TreeNet: Multi-Loss Deep Learning Network to Predict Branch Direction for Extracting 3D Anatomical Trees
|
|
|
- Paulina Parks
- 5 years ago
- Views:
Transcription
1 TreeNet: Multi-Loss Deep Learning Network to Predict Branch Direction for Extracting 3D Anatomical Trees Mengliu Zhao and Ghassan Hamarneh {mengliuz, School of Computing Science, Simon Fraser University, Canada Abstract. Calculation of blood vessel or airway direction is important for the task of tree tracking in 3D medical images. However, most existing works treat branch direction estimation as only a by-product of vesselness or tubularness computation. In this work, we propose a deep learning framework for predicting tracking directions of anatomical tree structures. We modify the deep V-Net architecture with extra layers and leverage a novel multi-loss function that encodes direction as well as cross sectional plane information. We evaluate our method on both 3D synthetic and 3D clinical pulmonary CT datasets. On the synthetic dataset, we outperform state of the art methods by at least 10% in direction estimation accuracy. For the clinical dataset, we outperform competing methods by 1 4% in mean direction accuracy and 4 10% in corresponding standard deviation. 1 Introduction Tree extraction is a crucial task in 3D medical image analysis, and accurately extracted circulatory and respiratory trees can be further utilized in surgery planning, registration and tree space analysis [1,2,3]. However, the automation and accuracy of tree extraction still remains an open problem due to the natural complexity and variability of the topologies of anatomical tree structures [4], the various imaging reconstruction artifacts [5], varying image intensities along branches, the similarity between tubular structure lumen and background tissue lumen, and the changing geometry due to different pathologies [6,7,8]. One major category of tree extraction algorithms is based on iterative tracking, which usually starts from a given seed point near the root of the tree, predicts the direction of the branch to track along it, detects bifurcations to spawn children trackers, and progresses down the tree hierarchy to smaller branches until some stopping criteria are met [9]. Although there have been several works that tackle the important bifurcation detection step of the tracker [10,11], very few are specifically designed to determine the direction of the branch at a given point. Most works simply treat the problem of direction estimation as a by-product of vesselness or tubularness calculation [12,13,14]. Most existing methods on vesselness (with explicit/implicit direction prediction) rely on certain assumptions made on the geometry of tubular structures.
2 2 Most Hessian based vessel enhancement filters, e.g., Frangi et al. [15] and Jerman et al. [13,14], assumed the vessels were elongated structures. Cetin et al. [10] measured intensity distribution within an oriented cylinder-sphere combined model and constructed a corresponding tensor representation to optimize for vessel directions, however, their success relied on a good match between the cylinder and the actual vessel shape. Law et al. [12] used a gradient based tensor to model oriented flux flow and the vessel direction was also approximated by the eigenvector intrinsically their assumption of vessel shapes were still straight tubes. However, in clinical datasets, especially those exhibiting various pathologies or abnormalities such as narrowing (e.g., in COPD [6]), aneurysms [7], and high tortuosity (which might indicate diseases like arterial hypertension and strokes [8]), the aforementioned shape assumptions might no longer hold true, which results in incorrect direction estimates. In contrast to the above deterministic methods, stochastic and learning based tracking methods provide more flexibility by adjusting the prediction retrospectively during the tracking process, or by using prior information learnt from the training data [16]. Lee et al. [17] proposed to use particle filtering to track vessel contours slice by slice, with the per-slice contour obtained by the Chan-Vese model. Lesage et al. [18] also proposed to use particle filtering method, but in contrast, utilized geometric flow, image features, as well as radius and direction prior distributions to perform Bayesian inference. In these tracking processes, vessel directions were found implicitly by subtracting neighboring points along the detected centerlines. On the other hand, a significant number of machine learning based methods ignore directional information completely by focusing on pixel-wise classification [19,9,20]. The fast development of deep learning methods provides vast opportunities in exploring the structures in 3D images from coarse to fine scales [21], however, limited work has been done on analyzing 3D vasculature images, and none of them estimated tree branch directions. Mirikharaji et al. [22] proposed to use an artificial neural network trained on 2D patches to learn the probability map of airway bifurcation locations; instead of tracking new branches, they connected the bifurcations by minimal paths to form the whole tree. Fu et al. [23] proposed to combine a convolutional neural network architecture with a conditional random field model to achieve a smooth binary segmentation for retinal vessels, but their method was only performed on 2D retinal images and no vessel direction was estimated. Chen et al. [24] proposed to use a convolutional autoencoder for voxel-wise cerebral arteries segmentation while completely ignoring directional information. We claim the following contributions are made in this paper: i) The proposed method, which extends V-Net [21], is the first tree branch direction prediction deep learning method; ii) The proposed multi-loss function is novel and specially designed for tracking 3D tubular structures; iii) The proposed model is trained and tested in a branch-specific way, which takes advantage of the anatomical tree statistics [25,16] and fully utilizes statistical and geometrical information.
3 3 2 Methodology Architecture: Our proposed deep learning architecture is an improvement of V-Net [21]. The choice of V-Net is two-fold: i) its encoding-decoding process propagates contextual information into higher resolution layers in our case, the context information is the tubularity of the neighboring points; and, ii) our multiloss function (introduced below) relies on cross sectional plane information, so the prediction process implicitly involves plane segmentation and reconstruction. We rescale all input volumes to voxels with histogram equalization. We use batch normalization and add three extra fully connected layers (FCs, with output channels 1024, 64 and 4) at the end of the V-Net and output a 4-element vector < v, R > where v predicts the direction of the vessel in the center of the input cube, and a radius R that serves as intermediate input in training the loss layer. The overall methodology is illustrated in Fig. 1a. At testing time: a region of interest (ROI, or 3D patch, which we use in the context interchangeably) is generated and input into the network (as shown in red dotted box in Fig. 1a), and the output is the predicted vector of the corresponding branch (shown as v gt in Fig. 1b). (a) (b) Fig. 1: (a): The proposed architecture and tracking process. (b): Illustration of B gt, v gt and I gt in Equation 1. Loss Function: We define the following multi-loss function: L(v i dt, R i dt) = ω 1 L dir + ω 2 L mask + ω 3 L image + ω 4 L radius (1) L dir (v i dt, Rdt) i = v i gt v i dt 2, L mask (v i dt, Rdt) i = Bgt i Bdt i 2 L 2 L image (v i dt, Rdt) i = Igt i Idt i 2 L 2, L radius (v i dt, Rdt) i = Rgt i Rdt i 2 where i is the training index, gt refers to ground truth value, dt refers to model prediction. Igt i and Idt i are corresponding gt and predicted cross sectional planes (going through the center voxel). Bgt i and Bdt i are the ground truth and predicted (using radius Rdt i and circular expression) branch masks on the cross sectional
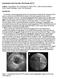
4 4 planes, Rgt i and Rdt i are the ground truth and predicted radii, as illustrated in Fig. 1b. The four terms L dir, L image, L mask, L radius capture the errors in, respectively, direction estimation, cross sectional image plane reconstruction, branch internal area estimation and radius estimation. We normalize L image and L mask by the patch cube size and set the weights empirically to ω 1 = ω 2 = ω 3 = ω 4 = 1. The total loss is optimized over vdt i and Ri dt. Since accurate direction prediction leads to accurate cross section plane prediction, using multiple loss terms should theoretically increase the direction prediction accuracy. Tree Tracking: We follow the tracking procedure in [9], which starts from a given seed point in a branch and tracks along vessel/airway detected by the proposed architecture. Additional tracking details are given in Section 3. 3 Evaluation Synthetic Dataset: We use three different types of synthetic dataset to mimic pathologies such as narrowing and aneurysms, and high tortuosity [6,7,8]: i) Occlusion, ii) Torus and iii) Leakage. Examples are shown in Fig. 2. We augment the data by rotating the volumes along each axis randomly between [0, 60 ], using two radius values, translating along each dimension separately by three values ([- 1, 0, 1], so 9 cases in total) and adding Gaussian noise with standard deviation from to 0.1 (20 cases). This brings the total number of image volumes per each category to 360. We then run a 3-fold cross validation ensuring that augmentations of any volume are not split across the train and test sets. Fig. 2: Synthetic examples (noise free). Clinical Dataset: The clinical dataset is from the Extraction of Airways from CT (EXACT) 2009 challenge 1. Sixteen training volumes with binary segmentations were provided by the organizers. We extracted two categories of data: 1) ROIs, each is a cuboid containing one of the following 7 anatomical structures: trachea, right main bronchi (RMB), left main bronchi (LMB), right superior lobar bronchus (RSLB), right intermediate bronchus (RIB), left superior lobar bronchus (LSLB) and left inferior lobar bronchus (LILB); 2) patches, each is a cube randomly sampled from the branch centerlines, with radii twice the mean radii of the branch, intensities normalized to [0, 1], and augmented by adding Gaussian noise with standard deviation [0.01, 0.04] with step size= red We perform a 4-fold cross validation on the patients for training and testing. 1
5 5 Competing Methods: We compare with 4 state-of-the-art algorithms: i) OOF [12]; ii) Tensor [10]; iii) Jerman [13,14]; 4) Particle filtering [17]. Since particle filtering doesn t directly predict the vessel direction, we use a primitive tracking method to first trace the branch centerline, then estimate the directions. For multiscale methods, the scale ranges are set according to mean branch scales learnt from the dataset, and all other parameters are set according to the original paper (for airways, i.e., dark-on-bright, some parameters are inverted accordingly). Note that although i) and ii) are not direction prediction methods per se, they leverage direction estimates to filter branches, which makes the comparison fair. Tracking Details: Both the proposed method and the competing ones use the same initial seeds, which are selected from the ground truth branch centerlines. By calculating the mean radii R of the corresponding branch, the ROI radii are set automatically as 2 R. Evaluation Metrics: Two metrics are used to evaluate the results. For the tracking method, we use the asymmetric distance function proposed in [9] to compare the ground truth centerline to the extracted centerline: D(C 1, C 2 ) = { min s 2 C 2 dist(s 1, s 2 ) s 1 C 1 } (2) where dist(s 1, s 2 ) is 3D Euclidean distance between voxels s 1 and s 2, C 1 the ground truth centerline and C 2 the detected centerline (by either the proposed method or particle filtering). D(C 1, C 2 ) returns a set of distance values for all the points on C 1, so a smaller mean value and standard deviation would indicate a better result. For other competing methods, since they return a per-voxel direction estimate, we use the following symmetric accuracy metric: accu(v 1, v 2 ) = v 1 v 2 (3) where v 1 and v 2 are the branch direction vectors to be compared. Experiments: The evaluation result on the synthetic dataset is shown in Table 1. We can see a marked improvement in the proposed method over the competing ones by at least 10% in mean direction accuracy. For the Occlusion category, all competing methods performed poorly, since all these methods assume that the foreground is always luminous. For the Torus category, we can see the Tensor method [10] performing the worst, as it modeled the blood vessel as cylindrical tubes, which were very different from the torus shapes in the given images. On the contrary, the Tensor method performed much better than other competing methods on the Leakage category, as a long cylinder might overcome the small leakage (but not good enough to overcome the occlusion) and found the correct direction. It is worth noting that, although the Jerman filter could achieve good enhancement results at tortuous and bulging branches [13,14], the filter was not designed to deal with the direction estimation task. We observe that by removing only one of the loss terms (other than L dir ) actually performs worse than using only L dir. This is not surprising when we remember that the cross sectional plane and the direction together serve as the Frenet frame, so removing one term would invalidate the frame representation.
6 6 Since L image contains the most information on the cross sectional plane, removing it leads to the worst performance. The improvement in prediction accuracy of the multi-loss function supports our hypothesis that all four terms contribute to the result, given their complementary nature. Fig. 3a shows an example where an airway centerline tree is extracted using our proposed method (red curves) and compared to the ground truth tree centerlines (yellow curves). Fig. 3b shows a qualitative comparison between the tracking result, along branch LIB, between the particle filtering and the proposed method. The mean and standard deviation of distance errors of each branch are shown in Fig. 4. The proposed method achieves a lower error mean and standard deviation on every anatomical branch. The results in Table 2 are consistent with the synthetic data results. The proposed method outperforms all the competing methods on all branches. We run our experiments on a Nvidia GTX GeForce 12 GB TITAN GPU, and the processing time per patch at testing stage is 0.04 second. Year Occlusion Torus Leakage OOF [12] (0.07) 0.69(0.28) 0.21(0.12) Tensor [10] (0.1) 0.48(0.037) 0.89(0.03) Jerman [13,14] (0.46) 0.62(0.48) 0.45(0.07) Proposed with L dir only 0.95(0.06) 0.96(0.09) 0.97(0.04) Proposed w/o L image 0.90(0.07) 0.95(0.12) 0.94(0.04) Proposed w/o L mask 0.93(0.07) 0.93(0.13) 0.97(0.04) Proposed w/o L radius 0.94(0.07) 0.93(0.10) 0.97(0.05) Proposed 0.97(0.02) 0.97(0.06) 0.99(0.04) Table 1: Three fold cross validation result on synthetic dataset. (a) (b) Fig. 3: (a): Whole tree extracted. (b): Centerlines tracked by proposed algorithm and competing particle filtering algorithm on LIB. 4 Conclusion and future work We proposed the first deep learning architecture for estimating anatomical tree branch directions, which is a critical step for the common tracking-based
7 7 Fig. 4: Distance error bar between GT centerlines and detected centerlines. Level 1 Level 2 Level 3 Trachea LMB RMB LILB LSLB RSLB RIB OOF [12] 0.19(0.18) 0.24(0.24) 0.29(0.26) 0.30(0.24) 0.42(0.31) 0.40(0.31) 0.43(0.28) Tensor [10] 0.86(0.15) 0.60(0.20) 0.78(0.15) 0.68(0.22) 0.34(0.29) 0.33(0.26) 0.82(0.13) Jerman [13,14] 0.91(0.17) 0.93(0.16) 0.90(0.18) 0.87(0.22) 0.87(0.17) 0.86(0.21) 0.88(0.18) Proposed 0.92(0.11) 0.95(0.07) 0.93(0.08) 0.89(0.15) 0.91(0.08) 0.90(0.11) 0.90(0.10) Table 2: Direction accuracy (mean and standard deviation) on airway branches of different levels. tree extraction methods. Our proposed loss function is unique in that it follows the geometry of the target structure (i.e. the curvilinear tree branches) by using branch direction agreement and cross sectional image information, based on a Frenet frame of reference. In future work, we intend to apply our model on other anatomical trees, such as cerebral vasculature and coronary vessels. Acknowledgments. We thank NVIDIA Corporation for the donation of Titan X GPUs used in this research and the Natural Sciences and Engineering Research Council of Canada (NSERC) for partial funding. References 1. Foruzan et al.: Analysis of CT images of liver for surgical planning. Analysis 2(2) (2012) Baka et al.: Oriented Gaussian mixture models for nonrigid 2D/3D coronary artery registration. TMI 33(5) (2014) Feragen et al.: Tree-space statistics and approximations for large-scale analysis of anatomical trees. In: IPMI. (2013) Kelch et al.: Organ-wide 3D-imaging and topological analysis of the continuous microvascular network in a murine lymph node. Scientific Reports 5 (2015)
8 8 5. Rodriguez et al.: CT reconstruction techniques for improved accuracy of lung CT airway measurement. Medical Physics 41(11) (2014) 1 6. Wiggs et al.: A model of airway narrowing in asthma and in chronic obstructive pulmonary disease. Am Rev Respir Dis 145 (1992) , 2, 4 7. Baráth et al.: Anatomically shaped internal carotid artery aneurysm in vitro model for flow analysis to evaluate stent effect. American Journal of Neuroradiology 25(10) (2004) , 2, 4 8. Han, H.C.: Twisted blood vessels: symptoms, etiology and biomechanical mechanisms. Journal of Vascular Research 49(3) (2012) , 2, 4 9. Zhao, M., Hamarneh, G.: Bifurcation detection in 3D vascular images using novel features and random forest. In: ISBI. (2014) , 2, 4, Cetin, S., Unal, G.: A higher-order tensor vessel tractography for segmentation of vascular structures. TMI 34(10) (2015) , 2, 5, 6, McIntosh, C., Hamarneh, G.: Vessel crawlers: 3D physically-based deformable organisms for vasculature segmentation and analysis. In: CVPR. Volume 1. (2006) Law, M., Chung, A.: Three dimensional curvilinear structure detection using optimally oriented flux. In: European Conference on Computer Vision. (2008) , 2, 5, 6, Jerman et al.: Enhancement of vascular structures in 3D and 2D angiographic images. TMI 35(9) (2016) , 2, 5, 6, Jerman et al.: Blob enhancement and visualization for improved intracranial aneurysm detection. IEEE Trans on Visualization and Computer Graphics 22(6) (2016) , 2, 5, 6, Frangi et al.: Multiscale vessel enhancement filtering. In: MICCAI. (1998) Zhao et al.: Leveraging tree statistics for extracting anatomical trees from 3D medical images. In: CRV. (2017) Lee et al.: Enhanced particle-filtering framework for vessel segmentation and tracking. Computer Methods and Programs in Biomedicine 148 (2017) , Lesage et al.: Medial-based Bayesian tracking for vascular segmentation: Application to coronary arteries in 3D CT angiography. In: ISBI. (2008) Khorshed, R., Celso, C.: Machine learning classification of complex vasculature structures from in-vivo bone marrow 3D data. In: ISBI. (2016) Zhou et al.: Vascular structure segmentation and bifurcation detection. In: ISBI. (2007) Field et al.: V-Net: A framework for a versatile network architecture to support real-time communication performance guarantees. In: Fourteenth Annual Joint Conference of the IEEE Computer and Communications Societies. Volume 3. (1995) , BenTaieb, A., Hamarneh, G.: Uncertainty driven multi-loss fully convolutional networks for histopathology. In: Intravascular Imaging and Computer Assisted Stenting, and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. Springer (2017) Fu et al.: DeepVessel: Retinal vessel segmentation via deep learning and conditional random field. In: MICCAI. (2016) Chen et al.: 3D intracranial artery segmentation using a convolutional autoencoder. In: IEEE International Conference on Bioinformatics and Biomedicine. (2017) Mirikharaji et al.: Globally-optimal anatomical tree extraction from 3D medical images using pictorial structures and minimal paths. In: MICCAI. (2017)
Globally-Optimal Anatomical Tree Extraction from 3D Medical Images using Pictorial Structures and Minimal Paths
 Globally-Optimal Anatomical Tree Extraction from 3D Medical Images using Pictorial Structures and Minimal Paths Zahra Mirikharaji, Mengliu Zhao, Ghassan Hamarneh Medical Image Analysis Lab, Simon Fraser
Globally-Optimal Anatomical Tree Extraction from 3D Medical Images using Pictorial Structures and Minimal Paths Zahra Mirikharaji, Mengliu Zhao, Ghassan Hamarneh Medical Image Analysis Lab, Simon Fraser
Probabilistic Tracking and Model-based Segmentation of 3D Tubular Structures
 Probabilistic Tracking and Model-based Segmentation of 3D Tubular Structures Stefan Wörz, William J. Godinez, Karl Rohr University of Heidelberg, BIOQUANT, IPMB, and DKFZ Heidelberg, Dept. Bioinformatics
Probabilistic Tracking and Model-based Segmentation of 3D Tubular Structures Stefan Wörz, William J. Godinez, Karl Rohr University of Heidelberg, BIOQUANT, IPMB, and DKFZ Heidelberg, Dept. Bioinformatics
Comparison of Vessel Segmentations using STAPLE
 Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
Human Heart Coronary Arteries Segmentation
 Human Heart Coronary Arteries Segmentation Qian Huang Wright State University, Computer Science Department Abstract The volume information extracted from computed tomography angiogram (CTA) datasets makes
Human Heart Coronary Arteries Segmentation Qian Huang Wright State University, Computer Science Department Abstract The volume information extracted from computed tomography angiogram (CTA) datasets makes
Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA.
 Shape prior integration in discrete optimization segmentation algorithms M. Freiman Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA. Email: moti.freiman@childrens.harvard.edu
Shape prior integration in discrete optimization segmentation algorithms M. Freiman Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA. Email: moti.freiman@childrens.harvard.edu
Comparison of Vessel Segmentations Using STAPLE
 Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
DETECTING and extracting blood vessels in magnetic resonance
 1224 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 9, SEPTEMBER 2007 Weighted Local Variance-Based Edge Detection and Its Application to Vascular Segmentation in Magnetic Resonance Angiography Max
1224 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 9, SEPTEMBER 2007 Weighted Local Variance-Based Edge Detection and Its Application to Vascular Segmentation in Magnetic Resonance Angiography Max
Vessel Centerline Tracking in CTA and MRA Images Using Hough Transform
 Vessel Centerline Tracking in CTA and MRA Images Using Hough Transform Maysa M.G. Macedo 1, Choukri Mekkaoui 2, and Marcel P. Jackowski 1 1 University of São Paulo, Department of Computer Science, Rua
Vessel Centerline Tracking in CTA and MRA Images Using Hough Transform Maysa M.G. Macedo 1, Choukri Mekkaoui 2, and Marcel P. Jackowski 1 1 University of São Paulo, Department of Computer Science, Rua
8/3/2017. Contour Assessment for Quality Assurance and Data Mining. Objective. Outline. Tom Purdie, PhD, MCCPM
 Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Deep Learning in Pulmonary Image Analysis with Incomplete Training Samples
 Deep Learning in Pulmonary Image Analysis with Incomplete Training Samples Ziyue Xu, Staff Scientist, National Institutes of Health Nov. 2nd, 2017 (GTC DC Talk DC7137) Image Analysis Arguably the most
Deep Learning in Pulmonary Image Analysis with Incomplete Training Samples Ziyue Xu, Staff Scientist, National Institutes of Health Nov. 2nd, 2017 (GTC DC Talk DC7137) Image Analysis Arguably the most
Automatic Cerebral Aneurysm Detection in Multimodal Angiographic Images
 Automatic Cerebral Aneurysm Detection in Multimodal Angiographic Images Clemens M. Hentschke, Oliver Beuing, Rosa Nickl and Klaus D. Tönnies Abstract We propose a system to automatically detect cerebral
Automatic Cerebral Aneurysm Detection in Multimodal Angiographic Images Clemens M. Hentschke, Oliver Beuing, Rosa Nickl and Klaus D. Tönnies Abstract We propose a system to automatically detect cerebral
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 9: Medical Image Segmentation (III) (Fuzzy Connected Image Segmentation)
 SPRING 2017 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 9: Medical Image Segmentation (III) (Fuzzy Connected Image Segmentation) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
SPRING 2017 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 9: Medical Image Segmentation (III) (Fuzzy Connected Image Segmentation) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
Application of level set based method for segmentation of blood vessels in angiography images
 Lodz University of Technology Faculty of Electrical, Electronic, Computer and Control Engineering Institute of Electronics PhD Thesis Application of level set based method for segmentation of blood vessels
Lodz University of Technology Faculty of Electrical, Electronic, Computer and Control Engineering Institute of Electronics PhD Thesis Application of level set based method for segmentation of blood vessels
Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach
 Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach Julien Jomier and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel
Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach Julien Jomier and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel
Modeling and preoperative planning for kidney surgery
 Modeling and preoperative planning for kidney surgery Refael Vivanti Computer Aided Surgery and Medical Image Processing Lab Hebrew University of Jerusalem, Israel Advisor: Prof. Leo Joskowicz Clinical
Modeling and preoperative planning for kidney surgery Refael Vivanti Computer Aided Surgery and Medical Image Processing Lab Hebrew University of Jerusalem, Israel Advisor: Prof. Leo Joskowicz Clinical
CHAPTER 3 RETINAL OPTIC DISC SEGMENTATION
 60 CHAPTER 3 RETINAL OPTIC DISC SEGMENTATION 3.1 IMPORTANCE OF OPTIC DISC Ocular fundus images provide information about ophthalmic, retinal and even systemic diseases such as hypertension, diabetes, macular
60 CHAPTER 3 RETINAL OPTIC DISC SEGMENTATION 3.1 IMPORTANCE OF OPTIC DISC Ocular fundus images provide information about ophthalmic, retinal and even systemic diseases such as hypertension, diabetes, macular
Weakly Supervised Fully Convolutional Network for PET Lesion Segmentation
 Weakly Supervised Fully Convolutional Network for PET Lesion Segmentation S. Afshari a, A. BenTaieb a, Z. Mirikharaji a, and G. Hamarneh a a Medical Image Analysis Lab, School of Computing Science, Simon
Weakly Supervised Fully Convolutional Network for PET Lesion Segmentation S. Afshari a, A. BenTaieb a, Z. Mirikharaji a, and G. Hamarneh a a Medical Image Analysis Lab, School of Computing Science, Simon
arxiv: v1 [cs.cv] 26 Apr 2013
![arxiv: v1 [cs.cv] 26 Apr 2013 arxiv: v1 [cs.cv] 26 Apr 2013](/thumbs/80/82264082.jpg) OAGM/AAPR Workshop 2013 (arxiv:1304.1876) 1 Pulmonary Vascular Tree Segmentation from Contrast-Enhanced CT Images M. Helmberger 1,2 M. Urschler 1,3 M. Pienn 2 Z. Bálint 2 A. Olschewski 2,4 H. Bischof 1
OAGM/AAPR Workshop 2013 (arxiv:1304.1876) 1 Pulmonary Vascular Tree Segmentation from Contrast-Enhanced CT Images M. Helmberger 1,2 M. Urschler 1,3 M. Pienn 2 Z. Bálint 2 A. Olschewski 2,4 H. Bischof 1
A Hybrid Method for Coronary Artery Stenoses Detection and Quantification in CTA Images
 A Hybrid Method for Coronary Artery Stenoses Detection and Quantification in CTA Images İlkay Öksüz 1, Devrim Ünay 2, Kamuran Kadıpaşaoğlu 2 1 Electrical and Electronics Engineering, Bahçeşehir University,
A Hybrid Method for Coronary Artery Stenoses Detection and Quantification in CTA Images İlkay Öksüz 1, Devrim Ünay 2, Kamuran Kadıpaşaoğlu 2 1 Electrical and Electronics Engineering, Bahçeşehir University,
2D Vessel Segmentation Using Local Adaptive Contrast Enhancement
 2D Vessel Segmentation Using Local Adaptive Contrast Enhancement Dominik Schuldhaus 1,2, Martin Spiegel 1,2,3,4, Thomas Redel 3, Maria Polyanskaya 1,3, Tobias Struffert 2, Joachim Hornegger 1,4, Arnd Doerfler
2D Vessel Segmentation Using Local Adaptive Contrast Enhancement Dominik Schuldhaus 1,2, Martin Spiegel 1,2,3,4, Thomas Redel 3, Maria Polyanskaya 1,3, Tobias Struffert 2, Joachim Hornegger 1,4, Arnd Doerfler
Pearling: Medical Image Segmentation with Pearl Strings
 Pearling: Medical Image Segmentation with Pearl Strings Jarek Rossignac 1, Brian Whited 1, Greg Slabaugh 2, Tong Fang 2, Gozde Unal 2 1 Georgia Institute of Technology Graphics, Visualization, and Usability
Pearling: Medical Image Segmentation with Pearl Strings Jarek Rossignac 1, Brian Whited 1, Greg Slabaugh 2, Tong Fang 2, Gozde Unal 2 1 Georgia Institute of Technology Graphics, Visualization, and Usability
TUBULAR SURFACES EXTRACTION WITH MINIMAL ACTION SURFACES
 TUBULAR SURFACES EXTRACTION WITH MINIMAL ACTION SURFACES XIANGJUN GAO Department of Computer and Information Technology, Shangqiu Normal University, Shangqiu 476000, Henan, China ABSTRACT This paper presents
TUBULAR SURFACES EXTRACTION WITH MINIMAL ACTION SURFACES XIANGJUN GAO Department of Computer and Information Technology, Shangqiu Normal University, Shangqiu 476000, Henan, China ABSTRACT This paper presents
Three Dimensional Segmentation of Intravascular Ultrasound Data
 Three Dimensional Segmentation of Intravascular Ultrasound Data Marc Wennogle 1 and William Hoff 2 1 Veran Medical Technologies, Nashville, Tennessee marc.wennogle@veranmedical.com 2 Colorado School of
Three Dimensional Segmentation of Intravascular Ultrasound Data Marc Wennogle 1 and William Hoff 2 1 Veran Medical Technologies, Nashville, Tennessee marc.wennogle@veranmedical.com 2 Colorado School of
Medical Image Segmentation
 Medical Image Segmentation Xin Yang, HUST *Collaborated with UCLA Medical School and UCSB Segmentation to Contouring ROI Aorta & Kidney 3D Brain MR Image 3D Abdominal CT Image Liver & Spleen Caudate Nucleus
Medical Image Segmentation Xin Yang, HUST *Collaborated with UCLA Medical School and UCSB Segmentation to Contouring ROI Aorta & Kidney 3D Brain MR Image 3D Abdominal CT Image Liver & Spleen Caudate Nucleus
SPNet: Shape Prediction using a Fully Convolutional Neural Network
 SPNet: Shape Prediction using a Fully Convolutional Neural Network S M Masudur Rahman Al Arif 1, Karen Knapp 2 and Greg Slabaugh 1 1 City, University of London 2 University of Exeter Abstract. Shape has
SPNet: Shape Prediction using a Fully Convolutional Neural Network S M Masudur Rahman Al Arif 1, Karen Knapp 2 and Greg Slabaugh 1 1 City, University of London 2 University of Exeter Abstract. Shape has
Simultaneous Multiple Surface Segmentation Using Deep Learning
 Simultaneous Multiple Surface Segmentation Using Deep Learning Abhay Shah 1, Michael D. Abramoff 1,2 and Xiaodong Wu 1,3 Department of 1 Electrical and Computer Engineering, 2 Radiation Oncology, 3 Department
Simultaneous Multiple Surface Segmentation Using Deep Learning Abhay Shah 1, Michael D. Abramoff 1,2 and Xiaodong Wu 1,3 Department of 1 Electrical and Computer Engineering, 2 Radiation Oncology, 3 Department
Automatic Vascular Tree Formation Using the Mahalanobis Distance
 Automatic Vascular Tree Formation Using the Mahalanobis Distance Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, Department of Radiology The University
Automatic Vascular Tree Formation Using the Mahalanobis Distance Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, Department of Radiology The University
GPU Accelerated Segmentation and Centerline Extraction of Tubular Structures from Medical Images
 Noname manuscript No. (will be inserted by the editor) GPU Accelerated Segmentation and Centerline Extraction of Tubular Structures from Medical Images Erik Smistad Anne C. Elster Frank Lindseth the date
Noname manuscript No. (will be inserted by the editor) GPU Accelerated Segmentation and Centerline Extraction of Tubular Structures from Medical Images Erik Smistad Anne C. Elster Frank Lindseth the date
An Advanced Automatic Fuzzy Rule-Based Algorithm for 3D Vessel Segmentation
 An Advanced Automatic Fuzzy Rule-Based Algorithm for 3D Vessel Segmentation Mohamed A. Abdou *1, Ashraf El-Sayed 2, Eman Ali 2 1 Informatics Research Institute, City for Scientific, Research & Technology
An Advanced Automatic Fuzzy Rule-Based Algorithm for 3D Vessel Segmentation Mohamed A. Abdou *1, Ashraf El-Sayed 2, Eman Ali 2 1 Informatics Research Institute, City for Scientific, Research & Technology
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008:
 Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Nearly automatic vessels segmentation using graph-based energy minimization
 Nearly automatic vessels segmentation using graph-based energy minimization Release 1.00 M. Freiman 1, J. Frank 1, L. Weizman 1 E. Nammer 2, O. Shilon 2,, L. Joskowicz 1 and J. Sosna 3 July 16, 2009 1
Nearly automatic vessels segmentation using graph-based energy minimization Release 1.00 M. Freiman 1, J. Frank 1, L. Weizman 1 E. Nammer 2, O. Shilon 2,, L. Joskowicz 1 and J. Sosna 3 July 16, 2009 1
Visual Medicine: Part Two Advanced Topics in Visual Medicine. Advanced Topics in Visual Medicine
 Visual Medicine: Part Two Advanced Topics in Visual Medicine Visualization of Vasculature Steffen Oeltze Visualization Research Group University of Magdeburg, Germany stoeltze@isg.cs.uni-magdeburg.de Outline
Visual Medicine: Part Two Advanced Topics in Visual Medicine Visualization of Vasculature Steffen Oeltze Visualization Research Group University of Magdeburg, Germany stoeltze@isg.cs.uni-magdeburg.de Outline
Vessel Segmentation Using A Shape Driven Flow
 Vessel Segmentation Using A Shape Driven Flow Delphine Nain, Anthony Yezzi and Greg Turk Georgia Institute of Technology, Atlanta GA 30332, USA {delfin, turk}@cc.gatech.edu, ayezzi@ece.gatech.edu Abstract.
Vessel Segmentation Using A Shape Driven Flow Delphine Nain, Anthony Yezzi and Greg Turk Georgia Institute of Technology, Atlanta GA 30332, USA {delfin, turk}@cc.gatech.edu, ayezzi@ece.gatech.edu Abstract.
Phantom-based evaluation of a semi-automatic segmentation algorithm for cerebral vascular structures in 3D ultrasound angiography (3D USA)
 Phantom-based evaluation of a semi-automatic segmentation algorithm for cerebral vascular structures in 3D ultrasound angiography (3D USA) C. Chalopin¹, K. Krissian², A. Müns 3, F. Arlt 3, J. Meixensberger³,
Phantom-based evaluation of a semi-automatic segmentation algorithm for cerebral vascular structures in 3D ultrasound angiography (3D USA) C. Chalopin¹, K. Krissian², A. Müns 3, F. Arlt 3, J. Meixensberger³,
Detecting Bone Lesions in Multiple Myeloma Patients using Transfer Learning
 Detecting Bone Lesions in Multiple Myeloma Patients using Transfer Learning Matthias Perkonigg 1, Johannes Hofmanninger 1, Björn Menze 2, Marc-André Weber 3, and Georg Langs 1 1 Computational Imaging Research
Detecting Bone Lesions in Multiple Myeloma Patients using Transfer Learning Matthias Perkonigg 1, Johannes Hofmanninger 1, Björn Menze 2, Marc-André Weber 3, and Georg Langs 1 1 Computational Imaging Research
THE quantification of vessel features, such as length, width,
 1200 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 25, NO. 9, SEPTEMBER 2006 Segmentation of Retinal Blood Vessels by Combining the Detection of Centerlines and Morphological Reconstruction Ana Maria Mendonça,
1200 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 25, NO. 9, SEPTEMBER 2006 Segmentation of Retinal Blood Vessels by Combining the Detection of Centerlines and Morphological Reconstruction Ana Maria Mendonça,
RSRN: Rich Side-output Residual Network for Medial Axis Detection
 RSRN: Rich Side-output Residual Network for Medial Axis Detection Chang Liu, Wei Ke, Jianbin Jiao, and Qixiang Ye University of Chinese Academy of Sciences, Beijing, China {liuchang615, kewei11}@mails.ucas.ac.cn,
RSRN: Rich Side-output Residual Network for Medial Axis Detection Chang Liu, Wei Ke, Jianbin Jiao, and Qixiang Ye University of Chinese Academy of Sciences, Beijing, China {liuchang615, kewei11}@mails.ucas.ac.cn,
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2014 October 07.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
REAL-TIME ADAPTIVITY IN HEAD-AND-NECK AND LUNG CANCER RADIOTHERAPY IN A GPU ENVIRONMENT
 REAL-TIME ADAPTIVITY IN HEAD-AND-NECK AND LUNG CANCER RADIOTHERAPY IN A GPU ENVIRONMENT Anand P Santhanam Assistant Professor, Department of Radiation Oncology OUTLINE Adaptive radiotherapy for head and
REAL-TIME ADAPTIVITY IN HEAD-AND-NECK AND LUNG CANCER RADIOTHERAPY IN A GPU ENVIRONMENT Anand P Santhanam Assistant Professor, Department of Radiation Oncology OUTLINE Adaptive radiotherapy for head and
Evaluation of Diffusion Techniques for Improved Vessel Visualization and Quantification in Three-Dimensional Rotational Angiography
 Evaluation of Diffusion Techniques for Improved Vessel Visualization and Quantification in Three-Dimensional Rotational Angiography Erik Meijering 1, Wiro Niessen 1, Joachim Weickert 2, and Max Viergever
Evaluation of Diffusion Techniques for Improved Vessel Visualization and Quantification in Three-Dimensional Rotational Angiography Erik Meijering 1, Wiro Niessen 1, Joachim Weickert 2, and Max Viergever
Deep Tracking: Biologically Inspired Tracking with Deep Convolutional Networks
 Deep Tracking: Biologically Inspired Tracking with Deep Convolutional Networks Si Chen The George Washington University sichen@gwmail.gwu.edu Meera Hahn Emory University mhahn7@emory.edu Mentor: Afshin
Deep Tracking: Biologically Inspired Tracking with Deep Convolutional Networks Si Chen The George Washington University sichen@gwmail.gwu.edu Meera Hahn Emory University mhahn7@emory.edu Mentor: Afshin
Reconstruction of Blood Vessels from Neck CT Datasets using Stable 3D Mass-Spring Models
 Eurographics Workshop on Visual Computing for Biomedicine (2008) C. P. Botha, G. Kindlmann, W. J. Niessen, and B. Preim (Editors) Reconstruction of Blood Vessels from Neck CT Datasets using Stable 3D Mass-Spring
Eurographics Workshop on Visual Computing for Biomedicine (2008) C. P. Botha, G. Kindlmann, W. J. Niessen, and B. Preim (Editors) Reconstruction of Blood Vessels from Neck CT Datasets using Stable 3D Mass-Spring
Predicting Cancer with a Recurrent Visual Attention Model for Histopathology Images
 Predicting Cancer with a Recurrent Visual Attention Model for Histopathology Images Aïcha BenTaieb and Ghassan Hamarneh School of Computing Science, Simon Fraser University, Canada {abentaie, hamarneh}@sfu.ca
Predicting Cancer with a Recurrent Visual Attention Model for Histopathology Images Aïcha BenTaieb and Ghassan Hamarneh School of Computing Science, Simon Fraser University, Canada {abentaie, hamarneh}@sfu.ca
Three-Dimensional Blood Vessel Modeling Method Considering IVUS Catheter Insertion
 Three-Dimensional Blood Vessel Modeling Method Considering IVUS Catheter Insertion Jinwon Son School of Mechanical Engineering, Chung-Ang University jinwon.son@cau.ac.kr Young Choi School of Mechanical
Three-Dimensional Blood Vessel Modeling Method Considering IVUS Catheter Insertion Jinwon Son School of Mechanical Engineering, Chung-Ang University jinwon.son@cau.ac.kr Young Choi School of Mechanical
Robust extraction of tumorous vascular networks in high resolution 3D images
 Robust extraction of tumorous vascular networks in high resolution 3D images Laurent Risser Institut de mathématiques de Toulouse lrisser@math.univ-toulouse.fr Joint work with: 2006 2008: F. Plouraboué
Robust extraction of tumorous vascular networks in high resolution 3D images Laurent Risser Institut de mathématiques de Toulouse lrisser@math.univ-toulouse.fr Joint work with: 2006 2008: F. Plouraboué
arxiv: v1 [cs.cv] 6 Jun 2017
![arxiv: v1 [cs.cv] 6 Jun 2017 arxiv: v1 [cs.cv] 6 Jun 2017](/thumbs/96/128307097.jpg) Volume Calculation of CT lung Lesions based on Halton Low-discrepancy Sequences Liansheng Wang a, Shusheng Li a, and Shuo Li b a Department of Computer Science, Xiamen University, Xiamen, China b Dept.
Volume Calculation of CT lung Lesions based on Halton Low-discrepancy Sequences Liansheng Wang a, Shusheng Li a, and Shuo Li b a Department of Computer Science, Xiamen University, Xiamen, China b Dept.
Lung and Lung Lobe Segmentation Methods by Fraunhofer MEVIS
 Lung and Lung Lobe Segmentation Methods by Fraunhofer MEVIS Bianca Lassen, Jan-Martin Kuhnigk, Michael Schmidt, and Heinz-Otto Peitgen Fraunhofer MEVIS, Universitaetsallee 29, 28359 Bremen, Germany http://www.mevis.fraunhofer.de
Lung and Lung Lobe Segmentation Methods by Fraunhofer MEVIS Bianca Lassen, Jan-Martin Kuhnigk, Michael Schmidt, and Heinz-Otto Peitgen Fraunhofer MEVIS, Universitaetsallee 29, 28359 Bremen, Germany http://www.mevis.fraunhofer.de
Tutorial Syllabus. Surface Visualization. (20 min.) Direct Volume Visualization. (30 min.) Medical Training and Surgical Planning
 Tutorial Syllabus Surface Visualization - Marching Cubes and its improvements - Smoothing of surface visualizations Direct Volume Visualization - Ray casting and texture-based approaches - Projection methods
Tutorial Syllabus Surface Visualization - Marching Cubes and its improvements - Smoothing of surface visualizations Direct Volume Visualization - Ray casting and texture-based approaches - Projection methods
Robust and Accurate Coronary Artery Centerline Extraction in CTA by Combining Model-Driven and Data-Driven Approaches
 Robust and Accurate Coronary Artery Centerline Extraction in CTA by Combining Model-Driven and Data-Driven Approaches Yefeng Zheng, Huseyin Tek, and Gareth Funka-Lea Imaging and Computer Vision, Siemens
Robust and Accurate Coronary Artery Centerline Extraction in CTA by Combining Model-Driven and Data-Driven Approaches Yefeng Zheng, Huseyin Tek, and Gareth Funka-Lea Imaging and Computer Vision, Siemens
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information
 Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Calculating the Distance Map for Binary Sampled Data
 MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Calculating the Distance Map for Binary Sampled Data Sarah F. Frisken Gibson TR99-6 December 999 Abstract High quality rendering and physics-based
MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Calculating the Distance Map for Binary Sampled Data Sarah F. Frisken Gibson TR99-6 December 999 Abstract High quality rendering and physics-based
A Geometric Flow for Segmenting Vasculature in Proton-Density Weighted MRI
 A Geometric Flow for Segmenting Vasculature in Proton-Density Weighted MRI Maxime Descoteaux a D. Louis Collins b Kaleem Siddiqi c, a Odyssée Project Team, INRIA, Sophia-Antipolis, France b McConnell Brain
A Geometric Flow for Segmenting Vasculature in Proton-Density Weighted MRI Maxime Descoteaux a D. Louis Collins b Kaleem Siddiqi c, a Odyssée Project Team, INRIA, Sophia-Antipolis, France b McConnell Brain
3D Visualization of Vascular Structures
 3D Visualization of Vascular Structures Bernhard Preim, University of Magdeburg, Visualization Research Group Outline Methods for 3D Visualization of Vasculature Model-based Surface Visualization Explicit
3D Visualization of Vascular Structures Bernhard Preim, University of Magdeburg, Visualization Research Group Outline Methods for 3D Visualization of Vasculature Model-based Surface Visualization Explicit
Model-Based Respiratory Motion Compensation for Image-Guided Cardiac Interventions
 Model-Based Respiratory Motion Compensation for Image-Guided Cardiac Interventions February 8 Matthias Schneider Pattern Recognition Lab Friedrich-Alexander-University Erlangen-Nuremberg Imaging and Visualization
Model-Based Respiratory Motion Compensation for Image-Guided Cardiac Interventions February 8 Matthias Schneider Pattern Recognition Lab Friedrich-Alexander-University Erlangen-Nuremberg Imaging and Visualization
Automatic Thoracic CT Image Segmentation using Deep Convolutional Neural Networks. Xiao Han, Ph.D.
 Automatic Thoracic CT Image Segmentation using Deep Convolutional Neural Networks Xiao Han, Ph.D. Outline Background Brief Introduction to DCNN Method Results 2 Focus where it matters Structure Segmentation
Automatic Thoracic CT Image Segmentation using Deep Convolutional Neural Networks Xiao Han, Ph.D. Outline Background Brief Introduction to DCNN Method Results 2 Focus where it matters Structure Segmentation
Interactive segmentation of vascular structures in CT images for liver surgery planning
 Interactive segmentation of vascular structures in CT images for liver surgery planning L. Wang¹, C. Hansen¹, S.Zidowitz¹, H. K. Hahn¹ ¹ Fraunhofer MEVIS, Institute for Medical Image Computing, Bremen,
Interactive segmentation of vascular structures in CT images for liver surgery planning L. Wang¹, C. Hansen¹, S.Zidowitz¹, H. K. Hahn¹ ¹ Fraunhofer MEVIS, Institute for Medical Image Computing, Bremen,
Segmentation of hepatic artery in multi-phase liver CT using directional dilation and connectivity analysis
 Segmentation of hepatic artery in multi-phase liver CT using directional dilation and connectivity analysis Lei Wang a, Alena-Kathrin Schnurr b, Yue Zhao c, Stephan Zidowitz a, Joachim Georgii a, Horst
Segmentation of hepatic artery in multi-phase liver CT using directional dilation and connectivity analysis Lei Wang a, Alena-Kathrin Schnurr b, Yue Zhao c, Stephan Zidowitz a, Joachim Georgii a, Horst
Introduction to Medical Image Processing
 Introduction to Medical Image Processing Δ Essential environments of a medical imaging system Subject Image Analysis Energy Imaging System Images Image Processing Feature Images Image processing may be
Introduction to Medical Image Processing Δ Essential environments of a medical imaging system Subject Image Analysis Energy Imaging System Images Image Processing Feature Images Image processing may be
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
A Generic Lie Group Model for Computer Vision
 A Generic Lie Group Model for Computer Vision Within this research track we follow a generic Lie group approach to computer vision based on recent physiological research on how the primary visual cortex
A Generic Lie Group Model for Computer Vision Within this research track we follow a generic Lie group approach to computer vision based on recent physiological research on how the primary visual cortex
Fuzzy-Based Extraction of Vascular Structures from Time-of-Flight MR Images
 816 Medical Informatics in a United and Healthy Europe K.-P. Adlassnig et al. (Eds.) IOS Press, 2009 2009 European Federation for Medical Informatics. All rights reserved. doi:10.3233/978-1-60750-044-5-816
816 Medical Informatics in a United and Healthy Europe K.-P. Adlassnig et al. (Eds.) IOS Press, 2009 2009 European Federation for Medical Informatics. All rights reserved. doi:10.3233/978-1-60750-044-5-816
Image Analysis, Geometrical Modelling and Image Synthesis for 3D Medical Imaging
 Image Analysis, Geometrical Modelling and Image Synthesis for 3D Medical Imaging J. SEQUEIRA Laboratoire d'informatique de Marseille - FRE CNRS 2246 Faculté des Sciences de Luminy, 163 avenue de Luminy,
Image Analysis, Geometrical Modelling and Image Synthesis for 3D Medical Imaging J. SEQUEIRA Laboratoire d'informatique de Marseille - FRE CNRS 2246 Faculté des Sciences de Luminy, 163 avenue de Luminy,
CHAPTER-4 LOCALIZATION AND CONTOUR DETECTION OF OPTIC DISK
 CHAPTER-4 LOCALIZATION AND CONTOUR DETECTION OF OPTIC DISK Ocular fundus images can provide information about ophthalmic, retinal and even systemic diseases such as hypertension, diabetes, macular degeneration
CHAPTER-4 LOCALIZATION AND CONTOUR DETECTION OF OPTIC DISK Ocular fundus images can provide information about ophthalmic, retinal and even systemic diseases such as hypertension, diabetes, macular degeneration
GPU-Based Airway Segmentation and Centerline Extraction for Image Guided Bronchoscopy
 GPU-Based Airway Segmentation and Centerline Extraction for Image Guided Bronchoscopy Erik Smistad 1, Anne C. Elster 1, Frank Lindseth 1,2 1) Norwegian University of Science and Technology 2) SINTEF Medical
GPU-Based Airway Segmentation and Centerline Extraction for Image Guided Bronchoscopy Erik Smistad 1, Anne C. Elster 1, Frank Lindseth 1,2 1) Norwegian University of Science and Technology 2) SINTEF Medical
Overview. Related Work Tensor Voting in 2-D Tensor Voting in 3-D Tensor Voting in N-D Application to Vision Problems Stereo Visual Motion
 Overview Related Work Tensor Voting in 2-D Tensor Voting in 3-D Tensor Voting in N-D Application to Vision Problems Stereo Visual Motion Binary-Space-Partitioned Images 3-D Surface Extraction from Medical
Overview Related Work Tensor Voting in 2-D Tensor Voting in 3-D Tensor Voting in N-D Application to Vision Problems Stereo Visual Motion Binary-Space-Partitioned Images 3-D Surface Extraction from Medical
LUNG NODULE DETECTION IN CT USING 3D CONVOLUTIONAL NEURAL NETWORKS. GE Global Research, Niskayuna, NY
 LUNG NODULE DETECTION IN CT USING 3D CONVOLUTIONAL NEURAL NETWORKS Xiaojie Huang, Junjie Shan, and Vivek Vaidya GE Global Research, Niskayuna, NY ABSTRACT We propose a new computer-aided detection system
LUNG NODULE DETECTION IN CT USING 3D CONVOLUTIONAL NEURAL NETWORKS Xiaojie Huang, Junjie Shan, and Vivek Vaidya GE Global Research, Niskayuna, NY ABSTRACT We propose a new computer-aided detection system
Adaptive Branch Tracing and Image Sharpening for Airway Tree Extraction in 3-D Chest CT
 Adaptive Branch Tracing and Image Sharpening for Airway Tree Extraction in 3-D Chest CT Marco Feuerstein 1, Takayuki Kitasaka 2,3, Kensaku Mori 1,3 1 Graduate School of Information Science, Nagoya University,
Adaptive Branch Tracing and Image Sharpening for Airway Tree Extraction in 3-D Chest CT Marco Feuerstein 1, Takayuki Kitasaka 2,3, Kensaku Mori 1,3 1 Graduate School of Information Science, Nagoya University,
The MAGIC-5 CAD for nodule detection in low dose and thin slice lung CT. Piergiorgio Cerello - INFN
 The MAGIC-5 CAD for nodule detection in low dose and thin slice lung CT Piergiorgio Cerello - INFN Frascati, 27/11/2009 Computer Assisted Detection (CAD) MAGIC-5 & Distributed Computing Infrastructure
The MAGIC-5 CAD for nodule detection in low dose and thin slice lung CT Piergiorgio Cerello - INFN Frascati, 27/11/2009 Computer Assisted Detection (CAD) MAGIC-5 & Distributed Computing Infrastructure
3D Vascular Segmentation using MRA Statistics and Velocity Field Information in PC-MRA
 3D Vascular Segmentation using MRA Statistics and Velocity Field Information in PC-MRA Albert C. S. Chung 1, J. Alison Noble 1, Paul Summers 2 and Michael Brady 1 1 Department of Engineering Science, Oxford
3D Vascular Segmentation using MRA Statistics and Velocity Field Information in PC-MRA Albert C. S. Chung 1, J. Alison Noble 1, Paul Summers 2 and Michael Brady 1 1 Department of Engineering Science, Oxford
Geodesic atlas-based labeling of anatomical trees: Application and evaluation on airways extracted from CT
 IEEE TRANSACTIONS ON MEDICAL IMAGING 1 Geodesic atlas-based labeling of anatomical trees: Application and evaluation on airways extracted from CT Aasa Feragen, Jens Petersen, Megan Owen, Pechin Lo, Laura
IEEE TRANSACTIONS ON MEDICAL IMAGING 1 Geodesic atlas-based labeling of anatomical trees: Application and evaluation on airways extracted from CT Aasa Feragen, Jens Petersen, Megan Owen, Pechin Lo, Laura
AN ADAPTIVE REGION GROWING SEGMENTATION FOR BLOOD VESSEL DETECTION FROM RETINAL IMAGES
 AN ADAPTIVE REGION GROWING SEGMENTATION FOR BLOOD VESSEL DETECTION FROM RETINAL IMAGES Alauddin Bhuiyan, Baikunth Nath and Joselito Chua Computer Science and Software Engineering, The University of Melbourne,
AN ADAPTIVE REGION GROWING SEGMENTATION FOR BLOOD VESSEL DETECTION FROM RETINAL IMAGES Alauddin Bhuiyan, Baikunth Nath and Joselito Chua Computer Science and Software Engineering, The University of Melbourne,
Generalizing vesselness with respect to dimensionality and shape Release 1.00
 Generalizing vesselness with respect to dimensionality and shape Release 1.00 Luca Antiga 1 August 3, 2007 1 Medical Imaging Unit Mario Negri Institute, Bergamo, Italy email: antiga at marionegri.it Abstract
Generalizing vesselness with respect to dimensionality and shape Release 1.00 Luca Antiga 1 August 3, 2007 1 Medical Imaging Unit Mario Negri Institute, Bergamo, Italy email: antiga at marionegri.it Abstract
arxiv: v1 [cs.cv] 4 Apr 2019
![arxiv: v1 [cs.cv] 4 Apr 2019 arxiv: v1 [cs.cv] 4 Apr 2019](/thumbs/96/127096255.jpg) mproved nference via Deep nput Transfer Saied Asgari Taghanaki, Kumar Abhishek, and Ghassan Hamarneh School of Computing Science, Simon Fraser University, Canada {sasgarit, kabhishe, hamarneh}@sfu.ca arxiv:194.237v1
mproved nference via Deep nput Transfer Saied Asgari Taghanaki, Kumar Abhishek, and Ghassan Hamarneh School of Computing Science, Simon Fraser University, Canada {sasgarit, kabhishe, hamarneh}@sfu.ca arxiv:194.237v1
Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching
 Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching Michael Schwenke 1, Anja Hennemuth 1, Bernd Fischer 2, Ola Friman 1 1 Fraunhofer MEVIS, Institute
Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching Michael Schwenke 1, Anja Hennemuth 1, Bernd Fischer 2, Ola Friman 1 1 Fraunhofer MEVIS, Institute
Learning and Inferring Depth from Monocular Images. Jiyan Pan April 1, 2009
 Learning and Inferring Depth from Monocular Images Jiyan Pan April 1, 2009 Traditional ways of inferring depth Binocular disparity Structure from motion Defocus Given a single monocular image, how to infer
Learning and Inferring Depth from Monocular Images Jiyan Pan April 1, 2009 Traditional ways of inferring depth Binocular disparity Structure from motion Defocus Given a single monocular image, how to infer
Characterizing Vascular Connectivity from microct Images
 Characterizing Vascular Connectivity from microct Images Marcel Jackowski 1, Xenophon Papademetris 1,2,LawrenceW.Dobrucki 1,3, Albert J. Sinusas 1,3, and Lawrence H. Staib 1,2 1 Departments of Diagnostic
Characterizing Vascular Connectivity from microct Images Marcel Jackowski 1, Xenophon Papademetris 1,2,LawrenceW.Dobrucki 1,3, Albert J. Sinusas 1,3, and Lawrence H. Staib 1,2 1 Departments of Diagnostic
MRI Tumor Segmentation with Densely Connected 3D CNN. Lele Chen, Yue Wu, Adora M. DSouze, Anas Z. Abidin, Axel Wismüller, and Chenliang Xu
 1 MRI Tumor Segmentation with Densely Connected 3D CNN Lele Chen, Yue Wu, Adora M. DSouze, Anas Z. Abidin, Axel Wismüller, and MRI Brain Tumor Segmentation 2 Image source: https://github.com/naldeborgh7575/brain_segmentation
1 MRI Tumor Segmentation with Densely Connected 3D CNN Lele Chen, Yue Wu, Adora M. DSouze, Anas Z. Abidin, Axel Wismüller, and MRI Brain Tumor Segmentation 2 Image source: https://github.com/naldeborgh7575/brain_segmentation
Quantitative IntraVascular UltraSound (QCU)
 Quantitative IntraVascular UltraSound (QCU) Authors: Jouke Dijkstra, Ph.D. and Johan H.C. Reiber, Ph.D., Leiden University Medical Center, Dept of Radiology, Leiden, The Netherlands Introduction: For decades,
Quantitative IntraVascular UltraSound (QCU) Authors: Jouke Dijkstra, Ph.D. and Johan H.C. Reiber, Ph.D., Leiden University Medical Center, Dept of Radiology, Leiden, The Netherlands Introduction: For decades,
Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data
 Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data An Experimental Comparison Thomas Lange 1, Stefan Wörz 2, Karl Rohr 2, Peter M. Schlag 3 1 Experimental and Clinical Research
Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data An Experimental Comparison Thomas Lange 1, Stefan Wörz 2, Karl Rohr 2, Peter M. Schlag 3 1 Experimental and Clinical Research
Open Topology: A Toolkit for Brain Isosurface Correction
 Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Structured Light II. Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov
 Structured Light II Johannes Köhler Johannes.koehler@dfki.de Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov Introduction Previous lecture: Structured Light I Active Scanning Camera/emitter
Structured Light II Johannes Köhler Johannes.koehler@dfki.de Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov Introduction Previous lecture: Structured Light I Active Scanning Camera/emitter
Learning Algorithms for Medical Image Analysis. Matteo Santoro slipguru
 Learning Algorithms for Medical Image Analysis Matteo Santoro slipguru santoro@disi.unige.it June 8, 2010 Outline 1. learning-based strategies for quantitative image analysis 2. automatic annotation of
Learning Algorithms for Medical Image Analysis Matteo Santoro slipguru santoro@disi.unige.it June 8, 2010 Outline 1. learning-based strategies for quantitative image analysis 2. automatic annotation of
3D Guide Wire Navigation from Single Plane Fluoroscopic Images in Abdominal Catheterizations
 3D Guide Wire Navigation from Single Plane Fluoroscopic Images in Abdominal Catheterizations Martin Groher 2, Frederik Bender 1, Ali Khamene 3, Wolfgang Wein 3, Tim Hauke Heibel 2, Nassir Navab 2 1 Siemens
3D Guide Wire Navigation from Single Plane Fluoroscopic Images in Abdominal Catheterizations Martin Groher 2, Frederik Bender 1, Ali Khamene 3, Wolfgang Wein 3, Tim Hauke Heibel 2, Nassir Navab 2 1 Siemens
A method for quantitative measurement of gas volume changes in upper airway
 A method for quantitative measurement of gas volume changes in upper airway TAO WANG AND ANUP BASU DEPARTMENT OF COMPUTING SCIENCE, UNIVERSITY OF ALBERTA Abstract A method for quantitative measurement
A method for quantitative measurement of gas volume changes in upper airway TAO WANG AND ANUP BASU DEPARTMENT OF COMPUTING SCIENCE, UNIVERSITY OF ALBERTA Abstract A method for quantitative measurement
Automated segmentation methods for liver analysis in oncology applications
 University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
Octree Generating Networks: Efficient Convolutional Architectures for High-resolution 3D Outputs Supplementary Material
 Octree Generating Networks: Efficient Convolutional Architectures for High-resolution 3D Outputs Supplementary Material Peak memory usage, GB 10 1 0.1 0.01 OGN Quadratic Dense Cubic Iteration time, s 10
Octree Generating Networks: Efficient Convolutional Architectures for High-resolution 3D Outputs Supplementary Material Peak memory usage, GB 10 1 0.1 0.01 OGN Quadratic Dense Cubic Iteration time, s 10
Frequency split metal artifact reduction (FSMAR) in computed tomography
 The Johns Hopkins University Advanced Computer Integrated Surgery Group 4 Metal Artifact Removal in C-arm Cone-Beam CT Paper Seminar Critical Review of Frequency split metal artifact reduction (FSMAR)
The Johns Hopkins University Advanced Computer Integrated Surgery Group 4 Metal Artifact Removal in C-arm Cone-Beam CT Paper Seminar Critical Review of Frequency split metal artifact reduction (FSMAR)
Iterative fully convolutional neural networks for automatic vertebra segmentation
 Iterative fully convolutional neural networks for automatic vertebra segmentation Nikolas Lessmann Image Sciences Institute University Medical Center Utrecht Pim A. de Jong Department of Radiology University
Iterative fully convolutional neural networks for automatic vertebra segmentation Nikolas Lessmann Image Sciences Institute University Medical Center Utrecht Pim A. de Jong Department of Radiology University
A 3-D Modeling Scheme for Cerebral Vasculature from MRA Datasets
 3-D Modeling Scheme for Cerebral Vasculature from MR Datasets Zhongyuan Qin, Xuanqin Mou Institute of Image Processing Xi'an Jiaotong University Xi'an, Shaanxi 710049, P. R. China Ruofei Zhang Department
3-D Modeling Scheme for Cerebral Vasculature from MR Datasets Zhongyuan Qin, Xuanqin Mou Institute of Image Processing Xi'an Jiaotong University Xi'an, Shaanxi 710049, P. R. China Ruofei Zhang Department
arxiv: v1 [cs.cv] 31 Mar 2016
![arxiv: v1 [cs.cv] 31 Mar 2016 arxiv: v1 [cs.cv] 31 Mar 2016](/thumbs/92/108399479.jpg) Object Boundary Guided Semantic Segmentation Qin Huang, Chunyang Xia, Wenchao Zheng, Yuhang Song, Hao Xu and C.-C. Jay Kuo arxiv:1603.09742v1 [cs.cv] 31 Mar 2016 University of Southern California Abstract.
Object Boundary Guided Semantic Segmentation Qin Huang, Chunyang Xia, Wenchao Zheng, Yuhang Song, Hao Xu and C.-C. Jay Kuo arxiv:1603.09742v1 [cs.cv] 31 Mar 2016 University of Southern California Abstract.
Segmentation of Airways Based on Gradient Vector Flow
 EXACT'09-191- Segmentation of Airways Based on Gradient Vector Flow Christian Bauer 1,2, Horst Bischof 1, and Reinhard Beichel 2,3,4 1 Inst. for Computer Graphics and Vision, Graz University of Technology,
EXACT'09-191- Segmentation of Airways Based on Gradient Vector Flow Christian Bauer 1,2, Horst Bischof 1, and Reinhard Beichel 2,3,4 1 Inst. for Computer Graphics and Vision, Graz University of Technology,
The VesselGlyph: Focus & Context Visualization in CT-Angiography
 The VesselGlyph: Focus & Context Visualization in CT-Angiography Matúš Straka M. Šrámek, A. La Cruz E. Gröller, D. Fleischmann Contents Motivation:» Why again a new visualization method for vessel data?
The VesselGlyph: Focus & Context Visualization in CT-Angiography Matúš Straka M. Šrámek, A. La Cruz E. Gröller, D. Fleischmann Contents Motivation:» Why again a new visualization method for vessel data?
Visual Working Efficiency Analysis Method of Cockpit Based On ANN
 Visual Working Efficiency Analysis Method of Cockpit Based On ANN Yingchun CHEN Commercial Aircraft Corporation of China,Ltd Dongdong WEI Fudan University Dept. of Mechanics an Science Engineering Gang
Visual Working Efficiency Analysis Method of Cockpit Based On ANN Yingchun CHEN Commercial Aircraft Corporation of China,Ltd Dongdong WEI Fudan University Dept. of Mechanics an Science Engineering Gang
Automated 3D Segmentation of the Lung Airway Tree Using Gain-Based Region Growing Approach
 Automated 3D Segmentation of the Lung Airway Tree Using Gain-Based Region Growing Approach Harbir Singh 1, Michael Crawford, 2, John Curtin 2, and Reyer Zwiggelaar 1 1 School of Computing Sciences, University
Automated 3D Segmentation of the Lung Airway Tree Using Gain-Based Region Growing Approach Harbir Singh 1, Michael Crawford, 2, John Curtin 2, and Reyer Zwiggelaar 1 1 School of Computing Sciences, University
SEPARATION AND SEGMENTATION OF THE HEPATIC VASCULATURE IN CT IMAGES. Qingyang Shang. Dissertation. Submitted to the Faculty of the
 SEPARATION AND SEGMENTATION OF THE HEPATIC VASCULATURE IN CT IMAGES By Qingyang Shang Dissertation Submitted to the Faculty of the Graduate School of Vanderbilt University in partial fulfillment of the
SEPARATION AND SEGMENTATION OF THE HEPATIC VASCULATURE IN CT IMAGES By Qingyang Shang Dissertation Submitted to the Faculty of the Graduate School of Vanderbilt University in partial fulfillment of the
Evaluation of Hessian-based filters to enhance the axis of coronary arteries in CT images
 International Congress Series 1256 (2003) 1191 1196 Evaluation of Hessian-based filters to enhance the axis of coronary arteries in CT images S.D. Olabarriaga a, *, M. Breeuwer b, W.J. Niessen a a University
International Congress Series 1256 (2003) 1191 1196 Evaluation of Hessian-based filters to enhance the axis of coronary arteries in CT images S.D. Olabarriaga a, *, M. Breeuwer b, W.J. Niessen a a University
SEGMENTATION AND RECONSTRUCTION OF 3D ARTERY MODELS FOR SURGICAL PLANNING
 SEGMENTATION AND RECONSTRUCTION OF 3D ARTERY MODELS FOR SURGICAL PLANNING QI YINGYI (B.Sc., FUDAN UNIVERSITY, 2005 ) A THESIS SUBMITTED FOR THE DEGREE OF MASTER OF SCIENCE DEPARTMENT OF COMPUTER SCIENCE
SEGMENTATION AND RECONSTRUCTION OF 3D ARTERY MODELS FOR SURGICAL PLANNING QI YINGYI (B.Sc., FUDAN UNIVERSITY, 2005 ) A THESIS SUBMITTED FOR THE DEGREE OF MASTER OF SCIENCE DEPARTMENT OF COMPUTER SCIENCE
Laurent D. COHEN.
 for biomedical image segmentation Laurent D. COHEN Directeur de Recherche CNRS CEREMADE, UMR CNRS 7534 Université Paris-9 Dauphine Place du Maréchal de Lattre de Tassigny 75016 Paris, France Cohen@ceremade.dauphine.fr
for biomedical image segmentation Laurent D. COHEN Directeur de Recherche CNRS CEREMADE, UMR CNRS 7534 Université Paris-9 Dauphine Place du Maréchal de Lattre de Tassigny 75016 Paris, France Cohen@ceremade.dauphine.fr
Computational Medical Imaging Analysis Chapter 4: Image Visualization
 Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
arxiv: v1 [cs.cv] 11 Apr 2018
![arxiv: v1 [cs.cv] 11 Apr 2018 arxiv: v1 [cs.cv] 11 Apr 2018](/thumbs/83/88598484.jpg) Unsupervised Segmentation of 3D Medical Images Based on Clustering and Deep Representation Learning Takayasu Moriya a, Holger R. Roth a, Shota Nakamura b, Hirohisa Oda c, Kai Nagara c, Masahiro Oda a,
Unsupervised Segmentation of 3D Medical Images Based on Clustering and Deep Representation Learning Takayasu Moriya a, Holger R. Roth a, Shota Nakamura b, Hirohisa Oda c, Kai Nagara c, Masahiro Oda a,
