(a) (b) Penalty Prior CS BIC Likelihood -600 Likelihood
|
|
|
- Kerrie Clarke
- 6 years ago
- Views:
Transcription
1 A Bayesian Approach to Temporal Data Clustering using Hidden Markov Models Cen Li Gautam Biswas Department of EECS, Box 1679 Station B, Vanderbilt University, Nashville, TN USA Abstract This paper presents clustering techniques that partition temporal data into homogeneous groups, and constructs state based profiles for each group in the hidden Markov model (HMM) framework. We propose a Bayesian HMM clustering methodology that improves upon existing HMM clustering by incorporating HMM model size selection into clustering control structure to derive better cluster models and partitions. Experimental results indicate the effectiveness of our methodology. 1. Introduction Unsupervised classification, or clustering, derives structure from data by using objective criteria to partition the data into homogeneous groups so that the within group object similarity and the between group object dissimilarity are optimized simultaneously. Categorization and interpretation of structure are achieved by analyzing the models constructed in terms of the feature value distributions within each group. In the past, cluster analysis techniques have focused on data described by static features. In many real applications, the dynamic characteristics, i.e., how a system interacts with the environment and evolves over time, are of interest. Such behavior or characteristic of these systems is best described by temporal features whose values change significantly during the observation period. Our goal for temporal data clustering is to construct profiles of dynamic processes by constructing and analyzing well defined, parsimonious models of data. Clustering temporal data is inherently more complex than clustering static data because: (i) the dimensionality of data is significantly larger in the temporal case, and (ii) the complexity of the models that describe the cluster structures, and their interpretation is much more complex. We assume that the temporal data sequences that define the dynamic characteristics of the phenomenon under study satisfy the Markov property, and the data generation may be viewed as a probabilistic walk through a fixed set of states. When state definitions directly correspond to feature values, a Markov chain model representation of the data may be appropriate (Sebastiani et al., 1999). When the state definitions are not directly observable, or it is not feasible to define states by exhaustive enumeration of feature values, i.e., data is described with multiple continuous valued temporal features, the hidden Markov model (HMM) is appropriate. In this paper, we focus on temporal data clustering using the HMM representation. Our ultimate goal is to develop through the extracted HMM models, an accurate and explainable representation of the system dynamics. It is important for our clustering system to determine the best number of clusters to partition the data, and the best model structure, i.e., the number of states in a model, to characterize the dynamics of the homogeneous data within each cluster. We approach these tasks by (i) developing an explicit HMM model size selection procedure that dynamically modifies the size of the HMMs during the clustering process, and (ii) casting the HMM model size selection and partition selection problems in terms of a Bayesian model selection problem. 2. Modeling with HMMs A HMM is a non-deterministic stochastic Finite State Automata. The basic structure of a HMM consists of a connected set of states, S =(S 1 ;S 2 ; :::; S n ). We use first order HMMs, where the state of a system at a particular time t is only dependent on the state of the system at the immediate previous time point, i.e., P (S t js t 1 ;S t 2 ;:::;S 1 )=P(S t js t 1 ). In addition, we assume all the temporal feature values are continuous, therefore, we use the continuous density HMM
2 (CDHMM) representation where all temporal features have continuous values. A CDHMM of n states for data having m temporal features can be characterized 1 in terms of three sets of probabilities (Rabiner, 1989): the initial state probabilities, the transition probability, and the emission probabilities. The initial state probabilities, ~ß of size n, defines the probability of any of the given states being the initial state of the given sequence. The transition probability matrix, A of size n n, defines the probability of transition from state i at time t, to state j at the next time step. And the emission probability matrix,b of size n m, defines the probability of generating feature values at any given state. For CDHMM, the emission probability density function of each state is defined by a multivariate Gaussian distribution. There are a numberofadvantages to using the HMM representation for the data profiling. First, the hidden states of a HMM can be used to effectively model the set of potentially valid states in a dynamic process. While the complete set of states and the exact sequence of states a system goes through may not be observable, it can be estimated by studying the observed behaviors of the system. Second, HMMs represent a well-defined probabilistic model. The parameters of a HMM can be determined in a well-defined manner. Furthermore, HMMs provide a graphical state based representation of the underlying dynamic processes that govern system behavior. Graphical models may aid the cluster analysis and model interpretation tasks. 3. The HMM Clustering Problem HMM clustering incorporating HMM model size selection can be described in terms of four nested search steps: Step 1: the number of clusters in a partition; Step 2: the object distribution to clusters in a given partition size; Step 3: the HMM model sizes for individual clusters in the partition; and Step 4: the HMM parameter configuration for the individual clusters. One primary limitation of the earlier work on HMM clustering (Rabiner et al., 1989), (Dermatas & Kokkinakis, 1996), (Kosaka etal., 1995), (Smyth, 1997) is that for search step 1, no objective criterion measure is used to automatically select the cluster partition based on data. A pre-determined threshold value on 1 We assume the continuous features are sampled at a pre-defined rate, and the temporal feature values are defined as a sequence of values. data likelihood, or a post-clustering Monte-Carlo simulation, is used instead, Another limitation is that they assume a pre-specified and uniform HMM size for all models in the intermediate and final clusters in a partition. Therefore, search step 3 does not exist in those systems. There is a separate line of work that derives the HMM model structure from homogeneous data (Stolcke & Omohundro, 1994) (Ostendorf & Singer, 1997), i.e., no clustering is involved. The main difficulty of directly applying their methods in our HMM clustering problem is that these methods either do not scale to continuous valued temporal data, or they do not have an objective way of comparing quality of models of different sizes. Once a model size (i.e., the number of states in the HMM model) is selected, step 4 is invoked to select model parameters that optimize a chosen criterion. A well known Maximum Likelihood (ML) parameter estimation method, the Baum-Welch procedure (Rabiner, 1989), is a variation of the more general EM algorithm (Dempster et al., 1977). It iterates between an expectation step (E-step) and a maximization step (M-step). The E-step assumes the current parameter configuration of the model and computes the expected values of necessary statistics. The M-step uses these statistics to update the model parameters so as to maximize the expected likelihood of the parameters. The iterative process continues until the parameter configuration converges Bayesian Clustering Methodology 4.1 Bayesian Clustering In model-based clustering, it is assumed that data is generated by a mixture of underlying probability distributions. The mixture model, M, is represented by K component models and a hidden, independent discrete variable C, where each value i of C represents a component cluster, modeled by i. Given observations X =(x 1 ; ;x N ), let f k (x i j k ; k ) be the density of an observation x i from the kth component model, k, where k is the corresponding parameters of the model. The likelihood of the mixture model given data is expressed as: P (Xj 1; ; K;P 1; ;P K) = Q N P K P i=1 k=1 k f k (x ij k ; k ); where P k is the probability that an observation belongs to the kth component(p k 0; P K k=1 P k =1). Bayesian clustering casts the model-based clustering problem into the Bayesian model selection problem. Given partitions with different component clusters, the goal is to se- 2 A HMM converges when the log data likelihood given two consecutive model configurations differ less than 10 6.
3 lect the best overall model, M, that has the highest posterior probability, P (M jx). 4.2 Model Selection Criterion From Bayes theorem, the posterior probability of the P (M)P (XjM) model, P (M jx), is given by: P (M jx) = P ; (X) where P (X) and P (M) are prior probabilities of the data and the model respectively, andp (XjM) is the marginal likelihood of the data. For the purpose of comparing alternate models, we have P (M jx) / P (M)P (XjM). Assuming none of the models considered is favored a priori, P (M jx) / P (XjM). That is, the posterior probability of a model is directly proportional to the marginal likelihood. Therefore, the goal is to select the mixture model that gives the highest marginal likelihood. Given the parameter configuration,, of a model M, the marginal likelihood of the data is computed as P (XjM) = R P (Xj ; M)P ( jm)d : When parameters involved are continuous valued, as in the case of CDHMM, the integration computation often becomes too complex to obtain a closed form solution. Common approaches include Monte-Carlo methods, i.e., Gibbs sampling methods (Chib, 1995), and various approximation methods, such as the Laplace approximation (Kass & Raftery, 1995). It has been well documented that although the Monte-Carlo methods and the Laplace approximation are quite accurate, they are computationally intense. Next, we lookattwo efficient approximation methods: the Bayesian information criterion (BIC) (Heckerman et al., 1995), and the Cheeseman-Stutz (CS) approximation (Cheeseman & Stutz, 1996) Bayesian Information Criterion BIC is derived from the Laplace approximation(heckerman et al., 1995): logp(m jx) ß logp(xjm; ^ ) d 2 logn; where d is the number of parameters in the model, N is the number of data objects, and ^ is the ML parameter configuration of model M. logp(xjm; ^ ), the data likelihood, tends to promote larger and more detailed models of data, whereas the second term, d 2 logn, is the penalty term which favors smaller models with less parameters. BIC selects the best model for the data by balancing these two terms Cheeseman-Stutz Approximation Cheeseman and Stutz (Cheeseman & Stutz, 1996) first proposed the CS approximation method for their Bayesian clustering system, AUTOCLASS. P (XjM) = P (X 0 jm) P (XjM) P (X0 jm) ; where X 0 represents complete data, i.e., data with known cluster labels. The first term is the marginal likelihood of the complete data. The exact integration is approximated by a summation over a set of local maximum P parameter configurations, s : P (X 0 jm) ß 2 s P ( jm)p (X 0 j ; M): To further reduce the computation burden, in our work, we have taken this approximation further by using a single maximum likelihood configuration, ^, ^ 2 s, to approximate the summation, i.e., P (X 0 jm) ß P (^ jm)p (X 0 j^ ; M). The second term in the CS approximation is a gross adjustment term. Both its numerator and denominator are expanded using the BIC measure. Ignoring differences between the penalty terms in the numerator and the denominator, we obtain: logp (XjM) ß logp (^ jm)+logp (Xj^ ; M); where X is the incomplete data and P (^ jm) is the prior probability of the ML model parameter values. 5. Bayesian HMM Clustering Recently, graphical models have been incorporated into the Bayesian clustering framework. Components of a Bayesian mixture model, which typically are modeled as multivariate normal distributions (Cheeseman & Stutz, 1996) are replaced with graphical models such asbayesian networks (Thiesson et al., 1998) and Markov chain models (Sebastiani et al., 1999). We have adapted Bayesian clustering to CDHMM clustering, so that: (i) components of a Bayesian mixture model are represented by CDHMMs, and (ii) f k (X i j k ; k )indatalikelihood computation computes the likelihood of a multi-feature temporal sequence given a CDHMM. First, we describe how the general Bayesian model selection criterion is adapted for the HMM model size selection and the cluster partition selection problems. Then we describe how the characteristics of these criterion functions are used to design our heuristic clustering search control structure. 5.1 Criterion Functions Criterion for HMM Size Selection The HMM model size selection process picks the HMM with the number of states that best describe the data. We use Bayesian model selection criterion to select the best HMM model size given data. From our discussion earlier, for a model trained on data X, thebest
4 model size is the one, when coupled with its ML configuration, gives the highest posterior probability. From section 4.2, we knowthatp ( jx) / P (Xj ), and the marginal likelihood can be efficiently approximated using the BIC and the CS measures. Applying the BIC approximation, marginal likelihood of the HMM, k, for cluster k is computed as: (a) 6 (b) 0 Penalty Prior Likelihood -600 Likelihood -800 CS BIC Model Size Model Size logp (X k j k ) ß P N k j=1 logp (X kjj k ; ^ k ) d k 2 logn k; where N k is the number of objects in cluster k, d k is the number of parameters 3 in k, and ^ k is the ML parameters in k. Applying the CS approximation, the marginal likelihood of a HMM, k, is computed as the sum of data likelihood and model prior probability. To compute model prior probability, we made certain assumptions about the prior probability distributions of the parameters. Given that the HMM for cluster k has m k states, the set of initial state probabilities, ß 0 s, and the transition probabilities from any single state i, a 0 ijs, can be regarded as following the Bernoulli distribution, i.e., ß i 0; P m k i=1 ß i =1,and a ij 0; P m k j=1 a ij =1,we assume that they follow the Dirichlet prior distribution: P (a i1; ;a imk j k ) = D(a i1; ;a imj k ) and P (ß 1; ;ß mk j k ) = D(ß 1; ;ß mj k ): For parameters defining the emission distributions, we assume that the feature mean values in each state, μ i, are uniformly distributed, and the standard deviations of each state,ff i, follow Jeffery's prior distribution (Cheeseman & Stutz, 1996), i.e., P (μ f j k ) = 1 P (ff f j k ) = ff 1 f μ fmax μ fmin ; [log ff fmax ff fmin ] 1 ; where μ f and ff f represents the mean and standard deviation value for the f th temporal feature, and μ fmax /μ fmin and ff fmax /ff fmin are the maximum/minimum mean and standard deviation values for feature f across all clusters. Therefore, logp ( ^ k j k ) = logp (ß 1; ;ß mk j k ) + P m k [logp (ff1; i=1 ;ffim k j k) + P F f log(p (μ =1 f j k ) P (ff f j k ))]: Figure 1 illustrates how BIC and CS work for HMM model size selection. Data generated on a 5-state HMM is modeled using HMMs of sizes ranging from 2 3 Significant parameters include all the parameters for emission probability definitions and only the initial probabilities and transition probabilities that are greater than a threshold value t, t is set to 10 6 for all experiments reported in this paper. Figure 1. HMM model size selection to 10. Results for the BIC and CS measures are plotted. The dashed lines show the likelihood of data for the different size HMMs. The dotted lines show the penalty (Figure 1 (a)) and the parameter prior probability (Figure 1 (b)) for each model. And the solid lines show BIC (Figure 1(a))and CS (Figure 1 (b)) as a combination of the above two terms. We observe that as the size of the model increases, the model likelihood also increases and the model penalty and parameter prior decreases monotonically. Both BIC and CS have their highest value corresponding to the size of the original HMM for data Criterion for Partition Selection In the Bayesian framework, the best clustering mixture model, M, has the highest partition posterior probability (PPP), P (M jx). We approximate PPP with the marginal likelihood of the mixture model, P (XjM). We compare the BIC and CS approximations of the marginal likelihood computation for cluster partition selection. For partition with K clusters, modeled as 1 ; ; K, the PPP computed using the BIC approximation is: logp(xjm) P N ß i=1 log[p K k=1 P k P (X i j ^ k ; k )] K+P K k=1 d k 2 logn; where ^ k and d k are the ML model parameter configuration and the number of significant model parameters of cluster k, respectively. P k is the likelihood of data given the model for cluster k. When computing the data likelihood, we assume that the data is complete, i.e., each object is assigned to one known cluster in the partition. Therefore, P k = 1 if object X i is in cluster k, and P k = 0 otherwise. The best model is the one that balances the overall data likelihood and the complexity of the entire cluster partition. In the CS approximation, the independent variable C can be regarded as following a Bernoulli distribution, i.e., P k 0, P K k=1 P k = 1, again making the assumption that the prior distribution of C follows the Dirichlet distribution, we get
5 Penalty Model Prior Likelihood BIC Likelihood CS Partition size Partition size Figure 2. Cluster partition selection Q K logp (XjM) ß log(p (P 1; ;P Kj k ) P ( ^ k=1 k j k )) + P N log(p K P i=1 k=1 k P (X ij ^ k ; k )); where logp( ^ k j k )s, are the prior probability of the ML parameter configuration for individual cluster models. Figure 2 illustrates how the BIC and the CS criteria work for cluster partition selection: given data consisting of an equal number of objects from four randomly generated HMMs, the BIC and the CS scores are measured when data is partitioned into 1 to 10 clusters. At first when the number of clusters is small, because the improvements of data likelihood dominates the change of the BIC and the CS values, values of both criteria monotonically increase as the size of the partition increases. Both criteria reach the peak value when the size of the partition corresponds to the true partition size, four. Subsequently, the improvements of data likelihood becomes less and less significant, the penalty on model complexity and the model prior terms dominate the change of the BIC and the CS measures, and both decrease monotonically as the size of the partition continues to increase. 5.2 The Clustering Search Control Structure The exponential complexity associated with the four nested search steps for our HMM clustering prompts us to introduce heuristics into the search process. Figures 1 & 2 illustrated the characteristics of the BIC and the CS criterion in partition selection (step 1) and HMM model size selection (step 3). They both monotonically increase as the size of the HMM model or the cluster partition increase, until they reach a peak, and then begin to decrease because the penalty termdom- inates. The peak corresponds to the optimal model size. Given this characteristic, we employ the same sequential search strategy for both search steps 1 and 3. We start with the simplest model, i.e., a one cluster partition for step 1 and a one state HMM for step 3. Then, we gradually increase the size of the model, i.e., adding one cluster to the partition or adding one state to the HMM, and re-estimate the model. After each expansion, we evaluate the model using the BIC or the CS measure. If the score of the current model decreases from that of the previous model, we may conclude that we have just passed the peak point, and accept the previous model as our final model. Otherwise, we continue with the model expansion process. To expand a partition, we first select a seed based on the cluster models in the current partition. A seed includes a set of k objects, (k = 3 for all experiments shown here). The purpose of including more than one object in each seed is to ensure that there is sufficient data to build a reliable initial HMM. The first object in a seed is selected by choosing an object that has the least likelihood given all cluster models in the current partition. Then the remaining objects in the seed are the ones have the highest likelihood given the HMM model built based on the first object. A seed selected in this way is more likely to correspond to the centroid of a new cluster in the given partition. We apply HMM model size selection for each chosen seed. Once a partition is expanded with a seed, search step 2 distributes objects to individual clusters such that the overall data likelihood given the partition is maximized. We assign object, x i, to cluster, ( ^ k ; k ), based on its sequence-to-hmm likelihood measure (Rabiner, 1989), P (x i j ^ k ; k ). Individual objects are assigned to clusters whose model provides the highest data likelihood. If after one round of object distribution, any object changes its cluster membership, models for all clusters are updated to reflect the current data in the clusters. Then, all objects are redistributed based on the set of new models. Otherwise, the distribution is accepted. After the objects are distributed into clusters, for a HMM model size, step 4 estimates the model parameters for each cluster using the Baum-Welch procedure discussed in section 2.2. Table 1 gives the complete description of the sequential Bayesian HMM clustering (BHMMC) algorithm. In this algorithm, the HMM model size selection is not applied during object redistribution. This is because: when a single HMM is built based on data generated from k different HMMs, if there is no limit on the size of the model, the best model for the data may be a k-composite model, i.e., one that has the set of states equivalent to the combination of states from all k models. During BHMMC clustering, the intermediate clusters tend to include data from multiple models.
6 Table 1. The sequential BHMMC control structure Initial partition construction: Select first seed Apply HMM model construction based on the seed Form the initial partition with the HMM do Partition expansion: Select one additional seed Apply HMM model construction based on the seed Add HMM to the current partition Object redistribution: do Distribute objects to clusters with the highest likelihood Apply HMM configuration for all clusters while there are objects change cluster memberships Compute PPP of the current partition while PPP of the current partition > PPP of the previous partition Accept the previous partition as the final cluster partition Apply HMM model construction to final clusters To apply the HMM model construction for these intermediate clusters may create composite models that can give high data likelihood compared to HMM models built based on data from a seed. These composite models can be hard to split up in subsequent partitioning iterations. Because our goal is to learn the number of patterns exhibited in data and to study models characterizing individual patterns, in general, if k models are significantly different fromeach other, we prefer to build k seperate, smaller, models, than to build one complex model that tries to include all k models. This is especially true when some of the models involved share a number of states, i.e., states appear in different models havevery similar state definitions. In this case, given the complex, composite model, it is difficult to determine the set of submodels because the transition probabilities in and out of these shared states represent composite statistics accumulated from all models involved. 6. Experimental Results First, we describe how synthetic models and data are generated for the experiments. Then, we give the performance indices we use to evaluate the experimental results. Finally, we analyze the experimental results using the proposed performance indices. 6.1 Data To construct HMM models of different sizes, first, we assign state definitions by randomly selecting mean and variance values from value ranges [0, 100] and [0,25] respectively. Then we assign state transition probabilities and initial probabilities by randomly sampling from value range [0, 1], and then normalize the probabilities. Based on each model, we randomly generated 30 objects, each object is described by two temporal features, and the sequence length of each feature is set to 100. For experiment 1,we generated five different HMMs for each of the three model sizes: 5, 10, and 15 states. Then, a separate data set is created based on each of these 15 HMMs. For experiment 2,we constructed five sets of HMMs, each set consists of four HMMs of different sizes, i.e., 3, 6, 9, and 12 states. For each set of HMMs, one data set is created by combining data objects generated from the 4 different HMMs. For these combined data sets, we know the number of models involved, and the model size and parameter configuration of each. 6.2 Performance Indices In addition to the partition posterior probability, we propose two other performance indices to evaluate the quality of the cluster partitions generated: Partition Misclassification Count (PMC) computes the number of object misclassification in the C i cluster assuming G i is the true cluster. C i and G i correspond when a majority of the C i objects have the G i label, and no other G k (k 6= i) group has a larger number of objects in C i. If more than three groups are formed, the additional smaller C groups are labeled as fragmented groups. The misclassification count is computed as follows: if an object falls into a fragmented C group, where its type (G label) is a majority, it is assigned a misclassification value of 1. If the object is a minority in a non-fragmented group, it is assigned a misclassification value of 2. Otherwise the misclassification value for an object is 0. When the true partition model is available, the smaller the sum of the misclassification counts for all objects in a partition, the better quality the partition in comparison. Between Partition Similarity (BPS) measures the similarity between two partitions in terms of the likelihood of temporal sequences generated by one partition given the other partition, and vice versa. Given two partitions P i and P j,each with N i and N j number of clusters and models: 1 ; ; k ; ; Ni, and 1 ; ; 0 k 0; ; N j respectively, the distance between the two partitions can be computed by: BPS(P P i;p j)= Ni k=1 Max P 0 k 0 2P j P (Sk i j 0 Nj k0 )+ k0=1 Max k 2P i P (Sj k j k ) N i +N j ; where Si k and Sj k0 are temporal sequences generated based on the kth HMM in partition i and the k 0 th HMM in partition j respectively. For each partition, one temporal sequence is generated from the HMM of each cluster. The likelihood of the temporal
7 Table 2. BIC and CS for HMM model size selection Criterion True HMM model size measure BIC 5(0) 10(0) 13.2(2.2) CS 5(0) 10(0) 13.2(2.2) BPS Varying size Fixed 15 Fixed 3 Fixed Trial PPP Varying size Fixed 15 Fixed 8 Fixed Trial Table 3. BIC and CS for cluster partition selection Criterion Fixed HMM size clustering Varying HMM measure size clustering BIC 48(24) 26(27) 70(22) 0(0) CS 26(27) 24(29) 84(29) 0(0) sequence given the other partition is approximated by the largest likelihood of the temporal sequence given all HMMs in the other partition. The overall BPS is the normalized sum of the likelihood of all temporal sequences from the two partitions. When computing the distance between one partition and the true partition, i.e., partition with the set of HMMs that generate the data, the larger the BPS, the more similar the partition in comparison to the true partition, thus the better the partition. 6.3 Experiments The first experiment compares the effectiveness of the BIC and the CS measures in selecting HMM model sizes based on data. Table 2 shows average model sizes and the standard deviations of the HMMs derived from data. In all cases, the BIC and the CS measures selected HMM models of exactly the same sizes. For 5-state and 10-state HMMs, both measures selected HMMs that have sizes identical to the generative HMMs. For 15-state generative HMMs, the sizes of the derived models differ among trials, and have an average size smaller than that of the true HMMs. This is attributed to the well known problem with the Baum-Welch ML parameter estimation procedure, i.e., it sometimes converges to locally maximum parameter configuration, which prematurely terminates the sequential HMM model size search process. The second experiment compares the performance of the BIC and the CS measures to cluster partition selection. It also studies the effect of the HMM model size selection on cluster partition generation. Two different clustering methods are used: (1) the sequential BHMMC which employs the dynamic HMM model size selection, and (2) fixed HMM size clustering which does not perform HMM model size selection. Instead, Figure 3. HMM cluster using HMM model size selection vs. using fixed HMM model size a pre-determined, fixed size HMM is used throughout clustering. Table 3shows the PMC for partitions generated for different data. The cluster partitions selected by the BIC and the CS measures are not always identical, but they generally agree with each other When model size selection is applied, both measures select the partitions that are perfect match to the partition with the set of generative models. When model size selection is not applied, for both measures, the partitions generated with too small a fixed HMM, i.e., a 3-state HMM, are considered better than those generated with too big a fixed HMM, i.e., a 15-state HMM. Both measures generated partitions of better quality when the fixed HMM size equals to the average size of the four generative HMMs. When the size of the HMMs are fixed and small, they do not possess the ability to discriminate among objects that are generated from multiple, more complex HMMs. Therefore, objects from different generative HMMs are grouped into the same cluster in the final partition. On the other hand, when using fixed size HMMs that are too big, adding one new cluster to the partition incurs a large model complexity penalty that sometimes can not be offset by the data likelihood gain. Also, the problem of forming composite models arises when the fixed HMM size used is too large. In both cases, objects from different generative HMMs are mixed into the same cluster in the partition generated. When the HMM model selection procedure is applied, individual clusters are modeled with HMMs of appropriate sizes to best fit data, and the complexity ofall HMMs in the partition and the overall data likelihood are carefully balanced. These lead to better quality cluster partitions. In addition to counting the misclassifications of each partition, we also compare the partition in terms of its PPP and BPS scores in Figure 3. The solid lines represent the sequential BHMMC, and the three dashed lines represent clustering with fixed 3-state HMM, 8-state HMM, and 15-state HMM. For all trials, partitions generated with HMM model size selection have higher
8 posterior model probability and larger between partition distance than those obtained from clustering with the fixed size HMMs. 7. Summary We have presented a Bayesian clustering methodology for temporal data using HMMs. The clustering process incorporates a HMM model size selection procedure which not only generates more accurate model structure for individual clusters, but also improves the quality of the partitions generated. This creates a tradeoff between improved quality oftheclustermod- els and the cluster partitions and increased computational complexity of the algorithm 4. From the experimental results, we believe thatthe improvements are significant enough to make the extra computation worth while. We cast both HMM model size selection and cluster partition selection problems into the Bayesian model selection framework and have experimentally shown that the BIC and the CS measures are comparable when applied in both tasks. The incorporation of heuristics into the search control structure have dramatically cut down the search space involved, but HMM clustering algorithm is still computationally complex. Therefore, we would like to employ incremental clustering strategies where we start with a cluster partition built based on small data, and gradually revise the size and structure of the partition as more and more data is collected. Also, wewould like to look into other partition evaluation criterion that is based on model prediction accuracy. Even though the purpose of our HMM clustering is not prediction per se, how well the set of models can predict may beused to evaluate the quality of the partition. References Cheeseman, P., & Stutz, J. (1996). Bayesian classification (autoclass): Theory and results. In U. M. Fayyad, G. Piatetsky-Shapiro, P. Smyth and R. Uthurusamy (Eds.), Advances in knowledge discovery and data mining, Cambridge, MA: MIT press. Chib, S. (1995). Marginal likelihood from the gibbs sampling. Journal of the American Statistical Association, 90, Dempster, A. P., Laird, N. M., & Rubin, D. B. (1977). Maximum likelihood from incomplete data via the 4 For data in experiment two, BHMMC with HMM model size selection takes an average of 30 hours on a Sparc Ultra machine. em algorithm. Journal of Royal Statistical Society Series B(methodological), 39, Dermatas, E., & Kokkinakis, G. (1996). Algorithm for clustering continuous density hmm by recognition error. IEEE Transactions on Speech and Audio Processing, 4, Heckerman, D., Geiger, D., & Chickering, D. M. (1995). A tutorial on learning with bayesian networks. Machine Learning, 20, Kass, R. E., & Raftery, A. E. (1995). Bayes factor. Journal of the American Statistical Association, 90, Kosaka, T., Masunaga, S., & Kuraoka, M. (1995). Speaker-independent phone modeling based on speaker-dependent hmm's composition and clustering. Proceedings of the Twentieth International Conference on Acoustics, Speech, and Signal Processing (pp ). Ostendorf, M., & Singer, H. (1997). Hmm topology design using maximum likelihood successive state splitting. Computer Speech and Language, 11, Rabiner, L. R. (1989). A tutorial on hidden markov models and selected applications in speech recognition. Proceedings of the IEEE, 77, Rabiner, L. R., Lee, C. H., Juang, B. H., & Wilpon, J. G. (1989). Hmm clustering for connected word recognition. Proceedings of the Fourteenth International Conference on Acoustics, Speech, and Signal Processing (pp ). Sebastiani, P., Ramoni, M., Cohen, P., Warwick, J., & Davis, J. (1999). Discovering dynamics using bayesian clustering. In D. J. Hand, J. N. Kok and M. R. Berthold (Eds.), Advances in intelligent data analysis, Berlin, Springer-Verlag. Smyth, P. (1997). Clustering sequences with hidden markov models. In M. C. Mozer, M. I. Jordan and T. Petsche (Eds.), Advances in neural information processing, Cambridge:MA, MIT Press. Stolcke, A., & Omohundro, S. M. (1994). Best-first model merging for hidden markov model induction (Technical Report TR ). International Computer Science Institute, 1947 Center St. Berkeley, CA. Thiesson, B., Meek, C., Chickering, D. M., & Heckerman, D. (1998). Learning mixtures of bayesian networks (Technical Report MSR-TR-97-30). Microsoft Research, One Microsoft Way, Redmond, WA.
Clustering Sequence Data using Hidden Markov Model. Representation. Cen Li and Gautam Biswas. Box 1679 Station B,
 Clustering Sequence Data using Hidden Markov Model Representation Cen Li and Gautam Biswas Box 1679 Station B, Department of Computer Science, Vanderbilt University, Nashville, TN 37235 USA ABSTRACT This
Clustering Sequence Data using Hidden Markov Model Representation Cen Li and Gautam Biswas Box 1679 Station B, Department of Computer Science, Vanderbilt University, Nashville, TN 37235 USA ABSTRACT This
Summary: A Tutorial on Learning With Bayesian Networks
 Summary: A Tutorial on Learning With Bayesian Networks Markus Kalisch May 5, 2006 We primarily summarize [4]. When we think that it is appropriate, we comment on additional facts and more recent developments.
Summary: A Tutorial on Learning With Bayesian Networks Markus Kalisch May 5, 2006 We primarily summarize [4]. When we think that it is appropriate, we comment on additional facts and more recent developments.
Clustering Sequences with Hidden. Markov Models. Padhraic Smyth CA Abstract
 Clustering Sequences with Hidden Markov Models Padhraic Smyth Information and Computer Science University of California, Irvine CA 92697-3425 smyth@ics.uci.edu Abstract This paper discusses a probabilistic
Clustering Sequences with Hidden Markov Models Padhraic Smyth Information and Computer Science University of California, Irvine CA 92697-3425 smyth@ics.uci.edu Abstract This paper discusses a probabilistic
A Model Selection Criterion for Classification: Application to HMM Topology Optimization
 A Model Selection Criterion for Classification Application to HMM Topology Optimization Alain Biem IBM T. J. Watson Research Center P.O Box 218, Yorktown Heights, NY 10549, USA biem@us.ibm.com Abstract
A Model Selection Criterion for Classification Application to HMM Topology Optimization Alain Biem IBM T. J. Watson Research Center P.O Box 218, Yorktown Heights, NY 10549, USA biem@us.ibm.com Abstract
Similarity-Based Clustering of Sequences using Hidden Markov Models
 Similarity-Based Clustering of Sequences using Hidden Markov Models Manuele Bicego 1, Vittorio Murino 1, and Mário A.T. Figueiredo 2 1 Dipartimento di Informatica, Università di Verona Ca Vignal 2, Strada
Similarity-Based Clustering of Sequences using Hidden Markov Models Manuele Bicego 1, Vittorio Murino 1, and Mário A.T. Figueiredo 2 1 Dipartimento di Informatica, Università di Verona Ca Vignal 2, Strada
Mixture Models and the EM Algorithm
 Mixture Models and the EM Algorithm Padhraic Smyth, Department of Computer Science University of California, Irvine c 2017 1 Finite Mixture Models Say we have a data set D = {x 1,..., x N } where x i is
Mixture Models and the EM Algorithm Padhraic Smyth, Department of Computer Science University of California, Irvine c 2017 1 Finite Mixture Models Say we have a data set D = {x 1,..., x N } where x i is
Geoff McLachlan and Angus Ng. University of Queensland. Schlumberger Chaired Professor Univ. of Texas at Austin. + Chris Bishop
 EM Algorithm Geoff McLachlan and Angus Ng Department of Mathematics & Institute for Molecular Bioscience University of Queensland Adapted by Joydeep Ghosh Schlumberger Chaired Professor Univ. of Texas
EM Algorithm Geoff McLachlan and Angus Ng Department of Mathematics & Institute for Molecular Bioscience University of Queensland Adapted by Joydeep Ghosh Schlumberger Chaired Professor Univ. of Texas
Hierarchical Mixture Models for Nested Data Structures
 Hierarchical Mixture Models for Nested Data Structures Jeroen K. Vermunt 1 and Jay Magidson 2 1 Department of Methodology and Statistics, Tilburg University, PO Box 90153, 5000 LE Tilburg, Netherlands
Hierarchical Mixture Models for Nested Data Structures Jeroen K. Vermunt 1 and Jay Magidson 2 1 Department of Methodology and Statistics, Tilburg University, PO Box 90153, 5000 LE Tilburg, Netherlands
Pattern Recognition. Kjell Elenius. Speech, Music and Hearing KTH. March 29, 2007 Speech recognition
 Pattern Recognition Kjell Elenius Speech, Music and Hearing KTH March 29, 2007 Speech recognition 2007 1 Ch 4. Pattern Recognition 1(3) Bayes Decision Theory Minimum-Error-Rate Decision Rules Discriminant
Pattern Recognition Kjell Elenius Speech, Music and Hearing KTH March 29, 2007 Speech recognition 2007 1 Ch 4. Pattern Recognition 1(3) Bayes Decision Theory Minimum-Error-Rate Decision Rules Discriminant
A Visualization Tool to Improve the Performance of a Classifier Based on Hidden Markov Models
 A Visualization Tool to Improve the Performance of a Classifier Based on Hidden Markov Models Gleidson Pegoretti da Silva, Masaki Nakagawa Department of Computer and Information Sciences Tokyo University
A Visualization Tool to Improve the Performance of a Classifier Based on Hidden Markov Models Gleidson Pegoretti da Silva, Masaki Nakagawa Department of Computer and Information Sciences Tokyo University
Machine Learning and Data Mining. Clustering (1): Basics. Kalev Kask
 Machine Learning and Data Mining Clustering (1): Basics Kalev Kask Unsupervised learning Supervised learning Predict target value ( y ) given features ( x ) Unsupervised learning Understand patterns of
Machine Learning and Data Mining Clustering (1): Basics Kalev Kask Unsupervised learning Supervised learning Predict target value ( y ) given features ( x ) Unsupervised learning Understand patterns of
Probabilistic Abstraction Lattices: A Computationally Efficient Model for Conditional Probability Estimation
 Probabilistic Abstraction Lattices: A Computationally Efficient Model for Conditional Probability Estimation Daniel Lowd January 14, 2004 1 Introduction Probabilistic models have shown increasing popularity
Probabilistic Abstraction Lattices: A Computationally Efficient Model for Conditional Probability Estimation Daniel Lowd January 14, 2004 1 Introduction Probabilistic models have shown increasing popularity
Note Set 4: Finite Mixture Models and the EM Algorithm
 Note Set 4: Finite Mixture Models and the EM Algorithm Padhraic Smyth, Department of Computer Science University of California, Irvine Finite Mixture Models A finite mixture model with K components, for
Note Set 4: Finite Mixture Models and the EM Algorithm Padhraic Smyth, Department of Computer Science University of California, Irvine Finite Mixture Models A finite mixture model with K components, for
Machine Learning (BSMC-GA 4439) Wenke Liu
 Machine Learning (BSMC-GA 4439) Wenke Liu 01-25-2018 Outline Background Defining proximity Clustering methods Determining number of clusters Other approaches Cluster analysis as unsupervised Learning Unsupervised
Machine Learning (BSMC-GA 4439) Wenke Liu 01-25-2018 Outline Background Defining proximity Clustering methods Determining number of clusters Other approaches Cluster analysis as unsupervised Learning Unsupervised
Machine Learning. Unsupervised Learning. Manfred Huber
 Machine Learning Unsupervised Learning Manfred Huber 2015 1 Unsupervised Learning In supervised learning the training data provides desired target output for learning In unsupervised learning the training
Machine Learning Unsupervised Learning Manfred Huber 2015 1 Unsupervised Learning In supervised learning the training data provides desired target output for learning In unsupervised learning the training
Constraints in Particle Swarm Optimization of Hidden Markov Models
 Constraints in Particle Swarm Optimization of Hidden Markov Models Martin Macaš, Daniel Novák, and Lenka Lhotská Czech Technical University, Faculty of Electrical Engineering, Dep. of Cybernetics, Prague,
Constraints in Particle Swarm Optimization of Hidden Markov Models Martin Macaš, Daniel Novák, and Lenka Lhotská Czech Technical University, Faculty of Electrical Engineering, Dep. of Cybernetics, Prague,
CS839: Probabilistic Graphical Models. Lecture 10: Learning with Partially Observed Data. Theo Rekatsinas
 CS839: Probabilistic Graphical Models Lecture 10: Learning with Partially Observed Data Theo Rekatsinas 1 Partially Observed GMs Speech recognition 2 Partially Observed GMs Evolution 3 Partially Observed
CS839: Probabilistic Graphical Models Lecture 10: Learning with Partially Observed Data Theo Rekatsinas 1 Partially Observed GMs Speech recognition 2 Partially Observed GMs Evolution 3 Partially Observed
Semi-Supervised Clustering with Partial Background Information
 Semi-Supervised Clustering with Partial Background Information Jing Gao Pang-Ning Tan Haibin Cheng Abstract Incorporating background knowledge into unsupervised clustering algorithms has been the subject
Semi-Supervised Clustering with Partial Background Information Jing Gao Pang-Ning Tan Haibin Cheng Abstract Incorporating background knowledge into unsupervised clustering algorithms has been the subject
Hidden Markov Model for Sequential Data
 Hidden Markov Model for Sequential Data Dr.-Ing. Michelle Karg mekarg@uwaterloo.ca Electrical and Computer Engineering Cheriton School of Computer Science Sequential Data Measurement of time series: Example:
Hidden Markov Model for Sequential Data Dr.-Ing. Michelle Karg mekarg@uwaterloo.ca Electrical and Computer Engineering Cheriton School of Computer Science Sequential Data Measurement of time series: Example:
Introduction to Machine Learning CMU-10701
 Introduction to Machine Learning CMU-10701 Clustering and EM Barnabás Póczos & Aarti Singh Contents Clustering K-means Mixture of Gaussians Expectation Maximization Variational Methods 2 Clustering 3 K-
Introduction to Machine Learning CMU-10701 Clustering and EM Barnabás Póczos & Aarti Singh Contents Clustering K-means Mixture of Gaussians Expectation Maximization Variational Methods 2 Clustering 3 K-
10. MLSP intro. (Clustering: K-means, EM, GMM, etc.)
 10. MLSP intro. (Clustering: K-means, EM, GMM, etc.) Rahil Mahdian 01.04.2016 LSV Lab, Saarland University, Germany What is clustering? Clustering is the classification of objects into different groups,
10. MLSP intro. (Clustering: K-means, EM, GMM, etc.) Rahil Mahdian 01.04.2016 LSV Lab, Saarland University, Germany What is clustering? Clustering is the classification of objects into different groups,
Supervised vs unsupervised clustering
 Classification Supervised vs unsupervised clustering Cluster analysis: Classes are not known a- priori. Classification: Classes are defined a-priori Sometimes called supervised clustering Extract useful
Classification Supervised vs unsupervised clustering Cluster analysis: Classes are not known a- priori. Classification: Classes are defined a-priori Sometimes called supervised clustering Extract useful
Unsupervised: no target value to predict
 Clustering Unsupervised: no target value to predict Differences between models/algorithms: Exclusive vs. overlapping Deterministic vs. probabilistic Hierarchical vs. flat Incremental vs. batch learning
Clustering Unsupervised: no target value to predict Differences between models/algorithms: Exclusive vs. overlapping Deterministic vs. probabilistic Hierarchical vs. flat Incremental vs. batch learning
Mixture models and frequent sets: combining global and local methods for 0 1 data
 Mixture models and frequent sets: combining global and local methods for 1 data Jaakko Hollmén Jouni K. Seppänen Heikki Mannila Abstract We study the interaction between global and local techniques in
Mixture models and frequent sets: combining global and local methods for 1 data Jaakko Hollmén Jouni K. Seppänen Heikki Mannila Abstract We study the interaction between global and local techniques in
Expectation Maximization (EM) and Gaussian Mixture Models
 Expectation Maximization (EM) and Gaussian Mixture Models Reference: The Elements of Statistical Learning, by T. Hastie, R. Tibshirani, J. Friedman, Springer 1 2 3 4 5 6 7 8 Unsupervised Learning Motivation
Expectation Maximization (EM) and Gaussian Mixture Models Reference: The Elements of Statistical Learning, by T. Hastie, R. Tibshirani, J. Friedman, Springer 1 2 3 4 5 6 7 8 Unsupervised Learning Motivation
Assignment 2. Unsupervised & Probabilistic Learning. Maneesh Sahani Due: Monday Nov 5, 2018
 Assignment 2 Unsupervised & Probabilistic Learning Maneesh Sahani Due: Monday Nov 5, 2018 Note: Assignments are due at 11:00 AM (the start of lecture) on the date above. he usual College late assignments
Assignment 2 Unsupervised & Probabilistic Learning Maneesh Sahani Due: Monday Nov 5, 2018 Note: Assignments are due at 11:00 AM (the start of lecture) on the date above. he usual College late assignments
ECE521: Week 11, Lecture March 2017: HMM learning/inference. With thanks to Russ Salakhutdinov
 ECE521: Week 11, Lecture 20 27 March 2017: HMM learning/inference With thanks to Russ Salakhutdinov Examples of other perspectives Murphy 17.4 End of Russell & Norvig 15.2 (Artificial Intelligence: A Modern
ECE521: Week 11, Lecture 20 27 March 2017: HMM learning/inference With thanks to Russ Salakhutdinov Examples of other perspectives Murphy 17.4 End of Russell & Norvig 15.2 (Artificial Intelligence: A Modern
Unsupervised Learning
 Unsupervised Learning Learning without Class Labels (or correct outputs) Density Estimation Learn P(X) given training data for X Clustering Partition data into clusters Dimensionality Reduction Discover
Unsupervised Learning Learning without Class Labels (or correct outputs) Density Estimation Learn P(X) given training data for X Clustering Partition data into clusters Dimensionality Reduction Discover
Modeling time series with hidden Markov models
 Modeling time series with hidden Markov models Advanced Machine learning 2017 Nadia Figueroa, Jose Medina and Aude Billard Time series data Barometric pressure Temperature Data Humidity Time What s going
Modeling time series with hidden Markov models Advanced Machine learning 2017 Nadia Figueroa, Jose Medina and Aude Billard Time series data Barometric pressure Temperature Data Humidity Time What s going
CS 543: Final Project Report Texture Classification using 2-D Noncausal HMMs
 CS 543: Final Project Report Texture Classification using 2-D Noncausal HMMs Felix Wang fywang2 John Wieting wieting2 Introduction We implement a texture classification algorithm using 2-D Noncausal Hidden
CS 543: Final Project Report Texture Classification using 2-D Noncausal HMMs Felix Wang fywang2 John Wieting wieting2 Introduction We implement a texture classification algorithm using 2-D Noncausal Hidden
Person Authentication from Video of Faces: A Behavioral and Physiological Approach Using Pseudo Hierarchical Hidden Markov Models
 Person Authentication from Video of Faces: A Behavioral and Physiological Approach Using Pseudo Hierarchical Hidden Markov Models Manuele Bicego 1, Enrico Grosso 1, and Massimo Tistarelli 2 1 DEIR - University
Person Authentication from Video of Faces: A Behavioral and Physiological Approach Using Pseudo Hierarchical Hidden Markov Models Manuele Bicego 1, Enrico Grosso 1, and Massimo Tistarelli 2 1 DEIR - University
Ambiguity Detection by Fusion and Conformity: A Spectral Clustering Approach
 KIMAS 25 WALTHAM, MA, USA Ambiguity Detection by Fusion and Conformity: A Spectral Clustering Approach Fatih Porikli Mitsubishi Electric Research Laboratories Cambridge, MA, 239, USA fatih@merl.com Abstract
KIMAS 25 WALTHAM, MA, USA Ambiguity Detection by Fusion and Conformity: A Spectral Clustering Approach Fatih Porikli Mitsubishi Electric Research Laboratories Cambridge, MA, 239, USA fatih@merl.com Abstract
Association Rule Mining and Clustering
 Association Rule Mining and Clustering Lecture Outline: Classification vs. Association Rule Mining vs. Clustering Association Rule Mining Clustering Types of Clusters Clustering Algorithms Hierarchical:
Association Rule Mining and Clustering Lecture Outline: Classification vs. Association Rule Mining vs. Clustering Association Rule Mining Clustering Types of Clusters Clustering Algorithms Hierarchical:
ALTERNATIVE METHODS FOR CLUSTERING
 ALTERNATIVE METHODS FOR CLUSTERING K-Means Algorithm Termination conditions Several possibilities, e.g., A fixed number of iterations Objects partition unchanged Centroid positions don t change Convergence
ALTERNATIVE METHODS FOR CLUSTERING K-Means Algorithm Termination conditions Several possibilities, e.g., A fixed number of iterations Objects partition unchanged Centroid positions don t change Convergence
10-701/15-781, Fall 2006, Final
 -7/-78, Fall 6, Final Dec, :pm-8:pm There are 9 questions in this exam ( pages including this cover sheet). If you need more room to work out your answer to a question, use the back of the page and clearly
-7/-78, Fall 6, Final Dec, :pm-8:pm There are 9 questions in this exam ( pages including this cover sheet). If you need more room to work out your answer to a question, use the back of the page and clearly
Data Mining Chapter 9: Descriptive Modeling Fall 2011 Ming Li Department of Computer Science and Technology Nanjing University
 Data Mining Chapter 9: Descriptive Modeling Fall 2011 Ming Li Department of Computer Science and Technology Nanjing University Descriptive model A descriptive model presents the main features of the data
Data Mining Chapter 9: Descriptive Modeling Fall 2011 Ming Li Department of Computer Science and Technology Nanjing University Descriptive model A descriptive model presents the main features of the data
Machine Learning A W 1sst KU. b) [1 P] Give an example for a probability distributions P (A, B, C) that disproves
![Machine Learning A W 1sst KU. b) [1 P] Give an example for a probability distributions P (A, B, C) that disproves Machine Learning A W 1sst KU. b) [1 P] Give an example for a probability distributions P (A, B, C) that disproves](/thumbs/93/111796408.jpg) Machine Learning A 708.064 11W 1sst KU Exercises Problems marked with * are optional. 1 Conditional Independence I [2 P] a) [1 P] Give an example for a probability distribution P (A, B, C) that disproves
Machine Learning A 708.064 11W 1sst KU Exercises Problems marked with * are optional. 1 Conditional Independence I [2 P] a) [1 P] Give an example for a probability distribution P (A, B, C) that disproves
Topics in Machine Learning-EE 5359 Model Assessment and Selection
 Topics in Machine Learning-EE 5359 Model Assessment and Selection Ioannis D. Schizas Electrical Engineering Department University of Texas at Arlington 1 Training and Generalization Training stage: Utilizing
Topics in Machine Learning-EE 5359 Model Assessment and Selection Ioannis D. Schizas Electrical Engineering Department University of Texas at Arlington 1 Training and Generalization Training stage: Utilizing
Clustering & Dimensionality Reduction. 273A Intro Machine Learning
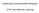 Clustering & Dimensionality Reduction 273A Intro Machine Learning What is Unsupervised Learning? In supervised learning we were given attributes & targets (e.g. class labels). In unsupervised learning
Clustering & Dimensionality Reduction 273A Intro Machine Learning What is Unsupervised Learning? In supervised learning we were given attributes & targets (e.g. class labels). In unsupervised learning
Clustering. SC4/SM4 Data Mining and Machine Learning, Hilary Term 2017 Dino Sejdinovic
 Clustering SC4/SM4 Data Mining and Machine Learning, Hilary Term 2017 Dino Sejdinovic Clustering is one of the fundamental and ubiquitous tasks in exploratory data analysis a first intuition about the
Clustering SC4/SM4 Data Mining and Machine Learning, Hilary Term 2017 Dino Sejdinovic Clustering is one of the fundamental and ubiquitous tasks in exploratory data analysis a first intuition about the
Machine Learning (BSMC-GA 4439) Wenke Liu
 Machine Learning (BSMC-GA 4439) Wenke Liu 01-31-017 Outline Background Defining proximity Clustering methods Determining number of clusters Comparing two solutions Cluster analysis as unsupervised Learning
Machine Learning (BSMC-GA 4439) Wenke Liu 01-31-017 Outline Background Defining proximity Clustering methods Determining number of clusters Comparing two solutions Cluster analysis as unsupervised Learning
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS
 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS CHAPTER 4 CLASSIFICATION WITH RADIAL BASIS AND PROBABILISTIC NEURAL NETWORKS 4.1 Introduction Optical character recognition is one of
ModelStructureSelection&TrainingAlgorithmsfor an HMMGesture Recognition System
 ModelStructureSelection&TrainingAlgorithmsfor an HMMGesture Recognition System Nianjun Liu, Brian C. Lovell, Peter J. Kootsookos, and Richard I.A. Davis Intelligent Real-Time Imaging and Sensing (IRIS)
ModelStructureSelection&TrainingAlgorithmsfor an HMMGesture Recognition System Nianjun Liu, Brian C. Lovell, Peter J. Kootsookos, and Richard I.A. Davis Intelligent Real-Time Imaging and Sensing (IRIS)
Classification. Vladimir Curic. Centre for Image Analysis Swedish University of Agricultural Sciences Uppsala University
 Classification Vladimir Curic Centre for Image Analysis Swedish University of Agricultural Sciences Uppsala University Outline An overview on classification Basics of classification How to choose appropriate
Classification Vladimir Curic Centre for Image Analysis Swedish University of Agricultural Sciences Uppsala University Outline An overview on classification Basics of classification How to choose appropriate
Effect of Initial HMM Choices in Multiple Sequence Training for Gesture Recognition
 Effect of Initial HMM Choices in Multiple Sequence Training for Gesture Recognition Nianjun Liu, Richard I.A. Davis, Brian C. Lovell and Peter J. Kootsookos Intelligent Real-Time Imaging and Sensing (IRIS)
Effect of Initial HMM Choices in Multiple Sequence Training for Gesture Recognition Nianjun Liu, Richard I.A. Davis, Brian C. Lovell and Peter J. Kootsookos Intelligent Real-Time Imaging and Sensing (IRIS)
Unsupervised Learning and Clustering
 Unsupervised Learning and Clustering Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Spring 2009 CS 551, Spring 2009 c 2009, Selim Aksoy (Bilkent University)
Unsupervised Learning and Clustering Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Spring 2009 CS 551, Spring 2009 c 2009, Selim Aksoy (Bilkent University)
Hidden Markov Models. Slides adapted from Joyce Ho, David Sontag, Geoffrey Hinton, Eric Xing, and Nicholas Ruozzi
 Hidden Markov Models Slides adapted from Joyce Ho, David Sontag, Geoffrey Hinton, Eric Xing, and Nicholas Ruozzi Sequential Data Time-series: Stock market, weather, speech, video Ordered: Text, genes Sequential
Hidden Markov Models Slides adapted from Joyce Ho, David Sontag, Geoffrey Hinton, Eric Xing, and Nicholas Ruozzi Sequential Data Time-series: Stock market, weather, speech, video Ordered: Text, genes Sequential
Mixture Models and EM
 Mixture Models and EM Goal: Introduction to probabilistic mixture models and the expectationmaximization (EM) algorithm. Motivation: simultaneous fitting of multiple model instances unsupervised clustering
Mixture Models and EM Goal: Introduction to probabilistic mixture models and the expectationmaximization (EM) algorithm. Motivation: simultaneous fitting of multiple model instances unsupervised clustering
Parameter Selection for EM Clustering Using Information Criterion and PDDP
 Parameter Selection for EM Clustering Using Information Criterion and PDDP Ujjwal Das Gupta,Vinay Menon and Uday Babbar Abstract This paper presents an algorithm to automatically determine the number of
Parameter Selection for EM Clustering Using Information Criterion and PDDP Ujjwal Das Gupta,Vinay Menon and Uday Babbar Abstract This paper presents an algorithm to automatically determine the number of
Supplementary Material: The Emergence of. Organizing Structure in Conceptual Representation
 Supplementary Material: The Emergence of Organizing Structure in Conceptual Representation Brenden M. Lake, 1,2 Neil D. Lawrence, 3 Joshua B. Tenenbaum, 4,5 1 Center for Data Science, New York University
Supplementary Material: The Emergence of Organizing Structure in Conceptual Representation Brenden M. Lake, 1,2 Neil D. Lawrence, 3 Joshua B. Tenenbaum, 4,5 1 Center for Data Science, New York University
Mix-nets: Factored Mixtures of Gaussians in Bayesian Networks with Mixed Continuous And Discrete Variables
 Mix-nets: Factored Mixtures of Gaussians in Bayesian Networks with Mixed Continuous And Discrete Variables Scott Davies and Andrew Moore School of Computer Science Carnegie Mellon University Pittsburgh,
Mix-nets: Factored Mixtures of Gaussians in Bayesian Networks with Mixed Continuous And Discrete Variables Scott Davies and Andrew Moore School of Computer Science Carnegie Mellon University Pittsburgh,
Invariant Recognition of Hand-Drawn Pictograms Using HMMs with a Rotating Feature Extraction
 Invariant Recognition of Hand-Drawn Pictograms Using HMMs with a Rotating Feature Extraction Stefan Müller, Gerhard Rigoll, Andreas Kosmala and Denis Mazurenok Department of Computer Science, Faculty of
Invariant Recognition of Hand-Drawn Pictograms Using HMMs with a Rotating Feature Extraction Stefan Müller, Gerhard Rigoll, Andreas Kosmala and Denis Mazurenok Department of Computer Science, Faculty of
Probabilistic Graphical Models
 Overview of Part Two Probabilistic Graphical Models Part Two: Inference and Learning Christopher M. Bishop Exact inference and the junction tree MCMC Variational methods and EM Example General variational
Overview of Part Two Probabilistic Graphical Models Part Two: Inference and Learning Christopher M. Bishop Exact inference and the junction tree MCMC Variational methods and EM Example General variational
Introduction to Pattern Recognition Part II. Selim Aksoy Bilkent University Department of Computer Engineering
 Introduction to Pattern Recognition Part II Selim Aksoy Bilkent University Department of Computer Engineering saksoy@cs.bilkent.edu.tr RETINA Pattern Recognition Tutorial, Summer 2005 Overview Statistical
Introduction to Pattern Recognition Part II Selim Aksoy Bilkent University Department of Computer Engineering saksoy@cs.bilkent.edu.tr RETINA Pattern Recognition Tutorial, Summer 2005 Overview Statistical
An Efficient Model Selection for Gaussian Mixture Model in a Bayesian Framework
 IEEE SIGNAL PROCESSING LETTERS, VOL. XX, NO. XX, XXX 23 An Efficient Model Selection for Gaussian Mixture Model in a Bayesian Framework Ji Won Yoon arxiv:37.99v [cs.lg] 3 Jul 23 Abstract In order to cluster
IEEE SIGNAL PROCESSING LETTERS, VOL. XX, NO. XX, XXX 23 An Efficient Model Selection for Gaussian Mixture Model in a Bayesian Framework Ji Won Yoon arxiv:37.99v [cs.lg] 3 Jul 23 Abstract In order to cluster
CSE 5243 INTRO. TO DATA MINING
 CSE 5243 INTRO. TO DATA MINING Cluster Analysis: Basic Concepts and Methods Huan Sun, CSE@The Ohio State University Slides adapted from UIUC CS412, Fall 2017, by Prof. Jiawei Han 2 Chapter 10. Cluster
CSE 5243 INTRO. TO DATA MINING Cluster Analysis: Basic Concepts and Methods Huan Sun, CSE@The Ohio State University Slides adapted from UIUC CS412, Fall 2017, by Prof. Jiawei Han 2 Chapter 10. Cluster
CSE 5243 INTRO. TO DATA MINING
 CSE 5243 INTRO. TO DATA MINING Cluster Analysis: Basic Concepts and Methods Huan Sun, CSE@The Ohio State University 09/25/2017 Slides adapted from UIUC CS412, Fall 2017, by Prof. Jiawei Han 2 Chapter 10.
CSE 5243 INTRO. TO DATA MINING Cluster Analysis: Basic Concepts and Methods Huan Sun, CSE@The Ohio State University 09/25/2017 Slides adapted from UIUC CS412, Fall 2017, by Prof. Jiawei Han 2 Chapter 10.
Nested Sampling: Introduction and Implementation
 UNIVERSITY OF TEXAS AT SAN ANTONIO Nested Sampling: Introduction and Implementation Liang Jing May 2009 1 1 ABSTRACT Nested Sampling is a new technique to calculate the evidence, Z = P(D M) = p(d θ, M)p(θ
UNIVERSITY OF TEXAS AT SAN ANTONIO Nested Sampling: Introduction and Implementation Liang Jing May 2009 1 1 ABSTRACT Nested Sampling is a new technique to calculate the evidence, Z = P(D M) = p(d θ, M)p(θ
A noninformative Bayesian approach to small area estimation
 A noninformative Bayesian approach to small area estimation Glen Meeden School of Statistics University of Minnesota Minneapolis, MN 55455 glen@stat.umn.edu September 2001 Revised May 2002 Research supported
A noninformative Bayesian approach to small area estimation Glen Meeden School of Statistics University of Minnesota Minneapolis, MN 55455 glen@stat.umn.edu September 2001 Revised May 2002 Research supported
Clustering Relational Data using the Infinite Relational Model
 Clustering Relational Data using the Infinite Relational Model Ana Daglis Supervised by: Matthew Ludkin September 4, 2015 Ana Daglis Clustering Data using the Infinite Relational Model September 4, 2015
Clustering Relational Data using the Infinite Relational Model Ana Daglis Supervised by: Matthew Ludkin September 4, 2015 Ana Daglis Clustering Data using the Infinite Relational Model September 4, 2015
k-means demo Administrative Machine learning: Unsupervised learning" Assignment 5 out
 Machine learning: Unsupervised learning" David Kauchak cs Spring 0 adapted from: http://www.stanford.edu/class/cs76/handouts/lecture7-clustering.ppt http://www.youtube.com/watch?v=or_-y-eilqo Administrative
Machine learning: Unsupervised learning" David Kauchak cs Spring 0 adapted from: http://www.stanford.edu/class/cs76/handouts/lecture7-clustering.ppt http://www.youtube.com/watch?v=or_-y-eilqo Administrative
Outline. Advanced Digital Image Processing and Others. Importance of Segmentation (Cont.) Importance of Segmentation
 Advanced Digital Image Processing and Others Xiaojun Qi -- REU Site Program in CVIP (7 Summer) Outline Segmentation Strategies and Data Structures Algorithms Overview K-Means Algorithm Hidden Markov Model
Advanced Digital Image Processing and Others Xiaojun Qi -- REU Site Program in CVIP (7 Summer) Outline Segmentation Strategies and Data Structures Algorithms Overview K-Means Algorithm Hidden Markov Model
Clustering Documents in Large Text Corpora
 Clustering Documents in Large Text Corpora Bin He Faculty of Computer Science Dalhousie University Halifax, Canada B3H 1W5 bhe@cs.dal.ca http://www.cs.dal.ca/ bhe Yongzheng Zhang Faculty of Computer Science
Clustering Documents in Large Text Corpora Bin He Faculty of Computer Science Dalhousie University Halifax, Canada B3H 1W5 bhe@cs.dal.ca http://www.cs.dal.ca/ bhe Yongzheng Zhang Faculty of Computer Science
Norbert Schuff VA Medical Center and UCSF
 Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Unsupervised Learning
 Unsupervised Learning Unsupervised learning Until now, we have assumed our training samples are labeled by their category membership. Methods that use labeled samples are said to be supervised. However,
Unsupervised Learning Unsupervised learning Until now, we have assumed our training samples are labeled by their category membership. Methods that use labeled samples are said to be supervised. However,
CHAPTER 4: CLUSTER ANALYSIS
 CHAPTER 4: CLUSTER ANALYSIS WHAT IS CLUSTER ANALYSIS? A cluster is a collection of data-objects similar to one another within the same group & dissimilar to the objects in other groups. Cluster analysis
CHAPTER 4: CLUSTER ANALYSIS WHAT IS CLUSTER ANALYSIS? A cluster is a collection of data-objects similar to one another within the same group & dissimilar to the objects in other groups. Cluster analysis
Machine Learning for Signal Processing Clustering. Bhiksha Raj Class Oct 2016
 Machine Learning for Signal Processing Clustering Bhiksha Raj Class 11. 13 Oct 2016 1 Statistical Modelling and Latent Structure Much of statistical modelling attempts to identify latent structure in the
Machine Learning for Signal Processing Clustering Bhiksha Raj Class 11. 13 Oct 2016 1 Statistical Modelling and Latent Structure Much of statistical modelling attempts to identify latent structure in the
Motivation. Technical Background
 Handling Outliers through Agglomerative Clustering with Full Model Maximum Likelihood Estimation, with Application to Flow Cytometry Mark Gordon, Justin Li, Kevin Matzen, Bryce Wiedenbeck Motivation Clustering
Handling Outliers through Agglomerative Clustering with Full Model Maximum Likelihood Estimation, with Application to Flow Cytometry Mark Gordon, Justin Li, Kevin Matzen, Bryce Wiedenbeck Motivation Clustering
Probabilistic Graphical Models
 Overview of Part One Probabilistic Graphical Models Part One: Graphs and Markov Properties Christopher M. Bishop Graphs and probabilities Directed graphs Markov properties Undirected graphs Examples Microsoft
Overview of Part One Probabilistic Graphical Models Part One: Graphs and Markov Properties Christopher M. Bishop Graphs and probabilities Directed graphs Markov properties Undirected graphs Examples Microsoft
Random projection for non-gaussian mixture models
 Random projection for non-gaussian mixture models Győző Gidófalvi Department of Computer Science and Engineering University of California, San Diego La Jolla, CA 92037 gyozo@cs.ucsd.edu Abstract Recently,
Random projection for non-gaussian mixture models Győző Gidófalvi Department of Computer Science and Engineering University of California, San Diego La Jolla, CA 92037 gyozo@cs.ucsd.edu Abstract Recently,
the number of states must be set in advance, i.e. the structure of the model is not t to the data, but given a priori the algorithm converges to a loc
 Clustering Time Series with Hidden Markov Models and Dynamic Time Warping Tim Oates, Laura Firoiu and Paul R. Cohen Computer Science Department, LGRC University of Massachusetts, Box 34610 Amherst, MA
Clustering Time Series with Hidden Markov Models and Dynamic Time Warping Tim Oates, Laura Firoiu and Paul R. Cohen Computer Science Department, LGRC University of Massachusetts, Box 34610 Amherst, MA
Online Pattern Recognition in Multivariate Data Streams using Unsupervised Learning
 Online Pattern Recognition in Multivariate Data Streams using Unsupervised Learning Devina Desai ddevina1@csee.umbc.edu Tim Oates oates@csee.umbc.edu Vishal Shanbhag vshan1@csee.umbc.edu Machine Learning
Online Pattern Recognition in Multivariate Data Streams using Unsupervised Learning Devina Desai ddevina1@csee.umbc.edu Tim Oates oates@csee.umbc.edu Vishal Shanbhag vshan1@csee.umbc.edu Machine Learning
INF4820, Algorithms for AI and NLP: Evaluating Classifiers Clustering
 INF4820, Algorithms for AI and NLP: Evaluating Classifiers Clustering Erik Velldal University of Oslo Sept. 18, 2012 Topics for today 2 Classification Recap Evaluating classifiers Accuracy, precision,
INF4820, Algorithms for AI and NLP: Evaluating Classifiers Clustering Erik Velldal University of Oslo Sept. 18, 2012 Topics for today 2 Classification Recap Evaluating classifiers Accuracy, precision,
Quantitative Biology II!
 Quantitative Biology II! Lecture 3: Markov Chain Monte Carlo! March 9, 2015! 2! Plan for Today!! Introduction to Sampling!! Introduction to MCMC!! Metropolis Algorithm!! Metropolis-Hastings Algorithm!!
Quantitative Biology II! Lecture 3: Markov Chain Monte Carlo! March 9, 2015! 2! Plan for Today!! Introduction to Sampling!! Introduction to MCMC!! Metropolis Algorithm!! Metropolis-Hastings Algorithm!!
Unsupervised naive Bayes for data clustering with mixtures of truncated exponentials
 Unsupervised naive Bayes for data clustering with mixtures of truncated exponentials José A. Gámez Computing Systems Department Intelligent Systems and Data Mining Group i 3 A University of Castilla-La
Unsupervised naive Bayes for data clustering with mixtures of truncated exponentials José A. Gámez Computing Systems Department Intelligent Systems and Data Mining Group i 3 A University of Castilla-La
HIDDEN MARKOV MODELS AND SEQUENCE ALIGNMENT
 HIDDEN MARKOV MODELS AND SEQUENCE ALIGNMENT - Swarbhanu Chatterjee. Hidden Markov models are a sophisticated and flexible statistical tool for the study of protein models. Using HMMs to analyze proteins
HIDDEN MARKOV MODELS AND SEQUENCE ALIGNMENT - Swarbhanu Chatterjee. Hidden Markov models are a sophisticated and flexible statistical tool for the study of protein models. Using HMMs to analyze proteins
MultiDimensional Signal Processing Master Degree in Ingegneria delle Telecomunicazioni A.A
 MultiDimensional Signal Processing Master Degree in Ingegneria delle Telecomunicazioni A.A. 205-206 Pietro Guccione, PhD DEI - DIPARTIMENTO DI INGEGNERIA ELETTRICA E DELL INFORMAZIONE POLITECNICO DI BARI
MultiDimensional Signal Processing Master Degree in Ingegneria delle Telecomunicazioni A.A. 205-206 Pietro Guccione, PhD DEI - DIPARTIMENTO DI INGEGNERIA ELETTRICA E DELL INFORMAZIONE POLITECNICO DI BARI
Clustering CS 550: Machine Learning
 Clustering CS 550: Machine Learning This slide set mainly uses the slides given in the following links: http://www-users.cs.umn.edu/~kumar/dmbook/ch8.pdf http://www-users.cs.umn.edu/~kumar/dmbook/dmslides/chap8_basic_cluster_analysis.pdf
Clustering CS 550: Machine Learning This slide set mainly uses the slides given in the following links: http://www-users.cs.umn.edu/~kumar/dmbook/ch8.pdf http://www-users.cs.umn.edu/~kumar/dmbook/dmslides/chap8_basic_cluster_analysis.pdf
Clustering. CS294 Practical Machine Learning Junming Yin 10/09/06
 Clustering CS294 Practical Machine Learning Junming Yin 10/09/06 Outline Introduction Unsupervised learning What is clustering? Application Dissimilarity (similarity) of objects Clustering algorithm K-means,
Clustering CS294 Practical Machine Learning Junming Yin 10/09/06 Outline Introduction Unsupervised learning What is clustering? Application Dissimilarity (similarity) of objects Clustering algorithm K-means,
An Introduction to Pattern Recognition
 An Introduction to Pattern Recognition Speaker : Wei lun Chao Advisor : Prof. Jian-jiun Ding DISP Lab Graduate Institute of Communication Engineering 1 Abstract Not a new research field Wide range included
An Introduction to Pattern Recognition Speaker : Wei lun Chao Advisor : Prof. Jian-jiun Ding DISP Lab Graduate Institute of Communication Engineering 1 Abstract Not a new research field Wide range included
A Deterministic Global Optimization Method for Variational Inference
 A Deterministic Global Optimization Method for Variational Inference Hachem Saddiki Mathematics and Statistics University of Massachusetts, Amherst saddiki@math.umass.edu Andrew C. Trapp Operations and
A Deterministic Global Optimization Method for Variational Inference Hachem Saddiki Mathematics and Statistics University of Massachusetts, Amherst saddiki@math.umass.edu Andrew C. Trapp Operations and
Evaluation of Model-Based Condition Monitoring Systems in Industrial Application Cases
 Evaluation of Model-Based Condition Monitoring Systems in Industrial Application Cases S. Windmann 1, J. Eickmeyer 1, F. Jungbluth 1, J. Badinger 2, and O. Niggemann 1,2 1 Fraunhofer Application Center
Evaluation of Model-Based Condition Monitoring Systems in Industrial Application Cases S. Windmann 1, J. Eickmeyer 1, F. Jungbluth 1, J. Badinger 2, and O. Niggemann 1,2 1 Fraunhofer Application Center
9.1. K-means Clustering
 424 9. MIXTURE MODELS AND EM Section 9.2 Section 9.3 Section 9.4 view of mixture distributions in which the discrete latent variables can be interpreted as defining assignments of data points to specific
424 9. MIXTURE MODELS AND EM Section 9.2 Section 9.3 Section 9.4 view of mixture distributions in which the discrete latent variables can be interpreted as defining assignments of data points to specific
Skill. Robot/ Controller
 Skill Acquisition from Human Demonstration Using a Hidden Markov Model G. E. Hovland, P. Sikka and B. J. McCarragher Department of Engineering Faculty of Engineering and Information Technology The Australian
Skill Acquisition from Human Demonstration Using a Hidden Markov Model G. E. Hovland, P. Sikka and B. J. McCarragher Department of Engineering Faculty of Engineering and Information Technology The Australian
Probabilistic Graphical Models
 Probabilistic Graphical Models Lecture 17 EM CS/CNS/EE 155 Andreas Krause Announcements Project poster session on Thursday Dec 3, 4-6pm in Annenberg 2 nd floor atrium! Easels, poster boards and cookies
Probabilistic Graphical Models Lecture 17 EM CS/CNS/EE 155 Andreas Krause Announcements Project poster session on Thursday Dec 3, 4-6pm in Annenberg 2 nd floor atrium! Easels, poster boards and cookies
Part I. Hierarchical clustering. Hierarchical Clustering. Hierarchical clustering. Produces a set of nested clusters organized as a
 Week 9 Based in part on slides from textbook, slides of Susan Holmes Part I December 2, 2012 Hierarchical Clustering 1 / 1 Produces a set of nested clusters organized as a Hierarchical hierarchical clustering
Week 9 Based in part on slides from textbook, slides of Susan Holmes Part I December 2, 2012 Hierarchical Clustering 1 / 1 Produces a set of nested clusters organized as a Hierarchical hierarchical clustering
Function approximation using RBF network. 10 basis functions and 25 data points.
 1 Function approximation using RBF network F (x j ) = m 1 w i ϕ( x j t i ) i=1 j = 1... N, m 1 = 10, N = 25 10 basis functions and 25 data points. Basis function centers are plotted with circles and data
1 Function approximation using RBF network F (x j ) = m 1 w i ϕ( x j t i ) i=1 j = 1... N, m 1 = 10, N = 25 10 basis functions and 25 data points. Basis function centers are plotted with circles and data
What is machine learning?
 Machine learning, pattern recognition and statistical data modelling Lecture 12. The last lecture Coryn Bailer-Jones 1 What is machine learning? Data description and interpretation finding simpler relationship
Machine learning, pattern recognition and statistical data modelling Lecture 12. The last lecture Coryn Bailer-Jones 1 What is machine learning? Data description and interpretation finding simpler relationship
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
Empirical Bayesian Motion Segmentation
 1 Empirical Bayesian Motion Segmentation Nuno Vasconcelos, Andrew Lippman Abstract We introduce an empirical Bayesian procedure for the simultaneous segmentation of an observed motion field estimation
1 Empirical Bayesian Motion Segmentation Nuno Vasconcelos, Andrew Lippman Abstract We introduce an empirical Bayesian procedure for the simultaneous segmentation of an observed motion field estimation
Unsupervised Learning and Clustering
 Unsupervised Learning and Clustering Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Spring 2008 CS 551, Spring 2008 c 2008, Selim Aksoy (Bilkent University)
Unsupervised Learning and Clustering Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Spring 2008 CS 551, Spring 2008 c 2008, Selim Aksoy (Bilkent University)
MCMC Methods for data modeling
 MCMC Methods for data modeling Kenneth Scerri Department of Automatic Control and Systems Engineering Introduction 1. Symposium on Data Modelling 2. Outline: a. Definition and uses of MCMC b. MCMC algorithms
MCMC Methods for data modeling Kenneth Scerri Department of Automatic Control and Systems Engineering Introduction 1. Symposium on Data Modelling 2. Outline: a. Definition and uses of MCMC b. MCMC algorithms
Clustering Lecture 5: Mixture Model
 Clustering Lecture 5: Mixture Model Jing Gao SUNY Buffalo 1 Outline Basics Motivation, definition, evaluation Methods Partitional Hierarchical Density-based Mixture model Spectral methods Advanced topics
Clustering Lecture 5: Mixture Model Jing Gao SUNY Buffalo 1 Outline Basics Motivation, definition, evaluation Methods Partitional Hierarchical Density-based Mixture model Spectral methods Advanced topics
Clustering. k-mean clustering. Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein
 Clustering k-mean clustering Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein A quick review The clustering problem: homogeneity vs. separation Different representations
Clustering k-mean clustering Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein A quick review The clustering problem: homogeneity vs. separation Different representations
Introduction to Mobile Robotics
 Introduction to Mobile Robotics Clustering Wolfram Burgard Cyrill Stachniss Giorgio Grisetti Maren Bennewitz Christian Plagemann Clustering (1) Common technique for statistical data analysis (machine learning,
Introduction to Mobile Robotics Clustering Wolfram Burgard Cyrill Stachniss Giorgio Grisetti Maren Bennewitz Christian Plagemann Clustering (1) Common technique for statistical data analysis (machine learning,
Gene Clustering & Classification
 BINF, Introduction to Computational Biology Gene Clustering & Classification Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Introduction to Gene Clustering
BINF, Introduction to Computational Biology Gene Clustering & Classification Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Introduction to Gene Clustering
Lesson 3. Prof. Enza Messina
 Lesson 3 Prof. Enza Messina Clustering techniques are generally classified into these classes: PARTITIONING ALGORITHMS Directly divides data points into some prespecified number of clusters without a hierarchical
Lesson 3 Prof. Enza Messina Clustering techniques are generally classified into these classes: PARTITIONING ALGORITHMS Directly divides data points into some prespecified number of clusters without a hierarchical
10701 Machine Learning. Clustering
 171 Machine Learning Clustering What is Clustering? Organizing data into clusters such that there is high intra-cluster similarity low inter-cluster similarity Informally, finding natural groupings among
171 Machine Learning Clustering What is Clustering? Organizing data into clusters such that there is high intra-cluster similarity low inter-cluster similarity Informally, finding natural groupings among
A Performance Evaluation of HMM and DTW for Gesture Recognition
 A Performance Evaluation of HMM and DTW for Gesture Recognition Josep Maria Carmona and Joan Climent Barcelona Tech (UPC), Spain Abstract. It is unclear whether Hidden Markov Models (HMMs) or Dynamic Time
A Performance Evaluation of HMM and DTW for Gesture Recognition Josep Maria Carmona and Joan Climent Barcelona Tech (UPC), Spain Abstract. It is unclear whether Hidden Markov Models (HMMs) or Dynamic Time
Supervised vs. Unsupervised Learning
 Clustering Supervised vs. Unsupervised Learning So far we have assumed that the training samples used to design the classifier were labeled by their class membership (supervised learning) We assume now
Clustering Supervised vs. Unsupervised Learning So far we have assumed that the training samples used to design the classifier were labeled by their class membership (supervised learning) We assume now
