Automated Data Pipelines & Intelligent Microscopy
|
|
|
- Ann Allen
- 5 years ago
- Views:
Transcription
1 Galaxy Workshop Freiburg University Automated Data Pipelines & Intelligent Microscopy RNAi Screening Facility BioQuant, Heidelberg University Jürgen Reymann
2 Scientific Infrastructure Automated Data Pipelines Intelligent Automated Imaging
3 BioQuant Center for Systems Biology Quantitative Analysis of Molecular and Cellular Biosystems Interaction of viral and cellular Systems Modeling Platform Technology Platform Large Scale Data Facility
4 Data Analytics Infrastructure Super resolution microscopy (>100 TB p.a.) Light sheet microscopy (>100 TB p.a.) hard drives 6 PB total capacity 500 TB p.a. Large Scale Data Facility M. Hemberger 5 PB p.a. 1 PB p.a. J. Eils, C. Lawerenz
5 RNAi Screening Facility Support
6 RNAi Solid Phase Reverse Transfection sirna mirna cdna complexed with tranfection solution Gene specific degradation of target mrna in the cells Loss of function phenotype INCENP -phenotype sirna spot distribution on cell array Spot diameter: 400µm Ziauddin J. and Sabatini D.M., Nature 411, (2001)
7 Assay Automation 8er head printing robot 384er well plates LabTeks (384er cell arrays) Cell arrays (whole genome) 384er head printing robot Erfle et al., JBS, 2008 Erfle et al., Nat. Prot. (2), 2007 Reymann et al., BioTech, 2009
8 Object Scanning 2D low resolution: Fully automated [ 10x/20x objective lens ] 1 scan field: 1-3s (1 channel auto-focusing) 1000 scan fields: 1 hour (3 channels auto-focusing stage moving) Wide field [ Olympus IX81 ScanR ] 3D high resolution: Fully automated [ 60x/63x objective lens ] 1 scan field: 30s (1 channel 30 optical sections auto-focusing) 1000 scan fields: 1 day (3 channels auto-focusing stage moving) Confocal [ Leica TCS SP5 ] 2D super resolution: Semi-automated [ 63x/100x objective lens ] 1 scan field: 3-10min (1 channel) 1000 scan fields: - Confocal [ Leica TCS SP5 ] Integrated: Wide field Super resolution
9 Screening Workflow & Timescales
10 Scientific Infrastructure Automated Data Pipelines Intelligent Automated Imaging
11 Biological Question Data Mining Assay Development Imaging Experiment Setup
12 Computing power Software platforms Algorithms Basic user availability Data Mining Biological Question Preparation methods Labeling strategies Fluorophores Parallel assays Assay Development Automation Correlative Throughput Resolution Feedback options Imaging Experiment Setup
13 Computing power Software platforms Algorithms Basic user availability Biological Question Preparation methods Labeling strategies Fluorophores Parallel assays Data Mining Increase of complexity Increase of information content Extraction / Interpretation of information Assay Development Automation Correlative Throughput Resolution Feedback options Imaging Experiment Setup
14 Computing power Software platforms Algorithms Basic user availability Biological Question Preparation methods Labeling strategies Fluorophores Parallel assays Data Mining Increase of complexity Increase of information content Extraction / Interpretation of information Assay Development Automation Correlative Throughput Resolution Feedback options Imaging Experiment Setup
15 Data Pipelines Screening Workflow :: Biological Assay Genome wide RNAi screening platform to identify host factors involved in HIV-1 (HCV, DV, AAV, PV) replication Boerner K. et al Biotechnol. J, 2010
16 Data Pipelines Information Workflow :: Data Mining Visualisation of raw data, meta data, intermediate and ultimate results Transfection protocol sirna library Sample preparation Assay automation Plate layouts Usability for basic user Data acquisition High-throughput High-content Correlative Data storage Image Processing Quality control Normalisation Segmentation Features Modular structure Statistics Normalisation (per plate/assay) Spatial normalisation Hit calling High-throughput capable, i.e. handling of large data sets, parallel computing, serverbased functionalities Bioinformatics Data integration Processes Pathways Interaction networks Extensive integrations Libraries, format handling, data manipulation, Screen meta data Screen meta data Image raw data Image meta data Analysis results Analysis results Analysis results Usability / Easy Integration of established algorithms Possibility to link to external data bases [ Visualisation (plate viewer, image viewer, plots) Tables Meta data ] Base Data
17 Data Pipelines Information Workflow :: Data Mining [ ] Visualisation of raw data, meta data, intermediate and ultimate results Transfection protocol sirna library Sample preparation Assay automation Plate layouts Usability for basic user Data acquisition High-throughput High-content Correlative Data storage Image Processing Quality control Normalisation Segmentation Features Modular structure Statistics Normalisation (per plate/assay) Spatial normalisation Hit calling High-throughput capable, i.e. handling of large data sets, parallel computing, serverbased functionalities Bioinformatics Data integration Processes Pathways Interaction networks Extensive integrations Libraries, format handling, data manipulation, Screen meta data Screen meta data Image raw data Image meta data Analysis results Analysis results Analysis results Usability / Easy Integration of established algorithms Possibility to link to external data bases [ Visualisation (plate viewer, image viewer, plots) Tables Meta data ] Base Data
18 Data Pipelines Information Workflow :: Data Mining [ ] Modular structure Visualisation of raw data, meta data, intermediate and ultimate results Usability for basic user High-throughput capable, i.e. handling of large data sets, parallel computing, serverbased functionalities Open-source / Platform independant Extensive integrations Libraries, format handling, data manipulation, Sustainability Supported by a large (developer) community / Usage of universal standards Usability / Easy Integration of established algorithms Possibility to link to external data bases
19 Data Pipelines Information Workflow :: Data Mining Integrations / Libraries Data Handling / Data Views ImageJ / ImageJ2 ImgLib2 (Fiji) WEKA R SVM s Clustering Classifiers Network Mining Normalisations Microscope Control Target identification FeedBack Data Base File reader / writer BioFormats (image data) OME-XML (image data) XML support Histograms / Plots Image / Table / Plate viewer Data manipulation Loop control OMERO
20 Data Pipelines Information Workflow :: Data Mining Workflow Repository Workflow Configuration Node Description Node Repository
21 Data Pipelines Information Workflow :: Data Mining Workflow Configuration Nuclei registration
22 Data Pipelines Information Workflow :: Data Mining Workflow Configuration Nuclei registration
23 Data Pipelines Information Workflow :: Data Mining Workflow Configuration Nuclei registration
24 Data Pipelines Information Workflow :: Data Mining Workflow Configuration Nuclei registration
25 Data Pipelines Information Workflow :: Data Mining Workflow Configuration Nuclei registration 02/10/2014 Galaxy Workshop Freiburg University
26 Data Pipelines
27 Data Pipelines Experiment Meta Data 96er head pipetting robot Well plates Printing robots 384 cell arrays (LabTeks) Whole genome cell arrays
28 Data Pipelines Image Raw Data Wide field [ 3x Olympus IX81 ScanR ] Confocal [ 2x (exp.) Leica TCS SP5 ] Integrated: Wide field Super resolution Spinning disc [ Perkin Elmer Opera LX ]
29 Data Pipelines Image Processing 2D high-throughput images Nuclei - NO phenotypic penetration Wide field Object-registration
30 Data Pipelines Statistics 2D high-throughput images Nuclei - NO phenotypic penetration sirna INCENP phenotypes Wide field Object-registration Hit-calling
31 Data Pipelines Statistics n = 8 Rel. distance of object to INCENP feature space Positive classified (Rel. to Incenp feature space) Negative classified (Rel. to Incenp feature space) Object
32 Data Pipelines Statistics n = 8 Rel. distance of object to INCENP feature space Positive classified (Rel. to Incenp feature space) n = 32 Erb n = 35 KIF11 n = 8 INCENP n = 35 PLK Negative classified (Rel. to Incenp feature space) Object
33 Data Pipelines Workflow Repository
34 Scientific Infrastructure Automated Data Pipelines Intelligent Automated Imaging
35 Computing power Software platforms Algorithms Basic user availability Data Mining Biological Question Preparation methods Labeling strategies Fluorophores Parallel assays Assay Development Automation Correlative Throughput Resolution Feedback options Imaging Experiment Setup
36 Computing power Software platforms Algorithms Basic user availability Data Mining Biological Question Preparation methods Labeling strategies Fluorophores Parallel assays Assay Development integrated Data Analysis Automation Correlative Throughput Resolution Feedback options i Imaging Experiment Setup
37 Computing power Software platforms Algorithms Basic user availability Data Mining Biological Question Preparation methods Labeling strategies Fluorophores Parallel assays Assay Development integrated Data Analysis Automation Correlative Throughput Resolution Feedback options i Imaging Experiment Setup
38 General Considerations Why Automated DAQ? Screening Large scale experiment number (up to genome wide) e.g.: human genes *3 sirnas = experiments Automated DAQ in general Automation NOT necessarily restricted to screening procedures Biological variability Increase amount of data / # Objects to be scanned Statistical relevance Reproducibility Increase image quality (e.g. quality controls) Increase information content (e.g. correlative microscopy / time lapse) Simplify data acquisition procedures
39 General Considerations Why Automated DAQ? Screening Large scale experiment number (up to genome wide) e.g.: human genes *3 sirnas = experiments Junk data Automated DAQ in general Automation NOT necessarily restricted to screening procedures Biological variability Increase amount of data / # Objects to be scanned Statistical relevance Reproducibility Increase image quality (e.g. quality controls) Increase information content (e.g. correlative microscopy / time lapse) Simplify data acquisition procedures Junk data Hits
40 General Considerations FeedBack -driven DAQ n-dim feature space of observables [ cells ]
41 General Considerations FeedBack -driven DAQ n-dim feature space of observables [ cells ] Shape Intensity Osterwald S. et al, Biotechnology J Co-localisations
42 General Considerations FeedBack -driven DAQ n-dim feature space of observables [ cells ] Shape PROBLEM: General measurement provides cloud of data points containing every feature! Intensity BUT: Every measurement aims at collecting the information content of specified features. Osterwald S. et al, Biotechnology J Co-localisations
43 General Considerations FeedBack -driven DAQ n-dim feature space of observables [ cells ] Shape PROBLEM: General measurement provides cloud of data points containing every feature! Intensity BUT: Every measurement aims at collecting the information content of specified features. Co-localisations Pre-Processing Identify objects of interest FeedBack to measuring system Acquire only objects of interest
44 General Considerations FeedBack -driven DAQ n-dim feature space of observables [ cells ] Shape Pure screening time: 6 months PROBLEM: Junk data: General measurement ~40% provides cloud of data points containing every feature! Intensity BUT: Every measurement aims at collecting the information content of specified features. Osterwald Osterwald S. et al, S. et Biotechnology al, J. J Co-localisations objects of interest Pre-Processing Identify objects of interest FeedBack to measuring system Acquire only objects of interest
45 Automated Imaging Combine/Link DAQ IP Transfection protocol Sample preparation Data acquisition Image Processing Statistics Bioinformatics sirna library Assay automation Plate layouts High-throughput High-content Correlative Data storage Quality control Normalisation Segmentation Features Normalisation (per plate/assay) Spatial normalisation Hit calling Data integration Processes Pathways Interaction networks Use information from Image Pre- Processing in order to optimise / upgrade DAQ
46 Automated Imaging Combine/Link DAQ IP Transfection protocol Sample preparation Data acquisition Image Processing Statistics Bioinformatics sirna library Assay automation Plate layouts High-throughput High-content Correlative Data storage Quality control Normalisation Segmentation Features Normalisation (per plate/assay) Spatial normalisation Hit calling Data integration Processes Pathways Interaction networks Use information from Image Pre- Processing in order to optimise / upgrade DAQ Junk data Costs Feasibility of experiments Bleaching Trigger (time lapse experiments) Pick only objects of interest Quality / Information content of image raw data Screening time Pick representative cells for high/super -resolution
47 Automated Imaging Data acquisition High-throughput High-content Correlative Data storage Image Pre-Processing Targets Trigger Hit calling Image Processing Quality control Normalisation Segmentation Features
48 Automated Imaging Data acquisition High-throughput High-content Correlative Data storage Image Pre-Processing Targets Trigger Hit calling Objects of interest Image Processing Quality control Normalisation Segmentation Features
49 Scientific Infrastructure Data Pipelines & Data Analysis Cross-Platform Solution
50 Cross-Platform Solution Idea: Acquire images of the same sample/target at multiple systems, in order to combine the advantages of different microscopic techniques. Examples:
51 Cross-Platform Solution Problem: Transfer System Coordinate transfer by 1.) Reference markers 2.) (SIFT) image registration
52 Cross-Platform Solution Problem: Transfer System Coordinate transfer by 1.) Reference markers 2.) (SIFT) image registration High-resolution Leica SP5 10 µm Image Processing Target identification Super-resolution dstorm 1 µm dstorm setup: Mike Heilemann Gunkel et al., Histochem & Cell Biol 2014
53 Cross-Platform Solution Integrated Correlative Microscopy High-throughput screening [ Target identification ] High-resolution screening Super-resolution screening C Collab.: Mike Heilemann, Fred Hamprecht, Vytaute Starkuviene Flottmann et al., Biotechniques 2013
54 Scientific Infrastructure Data Pipelines & Data Analysis Automated Microscopy
55 Automated Microscopy Leica Microsystems Computer Aided Microscopy (CAM) -Interface Enables to communicate with the microscope control software [ Retrieve / Send commands ] CAM Data storage (Pre-Screen) Image raw data Targets / Trigger Collab.:
56 Automated Microscopy Integrated Data Pipelines Collab.:
57 Automated Microscopy Integrated Data Pipelines Pre-Screen Configuration 1 2D low resolution imaging of full sample CAM Collab.:
58 Automated Microscopy Integrated Data Pipelines Pre-Screen Configuration 1 2D low resolution imaging of full sample CAM Image Pre-Processing Identify objects of interest (candidates) Collab.:
59 Automated Microscopy Integrated Data Pipelines Pre-Screen Configuration 1 2D low resolution imaging of full sample CAM Image Pre-Processing Identify objects of interest (candidates) Collab.:
60 Automated Microscopy Integrated Data Pipelines Target Screen Configuration 2 3D high resolution imaging of candidates CAM High resolution 3D information of sub-nuclear objects Collab.:
61
62
63
64
65
66
67
68
69
70
71
72
73 Automated Microscopy Highly accurate positioning of targets in the center of field of views Experiment setup Overall sample size: 8 * 12 = 96 scan fields Standard DAQ Config 2» Screening time: ~70h» junk data / less targets (NOT centered!) Automated Imaging Pre-Screen job1 Target Screen job2» Screening time: ~7h» 146 target regions (in center of field of view!) Overall acquisition time: 7 h 17 min Decreased overall screening time (factor 10) Increased number and quality (centered) of acquired target regions
74 Automated Microscopy Highly accurate positioning of targets in the center of field of views Experiment setup Overall sample size: 8 * 12 = 96 scan fields Standard DAQ Config 2» Screening time: ~70h» junk data / less targets (NOT centered!) Automated Imaging Pre-Screen Config 1 Target Screen Config 2» Screening time: ~7h» 146 target regions (in center of field of view!) Overall acquisition time: 7 h 17 min Decreased overall screening time (factor 10) Increased number and quality (centered) of acquired target regions
75 Automated Microscopy Some Numbers Costs confocal microscopy: 40 per hour Decreased overall screening time (factor 10) Increased number and quality (centered) of acquired target regions Overall sample size: 96 positions Standard DAQ job2» Screening time: ~70h Automated Imaging Pre-Screen job1 Target Screen job2» Screening time: ~7h Overall sample size: positions Standard DAQ job2» Screening time: 6 months» ~40% junk data Automated Imaging Pre-Screen job1 Target Screen job2» Screening time: 3,6 months ~ junk data
76 Perspectives Usability for basic user Data Analysis Data Management Visualisation Image Processing Statistics Bioinformatics Data storage File access File formats (Image) Raw data (Intermediate) Results Hists / Plots / Viewer Design automated workflow solutions Integrated Data Analysis Meta Data Data Bases Workflow repository Quality control Hit/Event identification Optical configuration Experiment Data Analysis Annotations Internal access External access Correlations Biological Question Data Acquisition Data Analysis Modeling Assay automation Plate layouts High-throughput High-content Correlative Data storage Quality control Normalisation Segmentation Features Normalisation (per plate/assay) Spatial normalisation Hit calling
77 Perspectives Usability for basic user Data Analysis Data Management Visualisation Image Processing Statistics Bioinformatics Data storage File access File formats (Image) Raw data (Intermediate) Results Hists / Plots / Viewer Design automated workflow solutions Integrated Data Analysis Meta Data Data Bases Workflow repository Quality control Hit/Event identification Optical configuration Experiment Data Analysis Annotations Internal access External access Correlations Biological Question Assay automation Plate layouts Data Acquisition i High-throughput High-content Correlative Imaging Data storage Data Analysis Quality control Normalisation Segmentation Features Modeling Normalisation (per plate/assay) Spatial normalisation Hit calling Integrated Data Analysis
78 Acknowledgements Holger Erfle Nina Beil Jürgen Beneke Ruben Bulkescher Manuel Gunkel Tautvydas Lisauskas Bastian Schumacher Michael Berthold Christian Dietz Thomas Gabriel Martin Horn Michael Zinsmeier Frank Sieckmann Constantin Kappel Werner Knebel Mike Heilemann Benjamin Flottmann
79 Perspectives Bioinformatics How could Galaxy help?
80 Perspectives Bioinformatics How could Galaxy help? How could Galaxy contribute?
Multiple Usage of KNIME in a Screening Laboratory Environment
 Multiple Usage of KNIME in a Screening Laboratory Environment KNIME UGM Zürich, 02.02.2012 Marc Bickle HT-TDS, MPI-CBG Outline Presentation of TDS Our problem: large complex datasets KNIME as data mining
Multiple Usage of KNIME in a Screening Laboratory Environment KNIME UGM Zürich, 02.02.2012 Marc Bickle HT-TDS, MPI-CBG Outline Presentation of TDS Our problem: large complex datasets KNIME as data mining
BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening. Alex Laude The BioImaging Unit
 BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening Alex Laude The BioImaging Unit Multi-dimensional, multi-modal imaging at the sub-cellular
BioImaging facility update: from multi-photon in vivo imaging to highcontent high-throughput image-based screening Alex Laude The BioImaging Unit Multi-dimensional, multi-modal imaging at the sub-cellular
Image Processing with KNIME
 Image Processing with KNIME Who we are?! Martin Horn Martin.horn@uni-konstanz.de (+49) 07531 88-5017 Z815 Active Segmentation Christian Dietz Christian.dietz@uni-konstanz.de (+49) 07531 88-3641 Z815 Active
Image Processing with KNIME Who we are?! Martin Horn Martin.horn@uni-konstanz.de (+49) 07531 88-5017 Z815 Active Segmentation Christian Dietz Christian.dietz@uni-konstanz.de (+49) 07531 88-3641 Z815 Active
Technology Development Studio (TDS) MPI-CBG, Dresden, Germany. Marc Bickle HT-TDS, MPI-CBG
 Technology Development Studio (TDS) MPI-CBG, Dresden, Germany Marc Bickle HT-TDS, MPI-CBG TDS: Core Screening Facility Core Screening facility of the MPI-CBG specialized in high content imaging (es 2003)
Technology Development Studio (TDS) MPI-CBG, Dresden, Germany Marc Bickle HT-TDS, MPI-CBG TDS: Core Screening Facility Core Screening facility of the MPI-CBG specialized in high content imaging (es 2003)
COLOCALISATION. Alexia Ferrand. Imaging Core Facility Biozentrum Basel
 COLOCALISATION Alexia Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to analyse
COLOCALISATION Alexia Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to analyse
COLOCALISATION. Alexia Loynton-Ferrand. Imaging Core Facility Biozentrum Basel
 COLOCALISATION Alexia Loynton-Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to
COLOCALISATION Alexia Loynton-Ferrand Imaging Core Facility Biozentrum Basel OUTLINE Introduction How to best prepare your samples for colocalisation How to acquire the images for colocalisation How to
Multi-channel Deep Transfer Learning for Nuclei Segmentation in Glioblastoma Cell Tissue Images
 Multi-channel Deep Transfer Learning for Nuclei Segmentation in Glioblastoma Cell Tissue Images Thomas Wollmann 1, Julia Ivanova 1, Manuel Gunkel 2, Inn Chung 3, Holger Erfle 2, Karsten Rippe 3, Karl Rohr
Multi-channel Deep Transfer Learning for Nuclei Segmentation in Glioblastoma Cell Tissue Images Thomas Wollmann 1, Julia Ivanova 1, Manuel Gunkel 2, Inn Chung 3, Holger Erfle 2, Karsten Rippe 3, Karl Rohr
Cellular Imaging Solutions Imaging with a vision
 Cellular Imaging Solutions Imaging with a vision www.moleculardevices.com High content screening (HCS) utilizes automated, high-resolution microscopy systems to assay and visualize phenotypic responses
Cellular Imaging Solutions Imaging with a vision www.moleculardevices.com High content screening (HCS) utilizes automated, high-resolution microscopy systems to assay and visualize phenotypic responses
GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow.
![GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow. GoTo [ File ] [ Import KNIME workflow ] Select archive file: and import the CountNuclei.zip example workflow.](/thumbs/75/71593469.jpg) Manual KNIME Image Processing :: First steps This manual aims to provide first insights in KNIME Image Processing. The image processing workflow Count nuclei is described in detail and can be downloaded
Manual KNIME Image Processing :: First steps This manual aims to provide first insights in KNIME Image Processing. The image processing workflow Count nuclei is described in detail and can be downloaded
HCS Tools Scripting Integration. Antje Niederlein Technology Development Studio / MPI-CBG Dresden
 HCS Tools Scripting Integration Antje Niederlein Technology Development Studio / MPI-CBG Dresden Who we are Screening service for academic laboratories Cells, model organisms RNAi, chemical screens Assay
HCS Tools Scripting Integration Antje Niederlein Technology Development Studio / MPI-CBG Dresden Who we are Screening service for academic laboratories Cells, model organisms RNAi, chemical screens Assay
GE Healthcare. Visualize Analyze. Realize. IN Cell Miner HCM Data management for high-content analysis and screening
 GE Healthcare Visualize Analyze Realize IN Cell Miner HCM Data management for high-content analysis and screening Managing the data mountain High-content analysis (HCA) provides quantitative insights in
GE Healthcare Visualize Analyze Realize IN Cell Miner HCM Data management for high-content analysis and screening Managing the data mountain High-content analysis (HCA) provides quantitative insights in
TUTORIAL. HCS- Tools + Scripting Integrations
 TUTORIAL HCS- Tools + Scripting Integrations HCS- Tools... 3 Setup... 3 Task and Data... 4 1) Data Input Opera Reader... 7 2) Meta data integration Expand barcode... 8 3) Meta data integration Join Layout...
TUTORIAL HCS- Tools + Scripting Integrations HCS- Tools... 3 Setup... 3 Task and Data... 4 1) Data Input Opera Reader... 7 2) Meta data integration Expand barcode... 8 3) Meta data integration Join Layout...
THE VERTICAL TURN. Leica TCS SP8 DLS Confocal and Digital LightSheet Microscope
 THE VERTICAL TURN Leica TCS SP8 DLS Confocal and Digital LightSheet Microscope Color coded three-dimensional image of eye specific GFP expression (3P3 promoter) in Drosophila melanogaster. Specimen courtesy
THE VERTICAL TURN Leica TCS SP8 DLS Confocal and Digital LightSheet Microscope Color coded three-dimensional image of eye specific GFP expression (3P3 promoter) in Drosophila melanogaster. Specimen courtesy
Textbook de.nbi Bioimage Analysis Workshop
 BIOIMAGE ANALYSIS WORKSHOP WITHIN THE FRAMEWORK OF HD-HUB Textbook de.nbi Bioimage Analysis Workshop An introduction into KNIME image analysis for life scientists Jürgen Reymann ViroQuant-CellNetworks
BIOIMAGE ANALYSIS WORKSHOP WITHIN THE FRAMEWORK OF HD-HUB Textbook de.nbi Bioimage Analysis Workshop An introduction into KNIME image analysis for life scientists Jürgen Reymann ViroQuant-CellNetworks
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences
 IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
IMARIS 3D and 4D interactive analysis and visualization solutions for the life sciences IMARIS A Brief History For over 20 years Bitplane has offered enabling scientific software tools for the life science
Detailed Program Image Processing Summer School 2010
 Detailed Program Image Processing Summer School 2010 Monday 08.30-09.00: Registration 09.00-10.00: Introduction to image processing (Peter Horvath) Basic definitions (digital image, bit depth, sampling,
Detailed Program Image Processing Summer School 2010 Monday 08.30-09.00: Registration 09.00-10.00: Introduction to image processing (Peter Horvath) Basic definitions (digital image, bit depth, sampling,
ONBI Practical 7: Comparison of techniques
 ONBI Practical 7: Comparison of techniques RM Parton 2014 Aims of practical 7: One of the most common issues confronting people new to microscopy is the confusing array of different techniques available.
ONBI Practical 7: Comparison of techniques RM Parton 2014 Aims of practical 7: One of the most common issues confronting people new to microscopy is the confusing array of different techniques available.
LOCI, Fiji & ImageJ2
 LOCI, Fiji & ImageJ2 Philosophy of optical research In vivo developmental biology Capture everything possible about a sample Multiple dimensions Emission spectra Lifetime Cell polarization Greater resolution
LOCI, Fiji & ImageJ2 Philosophy of optical research In vivo developmental biology Capture everything possible about a sample Multiple dimensions Emission spectra Lifetime Cell polarization Greater resolution
APPROACHES IN QUANTITATIVE, MULTI-DIMENSIONAL MICROSCOPY
 APPROACHES IN QUANTITATIVE, MULTI-DIMENSIONAL MICROSCOPY Profs. Zvi Kam and Benjamin Geiger Department of Molecular Cell Biology The Weizmann Institute of Science Rehovot, Israel In this presentation we
APPROACHES IN QUANTITATIVE, MULTI-DIMENSIONAL MICROSCOPY Profs. Zvi Kam and Benjamin Geiger Department of Molecular Cell Biology The Weizmann Institute of Science Rehovot, Israel In this presentation we
Installation KNIME AG. All rights reserved. 1
 Installation 1. Install KNIME Analytics Platform (from thumb drive) 2. Help > Install New Software > Add (> Archive): 00_InstallationFiles/CommunityContributions_trunk.zip https://update.knime.org/community-contributions/trunk
Installation 1. Install KNIME Analytics Platform (from thumb drive) 2. Help > Install New Software > Add (> Archive): 00_InstallationFiles/CommunityContributions_trunk.zip https://update.knime.org/community-contributions/trunk
Fiji Is Just ImageJ an open source platform for biological image analysis
 Fiji Is Just ImageJ an open source platform for biological image analysis Pavel Tomancak Max Planck Institute of Molecular Cell Biology and Genetics in Dresden (MPI-CBG) OME meeting, Paris June 15th Why
Fiji Is Just ImageJ an open source platform for biological image analysis Pavel Tomancak Max Planck Institute of Molecular Cell Biology and Genetics in Dresden (MPI-CBG) OME meeting, Paris June 15th Why
Goal-oriented Schema in Biological Database Design
 Goal-oriented Schema in Biological Database Design Ping Chen Department of Computer Science University of Helsinki Helsinki, Finland 00014 EMAIL: pchen@cs.helsinki.fi Abstract In this paper, I reviewed
Goal-oriented Schema in Biological Database Design Ping Chen Department of Computer Science University of Helsinki Helsinki, Finland 00014 EMAIL: pchen@cs.helsinki.fi Abstract In this paper, I reviewed
Introduction. Loading Images
 Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: http://www.cellprofiler.org/. CellProfiler
Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: http://www.cellprofiler.org/. CellProfiler
Virtual Frap User Guide
 Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
LAMBDA The LSDF Execution Framework for Data Intensive Applications
 LAMBDA The LSDF Execution Framework for Data Intensive Applications Thomas Jejkal 1, V. Hartmann 1, R. Stotzka 1, J. Otte 2, A. García 3, J. van Wezel 3, A. Streit 3 1 Institute for Dataprocessing and
LAMBDA The LSDF Execution Framework for Data Intensive Applications Thomas Jejkal 1, V. Hartmann 1, R. Stotzka 1, J. Otte 2, A. García 3, J. van Wezel 3, A. Streit 3 1 Institute for Dataprocessing and
Bosch Institute Advanced Microscopy Facility Workshops Summer/Autumn 2016
 Bosch Institute Advanced Microscopy Facility Workshops Summer/Autumn 2016 Presented by Dr Louise Cole and Dr Cathy Payne, Advanced Microscopy Facility, Bosch Institute, School of Medical Sciences, The
Bosch Institute Advanced Microscopy Facility Workshops Summer/Autumn 2016 Presented by Dr Louise Cole and Dr Cathy Payne, Advanced Microscopy Facility, Bosch Institute, School of Medical Sciences, The
Model-Based Segmentation and Colocalization Quantification in 3D Microscopy Images
 Model-Based Segmentation and Colocalization Quantification in 3D Microscopy Images Stefan Wörz 1, Petra Sander 2, Martin Pfannmöller 1, Ralf J. Rieker 3,4, Stefan Joos 2, Gunhild Mechtersheimer 3, Petra
Model-Based Segmentation and Colocalization Quantification in 3D Microscopy Images Stefan Wörz 1, Petra Sander 2, Martin Pfannmöller 1, Ralf J. Rieker 3,4, Stefan Joos 2, Gunhild Mechtersheimer 3, Petra
Workflow 1. Description
 Workflow 1 Description Determine protein staining intensities and distances in mitotic apparatus in z-stack intensity images, which were stained for the inner-centromere protein INCENP, and compare the
Workflow 1 Description Determine protein staining intensities and distances in mitotic apparatus in z-stack intensity images, which were stained for the inner-centromere protein INCENP, and compare the
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences
 Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris Imaris State of the Art Image Visualization and Analysis Over the last 25 years Imaris has continuously improved
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris Imaris State of the Art Image Visualization and Analysis Over the last 25 years Imaris has continuously improved
Confocal Application Notes April 2010
 April 2010 Motorized Stage Applications Mark & Find Prepared by Louise Bertrand Applications and Technical Support Group, Leica Microsystems, Inc In this issue of Leica Confocal Application Notes, we describe
April 2010 Motorized Stage Applications Mark & Find Prepared by Louise Bertrand Applications and Technical Support Group, Leica Microsystems, Inc In this issue of Leica Confocal Application Notes, we describe
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences
 Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris A Brief History Imaris was created 25 years ago to focus purely on the visualization of 3D fluorescence images
Imaris 3D and 4D interactive analysis and visualization solutions for the life sciences Imaris A Brief History Imaris was created 25 years ago to focus purely on the visualization of 3D fluorescence images
SENSITIVITY DIFFERENTLY TO SEE THINGS THE SPEED AND. Opera Phenix High Content Screening System
 THE SPEED AND SENSITIVITY TO SEE THINGS DIFFERENTLY Opera Phenix High Content Screening System For research use only. Not for use in diagnostic procedures. HIGH CONTENT SCREENING WITHOUT THE COMPROMISE
THE SPEED AND SENSITIVITY TO SEE THINGS DIFFERENTLY Opera Phenix High Content Screening System For research use only. Not for use in diagnostic procedures. HIGH CONTENT SCREENING WITHOUT THE COMPROMISE
DiSPIM A Flexible Dual-View Light Sheet Microscope Platform
 DiSPIM A Flexible Dual-View Light Sheet Microscope Platform ASI s DiSPIM Team: John Zemek Gary Rondeau Jon Daniels President Technical Director DiSPIM Lead Engineer NIH Collaborators and Inventors: Hari
DiSPIM A Flexible Dual-View Light Sheet Microscope Platform ASI s DiSPIM Team: John Zemek Gary Rondeau Jon Daniels President Technical Director DiSPIM Lead Engineer NIH Collaborators and Inventors: Hari
Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment
 Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment Harry Hochheiser and Ilya G. Goldberg Image Informatics and Computational Biology Unit, Laboratory
Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment Harry Hochheiser and Ilya G. Goldberg Image Informatics and Computational Biology Unit, Laboratory
Analysis of multi-channel cell-based screens
 Analysis of multi-channel cell-based screens Lígia Brás, Michael Boutros and Wolfgang Huber August 6, 2006 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
Analysis of multi-channel cell-based screens Lígia Brás, Michael Boutros and Wolfgang Huber August 6, 2006 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
Arena. Managing, grouping and searching images, controlling Batch runs, compiling results for Vantage
 1 Arena Managing, grouping and searching images, controlling Batch runs, compiling results for Vantage Imaris 8 Arena Search field (files, tags ) Arena tree provides direct access to all the components
1 Arena Managing, grouping and searching images, controlling Batch runs, compiling results for Vantage Imaris 8 Arena Search field (files, tags ) Arena tree provides direct access to all the components
Segmentation and descriptors for cellular immunofluoresence images
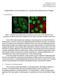 Christopher Probert cprobert@stanford.edu CS231A Project Report Segmentation and descriptors for cellular immunofluoresence images 1: Introduction Figure 1: Example immunoflouresence images of human cancer
Christopher Probert cprobert@stanford.edu CS231A Project Report Segmentation and descriptors for cellular immunofluoresence images 1: Introduction Figure 1: Example immunoflouresence images of human cancer
Technology and equipment at the Bioimaging Center
 Technology and equipment at the Bioimaging Center Light microscopy / Widefields Name Type Applications and equipment LEICA AF6000LX Nikon BioStation Timelaps imaging, CO2 and temperature controlled (14
Technology and equipment at the Bioimaging Center Light microscopy / Widefields Name Type Applications and equipment LEICA AF6000LX Nikon BioStation Timelaps imaging, CO2 and temperature controlled (14
2, the coefficient of variation R 2, and properties of the photon counts traces
 Supplementary Figure 1 Quality control of FCS traces. (a) Typical trace that passes the quality control (QC) according to the parameters shown in f. The QC is based on thresholds applied to fitting parameters
Supplementary Figure 1 Quality control of FCS traces. (a) Typical trace that passes the quality control (QC) according to the parameters shown in f. The QC is based on thresholds applied to fitting parameters
Quantitative analysis of Alzheimer plaques in mice using virtual microscopy
 Quantitative analysis of Alzheimer plaques in mice using virtual microscopy Monika Bieri, André Wethmar and Norbert Wey Technical- and Mediasupport Department of Pathology University Hospital Zurich, Switzerland
Quantitative analysis of Alzheimer plaques in mice using virtual microscopy Monika Bieri, André Wethmar and Norbert Wey Technical- and Mediasupport Department of Pathology University Hospital Zurich, Switzerland
a measurable difference
 Thermo Scientific Cell Analysis Software and Informatics Products a measurable difference for everyone Thermo Scientific HCS Studio Cell Analysis Software Thermo Scientific Store Image and Database Management
Thermo Scientific Cell Analysis Software and Informatics Products a measurable difference for everyone Thermo Scientific HCS Studio Cell Analysis Software Thermo Scientific Store Image and Database Management
Clustering Techniques
 Clustering Techniques Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 16 Lopresti Fall 2007 Lecture 16-1 - Administrative notes Your final project / paper proposal is due on Friday,
Clustering Techniques Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 16 Lopresti Fall 2007 Lecture 16-1 - Administrative notes Your final project / paper proposal is due on Friday,
A COMMON SOFTWARE FRAMEWORK FOR FEL DATA ACQUISITION AND EXPERIMENT MANAGEMENT AT FERMI
 A COMMON SOFTWARE FRAMEWORK FOR FEL DATA ACQUISITION AND EXPERIMENT MANAGEMENT AT FERMI R. Borghes, V. Chenda, A. Curri, G. Kourousias, M. Lonza, G. Passos, M. Prica, R. Pugliese 1 FERMI overview FERMI
A COMMON SOFTWARE FRAMEWORK FOR FEL DATA ACQUISITION AND EXPERIMENT MANAGEMENT AT FERMI R. Borghes, V. Chenda, A. Curri, G. Kourousias, M. Lonza, G. Passos, M. Prica, R. Pugliese 1 FERMI overview FERMI
Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences
 Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences W. J. Godinez 1, M. Lampe 2, S. Wörz 1, B. Müller 2, R. Eils 1 and K. Rohr 1 1 University of Heidelberg, IPMB, and DKFZ
Tracking of Virus Particles in Time-Lapse Fluorescence Microscopy Image Sequences W. J. Godinez 1, M. Lampe 2, S. Wörz 1, B. Müller 2, R. Eils 1 and K. Rohr 1 1 University of Heidelberg, IPMB, and DKFZ
Extract, Analyze, and Report Motion from Video
 Extract, Analyze, and Report Motion from Video Extract ProAnalyst provides motion analysis tools that can be applied to any video or image sequence, regardless of content or acquisition method. With ProAnalyst,
Extract, Analyze, and Report Motion from Video Extract ProAnalyst provides motion analysis tools that can be applied to any video or image sequence, regardless of content or acquisition method. With ProAnalyst,
PATVIS APA. Process analytical technology visual inspection system for automated particle analysis. Computer Vision Systems
 PATVIS APA Process analytical technology visual inspection system for automated particle analysis in-line or at-line process measurements simple installation in r & d or production of solid dosage forms
PATVIS APA Process analytical technology visual inspection system for automated particle analysis in-line or at-line process measurements simple installation in r & d or production of solid dosage forms
Live-cell 3D super-resolution imaging in thick biological samples
 Nature Methods Live-cell 3D super-resolution imaging in thick biological samples Francesca Cella Zanacchi, Zeno Lavagnino, Michela Perrone Donnorso, Alessio Del Bue, Lauria Furia, Mario Faretta & Alberto
Nature Methods Live-cell 3D super-resolution imaging in thick biological samples Francesca Cella Zanacchi, Zeno Lavagnino, Michela Perrone Donnorso, Alessio Del Bue, Lauria Furia, Mario Faretta & Alberto
Groovy in Jenkins. Ioannis K. Moutsatsos. Repurposing Jenkins for Life Sciences Data Pipelining
 Groovy in Jenkins Ioannis K. Moutsatsos Repurposing Jenkins for Life Sciences Data Pipelining Who Am I? Research scientist at local pharmaceutical company Software engineer Open Source advocate and contributor
Groovy in Jenkins Ioannis K. Moutsatsos Repurposing Jenkins for Life Sciences Data Pipelining Who Am I? Research scientist at local pharmaceutical company Software engineer Open Source advocate and contributor
MetaMorph Super-Resolution Software. Localization-based super-resolution microscopy
 MetaMorph Super-Resolution Software Localization-based super-resolution microscopy KEY BENEFITS Lateral Resolution up to 20 nm & Axial Resolution up to 40nm Real-time and offline superresolution image
MetaMorph Super-Resolution Software Localization-based super-resolution microscopy KEY BENEFITS Lateral Resolution up to 20 nm & Axial Resolution up to 40nm Real-time and offline superresolution image
How to store and visualize RNA-seq data
 How to store and visualize RNA-seq data Gabriella Rustici Functional Genomics Group gabry@ebi.ac.uk EBI is an Outstation of the European Molecular Biology Laboratory. Talk summary How do we archive RNA-seq
How to store and visualize RNA-seq data Gabriella Rustici Functional Genomics Group gabry@ebi.ac.uk EBI is an Outstation of the European Molecular Biology Laboratory. Talk summary How do we archive RNA-seq
Technical Computing with MATLAB
 Technical Computing with MATLAB University Of Bath Seminar th 19 th November 2010 Adrienne James (Application Engineering) 1 Agenda Introduction to MATLAB Importing, visualising and analysing data from
Technical Computing with MATLAB University Of Bath Seminar th 19 th November 2010 Adrienne James (Application Engineering) 1 Agenda Introduction to MATLAB Importing, visualising and analysing data from
Extract Features. 2-D and 3-D Analysis. Markerless Tracking. Create Data Reports
 Extract Features 2-D and 3-D Analysis Markerless Tracking Create Data Reports Extract ProAnalyst provides motion analysis tools that can be applied to any video or image sequence, regardless of content
Extract Features 2-D and 3-D Analysis Markerless Tracking Create Data Reports Extract ProAnalyst provides motion analysis tools that can be applied to any video or image sequence, regardless of content
LIFE SCIENCE APPLICATIONS Image-Pro Premier. Image Analysis Starts Here
 LIFE SCIENCE APPLICATIONS Image-Pro Premier Image Analysis Starts Here Image Analysis Starts with Image-Pro Incorporating more than 25 years of user input, Image-Pro Premier provides intuitive tools that
LIFE SCIENCE APPLICATIONS Image-Pro Premier Image Analysis Starts Here Image Analysis Starts with Image-Pro Incorporating more than 25 years of user input, Image-Pro Premier provides intuitive tools that
TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy
 TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy Introduction The OptiGrid converts the illumination system of a conventional wide
TN425: A study of fluorescence standards confirms that OptiGrid confocal images are suitable for quantitative microscopy Introduction The OptiGrid converts the illumination system of a conventional wide
Lecture 19 - Applied Image Analysis
 Lecture 19 - Applied Image Analysis Graeme Ball Three Questions 1. Why should you be interested in image processing & analysis? 2. What are the key concepts & techniques? 3. How can you apply them most
Lecture 19 - Applied Image Analysis Graeme Ball Three Questions 1. Why should you be interested in image processing & analysis? 2. What are the key concepts & techniques? 3. How can you apply them most
Overview. Experiment Specifications. This tutorial will enable you to
 Defining a protocol in BioAssay Overview BioAssay provides an interface to store, manipulate, and retrieve biological assay data. The application allows users to define customized protocol tables representing
Defining a protocol in BioAssay Overview BioAssay provides an interface to store, manipulate, and retrieve biological assay data. The application allows users to define customized protocol tables representing
Bioimage Informatics. Lecture 8, Spring Bioimage Data Analysis (II): Point Feature Detection
 Bioimage Informatics Lecture 8, Spring 2012 Bioimage Data Analysis (II): Point Feature Detection Lecture 8 February 13, 2012 1 Outline Project assignment 02 Comments on reading assignment 01 Review: pixel
Bioimage Informatics Lecture 8, Spring 2012 Bioimage Data Analysis (II): Point Feature Detection Lecture 8 February 13, 2012 1 Outline Project assignment 02 Comments on reading assignment 01 Review: pixel
Introduction to GE Microarray data analysis Practical Course MolBio 2012
 Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
SETTING UP AN HCS DATA ANALYSIS SYSTEM
 A WHITE PAPER FROM GENEDATA JANUARY 2010 SETTING UP AN HCS DATA ANALYSIS SYSTEM WHY YOU NEED ONE HOW TO CREATE ONE HOW IT WILL HELP HCS MARKET AND DATA ANALYSIS CHALLENGES High Content Screening (HCS)
A WHITE PAPER FROM GENEDATA JANUARY 2010 SETTING UP AN HCS DATA ANALYSIS SYSTEM WHY YOU NEED ONE HOW TO CREATE ONE HOW IT WILL HELP HCS MARKET AND DATA ANALYSIS CHALLENGES High Content Screening (HCS)
Discovery Net : A UK e-science Pilot Project for Grid-based Knowledge Discovery Services. Patrick Wendel Imperial College, London
 Discovery Net : A UK e-science Pilot Project for Grid-based Knowledge Discovery Services Patrick Wendel Imperial College, London Data Mining and Exploration Middleware for Distributed and Grid Computing,
Discovery Net : A UK e-science Pilot Project for Grid-based Knowledge Discovery Services Patrick Wendel Imperial College, London Data Mining and Exploration Middleware for Distributed and Grid Computing,
FLUOVIEW FV1000/FV1200
 FLUOVIEW FV1000/FV1200 UPGRADE TO 3D NANOIMAGING AND SINGLE MOLECULE TRACKING FOR OLYMPUS FLUOVIEW FV1000/FV1200 Within the past few years, several methods have been devised in order to obtain images with
FLUOVIEW FV1000/FV1200 UPGRADE TO 3D NANOIMAGING AND SINGLE MOLECULE TRACKING FOR OLYMPUS FLUOVIEW FV1000/FV1200 Within the past few years, several methods have been devised in order to obtain images with
ATFM 2016 ATFM Software installation guide
 ATFM 2016 Software installation guide The workshop is organized in the framework of the Czech-BioImaging research infrastructure supported by MEYS (LM2015062) 1 Software installation guide: Because the
ATFM 2016 Software installation guide The workshop is organized in the framework of the Czech-BioImaging research infrastructure supported by MEYS (LM2015062) 1 Software installation guide: Because the
Analysis of two-way cell-based assays
 Analysis of two-way cell-based assays Lígia Brás, Michael Boutros and Wolfgang Huber April 16, 2015 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
Analysis of two-way cell-based assays Lígia Brás, Michael Boutros and Wolfgang Huber April 16, 2015 Contents 1 Introduction 1 2 Assembling the data 2 2.1 Reading the raw intensity files..................
NDD FLIM Systems for Leica SP2 MP and SP5 MP Multiphoton Microscopes
 NDD FLIM Systems for Leica SP2 MP and SP5 MP Multiphoton Microscopes bh FLIM systems for the confocal and the multiphoton versions of the Leica SP2 and SP5 microscopes are available since 2002 [4]. These
NDD FLIM Systems for Leica SP2 MP and SP5 MP Multiphoton Microscopes bh FLIM systems for the confocal and the multiphoton versions of the Leica SP2 and SP5 microscopes are available since 2002 [4]. These
A Reliable and Distributed LIMS for Efficient Management of the Microarray Experiment Environment
 A Reliable and Distributed LIMS for Efficient Management of the Microarray Experiment Environment Hee-Jeong Jin BK Center for U-Port IT Research Education, Pusan National University, Busan, South Korea,
A Reliable and Distributed LIMS for Efficient Management of the Microarray Experiment Environment Hee-Jeong Jin BK Center for U-Port IT Research Education, Pusan National University, Busan, South Korea,
Data Life Cycle. Research. Access Collaborate. Acquire. Analyse. Comprehend. Plan. Manage Archive. Publish Reuse
 Automated ingest and management Access Collaborate Dataset transfer Databases Web-based file sharing Collaborative sites Acquire Analyse Technical advice Costing Grant assistance Plan Research Data Life
Automated ingest and management Access Collaborate Dataset transfer Databases Web-based file sharing Collaborative sites Acquire Analyse Technical advice Costing Grant assistance Plan Research Data Life
Metadata Models for Experimental Science Data Management
 Metadata Models for Experimental Science Data Management Brian Matthews Facilities Programme Manager Scientific Computing Department, STFC Co-Chair RDA Photon and Neutron Science Interest Group Task lead,
Metadata Models for Experimental Science Data Management Brian Matthews Facilities Programme Manager Scientific Computing Department, STFC Co-Chair RDA Photon and Neutron Science Interest Group Task lead,
Find the Correspondences
 Advanced Topics in BioImage Analysis - 3. Find the Correspondences from Registration to Motion Estimation CellNetworks Math-Clinic Qi Gao 03.02.2017 Math-Clinic core facility Provide services on bioinformatics
Advanced Topics in BioImage Analysis - 3. Find the Correspondences from Registration to Motion Estimation CellNetworks Math-Clinic Qi Gao 03.02.2017 Math-Clinic core facility Provide services on bioinformatics
Using Galaxy to provide a NGS Analysis Platform
 11/15/11 Using Galaxy to provide a NGS Analysis Platform Friedrich Miescher Institute - part of the Novartis Research Foundation - affiliated institute of Basel University - member of Swiss Institute of
11/15/11 Using Galaxy to provide a NGS Analysis Platform Friedrich Miescher Institute - part of the Novartis Research Foundation - affiliated institute of Basel University - member of Swiss Institute of
Marcel Worring Intelligent Sensory Information Systems
 Marcel Worring worring@science.uva.nl Intelligent Sensory Information Systems University of Amsterdam Information and Communication Technology archives of documentaries, film, or training material, video
Marcel Worring worring@science.uva.nl Intelligent Sensory Information Systems University of Amsterdam Information and Communication Technology archives of documentaries, film, or training material, video
Galaxy. Daniel Blankenberg The Galaxy Team
 Galaxy Daniel Blankenberg The Galaxy Team http://galaxyproject.org Overview What is Galaxy? What you can do in Galaxy analysis interface, tools and datasources data libraries workflows visualization sharing
Galaxy Daniel Blankenberg The Galaxy Team http://galaxyproject.org Overview What is Galaxy? What you can do in Galaxy analysis interface, tools and datasources data libraries workflows visualization sharing
Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform
 Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform 1. Exploring the IDR This current IDR web user interface (WUI) is based on the open source
Supplementary Note-- Williams et al The Image Data Resource: A Bioimage Data Integration and Publication Platform 1. Exploring the IDR This current IDR web user interface (WUI) is based on the open source
The walkthrough is available at /
 The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
The walkthrough is available at https://downloads.openmicroscopy.org/presentations/2018/gbi-sydney / Description We will demonstrate a number of features of the OMERO platform using an OMERO server based
Drug versus Disease (DrugVsDisease) package
 1 Introduction Drug versus Disease (DrugVsDisease) package The Drug versus Disease (DrugVsDisease) package provides a pipeline for the comparison of drug and disease gene expression profiles where negatively
1 Introduction Drug versus Disease (DrugVsDisease) package The Drug versus Disease (DrugVsDisease) package provides a pipeline for the comparison of drug and disease gene expression profiles where negatively
Kobe Open Workshop 25/10/2018. Software versions used for the workshop: Workshop content. Resources:
 Kobe Open Workshop 25/10/2018 The presentation and a PDF version of the workshop are available at https://downloads.openmicroscopy.org/presentations/2018/oow-kobe/ Software versions used for the workshop:
Kobe Open Workshop 25/10/2018 The presentation and a PDF version of the workshop are available at https://downloads.openmicroscopy.org/presentations/2018/oow-kobe/ Software versions used for the workshop:
Probabilistic Tracking of Virus Particles in Fluorescence Microscopy Image Sequences
 Probabilistic Tracking of Virus Particles in Fluorescence Microscopy Image Sequences W. J. Godinez 1,2, M. Lampe 3, S. Wörz 1,2, B. Müller 3, R. Eils 1,2, K. Rohr 1,2 1 BIOQUANT, IPMB, University of Heidelberg,
Probabilistic Tracking of Virus Particles in Fluorescence Microscopy Image Sequences W. J. Godinez 1,2, M. Lampe 3, S. Wörz 1,2, B. Müller 3, R. Eils 1,2, K. Rohr 1,2 1 BIOQUANT, IPMB, University of Heidelberg,
The quantitative analysis of interactions takes bioinformatics to the next higher dimension: we go from 1D to 2D with graph theory.
 1 The human protein-protein interaction network of aging-associated genes. A total of 261 aging-associated genes were assembled using the GenAge Human Database. Protein-protein interactions of the human
1 The human protein-protein interaction network of aging-associated genes. A total of 261 aging-associated genes were assembled using the GenAge Human Database. Protein-protein interactions of the human
Extract. Revolutionary Motion Analysis Software. Flexibility
 Extract ProAnalyst provides motion analysis tools that can be applied to any video or image sequence, regardless of content or acquisition method. With ProAnalyst, any digital video camera becomes a measurement
Extract ProAnalyst provides motion analysis tools that can be applied to any video or image sequence, regardless of content or acquisition method. With ProAnalyst, any digital video camera becomes a measurement
InVivo Analyzer Suite
 InVivo Analyzer Suite Live Cell Imaging Software for Fast Acquisition and Complete Analysis The Complete Live Cell Imaging Solution Events happen fast in live cell experimentation. InVivo InVivo Analyzer
InVivo Analyzer Suite Live Cell Imaging Software for Fast Acquisition and Complete Analysis The Complete Live Cell Imaging Solution Events happen fast in live cell experimentation. InVivo InVivo Analyzer
Leica TCS SPE. Simply Sophisticated Your Personal Confocal
 Leica TCS SPE Simply Sophisticated Your Personal Confocal 3 Leica TCS SPE Simply Sophisticated Many exciting discoveries begin with a confocal from Leica Microsystems. Here is one to start with the Leica
Leica TCS SPE Simply Sophisticated Your Personal Confocal 3 Leica TCS SPE Simply Sophisticated Many exciting discoveries begin with a confocal from Leica Microsystems. Here is one to start with the Leica
High-Resolution 3D Reconstruction of the Surface of Live Early-Stage Toad Embryo
 BIOL 507 PROJECT REPORT High-Resolution 3D Reconstruction of the Surface of Live Early-Stage Toad Embryo PLEDGE As my honor, I pledge that I have neither received nor given help in this homework. SIGNATURE
BIOL 507 PROJECT REPORT High-Resolution 3D Reconstruction of the Surface of Live Early-Stage Toad Embryo PLEDGE As my honor, I pledge that I have neither received nor given help in this homework. SIGNATURE
Use stage 16 embryos see Pereanu et al., 2007 for morphological staging criteria. First start Vaa3d (http://vaa3d.org/; Peng et al., 2010).
 Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
Image acquisition. Co- stain with anti- Even- skipped 2B8 (1:50, often this needs to be pre- absorbed several times for optimal staining], or 3C10 (1:50) available at DSHB. Use stage 16 embryos see Pereanu
Leica Microsystems Intelligent Structured Illumination Microscopy
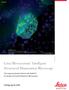 Widefield Mouse kidney section. Maximum projection of a stack containing 65 planes. Green: glomeruli and convoluted tubules (wheat germ agglutinin Alexa Fluor 488) Blue: Nuclei (DAPI). Structured Illumination
Widefield Mouse kidney section. Maximum projection of a stack containing 65 planes. Green: glomeruli and convoluted tubules (wheat germ agglutinin Alexa Fluor 488) Blue: Nuclei (DAPI). Structured Illumination
miscript mirna PCR Array Data Analysis v1.1 revision date November 2014
 miscript mirna PCR Array Data Analysis v1.1 revision date November 2014 Overview The miscript mirna PCR Array Data Analysis Quick Reference Card contains instructions for analyzing the data returned from
miscript mirna PCR Array Data Analysis v1.1 revision date November 2014 Overview The miscript mirna PCR Array Data Analysis Quick Reference Card contains instructions for analyzing the data returned from
MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS
 Mathematical and Computational Applications, Vol. 5, No. 2, pp. 240-247, 200. Association for Scientific Research MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS Volkan Uslan and Đhsan Ömür Bucak
Mathematical and Computational Applications, Vol. 5, No. 2, pp. 240-247, 200. Association for Scientific Research MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS Volkan Uslan and Đhsan Ömür Bucak
WHITE PAPER LIGHTNING IMAGE INFORMATION EXTRACTION BY ADAPTIVE DECONVOLUTION. LIGHTNING White Paper, September
 WHITE PAPER LIGHTNING IMAGE INFORMATION EXTRACTION BY ADAPTIVE DECONVOLUTION LIGHTNING White Paper, September 2018 1 LIGHTNING Image Information Extraction Seeing more than just the Image Author: Jürgen
WHITE PAPER LIGHTNING IMAGE INFORMATION EXTRACTION BY ADAPTIVE DECONVOLUTION LIGHTNING White Paper, September 2018 1 LIGHTNING Image Information Extraction Seeing more than just the Image Author: Jürgen
LEICA EM CRYO CLEM. One system to serve your workflow needs in Correlative Light and Electron Microscopy
 LEICA EM CRYO CLEM One system to serve your workflow needs in Correlative Light and Electron Microscopy CORRELATIVE LIGHT AND ELECTRON MICROSCOPY Researchers can only optimise results when their samples
LEICA EM CRYO CLEM One system to serve your workflow needs in Correlative Light and Electron Microscopy CORRELATIVE LIGHT AND ELECTRON MICROSCOPY Researchers can only optimise results when their samples
/ Computational Genomics. Normalization
 10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
Chapter 3. Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C.
 Chapter 3 Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C. elegans Embryos Abstract Differential interference contrast (DIC) microscopy is
Chapter 3 Automated Segmentation of the First Mitotic Spindle in Differential Interference Contrast Microcopy Images of C. elegans Embryos Abstract Differential interference contrast (DIC) microscopy is
From microarray images to Biological knowledge. Junior Barrera BIOINFO-USP DCC/IME-USP
 From microarray images to Biological knowledge Junior Barrera BIOINFO-USP DCC/IME-USP Team Hugo A. Armelin Junior Barrera Helena Brentaini Marcel Brun Y. Chen Edward R. Dougherty Roberto M. Cesar Jr. Daniel
From microarray images to Biological knowledge Junior Barrera BIOINFO-USP DCC/IME-USP Team Hugo A. Armelin Junior Barrera Helena Brentaini Marcel Brun Y. Chen Edward R. Dougherty Roberto M. Cesar Jr. Daniel
Galaxy. Data intensive biology for everyone. / #usegalaxy
 Galaxy Data intensive biology for everyone. www.galaxyproject.org @jxtx / #usegalaxy Engineering Dannon Baker Dan Blankenberg Dave Bouvier Nate Coraor Carl Eberhard Jeremy Goecks Sam Guerler Greg von Kuster
Galaxy Data intensive biology for everyone. www.galaxyproject.org @jxtx / #usegalaxy Engineering Dannon Baker Dan Blankenberg Dave Bouvier Nate Coraor Carl Eberhard Jeremy Goecks Sam Guerler Greg von Kuster
Correlative Light/Electron Microscopy
 Correlative Light/Electron Microscopy Dr. Guenter Resch Leica Beijing HPF Workshop, October 2014 Contents Overview Leica EM PACT2 CLEM Workflow Leica EM HPM100 CLEM Workflow Leica Cryo-LM Stage Section
Correlative Light/Electron Microscopy Dr. Guenter Resch Leica Beijing HPF Workshop, October 2014 Contents Overview Leica EM PACT2 CLEM Workflow Leica EM HPM100 CLEM Workflow Leica Cryo-LM Stage Section
4.2 Megapixel Scientific CMOS Camera
 CMOS, EMCCD AND CCD CAMERAS FOR LIFE SCIENCES 4.2 Megapixel Scientific CMOS Camera Photometrics Prime is the first intelligent scientific CMOS (scmos) camera to incorporate a powerful FPGA-based Embedded
CMOS, EMCCD AND CCD CAMERAS FOR LIFE SCIENCES 4.2 Megapixel Scientific CMOS Camera Photometrics Prime is the first intelligent scientific CMOS (scmos) camera to incorporate a powerful FPGA-based Embedded
TOPIM-2018 schedule. Session 1: Introduction to OMERO & CellProfiler
 Presentations are available at https://downloads.openmicroscopy.org/presentations/2018/topim-crete/ TOPIM-2018 schedule Thursday 12th July: 19:40-20:15 (35 mins) OMERO workshop introduction Friday 13th
Presentations are available at https://downloads.openmicroscopy.org/presentations/2018/topim-crete/ TOPIM-2018 schedule Thursday 12th July: 19:40-20:15 (35 mins) OMERO workshop introduction Friday 13th
Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays.
 Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays. http://www.nexus.ethz.ch/ -> Software -> TMARKER The plugin Intensity Clustering starts on step 7. Steps 1-6
Tutorial for cell nucleus intensity clustering on histochemically stained tissue microarrays. http://www.nexus.ethz.ch/ -> Software -> TMARKER The plugin Intensity Clustering starts on step 7. Steps 1-6
Image Analysis and Morphometry
 Image Analysis and Morphometry Lukas Schärer Evolutionary Biology Zoological Institute University of Basel 1 13. /15.3.2013 Zoology & Evolution Block Course Summary Quantifying morphology why do we need
Image Analysis and Morphometry Lukas Schärer Evolutionary Biology Zoological Institute University of Basel 1 13. /15.3.2013 Zoology & Evolution Block Course Summary Quantifying morphology why do we need
Software review. Biomolecular Interaction Network Database
 Biomolecular Interaction Network Database Keywords: protein interactions, visualisation, biology data integration, web access Abstract This software review looks at the utility of the Biomolecular Interaction
Biomolecular Interaction Network Database Keywords: protein interactions, visualisation, biology data integration, web access Abstract This software review looks at the utility of the Biomolecular Interaction
Digital Volume Correlation for Materials Characterization
 19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
MilkyWay Proteomics Documentation
 MilkyWay Proteomics Documentation Release alpha1 William D. Barshop, Hee Jong Kim, James A. Wohlschlegel Mar 25, 2018 Contents: 1 Uploading files to MilkyWay via the R/Shiny interface 1 1.1 Accessing
MilkyWay Proteomics Documentation Release alpha1 William D. Barshop, Hee Jong Kim, James A. Wohlschlegel Mar 25, 2018 Contents: 1 Uploading files to MilkyWay via the R/Shiny interface 1 1.1 Accessing
Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data
 Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data Mark-Anthony Bray, Ph.D Imaging Platform, Broad Institute Cambridge, Massachusetts, USA mbray@broadinstitute.org
Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data Mark-Anthony Bray, Ph.D Imaging Platform, Broad Institute Cambridge, Massachusetts, USA mbray@broadinstitute.org
