft Office 365 Contents What is Office 365?... 2 What is OneDrive?... 2 What if you already have a Microsoft Account?... 2 Download Office for FREE... 3 How to Access OneDrive... 4 Office Online...
MiniBase Workbook. Schoolwires Centricity2
 MiniBase Workbook Schoolwires Centricity2 Table of Contents Introduction... 1 Create a New MiniBase... 2 Add Records to the MiniBase:... 3 Add Records One at a Time... 3 Import Records:... 4 Deploy the
MiniBase Workbook Schoolwires Centricity2 Table of Contents Introduction... 1 Create a New MiniBase... 2 Add Records to the MiniBase:... 3 Add Records One at a Time... 3 Import Records:... 4 Deploy the
Tutorial: How to use the Wheat TILLING database
 Tutorial: How to use the Wheat TILLING database Last Updated: 9/7/16 1. Visit http://dubcovskylab.ucdavis.edu/wheat_blast to go to the BLAST page or click on the Wheat BLAST button on the homepage. 2.
Tutorial: How to use the Wheat TILLING database Last Updated: 9/7/16 1. Visit http://dubcovskylab.ucdavis.edu/wheat_blast to go to the BLAST page or click on the Wheat BLAST button on the homepage. 2.
FrontPage 98 Quick Guide. Copyright 2000 Peter Pappas. edteck press All rights reserved.
 Master web design skills with Microsoft FrontPage 98. This step-by-step guide uses over 40 full color close-up screen shots to clearly explain the fast and easy way to design a web site. Use edteck s QuickGuide
Master web design skills with Microsoft FrontPage 98. This step-by-step guide uses over 40 full color close-up screen shots to clearly explain the fast and easy way to design a web site. Use edteck s QuickGuide
Using the GEMM Applications System. Signing in When you first access the GEMM portal, you will be presented with this login screen.
 Using the GEMM Applications System Signing in When you first access the GEMM portal, you will be presented with this login screen. If this is your first time accessing the GEMM portal, then click the registration
Using the GEMM Applications System Signing in When you first access the GEMM portal, you will be presented with this login screen. If this is your first time accessing the GEMM portal, then click the registration
Beginning Excel. Revised 4/19/16
 Beginning Excel Objectives: The Learner will: Become familiar with terminology used in Microsoft Excel Create a simple workbook Write a simple formula Formatting Cells Adding Columns Borders Table of Contents:
Beginning Excel Objectives: The Learner will: Become familiar with terminology used in Microsoft Excel Create a simple workbook Write a simple formula Formatting Cells Adding Columns Borders Table of Contents:
The software comes with 2 installers: (1) SureCall installer (2) GenAligners (contains BWA, BWA- MEM).
 Release Notes Agilent SureCall 4.0 Product Number G4980AA SureCall Client 6-month named license supports installation of one client and server (to host the SureCall database) on one machine. For additional
Release Notes Agilent SureCall 4.0 Product Number G4980AA SureCall Client 6-month named license supports installation of one client and server (to host the SureCall database) on one machine. For additional
Genome Browsers Guide
 Genome Browsers Guide Take a Class This guide supports the Galter Library class called Genome Browsers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
Genome Browsers Guide Take a Class This guide supports the Galter Library class called Genome Browsers. See our Classes schedule for the next available offering. If this class is not on our upcoming schedule,
Annotating a single sequence
 BioNumerics Tutorial: Annotating a single sequence 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn how
BioNumerics Tutorial: Annotating a single sequence 1 Aim The annotation application in BioNumerics has been designed for the annotation of coding regions on sequences. In this tutorial you will learn how
SPREADSHEETS. (Data for this tutorial at
 SPREADSHEETS (Data for this tutorial at www.peteraldhous.com/data) Spreadsheets are great tools for sorting, filtering and running calculations on tables of data. Journalists who know the basics can interview
SPREADSHEETS (Data for this tutorial at www.peteraldhous.com/data) Spreadsheets are great tools for sorting, filtering and running calculations on tables of data. Journalists who know the basics can interview
Tutorial. Variant Detection. Sample to Insight. November 21, 2017
 Resequencing: Variant Detection November 21, 2017 Map Reads to Reference and Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Resequencing: Variant Detection November 21, 2017 Map Reads to Reference and Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
The UCSC Gene Sorter, Table Browser & Custom Tracks
 The UCSC Gene Sorter, Table Browser & Custom Tracks Advanced searching and discovery using the UCSC Table Browser and Custom Tracks Osvaldo Graña Bioinformatics Unit, CNIO 1 Table Browser and Custom Tracks
The UCSC Gene Sorter, Table Browser & Custom Tracks Advanced searching and discovery using the UCSC Table Browser and Custom Tracks Osvaldo Graña Bioinformatics Unit, CNIO 1 Table Browser and Custom Tracks
Tour Guide for Windows and Macintosh
 Tour Guide for Windows and Macintosh 2011 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Suite 100A, Ann Arbor, MI 48108 USA phone 1.800.497.4939 or 1.734.769.7249 (fax) 1.734.769.7074
Tour Guide for Windows and Macintosh 2011 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Suite 100A, Ann Arbor, MI 48108 USA phone 1.800.497.4939 or 1.734.769.7249 (fax) 1.734.769.7074
Gegenees genome format...7. Gegenees comparisons...8 Creating a fragmented all-all comparison...9 The alignment The analysis...
 User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
Tutorial 1: Exploring the UCSC Genome Browser
 Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Getting Started. Copyright statement
 Getting Started Copyright statement Copyright 2001 Accelrys, a subsidiary of Pharmacopeia Inc. All rights reserved. This document contains proprietary information of Accelrys and its licensors. It is their
Getting Started Copyright statement Copyright 2001 Accelrys, a subsidiary of Pharmacopeia Inc. All rights reserved. This document contains proprietary information of Accelrys and its licensors. It is their
Put the Graphs for Each Health Plan on the Same Graph
 At the conclusion of the technology assignment on graphing the total annual cost, you had a graph of each of health insurance plans you are examining. In this technology assignment, you ll combine those
At the conclusion of the technology assignment on graphing the total annual cost, you had a graph of each of health insurance plans you are examining. In this technology assignment, you ll combine those
ChIP-seq (NGS) Data Formats
 ChIP-seq (NGS) Data Formats Biological samples Sequence reads SRA/SRF, FASTQ Quality control SAM/BAM/Pileup?? Mapping Assembly... DE Analysis Variant Detection Peak Calling...? Counts, RPKM VCF BED/narrowPeak/
ChIP-seq (NGS) Data Formats Biological samples Sequence reads SRA/SRF, FASTQ Quality control SAM/BAM/Pileup?? Mapping Assembly... DE Analysis Variant Detection Peak Calling...? Counts, RPKM VCF BED/narrowPeak/
A short Introduction to UCSC Genome Browser
 A short Introduction to UCSC Genome Browser Elodie Girard, Nicolas Servant Institut Curie/INSERM U900 Bioinformatics, Biostatistics, Epidemiology and computational Systems Biology of Cancer 1 Why using
A short Introduction to UCSC Genome Browser Elodie Girard, Nicolas Servant Institut Curie/INSERM U900 Bioinformatics, Biostatistics, Epidemiology and computational Systems Biology of Cancer 1 Why using
Uploading Journal Entries from Excel
 Uploading Journal Entries from Excel Using Excel, you may create journal entries for upload to the AS400. The uploaded entries may add to an existing batch of journal entries, overwrite an existing batch,
Uploading Journal Entries from Excel Using Excel, you may create journal entries for upload to the AS400. The uploaded entries may add to an existing batch of journal entries, overwrite an existing batch,
Tutorial: Resequencing Analysis using Tracks
 : Resequencing Analysis using Tracks September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : Resequencing
: Resequencing Analysis using Tracks September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : Resequencing
A manual for the use of mirvas
 A manual for the use of mirvas Authors: Sophia Cammaerts, Mojca Strazisar, Jenne Dierckx, Jurgen Del Favero, Peter De Rijk Version: 1.0.2 Date: July 27, 2015 Contact: peter.derijk@gmail.com, mirvas.software@gmail.com
A manual for the use of mirvas Authors: Sophia Cammaerts, Mojca Strazisar, Jenne Dierckx, Jurgen Del Favero, Peter De Rijk Version: 1.0.2 Date: July 27, 2015 Contact: peter.derijk@gmail.com, mirvas.software@gmail.com
Nevada Genomics Center
 Nevada Genomics Center These are general instructions on how to use dnatools to submit Sanger sequencing samples to be run on the ABI Prism 3730 DNA analyzer. We here at the Nevada Genomics Center feel
Nevada Genomics Center These are general instructions on how to use dnatools to submit Sanger sequencing samples to be run on the ABI Prism 3730 DNA analyzer. We here at the Nevada Genomics Center feel
- G T G T A C A C
 Name Student ID.. Sequence alignment 1. Globally align sequence V (GTGTACAC) and sequence W (GTACC) by hand using dynamic programming algorithm. The alignment will be performed based on match premium of
Name Student ID.. Sequence alignment 1. Globally align sequence V (GTGTACAC) and sequence W (GTACC) by hand using dynamic programming algorithm. The alignment will be performed based on match premium of
Orientation Assignment for Statistics Software (nothing to hand in) Mary Parker,
 Orientation to MINITAB, Mary Parker, mparker@austincc.edu. Last updated 1/3/10. page 1 of Orientation Assignment for Statistics Software (nothing to hand in) Mary Parker, mparker@austincc.edu When you
Orientation to MINITAB, Mary Parker, mparker@austincc.edu. Last updated 1/3/10. page 1 of Orientation Assignment for Statistics Software (nothing to hand in) Mary Parker, mparker@austincc.edu When you
INTRODUCTION TO BIOINFORMATICS
 Molecular Biology-2017 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Molecular Biology-2017 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Shopping Cart: Queries, Personalizations, Filters, and Settings
 Shopping Cart: Queries, Personalizations, Filters, and Settings on the Shopping Cart Home Page Use this Job Aid to: Learn how to organize the Shopping Cart home page so that it is easier to use. BEFORE
Shopping Cart: Queries, Personalizations, Filters, and Settings on the Shopping Cart Home Page Use this Job Aid to: Learn how to organize the Shopping Cart home page so that it is easier to use. BEFORE
Service Line Export and Pivot Table Report (Windows Excel 2010)
 Service Line Export and Pivot Table Report (Windows Excel 2010) In this tutorial, we will take the Service Lines of the Active Students only and only the most recent record to take a snapshot look at approximate
Service Line Export and Pivot Table Report (Windows Excel 2010) In this tutorial, we will take the Service Lines of the Active Students only and only the most recent record to take a snapshot look at approximate
Comparative Sequencing
 Tutorial for Windows and Macintosh Comparative Sequencing 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh Comparative Sequencing 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Using Adobe Contribute 4 A guide for new website authors
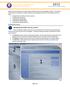 Using Adobe Contribute 4 A guide for new website authors Adobe Contribute allows you to easily update websites without any knowledge of HTML. This handout will provide an introduction to Adobe Contribute
Using Adobe Contribute 4 A guide for new website authors Adobe Contribute allows you to easily update websites without any knowledge of HTML. This handout will provide an introduction to Adobe Contribute
Importing from Blackboard Learn Grade Center Data to Banner 9 User Learning Scenarios
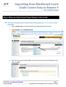 Importing from Blackboard Learn Grade Center Data to Banner 9 User Learning Scenarios Step 1: Make sure Final Grade Column Displays Letter Grade Ensure your final grade column in Grade Center has letter
Importing from Blackboard Learn Grade Center Data to Banner 9 User Learning Scenarios Step 1: Make sure Final Grade Column Displays Letter Grade Ensure your final grade column in Grade Center has letter
HOW TO Google Online Fillable Forms for Kiwanis Club Websites
 KIWANIS INTERNATIONAL HOW TO Google Online Fillable Forms for Kiwanis Club Websites Publish via email, website or blog Mandy Grover User instructions for creating free online fillable forms with Google
KIWANIS INTERNATIONAL HOW TO Google Online Fillable Forms for Kiwanis Club Websites Publish via email, website or blog Mandy Grover User instructions for creating free online fillable forms with Google
Shortest Path Algorithm
 Shortest Path Algorithm C Works just fine on this graph. C Length of shortest path = Copyright 2005 DIMACS BioMath Connect Institute Robert Hochberg Dynamic Programming SP #1 Same Questions, Different
Shortest Path Algorithm C Works just fine on this graph. C Length of shortest path = Copyright 2005 DIMACS BioMath Connect Institute Robert Hochberg Dynamic Programming SP #1 Same Questions, Different
Numbers Basics Website:
 Website: http://etc.usf.edu/te/ Numbers is Apple's new spreadsheet application. It is installed as part of the iwork suite, which also includes the word processing program Pages and the presentation program
Website: http://etc.usf.edu/te/ Numbers is Apple's new spreadsheet application. It is installed as part of the iwork suite, which also includes the word processing program Pages and the presentation program
Database Searching Using BLAST
 Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Computational Molecular Biology
 Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
Drupal Cloud Getting Started Guide Creating a Lab site with the MIT DLC Theme
 Introduction Drupal Cloud Getting Started Guide Creating a Lab site with the MIT DLC Theme In this Getting Started Guide, you can follow along as a website is built using the MIT DLC Theme. Whether you
Introduction Drupal Cloud Getting Started Guide Creating a Lab site with the MIT DLC Theme In this Getting Started Guide, you can follow along as a website is built using the MIT DLC Theme. Whether you
Assignment 7: Single-cell genomics. Bio /02/2018
 Assignment 7: Single-cell genomics Bio5488 03/02/2018 Assignment 7: Single-cell genomics Input Genotypes called from several exome-sequencing datasets derived from either bulk or small pools of cells (VCF
Assignment 7: Single-cell genomics Bio5488 03/02/2018 Assignment 7: Single-cell genomics Input Genotypes called from several exome-sequencing datasets derived from either bulk or small pools of cells (VCF
Sequence alignment algorithms
 Sequence alignment algorithms Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, February 23 rd 27 After this lecture, you can decide when to use local and global sequence alignments
Sequence alignment algorithms Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, February 23 rd 27 After this lecture, you can decide when to use local and global sequence alignments
Excel Intermediate
 Excel 2013 - Intermediate (103-124) Multiple Worksheets Quick Links Manipulating Sheets Pages EX16 EX17 Copying Worksheets Page EX337 Grouping Worksheets Pages EX330 EX332 Multi-Sheet Cell References Page
Excel 2013 - Intermediate (103-124) Multiple Worksheets Quick Links Manipulating Sheets Pages EX16 EX17 Copying Worksheets Page EX337 Grouping Worksheets Pages EX330 EX332 Multi-Sheet Cell References Page
4.1. Access the internet and log on to the UCSC Genome Bioinformatics Web Page (Figure 1-
 1. PURPOSE To provide instructions for finding rs Numbers (SNP database ID numbers) and increasing sequence length by utilizing the UCSC Genome Bioinformatics Database. 2. MATERIALS 2.1. Sequence Information
1. PURPOSE To provide instructions for finding rs Numbers (SNP database ID numbers) and increasing sequence length by utilizing the UCSC Genome Bioinformatics Database. 2. MATERIALS 2.1. Sequence Information
1 Introduction to Using Excel Spreadsheets
 Survey of Math: Excel Spreadsheet Guide (for Excel 2007) Page 1 of 6 1 Introduction to Using Excel Spreadsheets This section of the guide is based on the file (a faux grade sheet created for messing with)
Survey of Math: Excel Spreadsheet Guide (for Excel 2007) Page 1 of 6 1 Introduction to Using Excel Spreadsheets This section of the guide is based on the file (a faux grade sheet created for messing with)
Lab 8: Using POY from your desktop and through CIPRES
 Integrative Biology 200A University of California, Berkeley PRINCIPLES OF PHYLOGENETICS Spring 2012 Updated by Michael Landis Lab 8: Using POY from your desktop and through CIPRES In this lab we re going
Integrative Biology 200A University of California, Berkeley PRINCIPLES OF PHYLOGENETICS Spring 2012 Updated by Michael Landis Lab 8: Using POY from your desktop and through CIPRES In this lab we re going
Analyzing ChIP- Seq Data in Galaxy
 Analyzing ChIP- Seq Data in Galaxy Lauren Mills RISS ABSTRACT Step- by- step guide to basic ChIP- Seq analysis using the Galaxy platform. Table of Contents Introduction... 3 Links to helpful information...
Analyzing ChIP- Seq Data in Galaxy Lauren Mills RISS ABSTRACT Step- by- step guide to basic ChIP- Seq analysis using the Galaxy platform. Table of Contents Introduction... 3 Links to helpful information...
GenomeStudio Software Release Notes
 GenomeStudio Software 2009.2 Release Notes 1. GenomeStudio Software 2009.2 Framework... 1 2. Illumina Genome Viewer v1.5...2 3. Genotyping Module v1.5... 4 4. Gene Expression Module v1.5... 6 5. Methylation
GenomeStudio Software 2009.2 Release Notes 1. GenomeStudio Software 2009.2 Framework... 1 2. Illumina Genome Viewer v1.5...2 3. Genotyping Module v1.5... 4 4. Gene Expression Module v1.5... 6 5. Methylation
How to Create Student Accounts and Assignments
 How to Create Student Accounts and Assignments From the top navigation, select My Classes and click My Students Carolina Science Online will allow you to either create a single student account, one at
How to Create Student Accounts and Assignments From the top navigation, select My Classes and click My Students Carolina Science Online will allow you to either create a single student account, one at
SAM and VCF formats. UCD Genome Center Bioinformatics Core Tuesday 14 June 2016
 SAM and VCF formats UCD Genome Center Bioinformatics Core Tuesday 14 June 2016 File Format: SAM / BAM / CRAM! NEW http://samtools.sourceforge.net/ - deprecated! http://www.htslib.org/ - SAMtools 1.0 and
SAM and VCF formats UCD Genome Center Bioinformatics Core Tuesday 14 June 2016 File Format: SAM / BAM / CRAM! NEW http://samtools.sourceforge.net/ - deprecated! http://www.htslib.org/ - SAMtools 1.0 and
Positive Pay. Quick Start Set-up Guide
 Positive Pay Quick Start Set-up Guide 1-2-3 EASY STEPS TO FRAUD PROTECTION The following information will help you get up and running with Positive Pay quickly and easily. With Positive Pay, you provide
Positive Pay Quick Start Set-up Guide 1-2-3 EASY STEPS TO FRAUD PROTECTION The following information will help you get up and running with Positive Pay quickly and easily. With Positive Pay, you provide
Tutorial for Windows and Macintosh SNP Hunting
 Tutorial for Windows and Macintosh SNP Hunting 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh SNP Hunting 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
